
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 17 August 2021
Sec. Disease Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.694333
This article is part of the Research Topic New Trends in Integrated Plant Disease Management View all 5 articles
Phytopathogens and pests are two major factors that limit the growth of plants. The expression of a flavonoid regulator gene, AtMYB12(AT2G47460), has been reported to increase the endogenous flavonoid content of tobacco and tomato. Previous research has only focused on the regulation mechanism of v-myb avian myeloblastosis viral oncogene homolog (MYB) transcription factors under single stress conditions. Here, research showed that AtMYB12 was involved in regulating the resistance of tobacco to multiple biological stresses such as phytopathogens and aphid. We reported that transgenic tobacco carrying AtMYB12 was more resistant to Ralstonia solanacearum when the up-regulated expression of several defense-related markers, such as NbPR1a, NbNOA1, and NbrbohB, was activated, suggesting that the priming defense of a plant may contribute to bacterial disease resistance. The improvement of the resistance of AtMYB12-expressing transgenic tobacco is achieved by promoting the production of ROS, H2O2, and NO. AtMYB12-expressing transgenic tobacco also has resistance to fungal pathogens, such as Colletotrichum nicotianae Averna and Alternaria alternate. The enrichment of flavonols components, such as rutin, which directly inhibit the growth of C. nicotianae and A. alternate, may also contribute to the defense mediated by AtMYB12 over-expression. At the same time, the results also confirm that AtMYB12-expressing transgenic tobacco enhanced plant resistance to aphid-infested (Aphidoidea) pests. These results suggest that the AtMYB12 gene is a good candidate for pest and disease control, with limited resistance costs and enrichment in flavonols, and that AtMYB12 has a potential in the breeding of disease-resistant tobacco crops.
Flavonoids are a large class of secondary metabolites that are widely present in the plant kingdom and encompass over 9,000 molecules (Cheynier et al., 2013). They are suggested to be divided into diverse subgroups, such as anthocyanidins, flavonols, flavones, and flavanols (Nayak et al., 2015). Most flavonoids are well-known antioxidants because of their powerful capacity to donate electrons or hydrogen atoms (Chen et al., 2014). In vitro assays have shown that flavonoids affect the activity of many mammalian cell enzyme systems, such as kinases, phospholipase A2, phospholipase C, ATPases, topoisomerase, and others (Hoensch and Oertel, 2015; Wang et al., 2017).
In the vegetable kingdom, flavonoids are reported to play a significant role in plant resistance, both in terms of abiotic and biotic stresses (Nayak et al., 2015), such as drought, UV radiation, and heavy metal contamination (Izaguirre et al., 2007; Lei et al., 2019). In addition to their function in abiotic stress resistance, flavonoids have also been found to be involved in plant protection. Some flavonoids, such as naringenin, kaempferol, quercetin, and dihydroquercetin, show an inhibition effect on the gingipain activity of Porphyromonas gingivalis (Kariu et al., 2017).
Flavonoids, such as rutin, were found to be moderately effective at inhibiting aerobic bacteria and Pectobacterium atrosepticum, causing plant diseases (Taguri et al., 2006; Maddox et al., 2010; Kröner et al., 2012). Moreover, flavonoids are reported to have a main role in the postharvest resistance of fruits and vegetables (Li et al., 2007). They have also been shown to have antimicrobial effects on phytopathogens. For example, flavonoids have been reported to have an antifeedant activity against specific insects (Thoison et al., 2004; Napal et al., 2009). They have also been reported to be involved in modulating plant immunity. In another study, a handful of flavonoids, as signals, were exuded from plant roots to induce the transcription of virulent genes, that have been proved to be required for the infection process of rhizospheric microorganisms and mycorrhizal fungus (Nayak et al., 2015). In addition, spraying quercetin, fruit flavonol, has been shown to prime H2O2 burst and the SA signaling pathway to resist the virulent strain Pseudomonassyringae pv. tomato DC3000 (PstDC3000) in Arabidopsis thaliana (Jia et al., 2010). Rutin, as a major kind of flavonoid, can increase the resistance of rice, tobacco and Arabidopsis thaliana to pathogens with a concentration of 2 mM consistency pretreated: Xanthomonas oryzae pv. oryzae in rice, Ralstonia solanacearum in tobacco, and Pst DC3000 in Arabidopsis thaliana (Yang et al., 2016).
Most flavonoids are synthesized through the phenylpropanoid pathway, which is controlled by a series of enzymes in plants (Luo et al., 2008). Copious biology tactics have been utilized to ameliorate the production of polyphenolic compounds in plants (Broun, 2004). AtMYB12 is known as a flavonol-specific transcription factor in Arabidopsis (Stracke et al., 2007). This transcription factor has been heterologously expressed in a handful of plants, such as tobacco, tomatoes, potatoes, and buckwheat, resulting in a significant level of polyphenolic compound accumulation, especially flavonols (Li et al., 2015). Tobacco is one of the most important cash crops in the world. The occurrence of pests and diseases is an important factor that plagues tobacco production, which greatly hinders the improvement of tobacco yield and quality. Searching for resistance genes and using molecular breeding techniques to breed resistant tobacco are effective ways to prevent and control diseases and pests (Kakar et al., 2020). The transgenic tobacco with ameliorated accumulation of rutin resulted in boosted resistance against the insects Spodoptera litura and Helicoverpa armigera (two major insect pests to tobacco) (Misra et al., 2010; Pandey et al., 2012). However, it is still unknown whether the accumulation of the compound in plants is beneficial for themselves to increase resistance to pathogens and insects.
In this study, we evaluated the resistance of AtMYB12 transgenic plants to insect and phytopathogens to reveal whether overexpressing AtMYB12 could enhance the immunity of a plant. The results showed that the overexpression of AtMYB12 can increase the resistance of plants to aphids and phytopathogens by increasing the content of rutin and activating related immune response. It indicated that the regulation of the AtMYB12 expression can be used to manage plant diseases and pests in the future. This article focuses on the function of MYB transcription factors and analyzes their mechanisms involved in biological stresses. The mystery of MYB transcription factors, which regulate plant resistance to pathogens and insects, is revealed.
As previously reported, two different blended transgenic lines of T6 generation seeds from Nicotiana tabacum var. samsuncanik (35S::AtMYB12 OE-1, 35S::AtMYB12 OE-2) and a control (samsun) were grown in a greenhouse at 25°C, with 70% relative humidity and 16 h of photoperiod (Luo et al., 2008).
Ralstonia solanacearum strains of Shandong isolates (SD1) that cause tobacco bacterial wilt were cultured in an NA solid medium (5 g·L−1 tryptone, 1 g·L−1 yeast extract, 10 g·L−1 sugar, and 15 g·L−1 agar; pH 6.8–7) at 28°C in the dark for 2 days, and the medium was supplemented with triphenyltetrazolium chloride (TTC, 0.05% final concentration). The harvested pathogen was adjusted to 108 cfu·ml−1 (approximately OD600 = 0.2) with distilled water followed by a ten-fold dilution and then cultured in the NA solid medium at 28°C in the dark for 1 day. A. alternata and C. nicotianae Averna were cultured in a solid potato dextrose agar (PDA) medium at 25°C for a week, and plugs that were 0.5 cm in diameter were taken from the edge of each colony using a puncher and then cultured in the solid PDA medium supplemented with 10, 20, 30, and 40 g·L−1 concentrations of tobacco extracts. The colony morphology and colony diameter were measured 1 week after culturing at 25°C in the dark.
Leaves of 4-to-6-week-old tobacco were inoculated with R. solanacearum by syringe infiltration at a concentration of 105 cfu·ml−1. The bacterial growth on the leaves was counted by determining the cfu from 1 g of leaves (fresh weight basis) of either wild-type or transgenic tobacco in the NA medium (Jia et al., 2010). Five plants from each time point were inoculated. The same experiment was repeated twice with five replicates. A. alternata and C. nicotianae Averna plugs (0.5 cm in diameter) were taken from the edge of the colony using a puncher and then cultured (at 28°C, 70% relative humidity and with 12-h photoperiod) in the middle of each half of the AtMYB12-expressing transgenic tobacco and wild-type tobacco detached leaves, which were 4 weeks old. The diameter of lesions from each half of the leaves was measured 5 days post-inoculation. A. alternata (3 × 105 spores·ml−1) and C. nicotianae Averna (1 × 105 spores· ml−1) spore suspensions (20 μl) were dripped in the middle of each half of the OE-1 and WT tobacco detached leaves (at 28°C, 70% relative humidity, and with 12-h photoperiod). The spores of each half of the leaves were counted with a hemocytometer (Bright-Line, United States), 0, 12, 24, 36, 48 h, and 4 days post inoculation. The same experiment was repeated twice with five replicates. The infected tissues around the inoculation sites were immersed in a 70% trypan blue solution (2.5 mg·ml−1 trypan blue, 25% lactate, 25% phenol water, and 25% glycerol) for 24 h, and then the stained leaves were decolorized for 24 h with chloral hydrate (25 g/10 ml water). The spores inside the tissues were observed with a microscope (Nikon Eclipse 90i, Nikon Corporation, Tokyo, Japan). The same experiment was repeated twice with three replicates. The strains mentioned above are all preserved in the laboratory.
Total RNA was isolated from 100 mg of plant tissue in about 6 weeks with RNAiso Plus according to the instructions of the manufacturer (TaKaRa, Dalian, China). The first strand cDNA was synthesized from 1 μg of total RNA with a PrimeScriptTM RT reagent Kit with gDNA Eraser (cat No. RR047, TaKaRa, Dalian, China). Quantitative PCR was performed with SYBR® Premix Ex TaqTM (TliRNaseH Plus, Takara, Dalian, China) in an IQ5 Real-Time PCR system (Bio-Rad Laboratories, Hercules, CA, United States). The PCR program, following the reference (Li et al., 2013, 2015), was as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 55°C for 20 s, and 72°C for 30 s. A heat dissociation curve (55–95°C) following the final PCR cycle was used to verify the specificity of the PCR amplification. A relative quantification analysis was performed using the relative standard curve according to the threshold values (Ct) generated. The NbEF1α gene of tobacco was used as an internal control to standardize the results. For each gene, qRT-PCR assays were repeated at least twice with triplicate runs separately. The sequences of each primer for all the detected genes are listed in Table 1.
The reactions were performed using a hydrogen peroxide assay kit and a nitric oxide assay kit (Beyotime, Jiangsu, China). The experiment used tobacco that was about 4–6 weeks old. First, a standard curve was produced for each assay. To measure H2O2, 100 mg of tissue (six different leaves from diverse plants) was ground with liquid nitrogen, and 10 mg of dry-off powder was dissolved in 200 μl of schizolysis buffer and centrifuged. A 50-μl volume of supernatant was mixed with 100 μl of the test solution and incubated for 30 min at room temperature. A560 absorbance value was measured to calculate the concentration of H2O2 after comparing it with a standard curve (Qian et al., 2009). For NO quantification, the accumulation of NO2− was determined as an indicator of NO production in the medium. In brief, the sample (six different leaves from diverse plants) was collected and stored at −80°C, ground under liquid nitrogen, and centrifuged. The 50-μl supernatant was mixed with Griess reagents I and II at a 1:1 ratio. The absorbance was measured at a 540-nm wavelength, and the concentration was calculated according to the standard curve. The same experiment was repeated twice.
The extract from 100-g leaves of 6-week-old tobacco was purified using 1 L of methanol (Luo et al., 2008). The extract was shaken in an ice bath for 2 h and filtered with an organic filter membrane (diameter 0.22 μm). The filtrate was vacuum-concentrated with ScanSpeed 40 (Gene Company Limited, Hong Kong) by centrifugation at 6,000 rpm speed at room temperature. The extract was dissolved with 10 ml of dimethyl sulfoxide (DMSO). A 250-μl volume of the extract was added to 250 ml of PDA and defined as a concentration of 10 g·L−1. The wild-type extract (as described above) plus 12 mg rutin was defined as WT + rutin. The same experiment was repeated twice with five replicates each of the different transgenic lines (OE-1, OE-2).
In total, 12 seedlings each from wild-type and transgenic tobacco were mix-planted in a 6 × 6 array and surrounded with aphid-infested (Aphidoidea) tobacco plants. The number of aphids was counted for each plant and leaf after 1 month of co-culture (Jakobs et al., 2019). For the rutin spray assay, the wild-type seedlings were pretreated by spraying with 1, 2, and 4 mM rutin or water (control) before being moved into an insect nursery. Rutin was regularly sprayed, once a week, until the data were collected. The same experiment was replicated twice in the greenhouse.
Phenylpropanoids were extracted from fresh samples with 100% methanol and analyzed by high-performance liquid chromatography (HPLC) (Luo et al., 2008). These compounds were quantified by calculating the area of each individual peak and comparing this area with the standard curves obtained from the pure compounds. Rutin and chlorogenic acid were purchased from Sigma-Aldrich (St. Louis, MO, United States, http://www.sigmaaldrich.com). Kaempferol rutinoside was purchased from Extrasynthese (Genay, France, http://www.extrasynthese.com).
Each value represents repeated independent experiments, and the vertical bars expressed the arithmetic means ± standard deviations (SD). Tukey's test was performed to calculate statistical significance, and the significant differences between treatments and the untreated control are represented by ‘*' at p < 0.05 and ‘**' at p < 0.01. We used the SPSS software (https://www.ibm.com/cn-zh/analytics/spss-statistics-software) to verify.
High-performance liquid chromatography was performed to detect the flavonol content of the transgenic and wild-type tobacco plants. In AtMYB12-expressing transgenic tobacco, rutin was the predominant flavonol in the methanolic extract. The fresh weight of 0.692 to 1.142 mg·g−1rutin was detected in the AtMYB12 transgenic lines. The leaves of the two homozygous lines carrying AtMYB12 displayed a 12.58- to 20.76-fold increase in rutin relative to the wild type (Table 2). Although kaempferol rutinoside was not detected in the wild-type plants, its fresh weight increased by up to 0.089–0.221 mg·g−1 in the transgenic lines. The increased level of flavonol in tobacco was substantially higher than those demonstrated for AtMYB12 overexpression in Arabidopsis, which showed a three- to four-fold increase in the transgenic plants (Mehrtens et al., 2005), but less than the original report of a 46-fold increase for rutin in tobacco (Luo et al., 2008). Additionally, the increased concentration of chlorogenic acid in transgenic tobacco was less than the originally reported (Table 2).
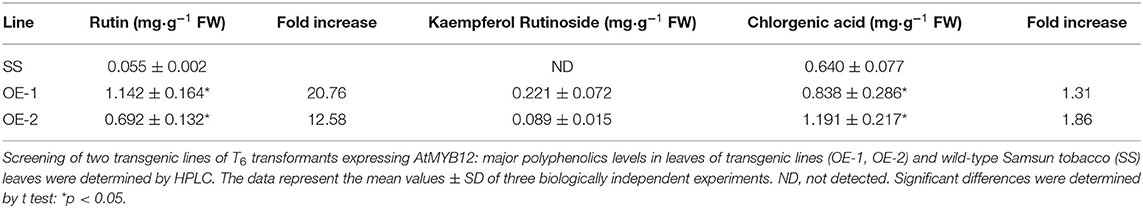
Table 2. Quantification of major polyphenolics in wild-type and AtMYB12 overexpression tobacco leaves.
Tobacco was subjected to many other diseases in the general categories of bacteria and fungi/molds except for insect pests, such as R. solanacearum, which causes bacterial wilt; C. nicotianae Averna, which causes anthracnose, and A. alternata, which causes brown spot disease. To understand how the AtMYB12 transcription factor affects tobacco resistance to diseases, we studied the sensitivity of the transgenic tobacco to R. Solanacearum SD1. The resistance responses of the two lines of transgenic tobacco (OE-1, OE-2) against R. solanacearum SD1 were detectable when compared with wild-type tobacco (Figure 1A). Wilt symptoms were only observed in inoculated half-leaves of the transgenic plants, and they expanded to the opposite half of the leaves and petiole of the wild-type plants 3 days post inoculation (Figure 1A). The bacterial growth curve also supported the observed symptoms, and the growth rate of SD1 on transgenic tobacco (average log = 5.405 at 96 h) was obviously lower than that of the wild type (log = 5.92 at 96 h) (Figure 1B). The number of bacteria in the wild type was 3.27-fold higher than in the transgenic plants 96 h post inoculation (Figure 1B).
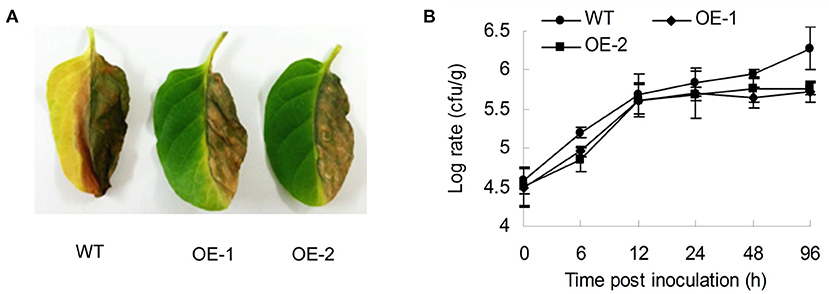
Figure 1. Characterization of resistance to R. solanacearum for AtMYB12-expressing transgenic tobacco. (A) Performance of the wild type (WT) and two lines of AtMYB12 overexpression tobacco (OE-1, OE-2) after inoculation with R. solanacearum (OD600 =0.5) for 3 days. (B) Growth curve of the R. solanacearum SD1 after the inoculation of 105cfu in tobacco leaves. Bacterial numbers were counted 0, 6, 12, 24, 48, and 96 h after infiltration. The values are the means ± SD (n = 5).
To understand the mechanisms involved in AtMYB12 induced resistance of transgenic tobacco, we compared the expression patterns of defense-related genes, such as NbPR1a, NbNOA1, and NbrbohB (Hyodo et al., 2017), in the wild-type and transgenic lines. In transgenic tobacco leaves (OE-1), we observed more than 187-fold induction of the NbPR1a gene in the transgenic plants, which was 52-fold higher induction relative to the wild-type leaves 24 h after inoculation (p < 0.01), and four to five-fold induction of NbrbohB (p < 0.01) and NbNOA1 (p < 0.01) in the transgenic plants, which were approximately 14- and 5.3-fold higher induction than in the wild-type leaves 6 h after inoculation. These results showed that the transgenic tobacco activated the stronger expression of defense-responsive genes than the wild type after the inoculation. Interestingly, in the absence of R. solanacearum, the expression level of defense-responsive genes was lower in the transgenic plant than in the wild type (Figure 2A).
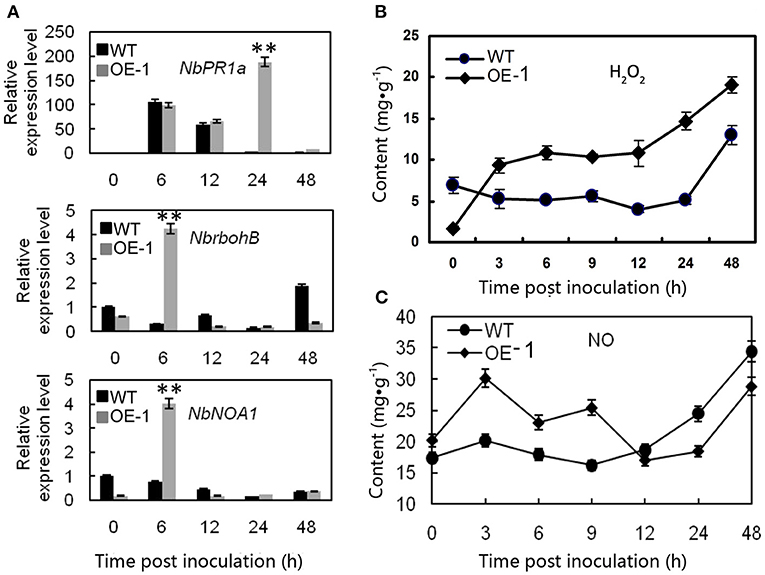
Figure 2. Different effects of the gene expression and the H2O2 and NO contents after R. solanacearum infection between wild-type (WT) and AtMYB12 overexpression tobacco (OE-1). (A) Relative expression analysis of NbPR1a, NbrbohB, and NbNOA1 after inoculation with the R. solanacearum SD1, which was performed 0, 6, 12, 24, and 48 h post-inoculation. The values represent the average of three technical replicates. The values are the means ± SD (n = 3, *p < 0.05, **p < 0.01). (B) Detection of the concentrations of H2O2. (C) Detection of the concentrations of NO, which were determined 0, 3, 6, 9, 12, 24, and 48 h post-inoculation. The values are the means ± SD (n = 6).
Because H2O2 and NO are a concern in diverse abiotic stress responses and are utilized to resist pathogen infections in plants, the H2O2 and NO contents of the tobacco leaves were quantified. The result showed that the background H2O2 level was lower in the transgenic tobacco (1.73 mg·g−1) than in the wild-type plants (6.91 mg·g−1) at 0 h (p < 0.01) (Figure 2B). Following the infiltration of R. solanacearum, the peroxide level increased rapidly and exceeded the content in the wild type at 3–6 h, and the maximum difference between the wild-type (5.16 mg·g−1) and transgenic tobacco (14.7 mg·g−1) occurred at 24 h significantly (p < 0.01). The peroxide level was consistent with the superoxide anions detected by staining with Nitrotetrazolium Blue chloride (NBT) (Supplementary Figure 1).
In the absence of pathogen inoculation, the NO content was a bit higher in the transgenic plants than in the wild type (Figure 2C). After pathogen inoculation, the NO concentration quickly increased to its peak value 3 h post inoculation. The result showed that the NO content of the transgenic leaves was much higher after being treated with R. solanacearum (30.14 mg·g−1) compared with that of the wild-type leaves (20.14 mg·g−1) (p < 0.01) at 3 h. There were no significant changes in the wild type at the early stage, and the NO level was dramatically increased 9 h post inoculation (Figure 2C).
Fungi also cause multiple serious tobacco diseases. In this experiment, tobacco was inoculated with A. alternata, which causes brown spot disease; and C. nicotianae Averna, which causes anthracnose disease, and infection was verified by detached leaf assay. In this study, the lesion diameter, lesion area percentage, spore number, and trypan blue staining were investigated. The transgenic tobacco showed significant resistance against A. alternata and C. nicotianae Averna relative to the wild type (Figures 3A,D). The diameter of the lesions caused by A. alternata in two different transgenic lines (OE-1, OE-2) for the tobacco leaves were 1.14 ± 0.08 and 1.19 ± 0.1 cm (Figure 3B) compared with 1.43 ± 0.05 cm (p < 0.01) in the wild type. Using the Photoshop (http://www.photoshop.com/) software, we also observed that the area percentage of lesions caused by A. alternata on the transgenic leaves was 43.67 ± 3.21%, which is dramatically lower than that on the leaves of the wild type, which was 77.02 ± 3.61% 9 days post inoculation (Supplementary Figure 2A). This resistant phenotype was also supported by pathological histology observation, which showed more conidia spores in the wild-type tobacco leaves than in transgenic leaves after inoculation with A. alternata (Figure 3C). Moreover, the hypersensitivity reaction (HR) response can be found in transgenic plants with small area 24 h post inoculation with A. alternata, and the leaves showed an intense HR response with wide area post 36 h post inoculation. In contrast, the wild-type plants showed no HR response, but had an increasing number of spores (Figure 4A).
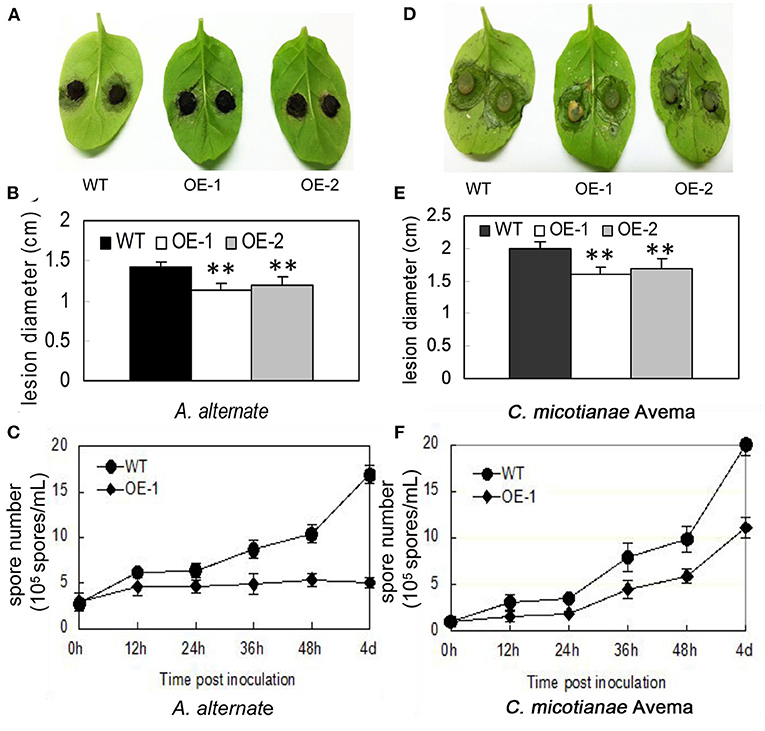
Figure 3. Characterization of resistance to A. alternata and C. nicotianae Averna for AtMYB12-expressing transgenic tobacco. (A) Performance of the wild-type (WT) and two lines of AtMYB12 overexpression tobacco (OE-1, OE-2) after inoculation with A. alternata for 7 days. (B) Lesion diameter from A. alternata after inoculation in tobacco leaves. The values represent the average of three technical replicates. (C) Growth curve of the A. alternata isolate after the inoculation of 3 × 106 spores·ml−1 in tobacco leaves. The spore numbers were counted 0, 12, 24, 36, 48 h, and 4 days after infiltration. The values are the means ± SD (n = 5). (D) Performance of the wild-type (WT) and two lines of AtMYB12 overexpression tobacco (OE-1, OE-2) after inoculation with C. nicotianae Averna for 7 days. (E) Lesion diameter of C. nicotianae Averna after inoculation in tobacco leaves. The values represent the average of three technical replicates. The values are the means ± SD (n = 3, *p < 0.05, **p < 0.01). (F) Growth curve of the C. nicotianae Averna isolate after the inoculation of 106 spores·ml−1 in tobacco leaves. Spore numbers were counted 0, 12, 24, 36, 48 h, and 4 days after infiltration. The values are the means ± SD (n = 5).
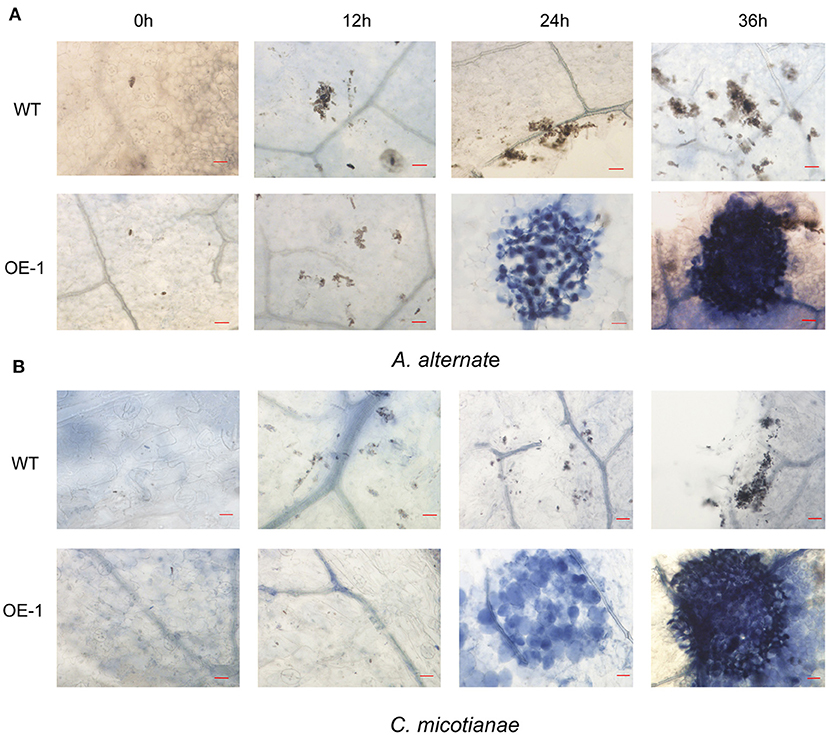
Figure 4. Infection phenotypes of wild-type (WT) and transgenic plant (OE-1) leaves inoculated with fungi. (A) WT and transgenic plants (OE-1) leaves were inoculated with A. alternata (3 × 106 spores·ml−1) and stained with trypan blue at 0, 12, 24, 36 h to reveal spores, mycelium, and dead plant cells. (B) WT and transgenic plant (OE-1) leaves were inoculated with C. nicotianae Averna (106 spores·ml−1) and stained with trypan blue at 0, 12, 24, 36 h to reveal spores, mycelium, and dead plant cells. Bar corresponds to 20 μm.
Using the same protocol as for A. alternata, the lesion area percentage was examined 5 days after inoculation with C. nicotianae Averna. The diameter of lesions caused by C. nicotianae Averna in the two different transgenic lines (OE-1,OE-2) for the tobacco leaves were 1.65 ± 0.04 and 1.72 ± 0.03 cm (Figure 3E) compared with 2.03 ± 0.02 cm (p < 0.01) in the wild type. This organism caused an average of 34.67 ± 2.31 and 27.67 ± 2.08% lesion areas in the wild-type and AtMYB12-overexpressing plants, respectively (Supplementary Figure 2B). In addition, fewer conidia spores and slower multiplication rate were observed in the transgenic tobacco leaves than in wild-type tobacco leaves (Figure 3F). Similar to inoculation with A. alternata, induced HR response was observed in the transgenic tobacco leaves 24 h after inoculation with C. nicotianae Averna (Figure 4B).
We analyzed the transcription level of pathogenesis-related (PR) genes: NbPR1a, NbNOA1 (nitric oxide-associated 1) which is related with NO production and defense responses, and NbrbohB (respiratory burst oxidase homolog B) which is related with active oxygen species generation. In the transgenic tobacco leaves (OE-1), we observed a six-fold induction of NbNOA1 (p < 0.01) in the transgenic plants, which was approximately five-fold higher than in the wild-type leaves at 48 h after inoculation with A. alternate (Figure 5A); more than 55-fold induction of the NbPR1a gene in the transgenic plants, which was seven-fold higher relative to the wild-type leaves 48 h after inoculation (p < 0.01) (Figure 5B); and 36-fold induction of NbrbohB (p < 0.01) which was 3.6-fold higher relative to the wild-type leaves 12 h after inoculation (p < 0.01) (Figure 5C). We also observed that there was four to six-fold induction of NbNOA1 (p < 0.01), NbrbohB (p < 0.01), and NbPR1a (p < 0.01) in the transgenic plants, which was approximately three- to four-fold higher than in the wild-type leaves 24 h after inoculation with C. nicotianae Averna (Figures 5D–F). These results showed that after inoculation, the transgenic tobacco activated the expression of defense-responsive genes better than the wild type.
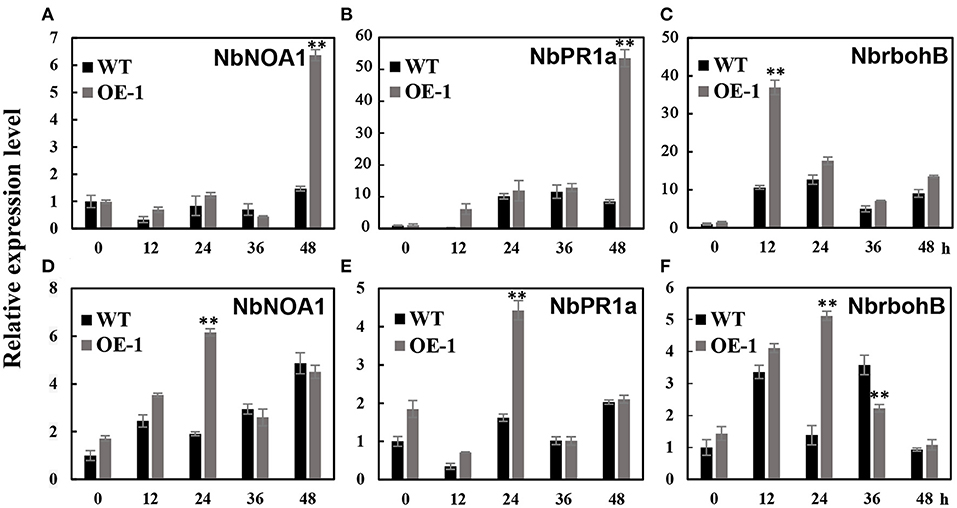
Figure 5. Different effects of the gene expression after A. alternate and C. nicotianae Averna infection between wild-type (WT) and AtMYB12 overexpression tobacco (OE-1). (A) Relative expression analysis of NbNOA1 after inoculation with the A. alternata isolate, which was performed 0, 12, 24, 36, and 48 h post-inoculation. The values represent the average of three technical replicates, and are the means ± SD (n = 3, *p < 0.05, **p < 0.01). (B) Relative expression analysis of NbPR1a after inoculation with the A. alternata isolate. (C) Relative expression analysis of NbrbohB after inoculation with the A. alternata isolate. (D) Relative expression analysis of NbNOA1 after inoculation with C. nicotianae Averna. (E) Relative expression analysis of NbPR1a after inoculation with C. nicotianae Averna. (F) Relative expression analysis of NbrbohB after inoculation with C. nicotianae Averna.
To explore the activation of plant defense by AtMYB12 overexpression, or directly with enriched flavonol components, we analyzed the growth of R. solanacearum in the medium with extracts from the wild-type and transgenic tobacco leaves. No visible differences in R. solanacearum growth in the culture medium with extracts from the wild-type and transgenic tobacco leaves were observed (data not shown).
We also tested the growth of the fungi C. nicotianae Averna and A. alternata in the culture medium with extracts from the wild-type and transgenic tobacco leaves, respectively. The in vitro assay showed that the growth-inhibiting effects on C. nicotianae Averna and A. alternata were almost the same. The colony diameter was significantly inhibited in the culture medium with extracts of transgenic tobacco or rutin only, and the inhibitory effects was positively correlated with the concentration of extract contents in the culture medium (Figures 6A,B). The average colony diameter was reduced to 7.60 cm and 6.87 cm for C. nicotianae Averna after adding a concentration of 10 g·L−1 leaf extract from the wild type and AtMYB12 OE-1, respectively (Figure 6C). These findings were approximately the same for added rutin to the wild-type extract, in which the colony diameter averaged 6.7 cm. As with C. nicotianae Averna, the colony diameter for A. alternata was reduced with the extract from the transgenic plant, and it averaged 5.87 cm for PDA culture plus with a 10 g·L−1 concentration of leaf extract from the wild type and 4.87 cm for the treatments with leaf extract from AtMYB12-overexpressing plants (OE-1). The reduced colony diameter could result from the addition of purified rutin (12 mg) to the wild-type extract (Figure 6D). These results indicate that enriched flavonol components (such as rutin) may have the ability to inhibit the growth of C. nicotianae Averna and A. alternata, and the correlation between rutin and inhibition was at a certain degree, but not absolute. However, the inhibitory effects of rutin on C. nicotianae Averna and A. alternata was significantly lower than that of extracts from the transgenic tobacco leaves. The difference between them was partially attributed to the defense mediated by AtMYB12 overexpression.
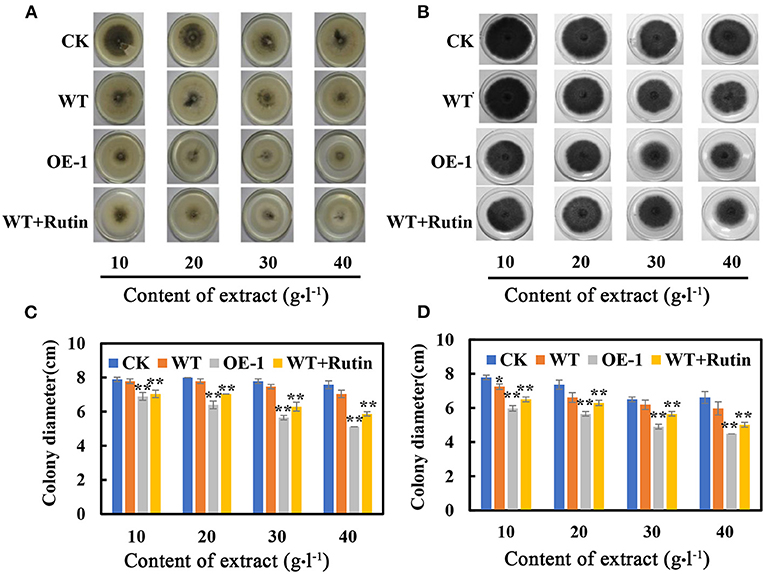
Figure 6. Effect of tobacco extract on C. nicotianae Averna and A. alternata. (A) Photographs were taken after the samples were cultured for 6 days on the PDA medium with different contents of C. nicotianae Averna extract. The extracts and rutin were dissolved in dimethyl sulfoxide (DMSO). (B) Photographs of A. alternata were taken after culturing the samples for 6 days on the culture medium with different extract contents. (C) Colony diameter of C. nicotianae Averna after culturing for 6 days. (D) Colony diameters of A. alternata after culturing for 6 days. CK, PDA plus DMSO. WT, PDA with the extract of wild-type tobacco leaves. OE-1, PDA with the extract of transgenic tobacco leaves. WT + rutin, PDA with the extract of wild-type tobacco leaves and rutin corresponding to transgenic leaf extracts. The values are the means ± SD (n = 5, *p < 0.05, **p < 0.01).
The accumulation of certain secondary metabolic components, such as flavonoids, has been demonstrated to enhance insect resistance to plants (Thoison et al., 2004; Sharma et al., 2018). The transgenic lines of AtMYB12 tobacco have greater significant resistance to insects compared with the control, with more than 70% mortality for S. litura and H. armigera (Misra et al., 2010). In this study, we also counted the average number of aphids (MyzusPersicae Sulzer) (371.32 ± 34.56 and 270.13 ± 25.45 per plant; 79.73 ± 9.7 and 51.39 ± 5.15 per leaf) on the leaves of the control tobacco and the transgenic tobacco that was carrying AtMYB12, respectively. The data showed that the transgenic tobacco has increased resistance against aphid insect pests (p < 0.01) (Figure 7A). We found that the insect suppression efficiency of the transgenic lines (OE-1, OE-2) was 27.3% per plant and 35.5% per leaf for aphids in contrast to the wild type (WT). This resistance was also observed through an insect bioassay that was performed with purified rutin sprayed on the tobacco plants. We counted the average number of aphids (307.13 ± 27.38, 298.13 ± 25.08, 291.13 ± 24.75, and 391.89 ± 30.16 per plant) on the leaves of the wild-type tobacco pre-sprayed with 1, 2, 4 mM rutin and water. The number of aphids on 4 mM rutin-sprayed plants was dramatically reduced to 74.3% of the control values; this effect reached the level of resistance comparable to that of the AtMYB12 transgenic plants (Figure 7B).
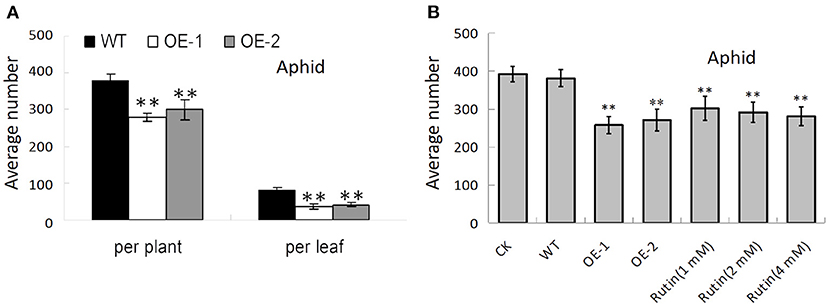
Figure 7. AtMYB12 overexpression enhanced insect resistance in tobacco plants. (A) Insect bioassay for aphids on wild-type (WT), AtMYB12 overexpression tobacco (OE-1 and OE-2). (B) Insect bioassay for aphids on WT, AtMYB12 overexpression tobacco (OE-1 and OE-2), WT tobacco pre-sprayed with water (CK) or 1, 2, 4 mM rutin. The values are the means ± SD (n = 12, *p < 0.05, **p < 0.01).
The appropriate accumulation of plant secondary substances, such as flavonoids, has been illustrated to demonstrate insect resistance in plants (Thoison et al., 2004; Sharma et al., 2018). The AtMYB12-expressing transgenic tobacco lines have been shown to develop resistance against the tobacco major insect pests S. litura and H. armigera because of the ameliorated content of rutin. In addition, artificial microRNA suppressed flavonol biosynthesis, which brought about the reversion of insect resistance in the AtMYB12-expressing tobacco (Misra et al., 2010; Pandey et al., 2012). This study showed that tobacco plants are subject to attack by numerous different microbial pathogens, such as bacteria and fungi. The transgenic tobacco lines showed enhanced resistance against R. solanacearum, A. alternata, and C. nicotianae Averna.
Studies have recently demonstrated that rutin plays a critical role in the protective mechanism of a plant; it has been shown that rutin is involved in herbivore resistance, and that there is a positive correlation between the insects on tobacco leaves (Hyodo et al., 2017) and the inter-specific derivatives of groundnut (Mallikarjuna et al., 2004). Rutin also inhibits P. atrosepticum pathogen growth in vitro at physiological concentrations (Kröner et al., 2012). Rutin was also one of the flavanoids that increase the susceptibility to B. cinerea in both tomato and grape (Zhang et al., 2015). For the recently research, rutin was defined as an resistance inducer which can induce plant resistance to pathogens. The activation of the immune mechanism is usually accompanied by the up-regulated expression of PR genes. At the same time, the expression of nitric oxide pathway genes and reactive oxygen species will also increase separately or coordinately (Yang et al., 2016), this results is equivalent to our results mentioned above. NtMYB12 is a highly homologous gene of AtMYB12 in tobacco and has been shown to be involved in regulating tobacco tolerance to abiotic stress by increasing the content of tobacco flavonoids (Song et al., 2020). Therefore, NtMYB12 may also play a role in the regulation of tobacco resistance to pests and diseases.
High flavonoid content can act as physical barriers, antimicrobials, or signals (Chen et al., 2014; Nayak et al., 2015). In this study, we showed that AtMYB12-overexpressing tobacco increased resistance to pathogens, and this plant defense may be caused by the high accumulation of rutin. However, the way in which its underlying molecular mechanism is controlled by rutin is a complicated issue. Due to various enemies of varied biotypes, such as microbial pathogens and insects, plants have countermeasures of evolution to protect themselves from potential invaders. As a result of the recognition of pathogen, plants activate copious defensive molecules to act as early responses (Jones and Dangl, 2006). In addition, plants also evolved into a systemic immune mechanism to resist the attack of pathogenic bacteria. Plants express a set of PR proteins that are correlated with the onset of systemic acquired resistance (SAR), and it has been indicated when some of these proteins were over-expressed in plants, the plant antimicrobial activity in vitro or the conferred resistance could be developed (Finkina et al., 2017).
Promoting plant defense gene expression, directly or indirectly, has been shown to control plant diseases. Many plant genes have been reported to be involved in enhanced resistance (Kou and Wang, 2009), most of which are reported to auto-activate the expression of the plant defense gene ahead of pathogen attack, which also mediates economic loss. In AtMYB12 transgenic plants, the expression levels of defense-related genes, such as NbrothB and NbNOA1, were even lower than that in the control plants when biotic stress was absent (Figure 2A). The NbPR1a expression was almost the same as it was in the control plant without inoculation. After inoculation with R. solanacearum, NbrothB and NbNOA1 were up-regulated only in the AtMYB12 transgenic plants, and NbPR1a was up-regulated approximately 100-fold in both the transgenic plants and the wild type 6 and 12 h post inoculation. This resistance was associated with different NbPR1 expressions 24 h post inoculation, and the transgenic plants showed 187-fold more expression level compared with the level in the wild-type leaves. The expression pattern of plant defense-related gene was highly similar to the pattern in leaves sprayed with quercetin; this pattern has been shown to prime a plant defense mechanism (Jia et al., 2010). This type of priming defense was regarded as a way to safely increase resistance ability in crops (Taheri and Tarighi, 2009; Zhang et al., 2019; Zhao et al., 2020). The flavonoid regulator AtMYB12 is a good candidate for breeding multifunctional resistance crops in the future.
The current perception about plant defense-related biochemical aspects, especially the roles of ROS and NO, can be found in some excellent reviews (Yoshioka et al., 2009; Kreslavski et al., 2012). When plants are under aerobic conditions, or attacked by biotic and abiotic elements, ROS will be produced continuously. ROS can be regarded as signaling molecules involved in a handful of developmental processes belonging to all organisms, in the basis of toxic by-products of aerobic elements (Nanda et al., 2010; Wang et al., 2012; Ma et al., 2017). In this study, the AtMYB12-expressing transgenic tobacco-enhanced resistance to R. solanacearum was accompanied by the accumulation of H2O2, an ROI species. The first burst of H2O2 occurred 3 h after inoculation, and the second burst occurred 24 h post inoculation (Figure 2B). This finding was similar to the non-host interaction between wheat and Uromyces fabae in which the first burst occurred at the location of attempted fungal penetration; a larger broad secondary burst happened at the whole cell level and coincided with a single-cell HR (Kong and Li, 2011; Zhang et al., 2011). According to current research on plants, NO is demonstrated to be a key signaling molecule; moreover, it orchestrates plethorical cellular activities to execute other progress, such as growth or development. It also plays important roles in triggering hypersensitive cell death and disease resistance in plants (Delledonne et al., 1998; Zhao et al., 2009; Du et al., 2012). In this study, the NO content of the transgenic leaves was much higher than that of the wild-type leaves after they were treated with R. solanacearum, and we observed approximately four- and five-fold induction of NbrbohB and NbNOA1 in the transgenic AtMYB12 tobacco leaves 6 h after inoculation compared with the levels in wild-type leaves (Figure 2A). We hypothesized that after inoculation with R. solanacearum, the AtMYB12 expression improved the NO and H2O2 contents and induced the expression of plant defense genes NbrbohB and NbNOA1constituting an aerobic metabolism system that results in an intense and faster response to biotic stresses from pathogen attacks compared with the wild-type tobacco. In addition, this study showed that the transgenic tobacco lines developed resistance against aphids (Figure 7A). Thus, AtMYB12 will be very beneficial for controlling insect pests in the future.
The AtMYB12 expression in tobacco not only increased the concentrations of flavonoids but also enhanced the defense against R. solanacearum, A. alternata, C. nicotianae Averna, and insect aphid; these results imply a route for the industrial production of polyphenols and high-resistance tobacco for agricultural production and for crop varieties. In conclusion, the flavonoid regulator AtMYB12 is a good candidate for breeding multifunctional resistance crops in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YL and XD designed the project and collected the samples. XD and HZ performed the experiments. ML and XZ analyzed the data. XD and HZ wrote the manuscript with the help of ZC, XZ, and XD. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (31801867 and 31872925), Shandong Provincial Key Research and Development Plan (2019JZZY020608, 2019GNC106152, and 2020CXGC010803), the Natural Science Fund for Outstanding Young Scholars of Shandong Province (JQ201807), and the Youth Innovation and Technology Support Program (2019KJF023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.694333/full#supplementary-material
Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7, 202–209. doi: 10.1016/j.pbi.2004.01.013
Chen, X., Mukwaya, E., Wong, M. S., and Zhang, Y. (2014). A systematic review on biological activities of prenylated flavonoids. Pharm. Biol. 52, 655–660. doi: 10.3109/13880209.2013.853809
Cheynier, V., Comte, G., Davies, K. M., Lattanzio, V., and Martens, S. (2013). Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 72, 1–20. doi: 10.1016/j.plaphy.2013.05.009
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
Du, Z. K.;, Zhu, L. S.;, Dong, M., Wang, J. H., Wang, J., Xie, H., and Zhu, S. (2012). Effects of the ionic liquid [Omim] PF6 on antioxidant enzyme systems, ROS and DNA damage in zebrafish (Danio rerio). Aquat. Toxicol. 124–125, 474–480. doi: 10.1016/j.aquatox.2012.08.002
Finkina, E. I., Melnikova, D. N., Bogdanov, I. V., and Ovchinnikova, T. V. (2017). Plant pathogenesis-related proteins PR-10 and PR-14 as components of innate immunity system and ubiquitous allergens. Curr. Med. Chem. 24, 1772–1787. doi: 10.2174/0929867323666161026154111
Hoensch, H. P., and Oertel, R. (2015). The value of flavonoids for the human nutrition: short review and perspectives. Clin. Nutr. Exp. 3, 86–99. doi: 10.1016/j.yclnex.2015.09.001
Hyodo, K., Suzuki, N., Mise, K., and Okuno, T. (2017). Roles of superoxide anion and hydrogen peroxide during replication of two unrelated plant RNA viruses in Nicotiana benthamiana. Plant Signal. Behav. 12:e1338223. doi: 10.1080/15592324.2017.1338223
Izaguirre, M. M., Mazza, C. A., Svatos, A., Baldwin, I. T., and Ballaré, C. L. (2007). Solar ultraviolet-B radiation and insect herbivory trigger partially overlapping phenolic responses in Nicotiana attenuata and Nicotiana longiflora. Ann. Bot. 99, 103–109. doi: 10.1093/aob/mcl226
Jakobs, R., Schweiger, R., and Müller, C. (2019). Aphid infestation leads to plant part-specific changes in phloem sap chemistry, which may indicate niche construction. New Phytol. 221, 503–514. doi: 10.1111/nph.15335
Jia, Z. H., Zou, B. H., Wang, X. M., Qiu, J., Ma, H., Gou, Z. H., et al. (2010). Quercetin-induced H2O2 mediates the pathogen resistance against Pseudomonas syringae pv. Tomato DC3000 in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 396, 522–527. doi: 10.1016/j.bbrc.2010.04.131
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kakar, K. U., Nawaz, Z., Cui, Z., Ahemd, N., and Ren, X. (2020). Molecular breeding approaches for production of disease-resilient commercially important tobacco. Brief. Funct. Genomics. 19, 10–25. doi: 10.1093/bfgp/elz038
Kariu, T., Nakao, R., Ikeda, T., Nakashima, K., Potempa, J., and Imamura, T. (2017). Inhibition of gingipains and Porphyromonas gingivalis growth and biofilm formation by prenyl flavonoids. J. Periodontal. Res. 52, 89–96. doi: 10.1111/jre.12372
Kong, X. P., and Li, D. Q. (2011). Hydrogen peroxide is not involved in HrpN from Erwinia amylovora-induced hypersensitive cell death in maize leaves. Plant Cell Rep. 30, 1273–1279. doi: 10.1007/s00299-011-1038-6
Kou, Y. J., and Wang, S. P. (2009). Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185. doi: 10.1016/j.pbi.2009.12.010
Kreslavski, V. D., Los, D. A., Allakhverdiev, S. I., and Kuznetsov, V. V. (2012). Signaling role of reactive oxygen species in plants under stress. Russ. J. Plant Phys. 59, 141–154. doi: 10.1134/S1021443712020057
Kröner, A., Marnet, N., Andrivon, D., and Val, F. (2012). Nicotiflorin, rutin and chlorogenic acid: phenylpropanoids involved differently in quantitative resistance of potato tubers to biotrophic and necrotrophic pathogens. Plant Physiol. Biochem. 57, 23–31. doi: 10.1016/j.plaphy.2012.05.006
Lei, Z. T., Sumner, B. W., Bhatia, A., Sarma, S. J., and Sumner, L. W. (2019). UHPLC-MS analyses of plant flavonoids. Curr. Protoc. Plant Biol. 4:e20085. doi: 10.1002/cppb.20085
Li, H., Flachowsky, H., Fischer, T. C., Hanke, M. V., Forkmann, G., Treutter, D., et al. (2007). Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226, 1243–1254. doi: 10.1007/s00425-007-0573-4
Li, N., Kong, L. G., Zhou, W. H., Zhang, X., Wei, S. T., Ding, X. H., et al. (2013). Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant CellTiss. Org. 115, 429–441. doi: 10.1007/s11240-013-0374-3
Li, Y., Chen, M., Wang, S. L., Ning, J., Ding, X. H., and Chu, Z. H. (2015). AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco. Plant Cell Tiss. Org. 122, 309–319. doi: 10.1007/s11240-015-0767-6
Luo, J., Butelli, E., Hill, L., Parr, A., Niggeweg, R., Bailey, P., et al. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326. doi: 10.1111/j.1365-313X.2008.03597.x
Ma, Q. J., Sun, M. H., Lu, J., Zhu, X. P., Gao, W. S., and Hao, Y. J. (2017). Ectopic expression of apple MdSUT2 gene influences development and abiotic stress resistance in tomato. Sci. Hortic. 220, 259–266. doi: 10.1016/j.scienta.2017.04.013
Maddox, C. E., Laur, L. M., and Tian, L. (2010). Antibacterial activity of phenolic compounds against the phytopathogen Xylella fastidiosa. Curr. Microbiol. 60, 53–58. doi: 10.1007/s00284-009-9501-0
Mallikarjuna, N., Kranthi, K. R., Jadhav, D. R., Kranthi, S., and Chandra, S. (2004). Influence of foliar chemical compounds on the development of Spodoptera litura (Fab.) in interspecific derivatives of groundnut. J. Appl. Entomol. 128, 321–328. doi: 10.1111/j.1439-0418.2004.00834.x
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Misra, P., Pandey, A., Tiwari, M., Chandrashekar, K., Sidhu, O. P., Asif, M. H., et al. (2010). Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 152, 2258–2268. doi: 10.1104/pp.109.150979
Nanda, A. K., Andrio, E., Marino, D., Pauly, N., and Dunand, C. (2010). Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 52, 195–204. doi: 10.1111/j.1744-7909.2010.00933.x
Napal, G. N. D., Carpinella, M. C., and Palacios, S. M. (2009). Antifeedant activity of ethanolic extract from flourensia oolepis and isolation of pinocembrin as its active principle compound. Bioresour. Technol. 100, 3669–3673. doi: 10.1016/j.biortech.2009.02.050
Nayak, B., Liu, R. H., and Tang, J. (2015). Effect of processing on phenolic antioxidants of fruits, vegetables, and grains-a review. Crit. Rev. Food Sci. Nutr. 55, 887–918. doi: 10.1080/10408398.2011.654142
Pandey, A., Misra, P., Chandrashekar, K., and Trivedi, P. K. (2012). Development of AtMYB12-expressing transgenic tobacco callus culture for production of rutin with biopesticidal potential. Plant Cell Rep. 31, 1867–1876. doi: 10.1007/s00299-012-1300-6
Qian, H. F., Chen, W., Li, J. J., Wang, J., Zhou, Z., Liu, W., et al. (2009). The effect of exogenous nitric oxide on alleviating herbicide damage in Chlorella vulgaris. Aquat. Toxicol. 92, 250–257. doi: 10.1016/j.aquatox.2009.02.008
Sharma, A., Verma, P., Mathur, A., and Mathur, A. K. (2018). Genetic engineering approach using early Vinca alkaloid biosynthesis genes led to increased tryptamine and terpenoid indole alkaloids biosynthesis in differentiating cultures of Catharanthus roseus. Protoplasma 255, 2070–2072. doi: 10.1007/s00709-017-1151-7
Song, Z. P., Luo, Y., Wang, W. F., Fan, N. B., Wang, D. B., Yang, C., et al. (2020). NtMYB12 positively regulates flavonol biosynthesis and enhances tolerance to low Pi stress in Nicotiana tabacum. Front. Plant Sci. 24:1683. doi: 10.3389/fpls.2019.01683
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Taguri, T., Tanaka, T., and Kouno, I. (2006). Antibacterial spectrum of plant polyphenols and extracts depending upon hydroxyphenyl structure. Biol. Pharm. Bull. 29, 2226–2235. doi: 10.1248/bpb.29.2226
Taheri, P., and Tarighi, S. (2009). Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J. Plant Physiol. 167, 201–208. doi: 10.1016/j.jplph.2009.08.003
Thoison, O., Sévenet, T., Niemeyer, H. M., and Russell, G. B. (2004). Insect antifeedant compounds from Nothofagusdombeyi and N. pumilio. Phytochemistry 65, 2173–2176. doi: 10.1016/j.phytochem.2004.04.002
Wang, N., Xu, H. F., Jiang, S. H., Zhang, Z. Y., Lu, N. N., Qiu, H. R., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wang, R. K., Li, L. L., Cao, Z. H., Zhao, Q., Li, M., Zhang, L. Y., et al. (2012). Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Plant Mol. Biol. 79, 123–135. doi: 10.1007/s11103-012-9899-9
Yang, W., Xu, X. N., Li, Y., Wang, Y. Z., Li, M., Wang, Y., et al. (2016). Rutin-mediated priming of plant resistance to three bacterial pathogens initiating the aarly SA signal pathway. PLoS ONE 11:e0146910. doi: 10.1371/journal.pone.0146910
Yoshioka, H., Asai, S., Yoshioka, M., and Kobayashi, M. (2009). Molecular mechanisms of generation for nitric oxide and reactive oxygen species, and role of the radical burst in plant immunity. Mol. Cells 28, 321–329. doi: 10.1007/s10059-009-0156-2
Zhang, H. C., Wang, C. F., Cheng, Y. L., Wang, X. J., Li, F., Han, Q. M., et al. (2011). Histological and molecular studies of the non-host interaction between wheat and Uromyces fabae. Planta 234, 979–991. doi: 10.1007/s00425-011-1453-5
Zhang, Y., Stefano, R. D., Robine, M., Butelli, E., Bulling, K., Hill, L., et al. (2015). Different reactive oxygen species scavenging properties of flavonoids determine their abilities to extend the shelf life of tomato. Plant Physiol. 169, 1568–1583. doi: 10.1104/pp.15.00346
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., Qi, C. H., You, C. X., et al. (2019). Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643. doi: 10.1007/s00425-019-03115-4
Zhao, J. P., Jiao, H. C., Song, Z. G., and Lin, H. (2009). Effects of l-arginine supplementation on glucose and nitric oxide (NO) levels and activity of NO synthase in corticosterone-challenged broiler chickens (Gallus gallus). Comp. Biochem. Phys. C. 150, 91–93. doi: 10.1016/j.cbpc.2009.07.003
Keywords: flavonols, plant defense, secondary metabolism, tobacco, transcription factor
Citation: Ding X, Zhang H, Li M, Yin Z, Chu Z, Zhao X, Li Y and Ding X (2021) AtMYB12-Expressing Transgenic Tobacco Increases Resistance to Several Phytopathogens and Aphids. Front. Agron. 3:694333. doi: 10.3389/fagro.2021.694333
Received: 13 April 2021; Accepted: 22 July 2021;
Published: 17 August 2021.
Edited by:
Ioannis Vagelas, University of Thessaly, GreeceReviewed by:
Sreepriya Pramod, Altria, United StatesCopyright © 2021 Ding, Zhang, Li, Yin, Chu, Zhao, Li and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Li, eWFuZ2xpMTk4OEBzZGF1LmVkdS5jbg==; Xinhua Ding, eGhkaW5nQHNkYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.