- Mycology and Plant Pathology Laboratory, Department of Botany, Visva Bharati University, Santiniketan, India
Over the past few decades, the massive increase in anthropogenic activity and industrialization processes has increased new pollutants in the environment. The effects of such toxic components (heavy metals, pesticides, etc.) in our ecosystem vary significantly and are of significant public health and economic concern. Because of this, environmental consciousness is increasing amongst consumers and industrialists, and legal constraints on emissions are becoming progressively stricter; for the ultimate aim is to achieve cost-effective emission control. Fortunately, certain taxonomically and phylogenetically diverse microorganisms (e.g., sulfur oxidizing/reducing bacteria) are endowed with the capability to remediate such undesired components from diverse habitats and have diverse plant-growth-promoting abilities (auxin and siderophore production, phosphate solubilization, etc.). However, the quirk of fate for pollutant and plant-growth-promoting microbiome research is that, even with an early start, genetic knowledge on these systems is still considered to be in its infancy due to the unavailability of in-depth functional genomics and population dynamics data from various ecosystems. This knowledge gap can be breached if we have adequate information concerning their genetic make-up, so that we can use them in a targeted manner or with considerable operational flexibility in the agricultural sector. Amended understanding regarding the genetic basis of potential microbes involved in such processes has led to the establishment of novel or advanced bioremediation technologies (such as the detoxification efficiency of heavy metals), which will further our understanding of the genomic/genetic landscape in these potential organisms. Our review aimed to unravel the hidden genomic basis and eco-physiological properties of such potent bacteria and their interaction with plants from various ecosystems.
Introduction
Delineating the genetic landscape of pollutant-degrading microbiomes and their use in eliminating environmental pollutants has always been preferred over physicochemical treatment strategies. Nevertheless, use of a microbe-assisted bioremediation process started a century ago when it's in the late 1960's it was used to remove contaminated oil mixed with water (Pieper and Reineke, 2000). Later, this new treatment approach emerged as one of the most promising methods for bioremediation in various areas for petroleum hydrocarbon degradation as well as in other contaminated habitats (Antizar-Ladislao, 2010). Among the major environmental pollutants, heavy metals and organic compounds, such as mineral oil hydrocarbons, polyaromatic and halogenated hydrocarbons, and benzene derivatives, severely affected the terrestrial as well as aquatic ecosystem or niches (Dvorák et al., 2017). However, among the existing environmental pollutants or contaminants, heavy metals achieved momentous attention due to their deadly long-term consequences on our society. In general, the toxic class of heavy metals includes Hg, Cr, Pb, Zn, Cu, Ni, Cd, As, Co, and Sn, which are released from the fertilizer and pesticide industry, sewage sludge, energy and fuel production, mining, combustion of fossil fuels, electroplating, photography, and electric appliance manufacturing. Notably, their long-persistent or recalcitrant chemical nature eventually has a hazardous impact on plant and human health (see Table 1 for detailed information) as well as on other living organisms; this simultaneously damages our eco-friendly framework and the ecological dynamics of their native beneficial microbial community (Ahemad, 2019). In the context of the present scenario, it has become imperative for us to concentrate our efforts toward creating novel bio-based and cost-effective sustainable remediation technology. Opportunely, certain taxonomically and phylogenetically diverse microorganisms (e.g., sulfur oxidizing/reducing bacteria) are endowed with the metabolic potential to remediate such undesired components from various habitats, along with having diverse plant-growth-promoting abilities (phytohormones and siderophore production, phosphate solubilization, etc.) (Zhuang et al., 2007; Bashan et al., 2008; Das et al., 2014). Some of the well-known bacterial genera that are encompassed with a dual potential, such as metal detoxification and plant growth promotion (PGP), are as follows: Pseudomonas spp., Psychrobacter spp., Bacillus sp., Bradyrhizobium spp., Achromobacter spp., Rhizobium spp., Methylobacterium spp., Azotobacter spp., Variovox spp., and Ochrobactrum spp. (Ahemad, 2019). Although there are ample reports that describe the isolation and characterization of potential microbial candidates with desired traits from various habitats, in depth knowledge regarding the genomic basis of the aforementioned physiologic potential is still lacking. It is noteworthy to mention that, with the rapid development of industrialization processes and anthropogenic activity, the increase of new pollutants will occur endlessly along with existing ones in our ecosystem. Thus, improved understanding regarding the genetic make-up of potential microbial candidates will help us to design engineered recombinant microbial strains with trait-specific desired properties, encompassed with better efficacy.
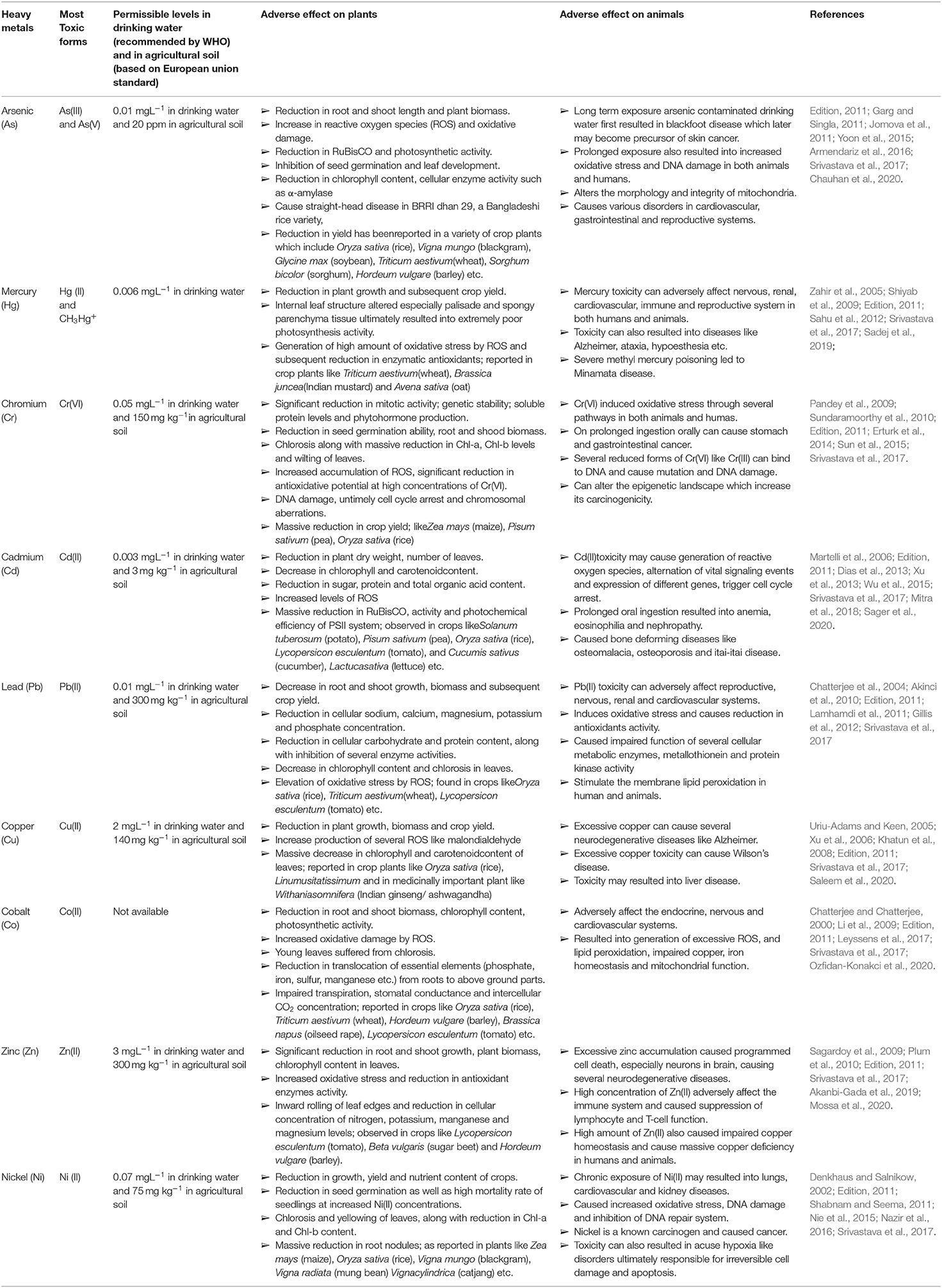
Table 1. Detailed information regarding permissible limit and adverse effect of different studied heavy metals/metalloid on plants and animals.
In this review, we aimed to unravel the comprehensive molecular mechanistic details or metabolic capacities of heavy-metal-metabolizing microbiota isolated thus far from diverse habitats and their potential use in the agroeconomic sector in the light of plant-growth-promoting attributes. Furthermore, we also tried to summarize the information on how modern meta-omics-based approaches are used in the revelation of bacterial community architecture and complex interactions of bioremediating microbiome. Further discussion has also been extended in relation to the prospects of microbe-mediated phytoremediation processes with special references to metallophytes.
Overview of the Physiological Strategy of Metal Detoxification or Immobilization Used by Potential Microbial Members
High concentrations of different heavy metals, like arsenic (As), mercury (Hg), cadmium (Cd), chromium (Cr), and lead (Pb), pose a very serious threat to our environment, causing different toxic effects both at the physiological and cellular/molecular level (see Table 1; for detailed information) viz., inhibition of several enzyme actions due to the binding of Hg2+, Cd2+, and Ag2+ with SH groups of concerned enzymes, generation of oxidative stress, and inhibition of activity of sulfate and phosphate groups containing compounds by structurally-related chromate and arsenate (Cervantes and Campos-García, 2007; Ventura-Lima et al., 2011). Although metals like manganese (Mn) Zinc (Zn), copper (Cu), nickel (Ni), molybdenum (Mo), and cobalt (Co) participate in the regulation of some important biochemical metabolic pathways in both prokaryotes and eukaryotes, higher concentrations of these heavy metals exert toxicity on humans, animals, and plants (see Table 1). Commonly, six widely known heavy metal resistance mechanisms have been characterized so far in different bacterial members (see Figure 1).
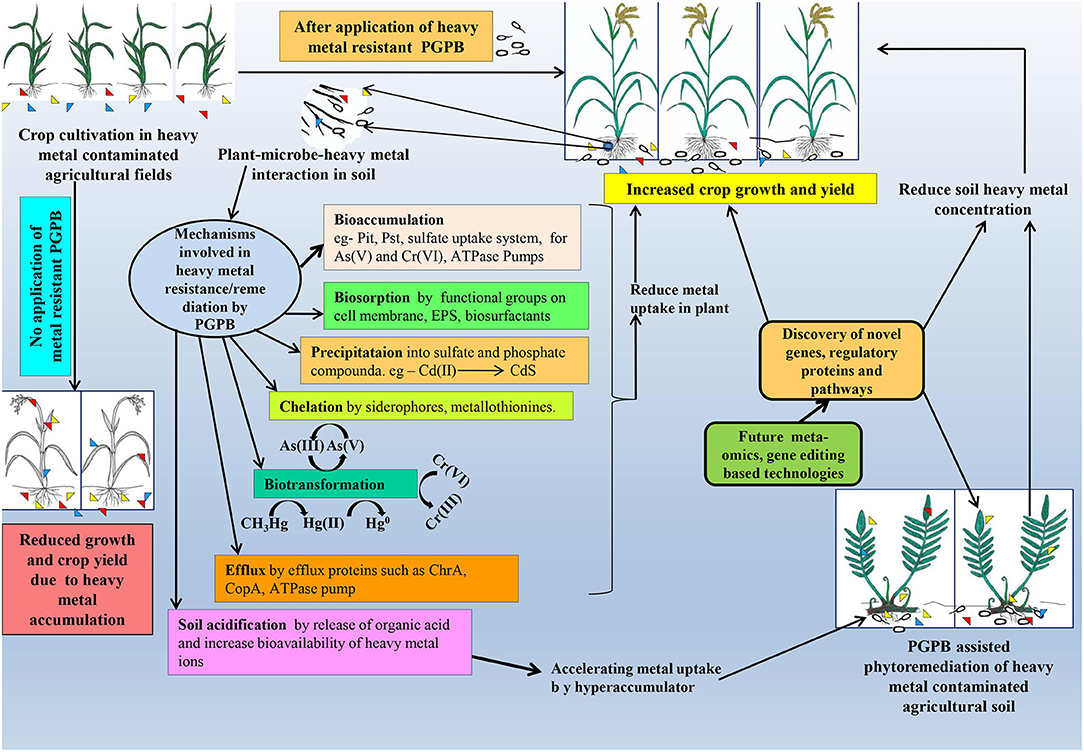
Figure 1. Thematic representation of microbe-assisted heavy metal remediation and its consequence on plant systems.
Exclusion of Heavy Metals by Permeability Barriers
Several bacteria (strains of Klebsiella aerogenes, Pseudomonas putida, and Arthrobacter viscosus) undergo structural alteration of the cell envelop, cell wall, plasma membrane, and surface layer (S-layer) in response to higher concentrations of certain heavy metals in their extracellular milieu. These changes ultimately reduce the permeability of different heavy metals (Bruins et al., 2000). Upon exposure to metal stress, such microbes started to secrete numerous components like exopolysaccharides (EPS), extracellular proteins, carbohydrates, and humic acids. These components in turn interact with heavy metals and prevent them from entering into the cell. Moreover, anionic functional groups like hydroxyl, carboxyl, amine, sulfhydryl, or sulfonate amide, which are the major constituents of EPS/S-layer and cell envelop, also caused unspecific entrapment of different heavy metals (Bruins et al., 2000; Etesami, 2018). A recent study by Wani et al. (2020) reported the biosorption of Cr(VI) by EPS and the biofilm-producing strain of Paenibacillus thiaminolyticus. Similarly, Upadhyay et al. (2017) also reported the involvement of EPS and biofilm in Zn(II) adsorption as well as Zn(II) resistance by plant-growth-promoting members of Pseudomonas sp. Apart from extracellular adsorption, a few earlier studies also reported the periplasmic binding of heavy metals such as for Cu(II) (Cha and Cooksey, 1991; Silver and Walderhaug, 1992; Silver and Ji, 1994; Bruins et al., 2000). However, both of these extracellular and periplasmic adsorptions strategy ultimately immobilized heavy metals in diverse environments (see Figure 1).
Extra and Intracellular Sequestration
An extracellular sequestration mechanism detoxifies heavy metals by forming stable complexes with them and ultimately reducing their bioavailability (Etesami, 2018). For instance, siderophore (Pyridine-2,6-bis thiocarboxylic acid type; pyoverdine, pyochelin, hydroxamate) of bacterial origin (Pseudomonas spp., Streptomyces spp. etc.) is able to form such complexes with several heavy metals (As, Cd, Hg, Cu, Ni, Zn, and Pb) and reduce their toxicity (Zawadzka et al., 2007; Dimkpa et al., 2008). Notably, a study conducted by Złoch et al. (2016) reported increased production of three different types of siderophore (hydroxymates, catecholates, and phenolates) by members of Streptomyces spp. under cadmium stress. They also suggested a possible role of ferrioxamine B (a hydroxamate type siderophore) in enhancing cadmium tolerance in Streptomyces sp. Apart from siderophore, several microbes (such as Bacillus spp. and Pseudomonas spp.) are able to produce different types of biosurfactants (surfactin, rhamnolipids, etc.) which can bind with diverse heavy metals (such as Cu or Zn) and reduce their bioavailability (Mulligan et al., 2001; Yang et al., 2018; Md Badrul Hisham et al., 2019; Sun et al., 2021).
However, in the case of intracellular sequestration, cytoplasmic hyperaccumulation of heavy metals involved several metal binding proteins like metallothioneins, metallo-chaperons, and many other low-molecular-weight cystine-rich proteins. In general, metallothioneins are low-molecular-weight cystine-rich proteins which reduce the bioavailability of noxious heavy metals by binding with them (Bruins et al., 2000; Blindauer et al., 2002; Naik et al., 2012). Several divalent cations of Cu, Cd, Zn, Pb, and As are found to be sequestrated intracellularly in this manner (Bruins et al., 2000). For example, Synechococcus PCC 7942 strain-derived SmtA protein is able to sequester four zinc ions per molecules (Blindauer et al., 2002). Similarly another plant growth-promoting strain of Pseudomonas was found to produce metallothioneins for extracellular sequestration of Pb(II) (Naik et al., 2012). Moreover, several bacteria are also able to accumulate large amounts of heavy metal ions intracellularly by compartmentalization or by cytoplasmic sequestration where various cellular proteins were involved for this (glutathione, metallothioneins, cystein rich proteins, etc.), thereby reducing the bioavailability of free metal ions (Etesami, 2018). A similar kind of observation by Kotoky et al. (2019) reported the involvement of glutathione S-transferase enzymes mediated Cd(II) detoxification system in members of Serratia marcescens.
Enzymatic Detoxification and Active Transport of Heavy Metals
Enzyme-mediated metal-detoxification mechanisms involve oxidation and/or reduction and demethylation and/or methylation reactions. This mechanism was found to be very effective in a wide range of bacteria for certain heavy metals e.g., Hg(II), As(V), and Cr(VI) (Bruins et al., 2000). For instance, organomercurial lyase and mercuric ion reductase are two such enzymes which can bind to the active site of Hg(II) (containing cystine sulfhydryl residue) and participate in hydrolyzing the stable carbon-mercury bond, finally forming a mercury thiolate adduct. This mercury thiolate adduct in turn was used as a substrate for mercuric ion reductase that finally reduced the Hg(II) into metallic mercury (Hg0) and forced it to diffuse back (mercury volatilization) into its natural environment (Bruins et al., 2000; Etesami, 2018). Moreover, Calzada Urquiza et al. (2017) reported that several plant growth-promoting members of Bacillus spp. were also capable of mercury volatilization. Enzymatic detoxification of arsenic involves oxidation of the more toxic As(III) into the less toxic As(V) by arsenite oxidase and conversion of arsenate (As5+) to the more toxic form aresenite (As3+) by the enzyme arsenate reductase, which may be exported out through some efflux system in response to the chemiosmotic gradient across the plasma membrane (Nies, 1999). So far as the other mechanisms are concerned, formation of metal sulfides and phosphate by anaerobic sulfur reducing bacteria (e.g., production of CdS and PbS by Klebsiella spp. or Alishewanella sp.), bacteria assisted heavy metal volatilization (conversion of arsenic into volatile derivatives by Clostridium spp.), and formation of gaseous substances has also been reported in several cases (Sharma et al., 2000; Meyer et al., 2007; Shi et al., 2018). Enzymatic detoxification system of Cr(VI) involves reduction of highly reactive Cr(VI) into less toxic Cr(III) by chromate reductase that has been observed in many bacteria like Sphingomonas spp. or Variovorax spp (Han et al., 2011; Bilal et al., 2018). Additionally, Wani et al. (2020) also reported an NADPH-dependent chromium reductase in Paenibacillus thiaminolyticus PS 5, having chromium resistance potential.
Notably, in certain cases, active transport of heavy metals is reported as a dominant mechanism in several bacteria (Bruins et al., 2000). These efflux systems can be linked to ATPase or not, but always have higher specificity for relevant cation or anion (Bruins et al., 2000). These efflux systems include Cd, Zn, and Pb by Cad-A system or active transport of Cd, Zn, and Co by Czc system, which have been reported in various bacteria such as Bacillus spp. or Pseudomonas spp. (Nies, 2003; Silver and Phung, 2005a). Furthermore, several plant growth-promoting bacteria are also known to have such an efflux system mediating their heavy metal resistance mechanism (see Supplementary Table 2 for detailed information).
On several occasions it has been observed that heavy-metal-induced oxidative stress can cause DNA damage which can in turn be countered by an indigenous DNA repair mechanism to some extent (Bruins et al., 2000). These resistant mechanisms may serve as potential plots for several remediation as well as industrial strategies, starting from metal recovery of industrial waste to remediation of metal contaminated soil. However, to achieve these, a comprehensive and improved understanding regarding the complex molecular mechanistic details of metal resistance in bacteria is very much necessary.
Molecular Mechanistic Insight Into Heavy Metal Resistance in Bacteria
Though there are no generalized resistance patterns for different heavy metals in bacteria, previous research has pointed out some major mechanisms which either solely or in combination are operating in diverse organisms. However, the genetic determinants for any heavy metal resistance in bacteria are either plasmid-born or a genome-integrated feature. Several factors, like concentration of heavy metals in the environment, types and toxicities of heavy metals, exposure time of organisms against those metals, and horizontal gene transfer through evolutionary time scale, play an imperative role in the development of their indigenous resistance property (Gadd, 1990). Here, we discuss the various molecular mechanisms of metal resistance in bacteria along with their utility in bioremediation purposes.
Resistance for Arsenic (As)
Arsenic is one of the most abundant elements on earth, often categorized as a heavy metalloid due to the presence of both metal and nonmetal moiety (Garg and Singla, 2011). It can exist in over 200 different forms, of which arsenopyrite is the most abundant (Mandal and Suzuki, 2002; Garg and Singla, 2011). Arsenic is a non-essential toxic element which mostly have two highly toxic forms, arsenite or As(III) and arsenate As(V), that can cause serious health hazards both in plants and animals (see Table 1 for detailed information). Among As(III) and arsenate As(V), As(V) is most abundant on the earth's crust whereas As(III) is mostly predominant in reducing environments (Nies, 1999; Mandal and Suzuki, 2002; Garg and Singla, 2011). Between them, As(III) has been found to be more soluble and toxic compared to As(V) (Mandal and Suzuki, 2002; Jomova et al., 2011). Arsenic, when present in combination with sulfur, oxygen, and chlorine, is generally referred to as inorganic arsenic; when in combination with carbon and hydrogen, it is commonly referred to as organic arsenic which is much less toxic than its inorganic counterparts (Mandal and Suzuki, 2002; Jomova et al., 2011). Having close structural similarities with (PO4)3−, As(V) frequently accumulates into cells via (PO4)3− transporting Pit or Pst system and results in 100-fold higher accumulation (Nies and Silver, 1995). Thus, arsenate toxicity often causes the inhibition of different phosphate transport systems (Nies and Silver, 1995; Nies, 1999). Bacteria had several arsenic resistance mechanisms viz. reduction of arsenate to arsenite and efflux of arsenite, oxidation of arsenite to arsenate, or intracellular accumulation and methylation of this metal (Andres and Bertin, 2016). In this context, operons like arsRBC and arsRDABC are well-characterized in diverse bacteria and located either in chromosome or in plasmid. For instance, arsRBC contain three genes, arsR, arsB, and arsC, which were discovered in the chromosome of E. coli (Diorio et al., 1995) and in plasmids pI258 (Ji and Silver, 1992) and pSX267 (Rosenstein et al., 1992) of Staphylococcus aureus and S. xylosus, respectively. On the contrary, arsRDABC operon contain two extra set of genes, arsA and arsD, that were initially discovered in plasmids R773 (Figure 2) and R46 of strains of E. coli (Chen et al., 1986; Bruhn et al., 1996). Both aforementioned operons are As(III) inducible. ArsR is a repressor protein which, in the absence of As(III), constitutively produce and bind to the operator region of both operons and prevents the transcription initiation process (Wu and Rosen, 1991). ArsD protein serves as a co-repressor of arsRDABC operon (Wu and Rosen, 1993). ArsC is an intracellular arsenate reductase which reduced As(V) into As(III) (Ji and Silver, 1992). Though arsenic reductase performs the same function in diverse organisms, their sequences vary greatly (Silver and Phung, 1996). ArsB is a transmembrane arsenite efflux pump protein that extruded out the As(III) from cytoplasm to cell surface in response to a chemiosmotic gradient (Chen et al., 1986), while ArsA is an AS(III) stimulated ATPase (Chen et al., 1986). According to Dey and Rosen (1995), both ArsA and ArsB formed a heterodimer (ArsA2B) which served as an ATP dependent efflux system of As(III). Moreover, in Microbacterium sp., arsenic resistance is also conferred by both ars and acr systems (see Figure 2), whereas arsC3 and acr3 are reported to transcribe in the opposite direction (Achour-Rokbani et al., 2010). Moreover, another arsenite efflux protein, namely Acr3, was found in various plant growth-promoting bacterial isolates like Pantoea agglomerans C1, Azospirillum brasilense Az39, Variovorax paradoxus S110, and Pseudompnas koreensis AGB-1 (Han et al., 2011; Armendariz et al., 2015; Babu et al., 2015; Luziatelli et al., 2020).
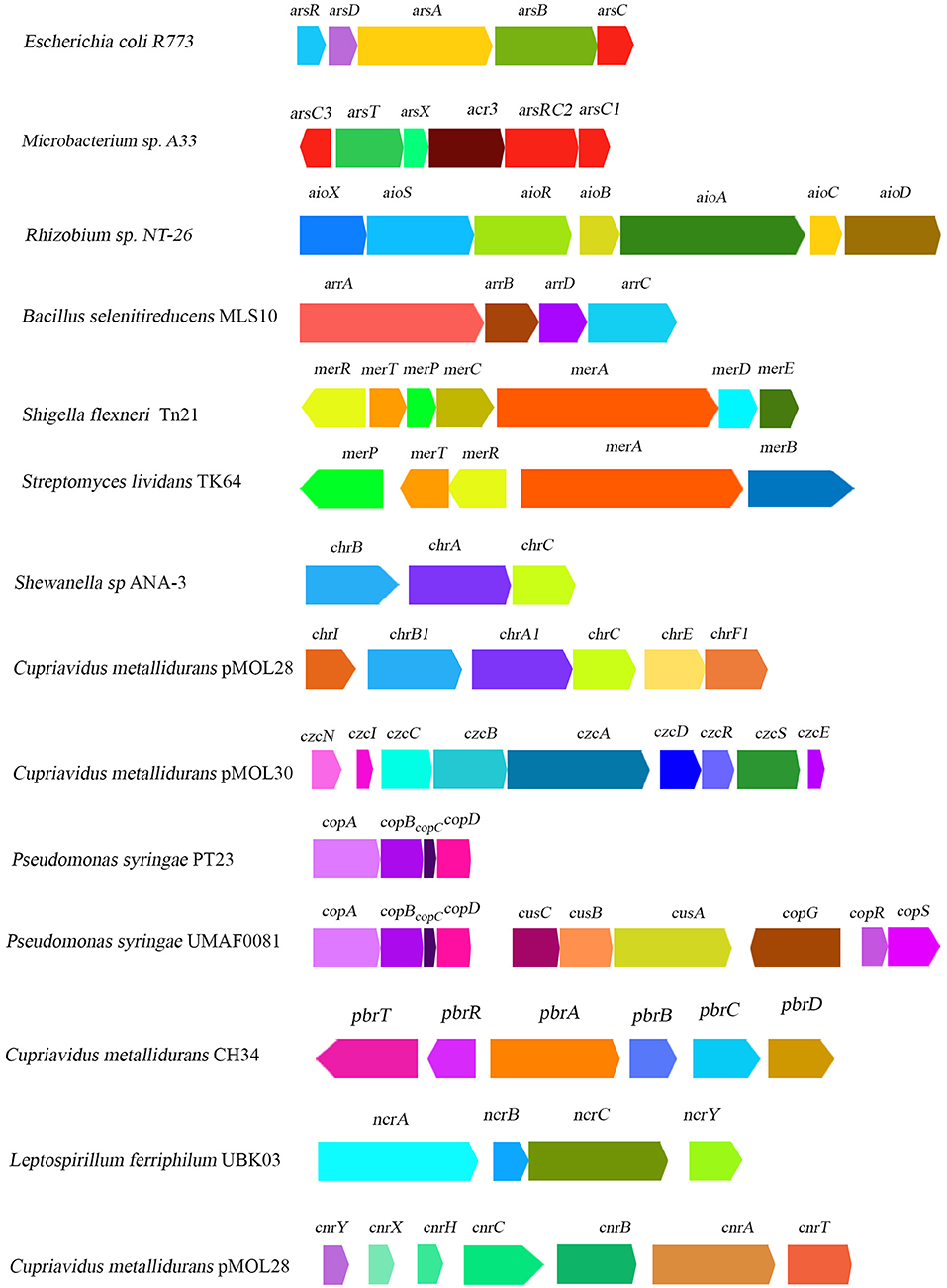
Figure 2. Schematic diagram illustrating the organization of heavy-metal-resistant related gene clusters reported from some major representative bacterial genera.
Notably, two other respiratory periplasmic, arsenite and arsenate oxidase (Anderson et al., 1992; Ahmann et al., 1994) enzymes, are known to be involved in arsenic resistance in bacteria (Silver and Phung, 2005a). Among all other bacterial isolates, arsenite oxidation system (another prime mechanism for arsenic resistance) is well-characterized in Alcaligenes faecalis where the first “arsenic gene island” was reported (Silver and Phung, 2005b; Phung et al., 2012). Arsenite oxidase is a heterodimeric protein that consists of two subunits: one large molybdopterin-containing subunit with an iron (3Fe-4S) center encoded by asoA/aoxA and another small rieske active subunit (Anderson et al., 1992) encoded asoB/aoxB genes (Silver and Phung, 2005b; Phung et al., 2012; Mallick et al., 2015). Upstream of the aoxA, there is a Twin Arginine Translocation (TAT) leader sequence which is involved in assemblage of arsenite oxidase and its translocation from cytoplasm to periplasm (Mallick et al., 2015). On the other hand, an anaerobic form of arsenate reductase (Arr) is a heterodimer (Mallick et al., 2015) and is encoded by arrA and arrB genes that were primarily characterized in Shewanella spp. (Saltikov and Newman, 2003) and in Bacillus selenitireducens (see Figure 2) (Afkar et al., 2003). Both cytoplasmic and periplasmic arsenate reductase, encoded by two different operons, i.e., arsDABC and arrAB respectively, were characterized in members of Shewanella (Saltikov et al., 2003). A few other arsenite oxidase encoding genes (e.g., aoxA, aoxB, aroB, and aroA) were also reported in different bacterial isolates, like Centibacterium arsenoxidans, and in Chemolithoautotrophic strain NT-26 (Muller et al., 2003; Santini and vanden Hoven, 2004; Silver and Phung, 2005b). Another gene, arsH encoded NADPH:FMN oxidase (ArsH) that oxidizes As(III) into As(V), has been reported in a few plant growth-promoting bacterial isolates (Pantoea agglomerans and Azospirillum brasilense; see Supplementary Table 2 for detailed information) (Armendariz et al., 2015; Luziatelli et al., 2020).
Both arsenite and arsenate reductase are coupled with inner membrane respiratory chain (Silver and Phung, 2005a), where arsenite oxidase serves as an electron donor while arsenate reductase serves as an electron acceptor (Ahmann et al., 1994). So far as the other mechanisms are concerned, biosorption of As(V) and As(III) on bacterial cell surface and methylation of inorganic arsenic in organic compounds (Thomas et al., 2004) are some of the most vital strategies for arsenic resistance, which have been reported in various bacteria. For example, Babu et al. (2015) reported the presence of gene bmtA (encoding metallothionein) in Pseudompnas koreensis, along with other arsenic resistance genes. On the other hand, methylation of arsenic and its subsequent volatilization is a multistep complex process and information is significantly lacking in this regard (Mallick et al., 2015). Another gene, namely arsM, was found to be responsible for arsenic methylation (Bertin et al., 2011). Mostly, arsenic resistance plant growth-promoting gram positive bacterial isolates belong to the phyla firmicutes and actinobacteria where Bacillus spp. was found to be the most abundant member. On the other hand, in the case of gram-negative bacterial isolates, they mostly belong to the phylum proteobacteria where Pseudomonas spp. holds a significant position. Among several bioremediation strategies used by bacterial isolates, biotransformation of As(V) and As (III) by enzymatic reduction and oxidation are found to be predominant, followed by arsenite efflux, bioaccumulation, and bio-adsorption strategy.
However, a few recent proteome-level investigations in Brevibacterium spp. and Exiguobacterium spp. shows that under metal stress [in the presence of 750 mg L−1 arsenic; As(III) and As(V)] conditions, various housekeeping proteins (ribosomal proteins or lipid metabolism-related proteins) are downregulated while proteins like superoxide dismutase, thioredoxin disulfide reductase, lipid hydroperoxide reductase, ArsR, ArsA, Cdr (encoding Co- enzyme A disulfide reductase), and ABC transporter are be upregulated (Castro-Severyn et al., 2019; Shah and Damare, 2020). Additionally, transcriptome analysis by Liu et al. (2020) also revealed the upregulation of areB gene in plant growth-promoting Paenibacillus polymyxa YCO136 in the presence of root exudates of tobacco.
Resistance for Mercury (Hg)
The most toxic bioavailable forms of mercury (Hg) are mercuric ions (Hg2+) and methyl mercury (CH3Hg+) (Nies, 1999). In general, microbe-assisted mercury detoxification mechanism involves enzymatic reduction of highly toxic mercuric ions (Hg2+) into less toxic volatile metallic mercury (Hg0), which passively diffuses from bacterial cells upon aeration (Silver and Walderhaug, 1992). A previous study identified that mercury resistance in microbes is conferred by mer operon that is located either in plasmid or in chromosomal DNA, and occasionally in the form of a transposons (see Figure 2) such as Tn21, characterized from plasmid NR1 in Shigella flexneri (Liebert et al., 1999; Silver and Phung, 2005a). This mer operon comprised of several functional genes, such as merRTPCADB, although their copy number and gene orientation synteny varied widely in different organisms (Silver and Walderhaug, 1992; Silver and Phung, 1996, 2005a). For instance, in Thiobacillus ferrooxidans, instead of one, two merR genes, merR1 and merR2, are present at a separate genetic locus (Inoue et al., 1991). MerR is a regulatory protein which contains a DNA binding region at carboxy terminal and a Hg2+ binding region at amino terminal (Silver and Phung, 2005a). MerR can both repress (in absence of Hg2+) and activate (in presence of Hg2+) the transcription initiation process of mer operon in taxonomically distantly related organisms (Silver and Phung, 1996).
MerT is an inner membrane Hg2+ transport protein encoded by merT gene, while merP and merC genes encode a periplasmic Hg2+ binding and transport protein respectively (Kusano et al., 1990). Notably, mer operon of gram-positive Streptomyces lividans is comprised of two divergent gene clusters (see Figure 2), out of which one cluster is encompassed with genetic determinants for MerR and MerT that are transcribed from right to left and another cluster contained gene merA and merB, transcribed from left to right (Sedlmeier and Altenbuchner, 1992). Mercuric reductase and organomercurial lyase are encoded by merA and merB, respectively (Silver and Phung, 1996). merB gene can also be found in bacterial isolates such as in plasmid pDU1358 of Serratia marcescens (Griffin et al., 1987) and in mer operon of several members of Pseudomonas spp. This operon-derived resistance sometimes represents the “narrow spectrum” that only contain merA gene and provides resistance for mercuric ions only, or it can be a “broad spectrum” which contains genes merA and merB that provide resistance against both Hg2+ and CH3Hg+ (Silver and Phung, 1996). Often, there is another genetic determinant, merD (encoding secondary regulatory protein), present at the end of mer operon that has been reported in many organisms (Mukhopadhyay et al., 1991).
Under mercury-stressed conditions, bacterial mer operon activates its genes. A small periplasmic protein MerP binds with the Hg2+ in the periplasm. Eventually, MerP protein is transferred from the Hg2+ to inner membrane protein MerT (Silver and Phung, 2005a). In this transfer event, no outer membrane protein is involved; therefore it can be considered as a passive process (Silver and Phung, 1996, 2005a). However, sometimes inner membrane proteins like MerC and MerF are also found to play similar roles in Hg2+ transport in some members (Kusano et al., 1990). Once on the inner surface of the cell membrane, Hg2+ is transferred to the homodimeric mercuric reductase, and then reduces Hg2+ into Hg0. Being volatile, Hg0 passively diffused out from an intracellular environment. On the other hand, more toxic organomercurials are detoxified by organomercurial lyase which follow a SE2 reaction pathway that break down the Hg-C bond by concerted proton attack. Subsequently, Hg2+ ions and reduced organic compounds are formed, such as methane after lysis of methyl mercury or benzene after lysis of phenylmercury (Silver and Phung, 1996; Begley and Ealick, 2004). A recent work related to Hg2+ resistance shed new light on MerT/MerP/MerA-mediated bioremediation potency in a marine strain of Pseudomonas pseudoalcaligenes (Zhang J. et al., 2020).
In recent years, a number of plant growth-promoting bacterial isolates (see Supplementary Table 2 for detailed information) were found to use mer operon encoded gene-mediated detoxification mechanisms (e.g., biotransformation). Notably, some other important mechanisms include merE gene-mediated detoxification of Hg2+ and CH3Hg+ (found in members of Bacillus) (Amin and Latif, 2016) and conversion of Hg2+ into less toxic and soluble HgS by plant growth-promoting members of Pseudomonas spp. or Enterobacter spp. (Tariq et al., 2015). Interestingly, gram positive bacterial isolates are mostly used for bioremediation of both Hg2+ and CH3Hg+ due to the existence of functional merA and merB genes, while gram-negative members mostly have a mercuric ion detoxification system, but lack an organomercurial system, except for a few Pseudomonas spp. which have merB gene for organomercurial lyase. However, a recent study by Calzada Urquiza et al. (2017) reported the presence of a merR gene for the first time in plant growth-promoting Bacillus spp. along with merA gene.
Resistance for Chromium (Cr)
Chromium (Cr) is one of the most abundant metals in the earth's crust, with an average concentration of 125 mg kg−1, while Cr(VI) or chromate and Cr(III), the trivalent cation, are two copious bioavailable forms of chromium, of which Cr(VI) is far more toxic than Cr(III) to all living cell (Nies, 1999; Sun et al., 2015). In bacteria, Cr(VI) is generally accumulated in the cell by sulfate uptake system which used Cr(VI) as an alternative substrate (Silver and Walderhaug, 1992). Reduction of intracellular Cr(VI) concentration by reducing its uptake system or by extrusion of Cr(VI) using membrane efflux pump or enzymatic reduction of Cr(VI) into Cr(III) are some of the imperative mechanisms of chromate resistance that have been reported in bacteria. The two most important genes, namely chrB and chrA (encode chromate efflux protein), for chromate resistance were initially sequenced and characterized from plasmid pUM505 (Cervantes et al., 1990) and pMOL28 (Nies et al., 1990) in Pseudomonas aeruginosa and Alcaligenes eutrophus CH34 (later named as Cupriavidus metallidurans CH34) respectively (Figure 2). The gene (chrB and chrA) expression system in Pseudomonas was found to be constitutive, while in Alcaligenes it is inducible nature (Cervantes et al., 1990; Nies et al., 1990). Viti et al. (2014) briefly reviewed genetic determinants of chromate detoxification system in bacteria (e.g., Cupriavidus metallidurans and Ochrobactrum tritici) which include several chr genes located in chr operons (chrFECAB) and encode proteins that belong to CHR superfamily (Branco et al., 2008; Monsieurs et al., 2011). ChrB served as a regulatory protein and was only activated by Cr(VI) or Cr(III) and sulfate in various organisms (Juhnke et al., 2002). However, chrC gene encoded a superoxide dismutase (SOD) (Juhnke et al., 2002). Gene clusters like chrABC are also found in plasmid of Shewanella sp. (Figure 2) (Aguilar-Barajas et al., 2008; Viti et al., 2014).
Notably, reduction of Cr(VI) to Cr(III) catalyzed by chromate reductase (ChrR), which was encoded by chrR or bio-adsorption of Cr(VI), is an alternate chromate resistance mechanism identified and characterized from several PGP bacteria (Sphingomonas sp.; Variovorax sp etc.) (Han et al., 2011; Bilal et al., 2018). Among different chromate bioremediating PGP bacterial members (see Supplementary Table 2 for detailed information), most are members of firmicutes, followed by actinobacteria. Whereas, gram negative isolates mostly belong to phylum proteobacteria where γ-proteobacteria were found to be the predominant class, followed by α-proteobacteria and other subsequent classes.
Resistance for Cadmium (Cd)
Cadmium is not an abundant metal in the Earth's crust and is mostly found in association with other elements such as Zinc, copper, and lead. The most toxic bioavailable redox form of cadmium is Cd2+ which is often taken up by Mg(II) uptake system (Nies, 1999). In bacteria, enzymatic detoxification of cadmium resistance mechanism is completely absent, because several divalent cations, like Cd(II), Zn(II), Co(II), and Ni(II), are quickly reoxidized into their former highly reactive toxic states, unlike passive removal of volatile Hg0 (Bruins et al., 2000). Efflux-pump-mediated resistance mechanism is the most predominant one for Cd(II) (Nies, 1999). Silver and Phung (2005a) summarized seven different types of metal ion efflux systems, of which CadA-, CzcCBA-, and CzcD-mediated efflux systems were responsible for Cd(II) efflux in various organisms. The CadA system (encode P-type ATPase protein) of Cd(II) efflux are well-characterized and were first discovered in plasmid pI258 of gram positive Staphylococcus aureus (Novick et al., 1979). Moreover, cadC encodes for a DNA binding repressor protein (able to bind at the operator-promoter site of DNA and prevent transcription) (Yoon et al., 1991) for transcriptional regulation of inducible cad operon. This cad operon was also found in a transposon element like Tn5422 in Listeria monocytogenes (Lebrun et al., 1994). When divalent metal ions like Zn2+/ Pb2+/Co2+ and Ni2+ bind with CadC protein, it is released from DNA and a transcription event occurs (Silver and Phung, 2005a).
Another well-characterized chemiosmotic efflux system is CzcCBA, driven by proton motive force. Necessary genes for Czc system are located in czcCBAD operon, either in chromosome or in plasmids of various organisms (e.g., plasmid pMOL30 of Cupriavidus metallidurans CH34, see Figure 2) (Nies et al., 1989; Gutiérrez-Barranquero et al., 2013). CzcD protein is a single membrane polypeptide efflux pump that belongs to the cation diffusion facilitator family (CDF) (Haney et al., 2005; Silver and Phung, 2005a). Whereas, CzcCBA protein complex is comprised of three polypeptides, of which a large inner membrane protein is CzcA, a smaller outer membrane protein is CzcC, and CzcB is a periplasmic protein. These CzcA and CzcB together formed a continuous channel from cytoplasm to the outside of the plasma membrane (Rensing et al., 1997; Silver and Phung, 2005a). Sometimes, CzcA alone can provide low levels of resistance for Cd(II), Co(II), and Zn(II), while CzcB creates a pathway for the removal of metal ions. CzcC creates a hole on the outer membrane surface for the release of those metal ions (Rensing et al., 1997). Moreover, adsorption of Cd(II) on cell surface by physical entrapment, ion exchange, or complexation prevents metals from entering inside the cell, and is also functional in several organisms (Bruins et al., 2000). However, resistance by Czc Cd(II) efflux pump and bio-adsorption on cell surface was found to be the predominant mechanism for cadmium resistance in different PGP bacterial isolates (e.g., Bacillus spp., Pseudomonas spp., Sphingomonas spp., Klebsiella spp., Enterobacter spp., Serratia spp., and Rhizobium spp. See Supplementary Table 2 for detailed information). Notably, a recent proteome-level investigation in Pseudomonas aeruginosa revealed that one third of the total upregulated proteins in cadmium amended culture are metalloproteins, suggesting their role in cadmium resistance (Izrael-Živković et al., 2018). Another separate proteomics-based study conducted by Zhang et al. (2019) shows that upregulation of Cd2+/Zn2+ exporting ATPase, type-VI protein secretion system, and glutathione-S-transferase under cadmium stress, which might be responsible for cadmium homeostasis in Burkhoderia cepacia strain GYP1 (Zhang et al., 2019). In this connection, a recent comparative transcriptome analysis by Wang H. et al. (2020) revealed a unique strategy: using montmorillonite(Mt) for enhanced cadmium tolerance in Chryseobacterium sp. WAL2. They put forward the fact that, in the presence of Mt and Cd(II) stress, the expression of genes related to efflux systems in strain WAL2 are found to be up-regulated. It has been observed that in the presence of Mt, expression of certain genes like czcA, czcB, and zntA increased several fold in Chryseobacterium sp. WAL2.
Resistance for Lead (Pb)
Divalent cation Pb2+ or Pb(II) is the most common bioavailable form, thus resistance against Pb(II) is mostly conferred by different efflux systems (Bruins et al., 2000). The Pb(II) resistance genetic determinants which are responsible for uptake, efflux, and accumulation of Pb(II) were first discovered in plasmid of Cupriavidus metallidurans strain CH34 (formerly known as Ralstonia metallidurans) (Borremans et al., 2001). All six genes for Pb(II) resistance are organized in two different operons: pbrABCD and pbrRT (Borremans et al., 2001). An earlier study revealed that the entire pbrABCD operon is co-transcribed into a single mRNA (Borremans et al., 2001; Silver and Phung, 2005a). However, pbr operon is an inducible one which is regulated by PbrR, a metal-sensing regulatory protein that belongs to Mer family (Chen et al., 2005). Another permease type protein, PbrT, is also involved in uptake of Pb(II). Gene pbrA encoded a P-type Pb(II) efflux ATPase of Cys-Pro-Cys soft metal efflux family and shared general sequence features with other P-type ATPase (Solioz and Vulpe, 1996). PbrB is an outer membrane protein involved in Pb(II) removal from periplasmic space to the extracellular side, whereas PbrC is a signal peptidase that cleaved off the signal peptide from a protein in periplasm (Borremans et al., 2001). Protein PbrD has Cys-rich metal binding sites and an important role in intracellular sequestration of Pb(II). Some other notable Pb(II) resistance mechanisms are adsorption of Pb(II) by EPS or by functional groups on cell wall or intracellular accumulation of Pb(II) as lead phosphate, reported in several organisms like Citrobacter freundii or Staphylococcus aureus (Levinson and Mahler, 1998). Notably, several reports revealed the involvement of pbrA gene in Pb(II) resistance in different plant growth-promoting bacteria which include members of Agrobacterium sp., Rahnella spp. Pseudomonas spp., Rhizobium spp., and Bacillus spp. (Jebara et al., 2015; Abdelkrim et al., 2018) Naik et al. (2012) reported another gene, namely bmtA, which encodes metallothionein and is involved in Pb(II) sequestration in several PGP bacteria like Pseudomonas aeruginosa. The occurrence of bio-adsorption was found to be key to the Pb(II)-resistant mechanism among PGP bacteria, followed by bioaccumulation and efflux by P-type ATPase.
Mechanism of Copper (Cu) Resistance
Unlike other heavy metals, Cu served as a vital micronutrient to all life forms. Cupric oxide [Cu(II)] and cuprous oxide [Cu(I)] are found to be the most abundant chemical forms that exist in the natural environment. Although it is necessary for certain physiological functions, excessive concentration of Cu(II) often imparts toxic side effects on various living entities. During the course of evolution, bacteria developed different mechanisms for Cu(II) resistance in response to high Cu(II) contaminants. Several such well-characterized systems include copYZAB system in Enterococcus hirae (Odermatt et al., 1992), cus (Franke et al., 2003; Gutiérrez-Barranquero et al., 2013), pco operon in plasmid pRJ1004 (Brown et al., 1995), cut systems in E. coli (Silver and Phung, 1996), and cop operon in Pseudomonas syringae (see Figure 2) (Gutiérrez-Barranquero et al., 2013).
For the last few decades, a number of seminal papers have been published in this regard which critically bring to the light the molecular tuning of Cu-metabolizing genes. For instance, in Enterococcus hirae, copper resistant gene cluster CopYZAB is organized in cop operon which activates upon copper starvation as well as in the presence of excessive copper concentration (Odermatt et al., 1992). In another separate study, deletion mutation of copA results in higher resistance in E. hirae, indicating that CopA might be involved in copper ions' uptake, especially Cu(I), while deletion of copB resulted in copper sensitivity (Silver and Phung, 1996). The expression of copA and copB are regulated by proteins CopY and CopZ ptotein, where CopY is an apo-repressor that is unable to bind directly with the DNA; instead, it forms CopY-Cu+ complex which in turn binds to the operator region and prevents transcription initiation process of cop operon (Odermatt and Solioz, 1995). On the other hand, CopZ is an anti-repressor protein which forms a CopZ- Cu+ protein complex and interacts with the CopY-Cu+ and finally results in removal of the repressor (Odermatt and Solioz, 1995). Similar to copYZAB in E. hirae, another operon, namely tcrYZAB, has also been reported to be involved in copper homeostasis in E. faecium and E. faecalis (Hasman, 2005).
In Pseudomonas syringae, cop operon encompassed with copABCD genes to encode four structural proteins: periplasmic copper binding protein (CopA), outer membrane protein (CopB), periplasmic copper binding protein (CopC), and inner membrane protein (CopD) (Silver and Phung, 1996). The regulation of this operon is solely dependent on two regulatory proteins, copR and copS; together they form two component regulation systems (Silver and Phung, 1996). Both CopA and CopC proteins are able to bind with the Cu(II) in the periplasmic site and reduce the Cu(II) toxicity. In this context, several earlier works suggested that bacteria can undergo colony color change under Cu stress. For instance, colonies of Pseudomonas turned to a blue color from a whitish one when they were grown in Cu(II)-containing medium (Cooksey, 1994; Silver and Phung, 1996). However, Cu resistance also came from some other operons like pco (encompassed with genes like pcoABCD and pcoRS) that were initially characterized in plasmids of E. coli (Cooksey, 1994). Although, gene clusters of pco operon encode cop operon-related proteins; only the former contain an additional genetic determinant pcoE, that is located downstream of other regulatory genes (Brown et al., 1995). Notably, cus system, which is characterized in E.coli, potentially encodes an RND efflux chemiosmotic carrier protein complex CusCFBA and cusRS (cusS encodes histidine kinase while cusR is a response regulator) involved in Cu(II) resistance only under anaerobic conditions (Franke et al., 2003). CusA is a member of the resistance-nodulation-division related (RND) protein and CusB is a membrane fusion protein (MFP). CusF is a copper-binding protein (Franke et al., 2003). Flow of Cu(II) and other heavy metals back and forth from the natural environment to inside the bacterial cell through proper uptake and efflux system is a vital eco-strategy that engaged a large portion of bioavailable heavy metals and ultimately reduced the possibility of their entry from soil to plant systems and eventually other living forms. The abovementioned Cu(II) resistance mechanisms can also be observed in different plant growth-promoting bacterial isolates (see Supplementary Table 2 for detailed information) and served as bioremediation agents and reduced Cu(II) toxicity in agricultural fields. Amongst such isolates, abundance of gram-negative bacteria occupied a higher position and mostly belonged to class α and γ proteobacteria, where members of Pseudomonas spp. holds a significant position. Use of P-type ATPase, which is either CopA- or CueA- (Adaikkalam and Swarup, 2002; Zhang and Rainey, 2007; Wei et al., 2009; Asaf et al., 2018) based efflux system of Cu(II), is found to be predominant in the Cu-remediating mechanism in PGP members. However, a few recent reports also exhibited the functionality of Cu(II) resistance by CusAB system in plant growth-promoting rhizobacteria i.e., Mesorhizobium amorphae 186 (Hao et al., 2015). Notably, Glibota et al. (2020) detected several genetic determinants (copA, copB, copC, pcoR, pcoA, and pcoD) that were responsible for conferring copper resistance in a large number of bacterial genera that include Bacillus spp., Chryseobacterium spp., Pseudomonas spp. Variovorax spp., and Stenotrophomonas spp. These were initially isolated from copper-compound-treated soil of an olive tree farm. Additionally, more copper resistance genetic determinants, tcrB (encoding copper export ATPase) and silE (encoding silver/copper periplasmic metal binding protein), were also detected in members of Enterobacter aerogenes and Pseudomonas sp.
Mechanism of Co(II) Resistance
The uptake of Cd(II) inside the cell by CorA system caused rapid accumulation of Co(II) (Smith et al., 1993). Mostly, Co(II) resistance occurred as a by-product of other resistance systems such as Cd(II), Zn(II), and Ni(II) (Nies, 1999). In gram-negative bacteria, Co(II) resistance is driven by trans envelop efflux system by RND protein (Nies, 1999). However, a widely reported Co(II) resistance system is the cation antiporter CzcCAB system, responsible for resistance to Cd(II), Co(II), and Zn(II), whereas CnrCBA and NccCBA efflux systems are responsible for Cd(II), Ni(II), Co(II), and Cd(II) resistance (Nanda et al., 2019). Notably, in Staphylococcus aureus, ZntA protein is responsible for Co(II) efflux (Xiong and Jayaswal, 1998) whereas in E. coli, gene yohM (rcnA) encodes a protein, RcnA/YohM, involved in Co(II) an Ni(II) resistance (Eitinger et al., 2005). There are several PGP bacteria that exhibited Co(II) resistance through different mechanisms. Out of these, CzcCAB-mediated efflux system was found to be the most prevalent and significant, followed by bio-adsorption and bioaccumulation mechanisms.
Mechanism of Zn(II) Resistance
Zinc is an important micronutrient required in trace amounts to maintain cellular function, thus zinc solubilization by bacteria is considered important event in nature (Kamran et al., 2017). However, at higher concentrations, Zn(II) can also cause severe health hazards in plants and animals (see Table 1). A study performed by Islam et al. (2014) exhibited reduction in the biomass of maize in the presence of 50 μM of Zinc. Generally, several unspecific cation transporters are responsible for the accumulation of Zn(II) in an intercellular environment. Among many, two such central Zn(II) uptake systems are CorA (a chemiosmotically driven transporter from the MgtE family) and MgtA type zinc transporter (a P-type ATPase transporter) (Nies, 1999). Bacterial Zn(II) resistance potency came from czc operon that encodes for chemiosmotic Zn(II) efflux pump. Another important efflux system is P type ATPase ZntCAB encoded by znt operon (Silver and Phung, 2005a). Glibota et al. (2020) reported the frequent presence of zntA gene among several bacterial isolates that belong to genera Chryseobacterium spp., Pseudomonas spp. Variovorax spp., and Stenotrophomonas spp. Another study by Hao et al. (2012) using genomic reverse transcriptase PCR and a transposon-based gene disruption approach revealed the zntA and zntR genes mediated resistance of Zn(II) and Cd(II) in PGPB members like Agrobacterium tumefaciens. Apart from this, several microbes also have the ability to produce metallothionein (a cytoplasmic metal cation binding protein) which can effectively reduce metal toxicity (Silver and Phung, 2005a). Nonetheless, several plant growth-promoting bacterial isolates (Pseudomonas spp., Sphingomonas spp., Serratia spp., and Rhizobium spp. See Supplementary Table 2 for detailed information) are also known to exhibit Zn(II) resistance potency via the czc operon (Arroyo-Herrera et al., 2020; Kang et al., 2020).
Another transcriptomic study in Pseudomonas putida strain KT2440 revealed that under Zn(II) stress, along with zinc resistance genes (e.g., cadA1, cadA2, czcCBA1, and czzD), several arsenic resistance genes belonging to two arsenic operons - arsRBCH1 and arsRBCH2- are significantly upregulated. A downstream proteomic study in that organism also exhibited that, under zinc stress, the multidrug and metal efflux system related genes were upregulated whereas genes related to membrane porins were found to be downregulated (Peng et al., 2018). Comparative transcriptomic analysis by Wang H. et al. (2020) also revealed the upregulated expression of gene zntA in Chryseobacterium sp. WAL2 in the presence of montmorillonite. This study hinted toward the role of montmorillonite as a positive regulator of metal detoxification related genes in diverse microbial groups, but requires further in-depth investigation in future.
Mechanism of Ni(II) Resistance
Intercellular accumulation of Ni(II) followed two different uptake systems. These include either a single polypeptide chemiosmotic system or a five component ABC uptake system (Mulrooney and Hausinger, 2003). An earlier study by Navarro et al. (1993) also reported the presence of two separate energy dependent Ni(II) uptake systems in E. coli, which include a non-specific high affinity magnesium uptake system and another highly specific, low capacity system encoded by nik operon. The nik operon (comprised of five genes: nikA, nikB, nikC, nikD, and nikE) remain activated at Ni(II) limiting conditions, whereas at high Ni(II) concentrations, the transcription is ceased and Ni(II) is found to be transported using magnesium uptake system. Another study by Liu et al. (2020) reported the upregulation of nikB and nikc genes in PGPB isolates Paenibacillus polymyxa YCO136 in the presence of root exudates of tobacco. So far, Ni(II) resistance in several bacteria involve cnr/ncr/nreB like tolerance system (Stoppel and Schlegel, 1995) which encode Ni(II) efflux proteins that extruded out Ni2+ from cytoplasm to the extracellular environment. Generally, cnr or ncr gene clusters responsible for the synthesis of efflux protein belong to RND type transporter whereas efflux systems encoded by nreB belong to MFS superfamily (Nanda et al., 2019). However, ncr system in Leptospirillum ferriphilum and cnr system in Cupriavidus metallidurans pMOL-28 are some well-characterized Ni(II) detoxification systems in prokaryotes (see Figure 2) (Tian et al., 2007; Mazhar et al., 2020) Nonetheless, several PGP Ni(II)-resistant bacterial isolates are reported where bio-adsorption of Ni(II) on the cell surface is found to be the most predominant resistance mechanism. Notably, metal-stress-induced transcriptome analysis of Sphingobium spp. revealed the upregulated expression of genes [NerB-like permease, a specific protein of secondary metal transporter family (HupE/UreJ)] coding for nickel resistance and other metal ion efflux systems along with overexpression of CopA, CopB, and CopC proteins that are involved in Copper resistance. This indicates the interlinking metal resistance potential in this organism (Volpicella et al., 2017).
Summarizing the Genomics of Major Plant-Growth-Promoting Attributes, Characterized so Far in Diverse Bacteria
Organisms with physiological potential, such as production and distribution of plant growth-promoting hormones like IAA/GAs, phosphate solubilization, nitrogen fixation, siderophore production, ACC deaminase production, and biofilm formation, are usually considered as potential plant growth-promoting candidates. Current advancements and the emerging picture of high-throughput genomic data helps in decoding a number of PGP bacterial genomes and relevant gene encoding proteins in distantly related microbial group isolated from diverse habitats.
Although microbes have the ability to produce multiple phytohormone, here we restricted our discussion to auxin. However, so far as the auxin-producing genes are concerned, mostly it has been characterized from bacteria thriving in rhizosphere (Spaepen and Vanderleyden, 2011); however, we detected a number of auxin-producing genes in the genome of several gut bacteria with high auxin production efficiency (Cox et al., 2018). A study by Indiragandhi et al. (2008) also revealed the presence of auxin-producing genes in several PGPB members isolated from gut environment, however delineation of the genetic basis of such attributes were not reported in that study. Microbes with such auxin production efficiency can significantly influence the plant growth and root architecture. In this context, the trp gene cluster plays a critical role in the biosynthesis of tryptophan (central metabolic precursor of auxin) from the chorismite. There are at least five different biochemical pathways for auxin production characterized so far in various PGP bacteria (e.g., members of Azospirillum spp., Bacillus spp., Bradyrhizobium spp., Enterobacter cloacae, Paenibacillus spp., Pseudomonas spp., Rhizobium spp., Acinetobacter spp., and Aeromonas spp.) (Spaepen et al., 2007; Yadav et al., 2017). For instance, pathways like indole-3-acetaldehyde (IAAld), indole-3-acetaldehyde (IAM), indole-3-acetamide (IPDC), indole-3- pyruvate decarboxylase (IPA;), and tryptophan (Trp) were found to be operative either singly or in combination (Spaepen et al., 2007). Relevant genes for auxin production are either found to be present in plasmid (Agrobacterium spp.) or in chromosome (P. syringae, Pantoea agglomerans) of diverse bacteria (Mole et al., 2007). High-throughput genetic and proteome level investigation emphasized the involvement of some major auxin-producing enzymes: Amino transferase, Trp monooxygenase, Nitrilase, Nitrile hydratase, IAM-hydrolase, Trp decarboxylase, Amine-oxidase, tryptophanase, tryptophan, and synthase (Spaepen et al., 2007). Transcriptomic study of PGPB members of Paenibacillus polymyxa YCO136 showed the upregulation of several metal resistance genes along with genes that are related to auxin production as well as phosphate solubilization (Liu et al., 2020). Due to the presence of multiple biochemical pathways, in-depth knowledge regarding genetic systems of auxin production still requires some more attention as these organisms could be used as important bioresources toward sustainable agriculture in the near future.
From the perspective of PGP attributes, microbe-mediated phosphate solubilization (conversion of insoluble phosphate into bioavailable form to the plants) potential holds substantial importance in the agricultural sector. Bacteria with such potential are being characterized and isolated from diverse ecosystems/or niche (soil ecosystem/ ectorhizospheric region/ as an endosymbionts). These currently include Acinetobacter spp., Pseudomonas spp., Enterobacter spp., Arthrobacter spp., Cellulosimicrobium spp., Desemzia spp., Flavobacterium spp., Providencia spp., Bacillus spp., Serratia spp., Burkholderia spp., and Erwinia spp (Goswami et al., 2016; Behera et al., 2017; Yadav et al., 2017; also see Supplementary Table 2 for detailed information). Acidification, chelation, and polymeric substances formation are some of the important mechanisms via which phosphate-solubilizing bacteria (PSB) act on mineral phosphates. However, the most common mechanism shown by maximum PSB is the production of low molecular weight organic acids (e.g., gluconic acid, oxalic acid, citric acid, acetic acid, succinic acid, etc.) with substantial lowering of the soil pH (Suleman et al., 2018; Wei et al., 2018). Genomic surveys of PSB highlighted the presence of several essential genes/enzymes involved in P-solubilization, such as glucose dehydrogenase (gcd), acid phosphatase (AphA), phosphatase, inorganic diphosphatase, alkaline phosphatase (AP), and sugar-phosphatase. Nonetheless, a few other accessory enzymes like phosphoenolpyruvate carboxylase (Pepc) have also been identified in several organisms responsible for the increase in supply of oxaloacetate, a vital anabolic precursor and an intermediate in biosynthesis of organic acids in phosphate (P) solubilization (Buch et al., 2010). Moreover, a previous study reported that enzymes like exopolyphosphatase (Ppx) and polyphosphate kinase (Ppk) could also be involved in the hydrolysis of inorganic polyphosphate P to release orthophosphate (Kaur et al., 2016).
Conventionally, siderophores are iron chelating low molecular weight (<1 kDa) compounds that are synthesized by various groups of microbial genera in response to low iron availability in their native environment (Goswami et al., 2016). These components play a considerable role in improving plant health through several mechanisms, i.e., mitigating the iron nutrition deficiency of host plant or preventing the growth of deadliest phytopathogens by limiting their iron availability. Multiple bacterial genera, such as Rhizobium spp., Mesorhizobium spp., Streptomyces spp., Burkholderia spp., Bacillus spp., and Pseudomonas spp. have been isolated with such siderophore production efficiency (Saharan and Nehra, 2011). Among many, some of the well-studied microbe-derived siderophore components are aerobactin, vibriobactin, bacillibactin, and enterobactin. Although functionally all siderophore components are the same, their chemical nature [catecholates (phenolates), hydroxamates, and carboxylates (e.g., derivatives of citric acid)] and genetic determinant vary significantly. For instance, aerobactin (hydroxamate-type siderophore) synthesis is tightly regulated by iuc (iron uptake chelate; encompassed with iucABCD genes) and iut (iron uptake transport; having iutAC genes) operon, identified and characterized in various bacteria such as E. coli, Gluconacetobacter spp., Micromonospora spp., and Acinetobacter spp. (Mihara et al., 2004; Logeshwaran et al., 2009; Ortúzar et al., 2020). On the contrary, genes [viuPDGC _ ABF (utilization related genes); vibABCDEH (biosynthesis related genes)] involved in vibriobactin biosynthesis/utilization are not clustered in a single locus; rather they are scattered throughout the genome (Crosa and Walsh, 2002). In the case of enterobactin biosynthesis, genes such as entA (encode for 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase), entB (encode for 2,3-dihydro-2,3-dihydroxybenzoate synthetase), entC (encode for isochorismate synthetase), and EntEFB are found to be involved (Crosa and Walsh, 2002). Notably, some other siderophore components like anguibactin (encoded by angCNHUTR genes), pyochelin (encoded by pchDCBAEF genes), yersiniabactin (encoded by YbtEUTSXQP genes), and mycobactin (encoded by mbtABCDEFGHIJ genes) are also known to play important roles in iron acquisition/uptake/chelation in various organisms.
In the context of potential PGP traits, 1-aminocyclopropane-1-carboxylic acid deaminase (ACC deaminase; encoded by acdS gene)-producing bacteria perform a vital role by blocking the synthesis of ethylene, thereby improving plant-fitness potentiality under many biotic stresses such as drought, salinity, or floods (Glick 2005). So far as its structural features are concerned, it is a multimeric enzyme (homodimer or homotrimer that uses pyridoxal 5-phosphate per subunit as a critical cofactor) with a molecular weight ranging between 35 and 42 kDa (Ali and Kim, 2018). Interestingly, several previous works report that production of this enzyme can be triggered in bacteria (Pseudomonas spp., Achromobacter spp., Serratia spp., Herbaspirillum spp., Ochrobactrum spp., Mesorhizobium spp., etc.) by inducing stressed conditions (Ali and Kim, 2018). Notably, several studies reported an LRP-like protein-coding region present in the close vicinity of many acdS genes in diverse bacteria (Rhizobium spp., Azospirillum spp. etc), termed acdR (ACC deaminase regulatory protein), which is required for optimum ACC deaminase production (Prigent-Combaret et al., 2008). Nevertheless, the presence of ACC deaminase is not only restricted within bacteria; rather their distribution is also reported in plant and fungal kingdoms, although their sequence composition varied greatly (Nascimento et al., 2014).
Moreover, in the context of PGP traits, nitrogen fixation ability of different PGPBs is considered as a key trait involved in plant growth promotion under heavy metal stress. Vicente and Dean (2017) exhibited the simplest form of molecular mechanism of nitrogen fixation by diazotrophs, while highlighted the promising research endeavor for developing genetically modified cereal plants capable of producing their own functional nitrogenase and subsequently able to perform nitrogen fixation function. Bacteria from a wide range of habitats are found to be capable of nitrogen fixation using oxygen sensitive metallozyme nitrogenase, which share common features of catalytic mechanism and assembly of metal-containing cofactors, which are important for its function. It has been observed that nitrogenase contain two catalytic parts: one is dinitrogenase reductase that serves as a nucleotide dependent system of electron delivery to another part of nitrogenase which, after receiving a sufficient amount of electrons from reductase, binds with nitrogen (N2) and converts it into ammonia (Vicente and Dean, 2017). Moreover, nif genes are found to be responsible for governing such nitrogen fixation. Nonetheless, diversity of diazotrophs is also responsible for altering soil fertility and optimizing plant growth, as it links directly/indirectly with the expression of nifH gene. Notably, nifH gene being highly conserved among diazotrophs serves as an indicator organism of soil (Chakraborty and Islam, 2018). The study by Chakraborty and Islam (2018) pointed out the fluctuation pattern in the abundance of proteobacteria and diazotrophs under arsenic stress. However, qPCR-based analysis of highly conserved 16S rRNA gene and nifH gene at higher arsenic concentration (10 ppm) showed significant reduction in the copy number of nifH gene, which indicated the decrease in diazotrophic population, whereas it remained unaltered where arsenic stress was lower (6 ppm). They have also suggested that factors like pH, arsenic, and (PO4)− is also linked with the fluctuation of nifH gene abundance. On the other hand, 16S rRNA gene-based diversity analysis revealed an increase in proteobacterial abundance with increased concentration of arsenic (Chakraborty and Islam, 2018). A similar finding was also reported by Chakraborty et al. (2019); they showed the decrease in expression of nifH gene and EPS production of arsenic oxidizing free living diazotrophs by members of Bacillus spp., Klebsiella spp., and Burkholderia spp. at higher arsenic concentrations.
Interrelation Between Bacterial Plant Growth Promoting Traits and Metal Resistance Mechanism for Stress Amelioration in Plants
Association of PGPB with their native host plant plays an instrumental role in their survival, especially during stressed conditions. Out of many components, heavy metals pose serious stress to plants that ultimately causes severe irreversible damage (see Table 1 for detailed information). To combat such issues, PGPB was found to play a critical role and remain associated with their host in metal-contaminated soil. A large number of PGPB have been isolated and characterized from diverse habitats which include rhizospheric soil, phyllosphere, costal sediments, heavy metal mine areas, sewage sludge, tannery effluents, endophytes, and root nodules (see Supplementary Table 2). PGPB have several traits that play important roles in plant growth promotion during heavy metal stress as well as metal detoxification. These are as follows.
Role of Phytohormones Under Heavy Metal Stress
It has been known for a long time that a large number of bacterial candidates have the ability to produce different plant growth hormones (gibberellic acid, cytokines, auxins, etc.). Of these, bacterial-derived auxin, especially IAA, gained significant attention as it plays a major role in plant growth promotion under such heavy metal stress (see Supplementary Table 2). Bacteria with such potency thus far include members of Pseudomonas spp., Bacillus spp., Serratia spp., Rhizobium spp., Mesorhizobium spp., Sinorhizobium spp., Burkholderia spp., Achromobacter spp., Azotobacter spp., and Paenibacillus spp. (see Supplementary Table 2). IAA not only helps plants to sustain metal tolerance by physiological changes but also enhances branching of plant roots and thereby aids nutrient uptake capacity under heavy metal stress (Etesami, 2018). Several reports also suggested that, during plant growth, IAA also loosened the cell wall of root cells which resulted in excess secretion of nutrient rich root-exudates in the surrounding environment, which eventually attracted native microbial populations of root rhizosphere (Bacilio-Jiménez et al., 2003). Root-exudates secreted in such a way contained components like phytochelatins and organic acids that subsequently prevent the entry of heavy metals in symplastic path (Manara, 2012).
Roll of ACC Deaminase Under Heavy Metal Stress
Under heavy metal stress, ACC deaminase promotes plant growth by inhibiting the generation of plant hormone ethylene which forms during different biotic and abiotic stresses, causing early senescence in plants (Glick, 2005). ACC deaminase cleaves the 1-aminocyclopropane 1-carboxilic acid, which is a precursor of ethylene, into α-ketobutarate and ammonia. (Carlos et al., 2016) reported the positive plant growth promotion effect of two ACC deaminase-producing bacteria Klebsiella Mc173 and Serratia K120 on Helianthus annuus under Ni(II) and Cd(II) stress. It has been observed that ACC deaminase activity of both strains remained unaffected in the presence of Ni(II) and Cd(II). A large number of heavy metal resistant plant growth-promoting bacteria has been isolated and reported to produce ACC deaminase (see Supplementary Table 2 for detailed information).
Role of Siderophore Under Heavy Metal Stress
Although plants reportedly produce phyto-siderophore to acquire Fe, it has been reported that bacterial-derived siderophore have a greater affinity toward iron (Etesami, 2018). Additionally, bacterial siderophores are reported to have the ability to bind with a number of different heavy metals, thereby immobilizing them and reducig their bioavailability to plants under heavy metal stress. A large number of PGPB isolates were reported to produce siderophores under such heavy metal stress (Złoch et al., 2016; Yang et al., 2018; Md Badrul Hisham et al., 2019; Sun et al., 2021).
Role of Nitrogen Fixation Under Heavy Metal Stress
Nitrogen fixation has been reported in a number of plant growth-promoting diazotrophs isolated from diverse habitats, many of which were found to have multi-metal tolerant potency. Nowadays, nitrogen fixing bacteria serve as key players as bioindicators of heavy metal contamination in soil. In this connection, two important studies by Chakraborty and Islam (2018) and Chakraborty et al. (2019) reported a decrease in population of nitrogen-fixing diazotrophs at higher concentrations of arsenic while the abundance of proteobacteria was found to be increased. Moreover, they also reported a decrease in EPS production among free living diazotrophs under high arsenic stress. However, in general, EPS production was found to be increased along with increased metal concentration to protect bacteria from metal stress. This metal resistance endeavor eventually created a microaerobic environment where free living diazotrophs were able to fix N2 and subsequentially support plant growth under heavy metal stress. This is another classic example of an interdependency relationship between heavy metal resistance and PGP traits in PGPB members.
Role of Phosphate Solubilization Under Heavy Metal Stress
Phosphate solubilization by PGPB or other microbes is a major plant growth promoting phenomenon, where the phosphate solubilizing bacteria (PSB) solubilized the mineral phosphate in rock and created a pool of bioavailable soluble phosphate in soil, which is further taken up by plants. Several bacterial-derived organic acids by PSB and lowering of pH and/or phosphatase enzyme activity are thought to be responsible for phosphate solubilization (Chen et al., 2006; Wei et al., 2018). These bioavailable phosphates eventually immobilized different heavy metals by complexation and reduced their bioavailability to plants. For example, immobilization of Pb(II) by formation of pyromorphite has been observed in Enterobacter cloacae (Park et al., 2011). Additionally, binding of heavy metals to the cell surface by forming phosphate-heavy-metal-complex has been reported for Pb(II) and Cd(II) adsorption by amide and carboxyl groups on cell surface of Enterobacter sp., Bacillus sp., and Lactococcus sp. (Yuan et al., 2017). For both instances, concerned heavy metals were found to be immobilized and insoluble, thus reducing the bioavailability and its stress generation capacity. A number of multi-metal tolerant PGPB from diverse habitats with phosphate solubilization ability has been reported and characterized over the past two decades (see Supplementary Table 2 for detailed information). Moreover, organic acid produced by PGPB/PSB members has a higher affinity for heavy metals and was also found to be involved in immobilization of such metals by complexation, thereby reducing heavy metal stress on plants (Najeeb et al., 2009; Gadd, 2010; Islam et al., 2016).
Role of Antioxidants Under Heavy Metal Stress
In the generation of reactive oxygen species, superoxide during heavy metal stress is one of the most expected events reported for both plants and animals (for details see Table 1). This heavy-metal-induced oxidative stress was reported to cause massive irreversible damage to DNA and cellular proteins (Islam et al., 2014). Furthermore, heavy-metal-induced oxidative stress resulted in a massive reduction in yield of several economically important crops, including Oryza sativa (rice), Triticum aestivum (wheat), and Zea mays (maize) (already mentioned in brief in Table 1). To combat such oxidative stress, plant cells produce several antioxidant enzymes like superoxide dismutase, catalase, peroxidise, and glutathione reductase. Several reports indicated that PGPBs like Azotobacter chroococcum and Proteus mirabilis can induce antioxidant systems in Zea mays so that the plant becomes more tolerant toward metals like Cu, Zn, and Pb (Islam et al., 2014; Rizvi and Khan, 2018). Additionally, PGPB was also found to produce several such enzymatic (such as catalyze and peroxidise) (Karthik and Arulselvi, 2017) and non-enzymatic (such as proline and glutathione) defense mechanisms that were eventually found to be involved in lowering the heavy-metal-induced oxidative stresses in plants (Islam et al., 2014; Rizvi and Khan, 2018).
Meta-Omics Perspective in Exploring the Structure-Function of Metal-Remediating Microbiota
Nowadays, the “meta-omics”-based approach offered a number of advantageous strategies in exploring microbiome framework, community dynamics, and metabolic networking in diverse environments (metagenomics, metatranscriptomics, metaproteomics, and metabolomics) that curtail different constraints posed by conventional culture dependent approaches. In the context of arsenic remediating microbiome, the work of Bertin et al. (2011) highlighted the use of the genome-binning approach that exhibited the prevalence of seven bacterial strains (contigs affiliated to novel bacterial phylum Candidatus “Fodinabacter communificans” β-Proteobacterium Thiomonas genus, Acidobacteria clade and A. ferrooxidans γ-Proteobacterium) in extreme habitats (e.g., acid mine drainage) with genetic potential for arsenic resistance [oxidizing As(III) into As(V) and their subsequent co-precipitation with sulfur] (Bertin et al., 2011). Among the seven genome bins reconstructed from the metagenomes, CARN6 contained the gene arsM encoding arsenite S-adenosylmethyltransferase which was further validated by the presence of monomethylarsonate and dimethylarsinate in their native habitat (Bertin et al., 2011). Subsequent metaproteomic-based investigations by the same group revealed the presence of Pst system which potentially influences the controlled uptake of As(V) in their inherent environment. Moreover, this study also unraveled the high-level expression of genes that were potentially involved in biosynthesis of biofilm, capsule, flagella, and pili, indicating that these components indeed have persuasive connection with metal-resistant genes (Bertin et al., 2011). In this connection, the functional importance between biofilm formation and immobilization of different heavy metals in metal-contaminated mine waters is further enlightened by the work of Drewniak et al. (2016). In total, 2% sequence reads were found to be ascribed for genes related to biofilm formation which indicates that biosorption might be a major process for sequestration of those heavy metals (Drewniak et al., 2016). Notably, another omics-based diversity study by Li et al. (2015) revealed that microbial community composition caused significant fluctuation between low and high arsenic containing samples collected from groundwater and sediments. Bacterial genera like Acinetobacter, Pseudomonas, Psychrobacter, and Alishewanella were found to be most abundant in high arsenic-containing groundwater systems, while Thiobacillus, Pseudomonas, Hydrogenophaga, Enterobacteriaceae, Sulfuricurvum, and Arthrobacter were found to be prevalent in high arsenic-containing sediment systems. Moreover, another separate metagenomic study by Wang et al. (2016) reveled the selective prevalence of certain bacterial genera (such as Alishewanella, Psychrobacter, Methylotenera, and Crenothrix) in high arsenic-containing groundwater systems of diverse geographical distribution, while Rheinheimera and the unidentified OP3 group were found to be abundant in low arsenic-containing groundwater systems. In this context, another study by Han et al. (2017) examined the impacts of As on bacterial communities' structure and their functional genetic determinants in the rhizosphere of As-hyperaccumulator plant Pteris vittate and reported that aroA-like genes were found to be more prevalent than arsC genes where As(V) was present in high concentrations in the soil. These studies actually exhibited how arsenic influences the bacterial community architecture in diverse environments. Moreover, another important study by Yu et al. (2020) revealed that cyanobacterial population (85.89%) was found to be present as dominant taxa among the diazotrophic microbial community studied in the citrus orchard where heavy metals like Pb, Cd, Cu, and Zn are present in higher concentrations. However, they also mentioned the dominance of other phylum types, which includes phylum like Proteobacteria, Firmicutes, and Verrucomicrobia, in different habitat types contaminated with the aforementioned heavy metals. High abundance of such candidates in metal-contaminated habitats sheds new light on their roles as key players for natural metal-remediation while maintaining the ecosystem balance. Omics-based functional gene analysis in other habitats, like activated sludge, coastal sediments, mangrove sediments, and contaminated ground water,. revealed the functionality of various metal-resistant genes such as ChrA (encoding chromate transport protein), ACR3 (encoding arsenic resistant protein), CusA (cation efflux system protein), CzcA (cobalt-zinc-cadmium resistance protein) aioA (arsenite oxidation), arrA, ACR3, arsM (arsenite methylation), aroA, arrA, arsB, and arsC (Cai et al., 2013; Luo et al., 2014; Cabral et al., 2019; Cavalca et al., 2019). However, so far as the metal-remediating microbiome architectures are concerned, microbial taxa like members of Firmicutes, Clostridiales, Verrucomicrobia, Methylococcaceae, Methylophilace, Rhizobiaceae, Burkholderia, Leptothrix, Thiothrix, Geobacter, Rhodococcus, Azoarcus, Rhodanobacter, Pseudomonas, and Beggiatoa are reported as predominant groups (Cai et al., 2013; Cabral et al., 2019; Cavalca et al., 2019; Wang L. et al., 2020) in most arsenic-contaminated ecological diverse environments. For the revelation of any pollutant-degrading novel microbial diversity, an amplicon sequencing (sequencing the hypervariable regions of the 16S rRNA gene) based approach is equally as important (cost-effective) as a metagenome approach. Ample work has been published in this regard that highlighted the presence of certain bacterial genera (e.g., Pseudomonas spp.; Methylobacillus spp.; Solirubrobacter spp., Ohtaekwangia etc.) in arsenic-contaminated sites as a major group (Kou et al., 2018; Espinosa et al., 2020). Notably, along with metagenomics, the use of stable isotope probe technique is also found to be effective in the revelation of nitrate dependent As(III) oxidizing bacterial members like Azoarcus, Rhodanobacter, and Pseudomonas, from arsenic-contaminated soil systems (Zhang M. et al., 2020). In meta-omics connection, another term “Fluxomics” has recently been introduced in the scientific community (Chandran et al., 2020). This approach is concerned with the estimating fluctuation and/or rate of metabolic reactions and predicts the fate of different metabolic process as well as intermediates in biological organisms. For instance, fluxome profiling approach was successfully used in the case of one marine strain of Shewanella for deciphering the co-metabolic pathways for the biodegradation of lethal metals, halogenated organic compounds, and radionucleotides (Tang et al., 2007). Nonetheless, using a metagenomic survey, several groups also reported the presence of potential arsenic reduction (80% metagenomic reads ascribed for this)/methylation/efflux related genes (6–9% metagenomic reads ascribed for this) in wetland environments like paddy soil (Xiao et al., 2016; Imchen et al., 2018). Notably, in a separate study conducted by Shi et al. (2019), the As(V) and Hg(II) bioremediation efficiency in methanotrophic bacterial members such as Methylocystis sp. HL18, whose assembled genome encompassed with operons like mer (having genes like merR, merT, merP, merC and merA), arsRCB, and arsRCCB, were reported (Shi et al., 2019). In this connection it is noteworthy to mention that functional-protein-based phylogenetic analysis (MerA, arsRCCB) indicates acquisition of metal-remediating genes in this organism follow a horizontal path of gene transfer, a phenomenon that hints toward the new possibilities for getting bioremediation potency in novel taxonomically diverse bacterial groups that has not been explored till date.
On some occasions it has been observed that bioremediating bacteria also has influence in other biogeochemical cycles. For instance, a functional marker gene and metagenome-based study that dealt with bacteria (Cupriavidus metallidurans MSR33) assisted mercury bioremediation efficiency and microbial community dynamics in a soil ecosystem unveiled that the removal of mercury (82%) from the soil system significantly increased the nitrogen fixing and nitrifying bacterial population (Bravo et al., 2020). On the contrary, a few works also used biomarker genes and metagenomics as ecological-indicator tools to detect some toxic pollutants like “methylmercury.” Genes like hgcAB that participate in Hg-methylation process [conversion of mercury (Hg) to MeHg] were efficiently used as biomarkers of diverse methylmercury contaminated environments that includes riverine areas, tidal marshes, and arctic permafrost (Christensen et al., 2019). A functional omics approach was also successfully used to explore the metallothionein-related genes [responsible for Cu(II) resistance] in a Cu-contaminated soil environment. Later on, downstream cloning and overexpression of these metallothionein-related genes were also used to develop recombinant strains that had better survival efficacy under metal stress along with greater absorption capacity for certain metals like cadmium, copper, and zinc (Liu et al., 2019). Notably, another important study by Wang et al. (2007) pointed out the effect of heavy metal stress in regulating microbial community composition and enzyme activity in soil environment. Recently, another comparative metatranscriptomic and metagenomic study in industrial wastewater discovered the presence of six novel genes potentially involved in Cr(VI) resistance. Out of six, two genes, gsr (general stress response protein) and mcr (membrane chromate reductase), when cloned and expressed in E. coli efficiently remove 50% of Cr(VI) out of 200–600 μm Cr(VI) (Pei et al., 2020). In this context, a recent finding by Ma et al. (2020) sheds new light on the role of minerals like phosphorus in changing bacterial population dynamics and subsequent Cr(VI) reduction efficiency in chromium-contaminated environments (Ma et al., 2020). They used an amplicon sequencing approach for microbial diversity analysis both in the presence and absence of liquid phosphorus (LP) and phosphorus (NP). In an LP system, Aeromonas spp. and Acinetobacteri spp. etc. occurred as a prevalent microbial group, whereas Alcaligenes spp. and Parabacteroides spp. etc. were found to be abundant in an NP system. Interestingly, genera like Delftia, Stenotrophonas, and Comamonas disappeared in the presence of phosphorus minerals, while genera like Pseudomonas and Clostridium were found to be enriched in the presence of different types of phosphorus components (Ma et al., 2020).
Potency and Prospect of Microbe Assisted Phytoremediation of Heavy Metals
Although plants are prime colonizers, they become highly susceptible when they are exposed to high heavy metal containing environments. Interestingly, it has often been observed that several socio-economically important crops have the ability to accumulate high concentrations of heavy metals in metal-contaminated sites. Reasonably, such phenomenon ultimately resulted in severe health hazards through the process of biomagnifications. Nevertheless, there are several non-crop plant species encompassed with high metal resistance/accumulation efficiency that also exist in nature, commonly regarded as metallophytes (e.g., Thlaspi caerulescens, Psycotria vanhermanni, Viola calaminaria, and Pteris vitata). Surprisingly, these plant systems have been undervalued since their discovery in remediation purposes (Khan, 2005; Marques et al., 2009). Notably, a number of heavy metal resistant plant growth-promoting bacteria were reported to enhance metal accumulation (e.g., Cr, As, Cd, Cu, Hg, Pb, Ni, and Zn) in large numbers of crop/ fodder plants (such as maize, tomato, black mustard, rapeseed, and Indian pea) (Ma et al., 2009; Cruz Medina et al., 2013; Ahmad et al., 2016; Kamran et al., 2016; Franchi et al., 2017; Abdelkrim et al., 2018). In this context, use of those microbes as bioinoculant/biofertilizer was obviously subjected to objectionable concern when they can be employed safely for the growth of hyperaccumulator plants, which are able to remediate agricultural soil due to their massive accumulation potency. Additionally, these plant biomasses can also then be used in energy production purposes (Ma et al., 2010). Therefore, phytoextraction can be considered as a most pertinent approach in the domain of bacteria-assisted phytoremediation strategies. Notably, a few studies also reported the enhanced accumulation of different heavy metals in plant roots only in the presence of plant growth-promoting bacteria, which suggests their probable future application in remediation strategies like phytostabilization (Moreira et al., 2014; Rojjanateeranaj et al., 2017). Even though these are very promising strategies for remediation of agriculturally important soil, improved understanding on molecular-genomic basis of plant-microbe-metal interaction is a prerequisite before its vast environment application of agronomic benefits for farmers.
Discussion and Conclusive Remarks
Along with genomics/molecular mechanism-based discussion, here we also tried to summarize the global scenario of bacteria-assisted metal remediation potential using a heat-map approach. Accordingly, Figure 3 exhibited the bioremediation potency of different toxic heavy metals by phylogenetically and ecologically diverse bacteria with plant growth-promoting traits. Remarkably, multi-metal tolerance ability was only found in the case of genera like Bacillus (belong to the phylum Firmicutes), Acinetobacter, Pseudomonas, and Rhizobium (three genera that belong to the phylum Proteobacteria). However, among the studied bacteria, Hg(II) tolerance potentiality was only shown by certain bacterial genera, out of which Bacillus shows very promising results [4,011 mg/l Hg(II)]. Notably, various PGP members of Bacillus, Azotobacter, and Paenibacillus reported so far have high Pb(II) tolerance ability (≈5,175 mg/l) while members of Acinetobacter have the highest Pb(II) capability (16,000 mg/l). So far as the As(III) and As(v) metalloids are concerned, the highest tolerance ability was found in Exiguobacterium [13,485 and 52,444 mg/l for As(III) and As(v) respectively], Pseudomonas [59,936 mg/l for As(V)] along with Rhodococcus [52,444 mg/l for As(V)], and Acinetobacter [50,000 mg/l for As(V)]. Notably, several PGP members of proteobacteria were found to be competent for tolerance of other heavy metals. For instance, Sphingomonas efficiently tolerates high concentration of heavy metals like Cd(II), Zn(II), and Co(II) (10,000 mg/l); Cupriavidus can tolerate the highest Ni(II) concentration (2,948 mg/l). Providencia had the highest Cu(II) tolerance ability (63,540 mg/l), whereas Cellulosimicrobium had the ability to tolerate Cr(VI) at a concentration of 23,395 mg/l. Considering the current state of knowledge, it has been observed that most of the bacterial-heavy metal tolerance mechanisms have been reported for As, Pb, Zn, and Cd. In this context, it is noteworthy to mention that other heavy metals, particularly Hg and Cr, needs special attention and indeed require further investigation. From the perspective of socio-economic progress, we cannot overlook the importance of industrial development; at the same time, the necessity of a sustainable pollution-free ecosystem cannot be ruled out. With the rapid development of modern meta-omics, comparative genomics, and molecular and/or system biology techniques, various groups have already made significant progress in the field of pollutant microbiome research. This will help us to combat nefarious heavy metal pollution issues and unravel the complex microbial population dynamics and their trophic interaction. Throughout this review, we have made an effort to decode the molecular genetic basis of heavy metal detoxification/resistance potential (having PGP traits) from bacteria with diverse taxonomical and ecological spectrums. Deeper molecular-genetic understanding of these potential microbes involved in such processes brings to the fore novel or advanced bioremediation technologies that could be used to polish our understanding in this regard.
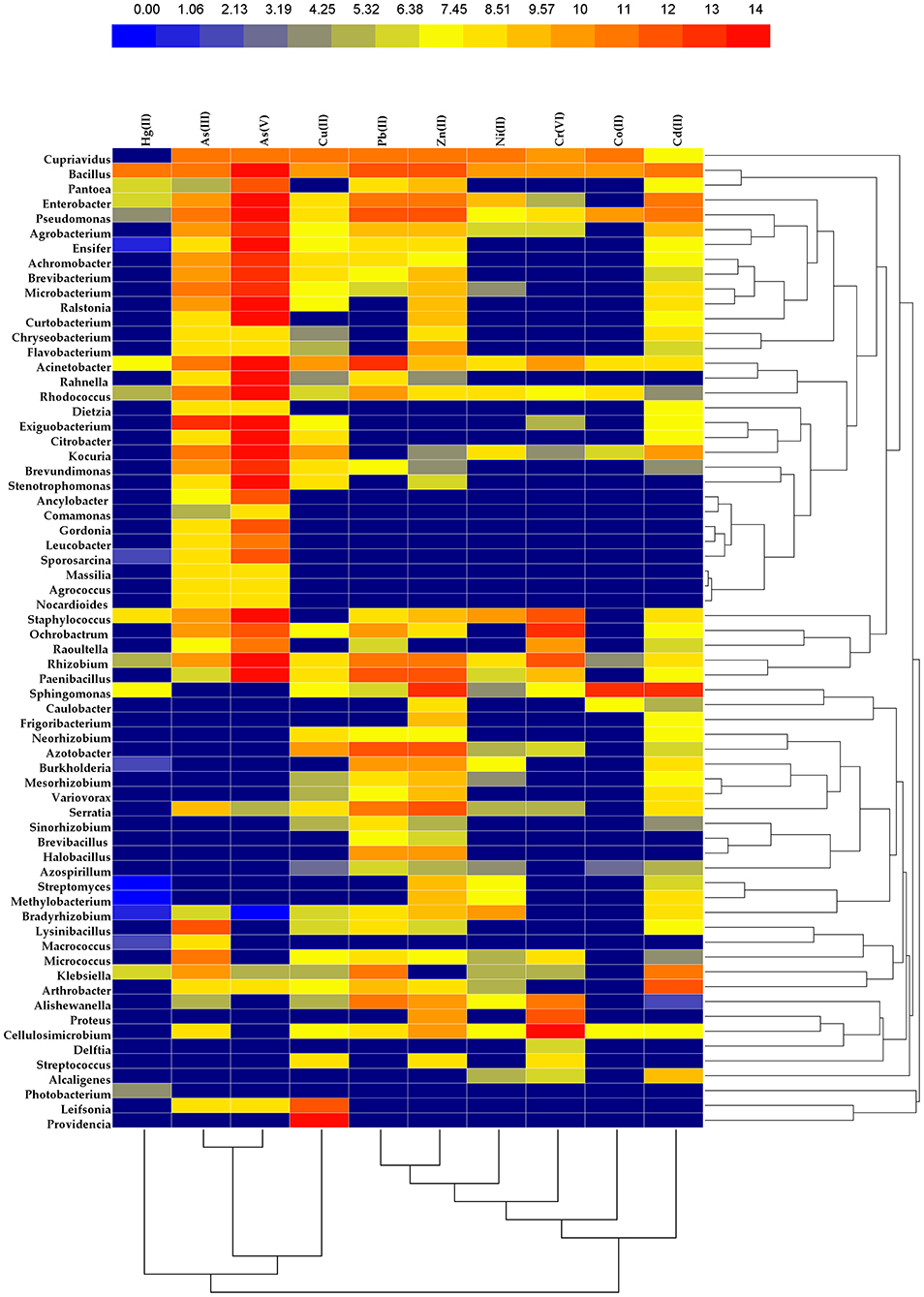
Figure 3. Heat map comparing the different levels of heavy metal resistant efficiency in several majorly studied bacterial genera having plant-growth-promotion attributes. Two-dimensional clustering is also shown involving the 10 major heavy metals on one hand and 67 representative bacterial genera on the other. Concentration of heavy metals are calculated and expressed using mg/L unit.
Author Contributions
SM conceived the idea of the review, provided the general concept and inputs for each specific section, drafted part of the manuscript, and wrote the review in conjunction with NM. SM and NM edited, compiled, and finalized the draft. KS participated in manuscript writing and extensive data mining. All authors contributed to the article and vetted the submitted version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.664126/full#supplementary-material
References
Abdelkrim, S., Jebara, S. H., Saadani, O., Chiboub, M., Abid, G., and Jebara, M. (2018). Effect of Pb-resistant plant growth-promoting rhizobacteria inoculation on growth and lead uptake by Lathyrus sativus. J. Basic Microbiol. 58, 579–589. doi: 10.1002/jobm.201700626
Achour-Rokbani, A., Cordi, A., Poupin, P., Bauda, P., and Billard, P. (2010). Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl. Environ. Microbiol. 76, 948–955. doi: 10.1128/AEM.01738-09
Adaikkalam, V., and Swarup, S. (2002). Molecular characterization of an operon, cueAR, encoding a putative P1-type ATPase and a MerR-type regulatory protein involved in copper homeostasis in Pseudomonas putida The GenBank accession number for the sequence reported in this paper is AF390440. Microbiology 148, 2857–2867. doi: 10.1099/00221287-148-9-2857
Afkar, E., Lisak, J., Saltikov, C., Basu, P., Oremland, R. S., and Stolz, J. F. (2003). The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 226, 107–112. doi: 10.1016/S0378-1097(03)00609-8
Aguilar-Barajas, E., Paluscio, E., Cervantes, C., and Rensing, C. (2008). Expression of chromate resistance genes from Shewanella sp. strain ANA-3 in Escherichia coli. FEMS Microbiol. Lett. 285, 97–100. doi: 10.1111/j.1574-6968.2008.01220.x
Ahemad, M. (2019). Remediation of metalliferous soils through the heavy metal resistant plant growth promoting bacteria: paradigms and prospects. Arab. J. Chem. 12, 1365–1377. doi: 10.1016/j.arabjc.2014.11.020
Ahmad, I., Akhtar, M. J., Asghar, H. N., Ghafoor, U., and Shahid, M. (2016). Differential effects of plant growth-promoting rhizobacteria on maize growth and cadmium uptake. J. Plant Growth Regul. 35, 303–315. doi: 10.1007/s00344-015-9534-5
Ahmann, D., Roberts, A. L., Krumholz, L. R., and Morel, F. M. (1994). Microbe grows by reducing arsenic. Nature 371:750. doi: 10.1038/371750a0
Akanbi-Gada, M. A., Ogunkunle, C. O., Vishwakarma, V., Viswanathan, K., and Fatoba, P. O. (2019). Phytotoxicity of nano-zinc oxide to tomato plant (Solanum lycopersicum L.): Zn uptake, stress enzymes response and influence on non-enzymatic antioxidants in fruits. Environ. Technol. Innov. 14:100325. doi: 10.1016/j.eti.2019.100325
Akinci, I. E., Akinci, S., and Yilmaz, K. (2010). Response of tomato (Solanum lycopersicum L.) to lead toxicity: growth, element uptake, chlorophyll and water content. Afr. J. Agric. Res. 5, 416–423. doi: 10.5897/AJAR10
Ali, S., and Kim, W. C. (2018). Plant growth promotion under water: decrease of waterlogging-induced ACC and ethylene levels by ACC deaminase-producing bacteria. Front. Microbiol. 9:1096. doi: 10.3389/fmicb.2018.01096
Amin, A., and Latif, Z. (2016). Screening of mercury-resistant and indole-3-acetic acid producing bacterial-consortium for growth promotion of Cicer arietinum L. J. Basic Microbiol. 57, 204–217. doi: 10.1002/jobm.201600352
Anderson, G. L., Williams, J., and Hille, R. (1992). The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J. Biol. Chem. 267, 23674–23682. doi: 10.1016/S0021-9258(18)35891-5
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40, 299–322. doi: 10.1093/femsre/fuv050
Antizar-Ladislao, B. (2010). Bioremediation: working with bacteria. Elements 6, 389–394. doi: 10.2113/gselements.6.6.389
Armendariz, A. L., Talano, M. A., Oller, A. L. W., Medina, M. I., and Agostini, E. (2015). Effect of arsenic on tolerance mechanisms of two plant growth-promoting bacteria used as biological inoculants. J. Environ. Sci. 33, 203–210. doi: 10.1016/j.jes.2014.12.024
Armendariz, A. L., Talano, M. A., Travaglia, C., Reinoso, H., Oller, A. L. W., and Agostini, E. (2016). Arsenic toxicity in soybean seedlings and their attenuation mechanisms. Plant Physiol. Biochem. 98, 119–127. doi: 10.1016/j.plaphy.2015.11.021
Arroyo-Herrera, I., Rojas-Rojas, F. U., Lozano-Cervantes, K. D., Larios-Serrato, V., Vásquez-Murrieta, M. S., Whtiman, W. B., et al. (2020). Draft genome of five Cupriavidus plantarum strains: agave, maize and sorghum plant-associated bacteria with resistance to metals. Biotech 10, 1–10. doi: 10.1007/s13205-020-02210-8
Asaf, S., Khan, A. L., Khan, M. A., Al-Harrasi, A., and Lee, I. J. (2018). Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. Biotech 8, 1–14. doi: 10.1007/s13205-018-1403-z
Babu, A. G., Shea, P. J., Sudhakar, D., Jung, I. B., and Oh, B. T. (2015). Potential use of Pseudomonas koreensis AGB-1 in association with Miscanthus sinensis to remediate heavy metal (loid)-contaminated mining site soil. J. Environ. Manage. 151, 160–166. doi: 10.1016/j.jenvman.2014.12.045
Bacilio-Jiménez, M., Aguilar-Flores, S., Ventura-Zapata, E., Pérez-Campos, E., Bouquelet, S., and Zenteno, E. (2003). Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 249, 271–277. doi: 10.1023/A:1022888900465
Bashan, Y., Puente, M. E., de-Bashan, L. E., and Hernandez, J. P. (2008). Environmental uses of plant growth-promoting bacteria. Plant-Microbe Interact. 661, 69–93.
Begley, T. P., and Ealick, S. E. (2004). Enzymatic reactions involving novel mechanisms of carbanion stabilization. Curr Opin Chem Biol. 8, 508–515. doi: 10.1016/j.cbpa.2004.08.004
Behera, B. C., Yadav, H., Singh, S. K., Mishra, R. R., Sethi, B. K., Dutta, S. K., et al. (2017). Phosphate solubilization and acid phosphatase activity of Serratia sp. isolated from mangrove soil of Mahanadi river delta, Odisha, India. J. Genet. Eng. Biotechnol. 15, 169–178. doi: 10.1016/j.jgeb.2017.01.003
Bertin, P. N., Heinrich-Salmeron, A., Pelletier, E., Goulhen-Chollet, F., Arsène-Ploetze, F., Gallien, S., et al. (2011). Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta-and proteo-genomics. ISME J. 5, 1735–1747. doi: 10.1038/ismej.2011.51
Bilal, S., Shahzad, R., Khan, A. L., Kang, S. M., Imran, Q. M., Al-Harrasi, A., et al. (2018). Endophytic microbial consortia of phytohormones-producing fungus Paecilomyces formosus LHL10 and bacteria Sphingomonas sp. LK11 to Glycine max L. regulates physio-hormonal changes to attenuate aluminum and zinc stresses. Front. Plant Sci. 9:1273. doi: 10.3389/fpls.2018.01273
Blindauer, C. A., Harrison, M. D., Robinson, A. K., Parkinson, J. A., Bowness, P. W., Sadler, P. J., et al. (2002). Multiple bacteria encode metallothioneins and SmtA-like zinc fingers. Mol. Microbiol. 45, 1421–1432. doi: 10.1046/j.1365-2958.2002.03109.x
Borremans, B., Hobman, J. L., Provoost, A., Brown, N. L., and van Der Lelie, D. (2001). Cloning and functional analysis of thepbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 183, 5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001
Branco, R., Chung, A. P., Johnston, T., Gurel, V., Morais, P., and Zhitkovich, A. (2008). The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium (VI) and superoxide. J. Bacteriol. 190, 6996–7003. doi: 10.1128/JB.00289-08
Bravo, G., Vega-Celedón, P., Gentina, J. C., and Seeger, M. (2020). Bioremediation by Cupriavidus metallidurans strain MSR33 of mercury-polluted agricultural soil in a rotary drum bioreactor and its effects on nitrogen cycle microorganisms. Microorganisms 8:1952. doi: 10.3390/microorganisms8121952
Brown, N. L., Barrett, S. R., Camakaris, J., Lee, B. T., and Rouch, D. A. (1995). Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17, 1153–1166. doi: 10.1111/j.1365-2958.1995.mmi_17061153.x
Bruhn, D. F., Li, J., Silver, S., Roberta, F., and Rosen, B. P. (1996). The arsenical resistance operon of IncN plasmid R46. FEMS Microbiol. Lett. 139, 149–153. doi: 10.1111/j.1574-6968.1996.tb08195.x
Bruins, M. R., Kapil, S., and Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 45, 198–207. doi: 10.1006/eesa.1999.1860
Buch, A., Archana, G., and Kumar, G. N. (2010). Heterologous expression of phosphoenolpyruvate carboxylase enhances the phosphate solubilizing ability of fluorescent pseudomonads by altering the glucose catabolism to improve biomass yield. Bioresour. Technol. 101, 679–687. doi: 10.1016/j.biortech.2009.08.075
Cabral, L., Noronha, M. F., de Sousa, S. T. P., Lacerda-Júnior, G. V., Richter, L., Fostier, A. H., et al. (2019). The metagenomic landscape of xenobiotics biodegradation in mangrove sediments. Ecotoxicol. Environ. Saf. 179, 232–240. doi: 10.1016/j.ecoenv.2019.04.044
Cai, L., Yu, K., Yang, Y., Chen, B. W., Li, X. D., and Zhang, T. (2013). Metagenomic exploration reveals high levels of microbial arsenic metabolism genes in activated sludge and coastal sediments. Appl. Microbiol. Biotechnol. 97, 9579–9588. doi: 10.1007/s00253-012-4678-8
Calzada Urquiza, C., Arvizu Hernández, I., Cruz Medina, J. A., Ramos Lopez, M. A., Perez Cruz, J., Rivera Zarazúa, R. L., et al. (2017). Identification by MALDI-TOF mass spectrometry of mercury-resistant bacteria associated with the rhizosphere of an apple orchard. Geomicrobiol. J. 34, 176–182. doi: 10.1080/01490451.2016.1165316
Carlos, M. H. J., Stefani, P. V. Y., Janette, A. M., Melani, M. S. S., and Gabriela, P. O. (2016). Assessing the effects of heavy metals in ACC deaminase and IAA production on plant growth-promoting bacteria. Microbiol. Res. 188, 53–61. doi: 10.1016/j.micres.2016.05.001
Castro-Severyn, J., Pardo-Esté, C., Sulbaran, Y., Cabezas, C., Gariazzo, V., Briones, A., et al. (2019). Arsenic response of three altiplanic Exiguobacterium strains with different tolerance levels against the metalloid species: a proteomics study. Front. Microbiol. 10:2161. doi: 10.3389/fmicb.2019.02161
Cavalca, L., Zecchin, S., Zaccheo, P., Abbas, B., Rotiroti, M., Bonomi, T., et al. (2019). Exploring biodiversity and arsenic metabolism of microbiota inhabiting arsenic-rich groundwaters in Northern Italy. Front. Microbiol. 10:1480. doi: 10.3389/fmicb.2019.01480
Cervantes, C., and Campos-García, J. (2007). “Reduction and efflux of chromate by bacteria,” in Molecular Microbiology of Heavy Metals, eds D. H. Nies and S. Silver (Berlin, Heidelberg: Springer), 407–419. doi: 10.1007/7171_2006_087
Cervantes, C. A. R. L. O. S., Ohtake, H., Chu, L. I. E. N., Misra, T. K., and Silver, S. (1990). Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J. Bacteriol. 172, 287–291. doi: 10.1128/JB.172.1.287-291.1990
Cha, J. S., and Cooksey, D. A. (1991). Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 88, 8915–8919. doi: 10.1073/pnas.88.20.8915
Chakraborty, A., Chowdhury, A. A., Bhakat, K., and Islam, E. (2019). Elevated level of arsenic negatively influences nifH gene expression of isolated soil bacteria in culture condition as well as soil system. Environ. Geochem. Health. 41, 1953–1966. doi: 10.1007/s10653-019-00261-2
Chakraborty, A., and Islam, E. (2018). Temporal dynamics of total and free-living nitrogen-fixing bacterial community abundance and structure in soil with and without history of arsenic contamination during a rice growing season. Environ. Sci. Pollut. Res. 25, 4951–4962. doi: 10.1007/s11356-017-0858-5
Chandran, H., Meena, M., and Sharma, K. (2020). Microbial biodiversity and bioremediation assessment through omics approaches. Front. Environ. Chem. 1:570326. doi: 10.3389/fenvc.2020.570326
Chatterjee, C., Dube, B. K., Sinha, P., and Srivastava, P. (2004). Detrimental effects of lead phytotoxicity on growth, yield, and metabolism of rice. Commun. Soil Sci. Plant Anal. 35, 255–265. doi: 10.1081/CSS-120027648
Chatterjee, J., and Chatterjee, C. (2000). Phytotoxicity of cobalt, chromium and copper in cauliflower. Environ. Poll. 109, 69–74. doi: 10.1016/S0269-7491(99)00238-9
Chauhan, R., Awasthi, S., Indoliya, Y., Chauhan, A. S., Mishra, S., Agrawal, L., et al. (2020). Transcriptome and proteome analyses reveal selenium mediated amelioration of arsenic toxicity in rice (Oryza sativa L.). J. Hazard. Mater. 390:122122. doi: 10.1016/j.jhazmat.2020.122122
Chen, C. M., Misra, T. K., Silver, S., and Rosen, B. P. (1986). Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J. Biol. Chem. 261, 15030–15038. doi: 10.1016/S0021-9258(18)66824-3
Chen, P., Greenberg, B., Taghavi, S., Romano, C., van der Lelie, D., and He, C. (2005). An exceptionally selective lead (II)-regulatory protein from Ralstonia metallidurans: development of a fluorescent lead (II) Probe. Angew. Chem. 117, 2775–2779. doi: 10.1002/ange.200462443
Chen, Y. P., Rekha, P. D., Arun, A. B., Shen, F. T., Lai, W. A., and Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 34, 33–41. doi: 10.1016/j.apsoil.2005.12.002
Christensen, G. A., Gionfriddo, C. M., King, A. J., Moberly, J. G., Miller, C. L., Somenahally, A. C., et al. (2019). Determining the reliability of measuring mercury cycling gene abundance with correlations with mercury and methylmercury concentrations. Environ. Sci. Technol. 53, 8649–8663. doi: 10.1021/acs.est.8b06389
Cooksey, D. A. (1994). Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol. Rev. 14, 381–386. doi: 10.1111/j.1574-6976.1994.tb00112.x
Cox, C. E., Brandl, M. T., de Moraes, M. H., Gunasekera, S., and Teplitski, M. (2018). Production of the plant hormone auxin by Salmonella and its role in the interactions with plants and animals. Front. Microbiol. 8:2668. doi: 10.3389/fmicb.2017.02668
Crosa, J. H., and Walsh, C. T. (2002). Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66, 223–249. doi: 10.1128/MMBR.66.2.223-249.2002
Cruz Medina, J. A., Farias, J. E., Cruz Hernández, A., Martinez, R. G., Valdes, S. S., Silva, G. H., et al. (2013). Isolation and characterization of mercury resistant Bacillus sp. from soils with an extensive history as substrates for mercury extraction in Mexico. Geomicrobiol. J. 30, 454–461. doi: 10.1080/01490451.2012.705229
Das, S., Jean, J. S., Kar, S., Chou, M. L., and Chen, C. Y. (2014). Screening of plant growth-promoting traits in arsenic-resistant bacteria isolated from agricultural soil and their potential implication for arsenic bioremediation. J. Hazard. Mater. 272, 112–120. doi: 10.1016/j.jhazmat.2014.03.012
Denkhaus, E., and Salnikow, K. (2002). Nickel essentiality, toxicity, and carcinogenicity. Crit. Rev. Oncol. Hematol. 42, 35–56. doi: 10.1016/S1040-8428(01)00214-1
Dey, S., and Rosen, B. P. (1995). Dual mode of energy coupling by the oxyanion-translocating ArsB protein. J. Bacteriol. 177, 385–389. doi: 10.1128/jb.177.2.385-389.1995
Dias, M. C., Monteiro, C., Moutinho-Pereira, J., Correia, C., Gonçalves, B., and Santos, C. (2013). Cadmium toxicity affects photosynthesis and plant growth at different levels. Acta Physiol. Plant. 35, 1281–1289. doi: 10.1007/s11738-012-1167-8
Dimkpa, C., Svatoš, A., Merten, D., Büchel, G., and Kothe, E. (2008). Hydroxamate siderophores produced by Streptomyces acidiscabies E13 bind nickel and promote growth in cowpea (Vigna unguiculata L.) under nickel stress. Can. J. Microbiol. 54, 163–172. doi: 10.1139/W07-130
Diorio, C., Cai, J., Marmor, J., Shinder, R., and DuBow, M. S. (1995). An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J. Bacteriol. 177, 2050–2056. doi: 10.1128/JB.177.8.2050-2056.1995
Drewniak, L., Krawczyk, P. S., Mielnicki, S., Adamska, D., Sobczak, A., Lipinski, L., et al. (2016). Physiological and metagenomic analyses of microbial mats involved in self-purification of mine waters contaminated with heavy metals. Front. Microbiol. 7:1252. doi: 10.3389/fmicb.2016.01252
Dvorák, P., Nikel, P. I., Damborský, J., and de Lorenzo, V. (2017). Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 35, 845–866. doi: 10.1016/j.biotechadv.2017.08.001
Eitinger, T., Suhr, J., Moore, L., and Smith, J. A. C. (2005). Secondary transporters for nickel and cobalt ions: theme and variations. Biometals 18, 399–405. doi: 10.1007/s10534-005-3714-x
Erturk, F. A., Agar, G., Arslan, E., Nardemir, G., and Sahin, Z. (2014). Determination of genomic instability and DNA methylation effects of Cr on maize (Zea mays L.) using RAPD and CRED-RA analysis. Acta Physiol. Plant. 36, 1529–1537. doi: 10.1007/s11738-014-1529-5
Espinosa, M. J. C., Blanco, A. C., Schmidgall, T., Atanasoff-Kardjalieff, A. K., Kappelmeyer, U., Tischler, D., et al. (2020). Toward biorecycling: isolation of a soil bacterium that grows on a polyurethane oligomer and monomer. Front. Microbiol. 11:404. doi: 10.3389/fmicb.2020.00404
Etesami, H. (2018). Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues: mechanisms and future prospects. Ecotoxicol. Environ. Saf. 147, 175–191. doi: 10.1016/j.ecoenv.2017.08.032
Franchi, E., Rolli, E., Marasco, R., Agazzi, G., Borin, S., Cosmina, P., et al. (2017). Phytoremediation of a multi contaminated soil: mercury and arsenic phytoextraction assisted by mobilizing agent and plant growth promoting bacteria. J. Soils Sediment. 17, 1224–1236. doi: 10.1007/s11368-015-1346-5
Franke, S., Grass, G., Rensing, C., and Nies, D. H. (2003). Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185, 3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003
Gadd, G. M. (1990). Heavy metal accumulation by bacteria and other microorganisms. Experientia 46, 834–840. doi: 10.1007/BF01935534
Gadd, G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156, 609–643. doi: 10.1099/mic.0.037143-0
Garg, N., and Singla, P. (2011). Arsenic toxicity in crop plants: physiological effects and tolerance mechanisms. Environ. Chem. Lett. 9, 303–321. doi: 10.1007/s10311-011-0313-7
Gillis, B. S., Arbieva, Z., and Gavin, I. M. (2012). Analysis of lead toxicity in human cells. BMC Genom. 13, 1–12. doi: 10.1186/1471-2164-13-344
Glibota, N., Grande, M., Galvez, A., and Ortega, E. (2020). Genetic determinants for metal tolerance and antimicrobial resistance detected in bacteria isolated from soils of olive tree farms. Antibiotics 9:476. doi: 10.3390/antibiotics9080476
Glick, B. R. (2005). Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7. doi: 10.1016/j.femsle.2005.07.030
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2016). Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2:1127500. doi: 10.1080/23311932.2015.1127500
Griffin, H. G., Foster, T. J., Silver, S., and Misra, T. K. (1987). Cloning and DNA sequence of the mercuric-and organomercurial-resistance determinants of plasmid pDU1358. Proc. Natl. Acad. Sci. U. S. A. 84, 3112–3116. doi: 10.1073/pnas.84.10.3112
Gutiérrez-Barranquero, J. A., de Vicente, A., Carrión, V. J., Sundin, G. W., and Cazorla, F. M. (2013). Recruitment and rearrangement of three different genetic determinants into a conjugative plasmid increase copper resistance in Pseudomonas syringae. Appl. Environ. Microbiol. 79, 1028–1033. doi: 10.1128/AEM.02644-12
Han, J. I., Choi, H. K., Lee, S. W., Orwin, P. M., Kim, J., LaRoe, S. L., et al. (2011). Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 193, 1183–1190. doi: 10.1128/JB.00925-10
Han, Y. H., Fu, J. W., Xiang, P., Cao, Y., Rathinasabapathi, B., Chen, Y., et al. (2017). Arsenic and phosphate rock impacted the abundance and diversity of bacterial arsenic oxidase and reductase genes in rhizosphere of As-hyperaccumulator Pteris vittata. J. Hazard. Mater. 321, 146–153. doi: 10.1016/j.jhazmat.2016.08.079
Haney, C. J., Grass, G., Franke, S., and Rensing, C. (2005). New developments in the understanding of the cation diffusion facilitator family. J. Ind. Microbiol. Biotechnol. 32, 215–226. doi: 10.1007/s10295-005-0224-3
Hao, X., Xie, P., Johnstone, L., Miller, S. J., Rensing, C., and Wei, G. (2012). Genome sequence and mutational analysis of plant-growth-promoting bacterium Agrobacterium tumefaciens CCNWGS0286 isolated from a zinc-lead mine tailing. Appl. Environ. Microbiol. 78, 5384–5394. doi: 10.1128/AEM.01200-12
Hao, X., Xie, P., Zhu, Y. G., Taghavi, S., Wei, G., and Rensing, C. (2015). Copper tolerance mechanisms of Mesorhizobium amorphae and its role in aiding phytostabilization by Robinia pseudoacacia in copper contaminated soil. Environ. Sci. Technol. 49, 2328–2340. doi: 10.1021/es504956a
Hasman, H. (2005). The tcrB gene is part of the tcrYAZB operon conferring copper resistance in Enterococcus faecium and Enterococcus faecalis. Microbiology 151, 3019–3025. doi: 10.1099/mic.0.28109-0
Imchen, M., Kumavath, R., Barh, D., Vaz, A., Góes-Neto, A., Tiwari, S., et al. (2018). Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Sci. Rep. 8, 1–15. doi: 10.1038/s41598-018-29521-4
Indiragandhi, P., Anandham, R., Madhaiyan, M., and Sa, T. M. (2008). Characterization of plant growth–promoting traits of bacteria isolated from larval guts of diamondback moth Plutella xylostella (Lepidoptera: Plutellidae). Curr. Microbiol. 56, 327–333. doi: 10.1007/s00284-007-9086-4
Inoue, C., Sugawara, K., and Kusano, T. (1991). The merR regulatory gene in Thiobacillus ferrooxidans is spaced apart from the mer structural genes. Mol. Microbiol. 5, 2707–2718. doi: 10.1111/j.1365-2958.1991.tb01979.x
Islam, F., Yasmeen, T., Arif, M. S., Riaz, M., Shahzad, S. M., Imran, Q., et al. (2016). Combined ability of chromium (Cr) tolerant plant growth promoting bacteria (PGPB) and salicylic acid (SA) in attenuation of chromium stress in maize plants. Plant Physiol. Biochem. 108, 456–467. doi: 10.1016/j.plaphy.2016.08.014
Islam, F., Yasmeen, T., Riaz, M., Arif, M. S., Ali, S., and Raza, S. H. (2014). Proteus mirabilis alleviates zinc toxicity by preventing oxidative stress in maize (Zea mays) plants. Ecotoxicol. Environ. Saf. 110, 143–152. doi: 10.1016/j.ecoenv.2014.08.020
Izrael-Živković, L., Rikalović, M., Gojgić-Cvijović, G., Kazazić, S., Vrvić, M., Brčeski, I., et al. (2018). Cadmium specific proteomic responses of a highly resistant Pseudomonas aeruginosa san ai. RSC Adv. 8, 10549–10560. doi: 10.1039/C8RA00371H
Jebara, S. H., Saadani, O., Fatnassi, I. C., Chiboub, M., Abdelkrim, S., and Jebara, M. (2015). Inoculation of Lens culinaris with Pb-resistant bacteria shows potential for phytostabilization. Environ. Sci. Pollut. Res. 22, 2537–2545. doi: 10.1007/s11356-014-3510-7
Ji, G., and Silver, S. (1992). Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc. Natl. Acad. Sci. U. S. A. 89, 9474–9478. doi: 10.1073/pnas.89.20.9474
Jomova, K., Jenisova, Z., Feszterova, M., Baros, S., Liska, J., Hudecova, D., et al. (2011). Arsenic: toxicity, oxidative stress and human disease. J. Appl. Toxicol. 31, 95–107. doi: 10.1002/jat.1649
Juhnke, S., Peitzsch, N., Hübener, N., Große, C., and Nies, D. H. (2002). New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch. Microbiol. 179, 15–25. doi: 10.1007/s00203-002-0492-5
Kamran, M., Eqani, S. A. M. A. S., Katsoyiannis, A., Xu, R., Bibi, S., Benizri, E., et al. (2016). Phytoextraction of chromium (Cr) and influence of Pseudomonas putida on Eruca sativa growth. J. Geochem. Explor. 182, 269–274. doi: 10.1016/j.gexplo.2016.09.005
Kamran, S., Shahid, I., Baig, D. N., Rizwan, M., Malik, K. A., and Mehnaz, S. (2017). Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 8:2593. doi: 10.3389/fmicb.2017.02593
Kang, S. M., Asaf, S., Khan, A. L., Khan, A., Mun, B. G., Khan, M. A., et al. (2020). Complete genome sequence of jgexplo201609005 Pseudomonas psychrotolerans CS51, a plant growth-promoting bacterium, under heavy metal stress conditions. Microorganisms 8:382. doi: 10.3390/microorganisms8030382
Karthik, C., and Arulselvi, P. I. (2017). Biotoxic effect of chromium (VI) on plant growth-promoting traits of novel Cellulosimicrobium funkei strain AR8 isolated from Phaseolus vulgaris rhizosphere. Geomicrobiol. J. 34, 434–442. doi: 10.1080/01490451.2016.1219429
Kaur, C., Selvakumar, G., and Ganeshamurthy, A. N. (2016). Draft genome sequence of phosphate-solubilizing bacterium Paraburkholderia tropica strain P-31 isolated from pomegranate (Punica granatum) rhizosphere. Genome Announc. 4:16. doi: 10.1128/genomeA.00844-16
Khan, A. G. (2005). Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J. Trace Elem. Med. Biol. 18, 355–364. doi: 10.1016/j.jtemb.2005.02.006
Khatun, S., Ali, M. B., Hahn, E. J., and Paek, K. Y. (2008). Copper toxicity in Withania somnifera: Growth and antioxidant enzymes responses of in vitro grown plants. Environ. Exp. Bot. 64, 279–285. doi: 10.1016/j.envexpbot.2008.02.004
Kotoky, R., Nath, S., Maheshwari, D. K., and Pandey, P. (2019). Cadmium resistant plant growth promoting rhizobacteria Serratia marcescens S2I7 associated with the growth promotion of rice plant. Environ. Sustain. 2, 135–144. doi: 10.1007/s42398-019-00055-3
Kou, S., Vincent, G., Gonzalez, E., Pitre, F. E., Labrecque, M., and Brereton, N. J. (2018). The response of a 16S ribosomal RNA gene fragment amplified community to lead, zinc, and copper pollution in a Shanghai field trial. Front. Microbiol. 9:366. doi: 10.3389/fmicb.2018.00366
Kusano, T., Ji, G. Y., Inoue, C., and Silver, S. (1990). Constitutive synthesis of a transport function encoded by the Thiobacillus ferrooxidans merC gene cloned in Escherichia coli. J. Bacteriol. 172, 2688–2692. doi: 10.1128/JB.172.5.2688-2692.1990
Lamhamdi, M., Bakrim, A., Aarab, A., Lafont, R., and Sayah, F. (2011). Lead phytotoxicity on wheat (Triticum aestivum L.) seed germination and seedlings growth. Comptes Rendus Biol. 334, 118–126. doi: 10.1016/j.crvi.2010.12.006
Lebrun, M., Audurier, A., and Cossart, P. (1994). Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J. Bacteriol. 176, 3049–3061. doi: 10.1128/jb.176.10.3049-3061.1994
Levinson, H. S., and Mahler, I. (1998). Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus. FEMS Microbiol. Lett. 161, 135–138. doi: 10.1111/j.1574-6968.1998.tb12939.x
Leyssens, L., Vinck, B., Van Der Straeten, C., Wuyts, F., and Maes, L. (2017). Cobalt toxicity in humans—a review of the potential sources and systemic health effects. Toxicology 387, 43–56. doi: 10.1016/j.tox.2017.05.015
Li, H. F., Gray, C., Mico, C., Zhao, F. J., and McGrath, S. P. (2009). Phytotoxicity and bioavailability of cobalt to plants in a range of soils. Chemosphere 75, 979–986. doi: 10.1016/j.chemosphere.2008.12.068
Li, P., Wang, Y., Dai, X., Zhang, R., Jiang, Z., Jiang, D., et al. (2015). Microbial community in high arsenic shallow groundwater aquifers in Hetao Basin of Inner Mongolia, China. PLoS ONE 10:e0125844. doi: 10.1371/journal.pone.0125844
Liebert, C. A., Hall, R. M., and Summers, A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522. doi: 10.1128/MMBR.63.3.507-522.1999
Liu, C., Lin, H., Dong, Y., Li, B., and Wang, L. (2019). Identification and characterization of plant growth–promoting endophyte RE02 from Trifolium repens L. in mining smelter. Environ. Sci. Pollut. Res. 26, 17236–17247. doi: 10.1007/s11356-019-04904-w
Liu, H., Wang, J., Sun, H., Han, X., Peng, Y., Liu, J., et al. (2020). Transcriptome profiles reveal the growth-promoting mechanisms of Paenibacillus polymyxa YC0136 on Tobacco (Nicotiana tabacum L.). Front. Microbiol. 11:2384. doi: 10.3389/fmicb.2020.584174
Logeshwaran, P., Thangaraju, M., and Rajasundari, K. (2009). Hydroxamate siderophores of endophytic bacteria Gluconacetobacter diazotrophicus isolated from sugarcane roots. Aust. J. Basic Appl. Sci. 3, 3564–3567.
Luo, J., Bai, Y., Liang, J., and Qu, J. (2014). Metagenomic approach reveals variation of microbes with arsenic and antimony metabolism genes from highly contaminated soil. PLoS ONE 9:e108185. doi: 10.1371/journal.pone.0108185
Luziatelli, F., Ficca, A. G., Cardarelli, M., Melini, F., Cavalieri, A., and Ruzzi, M. (2020). Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 8:153. doi: 10.3390/microorganisms8020153
Ma, L., Chen, N., Feng, C., Li, M., Gao, Y., and Hu, Y. (2020). Coupling enhancement of Chromium (VI) bioreduction in groundwater by phosphorus minerals. Chemosphere 240:124896. doi: 10.1016/j.chemosphere.2019.124896
Ma, Y., Rajkumar, M., and Freitas, H. (2009). Inoculation of plant growth promoting bacterium Achromobacter xylosoxidans strain Ax10 for the improvement of copper phytoextraction by Brassica juncea. J. Environ. Manage. 90, 831–837. doi: 10.1016/j.jenvman.2008.01.014
Ma, Y., Rajkumar, M., Vicente, J. A. F., and Freitas, H. (2010). Inoculation of Ni-resistant plant growth promoting bacterium Psychrobacter sp. strain SRS8 for the improvement of nickel phytoextraction by energy crops. Int. J. Phytoremediation. 13, 126–139. doi: 10.1080/15226511003671403
Mallick, I., Islam, E., and Kumar Mukherjee, S. (2015). Fundamentals and application potential of arsenic-resistant bacteria for bioremediation in rhizosphere: a review. Soil. Sediment. Contam. 24, 704–718. doi: 10.1080/15320383.2015.1010072
Manara, A. (2012). “Plant responses to heavy metal toxicity,” in Plants and Heavy Metals, ed A. Furini (Dordrecht: Springer), 27–53. doi: 10.1007/978-94-007-4441-7_2
Mandal, B. K., and Suzuki, K. T. (2002). Arsenic round the world: a review. Talanta 58, 201–235. doi: 10.1016/S0039-9140(02)00268-0
Marques, A. P., Rangel, A. O., and Castro, P. M. (2009). Remediation of heavy metal contaminated soils: phytoremediation as a potentially promising clean-up technology. Crit. Rev. Environ. Sci. Technol. 39, 622–654. doi: 10.1080/10643380701798272
Martelli, A., Rousselet, E., Dycke, C., Bouron, A., and Moulis, J. M. (2006). Cadmium toxicity in animal cells by interference with essential metals. Biochimie 88, 1807–1814. doi: 10.1016/j.biochi.2006.05.013
Mazhar, S. H., Herzberg, M., Ben Fekih, I., Zhang, C., Bello, S. K., Li, Y. P., et al. (2020). Comparative insights into the complete genome sequence of highly metal resistant Cupriavidus metallidurans strain BS1 isolated from a gold–copper mine. Front. Microbiol. 11:47. doi: 10.3389/fmicb.2020.00047
Md Badrul Hisham, N. H., Ibrahim, M. F., Ramli, N., and Abd-Aziz, S. (2019). Production of biosurfactant produced from used cooking oil by Bacillus sp. HIP3 for heavy metals removal. Molecules 24:2617. doi: 10.3390/molecules24142617
Meyer, J., Schmidt, A., Michalke, K., and Hensel, R. (2007). Volatilisation of metals and metalloids by the microbial population of an alluvial soil. Syst. Appl. Microbiol. 30, 229–238. doi: 10.1016/j.syapm.2006.05.001
Mihara, K., Tanabe, T., Yamakawa, Y., Funahashi, T., Nakao, H., Narimatsu, S., et al. (2004). Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 150, 2587–2597. doi: 10.1099/mic.0.27141-0
Mitra, S., Pramanik, K., Ghosh, P. K., Soren, T., Sarkar, A., Dey, R. S., et al. (2018). Characterization of Cd-resistant Klebsiella michiganensis MCC3089 and its potential for rice seedling growth promotion under Cd stress. Microbiol. Res. 210, 12–25. doi: 10.1016/j.micres.2018.03.003
Mole, B. M., Baltrus, D. A., Dangl, J. L., and Grant, S. R. (2007). Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 15, 363–371. doi: 10.1016/j.tim.2007.06.005
Monsieurs, P., Moors, H., Van Houdt, R., Janssen, P. J., Janssen, A., Coninx, I., et al. (2011). Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals 24, 1133–1151. doi: 10.1007/s10534-011-9473-y
Moreira, H., Marques, A. P., Franco, A. R., Rangel, A. O., and Castro, P. M. (2014). Phytomanagement of Cd-contaminated soils using maize (Zea mays L.) assisted by plant growth-promoting rhizobacteria. Environ. Sci. Pollut. Res. 21, 9742–9753. doi: 10.1007/s11356-014-2848-1
Mossa, A. W., Young, S. D., and Crout, N. M. (2020). Zinc uptake and phyto-toxicity: Comparing intensity-and capacity-based drivers. Sci. Total Environ. 699:134314. doi: 10.1016/j.scitotenv.2019.134314
Mukhopadhyay, D., Yu, H. R., Nucifora, G., and Misra, T. K. (1991). Purification and functional characterization of MerD. A coregulator of the mercury resistance operon in gram-negative bacteria. J. Biol. Chem. 266, 18538–18542. doi: 10.1016/S0021-9258(18)55095-X
Muller, D., Lièvremont, D., Simeonova, D. D., Hubert, J. C., and Lett, M. C. (2003). Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J. Bacteriol. 185, 135–141. doi: 10.1128/JB.185.1.135-141.2003
Mulligan, C. N., Yong, R. N., and Gibbs, B. F. (2001). Heavy metal removal from sediments by biosurfactants. J. Hazard. Mater. 85, 111–125. doi: 10.1016/S0304-3894(01)00224-2
Mulrooney, S. B., and Hausinger, R. P. (2003). Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27, 239–261. doi: 10.1016/S0168-6445(03)00042-1
Naik, M. M., Pandey, A., and Dubey, S. K. (2012). Pseudomonas aeruginosa strain WI-1 from Mandovi estuary possesses metallothionein to alleviate lead toxicity and promotes plant growth. Ecotoxicol. Environ. Saf. 79, 129–133. doi: 10.1016/j.ecoenv.2011.12.015
Najeeb, U., Xu, L., Ali, S., Jilani, G., Gong, H. J., Shen, W. Q., et al. (2009). Citric acid enhances the phytoextraction of manganese and plant growth by alleviating the ultrastructural damages in Juncus effusus L. J. Hazard. Mater. 170, 1156–1163. doi: 10.1016/j.jhazmat.2009.05.084
Nanda, M., Kumar, V., and Sharma, D. K. (2019). Multimetal tolerance mechanisms in bacteria: the resistance strategies acquired by bacteria that can be exploited to ‘clean-up’ heavy metal contaminants from water. Aquat. Toxicol. 212, 1–10. doi: 10.1016/j.aquatox.2019.04.011
Nascimento, F. X., Rossi, M. J., Soares, C. R., McConkey, B. J., and Glick, B. R. (2014). New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE 9:e99168. doi: 10.1371/journal.pone.0099168
Navarro, C., Wu, L. F., and Mandrand-Berthelot, M. A. (1993). The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9, 1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x
Nazir, H., Asghar, H. N., Zahir, Z. A., Akhtar, M. J., and Saleem, M. (2016). Judicious use of kinetin to improve growth and yield of rice in nickel contaminated soil. Int. J. Phytoremediat. 18, 651–655. doi: 10.1080/15226514.2015.1094444
Nie, J., Pan, Y., Shi, J., Guo, Y., Yan, Z., Duan, X., et al. (2015). A comparative study on the uptake and toxicity of nickel added in the form of different salts to maize seedlings. Int. J. Environ. Res. Public Health 12, 15075–15087. doi: 10.3390/ijerph121214972
Nies, A., Nies, D. H., and Silver, S. (1990). Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J. Biol. Chem. 265, 5648–5653. doi: 10.1016/S0021-9258(19)39411-6
Nies, D. H. (1999). Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51, 730–750. doi: 10.1007/s002530051457
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
Nies, D. H., Nies, A., Chu, L., and Silver, S. (1989). Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. U. S. A. 86, 7351–7355. doi: 10.1073/pnas.86.19.7351
Nies, D. H., and Silver, S. (1995). Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14, 186–199. doi: 10.1007/BF01569902
Novick, R. P., Murphy, E., Gryczan, T. J., Baron, E., and Edelman, I. (1979). Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid 2, 109–129. doi: 10.1016/0147-619X(79)90010-6
Odermatt, A., and Solioz, M. (1995). Two trans-acting metallo-regulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J. Biol. Chem. 270, 4349–4354. doi: 10.1074/jbc.270.9.4349
Odermatt, A., Suter, H., Krapf, R., and Solioz, M. (1992). An ATPase operon involved in copper resistance by Enterococcus hiraea. Ann. N. Y. Acad. Sci. 671, 484–486. doi: 10.1111/j.1749-6632.1992.tb43836.x
Ortúzar, M., Trujillo, M. E., Román-Ponce, B., and Carro, L. (2020). Micromonospora metallophores: a plant growth promotion trait useful for bacterial-assisted phytoremediation? Sci. Total Environ. 739:139850. doi: 10.1016/j.scitotenv.2020.139850
Ozfidan-Konakci, C., Yildiztugay, E., Elbasan, F., Kucukoduk, M., and Turkan, I. (2020). Hydrogen sulfide (H2S) and nitric oxide (NO) alleviate cobalt toxicity in wheat (Triticum aestivum L.) by modulating photosynthesis, chloroplastic redox and antioxidant capacity. J. Hazard. Mater. 388:122061. doi: 10.1016/j.jhazmat.2020.122061
Pandey, V., Dixit, V., and Shyam, R. (2009). Chromium effect on ROS generation and detoxification in pea (Pisum sativum) leaf chloroplasts. Protoplasma 236, 85–95. doi: 10.1007/s00709-009-0061-8
Park, J. H., Bolan, N., Megharaj, M., and Naidu, R. (2011). Concomitant rock phosphate dissolution and lead immobilization by phosphate solubilizing bacteria (Enterobacter sp.). J. Environ. Manage. 92, 1115–1120. doi: 10.1016/j.jenvman.2010.11.031
Pei, Y., Tao, C., Ling, Z., Yu, Z., Ji, J., Khan, A., et al. (2020). Exploring novel Cr (VI) remediation genes for Cr (VI)-contaminated industrial wastewater treatment by comparative metatranscriptomics and metagenomics. Sci. Total Environ. 742:140435. doi: 10.1016/j.scitotenv.2020.140435
Peng, J., Miao, L., Chen, X., and Liu, P. (2018). Comparative transcriptome analysis of Pseudomonas putida KT2440 revealed its response mechanisms to elevated levels of zinc stress. Front. Microbiol. 9:1669. doi: 10.3389/fmicb.2018.01669
Phung, L. T., Trimble, W. L., Meyer, F., Gilbert, J. A., and Silver, S. (2012). Draft genome sequence of Alcaligenes faecalis subsp. faecalis NCIB 8687 (CCUG 2071). J. Bacteriol. 194:18. doi: 10.1128/JB.01185-12
Pieper, D. H., and Reineke, W. (2000). Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11, 262–270. doi: 10.1016/S0958-1669(00)00094-X
Plum, L. M., Rink, L., and Haase, H. (2010). The essential toxin: impact of zinc on human health. Int. J. Environ. Res. Public Health 7, 1342–1365. doi: 10.3390/ijerph7041342
Prigent-Combaret, C., Blaha, D., Pothier, J. F., Vial, L., Poirier, M. A., Wisniewski-Dy,é, F., et al. (2008). Physical organization and phylogenetic analysis of acdR as leucine-responsive regulator of the 1-aminocyclopropane-1-carboxylate deaminase gene acdS in phytobeneficial Azospirillum lipoferum 4B and other Proteobacteria. FEMS Microbiol. Ecol. 65, 202–219. doi: 10.1111/j.1574-6941.2008.00474.x
Rensing, C., Pribyl, T., and Nies, D. H. (1997). New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 179, 6871–6879. doi: 10.1128/JB.179.22.6871-6879.1997
Rizvi, A., and Khan, M. S. (2018). Heavy metal induced oxidative damage and root morphology alterations of maize (Zea mays L.) plants and stress mitigation by metal tolerant nitrogen fixing Azotobacter chroococcum. Ecotoxicol. Environ. Saf. 157, 9–20. doi: 10.1016/j.ecoenv.2018.03.063
Rojjanateeranaj, P., Sangthong, C., and Prapagdee, B. (2017). Enhanced cadmium phytoremediation of Glycine max L. through bioaugmentation of cadmium-resistant bacteria assisted by biostimulation. Chemosphere 185, 764–771. doi: 10.1016/j.chemosphere.2017.07.074
Rosenstein, R., Peschel, A., Wieland, B., and Götz, F. (1992). Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J. Bacteriol. 174, 3676–3683. doi: 10.1128/JB.174.11.3676-3683.1992
Sadej, W., Zołnowski, A. C., Ciećko, Z., Grzybowski, Ł., and Szostek, R. (2019). Evaluation of the impact of soil contamination with mercury and application of soil amendments on the yield and chemical composition of Avena sativa L. J. Environ. Sci. Health A 55, 82–96. doi: 10.1080/10934529.2019.1667671
Sagardoy, R. U. T. H., Morales, F. E. R. M. Í. N., López-Millán, A. F., Abadía, A. N. U. N. C. I. A. C. I. Ó. N., and Abadía, J. A. V. I. E. R. (2009). Effects of zinc toxicity on sugar beet (Beta vulgaris L.) plants grown in hydroponics. Plant Biol. 11, 339–350. doi: 10.1111/j.1438-8677.2008.00153.x
Sager, S. M. A., Wijaya, L., Alyemeni, M. N., Hatamleh, A. A., and Ahmad, P. (2020). Impact of different cadmium concentrations on two Pisum sativum l. genotypes. Pak. J. Bot. 52, 821–829. doi: 10.30848/PJB2020-3(10)
Saharan, B. S., and Nehra, V. (2011). Plant growth promoting rhizobacteria: a critical review. Life Sci. Med. Res. 21:30.
Sahu, G. K., Upadhyay, S., and Sahoo, B. B. (2012). Mercury induced phytotoxicity and oxidative stress in wheat (Triticum aestivum L.) plants. Physiol. Mol. Biol. Plants 18, 21–31. doi: 10.1007/s12298-011-0090-6
Saleem, M. H., Fahad, S., Khan, S. U., Din, M., Ullah, A., Sabagh, A. E., et al. (2020). Copper-induced oxidative stress, initiation of antioxidants and phytoremediation potential of flax (Linum usitatissimum L.) seedlings grown under the mixing of two different soils of China. Environ. Sci. Poll. Res. 27, 5211–5221. doi: 10.1007/s11356-019-07264-7
Saltikov, C. W., Cifuentes, A., Venkateswaran, K., and Newman, D. K. (2003). The ars detoxification system is advantageous but not required for As (V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl. Environ. Microbiol. 69, 2800–2809. doi: 10.1128/AEM.69.5.2800-2809.2003
Saltikov, C. W., and Newman, D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U. S. A. 100, 10983–10988. doi: 10.1073/pnas.1834303100
Santini, J. M., and vanden Hoven, R. N. (2004). Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J. Bacteriol. 186, 1614–1619. doi: 10.1128/JB.186.6.1614-1619.2004
Sedlmeier, R., and Altenbuchner, J. (1992). Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol. Gen. Genet. 236, 76–85. doi: 10.1007/BF00279645
Shabnam, I., and Seema, M. (2011). Phytotoxicity of nickel and its accumulation in tissues of three Vigna species at their early growth stages. J. Appl. Bot. Food Qual. 84, 223–228.
Shah, S., and Damare, S. (2020). Cellular response of Brevibacterium casei NIOSBA88 to arsenic and chromium—a proteomic approach. Braz. J. Microbiol. 51, 1885–1895. doi: 10.1007/s42770-020-00353-7
Sharma, P. K., Balkwill, D. L., Frenkel, A., and Vairavamurthy, M. A. (2000). A new Klebsiella planticola strain (Cd-1) grows anaerobically at high cadmium concentrations and precipitates cadmium sulfide. Appl. Environ. Microbiol. 66, 3083–3087. doi: 10.1128/AEM.66.7.3083-3087.2000
Shi, L. D., Chen, Y. S., Du, J. J., Hu, Y. Q., Shapleigh, J. P., and Zhao, H. P. (2019). Metagenomic evidence for a Methylocystis species capable of bioremediation of diverse heavy metals. Front. Microbiol. 9:3297. doi: 10.3389/fmicb.2018.03297
Shi, X., Zhou, G., Liao, S., Shan, S., Wang, G., and Guo, Z. (2018). Immobilization of cadmium by immobilized Alishewanella sp. WH16-1 with alginate-lotus seed pods in pot experiments of Cd-contaminated paddy soil. J. Hazard. Mater. 357, 431–439. doi: 10.1016/j.jhazmat.2018.06.027
Shiyab, S., Chen, J., Han, F. X., Monts, D. L., Matta, F. B., Gu, M., et al. (2009). Phytotoxicity of mercury in Indian mustard (Brassica juncea L.). Ecotoxicol. Environ. Safety 72, 619–625. doi: 10.1016/j.ecoenv.2008.06.002
Silver, S., and Ji, G. (1994). Newer systems for bacterial resistances to toxic heavy metals. Environ. Health. Perspect. 102, 107–113. doi: 10.1289/ehp.94102s3107
Silver, S., and Phung, L. T. (1996). Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50, 753–789. doi: 10.1146/annurev.micro.50.1.753
Silver, S., and Phung, L. T. (2005a). A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J. Ind. Microbiol. Biotechnol. 32, 587–605. doi: 10.1007/s10295-005-0019-6
Silver, S., and Phung, L. T. (2005b). Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71, 599–608. doi: 10.1128/AEM.71.2.599-608.2005
Silver, S. I. M. O. N., and Walderhaug, M. A. R. K. (1992). Gene regulation of plasmid-and chromosome-determined inorganic ion transport in bacteria. Microbiol. Mol. Biol. Rev. 56, 195–228. doi: 10.1128/MMBR.56.1.195-228.1992
Smith, D. L., Tao, T., and Maguire, M. E. (1993). Membrane topology of a P-type ATPase. The MgtB magnesium transport protein of Salmonella typhimurium. J. Biol. Chem. 268, 22469–22479. doi: 10.1016/S0021-9258(18)41553-0
Solioz, M., and Vulpe, C. (1996). CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends. Biochem. Sci. 21, 237–241. doi: 10.1016/S0968-0004(96)20016-7
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signalling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Srivastava, V., Sarkar, A., Singh, S., Singh, P., de Araujo, A. S., and Singh, R. P. (2017). Agroecological responses of heavy metal pollution with special emphasis on soil health and plant performances. Front. Environ. Sci. 5:64. doi: 10.3389/fenvs.2017.00064
Stoppel, R. D., and Schlegel, H. G. (1995). Nickel-resistant bacteria from anthropogenically nickel-polluted and naturally nickel-percolated ecosystems. Appl. Environ. Microbiol. 61, 2276–2285. doi: 10.1128/AEM.61.6.2276-2285.1995
Suleman, M., Yasmin, S., Rasul, M., Yahya, M., Atta, B. M., and Mirza, M. S. (2018). Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS ONE 13:e0204408. doi: 10.1371/journal.pone.0204408
Sun, H., Brocato, J., and Costa, M. (2015). Oral chromium exposure and toxicity. Curr. Environ. Health Rep. 2, 295–303. doi: 10.1007/s40572-015-0054-z
Sun, W., Zhu, B., Yang, F., Dai, M., Sehar, S., Peng, C., et al. (2021). Optimization of biosurfactant production from Pseudomonas sp. CQ2 and its application for remediation of heavy metal contaminated soil. Chemosphere 265:129090. doi: 10.1016/j.chemosphere.2020.129090
Sundaramoorthy, P., Chidambaram, A., Ganesh, K. S., Unnikannan, P., and Baskaran, L. (2010). Chromium stress in paddy:(i) nutrient status of paddy under chromium stress;(ii) phytoremediation of chromium by aquatic and terrestrial weeds. Comptes Rendus Biol. 333, 597–607. doi: 10.1016/j.crvi.2010.03.002
Tang, Y. J., Hwang, J. S., Wemmer, D. E., and Keasling, J. D. (2007). Shewanella oneidensis MR-1 fluxome under various oxygen conditions. Appl. Environ. Microbiol. 73, 718–729. doi: 10.1128/AEM.01532-06
Tariq, S., Amin, A., and Latif, Z. (2015). PCR based DNA fingerprinting of mercury resistant and nitrogen fixing Pseudomonas spp. Pure Appl. Biol. 4:129. doi: 10.19045/bspab.2015.41018
Thomas, D. J., Waters, S. B., and Styblo, M. (2004). Elucidating the pathway for arsenic methylation. Toxicol. Appl. Pharmacol. 198, 319–326. doi: 10.1016/j.taap.2003.10.020
Tian, J., Wu, N., Li, J., Liu, Y., Guo, J., Yao, B., et al. (2007). Nickel-resistant determinant from Leptospirillum ferriphilum. Appl. Environ. Microbiol. 73, 2364–2368. doi: 10.1128/AEM.00207-07
Upadhyay, A., Kochar, M., Rajam, M. V., and Srivastava, S. (2017). Players over the surface: unraveling the role of exopolysaccharides in zinc biosorption by fluorescent Pseudomonas strain psd. Front. Microbiol. 8:284. doi: 10.3389/fmicb.2017.00284
Uriu-Adams, J. Y., and Keen, C. L. (2005). Copper, oxidative stress, and human health. Mol. Aspects Med. 26, 268–298. doi: 10.1016/j.mam.2005.07.015
Ventura-Lima, J., Bogo, M. R., and Monserrat, J. M. (2011). Arsenic toxicity in mammals and aquatic animals: a comparative biochemical approach. Ecotoxicol. Environ. Saf. 74, 211–218. doi: 10.1016/j.ecoenv.2010.11.002
Vicente, E. J., and Dean, D. R. (2017). Keeping the nitrogen-fixation dream alive. Proc. Natl. Acad. Sci. U. S. A. 114, 3009–3011. doi: 10.1073/pnas.1701560114
Viti, C., Marchi, E., Decorosi, F., and Giovannetti, L. (2014). Molecular mechanisms of Cr (VI) resistance in bacteria and fungi. FEMS Microbiol. Rev. 38, 633–659. doi: 10.1111/1574-6976.12051
Volpicella, M., Leoni, C., Manzari, C., Chiara, M., Picardi, E., Piancone, E., et al. (2017). Transcriptomic analysis of nickel exposure in Sphingobium sp. ba1 cells using RNA-seq. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-08934-7
Wang, H., Wu, P., Liu, J., Yang, S., Ruan, B., Rehman, S., et al. (2020). The regulatory mechanism of Chryseobacterium sp. resistance mediated by montmorillonite upon cadmium stress. Chemosphere 240:124851. doi: 10.1016/j.chemosphere.2019.124851
Wang, L., Yin, Z., and Jing, C. (2020). Metagenomic insights into microbial arsenic metabolism in shallow groundwater of Datong basin, China. Chemosphere 245:125603. doi: 10.1016/j.chemosphere.2019.125603
Wang, Y., Li, P., Jiang, Z., Sinkkonen, A., Wang, S., Tu, J., et al. (2016). Microbial community of high arsenic groundwater in agricultural irrigation area of Hetao Plain, Inner Mongolia. Front. Microbiol. 7:1917. doi: 10.3389/fmicb.2016.01917
Wang, Y., Shi, J., Wang, H., Lin, Q., Chen, X., and Chen, Y. (2007). The influence of soil heavy metals pollution on soil microbial biomass, enzyme activity, and community composition near a copper smelter. Ecotoxicol. Environ. Saf. 67, 75–81. doi: 10.1016/j.ecoenv.2006.03.007
Wani, P. A., Wahid, S., Rafi, N., and Wani, U. (2020). Role of NADH-dependent chromium reductases, exopolysaccharides and antioxidants by Paenibacillus thiaminolyticus PS 5 against damage induced by reactive oxygen species. Chem. Ecol. 36, 663–684. doi: 10.1080/02757540.2020.1770736
Wei, G., Fan, L., Zhu, W., Fu, Y., Yu, J., and Tang, M. (2009). Isolation and characterization of the heavy metal resistant bacteria CCNWRS33-2 isolated from root nodule of Lespedeza cuneata in gold mine tailings in China. J. Hazard. Mater. 162, 50–56. doi: 10.1016/j.jhazmat.2008.05.040
Wei, Y., Zhao, Y., Shi, M., Cao, Z., Lu, Q., Yang, T., et al. (2018). Effect of organic acids production and bacterial community on the possible mechanism of phosphorus solubilization during composting with enriched phosphate-solubilizing bacteria inoculation. Bioresour. Technol. 247, 190–199. doi: 10.1016/j.biortech.2017.09.092
Wu, J., Guo, J., Hu, Y., and Gong, H. (2015). Distinct physiological responses of tomato and cucumber plants in silicon-mediated alleviation of cadmium stress. Front. Plant Sci. 6:453. doi: 10.3389/fpls.2015.00453
Wu, J., and Rosen, B. P. (1991). The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5, 1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x
Wu, J., and Rosen, B. P. (1993). The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol. Microbiol. 8, 615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x
Xiao, K. Q., Li, L. G., Ma, L. P., Zhang, S. Y., Bao, P., Zhang, T., et al. (2016). Metagenomic analysis revealed highly diverse microbial arsenic metabolism genes in paddy soils with low-arsenic contents. Environ. Pollut. 211, 1–8. doi: 10.1016/j.envpol.2015.12.023
Xiong, A., and Jayaswal, R. K. (1998). Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J. Bacteriol. 180, 4024–4029. doi: 10.1128/JB.180.16.4024-4029.1998
Xu, D., Chen, Z., Sun, K., Yan, D., Kang, M., and Zhao, Y. (2013). Effect of cadmium on the physiological parameters and the subcellular cadmium localization in the potato (Solanum tuberosum L.). Ecotoxicol. Environ. Saf. 97, 147–153. doi: 10.1016/j.ecoenv.2013.07.021
Xu, J., Yang, L., Wang, Z., Dong, G., Huang, J., and Wang, Y. (2006). Toxicity of copper on rice growth and accumulation of copper in rice grain in copper contaminated soil. Chemosphere 62, 602–607. doi: 10.1016/j.chemosphere.2005.05.050
Yadav, A. N., Verma, P., Kour, D., Rana, K. L., Kumar, V., Singh, B., et al. (2017). Plant microbiomes and its beneficial multifunctional plant growth promoting attributes. Int. J. Environ. Sci. Nat. Resour. 3, 1–8. doi: 10.19080/IJESNR.2017.03.555601
Yang, Z., Shi, W., Yang, W., Liang, L., Yao, W., Chai, L., et al. (2018). Combination of bioleaching by gross bacterial biosurfactants and flocculation: a potential remediation for the heavy metal contaminated soils. Chemosphere 206, 83–91. doi: 10.1016/j.chemosphere.2018.04.166
Yoon, K. P., Misra, T. K., and Silver, S. (1991). Regulation of the cadA cadmium resistance determinant of Staphylococcus aureus plasmid pI258. J. Bacteriol. 173, 7643–7649. doi: 10.1128/JB.173.23.7643-7649.1991
Yoon, Y., Lee, W. M., and An, Y. J. (2015). Phytotoxicity of arsenic compounds on crop plant seedlings. Environ. Sci. Poll. Res. 22, 11047–11056. doi: 10.1007/s11356-015-4317-x
Yu, F., Lin, J., Xie, D., Yao, Y., Wang, X., Huang, Y., et al. (2020). Soil properties and heavy metal concentrations affect the composition and diversity of the diazotrophs communities associated with different land use types in a mining area. Appl. Soil Ecol. 155:103669. doi: 10.1016/j.apsoil.2020.103669
Yuan, Z., Yi, H., Wang, T., Zhang, Y., Zhu, X., and Yao, J. (2017). Application of phosphate solubilizing bacteria in immobilization of Pb and Cd in soil. Environ. Sci. Pollut. Res. 24, 21877–21884. doi: 10.1007/s11356-017-9832-5
Zahir, F., Rizwi, S. J., Haq, S. K., and Khan, R. H. (2005). Low dose mercury toxicity and human health. Environ. Toxicol. Pharmacol. 20, 351–360. doi: 10.1016/j.etap.2005.03.007
Zawadzka, A. M., Crawford, R. L., and Paszczynski, A. J. (2007). Pyridine-2, 6-bis (thiocarboxylic acid) produced by Pseudomonas stutzeri KC reduces chromium (VI) and precipitates mercury, cadmium, lead and arsenic. Biometals 20, 145–158. doi: 10.1007/s10534-006-9022-2
Zhang, J., Li, Q., Zeng, Y., Zhang, J., Lu, G., Dang, Z., et al. (2019). Bioaccumulation and distribution of cadmium by Burkholderia cepacia GYP1 under oligotrophic condition and mechanism analysis at proteome level. Ecotoxicol. Environ. Saf. 176, 162–169. doi: 10.1016/j.ecoenv.2019.03.091
Zhang, J., Zeng, Y., Liu, B., and Deng, X. (2020). MerP/MerT-mediated mechanism: a different approach to mercury resistance and bioaccumulation by marine bacteria. J. Hazard. Mater. 388:122062. doi: 10.1016/j.jhazmat.2020.122062
Zhang, M., Li, Z., Haggblom, M. M., Young, L., He, Z., Li, F., et al. (2020). Characterization of nitrate-dependent As (III)-oxidizing communities in arsenic-contaminated soil and investigation of their metabolic potentials by the combination of DNA-stable isotope probing and metagenomics. Environ. Sci. Technol. 54, 7366–7377. doi: 10.1021/acs.est.0c01601
Zhang, X. X., and Rainey, P. B. (2007). The role of a P1-type ATPase from Pseudomonas fluorescens SBW25 in copper homeostasis and plant colonization. Mol. Plant Microbe Interact. 20, 581–588. doi: 10.1094/MPMI-20-5-0581
Zhuang, X., Chen, J., Shim, H., and Bai, Z. (2007). New advances in plant growth-promoting rhizobacteria for bioremediation. Environ. Int. 33, 406–413. doi: 10.1016/j.envint.2006.12.005
Keywords: plant growth promotion, heavy metal resistance, meta-omics approaches, molecular mechanism, eco-physiological properties
Citation: Mandal S, Saha KK and Mandal NC (2021) Molecular Insight Into Key Eco-Physiological Process in Bioremediating and Plant-Growth-Promoting Bacteria. Front. Agron. 3:664126. doi: 10.3389/fagro.2021.664126
Received: 04 February 2021; Accepted: 31 March 2021;
Published: 11 May 2021.
Edited by:
Avishek Banik, Presidency University, IndiaReviewed by:
Vikram Hiren Raval, Gujarat University, IndiaArindam Chakraborty, Jadavpur University, India
Copyright © 2021 Mandal, Saha and Mandal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subhrangshu Mandal, cy5tYW5kYWxib2xwdXJAZ21haWwuY29t; Narayan Chandra Mandal, bWFuZGFsbmNAdmlzdmEtYmhhcmF0aS5hYy5pbg==
†These authors have contributed equally to this work
 Subhrangshu Mandal
Subhrangshu Mandal Kunal Kumar Saha
Kunal Kumar Saha Narayan Chandra Mandal*
Narayan Chandra Mandal*