
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron., 11 May 2021
Sec. Plant-Soil Interactions
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.661295
This article is part of the Research TopicMaximizing Nitrogen Fixation in Legumes as a Tool for Sustainable Agriculture IntensificationView all 17 articles
 Mustapha Missbah El Idrissi1*
Mustapha Missbah El Idrissi1* Omar Bouhnik1
Omar Bouhnik1 Salma ElFaik1
Salma ElFaik1 Soufiane Alami1
Soufiane Alami1 Hanane Lamin1
Hanane Lamin1 Eulogio J. Bedmar2
Eulogio J. Bedmar2 Hanaa Abdelmoumen1
Hanaa Abdelmoumen1In this work, we analyzed the diversity of the nodule-forming bacteria associated with Lupinus luteus and Lupinus cosentinii grown in the Maamora Cork oak forest acidic soils in Morocco. The phenotypic analysis showed the high diversity of the strains nodulating the two lupine's species. The strains were not tolerant to acidity or high alkalinity. They do not tolerate salinity or high temperatures either. The strains isolated from L. luteus were more tolerant to antibiotics and salinity than those isolated from L. cosentinii. The plant growth promoting (PGP) activities of our strains are modest, as among the 28 tested isolates, only six produced auxins, six produced siderophores, whereas three solubilized phosphates. Only two strains possess the three activities. The rrs gene sequences from eight representative strains selected following ARDRA and REP-PCR results revealed that they were members of the genus Bradyrhizobium. Six strains were then retained for further molecular analysis. The glnII, recA, gyrB, dnaK, and rpoB housekeeping gene sequence phylogeny showed that some strains were close to B. lupini LMG28514T whereas others may constitute new genospecies in the genus Bradyrhizobium. The strains were unable to nodulate Glycine max and Phaseolus vulgaris and effectively nodulated L. luteus, L. cosentinii, L. angustifolius, Chamaecytisus albidus, and Retama monosperma. The nodC and nodA symbiotic gene phylogenies showed that the strains are members of the genistearum symbiovar.
Legumes (Fabaceae) are of great importance in different aspects, whether in agriculture, medicine, or ecology. Some species of this plant family can establish a specific symbiosis with rhizobia. The latter are soil bacteria normally found in the rhizosphere and in large numbers on the root surface, because of the nutrients secreted by the plant as root exudates (Sessitsch et al., 2002). In the rhizobia–legume symbiosis, bacteria provide plants with nitrogen compounds and plants provide them with carbon sources. A successful symbiotic interaction is based on a molecular dialogue between the two partners and generally involves rhizobial lipo-chitooligosaccharide signals called Nod factors (NF) (Wang et al., 2018; Lindström and Mousavi, 2020). These nodulation factors secreted by the rhizobia and perceived by the root cell receptors will initiate molecular and physiological responses in the plant, leading to the formation of the nodule, which is the site of fixation of the nitrogen (Clúa et al., 2018). Nitrogen fixation and uptake contributes to the high seed protein content and also provides residual N for the following crops, particularly cereals (Kermah et al., 2018).
Rhizobia are also collectively called root or shoot nodule bacteria, comprise more than 238 species in 18 genera regrouped in two clades; however, the description of rhizobial species included only 23% of the 19,000 legume species that exist throughout the world (Shamseldin et al., 2017).
Lupinus is one of the most important genera in Fabaceae with a rich diversity of species divided into Mediterranean “Old World” species and American “New World” species. Wild lupines are distributed over a wide range of climates around the world, from the coldest to the warmest and from the driest to the most humid (Wolko et al., 2011). On the other hand, the centers of diversity of Lupinus are the Americas, the Mediterranean region, and the regions of North and East Africa (Susek et al., 2016).
Wild lupines are toxic to humans and unpalatable to animals because their seeds contain alkaloids. Lupine cultivation improved in the 1930's, when breeders discovered sweet varieties of white lupine. Thus, its culture was stimulated in different European countries (Kohajdova et al., 2011). Nowadays, lupines are mainly consumed by ruminants. They are used in human food for the manufacture of ingredients used in the bakery and pastry industries in Europe and Australia (Hall et al., 2005; Smith et al., 2006). The current cultivated varieties of lupines, known as sweet varieties, are devoid of bitterness such as white and yellow lupines, which are used for human consumption. The white lupine is exploited generally as flour, recommended in low glycemic index protein, and gluten-free diets (Kohajdova et al., 2011; Yorgancilar and Bilgiçli, 2014).
The extensive and ongoing development of molecular biology tools over the last 20 years has facilitated the identification of new nodulating bacteria and has resulted in significant changes in the classification and proposition of new and different species. Lupines were first thought to be mainly nodulated by slow-growing rhizobia of the genus Bradyrhizobium, although fast-growing strains associated with lupines have been identified since 1988 (Miller and Pepper, 1988). Lupinus is currently reported as a promiscuous host that can be nodulated by different symbiotic bacteria belonging to the genera Bradyrhizobium, Ochrobactrum, Microvirga, Phyllobacterium, Neorhizobium, and Rhizobium (Jarabo-Lorenzo et al., 2003; Trujillo et al., 2005; Velázquez et al., 2010; Ardley et al., 2012; Bourebaba et al., 2016; Msaddak et al., 2018; Tounsi-Hammami et al., 2019; Missbah El Idrissi et al., 2020).
There has probably been a horizontal transfer of symbiotic genes between different Bradyrhizobium spp. associated with Lupinus spp. and other legumes of the Genisteae tribe, and it is very likely that the Microvirga and Ochrobactrum species isolated from different lupins have obtained their symbiotic genes from other more common rhizobial genera (Andrews et al., 2018).
Lateral transfer of specific symbiosis genes into rhizobial genera is an important mechanism for legumes to form symbiosis with selected rhizobia more adapted to particular soils and a legume-specific rhizobium strain symbiosis can develop in specific habitats.
In this work, we isolated 36 bacteria from the root nodules of two Lupine species, Lupinus cosentinii and Lupinus luteus, grown in soils of the Maamora forest, one of the most important Cork Oak Forest in the world (Aafi et al., 2005). The bacteria were first screened for their nodulation gene possession as a marker for their belonging to symbiotic microsymbionts and then characterized by molecular and phenotypic analysis.
All the 36 strains were isolated from different soils in Maamora forest in the region of Rabat known by its mild climate (Aafi et al., 2005) (Supplementary Table 1). Twenty isolates were isolated from L. cosentinii and 16 were isolated from L. luteus plants root nodules, according to the method of Howieson and Dilworth (2016). The root segments bearing nodules were collected, washed under running tap water and surface sterilized by immersion in 5% sodium hypochlorite for 3 min, and finally washed seven times with sterile distilled water. Each nodule was then crushed, and the extract was streaked onto plates of YMA. Plates were incubated for 15 days at 28°C, and then the single colonies obtained were checked for purity by repeated streaking on YMA medium, supplemented with Congo red dye. The incubation period of 2 weeks was sufficient to check for the growth of any slow-growing symbiotic bacteria on YEM medium. Pure isolates were maintained at −20°C in 50% (m/v) glycerol and at 4°C in YMA that was routinely used for rhizobial culture.
The seeds of the two lupines were surface disinfected with 70% ethanol for 1 min and later sterilized with 5% sodium hypochlorite for 15 min. The seeds were scarified with concentrated sulfuric acid for 10 min and germinated in water agar (0.6% w/v) containing Petri plates. The seedlings were then transferred to Gibson tubes (Howieson and Dilworth, 2016) and inoculated with 1 ml per plant of a suspension containing ~108 isolates CFU ml−1. The plants were cultivated at 26°C for 60 days under a 16.0/8.0 h light/dark photoperiod. Eight weeks after inoculation, the plants were checked for nodules appearance and plant aspect. Indirect effectiveness of the nodules for N2 fixation was estimated by visual assay of red leghemoglobin presence in cross-sections and by the dark green intensity of the leaves compared to uninoculated control plants.
The selected strains Lcos6, Lcos7.2.1, Lcos8.1, Lcos10.2, Llut4, Llut5, Llut6, and Llut8 were also tested for nodulation on Lupinus angustifolius, Chamaecytisus albidus, Retama monosperma, Cytisus monspesulanus, Glycine max, and Phaseolus vulgaris in the same conditions.
Bacteria were grown on solid Tryptone-Yeast Extract (TY) slants (Beck et al., 1993) for 96 h at 28°C, and colonies were suspended in 2 ml of distilled water. The suspensions were then centrifuged twice at 3,000 r/min for 15 min. The resulting pellet was treated with proteinase K (20 mg/ml), and total DNA was extracted as previously described by Guerrouj et al. (2013). Rep-PCR (Repetitive Extragenic Palindromic Polymerase Chain Reaction) using ERIC1R and ERIC2 primers (De Bruijn, 1992) was used for amplification of the DNA. The DNA template was denatured for 5 min at 95°C, and PCR was carried out for 35 cycles (94°C for 30 s, 52°C for 1 min, and 72°C for 1 min), with an elongation step at 72°C for 7 min. PCR products were analyzed by horizontal electrophoresis in 2% agarose gels in TAE buffer at 55 V for 4 h. Gels were stained with ethidium bromide, visualized under UV radiation, and photographed with a digital camera. Cluster analysis of the fingerprints obtained was performed with Gelcompar II program (version 2.15) (Supplementary Figure 1).
PCR amplification was performed as described by Lamrabet et al. (2020) in 25-μl reaction mixtures containing 2.5 μ DNA extract, 10 mM Tris/HCl pH 8.3, 1.5 mM MgCl2, 50 mM KCl, 0.001% (w/v) gelatin, 1 U fastTaq DNA polymerase (Applied Biosystems), 200 mM of each deoxyribonucleotide, and 50 ng of each primer in Applied Biosystems 2720 Thermal Cycler.
All the amplifications consisted in an initial 3-min denaturation at 94°C and 35 cycles of 30-s denaturation at 94°C, 30 s of annealing depending on the primers used, and an extension at 72°C for 45 s. The final extension is performed at 72°C for 3 min. PCR amplification of 16S rRNA gene fragments was done using the two opposing primers fD1 and rD1 and the primer pairs nodCFn–nodCI and nodA1f–nodAb1r were used for amplification of the nodC and nodA gene (Chaintreuil et al., 2001; Laguerre et al., 2001).
Primers dnaK 1466F/1777R, glnII 12F/689R, gyrB 343F/1034R, TSrecA F/R, and rpoB 83F/1061R were used to amplify the chaperone protein DnaK (dnaK), the glutamine synthase (glnII), DNA gyrase subunit B (gyrB) genes, RecA (recA), and RNA polymerase β subunit gene (rpoB), using the annealing temperatures indicated in Supplementary Table 2. The PCR products and their concentration were assessed by electrophoresis of 6 μl of product on a 1% agarose gel and staining with ethidium bromide. A molecular mass marker (Promega 1-kb ladder) was included to estimate the length of the amplicons.
The amplification products were purified using a Qiaquick PCR purification kit (Qiagen) and sequenced using the same primers as for PCR amplification using the dideoxynucleotide chain-termination method with fluorescent ddNTPs (Applied Biosystems), with ABI Prism Dye Chemistry, and analyzed with a 3130xl automatic sequencer at the sequencing facilities of UATRS (Unités d'Appui Technique à la Recherche Scientifique, CNRST, Rabat, Morocco) and the CSIC sequencing facilities in Zaidin experimental station, CSIC, Granada, Spain. The sequences obtained were compared with those from GenBank using the BLASTN program (Altschul et al., 1990).
Sequences were aligned using MEGA 7 software (Kumar et al., 2016), and the distances calculated according to Kimura's two-parameter model (Kimura, 1980) were used to infer phylogenetic trees with the neighbor-joining analysis (Saitou and Nei, 1987) with MEGA 7 software (Kumar et al., 2016). Phylogenetic trees were subjected to 1000 bootstrap replications, and preferred topologies were plotted.
An in silico ARDRA of the 16S rDNA gene using 30 restriction enzymes (AccII, AcuI, AcvI, AgeI, AloI, AspLEI, BccI, BpmI, BsaXI, BslI, Bsp143I, Bst6I, BstC8I, BstF5I, Cac8I, Cfr9I, Eco91I, FaqI, FokI, HaeIII, HgaI, HhaI, Hinf I, LweI, MlyI, MspI, PspN4I, RsaI, SfaNI, and TaqI) was performed using the Silent Mutator program (Cermak, 2018b), which scans through the sequences and recognizes sites for each restriction enzyme. The enzymes were selected following the results obtained with the Restriction Comparator program (Cermak, 2018a). Finally, a dendrogram of similarities based on the concatenated sequences database was generated using Statistica software version 7.0 (StatSoft Inc, 2004).
The analysis was pursued by determining some phenotypic properties of the isolated strains. Tests were performed in broth tubes or agar plates inoculated with an exponentially growing liquid culture. The tolerance of the rhizobial isolates to high temperatures was tested on TY broth medium (Beringer, 1974) at 30, 35, 40, and 45°C. The ability of isolates to grow in acidic or basic media was determined on YMA Petri dishes for which the pH has been adjusted and buffered to 5.0, 5.5, 6.0, 7.0, 8.0, 9.0, 9.5, or 10.0, as described by Zerhari et al. (2000). The salt tolerance of the isolates was tested at 0, 86, 171, 342, 510, and 685 mM.
Utilization of 12 amino acids as sole nitrogen source was investigated on a modified YMB medium in which the yeast extract was replaced by the amino acid to test. The amino acids were sterilized separately by filtration and added to the medium at a concentration of at 1% (w/v). Carbohydrate assimilation was carried out on solid YEM containing 0.005% YEM and 0.4% (w/v) of glucose, fructose, sucrose, maltose, lactose or starch. The carbohydrates were sterilized separately by filtration.
The antibiotics resistance of the isolates was determined on solid YEM supplemented with the following antibiotics (μg/ml): kanamycin (20), ampicillin (20), bacitracin (20), tetracycline (10), vancomycin (20), tetracycline (10), streptomycin (25), trimethoprim (10), nalidixic acid (20), gentamicin (20), and chloramphenicol (20).
The intrinsic heavy metal resistance of the isolates was determined on TY agar plates containing the following heavy metals (μg/ml): CuSO4, 5H2O (500), AlCl3 (100), AlCl3 (450), HgCl2 (5), CdSO4 (50), ZnSO4 (250), Pb-acetate (1000), MnCl2, 4H2O (500), MnSO4 (500), MgCl2 (1000), MgSO4 (1000), BaCl2, 2H2O (1000), CoSO4 (150), FeCl3 (500), and NiCl2 (500).
To evaluate oxidase activity, a loopful of bacterial mass from the colonies of the isolates was spread out over cellulose paper soaked in a solution containing 1% N,N-dimethyl-p-phenylenediamine oxalate 98%. The appearance of blue color is indicative of oxidase activity.
We used the PVK solid medium containing 2.5 g L−1 of (Ca3PO4)2 to determine the ability of the isolates to solubilize phosphates. To this purpose, 5 μl of the bacterial suspension was plated onto PVK medium in Petri dishes and incubated at 28°C for 10 days. The size of the halos that appeared around the colonies and the size of the colonies were measured to estimate the ability of the isolates to solubilize phosphate. Siderophore-producing isolates were identified by the development of orange halos around the colonies according to Lakshmanan et al. (2015). To assess the ability of the isolates to produce indole acetic acid (IAA) and IAA-related compounds, a volume of 5 μl of the bacterial suspension was placed on the solid YEM medium supplemented with 0.5 g L−1 tryptophan, and the plates were then incubated at 28°C for 10 days. The production of IAA was detected using the Salkowski reagent followed by the appearance of a pink halo around the colonies after incubation for 30 to 60 min at 28°C.
Thirty-six bacterial cells were isolated from root nodules of L. cosentinii (20) and L. luteus (16) grown in Maamora cork oak forest soils. Among the strains, 28 were able to re-nodulate their original host under axenic conditions using Gibson's tubes. REP-PCR using primers REP1R-I and REP2-I grouped the strains into six clusters and some single strains (Supplementary Figure 1). The REP PCR technique is employed for analysis of the genetic diversity of rhizobia (Laguerre et al., 1997; Menna et al., 2009).
Eight strains representing the different groups were randomly selected and used for in vitro ARDRA, based on 1,450 nucleotides of rrs gene, with the restriction enzymes cfoI, HaeIII, and MspI, which distributed the strains in three ribotypes (Supplementary Table 3). Furthermore, the in silico ARDRA of the eight representative strains, together with other 45 bradyrhizobial species and the outgroup R. gallicum R602, using 30 restriction enzymes, including MspI, HaeIII, and RsaI, showed that the strains from the two lupines clustered in three main groups, with B. lupini–B. canariense and B. cytisi (Supplementary Figure 2).
The rrs gene sequences of strains Llut4 (MT468660), Llut5 (MT468656), Llut6 (MT468653), Llut8 (MT468657), Lcos6 (MT468659), Lcos7.2 (MW494674), Lcos8.1 (MW494669), and Lcos102 (MT468658) revealed they were members of the genus Bradyrhizobium of the Alphaproteobacteria and share 99 to 99.93% of similitude. The strains Llu4, Lut6, Lcos6, Lcos7.2, and Lcos102 have 99.43–100% of similitude with B. lupini USDA 3051T and share similarities from 99.55 to 100% with strains nodulating L. angustifolius, L micranthus, and L. luteus in Algeria and Tunisia (Msaddak et al., 2017; Mellal et al., 2019).
The strains Lcos102 and Llut5 have similarity values of 99.79 and 99.86% with B. canariense BTA-1T. They have similarities of 99.08–99.85% with the North African strains tested (Supplementary Table 4). The strains Lcos8.1 and Llut8 share similarities of 99.64 and 99.79% with B. cytisi CTAW11T and share also similarities of 98.93–99.85% with the strains isolated from the Algerian and Tunisian lupines.
The phylogenetic tree based on the 16S rRNA sequences showed that the strains Lut6, Lcos6, and Lcos7.2 clustered together with different strains isolated from North African lupines, in Algeria and Tunisia in group I, along with B. lupini USDA 3051T. The strains Lcos102 and Llut5 clustered in group II with B. canariense LMG 22265T, whereas strains Lcos8.1 and Llut8 clustered in group III with B. cytisi CTAWT and some strains isolated from Algerian and Tunisian lupines. The strain Llut4 grouped further into the cluster composed of groups 1 and 2 (Figure 1). The rrs sequences of strains Lcos6, Lcos7.2, and Llut6 are 100% similar, and thus, we selected one strain (Llcos6) to represent the three. The strains retained as representatives for further molecular analyses are Lcos6, Llut4, Llut5, Lcos102, Lcos8.1, and Llut8.
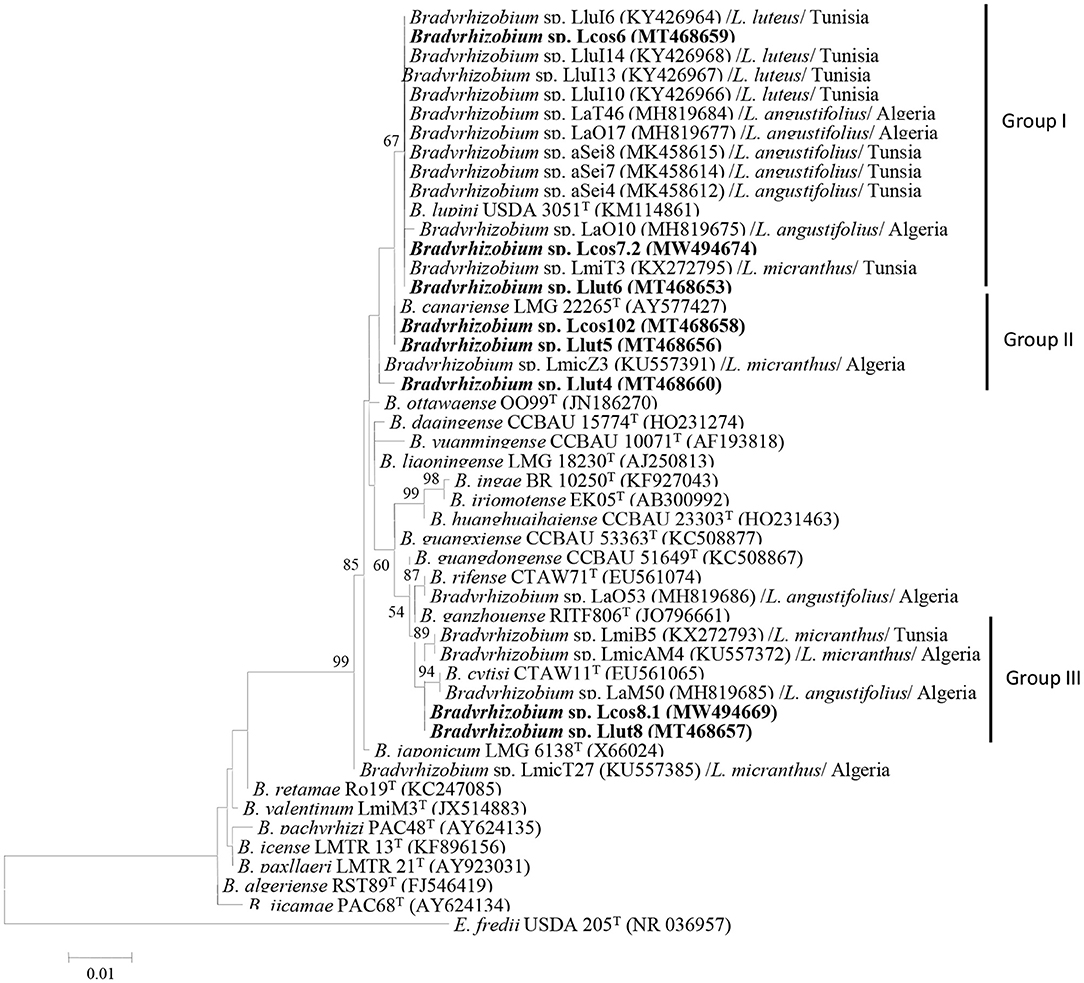
Figure 1. Maximum likelihood phylogenetic tree based on 16S rRNA sequences of rhizobial strains from nodules L. cosentinii and L. luteus grown in the Maamora forest and phylogenetically related species within the genus Bradyrhizobium. The analysis was based on 1,400 nucleotides. Isolates from this study are denoted in bold. Bootstrap values are indicated as percentages derived from 1,000 replications. Values lower than 50% are not shown. Bar, 1 nucleotide substitution per 100 nucleotides. The tree is rooted with Ensifer fredii USDA 205T.
Five housekeeping genes, dnaK, glnII, gyrB, recA, and rpoB of the six representative strains were analyzed in this study. The recA and glnII sequences were analyzed and compared with other strains isolated from North African lupines root nodules. All the sequence accession numbers are reported on the different figures. We could not compare our sequences with the dnaK, gyrB, and rpoB sequences of Algerian and Tunisian strains since they are not published.
The sequence analyses showed that the recA genes of strains Llut5 (MK346962) and Lcos6 (MK346959) are 100% similar and have 99.80% of similarity with B. lupini 3051T. The two strains share 99.58–100% similarity with strain LaO10, LaO17, and Lat46 isolated from L. angustifolius in Algeria (Mellal et al., 2019), and 99.59% with strain LmiT3 isolated from the root nodules of L. micranthus in Tunisia (Msaddak et al., 2017). The strains Lcos8.1 (MW527003), Llut8 (MK346964), and Lcos102 (MK346960) share 99.23 to 99.60% and have similarity percentages of 94.43 to 95.23% with Llut4 (MK346961). They have percentages of similarities ranging from 93.51 to 94.30 with the different Algerian and Tunisian strains. The phylogenetic tree based on the recA sequences (Supplementary Figure 3) shows that strains Lcos6 and Llut5 clustered with strains nodulating L. micranthus and L. angustifolius in Algeria all together with the type strain B. lupini USDA3051. The strain Llut4 was closer to B. cytisi LMG25866T with which it constituted a group containing B. japonicum LMG6138T and B. diazoefficiens USDA110T. The strains Llut8, Lcos8.1, and Lcos102 were regrouped in a different cluster with B. ganzhouense RITF806T.
The glnII sequences of strains Lcos6 (MK346977) and Llut5 (MW565871) share 98.98% similarity and have similarity percentages of 98.81 and 99.84 with B. lupini 3051T, respectively. They have also percentages of similarity ranging between 98.45 and 99.83 with strains LaO10, LaO17, LaT46 (Mellal et al., 2019), and LmiT3 (Msaddak et al., 2017).
The glnII neighbor-joining phylogenetic tree (Supplementary Figure 3) shows that strains Lcos6 and Llut5 are close to B. lupini 3051T with which they were regrouped along with strains LaO10, LaO17, and LaT46 isolated from L. angustifolius in Algeria and strain LmiT3 isolated from L. micranthus in Tunisia.
Similar results were obtained with the gyrB, dnaK, and rpoB sequences (Supplementary Figures 4–8). However, we could not compare our sequences with those of other North African lupines' microsymbionts.
The concatenation of the glnII, gyrB, and recA genes showed that the strains Llut8, Llcos81, and Llcos102 have identity percentages of 95.8, 95.73, and 95.59% with B. ganzhouense RITF806 T and share similarities of 99.43–99.57%. The strains Lcos6 and Llut5 share 99.70 similarities and have identity percentages of 99.43 and 99.50% with B. lupini LMG28514T, respectively. The strain Llut4 has low similarities with all the strains tested and shares a maximum identity of 94% with B. cytisi LMG25866 T (Supplementary Table 4). The phylogenetic tree based on the concatenation of the three genes (Figure 2) revealed also that B. cytisi LMG25866T is the closest parent to strain Llut4, whereas the strains Lut5 and Lcos6 clustered with B. lupini LMG28514T, and strains Llut8, Llcos81, and Llcos102 were regrouped with B. ganzhouense RITF806T.
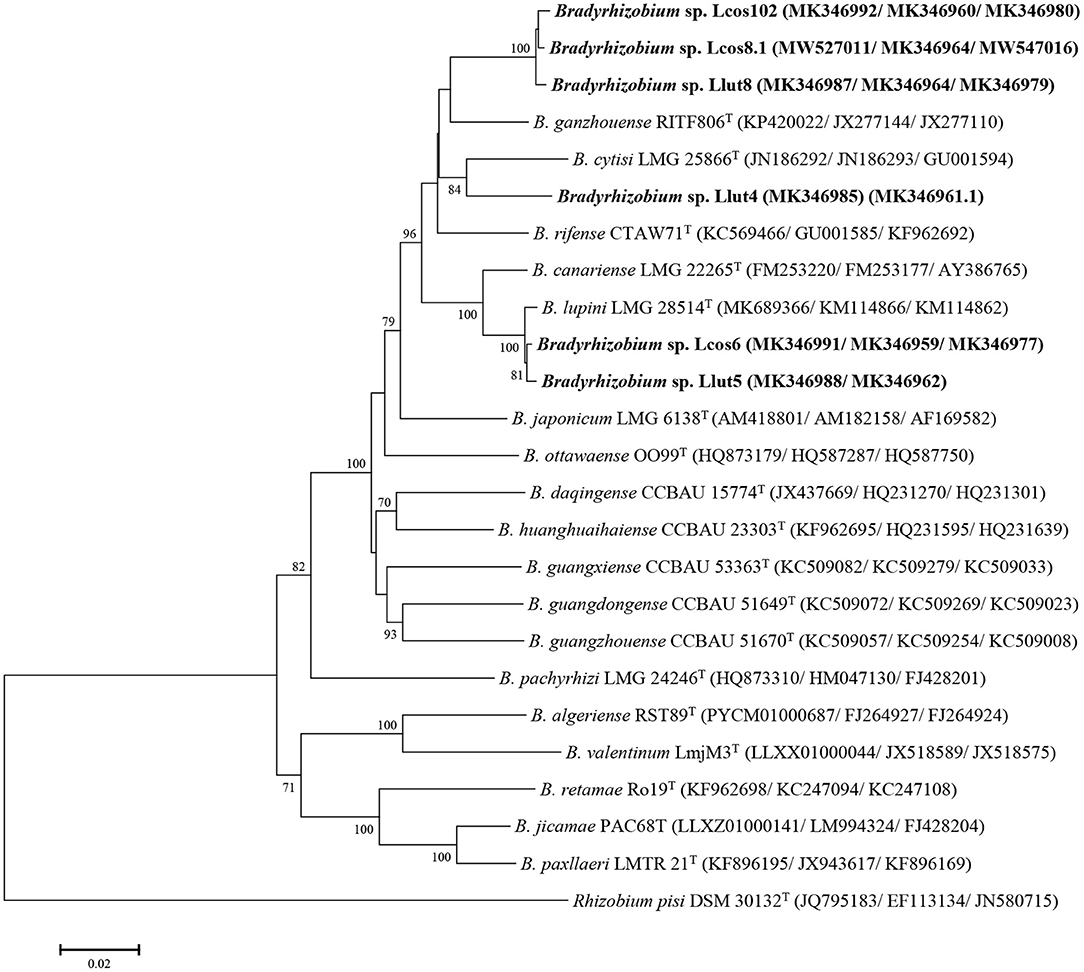
Figure 2. Maximum likelihood phylogenetic tree based on concatenated gyrB + recA + glnII sequences (1,430 nt) of strains from nodules of L. cosentinii and L. luteus and phylogenetically related species within the genus Bradyrhizobium. The analysis was based on 1,405 nucleotides. Isolates are denoted in bold. Bootstrap values are indicated as percentages derived from 1,000 replications. Values lower than 50 are not shown. Bar, 2 nucleotides substitution per 100 nucleotides. The tree is rooted with Rhizobium pisi DSM 30132T.
The nodC sequences of the strains Lut5, Llut8, and Lcos6 share 99.9–100% similarity. The nodC-based phylogenetic tree (Figure 3) showed that they were regrouped in a same cluster with strain LAM15, isolated from the nodules of L. angustifolius in Algeria (Mellal et al., 2019), with which they have a similarity percentage of 92.1%, within the genistearum symbiovar. The strain Llut4 has a 95.9% percentage similarity with the three other representatives and was regrouped with B. cytisi LMG25866T with which it shares 97% similarity.
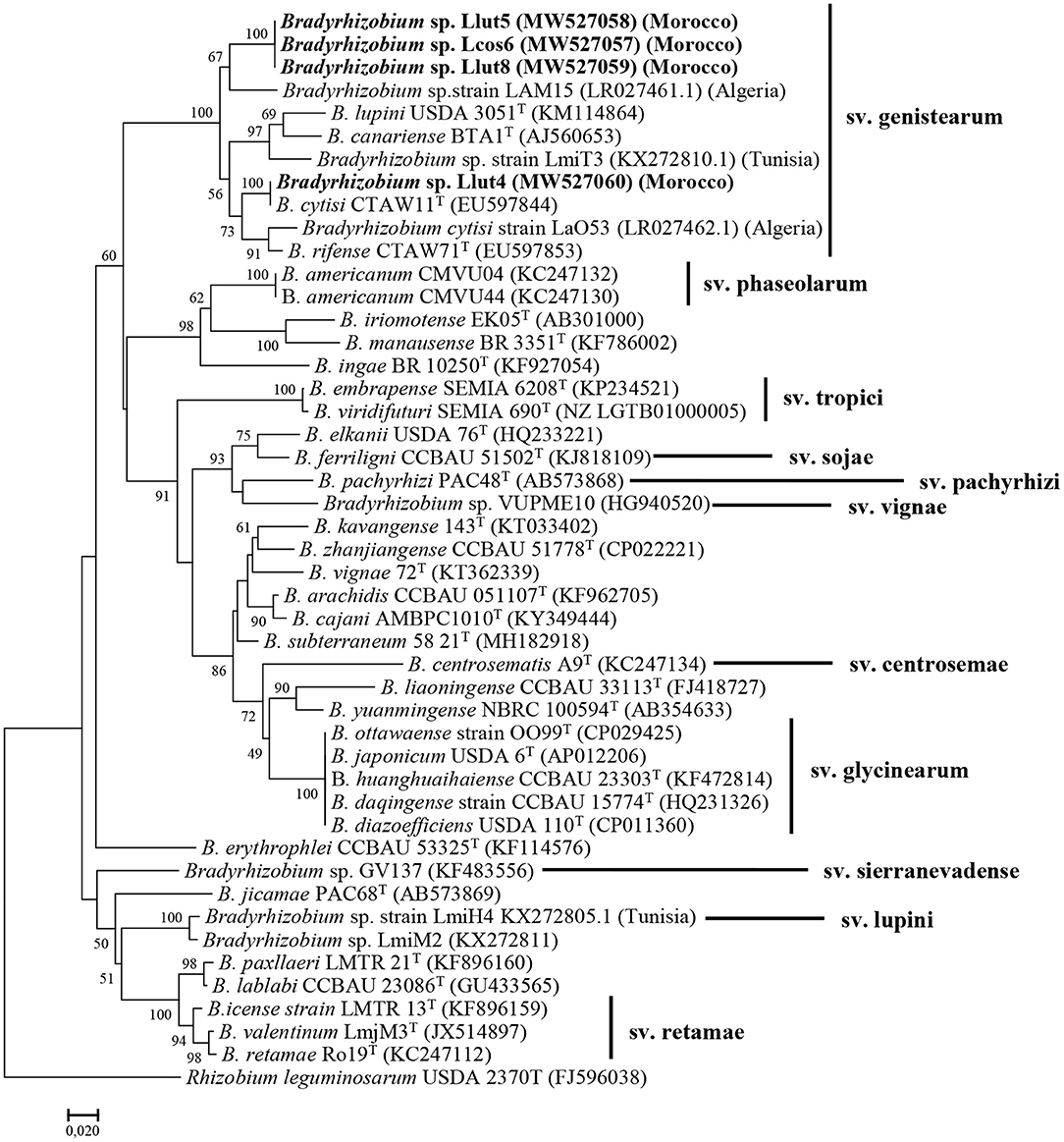
Figure 3. Maximum likelihood phylogenetic tree based on concatenation of the nodC gene partial sequences of the strains Lut4, Llut5, Lcos6, and Llut8, isolated from nodules of L. cosentinii and L. luteus and related species within the genus Bradyrhizobium. Isolates are denoted in bold. Bootstrap values are indicated as percentages derived from 1000 replications. Values lower than 50 are not shown. Bar, 2 nucleotide substitution per 100 nucleotides. The original geographical origin of the species is shown in brackets. The tree is rooted with R. leguminosarum USDA 2370T.
The analysis of the nodA gene sequences showed also that our strains were very close and share 99.6 to 100% similarity. The phylogenetic tree based on the nodA sequences (Supplementary Figure 9) showed also that they formed a single sub-cluster in the symbiovar genistearum. The phylogenetic analysis of the two symbiotic genes' concatenated sequences also produced similar results (Supplementary Figure 10).
The Moroccan as well as the Algerian and the Tunisian strains were clustered together in the same clade with B. lupini, B. canariense, B. cytisi, and B. rifense, members of the genistearum symbiovar.
All the strains selected for the host range experiments were able to nodulate L. angustifolius, L. cosentinii, L. luteus, C. monspessilanus, C. albidus, and R. monosperma, but not L. albus, P. vulgaris, or G. max (Supplementary Table 5).
All the isolated strains were slow growing, as their colonies appeared on YEM solid medium after 7 days incubation time at 28°C. The phenotypic results reported in Supplementary Table 6 show that the strains grow in media with different pH values ranging from 6 to 8. All strains nodulating L. cosentinii use trehalose as sole carbon source; 93% assimilate fructose and myoinositol, and 64% grew in the presence of arabinose, while only 7% can assimilate maltose. The latter cannot be assimilated by strains nodulating L. luteus. The results also showed that all strains of L. luteus assimilate arabinose and fructose, while 79% of these strains use myo-inositol and 36% develop in the presence of trehalose. None of the two lupine species microsymbionts is capable to use lactose, starch, or carboxy-methyl-cellulose (CMC).
The strains nodulating lupines in the Maâmora forest are able to use a wide range of amino acids as sole nitrogen source. All the strains nodulating L. luteus assimilate 10 amino acids, arginine, tyrosine, proline, asparagine, histidine, valine, serine, tryptophan, leucine, and phenylalanine, while 93% of these strains use methionine, and 64% use alanine, whereas only 14% are able to use glycine, lysine, and aspartic acid. On the other hand, all strains that nodulate L. cosentinii are able to use tyrosine, serine, and phenylalanine as the sole source of nitrogen, while 93% assimilate arginine, proline, valine, tryptophan, and leucine, whereas 86% can assimilate asparagine and histidine while 57% are able to assimilate methionine. Glycine, lysine, and aspartic acid are used by only 21% of these strains.
We also tested some enzymatic activities in the strains studied. Thus, 18% of strains nodulating L. cosentinii and 9% among strains of L. luteus produce gelatinase, and all the strains isolated from the two lupine species produce catalase.
Tolerance to NaCl varies considerably between strains nodulating the two species. The results show that all strains tested can grow in the presence of 85 mM NaCl, while 21% of L. cosentinii strains and 14% of L. luteus strains develop in salt concentrations between 170 mM and 680 mM, while only 7% of the strains nodulating L. cosentinii and 7% of those of L. luteus continue to develop at 854 mM NaCl.
Strains react differently when grown in YEM media with different pH values. All the strains nodulating the two species of lupine have the ability to grow in a slightly acidic to neutral pH (6 to 7). All the strains nodulating L. luteus grow in a slightly alkaline pH (pH 7.5) and 93% grow at pH 8. They are sensitive to more alkaline pH, with only 50% of the strains able to grow at pH 8.5 and 14% at pH 9. The sensitivity of these strains was also noted with respect to acidic pH; thus, no strain tolerated pH 5; however, 14% of these strains were able to grow at pH 5.5. The results also show that the majority of strains nodulating L. cosentinii (86%) can develop at pH 7.5 and that with the increase in pH, the number of strains that manage to grow there decreases, 36% grow at pH 8, 29% at pH 8.5, and only 21% at pH 9. In acidic environments, 21% of the strains manage to develop at pH 5.5 and 7% at pH 5.
Among all the heavy metals tested, barium, manganese chloride, manganese sulfate, aluminum, and cadmium (at 100 μg/ml) were the least harmful for the growth of the strains nodulating L. luteus with 79–93% that resist. On the other hand, the presence of cobalt and copper inhibited the growth of half of the strains. The heavy metals most toxic to strains of L. luteus are mercury, magnesium, zinc, and lead, and only 21% could grow in their presence. The results showed also that 50% of strains nodulating L. cosentinii develop in the presence of manganese chloride and 43% are resistant to aluminum. On the other hand, in presence of barium, cobalt, and manganese sulfate, 36% of the strains were able to resist, and 28% of the strains were able to develop in the presence of copper. Mercury, magnesium, zinc at 500 μg/ml, and lead at 1,000 μg/ml were found to be more toxic with only 21% of strains that were able to tolerate their presence.
The tolerance to antibiotic is one of the defining criteria in the analysis of rhizobial biodiversity. In this work, we tested 11 antibiotics on the different strains isolated from the two lupines and the results showed that there is variability in tolerance. Thus, all the strains nodulating L. luteus were resistant to several families of antibiotics: beta-lactams (ampicillin and penicillin), quinolones (nalidixic acid), glucopeptides (vancomycin), aminoglycosides (spectinomycin), gramicidins (bacitracin), and trimethoprims. They grew also in presence of tetracyclines and aminoglycosides (gentamycins and kanamycin) with high percentages between 70 and 86%. The chloramphenicol affects the growth of these bacteria since only 36% were able to grow in its presence, at the concentrations used. The results also showed that 43% of the strains nodulating L. cosentinii tolerated the majority of antibiotics: nalidixic acid, vancomycin, spectinomycin, bacitracin, trimethoprim, and beta-lactams, and 36% developed in the presence of chloramphenicol. These strains are sensitive to aminoglycosides, since only 21% are resistant to kanamycin and 14% are resistant to gentamycin.
Temperature also has a differential effect on strains' growth. Thus, all the strains grew at 30°C, whereas at 35°C, the strains showed a fluctuation in their responses, with 79% of the strains nodulating L. cosentinii, and 71% for the strains nodulating L. luteus, which were able to grow at this temperature. No strain can grow above 40°C.
Out of the 28 strains tested, six strains produced the IAA, four strains from L. cosentinii and two from L. luteus. Only three strains were able to solubilize the inorganic phosphates, two strains from L. cosentinii and one strain among L. luteus microsymbionts. Two strains were hence very effective with a solubilization index of 275% for lut5 and 375% for cos10.2. Six strains produced siderophores, four strains from L. cosentinii and two from L. luteus. The two strains Lcos10.2 and Llut5 have the capacity to solubilize phosphates and produce siderophores and IAA.
In this work, we aimed to characterize and identify some bacterial strains isolated from the root nodules of two lupines grown in different soils of the Maamora forest, in the vicinity of Rabat.
We started the molecular characterization by the analysis of the eight representative strains' rrs gene sequences, and their phylogeny revealed that they are more related to B. cytisi and B. lupini. The rrs gene was considered as a preliminary genetic marker to situate a strain taxonomic position at the genus level, although it does not provide clear phylogenetic diversity for species of the Bradyrhizobium genus (Martinez-Romero and Ormeño-Orrillo, 2019). To this purpose, an MLSA of different housekeeping genes as additional phylogenetic markers is frequently used for accurate identification of Bradyrhizobium species and strains (Tampakaki et al., 2017; Wójcik and Kalita, 2019).
Six strains were then selected from the eight representatives, as the rrs sequences of strains Llut6 and Lcos7.2 were identical (100%) to strain Lcos6 rrs sequence. The phylogenies of the three individual genes glnII, gyrB, and recA showed that strains Lcos6 and Llut5 were closely related to B. lupini LMG 28514T. In fact, we sequenced also the dnaK and rpoB genes of most strains, but we failed to amplify them in some. Furthermore, data concerning these two genes in B. lupini are not available in the databases, and the dnaK (JACJNR010000003.1) and rpoB (JACJNR010000001.1) sequences of the strain B. lupini DSM 30140 extracted from the whole genome sequence available in the NCBI database are more related to B. japonicum LMG6138T (Supplementary Figures 5, 6). Consequently, we preferred to continue with only three genes.
The concatenated housekeeping genes' sequences phylogeny confirmed that the closest parent of the strains Lcos6 and Llut5 is B. lupini LMG 28514T, whereas strain Llut4 was more related to B. cytisi LMG 25866T. The three strains Lcos8.1, Lcos102 and Llut8 were regrouped in a single cluster more related to B. ganzhouense RITF806T, isolated from the rot nodules of Acacia melanoxylon in China (Lu et al., 2014). The two strains Lcos8.1 and Lcos102 were isolated from L. cosentinii, whereas strain Llut8 was isolated from L. luteus. None of the three strains grew in YEMA medium in the presence of 1% NaCl. The strain Lcos6 grows between pH 6 and 7.5, while cos8.1 grows between pH 6 and 8, and the Lut8 strain grows in pH 6–8.5, but do not grow at 40°C. They use glucose, fructose, mannose, and sucrose as sole carbon sources. Inversely, the type strain of B. ganzhouense does not assimilate sucrose; it grows at pH 5–12 and develops in the presence of 3% NaCl (Lu et al., 2014). Their low percentages of similarity with their closest parent suggest that they may constitute a putative new genospecies in the genus Bradyrhizobium.
Plant species of the genus Lupinus were considered as nodulated exclusively by Bradyrhizobium spp. for a long time until Ardley et al. (2012) reported the isolation and identification of three symbiotic Microvirga species from L. texensis and Listia angolensis root nodules. More recently, lupines were found to be nodulated by Microvirga strains in Tunisian, Moroccan, and American soils (Beligala et al., 2017; Msaddak et al., 2017, 2019; Rejili et al., 2019; Missbah El Idrissi et al., 2020). Msaddak et al. (2017) suggested that lupines would prefer Bradyrhizobium spp. strains as microsymbionts in acidic soils and Microvirga spp. in the alkaline soils. Lamrabet et al. (2020) also reported that in alkaline soils, some Genistae such as Retama spp. would prefer members of Microvirga as microsymbiont instead of Bradyrhizobium, although Tounsi-Hammami et al. (2019) found that strains isolated from L. albus in alkaline soils were affiliated to Rhizobium and Neorhizobium. Furthermore, Durán et al. (2014) reported the nodulation of the endemic Lupinus mariae-josephae by Bradyrhizobium valentinum in alkaline-limed soils in Eastern Spain.
The phenotypic analysis showed that all the strains assimilate different carbon sources including glucose, mannose, galactose, and sucrose, while no strain used starch or lactose; this result agrees with previously reported results (Rome et al., 1996; Missbah El Idrissi et al., 2020). Bradyrhizobium canariense BTA-1T does not use sucrose or lactose either (Vinuesa et al., 2005).
There was diversity in the utilization of amino acids, but all the strains were able to assimilate tyrosine and phenylalanine as sole nitrogen source, which contradicts results of Miller and Pepper (1988) who reported that the asparagine was the most preferred amino acid for the growth of rhizobia isolated from lupines in the desert of Mexico. Vinuesa et al. (2005) reported also that B. canariense and B. yuanmingense do not use glycine. Some strains possess the gelatinase activity, an enzyme that hydrolyzes collagen into amino acids or peptides, which allows them to persist in in the rhizosphere while waiting for the plant partner.
We analyzed the effect of environmental factors including salinity, drought, acidity, and alkalinity as well as the behavior of strains toward heavy metals and antibiotics, factors that compromise survival, growth, and nitrogen fixing ability of rhizobia strains (Zahran et al., 1994). We noticed a low tolerance to salinity in the majority of the strains as reported by Vinuesa et al. (2005), but some strains tolerated salt concentrations as high as 854 mM. Many studies have reported very interesting resistance profiles in strains isolated from lupines in Egypt that can tolerate up to 1,700 mM NaCl (Zahran et al., 1994). However, the limits of salinity tolerance between rhizobia can vary considerably from one species to another (Elsheikh and Wood, 1989), and even between strains of the same species (Boukhatem et al., 2012).
Peix et al. (2015) found that among 11 type strains of Bradyrhizobium species tested, only B. betae, B. japonicum, and B. ganzhouense grew at 171 mM of NaCl. In fact, in saline environments, the symbiosis between rhizobia and lupines depends not only on the bacteria but on the host plant also. Some strains were able to grow at pH 9, while few strains tolerated pH 5. Our strains tolerate more alkaline than acidic pHs. The type strains of B. cytisi and B. rifense are reported to grow at pH 4.5 (Peix et al., 2015). All the isolates grow up to 35°C, while no strain grew at 40°C, which is very common in bradyrhizobia (Peix et al., 2015). Bradyrhizobium sp. strains isolated from L. luteus in Tunisia were not able to grow at 37°C, pH 12, and 1% NaCl (Msaddak et al., 2017). Other studies have shown that certain rhizobial strains can tolerate higher temperatures, but it has been suggested that rhizobia may be protected against different soil constraints by living in particular niches in the rhizosphere and inside nodules (Boukhatem et al., 2012).
L. luteus nodulating strains are more resistant to antibiotics (ATB) and heavy metals than strains isolated from L. cosentinii, among which only 50% are able to resist to some antibiotics. The resistance level varies between strains, depending on the type of ATB and its mechanism of action. Similar results were obtained by (Msaddak et al., 2017).
The majority of our strains showed modest activities of phosphate solubilization and production of siderophores or auxins. Sarkar and Laha (2013) reported that 80% of rhizobia have the capacity to produce IAA and IAA-related compounds. However; siderophore production is usually low and phosphate solubilization is variable in rhizobia (Lamrabet et al., 2020). Few rhizobia may possess all the plant growth promoting (PGP) activities and the plants need hence to interact with different beneficial strains in their rhizosphere. To sustain their development, the most healthy plants attract the best PGPRs (Backer et al., 2018; Gouda et al., 2018).
It is evident that environmental factors such as salinity, drought, acidity, alkalinity, heavy metals, and the presence of antibiotics compromise the survival, growth, and the ability to fix nitrogen of strains of rhizobia (De la Peña and Pueyo, 2012), which shows the interest of knowing their phenotypic and biochemical characteristics in advance before being introduced and inoculated to the plants in the field, to ensure their survival, their adaptability, and their competitiveness (Boukhatem et al., 2012).
The nodC symbiotic gene (encoding the N-acetylglucosaminyl transferase protein) is essential for nodulation of compatible host legumes (Laguerre et al., 2001). However, although its phylogeny is not congruent with the core genes' phylogeny (Andrews and Andrews, 2017), it is used to characterize the new isolates at the symbiovar level (Rogel et al., 2011).
Analysis of the individual and concatenated nodA and nodC gene sequences and their phylogeny showed that the strains Llut5, Lcos6, Llut4, and Llut8 are members of the genistearum symbiovar.
All the strains are able to nodulate R. monosperma, L. luteus, L. albus, L. cosentinii, C. albidus, and C. monspessulanus but not G. max or Phaseolus vulgaris. It is known that the Genisteae microsymbionts do not nodulate soybean (G. max or G. soya: Phaseoleae tribe). However, they are able to nodulate different genera and species of the tribe Genisteae (Vinuesa et al., 2005). The fact that the strains are able to nodulate and fix nitrogen with different Genisteae is an advantage to members of this tribe. It is important to note that we observed no significant visible differences between the two lupine plants inoculated with the Lcos strains (isolated from L. cosentinii) or the Llut strains (isolated from L. luteus). All the plants were in good health as shown by the leaves' green color after 3 months. This represents an advantage for these legume plants, because they can establish a nitrogen fixing symbiosis with different strains with different PGP potentialities, which contributes to their resilience in the case of any edaphoclimatic changes.
In this work, we report on the characterization of some strains isolated from the root nodules of two lupines grown in the acidic soils of the Maamora cork forest, in Morocco.
We found that L. cosentinii is nodulated by members of the genus Bradyrhizobum, whereas the species has been reported as nodulated by Microvirga sp. in alkaline soils of the same forest. As already reported, the selection of lupine microsymbionts in the south Mediterranean would depend on the type and pH of the soil. Some Bradyrhizobium spp. strains newly isolated from L. luteus and L. cosentinii may constitute a new genospecies, and this needs confirmation by more sophisticated research such as whole genome sequencing or DNA–DNA hybridization.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank [accession: MW526996-MW527060, MW494669-MW494676, MW547016, and MW565871-MW565872].
MM participated in the conception and design of the experimentations and discussion of the results. OB participated in the analysis of genomic data and phylogenies. SE, HL, and SA participated in the isolation and characterization of isolates. EB participated in the sequencing experiments. HA participated in the conception of the project. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.661295/full#supplementary-material
Aafi, A., El Kadmiri, A. A., Benabid, A., and Rochdi, M. (2005). Richesse et diversité floristique de la suberaie de la Mamora (Maroc). Acta Bot. Malacit. 30, 127–138. doi: 10.24310/abm.v30i0.7187
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andrews, M., and Andrews, M. E. (2017). Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 18, 705. doi: 10.3390/ijms18040705
Andrews, M., De Meyer, S., James, E. K., Stepkowski, T., Hodge, S., Simon, M. F., et al. (2018). Horizontal transfer of symbiosis genes within and between rhizobial genera: occurrence and importance. Genes. 9:321. doi: 10.3390/genes9070321
Ardley, J. K., Parker, M. A., De Meyer, S. E., Trengove, R. D., O'Hara, G. W., Reeve, W. G., et al. (2012). Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int. J. Syst. Evol. Microbiol. 62, 2579–2588. doi: 10.1099/ijs.0.035097-0
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Beck, D. P., Materon, L. A., and Afandi, F. (1993). Practical rhizobium-legume technology manual. Technical Manual. ICARDA. 19.
Beligala, D. H., Michaels, H. J., Devries, M., and Phuntumart, V. (2017). Multilocus sequence analysis of root nodule bacteria associated with Lupinus spp. and glycine max. Adv. Microbiol. 7, 790–812. doi: 10.4236/aim.2017.711063
Beringer, J. E. (1974). R factor transfer in Rhizobium leguminosarum. Microbiology 84, 188–198. doi: 10.1099/00221287-84-1-188
Boukhatem, Z. F., Domergue, O., Bekki, A., Merabet, C., Sekkour, S., Bouazza, F., et al. (2012). Symbiotic characterization and diversity of rhizobia associated with native and introduced acacias in arid and semi-arid regions in Algeria. FEMS Microbiol. Ecol. 80, 534–547. doi: 10.1111/j.1574-6941.2012.01315.x
Bourebaba, Y., Durán, D., Boulila, F., Ahnia, H., Boulila, A., Temprano, F., et al. (2016). Diversity of Bradyrhizobium strains nodulating Lupinus micranthus on both sides of the Western Mediterranean: Algeria and Spain. Syst. Appl. Microbiol. 39, 266–274. doi: 10.1016/j.syapm.2016.04.006
Cermak, V. (2018a). Restriction Comparator. Available online at: http://www.molbiotools.com/restrictioncomparator.html (accessed November, 2010).
Cermak, V. (2018b). Silent Mutator. Available online at: http://www.molbiotools.com/silentmutator.html
Chaintreuil, C., Boivin, C., Dreyfus, B., and Giraud, E. (2001). Characterization of the common nodulation genes of the photosynthetic Bradyrhizobium sp. ORS285 reveals the presence of a new insertion sequence upstream of nodA. FEMS Microbiol. Lett. 194, 83–86. doi: 10.1111/j.1574-6968.2001.tb09450.x
Clúa, J., Roda, C., Zanetti, M. E., and Blanco, F. A. (2018). Compatibility between legumes and rhizobia for the establishment of a successful nitrogen-fixing symbiosis. Genes. 9:125. doi: 10.3390/genes9030125
De Bruijn, F. J. (1992). Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58, 2180–2187. doi: 10.1128/AEM.58.7.2180-2187.1992
De la Peña, T. C., and Pueyo, J. J. (2012). Legumes in the reclamation of marginal soils, from cultivar and inoculant selection to transgenic approaches. Agron. Sustain. Dev. 32, 65–91. doi: 10.1007/s13593-011-0024-2
Durán, D., Rey, L., Navarro, A., Busquets, A., Imperial, J., and Ruiz-Argüeso, T. (2014). Bradyrhizobium valentinum sp. nov., isolated from effective nodules of Lupinus mariae-josephae, a lupine endemic of basic-lime soils in Eastern Spain. Syst. Appl. Microbiol. 37, 336–341. doi: 10.1016/j.syapm.2014.05.002
Elsheikh, E. A. E., and Wood, M. (1989). Response of chickpea and soybean rhizobia to salt: osmotic and specific ion effects of salts. Soil Biol. Biochem. 21, 889–895. doi: 10.1016/0038-0717(89)90077-1
Gouda, S., Kerry, R. G., Das, G., Paramithiotis, S., Shin, H.-S., and Patra, J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 206, 131–140. doi: 10.1016/j.micres.2017.08.016
Guerrouj, K., Pérez-Valera, E., Chahboune, R., Abdelmoumen, H., Bedmar, E. J., and El Idrissi, M. M. (2013). Identification of the rhizobial symbiont of Astragalus glombiformis in Eastern Morocco as Mesorhizobium camelthorni. Antonie Van Leeuwenhoek 104, 187–198. doi: 10.1007/s10482-013-9936-y
Hall, R. S., Thomas, S. J., and Johnson, S. K. (2005). Australian sweet lupin flour addition reduces the glycaemic index of a white bread breakfast without affecting palatability in healthy human volunteers. Asia Pac. J. Clin. Nutr. 14:91.
Howieson, J. G., and Dilworth, M. J., (Eds.). (2016). Working with Rhizobia. ACIAR Monograph No. 173. Canberra: Australian Centre for International Agricultural Research (ACIAR). p. 312.
Jarabo-Lorenzo, A., Pérez-Galdona, R., Donate-Correa, J., Rivas, R., Velázquez, E., Hernández, M., et al. (2003). Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst. Appl. Microbiol. 26, 611–623. doi: 10.1078/072320203770865927
Kermah, M., Franke, A. C., Adjei-Nsiah, S., Ahiabor, B. D. K., Abaidoo, R. C., and Giller, K. E. (2018). N2-fixation and N contribution by grain legumes under different soil fertility status and cropping systems in the Guinea savanna of northern Ghana. Agric. Ecosyst. Environ. 261, 201–210. doi: 10.1016/j.agee.2017.08.028
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kohajdova, Z., KaroVičoVá, J., and Schmidt, Š. (2011). Lupin composition and possible use in bakery-a review. Czech J. Food Sci. 29, 203–211. doi: 10.17221/252/2009-CJFS
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laguerre, G., Nour, S. M., Valérie, M., Sanjuan, J., Drouin, P., and Amarger, N. (2001). Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147, 981–993. doi: 10.1099/00221287-147-4-981
Laguerre, G., van Berkum, P., Amarger, N., and Prévost, D. (1997). Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl. Environ. Microbiol. 63, 4748–4758. doi: 10.1128/AEM.63.12.4748-4758.1997
Lakshmanan, V., Shantharaj, D., Li, G., Seyfferth, A. L., Janine Sherrier, D., and Bais, H. P. (2015). A natural rice rhizospheric bacterium abates arsenic accumulation in rice (Oryza sativa L.). Planta 242, 1037–1050. doi: 10.1007/s00425-015-2340-2
Lamrabet, M., Lamin, H., Bouhnik, O., Bennis, M., Abdelmoumen, H., Eulogio, J. B., et al. (2020). Nodulation of Retama species by members of the genus Microvirga in Morocco. Symbiosis. 82, 249–258. doi: 10.1007/s13199-020-00725-5
Lindström, K., and Mousavi, S. A. (2020). Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 13, 1314–1335. doi: 10.1111/1751-7915.13517
Lu, J. K., Dou, Y. J., Zhu, Y. J., Wang, S. K., Sui, X. H., and Kang, L. H. (2014). Bradyrhizobium ganzhouense sp. nov., an effective symbiotic bacterium isolated from Acacia melanoxylon R. Br. nodules. Int. J. Syst. Evol. Microbiol. 64:1900. doi: 10.1099/ijs.0.056564-0
Martinez-Romero, E., and Ormeño-Orrillo, E. (2019). A genomotaxonomy view of the Bradyrhizobium genus. Front. Microbiol. 10:1334. doi: 10.3389/fmicb.2019.01334
Mellal, H., Yacine, B., Boukaous, L., Khouni, S., Benguedouar, A., Castellano-Hinojosa, A., et al. (2019). Phylogenetic diversity of Bradyrhizobium strains isolated from root nodules of Lupinus angustifolius grown wild in the North East of Algeria. Syst. Appl. Microbiol. 42, 397–402. doi: 10.1016/j.syapm.2019.01.003
Menna, P., Pereira, A. A., Bangel, E. V., and Hungria, M. (2009). Rep-PCR of tropical rhizobia for strain fingerprinting, biodiversity appraisal and as a taxonomic and phylogenetic tool. Symbiosis 48, 120–130. doi: 10.1007/BF03179991
Miller, M. S., and Pepper, I. L. (1988). Physiological and biochemical characteristics of a fast-growing strain of lupin rhizobia isolated from the Sonoran Desert. Soil Biol. Biochem. 20, 319–322. doi: 10.1016/0038-0717(88)90011-9
Missbah El Idrissi, M., Lamin, H., ElFaik, S., Tortosa, G., Peix, A., Bedmar, E. J., et al. (2020). Microvirga sp. symbiovar mediterranense nodulates Lupinus cosentinii grown wild in Morocco. J. Appl. Microbiol. 128, 1109–1118. doi: 10.1111/jam.14526
Msaddak, A., Rejili, M., Durán, D., Mars, M., Palacios, J. M., Ruiz-Argüeso, T., et al. (2019). Microvirga tunisiensis sp. nov., a root nodule symbiotic bacterium isolated from Lupinus micranthus and L. luteus grown in Northern Tunisia. Syst. Appl. Microbiol. 42:126015. doi: 10.1016/j.syapm.2019.126015
Msaddak, A., Rejili, M., Durán, D., Rey, L., Imperial, J., Palacios, J. M., et al. (2017). Members of Microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in Northern Tunisian soils. FEMS Microbiol. Ecol. 93, 1–13. doi: 10.1093/femsec/fix068
Msaddak, A., Rejili, M., Durán, D., Rey, L., Palacios, J. M., Imperial, J., et al. (2018). Definition of two new symbiovars, sv. lupini and sv. mediterranense, within the genera Bradyrhizobium and Phyllobacterium efficiently nodulating Lupinus micranthus in Tunisia. Syst. Appl. Microbiol. 41, 487–493. doi: 10.1016/j.syapm.2018.04.004
Peix, A., Ramírez-Bahena, M. H., Flores-Félix, J. D., Alonso de la Vega, P., Rivas, R., Mateos, P. F., et al. (2015). Revision of the taxonomic status of the species Rhizobium lupini and reclassification as Bradyrhizobium lupini comb. nov. Int. J. Syst. Evol. Microbiol. 65, 1213–1219. doi: 10.1099/ijs.0.000082
Rejili, M., Msaddak, A., Filali, I., Benabderrahim, M. A., Mars, M., and Marín, M. (2019). New chromosomal lineages within Microvirga and Bradyrhizobium genera nodulate Lupinus angustifolius growing on different Tunisian soils. FEMS Microbiol. Ecol. 95:fiz118. doi: 10.1093/femsec/fiz118
Rogel, M. A., Ormeño-Orrillo, E., and Martinez Romero, E. (2011). Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst. Appl. Microbiol. 34, 96–104. doi: 10.1016/j.syapm.2010.11.015
Rome, S., Fernandez, M. P., Brunel, B., Normand, P., and Cleyet-Marel, J.-C. (1996). Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int. J. Syst. Evol. Microbiol. 46, 972–980. doi: 10.1099/00207713-46-4-972
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sarkar, D., and Laha, S. (2013). Production of phytohormone auxin (IAA) from soil born Rhizobium sp, isolated from different leguminous plant. Int. J. Appl. Environ. Sci. 8, 521–528.
Sessitsch, A., Howieson, J. G., Perret, X., Antoun, H., and Martinez-Romero, E. (2002). Advances in rhizobium research. CRC Crit. Rev. Plant Sci. 21, 323–378. doi: 10.1080/0735-260291044278
Shamseldin, A., Abdelkhalek, A., and Sadowsky, M. J. (2017). Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: a review. Symbiosis 71, 91–109. doi: 10.1007/s13199-016-0462-3
Smith, S. C., Choy, R., Johnson, S. K., Hall, R. S., Wildeboer-Veloo, A. C. M., and Welling, G. W. (2006). Lupin kernel fiber consumption modifies fecal microbiota in healthy men as determined by rRNA gene fluorescent in situ hybridization. Eur. J. Nutr. 45, 335–341. doi: 10.1007/s00394-006-0603-1
StatSoft Inc (2004). STATISTICA (Data Analysis Software System), Version 7. Available online at: www.statsoft.com (accessed November, 2010).
Susek, K., Bielski, W. K., Hasterok, R., Naganowska, B., and Wolko, B. (2016). A first glimpse of wild lupin karyotype variation as revealed by comparative cytogenetic mapping. Front. Plant Sci. 7:1152. doi: 10.3389/fpls.2016.01152
Tampakaki, A. P., Fotiadis, C. T., Ntatsi, G., and Savvas, D. (2017). Phylogenetic multilocus sequence analysis of indigenous slow-growing rhizobia nodulating cowpea (Vigna unguiculata L.) in Greece. Syst. Appl. Microbiol. 40, 179–189. doi: 10.1016/j.syapm.2017.01.001
Tounsi-Hammami, S., Le Roux, C., Dhane-Fitouri, S., De Lajudie, P., Duponnois, R., Jeddi, F., and Ben (2019). Genetic diversity of rhizobia associated with root nodules of white lupin (Lupinus albus L.) in Tunisian calcareous soils. Syst. Appl. Microbiol. 42, 448–456. doi: 10.1016/j.syapm.2019.04.002
Trujillo, M. E., Willems, A., Abril, A., Planchuelo, A.-M., Rivas, R., Ludena, D., et al. (2005). Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl. Environ. Microbiol. 71, 1318–1327. doi: 10.1128/AEM.71.3.1318-1327.2005
Velázquez, E., Valverde, A., Rivas, R., Gomis, V., Peix, Á., Gantois, I., et al. (2010). Strains nodulating Lupinus albus on different continents belong to several new chromosomal and symbiotic lineages within Bradyrhizobium. Antonie Van Leeuwenhoek 97, 363–376. doi: 10.1007/s10482-010-9415-7
Vinuesa, P., Silva, C., Werner, D., and Martínez-Romero, E. (2005). Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34, 29–54. doi: 10.1016/j.ympev.2004.08.020
Wang, J., Andersen, S. U., and Ratet, P. (2018). Molecular and cellular mechanisms of the legume-rhizobia symbiosis. Front. Plant Sci. 9:1839. doi: 10.3389/fpls.2018.01839
Wójcik, M., and Kalita, M. (2019). Numerical analysis of phenotypic properties, genomic fingerprinting, and multilocus sequence analysis of Bradyrhizobium strains isolated from root nodules of Lembotropis nigricans of the tribe Genisteae. Ann. Microbiol. 69, 1123–1134. doi: 10.1007/s13213-019-01491-6
Wolko, B., Clements, J. C., Naganowska, B., and Nelson, M. N. (2011). “Lupinus,” in Wild Crop Relatives: Genomic And Breeding Resources, Ed C. Kole (Verlag Berlin Heidelberg: Springer), 153–206. doi: 10.1007/978-3-642-14387-8_9
Yorgancilar, M., and Bilgiçli, N. (2014). Chemical and nutritional changes in bitter and sweet lupin seeds (Lupinus albus L.) during bulgur production. J. Food Sci. Technol. 51, 1384–1389. doi: 10.1007/s13197-012-0640-0
Zahran, H. H., Räsänen, L. A., Karsisto, M., and Lindström, K. (1994). Alteration of lipopolysaccharide and protein profiles in SDS-PAGE of rhizobia by osmotic and heat stress. World J. Microbiol. Biotechnol. 10, 100–105. doi: 10.1007/BF00357572
Keywords: Lupinus, Bradyrhizobium, symbiosis, nodulation, diversity, MLSA
Citation: Missbah El Idrissi M, Bouhnik O, ElFaik S, Alami S, Lamin H, Bedmar EJ and Abdelmoumen H (2021) Characterization of Bradyrhizobium spp. Nodulating Lupinus cosentinii and L. luteus Microsymbionts in Morocco. Front. Agron. 3:661295. doi: 10.3389/fagro.2021.661295
Received: 30 January 2021; Accepted: 22 March 2021;
Published: 11 May 2021.
Edited by:
Jose Palacios, Polytechnic University of Madrid, SpainReviewed by:
Luis Rey, Polytechnic University of Madrid, SpainCopyright © 2021 Missbah El Idrissi, Bouhnik, ElFaik, Alami, Lamin, Bedmar and Abdelmoumen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mustapha Missbah El Idrissi, bWlzc2JhaDQ5QHlhaG9vLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.