- National Biological Control Laboratory, Biological Control of Pests Research Unit, US Department of Agriculture, Agricultural Research Service, Stoneville, MS, United States
RNA interference (RNAi) has inspired insect scientists to aim toward the development of this technology for protection against insect pests. The RNAi mechanism works at the intracellular level by exploiting a mode of action specific to the expression of genes, interrupting the transcription to translation process. Many of the limitations of RNAi technology are being addressed to adapt it for insect pest application. However, most of the insect pest problems for which RNAi is being developed involve direct plant-insect interactions, primarily in monocultures. Ants (Hymenoptera: Formicidae) are important agricultural pests that generally do not feed directly on crops, yet have dramatic impact on agroecosystems such as pastures, orchards, and nurseries. The application of RNAi to pest ants is complicated by the social nature of ants. Here the goal is to examine the potential application of RNAi to ant pests, especially invasive ants, which present distinct challenges with regard to delivery, targeting, efficacy, and risks.
Introduction
RNA interference (RNAi) is a cellular process that degrades messenger RNA (mRNA), the key to protein synthesis, and hence gene function. The RNAi process is conserved in eukaryotic organisms including insects (Agrawal et al., 2003; Dowling et al., 2016). The overall process is triggered by double-stranded RNA (dsRNA) inside the cell. In a generalized model eukaryotic cell, the presence of dsRNA signals invasion by viral molecules or activity of transposable elements. Either of these conditions represent a threat to the cell and triggers a defensive RNAi mechanism. A chain of events within the cell is triggered by recognition of dsRNA, and genes are activated that produce the RNAi core machinery: sets of proteins that assemble into an enzymatic apparatus called the RNA-induced silencing complex (RISC) [reviewed in Cooper et al. (2019) and Zhu and Palli (2020)]. This apparatus disassembles the dsRNA and enables the degradation of any mRNA that is precisely identical to the dsRNA sequence. The generalized RNAi mechanism relies on several coordinated enzymes; both the mechanism and enzymes have been characterized in model organisms. Specific enzymatic pathways that enable RNAi within a single cell and among tissues within a single insect vary among insect species. In social insects, the specific RNAi genes and pathways, while predicted as present by homology are mostly undeciphered so far.
The precise specificity of RNAi to a unique sequence has inspired molecular genetic techniques and led to exploitation of the RNAi process and mechanisms for experimental and applied designs. One exciting potential application of RNAi is insect control (Baum et al., 2007; Price and Gatehouse, 2008; Scott et al., 2013). Ideally, a dsRNA could be designed that interrupts one or more vital insect species' genes so specifically and effectively that only the target insect species would be killed.
Delivery of RNAi products to combat crop pests through transgenic plants has been enthusiastically pursued by science and the agricultural biotechnology industry (Baum et al., 2007; Cagliari et al., 2019) and has entered the commercial marketplace. The recent development of a transgenic crop product, SmartStax PRO maize, approved for use in North America, is an example of an application of RNAi technology that combats a destructive beetle, the corn rootworm Diabrotica spp. (Coleoptera: Chrysomelidae) (Head et al., 2017). Non-transgenic plant methods to deliver RNAi pest control products have also been theorized, designed, and tested [reviewed in Cagliari et al. (2019) and Taning et al. (2020)].
In this discussion of RNAi and insect pest control, ants are considered as potential targets of RNAi technology. Generalized barriers and limitations to dsRNA delivery and efficiency are examined in light of progress that has been made to date.
RNAi in Insects
The RNAi mechanism in general and in insects specifically has been reviewed (Vélez and Fishilevich, 2018; Vogel et al., 2018; Christiaens et al., 2020). In a broad study of insect transcriptomes, many taxonomic groups of insects were examined and found to have a suite of genes encoding core RNAi enzymes; unfortunately ants were not included (Dowling et al., 2016). An ant genome project (Smith et al., 2011) found both nuclear and cytoplasmic RNAi genes in a harvester ant, Pogonomyrmex barbatus. RNAi components were described in the genomes of Jerdon's jumping ant Harpegnathos saltator and the Florida carpenter ant Camponotus floridanus, and injection of dsRNA resulted in decreased measures of gene transcription (Bonasio et al., 2010). To illustrate that other members of the Formicidae are no exception to the apparent presence of RNAi core enzyme genes, three species of invasive ant were queried in NCBI (BLASTp) with proteins of core and systemic RNAi enzymes, and the resulting homologs are listed in Supplementary Table 1.
Insects are multicellular organisms, and application of RNAi relies on the propagation of the mechanism throughout cells and tissues. In isolated insect cells derived from the model insect Drosophila melanogaster, a protein called systemic RNA interference–deficient (SID) identified from the model nematode Caenorhabditis elegans (Winston et al., 2002) was shown to improve intercellular distribution of dsRNA (Feinberg and Hunter, 2003). A second SID protein was required for intestinal absorption in C. elegans (Winston et al., 2007). Ant genomes have genes similar to sid (Supplementary Table 1).
RNAi evaluation in insects is most often initiated by microinjection studies that are aimed at characterizing gene function or identifying RNAi gene targets. This is the obvious method for delivering dsRNA to cells; ideally a fertilized or viable haploid insect egg in pre-cellularization stage is injected. This maximizes likelihood of delivery of the dsRNA effector construct to each cell, bypassing the cell membrane barrier to intercellular RNAi distribution. Cell fates in insects are determined during oogenesis and early in embryogenesis. In later stages of development the integument and cuticle are formidable barriers for both the insect's exterior and most of the cells within the insect gut. In a major agricultural pest, Helicoverpa zea (Lepidoptera: Noctuidae), four candidate genes were evaluated for RNAi effect, using egg microinjection, egg soaking, and feeding as delivery strategies. Only egg microinjection resulted in decreased transcript levels, demonstrating the recalcitrance of some insect species to diet-delivered dsRNA (Wang et al., 2018). Thus, far, due to their delicate nature as components of a superorganism, ant eggs have not been injected with dsRNA. Testing RNAi candidate genes in cell lines is also preferable, however there is a dearth of insect cell lines available for research, and none that are derived from ants (Arunkarthick et al., 2017; Guo et al., 2020). In the absence of cell lines or egg microinjection methods, dsRNA delivery for the purpose of biopesticide development in insects is most often achieved via haemocoelic microinjection or through feeding.
The Nature of Ants
While many members of Hymenoptera are social insects, all members of the family Formicidae are eusocial, the most advanced form of sociality (Branstetter et al., 2017). The definition of eusociality includes three characteristics: cooperative care of juvenile forms, overlapping generations in the colony, and development of differing adult forms (castes) that include both sterile and reproductive forms. Ants do not function as individuals but as members of a colony that includes at least one reproductive queen and sterile female workers (Hölldobler and Wilson, 1990). Because of the interdependency of the diverse members of the colony, ants are described as a superorganism (Hölldobler and Wilson, 2009; Pull and McMahon, 2020). While many insects exhibit social behaviors, the ants certainly exhibit the most social complexity. Termites, another insect that might be considered an apex of social complexity, are in another phylogenetic order evolutionarily distant from the ants and are adapted to degrade woody materials, a quite different trophic niche from the ants. Only the family Formicidae are considered here.
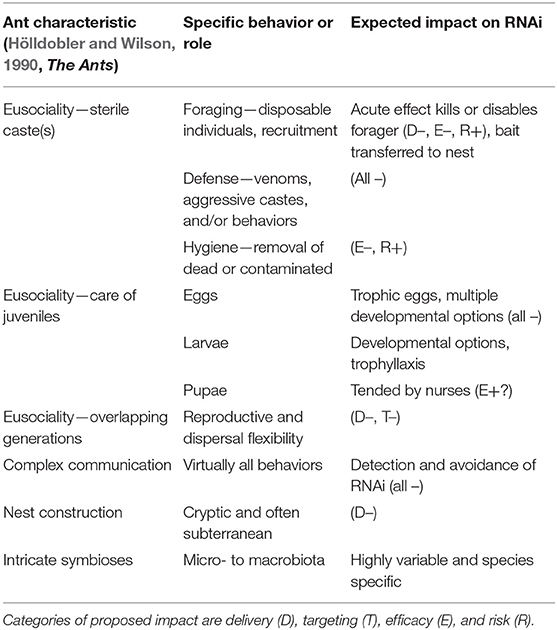
Division of labor among workers is pronounced in most ant species and is important in their ecological and invasion success. In many social insects, division of labor is based on age (temporal castes). Some ant workers show morphological specialization for particular tasks (physical castes); these are often those major workers with adaptation of the head and mandibles for colony defense. For example, the genus Pheidole includes species that produce soldier ants with greatly enlarged heads that can be manipulated using RNAi (Rajakumar et al., 2018). Other genera have similarly differentiated soldier castes that are enlarged overall and have heads and mandibles adapted for defense. Other soldier caste morphological adaptations include those adapted for blocking nest entrances and those with mandibles adapted for grinding grain. These highly visible adaptations are in sterile forms that may be useful for study but are not useful for designing colony-level targeted RNAi biopesticides. Subtle behavioral castes are currently being identified (Valentini et al., 2020) but generalized adult roles that are vital to the success of the colony, and thus logically critical to RNAi biopesticide success, are the forager and nurse worker. Foraging workers are often numerous and somewhat expendable, ranging outside of the colony and exposed to dangers. Nurses stay within the colony and tend juveniles, collectively referred to as “brood.” Other adult worker roles are defense, hygiene, food processing, queen tending, and others. While every role in the colony is important, logical targets for pest control focused RNAi, those vital to the overall and immediate colony survival, should involve central processes of brood care and reproduction. The basic function of communication within the colony, if disrupted, could also lead to colony death.
After an egg has been produced by the ant queen, it is cared for by worker adults of the nurse caste. Eggs are collected and incorporated into the brood pile that often includes other juvenile stages, which require constant tending by workers to survive. An ant egg may develop into a sterile female, a fertile female, or a male, or may not develop at all; ants are known to produce “trophic eggs” that provide nutrition for the colony (Hölldobler and Wilson, 1990; Crespi, 1992; Perry and Roitberg, 2006). As larvae, those destined to be reproductives—males or females—often become substantially larger (or otherwise differentiated) than the sterile females, the workers. Reproductive adults are generally winged, although the female reproductive wings are deciduous, while workers are wingless. The constant colony care required for juvenile ants makes handling and experimenting with isolated stages in a laboratory environment nearly impossible; this is an impediment to developing RNAi tests.
Importantly, juveniles play a vital nutritional role in the ant colony; the larvae serve as the colony digestive organ (Dussutour and Simpson, 2009; Allen et al., 2018). The morphology of adult ants includes a narrow constriction, the petiole, between the mouth and the internal digestive organs, the crop and the midgut. This mechanically prevents passage of large food particles to the gut and is supplemented in some species by an internal filter (Hölldobler and Wilson, 1990; Lanan et al., 2016). Only liquids can pass into the crop or midgut of adult ants. Larvae, on the other hand, have no such constriction, and they produce digestive enzymes adults lack (Hölldobler and Wilson, 1990; Lanan et al., 2016; Allen et al., 2018), and are able to digest food solids. Through trophallaxis, regurgitation of stomach contents, larvae provide digested liquids to feed all members of the colony (Cassill et al., 2005). Thus, digestion and nutrition in the colony is a distributed activity; food particles are collected by adult foragers, digested by larvae, and passed to other colony members through trophallaxis. In many species food processing involves much more complex steps, notably in fungus-gardening ants (Hölldobler and Wilson, 1990). For the purposes of this discussion all pre-feeding activities will be called “processing,” and yet those very activities may be critical to success or failure of an RNAi ant control product. The complexity of diverse ant food collection and processing is the first barrier to RNAi ant control implementation. Cooper et al. (2019) defines five impediments to RNAi efficiency in insect systems: (1) dsRNA instability, (2) problems with dsRNA internalization, (3) problems with core RNAi machinery (4) problems with spreading of RNAi signal/mechanism within the insect body, and (5) target gene associated problems. In an ant colony, all five of these potential issues must first overcome the social behaviors and mechanisms associated with feeding, presenting further difficult entry into the cellular RNAi environment within the target individual within the colony. Better understanding of the food collection and processing steps performed by each pest ant species will reduce barriers and present opportunities for delivery of RNAi biopesticides to the colony if the five impediments can be overcome. The ant colony presents additional impediments, and opportunities, associated with the nature of ants as a superorganism. The adult worker ants, the foragers, collect materials to feed the colony but do not ingest them directly nor immediately. The foragers first mechanically disassemble food into bits that can be transported back to the colony. Food sources may be covered with particulate matter prior to disassembly and transport (Wang et al., 2016; Qin et al., 2019). Food sources may be cut into small pieces and stored within the colony (Haack et al., 1995; Gayahan and Tschinkel, 2008). Solid particles are then passed to larvae for digestion of complex and high molecular weight materials: proteinaceous, complex carbohydrate, or other macromolecules. Liquids are collected in the forager's crop. After various “processing” steps, nutrients are distributed throughout the colony via trophallaxis. All of these food handling steps may be viewed as additional barriers/opportunities between presentation of a potentially effective dsRNA molecule and the interior of the ant cell in which it is intended to interrupt gene function. Some genes that may be involved in dsRNA degradation in ants (Supplementary Table 2) show homology to orthopteran genes (Song et al., 2019). Beyond being degraded by the gut contents or hemolymph, similar to most insects (Cooper et al., 2019), dsRNA targeting ant genes may be degraded by the pre-ingestion environment, by ant forager exudates in saliva or venom gland secretions during collection or storage of solids. Although ant saliva and venoms have not yet been tested for dsRNA-degrading activity, ant venom is antimicrobial (Tragust, 2016; Aanen, 2018; Cremer et al., 2018), and some ants produce an unidentified RNA-degrading compound (Valles et al., 2012). DsRNA may also be degraded within the crop of the forager ant in the case of liquids. Additionally, soil microorganisms may interact with the dsRNA. Figure 1 presents a generalized diagrammatic representation of the potential ant superorganism barriers to RNAi application considered in this text, many of which have yet to be examined experimentally. Potential barriers are also listed in Table 1.
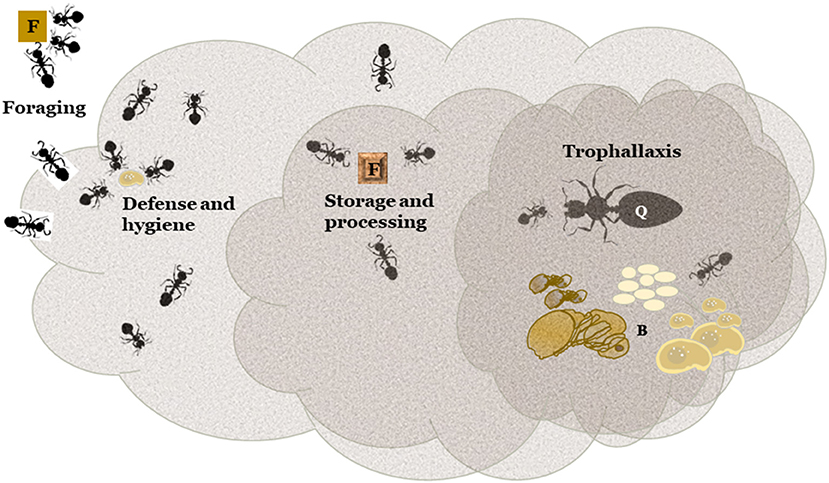
Figure 1. Impediments to RNAi efficacy in ant colonies based on their social nature. Preparations of RNAi effector molecules (square labeled F for “food”) must be collected by foraging workers. Workers dedicated to defending the colony may reject contaminated food or foragers. Furthermore, worker or brood that are defective or contaminated may be discarded by worker ants functioning in a hygiene role (social immunity). Food may be stored and/or processed in sites within the nest or within workers. Modified food is represented by the square labeled F with a modified shape effect. When food is needed it is digested and converted to liquid form by larvae and distributed among juveniles, workers, and queen(s) by trophallaxis. Q, queen; B, brood.
Ants as Pests
The agronomic impact of ants is usually indirect, because very few ants attack growing plants. A notable exception is fungus farming ants that may defoliate vast territories to support enormous colonies (Zanetti et al., 2003). Many ants defend sucking pests, improving their success and thus increasing crop damage. More commonly, the nest-building activities of ants, particularly invasive ants, disrupt infrastructures such as roads, sidewalks, or other stone or cement foundations (Vinson, 2013). Ants also invade commercial beekeeping operations (Weeks and Drees, 2002). Nests may be made on or near farm equipment and electrical infrastructure, and result in damage to the equipment itself or very often contact with humans or livestock that results in bites, stings, and in extreme cases death (Goddard, 2004; Tankersley, 2008). Certain ants have become some of the most successful invasive pests, notably the Argentine ant, Linepithema humile, and the red imported fire ant, Solenopsis invicta, which has been extensively studied as an ant, as a pest, and as a target for RNAi.
RNAi Applied to Ants
In the first demonstration of RNAi in an ant, the vitellogenin receptor (VgR) gene of the red imported fire ant, S. invicta, was targeted. Transfer of the protein vitellogenin via VgR is pivotal to the development of insect oocytes in the ovary. Injection of VgR dsRNA into the abdomens of queen pupae while the cuticle was still white but eyespots were visible resulted in silencing of the gene and delayed oocyte development (Lu et al., 2009). Interestingly, injection of more advanced queen pupae in which cuticle darkening had progressed did not result in silencing (Lu et al., 2009). To demonstrate a mechanism for social insect communication, a pheromone biosynthesis activating neuropeptide (PBAN) and its receptor were targeted with RNAi, again in S. invicta. Silencing was demonstrated by decreased transcription, differences in pheromone production, and the inability of dsRNA-injected ants to produce functional trail pheromones (Choi and Vander Meer, 2012). The same PBAN dsRNA interfered with melanization (cuticle darkening) of pupae when injected, was shown to result in increased mortality when injected, and importantly increased mortality of larvae when dsRNA was fed in a sucrose solution to nurse ants (Choi et al., 2012). This dsRNA feeding experiment resulted in multiple patent awards (US Patent 8,575,328, 2013; US Patent 9,000,145, 2015; US Patent 9,617,542, 2017). Another feeding study in S. invicta utilized small dsRNA to test a chemosensory gene, CSP9 and a protein kinase gene. Feeding resulted in gene silencing after 48 h and the larvae fed by nurse ants provided with the dsRNA/sucrose solution had increased mortality and failed to molt normally (Cheng et al., 2015). Similarly, feeding dsRNA based on the sequence of a short neuropeptide receptor gene may have affected larval mortality, although supporting data from multiple feeding studies varied (Castillo Bravo, 2015). In another feeding study, dsRNA complementary to sequences of the capsid protein gene of an ant-infecting virus, Solenopis invicta virus 1 (SINV-1) was provided to colonies of ants intentionally infected with the virus. The results showed a leveling-off of the effects of viral infection (death of colony members) and a decrease in viral titre after dsRNA treatment (Rydzak and Bextine, 2016).
In the sexually dimorphic ant genus, Diacamma, microinjected dsRNA silenced expression of the gene yellow which is involved in melanization. After injecting the pre-darkened pupal stage twice with a 666 bp preparation of dsRNA, individual specimens were observed for color changes. Expression levels of the yellow gene were also measured and treated and control specimens had significantly different levels overall. Color results were somewhat inconsistent among the treated specimens but color changes were documented and significant in females, which normally are black; the yellow dsRNA treated specimens were brown. In males, an increase in mortality was noted in the dsRNA treated group. The males were normally brown so the dsRNA-based color effect was not apparent (Miyazaki et al., 2014).
In another ant, C. floridanus, two genes encoding peptidoglycan recognition proteins thought to be involved in the regulation of the intracellular endosymbionts were used as targets for RNAi (Ratzka et al., 2013). Earlier work on this species had shown that injection of dsRNA resulted in decreased measured gene transcription (Bonasio et al., 2010). DsRNA was fed to insects and decreased gene expression was demonstrated for one of the genes, but only in adult insects fed dsRNA, not in the larvae fed by the adults. Additionally, and counterintuitively, the treated adult midgut tissue did not show silencing while the dsRNA was shown to persist in the adult crop (Ratzka et al., 2013).
Additional studies focused on the physiological function of estrogen receptor and associated pathway genes in the ant Polyrhachis vicina. DsRNA was used in feeding bioassays and results depended on the caste and time following dsRNA exposure (Zhang and Xi, 2018; Xi et al., 2019).
Injection of dsRNA in ants have yielded more dramatic results in Harpegnathos saltator and Pheidole hyatti (Gospocic et al., 2017; Rajakumar et al., 2018), reviewed in Arsenault et al. (2019). In H. saltator, an ant that has “unusual reproductive flexibility” in that a sterile worker of a colony that has lost a queen is able to convert to a reproductive status, RNAi facilitated the identification of a gene critical to caste identity, corazonin (Gospocic et al., 2017). The RNAi component of the study utilized small interfering RNAs (siRNA) designed to knock down transcription of the receptor gene for the corazonin peptide. The siRNA was injected into the heads of living adult ants, with behavioral and transcriptional effects. Counts of ants interacting with crickets in a confined space were categorized as hunting behavior, and transcription was measured using both qRT-PCR and RNA-Seq comparisons. The results indicated convincingly that the siRNA treatment was effective, and supported the study conclusions: corazonin receptor knockdown decreased hunting. In P. hyatti RNAi was used to show how a specific gene vestigial was related to differentiation of a soldier subcaste phenotype, characterized by an oversized head (Rajakumar et al., 2018). This RNAi experiment was part of the larger study of caste differentiation in a species with highly dimorphic workers. A very concentrated (3.5 mg/ml) solution of dsRNA was used and microinjected into larvae at the wing bud region where the vestigial gene is expressed. This study used a relatively long, over 600 base pairs, dsRNA construct. Measurements of the RNAi effect included overall size, head size, wing development in males, and semiquantitative RT-PCR. While the overall results reported were compelling with respect to the topic of gene function in caste differentiation, the range of recorded RNAi results overlap in the treated and untreated groups, so the knockdown was far from efficient, even using the very stringent methods.
The most recent study of RNAi in ants focused on feeding dsRNA based on six highly conserved housekeeping genes to the invasive pest ant Nylanderia fulva, another highly invasive ant. Heat-inactivated bacterial expression vectors were incorporated into artificial diet prepared from expression vector cells, glucose solution, and agar, and fed to worker ants. Reduced gene expression was measured after feeding, and two sequences resulted in modestly higher ant mortality (Meng et al., 2020).
These studies have little in common, other than ants as target organisms. Target genes, control genes, methods of introduction and measurement were dramatically different and focused on very different experimental goals (see Supplementary Table 3). The studies that used feeding as a delivery method did not use similar methods for feeding. Taken together, however, these studies demonstrate that RNAi is functional in ants, but that effectiveness is highly variable. One implication that can be gleaned from injection experiments is that direct application of either long or siRNA constructs can decrease transcription of a target gene if delivered in close proximity to the desired tissue and at the specific developmental stage of gene expression. However, there is substantial variation among treated individuals. Routine procedures and standardized results need to be established. Feeding experiments reach similar conclusions: results vary depending on the gene, delivery, stage, and tissue.
Prospects and Recommendations
No experiments have yet examined whole colony impacts. To produce RNAi-based pest control products for ants, the interfering molecule, dsRNA, must be delivered to the organism, and further to the superorganism, the colony as a whole, overcoming numerous barriers (Table 1). Products used to control ant pests are most often formulated as baits. Large scale production of dsRNA and other RNAi materials is progressing to a point that testing bait formulations for ant control should become possible (Taning et al., 2020). And yet, basic questions about applying RNAi to control ants persist. Will RNAi baits effectively reach the cells of the vital stage of the vital caste of the targeted species? Once the dsRNA molecule is internalized by an insect in the colony, having passed potentially through or by several individuals, the RNAi machinery may be activated and the signal spread throughout the individual. But will killing or changing that individual impact the entire colony? The eusocial characteristics of ants require special consideration in designing an RNAi biopesticide strategy. Table 1 summarizes the ant characteristics and potential impacts; further research is needed to elucidate solutions and overcome challenges in specific pest species. In general, sterile foraging individuals must be attracted to the product and transport it to the colony. The product must withstand any food handling performed by the colony workers and resist degradation by the environment of the colony and any microbiota within the nest. The product must not trigger any defensive or hygiene behaviors. Once these challenges have been met, the product should be distributed throughout the colony by trophallaxis, enhancing the potential for the biopesticide to reach all colony members, both juvenile and adult. Target genes may be aimed at disrupting basic cellular function, digestive, reproductive, or communications systems. Choosing an appropriate target gene or genes must be done in the context of the colony, not the individual (Rodovalho et al., 2011; Allen et al., 2018). Progress to date, and indications from genome sequencing (Supplementary Tables 1–3) indicate likelihood of success. However, efficiency is still a problem (Meng et al., 2020), and the possibility that dsRNA is degraded within each pest species must be addressed. DsRNA stability and RNAi signal propagation have been discussed as limitations to effective RNAi pest control strategies (Terenius et al., 2011; Allen and Walker, 2012; Shukla et al., 2016; Vélez and Fishilevich, 2018) and dsRNA degrading enzymes have recently been discovered and described as problematic (Guan et al., 2018; Song et al., 2019). Many genes are being identified through recent and ongoing sequencing efforts (Boomsma et al., 2017). None of the studies performed so far provide substantial insight to these questions, although the implications of overall results are tantalizing. While the number of ant genomes continues to increase, the functional studies of ant genes specific to the diverse and fascinating behaviors of ants are relatively few. Studies that focus on effective delivery methods for colony-level effects are needed. New technologies for assisting and packaging RNAi biopesticides, as discussed in Zhu and Palli (2020), should be assessed in ant bioassays, which also need to be established, independently verified, and standardized as much as possible.
The core RNAi components appear to be present in ants (Supplementary Table 1). Detailed transcription studies identifying the timing, castes, developmental stages, and tissues in which RNAi enzymes and RNA-degrading enzymes are present in ants in the presence and absence of dsRNA would be extremely useful to the implementation of RNAi both for study purposes and for development of RNAi biopesticides. Ant-derived cell lines would also be extremely helpful for studying gene interactions in ants.
RNAi detection in ants is a challenge. Visible markers have been enormously helpful; eye color knockouts have been demonstrated in Orthoptera (Dong and Friedrich, 2005) and Lepidoptera (Fabrick et al., 2004), but have not been demonstrated in ants. As primarily subterranean organisms, ants generally do not have well-developed eyes. Still, eye pigments can be visualized in the developing pupae of many ant species, so a knockout phenotype study could prove valuable, and is recommended. Experimentally, knockout of visual pigment or transport genes may be more efficiently conducted initially using CRISPR tools (Trible et al., 2017; Yan et al., 2017) rather than RNAi. Sadly, some of the visible phenotype studies, such as the Pheidole supersoldier head phenotype, are not applicable to most ant species. Thus, for ants in general, there is a lack of a broadly useful positive control. The sort of cuticle color RNAi applications that were successful in the genus Diacamma (Miyazaki et al., 2014) have proven difficult in other ant genera (unpublished observations by this author). Thus, for ants in general, there is a lack of a broadly useful positive control for phenotype.
Certainly ants, as most other broad taxonomic groups, are not solely composed of invasives and/or pests. Furthermore, those ants that are pests in one context may perform environmental services in another (Helms et al., 2016; Baldwin et al., 2020). Therefore, it is essential to design novel ant control materials that have few off-target effects, to protect other ants and other benign organisms that might contact a preparation of ant RNAi materials. The high target specificity due to the complementary nature of gene-coding molecules makes RNAi technology especially appealing to insects in light of the many ecosystem services provided by ants and other insects as pollinators, nutrient and soil cyclers, predators, and food for wildlife (Baldwin et al., 2020; Elizalde et al., 2020).
So many aspects of ant biology, while fascinating, present concerns with respect to the development of RNAi ant control products. All of the facets of ant biology, from behavior to biochemistry, await study and illumination so that we can develop and apply this powerful molecular technology now available to science and agriculture.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by USDA ARS In-House Appropriated Research Project: Biology and Control of Invasive Ants, Project Number: 6066-22320-010-00-D.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Jian Chen and Yuzhe Du kindly provided internal (USDA) review and suggestions for an earlier version of this manuscript. I thank the reviewers and the editor who provided valuable guidance. The USDA/ARS is an equal opportunity employer and provider.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.591539/full#supplementary-material
References
Aanen, D. K. (2018). Social immunity: the disposable individual. Curr. Biol. 28, R322–R324. doi: 10.1016/j.cub.2018.02.050
Agrawal, N., Dasaradhi, P., Mohmmed, A., Malhotra, P., Bhatnagar, R. K., and Mukherjee, S. K. (2003). RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. doi: 10.1128/MMBR.67.4.657-685.2003
Allen, M. L., Rhoades, J. H., Sparks, M. E., and Grodowitz, M. J. (2018). Differential gene expression in red imported fire ant (Solenopsis invicta) (Hymenoptera: Formicidae) larval and pupal stages. Insects 9:185. doi: 10.3390/insects9040185
Allen, M. L., and Walker, W. B. III. (2012). Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 58, 391–396. doi: 10.1016/j.jinsphys.2011.12.014
Arsenault, S. V., Glastad, K. M., and Hunt, B. G. (2019). Leveraging technological innovations to investigate evolutionary transitions to eusociality. Curr. Opin. Insect Sci. 34, 27–32. doi: 10.1016/j.cois.2019.03.003
Arunkarthick, S., Asokan, R., Aravintharaj, R., Niveditha, M., and Kumar, N. K. (2017). A review of insect cell culture: establishment, maintenance and applications in entomological research. J. Entomol. Sci. 52, 261–273. doi: 10.18474/JES17-02PT.1
Baldwin, J., Paula-Moraes, S. V., and Pereira, R. (2020). The good side of the bad guys: predation of lepidopteran pests by Solenopsis invicta Buren (Hymenoptera: Formicidae) in the Florida Panhandle. Florida Entomol. 103, 68–71. doi: 10.1653/024.103.0411
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Bonasio, R., Zhang, G., Ye, C., Mutti, N. S., Fang, X., Qin, N., et al. (2010). Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071. doi: 10.1126/science.1192428
Boomsma, J. J., Brady, S. G., Dunn, R. R., Gadau, J., Heinze, J., Keller, L., et al. (2017). The global ant genomics alliance (GAGA). Myrmecol. News 25, 61–66. Available online at: https://myrmecologicalnews.org/cms/index.php?option=com_content&view=category&id=1486&Itemid=407
Branstetter, M. G., Danforth, B. N., Pitts, J. P., Faircloth, B. C., Ward, P. S., Buffington, M. L., et al. (2017). Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr. Biol. 27, 1019–1025. doi: 10.1016/j.cub.2017.03.027
Cagliari, D., Dias, N. P., Galdeano, D., dos Santos, E. Á., Smagghe, G., and Zotti, M. J. (2019). Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 10:1319. doi: 10.3389/fpls.2019.01319
Cassill, D. L., Butler, J., Vinson, S. B., and Wheeler, D. E. (2005). Cooperation during prey digestion between workers and larvae in the ant, Pheidole spadonia. Insectes Soc. 52, 339–343. doi: 10.1007/s00040-005-0817-x
Castillo Bravo, P. A. (2015). Short Neuropeptide F Receptor in the Worker Brain of the Red Imported Fire Ant (Solenopsis invicta Buren) and Methodology for RNA Interference. MS, TAMU.
Cheng, D., Lu, Y., Zeng, L., Liang, G., and He, X. (2015). Si-CSP9 regulates the integument and moulting process of larvae in the red imported fire ant, Solenopsis invicta. Sci. Rep. 5:9245. doi: 10.1038/srep09245
Choi, M. Y., and Vander Meer, R. K. (2012). Ant trail pheromone biosynthesis is triggered by a neuropeptide hormone. PLoS ONE 7:e50400. doi: 10.1371/journal.pone.0050400
Choi, M. Y., Vander Meer, R. K., Coy, M., and Scharf, M. E. (2012). Phenotypic impacts of PBAN RNA interference in an ant, Solenopsis invicta, and a moth, Helicoverpa zea. J. Insect Physiol. 58, 1159–1165. doi: 10.1016/j.jinsphys.2012.06.005
Christiaens, O., Whyard, S., Velez, A. M., and Smagghe, G. (2020). Double-stranded RNA technology to control insect pests: current status and challenges. Front. Plant Sci. 11:451. doi: 10.3389/fpls.2020.00451
Cooper, A. M., Silver, K., Zhang, J., Park, Y., and Zhu, K. Y. (2019). Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manage. Sci. 75, 18–28. doi: 10.1002/ps.5126
Cremer, S., Pull, C. D., and Fuerst, M. A. (2018). Social immunity: emergence and evolution of colony-level disease protection. Annual Rev. Entomol. 63, 105–123. doi: 10.1146/annurev-ento-020117-043110
Crespi, B. J. (1992). “Cannibalism and trophic eggs in subsocial and eusocial insects,” in Cannibalism: Ecology and Evolution Among Diverse Taxa, eds B. J. Crespi and M. A. Elgar (Oxford: Oxford University Press), 176–213.
Dong, Y., and Friedrich, M. (2005). Nymphal RNAi: systemic RNAi mediated gene knockdown in juvenile grasshopper. BMC Biotechnol. 5:25. doi: 10.1186/1472-6750-5-25
Dowling, D., Pauli, T., Donath, A., Meusemann, K., Podsiadlowski, L., Petersen, M., et al. (2016). Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol. Evol. 8, 3784–3793. doi: 10.1093/gbe/evw281
Dussutour, A., and Simpson, S. J. (2009). Communal nutrition in ants. Curr. Biol. 19, 740–744. doi: 10.1016/j.cub.2009.03.015
Elizalde, L., Arbetman, M., Arnan, X., Eggleton, P., Leal, I. R., Lescano, M. N., et al. (2020). The ecosystem services provided by social insects: traits, management tools and knowledge gaps. Biol. Rev. 95, 1418–1441. doi: 10.1111/brv.12616
Fabrick, J. A., Kanost, M. R., and Baker, J. E. (2004). RNAi-induced silencing of embryonic tryptophan oxygenase in the Pyralid moth, Plodia interpunctella. J. Insect Sci. 4:15. doi: 10.1093/jis/4.1.15
Feinberg, E. H., and Hunter, C. P. (2003). Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547. doi: 10.1126/science.1087117
Gayahan, G. G., and Tschinkel, W. R. (2008). Fire ants, Solenopsis invicta, dry and store insect pieces for later use. J. Insect Sci. 8:39. doi: 10.1673/031.008.3901
Goddard, J. (2004). Fire ant attacks on humans. Infect. Med. 21, 587–599. doi: 10.1007/978-3-319-75874-9_11
Gospocic, J., Shields, E. J., Glastad, K. M., Lin, Y., Penick, C. A., Yan, H., et al. (2017). The neuropeptide corazonin controls social behavior and caste identity in ants. Cell 170, 748–759.e712. doi: 10.1016/j.cell.2017.07.014
Guan, R. B., Li, H. C., Fan, Y. J., Hu, S. R., Christiaens, O., Smagghe, G., et al. (2018). A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 293, 6011–6021. doi: 10.1074/jbc.RA117.001553
Guo, Y., Goodman, C. L., Stanley, D. W., and Bonning, B. C. (2020). Cell lines for honey bee virus research. Viruses 12:236. doi: 10.3390/v12020236
Haack, K. D., Vinson, S. B., and Olson, J. K. (1995). Food distribution and storage in colonies of Monomorium pharaonis (L.) (Hymenoptera: Formicidae). J. Entomol. Sci. 30, 70–81. doi: 10.18474/0749-8004-30.1.70
Head, G. P., Carroll, M. W., Evans, S. P., Rule, D. M., Willse, A. R., Clark, T. L., et al. (2017). Evaluation of SmartStax and SmartStax PRO maize against western corn rootworm and northern corn rootworm: efficacy and resistance management. Pest Manage. Sci. 73, 1883–1899. doi: 10.1002/ps.4554
Helms, J. A., Godfrey, A. P., Ames, T., and Bridge, E. S. (2016). Predator foraging altitudes reveal the structure of aerial insect communities. Sci. Rep. 6:28670. doi: 10.1038/srep28670
Hölldobler, B., and Wilson, E. O. (1990). The Ants. Cambridge, MA: Harvard University Press. doi: 10.1007/978-3-662-10306-7
Hölldobler, B., and Wilson, E. O. (2009). The Superorganism: The Beauty, Elegance, and Strangeness of Insect Societies. New York, NY: WW Norton & Company.
Lanan, M. C., Rodrigues, P. A. P., Agellon, A., Jansma, P., and Wheeler, D. E. (2016). A bacterial filter protects and structures the gut microbiome of an insect. ISME J. 10, 1866–1876. doi: 10.1038/ismej.2015.264
Lu, H. L., Vinson, S., and Pietrantonio, P. V. (2009). Oocyte membrane localization of vitellogenin receptor coincides with queen flying age, and receptor silencing by RNAi disrupts egg formation in fire ant virgin queens. FEBS J. 276, 3110–3123. doi: 10.1111/j.1742-4658.2009.07029.x
Meng, J., Lei, J., Davitt, A., Holt, J. R., Huang, J., Gold, R., et al. (2020). Suppressing tawny crazy ant (Nylanderia fulva) by RNAi technology. Insect Sci. 27, 113–121. doi: 10.1111/1744-7917.12604
Miyazaki, S., Okada, Y., Miyakawa, H., Tokuda, G., Cornette, R., Koshikawa, S., et al. (2014). Sexually dimorphic body color is regulated by sex-specific expression of yellow gene in ponerine ant, Diacamma sp. PLoS ONE 9:e92875. doi: 10.1371/journal.pone.0092875
Perry, J. C., and Roitberg, B. D. (2006). Trophic egg laying: hypotheses and tests. Oikos 112, 706–714. doi: 10.1111/j.0030-1299.2006.14498.x
Price, D. R., and Gatehouse, J. A. (2008). RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400. doi: 10.1016/j.tibtech.2008.04.004
Pull, C. D., and McMahon, D. P. (2020). Superorganism immunity: a major transition in immune system evolution. Front. Ecol. Evol. 8:186. doi: 10.3389/fevo.2020.00186
Qin, W., Chen, X., Hooper-Bùi, L. M., Cai, J., Wang, L., Sun, Z., et al. (2019). Food-burying behavior in red imported fire ants (Hymenoptera: Formicidae). PeerJ 7:e6349. doi: 10.7717/peerj.6349
Rajakumar, R., Koch, S., Couture, M., Favé, M. J., Lillico-Ouachour, A., Chen, T., et al. (2018). Social regulation of a rudimentary organ generates complex worker-caste systems in ants. Nature 562, 574–577. doi: 10.1038/s41586-018-0613-1
Ratzka, C., Gross, R., and Feldhaar, H. (2013). Systemic gene knockdown in Camponotus floridanus workers by feeding of dsRNA. Insectes Soc. 60, 475–484. doi: 10.1007/s00040-013-0314-6
Rodovalho, C. M., Ferro, M., Fonseca, F. P., Antonio, E. A., Guilherme, I. R., Henrique-Silva, F., et al. (2011). Expressed sequence tags from Atta laevigata and identification of candidate genes for the control of pest leaf-cutting ants. BMC Res. Notes 4:203. doi: 10.1186/1756-0500-4-203
Rydzak, P. M., and Bextine, B. R. (2016). Manipulation of viral titers of Solenopsis invicta Virus-1 by RNA interference in laboratory colonies of red imported fire ant. Southwest. Entomol. 41, 379–388. doi: 10.3958/059.041.0203
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Shukla, J. N., Kalsi, M., Sethi, A., Narva, K. E., Fishilevich, E., Singh, S., et al. (2016). Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 13, 656–669. doi: 10.1080/15476286.2016.1191728
Smith, C. R., Smith, C. D., Robertson, H. M., Helmkampf, M., Zimin, A., Yandell, M., et al. (2011). Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl. Acad. Sci. U.S.A. 108, 5667–5672. doi: 10.1073/pnas.1007901108
Song, H., Fan, Y., Zhang, J., Cooper, A. M., Silver, K., Li, D., et al. (2019). Contributions of dsRNases to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest Manage. Sci. 75, 1707–1717. doi: 10.1002/ps.5291
Taning, C. N., Arpaia, S., Christiaens, O., Dietz-Pfeilstetter, A., Jones, H., Mezzetti, B., et al. (2020). RNA-based biocontrol compounds: current status and perspectives to reach the market. Pest Manage. Sci. 76, 841–845. doi: 10.1002/ps.5686
Tankersley, M. S. (2008). The stinging impact of the imported fire ant. Curr. Opin. Allergy Clin. Immunol, 8, 354–359. doi: 10.1097/ACI.0b013e3283073b48
Terenius, O., Papanicolaou, A., Garbutt, J. S., Eleftherianos, I., Huvenne, H., Kanginakudru, S., et al. (2011). RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 57, 231–245. doi: 10.1016/j.jinsphys.2010.11.006
Tragust, S. (2016). External immune defence in ant societies (Hymenoptera: Formicidae): the role of antimicrobial venom and metapleural gland secretion. Myrmecol. News 23, 119–128. doi: 10.25849/myrmecol.news_023:119
Trible, W., Olivos-Cisneros, L., McKenzie, S. K., Saragosti, J., Chang, N. C., Matthews, B. J., et al. (2017). Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 170, 727–735.e10. doi: 10.1016/j.cell.2017.07.001
Valentini, G., Masuda, N., Shaffer, Z., Hanson, J. R., Sasaki, T., Walker, S. I., et al. (2020). Division of labour promotes the spread of information in colony emigrations by the ant Temnothorax rugatulus. Proc. R. Soc. B 287:20192950. doi: 10.1098/rspb.2019.2950
Valles, S. M., Strong, C. A., Buss, E. A., and Oi, D. H. (2012). Non-enzymatic hydrolysis of RNA in workers of the ant Nylanderia pubens. J. Insect Sci. 12:146. doi: 10.1673/031.012.14601
Vélez, A. M., and Fishilevich, E. (2018). The mysteries of insect RNAi: a focus on dsRNA uptake and transport. Pestic. Biochem. Physiol. 151, 25–31. doi: 10.1016/j.pestbp.2018.08.005
Vinson, S. B. (2013). Impact of the invasion of the imported fire ant. Insect Sci. 20, 439–455. doi: 10.1111/j.1744-7917.2012.01572.x
Vogel, E., Santos, D., Mingels, L., Verdonckt, T. W., and Broeck, J. V. (2018). RNA interference in insects: protecting beneficials and controlling pests. Front. Physiol. 9:1912. doi: 10.3389/fphys.2018.01912
Wang, C., Chen, X., Strecker, R., Henderson, G., Wen, X., and Hooper-Bùi, L. M. (2016). Individual and cooperative food transport of the red imported fire ant (Hymenoptera: Formicidae): laboratory observations. J. Insect Behav. 29, 99–107. doi: 10.1007/s10905-016-9546-4
Wang, J., Gu, L., and Knipple, D. C. (2018). Evaluation of some potential target genes and methods for RNAi-mediated pest control of the corn earworm Helicoverpa zea. Pestic. Biochem. Physiol. 149, 67–72. doi: 10.1016/j.pestbp.2018.05.012
Weeks, R. D., and Drees, B. M. (2002). Barrier treatments for red imported fire ants Solenpopsis invicta (Hymenoptera, Formicidae) in commercial bee colonies. Southwestern Entomologist, 27, 185–189.
Winston, W. M., Molodowitch, C., and Hunter, C. P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459. doi: 10.1126/science.1068836
Winston, W. M., Sutherlin, M., Wright, A. J., Feinberg, E. H., and Hunter, C. P. (2007). Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. U.S.A. 104, 10565–10570. doi: 10.1073/pnas.0611282104
Xi, G., Ying, Q., and Xie, H. (2019). The functional relevance of oestrogen-related receptor gene to ultraspiracle gene in regulating the development of Polyrhachis vicina (Hymenoptera: Formicidae). Orient. Insects 53, 362–378. doi: 10.1080/00305316.2018.1508520
Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., et al. (2017). An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170, 736–747.e9. doi: 10.1016/j.cell.2017.06.051
Zanetti, R., Zanuncio, J. C., Vilela, E. F., Leite, H. G., Jaffé, K., and Oliveira, A. C. (2003). Level of economic damage for leaf-cutting ants (Hymenoptera: Formicidae) in eucalyptus plantations in Brazil. Sociobiology 42, 433–442.
Zhang, J. J., and Xi, G. S. (2018). Morphological changes in different caste adult ant specificities of Polyrhachis vicina Roger (Hymenoptera, Formicidae) caused in estrogen-related receptor. Gen. Comp. Endocrinol. 266, 29–37. doi: 10.1016/j.ygcen.2018.04.002
Keywords: Formicidae, invasive species, core enzymes, dsRNA -degrading enzymes, social immunity, trophallaxis
Citation: Allen ML (2021) Prospects for Using RNAi as Control for Ants. Front. Agron. 3:591539. doi: 10.3389/fagro.2021.591539
Received: 04 August 2020; Accepted: 30 March 2021;
Published: 07 May 2021.
Edited by:
Lynne K. Rieske-Kinney, University of Kentucky, United StatesReviewed by:
Rob Morrison, Center for Grain and Animal Health, Agricultural Research Service, United States Department of Agriculture, United StatesDomenico Bosco, University of Turin, Italy
Olivier Christiaens, Ghent University, Belgium
Copyright © 2021 Allen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margaret L. Allen, bWVnLmFsbGVuQHVzZGEuZ292
 Margaret L. Allen
Margaret L. Allen