- 1Soil Microbiology Division, CSIR-Soil Research Institute, Kumasi, Ghana
- 2Department of Crop and Soil Sciences, Faculty of Agriculture, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
- 3Department of Theoretical and Applied Biology, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
- 4Soil Microbiology Section, International Institute of Tropical Agriculture, Ibadan, Nigeria
- 5Embrapa Agrobiologia, Rio de Janeiro, Brazil
- 6Soil Microbiology Section, CSIR-Savannah Agricultural Research Institute, Tamale, Ghana
The existence of large population of ineffective native rhizobia and inconsistent performance of exotic strains in Ghanaian soils necessitate the need to identify effective and locally adapted elite strains for enhanced legume-rhizobium symbiosis. This study was designed to test the suitability of two previously selected potential elite Bradyrhizobium strains for use as inoculants on cowpea (Vigna unguiculata L. Walp) and groundnut (Arachis hypogaea L.) in multilocation experiments. Field experiments were set up at 26 locations (12 planted with cowpea and 14 planted with groundnut) in the Northern region of Ghana. Four treatments were applied at each location: inoculation with Bradyrhizobium strains KNUST 1002 and KNUST 1006, a positive nitrogen (+N) control and a negative control (without nitrogen or inoculation) arranged in randomized complete blocks with four replications. The results showed that inoculation with strains KNUST 1002 and KNUST 1006 promoted significant increases in grain yields of both cowpea and groundnut. On average, inoculating cowpea with strains KNUST 1002 and KNUST 1006 resulted in 63 and 52% increases in grain yield when compared to the negative control. Pod yields of groundnut, on the other hand, were significantly (p < 0.05) increased at 57% of the study locations with one or both test strains. Responses to inoculation were, however, highly variable across the different study locations (i.e., significant Treatment × Location Interaction, TLI). A detailed analysis of this significant TLI based on the genotype main effect (G) plus genotype-by-environment (GE) interaction (GGE) biplot analysis revealed that location contributed 71 and 88% of the variation observed in cowpea and groundnut, respectively, and grouped the locations into mega-environments for cowpea. These results demonstrate that native elite Bradyrhizobium strains KNUST 1002 and KNUST 1006 have potential for use as inoculants to increase cowpea and groundnut production in Northern Ghana.
Introduction
Cowpea and groundnut form a vital component of the traditional cropping system in most countries in Africa including Ghana due to the high protein contents of these grains (Ajeigbe et al., 2012). Other advantages of these crops are their contribution to soil fertility improvement and assistance in reducing the incidence of pests and diseases (Graham and Vance, 2000). Until recently, legumes were cultivated as secondary crops to cereals by most smallholder farmers and, as such, received little or no fertilizer (Naab et al., 2009). Farmers nowadays cultivate these legumes on a large scale as cash crops but still without the application of adequate quantities of fertilizer. It is also evident that yields obtained are just a fraction of their maximum potential. For instance, average groundnut yields obtained on farmers' fields in Ghana are estimated at 900 kg ha−1 while potential yields of the improved varieties have been tagged between 1,800 and 2,800 kg ha−1 (Adu-Dapaah et al., 2004). Additionally, a yield gap of about 1,560 kg ha−1 has been reported for cowpea (Karikari et al., 2015). Although legumes are characterized by the ability to partially or fully meet their nitrogen (N) needs through biological nitrogen fixation (BNF) (Hungria and Kaschuk, 2014), the process is often limited by nutrient deficiencies in soils, particularly phosphorus (P). Such nutrient deficiencies in soils are predominant in the legume growing areas of the savanna zones of Ghana (Adjei-Nsiah et al., 2018).
For the past decade, several approaches have been recommended as means to improve BNF and subsequently yields of legumes. Among these approaches are the use of high yielding varieties (Okogun et al., 2005; Buruchara et al., 2011), P-based fertilizers (Kamara et al., 2007; Kamanga et al., 2010; Kolawole, 2012), rhizobium inoculants (Sanginga et al., 2000; Osunde et al., 2003; Thuita et al., 2012), or a combination of these approaches (Ndakidemi et al., 2006; Ronner et al., 2016). The International Institute of Tropical Agriculture (IITA) bred promiscuous legumes, particularly soybeans, with the aim of improving BNF (Vanlauwe and Giller, 2006). Consequently, smallholder farmers in Zimbabwe and South Africa, for instance, preferred these promiscuous varieties to the high yielding ones due to limited access to inoculants (Mpepereki et al., 2000; Musiyiwa et al., 2005). However, the performance of promiscuous soybeans in terms of nodulation and BNF has been erratic due to the diverse nature of native rhizobia present in soils (Mpepereki et al., 2000; Sanginga, 2003). In effect, it was concluded to be impossible to obtain a promiscuous soybean variety that nodulates in all environments. Hence, the need for rhizobium inoculation remains paramount in the subregion.
Recent studies that employed the use of rhizobium inoculants in combination with phosphorus fertilizers revealed clearly that this approach results in increased yields of legumes (Ronner et al., 2016; Ulzen et al., 2016; Kyei-Boahen et al., 2017). However, the widespread use of rhizobium inoculants by smallholder farmers in Ghana is historically rare owing to limited access. In addition, the variable quality of some imported inoculants has resulted in inconsistent responses (Brockwell and Bottomley, 1995; Singleton et al., 1997), creating doubt among farmers. Until recently, there were no facilities in the region to produce and distribute inoculants, and as such, all the inoculants available on the market were imported. However, since 2017, the first inoculant production facility established in the Council for Scientific and Industrial Research (CSIR)-Savanna Agricultural Research Institute in Tamale, Ghana, has been producing rhizobium inoculants for the local market using exotic strains. Promiscuous native rhizobium, on the other hand, also competes with introduced exotic strains precluding inoculation responses in some cases (McInnes and Haq, 2007).
As many strains used in inoculant formulations originated from indigenous pools of their country of origin, isolation and characterization to identify local elite strains present a strategy to improve BNF. Studies that focused on the characterization of indigenous rhizobium have revealed them as important sources of inoculant strains that are able to improve nodulation and BNF in grain legumes (Fening and Danso, 2002; Bogino et al., 2006; Ampomah et al., 2008; Grönemeyer et al., 2014). Furthermore, effective native strains, when identified, will serve as an important source of local inoculants to enhance BNF and subsequently yields of target legumes due to inherent adaptability to local conditions.
In this context, the present study was performed to assess the symbiotic potential of two previously selected native Bradyrhizobium strains to improve grain yields of cowpea and groundnut in multiple environments in Northern Ghana.
Materials and Methods
General Description of Study Locations
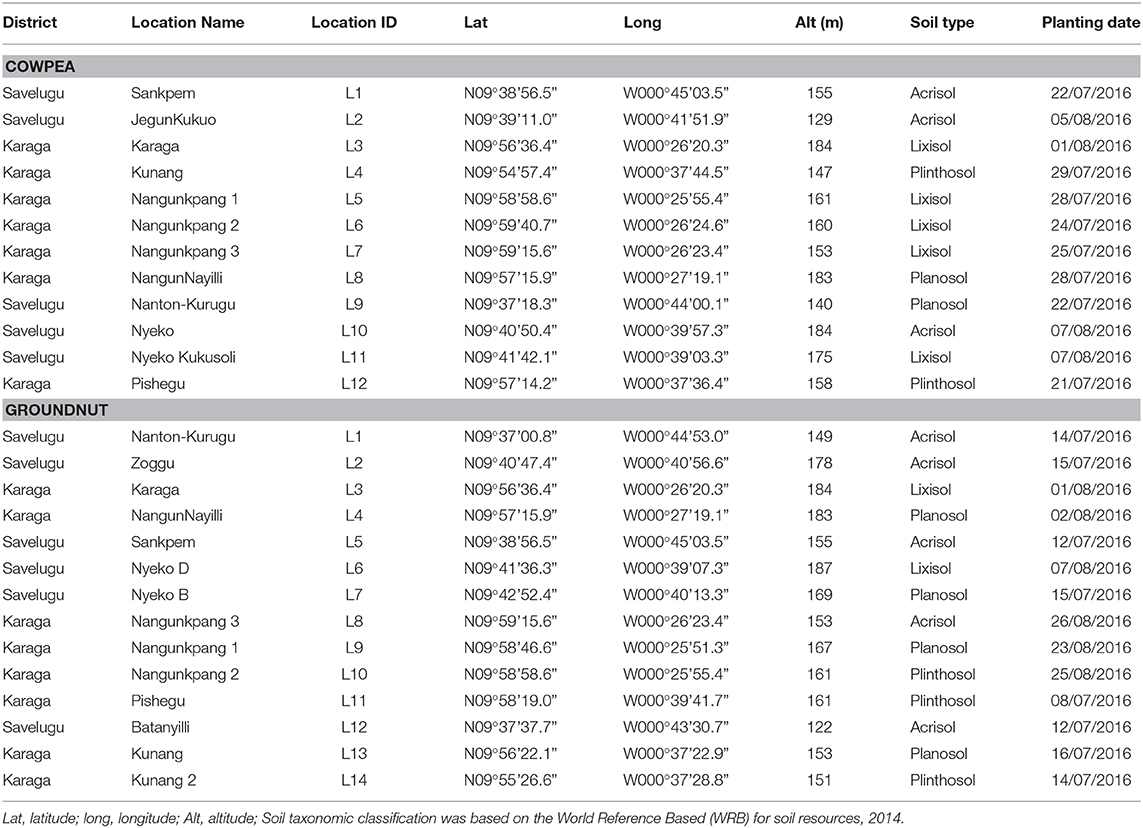
The experiments were conducted during the 2016 cropping season in two districts in the Northern region of Ghana. The region falls within the Guinea savannah agroecological zone with a unimodal rainfall pattern and an annual rainfall distribution of 1,000–1,200 mm. Mean temperature for the area ranges from 26 to 30°C with slight variations within a year (MOFA, 2016). Twenty-six locations with no known history of recent legume cultivation were selected for the field experiments. The geographical positions and other information about the study locations are presented in Table 1.
Physicochemical Analyses of Soils From Study Locations
Prior to the establishment of the experiments, soils were sampled at a depth of 0–20 cm from each of the study sites. At each site, eight soil core samples were taken, thoroughly mixed, and composite samples taken. A portion of the composite sample for each field was air-dried, sieved through a 2 mm wire mesh, and kept for physicochemical analysis. The hydrometer method based on the principle of dispersion and sedimentation was used to determine soil texture (Bouyoucos, 1962). Soil pH was measured by inserting the electrode of a pH meter (Jenway 3510, England) into partly settled 1:2.5 (w/v) soil:water suspension. For organic carbon determination, the modified Walkley and Black method described by Nelson and Sommers (1982), which involves a wet combustion of organic matter using a mixture of potassium dichromate (K2Cr2O7) solution and sulfuric acid, was used. Total N was determined by the Kjeldahl method with samples initially digested to convert organic N to ammonium–N, followed by the determination of ammonia in the digested samples (Bremner and Mulvaney, 1982). Available phosphorus was extracted using the Bray's No 1 solution (Bray and Kurtz, 1945), after which phosphorus in the extracts was measured with the spectrophotometer using blue ammonium molybdate with ascorbic acid as the reducing agent. Exchangeable cations were extracted using the procedure by Black (1965) with 1.0 M ammonium acetate extract.
Estimation of Indigenous Rhizobia in Soils
The rhizobium population in each composite soil sample was estimated using the most probable number (MPN) method described by Yates et al. (2016) with cowpea (variety, Asontem) as the trap host. Pregerminated cowpea seeds of equal radicle length were aseptically transferred into growth pouches (Mega International, USA). A week after transplanting, growth pouches with healthy seedlings were selected and inoculated with serial dilutions of the soil samples. The soil dilutions were prepared using the six steps, 5-fold serial dilution approach. One milliliter of each dilution was used to inoculate the seedlings with four replicates. The setup was monitored for 28 days, after which the MPN of rhizobium in each soil was assessed using the MPNES software (Woomer et al., 1990).
Preparation of Planting Materials
For the field experiments, groundnut (Arachis hypogaea L.) variety Shitaochi commonly known as “Chinese” was used, while the cowpea (Vigna unguiculata L. Walp) variety “Songotra” was used for the cowpea experiments. These varieties were selected based on farmers' preference. The two elite Bradyrhizobium strains (KNUST 1002 and KNUST 1006), selected from preliminary controlled experiments based on their symbiotic effectiveness (Osei et al., 2018), were processed into inoculant formulations under aseptic conditions. Each strain was cultured in 50 ml yeast mannitol broth (YMB) until the late logarithm phase, and this broth was then mixed with 50 g of prepackaged and gamma-radiated (sterile) peat. The inoculated peat was gently massaged for even distribution of inoculum and incubated at 28°C for 2 weeks to cure and reach a cell concentration of at least 108 colony forming units (CFU) g peat−1 (Somasegaran and Hoben, 2012).
Field Preparation and Imposition of Treatment
Experimental fields were plowed, harrowed, and laid out in randomized complete blocks with four replications. The treatments were: (i) Bradyrhizobium strain KNUST 1002, (ii) Bradyrhizobium strain KNUST 1006, (iii) positive nitrogen control (i.e., uninoculated and fertilized with 100 kg N ha−1 in the form of urea) and iv) negative control (i.e., uninoculated and unfertilized). Each plot measured 5 m × 5 m spaced by 1 m alleys between blocks and treatments. The inoculants were applied to the seeds and planted at a spacing of 50 cm × 20 cm for both cowpea and groundnut. Prior to planting, the seeds were coated with 3% w/v gum arabic solution, followed by inoculation with each of the inoculant strains at a rate of 5 g kg seed−1. The seeds were mixed gently and air-dried for 30 min before planting. Each treatment received a basal application of 30 kg P ha−1 in the form of triple super phosphate. One hundred kg N ha−1 in the form of urea was applied as the +N treatment and was split during the application at the rate of 20 kg N ha−1 at 2 weeks after planting and the remaining 80 kg N ha−1 at 50 % flowering (R2 growth state).
Harvesting and Data Collection
Plants were harvested manually from the inner rows excluding the border rows at full maturity (R8 stage) (Fehr et al., 1971) and pods of both groundnut and cowpea separated. Cowpea pods were dried to constant weight, threshed, and winnowed for grain yield determination. Yield of groundnut, on the other hand, was determined unshelled. Grains and pods were weighed using a standard electronic balance and yields estimated on per hectare basis.
Statistical Analyses
Since two different legume hosts (groundnut and cowpea) were used, separate analysis was done for each crop. The data from all the locations were pooled together and checked for normal distribution and homogeneity of residuals variance before subjecting to analysis of variance (ANOVA) using R (R Core Team, 2017) and SISVAR (Ferreira, 2008), respectively. The ANOVA model included treatment, location, and their interaction. Significant mean differences were separated using Scott Knott method at 5% probability. Treatment × Location Interaction (TLI) was further evaluated using the Genotype (G) mean effect plus Genotype-by-Environment (GE) interaction (GGE) biplot function in GEA-R (Genotype x Environment Analysis with R for windows) software version 4.0 (Pacheco et al., 2015). GGE biplot analysis helps to address important concepts such as crossover Treatment × Location responses and specific adaptation of treatment in an easy to understand form (i.e., graphically) and is also very efficient especially when a few number of treatments are considered.
For genotype evaluation (herein referred to as treatment evaluation), treatment-focused singular value partitioning (SVP = 1) was used based on the Mean vs. Stability option in the software. The “which-won-where” option was used to identify the treatment with the best performance at specific locations and to delineate mega-environments. Yan and Tinker (2006) have explained in detail the statistical theory of the GGE methodology.
Results
Chemical and Biological Properties of Soil and Rainfall Distribution of the Study Sites
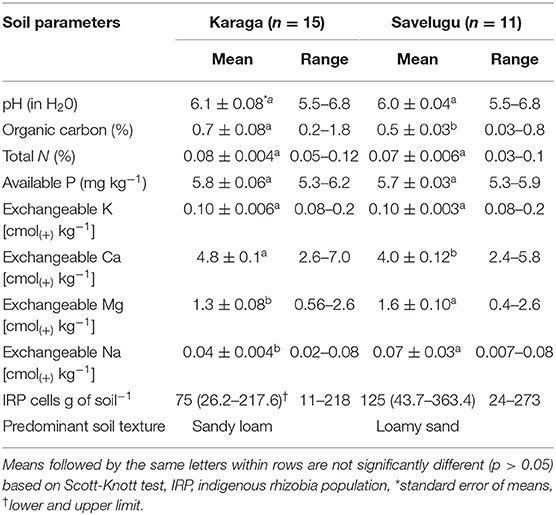
The soils from all the study locations ranged from moderately to slightly acidic (pH 5.5 to 6.8) (Table 2). The organic carbon and total N contents of soils ranged from 0.03 to 1.8% and 0.03 to 0.1%, respectively, and were considered as very low. The available P-values averaged 5.8 mg kg−1 (5.3–6.2 mg kg−1) and 5.7 mg kg−1 (5.3–5.9 mg kg−1) for locations under the Karaga and Savelugu districts, respectively, and were considered low. Estimates of native rhizobium population of soils at the study locations averaged 75 and 125 rhizobium cells g−1 of soil for Karaga and Savelugu districts, respectively. The textural class analysis revealed that soils of the sites in the Karaga district were predominately sandy loam while those in the Savelugu district were mostly loamy sand in nature. In general, the fertility status of the soils considered in this study were low (Table 2) based on the description of Landon (2014). The amount of rainfall within 3 days before or after planting varied per study location for both cowpea and groundnut. For the cowpea study locations, the rainfall amount within the period of planting was 9 mm or more for only 4 out of the 12 sites, while that of groundnut fields recorded was 11 mm or more for 4 out of the 14 sites.
Response of Target Legumes to Treatment
Cowpea Yield Response
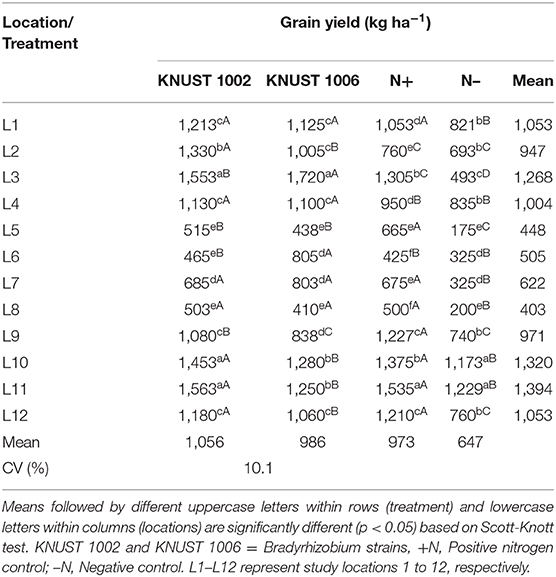
The grain yield response of cowpea to rhizobium inoculation was significantly positive compared to the negative control treatment for both strains at 8 of the 12 sites (Table 3). At all the sites except L6, KNUST 1002 promoted significant increase in grain yield compared to the negative control plots while with the exception of sites L9, L10, and L11 (see sites description in Table 1), inoculation with strain KNUST 1006 promoted significant increase in grain yield at all the other sites. The application of 100 kg N ha−1 at most sites promoted an increase in grain yield compared to the negative control. Only at two sites (L5 and L9) did the positive control treatment outyield both inoculated treatments with KNUST 1002 and KNUST 1006. There were large and significant differences in grain yield for the negative control plots ranging from 175 to 1,229 kg ha−1. At the sites with lowest yields in control plots, the increase in the grain yield due to inoculation was often very high. For example, strain KNUST 1002 resulted in increases of 194 and 111% at sites L5 and L7, respectively, over the negative control. For the negative control treatment, grain yield only exceeded 1,000 kg ha−1 at two sites (Table 3). The average yield increases of cowpea following inoculation with the rhizobium strains were 409 kg ha−1 and 339 kg ha−1 for KNUST 1002 and KNUST 1006, representing percentage increases of 63 and 52, respectively, over the negative control.
Groundnut (Unshelled) Yield Response
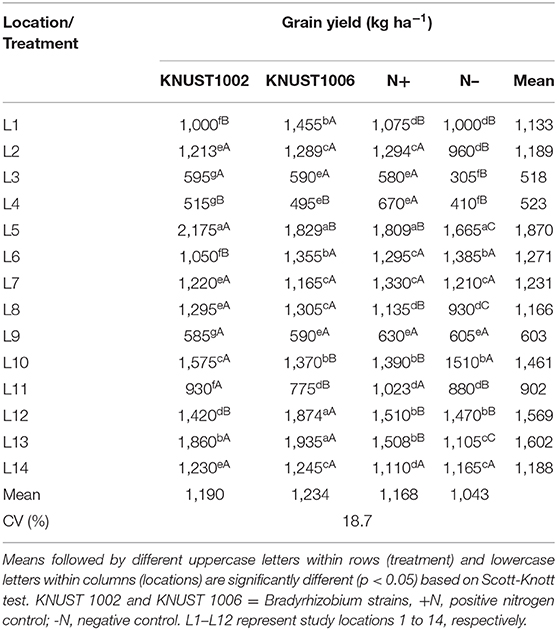
The yield response of groundnut to rhizobium inoculation was significantly positive compared to the negative control treatment, for both strains at only five sites (Table 4). At two sites (L1 and L12), only KNUST 1006 promoted a significant yield increase and at sites L5 and L11, only KNUST 1002 significantly increased yield over the control. Thus, there were statistically significant (p < 0.05) increases in yield responses to one or both rhizobium strains at 8 (L1, L2, L3, L5, L8, L11, L12, and L13) of the 14 sites compared to the negative control. The application of 100 kg N ha−1 promoted an increase in yield compared to the negative control treatment at seven sites. However, at two sites (L5 and L10), the yield of the plants inoculated with KNUST 1002 was significantly higher (p < 0.05) than those of plants receiving 100 kg N ha−1. At sites L8 and L13, both rhizobium strains promoted significantly higher yields than the positive control treatment. The positive N treatment promoted yield increases significantly at par with one or both strains at all the locations except L4. The mean yield increases of groundnut in relation to the negative control at all sites for KNUST 1002 and 1006 were 147 and 191 kg ha−1, respectively, but at some sites, yield increases were highly significant. For example, yield increased from 1,105 kg ha−1 in the negative control treatment to 1,860 kg ha−1 with strain KNUST 1002 and 1,935 kg ha−1 when groundnut was inoculated with KNUST 1006 at site L13. At the sites L1 and L13, inoculation promoted yield increases of over 400 kg ha−1.
Exploring the Interaction Between Treatment and Location
Which-Won-Where Pattern
The best treatment(s) for each location was identified based on the which-won-where function in the GGE biplot analysis. The analysis grouped the study locations into three mega-environments with KNUST 1002, KNUST 1006, and positive control treatment, producing the best yields in each of the mega-environments for cowpea. Thus, three locations (L5, L9, and L11) fell within mega-environment 1 delineated by the red lines in the biplot with the +N treatment at the polygon vertex. This indicates that cowpea yields were increased for plots that received N at these three locations when compared with the other treatments. Similarly, four locations, L1, L8, L10, and L12, formed the second mega-environment with KNUST 1002 producing comparatively higher yields at these locations than the other treatments. The remaining locations fell under mega-environment 3 with KNUST 1006, resulting in increased cowpea yields at these locations (Figure 1).
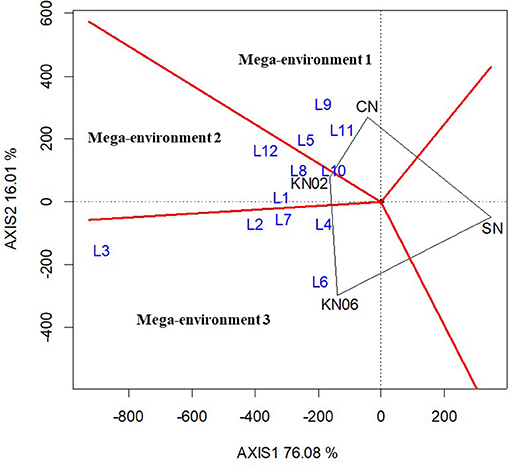
Figure 1. Polygon view of the Genotype main effect plus Genotype-by-Environment (GGE) biplot for treatment performance at different locations for cowpea based on the which-won-where pattern. KN02, KN06, CN, and SN represent Bradyrhizobium strains KNUST 1002, KNUST 1006, positive nitrogen, and negative controls, respectively and L1–L12 represent study locations 1 to 12. Mega-environment = group of locations sharing the same best set of treatment(s).
The biplot analysis of groundnut data resulted in the delineation of three mega-environments (Figure 2). The first mega-environment comprised six locations (L1, L2, L8, L12, L13, and L14) with KNUST 1006 promoting yield increases in these locations. The second mega-environment comprised only location L6 with the negative control treatment recording greater yield than the inoculated and positive control treatments. Seven locations (L3, L4, L5, L7, L9, L10, and L11) fell within the third mega-environment with KNUST 1002 producing high yields, which were comparable to +N treatment (Figure 2).
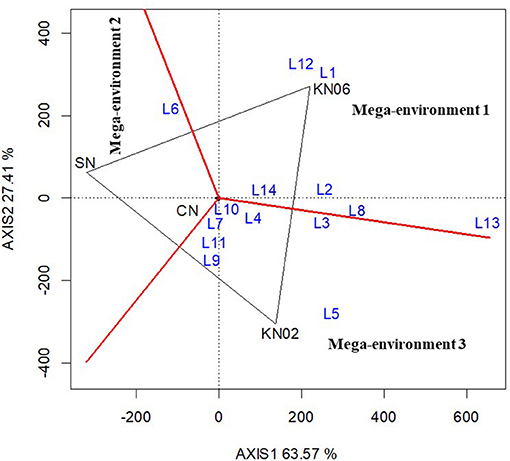
Figure 2. Polygon view of the GGE biplot for treatment performance at different locations for groundnut based on the which-won-where pattern. KN02 KN02, KN06, CN, and SN represent Bradyrhizobium strains KNUST 1002, KNUST 1006, positive nitrogen, and negative controls, respectively and L1–L14 represent study locations 1 to 14. Mega-environment = group of locations sharing the same best set of treatment(s).
Overall, the soil fertility status of the study locations was low. The native rhizobium population, however, varied for the delineated mega-environments. For instance, the cowpea study locations within mega-environment 3, where KNUST 1006 caused yield increases, all had rhizobium populations below 100 cells g soil−1. These populations were relatively lower than the populations at the locations within mega-environment 2 where KNUST 1002 produced greater yields. A similar but opposite trend was observed for the mega-environment delineation of groundnut locations. The locations in mega-environment 3, where KNUST 1002 was the best treatment, had relatively lower populations of native rhizobium than the locations in mega-environment 1, where KNUST 1006 produced greater yields (Table 5).
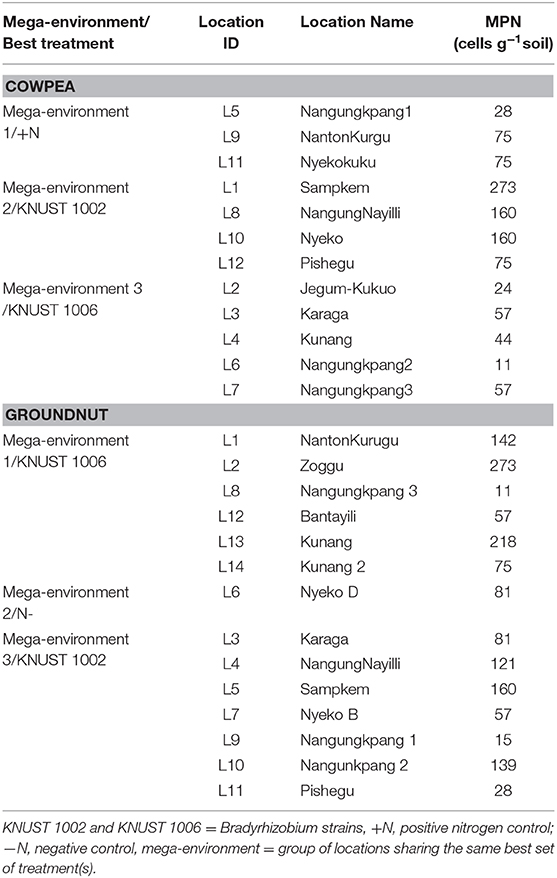
Table 5. Native rhizobia population for locations within each mega-environment for cowpea and groundnut.
Treatment-Focused Evaluation
The first two principal components (axis 1 and axis 2) explained 92.1% of the variation in grain yield for cowpea (Figure 3A). The consistency in performance of treatment across the cowpea study locations was in the order KNUST 1002 > +N > KNUST 1006. This is indicated by the shortness of the line projecting from the abscissa of the biplot for each treatment. KNUST 1002 thus consistently promoted significant yield increases across the cowpea study locations. The negative control treatment also had a very short projection from the abscissa of the biplot, which means that lower yields were recorded for this treatment across all the study locations. Evaluation of the ability of a treatment to perform similar to or better than an ideal treatment (i.e., a standard strain) showed that KNUST 1002 was an ideal strain for inoculating cowpea as it produced greater yields at almost all the study locations. This was depicted by its position in the concentric circles drawn to visualize the distance between each treatment and the hypothetical ideal treatment (Figure 4A).
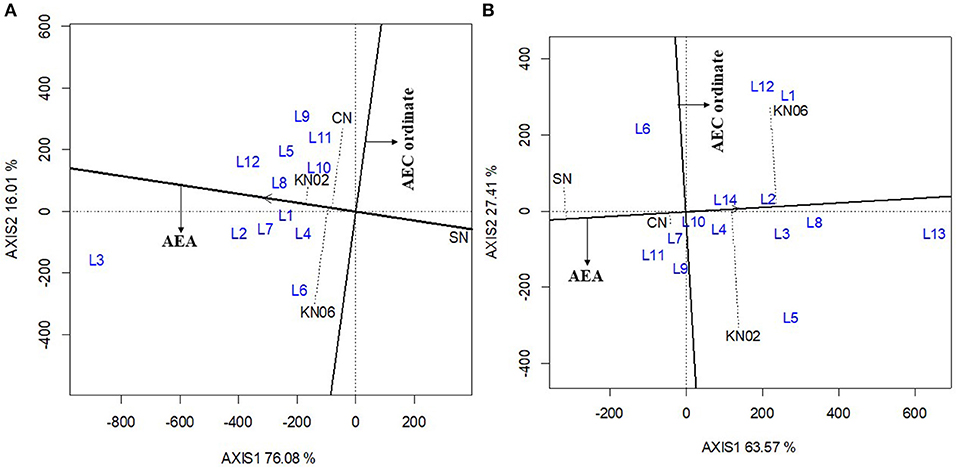
Figure 3. GGE biplot based on the genotype-focused view for evaluation of treatment performance on cowpea (A) and groundnut (B). KN02, KN06, CN, and SN represent Bradyrhizobium strains KNUST 1002, KNUST 1006, positive nitrogen, and negative controls, respectively. L1-L14 represent study locations. AEA, Average Environment Axis; AEC, Average Environment Coordination.
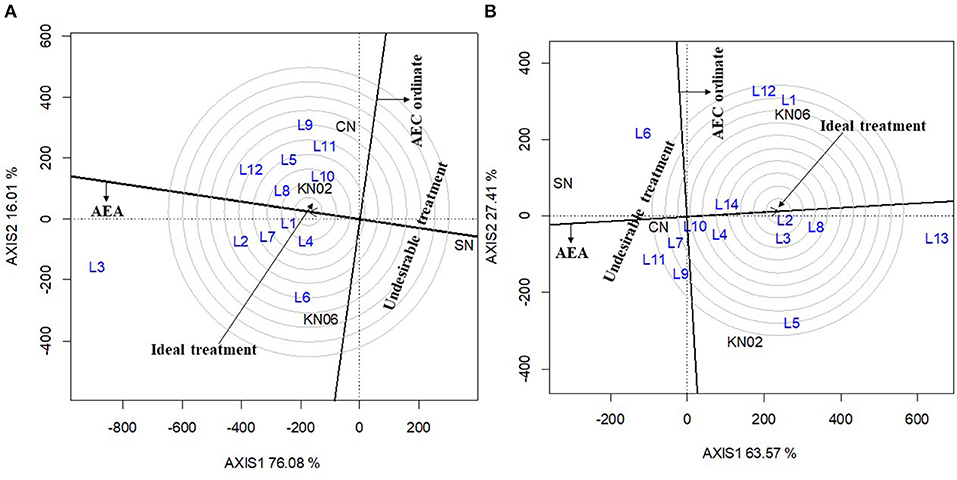
Figure 4. GGE biplot based on genotype-focused view for comparing treatment in this study with an ideal treatment cowpea (A) and groundnut (B). KN02, KN06, CN, and SN represent Bradyrhizobium strains KNUST 1002, KNUST 1006, positive nitrogen, and absolute controls, respectively. L1–L14 represent study locations. Ideal treatment = treatment with high mean performance and high stability, Undesirable treatment = treatment that is less stable and has below average yields.
The biplot for treatment-focused view for groundnut data explained 90.1% of the variability in performance observed with the +N treatment, having a short projection from the abscissa, followed by KNUST 1006 and KNUST 1002. The consistency in performance of treatment was relatively low on groundnut compared with cowpea. The negative control treatment consistently recorded the lowest yields across the groundnut study locations (Figure 3B). These results were confirmed by the output of the ideal treatment evaluation (Figure 4B).
Discussion
Although efficient exploration of BNF for enhanced grain yields, particularly in legumes, can be achieved via the use of effective native rhizobia, there are several factors that influence inoculation responses. Chiefly among these factors are the population of native rhizobia in soils and the soil N concentration. In this study, the general soil fertility and particularly N content of soils at the experimental sites were very low (Landon, 2014). This poor fertility status might have influenced the low average population estimates (i.e., between 75 and 125 cells g−1 soil) of rhizobia in these soils. This population of rhizobia is within the 300 cells g−1 soil threshold below which inoculation is likely to give beneficial results to farmers (Herridge, 2008). Furthermore, Thies et al. (1991) indicated that there must be a demand for fixed N not being met by soil N in order to achieve desired responses to inoculation. This was generally the case for soils in our study locations. Inoculation with native strains in this study led to increases in grain yields of cowpea and groundnut, which confirms the effectiveness of the strains in BNF under controlled conditions previously reported by Osei et al. (2018). Furthermore, strains isolated from a particular region have been reported to be the most effective on a given crop in the same region (Sattar et al., 1995).
Increases in grain yields following inoculation of legumes with native rhizobia in this study are remarkable and disprove earlier reports by some authors that these crops do not respond to inoculation with rhizobia (Awonaike et al., 1990; de Freitas et al., 2012; Mweetwa et al., 2014). The results, however, agree with the findings of several authors including Bogino et al. (2006), Boddey et al. (2016), Ulzen et al. (2016), and Martins et al. (2003) that inoculating cowpea and groundnut leads to yield improvements. Variations in yield response to inoculation of cowpea and groundnut at the different locations can be attributed to the differences in compatibility between the host plant genotype and specific rhizobium strains under the influence of environmental factors. Several authors have reported similar observations of variability in rhizobium strain performance on different plant genotypes (Tirichine et al., 2000; Simsek et al., 2007; Schumpp and Deakin, 2010). Such variability in performance of rhizobia on different legume hosts emphasizes the significance of evaluating strains on different legume hosts under varying edapho-climatic conditions (Zaman-Allah et al., 2007).
The GGE biplot analysis permitted a detailed interpretation of the observed significant interaction between treatment and the various locations considered in this study. Considering the contribution of each factor (i.e., treatment, location, and their interaction) to the variation observed, the results showed that location contributed most (71–88%) to the variation, which means that the treatment performance was highly influenced by the different environments. This observation could have resulted from differences in rhizobium population, weather conditions (mainly rainfall), and management practices of farmers at the different locations. For instance, the variable and insufficient amount of rainfall before and/or at planting and the planting and weeding dates could have affected the performance of the inoculated strains and the efficient use of N in the case of the +N treated plots (Tittonell et al., 2008). Since factors that affect the host also affect the microsymbiont (Thies et al., 1995), insufficient amounts of rainfall (below 15 mm) received at some locations prior to, or at planting, may have affected the survival of inoculated strains, resulting in the differences in yield responses observed (Subedi and Ma, 2009). The differential ranking of best treatment across study locations indicates existence of crossover TLI. This means that, although inoculation with either of the strains induced significant yield increases over the control, their adaptation to environmental conditions in the various location possibly differed, which influenced their performance and hence the delineation of mega-environments. In the case of cowpea, the positive yield responses induced by Bradyrhizobium strain KNUST 1002 at the locations within mega-environment 2, even in the presence of relatively large numbers of ineffective native rhizobia populations, signify that this strain was possibly more compatible with the legume host and possessed an inherent ability to outcompete native rhizobia during nodulation. Bradyrhizobium strain KNUST 1006, on the other hand, demonstrated high compatibility with groundnut and competitiveness against the native rhizobium populations in mega-environment 1. It can be deduced from the results that strain-specific legume rhizobia symbiosis may have occurred in the various locations across the mega-environments. According to Woomer et al. (2014), positive inoculation response is primarily influenced by the rhizobium/legume genotype, environmental conditions, and crop management. In addition, Berrada et al. (2012) observed that strains that exhibit a wide adaptation to their environment are able to elude limiting factors and maintain a higher capacity for BNF. These findings agree with the results of a previous study, which showed that inoculation responses could be highly variable and site specific (Singleton et al., 1992).
Another important trait that has to be considered when selecting native rhizobium for use as inoculant is the ability to constantly produce desired responses in a measured parameter. A treatment (rhizobium strain) that possesses such a trait is referred to as stable (Yan and Tinker, 2006). In this study, Bradyrhizobium strain KNUST 1002 was considered stable on cowpea, producing consistently higher yields than the other treatments, while Bradyrhizobium strain KNUST 1006 was stable on groundnut. Such stability in performance of test strains in this study makes it easy and convenient to recommend them for inoculant production to increase cowpea and groundnut yields, respectively, with little financial risks for farmers, which will subsequently boost their confidence in the use of the inoculant technology. Several studies have reported on positive yield responses following inoculation of cowpea and groundnut (Martins et al., 2003; Bogino et al., 2006; Boddey et al., 2016; Ulzen et al., 2018), but none of these have considered the stability in performance of the strains. The GGE biplot analyses have been widely employed in the field of plant breeding to identify superior genotypes and for delineation of mega-environments (Yan et al., 2000; Kaya et al., 2006). It is interesting to note that this is the first time the outcome of multilocational inoculation experiments is being reported based on GGE biplot analysis. This GGE biplot analysis has revealed the differences in relative performance of treatments within locations and the stability in performance of native elite strains across locations, which would not have been possible by just considering mean yields based on simple ANOVA alone (Samonte et al., 2005).
Conclusion
This study has shown that inoculation with native Bradyrhizobium strains KNUST 1002 and KNUST 1006 increases grain yields of field-grown cowpea and groundnut, respectively, in multiple locations within the Sudan savanna agroecological zone. Therefore, these strains have potential for use as inoculants to increase legume production of the study locations and environments of similar soil fertility and climatic conditions.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
OO, RA, and LR conceived the research. OO, RA, and AO planned the work and set up field experiments. OO and JR collected and analyzed the data. OO drafted the manuscript. RA, RB, LR, JR, BA, and AO edited, revised, and made significant contributions to the write-up.
Funding
This research activities were sponsored by the Bill & Melinda Gates Foundation through the Soil Health Project (Grant Number 2013 SHP025).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors express their gratitude to the rhizobiology team of KNUST for their assistance during the field studies. We also appreciate the technical support of Mr. Ayamah Azumah and the team of Agricultural Extension Agents (AEAs) headed by Mr. Jibreel Mohammed Basit during the laboratory work and field monitoring and Professor Charles Quansah of the Department of Crop and Soil Sciences, Faculty of Agriculture, KNUST for proofreading this document. We are grateful for the financial support from the N2 Africa Phase II project. LR and RB acknowledge the research fellowship from the Brazilian National Research Council (CNPq, process nos. 308898/2017-6 and 305047/2016-7). RB also acknowledges the invaluable support from the Rio de Janeiro State Research Council (FAPERJ – Process No. E-26/203.031/2017).
References
Adjei-Nsiah, S., Alabi, B., Ahiakpa, J., and Kanampiu, F. (2018). Response of grain legumes to phosphorus application in the guinea savanna agro-ecological zones of Ghana. Agron. J. 110, 1089–1096. doi: 10.2134/agronj2017.11.0667
Adu-Dapaah, H. K., Asibuo, J., Danquah, O., Asumadu, H., Haleegoah, J., and Agyei, B. (2004). “Farmer participation in groundnut rosette resistant varietal selection in Ghana”, in New Direction for a Diverse Planet: Proceedings for the 4th International Crop Science Congress (Brisbane, QLD).
Ajeigbe, H. A., Adamu, R. S., and Singh, B. B. (2012). Yield performance of cowpea as influenced by insecticide types and their combinations in the dry savannas of Nigeria. Afr. J. Agr. Res. 7, 5930–5938. doi: 10.5897/AJAR12.431
Ampomah, O. Y., Ofori-Ayeh, E., Solheim, B., and Svenning, M. M. (2008). Host range, symbiotic effectiveness and nodulation competitiveness of some indigenous cowpea bradyrhizobia isolates from the transitional savanna zone of Ghana. Afr. J. Biotechnol. 7, 988–996.
Awonaike, K., Kumarasinghe, K., and Danso, S. (1990). Nitrogen fixation and yield of cowpea (Vigna Unguiculata) as influenced by cultivar and Bradyrhizobium strain. Field Crops Res. 24, 163–171. doi: 10.1016/0378-4290(90)90035-A
Berrada, H., Nouioui, I., Houssaini, M. I., Ghachtouli, N., Gtari, M., and Benbrahim, K. (2012). Phenotypic and genotypic characterizations of rhizobia isolated from root nodules of multiple legume species native of fez, Morocco. Afr. J. Microbiol. Res. 6, 5314–5324. doi: 10.5897/AJMR11.1505
Black, C. A. (1965). Methods of Soil Analysis Part 2. Chemical and Microbiological Properties. 1st Edn. Madison, WI: Soil Science Society of America.
Boddey, R. M., Fosu, M., Atakora, W. K., Miranda, C. H. B., Boddey, L. H., Guimaraes, A. P., et al. (2016). Cowpea (Vigna Unguiculata) crops in Africa can respond to inoculation with rhizobium. Exp. Agric. 53, 578–587. doi: 10.1017/s0014479716000594
Bogino, P., Banchio, E., Rinaudi, L., Cerioni, G., Bonfiglio, C., and Giordano, W. (2006). Peanut (Arachis Hypogaea) response to inoculation with Bradyrhizobium sp. in soils of Argentina. Ann. Appl. Biol. 148, 207–212. doi: 10.1111/j.1744-7348.2006.00055.x
Bouyoucos, G. J. (1962). Hydrometer method improved for making particle size analyses of soils 1. Agron. J. 54, 464–465. doi: 10.2134/agronj1962.00021962005400050028x
Bray, R. H., and Kurtz, L. (1945). Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 59, 39–46. doi: 10.1097/00010694-194501000-00006
Bremner, J., and Mulvaney, C. (1982). Total nitrogen in: page, Al Miller, Rh and Keeney methods of soil analysis part 2. Agronomy 9, 149–157.
Brockwell, J., and Bottomley, P. J. (1995). Recent advances in inoculant technology and prospects for the future. Soil Biol. Biochem. 27, 683–697. doi: 10.1016/0038-0717(95)98649-9
Buruchara, R., Chirwa, R., Sperling, L., Mukankusi, C., Rubyogo, J. C., Mutonhi, R., et al. (2011). Development and delivery of bean varieties in Africa: the Pan-Africa bean research alliance (Pabra) model. Afr. Crop Sci. J. 19, 227–245.
de Freitas, A. D. S., Silva, A. F., and de Sá Barretto Sampaio, E. V. (2012). Yield and biological nitrogen fixation of cowpea varieties in the semi-arid region of Brazil. Biomass Bioenergy 45, 109–114. doi: 10.1016/j.biombioe.2012.05.017
Fehr, W., Caviness, C., Burmood, D., and Pennington, J. (1971). Stage of development descriptions for soybeans, Glycine Max (L.) merrill 1. Crop Sci. 11, 929–931. doi: 10.2135/cropsci1971.0011183X001100060051x
Fening, J., and Danso, S. (2002). Variation in symbiotic effectiveness of cowpea Bradyrhizobia indigenous to Ghanaian soils. Appl. Soil Ecol. 21, 23–29. doi: 10.1016/S0929-1393(02)00042-2
Ferreira, D. F. (2008). “Sisvar: Um Programa Para Análises E Ensino De Estatística,” in Revista Symposium (Lavras), 36–41.
Graham, P., and Vance, C. (2000). Nitrogen fixation in perspective: an overview of research and extension needs. Field Crops Res. 65, 93–106. doi: 10.1016/S0378-4290(99)00080-5
Grönemeyer, J. L., Kulkarni, A., Berkelmann, D., Hurek, T., and Reinhold-Hurek, B. (2014). Rhizobia indigenous to the Okavango region in Sub-Saharan Africa: diversity, adaptations, and host specificity. Appl. Environ. Microbiol. 80, 7244–7257. doi: 10.1128/AEM.02417-14
Herridge, D. (2008). “Inoculation technology for legumes,” in Nitrogen-Fixing Leguminous Symbioses, eds M. J. Dilworth, E. K. James, J. I. Sprent, and W. E. Newton (Dordrecht: Springer), 77–115.
Hungria, M., and Kaschuk, G. (2014). Regulation of N2 fixation and / assimilation in nodulated and N-fertilized Phaseolus Vulgaris L. exposed to high temperature stress. Environ. Exp. Bot. 98, 32–39. doi: 10.1016/j.envexpbot.2013.10.010
Kamanga, B., Whitbread, A., Wall, P., Waddington, S., Almekinders, C., and Giller, K. (2010). Farmer evaluation of phosphorus fertilizer application to annual legumes in chisepo, Central Malawi. Afr. J Agr. Res. 5, 668–680.
Kamara, A. Y., Abaidoo, R., Kwari, J., and Omoigui, L. (2007). Influence of phosphorus application on growth and yield of soybean genotypes in the tropical savannas of Northeast Nigeria. Arch. Agron. Soil Sci. 53, 539–552. doi: 10.1080/03650340701398452
Karikari, B., Arkorful, E., and Addy, S. (2015). Growth, nodulation and yield response of cowpea to phosphorus fertilizer application in Ghana. J. Agron. 14, 234–240. doi: 10.3923/ja.2015.234.240
Kaya, Y., Akçura, M., and Taner, S. (2006). Gge-biplot analysis of multi-environment yield trials in bread wheat. Turk. J. Agric. For. 30, 325–337.
Kolawole, G. O. (2012). Effect of phosphorus fertilizer application on the performance of maize/soybean intercrop in the southern guinea savanna of Nigeria. Arch. Agron. Soil Sci. 58, 189–198. doi: 10.1080/03650340.2010.512723
Kyei-Boahen, S., Savala, C. E., Chikoye, D., and Abaidoo, R. (2017). Growth and yield responses of cowpea to inoculation and phosphorus fertilization in different environments. Front. Plant Sci. 8:646. doi: 10.3389/fpls.2017.00646
Landon, J. R. (2014). Booker Tropical Soil Manual: A Handbook for Soil Survey and Agricultural Land Evaluation in the Tropics and Subtropics. Abingdon: Routledge.
Martins, L., Xavier, G., Rangel, F., Ribeiro, J., Neves, M., Morgado, L., et al. (2003). Contribution of biological nitrogen fixation to cowpea: a strategy for improving grain yield in the semi-arid region of Brazil. Biol. Fert. Soils 38, 333–339. doi: 10.1007/s00374-003-0668-4
McInnes, A., and Haq, K. (2007). “Contributions of rhizobia to soil nitrogen fertility,” in Soil Biological Fertility, eds L. K. Abbott and D. V. Murphy (Dordrecht: Springer), 99–128.
Mpepereki, S., Javaheri, F., Davis, P., and Giller, K. (2000). Soyabeans and sustainable agriculture: promiscuous soyabeans in Southern Africa. Field Crops Res. 65, 137–149. doi: 10.1016/S0378-4290(99)00083-0
Musiyiwa, K., Mpepereki, S., and Giller, K. (2005). Symbiotic effectiveness and host ranges of indigenous rhizobia nodulating promiscuous soyabean varieties in Zimbabwean soils. Soil Biol. Biochem. 37, 1169–1176. doi: 10.1016/j.soilbio.2004.12.004
Mweetwa, A. M., Mulenga, M., Mulilo, X., Ngulube, M., Banda, J. S., Kapulu, N., et al. (2014). Response of cowpea, soya beans and groundnuts to non-indigenous legume inoculants. Sustain. Agric. Res. 3:84. doi: 10.5539/sar.v3n4p84
Naab, J. B., Chimphango, S. M., and Dakora, F. D. (2009). N2 fixation in cowpea plants grown in farmers' fields in the upper west region of Ghana, measured using 15N natural abundance. Symbiosis 48, 37–46. doi: 10.1007/BF03179983
Ndakidemi, P., Dakora, F., Nkonya, E., Ringo, D., and Mansoor, H. (2006). Yield and economic benefits of common bean (Phaseolus Vulgaris) and soybean (Glycine Max) inoculation in Northern Tanzania. Anim. Prod. Sci. 46, 571–577. doi: 10.1071/EA03157
Nelson, D., and Sommers, L. E. (1982). “Total carbon, organic carbon, and organic matter 1,” in Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties, 2nd Edn, eds A. L. Page, R. H. Miller, and D. R. Keeney (Madison, WI: Soil Science Society of America), 539–579.
Okogun, J., Sanginga, N., Abaidoo, R., Dashiell, K., and Diels, J. (2005). On-farm evaluation of biological nitrogen fixation potential and grain yield of lablab and two soybean varieties in the Northern Guinea Savanna of Nigeria. Nutr. Cycl. Agroecosys. 73, 267–275. doi: 10.1007/s10705-005-3821-7
Osei, O., Abaidoo, R. C., Ahiabor, B. D., Boddey, R. M., and Rouws, L. F. (2018). Bacteria related to Bradyrhizobium yuanmingense from Ghana are effective groundnut micro-symbionts. Appl. Soil Ecol. 127 41–50. doi: 10.1016/j.apsoil.2018.03.003
Osunde, A., Gwam, S., Bala, A., Sanginga, N., and Okogun, J. (2003). Responses to rhizobial inoculation by two promiscuous soybean cultivars in soils of the southern guinea savanna zone of Nigeria. Biol. Fert. Soils 37, 274–279. doi: 10.1007/s00374-003-0609-2
Pacheco, A., Vargas, M., Alvarado, G., Rodriguez, F., Crossa, J., and Burgueño, J. (2015). Gea-R (Genotype X Environment Analysis with R for Windows) Version 2.0. Mexico: CIMMYT.
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ronner, E., Franke, A., Vanlauwe, B., Dianda, M., Edeh, E., Ukem, B., et al. (2016). Understanding variability in soybean yield and response to P-fertilizer and rhizobium inoculants on farmers' fields in Northern Nigeria. Field Crops Res. 186, 133–145. doi: 10.1016/j.fcr.2015.10.023
Samonte, S. O. P., Wilson, L. T., McClung, A. M., and Medley, J. C. (2005). Targeting cultivars onto rice growing environments using ammi and sreg gge biplot analyses. Crop Sci. 45, 2414–2424. doi: 10.2135/cropsci2004.0627
Sanginga, N. (2003). Role of biological nitrogen fixation in legume based cropping systems; a case study of West Africa farming systems. Plant Soil 252, 25–39. doi: 10.1023/A:1024192604607
Sanginga, N., Thottappilly, G., and Dashiell, K. (2000). Effectiveness of rhizobia nodulating recent promiscuous soyabean selections in the moist savanna of Nigeria. Soil Biol. Biochem. 32, 127–133. doi: 10.1016/S0038-0717(99)00143-1
Sattar, M., Quader, M., and Danso, S. (1995). Nodulation, N2 fixation and yield of chickpea as influenced by host cultivar and Bradyrhizobium strain differences. Soil Biol. Biochem. 27, 725–727. doi: 10.1016/0038-0717(95)98656-9
Schumpp, O., and Deakin, W. J. (2010). How inefficient rhizobia prolong their existence within nodules. Trends Plant Sci. 15, 189–195. doi: 10.1016/j.tplants.2010.01.001
Simsek, S., Ojanen-Reuhs, T., Stephens, S. B., and Reuhs, B. L. (2007). Strain-ecotype specificity in Sinorhizobium Meliloti-medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J. Bacteriol. 189, 7733–7740. doi: 10.1128/JB.00739-07
Singleton, P., Bohlool, B., and Nakao, P. (1992). Legume response to rhizobial inoculation in the tropics: myths and realities. Myths Sci.Soils Trop. 29, 135–155. doi: 10.2136/sssaspecpub29.c8
Singleton, P., Boonkerd, N., Carr, T., and Thompson, J. (1997). Technical and Market Constraints Limiting Legume Inoculant Use in Asia. Extending Nitrogen Fixation Research to Farmers' Fields. Patancheru: ICRISAT, 17–38.
Somasegaran, P., and Hoben, H. J. (2012). Handbook for Rhizobia: Methods in Legume-Rhizobium Technology. Berlin; Heidelberg: Springer Science & Business Media.
Subedi, K., and Ma, B. (2009). Assessment of some major yield-limiting factors on maize production in a humid temperate environment. Field Crops Res. 110, 21–26. doi: 10.1016/j.fcr.2008.06.013
Thies, J., Woomer, P., and Singleton, P. (1995). Enrichment of Bradyrhizobium spp populations in soil due to cropping of the homologous host legume. Soil Biol. Biochem. 27, 633–636. doi: 10.1016/0038-0717(95)98643-3
Thies, J. E., Singleton, P. W., and Bohlool, B. B. (1991). Influence of the size of indigenous rhizobial populations on establishment and symbiotic performance of introduced rhizobia on field-grown legumes. Appl. Environ. Microbiol. 57, 19–28. doi: 10.1128/AEM.57.1.19-28.1991
Thuita, M., Pypers, P., Herrmann, L., Okalebo, R. J., Othieno, C., Muema, E., et al. (2012). Commercial rhizobial inoculants significantly enhance growth and nitrogen fixation of a promiscuous soybean variety in Kenyan soils. Biol. Fert. Soils 48, 87–96. doi: 10.1007/s00374-011-0611-z
Tirichine, L., de Billy, F., and Huguet, T. (2000). Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago Truncatula. Plant Physiol. 123, 845–852. doi: 10.1104/pp.123.3.845
Tittonell, P., Vanlauwe, B., Corbeels, M., and Giller, K. E. (2008). Yield gaps, nutrient use efficiencies and response to fertilisers by maize across heterogeneous smallholder farms of Western Kenya. Plant Soil 313, 19–37. doi: 10.1007/s11104-008-9676-3
Ulzen, J., Abaidoo, R. C., Masso, C., Owusu-Ansah, E. D. J., and Ewusi-Mensah, N. (2018). Is there a need for Bradyrhizobium Yuanmingense and B. Japonicum reinoculation in subsequent cropping seasons under smallholder farmers' conditions? Appl. Soil Ecol. 128, 54–60. doi: 10.1016/j.apsoil.2018.04.003
Ulzen, J., Abaidoo, R. C., Mensah, N. E., Masso, C., and AbdelGadir, A. H. (2016). Bradyrhizobium inoculants enhance grain yields of soybean and cowpea in Northern Ghana. Front. Plant Sci. 7:1770. doi: 10.3389/fpls.2016.01770
Vanlauwe, B., and Giller, K. E. (2006). Popular myths around soil fertility management in Sub-Saharan Africa. Agric. Ecosyst. Environ. 116, 34–46. doi: 10.1016/j.agee.2006.03.016
Woomer, P., Bennett, J., and Yost, R. (1990). Overcoming the inflexibility of most-probable-number procedures. Agron. J. 82, 349–353. doi: 10.2134/agronj1990.00021962008200020035x
Woomer, P., Huising, J., Giller, K., Baijukya, F., Kantengwa, S., Vanlauwe, B., et al. (2014). N2africa: Final Report of the First Phase-2009-2013. N2Africa.
Yan, W., Hunt, L., Sheng, Q., and Szlavnics, Z. (2000). Cultivar evaluation and mega-environment investigation based on the Gge biplot. Crop Sci. 40, 597–605. doi: 10.2135/cropsci2000.403597x
Yan, W., and Tinker, N. A. (2006). Biplot analysis of multi-environment trial data: principles and applications. Can. J. Plant Sci. 86, 623–645. doi: 10.4141/P05-169
Yates, R., Howieson, J., Hungria, M., Bala, A., O'Hara, G., and Terpolilli, J. (2016). “Authentication of rhizobia and assessment of the legume symbiosis in controlled plant growth systems,” in Working with Rhizobia, eds J. G. Howieson and M. J. Dilworth (Canberra, ACT: Australian Centre for International Agricultural Research), 73–108.
Keywords: inoculation, multiple location experiment, GGE biplot, site-specific response, grain yield
Citation: Osei O, Abaidoo RC, Opoku A, Rouws JRC, Boddey RM, Ahiabor BDK and Rouws LFM (2020) Native Bradyrhizobium Strains From Ghana Can Enhance Grain Yields of Field-Grown Cowpea and Groundnut. Front. Agron. 2:2. doi: 10.3389/fagro.2020.00002
Received: 16 January 2020; Accepted: 17 March 2020;
Published: 16 April 2020.
Edited by:
Antonio Rafael Sánchez-Rodríguez, Universidad de Córdoba, SpainReviewed by:
Ana María García-López, University of Seville, SpainIsabel Díaz De La Torre, University of Seville, Spain
Copyright © 2020 Osei, Abaidoo, Opoku, Rouws, Boddey, Ahiabor and Rouws. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ophelia Osei, b29zZWk1MkB5YWhvby5jb20=
 Ophelia Osei
Ophelia Osei Robert C. Abaidoo
Robert C. Abaidoo Andrews Opoku
Andrews Opoku Janaina R. C. Rouws
Janaina R. C. Rouws Robert M. Boddey
Robert M. Boddey Benjamin D. K. Ahiabor6
Benjamin D. K. Ahiabor6 Luc F. M. Rouws
Luc F. M. Rouws