- 1Department of Veterans Affairs Health Care System, The HLA Research Group, Brain Sciences Center, Minneapolis, MN, United States
- 2Department of Neuroscience, University of Minnesota Medical School, Minneapolis, MN, United States
- 3Department of Psychiatry, University of Minnesota Medical School, Minneapolis, MN, United States
- 4Institute for Health Informatics, University of Minnesota Medical School, Minneapolis, MN, United States
Introduction: The human immune system is equipped to neutralize and eliminate viruses and other foreign antigens via binding of human leukocyte antigen (HLA) molecules with foreign antigen epitopes and presenting them to T cells. HLA is highly polymorphic, resulting in subtle differences in the binding groove that influence foreign antigen binding and elimination. Here we tested the hypothesis that certain HLA alleles may promote longevity by enhanced ability to counter virus antigens that may otherwise contribute to morbidity and mortality.
Methods: We utilized high-resolution genotyping to characterize HLA and apolipoprotein E in a large sample (N = 986) of participants (469 men, 517 women) ranging in age from 24 to 90+ years old (mean age: 58.10 years) and identified 244 HLA alleles that occurred in the sample. Since each individual carries 12 classical HLA alleles (6 alleles of each Class I and Class II), we determined in silico the median predicted binding affinity for each individual (across the 12 HLA alleles) and each of 13 common viruses (Human Herpes Virus 1 [HHV1], HHV2, HHV3, HHV4, HHV5, HHV6A, HHV6B, HHV7, HHV8, human papilloma virus [HPV], human polyoma virus [JCV], human endogenous retrovirus K [HERVK], and HERVW). Next, we performed a stepwise multiple linear regression where the age of the participant was the dependent variable and the 13 median predicted HLA-virus binding affinities were the independent variables.
Results: The analyses yielded only one statistically significant effect–namely, a positive association between age and HERVK (P = 0.005). Furthermore, we identified 13 HLA alleles (9 HLA-I and 4 HLA-II) that occurred at greater frequency in very old individuals (age ≥90 years) as compared to younger individuals. Remarkably, for those 13 alleles, the predicted binding affinities were significantly higher for HERVK than for the other viruses (P < 0.001). ApoE genotypes did not differ significantly between older and younger groups.
Discussion: Taken together, the results showed that HLA-HERVK binding affinity is a robust predictor of longevity and that HLA alleles that bind with high affinity to HERVK were enriched in very old individuals. The findings of the present study highlight the influence of interactions between host immunogenetics and virus exposure on longevity and suggest that specific HLA alleles may promote longevity via enhanced immune response to specific common viruses, notably HERVK.
1 Introduction
Exposure to viruses and other foreign antigens is associated with numerous and wide-ranging health conditions and sequelae (Chang et al., 2017; Mui et al., 2017; Krump and You, 2018; Houen and Trier, 2021; Hussein and Rahal, 2019; Lotz et al., 2021; Al-Haddad et al., 2019; Burgdorf et al., 2019). Under optimal conditions–that is, immunocompetence–the human immune system is equipped to neutralize and eliminate foreign antigens that may otherwise contribute to morbidity and mortality. Specifically, the Human Leukocyte Antigen (HLA) region of chromosome 6 codes for cell-surface glycoproteins that play a critical role in the host immune response to foreign antigens including viruses, bacteria, and cancer neoantigens. Each individual possesses 12 classical HLA alleles, inherited in a Mendelian fashion, including six each from Class I (HLA-A, B, C) and Class II (HLA-DPB1, DQB1, DRB1). The steps involved in antigen processing and presentation by HLA-I and HLA-II alleles are reviewed elsewhere (Pishesha et al., 2022). Briefly, HLA-I molecules, which are expressed on all nucleated cells, help clear foreign antigens via binding and transporting cytosolic foreign antigen epitopes to the cell surface for presentation to cytotoxic CD8+ T cells to signal destruction of an infected cell. HLA-II molecules, which are expressed on lymphocytes and professional antigen presenting cells, bind and present endocytosed exogenous antigen epitopes to CD4+ T cells to stimulate antibody production and adaptive immunity. The HLA region is the most highly polymorphic of the human genome (Trowsdale and Knight, 2013). Even single amino acid differences can alter HLA-antigen binding (Hov et al., 2011), thereby influencing foreign antigen elimination and disease susceptibility (Dendrou et al., 2018). A recent genome-wide association meta-analysis documented that the HLA-DRB1 region is also associated with longevity as is apolipoprotein E (apoE) (Joshi et al., 2017). Here we tested the hypothesis that certain HLA alleles may promote longevity by enhanced ability to eliminate virus antigens that may otherwise contribute to morbidity and mortality. Specifically, we utilized high-resolution genotyping to 1) characterize the composition of HLA in a large sample of participants, 2) evaluate in silico the binding affinity of their HLA alleles with common viruses, and 3) determine the association between HLA-virus antigen binding affinity and age. We also evaluated the influence of apoE on longevity.
2 Materials and methods
2.1 Participants
A total of 986 participants (469 men, 517 women), the majority (87%) of whom were United States (US) veterans, were included in the present analyses. Participants were excluded from participation if they had been diagnosed with medical or psychiatric conditions that could impair ability to provide informed consent or participate in the study (e.g., Alzheimer’s dementia, schizophrenia). All participants provided written informed consent and were compensated for their participation. The study was approved by the Minneapolis Veterans Affairs Healthcare System Institutional Review Board and all research was performed in accordance with relevant guidelines and regulations.
2.2 HLA genotyping
DNA isolation was carried out from whole blood or saliva samples using commercially available kits (blood: ArchivePure cat. 2300730 from 5Prime distributed by Fisher Scientific or VWR; saliva: Oragene-Discover cat. OGR-500 coupled with prepIT purifier reagent cat. PT-L2P/DNA Genotek Inc. Ottawa, ON, Canada). The purified DNA samples were sent to HistoGenetics (http://www.histogenetics.com/) for high-resolution HLA Sequence-based Typing (SBT; details are given in https://bioinformatics.bethematchclinical.org/HLA-Resources/HLA-Typing/High-Resolution-Typing-Procedures/ and https://bioinformatics.bethematchclinical.org/WorkArea/DownloadAsset.aspx?id=6482). Their sequencing DNA templates are produced by locus- and group-specific amplifications that include exon 2 and 3 for Class I (A, B, C) and exon 2 for Class II (DRB1, DRB3/4/5, DQB1, and DPB1) and reported as Antigen Recognition Site (ARS) alleles as per ASHI recommendation (Cano et al., 2007).
2.3 ApoE genotyping
DNA samples were genotyped using PCR amplification followed by restriction enzyme digestion (Reymer et al., 1995). Each amplification reaction contained PCR buffer with 15 mmol/L MgCl2 ng amounts of genomic DNA, 20 pmol apoE forward (5N TAA GCT TGG CAC GGC TGT CCA AGG A 3N) and reverse (5N ATA AAT ATA AAA TAT AAA TAA CAG AAT TCG CCC CGG CCT GGT ACA C 3N) primers, 1.25 mmol/L of each deoxynucleotide triphosphate, 10% dimethylsulfoxide, and 0.25 μL AmpliTaq DNA polymerase. Reaction conditions in a thermocycler included an initial denaturing period of 3 min at 95 C, 1 min at 60 C, and 2 min at 72 C; followed by 32 cycles of 1 min at 95 C, 1 min at 60 C, and 2 min at 72 C; and a final extension of 1 min at 95 C, 1 min at 60 C, and 3 min at 72 C. PCR products were digested with HhaI and separated on a 4% Agarose gel which was stained with ethidium bromide. Known apoE isoform standards were included in the analysis.
2.4 Viruses
Thirteen viruses implicated in various diseases in humans were investigated, including 9 human herpes viruses (HHV1, HHV2, HHV3, HHV4, HHV5, HHV6A, HHV6B, HHV7, HHV8), human polyoma JC virus (JCV), human papilloma virus 16 (HPV), human endogenous retrovirus K (HERVK), and human endogenous retrovirus W (HERVW). Details of the viral proteins used are given in Table 1.
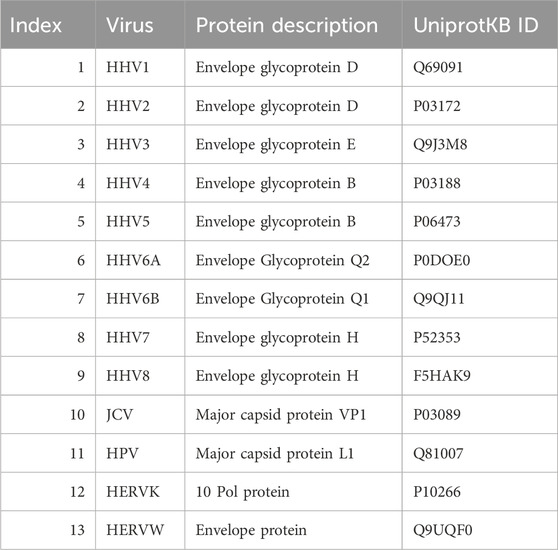
Table 1. Viral proteins used. HHV, human herpes virus; JCV, human polyomavirus JC; HPV, human papillomavirus; HERVK, human endogenous retrovirus K; HERVW, human endogenous retrovirus W.
2.5 In silico determination of predicted binding affinity of HLA-I and HLA-II alleles
Predicted binding affinities were obtained for viral protein epitopes using the Immune Epitope Database (IEDB) NetMHCpan (ver. 4.1) tool (Reynisson et al., 2020; IEDB, 2020). More specifically, we used the sliding window approach (Charonis et al., 2020a; Charonis et al., 2020b; Charonis et al., 2021) to test exhaustively all possible linear 9-mer (for HLA-I predictions) and 15-mer (for HLA-II predictions) AA residue epitopes of the 13 viral proteins analyzed (Table 1; Supplementary Table S1). The method is illustrated in Figure 1 for the HERVK virus protein. For each epitope-HLA molecule tested, this tool gives, as an output, the percentile rank of binding affinity of the HLA molecule and the epitope, among predicted binding affinities of the same HLA molecule to a large number of different peptides of the same AA length; the smaller the percentile rank, the better the binding affinity. Now, given a protein of N amino acid length and an epitope length of k AA, there are N-k binding affinity predictions, i.e., N-k percentile ranks. Of these predictions, for each viral protein and HLA molecule tested, we retained the lowest percentile rank (LPR) as the best binding affinity of the protein-HLA molecule pair. Finally, we took the inverse of LPR, so that higher values mean better binding affinities for more intuitive interpretation Equation 1:
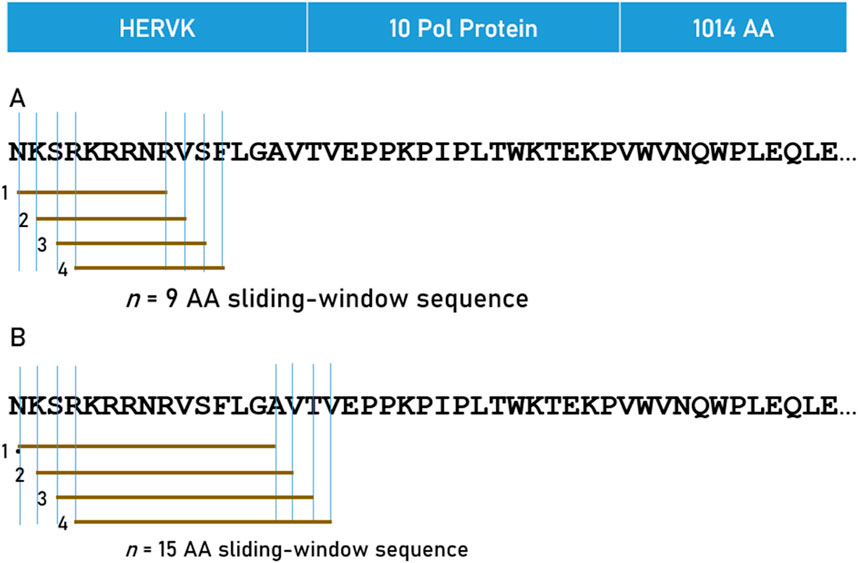
Figure 1. The 9-AA sliding window approach used for HLA Class I (A) and 15-AA window for HLA Class II (B) analyses are illustrated for HERVK.
2.6 Statistical analyses
Standard statistical methods were used to analyze the data using the IBM-SPSS statistical package (version 29). For multidimensional scaling (MDS), the ALSCAL procedure of the IBM-SPSS package was used (Level, ordinal; Condition, Matrix; Model, Euclid; Dimension [2,2]; Criteria: S-stress convergence = 0.001, minimum s-stress = 0.005, number of iterations = 30), and the K-means clustering procedure of the same package for identifying clusters in the MDS plot (Number of clusters = 3, Method: Iterate and Classify).
3 Results
3.1 Age
The frequency distribution of age is shown in Figure 2; mean = 58.10 y, SD = 13.77 y, median = 56.67 y, minimum = 24.08 y, maximum = 90 + y.
3.2 HLA alleles
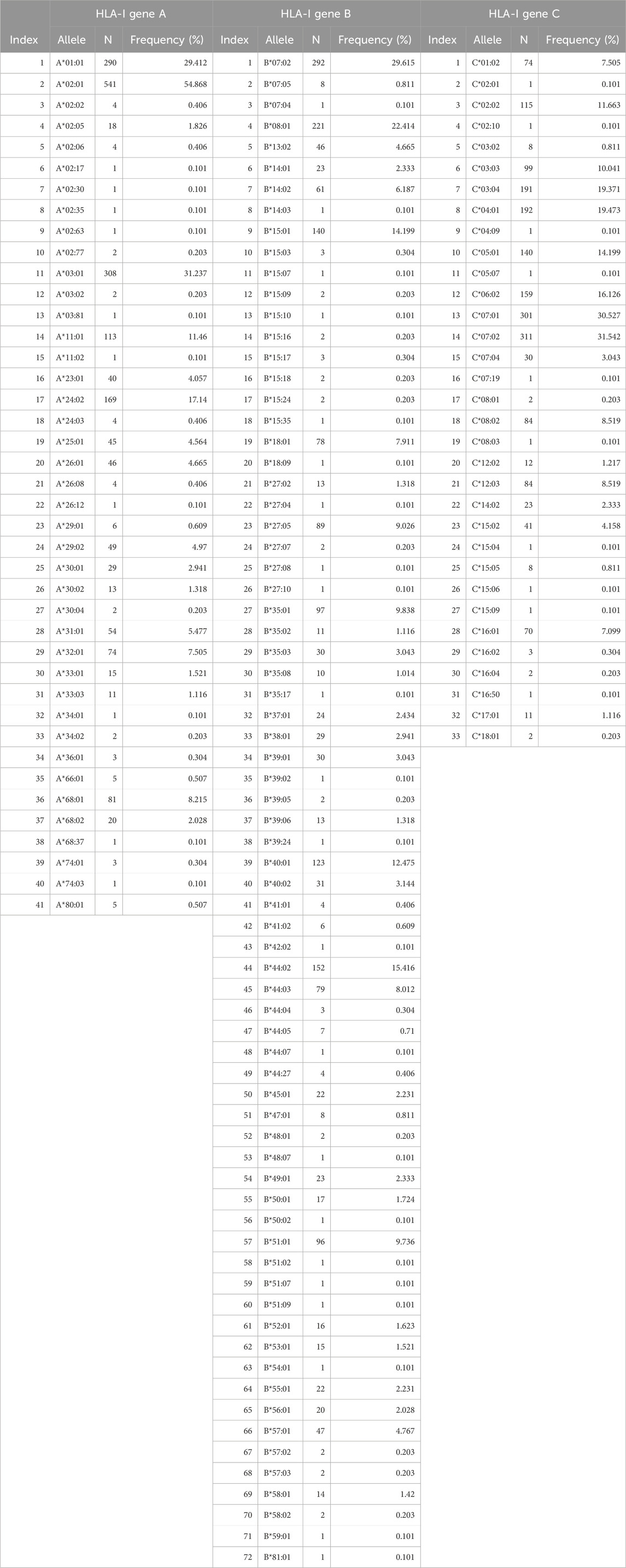
There were 244 distinct HLA alleles (142 of Class I and 93 of Class II) comprising the classical genes of Class I (A, B, C) and Class II (DPB1, DQB1, DRB1). The alleles and their frequencies (percentages) in our sample (N = 986) are given in Tables 2, 3.
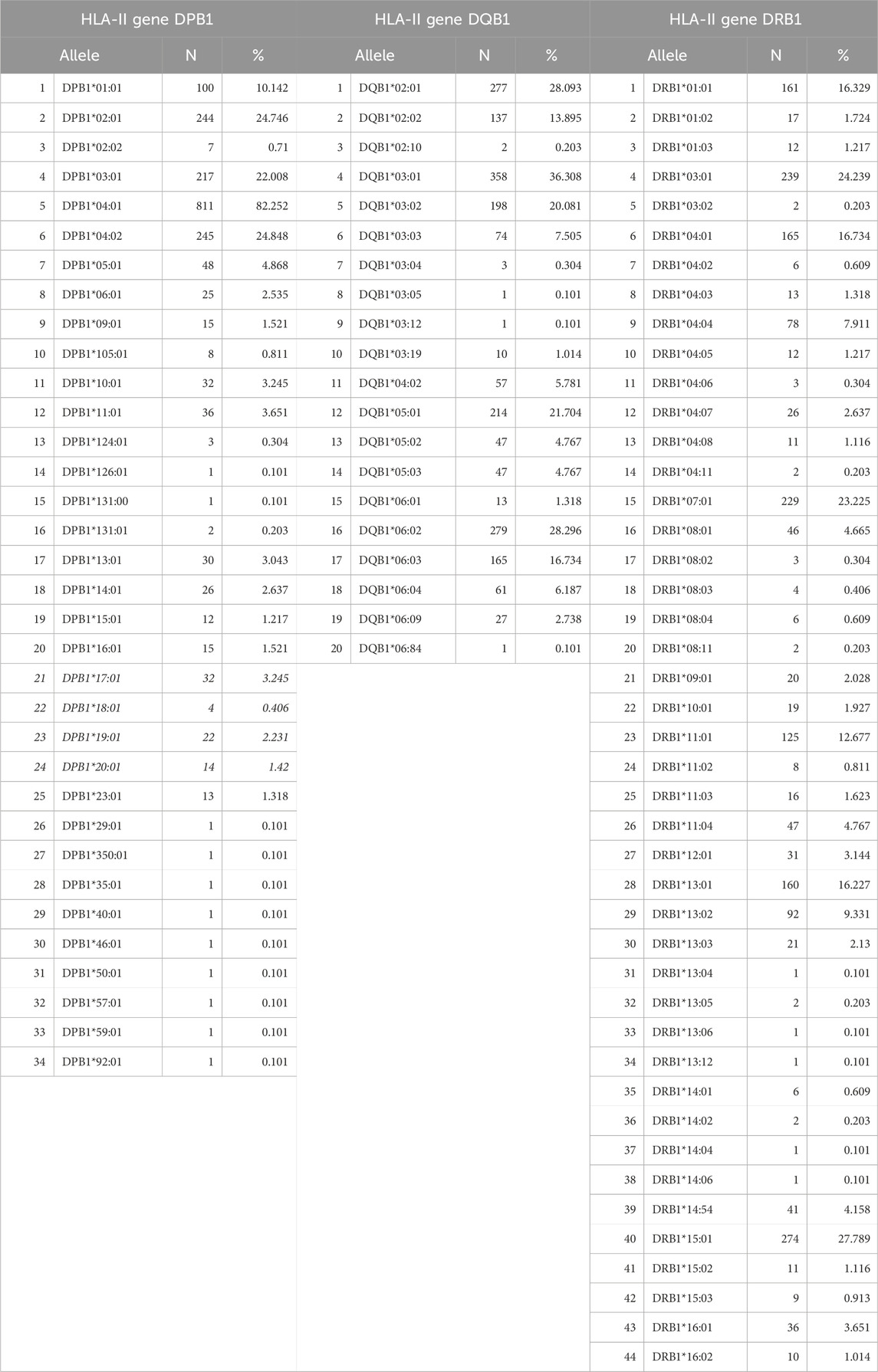
Table 3. Alleles of HLA Class II, Genes DPB1, DQB1, DRB1 found in our sample (N = 986 participants).
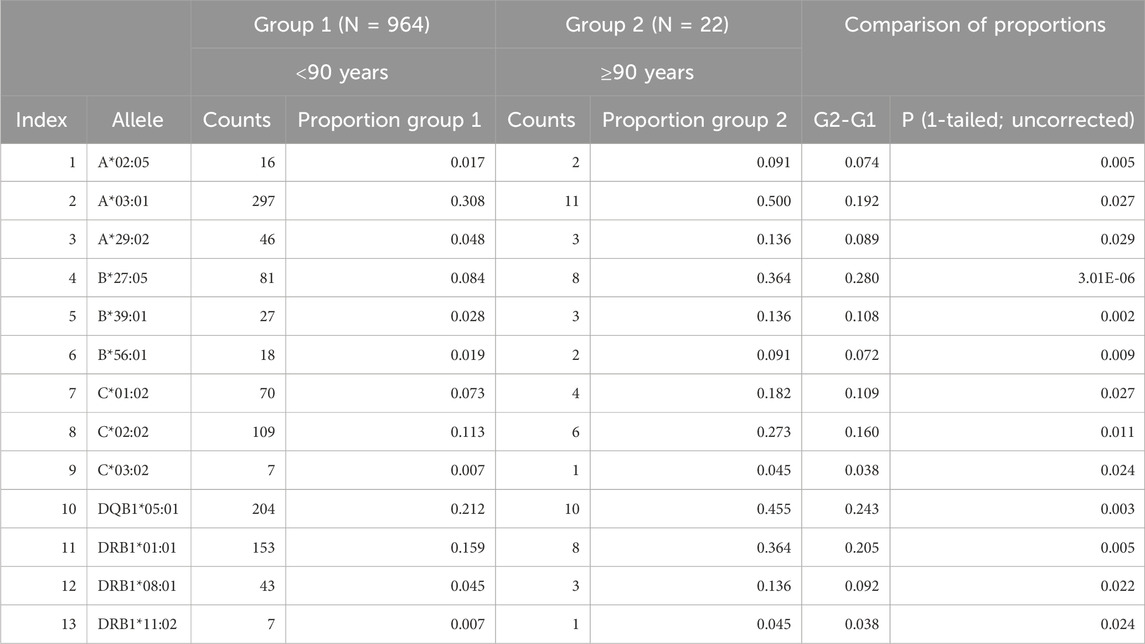
We used the age of 90 years as a conservative cut-off point to separate participants in 2 groups: Group1 (age <90 years, N = 964) and Group 2 (age ≥90 years, N = 22, “very old”). We hypothesized that surviving to very old age could/would be associated with the presence of specific HLA alleles. For that purpose, we searched for alleles with higher frequencies in Group 2 (as compared to Group 1) and found 13 alleles with significantly higher proportions in Group 2 than in Group 1 (Very Old Alleles, VOA); since we were testing the hypothesis of only higher allele frequencies in Group 2, we used a one-tailed test of proportions to obtain the statistical significance of the difference between the two proportions. These alleles with detailed statistics are shown in Table 4; they comprise 9 alleles of HLA Class I (3 of each A, B, and C genes) and 4 of Class II (1 of DPB1 gene, 1 of DQB1 gene, and 2 of DRB1 gene). The ratios of Group 2/Group 1 proportions ranged from 1.62 (allele A*03:01) to 6.43 (alleles C*03:02 and DRB1*11:02).
In a different analysis, we computed proportions of occurrence of the 13 VOAs across age starting at 51 years and moving forward every year–i.e., those younger than 51 vs those 51 years and older, those younger than 52 vs those 52 years and older, and so forth. The time course of the difference of proportions between the older and younger groups is shown in Figure 3. It can be seen that a systematic, monotonic increase in proportions (“enrichment”) of the 13 VOAs starts at age 74, marked by a thin vertical line in Figure 3.
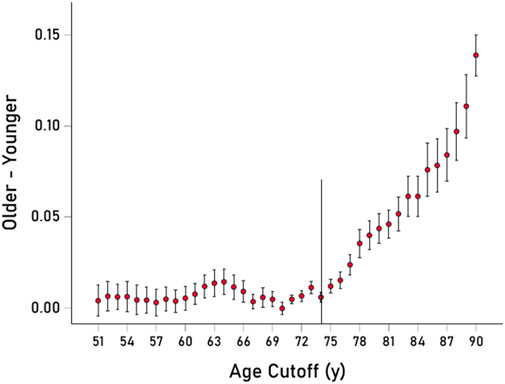
Figure 3. The mean ± SEM of the difference of proportions of the 13 VOAs (older–younger group samples) is plotted against the age cut-off threshold for younger/older group definitions. Thin vertical line is at 74 y. See text for details.
3.3 Association of virus PBA with longevity
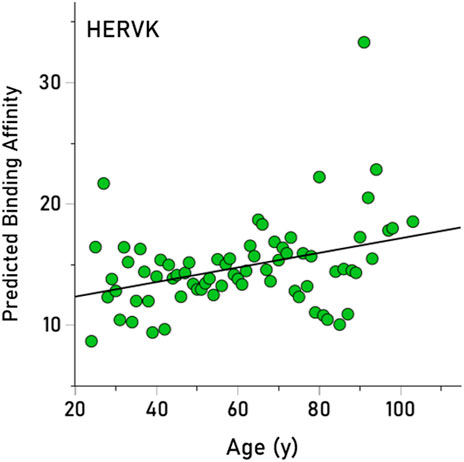
Here we tested the hypothesis that longevity could be associated with higher PBA for viruses that impede it, given that higher PBA entails a higher chance in eliminating a harmful virus and, hence, a higher chance of living longer. We obtained PBA estimates for 235/244 (96.3%) alleles, since 9 alleles could not be analyzed by the binding affinity tool (These alleles occurred in 10 participants, hence the number of participants carrying, as a pool, 235 alleles was 976.) We tested the hypothesis above as follows: (i) Given that there are 12 HLA alleles per participant (6 of HLA-I and 6 of HLA-II classical genes, namely, A, B, C for HLA-I and DPB1, DQB1, DRB1 for HLA-II) and 13 virus PBAs per allele, we first obtained the median PBA for each participant and virus, from the set of these12 alleles. This yielded a matrix of 976 participants x 13 median virus PBAs. (ii) Next, we performed a stepwise multiple linear regression where the age of the participant was the dependent variable and the 13 median virus PBAs were the independent variables. This analysis yielded only one statistically significant effect, namely, a positive association between age and HERVK (P = 0.005). (iii) This effect was further quantified and visualized using a simple linear regression between the annual averages of age and HERVK median PBA (across all participants). The result is plotted in Figure 4, illustrating the finding that HERVK PBA increases significantly with age (r = 0.357, P = 0.002) (It can be seen in Figure 4 that there is a high PBA value; if that is removed from the analysis, the positive association between and HERVK PBA remains strong and highly significant [r = 0.326, P = 0.005], attesting to the robustness of this effect).
3.4 HLA alleles in the very old and their association with virus PBA
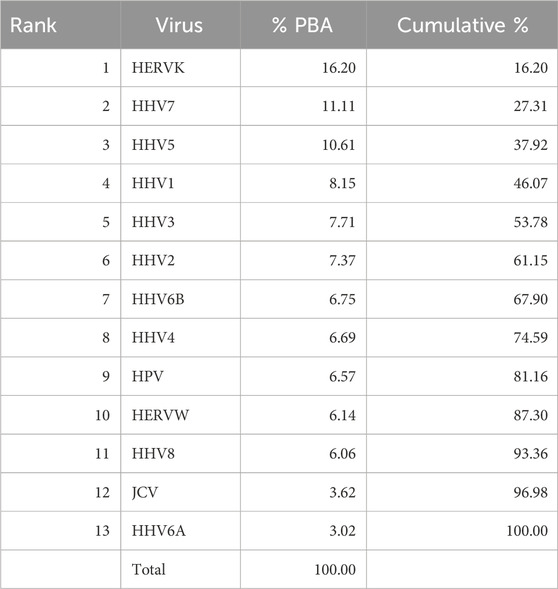
We then analyzed the PBAs of the 13 viruses in 11/13 VOA alleles (alleles C*02:02 and C*03:02 could not be modeled by the IEDB NetMHCpan affinity tool) using a repeated-measures analysis of variance (ANOVA), where virus PBAs were the dependent variables. We found that the PBA for HERVK was significantly higher than those of all the other viruses (P < 0.001, Bonferroni corrected for multiple comparisons) (Figure 5). The relative contribution of specific viral PBAs of the 11 VOAs, considered as an aggregate, is given in Table 5 (as percent of the sum of all 13 PBAs) and illustrated in Figure 6. It can be seen that HERVK PBA contributed the most (16.2%), followed by HHV7 (11.11%) and HHV5 (10.61%); collectively, the PBAs of these 3 viruses accounted for 37.92% of the total PBA of the 11 VOA alleles. Finally, the grouping of viral PBAs was evaluated using MDS and is shown in Figure 7. It can be seen that HERVK is farthest away from all other viruses, and that HHV7 and HHV5 are well separated from the remaining viruses. The apparent 3 cluster formation in the MDS plot of Figure 7 was documented by the result of a K-means clustering analysis of the 13 X-Y viral MDS coordinates, which assigned each viral PBA to one of the 3 distinct clusters, as demarcated in Figure 7. The cluster separation was further evaluated by performing a multivariate analysis of variance (MANOVA), where the MDS X and Y coordinates were the dependent variables and the cluster assignment was a fixed factor. This analysis yielded a highly statistically significant cluster separation (Hoteling’s Trace test = 6.019, P = 0.0001).
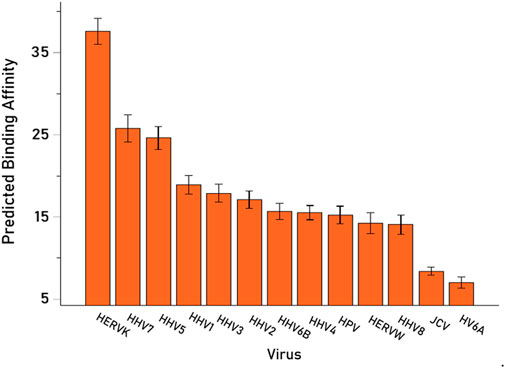
Figure 5. Mean (±95% Confidence Intervals) PBA of the 13 viruses studied for the 11 VOA alleles (alleles C*02:02 and C*03:02 could not by modeled by the IEDB NetMHCpan affinity tool).
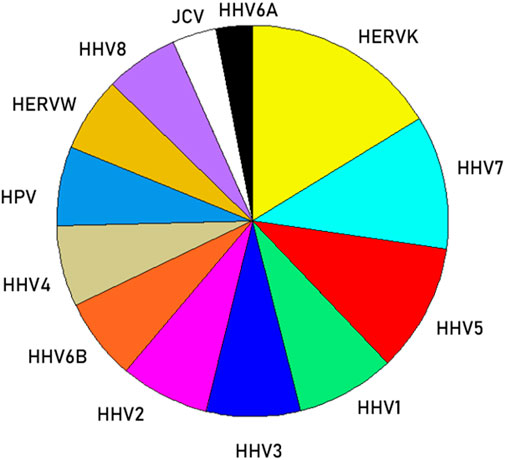
Figure 6. Pie plot to illustrate the relative (%) contribution of viral PBAs to VOAs (Table 5).
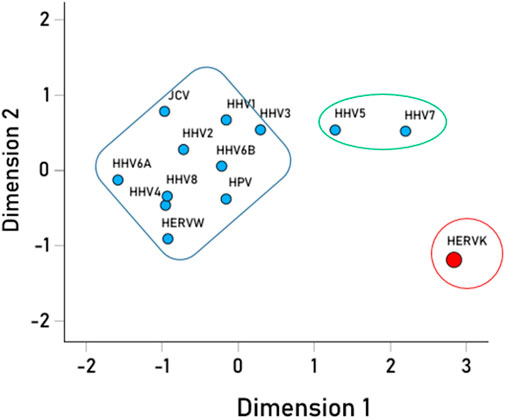
Figure 7. Multidimensional scaling plot of the viral PBAs. The clusters outlined were the outcomes of the K-means clustering analysis of the PBA X-Y coordinates.
3.5 ApoE
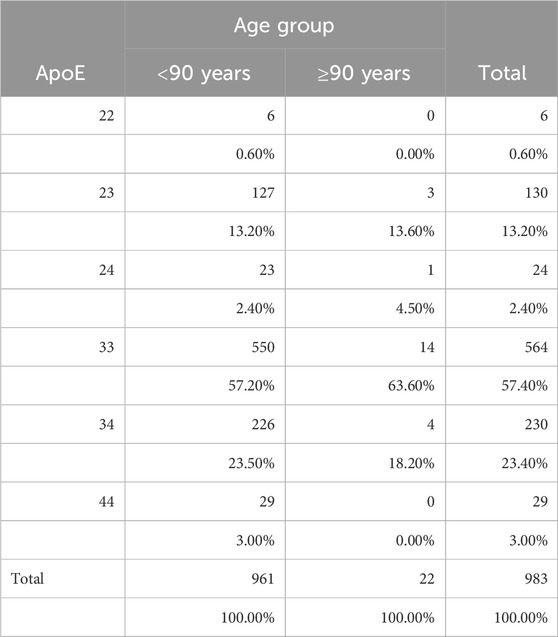
ApoE genotype is shown in Table 6. ApoE data were missing for three of the individuals in the younger group. The ApoE genotype distribution did not differ between the two groups (P = 0.898, chi-square test). There was also no difference in the proportion of the E2 allele (E2/E2, E2/E3) and E4 allele (E3/E4, E4/E4) between the two groups (E2: P = 0.978; E4: P = 0.379; Wilson test of two proportions).
4 Discussion
Here we evaluated longevity with respect to HLA and tested the hypothesis that longevity may be related to the enhanced ability to counter common viruses that have been implicated in morbidity and mortality. We first identified 244 HLA alleles in this sample and documented a highly significant positive association between the predicted binding affinity of those alleles to HERVK (but not any of the other viruses) and age. We then identified 13 HLA alleles (out of 244) that were more common among very old individuals than their younger counterparts, suggesting that those alleles may promote longevity. We documented that the set of very old alleles was associated with significantly higher binding affinity to HERVK, followed by HHV5 and HHV7, than to other common viruses. Taken together, the present findings suggest that specific HLA alleles may promote longevity via enhanced ability to mount immune responses to HERVK, an endogenous retrovirus that may otherwise contribute to morbidity and mortality.
4.1 Methodological considerations
With respect to HLA genotyping, we analyzed alleles at 2-field resolution, where the first field defines the allele group that corresponds to the serologically defined specificity of the HLA protein and the second field indicates differences in the DNA sequence that lead to a difference in the amino acid sequence of the resulting protein. The third field indicates synonymous DNA substitutions in the coding region, whereas the fourth field refers to differences in the non-coding regions. Since differences in the third and fourth fields do not have any influence on the resulting protein, which was the focus of this study, we did not use them in this analysis. However, ambiguities stemming from match/mismatches in the coding (third field) and/or non-coding (fourth field) DNA sequences (Voorter et al., 2014) are likely to be important with regard to successful transplantation (Mayor et al., 2021).
Finally, with respect to epitope length, although various lengths are possible, it is recommended that 9-mers for HLA-I (Lafuente and Reche, 2009) and 15-mers for HLA-II) (Rudensky et al., 1991) are most suitable. More specifically, for HLA-I, “… most MHCI-peptide ligands have nine residues (they are 9-mers), making models for the prediction of 9-mer binders preferable” (23, page 3211), as exemplified by the choice to use 9-mers in a detailed biophysical study of antigen presentation to HLA-I molecules (Garstka et al., 2015).
4.2 Viruses and disease associations
HERVK is part of a broad class of human endogenous retroviruses that are integrated into the human genome. Among the endogenous retroviruses, HERVK (particularly the HML-2 subgroup evaluated here) is the most transcriptionally active, possessing open reading frames allowing for coding of proteins (Garcia-Montojo et al., 2018) that are found in several tissues (Flockerzi et al., 2008). HERVK is minimally expressed in healthy adult cells but is upregulated and expressed, producing virus-like particles, in various conditions (Shin et al., 2023) and has been associated with cancers (Rivas et al., 2022; Curty et al., 2020), neurological conditions, diabetes, and autoimmune disorders (Garcia-Montojo et al., 2018; Xue et al., 2020). HERVK has also been associated with cellular senescence and tissue aging (Liu et al., 2023). Here, the predicted binding affinity of HERVK was highly significantly and positively associated with age. In addition, the 13 alleles identified in the very old group were shown to confer enhanced protection against HERVK, suggesting reduced HERVK-associated risks in the very old group. Notably, the 13 very old alleles were also significantly associated with the predicted binding affinities of HHV7 and HHV5. Several studies, reviewed elsewhere (Chen et al., 2019) have documented that infection with herpes viruses, among others, can induce HERV transactivation, contributing to virus-associated diseases and tumors. Thus, possessing the very old alleles identified here may promote longevity by facilitating not only an immune response to HERVK but also to HHV7 and HHV5 thereby reducing herpes virus-related HERV transactivation and long-term impacts on HERVK-associated morbidity and mortality. It should be noted that these 13 alleles were not absent in the younger group but were significantly less frequent; thus, members of the younger cohort who possess these 13 alleles would presumably be afforded greater longevity.
4.3 HLA and longevity
Previous research has documented an association between the HLA-DRB1 region and longevity (Joshi et al., 2017). Similarly, we found that 3 of the 13 alleles which were more frequent in the very old group belong to the Class II DRB1 gene. Here, however, several other HLA genes including Class I HLA-A (3 alleles), HLA-B (3 alleles), HLA-C (3 alleles), and Class II DQB1 (1 allele) were also more frequent in the very old group. Thus, the findings of our study which was based on high-resolution genotyping (vs imputation), confirm those of previous GWAS studies and document the relevance of additional HLA genes to longevity. Indeed, several of the very old alleles have been associated with disease protection. For instance, HLA-A*02:05 and HLA-A*03:01 are protective against severe clinical manifestations of SARS-CoV-2 (Littera et al., 2020; Shkurnikov et al., 2021), DRB1*01:01 is protective against MS (Mamedov et al., 2020), and DRB1*11:02 protects against rheumatoid arthritis (Cruz-Tapias et al., 2013). Perhaps the most well-investigated is HLA-B*27:05 which has been associated with risk for ankylosing spondylitis (Khan, 2023), yet superior immune control of HIV (International et al., 2010) and hepatitis C virus (Neumann-Haefelin et al., 2010). Thus, this group of alleles that are enhanced in very old individuals have been shown to protect against certain viruses and diseases, presumably promoting longevity.
4.4 Host-virus interactions
When considering protective or susceptibility effects related to HLA, it is crucial to keep in mind the role of HLA in host protection via signaling the immune system to eliminate foreign antigens. That is, protective or susceptibility effects are not conferred by HLA alone but by interactions between an individual’s HLA composition and exposure to viruses and other foreign antigens. Consequently, a given allele may preferentially bind and mount an immune response to certain pathogens over others. Indeed, this is what was documented for the 13 very old alleles here with regard to the 13 viruses we investigated. That is, as a group of alleles, those that were more common in the very old group had enhanced ability to eliminate HERVK relative to others.
4.5 ApoE and longevity
Notably, apolipoprotein E composition did not differ between the very old cohort and the younger cohort in the present study. Some previous research has found that the E2 allele is associated with increased longevity and the E4 allele with decreased longevity (Sebastiani et al., 2019), although the overall findings with regard to apoE and longevity are mixed (Abondio et al., 2019). ApoE is most prominently implicated in dementia (Serrano-Pozo et al., 2021); individuals who had been diagnosed with dementia were not included in the present study.
4.6 Conclusions and limitations
In summary, the findings of the present study suggest that longevity is associated with enhanced immune response to specific common viruses–particularly, HERVK–conferred by specific HLA alleles. These novel findings, which highlight the effect of interaction between host immunogenetics and virus exposure on longevity, need to be considered in light of several qualifications. First, the findings here are based on in silico analyses; future studies evaluating HLA and HERVK antigen-specific CD8 T cell receptor repertoire and in long-living individuals are warranted to further substantiate the conclusions herein. Second, we focused exclusively on the 244 alleles that were documented in our sample and particularly on 13 alleles that were enriched in the very old group of participants. That does not preclude the possibility of other alleles also mounting robust immune responses to HERVK. Indeed, in light of the role of HLA in host protection against viruses, it is likely that additional alleles that were equally present (or absent) in both groups bind with high affinity to the common viruses investigated here. In addition, it is worth noting that the current findings are specific to longevity and may not equate with overall health as only health conditions that would interfere with comprehension or participation were exclusionary. Finally, although the viruses and HLA alleles are relatively common globally, the frequencies of both are known to vary geographically (Looker et al., 2017; Adane and Getawa, 2021; Prugnolle et al., 2005; Singh et al., 2007). Although additional studies are warranted to determine whether the longevity alleles identified here generalize to other populations, all of the 13 alleles that were enriched in the very old group of participants here are common worldwide (Hurley et al., 2020), suggesting these alleles with high affinity to HERVK may promote global longevity.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Institutional Review Board of the Minneapolis VA Health Care System. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
LJ: Data curation, Investigation, Project administration, Writing–original draft, Writing–review and editing. AG: Conceptualization, Formal Analysis, Methodology, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Partial funding for this study was provided by the University of Minnesota (the Anita Kunin Chair in Women’s Healthy Brain Aging, the Brain and Genomics Fund, the McKnight Presidential Chair of Cognitive Neuroscience, and the American Legion Brain Sciences Chair) and the U.S. Department of Veterans Affairs. The sponsors had no role in the current study design, analysis or interpretation, or in the writing of this paper. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fragi.2025.1471202/full#supplementary-material
References
Abondio, P., Sazzini, M., Garagnani, P., Boattini, A., Monti, D., Franceschi, C., et al. (2019). The genetic variability of APOE in different human populations and its implications for longevity. Genes 10, 222. doi:10.3390/genes10030222
Adane, T., and Getawa, S. (2021). Cytomegalovirus seroprevalence among blood donors: a systematic review and meta-analysis. J. Int. Med. Res. 49, 03000605211034656. doi:10.1177/03000605211034656
Al-Haddad, B. J. S., Jacobsson, B., Chabra, S., Modzelewska, D., Olson, E. M., Bernier, R., et al. (2019). Long-term risk of neuropsychiatric disease after exposure to infection in utero. JAMA Psychiat. 76, 594–602. doi:10.1001/jamapsychiatry.2019.0029
Burgdorf, K. S., Trabjerg, B. B., Pedersen, M. G., Nissen, J., Banasik, K., Pedersen, O. B., et al. (2019). Large-scale study of Toxoplasma and Cytomegalovirus shows an association between infection and serious psychiatric disorders. Brain Behav. Immun. 79, 152–158. doi:10.1016/j.bbi.2019.01.026
Cano, P., Klitz, W., Mack, S. J., Maiers, M., Marsh, S. G. E., Noreen, H., et al. (2007). Common and well-documented HLA alleles: report of the ad-hoc committee of the American society for histocompatiblity and immunogenetics. Hum. Immunol. 68, 392–417. doi:10.1016/j.humimm.2007.01.014
Chang, Y., Moore, P. S., and Weiss, R. A. (2017). Human oncogenic viruses: nature and discovery. Philos. Trans. R. Soc. Lond B Biol. Sci. 372, 20160264. doi:10.1098/rstb.2016.0264
Charonis, S., James, L. M., and Georgopoulos, A. P. (2020a). In silico assessment of binding affinities of three dementia-protective Human Leukocyte Antigen (HLA) alleles to nine human herpes virus antigens. Curr. Res. Transl. Med. 68, 211–216. doi:10.1016/j.retram.2020.06.002
Charonis, S., Tsilibary, E. P., and Georgopoulos, A. P. (2020b). SARS-CoV-2 virus and Human Leukocyte Antigen (HLA) Class II: investigation in silico of binding affinities for COVID-19 protection and vaccine development. J. Immunol. Sci. 4, 12–23. doi:10.29245/2578-3009/2020/4.1198
Charonis, S. A., Tsilibary, E. P., and Georgopoulos, A. P. (2021). In silico investigation of binding affinities between human leukocyte antigen class I molecules and SARS-CoV-2 virus spike and ORF1ab proteins. Explor. Immunol. 1, 16–26. doi:10.37349/ei.2021.00003
Chen, J., Foroozesh, M., and Qin, Z. (2019). Transactivation of human endogenous retroviruses by tumor viruses and their functions in virus-associated malignancies. Oncogenesis 8, 6. doi:10.1038/s41389-018-0114-y
Cruz-Tapias, P., Castiblanco, J., and Anaya, J. M. (2013). “HLA association with autoimmune diseases,” in Autoimmunity: from bench to bedside. Editors J. M. Anaya, Y. Shoenfeld, A. Rojas-Villarraga, R. A. Levy, and R. Cervera (Bogota, Colombia: El Rosario University Press), 271–284.
Curty, G., Marston, J. L., de Mulder Rougvie, M., Leal, F. E., Nixon, D. F., Soares, M. A., et al. (2020). Human Endogenous Retrovirus K in cancer: a potential biomarker and immunotherapeutic target. Viruses 12, 726. doi:10.3390/v12070726
Dendrou, C. A., Petersen, J., Rossjohn, J., and Fugger, L. (2018). HLA variation and disease. Nat. Rev. Immunol. 18, 325–339. doi:10.1038/nri.2017.143
Flockerzi, A., Ruggieri, A., Frank, O., Sauter, M., Maldener, E., Kopper, B., et al. (2008). Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics 9, 354. doi:10.1186/1471-2164-9-354
Garcia-Montojo, M., Doucet-O’Hare, T., Henderson, L., and Nath, A. (2018). Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit. Rev. Microbiol. 44, 715–738. doi:10.1080/1040841X.2018.1501345
Garstka, M. A., Fish, A., Celie, P. H., Joosten, R. P., Janssen, G. M., Berlin, I., et al. (2015). The first step of peptide selection in antigen presentation by MHC class I molecules. Proc. Natl. Acad. Sci. U. S. A. 112, 1505–1510. doi:10.1073/pnas.1416543112
Houen, G., and Trier, N. H. (2021). Epstein-Barr virus and systemic autoimmune diseases. Front. Immunol. 11, 587380. doi:10.3389/fimmu.2020.587380
Hov, J. R., Kosmoliaptsis, V., Traherne, J. A., Olsson, M., Boberg, K. M., Bergquist, A., et al. (2011). Electrostatic modifications of the human leukocyte antigen-DR P9 peptide-binding pocket and susceptibility to primary sclerosing cholangitis. Hepatology 53, 1967–1976. doi:10.1002/hep.24299
Hurley, C. K., Kempenich, J., Wadsworth, K., Sauter, J., Hofmann, J. A., Schefzyk, D., et al. (2020). Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0.0. HLA 95, 516–531. doi:10.1111/tan.13811
Hussein, H. M., and Rahal, E. A. (2019). The role of viral infections in the development of autoimmune diseases. Crit. Rev. Microbiol. 45, 394–412. doi:10.1080/1040841X.2019.1614904
IEDB (2020). Analysis resource. Available at: http://tools.iedb.org/mhci/result/(Accessed April 2, 2024).
International, H. I. V., Pereyra, F., Jia, X., McLaren, P. J., Telenti, A., de Bakker, P. I. W., et al. (2010). The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557. doi:10.1126/science.1195271
Joshi, P. K., Piratsu, N., Kenitsou, K. A., Fischer, K., Hofer, E., Schraut, K. E., et al. (2017). Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat. Commun. 8, 910. doi:10.1038/s41467-017-00934-5
Khan, M. A. (2023). HLA-B*27 and ankylosing spondylitis: 50 years of insights and discoveries. Curr. Rheumatol. Rep. 25, 327–340. doi:10.1007/s11926-023-01118-5
Krump, N. A., and You, J. (2018). Molecular mechanisms of viral oncogenesis in humans. Nat. Rev. Microbiol. 16, 684–698. doi:10.1038/s41579-018-0064-6
Lafuente, E. M., and Reche, P. A. (2009). Prediction of MHC-peptide binding: a systematic and comprehensive overview. Curr. Pharm. Des. 15 (28), 3209–3220. doi:10.2174/138161209789105162
Littera, R., Campagna, M., Deidda, S., Angioni, G., Cipri, S., Melis, M., et al. (2020). Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. The Sardinian experience. Front. Immunol. 11, 605688. doi:10.3389/fimmu.2020.605688
Liu, X., Liu, Z., Wu, Z., Ren, J., Fan, Y., Sun, L., et al. (2023). Resurrection of endogenous retroviruses during aging reinforces senescence. Cell 186, 287–304.e26. doi:10.1016/j.cell.2022.12.017
Looker, K. J., Magaret, A. S., May, M. T., Turner, K. M. E., Vickerman, P., Newman, L. M., et al. (2017). First estimates of the global and regional incidence of neonatal herpes infection. Lancet Glob. Health 5, e300–e309. doi:10.1016/S2214-109X(16)30362-x
Lotz, S. K., Blackhurst, B. M., Reagin, K. L., and Funk, K. E. (2021). Microbial infections are a risk factor for neurodegenerative diseases. Front. Cell Neurosci. 15, 691136. doi:10.3389/fncel.2021.691136
Mamedov, A., Vorobyeva, N., Filimonova, I., Zakharova, M., Kiselev, I., Bashinskaya, V., et al. (2020). Protective allele for multiple sclerosis HLA-DRB1*01:01 provides kinetic discrimination of myelin and exogenous antigenic peptides. Front. Immunol. 10, 3088. doi:10.3389/fimmu.2019.03088
Mayor, N. P., Wang, T., Lee, S. J., Kuxhausen, M., Vierra-Green, C., Barker, D. J., et al. (2021). Impact of previously unrecognized HLA mismatches using ultrahigh resolution typing in unrelated donor hematopoietic cell transplantation. J. Clin. Oncol. 39, 2397–2409. doi:10.1200/JCO.20.03643
Mui, U. N., Haley, C. T., and Tyring, S. K. (2017). Viral oncology: molecular biology and pathogenesis. J. Clin. Med. 6, 111. doi:10.3390/jcm6120111
Neumann-Haefelin, C., Timm, J., Schmidt, J., Kersting, N., Fitzmaurice, K., Oniangue-Ndza, C., et al. (2010). Protective effect of human leukocyte antigen B27 in hepatitis C virus infection requires the presence of a genotype-specific immunodominant CD8+ T-cell epitope. Hepatology 51, 54–62. doi:10.1002/hep.23275
Pishesha, N., Harmand, T. J., and Ploegh, H. L. (2022). A guide to antigen processing and presentation. Nat. Rev. Immunol. 22, 751–764. doi:10.1038/s41577-022-00707-2
Prugnolle, F., Mnica, A., Charpentier, M., Guégan, J. F., Guernier, V., and Balloux, F. (2005). Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 15, 1022–1027. doi:10.1016/j.cub.2005.04.050
Reymer, P. W., Groenemeyer, B. E., Van de Burg, R., and Kastelein, J. J. (1995). Apolipoprotein E genotyping on agarose gels. Clin. Chem. 41, 1046–1047. doi:10.1093/clinchem/41.7.1046
Reynisson, B., Alvarez, B., Paul, S., Peters, B., and Nielsen, M. (2020). NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48 (W1), W449–W454. doi:10.1093/nar/gkaa379
Rivas, S. R., Valdez, M. J. M., Govindarajan, V., Seetharam, D., Doucet-O'Hare, T. T., Heiss, J. D., et al. (2022). The role of HERV-K in cancer stemness. Viruses 14, 2019. doi:10.3390/v14092019
Rudensky, Y., Preston-Hurlburt, P., Hong, S. C., Barlow, A., and Janeway, C. A. (1991). Sequence analysis of peptides bound to MHC class II molecules. Nature 353, 622–627. doi:10.1038/353622a0
Sebastiani, P., Gurinovich, A., Nygaard, M., Sasaki, T., Sweigart, B., Bae, H., et al. (2019). APOE alleles and extreme human longevity. J. Gerontol. A Biol. Sci. Med. Sci. 74, 44–51. doi:10.1093/gerona/gly174
Serrano-Pozo, A., Das, S., and Hyman, B. T. (2021). APOE and Alzheimer's disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 20, 68–80. doi:10.1016/S1474-4422(20)30412-9
Shin, W., Mun, S., and Han, K. (2023). Human endogenous retrovirus-K (HML-2)-Related genetic variation: human genome diversity and disease. Genes (Basel) 14 (12), 2150. doi:10.3390/genes14122150
Shkurnikov, M., Nersisyan, S., Jankevic, T., Galatenko, A., Gordeev, I., Vechorko, V., et al. (2021). Association of HLA class I genotypes with severity of coronavirus disease-19. Front. Immunol. 12, 641900. doi:10.3389/fimmu.2021.641900
Singh, R., Kaul, R., Kaul, A., and Khan, K. (2007). A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J. Gastron. 13, 1770–1787. doi:10.3748/wjg.v13.i12.1770
Trowsdale, J., and Knight, J. C. (2013). Major histocompatibility complex genomics and human disease. Ann. Rev. Genom Hum. Genet. 14, 301–323. doi:10.1146/annurev-genom-091212-153455
Voorter, C. E., Palusci, F., and Tilanus, M. G. (2014). Sequence-based typing of HLA: an improved group-specific full-length gene sequencing approach. Methods Mol. Biol. 1109, 101–114. doi:10.1007/978-1-4614-9437-9_7
Keywords: longevity, human leukocyte antigen (HLA), viruses, human endogenous retrovirus K (HERV-K), apolipoprotein E
Citation: James LM and Georgopoulos AP (2025) Immunogenetics of longevity and its association with human endogenous retrovirus K. Front. Aging 6:1471202. doi: 10.3389/fragi.2025.1471202
Received: 26 July 2024; Accepted: 20 January 2025;
Published: 04 February 2025.
Edited by:
Calogero Caruso, University of Palermo, ItalyReviewed by:
Elena Ciaglia, University of Salerno, ItalyMilena Ivanova Ivanova-Shivarova, Aleksandrovska University Hospital, Bulgaria
Copyright © 2025 James and Georgopoulos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa M. James, bG1qYW1lc0B1bW4uZWR1
 Lisa M. James
Lisa M. James Apostolos P. Georgopoulos
Apostolos P. Georgopoulos