
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Aging, 15 May 2023
Sec. Molecular Mechanisms of Aging
Volume 4 - 2023 | https://doi.org/10.3389/fragi.2023.1171850
This article is part of the Research TopicInsights in Molecular Mechanisms of Aging 2022View all 10 articles
Age-related loss of skeletal muscle mass leads to a reduction of strength. It is likely due to an inadequate stimulation of muscle protein synthesis (MPS) in response to anabolic stimuli, such as mechanical load. Ribosome biogenesis is a major determinant of translational capacity and is essential for the control of muscle mass. This mini-review aims to put forth the hypothesis that ribosome biogenesis is impaired by aging in response to mechanical load, which could contribute to the age-related anabolic resistance and progressive muscle atrophy. Recent animal studies indicate that aging impedes muscle hypertrophic response to mechanical overload. This is associated with an impaired transcription of ribosomal DNA (rDNA) by RNA polymerase I (Pol I), a limited increase in total RNA concentration, a blunted activation of AKT/mTOR pathway, and an increased phosphorylation of AMPK. In contrast, an age-mediated impairment of ribosome biogenesis is unlikely in response to electrical stimulations. In human, the hypertrophic response to resistance exercise training is diminished with age. This is accompanied by a deficit in long-term MPS and an absence of increased total RNA concentration. The results addressing the acute response to resistance exercise suggest an impaired Pol I-mediated rDNA transcription and attenuated activation/expression of several upstream regulators of ribosome biogenesis in muscles from aged individuals. Altogether, emerging evidence indicates that impaired ribosome biogenesis could partly explain age-related anabolic resistance to mechanical load, which may ultimately contribute to progressive muscle atrophy. Future research should develop more advanced molecular tools to provide in-depth analysis of muscle ribosome biogenesis.
Skeletal muscle plays a vital role in movement, body temperature regulation and inter-organ crosstalk. Aging can lead to sarcopenia, which is characterized by decreased muscle mass, strength, and physical performance (Cruz-Jentoft et al., 2019). Sarcopenia is associated with increased risk for adverse health events, including falls, loss of independence and chronic disability (Batsis et al., 2014; Cruz-Jentoft and Sayer, 2019). Muscle atrophy directly relates to loss of muscle strength (Nilwik et al., 2013). Thus, understanding the mechanisms behind age-related muscle atrophy is crucial to develop early preventive measures and mitigate sarcopenia risk in advanced aging.
The etiology of age-related atrophy is multifactorial but could mainly result from an imbalance between muscle protein synthesis (MPS) and muscle protein breakdown (Wilkinson et al., 2018). Cumulative evidence indicates that basal rates of MPS are not compromised in old compared with young adults (Volpi et al., 2001; Cuthbertson et al., 2005; Katsanos et al., 2005; Mayhew et al., 2009). Instead, an inadequate stimulation of MPS in response to anabolic stimuli [called anabolic resistance (AR)], including mechanical load associated with resistance exercise (RE), dietary amino acids (AA), and anabolic hormonal milieu could underlie age-related catabolic perturbations in protein homeostasis, leading to a progressive decline in muscle mass (Wilkinson et al., 2018). Recent reviews have analyzed the mechanisms linking AR and age-related muscle atrophy (Wilkinson et al., 2018; Prokopidis et al., 2021; Zhao et al., 2021; Kanova and Kohout, 2022).
Novel insights indicate that ribosome biogenesis is a major determinant of translational capacity and is involved in the control of skeletal muscle mass (Chaillou et al., 2014; Figueiredo and McCarthy, 2019; von Walden, 2019). To date, the impact of age-related AR to mechanical load on translational capacity is under debate. After providing a brief overview of the regulation of ribosome biogenesis, this mini-review will put forth the hypothesis that ribosome biogenesis is impaired by aging in response to mechanical load, which could contribute to the age-related AR and progressive muscle atrophy.
The human 80S ribosome, a 4.3-MDa ribonucleoprotein complex, is composed of one small 40S subunit (40S-SU) and one large 60S subunit (60S-SU). The 40S-SU contains 18S rRNA and 33 ribosomal proteins (RPs), while 60S-SU is composed of 5S, 5.8S, and 28S rRNAs, and 47 RPs (Khatter et al., 2015). During protein translation, 40S-SU decodes the mRNA by promoting the interaction between the mRNA codons and the transfer RNA (tRNA) anticodons (aminoacyl-tRNA), and each added amino acid is then incorporated into the newly translated polypeptide by 60S-SU. Ribosomes are crucial for protein translation, which is determined by both translational efficiency (i.e., protein synthesis rate per ribosome) and translational capacity (i.e., number of ribosomes per unit tissue or total number).
Ribosome biogenesis is a multistep process leading to the de novo synthesis of ribosomes [for more detailed information, see (Thomson et al., 2013; Chaillou et al., 2014; Kusnadi et al., 2015; Figueiredo and McCarthy, 2019)]. It includes numerous steps (Figure 1): the transcription of ribosomal DNA (rDNA), mRNAs encoding RPs, and tRNAs, the translation of RPs, the processing of the polycistronic 47S rRNA precursor (47S pre-rRNA) into smaller rRNA (18S, 5.8S and 28S rRNAs), the assembly of rRNAs and RPs to form 40S- and 60S-SU, and the nuclear export and maturation of these subunits. Ribosome biogenesis requires the coordinated actions of all three RNA polymerases (Pol): RNA Pol I is responsible for the transcription of 47 pre-rRNA, RNA Pol II transcribes mRNAs encoding RPs (as well as all other mRNAs), and RNA Pol III transcribes 5S rRNA gene (as well as tRNA genes).
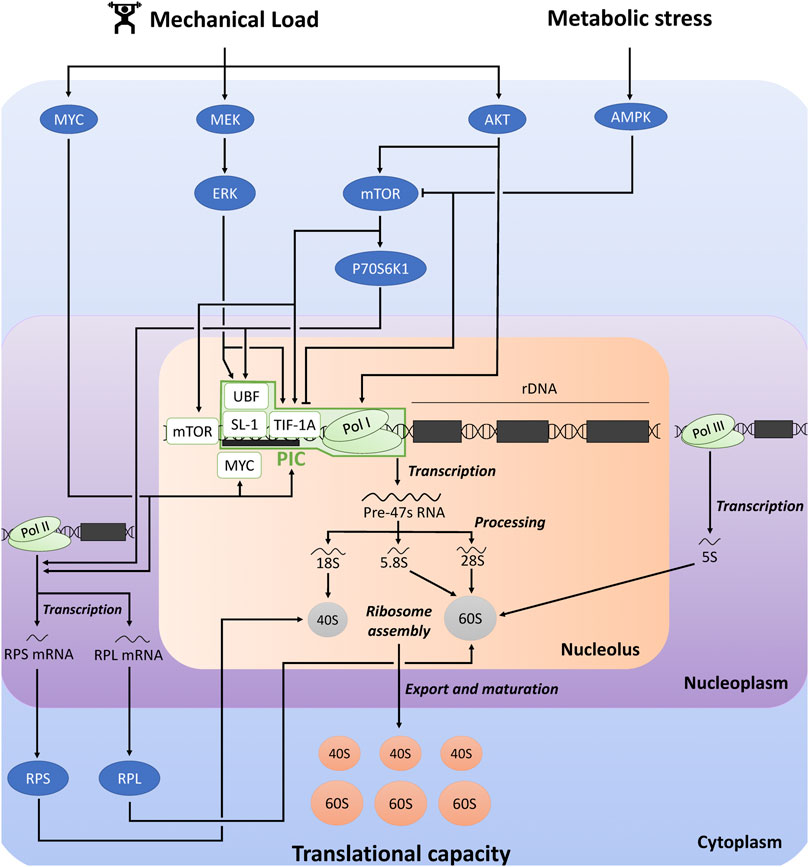
FIGURE 1. Schematic representation of the regulation of ribosome biogenesis and translational capacity in response to mechanical load and metabolic stress. The figure was adapted from previous reviews (Chaillou et al., 2014; Kusnadi et al., 2015; Figueiredo and McCarthy, 2019). AKT, protein kinase B; AMPK, AMP-activated protein kinase; ERK, extracellular signal-regulated kinase; mTOR, mechanistic target of rapamycin; PIC, pre-initiation complex; RPS, ribosomal protein small; RPL, ribosomal protein large; Pol, RNA polymerase; SL-1, selectively factor 1; TIF-1A, transcription initiation factor 1A; UBF, upstream binding factor.
Pol I-mediated rDNA transcription is the rate-limiting step in ribosome biogenesis, and 47S pre-rRNA production is considered as the major regulatory point of ribosome biogenesis (Kusnadi et al., 2015). Pol I-mediated rDNA transcription is largely dependent on the expression and activity of the pre-initiation complex (PIC) at the rDNA promoter. PIC is composed of RNA Pol I complex, and the Pol I-specific transcription factors transcription initiation factor 1A (TIF-1A), selectively factor 1 (SL-1) complex [formed by the TATA-binding protein (TBP) and four TBP-associated factors (TAFs)], and upstream binding factor (UBF) (Gorski et al., 2007; Goodfellow and Zomerdijk, 2013). UBF first binds to the rDNA promoter and recruits SL-1 complex. UBF/SL-1 complex facilitates DNA binding and TIF-1A promotes the connection between these factors and Pol I.
Pol I-mediated rDNA transcription and the other steps of ribosome biogenesis are controlled by several signaling pathways which receive inputs from different cellular stimuli, including mechanical load, growth factors, hormonal, nutritional and metabolic influences [for more detailed information, see (Chaillou et al., 2014; Kusnadi et al., 2015; Brook et al., 2019; Figueiredo and McCarthy, 2019; Kirby, 2019; von Walden, 2019)]. The MAPK-ERK kinase (MEK)/extracellular signal-regulated kinase (ERK) and AKT/mechanistic target of rapamycin (mTOR) signaling pathways plays a critical role in regulating rDNA transcription. For instance, ERK and mTOR complex 1 (mTORC1) activates TIF-1A, and ERK and ribosomal protein S6 kinase 1 (P70S6K1) activates UBF (Kusnadi et al., 2015). Moreover, mTOR stimulates rDNA transcription through its association with the rDNA promoter (von Walden et al., 2016). AKT also promotes rDNA transcription by improving Pol I loading on rDNA promoter and transcription elongation, which are mediated by mTORC1-dependent and independent mechanisms (Chan et al., 2011). The transcription factor MYC is another key regulator of ribosome biogenesis, through its direct and indirect influences on Pol I-mediated rDNA transcription (Kusnadi et al., 2015). MYC stimulates the transcription of several factors associated with Pol I-mediated rDNA transcription, including the core factors of PIC (UBF, TIF-1A and Pol I subunits). Furthermore, MYC can translocate into the nucleolus where it binds to the rDNA promoter, thereby activating Pol I transcription. It can also interact with SL-1, which facilitates the stabilization of UBF/SL-1 complex and the initiation of rDNA transcription. Furthermore, AMP-dependent protein kinase (AMPK) is a crucial sensor of metabolic stress, which is activated in response to depletion of adenosine triphosphate (ATP) and accumulation of adenosine monophosphate (AMP). AMPK can directly and indirectly inactivate mTORC1 by phosphorylation (Hardie, 2008), which thus potentially impair rDNA transcription and ribosome biogenesis (Kusnadi et al., 2015). AMPK could also prevent Pol I-mediated rDNA transcription by inhibiting the interaction between TIF-1A and Pol I complex with the rDNA promoter, which blunts PIC assembly (Hoppe et al., 2009).
Although Pol I-mediated rDNA transcription is considered as the rate-limiting step of ribosome biogenesis, other regulatory steps could be influenced. RPs and other components of the translational machinery are translated from 5′-terminal tract of pyrimidine (5′-TOP) mRNAs. P70S6K was proposed to promote the translation of 5′-TOP mRNAs (Jefferies et al., 1997), while AMPK could suppress the translation of specific 5′-TOP mRNAs (Reiter et al., 2008). MYC could also promote the transcription of several genes encoding RPs of 40S-SU (ribosomal protein small, RPS) and 60S-SU (ribosomal protein large, RPL) (Coller et al., 2000; Kim et al., 2000).
For the analysis of this mini-review, we selected studies comparing the level/expression of markers/regulators of skeletal muscle ribosome biogenesis between healthy young and healthy old humans/animals in response to mechanical load (RE in human studies; mechanical overload, and electrical stimulations in animal studies) (Table 1). We also included a few human studies combining RE and AA supplementation. These factors include markers of ribosome content/concentration (total RNA, rRNA, RPs), specific mediators of rDNA transcription by Pol I (TIF-1A, UBF, POL1s, TAFs, pre-47S rRNA) and important upstream regulators of ribosome biogenesis (MEK/ERK and AKT/mTOR pathways, MYC, AMPK) introduced in the previous section.
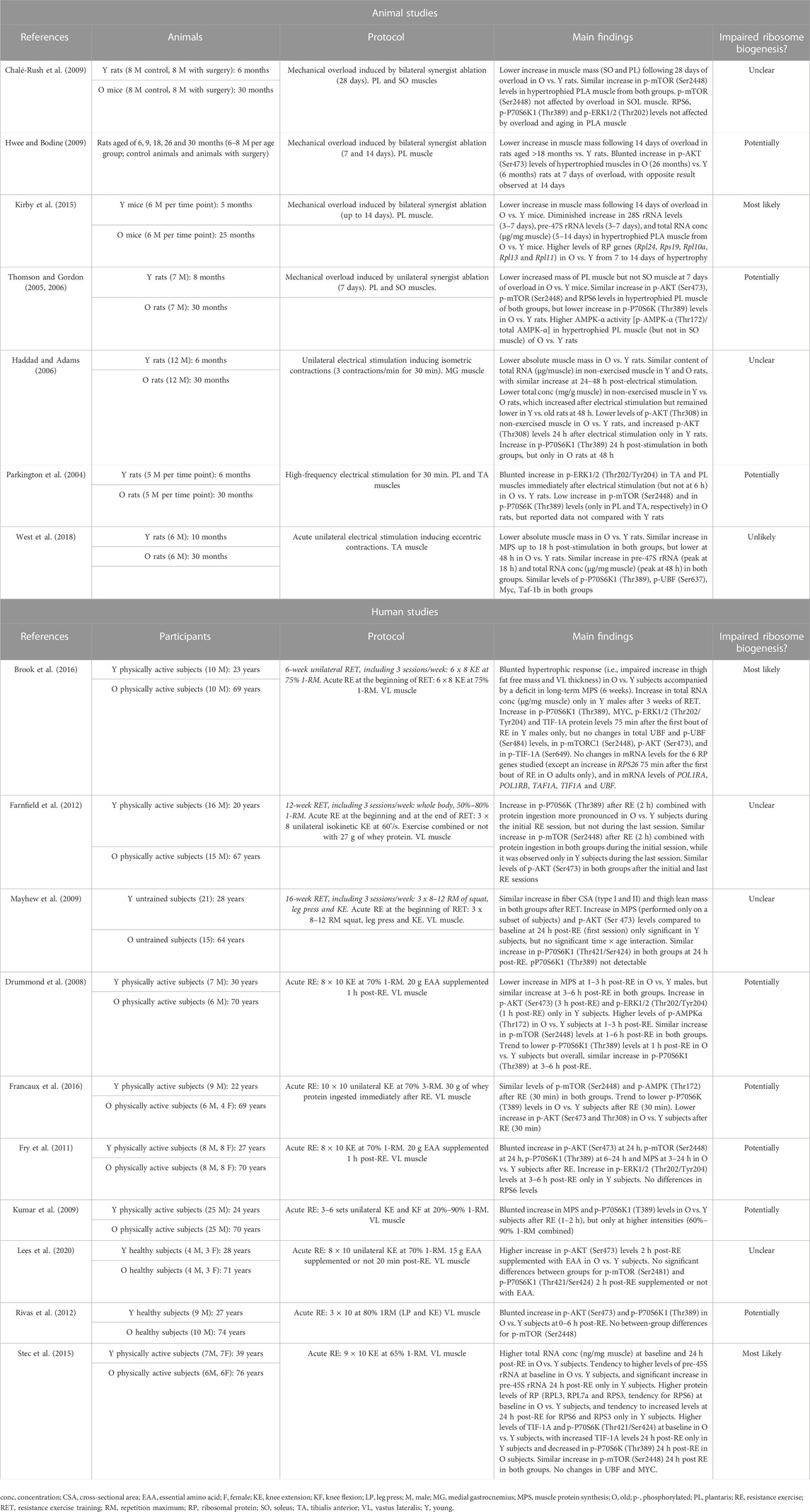
TABLE 1. Summary of the studies comparing the changes in ribosome biogenesis in response to mechanical load between young and old animals and subjects.
Several rodent studies indicate that aging impairs muscle hypertrophic response to mechanical overload induced by synergist ablation (Thomson and Gordon, 2005; Thomson and Gordon, 2006; Chalé-Rush et al., 2009; Hwee and Bodine, 2009; Kirby et al., 2015). This was accompanied by a lower increase in total RNA concentration in hypertrophied plantaris muscle from old compared with young mice (Kirby et al., 2015). This difference in total RNA concentration (µg/mg muscle) implies that total RNA content (µg/muscle) was reduced in old vs. young hypertrophied muscles due to the differences in muscle mass between the two groups. Given that rRNAs constitutes the largest proportion of total cellular RNA (ranging from 70% to 90%), reduced total RNA content indicates a decreased translational capacity mediated by impaired ribosome biogenesis (Chaillou et al., 2014; Figueiredo and McCarthy, 2019). Kirby et al., found that pre-47S rRNA and 28S rRNA levels paralleled the changes in total RNA concentration. In addition, they observed that the expression of numerous RP genes similarly increased in young and old hypertrophied muscles until 5 days of overload, after which some of these RP genes (especially Rpl11) remained only highly expressed in old muscles subjected to mechanical overload. This finding is consistent with previous results showing that the expression of numerous RP genes was negatively correlated with the increase in human lean mass after 20 weeks of resistance exercise training (RET) (Phillips et al., 2013). Although it remains to be investigated, impaired induction of ribosome biogenesis with age in response to mechanical load might be associated with changes in RP translation (all or specific RPs).
The activation of AKT/mTOR pathway is also attenuated by aging during the first week of overload (Thomson and Gordon, 2006; Hwee and Bodine, 2009). The increase in AKT phosphorylation in hypertrophied plantaris muscles (7 days of overload) was impaired in old compared with young rats (Hwee and Bodine, 2009), a result not confirmed in another study (Thomson and Gordon, 2006). However, the latter study observed that overload-mediated increase in P70S6K phosphorylation was attenuated in plantaris muscles from old vs. young rats. In addition, AMPK-α phosphorylation status (phosphorylated/total forms) was more elevated in overloaded plantaris muscles (but not in overloaded soleus muscles) from old than young rats, which is consistent with the diminished overload-induced hypertrophy by aging only observed in plantaris muscles (Thomson and Gordon, 2005). Furthermore, the attenuation of muscle hypertrophy by aging following prolonged overload (28 days), was not associated with age-differences in translational signaling (mTOR pathway and ERK1/2 phosphorylation) (Chalé-Rush et al., 2009). Altogether, aging blunts muscle hypertrophy induced by synergist ablation, and this corroborates with an altered ribosome biogenesis, as evidenced by a lower ribosome content (i.e., rRNA content), and an impaired rDNA transcription and blunted anabolic pathways during the early phase of muscle growth.
To our knowledge, no studies have investigated in a more physiological model inducing mild muscle growth, whether age-related attenuation of muscle hypertrophy is directly associated with impaired ribosome biogenesis. Some studies assessed markers/regulators of ribosome biogenesis following a single bout of electrical stimulations (Parkington et al., 2004; Haddad and Adams, 2006; West et al., 2018), a physiological model of RE known to stimulate MPS (Wong and Booth, 1990; West et al., 2018) and to moderately increase muscle mass (Baar and Esser, 1999). Following maximal eccentric contractions induced by electrical stimulations, MPS similarly increased in young and old rats at 18 h, while it remained only elevated in young animals at 48 h (West et al., 2018). No age-related differences were found for pre-47S rRNA, total RNA concentration, P70S6K1 and UBF phosphorylations, and Myc and Taf-1b mRNA. These findings suggest that ribosome biogenesis is not impaired by aging in response to electrical stimulation, and this cannot explain the age-mediated attenuation of MPS observed at 48 h. In another study using maximal isometric contractions induced by electrical stimulation (Haddad and Adams, 2006), total RNA content similarly increased at 24–48 h following exercise in young and old rats. Furthermore, the activation of several upstream regulators of ribosome biogenesis was impaired by aging (AKT, ERK1/2, mTOR, P70S6K) in response to electrical stimulation (Parkington et al., 2004; Haddad and Adams, 2006). Despite these results, the absence of specific impact of aging on ribosome content and Pol I-mediated rDNA transcription suggests that ribosome biogenesis is not altered by aging after a single bout of electrical stimulation. Further studies should investigate the effect of aging on muscle hypertrophy and translational capacity in response to chronic electrical stimulation.
Currently, only one study investigated the causes of age-related AR and attenuated muscle hypertrophic growth during a training intervention with a focus on ribosome biogenesis (Brook et al., 2016). Following 6 weeks of unilateral RET, muscle hypertrophy was blunted in old compared with young men, and this was accompanied by a deficit in long-term MPS. An increase in total RNA concentration was found in young but not in old males following 3 weeks of the training intervention, and similar results were observed for phosphorylated P70S6K1 and ERK1/2, MYC, and TIF-1A 75 min after the first RE session. Noteworthy, other factors associated with ribosome biogenesis were unaffected by aging in this study (phosphorylated forms of Akt, mTORC1, UBF, and TIF-1A, and 5 RP genes), while RPS26 mRNA only increased in old males 75 min after the first RE session (Table 1). Although blunted muscle hypertrophy and AR mediated by aging are certainly multifactorial, impaired ribosome biogenesis (and more likely rDNA transcription) seems to be one contributing factor of this age-related deficit. In another study, no clear age-differences in MPS and translational signaling (AKT and P70S6K1) were found 24 h following the first session of a 16-week RET program, which is consistent with the similar gains in lean mass and fiber size observed in young and old subjects after RET (Mayhew et al., 2009). Although type-II muscle fibers were smaller in old than in young individuals in this study, the absence of age-related differences in hypertrophic response is surprising and is in contrast to most studies published in the field (Welle et al., 1996; Kosek et al., 2006; Greig et al., 2011; Peterson et al., 2011; Mero et al., 2013; Brook et al., 2016). An age-related effect on translational signaling associated with ribosome biogenesis (AKT, mTOR and P70S6K) was also uncertain in another study where muscle biopsies were collected after the first and last sessions of a 12-week RET (Farnfield et al., 2012). Unfortunately, clear conclusions cannot be drawn from this study because changes in muscle size after RET were not assessed.
Recently, the magnitude of RET-induced muscle hypertrophy was assessed in relation to changes in ribosome biogenesis in old subjects (65 years of age) (Stec et al., 2016). Unfortunately, this study did not include a control group of young participants (study not presented in Table 1 for this reason). Following 4 weeks of RET (3 sessions/week), total RNA concentration increased in moderate and extreme responders, but not in nonresponders. RET led to a more robust increase in MYC levels in moderate and extreme responders compared with nonresponders, and rRNA abundance only increased in extreme responders after RET. Other studies have proposed that the extent of ribosome biogenesis may be predictive of the hypertrophic response irrespective of age (Kim et al., 2007; Figueiredo et al., 2015), while blunting Pol I-mediated rRNA transcription abolishes in vitro hypertrophy in human cells (Stec et al., 2016). Altogether, these findings support the idea that impaired ribosome biogenesis (especially Pol I rDNA transcription) may contribute to the limited hypertrophic response observed in aged populations displaying a chronic deficit in MPS during RET.
Currently, only one study compared Pol I-mediated rRNA transcription between old and young subjects following a single RE bout (Stec et al., 2015). Increased pre-45S rRNA levels 24 h following RE were observed in muscle from young but not old individuals. This was associated with a concomitant elevation of TIF-1A protein in the young group only, while UBF and MYC levels remained unchanged. Intriguingly, muscles from older participants had at baseline higher total RNA concentration, levels of several RPs and TIF-1A, and tended to present greater levels of pre-45S rRNA in comparison with younger participants. These results are in agreement with the higher muscle RNA concentration found in old than in young rats (Haddad and Adams, 2006) (Table 1), which according to Stec et al. (2015), might indicate an age-related accumulation of ribosome (potentially dysfunctional) at rest. Another possibility is that ribosome content (number/muscle) is not affected by aging in rested muscle, but its concentration (number/unit muscle) increases due to muscle atrophy (Figueiredo and McCarthy, 2019). This idea seems plausible since RNA content was similar in young and aged muscles at baseline, at least in rodents (Haddad and Adams, 2006). These two discordant hypotheses underline that accurate normalization of biological data in conditions of marked changes of muscle mass is crucial to properly interpret scientific results and avoid misleading conclusions (Chaillou et al., 2011).
Finally, other studies investigated the impact of aging on upstream regulators of muscle ribosome biogenesis after one RE session. An attenuated stimulation of MPS by aging is commonly observed following RE (Drummond et al., 2008; Kumar et al., 2009; Fry et al., 2011). This corroborates with an impaired activation of AKT/mTOR pathway and ERK1/2 (Drummond et al., 2008; Kumar et al., 2009; Fry et al., 2011; Rivas et al., 2012; Stec et al., 2015; Francaux et al., 2016), although one study did not confirm this result (Lees et al., 2020). AMPK phosphorylation was also higher in muscles from old compared with young participants at 1 and 3 h following RE (Drummond et al., 2008), which was not confirmed in another study ∼30 min after RE (Francaux et al., 2016). Factors such as protein supplementation (only applied in the latter study) and the time-points of tissue collection might explain the discrepancy between the two studies. In summary, the human studies focusing on the acute response to RE suggest that aging impairs Pol I rDNA transcription and limits the activation/expression of several upstream regulators of ribosome biogenesis, which may contribute to the attenuated stimulation of MPS.
Aging appears to impair muscle hypertrophic response to mechanical load in both animals and humans. This is associated with a deficit in long-term MPS (only studied in humans) and a blunted increase in RNA concentration. An attenuated acute stimulation of MPS and anabolic signaling by aging in response to mechanical load is commonly observed in animal and human studies. Although multifactorial, several lines of evidence indicate that AR to mechanical load partly results from the impaired induction of ribosome biogenesis (especially rDNA transcription by Pol I), which may contribute to the progressive decline in muscle mass with age.
The analysis presented in this mini-review focused on healthy aged humans and animals (Table 1). Aging is also associated with higher risks of developing diseases, such as cancer (White et al., 2014). Patients with advanced stage cancer are susceptible of suffering from cachexia, a wasting and multifactorial syndrome characterized by a severe loss of skeletal muscle mass with or without a reduction of body fat (Dodson et al., 2011). Skeletal muscle wasting is accompanied by a blunted protein synthetic capacity in preclinical models of cancer cachexia (Brown et al., 2018; Kim et al., 2021). This muscle anabolic deficit corroborates with an impaired ribosomal production in several models of cancer cachexia, including ovarian cancer (Kim et al., 2021), colorectal cancer (Kim et al., 2022), and lung cancer (Belcher et al., 2022). Specifically, a reduced rRNA concentration, and an impaired Pol I-mediated rDNA transcription initiation and/or elongation were found in these three types of cancer (Kim et al., 2021; Belcher et al., 2022; Kim et al., 2022). Nevertheless, some divergences in ribosomal capacity were observed in these studies, which were likely due to differences in tumor types (e.g., LP07 and LLC models of lung cancer) and tumor burdens (e.g., metastatic and xenograft tumors) (Belcher et al., 2022; Kim et al., 2022). Although these findings remain to be confirmed in clinical studies with cancer patients, impaired ribosome biogenesis could contribute to anabolic deficits associated with cancer cachexia. Chemotherapeutic agents, which are commonly used to treat cancer and blunt tumor progression, have also a detrimental effect on skeletal muscle mass (Barreto et al., 2016; Damrauer et al., 2018). A recent study showed in C2C12 myotubes that exposure to chemotherapeutic agents reduced protein synthesis, rRNA content, and Pol I-mediated rDNA transcription at the initiation step, suggesting that chemotherapy-induced anabolic deficits may partly result from impaired ribosome production (Guo et al., 2021).
Muscle disuse, which is commonly observed in older individuals due to frequent hospitalizations, immobilization, or bed rest, leads to a rapid loss of skeletal muscle mass and force (Urso et al., 2006; Kortebein et al., 2007). A recent study indicates in middle-aged rats (10 months) that muscle disuse (hindlimb suspension) reduces Pol I-mediated rDNA transcription, total RNA concentration and RNA synthesis, and increases RNA degradation (Figueiredo et al., 2021a). A reduced total RNA concentration was also observed in the latter study in healthy middle-aged men (50 years) after 2 weeks of knee immobilization. These findings provide strong evidence that translational capacity is impaired during muscle disuse, which could contribute to muscle atrophy. Since elderly population has higher risks of developing cancer and being hospitalized or immobilized, impaired ribosome biogenesis and translational capacity could be common mechanisms contributing to muscle wasting in aging population exhibiting sarcopenia, cachexia and muscle disuse.
The selected studies of our analysis essentially used standard methods (i.e., immunoblotting, quantitative Polymerase Chain Reaction, and spectrophotometry) to compare the level/expression of markers/regulators of ribosome biogenesis between young and old skeletal muscles in response to mechanical load. These methods are unfortunately limited to capture complex molecular events and provide an in-depth analysis of ribosome biogenesis. In addition, current knowledge on muscle ribosome biogenesis is recent (10–15 years) and has mainly focused on rDNA transcription. Future research should develop more advanced molecular tools and innovative technologies. For instance, proteomic analyses and translatomic methods such as ribosomal profiling could be used to assess changes in RP translation (Brar and Weissman, 2015). A recent study performed in skeletal muscle of older individuals (72 years) and combining ribosomal profiling and RNA-sequencing revealed a reduced translation of most mRNAs encoding for RPs in response to physical inactivity, while the opposite result was found after leucine ingestion (Mahmassani et al., 2021). Using identical methods, another study from the same group found in adult mice that leucine administration increased the translation of ribosomal transcripts (32 Rpl and 25 Rps) and the abundance of Snord22 and Snord82, two small nucleolar RNAs essential for rRNA maturation and ribosome biogenesis (Drummond et al., 2017). Future studies should use these innovative tools to investigate whether RP translation and other regulatory processes of ribosome biogenesis are impaired with age in response to mechanical load. Because muscle cells do not exclusively contribute to global gene expression in a baseline state and during skeletal muscle hypertrophy (Murach et al., 2022), combining ribosome profiling with a ribosome-tagging approach in animal models would be of interest to answer the latter research question specifically in muscle fibers during aging (Doroudgar et al., 2019).
The results from Mahmassani et al. (2021) indicate that changes in RP translation observed in skeletal muscle from aged adults in response to physical inactivity or leucine ingestion are not specific to the small or large ribosome subunit. Nevertheless, a recent study performed in Drosophila showed that the mRNA levels of RpS28a and RpS28-like declined in skeletal muscle during aging (Jiao et al., 2021). In addition, the results reported in Table 1 suggest that aging increases the level of some specific RP transcripts in response to mechanical load in both animals (Kirby et al., 2015) and humans (Brook et al., 2016), and increases the abundance of some ribosomal proteins at baseline in humans (Stec et al., 2015). Since manipulation of specific ribosome proteins appears to influence skeletal muscle growth, at least in vitro (Chaillou et al., 2015), the possibility of an age effect on muscle ribosome composition, in particular in response to anabolic stimuli such as mechanical load, deserves further consideration. The concepts of ribosome heterogeneity and functional ribosome specialization are relatively recent and currently poorly explored in skeletal muscle (Chaillou, 2019). The impact of aging on muscle RP composition and stoichiometry of both polysomes and free subunits could be investigated by employing selected reaction monitoring-based proteomics and mass spectrometry using tandem mass tag technology (Shi et al., 2017). Finally, some other emerging fields such as genetic and epigenetic regulation of ribosome biogenesis (Figueiredo et al., 2021b) and ribophagy (Figueiredo et al., 2021a) would also be of interest to explore ribosome turnover in aged muscle.
TC contributed to the development of the idea for the review. TC and DM-R contributed equally for the search on literature, discuss the main points of the theme, and wrote the review. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. All authors contributed to the article and approved the submitted version.
Publication fees were covered by Örebro University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baar, K., and Esser, K. (1999). Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am. J. Physiol. 276 (1), C120–C127. doi:10.1152/ajpcell.1999.276.1.C120
Barreto, R., Mandili, G., Witzmann, F. A., Novelli, F., Zimmers, T. A., and Bonetto, A. (2016). Cancer and chemotherapy contribute to muscle loss by activating common signaling pathways. Front. Physiol. 7, 472. doi:10.3389/fphys.2016.00472
Batsis, J. A., Mackenzie, T. A., Barre, L. K., Lopez-Jimenez, F., and Bartels, S. J. (2014). Sarcopenia, sarcopenic obesity and mortality in older adults: Results from the national health and nutrition examination survey III. Eur. J. Clin. Nutr. 68 (9), 1001–1007. doi:10.1038/ejcn.2014.117
Belcher, D. J., Guitart, M., Hain, B., Kim, H. G., Waning, D., Barreiro, E., et al. (2022). LP07 and LLC preclinical models of lung cancer induce divergent anabolic deficits and expression of pro-inflammatory effectors of muscle wasting. J. Appl. Physiol. (1985) 133(6), 1260–1272. doi:10.1152/japplphysiol.00246.2022
Brar, G. A., and Weissman, J. S. (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 16 (11), 651–664. doi:10.1038/nrm4069
Brook, M. S., Wilkinson, D. J., Mitchell, W. K., Lund, J. N., Phillips, B. E., Szewczyk, N. J., et al. (2016). Synchronous deficits in cumulative muscle protein synthesis and ribosomal biogenesis underlie age-related anabolic resistance to exercise in humans. J. Physiol. 594, 7399–7417. doi:10.1113/jp272857
Brook, M. S., Wilkinson, D. J., Smith, K., and Atherton, P. J. (2019). It's not just about protein turnover: The role of ribosomal biogenesis and satellite cells in the regulation of skeletal muscle hypertrophy. Eur. J. Sport Sci. 19, 952–963. doi:10.1080/17461391.2019.1569726
Brown, J. L., Lee, D. E., Rosa-Caldwell, M. E., Brown, L. A., Perry, R. A., Haynie, W. S., et al. (2018). Protein imbalance in the development of skeletal muscle wasting in tumour-bearing mice. J. Cachexia Sarcopenia Muscle 9 (5), 987–1002. doi:10.1002/jcsm.12354
Chaillou, T., Kirby, T. J., and McCarthy, J. J. (2014). Ribosome biogenesis: Emerging evidence for a central role in the regulation of skeletal muscle mass. J. Cell Physiol. 229, 1584–1594. doi:10.1002/jcp.24604
Chaillou, T., Malgoyre, A., Banzet, S., Chapot, R., Koulmann, N., Pugniere, P., et al. (2011). Pitfalls in target mRNA quantification for real-time quantitative RT-PCR in overload-induced skeletal muscle hypertrophy. Physiol. Genomics 43 (4), 228–235. doi:10.1152/physiolgenomics.00109.2010
Chaillou, T. (2019). Ribosome specialization and its potential role in the control of protein translation and skeletal muscle size. J. Appl. Physiol. (1985) 127(2), 599–607. doi:10.1152/japplphysiol.00946.2018
Chaillou, T., Zhang, X., and McCarthy, J. J. (2015). Expression of muscle-specific ribosomal protein L3-like impairs myotube growth. J. Cell Physiol. 231, 1894–1902. doi:10.1002/jcp.25294
Chalé-Rush, A., Morris, E. P., Kendall, T. L., Brooks, N. E., and Fielding, R. A. (2009). Effects of chronic overload on muscle hypertrophy and mTOR signaling in young adult and aged rats. J. Gerontol. A Biol. Sci. Med. Sci. 64 (12), 1232–1239. doi:10.1093/gerona/glp146
Chan, J. C., Hannan, K. M., Riddell, K., Ng, P. Y., Peck, A., Lee, R. S., et al. (2011). AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci. Signal 4 (188), ra56. doi:10.1126/scisignal.2001754
Coller, H. A., Grandori, C., Tamayo, P., Colbert, T., Lander, E. S., Eisenman, R. N., et al. (2000). Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. U. S. A. 97 (7), 3260–3265. doi:10.1073/pnas.97.7.3260
Cruz-Jentoft, A. J., Bahat, G., Bauer, J., Boirie, Y., Bruyère, O., Cederholm, T., et al. (2019). Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 48 (1), 16–31. doi:10.1093/ageing/afy169
Cruz-Jentoft, A. J., and Sayer, A. A. (2019). Sarcopenia. Lancet 393 (10191), 2636–2646. doi:10.1016/s0140-6736(19)31138-9
Cuthbertson, D., Smith, K., Babraj, J., Leese, G., Waddell, T., Atherton, P., et al. (2005). Anabolic signaling deficits underlie amino acid resistance of wasting, aging muscle. Faseb J. 19 (3), 422–424. doi:10.1096/fj.04-2640fje
Damrauer, J. S., Stadler, M. E., Acharyya, S., Baldwin, A. S., Couch, M. E., and Guttridge, D. C. (2018). Chemotherapy-induced muscle wasting: Association with NF-κB and cancer cachexia. Eur. J. Transl. Myol. 28 (2), 7590. doi:10.4081/ejtm.2018.7590
Dodson, S., Baracos, V. E., Jatoi, A., Evans, W. J., Cella, D., Dalton, J. T., et al. (2011). Muscle wasting in cancer cachexia: Clinical implications, diagnosis, and emerging treatment strategies. Annu. Rev. Med. 62, 265–279. doi:10.1146/annurev-med-061509-131248
Doroudgar, S., Hofmann, C., Boileau, E., Malone, B., Riechert, E., Gorska, A. A., et al. (2019). Monitoring cell-type-specific gene expression using ribosome profiling in vivo during cardiac hemodynamic stress. Circ. Res. 125 (4), 431–448. doi:10.1161/circresaha.119.314817
Drummond, M. J., Dreyer, H. C., Pennings, B., Fry, C. S., Dhanani, S., Dillon, E. L., et al. (2008). Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J. Appl. Physiol. (1985) 104 (5), 1452–1461. doi:10.1152/japplphysiol.00021.2008
Drummond, M. J., Reidy, P. T., Baird, L. M., Dalley, B. K., and Howard, M. T. (2017). Leucine differentially regulates gene-specific translation in mouse skeletal muscle. J. Nutr. 147, 1616–1623. doi:10.3945/jn.117.251181
Farnfield, M. M., Breen, L., Carey, K. A., Garnham, A., and Cameron-Smith, D. (2012). Activation of mTOR signalling in young and old human skeletal muscle in response to combined resistance exercise and whey protein ingestion. Appl. Physiol. Nutr. Metab. 37 (1), 21–30. doi:10.1139/h11-132
Figueiredo, V. C., Caldow, M. K., Massie, V., Markworth, J. F., Cameron-Smith, D., and Blazevich, A. J. (2015). Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. Am. J. Physiol. Endocrinol. Metab. 309 (1), E72–E83. doi:10.1152/ajpendo.00050.2015
Figueiredo, V. C., D'Souza, R. F., Van Pelt, D. W., Lawrence, M. M., Zeng, N., Markworth, J. F., et al. (2021a). Ribosome biogenesis and degradation regulate translational capacity during muscle disuse and reloading. J. Cachexia Sarcopenia Muscle 12 (1), 130–143. doi:10.1002/jcsm.12636
Figueiredo, V. C., and McCarthy, J. J. (2019). Regulation of ribosome biogenesis in skeletal muscle hypertrophy. Physiol. (Bethesda) 34 (1), 30–42. doi:10.1152/physiol.00034.2018
Figueiredo, V. C., Wen, Y., Alkner, B., Fernandez-Gonzalo, R., Norrbom, J., Vechetti, I. J., et al. (2021b). Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J. Physiol. 599 (13), 3363–3384. doi:10.1113/jp281244
Francaux, M., Demeulder, B., Naslain, D., Fortin, R., Lutz, O., Caty, G., et al. (2016). Aging reduces the activation of the mTORC1 pathway after resistance exercise and protein intake in human skeletal muscle: Potential role of REDD1 and impaired anabolic sensitivity. Nutrients 8 (1), 47. doi:10.3390/nu8010047
Fry, C. S., Drummond, M. J., Glynn, E. L., Dickinson, J. M., Gundermann, D. M., Timmerman, K. L., et al. (2011). Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skelet. Muscle 1 (1), 11. doi:10.1186/2044-5040-1-11
Goodfellow, S. J., and Zomerdijk, J. C. (2013). Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell. Biochem. 61, 211–236. doi:10.1007/978-94-007-4525-4_10
Gorski, J. J., Pathak, S., Panov, K., Kasciukovic, T., Panova, T., Russell, J., et al. (2007). A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. Embo J. 26 (6), 1560–1568. doi:10.1038/sj.emboj.7601601
Greig, C. A., Gray, C., Rankin, D., Young, A., Mann, V., Noble, B., et al. (2011). Blunting of adaptive responses to resistance exercise training in women over 75y. Exp. Gerontol. 46 (11), 884–890. doi:10.1016/j.exger.2011.07.010
Guo, B., Bennet, D., Belcher, D. J., Kim, H. G., and Nader, G. A. (2021). Chemotherapy agents reduce protein synthesis and ribosomal capacity in myotubes independent of oxidative stress. Am. J. Physiol. Cell Physiol. 321 (6), C1000–C1009. doi:10.1152/ajpcell.00116.2021
Haddad, F., and Adams, G. R. (2006). Aging-sensitive cellular and molecular mechanisms associated with skeletal muscle hypertrophy. J. Appl. Physiol. (1985) 100(4), 1188–1203. doi:10.1152/japplphysiol.01227.2005
Hardie, D. G. (2008). AMPK and raptor: Matching cell growth to energy supply. Mol. Cell 30 (3), 263–265. doi:10.1016/j.molcel.2008.04.012
Hoppe, S., Bierhoff, H., Cado, I., Weber, A., Tiebe, M., Grummt, I., et al. (2009). AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. U. S. A. 106 (42), 17781–17786. doi:10.1073/pnas.0909873106
Hwee, D. T., and Bodine, S. C. (2009). Age-related deficit in load-induced skeletal muscle growth. J. Gerontol. A Biol. Sci. Med. Sci. 64 (6), 618–628. doi:10.1093/gerona/glp026
Jefferies, H. B., Fumagalli, S., Dennis, P. B., Reinhard, C., Pearson, R. B., and Thomas, G. (1997). Rapamycin suppresses 5'TOP mRNA translation through inhibition of p70s6k. EMBO J. 16 (12), 3693–3704. doi:10.1093/emboj/16.12.3693
Jiao, J., Kavdia, K., Pagala, V., Palmer, L., Finkelstein, D., Fan, Y., et al. (2021). An age-downregulated ribosomal RpS28 protein variant regulates the muscle proteome. G3 (Bethesda) 11 (7), jkab165. doi:10.1093/g3journal/jkab165
Kanova, M., and Kohout, P. (2022). Molecular mechanisms underlying intensive care unit-acquired weakness and sarcopenia. Int. J. Mol. Sci. 23 (15), 8396. doi:10.3390/ijms23158396
Katsanos, C. S., Kobayashi, H., Sheffield-Moore, M., Aarsland, A., and Wolfe, R. R. (2005). Aging is associated with diminished accretion of muscle proteins after the ingestion of a small bolus of essential amino acids. Am. J. Clin. Nutr. 82 (5), 1065–1073. doi:10.1093/ajcn/82.5.1065
Khatter, H., Myasnikov, A. G., Natchiar, S. K., and Klaholz, B. P. (2015). Structure of the human 80S ribosome. Nature 520 (7549), 640–645. doi:10.1038/nature14427
Kim, H. G., Huot, J. R., Pin, F., Belcher, D. J., Bonetto, A., and Nader, G. A. (2022). Metastatic or xenograft colorectal cancer models induce divergent anabolic deficits and expression of pro-inflammatory effectors of muscle wasting in a tumor-type-dependent manner. J. Appl. Physiol. (1985) 133(6), 1273–1283. doi:10.1152/japplphysiol.00247.2022
Kim, H. G., Huot, J. R., Pin, F., Guo, B., Bonetto, A., and Nader, G. A. (2021). Reduced rDNA transcription diminishes skeletal muscle ribosomal capacity and protein synthesis in cancer cachexia. FASEB J. 35 (2), e21335. doi:10.1096/fj.202002257R
Kim, J. S., Petrella, J. K., Cross, J. M., and Bamman, M. M. (2007). Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: A cluster analysis. J. Appl. Physiol. (1985) 103(5), 1488–1495. doi:10.1152/japplphysiol.01194.2006
Kim, S., Li, Q., Dang, C. V., and Lee, L. A. (2000). Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl. Acad. Sci. U. S. A. 97 (21), 11198–11202. doi:10.1073/pnas.200372597
Kirby, T. J., Lee, J. D., England, J. H., Chaillou, T., Esser, K. A., and McCarthy, J. J. (2015). Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J. Appl. Physiol. (1985) 119 (4), 321–327. doi:10.1152/japplphysiol.00296.2015
Kirby, T. J. (2019). Mechanosensitive pathways controlling translation regulatory processes in skeletal muscle and implications for adaptation. J. Appl. Physiol. (1985) 127(2), 608–618. doi:10.1152/japplphysiol.01031.2018
Kortebein, P., Ferrando, A., Lombeida, J., Wolfe, R., and Evans, W. J. (2007). Effect of 10 days of bed rest on skeletal muscle in healthy older adults. JAMA 297 (16), 1772–1774. doi:10.1001/jama.297.16.1772-b
Kosek, D. J., Kim, J. S., Petrella, J. K., Cross, J. M., and Bamman, M. M. (2006). Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J. Appl. Physiol. (1985) 101 (2), 531–544. doi:10.1152/japplphysiol.01474.2005
Kumar, V., Selby, A., Rankin, D., Patel, R., Atherton, P., Hildebrandt, W., et al. (2009). Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. J. Physiol. 587 (1), 211–217. doi:10.1113/jphysiol.2008.164483
Kusnadi, E. P., Hannan, K. M., Hicks, R. J., Hannan, R. D., Pearson, R. B., and Kang, J. (2015). Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 556 (1), 27–34. doi:10.1016/j.gene.2014.11.010
Lees, M. J., Wilson, O. J., Webb, E. K., Traylor, D. A., Prior, T., Elia, A., et al. (2020). Novel essential amino acid supplements following resistance exercise induce aminoacidemia and enhance anabolic signaling irrespective of age: A proof-of-concept trial. Nutrients 12 (7), 2067. doi:10.3390/nu12072067
Mahmassani, Z. S., McKenzie, A. I., Petrocelli, J. J., de Hart, N. M., Fix, D. K., Kelly, J. J., et al. (2021). Reduced physical activity alters the leucine-stimulated translatome in aged skeletal muscle. J. Gerontol. A Biol. Sci. Med. Sci. 76 (12), 2112–2121. doi:10.1093/gerona/glab077
Mayhew, D. L., Kim, J. S., Cross, J. M., Ferrando, A. A., and Bamman, M. M. (2009). Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J. Appl. Physiol. (1985) 107(5), 1655–1662. doi:10.1152/japplphysiol.91234.2008
Mero, A. A., Hulmi, J. J., Salmijärvi, H., Katajavuori, M., Haverinen, M., Holviala, J., et al. (2013). Resistance training induced increase in muscle fiber size in young and older men. Eur. J. Appl. Physiol. 113 (3), 641–650. doi:10.1007/s00421-012-2466-x
Murach, K. A., Liu, Z., Jude, B., Figueiredo, V. C., Wen, Y., Khadgi, S., et al. (2022). Multi-transcriptome analysis following an acute skeletal muscle growth stimulus yields tools for discerning global and MYC regulatory networks. J. Biol. Chem. 298 (11), 102515. doi:10.1016/j.jbc.2022.102515
Nilwik, R., Snijders, T., Leenders, M., Groen, B. B., van Kranenburg, J., Verdijk, L. B., et al. (2013). The decline in skeletal muscle mass with aging is mainly attributed to a reduction in type II muscle fiber size. Exp. Gerontol. 48 (5), 492–498. doi:10.1016/j.exger.2013.02.012
Parkington, J. D., LeBrasseur, N. K., Siebert, A. P., and Fielding, R. A. (2004). Contraction-mediated mTOR, p70S6k, and ERK1/2 phosphorylation in aged skeletal muscle. J. Appl. Physiol. (1985) 97 (1), 243–248. doi:10.1152/japplphysiol.01383.2003
Peterson, M. D., Sen, A., and Gordon, P. M. (2011). Influence of resistance exercise on lean body mass in aging adults: A meta-analysis. Med. Sci. Sports Exerc 43 (2), 249–258. doi:10.1249/MSS.0b013e3181eb6265
Phillips, B. E., Williams, J. P., Gustafsson, T., Bouchard, C., Rankinen, T., Knudsen, S., et al. (2013). Molecular networks of human muscle adaptation to exercise and age. PLoS Genet. 9 (3), e1003389. doi:10.1371/journal.pgen.1003389
Prokopidis, K., Chambers, E., Ni Lochlainn, M., and Witard, O. C. (2021). Mechanisms linking the gut-muscle Axis with muscle protein metabolism and anabolic resistance: Implications for older adults at risk of sarcopenia. Front. Physiol. 12, 770455. doi:10.3389/fphys.2021.770455
Reiter, A. K., Bolster, D. R., Crozier, S. J., Kimball, S. R., and Jefferson, L. S. (2008). AMPK represses TOP mRNA translation but not global protein synthesis in liver. Biochem. Biophys. Res. Commun. 374 (2), 345–350. doi:10.1016/j.bbrc.2008.07.025
Rivas, D. A., Morris, E. P., Haran, P. H., Pasha, E. P., Morais Mda, S., Dolnikowski, G. G., et al. (2012). Increased ceramide content and NFκB signaling may contribute to the attenuation of anabolic signaling after resistance exercise in aged males. J. Appl. Physiol. (1985) 113 (11), 1727–1736. doi:10.1152/japplphysiol.00412.2012
Shi, Z., Fujii, K., Kovary, K. M., Genuth, N. R., Rost, H. L., Teruel, M. N., et al. (2017). Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67 (1), 71–83. doi:10.1016/j.molcel.2017.05.021
Stec, M. J., Kelly, N. A., Many, G. M., Windham, S. T., Tuggle, S. C., and Bamman, M. M. (2016). Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. Am. J. Physiol. Endocrinol. Metab. Ajpendo. 310, E652. doi:10.1152/ajpendo.00486.2015
Stec, M. J., Mayhew, D. L., and Bamman, M. M. (2015). The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J. Appl. Physiol. (1985) 119 (8), 851–857. doi:10.1152/japplphysiol.00489.2015
Thomson, D. M., and Gordon, S. E. (2005). Diminished overload-induced hypertrophy in aged fast-twitch skeletal muscle is associated with AMPK hyperphosphorylation. J. Appl. Physiol. 98 (2), 557–564. doi:10.1152/japplphysiol.00811.2004
Thomson, D. M., and Gordon, S. E. (2006). Impaired overload-induced muscle growth is associated with diminished translational signalling in aged rat fast-twitch skeletal muscle. J. Physiol. 574 (1), 291–305. doi:10.1113/jphysiol.2006.107490
Thomson, E., Ferreira-Cerca, S., and Hurt, E. (2013). Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126 (21), 4815–4821. doi:10.1242/jcs.111948
Urso, M. L., Clarkson, P. M., and Price, T. B. (2006). Immobilization effects in young and older adults. Eur. J. Appl. Physiol. 96 (5), 564–571. doi:10.1007/s00421-005-0109-1
Volpi, E., Sheffield-Moore, M., Rasmussen, B. B., and Wolfe, R. R. (2001). Basal muscle amino acid kinetics and protein synthesis in healthy young and older men. Jama 286 (10), 1206–1212. doi:10.1001/jama.286.10.1206
von Walden, F., Liu, C., Aurigemma, N., and Nader, G. A. (2016). mTOR signaling regulates myotube hypertrophy by modulating protein synthesis, rDNA transcription, and chromatin remodeling. Am. J. Physiol. Cell Physiol. 311 (4), C663–c672. doi:10.1152/ajpcell.00144.2016
von Walden, F. (2019). Ribosome biogenesis in skeletal muscle: Coordination of transcription and translation. J. Appl. Physiol. (1985) 127 (2), 591–598. doi:10.1152/japplphysiol.00963.2018
Welle, S., Totterman, S., and Thornton, C. (1996). Effect of age on muscle hypertrophy induced by resistance training. J. Gerontol. A Biol. Sci. Med. Sci. 51 (6), M270–M275. doi:10.1093/gerona/51a.6.m270
West, D. W. D., Marcotte, G. R., Chason, C. M., Juo, N., Baehr, L. M., Bodine, S. C., et al. (2018). Normal ribosomal biogenesis but shortened protein synthetic response to acute eccentric resistance exercise in old skeletal muscle. Front. Physiol. 9, 1915. doi:10.3389/fphys.2018.01915
White, M. C., Holman, D. M., Boehm, J. E., Peipins, L. A., Grossman, M., and Henley, S. J. (2014). Age and cancer risk: A potentially modifiable relationship. Am. J. Prev. Med. 46 (3), S7–S15. doi:10.1016/j.amepre.2013.10.029
Wilkinson, D. J., Piasecki, M., and Atherton, P. J. (2018). The age-related loss of skeletal muscle mass and function: Measurement and physiology of muscle fibre atrophy and muscle fibre loss in humans. Ageing Res. Rev. 47, 123–132. doi:10.1016/j.arr.2018.07.005
Wong, T. S., and Booth, F. W. (1990). Protein metabolism in rat tibialis anterior muscle after stimulated chronic eccentric exercise. J. Appl. Physiol. (1985) 69 (5), 1718–1724. doi:10.1152/jappl.1990.69.5.1718
Keywords: anabolic resistance, resistance exercise, elderly, translational capacity, sarcopenia, muscle atrophy, rDNA transcription
Citation: Chaillou T and Montiel-Rojas D (2023) Does the blunted stimulation of skeletal muscle protein synthesis by aging in response to mechanical load result from impaired ribosome biogenesis?. Front. Aging 4:1171850. doi: 10.3389/fragi.2023.1171850
Received: 22 February 2023; Accepted: 04 May 2023;
Published: 15 May 2023.
Edited by:
Marco P. Brotto, University of Texas at Arlington, United StatesReviewed by:
Arpit Sharma, Harvard University, United StatesCopyright © 2023 Chaillou and Montiel-Rojas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Chaillou, dGhvbWFzLmNoYWlsbG91QG9ydS5zZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.