- Max Planck Institute for Infection Biology, Berlin, Germany
Intestinal microbial communities participate in essential aspects of host biology, including nutrient acquisition, development, immunity, and metabolism. During host aging, dramatic shifts occur in the composition, abundance, and function of the gut microbiota. Although such changes in the microbiota are conserved across species, most studies remain descriptive and at most suggest a correlation between age-related pathology and particular microbes. Therefore, the causal role of the microbiota in host aging has remained a challenging question, in part due to the complexity of the mammalian intestinal microbiota, most of which is not cultivable or genetically amenable. Here, we summarize recent studies in the fruit fly Drosophila melanogaster that have substantially progressed our understanding at the mechanistic level of how gut microbes can modulate host aging.
Introduction
Biological aging is a complex, multi-factorial phenomenon regulated by a combination of genetic and environmental factors. Genetic studies in laboratory model organisms identified several conserved host pathways, including the insulin/insulin-like growth factor signaling (IIS) and mechanistic Target of Rapamycin (mTOR), which negatively regulate the lifespan of multiple species (Kapahi et al., 2010; Lapierre and Hansen, 2012; Fernandes and Demetriades, 2021). Environmental perturbations, such as changes in temperature, dietary restriction, and stress, can similarly significantly affect life expectancy, and aging in experimental organisms (Valenzano et al., 2006; Fontana et al., 2010; Tatar et al., 2014; Regan et al., 2016; Dutta et al., 2022; Vakkayil and Hoppe, 2022).
Residing at the interface between the host organisms and the environment, commensal microbial communities participate in multiple essential processes, including development, synthesis of essential vitamins, metabolism, immune system modulation, and defense against pathogens (Nicholson et al., 2012; Sommer and Bäckhed, 2013; Geva-Zatorsky et al., 2017; Pickard et al., 2017; Sannino et al., 2018; Ducarmon et al., 2019; Consuegra et al., 2020).
Not surprisingly, there is accumulating evidence that the gut microbiota also plays a significant role in longevity across species (O’Toole and Jeffery, 2015; Clark and Walker, 2018; Kim and Jazwinski, 2018; Valenzano and Seidel, 2018; Bana and Cabreiro, 2019). For example, studies using different human age cohorts identified differences in microbiota composition across various age groups (Biagi et al., 2016). Overall, several studies found that microbial diversity in the gut declines with age (Yatsunenko et al., 2012; Biagi et al., 2016; Leite et al., 2021). However, health-associated genera, such as Christensenella, Akkermansia, and Bifidobacterium, were consistently found in exceptionally long-lived individuals, like supercentenarians, suggesting their potential life span–promoting effects (Biagi et al., 2016).
Although the composition of microbiota varies among taxa, similar to what has been observed in humans, extensive remodeling of the gut microbial communities during aging has also been identified in several model organisms, such as the nematode Caenorhabditis elegans (Cabreiro and Gems, 2013; Han B. et al., 2017), the fly Drosophila melanogaster (Clark et al., 2015; Li et al., 2016), the fish Nothobranchius furzeri (Smith et al., 2017) and the mouse Mus musculus (Langille et al., 2014). Whether such age-related changes in the microbiota are causative or a consequence of the aging of the organism remains a challenging question to be answered.
This review summarizes the insights into the intricate relationship between the gut microbiome and host aging obtained using the Drosophila model.
Several features of Drosophila microbiota laid the foundations for the successful use of the fruit fly model in microbiome research. The simple taxonomic composition combined with the cultivability and genetic tractability of the members of the fly microbiota enables functional studies and deciphering of the molecular mechanisms of commensal influence on the host physiology (Douglas, 2018; Douglas, 2019; Grenier and Leulier, 2020; Lesperance and Broderick, 2020). A wealth of genetic, genomic, and molecular resources available in Drosophila helps in the study of host mechanisms of microbiota control and host factors targeted by the commensals (Hales et al., 2015; Ludington and Ja, 2020). The simplicity of generating and maintaining germ-free, also known as axenic, animals is another particular advantage of the Drosophila model. Furthermore, gnotobiotic animals colonized with a standardized microbiota can be easily obtained (Douglas, 2018; Ludington and Ja, 2020).
Not surprisingly, due to its exceptional amenability to the experimental manipulations of the microbiome, Drosophila has been extensively used to study the impact of microbiota on various physiological processes, including aging (Kuraishi et al., 2013; Trinder et al., 2017; Gould et al., 2018; Ludington and Ja, 2020; Kong et al., 2021; Hrdina and Iatsenko, 2022).
Composition and Maintenance of Drosophila Microbiota
Drosophila melanogaster harbors relatively simple microbial communities (2–30 species) in the laboratory and in the field, represented by only two phyla, Proteobacteria and Firmicutes, and is dominated by two prominent families, Acetobacteraceae and Lactobacillaceae and two minor families, Enterococceae and Enterobacteriaceae (Staubach et al., 2013; Wong et al., 2013; Adair et al., 2018). The most consistently associated species across different studies are Lactiplantibacillus plantarum, Levilactobacillus brevis, Acetobacter pomorum, A. pasteurianus, and Enterococcus faecalis (Broderick et al., 2014; Erkosar and Leulier, 2014; Grenier and Leulier, 2020; Lesperance and Broderick, 2020; Ludington and Ja, 2020). Such a community of lactic acid and acetic acid bacteria reflects the fermentative substrates on which flies feed (Chandler et al., 2011; Broderick and Lemaitre, 2012). Diet (substrate) plays an essential role in shaping the Drosophila microbiota as the establishment and maintenance of Drosophila intestinal commensals relies on the constant intake of microbes from the diet (Erkosar et al., 2014). The vast majority of the intestinal microbes of the fruit flies cannot stably persist in the gut and must be constantly re-ingested with the food (Blum et al., 2013; Broderick et al., 2014; Storelli et al., 2018). The gut of newly emerged flies is colonized with a low number of microbes. However, these flies acquire their microbiota within the first day of their adult life by ingesting bacteria from the food contaminated by the feces of their parents (Blum et al., 2013; Broderick et al., 2014). Additionally, females transmit their microbiota to the offspring by seeding the eggshells of their progenies. Hatching larvae get colonized by eating the contaminated eggshell and ingesting the microbe-rich food on which the bacteria thrive (Erkosar et al., 2014; Storelli et al., 2018). Such associations between D. melanogaster, microbiota, and nutrition likely contribute to the high variability in composition and density of microbiota observed between individual flies reared in the same culture vial. The bacterial load can vary by one log between individual co-housed flies (Broderick et al., 2014). Also, flies that are frequently transferred to sterile food, which prevents the re-ingestion of microbes from the substrate, can lose their microbiota, and become germ-free (Blum et al., 2013; Pais et al., 2018). The transitory nature of Drosophila microbiota was established using bacterial isolates from Drosophila laboratory stocks. Interestingly, some of the bacterial isolates from wild-caught D. melanogaster can stably persist and proliferate in the guts of fruit flies. Such stable association facilitated continuous bacterial spreading to new environment and colonization of the next generation of flies with beneficial bacterium which accelerated the growth of Drosophila larvae and enhanced the fertility of adult flies, thus conferring fitness advantages for both partners in an ecological context (Pais et al., 2018). Under laboratory conditions, where both microbes and flies can live perfectly fine independently, the evolutionary pressure to maintain a stable association has likely been lost. In line with this, it was found that the diet, rather than the host, is the major force driving the evolution of symbiotic properties of the prominent fly commensal L. plantarum (Martino et al., 2018). Such diet-driven evolution of improved symbiotic properties is an example of by-product mutualism, where the host benefits from the by-products of its bacterial symbiont.
Influence of Gut Microbiota on Drosophila Lifespan
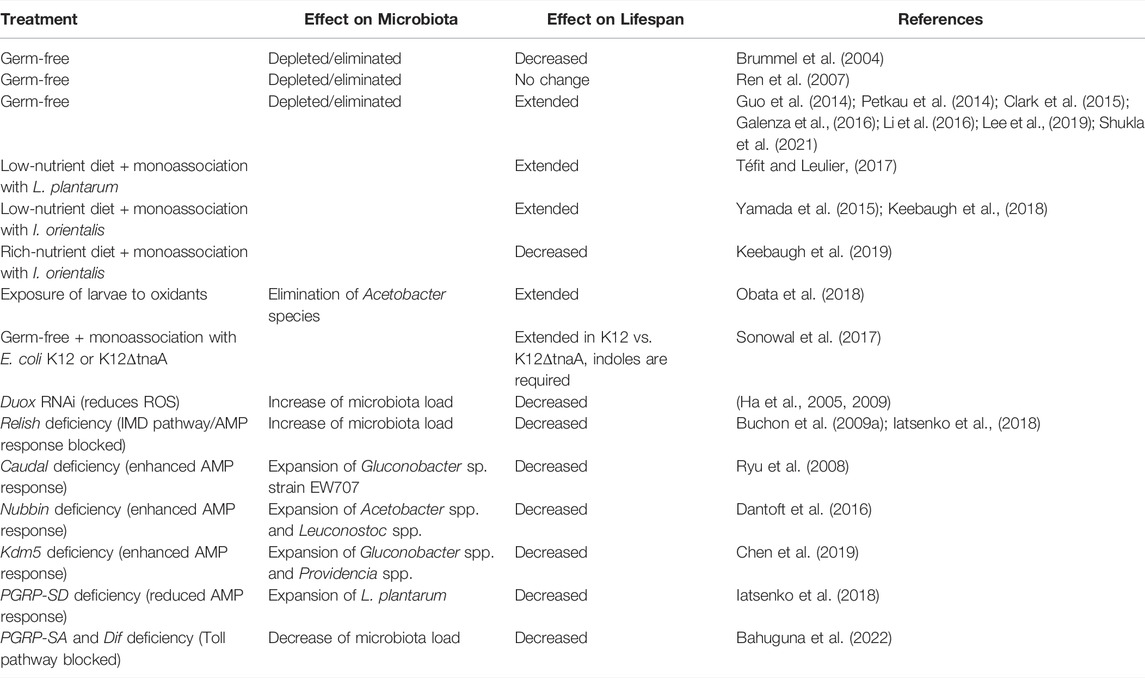
The microbiota of the fruit fly changes in abundance and composition throughout aging (Broderick et al., 2014; Guo et al., 2014; Clark et al., 2015; Li et al., 2016; Salazar et al., 2018). Old flies often harbor a higher bacterial load in the intestine than their young counterparts (Blum et al., 2013; Broderick et al., 2014; Marra et al., 2021a). Considering the reported microbiota perturbations, expectedly, the role of microbiota in Drosophila aging has been an active area of research. The most direct approach to examining the influence of the gut microbiota on host longevity is to investigate the impact of microbiota removal using axenic flies. Multiple studies that used such an approach reported conflicting results. The pioneering work of Brummel et al. reported that the lifespan of flies reared axenically was shorter than that of conventionally reared flies (Brummel et al., 2004). This effect could be rescued by exposing the flies to microbes within 2–3 days from eclosion. A follow-up study from Ren showed no impact of microbiota elimination on the lifespan (Ren et al., 2007).
Differences in nutrient conditions between laboratories may contribute to the inconsistent results on the impact of microbiota on fly lifespan. Indeed, the bacterial fly commensal L. plantarum (Téfit and Leulier, 2017) and yeast Issatchenkia orientalis (Yamada et al., 2015; Keebaugh et al., 2018) can promote fly longevity when flies are reared on an undernutrition diet. Remarkably, the same microbes (I. orientalis) that extended the lifespan of the fly under poor nutritional conditions shortened the lifespan when flies were reared on nutrient-rich diets (Keebaugh et al., 2019). This observation raises awareness that the nutritional environment and possibly other environmental factors are important determinants of the effect of commensals on the host physiology. Although Keebaugh et al. have not investigated the underlying mechanisms of the detrimental impact of fly commensals on the lifespan under nutrient-rich conditions, their observation provides experimental support for the “overfeeding hypothesis.” Namely, Lachnit et al. (2019) proposed that overfeeding changes functionality of the intestinal microbiota and increases its activity, resulting in the increase of microbial byproducts that promote disease development. Such a scenario could explain the detrimental effect of I. orientalis on Drosophila lifespan under a nutrient-rich diet. In addition to the nutritional value, dietary pH has also been shown to have an effect on life expectancy. In fact, flies live longer on an acidic diet, similar to what has been observed in axenic animals, suggesting that this effect may be independent of the microbiota (Deshpande et al., 2015). However, the authors observed a shift in the microbiota composition on alkaline pH diets, and this shift might have exacerbated the deleterious effect of alkaline foods on the lifespan (Deshpande et al., 2015). In addition, chemical stressors present in the food could have an impact on the lifespan of fruit flies via affecting the composition of microbiota. For example, transient exposure of Drosophila larvae to low concentrations of oxidants extended adult lifespan by selectively eliminating Acetobacter species which were causally linked to lifespan shortening (Obata et al., 2018).
Despite conflicting results of early studies, there is lots of evidence showing that the absence of microbes extends the lifespan of Drosophila. Numerous studies that utilized different fly genotypes, various diets (cornmeal, chemically-defined), and different ways of generating axenic flies (bleaching, antibiotic treatment) consistently reported an increase in the lifespan of axenic flies (Petkau et al., 2014; Clark et al., 2015; Galenza et al., 2016; Téfit and Leulier, 2017; Iatsenko et al., 2018; Obata et al., 2018; Lee et al., 2019; Shukla et al., 2021). Therefore, the presence of microbiota can be detrimental to the lifespan of fruit flies. Although the microbiota of Drosophila undergoes age-related changes in composition and some members of the microbiota appear to be particularly harmful to the host during aging, the extensive work of Lee e al. showed that the abundance of microbes is a stronger determinant of host lifespan than changes in the composition (Lee et al., 2019; Lee et al., 2022).
Several studies attempted to understand the impact of microbes on fruit fly aging and compared the differences between axenic and conventional flies. Shukla et al. took a transcriptomics approach and discovered that around 70% of age-associated changes in gene expression do not occur in germ-free flies (Shukla et al., 2021). Among the processes that did not occur in axenic flies were two hallmarks of aging conserved across animal species: the down-regulation of stress-response genes and the up-regulation of genes of the innate immune system. These processes therefore represent an adaptive response of the organism to microbes during aging.
Besides regulating gene expression, commensals also modify the fly metabolome during aging. Specifically, Yamauchi et al. discovered that allantoin, an end product of purine degradation pathway in Drosophila, levels increase with aging in a microbiota-dependent manner and contribute to the shortening of the lifespan (Yamauchi et al., 2020). Acetobacter persici was identified as a species responsible for increasing allantoin levels by activating the immune deficiency (IMD) pathway in Malpighian tubules. It remains to be discovered how increased allantoin shortens lifespan and how the IMD pathway regulates allantoin levels.
In addition to affecting the fly metabolome, commensals themselves produce metabolites that affect host aging. For example, indoles, aromatic heterocyclic organic intermediates in the biosynthesis of tryptophan, released by E. coli were shown to extend the lifespan of Drosophila by activating aryl hydrocarbon receptors (Sonowal et al., 2017). Although E. coli is not a typical fly gut commensal, a similar mechanism may operate in some of the Drosophila microbiota members that produce indoles, like Lactobacilli. Another bacterial metabolite, colonic acid, exhibited a similar longevity-promoting effect likely via the host’s mitochondrial dynamics regulation and unfolded protein response (UPRmt) (Han B. et al., 2017). Furthermore, a genetic approach in bacteria, a metagenome-wide association, identified bacterial methionine metabolism genes associated with the variation in Drosophila lifespan, suggesting a potential role of methionine in fruit fly longevity (Matthews et al., 2020).
Since the composition of the Drosophila microbiota changes with age and abundance increases, several studies investigated the potential causes, and consequences of such perturbations. For example, Guo et al. proposed a mechanism explaining the causes of age-related commensal dysbiosis and its impact on fly longevity (Guo et al., 2014). The authors discovered that there is a chronic activation of the FOXO transcription factor in the aging intestines. This leads to FOXO-mediated suppression of PGRP-SC2, a negative regulator of the IMD pathway, and thus to a deregulation of the activity of the IMD pathway and the induction of commensal dysbiosis in the form of increased microbial load and expansion of pathobionts (Guo et al., 2014). Consequently, such a dysbiotic microbial community caused intestinal stem cell (ISC) overproliferation, dysplasia, and reduced lifespan (Figure 1). However, how PGRP-SC2 controls microbiota, e.g., either via regulation of IMD pathway activity or by acting as an effector molecule remains to be investigated. In a subsequent study, Li et al. identified an additional mechanism behind commensal dysbiosis during aging (Li et al., 2016). Specifically, the authors found that the JAK-STAT pathway activity in the Drosophila gut increases with aging, likely due to the increased production of cytokines. This leads to metaplasia in the stomach-like copper cell region of the intestine. Due to its acidic pH, this region controls the distribution and composition of the microbiota. Therefore, age-related metaplasia due to increased JAK-STAT activation disrupts the copper cell region, leading to commensal dysbiosis, epithelial dysplasia, and reduced lifespan (Li et al., 2016). Thus, the decline in intestinal compartmentalization caused by age-related chronic inflammation is a crucial mechanism behind commensal dysbiosis and intestinal dysplasia.
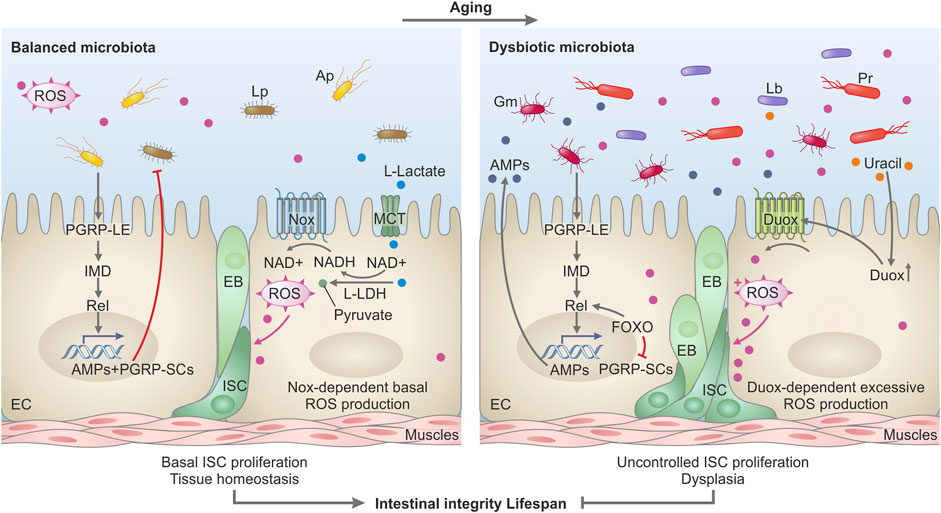
FIGURE 1. Model depicting the impact of microbiota on intestinal homeostasis during aging in Drosophila. In a healthy, young gut (left panel), the microbiota helps to sustain intestinal homeostasis, intestinal integrity, and optimal lifespan. Commensal bacteria like L. plantarum release L-lactate in the intestine. L-lactate enters the intestinal cells via monocarboxylate transporters (MCT) and is oxidized by the host lactate dehydrogenase (LDH), thus generating NADH. NADH is used by the NADPH-oxidase Nox to generate Reactive Oxygen Species (ROS) which stimulate basal intestinal stem cell (ISC) proliferation and differentiation in EnteroBlasts (EB) and Enterocytes (EC) to ensure tissue homeostasis. L. plantarum (Lp) and other commensals such as A. pomorum (Ap) stimulate basal IMD pathway activity by triggering the receptor PGRP-LE and the Relish transcription factor which consequently induces expression of PGRP-SCs and antimicrobial peptides (AMPs)—molecules that control the proliferation of the commensal bacteria. In the aging gut (right panel), the activated FOXO transcription factor boosts Relish activity and AMPs genes expression but suppresses PGRP-SCs expression. Such an environment with an excessive amount of AMPs and reduced production of PGRP-SCs favors the emergence and selection of pathobionts in the commensal community. Such pathobionts like G. morbifer (Gm), L. brevis (Lb), or P. rettgeri (Pr) are resistant to AMPs and release uracil which promotes excessive ROS production via activation of DUOX. DUOX-dependent ROS in turn stimulates uncontrolled ISC proliferation and defective differentiation resulting in gut dysplasia, loss of tissue integrity, and reduced lifespan.
Link Between Host Genetics, Microbiota Dysbiosis, and Lifespan
Host genetics is one of the determinants of microbiota composition and abundance (Chaston et al., 2016). Several mutations were identified that predispose fruit flies to a shorter lifespan by affecting the intestinal microbial communities. Those mutations predominantly disrupt the host pathways implicated in the control of intestinal commensals. Fruit flies rely on two inducible defense mechanisms to control pathogens and gut microbiota: antimicrobial peptides (AMPs) and reactive oxygen species (ROS) (Ha et al., 2005; Ryu et al., 2006; Buchon et al., 2009b; Marra et al., 2021a). The dual oxidase Duox produces high levels of microbicidal ROS in response to uracil released by pathogens and pathobionts (Ha et al., 2005; Lee et al., 2013). Flies lacking Duox activity cannot control mutualistic and pathogenic bacteria and have a short lifespan (Ha et al., 2005; Ha et al., 2009). Importantly, ROS are not only microbicidal but also can damage intestinal cells, thereby inducing the compensatory proliferation of ISCs to repair the damage (Buchon et al., 2009a; Hochmuth et al., 2011). However, excessive accumulation of stem cells results in dysplasia and intestinal dysfunction.
The activation of the IMD pathway by intestinal bacteria results in the translocation of the NF-kB-like transcription factor Relish to the nucleus, which promotes the expression of antimicrobial peptides (AMPs). The IMD pathway is initiated when bacterial peptidoglycan (PGN) is sensed by the transmembrane receptor PGRP-LC in the ectodermal parts of the gut or by the intracellular receptor PGRP- LE in the midgut (Bosco-Drayon et al., 2012; Neyen et al., 2012). IMD-induced AMPs act primarily against ROS-resistant microbes and complement Duox-mediated ROS defense (Ryu et al., 2006). Flies with disrupted IMD signaling, like Relish mutants or mutants lacking AMPs exhibit an excessive load of commensals and are short-lived (Buchon et al., 2009a; Broderick et al., 2014; Iatsenko et al., 2018; Marra et al., 2021a). Several additional mutants with affected ROS- or IMD-mediated responses were shown to have reduced longevity due to commensal dysbiosis. For example, flies with reduced expression of the transcription factor Caudal have elevated IMD pathway activity and increased expression of AMPs in the gut (Ryu et al., 2008). Such an inflamed intestinal environment favors the growth of the pathobiont Gluconobacter EW707, which drives host mortality. A subsequent study showed that Gluconobacter, in contrast to beneficial microbiota members, releases uracil, which triggers chronic DUOX activation and ROS production, causing intestinal damage, uncontrolled ISC proliferation, dysplasia, and mortality (Figure 1) (Lee et al., 2013).
Loss of function of another transcription factor, Nubbin, similarly leads to a constitutively active immune response in the gut and shift in microbiota abundance and composition. Consequently, flies lacking Nubbin are short-lived due to commensal dysbiosis and overproliferation of Acetobacter spp. and Leuconostoc spp (Dantoft et al., 2016).
Gluconobacter spp was implicated in the intestinal pathology in another mutant with deregulated IMD activity. Chen et al. reported that flies lacking histone demethylase KDM5 exhibit gut dysbiosis due to excessive IMD pathway activity and a reduced lifespan (Chen et al., 2019). Specifically, kdm mutants exhibit reduced abundance of Lactobacillus spp but increased abundance of Gluconobacter spp and Providencia spp (Chen et al., 2019). Interestingly, another genetic deficiency, namely that of transglutaminase, results in a similar phenotype—elevated IMD activity, reduced lifespan, and commensal dysbiosis with the expansion of Providencia spp. and Acetobacter spp (Sekihara et al., 2016). Such consistency suggests that excessive IMD pathway activity creates conditions optimal for pathobionts like Gluconobacter and Providencia. Notably, both Gluconobacter and Providencia are resistant to host AMPs and ROS, while both release uracil and induce ROS via DUOX, thus not only thriving in the inflamed intestine but also further exacerbating the inflammation, intestinal damage, and dysplasia.
Flies deficient for several negative regulators of the IMD pathway, PGRP-LB, PGRP-SCs, and Pirk, are short-lived similar to other mutants with overactive IMD pathway (Kleino et al., 2008; Lhocine et al., 2008; Paredes et al., 2011). The fact that their lifespan can be extended under germ-free conditions suggests that chronic immune activation to endogenous microbes likely causes intestinal dysfunction and early death. However, whether such flies lacking negative IMD pathway regulators experience any changes in commensal composition and abundance reported for other mutants with excessive IMD activity remains to be investigated. The same question applies to the big bang mutant, which, due to disrupted septate junctions in the gut, has a reduced lifespan caused by constitutive immune activation that is driven by intestinal commensals (Bonnay et al., 2013).
Beyond pathobionts, bacteria that are typically considered beneficial for flies, like Lactobacillus, can also cause intestinal pathologies during aging when their abundance is not controlled (Fast et al., 2018; Iatsenko et al., 2018). During aging, such uncontrolled growth was observed in flies lacking PGRP-SD—a secreted receptor upstream of the IMD pathway (Iatsenko et al., 2016, 2018). Overgrowth of L. plantarum in the intestines of PGRP-SD mutant flies was accompanied by the accumulation of lactic acid, which triggered the generation of elevated levels of ROS via the intestinal NADPH oxidase Nox. Nox-generated ROS consequently caused intestinal damage, compensatory overproliferation of ISCs, dysplasia, and shortened lifespan (Iatsenko et al., 2018). The fact that L. plantarum induces Nox-dependent ISC proliferation in the mammalian intestine (Jones et al., 2013) suggests a conserved mechanism of Nox activation by lactate which couples bacterial growth to ISC proliferation.
Additionally, commensals might affect the host aging by modulating the key longevity-controling pathways. Indeed, mTOR and insulin-like growth factor signaling are evolutionary conserved pathways known to be regulated by fruit fly commensals (Fan et al., 2018; Bana and Cabreiro, 2019). For example, pyrroloquinoline quinone–dependent alcohol dehydrogenase (PQQ-ADH) activity of a commensal, A. pomorum, modulates IIS in Drosophila and is necessary to promote larval growth on a low-yeast diet (Shin et al., 2011). Similarly, L. plantarum, promotes growth of larvae on low-nutrient diet via up-regulation of the TOR pathway. Flies overexpressing the inhibitor of TOR complex 1 are resistant to the effects of L. plantarum on growth (Storelli et al., 2011). Given that suppression of TOR and IIS is known to promote longevity (Bana and Cabreiro, 2019), it will be important to determine whether A. pomorum and L. plantarum might negatively affect aging by activating IIS and TOR pathways respectively. Interestingly, chemical inhibition of TOR by rapamycin treatment altered microbiome composition and extended the lifespan but both in CR and axenic flies, suggesting that rapamycin’s lifespan-promoting effect is independent of microbiota (Schinaman et al., 2019).
Although Toll pathway doesn’t play a major role in intestinal immunity (Broderick et al., 2014), its role in the microbiota control is emerging. For example, Toll-deficient flies lacking PGRP-SA or DIF have reduced bacterial load and shortened lifespan. Mechanistically, PGRP-SA regulates microbiota via metabolic rather than immune function. Specifically, Toll pathway activation in enterocytes following the recognition of bacteria by PGRP-SA receptor keeps increased transcription of translational regulation factor 4E-BP. Toll activated 4E-BP enables fat catabolism, thus sustaining the microbiota. In the absence of Toll pathway activity, TOR-mediated suppression of 4E-BP results in intestinal lipid accumulation and inaccessibility which correlates with loss of intestinal bacterial density (Bahuguna et al., 2022). However, whether the reduced lifespan of PGRP-SA and dif mutants is due to lipid accumulation or changes in microbiota communities is an open question.
Similar to overactivation of IMD pathway, chronic stimulation of Toll pathway by PGN released by Gram-positive microbiota results in the short lifespan of flies. Namely, flies deficient for Kruppel-like factor 15 (Klf15) lack nephrocytes and not able to filter the hemolymph, thus accumulating microbiota-derived PGN in the hemolymph. This creates a chronic state of systemic Toll pathway activation by the PGN and shortened lifespan (Troha et al., 2019). In summary, both constitutive activation and deficiency in the major immune pathways disrupts host-commensal homeostasis leading to premature death of the host (Table 1).
Discussion
In recent years substantial progress has been made in our understanding of the molecular mechanisms of microbiota influence on the fruit fly lifespan. While we have advanced our knowledge on host-microbiota interactions during aging, many essential questions remain to be addressed. For example, most studies focused on changes in bacterial communities during aging and the impact of particular bacteria on Drosophila lifespan. Yet, the role of other microbiome residents, like fungi and viruses, in the aging process has not been investigated and represents an exciting avenue for future studies. Similarly, fruit fly endosymbionts, like Wolbachia and Spiroplasma, have dramatic effect on the lifespan (Maistrenko et al., 2016; Marra et al., 2021b). The results, however, are inconsistent and lack mechanistic explanation. While Wolbachia is known to interfere with the longevity-modulating pathways (Ikeya et al., 2009; Maistrenko et al., 2016), it also modifies the intestinal microbiome via yet to be discovered mechanism (Simhadri et al., 2017). Whether such microbiome remodeling contributes to the effect of Wolbachia on Drosophila lifespan requires further investigation.
Another critical aspect that needs additional insights is the contribution of microbiota to sex differences in longevity. While it is known that Drosophila males live shorter than females and that there are differences between sexes in age-related intestinal pathologies and gene expression (Austad and Fischer, 2016; Hudry et al., 2016; Regan et al., 2016; Belmonte et al., 2020), the microbiome’s contribution to these differences is largely unknown. Although males and females differ in microbiota composition (Han G. et al., 2017; Leech et al., 2021), it has to be studied whether sexual dimorphism in microbiome composition and abundance or age-related microbial dysbiosis contributes to the observed pathologies and lifespan differences. An alternative possibility that needs to be tested is that the host might respond to the same commensal microbes in a sex-dimorphic manner. Thus, the same commensals might trigger different phenotypes in males and females. To address this complex question, future studies should include sex as a variable to investigate the impact of the microbiome on the aging process.
There is plenty of evidence that the composition of the diet affects various physiological processes in the host, including aging (Tatar et al., 2014; Stefana et al., 2017). Considering that the nutritional environment also dictates microbiome composition, the effect of diet on the host could also be indirectly mediated via an altered microbiome (Harris et al., 2019; Ghosh et al., 2020; Sanchez-Morate et al., 2020). Numerous studies investigated how macronutrients like proteins, lipids, carbohydrates, and their ratios affect the fruit fly physiology and commensal populations (Woodcock et al., 2015; Galenza et al., 2016; Jang and Lee, 2018; Evangelakou et al., 2019; Keebaugh et al., 2019). However, the role of micronutrients and particularly dietary transition metals remains understudied, despite their crucial importance in insect physiology (Dow, 2017; Missirlis, 2021) and interactions with pathogenic and symbiotic microbes (Iatsenko et al., 2020; Hrdina and Iatsenko, 2022). This raises the necessity to investigate how trace metals affect fly aging directly and indirectly via altering microbiome composition and function. Indeed, studies in mammals support the notion that trace metals modulate the microbiome and thus host-microbe interactions and animal health (Lopez and Skaar, 2018). Whether this is also applicable to the Drosophila microbiome and aging remains to be investigated.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the Max Planck Society and by the Deutsche Forschungsgemeinschaft (DFG) grant IA 81/2-1 to II.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Stanislava Chtarbanova for the invitation to contribute this manuscript, Diane Schad for preparing the figures, and Alexandra Hrdina for critical reading of the manuscript. The authors apologize to colleagues whose work could not be directly cited due to space restrictions.
References
Adair, K. L., Wilson, M., Bost, A., and Douglas, A. E. (2018). Microbial Community Assembly in Wild Populations of the Fruit Fly Drosophila melanogaster. ISME J. 12, 959–972. doi:10.1038/s41396-017-0020-x
Austad, S. N., and Fischer, K. E. (2016). Sex Differences in Lifespan. Cell. Metab. 23, 1022–1033. doi:10.1016/J.CMET.2016.05.019
Bahuguna, S., Atilano, M., Glittenberg, M., Lee, D., Arora, S., Wang, L., et al. (2022). Bacterial Recognition by PGRP-SA and Downstream Signalling by Toll/DIF Sustain Commensal Gut Bacteria in Drosophila. PLOS Genet. 18, e1009992. doi:10.1371/JOURNAL.PGEN.1009992
Bana, B., and Cabreiro, F. (2019). The Microbiome and Aging. Annu. Rev. Genet. 53, 239–261. doi:10.1146/annurev-genet-112618-043650
Belmonte, R. L., Corbally, M.-K., Duneau, D. F., and Regan, J. C. (2020). Sexual Dimorphisms in Innate Immunity and Responses to Infection in Drosophila melanogaster. Front. Immunol. 10, e03075. doi:10.3389/fimmu.2019.03075
Biagi, E., Franceschi, C., Rampelli, S., Severgnini, M., Ostan, R., Turroni, S., et al. (2016). Gut Microbiota and Extreme Longevity. Curr. Biol. 26, 1480–1485. doi:10.1016/J.CUB.2016.04.016
Blum, J. E., Fischer, C. N., Miles, J., and Handelsman, J. (2013). Frequent Replenishment Sustains the Beneficial Microbiome of Drosophila melanogaster. MBio 4, e00860–13. doi:10.1128/mBio.00860-13
Bonnay, F., Cohen-Berros, E., Hoffmann, M., Kim, S. Y., Boulianne, G. L., Hoffmann, J. A., et al. (2013). Big Bang Gene Modulates Gut Immune Tolerance in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 110, 2957–2962. doi:10.1073/pnas.1221910110
Bosco-Drayon, V., Poidevin, M., Boneca, I. G., Narbonne-Reveau, K., Royet, J., and Charroux, B. (2012). Peptidoglycan Sensing by the Receptor PGRP-LE in the Drosophila Gut Induces Immune Responses to Infectious Bacteria and Tolerance to Microbiota. Cell. Host Microbe 12, 153–165. doi:10.1016/j.chom.2012.06.002
Broderick, N. A., Buchon, N., and Lemaitre, B. (2014). Microbiota-induced Changes in drosophila Melanogaster Host Gene Expression and Gut Morphology. MBio 5, e01117–14. doi:10.1128/mBio.01117-14
Broderick, N. A., and Lemaitre, B. (2012). Gut-associated Microbes of Drosophila melanogaster. Gut Microbes 3, 307–321. doi:10.4161/gmic.19896
Brummel, T., Ching, A., Seroude, L., Simon, A. F., and Benzer, S. (2004). Drosophila Lifespan Enhancement by Exogenous Bacteria. Proc. Natl. Acad. Sci. U.S.A. 101, 12974–12979. doi:10.1073/PNAS.0405207101
Buchon, N., Broderick, N. A., Chakrabarti, S., and Lemaitre, B. (2009a). Invasive and Indigenous Microbiota Impact Intestinal Stem Cell Activity through Multiple Pathways in Drosophila. Genes Dev. 23, 2333–2344. doi:10.1101/gad.1827009
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S., and Lemaitre, B. (2009b). Drosophila Intestinal Response to Bacterial Infection: Activation of Host Defense and Stem Cell Proliferation. Cell. Host Microbe 5, 200–211. doi:10.1016/j.chom.2009.01.003
Cabreiro, F., and Gems, D. (2013). Worms Need Microbes Too: Microbiota, Health and Aging inCaenorhabditis Elegans. EMBO Mol. Med. 5, 1300–1310. doi:10.1002/emmm.201100972
Chandler, J. A., Morgan Lang, J., Bhatnagar, S., Eisen, J. A., and Kopp, A. (2011). Bacterial Communities of Diverse Drosophila Species: Ecological Context of a Host-Microbe Model System. PLOS Genet. 7, e1002272. doi:10.1371/JOURNAL.PGEN.1002272
Chaston, J. M., Dobson, A. J., Newell, P. D., and Douglas, A. E. (2016). Host Genetic Control of the Microbiota Mediates the Drosophila Nutritional Phenotype. Appl. Environ. Microbiol. 82, 671–679. doi:10.1128/AEM.03301-15
Chen, K., Luan, X., Liu, Q., Wang, J., Chang, X., Snijders, A. M., et al. (2019). Drosophila Histone Demethylase KDM5 Regulates Social Behavior through Immune Control and Gut Microbiota Maintenance. Cell. Host Microbe 25, 537–552. e8. doi:10.1016/j.chom.2019.02.003
Clark, R. I., Salazar, A., Yamada, R., Fitz-Gibbon, S., Morselli, M., Alcaraz, J., et al. (2015). Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell. Rep. 12, 1656–1667. doi:10.1016/J.CELREP.2015.08.004
Clark, R. I., and Walker, D. W. (2018). Role of Gut Microbiota in Aging-Related Health Decline: Insights from Invertebrate Models. Cell. Mol. Life Sci. 75, 93–101. doi:10.1007/s00018-017-2671-1
Consuegra, J., Grenier, T., Baa-Puyoulet, P., Rahioui, I., Akherraz, H., Gervais, H., et al. (2020). Drosophila-associated Bacteria Differentially Shape the Nutritional Requirements of Their Host during Juvenile Growth. PLoS Biol. 18, e3000681. doi:10.1371/journal.pbio.3000681
Dantoft, W., Lundin, D., Esfahani, S. S., and Engström, Y. (2016). The POU/Oct Transcription Factor Pdm1/nub Is Necessary for a Beneficial Gut Microbiota and Normal Lifespan of Drosophila. J. Innate Immun. 8, 412–426. doi:10.1159/000446368
Deshpande, S. A., Yamada, R., Mak, C. M., Hunter, B., Obando, A. S., Hoxha, S., et al. (2015). Acidic Food pH Increases Palatability and Consumption and Extends Drosophila Lifespan. J. Nutr. 145, 2789–2796. doi:10.3945/JN.115.222380
Douglas, A. E. (2019). Simple Animal Models for Microbiome Research. Nat. Rev. Microbiol. 17, 764–775. doi:10.1038/s41579-019-0242-1
Douglas, A. E. (2018). The Drosophila Model for Microbiome Research. Lab. Anim. 47, 157–164. doi:10.1038/S41684-018-0065-0
Dow, J. A. (2017). The Essential Roles of Metal Ions in Insect Homeostasis and Physiology. Curr. Opin. Insect Sci. 23, 43–50. doi:10.1016/J.COIS.2017.07.001
Ducarmon, Q. R., Zwittink, R. D., Hornung, B. V. H., van Schaik, W., Young, V. B., and Kuijper, E. J. (2019). Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 83, e00007. doi:10.1128/mmbr.00007-19
Dutta, N., Garcia, G., and Higuchi-Sanabria, R. (2022). Hijacking Cellular Stress Responses to Promote Lifespan. Front. Aging 3, 20. doi:10.3389/FRAGI.2022.860404
Erkosar, B., Leulier, F., Kasper, L. H., and Just, W. (2014). Transient Adult Microbiota, Gut Homeostasis and Longevity: Novel Insights from the Drosophila Model. FEBS Lett. 588, 4250–4257. doi:10.1016/J.FEBSLET.2014.06.041
Erkosar, B., and Leulier, F. (2014). Transient Adult Microbiota, Gut Homeostasis and Longevity: Novel Insights from the Drosophila Model. FEBS Lett. 588, 4250–4257. doi:10.1016/j.febslet.2014.06.041
Evangelakou, Z., Manola, M., Gumeni, S., and Trougakos, I. P. (2019). Nutrigenomics as a Tool to Study the Impact of Diet on Aging and Age-Related Diseases: the Drosophila Approach. Genes Nutr. 14, e0638. doi:10.1186/S12263-019-0638-6
Fan, X., Gaur, U., and Yang, M. (2018). “Intestinal Homeostasis and Longevity: Drosophila Gut Feeling,” in Advances in Experimental Medicine and Biology (Singapore: Springer), 157–168. doi:10.1007/978-981-13-1117-8_10
Fast, D., Duggal, A., and Foley, E. (2018). Monoassociation with Lactobacillus Plantarum Disrupts Intestinal Homeostasis in Adult Drosophila melanogaster. MBio 9, e01114–18. doi:10.1128/mBio.01114-18
Fernandes, S. A., and Demetriades, C. (2021). The Multifaceted Role of Nutrient Sensing and mTORC1 Signaling in Physiology and Aging. Front. Aging 2, 38. doi:10.3389/FRAGI.2021.707372
Fontana, L., Partridge, L., and Longo, V. D. (2010). Extending Healthy Life Span-From Yeast to Humans. Science 328, 321–326. doi:10.1126/SCIENCE.1172539/SUPPL_FILE/PLAYLIST
Galenza, A., Hutchinson, J., Campbell, S. D., Hazes, B., and Foley, E. (2016). Glucose Modulates Drosophila Longevity and Immunity Independent of the Microbiota. Biol. Open 5, 165–173. doi:10.1242/BIO.015016
Geva-Zatorsky, N., Sefik, E., Kua, L., Pasman, L., Tan, T. G., Ortiz-Lopez, A., et al. (2017). Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 168, 928–943. e11. doi:10.1016/j.cell.2017.01.022
Ghosh, T. S., Rampelli, S., Jeffery, I. B., Santoro, A., Neto, M., Capri, M., et al. (2020). Mediterranean Diet Intervention Alters the Gut Microbiome in Older People Reducing Frailty and Improving Health Status: the NU-AGE 1-year Dietary Intervention across Five European Countries. Gut 69, 1218–1228. doi:10.1136/GUTJNL-2019-319654
Gould, A. L., Zhang, V., Lamberti, L., Jones, E. W., Obadia, B., Korasidis, N., et al. (2018). Microbiome Interactions Shape Host Fitness. Proc. Natl. Acad. Sci. U.S.A. 115, E11951–E11960. doi:10.1073/pnas.1809349115
Grenier, T., and Leulier, F. (2020). How Commensal Microbes Shape the Physiology of Drosophila melanogaster. Curr. Opin. Insect Sci. 41, 92–99. doi:10.1016/J.COIS.2020.08.002
Guo, L., Karpac, J., Tran, S. L., and Jasper, H. (2014). PGRP-SC2 Promotes Gut Immune Homeostasis to Limit Commensal Dysbiosis and Extend Lifespan. Cell. 156, 109–122. doi:10.1016/j.cell.2013.12.018
Ha, E.-M., Lee, K.-A., Park, S. H., Kim, S.-H., Nam, H.-J., Lee, H.-Y., et al. (2009). Regulation of DUOX by the Gαq-Phospholipase Cβ-Ca2+ Pathway in Drosophila Gut Immunity. Dev. Cell. 16, 386–397. doi:10.1016/J.DEVCEL.2008.12.015
Ha, E.-M., Oh, C.-T., Bae, Y. S., and Lee, W.-J. (2005). A Direct Role for Dual Oxidase in Drosophila Gut Immunity. Science 310, 847–850. doi:10.1126/science.1117311
Hales, K. G., Korey, C. A., Larracuente, A. M., and Roberts, D. M. (2015). Genetics on the Fly: A Primer on the Drosophila Model System. Genetics 201, 815–842. doi:10.1534/GENETICS.115.183392
Han, B., Sivaramakrishnan, P., Lin, C.-C. J., Neve, I. A. A., He, J., Tay, L. W. R., et al. (2017a). Microbial Genetic Composition Tunes Host Longevity. Cell. 169, 1249–1262. e13. doi:10.1016/j.cell.2017.05.036
Han, G., Lee, H. J., Jeong, S. E., Jeon, C. O., and Hyun, S. (2017b). Comparative Analysis of Drosophila melanogaster Gut Microbiota with Respect to Host Strain, Sex, and Age. Microb. Ecol. 74, 207–216. doi:10.1007/S00248-016-0925-3/FIGURES/6
Harris, E. V., De Roode, J. C., and Gerardo, N. M. (2019). Diet-microbiome-disease: Investigating Diet's Influence on Infectious Disease Resistance through Alteration of the Gut Microbiome. PLoS Pathog. 15, e1007891. doi:10.1371/journal.ppat.1007891
Hochmuth, C. E., Biteau, B., Bohmann, D., and Jasper, H. (2011). Redox Regulation by Keap1 and Nrf2 Controls Intestinal Stem Cell Proliferation in Drosophila. Cell. Stem Cell. 8, 188–199. doi:10.1016/J.STEM.2010.12.006
Hrdina, A., and Iatsenko, I. (2022). The Roles of Metals in Insect-Microbe Interactions and Immunity. Curr. Opin. Insect Sci. 49, 71–77. doi:10.1016/j.cois.2021.12.004
Hudry, B., Khadayate, S., and Miguel-Aliaga, I. (2016). The Sexual Identity of Adult Intestinal Stem Cells Controls Organ Size and Plasticity. Nature 530, 344–348. doi:10.1038/nature16953
Iatsenko, I., Boquete, J.-P., and Lemaitre, B. (2018). Microbiota-Derived Lactate Activates Production of Reactive Oxygen Species by the Intestinal NADPH Oxidase Nox and Shortens Drosophila Lifespan. Immunity 49, 929–942. doi:10.1016/J.IMMUNI.2018.09.017
Iatsenko, I., Kondo, S., Mengin-Lecreulx, D., Lemaitre, B., Bischoff, V., Vignal, C., et al. (2016). PGRP-SD, an Extracellular Pattern-Recognition Receptor, Enhances Peptidoglycan-Mediated Activation of the Drosophila Imd Pathway. Immunity 45, 1013–1023. doi:10.1016/j.immuni.2016.10.029
Iatsenko, I., Marra, A., Boquete, J.-P., Peña, J., and Lemaitre, B. (2020). Iron Sequestration by Transferrin 1 Mediates Nutritional Immunity in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 117, 7317–7325. doi:10.1073/pnas.1914830117
Ikeya, T., Broughton, S., Alic, N., Grandison, R., and Partridge, L. (2009). The endosymbiontWolbachiaincreases insulin/IGF-like Signalling inDrosophila. Proc. R. Soc. B 276, 3799–3807. doi:10.1098/RSPB.2009.0778
Jang, T., and Lee, K. P. (2018). Comparing the Impacts of Macronutrients on Life-History Traits in Larval and Adult Drosophila melanogaster: the Use of Nutritional Geometry and Chemically Defined Diets. J. Exp. Biol. 221, 181115. doi:10.1242/JEB.181115
Jones, R. M., Luo, L., Ardita, C. S., Richardson, A. N., Kwon, Y. M., Mercante, J. W., et al. (2013). Symbiotic Lactobacilli Stimulate Gut Epithelial proliferationviaNox-Mediated Generation of Reactive Oxygen Species. EMBO J. 32, 3017–3028. doi:10.1038/emboj.2013.224
Kapahi, P., Chen, D., Rogers, A. N., Katewa, S. D., Li, P. W.-L., Thomas, E. L., et al. (2010). With TOR, Less Is More: A Key Role for the Conserved Nutrient-Sensing TOR Pathway in Aging. Cell. Metab. 11, 453–465. doi:10.1016/J.CMET.2010.05.001
Keebaugh, E. S., Yamada, R., and Ja, W. W. (2019). The Nutritional Environment Influences the Impact of Microbes on Drosophila melanogaster Life Span. MBio 10, e00885. doi:10.1128/mBio.00885-19
Keebaugh, E. S., Yamada, R., Obadia, B., Ludington, W. B., and Ja, W. W. (2018). Microbial Quantity Impacts Drosophila Nutrition, Development, and Lifespan. iScience 4, 247–259. doi:10.1016/J.ISCI.2018.06.004
Kim, S., and Jazwinski, S. M. (2018). The Gut Microbiota and Healthy Aging: A Mini-Review. Gerontology 64, 513–520. doi:10.1159/000490615
Kleino, A., Myllymäki, H., Kallio, J., Vanha-aho, L.-M., Oksanen, K., Ulvila, J., et al. (2008). Pirk Is a Negative Regulator of theDrosophilaImd Pathway. J. Immunol. 180, 5413–5422. doi:10.4049/jimmunol.180.8.5413
Kong, Y., Wang, L., and Jiang, B. (2021). The Role of Gut Microbiota in Aging and Aging Related Neurodegenerative Disorders: Insights from Drosophila Model. Life 11, 855. doi:10.3390/LIFE11080855
Kuraishi, T., Hori, A., and Kurata, S. (2013). Host-microbe Interactions in the Gut of Drosophila melanogaster. Front. Physiol. 4 (DEC), 375. doi:10.3389/FPHYS.2013.00375
Lachnit, T., Bosch, T. C. G., and Deines, P. (2019). Exposure of the Host-Associated Microbiome to Nutrient-Rich Conditions May Lead to Dysbiosis and Disease Development— an Evolutionary Perspective. MBio 10, e0035. doi:10.1128/MBIO.00355-19
Langille, M. G., Meehan, C. J., Koenig, J. E., Dhanani, A. S., Rose, R. A., Howlett, S. E., et al. (2014). Microbial Shifts in the Aging Mouse Gut. Microbiome 2, 50–12. doi:10.1186/S40168-014-0050-9/FIGURES/5
Lapierre, L. R., and Hansen, M. (2012). Lessons from C. elegans: Signaling Pathways for Longevity. Trends Endocrinol. Metabolism 23, 637–644. doi:10.1016/J.TEM.2012.07.007
Lee, H.-Y., Lee, S.-H., Lee, J.-H., Lee, W.-J., and Min, K.-J. (2019). The Role of Commensal Microbes in the Lifespan of Drosophila melanogaster. Aging 11, 4611–4640. doi:10.18632/aging.102073
Lee, H.-Y., Lee, S.-H., Min, K.-J., Lee, S.-H., and Min, K.-J. (2022). The Increased Abundance of Commensal Microbes Decreases Drosophila melanogaster Lifespan through an Age-Related Intestinal Barrier Dysfunction. Insects 13, 219. doi:10.3390/INSECTS13020219
Lee, K.-A., Kim, S.-H., Kim, E.-K., Ha, E.-M., You, H., Kim, B., et al. (2013). Bacterial-Derived Uracil as a Modulator of Mucosal Immunity and Gut-Microbe Homeostasis in Drosophila. Cell. 153, 797–811. doi:10.1016/j.cell.2013.04.009
Leech, T., McDowall, L., Hopkins, K. P., Sait, S. M., Harrison, X. A., and Bretman, A. (2021). Social Environment Drives Sex and Age‐specific Variation in Drosophila melanogaster Microbiome Composition and Predicted Function. Mol. Ecol. 30, 5831–5843. doi:10.1111/MEC.16149
Leite, G., Pimentel, M., Barlow, G. M., Chang, C., Hosseini, A., Wang, J., et al. (2021). Age and the Aging Process Significantly Alter the Small Bowel Microbiome. Cell. Rep. 36, 109765. doi:10.1016/J.CELREP.2021.109765
Lesperance, D. N., and Broderick, N. A. (2020). Microbiomes as Modulators of Drosophila melanogaster Homeostasis and Disease. Curr. Opin. Insect Sci. 39, 84–90. doi:10.1016/j.cois.2020.03.003
Lhocine, N., Ribeiro, P. S., Buchon, N., Wepf, A., Wilson, R., Tenev, T., et al. (2008). PIMS Modulates Immune Tolerance by Negatively Regulating Drosophila Innate Immune Signaling. Cell. Host Microbe 4, 147–158. doi:10.1016/J.CHOM.2008.07.004
Li, H., Qi, Y., and Jasper, H. (2016). Preventing Age-Related Decline of Gut Compartmentalization Limits Microbiota Dysbiosis and Extends Lifespan. Cell. Host Microbe 19, 240–253. doi:10.1016/j.chom.2016.01.008
Lopez, C. A., and Skaar, E. P. (2018). The Impact of Dietary Transition Metals on Host-Bacterial Interactions. Cell. Host Microbe 23, 737–748. doi:10.1016/J.CHOM.2018.05.008
Ludington, W. B., and Ja, W. W. (2020). Drosophila as a Model for the Gut Microbiome. PLOS Pathog. 16, e1008398. doi:10.1371/JOURNAL.PPAT.1008398
Maistrenko, O. M., Serga, S. V., Vaiserman, A. M., and Kozeretska, I. A. (2016). Longevity-modulating Effects of Symbiosis: Insights from Drosophila-Wolbachia Interaction. Biogerontology 17 (17), 785–803. doi:10.1007/S10522-016-9653-9
Marra, A., Hanson, M. A., Kondo, S., Erkosar, B., Lemaitre, B., and Strains, G. (2021a). Drosophila Antimicrobial Peptides and Lysozymes Regulate Gut Microbiota Composition and Abundance. mBio 12 (17), e008241. doi:10.1128/mBio.00824-21
Marra, A., Masson, F., and Lemaitre, B. (2021b). The Iron Transporter Transferrin 1 Mediates Homeostasis of the Endosymbiotic Relationship between Drosophila melanogaster and Spiroplasma Poulsonii. microLife 2, equab008. doi:10.1093/femsml/uqab008
Martino, M. E., Joncour, P., Leenay, R., Gervais, H., Shah, M., Hughes, S., et al. (2018). Bacterial Adaptation to the Host's Diet Is a Key Evolutionary Force Shaping Drosophila-Lactobacillus Symbiosis. Cell. Host Microbe 24, 109–119. e6. doi:10.1016/j.chom.2018.06.001
Matthews, M. K., Wilcox, H., Hughes, R., Veloz, M., Hammer, A., Banks, B., et al. (2020). Genetic Influences of the Microbiota on the Life Span of drosophila Melanogaster. Appl. Environ. Microbiol. 86. doi:10.1128/AEM.00305-20
Missirlis, F. (2021). Regulation and Biological Function of Metal Ions in Drosophila. Curr. Opin. Insect Sci. 47, 18–24. doi:10.1016/j.cois.2021.02.002
Neyen, C., Poidevin, M., Roussel, A., and Lemaitre, B. (2012). Tissue- and Ligand-specific Sensing of Gram-Negative Infection in drosophila by PGRP-LC Isoforms and PGRP-LE. J. Immunol. 189, 1886–1897. doi:10.4049/jimmunol.1201022
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut Microbiota Metabolic Interactions. Science 336, 1262–1267. doi:10.1126/science.1223813
Obata, F., Fons, C. O., and Gould, A. P. (2018). Early-life Exposure to Low-Dose Oxidants Can Increase Longevity via Microbiome Remodelling in Drosophila. Nat. Commun. 9, 975. doi:10.1038/s41467-018-03070-w
O’Toole, P. W., and Jeffery, I. B. (2015). Gut Microbiota and Aging. Science 350, 1214–1215. doi:10.1126/science.aac8469
Pais, I. S., Valente, R. S., Sporniak, M., and Teixeira, L. (2018). Drosophila melanogaster Establishes a Species-specific Mutualistic Interaction with Stable Gut-Colonizing Bacteria. PLOS Biol. 16, e2005710. doi:10.1371/journal.pbio.2005710
Paredes, J. C., Welchman, D. P., Poidevin, M., and Lemaitre, B. (2011). Negative Regulation by Amidase PGRPs Shapes the Drosophila Antibacterial Response and Protects the Fly from Innocuous Infection. Immunity 35, 770–779. doi:10.1016/j.immuni.2011.09.018
Petkau, K., Parsons, B. D., Duggal, A., and Foley, E. (2014). A Deregulated Intestinal Cell Cycle Program Disrupts Tissue Homeostasis without Affecting Longevity in Drosophila. J. Biol. Chem. 289, 28719–28729. doi:10.1074/JBC.M114.578708
Pickard, J. M., Zeng, M. Y., Caruso, R., and Núñez, G. (2017). Gut Microbiota: Role in Pathogen Colonization, Immune Responses, and Inflammatory Disease. Immunol. Rev. 279, 70–89. doi:10.1111/imr.12567
Regan, J. C., Khericha, M., Dobson, A. J., Bolukbasi, E., Rattanavirotkul, N., and Partridge, L. (2016). Sex Difference in Pathology of the Ageing Gut Mediates the Greater Response of Female Lifespan to Dietary Restriction. Elife 5, 10956. doi:10.7554/eLife.10956
Ren, C., Webster, P., Finkel, S. E., and Tower, J. (2007). Increased Internal and External Bacterial Load during Drosophila Aging without Life-Span Trade-Off. Cell. Metab. 6, 144–152. doi:10.1016/J.CMET.2007.06.006
Ryu, J.-H., Ha, E.-M., Oh, C.-T., Seol, J.-H., Brey, P. T., Jin, I., et al. (2006). An Essential Complementary Role of NF-Κb Pathway to Microbicidal Oxidants in Drosophila Gut Immunity. EMBO J. 25, 3693–3701. doi:10.1038/sj.emboj.7601233
Ryu, J.-H., Kim, S.-H., Lee, H.-Y., Bai, J. Y., Nam, Y.-D., Bae, J.-W., et al. (2008). Innate Immune Homeostasis by the Homeobox Gene Caudal and Commensal-Gut Mutualism in Drosophila. Science 319, 777–782. doi:10.1126/science.1149357
Salazar, A. M., Resnik-Docampo, M., Ulgherait, M., Clark, R. I., Shirasu-Hiza, M., Jones, D. L., et al. (2018). Intestinal Snakeskin Limits Microbial Dysbiosis during Aging and Promotes Longevity. iScience 9, 229–243. doi:10.1016/j.isci.2018.10.022
Sanchez-Morate, E., Gimeno-Mallench, L., Stromsnes, K., Sanz-Ros, J., Román-Domínguez, A., Parejo-Pedrajas, S., et al. (2020). Relationship between Diet, Microbiota, and Healthy Aging. Biomedicines 8, 287. doi:10.3390/BIOMEDICINES8080287
Sannino, D. R., Dobson, A. J., Edwards, K., Angert, E. R., and Buchon, N. (2018). The Drosophila melanogaster Gut Microbiota Provisions Thiamine to its Host. MBio 9. doi:10.1128/MBIO.00155-18
Schinaman, J. M., Rana, A., Ja, W. W., Clark, R. I., and Walker, D. W. (2019). Rapamycin Modulates Tissue Aging and Lifespan Independently of the Gut Microbiota in Drosophila. Sci. Rep. 9, 1. doi:10.1038/s41598-019-44106-5
Seidel, J., and Valenzano, D. R. (2018). The Role of the Gut Microbiome during Host Ageing. F1000Res 7, 1086. doi:10.12688/f1000research.15121.1
Sekihara, S., Shibata, T., Hyakkendani, M., and Kawabata, S.-i. (2016). RNA Interference Directed against the Transglutaminase Gene Triggers Dysbiosis of Gut Microbiota in Drosophila. J. Biol. Chem. 291, 25077–25087. doi:10.1074/jbc.M116.761791
Shin, S. C., Kim, S.-H., You, H., Kim, B., Kim, A. C., Lee, K.-A., et al. (2011). Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science 80334, 670–674. doi:10.1126/SCIENCE.1212782
Shukla, A. K., Johnson, K., and Giniger, E. (2021). Common Features of Aging Fail to Occur in Drosophila Raised without a Bacterial Microbiome. iScience 24, 102703. doi:10.1016/J.ISCI.2021.102703
Simhadri, R. K., Fast, E. M., Guo, R., Schultz, M. J., Vaisman, N., Ortiz, L., et al. (2017). The Gut Commensal Microbiome of Drosophila melanogaster Is Modified by the Endosymbiont Wolbachia. mSphere 2, e00287. doi:10.1128/MSPHERE.00287
Smith, P., Willemsen, D., Popkes, M., Metge, F., Gandiwa, E., Reichard, M., et al. (2017). Regulation of Life Span by the Gut Microbiota in the Short-Lived African Turquoise Killifish. Elife 6, 27014. doi:10.7554/eLife.27014
Sommer, F., and Bäckhed, F. (2013). The Gut Microbiota - Masters of Host Development and Physiology. Nat. Rev. Microbiol. 11, 227–238. doi:10.1038/nrmicro2974
Sonowal, R., Swimm, A., Sahoo, A., Luo, L., Matsunaga, Y., Wu, Z., et al. (2017). Indoles from Commensal Bacteria Extend Healthspan. Proc. Natl. Acad. Sci. U. S. A. 114, E7506–E7515. doi:10.1073/PNAS.1706464114/SUPPL_FILE/PNAS
Staubach, F., Baines, J. F., Künzel, S., Bik, E. M., and Petrov, D. A. (2013). Host Species and Environmental Effects on Bacterial Communities Associated with Drosophila in the Laboratory and in the Natural Environment. PLoS One 8, e70749. doi:10.1371/JOURNAL.PONE.0070749
Stefana, M. I., Driscoll, P. C., Obata, F., Pengelly, A. R., Newell, C. L., MacRae, J. I., et al. (2017). Developmental Diet Regulates Drosophila Lifespan via Lipid Autotoxins. Nat. Commun. 8, 1–13. doi:10.1038/s41467-017-01740-9
Storelli, G., Defaye, A., Erkosar, B., Hols, P., Royet, J., and Leulier, F. (2011). Lactobacillus Plantarum Promotes Drosophila Systemic Growth by Modulating Hormonal Signals through TOR-dependent Nutrient Sensing. Cell. Metab. 14, 403–414. doi:10.1016/J.CMET.2011.07.012
Storelli, G., Strigini, M., Grenier, T., Bozonnet, L., Schwarzer, M., Daniel, C., et al. (2018). Drosophila Perpetuates Nutritional Mutualism by Promoting the Fitness of its Intestinal Symbiont Lactobacillus Plantarum. Cell. Metab. 27, 362–377. doi:10.1016/j.cmet.2017.11.011
Tatar, M., Post, S., and Yu, K. (2014). Nutrient Control of Drosophila Longevity. Trends Endocrinol. Metabolism 25, 509–517. doi:10.1016/j.tem.2014.02.006
Téfit, M. A., and Leulier, F. (2017). Lactobacillus Plantarum Favors the Early Emergence of Fit and Fertile Adult Drosophila upon Chronic Undernutrition. J. Exp. Biol. 220, 900–907. doi:10.1242/JEB.151522
Trinder, M., Daisley, B. A., Dube, J. S., and Reid, G. (2017). Drosophila melanogaster as a High-Throughput Model for Host-Microbiota Interactions. Front. Microbiol. 8, 751. doi:10.3389/fmicb.2017.00751
Troha, K., Nagy, P., Pivovar, A., Lazzaro, B. P., Hartley, P. S., and Buchon, N. (2019). Nephrocytes Remove Microbiota-Derived Peptidoglycan from Systemic Circulation to Maintain Immune Homeostasis. Immunity 51, 625–637. doi:10.1016/j.immuni.2019.08.020
Vakkayil, K. L., and Hoppe, T. (2022). Temperature-Dependent Regulation of Proteostasis and Longevity. Front. Aging 3, 15. doi:10.3389/FRAGI.2022.853588
Valenzano, D. R., Terzibasi, E., Cattaneo, A., Domenici, L., and Cellerino, A. (2006). Temperature Affects Longevity and Age-Related Locomotor and Cognitive Decay in the Short-Lived Fish Nothobranchius Furzeri. Aging Cell. 5, 275–278. doi:10.1111/J.1474-9726.2006.00212.X
Wong, A. C.-N., Chaston, J. M., and Douglas, A. E. (2013). The Inconstant Gut Microbiota of Drosophila Species Revealed by 16S rRNA Gene Analysis. ISME J. 7 (7), 1922–1932. doi:10.1038/ismej.2013.86
Woodcock, K. J., Kierdorf, K., Pouchelon, C. A., Vivancos, V., Dionne, M. S., and Geissmann, F. (2015). Macrophage-Derived Upd3 Cytokine Causes Impaired Glucose Homeostasis and Reduced Lifespan in Drosophila Fed a Lipid-Rich Diet. Immunity 42, 133–144. doi:10.1016/j.immuni.2014.12.023
Yamada, R., Deshpande, S. A., Bruce, K. D., Mak, E. M., and Ja, W. W. (2015). Microbes Promote Amino Acid Harvest to Rescue Undernutrition in Drosophila. Cell. Rep. 10, 865–872. doi:10.1016/J.CELREP.2015.01.018
Yamauchi, T., Oi, A., Kosakamoto, H., Akuzawa-tokita, Y., Murakami, T., Mori, H., et al. (2020). Gut Bacterial Species Distinctively Impact Host Purine Metabolites during Aging in Drosophila. iScience 23, 101477. doi:10.1016/j.isci.2020.101477
Keywords: drosophila, aging, microbiota, lifespan, dysbiosis, dysplasia, intestinal immunity
Citation: Arias-Rojas A and Iatsenko I (2022) The Role of Microbiota in Drosophila melanogaster Aging. Front. Aging 3:909509. doi: 10.3389/fragi.2022.909509
Received: 31 March 2022; Accepted: 22 April 2022;
Published: 19 May 2022.
Edited by:
Stanislava Chtarbanova, University of Alabama, United StatesReviewed by:
Rupinder Kaur, Vanderbilt University, United StatesCopyright © 2022 Arias-Rojas and Iatsenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Igor Iatsenko, aWF0c2Vua29AbXBpaWItYmVybGluLm1wZy5kZQ==
 Aranzazu Arias-Rojas
Aranzazu Arias-Rojas Igor Iatsenko
Igor Iatsenko