1 Introduction
1.1 The challenge of understanding individual variation in immunosenescence
The global demographic is shifting towards an aged population (United Nations, 2019). Yet, it is our oldest members of society that are the most vulnerable to infectious disease, reflecting immune system ageing, or “immunosenescence” (Glynn and Moss, 2020; Chen et al., 2021). Immunosenescence has two defining features: heightened susceptibility to pathogens, and increased systemic, basal inflammation (Franceschi, 2000; Zerofsky et al., 2005; Benayoun et al., 2019), which is strongly linked to the development of age-related diseases such as neurodegeneration and cancer (Franceschi et al., 2018; Rea, 2018). Understanding the mechanisms of immunosenescence could offer therapeutic potential in reducing age-associated morbidity (Neves and Sousa-Victor, 2020; Borgoni et al., 2021). However, individuals do not age at the same rate, with variation arising from genotype (Burger and Promislow, 2006; Belsky et al., 2015), including sex (Sampathkumar et al., 2020; Xirocostas et al., 2020; Bronikowski et al., 2022), and from the environment. While most mechanistic biology research ignores this diversity, largely due to the practicality of using single genotypes and sexes, individual variation will determine the response to therapeutics targeting ageing pathologies (Meyer et al., 2013; Fuselli, 2019). Here, we champion the use of Drosophila melanogaster as an ideal model for the early stages of an approach that aims to leverage population variation to understand immunosenescence and how to treat it. We briefly describe the rich history of this model system in the fields of the biology of ageing, biogerontology and immunology, and their conceptual and technical convergence on the fly model. We outline the unique potential of Drosophila in elucidating variation in immunosenescence, its underlying mechanisms and its treatments.
1.2 Drosophila’s central role in formulating evolutionary theories of ageing—why do organisms age?
Drosophila have proven pivotal in the synthesis and empirical testing of the evolutionary theories of ageing, which provide a conceptual framework within which to understand the biology of ageing. Experimental evolution experiments highlighted a trade-off between longevity and early-life fecundity (Rose and Charlesworth, 1981; Luckinbill and Clare, 1985; Rose, 1991). Such fitness trade-offs are commonly seen in insects (Flatt et al., 2013) and give credence to Williams’ theory of antagonistic pleiotropy (Williams, 1957). These trade-offs may rely on resource allocation between germline and “disposable” soma (Kirkwood, 1977), and are not a universal, but rather a contextual, quid pro quo that depends on environmental factors. The range of phenotypic expression of a genotype across varying environments is called phenotypic plasticity. It is an important source of natural variation for life history traits. This has been demonstrated in multiple settings, such as temperature and commensal status, using Drosophila (e.g., Lee et al., 2019; Huang et al., 2020). An influential evolutionary theory, the resource reallocation hypothesis, suggests that natural selection may favour phenotypically plastic responses in allocation, prioritising somatic maintenance during scarcity, and reproduction in replete settings (Shanley and Kirkwood, 2000; Regan, 2020). Drosophila has offered an easy-to-manipulate empirical platform to understand these phenotypically plastic responses to nutritional resources, and their associated trade-offs (Lee et al., 2008; Jensen et al., 2015; Lee, 2015; Zanco et al., 2021). These studies have served to shape our understanding of key evolutionary concepts on the consequence of reproductive investment, and phenotypically plastic responses to the environment, in determining rates of ageing.
1.3 Drosophila’s central role in biogerontology—how can we slow down ageing?
The goal of Biogerontology, born out of evolutionary biology, is to enhance healthy ageing by limiting age-associated multi-morbidity through therapeutic intervention. Key to this goal is a thorough understanding of the underlying molecular mechanisms that govern the ageing process. Ageing has been studied using flies for over a century (Hyde, 1913; Piper and Partridge, 2018). One of the observations fundamental to modern-day biogerontology was the remarkably beneficial effects of dietary restriction on ageing across taxa. The use of invertebrate model organisms, such as flies and worms, has given insight into the dietary macronutrients, genes and pathways underpinning this effect (Grandison et al., 2009; Piper and Partridge, 2018). These studies have not only implicated environment-sensing pathways like IIS (Clancy et al., 2001; Tatar et al., 2001), and TOR (Vellai et al., 2003; Kapahi, 2004), but also numerous longevity-associated pathways including Toll, Ras-ERK-ETS, AMPK, and Myc pathways (Piper and Partridge, 2018). The extension of lifespan through targeted manipulation of pathways such as TOR was initially demonstrated in invertebrates (Vellai et al., 2003), including Drosophila (Bjedov et al., 2010), before it was shown to be effective in mammals (Harrison et al., 2009).
1.4 Drosophila’s central role in immunology—the genetics of innate immunity
The famous intersection of genetic tractability with immunology following the identification of Toll as an immune signalling molecule in Drosophila (Lemaitre et al., 1996), and later characterisation of mammalian Toll-like pathogen-recognition receptors (Medzhitov et al., 1997), sealed Drosophila’s fate as an indispensable model system in innate immunity research. Indeed, there are multiple conserved immune components of the model. Immune signalling culminates in expression of antimicrobial peptides (AMPs) and immune effectors via conserved NF-kB-like transcription factors (Lemaitre and Hoffmann, 2007; Buchon et al., 2014). Haematopoiesis, controlled by conserved transcription factors (Evans et al., 2003), gives rise to macrophage-like hemocytes which share a common superfamily of phagocytic receptors with humans (Kurucz et al., 2007). Epithelial immunity, such as the gut, is governed by conserved immune signalling, regulates commensal interactions, and is structurally similar to that of humans (Liu et al., 2017).
1.5 Power to the fly: Drosophila as a nexus for immunity, evolution and biogerontology to understand variation in immunosenescence
The rich history of Drosophila as a model system, spanning more than a century, has produced biological insights too numerous to mention. Here, we have briefly detailed the contributions of the model to three, interconnected disciplines. The power, we argue, in using Drosophila as a model in immunosenescence, which shares many similarities with that of mammals and has been reviewed elsewhere (Flatt and Garschall, 2018; Min and Tatar, 2018; Sciambra and Chtarbanova, 2021), comes from the conceptual and technical convergence of these fields on the system (Figure 1). Coupling the genetic and technical approaches possible in Drosophila with an evolutionary biology approach to address different forms of variation in immune responses will not only provide mechanistic insights into immunosenescence, but will also guide our ultimate goal of treating it.
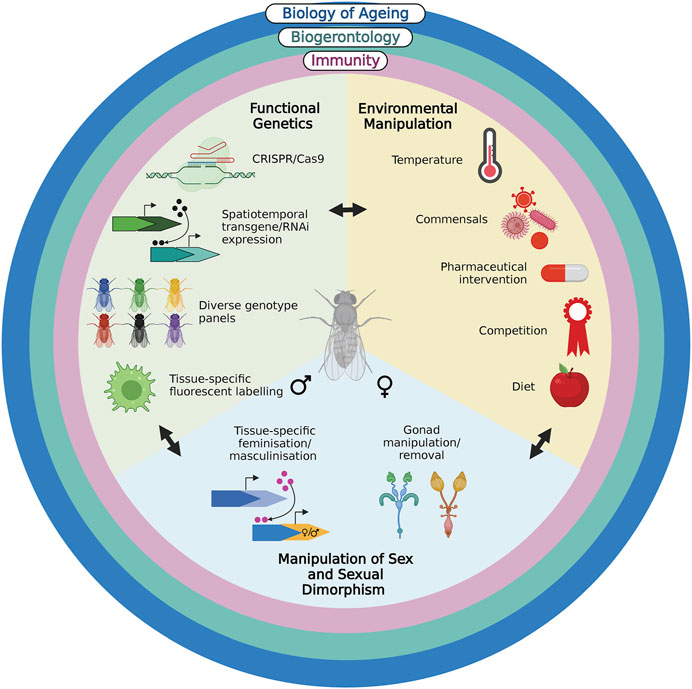
FIGURE 1. Drosophila’s rich history in the fields of evolutionary biology of ageing, biogerontology and immunity have endowed the model with multiple conceptual approaches and techniques. The functional genetic approaches, ease of environmental and sex manipulation render it invaluable in understanding natural variation in immunosenescence and its potential treatments. Created with BioRender.com.
1.5.1 Genetic variation
Genetic and phenotypic heterogeneity in immune responses are ubiquitous across species. Panels of fully-sequenced, isogenic lines derived from wild-caught Drosophila (Mackay, 2012; Grenier et al., 2015), or from extensive recombination (King, 2012), are an important community resource for understanding the genetic bases of complex phenotypic traits. These lines have been used to explore natural variation in, and genetics underpinning, susceptibility to pathogens. Susceptibility is determined by both resistance, which describes the ability to control pathogen burden (Wang et al., 2017; Palmer, 2018; Chapman et al., 2020) and disease tolerance (Howick and Lazzaro, 2017), which describes the ability to cope with infection and represents a less well-understood immune strategy (Soares et al., 2017; Schneider, 2021). Drosophila offers an ideal model to address the elusive processes determining disease tolerance, as they offer readily measurable fitness metrics and genetic tractability (e.g., Kutzer and Armitage, 2016; Duneau et al., 2017a; Gupta and Vale, 2017). Assessing variation in disease tolerance to Providencia rettgeri has implicated several biological processes, such as genes related to the endoplasmic reticulum (Howick and Lazzaro, 2017), which was further linked to tolerance in a transcriptomic analysis of infection response (Troha et al., 2018). These studies have granted some initial insight into the physiology underpinning this relatively elusive immune strategy, and genetic variation therein.
Use of isogenic panels could be extended to explore natural variation and plasticity in immunosenescence and the immune strategies affected by age. The role for declines in disease tolerance, and its variation, in driving immunosenescence is currently a black box, largely due to our lack of understanding of the mechanisms underpinning tolerance. There is a dearth of genotypically high-powered experiments assessing genetic variation in age-related immune dysregulation, despite prevalent genotype-by-age interactions (Lesser et al., 2006; Felix et al., 2012).
By performing genome wide association analysis (GWAS; e.g., King, 2012; Mackay, 2012) when assessing such variation in immunosenescence among the isogenic panels, candidate genes and pathways could potentially be identified, and ultimately validated. The ability to conditionally manipulate virtually any gene in the fly genome, through the spatiotemporal control of transgene expression via binary genetic systems (Brand and Perrimon, 1993; Osterwalder et al., 2001), makes validation of candidate genes possible in high throughput. These tools grant the model relative ease in assessing canonical and non-canonical components of immune defence and their impact on immunosenescence over ageing. Additionally, Drosophila are tractable to reverse genetic knock-out approaches using CRISPR/Cas9 technology, as has been demonstrated in the functional assessment of individual AMPs (Hanson et al., 2019). Recently, the potential of such CRISPR lines in the functional assessment of AMPs over ageing has been demonstrated. While ageing is associated with expansion of the AMP repertoire and loss of specificity, possibly indicative of immune dysregulation (Shit et al., 2022), their collective role in regulating microbiome dysbiosis was shown to be a critical determinant of lifespan (Hanson and Lemaitre, 2022), the control of which has previously been linked to lifespan and ageing pathologies (Guo et al., 2014; Clark et al., 2015; Li et al., 2016) Finally, fluorescent labelling of immune tissues, such as hemocytes (e.g., Sanchez Bosch et al., 2019; Krejčová, 2019; Chakrabarti and Visweswariah, 2020; Kierdorf, 2020; Coates, 2021), opens the possibility to assess their contribution to immune, and systemic, ageing.
Flies are highly amenable to experimental evolution, which when coupled with genomic sequencing, allows inferences to be made about the genetic response to a dominant selection pressure, such as delayed reproduction or longevity (McHugh and Burke, 2022). This technique can be used to explore trade-offs between immunity and life-history traits (McKean et al., 2008; Shahrestani et al., 2021), or lack thereof (Faria, 2015; Gupta, 2016). In lines selected for longevity, an improvement of measurable immune responses was observed (Fabian, 2018), whereas selection for resistance against a specific fungal pathogen was costly to lifespan (Shahrestani et al., 2021). It is possible, and remains to be tested, that improved early immunity has detrimental effects on the rate of immunosenescence, ultimately affecting ageing.
1.5.2 Plasticity in immunity
Immunology research has historically focused on the genetics underpinning immune responses within a controlled environment, but rarely assesses reaction norms under varying conditions (Martin, 2021). Drosophila has offered insight into such immune plasticity, including genotype-by-temperature (Lazzaro et al., 2008), and genotype-by-diet interactions using isogenic lines. The observed effects of diet on resistance (Unckless et al., 2015) underscore the importance of nutrition in determining immune responses (for e.g., Galenza, 2016; Ponton et al., 2020). Similarly, disease tolerance plasticity was observed in a diet- and pathogen-specific manner (Howick and Lazzaro, 2014; Kutzer and Armitage, 2016). Through the assessment of genotype-by-environment interactions, these investigations have highlighted an intrinsic link between immunity and metabolism, as has been confirmed elsewhere, for example, the induction of immune-response genes downstream of IIS and TOR nutrient-sensing pathways independently of Toll or Imd signalling (Becker et al., 2010; Varma et al., 2014). Thus, environmental factors are an important source of variation in immunity, as predicted by evolutionary theory.
Little is known about phenotypic plasticity in the manifestation of immunosenescence. Yet, the question “what kind of environments accelerate or slow down immunosenescence?” is crucial to an evolutionary biology approach in treating immune decline. Plasticity in immune responses in the face of varying environments early in life (Lazzaro and Little, 2009; Leech, 2019) is likely to have consequences on immunosenescence. While such plasticity is often ignored, or difficult to address, as in mouse studies (Martin, 2021), assessing the expression of immune responses across differing environments in flies is relatively tenable (McKean and Nunney, 2005; Lazzaro et al., 2008; Unckless et al., 2015).
Pharmacological intervention represents a highly relevant example of environmental manipulation, and we have barely scratched the surface in understanding genetic variation underpinning responses to geroprotective treatments (but see Lind, 2017; Rohde et al., 2021). Through robust genetic validation, flies have provided mechanistic insight into the action of multiple potential geroprotective drugs (for e.g., Bjedov et al., 2010; Slack et al., 2015; Castillo-Quan et al., 2019). Drosophila is the ideal candidate model to move towards high-powered studies addressing variation in response to either environmental (e.g., dietary restriction) or pharmacological (e.g., therapeutic mTOR attenuation) interventions targeting ageing, including immunosenescence. The crucial next step in biogerontology is to capture individual variation in responses to therapeutics, which, in contrast to mammalian systems, can be tested with relative ease in Drosophila.
1.5.3 Sex bias in immunity
Sex differences in immune responses, which have been observed across taxa (Kelly et al., 2018; Metcalf et al., 2020), are prevalent in Drosophila infectious disease models (Belmonte et al., 2020). Sex differences in the immune response appear contextual in Drosophila (Belmonte et al., 2020), mirroring the lack of ubiquitous sex differences across taxa (Kelly et al., 2018). Sex differences in infection outcome are likely to be dictated by a complex interplay of environmental factors, pathogen-specific consequences of infection, and the different life-histories of the sexes. Notwithstanding this complexity, fruit flies have offered mechanistic insight into pathogen-specific, dimorphic responses to infection, such as sex differences in the Toll pathway (D. Duneau et al., 2017a; Shahrestani et al., 2018), and sex-by-genotype interactions determining viral load and transmission potential of Drosophila C virus (Siva-Jothy and Vale, 2021).
The inclusion of both sexes when using isogenic, transgenic or outbred Drosophila lines will invariably provide insight into sex differences in immunosenescence. In particular, examining dimorphism across genetic panels could address the interaction of sex and ageing on immunosenescence. Furthermore, the ability to remove the gonads prior to next generation sequencing facilitates the examination of somatic, sex differential gene expression, as was done to compare the immune response to P. rettgeri (Duneau et al., 2017b). Coupling the cell autonomous sex determination system of Drosophila (Salz and Erickson, 2010) with their robust genetic toolkit allows the manipulation of sex in a tissue-specific manner, as has been demonstrated in exploration of sexual dimorphisms in physiology (Hudry et al., 2016; Millington et al., 2021) and in the infection response and pathology of the ageing gut (Regan et al., 2016). This offers an unparalleled system to tease apart how tissue-specific and systemic sex differences contribute to individual variation in immunosenescence. Considering the conserved dimorphism in both immune function and responses to drugs that delay ageing (Bjedov et al., 2010; Arriola Apelo and Lamming, 2016; Bitto et al., 2016; Regan et al., 2016; Regan et al., 2021; Strong et al., 2020); Drosophila genetic techniques could be leveraged for the early stages of developing sex-optimised drug treatments for immunosenescence.
2 Conclusion
Immune decline over ageing is linked to age-associated morbidity through increased pathogen susceptibility and dysregulated inflammation. Yet, immunosenescence remains relatively elusive where individual-to-individual variation in the onset and extent is virtually unknown, potentially determining responses to treatments targeting the decline. Steps to capturing population diversity in immune responses, arising from genotype, phenotypic plasticity and sex, discussed here, would provide mechanistic insight if applied to immunosenescence. We argue that Drosophila is uniquely situated to address the challenge of variation in ageing. Its vast history as a model in the fields of evolutionary biology, biogerontology, and immunity offers a foundation of knowledge about the species and a matchless system of tractable genetics, statistical power, and environmental manipulation. Using Drosophila to initiate an evolutionary biology approach to immunosenescence would address population diversity and, ultimately, aid the development of personalised therapeutics.
Author contributions
M-KC and JR conceived the article. M-KC wrote the manuscript with contribution and editing from JR. Both authors contributed to the article and approved the submitted version.
Funding
M-KC and JR were funded by The University of Edinburgh.
Acknowledgments
Thanks to David F. Duneau for valuable comments on the manuscript, and to members of the Regan lab for discussion.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arriola Apelo, S. I., and Lamming, D. W. (2016). Rapamycin: An InhibiTOR of aging emerges from the soil of easter island. J. Gerontol. A Biol. Sci. Med. Sci. 71 (7), 841–849. doi:10.1093/gerona/glw090
Becker, T., Loch, G., Beyer, M., Zinke, I., Aschenbrenner, A. C., Carrera, P., et al. (2010). FOXO-dependent regulation of innate immune homeostasis. Nature 463 (7279), 369–373. doi:10.1038/nature08698
Belmonte, R. L., Corbally, M. K., Duneau, D. F., and Regan, J. C. (2020). Sexual dimorphisms in innate immunity and responses to infection in Drosophila melanogaster. Front. Immunol. 10, 3075. doi:10.3389/FIMMU.2019.03075/BIBTEX
Belsky, D. W., Caspi, A., Houts, R., Cohen, H. J., Corcoran, D. L., Danese, A., et al. (2015). Quantification of biological aging in young adults. Proc. Natl. Acad. Sci. U. S. A. 112 (30), E4104–E4110. doi:10.1073/pnas.1506264112
Benayoun, B. A., Pollina, E. A., Priya Singh, P., Mahmoudi, S., Harel, I., Casey, K. M., et al. (2019). Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709. doi:10.1101/gr.240093.118
Bitto, A., Ito, T. K., Pineda, V. V., LeTexier, N. J., Huang, H. Z., Sutlief, E., et al. (2016). Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. eLife 5, e16351. doi:10.7554/eLife.16351
Bjedov, I., Toivonen, J. M., Kerr, F., Slack, C., Jacobson, J., Foley, A., et al. (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster, ” Cell metabolism. Cell Metab. 11 (1), 35–46. doi:10.1016/J.CMET.2009.11.010
Borgoni, S., Kudryashova, K. S., Burka, K., and de Magalhaes, J. P. (2021). Targeting immune dysfunction in aging. Ageing Res. Rev. 70, 101410. doi:10.1016/j.arr.2021.101410
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 (2), 401–415. doi:10.1242/dev.118.2.401
Bronikowski, A. M., Meisel, R. P., Peggy, R., Biga, R., Walters, J. R., Mank, J. E., et al. (2022). Sex-specific aging in animals: Perspective and future directions. Aging Cell 21, e13542e13542. doi:10.1111/acel.13542
Buchon, N., Silverman, N., and Cherry, S. (2014). Immunity in Drosophila melanogaster — From microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. doi:10.1038/nri3763
Burger, J. M. S., and Promislow, D. E. L. (2006). Are functional and demographic senescence genetically independent? Exp. Gerontol. 41 (11), 1108–1116. doi:10.1016/j.exger.2006.08.008
Castillo-Quan, J. I., Tain, L. S., Kinghorn, K. J., Li, L., Gronke, S., Hinze, Y., et al. (2019). A triple drug combination targeting components of the nutrient-sensing network maximizes longevity. Proc. Natl. Acad. Sci. U. S. A. 116 (42), 20817–20819. doi:10.1073/pnas.1913212116
Chakrabarti, S., and Visweswariah, S. S. (2020). Intramacrophage ROS primes the innate immune system via JAK/STAT and Toll activation. Cell Rep. 33 (6), 108368. doi:10.1016/J.CELREP.2020.108368
Chapman, J. R., Dowell, M. A., Chan, R., and Unckless, R. L. (2020). The genetic basis of natural variation in Drosophila melanogaster immune defense against Enterococcus faecalis. Genes 11, 234. doi:10.3390/GENES11020234
Chen, Y., Klein, S. L., Garibaldi, B. T., Li, H., Wu, C., Osevala, N. M., et al. (2021). Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res. Rev. 65, 101205. doi:10.1016/j.arr.2020.101205
Clancy, D. J., Gems, D., Harshman, L. G., Oldham, S., Stocker, H., Hafen, E., et al. (2001). Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein,” Science. Am. Assoc. Adv. Sci., 292, 104–106. doi:10.1126/SCIENCE.1057991/ASSET/04E1720A-7F71-4F12-A9DB-00508D22FF36/ASSETS/GRAPHIC/SE1219311004
Clark, R. I., Salazar, A., Yamada, R., Fitz-Gibbon, R., Morselli, R., Alcaraz, J., et al. (2015). Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep. 12, 1656–1667. doi:10.1016/J.CELREP.2015.08.004
Coates, J. A. (2021). Identification of functionally distinct macrophage subpopulations in drosophila eLife 10. doi:10.7554/ELIFE.58686
Duneau, D., Ferdy, J. B., Revah, J., Kondolf, H., Ortiz, G. A., Lazzaro, P. B., et al. (2017a). Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. eLife, 6, 28298. doi:10.7554/ELIFE.28298
Duneau, D., Kondolf, H. C., Im, J. H., Ortiz, G. A., Chow, C., Fox, M. A., et al. (2017b). The Toll pathway underlies host sexual dimorphism in resistance to both Gram-negative and Gram-positive bacteria in mated Drosophila. BMC Biol. 15 (1), 124. doi:10.1186/s12915-017-0466-3
Evans, C. J., Hartenstein, V., and Banerjee, U. (2003). Thicker than blood: Conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 5 (5), 673–690. doi:10.1016/S1534-5807(03)00335-6
Fabian, D. K. (2018). Evolution of longevity improves immunity in Drosophila. Evol. Lett. 2, 567–579. doi:10.1002/evl3.89
Faria, V. G. (2015). Evolution of Drosophila resistance against different pathogens and infection routes entails no detectable maintenance costs. Evol. Int. J. Org. Evol 69, 2799–2809. doi:10.1111/EVO.12782
Felix, T. M., Hughes, K. A., Stone, E. A., Drnevich, J. M., and Leips, J. (2012). Age-specific variation in immune response in Drosophila melanogaster has a genetic basis. Genetics 191 (3), 989–1002. doi:10.1534/genetics.112.140640
Flatt, T., Stone, E. A., Drnevich, J. M., and Leips, J. (2013). Life-history evolution and the polyphenic regulation of somatic maintenance and survival. Q. Rev. Biol. 88, 185–218. doi:10.1086/671484/ASSET/IMAGES/LARGE/FG6_ONLINE
Flatt, T., and Garschall, K. (2018). The interplay between immunity and aging in Drosophila. F1000Research 7, 1. doi:10.12688/F1000RESEARCH.13117.1
Franceschi, C. (2000). Inflamm-aging. An evolutionary perspective on immunosenescence,” Annals of the New York Academy of Sciences. Ann N Y Acad Sci 908, 244–254. doi:10.1111/J.1749-6632.2000.TB06651.X
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C., and Santoro, A. (2018). Inflammaging: A new immune–metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14 (10), 576–590. doi:10.1038/s41574-018-0059-4
Fuselli, S. (2019). Beyond drugs: The evolution of genes involved in human response to medications. Proc. Biol. Sci. 286, 20191716. doi:10.1098/rspb.2019.1716
Galenza, A (2016). Glucose modulates Drosophila longevity and immunity independent of the microbiota. Biol. open 5, 165–173. doi:10.1242/bio.015016
Glynn, J. R., and Moss, P. A. H. (2020). Systematic analysis of infectious disease outcomes by age shows lowest severity in school-age children. Sci. Data 7 (1), 329. doi:10.1038/s41597-020-00668-y
Grandison, R. C., Piper, M. D. W., and Partridge, L. (2009). Amino acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462 (7276), 1061–1064. doi:10.1038/NATURE08619
Grenier, J. K., Arguello, J. R., Moreira, M. C., Gottipati, S., Mohammed, J., Hackett, S. R., et al. (2015). Global diversity lines - a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 5 (4), 593–603. doi:10.1534/g3.114.015883
Guo, L., Karpac, J., Tran, S. L., and Jasper, H. (2014). PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 156 (0), 109–122. doi:10.1016/J.CELL.2013.12.018
Gupta, V. (2016). No apparent cost of evolved immune response in Drosophila melanogaster. Evol. Int. J. Org. Evol, 70, 934–943. doi:10.1111/EVO.12896
Gupta, V., and Vale, P. F. (2017). Nonlinear disease tolerance curves reveal distinct components of host responses to viral infection. R. Soc. Open Sci. 4, 170342. doi:10.1098/RSOS.170342
Hanson, M. A., Dostalova, A., Ceroni, C., Poidevin, M., Kondo, S., and Lemaitre, B. (2019). Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341. doi:10.7554/eLife.44341
Hanson, M. A., and Lemaitre, B. (2022).Antimicrobial peptides do not directly contribute to aging in Drosophila, but improve lifespan by preventing dysbiosis. bioRxiv. doi:10.1101/2022.08.24.504952
Harrison, D. E., Strong, R., Sharp, Z. D., Nelson, J. F., Astle, C. M., Flurkey, K., et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460 (7253), 392–395. doi:10.1038/nature08221
Howick, V. M., and Lazzaro, B. P. (2014). Genotype and diet shape resistance and tolerance across distinct phases of bacterial infection. BMC Evol. Biol. 14 (1), 56. doi:10.1186/1471-2148-14-56
Howick, V. M., and Lazzaro, B. P. (2017). The genetic architecture of defence as resistance to and tolerance of bacterial infection in Drosophila melanogaster. Mol. Ecol. 26, 1533–1546. doi:10.1111/MEC.14017
Huang, W., Campbell, T., Carbone, M. A., Jones, W. E., Unselt, D., Anholt, R. R. H., et al. (2020). Context-dependent genetic architecture of Drosophila life span. PLoS Biol. 18 (3), e3000645. doi:10.1371/JOURNAL.PBIO.3000645
Hudry, B., Khadayate, S., and Miguel-Aliaga, I. (2016). The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature 530 (7590), 344–348. doi:10.1038/nature16953
Jensen, K., McClure, C., Priest, N. K., and Hunt, J. (2015). Sex-specific effects of protein and carbohydrate intake on reproduction but not lifespan in Drosophila melanogaster. Aging Cell 14 (4), 605–615. doi:10.1111/acel.12333
Kapahi, P. (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. CB, 14, 885–890. doi:10.1016/J.CUB.2004.03.059
Kelly, C. D., Stoehr, A. M., Nunn, C., Smyth, K. N., and Prokop, Z. M. (2018). Sexual dimorphism in immunity across animals: A meta-analysis. Ecol. Lett. 21 (12), 1885–1894. doi:10.1111/ELE.13164
Kierdorf, K. (2020). Muscle function and homeostasis require cytokine inhibition of AKT activity in Drosophila. eLife 9, 51595. doi:10.7554/ELIFE.51595
King, E. G. (2012). Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22, 1558–1566. doi:10.1101/GR.134031.111
Krejčová, G. (2019). Drosophila macrophages switch to aerobic glycolysis to mount effective antibacterial defense. eLife 8, 50414. doi:10.7554/ELIFE.50414
Kurucz, É., Markus, R., Zsamboki, J., Folkl-Medzihradszky, K., Darula, Z., Vilmos, P., et al. (2007). Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes, ” Current biology: CB. Curr. Biol. 17 (7), 649–654. doi:10.1016/J.CUB.2007.02.041
Kutzer, M. A. M., and Armitage, S. A. O. (2016). The effect of diet and time after bacterial infection on fecundity, resistance, and tolerance in Drosophila melanogaster. Ecol. Evol. 6, 4229. doi:10.1002/ECE3.2185
Lazzaro, B. P., Flores, H. A., Lorigan, J. G., and Yourth, C. P. (2008). Genotype-by-Environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster, ” PLoS pathogens. PLoS Pathog. 4 (3), e1000025. doi:10.1371/JOURNAL.PPAT.1000025
Lazzaro, B. P., and Little, T. J. (2009). Immunity in a variable worldSeries B, Biological sciences. Philos. Trans. R. Soc. Lond B Biol. Sci. 364, 15–26. doi:10.1098/RSTB.2008.0141
Lee, H.-Y., Lee, S. H., Lee, J. H., Lee, W. J., and Min, K. J. (2019). The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging 11 (13), 4611–4640. doi:10.18632/aging.102073
Lee, K. P. (2015). Dietary protein:carbohydrate balance is a critical modulator of lifespan and reproduction in Drosophila melanogaster: A test using a chemically defined diet. J. Insect Physiol. 75, 12–19. doi:10.1016/j.jinsphys.2015.02.007
Lee, K. P., Simpson, S. J., Clissold, F. J., Brooks, R., Ballard, J. W. O., Taylor, P. W., et al. (2008). Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc. Natl. Acad. Sci. U. S. A. 105 (7), 2498–2503. doi:10.1073/pnas.0710787105
Leech, T. (2019). Interactive effects of social environment, age and sex on immune responses in Drosophila melanogaster. J. Evol. Biol. 32, 1082–1092. doi:10.1111/JEB.13509
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi:10.1146/ANNUREV.IMMUNOL.25.022106.141615
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M., and Hoffmann, J. A. (1996). The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults Cell 86 (6), 973–983. doi:10.1016/S0092-8674(00)80172-5
Lesser, K. J., Paiusi, I. C., and Leips, J. (2006). Naturally occurring genetic variation in the age-specific immune response of Drosophila melanogaster. Aging Cell 5 (4), 293–295. doi:10.1111/j.1474-9726.2006.00219.x
Li, H., Qi, Y., and Jasper, H. (2016). Preventing age-related decline of gut compartmentalization limits microbiota dysbiosis and extends lifespan. Cell Host Microbe 19 (2), 240–253. doi:10.1016/j.chom.2016.01.008
Lind, M. I. (2017). Rapamycin additively extends lifespan in short- and long-lived lines of the nematode Caenorhabditis remanei. Exp. Gerontol. 90, 79–82. doi:10.1016/j.exger.2017.01.017
Liu, X., Hodgson, J. J., and Buchon, N. (2017). Drosophila as a model for homeostatic, antibacterial, and antiviral mechanisms in the gut. PLoS Pathog. 13 (5), e1006277. doi:10.1371/journal.ppat.1006277
Luckinbill, L. S., and Clare, M. J. (1985). Selection for life span in Drosophila melanogaster, ” Heredity 1985 55:1. Heredity 55 (1), 9–18. doi:10.1038/hdy.1985.66
Mackay, T. F. C. (2012). The Drosophila melanogaster genetic reference panel. Nature 482, 173–178. doi:10.1038/nature10811
Martin, L. B. (2021). Genes, environments, and phenotypic plasticity in immunology. Trends Immunol. 42, 198–208. doi:10.1016/J.IT.2021.01.002
McHugh, K. M., and Burke, M. K. (2022). From microbes to mammals: The experimental evolution of aging and longevity across species. Evolution 76 (4), 692–707. doi:10.1111/evo.14442
McKean, K. A., and Nunney, L. (2005). BATEMAN’S principle and immunity: PHENOTYPICALLY plastic reproductive strategies predict changes in immunological sex differences, ” evolution; international journal of organic evolution. Evol. 59 (7), 1510–1517. doi:10.1554/04-657
McKean, K. A., Yourth, C. P., Lazzaro, B. P., and Clark, A. G. (2008). The evolutionary costs of immunological maintenance and deployment. BMC Evol. Biol. 8, 76. doi:10.1186/1471-2148-8-76
Medzhitov, R., Preston-Hurlburt, P., and Janeway, C. A. (1997). A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388 (6640), 394–397. doi:10.1038/41131
Metcalf, C. J. E., Roth, O., and Graham, A. L. (2020). Why leveraging sex differences in immune trade-offs may illuminate the evolution of senescence. Funct. Ecol. 34 (1), 129–140. doi:10.1111/1365-2435.13458
Meyer, U. A., Zanger, U. M., and Schwab, M. (2013). Omics and drug response. Annu. Rev. Pharmacol. Toxicol. 53, 475–502. doi:10.1146/annurev-pharmtox-010510-100502
Millington, J. W., Brownrigg, G. P., Chao, C., Sun, Z., Basner-Collins, P. J., Wat, L. W., et al. (2021). Female-biased upregulation of insulin pathway activity mediates the sex difference in Drosophila body size plasticity. eLife 10, e58341. doi:10.7554/eLife.58341
Min, K. J., and Tatar, M. (2018). Unraveling the molecular mechanism of immunosenescence in Drosophila. Int. J. Mol. Sci. 19, 9, doi:10.3390/IJMS19092472
Neves, J., and Sousa-Victor, P. (2020). Regulation of inflammation as an anti-aging intervention. FEBS J., 287, 43–52. doi:10.1111/febs.15061
Osterwalder, T., Yoon, K. S., White, B. H., and KesHisHian, H. (2001). A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U. S. A. 98 (22), 12596–12601. doi:10.1073/PNAS.221303298
Palmer, W. H. (2018). Isolation of a natural DNA virus of Drosophila melanogaster, and characterisation of host resistance and immune responses. Public Libr. Sci. 14, e1007050. doi:10.1371/journal.ppat.1007050
Piper, M. D. W., and Partridge, L. (2018). Drosophila as a model for ageing. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1864, 2707–2717. doi:10.1016/J.BBADIS.2017.09.016
Ponton, F., Morimoto, J., Robinson, K., Kumar, S. S., Cotter, S. C., Wilson, K., et al. (2020). Macronutrients modulate survival to infection and immunity in Drosophila. J. Anim. Ecol. 89 (2), 460–470. doi:10.1111/1365-2656.13126
Rea, I. M. (2018). Age and age-related diseases: Role of inflammation triggers and cytokines. Frontiers in immunology, 9, 586. doi:10.3389/fimmu.2018.00586
Regan, J. C. (2020). Dietary restriction and insulin-like signalling pathways as adaptive plasticity: A synthesis and re-evaluationFunct. Ecol. 34, 107–128. doi:10.1111/1365-2435.13418/SUPPINFO
Regan, J. C., Khericha, M., Dobson, A. J., Bolukbasi, E., Rattanavirotkul, N., and Partridge, L. (2016). Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. eLife 5, e10956. doi:10.7554/eLife.10956
Regan, J. C., Lu, Y. X., Urena, E., Meilenbrock, R., Catterson, J. H., KiBler, D., et al. (2021). Sexual identity of enterocytes regulates rapamycin-mediated intestinal homeostasis and lifespan extension. bioRxiv. doi:10.1101/2021.10.22.465415
Rohde, P. D., Bocker, A., Jensen, C. A. B., Bergstrom, A. L., Madsen, M. I. J., Christensen, S. L., et al. (2021). Genotype and trait specific responses to rapamycin intake in Drosophila melanogaster. Insects 12 (5), 474. doi:10.3390/insects12050474
Rose, M. R., and Charlesworth, B. (1981). Genetics of life history in DROSOPHILA MELANOGASTER. II. Exploratory selection experiments. Genetics 97 (1), 187–196. doi:10.1093/GENETICS/97.1.187
Salz, H. K., and Erickson, J. W. (2010). Sex determination in Drosophila: The view from the top. Fly 4 (1), 60–70. doi:10.4161/fly.4.1.11277
Sampathkumar, N. K., Bravo, J. I., Chen, Y., Danthi, P. S., Donahue, E. K., Lai, R. W., et al. (2020). Widespread sex dimorphism in aging and age-related diseases. Hum. Genet. 139 (3), 333–356. doi:10.1007/s00439-019-02082-w
Sanchez Bosch, P., Makhijani, K., Herboso, L., Gold, K. S., Baginsky, R., Woodcock, K. J., et al. (2019). Adult Drosophila Lack Hematopoiesis but Rely on a Blood Cell Reservoir at the Respiratory Epithelia to Relay Infection Signals to Surrounding Tissues. Dev. cell 51 (6), 787–803.e5. doi:10.1016/j.devcel.2019.10.017
Schneider, D. S. (2021). Immunology’s intolerance of disease tolerance. Nat. Rev. Immunol. 21 (10), 624–625. doi:10.1038/s41577-021-00619-7
Sciambra, N., and Chtarbanova, S. (2021). The impact of age on response to infection in Drosophila, ” microorganisms. Microorganisms 9 (5), 958. doi:10.3390/MICROORGANISMS9050958
Shahrestani, P., Chambers, M., Vandenberg, J., Garcia, K., Malaret, G., Chowdhury, P., et al. (2018). Sexual dimorphism in Drosophila melanogaster survival of Beauveria bassiana infection depends on core immune signaling, ” Scientific reports. Sci. Rep. 8 (1), 12501. doi:10.1038/s41598-018-30527-1
Shahrestani, P., King, E., Ramezan, R., Phillips, M., Riddle, M., Thronburg, M., et al. (2021). The molecular architecture of Drosophila melanogaster defense against Beauveria bassiana explored through evolve and resequence and quantitative trait locus mapping. G3 Genes, 11, 324. doi:10.1093/G3JOURNAL/JKAB324
Shanley, D. P., and Kirkwood, T. B. L. (2000). Calorie restriction and aging: A life-history analysis, ” evolution; international journal of organic evolution. Evolution 54 (3), 740–750. doi:10.1111/J.0014-3820.2000.TB00076.X
Shit, B., Prakash, A., Sarkar, S., Vale, P. F., and Khan, I. (2022). Ageing leads to nonspecific antimicrobial peptide responses in Drosophila melanogaster. bioRxiv. doi:10.1101/2022.06.25.497570
Siva-Jothy, J. A., and Vale, P. F. (2021). Dissecting genetic and sex-specific sources of host heterogeneity in pathogen shedding and spread. PLoS Pathog. 17 (1), e1009196. doi:10.1371/journal.ppat.1009196
Slack, C., Alic, N., Foley, A., Cabecinha, M., Hoddinott, M. P., and Partridge, L. (2015). The ras-erk-ETS-signaling pathway is a drug target for longevity. Cell 162 (1), 72–83. doi:10.1016/j.cell.2015.06.023
Soares, M. P., Teixeira, L., and Moita, L. F. (2017). Disease tolerance and immunity in host protection against infection. Nat. Rev. Immunol. 1717, 283–96. doi:10.1038/nri.2016.136
Strong, R., Miller, R. A., Bogue, M., Fernandez, E., Javors, M. A., Libert, S., et al. (2020). Rapamycin-mediated mouse lifespan extension: Late-life dosage regimes with sex-specific effects. Aging Cell 19 (11), e13269. doi:10.1111/acel.13269
Tatar, M., KopelmAn, A., Epstein, D., Tu, M. P., Yin, C. M., and Garofalo, R. S. (2001). A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function, ” science. Science 292 (5514), 107–110. doi:10.1126/science.1057987
Troha, K., Hyum, J., Lazzaro, B. P., and Buchon, N. (2018). Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. Public Libr. Sci. 14, e1006847. doi:10.1371/journal.ppat.1006847
Unckless, R. L., Rottschaefer, S. M., and Lazzaro, B. P. (2015). The complex contributions of genetics and nutrition to immunity in Drosophila melanogaster. PLoS Genet. 11 (3), e1005030. doi:10.1371/JOURNAL.PGEN.1005030
United Nations (2019). World population prospects 2019 highlights. Available at: https://www.un.org/development/desa/publications/world-population-prospects-2019-highlights.html.
Varma, D., Bülow, M. H., Pesch, Y. Y., Loch, G., and Hoch, M. (2014). Forkhead, a new cross regulator of metabolism and innate immunity downstream of TOR in Drosophila. J. insect physiology 69, 80–88. doi:10.1016/J.JINSPHYS.2014.04.006
Vellai, T., Takacs-Vellai, K., Zhang, Y., Kovacs, A. L., Orosz, L., and Muller, F. (2003). Influence of TOR kinase on lifespan in C. elegans, ” Nature. Nature 426 (6967), 620–620. doi:10.1038/426620a
Wang, J. B., Lu, H. L., and St. Leger, R. J. (2017). The genetic basis for variation in resistance to infection in the Drosophila melanogaster genetic reference panel. Public Libr. Sci. 13 (3), e1006260. doi:10.1371/JOURNAL.PPAT.1006260
Williams, G. C. (1957). Pleiotropy, natural selection, and the evolution of senescence, ” evolution; international journal of organic evolution. Evolution 11 (4), 398. doi:10.2307/2406060
Xirocostas, Z. A., Everingham, S. E., and Moles, A. T. (2020). The sex with the reduced sex chromosome dies earlier: A comparison across the tree of life. Biol. Lett. 16 (3), 20190867. doi:10.1098/RSBL.2019.0867
Zanco, B., Mirth, C. K., Sgro, C. M., and Piper, M. D. (2021). A dietary sterol trade-off determines lifespan responses to dietary restriction in Drosophila melanogaster females. eLife 10, e62335. doi:10.7554/eLife.62335
Keywords: Drosophila, immunosenescence, immunity, ageing, natural variation
Citation: Corbally M-K and Regan JC (2022) Fly immunity comes of age: The utility of Drosophila as a model for studying variation in immunosenescence. Front. Aging 3:1016962. doi: 10.3389/fragi.2022.1016962
Received: 11 August 2022; Accepted: 20 September 2022;
Published: 04 October 2022.
Edited by:
Susan Broughton, Lancaster University, United KingdomReviewed by:
Myriam Richaud, UMR5247 Institut des Biomolécules Max Mousseron (IBMM), FranceCopyright © 2022 Corbally and Regan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer C. Regan, amVubnkucmVnYW5AZWQuYWMudWs=
 Mary-Kate Corbally
Mary-Kate Corbally Jennifer C. Regan
Jennifer C. Regan