To fully understand how the acquisition, maintenance and recall of memories is achieved is one of the holy grails of neuroscience. Since the definition of the g-factor for “general cognitive ability” over a century ago (
Spearman, 1904
) and the characterization of the landmark patient H.M. in 1957 (
Scoville and Milner, 1957
), the field of cognitive neuropsychology has been attempting to elucidate the brain structures and molecular players involved in these processes. However, most of this progress has been reached stepwise, working forward from brain structure to structure and molecule to molecule in a hypothesis-driven fashion (
Lee and Silva, 2009
). Recently, the advent of high-throughput genomic polymorphism scanning technologies has revolutionized our ability to search for the common genetic drivers of memory and other cognitive domains in a hypothesis-free manner (
Goldberg and Weinberger, 2004
;
Fisher, 2006
;
Payton, 2006
,
2009
;
Potkin et al., 2009
). In the case of KIBRA (also known as WWC1 for WW and C2 domain containing 1), the topic of this review, evidence for a link to cognition occurred independently both from genetic association studies (
Papassotiropoulos et al., 2006
) and from biochemical work (
Büther et al., 2004
).
Beyond understanding the way memory works, one practical goal of studying genes with relation to cognitive performance is to identify new treatment strategies that cope with the various cognitive disabilities for which treatment options are urgently needed. The population aging in industrialized countries is expected to result in a strong increase in cognitive health problems in the next decades, both as a result of aging-associated cognitive decline, as well as increased prevalence of neuropsychiatric diseases such as Alzheimer’s disease. Here, novel discoveries in cognition are sought to broaden the spectrum of pharmacologically addressable targets and hypotheses. The KIBRA-pathway appears to present a novel promising inroad for this, since KIBRA has been both linked to normal cognitive performance and to Alzheimer’s disease.
Here, we highlight the most important functional aspects of KIBRA, and connect genetic and biochemical data to aid in the formation of hypotheses related to KIBRA’s cellular and molecular function.
The KIBRA Gene and Protein
KIBRA was first cloned and initially characterized by
Kremerskothen et al. (2003)
as a molecule that interacts with the postsynaptic protein dendrin. Human KIBRA, encoded by the
WWC1 gene, is located on chromosome 5q35.1, and contains 1113 amino acids and has a predicted size or 125.3 kDa. From N- to C-terminus a number of known protein domains have been identified which are depicted in Figure
1
. The WW-domains (aa 6–86), which cover a stretch of 35–40 amino acids contain two conserved tryptophan residues. These domains are responsible for the interaction with various proteins containing proline-rich sequences (PPxY). A putative nuclear localization signal was identified between amino acids 361–376 (
Rayala et al., 2006
). A C2 domain composed of two four-stranded β-sheets is located between amino acid 655 and 783. The 130 residues of the C2 domain are involved in binding phospholipids in a calcium-dependent manner. C2 domains are found in proteins with functions ranging from signal transduction to vesicular trafficking (
Rizo and Sudhof, 1998
). Calcium binding induces a change in the electrostatic potential, which enhances phospholipid binding. A glutamic acid-rich region is located between amino acids 845 and 873 (
Kremerskothen et al., 2003
;
Rayala et al., 2006
). Lastly, a putative class III PDZ-binding sequence has been identified between amino acids 1110 and 1113 (
Duning et al., 2008
).
Figure 1. Structural features of the human KIBRA protein. Shown are the identified domains in the KIBRA protein. WW domains are located between position 6 and 86, and cover about 40 amino acids containing two conserved tryptophan residues. WW domains are generally thought to be responsible for the interaction with various proteins that contain proline-rich sequences such as PPxY. The C2 domain is located between amino acids 655 and 783. This is a conserved membrane targeting motif composed of β-sheets. The 130 residues of C2 are involved in binding phospholipids in a calcium-dependent manner. The PKCζ binding region is located at amino acids 953–996 and contains two serine residues that can be phosphorylated by the kinase. The last four amino acids contain a PDZ binding motif.
Human Genetic Evidence for KIBRA’s Role in Cognition
KIBRA has come into the focus of the neurogenetics field following the publication of human evidence pointing to an involvement of the gene in memory performance and cognition (
Papassotiropoulos et al., 2006
). In that publication, the authors report that carriers of the KIBRA/
WWC1 (rs17070145) T allele or, to a lesser extent, the calsyntenin 2 (
CLSTN2) rs6439886 T allele performed significantly better on multiple episodic memory tasks than those homozygous for the C allele at either polymorphism. Furthermore, using functional magnetic resonance imaging (fMRI), they observed that brain activation (measured as oxygen extraction from blood) in key areas associated with memory retrieval was significantly greater in a selection of 15
WWC1 (rs17070145) T-allele-noncarriers than in 15 T-allele-carriers during an episodic memory task. After this initial finding, a considerable number of studies examined the
WWC1 polymorphism in different contexts of cognition and in different populations (summarized in Table
1 and reviewed below).
Table 1. Summary of the genetic association data available on the KIBRA polymorphism and memory and Alzheimer’s disease. Twelve studies that examine the KIBRA polymorphism in different populations are listed that were published before December 2009. Summarized are the citation and publication year, the population(s) examined, the study size, the effect(s) observed, and fi nally whether the study confi rmed the association of the rs17070145 SNP with memory performance.
A number of studies have examined the role of the rs17070145 polymorphism on the cognitive performance of non-demented individuals. The original finding was first confirmed in a small German cohort (
n = 64) of healthy elderly individuals with a mean age of approximately 67 (
Schaper et al., 2008
). A second study also confirmed the influence of the T-allele on memory performance in a cohort of 312 elderly individuals between the ages of 50–89 (
Almeida et al., 2008
). Of note, 136 individuals in their study met diagnostic criteria for mild cognitive impairment (MCI) and they were able to identify an excess risk for MCI among non-T-allele-carriers, although it failed to reach statistical significance. A study on individuals with subjective memory complaints found that family history of dementia was significantly elevated in KIBRA non-T-allele-carriers while these same individuals exhibited better performance on verbal episodic memory tests (
Nacmias et al., 2008
).
In mid-2008 a report of non-replication of the influence of rs17070145 on episodic memory was published (
Need et al., 2008
). In this study the authors utilized two cohorts of European genetic origin. The first cohort (
n = 300) was examined using a verbal recall memory test and the second cohort (
n = 365) was phenotyped with the exact same auditory verbal learning test (AVLT) that was used in the original publication (
Papassotiropoulos et al., 2006
). The authors failed to see any association of rs17070145 with memory performance in either cohort although they reported to be well powered to do so. Additionally, some tagging SNPs within the KIBRA genetic locus were tested and no significant association signals could be identified following multiple testing corrections. In the concluding paragraph of this work the authors indicate that their non-replication findings are likely a good indicator of the challenges facing the study of the genetics of human memory and specifically point to the need for consistency in phenotyping across any replication efforts.
Finally, the largest replication effort was published in 2009 (
Bates et al., 2009
). In two cohorts numbering over 2,500 individuals combined, association between rs17070145 and memory performance was confirmed. Using the AVLT and the Wechsler Logical Memory Test, it was possible to demonstrate an association between rs17070145 T-allele-carrier status and item-distinctiveness storage and/or recollection. The authors suggest that KIBRA action is involved primarily in the conscious recall of item-based information and underscore the importance of attention to phenotyping when attempting to replicate a genetic association. The most recent study in healthy individuals replicated the positive effects of the KIBRA T-allele on episodic memory in a cohort of 383 individuals and showed that the effect size of the KIBRA polymorphism was modulated by the associative components of the task (
Preuschhof et al., 2009
). Additionally, this work suggests an interaction between KIBRA and
CLSTN2 in episodic memory as the effect of carrying the rs17070145 T-allele was enhanced in carriers of the
CLSTN2 rs6439886 C-allele.
Recently, KIBRA was also examined in the context of developmental or current exposure to nicotine (
Jacobsen et al., 2009
;
Zhang et al., 2009
). One study assessed association of SNP rs17070145 with verbal and visuospatial memory and fMRI changes in adolescents and could not find any significant influence of the polymorphisms either alone or in interaction with smoking habits or smoking exposure (
Jacobsen et al., 2009
). However, this study only examined 101 subjects with various combinations of prenatal and current nicotine exposure. The most recent study analyzed cognitive flexibility measures with the Wisconsin Card Sorting Test, ethnicity, and smoking habits in relation to the rs17070145 polymorphism (
Zhang et al., 2009
). In European Americans homozygous for the C-allele, an association with better cognitive flexibility was found. Curiously, current smokers of European origin with the T-allele performed significantly better than past smokers with the T-allele while there was no difference for C-allele carriers. There was no difference in performance in subjects of African American ethnicity. Association of the C instead of the T allele with better cognitive performance in different studies may be caused by differential effects of the polymorphism itself or yet unknown functional polymorphisms in the KIBRA gene in linkage disequilibrium with SNP rs17070145 on different domains of cognitive performance (e.g., more frontally located functions versus hippocampal functions).
KIBRA and Alzheimer’s Disease: Clues from the Genome and Transcriptome
Based on the associations with memory performance in healthy subjects, the link between KIBRA and Alzheimer’s disease was also examined. One study demonstrated that the rs17070145 C-allele was significantly associated with increased risk for developing late-onset AD (
Corneveaux et al., 2008
). However, another study showed that the T-allele was associated with an increased risk for very-late-onset AD (
Rodriguez-Rodriguez et al., 2009
). These differential findings may be reflective of differences in the genetic background of the studied samples, in the case/control selection criteria, or simply in the statistical power differences between the studies. A study in early 2009 also noted an association between KIBRA and late-onset AD risk (
Beecham et al., 2009
). Utilizing imputation to compare their data set generated on the Illumina HumanHap beadchip with data generated on a similar density Affymetrix array in a large AD study of 1,411 cases and controls (
Reiman et al., 2007
), the authors reported on a SNP located within KIBRA (rs12514426) that was associated [joint analysis
p-value (uncorrected) = 0.000928] with late-onset AD. This SNP is in very low hypothetical linkage disequilibrium with rs17070145, but nonetheless represents a third independently reported genetic association between KIBRA and late-onset AD. Of note, the same analysis also yielded several hits in the SORL1 locus, a gene with strong biological and genetic links to Alzheimer’s disease (
Andersen et al., 2005
;
Rogaeva et al., 2007
;
Dodson et al., 2008
;
Mayeux and Hyslop, 2008
;
Ma et al., 2009
).
Two additional pieces of experimental evidence link KIBRA to AD (
Corneveaux et al., 2008
). First, the expression of KIBRA itself and the genes encoding several of its direct binding partners (
DYNLL1,
PRKCZ,
SNX4, and
KIAA0513) are significantly altered in non-tangle bearing neurons in key brain regions in AD patients versus matched controls. In neurons laser-dissected from the hippocampus, middle temporal gyrus, and posterior cingulate, KIBRA mRNA was found to be significantly increased between 2.4- and 3.0-fold, while in those same brain regions PKCζ mRNA was decreased by 2.3- to 3.3-fold. No significant changes were observed in the primary visual cortex, a region that is largely spared by AD pathology. Interestingly, only significant changes in PKCζ mRNA were detected in the hippocampus and middle temporal gyrus from patients with MCI (Huentelman lab communication, unpublished results), suggesting that perhaps the transcriptional alteration of these molecular species may follow a temporal pattern that could be related to dementia progression. Secondly, a link between rs17070145 T-allele carrier status and brain hypometabolism of fluorodeoxyglucose (FDG) as measured using positron emission tomography was described. It was shown that significant decreases in FDG uptake were noted in the precuneus and posterior/mid cingulate regions of neurologically normal individuals who were at elevated risk for developing AD. Some of these regions overlapped with previously identified hypometabolic changes in individuals stratified by
APOE E4 carrier status (
Reiman et al., 2004
;
Buckner et al., 2005
;
Reiman et al., 2005
).
In conclusion, several pieces of evidence point to a link between KIBRA and Alzheimer’s disease. At present, the genetic link appears not as clear-cut as with memory performance in healthy individuals and therefore requires further replication. An interesting question is whether this link represents an extension or consequence of the involvement of KIBRA in episodic memory performance or possibly that it represents an independent function of KIBRA in the pathophysiology of Alzheimer’s disease.
Expression Patterns of KIBRA mRNA and Protein
KIBRA is predominately expressed in the kidney and brain in the adult organism. In the rodent and human brain, KIBRA is expressed in memory-related structures (including the hippocampus and cortex), the cerebellum, and in the hypothalamus (
Johannsen et al., 2008
). During brain development, expression has been shown to decrease from juvenile postnatal stages to the adult animal (
Johannsen et al., 2008
). On a subcellular level within neurons, KIBRA shows a somatodendritic staining pattern with enrichment in the perinuclear region and in postsynaptic structures. In subcellular fractionation experiments, KIBRA was heavily enriched in the postsynaptic density (PSD) fraction (
Johannsen et al., 2008
). However, nuclear localization has also been noted (
Rayala et al., 2006
). So far, it is unclear if different localizations are due to the type of cells investigated, experimental conditions, or possible modified variants of KIBRA.
Interacting Partners of KIBRA
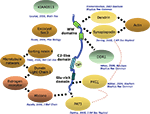
KIBRA has been shown to participate in a number of cellular functions such as cell polarity and migration, vesicle transport, transcriptional regulation and synaptogenesis. Most hints to its function have come from the study of interaction partners identified in yeast two hybrid screens. To date, 10 direct interaction partners have been described (Figure
2
). The two WW-domains, which cover a stretch of 35–40 amino acids containing two conserved tryptophan residues, appear responsible for many of the interactions with the identified proteins. The first interaction partner of KIBRA, identified by a yeast two hybrid screen, was the postsynaptic protein dendrin (
KIAA0749) (
Kremerskothen et al., 2003
). Dendrin was first characterized as a dendritic protein whose expression is altered by sleep deprivation in the forebrain of rats (
Neuner-Jehle et al., 1996
). Dendrin is translated from a dendritically localized mRNA and interacts with alpha-actinin and the synaptic scaffolding molecule S-SCAM (
Kremerskothen et al., 2006
).
Figure 2. Known interactors of KIBRA. Depicted are interaction partners of KIBRA identified by a number of laboratories with the respective citations given. Solid dotted lines: direct interactions that have been mapped to specific segments of the KIBRA protein; hatched broad lines: direct interactions where the location of binding is not known.
The proposal that KIBRA is involved in processes in the PSD was strengthened by the identification of two other postsynaptic binding proteins as KIBRA partners: synaptopodin (
Duning et al., 2008
) and PKCζ (
Büther et al., 2004
). Similar to dendrin, synaptopodin was also found to interact through the PPxY motif with the WW-domains of KIBRA. Both proteins are localized in dendrites and play a role in the organization of the cytoskeleton (
Kremerskothen et al., 2005
;
Duning et al., 2008
). The role of synaptopodin in dendritic spines has been demonstrated in knock-out mice where the animals suffer from impaired long-term synaptic plasticity and lack the dendritic spine apparatus (
Deller et al., 2003
;
Asanuma et al., 2005
).
Another interaction partner with a potential link to cognitive processing is
KIAA0513.
KIAA0513 expression is upregulated in the brain of schizophrenia patients, but the cellular function of the encoded protein is unknown (
Lauriat et al., 2006
). It is tempting to speculate that this could be an additional link to the cognitive deficits associated with the disease, a prominent aspect of the negative symptoms of schizophrenia. The identification of the protein PATJ (PALS1-associated tight junction protein) as an interaction partner of KIBRA suggests that it may play a role in cell polarity (
Duning et al., 2008
). PATJ interacts with the putative class III PDZ-binding site of KIBRA (ADDV). PATJ is a component of an evolutionarily conserved multiprotein complex, which regulates cell polarity in podocytes and neurons. Both cell types develop prominent cell processes and show highly dynamic actin filament generation. Intriguingly, PATJ regulates the subcellular localization of atypical PKC and Par3 during migration of epithelia (
Shin et al., 2005
,
2007
). Taken together, these similarities suggest overlapping functions of KIBRA and PATJ in the modulation of cell polarity and migration in cells, which are derived from the ectoderm. The identification of KIBRA in the PSD, the interaction with cell polarity proteins and its functional role in migration points toward a central role of KIBRA in rearrangements of the cellular cytoskeleton.
Another link of KIBRA to the cytoskeleton comes from a study providing evidence that KIBRA binds to the dynein-complex (
Traer et al., 2007
). The authors show that the sorting nexin 4 (SNX4) interacts with a complex of the microtubule motor dynein and KIBRA and that the complex regulates the sorting of transferrin receptor (TfnR) away from degradation and towards a recycling pathway (
Traer et al., 2007
). Downregulation of SNX4, KIBRA, or dynein proteins disrupts the trafficking of TfnR from the endosomes and into juxtanuclear endocytic recycling compartments thereby increasing the lysosomal-mediated degradation of the receptor. The role of KIBRA in vesicle-based transport processes was underlined by recent findings that KIBRA binds to sec3, a component of the exocyst complex (
Rosse et al., 2009
). The exocyst/KIBRA complex enables intracellular targeting of PKCζ, thus regulating localized signal cascades necessary for cell migration.
The interaction of KIBRA with the dynein complex was confirmed in a study describing the simultaneous interaction of KIBRA with histone H3 and dynein light chain 1 (DLC1) (
Rayala et al., 2006
). The binding of KIBRA to histone H3 appears to be mediated through the glutamic acid rich region of KIBRA near the
C-terminus. Deletion of the histone-binding region in KIBRA resulted in the inhibition of transactivation of the estrogen receptor-alpha in human breast cancer cells. Although KIBRA binds to histone and partially resides in the nucleus, the underlying mechanism of how KIBRA mediates ER transactivation remains unclear. The notion that KIBRA is also involved in transcriptional regulation is further supported in a study showing that KIBRA is upregulated through the application of progesterone and that KIBRA binds to discoidin domain receptor 1 (DDR1), a tyrosine kinase important for the development of the mammary glands (
Hilton et al., 2008
). KIBRA and DDR1 both interact with PKCζ in a molecular complex involved in the collagen-regulated stimulation of the MAPK cascade.
KIBRA Interacts with PKC/ PKMζ
The strongest indication of an involvement of KIBRA in cognition comes from the finding that KIBRA interacts with protein kinase C ζ (
Büther et al., 2004
), a molecule crucially involved in neuronal plasticity (
Sacktor, 2008
). This atypical PKC comprises four functional motifs, a PB1 domain at the N-terminus, a pseudosubstrate sequence, a C1 domain and the enzymatic active domain in the C-terminal end of the protein. PKCζ was originally identified by Nishizuka and coworkers in 1987 through homology screening (
Ono et al., 1987
,
1988
,
1989
;
Nishizuka, 1988
). Sacktor, Osten, and colleagues first described the presence of the constitutively active form, known as PKMζ, in the hippocampus and suggested a role for PKCζ/PKMζ in the maintenance of LTP (
Sacktor et al., 1993
). Importantly, PKMζ is the only PKM-form of all the PKCs expressed in the brain that is expressed in the hippocampus (
Sacktor et al., 1993
). The constitutive nature of PKMζ made it immediately attractive as being involved in the “synaptic tagging” machinery. Levels of PKMζ increase during LTP likely due to an increase in PKMζ synthesis (
Osten et al., 1996
;
Hernandez et al., 2003
). PKMζ can be generated by an independent promoter in the PKCζ gene, and one hypothesis is that PKMζ mRNA is locally translated in dendrites (
Muslimov et al., 2004
). PKMζ increases the LTP response (
Ling et al., 2002
) and blocking PKMζ abolishes LTP maintenance (
Serrano et al., 2005
). This occurs through the increase in the number of active AMPA receptors in the post-synaptic membrane (
Ling et al., 2006
) likely by influencing the trafficking of the receptors (
Yao et al., 2008
). A series of elegant papers finally demonstrated that PKMζ had an overwhelming influence on long-term memory maintenance in the hippocampus (
Drier et al., 2002
;
Pastalkova et al., 2006
;
Serrano et al., 2008
) and in the cortex (
Shema et al., 2007
,
2009
). It remains to be demonstrated how PKMζ is able to maintain memory for months with a protein half-life that is likely much shorter.
PKCζ interacts with a short stretch of amino acids near the carboxyterminal end of KIBRA (positions 953–996), and phosphorylates two serines at positions 975 and 978 (
Büther et al., 2004
). This observation was confirmed and extended, demonstrating that KIBRA co-localizes and interacts with PKMζ (
Yoshihama et al., 2009
). Both proteins were found to be co-localized in the hippocampus and dentate gyrus regions of fundamental importance for learning and memory processes (
Büther et al., 2004
;
Yoshihama et al., 2009
).
The significance of the interaction between KIBRA and PKMζ and the phosphorylation of KIBRA is unclear at this point. The overwhelming evidence for an indispensable functioning of PKMζ in the maintenance of long-term memory and the interaction with other postsynaptic proteins supports the notion that KIBRA serves a critical role in postsynaptic processes that might be regulated by phosphorylation.
KIBRA in Kidney and Brain: Parallels?
The name of the protein in question, KIBRA, also reflects the areas where work has been done on possible functions, the central nervous system and the kidney. Astonishingly, recent work suggests that kidney and brain might have more in common than previously thought. In both organs key physiological functions (synaptic transmission and urine filtration) are fulfilled by highly polarized cell types. It has been shown that KIBRA knock-down in podocytes impairs directed cell migration (
Duning et al., 2008
). The authors postulated that the motility of foot processes and flexibility of synaptic contacts could be regulated by an analogous set of proteins including KIBRA. The establishment of polarity in these cells is achieved by directed, vesicle-based transport of multi-protein complexes composed of proteins such as PATJ, Par3/Par6, PKC/Mζ and KIBRA (
Barnes and Polleux, 2009
;
Weide and Huber, 2009
). Furthermore, signal transmission in the brain and in the kidney are thought to be controlled by similar mechanisms including receptor clustering, vesicle exocytosis, and dynamic formation of cell–cell contacts (
Rastaldi et al., 2006
;
Weide and Huber, 2009
). Interestingly, a dense protein network at the cell periphery, the PSD in neurons and the slit membrane in podocytes, is crucial for memory formation in the brain and urine filtration in the kidney. KIBRA might control the composition and architecture of these protein networks through regulating the association, intracellular targeting, and posttranslational modification of key molecules such as Dendrin, Synaptopodin, and PKCζ.
We see a number of key areas where answers will likely lead to the fastest advancement of understanding KIBRA’s functions and their possible pharmacological exploitation. On the genetic side, we need more data on the association of KIBRA and Alzheimer’s disease or other states of cognitive impairment to clarify the role that the polymorphism may play. This includes further sequencing work within the associated haplotype to identify the likely biological functional variant(s). On the biochemical side, we need to understand the significance of the PKC/Mζ interaction with KIBRA. It will also be crucial to determine whether KIBRA plays a role in fast synaptic transmission or in long-term potentiation. Specifically, we need to test the hypothesis that KIBRA is involved in aspects of the maintenance of synaptic memory through constitutively active PKMζ. On the systems level, we need data from animals where KIBRA has been deleted, or its expression enhanced, and find out whether these animals show altered cognitive behavior. Finally, it will be necessary to establish or rule out whether there is another polymorphism in the KIBRA locus that would result in a clear alteration on the transcriptional or translational level. If it turns out that the rs17070145 polymorphism is specific to memory performance, it will be necessary to investigate the consequences of the C/T-Allele in detail. This should cover multiple parameters of gene function, including expression, transcript stability, or alternative splicing events.
A new player has entered the stage in the field of cognition and memory: KIBRA. In contrast to many other genes identified to play a role in cognition, KIBRA was identified in a hypothesis-free fashion by human genetic studies that have been largely replicated. From those studies, KIBRA appears to be involved in the ability to store episodic memory for at least 24 h but not in the acquisition of memory traces. Biochemistry and expression of KIBRA place it into a context highly compatible with a role in memory storage. Of special importance appears to be the link to PKC/Mζ a uniquely powerful player in maintaining long-term memory. The genetic link to KIBRA certainly gives credibility to relevance of this player in human memory performance. Moreover, KIBRA represents a new example of a cognition associated gene with a connection to Alzheimer’s disease. KIBRA therefore appears as a highly attractive novel target for pharmacological approaches to cognition and possibly dementia.
Conflict of Interest Statement
Matthew J. Huentelman is a co-inventor on patent applications related to KIBRA. Amnestix Inc. is a licensor of KIBRA patent applications from the Translational Genomics Research Institute, and is a subsidiary of SYGNIS Bioscience.
Almeida, O. P., Schwab, S. G., Lautenschlager, N. T., Morar, B., Greenop, K. R., Flicker, L., and Wildenauer, D. (2008). KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment.
J. Cell. Mol. Med. 12, 1672–1676.
Andersen, O. M., Reiche, J., Schmidt, V., Gotthardt, M., Spoelgen, R., Behlke, J., von Arnim, C. A., Breiderhoff, T., Jansen, P., Wu, X., Bales, K. R., Cappai, R., Masters, C. L., Gliemann, J., Mufson, E. J., Hyman, B. T., Paul, S. M., Nykjaer, A., and Willnow, T. E. (2005). Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein.
Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466.
Asanuma, K., Kim, K., Oh, J., Giardino, L., Chabanis, S., Faul, C., Reiser, J., and Mundel, P. (2005). Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner.
J. Clin. Invest. 115, 1188–1198.
Barnes, A. P., and Polleux, F. (2009). Establishment of axon-dendrite polarity in developing neurons.
Annu. Rev. Neurosci. 32, 347–381.
Bates, T. C., Price, J. F., Harris, S. E., Marioni, R. E., Fowkes, F. G., Stewart, M. C., Murray, G. D., Whalley, L. J., Starr, J. M., and Deary, I. J. (2009). Association of KIBRA and memory.
Neurosci. Lett. 458, 140–143.
Beecham, G. W., Martin, E. R., Li, Y. J., Slifer, M. A., Gilbert, J. R., Haines, J. L., and Pericak-Vance, M. A. (2009). Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease.
Am. J. Hum. Genet. 84, 35–43.
Buckner, R. L., Snyder, A. Z., Shannon, B. J., LaRossa, G., Sachs, R., Fotenos, A. F., Sheline, Y. I., Klunk, W. E., Mathis, C. A., Morris, J. C., and Mintun, M. A. (2005). Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory.
J. Neurosci. 25, 7709–7717.
Büther, K., Plaas, C., Barnekow, A., and Kremerskothen, J. (2004). KIBRA is a novel substrate for protein kinase Czeta.
Biochem. Biophys. Res. Commun. 317, 703–707.
Corneveaux, J. J., Liang, W. S., Reiman, E. M., Webster, J. A., Myers, A. J., Zismann, V. L., Joshipura, K. D., Pearson, J. V., Hu-Lince, D., Craig, D. W., Coon, K. D., Dunckley, T., Bandy, D., Lee, W., Chen, K., Beach, T. G., Mastroeni, D., Grover, A., Ravid, R., Sando, S. B., Aasly, J. O., Heun, R., Jessen, F., Kolsch, H., Rogers, J., Hutton, M. L., Melquist, S., Petersen, R. C., Alexander, G. E., Caselli, R. J., Papassotiropoulos, A., Stephan, D. A., and Huentelman, M. J. (2008). Evidence for an association between KIBRA and late-onset Alzheimer’s disease.
Neurobiol. Aging doi: 10.1016/j.neurobiolaging.2008.07.014
Deller, T., Korte, M., Chabanis, S., Drakew, A., Schwegler, H., Stefani, G. G., Zuniga, A., Schwarz, K., Bonhoeffer, T., Zeller, R., Frotscher, M., and Mundel, P. (2003). Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity.
Proc. Natl. Acad. Sci. U.S.A. 100, 10494–10499.
Dodson, S. E., Andersen, O. M., Karmali, V., Fritz, J. J., Cheng, D., Peng, J., Levey, A. I., Willnow, T. E., and Lah, J. J. (2008). Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer’s disease.
J. Neurosci. 28, 12877–12886.
Drier, E. A., Tello, M. K., Cowan, M., Wu, P., Blace, N., Sacktor, T. C., and Yin, J. C. (2002). Memory enhancement and formation by atypical PKM activity in
Drosophila melanogaster.
Nat. Neurosci. 5, 316–324.
Duning, K., Schurek, E. M., Schluter, M., Bayer, M., Reinhardt, H. C., Schwab, A., Schaefer, L., Benzing, T., Schermer, B., Saleem, M. A., Huber, T. B., Bachmann, S., Kremerskothen, J., Weide, T., and Pavenstadt, H. (2008). KIBRA modulates directional migration of podocytes.
J. Am. Soc. Nephrol. 19, 1891–1903.
Fisher, S. E. (2006). Tangled webs: tracing the connections between genes and cognition.
Cognition 101, 270–297.
Goldberg, T. E., and Weinberger, D. R. (2004). Genes and the parsing of cognitive processes.
Trends Cogn. Sci. 8, 325–335.
Hernandez, A. I., Blace, N., Crary, J. F., Serrano, P. A., Leitges, M., Libien, J. M., Weinstein, G., Tcherapanov, A., and Sacktor, T. C. (2003). Protein kinase M zeta synthesis from a brain mRNA encoding an independent protein kinase C zeta catalytic domain. Implications for the molecular mechanism of memory.
J. Biol. Chem. 278, 40305–40316.
Hilton, H. N., Stanford, P. M., Harris, J., Oakes, S. R., Kaplan, W., Daly, R. J., and Ormandy, C. J. (2008). KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling.
Biochim. Biophys. Acta 1783, 383–393.
Jacobsen, L. K., Picciotto, M. R., Heath, C. J., Mencl, W. E., and Gelernter, J. (2009). Allelic variation of calsyntenin 2 (CLSTN2) modulates the impact of developmental tobacco smoke exposure on mnemonic processing in adolescents.
Biol. Psychiatry 65, 671–679.
Johannsen, S., Duning, K., Pavenstadt, H., Kremerskothen, J., and Boeckers, T. M. (2008). Temporal–spatial expression and novel biochemical properties of the memory-related protein KIBRA.
Neuroscience 155, 1165–1173.
Kremerskothen, J., Kindler, S., Finger, I., Veltel, S., and Barnekow, A. (2003). Characterization of KIBRA, a novel WW domain-containing protein.
Biochem. Biophys. Res. Commun. 300, 862–867.
Kremerskothen, J., Plaas, C., Buther, K., Finger, I., Veltel, S., Matanis, T., Liedtke, T., and Barnekow, A. (2005). Synaptopodin, a molecule involved in the formation of the dendritic spine apparatus, is a dual actin/alpha-actinin binding protein.
J. Neurochem. 92, 597–606.
Kremerskothen, J., Plaas, C., Kindler, S., Frotscher, M., and Barnekow, A. (2006). Postsynaptic recruitment of dendrin depends on both dendritic mRNA transport and synaptic anchoring.
J. Neurochem. 96, 1659–1666.
Lauriat, T. L., Dracheva, S., Kremerskothen, J., Duning, K., Haroutunian, V., Buxbaum, J. D., Hyde, T. M., Kleinman, J. E., and McInnes, L. A. (2006). Characterization of KIAA0513, a novel signaling molecule that interacts with modulators of neuroplasticity, apoptosis, and the cytoskeleton.
Brain Res. 1121, 1–11.
Lee, Y. S., and Silva, A. J. (2009). The molecular and cellular biology of enhanced cognition.
Nat. Rev. Neurosci. 10, 126–140.
Ling, D. S., Benardo, L. S., Serrano, P. A., Blace, N., Kelly, M. T., Crary, J. F., and Sacktor, T. C. (2002). Protein kinase Mzeta is necessary and sufficient for LTP maintenance.
Nat. Neurosci. 5, 295–296.
Ling, D. S., Benardo, L. S., and Sacktor, T. C. (2006). Protein kinase Mzeta enhances excitatory synaptic transmission by increasing the number of active postsynaptic AMPA receptors.
Hippocampus 16, 443–452.
Ma, Q. L., Galasko, D. R., Ringman, J. M., Vinters, H. V., Edland, S. D., Pomakian, J., Ubeda, O. J., Rosario, E. R., Teter, B., Frautschy, S. A., and Cole, G. M. (2009). Reduction of SorLA/LR11, a sorting protein limiting beta-amyloid production, in Alzheimer disease cerebrospinal fluid.
Arch. Neurol. 66, 448–457.
Mayeux, R., and Hyslop, P. S. (2008). Alzheimer’s disease: advances in trafficking.
Lancet Neurol. 7, 2–3.
Muslimov, I. A., Nimmrich, V., Hernandez, A. I., Tcherepanov, A., Sacktor, T. C., and Tiedge, H. (2004). Dendritic transport and localization of protein kinase Mzeta mRNA: implications for molecular memory consolidation.
J. Biol. Chem. 279, 52613–52622.
Nacmias, B., Bessi, V., Bagnoli, S., Tedde, A., Cellini, E., Piccini, C., Sorbi, S., and Bracco, L. (2008). KIBRA gene variants are associated with episodic memory performance in subjective memory complaints.
Neurosci. Lett. 436, 145–147.
Need, A. C., Attix, D. K., McEvoy, J. M., Cirulli, E. T., Linney, K. N., Wagoner, A. P., Gumbs, C. E., Giegling, I., Moller, H. J., Francks, C., Muglia, P., Roses, A., Gibson, G., Weale, M. E., Rujescu, D., and Goldstein, D. B. (2008). Failure to replicate effect of Kibra on human memory in two large cohorts of European origin.
Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 667–668.
Neuner-Jehle, M., Denizot, J. P., Borbely, A. A., and Mallet, J. (1996). Characterization and sleep deprivation-induced expression modulation of dendrin, a novel dendritic protein in rat brain neurons.
J. Neurosci. Res. 46, 138–151.
Nishizuka, Y. (1988). The molecular heterogeneity of protein kinase C and its implications for cellular regulation.
Nature 334, 661–665.
Ono, Y., Fujii, T., Ogita, K., Kikkawa, U., Igarashi, K., and Nishizuka, Y. (1987). Identification of three additional members of rat protein kinase C family: delta-, epsilon- and zeta-subspecies.
FEBS Lett. 226, 125–128.
Ono, Y., Fujii, T., Ogita, K., Kikkawa, U., Igarashi, K., and Nishizuka, Y. (1988). The structure, expression, and properties of additional members of the protein kinase C family.
J. Biol. Chem. 263, 6927–6932.
Ono, Y., Fujii, T., Ogita, K., Kikkawa, U., Igarashi, K., and Nishizuka, Y. (1989). Protein kinase C zeta subspecies from rat brain: its structure, expression, and properties.
Proc. Natl. Acad. Sci. U.S.A. 86, 3099–3103.
Osten, P., Valsamis, L., Harris, A., and Sacktor, T. C. (1996). Protein synthesis-dependent formation of protein kinase Mzeta in long-term potentiation.
J. Neurosci. 16, 2444–2451.
Papassotiropoulos, A., Stephan, D. A., Huentelman, M. J., Hoerndli, F. J., Craig, D. W., Pearson, J. V., Huynh, K. D., Brunner, F., Corneveaux, J., Osborne, D., Wollmer, M. A., Aerni, A., Coluccia, D., Hanggi, J., Mondadori, C. R., Buchmann, A., Reiman, E. M., Caselli, R. J., Henke, K., and de Quervain, D. J. (2006). Common Kibra alleles are associated with human memory performance.
Science 314, 475–478.
Pastalkova, E., Serrano, P., Pinkhasova, D., Wallace, E., Fenton, A. A., and Sacktor, T. C. (2006). Storage of spatial information by the maintenance mechanism of LTP.
Science 313, 1141–1144.
Payton, A. (2006). Investigating cognitive genetics and its implications for the treatment of cognitive deficit.
Genes Brain Behav. 5(Suppl. 1), 44–53.
Payton, A. (2009). The impact of genetic research on our understanding of normal cognitive ageing: 1995 to 2009.
Neuropsychol. Rev. 19, 451–477.
Potkin, S. G., Turner, J. A., Guffanti, G., Lakatos, A., Torri, F., Keator, D. B., and Macciardi, F. (2009). Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations.
Cogn. Neuropsychiatry 14, 391–418.
Preuschhof, C., Heekeren, H. R., Li, S. C., Sander, T., Lindenberger, U., and Backman, L. (2009). KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory.
Neuropsychologia. 48, 402–408.
Rastaldi, M. P., Armelloni, S., Berra, S., Calvaresi, N., Corbelli, A., Giardino, L. A., Li, M., Wang, G. Q., Fornasieri, A., Villa, A., Heikkila, E., Soliymani, R., Boucherot, A., Cohen, C. D., Kretzler, M., Nitsche, A., Ripamonti, M., Malgaroli, A., Pesaresi, M., Forloni, G. L., Schlondorff, D., Holthofer, H., and D’Amico, G. (2006). Glomerular podocytes contain neuron-like functional synaptic vesicles.
FASEB J. 20, 976–978.
Rayala, S. K., den Hollander, P., Manavathi, B., Talukder, A. H., Song, C., Peng, S., Barnekow, A., Kremerskothen, J., and Kumar, R. (2006). Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells.
J. Biol. Chem. 281, 19092–19099.
Reiman, E. M., Chen, K., Alexander, G. E., Caselli, R. J., Bandy, D., Osborne, D., Saunders, A. M., and Hardy, J. (2004). Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia.
Proc. Natl. Acad. Sci. U.S.A. 101, 284–289.
Reiman, E. M., Chen, K., Alexander, G. E., Caselli, R. J., Bandy, D., Osborne, D., Saunders, A. M., and Hardy, J. (2005). Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism.
Proc. Natl. Acad. Sci. U.S.A. 102, 8299–8302.
Reiman, E. M., Webster, J. A., Myers, A. J., Hardy, J., Dunckley, T., Zismann, V. L., Joshipura, K. D., Pearson, J. V., Hu-Lince, D., Huentelman, M. J., Craig, D. W., Coon, K. D., Liang, W. S., Herbert, R. H., Beach, T., Rohrer, K. C., Zhao, A. S., Leung, D., Bryden, L., Marlowe, L., Kaleem, M., Mastroeni, D., Grover, A., Heward, C. B., Ravid, R., Rogers, J., Hutton, M. L., Melquist, S., Petersen, R. C., Alexander, G. E., Caselli, R. J., Kukull, W., Papassotiropoulos, A., and Stephan, D. A. (2007). GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers.
Neuron 54, 713–720.
Rizo, J., and Sudhof, T. C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain.
J. Biol. Chem. 273, 15879–15882.
Rodriguez-Rodriguez, E., Infante, J., Llorca, J., Mateo, I., Sanchez-Quintana, C., Garcia-Gorostiaga, I., Sanchez-Juan, P., Berciano, J., and Combarros, O. (2009). Age-dependent association of KIBRA genetic variation and Alzheimer’s disease risk.
Neurobiol. Aging 30, 322–324.
Rogaeva, E., Meng, Y., Lee, J. H., Gu, Y., Kawarai, T., Zou, F., Katayama, T., Baldwin, C. T., Cheng, R., Hasegawa, H., Chen, F., Shibata, N., Lunetta, K. L., Pardossi-Piquard, R., Bohm, C., Wakutani, Y., Cupples, L. A., Cuenco, K. T., Green, R. C., Pinessi, L., Rainero, I., Sorbi, S., Bruni, A., Duara, R., Friedland, R. P., Inzelberg, R., Hampe, W., Bujo, H., Song, Y. Q., Andersen, O. M., Willnow, T. E., Graff-Radford, N., Petersen, R. C., Dickson, D., Der, S. D., Fraser, P. E., Schmitt-Ulms, G., Younkin, S., Mayeux, R., Farrer, L. A., and St George-Hyslop, P. (2007). The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease.
Nat. Genet. 39, 168–177.
Rosse, C., Formstecher, E., Boeckeler, K., Zhao, Y., Kremerskothen, J., White, M. D., Camonis, J. H., and Parker, P. J. (2009). An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation.
PLoS Biol. 7, e1000235. doi: 10.1371/journal.pbio.1000235.
Sacktor, T. C. (2008). PKMzeta, LTP maintenance, and the dynamic molecular biology of memory storage.
Prog. Brain Res. 169, 27–40.
Sacktor, T. C., Osten, P., Valsamis, H., Jiang, X., Naik, M. U., and Sublette, E. (1993). Persistent activation of the zeta isoform of protein kinase C in the maintenance of long-term potentiation.
Proc. Natl. Acad. Sci. U.S.A. 90, 8342–8346.
Schaper, K., Kolsch, H., Popp, J., Wagner, M., and Jessen, F. (2008). KIBRA gene variants are associated with episodic memory in healthy elderly.
Neurobiol. Aging 29, 1123–1125.
Scoville, W. B., and Milner, B. (1957). Loss of recent memory after bilateral hippocampal lesions.
J. Neurol. Neurosurg. Psychiatr. 20, 11–21.
Serrano, P., Yao, Y., and Sacktor, T. C. (2005). Persistent phosphorylation by protein kinase Mzeta maintains late-phase long-term potentiation.
J. Neurosci. 25, 1979–1984.
Serrano, P., Friedman, E. L., Kenney, J., Taubenfeld, S. M., Zimmerman, J. M., Hanna, J., Alberini, C., Kelley, A. E., Maren, S., Rudy, J. W., Yin, J. C., Sacktor, T. C., and Fenton, A. A. (2008). PKMzeta maintains spatial, instrumental, and classically conditioned long-term memories.
PLoS Biol. 6, 2698–2706. doi: 10.1371/journal.pbio.0060318.
Shema, R., Sacktor, T. C., and Dudai, Y. (2007). Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKM zeta.
Science 317, 951–953.
Shema, R., Hazvi, S., Sacktor, T. C., and Dudai, Y. (2009). Boundary conditions for the maintenance of memory by PKMzeta in neocortex.
Learn. Mem. 16, 122–128.
Shin, K., Straight, S., and Margolis, B. (2005). PATJ regulates tight junction formation and polarity in mammalian epithelial cells.
J. Cell Biol. 168, 705–711.
Shin, K., Wang, Q., and Margolis, B. (2007). PATJ regulates directional migration of mammalian epithelial cells.
EMBO Rep. 8, 158–164.
Spearman, C. (1904). ‘General Intelligence’ objectively determined and measured.
Am. J. Psychol. 15, 201–293.
Traer, C. J., Rutherford, A. C., Palmer, K. J., Wassmer, T., Oakley, J., Attar, N., Carlton, J. G., Kremerskothen, J., Stephens, D. J., and Cullen, P. J. (2007). SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment.
Nat. Cell Biol. 9, 1370–1380.
Weide, T., and Huber, T. B. (2009). Signaling at the slit: podocytes chat by synaptic transmission.
J. Am. Soc. Nephrol. 20, 1862–1864.
Yao, Y., Kelly, M. T., Sajikumar, S., Serrano, P., Tian, D., Bergold, P. J., Frey, J. U., and Sacktor, T. C. (2008). PKM zeta maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors.
J. Neurosci. 28, 7820–7827.
Yoshihama, Y., Hirai, T., Ohtsuka, T., and Chida, K. (2009). KIBRA Co-localizes with protein kinase Mzeta (PKMzeta) in the mouse hippocampus.
Biosci. Biotechnol. Biochem. 73, 147–151.
Zhang, H., Kranzler, H. R., Poling, J., Gruen, J. R., and Gelernter, J. (2009). Cognitive flexibility is associated with KIBRA variant and modulated by recent tobacco use.
Neuropsychopharmacology 34, 2508–2516.