
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Aging Neurosci. , 16 January 2024
Sec. Alzheimer's Disease and Related Dementias
Volume 16 - 2024 | https://doi.org/10.3389/fnagi.2024.1332845
This article is part of the Research Topic Epigenetic Modifications in Neurological and Cognitive Disorders View all 7 articles
Background: Currently, the prevalence of Alzheimer’s disease (AD) is progressively rising, particularly in developed nations. There is an escalating focus on the onset and progression of AD. A mounting body of research indicates that epigenetics significantly contributes to AD and holds substantial promise as a novel therapeutic target for its treatment.
Objective: The objective of this article is to present the AD areas of research interest, comprehend the contextual framework of the subject research, and investigate the prospective direction for future research development.
Methods: ln Web of Science Core Collection (WOSCC), we searched documents by specific subject terms and their corresponding free words. VOSviewer, CiteSpace and Scimago Graphica were used to perform statistical analysis on measurement metrics such as the number of published papers, national cooperative networks, publishing countries, institutions, authors, co-cited journals, keywords, and visualize networks of related content elements.
Results: We selected 1,530 articles from WOSCC from January 2013 to June 2023 about epigenetics of AD. Based on visual analysis, we could get that China and United States were the countries with the most research in this field. Bennett DA was the most contributed and prestigious scientist. The top 3 cited journals were Journal of Alzheimer’s Disease, Neurobiology of Aging and Molecular Neurobiology. According to the analysis of keywords and the frequency of citations, ncRNAs, transcription factor, genome, histone modification, blood DNA methylation, acetylation, biomarkers were hot research directions in AD today.
Conclusion: According to bibliometric analysis, epigenetic research in AD was a promising research direction, and epigenetics had the potential to be used as AD biomarkers and therapeutic targets.
Alzheimer’s disease (AD), a slowly progressive and irreversible neurodegenerative disease, is the most common form of dementia (60–70%). It mainly affects the elderly, causing cognitive impairment and some changes in mood and behavior. In March 2023, according to the AD-related data released by the WHO, there were currently more than 550,000 people with dementia in the world, and there were nearly 100,000 new cases every year. There is doubtless that AD has become a public health priority (World Health Organization, 2023).
Epigenetics refers to epigenetic modifications associated with the regulation of gene expression and differentiation, as well as genetic changes in gene activity or cellular phenotype, without altering DNA sequence (Hernández-Romero et al., 2019). Current studies had shown that epigenetic mechanisms including DNA methylation and hydroxy methylation, histone modifications, and regulation of non-coding RNAs (ncRNAs) were closely linked to AD and helpful for the treatment of AD (Wang et al., 2020; Wu and Kuo, 2020; Xiao et al., 2020; Lauretti et al., 2021; Nikolac Perkovic et al., 2021). At present, the research hotspots in epigenetics of AD are still DNA methylation, regulation of miRNAs, histone acetylation, etc. Studies have shown that many microRNAs (miRNAs) can be used as markers for early diagnosis of AD, and epigenetic mechanisms regulate gene expression in preliminary AD (Gao et al., 2022). Therefore, studying the interaction between AD and epigenetics plays an important role in the treatment and prevention of AD. However, the pathogenesis of AD is complex, and existing drugs have extreme side effects, so there has been no considerable progress in the treatment of AD. Therefore, it is still very important to further elucidate the epigenetic mechanism of AD, find newer and more reliable biomarkers, propose new treatment strategies, and develop fresh research directions. Meanwhile, there is no bibliometric analysis in this field. Therefore, in order to facilitate researchers to better understand the current state and development trend of the link of epigenetics and AD, we used bibliometric methods to quantitatively analyze and visualize the literature in this field.
All articles from January 1, 2013 to June 12, 2023 were searched in WOSCC using File 1 (showed in the Supplementary materials) as the screening strategies. We restricted the language type of the article to English, and included papers and review papers. 3,743 articles were obtained. Then we excluded articles that did not fit the topic by reading the article title, abstract and even the full text of the paper, and finally we retained 1,530 papers. The specific retrieval strategy for this study could be found in Figure 1.
Data is imported into VOSviewer.1.6.18 and CiteSpace.6.2.3 to visualize the co-citation network maps of countries, institutions, authors, journals, keywords and articles and analyze relevant publication and citation data.
Bibliometrics is a discipline concerned with the study of distributional structure, quantitative relationship, change rule, and quantitative management of bibliometrics and data, and goes on to discuss some of the structures, characteristics and laws of science and technology using quantitative research methods such as numbers and statistics.
VOSviewer, developed by Leiden University in the Netherlands, is good at generating any type of text map, and can perform publication statistics, cooperative network analysis, co-occurrence analysis, and co-citation analysis on documents (Van Eck and Waltman, 2010).
CiteSpace is a visual bibliometric analysis software developed by Professor Chaomei Chen, which can be used for cooperative network analysis and clustering. In addition, the centrality and explosive display is its most prominent advantage (Synnestvedt et al., 2005). Centrality refers to the ability of an entity to connect the content of different groups together, revealing its role as a turning point in the entire network. The higher the centrality of a node, the more times it appears on the shortest path in the overall network, and the greater its influence and importance. “Citation bursts” refers to the sudden increase in the frequency of a specific type of event or the frequency of a large number of citations during a specific time period. Through centrality and bursting analysis, we can quickly understand the knowledge structure, research hotspots and frontiers, and the evolution of emerging topics. “Strength” indicated the intensity of the burst, “Begin” indicated the start year of the burst, and “End” indicated the end year of the burst. The red bar indicated the duration of the burst, and the blue bar indicated the entire period from 2013 to 2023 (Chen et al., 2014).
Through our search, we acquired 1,530 English-language articles in the field of epigenetics of AD including papers and review papers. As shown in Supplementary Figure 1, the number of publications experienced a steady increase from 66 in 2013 to 229 in 2022. 2013 to 2018 was the first stage of primary development, during which number of articles in this field grew with stability and was correspondingly few, demonstrating progress in this period had been slow and there was no further breakthrough. In 2019–2022, the publications on epigenetics of AD research went through a second phase of high-speed development. Despite a slight decline in publications in 2018 and 2021, the cumulative number of publications over the decade reached 1,442, averaging roughly 144 per year.
From January 1, 2013 to June 12, 2023, a total of 76 countries had participated in epigenetics of AD. The top 10 countries’ documents, citations, average citation/publication and centrality were showed in the Table 1. It showed that People’s Republic of China ranked first with 559 publications and followed by USA with 474 publications. USA had the most citations (20,321 citations) and the most average citations (Berson et al., 2018; Table 1). This can be inferred that China and USA were the countries with the most research related to epigenetics of AD and China, USA, Italy, Germany, Spain with centrality more than 0.1 suggested that they had strong academic influence in this field. Figure 2A showed collaborative networks between countries. A node in the figure represented a country and the size of node represented the corresponding country publication number and the lines between nodes represented collaborative relationships, which indicated China and USA were most closely linked in this regard. The colors represent the total intensity of cooperation between countries in this field. Figure 2B showed the national timeline diagram based on CiteSpace. The colored circles on one of the lines formed a cluster, and a total of five clusters were formed finally. A circle represented a country, the size of the circle represented the publication volume of the country, and the lines between the circles represented the partnership. The order of the circles represented the time when the country began research on epigenetics of AD. The purple ring in the circle represented the centrality of the country, and the thickness of the purple ring described the magnitude of the centrality. According to this, we could find that USA, Chinese and Spain had the greater centrality. England, Germany, Italy, USA, Spain, People’s Republic of China, India, South Korea, Canada started research earlier and Iran started later. It also reflected that the research on neurological disease and methylomics had lasted for a long time and they were hot topics.
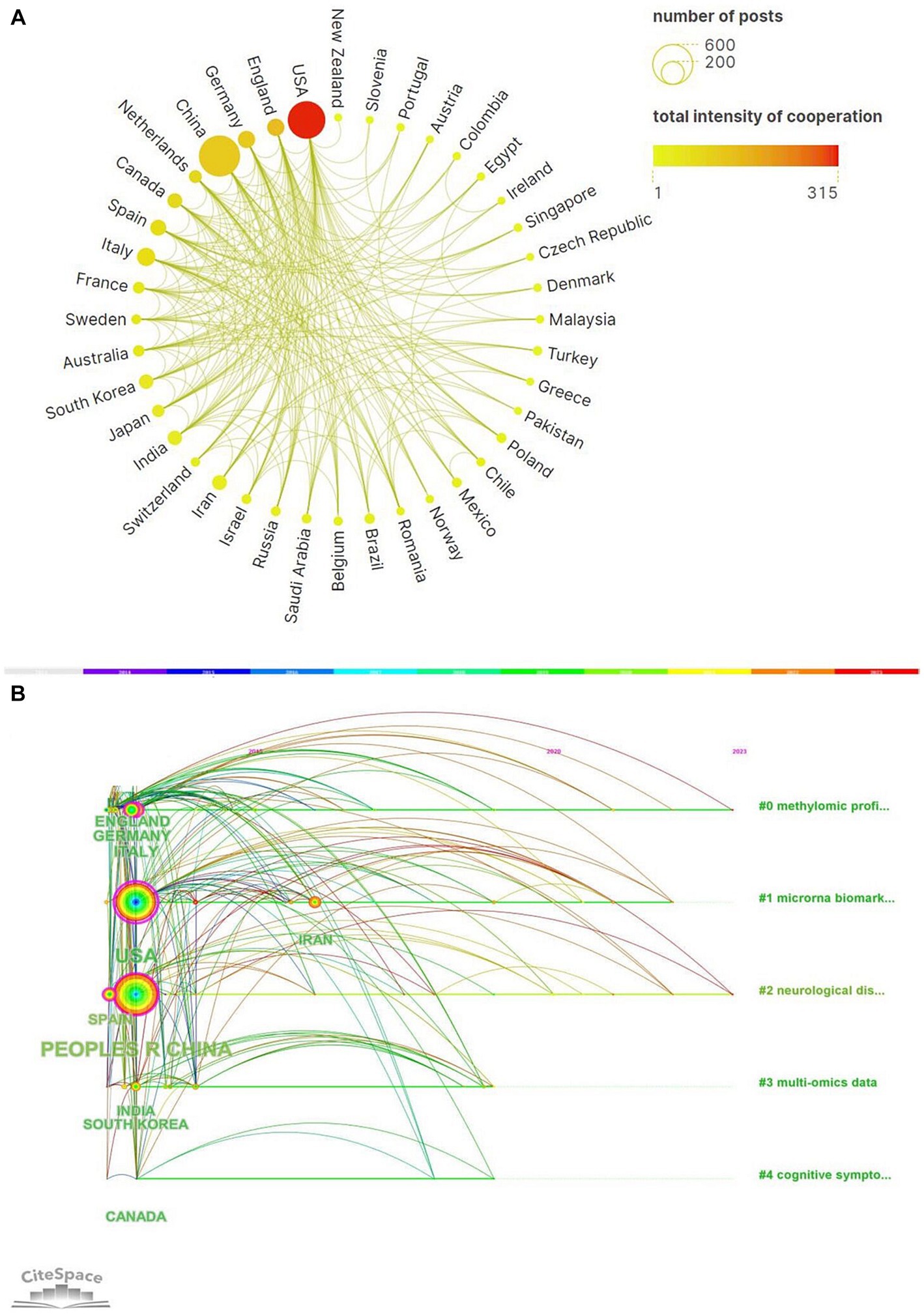
Figure 2. (A) A world map of the intensity of cooperation among 20 countries. (B) CiteSpace-based visualization of national timeline diagram.
We had analyzed productions of coming from 1899 institutions. We found that Capital Medical University’s documents accounted for the most. But Maastricht University had the most citations (Supplementary Figure 2A). It showed that Maastricht University’s documents were of high quality. Representing the 4 nodes of Harvard University, Shanghai Jiao Tong University, Rush University, Columbus University had been presented dense lines, which indicated that they cooperated with other institutions frequently (Supplementary Figure 2B). As shown in Figure 3, we could also find that among the clusters, the clusters represented by Harvard University had denser networks. Maybe the direction studied by Harvard University was recognized by most of institutions. The institutions were divided into 8 clusters according to title words and the colors of the clusters corresponded to the numbers #0–7 in the color bar in Figure 3.
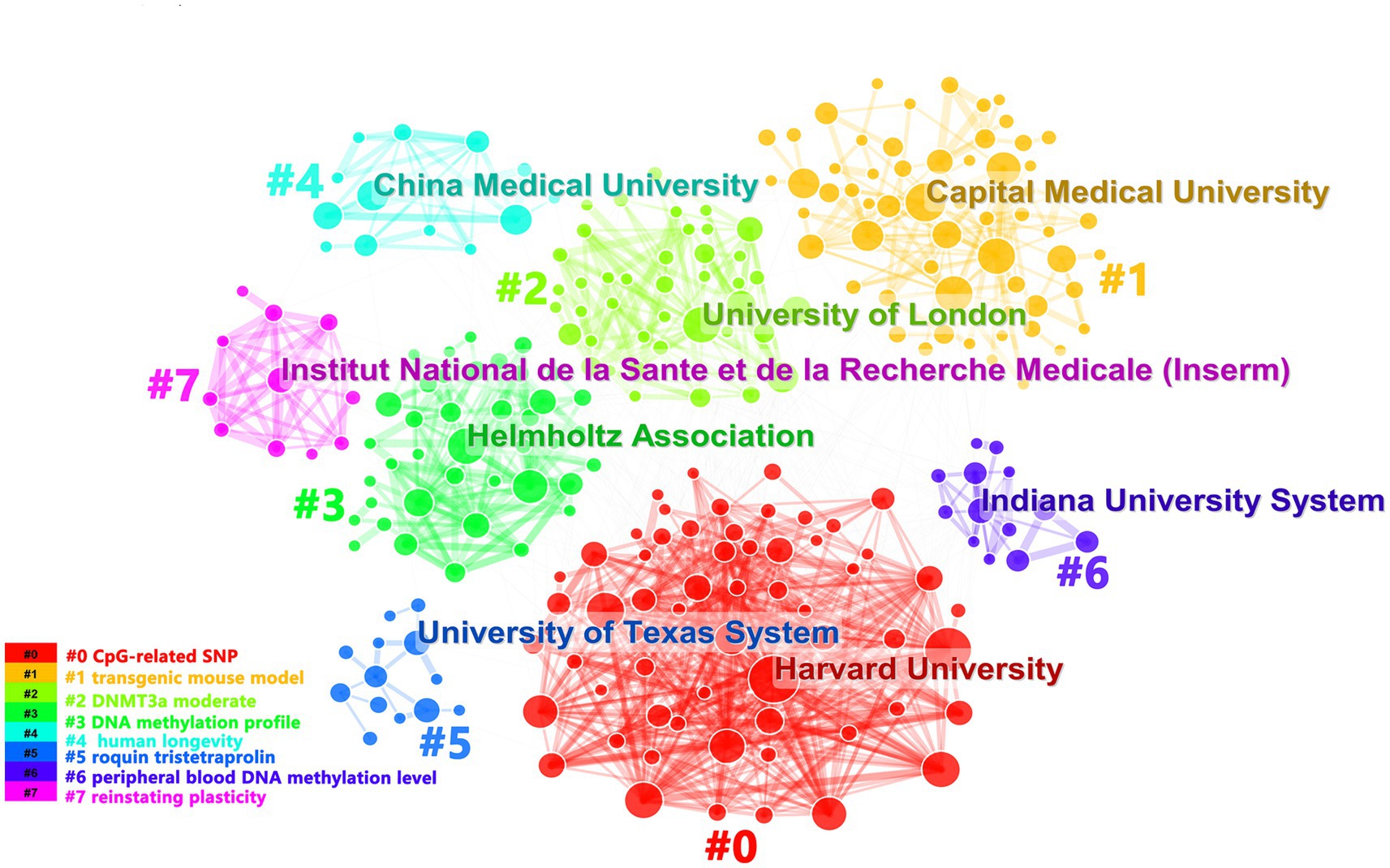
Figure 3. It showed organization cooperation clusters. Each cluster described the main institution and the topics of the cluster were shown in the corresponding legend.
A total of 44,213 authors had devoted to the research of epigenetics of AD over the past 10 years. We listed the top 11 most productive authors according to the documents, citations and average citation/publication (Table 2). The top 5 productive authors were Bennett DA, De Jager PL, Lukiw WJ, Lunnon K and Zhang W. The top 5 cited authors were Bennet DA, Lukiw WJ, Zhang W, Mill J and Zhao YH. As another parameter, average citation/publication was used to assess authors’ contribution, which was able to better reflect the quality of the author’s publications. Due to the highest number of posts and highest citation count, it had no doubt that Bennett DA was the most contributed and prestigious scientist in this field. Partnerships between authors were evaluated by collaboration network visualization of co-authors. As shown in Figure 4, the authors were divided into 6 clusters according to keyword relevance and cooperation strength. The color of the clusters corresponded to the numbers #0–5 in the color bar in Figure 4. Every node was regarded as a symbol of different author while the circle size reflected the number of articles published by each author. The thicker lines between the circles indicated the closer connection linking the writers.
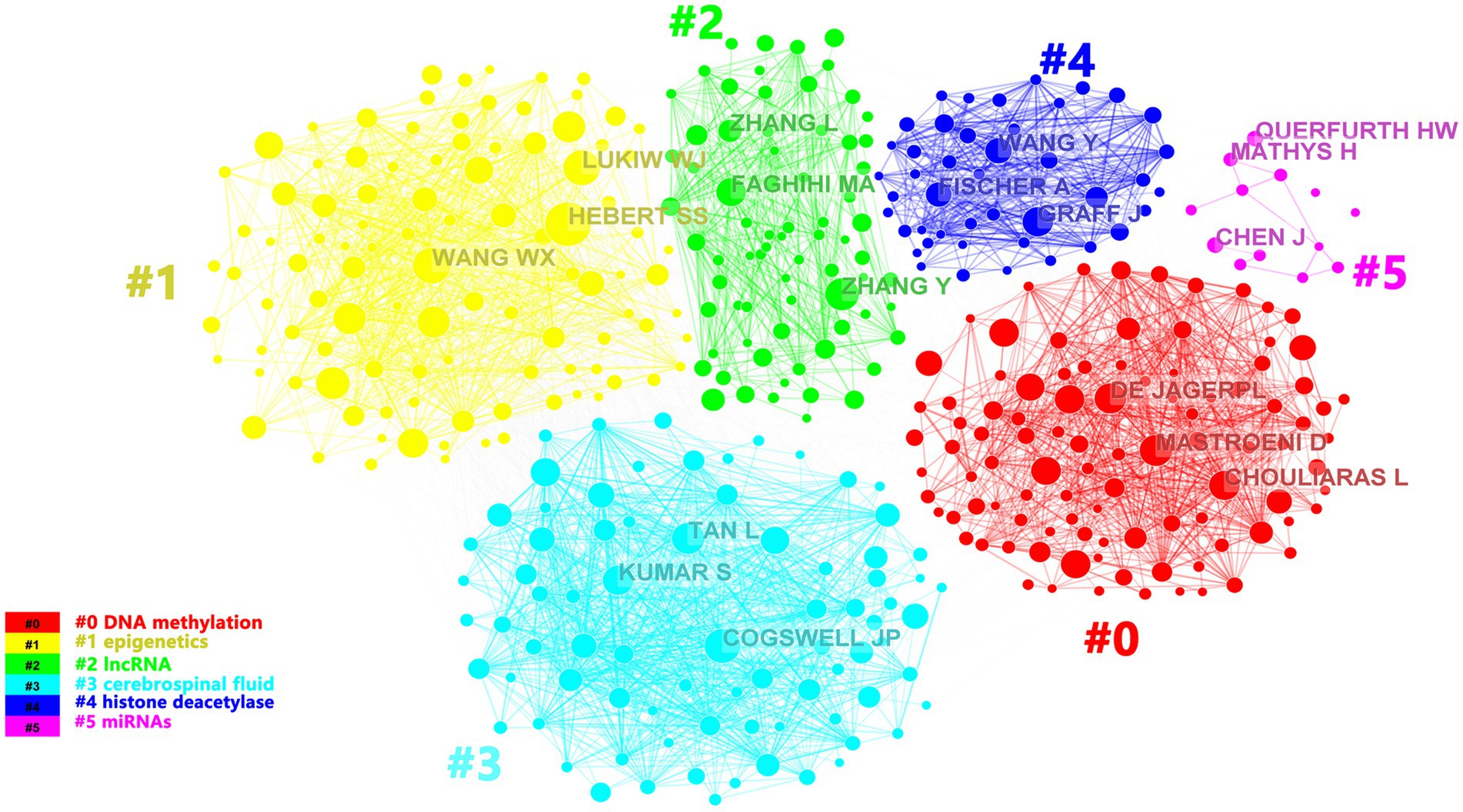
Figure 4. The cluster visualization map of topics for the authors. The colors of the clusters corresponded to the numbers #0–5 in the color bar and the topics of the cluster were shown in the corresponding legend.
All articles were distributed in 5,437 journals and Table 3 displayed the top 10 journals ranked by citation, documents and average citation/publication. In terms of the number of citations, the top 3 were Journal of Alzheimer’s Disease, Neurobiology of Aging and Molecular Neurobiology; In terms of the number of documents, the top 3 were Journal of Alzheimer’s Disease, International Journal of Molecular and Molecular Neurobiology; The impact factors (IF) of various journals in 2022 were also shown in Table 3, with the most influential journals being Aging-US, Cells. With no doubt that Journal of Alzheimer’s Disease was the most productive journal, but Aging-US was the most influential journal with the highest average citation/publication and IF.
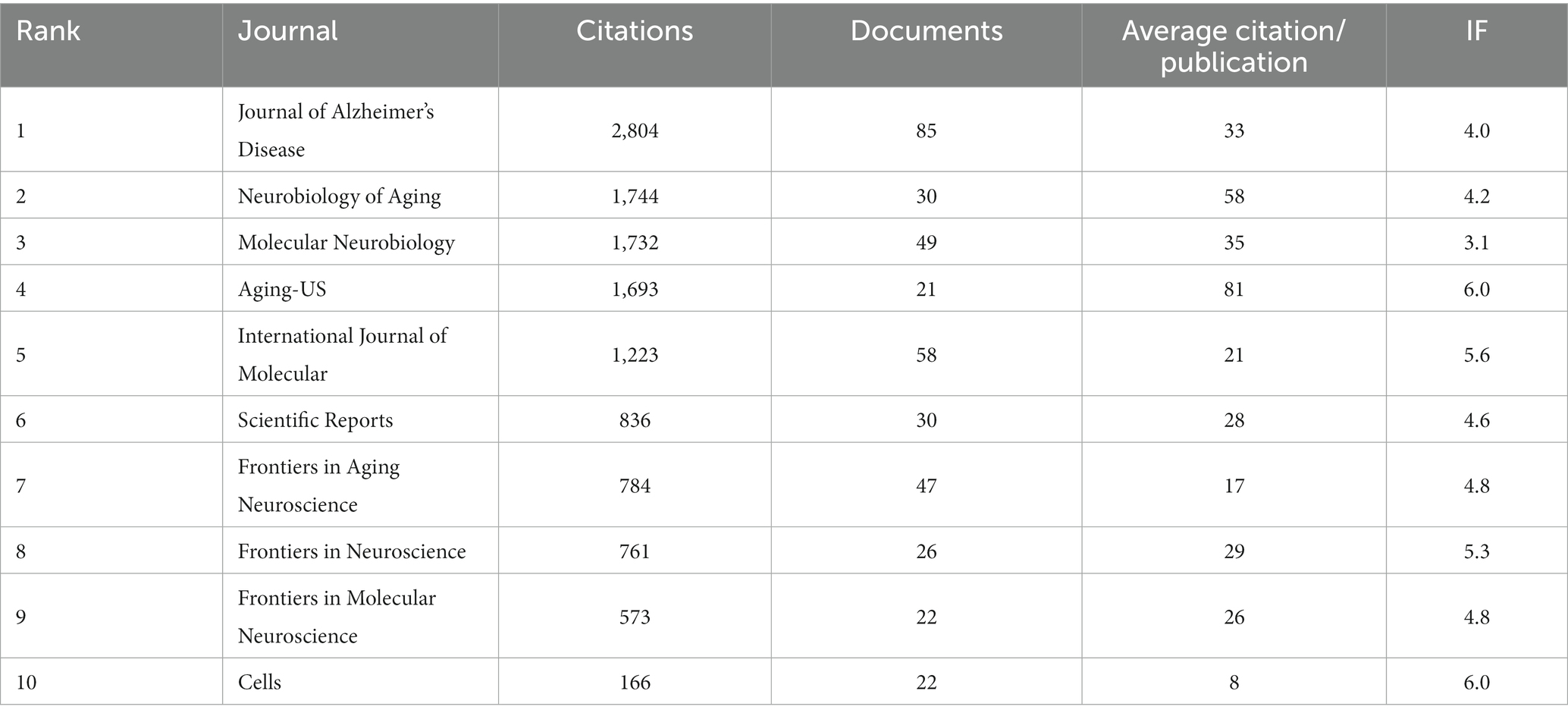
Table 3. The top 10 journals ranked by citations, documents, and average citation/publication and IF.
We used CiteSpace and VOSviewer to visualize the 5,533 keywords of 1,530 selected articles. The top 10 frequently occurring keywords were miRNAs (486), biomarkers (273), DNA methylation (257), amyloid beta (168), gene expression (165), amyloid precursor protein (160), oxidative stress (134), neurodegeneration (123), tau (116), cerebrospinal-fluid (110). And DNA methylation’s centrality (0.05) was the most, which indicated it had the greatest influence among them (Table 4). Based on the strength of the citation bursts, we got the top 25 keywords. CiteSpace conducted a burst detection of keywords. “Strength” indicated the intensity of the burst, “Begin” indicated the start year of the burst, and “End” indicated the end year of the burst. The red bar reflected the duration of the burst, and the blue bar reflected the entire period from 2013 to 2023. The strength of histone acetylation and histone deacetylase inhibitors’ (HDACI) citation bursts had been the highest in the past years. The figure also showed the current higher strength of deficits, tau, and receptor, indicating that they have been the focus of research in recent years (Figure 5A). According to the network visualization showed in the Figure 5B, the size of the circle indicated the frequency of the keyword, different colors represented different clusters, the lines between nodes showed the correlation between keywords, the more lines the keywords distribute, and the relationship between this keyword and other keywords was stronger, we found that apart from Alzheimer’s and epigenetics themselves, keywords related to miRNAs, biomarkers, DNA methylation appeared more. As shown in Figure 6, the keywords were grouped into 8 clusters by topic and labeled with different colors. The first group was epigenome-wide association studies (cluster #0). The keywords were DNA methylation, gene expression, cognitive impairment, apolipoprotein E and pathology. This group mainly studied the role of epigenetic gene modification in the pathogenesis of AD. The second group was memory formation (cluster #1) and involved keywords like transgenic mouse models, histone acetylation, synaptic plasticity and memory. This group mostly explored the effects and mechanisms of AD on memory. The third group was targeting β-secretase (BACE1) (cluster #2) and this group contained miRNAs, biomarkers, apoptosis and genes. It showed that epigenetic mechanisms apply to targeted therapy of AD. The fourth group was memory deficit (cluster #3), which also involved keywords such as tau, neurodegeneration, indicating that abnormal post-translational modifications of tau protein was related to memory deficit in AD. The fifth group was neurodegenerative disease (cluster #4) and the keywords were Parkinson’s disease and Huntington’s disease and illustrated the association between AD and other neurodegenerative diseases in terms of epigenetics. The sixth group was neuronal damage (cluster #5) and involved keywords like amyloid precursor protein, tau phosphorylation, neurotrophic factor and lncRNAs, which revealed the mechanism of neuronal damage. The seventh group’s keywords were mainly oxidative stress and amyloid beta (cluster #6). This group of studies revealed oxidative stress and neuroinflammation activation pathways in AD. The eighth group was systematic review (cluster #7).
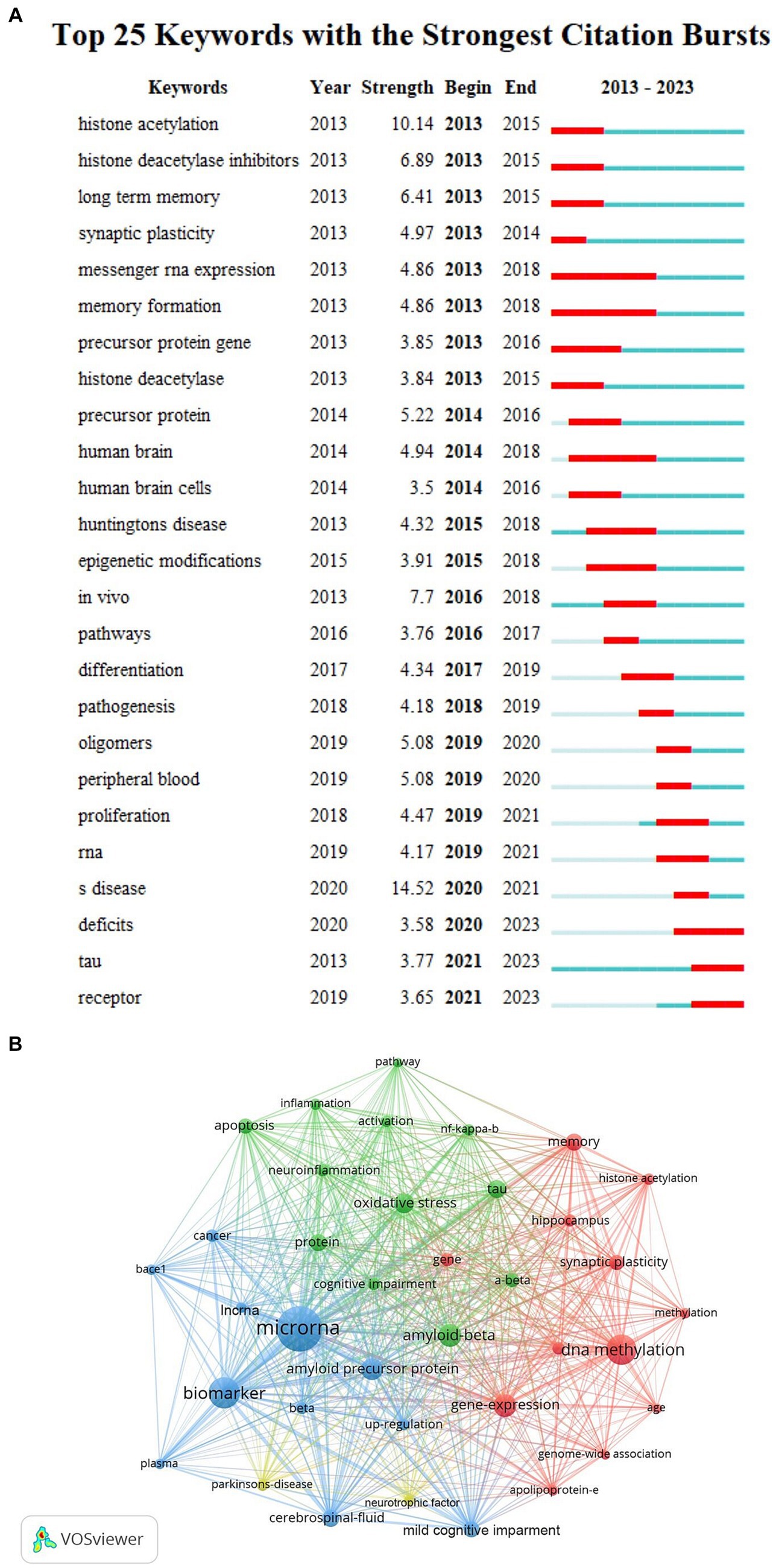
Figure 5. (A) Top 20 keywords with the strongest citation bursts. The strongest citation bursts time period was indicated in red. (B) The co-occurrence analysis of keywords based on VOSviewer. The size of the circle indicated the frequency of the keyword, different colors represented different clusters, the lines between nodes showed the correlation between keywords, the more lines the keywords distribute, and the relationship between this keyword and other keywords was stronger.
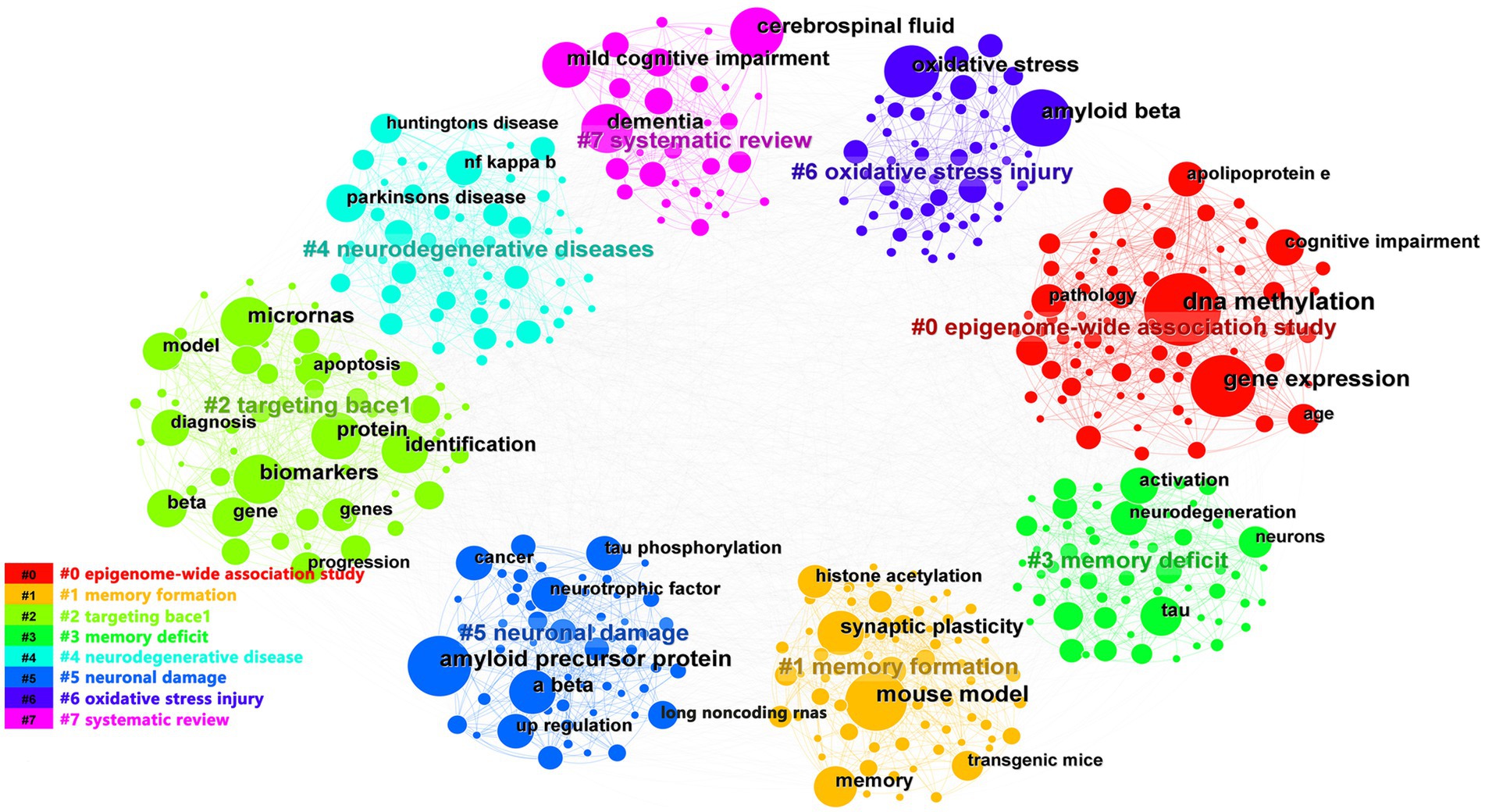
Figure 6. The co-occurrence analysis of keywords based on CiteSpace. The keywords were grouped into 8 clusters by topic and labeled with different colors. The topics of the cluster were shown in the corresponding legend.
In the past 10 years, a total of 1,530 articles have been cited in epigenetics of AD research field, with 67,138 citations and an average of 44 citations per document. As shown in Figure 7, references were divided into 8 clusters according to topics and marked with diverse colors. The color of the clusters corresponded to the numbers #0–7 in the color bar in Figure 7. #0, #1, and #2 had the largest clusters, indicating that the 3 areas of DNA methylation, exosome-based biomarker, and epigenome-wide association studies have been cited and studied the most in recent years.
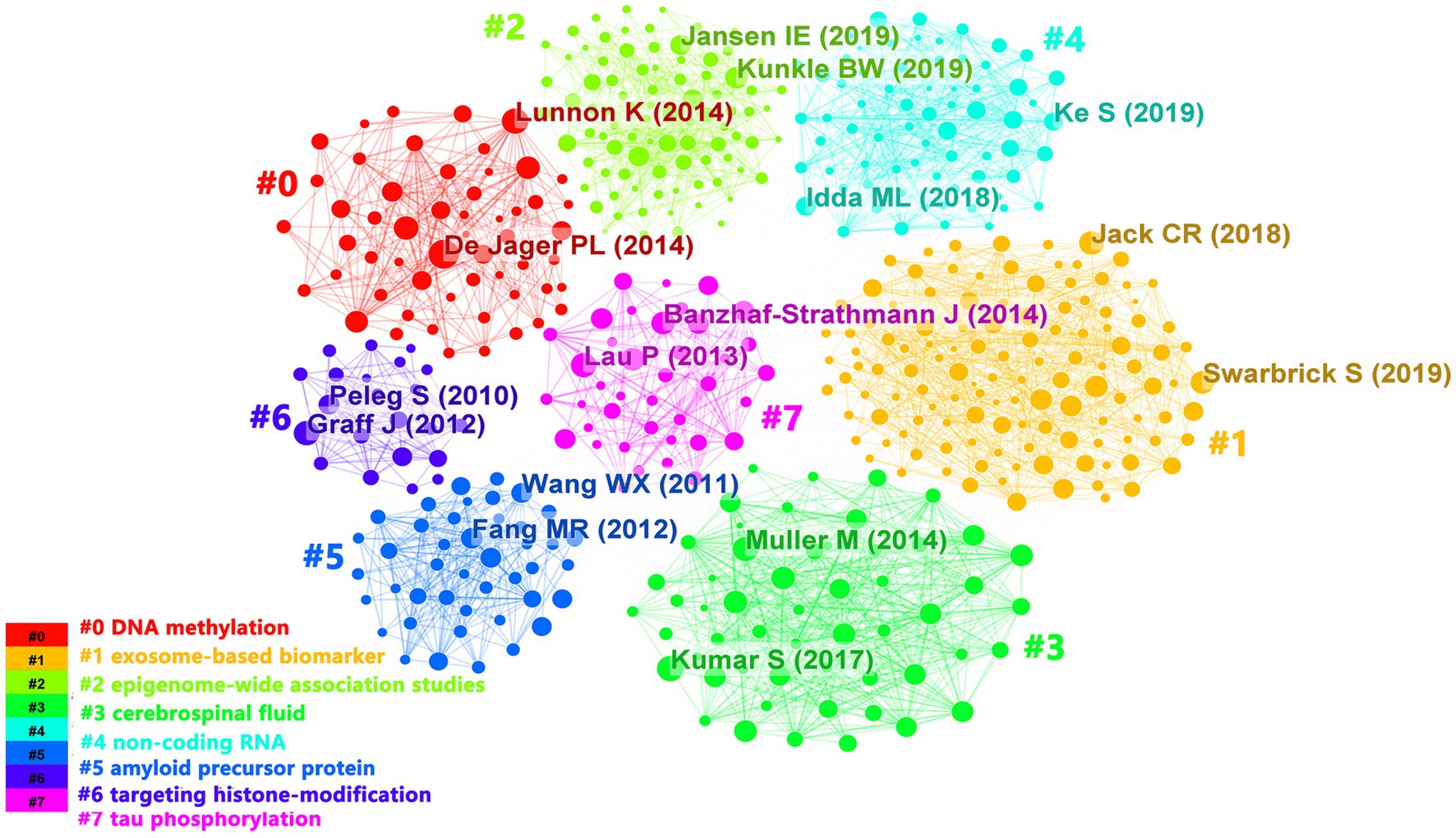
Figure 7. The clustering visualization map of topics for cited references. The colors of the clusters corresponded to the numbers #0–7 in the color bar and the topics of the cluster were shown in the corresponding legend.
Based on the co-occurrence analysis and keywords burst graph analysis of CiteSpace and VOSviewer, we found that keywords describing ncRNAs, transcription factor, genome, histone modification, blood DNA methylation, acetylation, biomarkers have been used most frequently in recent years. In addition, we should also pay attention to macroscopic mechanisms such as oxidative stress, synaptic plasticity, and apoptosis.
In terms of some correlative studies, several highly cited articles highlighted the main research directions in epigenetics of AD, which engrossed our attention. For example, Kumar et al. (2013) initially generated a compilation of prominent canonical pathways through the utilization of the Ingenuity Pathway Analysis platform. They discerned essential circulating miRNAs pathways that exhibited biological associations with the biology of AD. These pathways encompassed axonal guidance signaling, ephrin receptor signaling, actin cytoskeleton signaling, clathrin-mediated endocytosis signaling and rhoA signaling (Gallo, 2007; Lin et al., 2009; Wu and Yao, 2009; Chacon et al., 2011). Additionally, they demonstrated that a canonical pathway associated with TypeII Diabetes Mellitus signaling was notably enriched with targets of the signature miRNAs. “An epigenetic biomarker of aging for lifespan and healthspan (2018)” written by Levine mentioned they developed a new epigenetic biomarker of aging-DNAm Phenoage, using an innovative two-step process (Levine et al., 2018). The study conducted by Gjoneska et al. examined various aging outcomes, including all-cause mortality, cancer, health span, physical function, and AD. The researchers found that their evaluation method had notable advantages over previous measurements in predicting these outcomes. Specifically, they investigated the transcriptional and chromatin state dynamics in early and late hippocampal pathology in a mouse model of AD-like neurodegeneration (Gjoneska et al., 2015). Their study revealed a synergistic downregulation of genes and regulatory regions associated with synaptic plasticity, while immune response genes and regulatory regions were upregulated, suggesting that immune processes also play an important role in the progression of AD. When cells in the hippocampus, a region closely associated with AD, lose function or die, new connections can be made through healthy neurons within the population, and these connections can either exhibit parallel function (homologous sprouting) or fibers from the convergent pathway (heterologous sprouting) can maintain functional stability through enhanced or weakened signals, this result demonstrates the plasticity of synapses in the hippocampus (Cotman and Anderson, 1988). Epigenetic markers that are closely related to the development of AD, such as DNA and histone modifications, have been found in the peripheral blood of AD patients in clinical studies, and considering the reversibility of epigenetic modifications and the complexity of gene–environment interactions, we may need to combine epigenetic therapeutics with environmental strategies and drugs with multiple targets (Jeremic et al., 2023). The above studies indicate that based on the involvement of various epigenetic mechanisms in the development and pathogenesis of AD, the study of new AD biomarkers and targeted therapies is a major research direction in this field. To further advance, it is crucial to enhance our understanding of the fundamental biology of aging, particularly in terms of elucidating the interplay between cell type specificity and aging biology. Additionally, efforts such as the Model Biological Development and Evaluation of Late-Onset Alzheimer’s Disease should be undertaken to refine the model system (Oblak et al., 2020; Gonzales et al., 2022).
As shown in Figure 8, we will systematically elucidate the epigenetic mechanisms of AD from three aspects of DNA, histone modifications and ncRNAs.
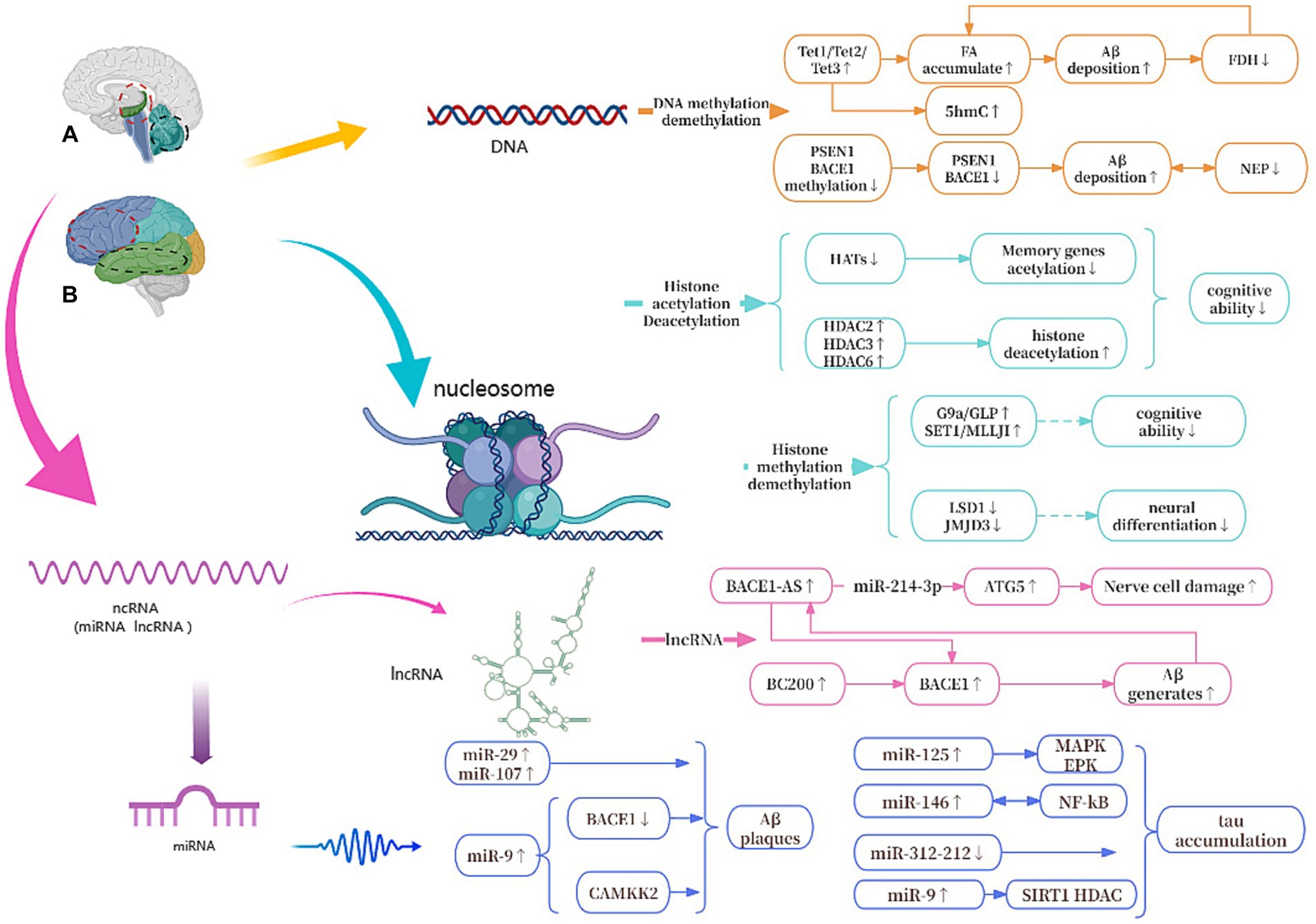
Figure 8. Epigenetic mechanisms of AD. (A) The red circle represents the hippocampus, and the black circle represents the cerebellum. (B) The red circle represents the frontal lobe, and the black circle represents the temporal lobe. Tet1, Tet2, and Tet3, All Tet proteins regulate DNA demethylation in the body; FA, formaldehyde; FDH, formaldehyde dehydrogenase; PSEN1, presenilin 1; BACE1, beta-site APP cleaving enzyme 1; NEP, neprilysin; G9a/GLP, Both are histone methyltransferase; LSD1/JMJD3, Both are histone demethylase; HATs, histone acetylases; HDACs, histone deacetylases; ATG5, autophagy-related genes 5; BC200, brain cytoplasmic 200; MAPK/ERK, protein kinase/extracellular signal-reduced kinases; NF-κB, nuclear factor κB; CAMKK2, calcium/calmodulin-dependent protein kinase 2.
Among all the known mechanisms of epigenetics, DNA methylation has been studied and known the most. During DNA methylation, a methyl group is added to the 5th atom of the cytosine ring, which forms 5-methylcytosine (5mC) (Prokhortchouk and Defossez, 2008). Oliveira et al. found that DNA methyltransferases3a2 (Dnmt3a2) reduced in the hippocampus of mice, and the restoration of Dnmt3a2 recovered the cognitive functions (Moore et al., 2013; Lee et al., 2014). This shows to some extent that methylation of DNA affects gene expression and may hinder the binding of transcription activators to DNA (Lee et al., 2014), its precise regulation is essential for maintaining normal cognitive function (Moore et al., 2013). There are three Tet proteins that regulate DNA demethylation in mammalian bodies and modify DNA by repeatedly oxidizing 5mC to produce 5-hydroxymethylcytosine (5hmC): Tet1, Tet2 and Tet3, which regulate the expression of different genes in different cells to control cellular behavior (Zhang et al., 2023). Tet1 is currently thought to play the most important role in AD. Disruption of Tet1 and subsequent disruption of the methylome can alter AD-associated hydroxy methylation, as well as the expression of key genes involved in maintaining the epigenome, neuronal health, and the activation and function of microglia (Armstrong et al., 2023). In addition, during DNA demethylation, the active substance formaldehyde (FA) is produced, and excessive exposure to FA or endogenous FA metabolism disorder can lead to abnormal accumulation of FA (Li et al., 2021). Excessive FA interacts with Aβ42 to accelerate Aβ aggregation, which in turn inhibits the activity of formaldehyde dehydrogenase (FDH), thereby reducing FA degradation and leading to FA accumulation (Fei et al., 2021). At the same time, Aβ plaque deposition can cause hypermethylation of Aβ-degrading enzymes such as neutral endopeptidase (NEP) and down-regulate the expression of NEP, while low expression of NEP will promote further deposition of Aβ (Lin et al., 2021). In addition, some studies have confirmed that S-adenosylmethionine via improving Presenilin-1 (PSEN1) and BACE1 expression reduces tau phosphorylation and Aβ in a mouse model (Cotman and Anderson, 1988; Zhang et al., 2023).
Relatively speaking, the study of DNA methylation is more mature, involving different areas of the blood, cerebrospinal fluid and brain. However, currently there is no evidence that DNA methylation in peripheral blood cells is associated with cerebral cortex samples (Yu et al., 2016), so the connection between blood and DNA methylation in the brain needs other clues. DNA methylation usually occurs in CpG islands within these gene promoter regions, which are long repeating sequences containing cytosine-guanine nucleotides (Xie et al., 2023). Studies have found that 5mC at certain CpG check points is highly correlated with age; therefore, biological age can be accurately predicted using relatively few CpG check points. Based on this, this modification can be used as an estimated indicator of an organism’s aging, called the epigenetic clock (Gao et al., 2022; Jeremic et al., 2023), which is also widely concerned.
Of course, there are also a small number of new studies that suggest that AD is associated with a decrease in bioenergy caused by mitochondrial dysfunction (Wallace, 2011; Coskun et al., 2012; Hroudová et al., 2014; Swerdlow et al., 2014), such as in one study, patients with low Aβ and high t-tau AD biomarkers and pre-symptomatic patients with PSEN1 mutation showed low levels of mitochondria in cerebrospinal fluid (Podlesniy et al., 2020). Mitochondrial DNA methylation is also regulated by epigenetic mechanisms, whether it is related to AD is also receiving increasing attention (Huang et al., 2022). However, research in the emerging field of mitochondrial DNA methylation is quite limited. For example, there are few specific displays of this epigenetic inheritance in the study of mitochondrial DNA. It is difficult to find the concentrated presentation area of mitochondrial DNA methylation, or to use means to present it, which will be a difficult problem (Bellizzi et al., 2013; Blanch et al., 2016).
Histone modification mainly refers to the modification that occurs at the amino tail end of histone translation, such as methylation, acetylation, phosphorylation, ubiquitination, etc., which not only occurs in nerve cell development, but also plays an important role in the aging brain, and in the pathogenesis of AD (Mastroeni et al., 2011; Berson et al., 2018; Geng et al., 2021). Among them, histone acetylation is the most studied, that is, under the catalysis of histone acetyltransferase (HAT), the acetyl group in cofactor acetyl-CoA is added to the amino acid residues (mainly lysine in H3 and H4) at the N-terminal tail of histone, and then transcription begins (Santana et al., 2023). During this process, changes in related enzymes reduce histone acetylation levels, lead to inactivation of memory-related genes, and cause abnormal phosphorylation of tau, which in turn leads to cognitive degeneration (Peixoto and Abel, 2013).
The inhibition of histone deacetylases (HDACs) can reverse these changes and alleviate cognitive impairment (Gao et al., 2022). For instance, sodium phenylbutyrate, an HDACI, can induce the expression of neurotrophic factors through the protein kinase C-cAMP-response element binding protein pathway, thereby alleviating memory impairment in transgenic AD mice (Yang et al., 2017). Previous research has shown that proteomic analysis revealed a higher number of changes in histone post-translational modifications in the brains of AD patients compared to healthy elderly brains. Specifically, a significant increase in Histone H3 lysine is acetylated at site 27 (H3K27ac) and Histone H3 lysine is acetylated at site 9 (H3K9ac) was observed (Creyghton et al., 2010; Lu et al., 2014). Disease-specific alterations enriched for single-nucleotide polymorphisms from AD Genome-Wide Association Studies (GWAS) and expression quantitative trait loci points to potential involvement of H3K27ac in AD, indicating a possible disease-specific increase (Lambert et al., 2013; Kunkle et al., 2019; Xiong et al., 2023). Integrated multi-omics data reveals that H3K27ac and H3K9ac may play a role as potential epigenetic drivers in AD (Nativio et al., 2020). At the same time, recent epigenome studies have revealed the potential relationship between epigenome information loss and apoptosis of neurons and other cells. Although GWAS has revealed more than 40 genetic loci associated with AD, because more than 90% of risk loci are located in non-coding regions, transcriptome data alone is difficult to pinpoint the disease-causing loci of AD and elucidate its molecular mechanisms (Bellenguez et al., 2022). For example, using this approach, the researchers revealed the global epigenomic information loss that occurs with H3K27ac histone modifications in patients with advanced AD; Runt-associated transcription factor 1, Spi-1 Proto-Oncogene and other transcription factors play an important role in the pathogenesis of microglia AD (Xiong et al., 2023). It can be seen that studies at the multi-omics level play an important role in revealing the combined effects at the cellular and molecular levels in AD. Therefore, integrating epigenome and transcriptome data may be the key to solving the difficulties of neurodegenerative diseases such as AD.
Histone methylation refers to the process of adding methyl groups to the lysine or arginine residues of histones, this process is catalyzed by histone methyltransferases (HMT) that catalyze the transfer of 1–3 methyl groups to histones (Zhou and Shao, 2021; Ding et al., 2023). The effect of histone methylation on transcription is determined by several factors, including the specific histone involved, the specific residues that are modified, and the overall degree of methylation at each site. Typically, methylation of H3K4, H3K36, and H3K79 is associated with active transcription, while methylation of H3K9, H3K27, and H4K20 is associated with inhibitory transcription (Yi and Kim, 2020).
G9a is a type of histone methyltransferase with a SET domain. They are involved in the in vivo modification of H3K9me2. H3K9me2 is the fourth lysine of histone H3 that is trimethylated HMT (Chaturvedi et al., 2012). Initially described as encoding a protein similar to G9a, G9a-like protein (GLP) has the same substrate specificity on histones as G9a. The G9a and GLP complex is associated with memory and learning, and participates in long-term potentiation, the maintenance of synaptic plasticity and upregulating Brain-derived neurotrophic factor (Sharma et al., 2017; Han et al., 2021). Treatment of transgenic mice with the G9a/GLP inhibitor UNC0642 reduces H3K9me2 and changes the overall levels of 5mC and 5hmC, increasing synaptic plasticity and reducing neuroinflammation, preventing Aβ plaque accumulation and enhancing cognitive function, thereby affecting the development of AD. Some research showed that inhibiting the activity of G9a/GLP may be a potential target for treating AD (Cao D. et al., 2020). Furthermore, the SET1 family of enzymes are well known for their involvement in H3K4 methylation. Thus, overexpression of the SET1/MLL family may be a potential cause of abnormally elevated H3K4me3 in the body. When the HMT function of SET1/MLL is inhibited, it can reduce the levels of H3K4me3 in the prefrontal cortex (PFC), restore the expression of glutamate receptor at synapses, increase PFC synaptic plasticity, and further improve cognitive impairment in AD mice. This suggests that targeting the SET1/MLL family of enzymes may also hold potential for the treatment of AD (Cao Q. et al., 2020). Proteins containing the Jumonji domain 3 (JMJD3) play a crucial role in activating neurogenesis in the adult brain ventricular zone by serving as histone H3K27 demethylases. High expression of JMJD3 leads to enhanced demethylation of H3K27me3 in related genes, such as DLX2, MLL1, and MASH1, thereby promoting neuronal differentiation (Cao D. et al., 2020). Previous studies showed inhibition of Lysine-specific histone demethylase 1 (LSD1) seems to be correlated with the activation of pluripotent genes and the induction of AD lesions. Moreover, research indicates that overexpression of LSD1 in hippocampal neurons can rescue cell death and inhibit the occurrence of inflammation (Engstrom et al., 2020).
ncRNAs are closely related to the epigenetic mechanisms of AD and are diverse. In this article, we focus on miRNAs and lncRNAs (He et al., 2023). So far, many different miRNAs have been widely studied and linked with the development of AD, and the end result of the action of miRNAs has been the translational inhibition and/or degradation of targeted miRNAs (Huntzinger and Izaurralde, 2011). Some of miRNAs can lead to synaptic and cognitive dysfunction, and others can suppress neuronal apoptosis and pathological damage and improve the intelligence of AD mice (Gao et al., 2022). Several studies have found that MAPK/ERK activation is associated with the formation of miR-125 bp-tau, and the expression of miR-146 is closely related to NF-κB. Increased levels of miR-125 and miR-146 in vivo lead to tau protein aggregation. While miR-9 is involved in regulating calcium/calmodulin-dependent protein kinase 2 (CAMKK2), a decrease in its content can lead to Aβ plaque deposition (Chang et al., 2014). Moreover, some studies have shown that certain miRNAs are potential in AD therapy, it is still a new direction in AD research.
lncRNAs refer to a type of endogenous non-coding RNA with a length exceeding 200 nucleotides (Li and Wang, 2023). These lncRNAs are expressed in brain tissue and are distributed in the cell nucleus, cytoplasm, and mitochondria (Bridges et al., 2021). One of the most well-studied lncRNAs is BACE1-AS, which is transcribed from the reverse strand of the BACE1 gene locus on chromosome 11. It has been shown that upregulation of BACE1-AS may promote the generation of Aβ. Moreover, in the brains of AD patients, sustained production of Aβ can induce upregulation of BACE1-AS, thereby creating a positive feedback mechanism that further promotes Aβ expression and worsening the pathological deterioration (Li et al., 2019). Furthermore, in mouse experiments, down-regulation of BACE1-AS has been shown to improve memory in mice (Zhang et al., 2018). In addition to its role in AD development, BACE1-AS is also of interest in the autophagy-lysosomal system, where it promotes neuronal damage mediated by autophagy (Zhou et al., 2021). Another lncRNA of interest is brain cytoplasmic 200 (BC200), which is mainly expressed in the cell bodies and dendrites of neurons in the hippocampus and neocortex (Ding et al., 2022). BC200 can inhibit the expression of BACE1 and increase cell survival rates in AD cell models. Overexpression of BC200 in brain tissue elevates BACE1 expression, accelerating Aβ deposition (Li et al., 2018).
Scientists believe that many natural compounds or their metabolites may have effects similar to those of known epigenetic regulators that can improve neurodegenerative diseases such as AD. For example, genistein, epigallocatechin gallate have the ability to inhibit DNMT and HDAC; lycopene and curcumin have antioxidant properties that delay aging (Castillo-Ordoñez et al., 2024). Moreover, postmenopausal women account for a higher proportion of women with AD (Barron and Pike, 2012). Loss of hormonal regulatory, synaptic, and neuroprotective effects likely contribute to AD pathology, and epigenetic regulation of hormones improved memory (Mills et al., 2023). This suggests that the epigenetic process of AD is closely related to estrogen, and estrogen receptors are widely distributed in nerve cells involved in learning and memory (Kumar et al., 2023).
The pathogenesis of most AD cases is generally considered to be a complex disease caused by a combination of multiple genetic and environmental factors, such as lifestyle, dietary habits, age, gender, education level, ethnicity, etc. The combination of these factors and their interaction may affect an individual’s sensitivity to AD risk. Therefore, through a comprehensive study of genetic and environmental factors, as well as an in-depth analysis of individual cases, a more comprehensive understanding of the pathogenesis of AD can provide more effective strategies for the prevention and treatment of AD (Sharma et al., 2020).
In the study of AD epigenetics, the correlation between DNA modification and histone modification and the combined effects of ncRNAs is not clear. Recent studies have shown that the wide application of omics technology in many fields such as epigenetics and genomics can collect many complex pathological data, and use its powerful computing and analysis capabilities to help discover more new biomarkers, which can eventually be used for AD research and related prediction and treatment (Pierouli et al., 2023; Tan et al., 2023). At present, it seems that the study of multiple omics data such as epigenomics, methylomics, transcriptomics will show an important role in determining the mechanism of epigenetics in AD in the future (Wang et al., 2023).
In recent years, the number of AD patients worldwide has been increasing, and with the development of new therapies, elucidating epigenetic mechanisms such as histone modification, DNA modification, and ncRNAs is crucial for the future development of safe and effective AD therapies (Zhang et al., 2022). The current clinical drugs for the treatment of AD are mainly pathological drugs targeting Aβ and tau proteins. This may be due to the lack of clinical trials of strategies such as neuroprotection, nerve cell suppression, immunomodulation, and protein homeostasis, and the high failure rate of drug clinical trials. In addition, many AD treatments are almost stuck in the experimental stage of animal models, and compounds recognized as disease-modifying pathways fail to enter clinical research without reason, thus hindering the development of clinical research (Dhapola et al., 2023; Jeremic et al., 2023). This requires re-examining the treatment of AD from various aspects. For example, first, to find new therapeutic targets. Second, to use multi-target therapy based on existing targets. In addition to classical Aβ and tau targets, a variety of targets such as cholinergic, glutaminergic, dopaminergic, GABAergic, adrenergic, and serotonergic signaling have been extensively studied as possible pathways for the treatment of AD (Kandimalla and Reddy, 2017). This trend of multi-target therapy provides new directions and possibilities for AD drug development, and it is hoped that more breakthroughs can be made in the future to bring more effective treatment options for AD patients. Third, to improve the current experimental technology, a variety of experimental methods such as multi-omics analysis, nanotechnology, etc. can be used for experiments (Dong et al., 2021; Cáceres et al., 2023). Generally speaking, in drug development related to epigenetics, advances in multi-target therapy should be the main focus of treatment for neurodegenerative diseases in the future (Ghosh and Saadat, 2023).
While there are many studies on the relationship between AD and epigenetics, there are still many challenges. Firstly, the epigenetic mechanism of AD has not been fully elucidated (Paniri et al., 2023). Although many studies have shown that AD is closely related to DNA methylation and hydroxy methylation, they have not really clarified the 5mC and 5hmC levels is the imbalance part of the cause of AD or just a consequence of AD (Nikolac Perkovic et al., 2021). Secondly, in the past 10 years, the number of AD patients has been increasing worldwide, but the treatment of AD has almost stayed in the experimental stage of animal models, and the trials in clinical application are insufficient. In the development of new drugs, the more detailed clarification of epigenetic mechanisms such as histone modification, DNA modification, and ncRNAs is crucial for the future development of safe and effective drugs for AD. Meanwhile, the current DNA methylation age estimation model lacks precision. The identification of a novel epigenetic clock capable of encompassing the entirety of human lifespan is a pressing concern within the field of DNA methylation research (Shireby et al., 2020). Additionally, it is important to acknowledge that epigenetic biomarkers, including miRNAs, do not comprehensively or accurately depict the pathological progression of AD (Ritchie et al., 2017). The primary challenge in utilizing miRNAs for external diagnosis lies in identifying specific miRNAs that can serve as markers for a diverse range of patients, as well as developing accurate, straightforward, and cost-effective methods to match them (Condrat et al., 2020).
This is the first bibliometric analysis of the epigenetics of AD, which has certain limitations. Firstly, the literature only sourced from the WOSCC, so the results obtained by the analysis are limited. Secondly, we have certain restrictions for screening the data. Recently published data and data other than papers and review papers published in English have not been analyzed. Finally, bibliometric analysis itself has certain limitations. It mainly quantitatively analyzes the characteristics of the literature and cannot analyze the details of the literature well, so the conclusions obtained are relatively superficial.
In order to explore the global development trend in the field of epigenetics of AD, we used bibliometrics to conclude that China is the most prolific country, Capital Medical University is the most prolific institution, Bennett is the most prolific and cited author. The current research focus is still to elucidate the epigenetic mechanism of AD in more detail from DNA modification, histone modification, ncRNAs, etc. and to find more specific biomarkers for the diagnosis of AD based on the above mechanisms. In terms of treatment, the comprehensive use of omics analysis technology to explore new therapeutic targets, and the development of drugs targeting HATs, TETs, HDACI, and RNA-based mechanisms for multi-target therapy should be the directions of future research.
YZ: Visualization, Writing – original draft. WA: Visualization, Writing – original draft. JZ: Visualization, Writing – original draft. XH: Funding acquisition, Supervision, Writing – review & editing. LZ: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Chengdu Medical College-Chengdu Eighth People’s Hospital Clinical Science Research Fund Project (YLLNZD2301); the Development and Regeneration Key Laboratory of Sichuan Province (SYS13-006). The National College Student Innovation and Entrepreneurship Training Program Project (202213705036); Sichuan Provincial College Student Innovation and Entrepreneurship Training Program Project (S202313705089).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2024.1332845/full#supplementary-material
Armstrong, M. J., Jin, Y., Vattathil, S. M., Huang, Y., Schroeder, J. P., Bennet, D. A., et al. (2023). Role of TET1-mediated epigenetic modulation in Alzheimer’s disease. Neurobiol. Dis. 185:106257. doi: 10.1016/j.nbd.2023.106257
Barron, A. M., and Pike, C. J. (2012). Sex hormones, aging, and Alzheimer’s disease. Front. Biosci. (Elite Ed.) 4, 976–997. doi: 10.2741/E434
Bellenguez, C., Küçükali, F., Jansen, I. E., Kleineidam, L., Moreno-Grau, S., Amin, N., et al. (2022). New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436. doi: 10.1038/s41588-022-01024-z
Bellizzi, D., D’Aquila, P., Scafone, T., Giordano, M., Riso, V., Riccio, A., et al. (2013). The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 20, 537–547. doi: 10.1093/dnares/dst029
Berson, A., Nativio, R., Berger, S. L., and Bonini, N. M. (2018). Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 41, 587–598. doi: 10.1016/j.tins.2018.05.005
Blanch, M., Mosquera, J. L., Ansoleaga, B., Ferrer, I., and Barrachina, M. (2016). Altered mitochondrial DNA methylation pattern in Alzheimer disease-related pathology and in Parkinson disease. Am. J. Pathol. 186, 385–397. doi: 10.1016/j.ajpath.2015.10.004
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell Biol. 220:e202009045. doi: 10.1083/jcb.202009045
Cáceres, C., Heusser, B., Garnham, A., and Moczko, E. (2023). The major hypotheses of Alzheimer’s disease: related nanotechnology-based approaches for its diagnosis and treatment. Cells 12:2669. doi: 10.3390/cells12232669
Cao, D., Jiang, D., Zhou, D., Yu, H., and Li, J. (2020). A comparative study on 5hmC targeting regulation of neurons in AD mice by several natural compounds. Biomed. Res. Int. 2020, 1–9. doi: 10.1155/2020/5016706
Cao, Q., Wang, W., Williams, J. B., Yang, F., Wang, Z. J., and Yan, Z. (2020). Targeting histone K4 trimethylation for treatment of cognitive and synaptic deficits in mouse models of Alzheimer’s disease. Sci. Adv. 6:50. doi: 10.1126/sciadv.abc8096
Castillo-Ordoñez, W. O., Cajas-Salazar, N., and Velasco-Reyes, M. A. (2024). Genetic and epigenetic targets of natural dietary compounds as anti-Alzheimer’s agents. Neural Regen. Res. 19, 846–854. doi: 10.4103/1673-5374.382232
Chacon, P. J., Garcia-Mejias, R., and Rodriguez-Tebar, A. (2011). Inhibition of RhoA GTPase and the subsequent activation of PTP1B protects cultured hippocampal neurons against amyloid β toxicity. Mol. Neurodegener. 6:14. doi: 10.1186/1750-1326-6-14
Chang, F. E. I., Zhang, L.-H., Xu, W.-P., Jing, P., and Zhan, P.-Y. (2014). microRNA-9 attenuates amyloidβ-induced synaptotoxicity by targeting calcium/calmodulin-dependent protein kinase kinase 2. Mol. Med. Rep. 9, 1917–1922. doi: 10.3892/mmr.2014.2013
Chaturvedi, C. P., Somasundaram, B., Singh, K., Carpenedo, R. L., Stanford, W. L., Dilworth, F. J., et al. (2012). Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc. Natl. Acad. Sci. U. S. A. 109, 18845–18850. doi: 10.1073/pnas.1213951109
Chen, C., Dubin, R., and Kim, M. C. (2014). Emerging trends and new developments in regenerative medicine: a scientometric update (2000 – 2014). Expert. Opin. Biol. Ther. 14, 1295–1317. doi: 10.1517/14712598.2014.920813
Condrat, C. E., Thompson, D. C., Barbu, M. G., Bugnar, O. L., Boboc, A., Cretoiu, D., et al. (2020). miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells 9:276. doi: 10.3390/cells9020276
Coskun, P., Wyrembak, J., Schriner, S. E., Chen, H. W., Marciniack, C., Laferla, F., et al. (2012). A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim. Biophys. Acta 1820, 553–564. doi: 10.1016/j.bbagen.2011.08.008
Cotman, C. W., and Anderson, K. J. (1988). Synaptic plasticity and functional stabilization in the hippocampal formation: possible role in Alzheimer’s disease. Adv. Neurol. 47, 313–335.
Creyghton, M. P., Cheng, A. W., Welstead, G. G., Kooistra, T., Carey, B. W., Steine, E. J., et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 107, 21931–21936. doi: 10.1073/pnas.1016071107
Dhapola, R., Kumari, S., Sharma, P., and HariKrishnaReddy, D. (2023). Insight into the emerging and common experimental in-vivo models of Alzheimer’s disease. Lab. Anim. Res. 39:33. doi: 10.1186/s42826-023-00184-1
Ding, Y., Liu, C., and Zhang, Y. (2023). Aging-related histone modification changes in brain function. Ibrain 9, 205–213. doi: 10.1002/ibra.12106
Ding, Y., Luan, W., Shen, X., Wang, Z., and Cao, Y. (2022). LncRNA BDNF-AS AS ceRNA regulates the miR-9-5p/BACE1 pathway affecting neurotoxicity in Alzheimer’s disease. Arch. Gerontol. Geriatr. 99:104614. doi: 10.1016/j.archger.2021.104614
Dong, X., Liu, C., and Dozmorov, M. (2021). Review of multi-omics data resources and integrative analysis for human brain disorders. Brief. Funct. Genomics 20, 223–234. doi: 10.1093/bfgp/elab024
Engstrom, A. K., Walker, A. C., Moudgal, R. A., Myrick, D. A., Kyle, S. M., Bai, Y., et al. (2020). The inhibition of LSD1 via sequestration contributes to tau-mediated neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 117, 29133–29143. doi: 10.1073/pnas.2013552117
Fei, X., Zhang, Y., Mei, Y., Yue, X., Jiang, W., Ai, L., et al. (2021). Degradation of FA reduces Abeta neurotoxicity and Alzheimer-related phenotypes. Mol. Psychiatry 26, 5578–5591. doi: 10.1038/s41380-020-00929-7
Gallo, G. (2007). Tau is actin up in Alzheimer’s disease. Nat. Cell Biol. 9, 133–134. doi: 10.1038/ncb0207-133
Gao, X., Chen, Q., Yao, H., Tan, J., Liu, Z., Zhou, Y., et al. (2022). Epigenetics in Alzheimer’s disease. Front. Aging Neurosci. 14:911635. doi: 10.3389/fnagi.2022.911635
Geng, H., Chen, H., Wang, H., and Wang, L. (2021). The histone modifications of neuronal plasticity. Neural Plast. 2021, 1–7. doi: 10.1155/2021/6690523
Ghosh, P., and Saadat, A. (2023). Neurodegeneration and epigenetics: a review. Neurologia 38, e62–e68. doi: 10.1016/j.nrl.2021.01.016
Gjoneska, E., Pfenning, A. R., Mathys, H., Quon, G., Kundaje, A., Tsai, L. H., et al. (2015). Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369. doi: 10.1038/nature14252
Gonzales, M. M., Garbarino, V. R., Pollet, E., Palavicini, J. P., Kellogg, D. L. Jr., Kraig, E., et al. (2022). Biological aging processes underlying cognitive decline and neurodegenerative disease. J. Clin. Invest. 132:e158453. doi: 10.1172/JCI158453
Han, J. L. T., Pang, K. K. L., Ang, S. R. X., Sharma, M., and Sajikumar, S. (2021). Inhibition of lysine methyltransferase G9a/GLP reinstates long-term synaptic plasticity and synaptic tagging/capture by facilitating protein synthesis in the hippocampal CA1 area of APP/PS1 mouse model of Alzheimer’s disease. Transl. Neurodegener. 10:23. doi: 10.1186/s40035-021-00247-0
He, C., Li, Z., Yang, M., Yu, W., Luo, R., Zhou, J., et al. (2023). Non-coding RNA in microglia activation and neuroinflammation in Alzheimer’s disease. J. Inflamm. Res. 16, 4165–4211. doi: 10.2147/JIR.S422114
Hernández-Romero, I. A., Guerra-Calderas, L., Salgado-Albarrán, M., Maldonado-Huerta, T., and Soto-Reyes, E. (2019). The regulatory roles of non-coding RNAs in angiogenesis and neovascularization from an epigenetic perspective. Front. Oncol. 9:1091. doi: 10.3389/fonc.2019.01091
Hroudová, J., Singh, N., and Fišar, Z. (2014). Mitochondrial dysfunctions in neurodegenerative diseases: relevance to Alzheimer’s disease. Biomed. Res. Int. 2014:175062, 1–9. doi: 10.1155/2014/175062
Huang, C. H., Chang, M. C., Lai, Y. C., Lin, C. Y., Hsu, C. H., Tseng, B. Y., et al. (2022). Mitochondrial DNA methylation profiling of the human prefrontal cortex and nucleus accumbens: correlations with aging and drug use. Clin. Epigenetics 14:79. doi: 10.1186/s13148-022-01300-z
Huntzinger, E., and Izaurralde, E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110. doi: 10.1038/nrg2936
Jeremic, D., Jiménez-Díaz, L., and Navarro-López, J. D. (2023). Targeting epigenetics: a novel promise for Alzheimer’s disease treatment. Ageing Res. Rev. 90:102003. doi: 10.1016/j.arr.2023.102003
Kandimalla, R., and Reddy, P. H. (2017). Therapeutics of neurotransmitters in Alzheimer’s disease. J. Alzheimer’s Dis. 57, 1049–1069. doi: 10.3233/JAD-161118
Kumar, P., Dezso, Z., MacKenzie, C., Oestreicher, J., Agoulnik, S., Byrne, M., et al. (2013). Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One 8:e69807. doi: 10.1371/journal.pone.0069807
Kumar, R., Fatima, F., Yadav, G., Singh, S., Haldar, S., and Alexiou, A. (2023). et al, Epigenetic modifications by estrogen and androgen in Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 22, 6–17. doi: 10.2174/1871527321666220225110501
Kunkle, B. W., Grenier-Boley, B., Sims, R., Bis, J. C., Damotte, V., Naj, A. C., et al. (2019). Author correction: genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 1423–1424. doi: 10.1038/s41588-019-0495-7
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458. doi: 10.1038/ng.2802
Lauretti, E., Dabrowski, K., and Praticò, D. (2021). The neurobiology of non-coding RNAs and Alzheimer’s disease pathogenesis: pathways, mechanisms and translational opportunities. Ageing Res. Rev. 71:101425. doi: 10.1016/j.arr.2021.101425
Lee, C. J., Evans, J., Kim, K., Chae, H., and Kim, S. (2014). Determining the effect of DNA methylation on gene expression in cancer cells. Methods Mol. Biol. 1101, 161–178. doi: 10.1007/978-1-62703-721-1_9
Levine, M. E., Lu, A. T., Quach, A., Chen, B. H., Assimes, T. L., Bandinelli, S., et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging Us 10, 573–591. doi: 10.18632/aging.101414
Li, K., and Wang, Z. (2023). lncRNA NEAT1: key player in neurodegenerative diseases. Ageing Res. Rev. 86:101878. doi: 10.1016/j.arr.2023.101878
Li, F., Wang, Y., Yang, H., Xu, Y., Zhou, X., Zhang, X., et al. (2019). The effect of BACE1-AS on β-amyloid generation by regulating BACE1 mRNA expression. BMC Mol. Biol. 20:23. doi: 10.1186/s12867-019-0140-0
Li, T., Wei, Y., Qu, M., Mou, L., Miao, J., Xi, M., et al. (2021). Formaldehyde and De/methylation in age-related cognitive impairment. Genes 12:913. doi: 10.3390/genes12060913
Li, H., Zheng, L., Jiang, A., Mo, Y., and Gong, Q. (2018). Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport 29, 1061–1067. doi: 10.1097/WNR.0000000000001057
Lin, L., Lesnick, T. G., Maraganore, D. M., and Isacson, O. (2009). Axon guidance and synaptic maintenance: preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 32, 142–149. doi: 10.1016/j.tins.2008.11.006
Lin, S., Pan, Z., Ma, Y., Gao, J., Shan, J., Chu, C., et al. (2021). Research Progress of epigenetic modification in the regulation of Alzheimer’s disease (in chinese). Prog. Biochem. Biophys. 49, 623–641. doi: 10.16476/j.pibb.2021.0252
Lu, T., Aron, L., Zullo, J., Pan, Y., Kim, H., Chen, Y., et al. (2014). REST and stress resistance in ageing and Alzheimer’s disease. Nature 507, 448–454. doi: 10.1038/nature13163
Mastroeni, D., Grover, A., Delvaux, E., Whiteside, C., Coleman, P. D., and Rogers, J. (2011). Epigenetic mechanisms in Alzheimer’s disease. Neurobiol. Aging 32, 1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017
Mills, Z. B., Faull, R. L. M., and Kwakowsky, A. (2023). Is hormone replacement therapy a risk factor or a therapeutic option for Alzheimer’s disease? Int. J. Mol. Sci. 24:3205. doi: 10.3390/ijms24043205
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi: 10.1038/npp.2012.112
Nativio, R., Lan, Y., Donahue, G., Sidoli, S., Berson, A., Srinivasan, A. R., et al. (2020). Author correction: an integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 52:1266. doi: 10.1038/s41588-020-00733-7
Nikolac Perkovic, M., Videtic Paska, A., Konjevod, M., Kouter, K., Svob Strac, D., Nedic Erjavec, G., et al. (2021). Epigenetics of Alzheimer’s disease. Biomol. Ther. 11:2. doi: 10.3390/biom11020195
Oblak, A. L., Forner, S., Territo, P. R., Sasner, M., Carter, G. W., Howell, G. R., et al. (2020). Model organism development and evaluation for late-onset Alzheimer’s disease: MODEL-AD. Alzheimer’s Dementia 6:e12110. doi: 10.1002/trc2.12110
Paniri, A., Hosseini, M. M., and Akhavan-Niaki, H. (2023). Alzheimer’s disease-related epigenetic changes: novel therapeutic targets. Mol. Neurobiol. doi: 10.1007/s12035-023-03626-y
Peixoto, L., and Abel, T. (2013). The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38, 62–76. doi: 10.1038/npp.2012.86
Pierouli, K., Papakonstantinou, E., Papageorgiou, L., Diakou, I., Mitsis, T., Dragoumani, K., et al. (2023). Role of non-coding RNAs as biomarkers and the application of omics technologies in Alzheimer’s disease (review). Int. J. Mol. Med. 51:5. doi: 10.3892/ijmm.2022.5208
Podlesniy, P., Llorens, F., Puigròs, M., Serra, N., Sepúlveda-Falla, D., Schmidt, C., et al. (2020). Cerebrospinal fluid mitochondrial DNA in rapid and slow progressive forms of Alzheimer’s disease. Int. J. Mol. Sci. 21:6298. doi: 10.3390/ijms21176298
Prokhortchouk, E., and Defossez, P. A. (2008). The cell biology of DNA methylation in mammals. Biochim. Biophys. Acta 1783, 2167–2173. doi: 10.1016/j.bbamcr.2008.07.015
Ritchie, C., Smailagic, N., Noel-Storr, A. H., Ukoumunne, O., Ladds, E. C., and Martin, S. (2017). CSF tau and the CSF tau/ABeta ratio for the diagnosis of Alzheimer’s disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst. Rev. 3:CD010803. doi: 10.1002/14651858.CD010803.pub2
Santana, D. A., Smith, M. A. C., and Chen, E. S. (2023). Histone modifications in Alzheimer’s disease. Genes 14:347. doi: 10.3390/genes14020347
Sharma, M., Dierkes, T., and Sajikumar, S. (2017). Epigenetic regulation by G9a/GLP complex ameliorates amyloid-beta 1-42 induced deficits in long-term plasticity and synaptic tagging/capture in hippocampal pyramidal neurons. Aging Cell 16, 1062–1072. doi: 10.1111/acel.12634
Sharma, V. K., Mehta, V., and Singh, T. G. (2020). Alzheimer’s disorder: epigenetic connection and associated risk factors. Curr. Neuropharmacol. 18, 740–753. doi: 10.2174/1570159X18666200128125641
Shireby, G. L., Davies, J. P., Francis, P. T., Burrage, J., Walker, E. M., Neilson, G. W. A., et al. (2020). Recalibrating the epigenetic clock: implications for assessing biological age in the human cortex. Brain 143, 3763–3775. doi: 10.1093/brain/awaa334
Swerdlow, R. H., Burns, J. M., and Khan, S. M. (2014). The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim. Biophys. Acta 1842, 1219–1231. doi: 10.1016/j.bbadis.2013.09.010
Synnestvedt, M. B., Chen, C., and Holmes, J. H. (2005). CiteSpace II: visualization and knowledge discovery in bibliographic databases. AMIA Annu. Symp. Proc. 2005, 724–728.
Tan, Y., Nie, D. R., Cao, Y., Ke, C., Pan, J., Shi, W. Y., et al. (2023). Trends in the application of “omics” to Alzheimer’s disease: a bibliometric and visualized study. Neurol. Sci. doi: 10.1007/s10072-023-07079-y
Van Eck, N. J., and Waltman, L. (2010). Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 84, 523–538. doi: 10.1007/s11192-009-0146-3
Wallace, D. C. (2011). Bioenergetic origins of complexity and disease. Cold Spring Harb. Symp. Quant. Biol. 76, 1–16. doi: 10.1101/sqb.2011.76.010462
Wang, E., Wang, M., Guo, L., Fullard, J. F., Micallef, C., Bendl, J., et al. (2023). Genome-wide methylomic regulation of multiscale gene networks in Alzheimer’s disease. Alzheimers Dement. 19, 3472–3495. doi: 10.1002/alz.12969
Wang, T., Zhang, J., and Xu, Y. (2020). Epigenetic basis of lead-induced neurological disorders. Int. J. Environ. Res. Public Health 17:4878. doi: 10.3390/ijerph17134878
World Health Organization (2023). Dementia. Available at: https://www.who.int/news-room/fact-sheets/detail/dementia.
Wu, Y. Y., and Kuo, H. C. (2020). Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 27:49. doi: 10.1186/s12929-020-00636-z
Wu, F., and Yao, P. J. (2009). Clathrin-mediated endocytosis and Alzheimer’s disease: an update. Ageing Res. Rev. 8, 147–149. doi: 10.1016/j.arr.2009.03.002
Xiao, X., Liu, X., and Jiao, B. (2020). Epigenetics: recent advances and its role in the treatment of Alzheimer’s disease. Front. Neurol. 11:538301. doi: 10.3389/fneur.2020.538301
Xie, J., Xie, L., Wei, H., Li, X. J., and Lin, L. (2023). Dynamic regulation of DNA methylation and brain functions. Biology 12:152. doi: 10.3390/biology12020152
Xiong, X., James, B. T., Boix, C. A., Park, Y. P., Galani, K., Victor, M. B., et al. (2023). Epigenomic dissection of Alzheimer’s disease pinpoints causal variants and reveals epigenome erosion. Cells 186, 4422–4437.e21. doi: 10.1016/j.cell.2023.08.040
Yang, S. S., Zhang, R., Wang, G., and Zhang, Y. F. (2017). The development prospection of HDAC inhibitors as a potential therapeutic direction in Alzheimer’s disease. Transl. Neurodegener. 6:19. doi: 10.1186/s40035-017-0089-1
Yi, S. J., and Kim, K. (2020). New insights into the role of histone changes in aging. Int. J. Mol. Sci. 21:8241. doi: 10.3390/ijms21218241
Yu, L., Chibnik, L. B., Yang, J., McCabe, C., Xu, J., Schneider, J. A., et al. (2016). Methylation profiles in peripheral blood CD4+ lymphocytes versus brain: the relation to Alzheimer’s disease pathology. Alzheimers Dement. 12, 942–951. doi: 10.1016/j.jalz.2016.02.009
Zhang, L., Liu, Y., Lu, Y., and Wang, G. (2022). Targeting epigenetics as a promising therapeutic strategy for treatment of neurodegenerative diseases. Biochem. Pharmacol. 206:115295. doi: 10.1016/j.bcp.2022.115295
Zhang, X., Zhang, Y., Wang, C., and Wang, X. (2023). TET (ten-eleven translocation) family proteins: structure, biological functions and applications. Signal Transduct. Target. Ther. 8:297. doi: 10.1038/s41392-023-01537-x
Zhang, W., Zhao, H., Wu, Q., Xu, W., and Xia, M. (2018). Knockdown of BACE1-AS by siRNA improves memory and learning behaviors in Alzheimer’s disease animal model. Exp. Ther. Med. 16, 2080–2086. doi: 10.3892/etm.2018.6359
Zhou, Y., Ge, Y., Liu, Q., Li, Y. X., Chao, X., Guan, J. J., et al. (2021). LncRNA BACE1-AS promotes autophagy-mediated neuronal damage through the miR-214-3p/ATG5 Signalling Axis in Alzheimer’s disease. Neuroscience 455, 52–64. doi: 10.1016/j.neuroscience.2020.10.028
Keywords: Alzheimer’s disease, epigenetics, bibliometric analysis, visualization, neurology
Citation: Zhao Y, Ai W, Zheng J, Hu X and Zhang L (2024) A bibliometric and visual analysis of epigenetic research publications for Alzheimer’s disease (2013–2023). Front. Aging Neurosci. 16:1332845. doi: 10.3389/fnagi.2024.1332845
Received: 07 November 2023; Accepted: 02 January 2024;
Published: 16 January 2024.
Edited by:
Mayur Doke, University of Miami Health System, United StatesReviewed by:
Alejandro Antón-Fernández, Spanish National Research Council (CSIC), SpainCopyright © 2024 Zhao, Ai, Zheng, Hu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: LuShun Zhang, emhhbmdsczIwMTJAY21jLmVkdS5jbg==; XianLiang Hu, NzQ5MDUyMDQ2QHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.