
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Aging Neurosci. , 20 October 2023
Sec. Alzheimer's Disease and Related Dementias
Volume 15 - 2023 | https://doi.org/10.3389/fnagi.2023.1287322
 Natalia Matveeva1,2†
Natalia Matveeva1,2† Ivan Kiselev1,2*†
Ivan Kiselev1,2*† Natalia Baulina1,2
Natalia Baulina1,2 Ekaterina Semina1
Ekaterina Semina1 Viktor Kakotkin1
Viktor Kakotkin1 Mikhail Agapov1
Mikhail Agapov1 Olga Kulakova1,2
Olga Kulakova1,2 Olga Favorova1,2
Olga Favorova1,2The severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) and the сoronavirus disease 2019 (COVID-19) have become a global health threat. At the height of the pandemic, major efforts were focused on reducing COVID-19-associated morbidity and mortality. Now is the time to study the long-term effects of the pandemic, particularly cognitive impairment associated with long COVID. In recent years much attention has been paid to the possible relationship between COVID-19 and Alzheimer’s disease, which is considered a main cause of age-related cognitive impairment. Genetic predisposition was shown for both COVID-19 and Alzheimer’s disease. However, the analysis of the similarity of the genetic architecture of these diseases is usually limited to indicating a positive genetic correlation between them. In this review, we have described intrinsic linkages between COVID-19 and Alzheimer’s disease, pointed out shared susceptibility genes that were previously identified in genome-wide association studies of both COVID-19 and Alzheimer’s disease, and highlighted a panel of SNPs that includes candidate genetic risk markers of the long COVID-associated cognitive impairment.
In recent years the SARS-CoV-2 and the COVID-19 have become a global health threat. The most common COVID-19 symptoms are nonspecific and occur in many other infectious diseases. The course of COVID-19 can vary from mild to severe, up to death (Umakanthan et al., 2020); as of July 19, 2023, there are 6,951,677 COVID-19 deaths worldwide according to WHO data1. The severity of COVID-19 is affected by age and the presence of comorbidities like oncology or diseases of the cardiovascular system, lungs, kidneys, etc. (Wang et al., 2020).
In a significant proportion of patients (up to 20%), COVID-19 develops into a long-term pathological condition characterized by symptoms lasting more than 12 weeks and initially called the post-COVID-19 syndrome. Now both the post-COVID-19 syndrome and ongoing symptomatic COVID-19 lasting 4 to 12 weeks are combined into another term, long COVID, which is increasingly being used in the literature (Greenhalgh et al., 2022). Clinical manifestations of the long COVID are varied and often include fatigue, breathlessness, impaired blood clotting, persistent immunosuppression, headache, chest pain, muscle and joint pain, impaired sense of smell and taste (Yong, 2021; Oronsky et al., 2023), as well as various neuropsychiatric symptoms including cognitive impairment (Yong, 2021; Efstathiou et al., 2022). Surprisingly, the development of the long COVID does not depend on the severity of COVID-19 itself; it has been described even in patients with mild or asymptomatic disease (Yong, 2021). The long COVID is multifaceted, it usually affects multiple systems or organs and results in dramatic decline in quality of life and increased mortality. At the height of the pandemic, major efforts were focused on reducing COVID-19-associated morbidity and mortality. Now is the time to study the long-term effects of the pandemic, particularly cognitive impairment in the long COVID.
The relationship between COVID-19 and cognitive impairment encompasses two main aspects: the first is the peculiarities of the course of COVID-19 in patients with psychiatric and neurodegenerative conditions and the second is a long COVID-associated cognitive impairment, which depends on the age of the patient (Herrera and González-Nosti, 2022). It has been convincingly shown that the infection affects the age of the first onset of various neurodegenerative diseases and their rate of progression (Rahmati et al., 2023). In the long term, the aspect of the long COVID-associated cognitive impairment is of particular importance, and we will focus on it in this review.
The main manifestations of cognitive dysfunction after COVID-19 are fatigue, “brain fog,” memory problems, attention deficit, sleep disturbances, anxiety, and depression (Premraj et al., 2022). Even 4 months after hospitalization, 30–40% of COVID-19 patients experience memory, attention, and sleep problems (Garrigues et al., 2020). Meta-analysis data show that patients who have recovered from COVID-19 experience a decline in overall cognitive abilities, when compared to healthy individuals, for a minimum of 7 months following the COVID-19 diagnosis (Crivelli et al., 2022). Even young adults who experienced a mild course of COVID-19, are at risk of developing cognitive symptoms (Gordon et al., 2022). Moreover, the mean age of patients who developed cognitive impairment after COVID-19 is significantly lower compared to those who did not (46.65 ± 9.08 vs. 50.05 ± 9.05, p < 0.001) (Matias-Guiu et al., 2023). The aforementioned facts emphasize high social significance of the long COVID and associated cognitive impairment.
Alzheimer’s disease (AD) is a severe neurodegenerative disorder of the central nervous system (CNS) characterized by irreversible and progressive loss of cognitive function due to metabolic dysfunction, accumulation of misfolded proteins – beta-amyloid and tau protein – in brain tissue, and progressive neuronal death (Kunkle et al., 2019). AD is considered the primary cause of cognitive impairment in older individuals, responsible for over two-thirds of all dementia cases in people over 65 years of age (Kumar et al., 2023).
COVID-19 may provoke the development of AD. Within 360 days following a COVID-19 diagnosis, individuals aged 65 and above exhibited a significantly increased risk of the first diagnosis of AD (Wang et al., 2022a). In several Mendelian randomized studies, hospitalization with COVID-19 has been shown to be significantly associated with an increased risk of developing AD (Li et al., 2022; Baranova et al., 2023). The topic of interactions and intrinsic linkages between COVID-19 and AD is the subject of reviews summarizing data up to 2023 (Axenhus and Winblad, 2023; Li et al., 2023).
Recently, the similarity of the long COVID and AD has attracted increasing attention. It was noted that the consequences of COVID-19 may resemble the decline of cognitive functions commonly seen in aging (Mavrikaki et al., 2022). Moreover, after COVID-19, pathologic changes in the CNS similar to certain changes in AD are detected. It has been shown that among hospitalized COVID-19 patients, levels of biomarkers associated with neurodegeneration (such as beta-amyloid protein, tau protein, glial fibrillary acidic protein, neurofilament light chain, and ubiquitin carboxy-terminal hydrolase L1), increase to levels observed in AD-induced dementia and correlate with older age and severity of COVID illness (Frontera et al., 2022).
Рossible mechanisms underlying neurological consequences of COVID-19 have been summarized in a number of reviews (Jha et al., 2021; Mannekote Thippaiah et al., 2023). It has been established that SARS-CoV-2 can penetrate CNS (Pezzini and Padovani, 2020). Upon neuroinvasion, the SARS-CoV-2 spike protein binds to angiotensin-converting enzyme 2 (ACE2) on capillary endothelium, which acts as a receptor for viral entry into the cell A systemic exacerbated release of pro-inflammatory cytokines (so-called “cytokine storm”), characteristic of SARS-CoV-2, leads to increase in blood–brain barrier (BBB) permeability and facilitates virus penetration into the CNS. Another way for the virus to enter brain tissues is through the cells of the olfactory epithelium and olfactory tract (Baig et al., 2020). In the CNS, SARS-CoV-2 can interact with neurons and glial cells, which highly express ACE2 (Jha et al., 2021) and trigger a neuroinflammatory cascade, including activation of microglia, local production of proinflammatory cytokines, activation of inflammatory signaling, and subsequent astroglial reactivity, all of which may lead to neurodegeneration (Steardo et al., 2020; Saucier et al., 2023).
ACE2 is a key player in maintaining the balance between the pressor axis and the tissue-protective axis of renin-angiotensin system (RAS). ACE2 converts Angiotensin II (Ang II) to an active heptopeptide Ang-(1–7) which induces vasorelaxation, suppresses inflammation, oxidative stress, apoptosis, fibrosis and coagulation [summarized in (Heyman et al., 2021a)]. Binding of the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein to ACE2 results in the internalization of the complex and depletion of ACE2 on the cell membrane. As a result, Ang-(1–7) production decreases and the activity of the pressor axis, potentiated by Ang II, rises. This eventually leads to inflammation, oxidative stress, tissue damage and coagulopathy typical for COVID-19 (Heyman et al., 2021b). In the CNS, the weakening of the protective RAS axis upon virus binding with ACE2 makes the neural tissue more vulnerable to damage (Baig et al., 2020). ACE2 depletion can cause an elevation in blood pressure and an increased risk of cerebral hemorrhage (Wu et al., 2020). Finally, ACE2 is involved in the beta-amyloid metabolism providing an additional link between COVID-19 and AD (Evans et al., 2020). Based on the body of experimental data reviewed, Reiss et al. distinguished three main mechanisms of the long-term neuropathological effects of SARS-CoV-2: (1) systemic inflammation leading to weakening of the blood–brain barrier (BBB), cytokine entry into the CNS, microglial activation, and production of neuroinflammatory mediators; (2) direct viral invasion of microvascular endothelium of the BBB and impairment of the blood flow in the brain; (3) entry of viral particles into the brain via the nasal epithelium and olfactory bulb, what causes neurotoxicity and neuronal loss (Reiss et al., 2023). Each of these mechanisms can impair neuronal function and thus cognition.
For both COVID-19 and different neurodegenerative diseases, numerous genome-wide association studies (GWASs) have been conducted, providing evidence of a genetic predisposition to these conditions. However, in only a few works genome-wide data were analyzed in order to search for shared genes involved in the pathogenesis of both COVID-19 and AD. Direct comparison of two GWASs allowed to identify locus chr19:44,744,108–46,102,697 containing APOE gene as a possible risk region for both COVID-19 and AD (Qiu et al., 2022). In the work (Baranova et al., 2023), a broader search for possible shared genes was carried out using GWAS catalog data. However, the database was accessed not later than 2022.05.20, when some important COVID-19 GWASs and meta-analyses had not yet been published. At that time, only for two genes, LUZP2 and RBFOX1, both COVID-19- and AD-associated SNPs were found at the genome-wide significance level p ≤ 5 × 10−8. For SNPs in 17 more genes (ANO3, CCDC171, CSMD1, DAB1, ECHDC3, EDAR, FAT1, GLIS3, GRIN2B, NAALADL2, NKAIN2, NTM, RBMS3, SHANK2, ST18, TCF7L2, and UNC5D) the association with COVID-19 and/or AD was observed on a significance level reaching only p ≤ 1 × 10−5. The APOE gene was not highlighted at all in this study.
In this study, we conducted a search for shared susceptibility genes for COVID-19 and AD in the GWAS Catalog database 13 months later than in Baranova et al. (2023), when the amount of information available in the database has significantly increased, especially for COVID-19. At the time we accessed the GWAS catalog, it contained 249 studies and 4,064 associations of SNPs with COVID-19 (p ≤ 1 × 10−5); for AD, 2588 associations at the same level of significance were found in 150 studies. Out of them, the number of SNPs that meet the genome-wide significance level (p ≤ 5 × 10−8) turned out to be 1,100 for COVID-19 and 1,550 for AD. By analyzing the entire dataset, we identified 39 protein-coding genes and seven non-protein-coding genes, which showed an association with both COVID-19 and AD; the list of these genes is presented in Table 1.
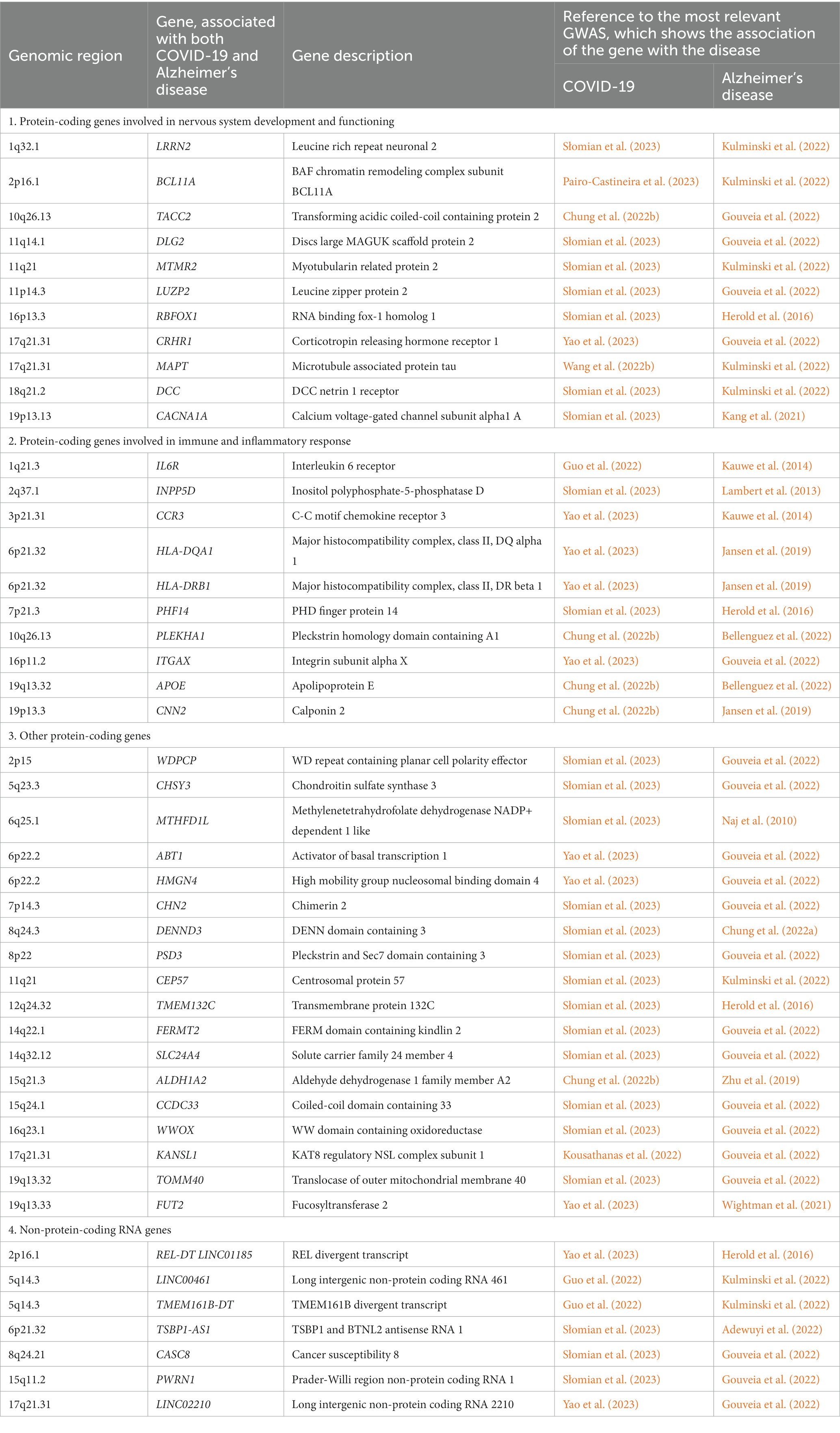
Table 1. Shared genes that carry one or the other SNP associated with both COVID-19 and Alzheimer’s disease at a genome-wide significance level (p ≤ 5 × 10−8).
Since neuronal dysfunction and inflammation have a critical role in the development of cognitive impairment associated with both COVID-19 (Gholami et al., 2021) and AD (Forloni and Balducci, 2018), we, using data from GeneCards database (Stelzer et al., 2016), identified two large subgroups of protein-coding genes from Table 1: the first subgroup includes genes involved in nervous system development and functioning and the second subgroup includes genes involved in immune and inflammatory response. Functions of these genes are discussed in more detail below.
The cognitive impairment in AD is known to be based on the dysfunction of lipid metabolism and metabolism of amyloid beta and tau protein, which eventually lead to accumulation of protein aggregates (Kunkle et al., 2019). Among the shared genes, belonging to the first subgroup, we found MAPT encoding precursor of the tau protein (Tabeshmehr and Eftekharpour, 2023), CRHR1 mediating tau hyperphosphorylation (Lemche, 2018) and CACNA1A involved in neuronal cell death and toxicity of amyloid beta (Demuro et al., 2010). In addition, four genes belonging to the first subgroup: DLG2, MTMR2, RBFOX1, and DCC, are involved in synapse formation and signal transduction (Lee et al., 2010; Alkallas et al., 2017; Wong et al., 2019; Prokopenko et al., 2022), what may indicate deregulation of these processes in the development of COVID-19 and AD-associated cognitive impairment.
Functions of the several highlighted genes (IL6R, INPP5D, CCR3, HLA-DQA1, HLA-DRB1, PHF14, PLEKHA1, ITGAX, APOE, CNN2), belonging to the second subgroup of shared protein-coding genes, relate to the development of the immune response and inflammation, what is in a good agreement with the current understanding of the critical role of inflammatory responses in the development of various neurodegenerative diseases, particularly AD (Hammond et al., 2019). Many genes involved in the regulation of the immune response and inflammation have been found among the risk genes for such diseases (Nott and Holtman, 2023).
Microglial cells are key regulators of inflammatory responses in the central nervous system (Kwon and Koh, 2020), and studies at the genomic and transcriptomic levels showed that neurodegeneration may be strongly associated with altering microglia function (Raj et al., 2014; Lopes et al., 2022). Among the genes of the second subgroup there are several genes involved in the functioning of microglia and the maintenance of its homeostasis; they are associated with inflammation, cell adhesion and migration (APOE, IL6R, CCR3, ITGAX, CNN2), as well as with regulation of microglial gene expression (INPP5D). It has been established that the development of COVID-19 is also associated with various inflammatory reactions; in severe cases an abrupt and uncontrolled release of proinflammatory cytokines (“cytokine storm”) develops leading to damage to many cells and tissues (Hu et al., 2021). The association of the APOE gene with the development of both diseases has been most well studied (see section 5).
In addition to the genes associated with innate immune response, the second subgroup includes MHC class II genes HLA-DQA1 and HLA-DRB1 and some genes that regulate the formation of germinal centers and the production of autoantibodies (PHF14 and PLEKHA1) (Sharma et al., 2018; Zhang et al., 2020), which suggests a possible involvement of adaptive immunity in cognitive impairment associated with both COVID-19 and AD.
According to the GeneCards database (Stelzer et al., 2016), functions of other protein-coding genes combined in the third subgroup relate to the chromosome organization and regulation of transcription (ABT1, HMGN4, CEP57, KANSL1), cell signaling (CHSY3, ALDH1A2, TMEM132C, FUT2, SLC24A4), metabolic processes (CHSY3, ALDH1A2, TMEM132C, FUT2), cytoskeleton organization and cell motility (WDPCP, FERMT2), vesicle-mediated transport (DENND3, PSD3) and mitochondria functioning (TOMM40, MTHFD1L).
Finally, non-protein-coding genes that make up the fourth subgroup remain largely unexplored, with only exceptions being the miRNA sponge LINC00461, involved in the regulation of MAPK/ERK and PI3K/AKT signaling pathways (Zhang et al., 2022), and imprinted PWRN1, which is located in the Prader-Willi syndrome region and associated with various cancers (Jiang et al., 2020).
The genetic architecture of COVID-19 and AD seems to exhibit certain similarities not only at the level of common genes involved in the same pathological processes, but also at the level of shared or linked particular polymorphic variants, what is confirmed by a positive genetic correlation between the diseases (Li et al., 2022; Qiu et al., 2022; Baranova et al., 2023). To date the only genetic variant known to be associated with both COVID-19 and AD is ε4 allele of the APOE gene (Xiong et al., 2021).
The APOE gene has three main alleles, ε2, ε3, and ε4, defined by a combination of variants of two SNPs: rs429358 and rs7412; these alleles encode three isoforms of the gene protein product, apolipoprotein E (ApoE), which differ from each other by amino acids at positions 112 and 158: ApoE2 (cys112, cys158), ApoE3 (cys112, arg158) and ApoE4 (arg112, arg158) (Weisgraber et al., 1981). ApoE4 stands apart from other ApoE isoforms, because its primary sequence causes formation of a more compact spatial structure and leads to the protein instability (Dong et al., 1994). ApoE is involved in transport of cholesterol and some other lipids. In the CNS it is mainly produced by astrocytes and microglial cells and involved in neuronal lipid metabolism and regulation of beta amyloid production, modifying risk of AD (Wang et al., 2021b). ApoE isoforms may affect microglial activation and neuroinflammatory state of the brain (Kloske et al., 2021) and thus modulate pathogenesis of both COVID-19 and AD. ApoE2 and ApoE3 isoforms were shown to inhibit beta amyloid aggregation and neurotoxicity, whereas ApoE4 lacks neuroprotective activity and facilitates AD (Drouet et al., 2001). ApoE4 seems to be involved in the neurological effects of COVID-19 by increasing rates of SARS-CoV-2 entry into neuronal cells and stimulating their apoptosis under infection (Wang et al., 2021a). The APOE ε4 allele is one of the primary genetic risk factors of AD (Raulin et al., 2022). It was also shown to significantly increase the chance to test positive for COVID-19 (Kuo et al., 2020a). The risk of symptomatic COVID-19, severe COVID-19 and disease-associated mortality were also elevated in APOE ε4 carriers (Kuo et al., 2020a,b; Del Ser et al., 2021; Hubacek et al., 2021).
In order to search for a panel of SNPs that can be used as genetic markers of the risk of the long COVID-associated cognitive impairment, we compared the lists of SNPs associated with COVID-19 or AD (p ≤ 5 × 10−8) and located in 46 shared genes listed in Table 1. Expectedly, APOE SNP rs429358, which, in combination with rs7412, is responsible for the formation of three main APOE alleles, was associated with both COVID-19 and AD (highest value of ps extracted from GWAS catalog were equal to 1 × 10−44 and 2 × 10−303, respectively); no significant shared SNPs were found for other genes.
Next, for each gene presented in the Тable 1 we have chosen the pairs of SNPs, in which one SNP is associated with COVID-19 and the other with AD, and extracted from LDlink database (Machiela and Chanock, 2015) data on linkage disequilibrium between these SNPs for the general population (complete dataset, ALL) and for the most represented in GWASs European populations (EUR) of the 1,000 Genomes Project. Regions of genes TSBP1-AS1, DLG2, PWRN1, CCDC33, and CACNA1A were excluded from this analysis because some of their SNPs were either absent in the 1,000 Genomes reference panel, or are not biallelic. If several SNPs of the same gene were associated with COVID-19 and/or AD, only the pair characterized by the highest R2 value was selected. As a result we pinpointed 19 pairs of SNPs associated with COVID-19 and AD and characterized by significant linkage disequilibrium (Table 2). Among them, 3 pairs of SNPs (in BCL11A, CHN2, and CNN2 genes) were in the linkage disequilibrium in European populations only. Three gene pairs (LINC00461 and TMEM161B-DT, HLA-DQA1, and HLA-DRB1, ABT1 and HMGN4) located in the same genomic loci were merged together in the Table 2 because they were assigned to the same SNPs. We believe that in addition to the APOE SNP rs429358, SNPs included in the Table 2 may be good candidate genetic risk markers of the long COVID-associated cognitive impairment. Unfortunately, the effects of alleles and genotypes of certain identified SNPs on the expression of genes, in which they are located, still remain unknown and require further research.
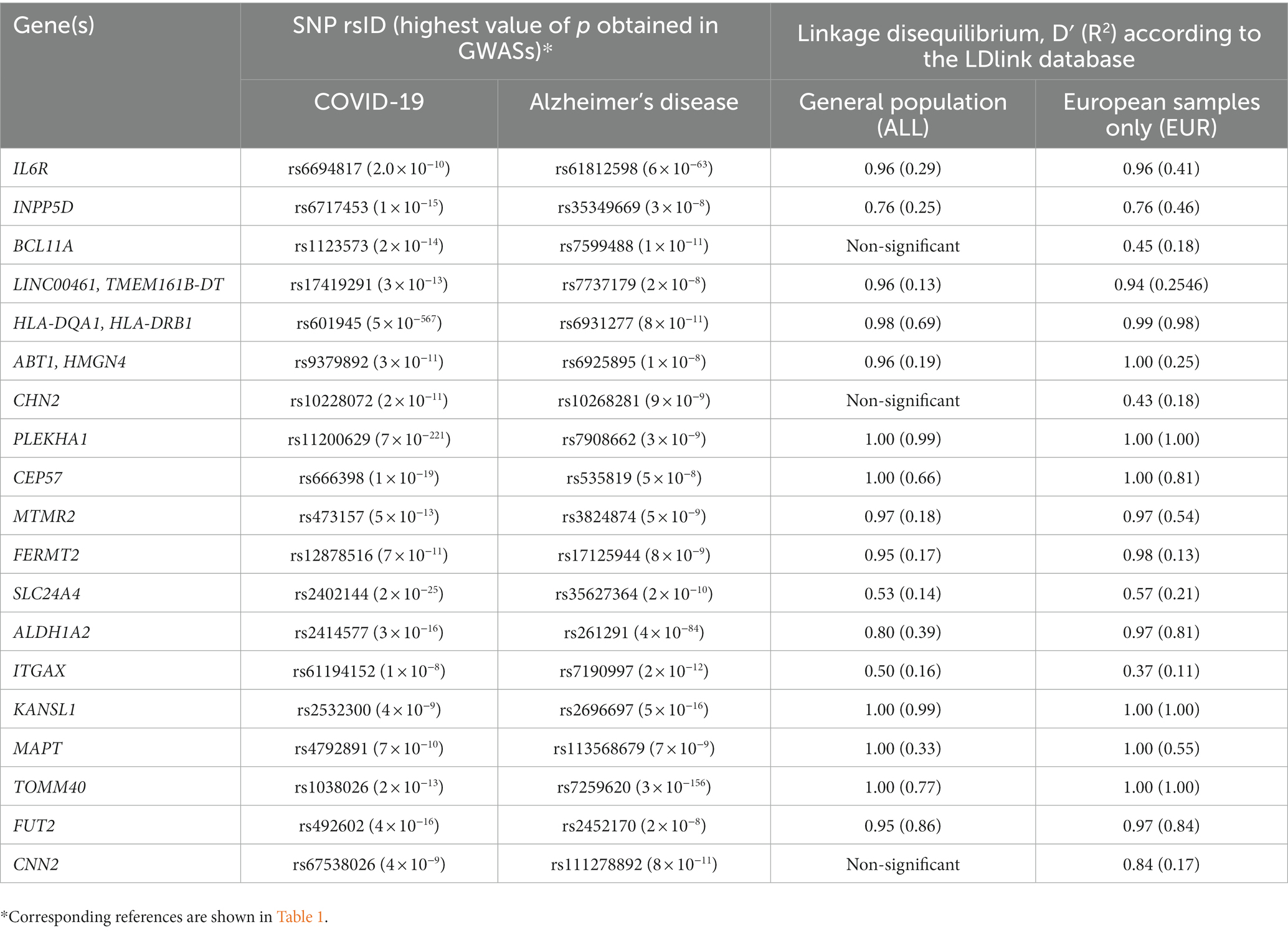
Table 2. SNPs in the shared genes, associated with COVID-19 and Alzheimer’s disease in GWASs, which are in linkage disequilibrium.
The assessment of the risk of developing cognitive impairment as a manifestation of the long COVID is of undoubted practical importance. Identification of high-risk groups among COVID-19 survivors may help to prevent cognitive impairment through a combination of lifestyle changes and medication interventions.
However, it is hindered due to a number of different factors. Only a small number of people undergo regular cognitive tests, which significantly limits the ability to assess cognitive impairment before and after COVID-19 (Nami et al., 2022). At the same time, the lack of unified protocols for the assessment of cognitive functions complicates the interpretation of the obtained data. In addition, hypoxia, endothelial dysfunction, intoxication, thrombosis, etc., which often accompany COVID-19, in some cases may act as nonspecific causes of cognitive impairment. And finally, the prolonged isolation, fear, and financial losses in COVID-19 can by themselves lead to depression, anxiety disorders, sleep disturbances, difficulties in concentration, and other symptoms of post-traumatic stress (Hossain et al., 2020).
Against the background of these modifiable factors, analysis of the genetic susceptibility may be a reliable method for predicting the risk of developing cognitive impairment due to COVID-19. Selection of the most informative risk markers among SNPs, which are localized in shared genes associated with COVID-19 and AD, assessment of their sensitivity and specificity for predicting the risk of developing cognitive impairment among COVID-19 survivors, and creation of an effective SNP panel to predict the individual risk is an urgent task of the near future.
NM: Conceptualization, Writing – original draft. IK: Conceptualization, Writing – original draft. NB: Writing – review & editing. ES: Writing – review & editing. VK: Writing – review & editing. MA: Writing – review & editing. OK: Supervision, Writing – review & editing. OF: Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported from the Russian Federal Academic Leadership Program Priority 2030 at the Immanuel Kant Baltic Federal University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adewuyi, E. O., O’Brien, E. K., Nyholt, D. R., Porter, T., and Laws, S. M. (2022). A large-scale genome-wide cross-trait analysis reveals shared genetic architecture between Alzheimer’s disease and gastrointestinal tract disorders. Commun Biol 5:691. doi: 10.1038/s42003-022-03607-2
Alkallas, R., Fish, L., Goodarzi, H., and Najafabadi, H. S. (2017). Inference of RNA decay rate from transcriptional profiling highlights the regulatory programs of Alzheimer’s disease. Nat. Commun. 8:909. doi: 10.1038/s41467-017-00867-z
Axenhus, M., and Winblad, B. (2023). Exploring the impact of coronavirus disease 2019 on dementia: a review. touchREVIEWS in neurology. Available at: https://touchneurology.com/alzheimers-disease-dementia/journal-articles/exploring-the-iimpact-of-coronavirus-disease-2019-on-dementia-a-review/ (Accessed August 16, 2023).
Baig, A. M., Khaleeq, A., Ali, U., and Syeda, H. (2020). Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 11, 995–998. doi: 10.1021/acschemneuro.0c00122
Baranova, A., Cao, H., and Zhang, F. (2023). Causal effect of COVID-19 on Alzheimer’s disease: a Mendelian randomization study. J. Med. Virol. 95:e28107. doi: 10.1002/jmv.28107
Bellenguez, C., Küçükali, F., Jansen, I. E., Kleineidam, L., Moreno-Grau, S., Amin, N., et al. (2022). New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436. doi: 10.1038/s41588-022-01024-z
Chung, J., Das, A., Sun, X., Sobreira, D. R., Leung, Y. Y., Igartua, C., et al. (2022a). Genome-wide association and multi-omics studies identify MGMT as a novel risk gene for Alzheimer’s disease among women. Alzheimers Dement. 19, 896–908. doi: 10.1002/alz.12719
Chung, J., Vig, V., Sun, X., Han, X., O’Connor, G. T., Chen, X., et al. (2022b). Genome-wide pleiotropy study identifies association of PDGFB with age-related macular degeneration and COVID-19 infection outcomes. J. Clin. Med. 12:109. doi: 10.3390/jcm12010109
Crivelli, L., Palmer, K., Calandri, I., Guekht, A., Beghi, E., Carroll, W., et al. (2022). Changes in cognitive functioning after COVID-19: a systematic review and meta-analysis. Alzheimers Dement. 18, 1047–1066. doi: 10.1002/alz.12644
Del Ser, T., Fernández-Blázquez, M. A., Valentí, M., Zea-Sevilla, M. A., Frades, B., Alfayate, E., et al. (2021). Residence, clinical features, and genetic risk factors associated with symptoms of COVID-19 in a cohort of older people in Madrid. Gerontology 67, 281–289. doi: 10.1159/000513182
Demuro, A., Parker, I., and Stutzmann, G. E. (2010). Calcium signaling and amyloid toxicity in Alzheimer disease. J. Biol. Chem. 285, 12463–12468. doi: 10.1074/jbc.R109.080895
Dong, L.-M., Wilson, C., Wardell, M. R., Simmons, T., Mahley, R. W., Weisgraber, K. H., et al. (1994). Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 269, 22358–22365. doi: 10.1016/S0021-9258(17)31797-0
Drouet, B., Fifre, A., Pinçon-Raymond, M., Vandekerckhove, J., Rosseneu, M., Guéant, J. L., et al. (2001). ApoE protects cortical neurones against neurotoxicity induced by the non-fibrillar C-terminal domain of the amyloid-beta peptide. J. Neurochem. 76, 117–127. doi: 10.1046/j.1471-4159.2001.00047.x
Efstathiou, V., Stefanou, M.-I., Demetriou, M., Siafakas, N., Makris, M., Tsivgoulis, G., et al. (2022). Long COVID and neuropsychiatric manifestations (review). Exp. Ther. Med. 23:363. doi: 10.3892/etm.2022.11290
Evans, C. E., Miners, J. S., Piva, G., Willis, C. L., Heard, D. M., Kidd, E. J., et al. (2020). ACE2 activation protects against cognitive decline and reduces amyloid pathology in the Tg2576 mouse model of Alzheimer’s disease. Acta Neuropathol. 139, 485–502. doi: 10.1007/s00401-019-02098-6
Forloni, G., and Balducci, C. (2018). Alzheimer’s disease, oligomers, and inflammation. J. Alzheimers Dis. 62, 1261–1276. doi: 10.3233/JAD-170819
Frontera, J. A., Boutajangout, A., Masurkar, A. V., Betensky, R. A., Ge, Y., Vedvyas, A., et al. (2022). Comparison of serum neurodegenerative biomarkers among hospitalized COVID-19 patients versus non-COVID subjects with normal cognition, mild cognitive impairment, or Alzheimer’s dementia. Alzheimers Dement. 18, 899–910. doi: 10.1002/alz.12556
Garrigues, E., Janvier, P., Kherabi, Y., Le Bot, A., Hamon, A., Gouze, H., et al. (2020). Post-discharge persistent symptoms and health-related quality of life after hospitalization for COVID-19. J. Infect. 81, e4–e6. doi: 10.1016/j.jinf.2020.08.029
Gholami, M., Safari, S., Ulloa, L., and Motaghinejad, M. (2021). Neuropathies and neurological dysfunction induced by coronaviruses. J. Neurovirol. 27, 380–396. doi: 10.1007/s13365-021-00977-x
Gordon, M. N., Heneka, M. T., Le Page, L. M., Limberger, C., Morgan, D., Tenner, A. J., et al. (2022). Impact of COVID-19 on the onset and progression of Alzheimer’s disease and related dementias: a roadmap for future research. Alzheimers Dement. 18, 1038–1046. doi: 10.1002/alz.12488
Gouveia, C., Gibbons, E., Dehghani, N., Eapen, J., Guerreiro, R., and Bras, J. (2022). Genome-wide association of polygenic risk extremes for Alzheimer’s disease in the UK biobank. Sci. Rep. 12:8404. doi: 10.1038/s41598-022-12391-2
Greenhalgh, T., Sivan, M., Delaney, B., Evans, R., and Milne, R. (2022). Long covid-an update for primary care. BMJ 378:e072117. doi: 10.1136/bmj-2022-072117
Guo, H., Li, T., and Wen, H. (2022). Identifying shared genetic loci between coronavirus disease 2019 and cardiovascular diseases based on cross-trait meta-analysis. Front. Microbiol. 13:993933. doi: 10.3389/fmicb.2022.993933
Hammond, T. R., Marsh, S. E., and Stevens, B. (2019). Immune Signaling in Neurodegeneration. Immunity 50, 955–974. doi: 10.1016/j.immuni.2019.03.016
Herold, C., Hooli, B. V., Mullin, K., Liu, T., Roehr, J. T., Mattheisen, M., et al. (2016). Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol. Psychiatry 21, 1608–1612. doi: 10.1038/mp.2015.218
Herrera, E., and González-Nosti, M. (2022). The effect of age on cognitive impairment associated with post COVID-19 syndrome. Psychiatry Res. 318:114945. doi: 10.1016/j.psychres.2022.114945
Heyman, S. N., Kinaneh, S., and Abassi, Z. (2021a). The duplicitous nature of ACE2 in COVID-19 disease. EBioMedicine 67:103356. doi: 10.1016/j.ebiom.2021.103356
Heyman, S. N., Walther, T., and Abassi, Z. (2021b). Angiotensin-(1-7)-a potential remedy for AKI: insights derived from the COVID-19 pandemic. J. Clin. Med. 10:1200. doi: 10.3390/jcm10061200
Hossain, M. M., Tasnim, S., Sultana, A., Faizah, F., Mazumder, H., Zou, L., et al. (2020). Epidemiology of mental health problems in COVID-19: a review. F1000Res 9:636. doi: 10.12688/f1000research.24457.1
Hu, B., Huang, S., and Yin, L. (2021). The cytokine storm and COVID-19. J. Med. Virol. 93, 250–256. doi: 10.1002/jmv.26232
Hubacek, J. A., Dlouha, L., Dusek, L., Majek, O., and Adamkova, V. (2021). Apolipoprotein E4 allele in subjects with COVID-19. Gerontology 67, 320–322. doi: 10.1159/000516200
Jansen, I. E., Savage, J. E., Watanabe, K., Bryois, J., Williams, D. M., Steinberg, S., et al. (2019). Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413. doi: 10.1038/s41588-018-0311-9
Jha, N. K., Ojha, S., Jha, S. K., Dureja, H., Singh, S. K., Shukla, S. D., et al. (2021). Evidence of coronavirus (CoV) pathogenesis and emerging pathogen SARS-CoV-2 in the nervous system: a review on neurological impairments and manifestations. J. Mol. Neurosci. 71, 2192–2209. doi: 10.1007/s12031-020-01767-6
Jiang, J., Wang, X., and Lu, J. (2020). PWRN1 suppressed Cancer cell proliferation and migration in glioblastoma by inversely regulating hsa-miR-21-5p. Cancer Manag. Res. 12, 5313–5322. doi: 10.2147/CMAR.S250166
Kang, S., Gim, J., Lee, J., Gunasekaran, T. I., Choi, K. Y., Lee, J. J., et al. (2021). Potential novel genes for late-onset Alzheimer’s disease in east-Asian descent identified by APOE-stratified genome-wide association study. J. Alzheimers Dis. 82, 1451–1460. doi: 10.3233/JAD-210145
Kauwe, J. S. K., Bailey, M. H., Ridge, P. G., Perry, R., Wadsworth, M. E., Hoyt, K. L., et al. (2014). Genome-wide association study of CSF levels of 59 alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet. 10:e1004758. doi: 10.1371/journal.pgen.1004758
Kloske, C. M., Dugan, A. J., Weekman, E. M., Winder, Z., Patel, E., Nelson, P. T., et al. (2021). Inflammatory pathways are impaired in Alzheimer disease and differentially associated with apolipoprotein E status. J. Neuropathol. Exp. Neurol. 80, 922–932. doi: 10.1093/jnen/nlab085
Kousathanas, A., Pairo-Castineira, E., Rawlik, K., Stuckey, A., Odhams, C. A., Walker, S., et al. (2022). Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 607, 97–103. doi: 10.1038/s41586-022-04576-6
Kulminski, A. M., Loiko, E., Loika, Y., and Culminskaya, I. (2022). Pleiotropic predisposition to Alzheimer’s disease and educational attainment: insights from the summary statistics analysis. Geroscience 44, 265–280. doi: 10.1007/s11357-021-00484-1
Kumar, A., Sidhu, J., Goyal, A., and Tsao, J. W. (2023). Alzheimer Disease. Available at: http://www.ncbi.nlm.nih.gov/books/NBK499922/ (Accessed August 16, 2023).
Kunkle, B. W., Grenier-Boley, B., Sims, R., Bis, J. C., Damotte, V., Naj, A. C., et al. (2019). Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430. doi: 10.1038/s41588-019-0358-2
Kuo, C.-L., Pilling, L. C., Atkins, J. L., Masoli, J. A. H., Delgado, J., Kuchel, G. A., et al. (2020a). APOE e4 genotype predicts severe COVID-19 in the UK biobank community cohort. J. Gerontol. A Biol. Sci. Med. Sci. 75, 2231–2232. doi: 10.1093/gerona/glaa131
Kuo, C.-L., Pilling, L. C., Atkins, J. L., Masoli, J. A. H., Delgado, J., Kuchel, G. A., et al. (2020b). ApoE e4e4 genotype and mortality with COVID-19 in UK biobank. J. Gerontol. A Biol. Sci. Med. Sci. 75, 1801–1803. doi: 10.1093/gerona/glaa169
Kwon, H. S., and Koh, S. H. (2020). Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl. Neurodegener 9:42. doi: 10.1186/s40035-020-00221-2
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458. doi: 10.1038/ng.2802
Lee, H. W., Kim, Y., Han, K., Kim, H., and Kim, E. (2010). The phosphoinositide 3-phosphatase MTMR2 interacts with PSD-95 and maintains excitatory synapses by modulating endosomal traffic. J. Neurosci. 30, 5508–5518. doi: 10.1523/JNEUROSCI.4283-09.2010
Lemche, E. (2018). Early life stress and epigenetics in late-onset Alzheimer’s dementia: a systematic review. Curr. Genomics 19, 522–602. doi: 10.2174/1389202919666171229145156
Li, C., Liu, J., Lin, J., and Shang, H. (2022). COVID-19 and risk of neurodegenerative disorders: a Mendelian randomization study. Transl. Psychiatry 12:283. doi: 10.1038/s41398-022-02052-3
Li, W., Sun, L., Yue, L., and Xiao, S. (2023). Alzheimer’s disease and COVID-19: interactions, intrinsic linkages, and the role of immunoinflammatory responses in this process. Front. Immunol. 14:1120495. doi: 10.3389/fimmu.2023.1120495
Lopes, K. D. P., Snijders, G. J. L., Humphrey, J., Allan, A., Sneeboer, M. A. M., Navarro, E., et al. (2022). Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat. Genet. 54, 4–17. doi: 10.1038/s41588-021-00976-y
Machiela, M. J., and Chanock, S. J. (2015). LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. doi: 10.1093/bioinformatics/btv402
Mannekote Thippaiah, S., Amanullah, S., Huang, Z. H., Goldschmidt, E., and Pradhan, B. (2023). Biological correlates of the neuropsychiatric symptoms in SARS-CoV-2 infection: an updated review. Egypt. J. Neurol. Psychiatr. Neurosurg. 59:101. doi: 10.1186/s41983-023-00705-8
Matias-Guiu, J. A., Herrera, E., González-Nosti, M., Krishnan, K., Delgado-Alonso, C., Díez-Cirarda, M., et al. (2023). Development of criteria for cognitive dysfunction in post-COVID syndrome: the IC-CoDi-COVID approach. Psychiatry Res. 319:115006. doi: 10.1016/j.psychres.2022.115006
Mavrikaki, M., Lee, J. D., Solomon, I. H., and Slack, F. J. (2022). Severe COVID-19 is associated with molecular signatures of aging in the human brain. Nat Aging 2, 1130–1137. doi: 10.1038/s43587-022-00321-w
Naj, A. C., Beecham, G. W., Martin, E. R., Gallins, P. J., Powell, E. H., Konidari, I., et al. (2010). Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 6:e1001130. doi: 10.1371/journal.pgen.1001130
Nami, M., Thatcher, R., Kashou, N., Lopes, D., Lobo, M., Bolanos, J. F., et al. (2022). A proposed brain-, spine-, and mental- health screening methodology (NEUROSCREEN) for healthcare systems: position of the society for brain mapping and therapeutics. J. Alzheimers Dis. 86, 21–42. doi: 10.3233/JAD-215240
Nott, A., and Holtman, I. R. (2023). Genetic insights into immune mechanisms of Alzheimer’s and Parkinson’s disease. Front. Immunol. 14:1168539. doi: 10.3389/fimmu.2023.1168539
Oronsky, B., Larson, C., Hammond, T. C., Oronsky, A., Kesari, S., Lybeck, M., et al. (2023). A review of persistent post-COVID syndrome (PPCS). Clin. Rev. Allergy Immunol. 64, 66–74. doi: 10.1007/s12016-021-08848-3
Pairo-Castineira, E., Rawlik, K., Bretherick, A. D., Qi, T., Wu, Y., Nassiri, I., et al. (2023). GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature 617, 764–768. doi: 10.1038/s41586-023-06034-3
Pezzini, A., and Padovani, A. (2020). Lifting the mask on neurological manifestations of COVID-19. Nat. Rev. Neurol. 16, 636–644. doi: 10.1038/s41582-020-0398-3
Premraj, L., Kannapadi, N. V., Briggs, J., Seal, S. M., Battaglini, D., Fanning, J., et al. (2022). Mid and long-term neurological and neuropsychiatric manifestations of post-COVID-19 syndrome: a meta-analysis. J. Neurol. Sci. 434:120162. doi: 10.1016/j.jns.2022.120162
Prokopenko, D., Lee, S., Hecker, J., Mullin, K., Morgan, S., Katsumata, Y., et al. (2022). Region-based analysis of rare genomic variants in whole-genome sequencing datasets reveal two novel Alzheimer’s disease-associated genes: DTNB and DLG2. Mol. Psychiatry 27, 1963–1969. doi: 10.1038/s41380-022-01475-0
Qiu, S., Hu, Y., and Cheng, L. (2022). A genome-wide cross-trait analysis highlights the shared genetic structure between COVID-19 and Alzheimer’s disease. J. Infect. 84, e1–e2. doi: 10.1016/j.jinf.2021.08.038
Rahmati, M., Yon, D. K., Lee, S. W., Soysal, P., Koyanagi, A., Jacob, L., et al. (2023). New-onset neurodegenerative diseases as long-term sequelae of SARS-CoV-2 infection: a systematic review and meta-analysis. J. Med. Virol. 95:e28909. doi: 10.1002/jmv.28909
Raj, T., Rothamel, K., Mostafavi, S., Ye, C., Lee, M. N., Replogle, J. M., et al. (2014). Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523. doi: 10.1126/science.1249547
Raulin, A.-C., Doss, S. V., Trottier, Z. A., Ikezu, T. C., Bu, G., and Liu, C.-C. (2022). ApoE in Alzheimer’s disease: pathophysiology and therapeutic strategies. Mol. Neurodegener. 17:72. doi: 10.1186/s13024-022-00574-4
Reiss, A. B., Greene, C., Dayaramani, C., Rauchman, S. H., Stecker, M. M., De Leon, J., et al. (2023). Long COVID, the brain, nerves, and cognitive function. Neurol. Int. 15, 821–841. doi: 10.3390/neurolint15030052
Saucier, J., Comeau, D., Robichaud, G. A., and Chamard-Witkowski, L. (2023). Reactive gliosis and neuroinflammation: prime suspects in the pathophysiology of post-acute neuroCOVID-19 syndrome. Front. Neurol. 14:1221266. doi: 10.3389/fneur.2023.1221266
Sharma, A., Liu, X., Hadley, D., Hagopian, W., Chen, W. M., Onengut-Gumuscu, S., et al. (2018). Identification of non-HLA genes associated with development of islet autoimmunity and type 1 diabetes in the prospective TEDDY cohort. J. Autoimmun. 89, 90–100. doi: 10.1016/j.jaut.2017.12.008
Słomian, D., Szyda, J., Dobosz, P., Stojak, J., Michalska-Foryszewska, A., Sypniewski, M., et al. (2023). Better safe than sorry-whole-genome sequencing indicates that missense variants are significant in susceptibility to COVID-19. PLoS One 18:e0279356. doi: 10.1371/journal.pone.0279356
Steardo, L., Steardo, L., Zorec, R., and Verkhratsky, A. (2020). Neuroinfection may contribute to pathophysiology and clinical manifestations of COVID-19. Acta Physiol (Oxf.) 229:e13473. doi: 10.1111/apha.13473
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1.30.1–1.30.33. doi: 10.1002/cpbi.5
Tabeshmehr, P., and Eftekharpour, E. (2023). Tau; one protein, so many diseases. Biology 12:244. doi: 10.3390/biology12020244
Umakanthan, S., Sahu, P., Ranade, A. V., Bukelo, M. M., Rao, J. S., Abrahao-Machado, L. F., et al. (2020). Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19). Postgrad. Med. J. 96, 753–758. doi: 10.1136/postgradmedj-2020-138234
Wang, L., Davis, P. B., Volkow, N. D., Berger, N. A., Kaelber, D. C., and Xu, R. (2022a). Association of COVID-19 with new-onset Alzheimer’s disease. J. Alzheimers Dis. 89, 411–414. doi: 10.3233/JAD-220717
Wang, T., Du, Z., Zhu, F., Cao, Z., An, Y., Gao, Y., et al. (2020). Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet 395:e52. doi: 10.1016/S0140-6736(20)30558-4
Wang, Y., Guga, S., Wu, K., Khaw, Z., Tzoumkas, K., Tombleson, P., et al. (2022b). COVID-19 and systemic lupus erythematosus genetics: a balance between autoimmune disease risk and protection against infection. PLoS Genet. 18:e1010253. doi: 10.1371/journal.pgen.1010253
Wang, H., Kulas, J. A., Wang, C., Holtzman, D. M., Ferris, H. A., and Hansen, S. B. (2021b). Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc. Natl. Acad. Sci. U. S. A. 118:e2102191118. doi: 10.1073/pnas.2102191118
Wang, C., Zhang, M., Garcia, G., Tian, E., Cui, Q., Chen, X., et al. (2021a). ApoE-isoform-dependent SARS-CoV-2 Neurotropism and cellular response. Cell Stem Cell 28, 331–342.e5. doi: 10.1016/j.stem.2020.12.018
Weisgraber, K. H., Rall, S. C., and Mahley, R. W. (1981). Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J. Biol. Chem. 256, 9077–9083. doi: 10.1016/S0021-9258(19)52510-8
Wightman, D. P., Jansen, I. E., Savage, J. E., Shadrin, A. A., Bahrami, S., Holland, D., et al. (2021). A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282. doi: 10.1038/s41588-021-00921-z
Wong, E. W., Glasgow, S. D., Trigiani, L. J., Chitsaz, D., Rymar, V., Sadikot, A., et al. (2019). Spatial memory formation requires netrin-1 expression by neurons in the adult mammalian brain. Learn. Mem. 26, 77–83. doi: 10.1101/lm.049072.118
Wu, Y., Xu, X., Chen, Z., Duan, J., Hashimoto, K., Yang, L., et al. (2020). Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 87, 18–22. doi: 10.1016/j.bbi.2020.03.031
Xiong, N., Schiller, M. R., Li, J., Chen, X., and Lin, Z. (2021). Severe COVID-19 in Alzheimer’s disease: APOE4’s fault again? Alzheimers Res. Ther. 13:111. doi: 10.1186/s13195-021-00858-9
Yao, M., Huang, X., Guo, Y., Zhao, J. V., and Liu, Z. (2023). Disentangling the common genetic architecture and causality of rheumatoid arthritis and systemic lupus erythematosus with COVID-19 outcomes: genome-wide cross trait analysis and bidirectional Mendelian randomization study. J. Med. Virol. 95:e28570. doi: 10.1002/jmv.28570
Yong, S. J. (2021). Long COVID or post-COVID-19 syndrome: putative pathophysiology, risk factors, and treatments. Infect Dis 53, 737–754. doi: 10.1080/23744235.2021.1924397
Zhang, L., Lu, Y., Wang, Y., Wang, F., Zhai, S., Chen, Z., et al. (2020). PHF14 is required for germinal center B cell development. Cell. Immunol. 358:104221. doi: 10.1016/j.cellimm.2020.104221
Zhang, Q., Zhong, C., Shen, J., Chen, S., Jia, Y., and Duan, S. (2022). Emerging role of LINC00461 in cancer. Biomed. Pharmacother. 152:113239. doi: 10.1016/j.biopha.2022.113239
Keywords: COVID-19, long Covid, Alzheimer’s disease, cognitive impairment, pathway analysis, genetic susceptibility, GWAS, SNP
Citation: Matveeva N, Kiselev I, Baulina N, Semina E, Kakotkin V, Agapov M, Kulakova O and Favorova O (2023) Shared genetic architecture of COVID-19 and Alzheimer’s disease. Front. Aging Neurosci. 15:1287322. doi: 10.3389/fnagi.2023.1287322
Received: 01 September 2023; Accepted: 09 October 2023;
Published: 20 October 2023.
Edited by:
Kevin Nicholas Hascup, Southern Illinois University Carbondale, United StatesReviewed by:
Christian Barbato, National Research Council (CNR), ItalyCopyright © 2023 Matveeva, Kiselev, Baulina, Semina, Kakotkin, Agapov, Kulakova and Favorova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivan Kiselev, a2lzZWxldi5pdmFuLjE5OTFAZ21haWwuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.