- 1Department of Biochemistry and Clinical Laboratories, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Department of Basic Medical Sciences, Khoy University of Medical Sciences, Khoy, Iran
Alzheimer's disease (AD) is characterized by the accumulation of misfolded amyloid-beta and tau proteins. Autophagy acts as a proteostasis process to remove protein clumps, although it progressively weakens with aging and AD, thus facilitating the accumulation of toxic proteins and causing neurodegeneration. This review examines the impact of impaired autophagy on the progression of AD disease pathology. Under normal circumstances, autophagy removes abnormal proteins and damaged organelles, but any dysfunction in this process can lead to the exacerbation of amyloid and tau pathology, particularly in AD. There is increasing attention to therapeutic tactics to revitalize autophagy, including reduced caloric intake, autophagy-stimulating drugs, and genetic therapy. However, the translation of these strategies into clinical practice faces several hurdles. In summary, this review integrates the understanding of the intricate role of autophagy dysfunction in Alzheimer's disease progression and reinforces the promising prospects of autophagy as a beneficial target for treatments to modify the course of Alzheimer's disease.
1. Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder that is the leading cause of dementia in the elderly population. This condition contributes to ~70% of the estimated 50 million cases of dementia worldwide (Morato et al., 2022; Alzheimer's Association, 2023). The hallmark pathological features of AD include the accumulation of amyloid beta (Aβ) plaques and the presence of neurofibrillary tangles of hyperphosphorylated tau protein in the brain (Hampel et al., 2021). These manifestations contribute to synaptic dysfunction and neuronal decline. The origins of sporadic or late-onset AD, which accounts for more than 95% of cases, remain largely unknown. However, they are likely influenced by a multifaceted interplay of genetic, epigenetic, and environmental factors (Gcwensa et al., 2021). Pathways implicated in the development of AD include deficiencies in Aβ clearance, tau-related pathology, neuroinflammation, mitochondrial dysfunction, and impaired proteostasis (Gadhave et al., 2020).
Proteostasis involves the regulatory processes that maintain the balance of the proteome through functions such as protein synthesis, folding, degradation, and catabolism, mediated by pathways such as the ubiquitin–proteasome system and autophagy. In situations of proteostatic imbalance, the heat shock response and the unfolded protein response step in to correct the situation, but their restorative efficiency declines with age (Lei et al., 2021; Sarkar and Nazir, 2022). The inability to restore proteostasis leads to proteotoxic stress and eventual cell death. Neurodegenerative disorders, such as AD, are characterized by ubiquitous proteostasis perturbations. However, whether these perturbations trigger or result from neurodegeneration remains a subject of ongoing debate (Hohn et al., 2020; Ruz et al., 2020).
Autophagy is an intracellular catabolic pathway involving the lysosomal degradation of cellular components from the cytoplasm. Autophagy begins with the formation of double-membrane vesicles called autophagosomes, which engulf materials such as misfolded proteins and damaged organelles. These autophagosomes then fuse with lysosomes to facilitate breakdown and recycling of the contents. Basal autophagy plays a critical role in maintaining cellular integrity by regulating quality control, and can be upregulated in response to stressors. Dysregulation of autophagy has been observed in several neurodegenerative diseases, and also shows declining efficacy over time, which has been implicated in cytotoxic protein accumulation in AD (Hughes et al., 2020; Senos Demarco and Jones, 2020).
In this review, we examine the complex interplay between aberrant proteostasis, autophagy dysfunction, and neuronal death in Alzheimer's disease pathogenesis. We explore proteostasis regulation and molecular details of the autophagy pathway. By evaluating evidence from AD models and human studies, we aim to elucidate this intricate relationship between proteostasis, autophagy, and neurodegeneration in AD, shedding light on mechanisms and revealing new therapeutic avenues.
2. Proteostasis and cellular homeostasis
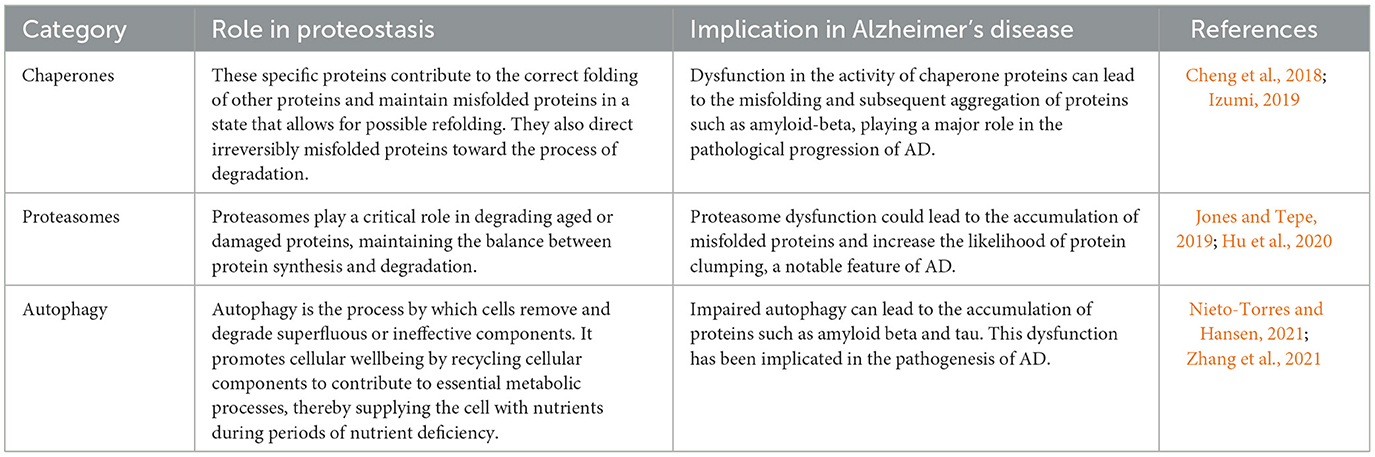
Proteostasis refers to a set of processes that maintain the structure and function of the proteome. It includes mechanisms that regulate protein synthesis, folding, unfolding, and degradation to ensure that proteins achieve the 3D shapes necessary for their biological roles (Sebastian and Shoulders, 2020; Powers and Gierasch, 2021). Disruptions in proteostasis are associated with diseases characterized by protein misfolding and aggregation, such as neurodegenerative disorders (Cortes Sanchon et al., 2021). The key components of proteostasis are molecular chaperones, which are proteins that mediate folding. Chaperones such as Hsp70 and Hsp90 recognize exposed hydrophobic regions on misfolded proteins. They maintain these proteins in a foldable state until proper folding is achieved through ATP hydrolysis and cochaperone action (Frumkin et al., 2014; Saibil, 2021). Notably, chaperones also direct misfolded proteins toward degradation. Aging impairs the effectiveness of the heat shock response, which activates chaperones to deal with proteotoxic stress, leading to proteostatic failure (Morimoto, 2020). In addition, the ubiquitin–proteasome system (UPS) is a major proteolytic pathway that directs the degradation of damaged ubiquitinated proteins. Through “proteasomal degradation”, the UPS removes defective oxidized, mutated, or misfolded proteins, thereby maintaining proteostasis (Fhu and Ali, 2021; Kong et al., 2021). UPS dysfunction leads to protein accumulation, a hallmark of neurodegenerative diseases such as Parkinson's disease, which is associated with reduced UPS function and aggregation of α-synuclein targeted for UPS degradation (Zheng et al., 2016; Al Mamun et al., 2020). In neurodegenerative diseases, protein aggregates accumulate in the neurons, in part due to impaired proteostatic mechanisms (Figure 1; Agius, 2016). Abnormal aggregation into insoluble fibrils can damage membranes, disrupt trafficking, and lead to synaptic dysfunction and neuronal death. Therefore, maintaining neuronal proteostasis is critical to mitigate neurotoxicity. Table 1 shows the components of proteostasis.
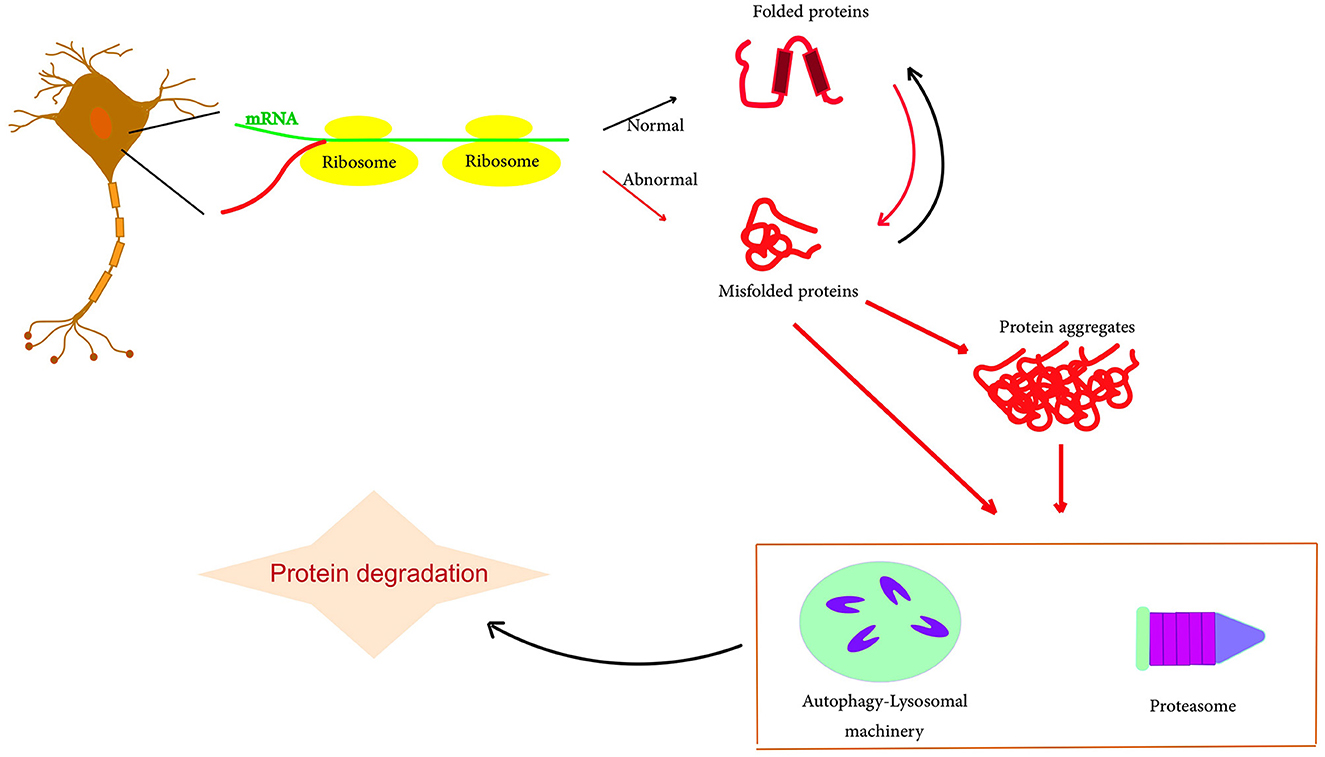
Figure 1. Schematic representation of protein misfolding and aggregation. The accumulation of misfolding proteins, including amyloid-beta and tau, due to an imbalance in protein production. Typically, these abnormal protein clusters are broken down through a process called autophagy or with the support of the cell's proteasome. However, if there is any malfunction in this degradation process, the accumulation of amyloid and tau can potentially escalate, particularly contributing to the progression of AD.
In summary, proteostasis regulates processes that manage protein lifecycles to maintain proteome integrity using specialized components such as chaperones, targeted proteolytic mechanisms, and stress responses. Elucidating the dysregulation of neuronal proteostasis in disease may reveal potential areas for therapeutic intervention.
3. Autophagy: mechanisms and regulation
Autophagy, a well-conserved lysosomal degradation pathway, performs the vital task of degrading and recycling cellular components (Schaefer and Dikic, 2021). This complex process consists of three primary types: macroautophagy, microautophagy, and chaperone-associated autophagy. Macroautophagy is the process by which cytoplasmic substances are packaged into vesicular structures known as autophagosomes, which then associate with lysosomes to facilitate degradation and recycling of the contents (Schuck, 2020; Wang L. et al., 2022). Microautophagy involves direct engulfment of the cytoplasm into lysosomes via invagination of the lysosomal membrane. The third type, chaperone-mediated autophagy, involves specific proteins called “chaperones” that recognize, guide, and transfer substrate proteins across the lysosomal membrane to the lumen for subsequent degradation (Kaushik and Cuervo, 2018; Schuck, 2020).
Among these types, macroautophagy has been the most thoroughly studied and defined. Effective execution of macroautophagy depends on ATG proteins, which regulate the sequential progression of autophagosomes from initiation to completion (Schuck, 2020; Metur and Klionsky, 2021). These processes are initiated by upstream signaling events that inhibit mTORC1, a nutrient sensor, which then facilitates the activation of the ULK1 kinase complex and the downstream generation of PI3P lipids, which are essential for the recruitment of other ATG proteins (Shimobayashi et al., 2013; Condon and Sabatini, 2019).
Autophagosome formation is highly dependent on two ubiquitin-like conjugation systems. These are the ATG12-ATG5-ATG16L1 system and the LC3 conjugation system. These systems determine the site of lipidation of the LC3 protein by the ATG12-ATG5-ATG16L1 complex, which then cleaves LC3 to form LC3-I. This LC3-I is then combined with phosphatidylethanolamine to form LC3-II, a characteristic marker of autophagosomal membranes (Runwal et al., 2019; Mizushima, 2020; Boehi et al., 2021; Vujic et al., 2021).
The mTORC1 complex plays a critical role in regulating autophagy by inhibiting its initiation under nutrient-rich conditions (Ramaian Santhaseela and Jayavelu, 2021). Another key regulator is AMP-activated protein kinase (AMPK), which senses the energy status of cells and triggers autophagy during periods of energy depletion (Gelinas et al., 2018). In addition, the transcription factor EB (TFEB) modulates the transcription of autophagy and lysosomal genes, thereby maintaining autophagic balance (Dang and Back, 2021).
Autophagy is a response mechanism to various stressors, allowing the use of intracellular nutrients and the removal of damaged components (Schaefer and Dikic, 2021). However, it also has a dark side. Dysregulation of autophagy has been implicated in several pathologies, including neurodegeneration, heart disease, and cancer. Understanding how this critical, multifaceted process is regulated could potentially provide new avenues for therapeutic intervention in various disease contexts (Kroemer and White, 2010; Markaki and Tavernarakis, 2020).
In summary, macroautophagy, enabled by ATG proteins, is a primary form of autophagy. mTORC1 inhibits autophagy under conditions of nutrient abundance, whereas AMPK induces it under conditions of energy deprivation. While autophagy typically plays an adaptive role, its dysregulation can wreak havoc and lead to disease pathology.
4. Autophagy dysregulation in Alzheimer's disease
AD is associated with progressive neuronal deterioration leading to dementia. The key pathological features include extracellular amyloid plaques containing aggregated Aβ peptide and intracellular neurofibrillary tangles containing aggregated hyperphosphorylated tau protein (Patil et al., 2010; Ciccone et al., 2020). Autophagy, a form of cellular degradation involving lysosomal turnover of cytoplasmic material, is increasingly implicated in AD pathogenesis (Schaefer and Dikic, 2021).
Autophagy is one of the primary pathways for the degradation of aggregated proteins. In the context of AD, impaired autophagy-mediated clearance of Aβ and tau aggregates is likely to contribute to their accumulation. Aβ peptides are derived from amyloid precursor protein (APP), which is proteolytically cleaved by β- and γ-secretases. Accumulation of Aβ, particularly the neurotoxic oligomeric forms, results in synaptic dysfunction, neuronal loss, and cognitive decline (Festa et al., 2021; Ntsapi and Loos, 2021). Tau is a microtubule-stabilizing protein that resides within axonal microtubules under typical conditions. Hyperphosphorylation forces tau to disengage from microtubules and form neurotoxic oligomeric aggregates that disrupt neuronal transport (Gyparaki et al., 2021; Wang D. et al., 2021). Impaired autophagic clearance accelerates the accumulation of Aβ and tau aggregates, exacerbating neurodegeneration (Festa et al., 2021).
Research indicates that dysfunctional autophagy impairs clearance of Aβ and tau aggregates, promotes their accumulation, and increases neurotoxicity in AD models. APP transgenic mice with knockout of an autophagy gene, Beclin-1 or ATG7, show increased extracellular Aβ deposition and intracellular Aβ accumulation (Carosi et al., 2021; Festa et al., 2021; Wu et al., 2021). Pharmacological autophagy has been shown to reduce Aβ levels and ameliorate cognitive deficits in APP mice (Wu et al., 2021). The restoration of autophagy has been found to exert an ameliorative effect on tau pathology when tau mutation or aggregation inhibitors are used (Anglada-Huguet et al., 2021). Autophagy stimulation has been found to result in tau clearance in primary neurons, whereas autophagy inhibition results in the accumulation of tau aggregates (Chinnathambi and Gorantla, 2021). Therefore, it is likely that autophagy impairment contributes to Aβ and tau pathology in AD.
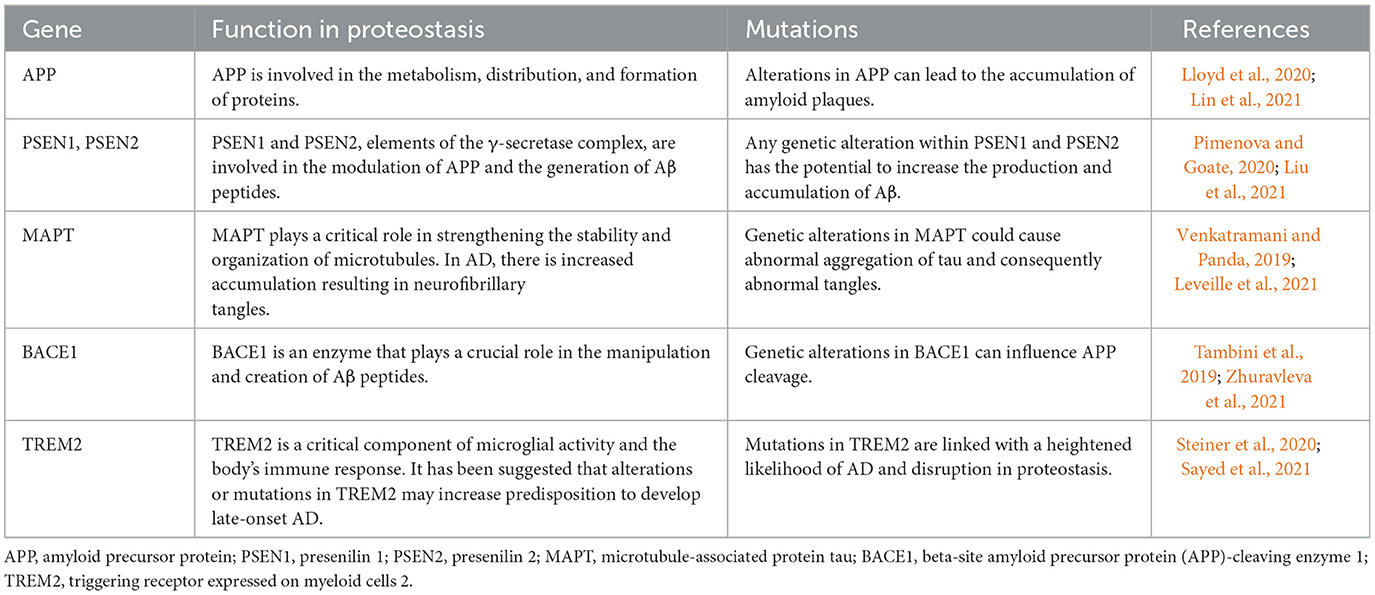
Several lines of evidence support a role for defective autophagy in the pathogenesis of AD. Indicators of autophagy, such as LC3-II and Beclin-1, are reduced, while defective lysosomal proteolysis and autophagosome accumulation are evident in the brains of AD patients (Mani et al., 2021). Increased AD risk is associated with variations in autophagy gene polymorphisms (Lipinski et al., 2010). The accumulation of autophagy-promoting proteins is caused by Aβ and tau oligomers, which impair autophagy function, setting up a damaging cycle (Feng et al., 2020). Autophagy is disrupted by risk factors for AD, such as apo E4 and presenilin 1 mutations (Yang et al., 2019). Autophagy stimulation reduces Aβ and tau pathology and improves cognition in AD models (Qi et al., 2021). Thus, mitigating autophagy dysfunction represents an attractive strategy for AD treatment, potentially modifying the course of the disease. Table 2 provides an overview of the key genes associated with proteostasis, while Figure 2 graphically illustrates the role of autophagy processes in the context of AD. In conclusion, impaired autophagic clearance of Aβ and tau aggregates likely contributes to their accumulation in AD. Enhancing autophagy helps to clear toxic protein aggregates and prevent neurodegeneration, providing strong evidence that autophagy dysfunction is an important factor in AD pathogenesis. Targeting the autophagy pathway may provide therapeutic opportunities for AD.
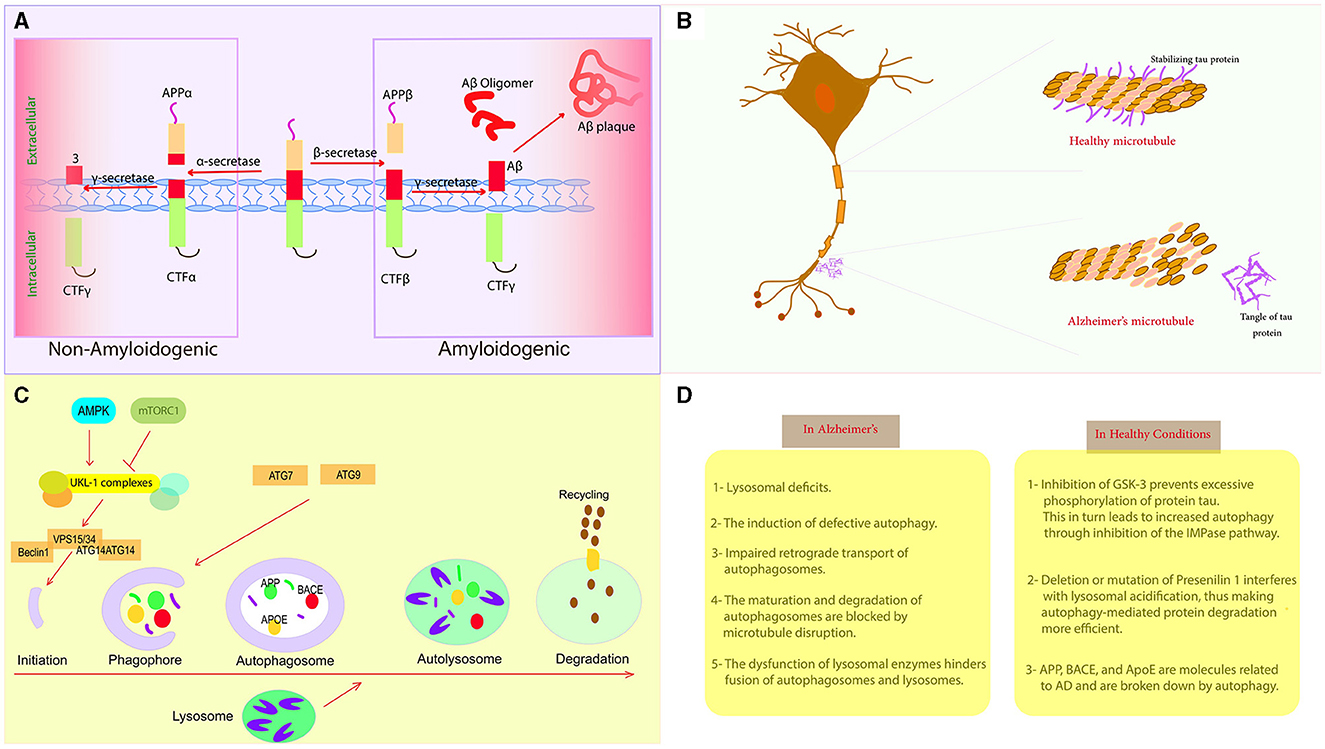
Figure 2. Autophagy and Alzheimer's disease. (A) AD development begins with the accumulation of abnormal Aβ peptides resulting from the proteolytic cleavage of APP by γ-secretase and β-secretase. (B) Tau is present in the axon of the neuron, but its hyperphosphorylated version is insoluble and unlikely to bind to microtubules, leading to axonal degeneration. (C) The primary processes that control different stages of autophagy. (D) Comparison between healthy conditions and the pathological conditions associated with AD.
5. Autophagy in animal models of AD
In animal models of AD, the introduction of bone marrow-derived mesenchymal stem cells (BMMSCs) has been shown to induce autophagy and reduce apoptosis, resulting in improvement of memory and cognitive deficits (Qin et al., 2022). The induction of autophagy is supported by the observed expression of signaling molecules, including Beclin-1, Atg5, LC3-II, and mTOR (Qin et al., 2022). Research beyond BMMSC transplantation has also investigated the function of autophagy in AD using various animal models. In one case, an APP transgenic mouse model with deletion of Beclin-1 showed a disruption in the basic level of autophagy, leading to escalation of the accumulation of Aβ in the cell (Liu and Li, 2019). This suggests that maintaining the flow of autophagy may be essential to prevent Aβ accumulation and subsequent neuronal death (Liu and Li, 2019). In addition, recent evidence suggests that specific perturbations of non-canonical autophagy in microglia may reduce the ability to clear β-amyloid, leading to progressive neurodegeneration in a mouse model of AD. These observations are consistent with previous findings in mice lacking autophagy-related gene 7 (atg7) (Wang Z. et al., 2022). Intriguingly, some research has suggested that boosting autophagy may have protective effects on the nervous system. For example, administration of rapamycin, a known mTOR inhibitor that triggers autophagy, has been shown to reduce Aβ levels and improve cognitive function in mouse models of AD (Liu and Li, 2019). In conclusion, these studies highlight the potential therapeutic role of autophagy modulation in AD. However, further research is needed to fully understand the complex interplay between autophagy and other cellular processes in the context of AD. The use of animal models will continue to be essential in this endeavor.
6. Autophagic biomarkers in AD
Recent research has aimed to explore the deep relationship between autophagy and AD, with the aim of discovering possible autophagy-related biomarkers for Alzheimer's disease. Researchers aim to do this by identifying critical autophagy genes that show striking differences in expression and investigating the potential roles of these genes (Li et al., 2023). One study has discovered ten DEAGs that play a role in the progression of AD. These include nine genes with increased activity (CAPNS1, GAPDH, IKBKB, LAMP1, LAMP2, MAPK1, PRKCD, RAB24, and RAF1) and one with decreased activity (CASP1). Correlational analysis has suggested possible links between these key DEAGs (Li et al., 2023). In addition to these genetic biomarkers, other studies have identified dysfunctional autophagic processes in the brains of Alzheimer's patients. For example, the accumulation of immature autophagic vacuoles (AVs) in dystrophic neurites in the brains of AD patients suggests that the autophagic process is disrupted (Liu and Li, 2019). In addition, presenilin has been proposed to play a necessary role in lysosomal calcium storage and release; without proper presenilin function, cells exhibit defective endosomal–lysosomal fusion, accompanied by the accumulation of endosomes and autophagosomes, and severely deficient autophagy (Orr and Oddo, 2013). The accumulation of Aβ protein fragments outside neurons, known as “Aβ plaques”, and the accumulation of an altered variant of the tau protein inside neurons, known as “tau tangles”, are two notable brain changes associated with AD4 (Wang Y. Y. et al., 2021). These pathological features could also be used as potential indicators of the presence of AD. In summary, these studies highlight the prospective importance of autophagy-related biomarkers for the detection and treatment of AD. However, further studies are essential to fully unravel the intricate relationship between autophagy and other cellular functions within the AD spectrum.
7. Crosstalk between autophagy and other cellular pathways
Recent research suggests that autophagy is involved in several other cellular pathways in AD, such as the ubiquitin–proteasome system (UPS), the endoplasmic reticulum (ER) stress response, and the unfolded protein response (UPR) (Kocaturk and Gozuacik, 2018). The UPS functions to degrade misfolded proteins and maintain protein balance in the cell. Recent research has shown that autophagy and the UPS undergo a complex interaction in the context of AD (Kocaturk and Gozuacik, 2018). One study found that the inhibition of autophagy led to the accumulation of ubiquitinated proteins and the activation of the UPS in AD animal models. The study also found that inhibition of the UPS led to the accumulation of autophagic vacuoles and the activation of autophagy in these models (Ke, 2017). These findings suggest that autophagy and the UPS interact in AD and that the regulation of these pathways may be a potential therapeutic target for the treatment of the disease. The ER stress response and the UPR are cellular pathways that respond to misfolded proteins and regulate protein balance. Recent research has shown that autophagy interacts with these pathways in AD (Chipurupalli et al., 2021). One study showed that when autophagy was inhibited, the ER stress response and the UPR were subsequently triggered in models of AD (Montibeller and de Belleroche, 2018). In addition, when autophagy was activated, there was a decrease in ER stress and the UPR in these models. These results suggest that autophagy interacts with the ER stress response and the UPR in AD, and that targeting these pathways could also be a potential therapeutic focus for managing the disease (Tooze et al., 2017). In addition to the UPS, the ER stress response, and the UPR, recent studies have shown that autophagy interacts with other cellular pathways in AD, including the immune response and the mitochondrial quality control system. One study found that activation of autophagy led to a reduction in neuroinflammation and an improvement in cognitive function in animal models of AD (Wang G. et al., 2019). The findings also indicated that the induction of autophagy led to an improvement in mitochondrial functionality and a reduction in oxidative stress in these models. These findings suggest that autophagy is involved in the immune response and the system that monitors mitochondrial quality in AD (Lee et al., 2012; Hyttinen et al., 2021). Control of these pathways may offer potential therapeutic strategies to combat the disease. In summary, autophagy dysregulation is a key feature of AD, and recent studies have shown that autophagy interacts with other cellular pathways in the disease, including the UPS, ER stress response, UPR, immune response, and mitochondrial quality control system. These findings suggest that regulation of these pathways may be potential therapeutic targets for the treatment of AD. Further research is needed to fully understand the crosstalk between autophagy and other cellular pathways in AD and to develop effective therapeutic strategies.
8. Therapeutic potential of modulating autophagy
Modulation of autophagy already offers a promising avenue for therapy given the contribution of autophagy dysfunction to AD pathology (Kaushik et al., 2021; Eisenberg et al., 2022).
Therapeutic approaches to modulation ofautophagy include a variety of strategies, including dietary control, pharmacological methods, and genetic therapies.
Of note is the potential of caloric restriction to induce autophagy by inhibiting mTOR signaling, a prominent inhibitor of autophagy. Stimulation of autophagy leads to significant improvements in cognitive function and reduced levels of amyloid-beta plaques and phosphorylated tau in mouse models of Alzheimer's under 30% dietary restriction. However, long-term dietary abstinence seems largely infeasible for AD patients (Ferreira-Marques et al., 2021; Muller et al., 2021). The feasibility tends toward clinical relevance in the case of the use of pharmacological inducers of autophagy such as mTOR inhibitors and lithium. Notably, the mTOR inhibitor rapamycin has been found to increase autophagy levels, reduce amyloid-beta and phosphorylated tau levels, and improve cognition in AD mouse models (Yang and Zhang, 2020). Lithium's similar enhancement of autophagy and attenuation of amyloid-beta and tau pathology enhances its therapeutic potential for AD (Morris and Berk, 2016; Uddin et al., 2021).
Evidence suggests that autophagy, along with its non-canonical processes such as LC3-associated phagocytosis (LAP) and LC3-associated non-canonical autophagy-dependent necroptosis (LANDO), plays a significant role in the development of AD (Hooper et al., 2021; Magne and Green, 2022). Recent evidence suggests an emerging role of LANDO in cytokine receptor signaling and innate immunity (Magne and Green, 2022). Research study by Martinez-Vicente et al. has shown that LC3-associated phagocytosis (LAP) plays an important role in Aβ plaque clearance. When LAP is inhibited in microglia, Aβ plaque clearance is reduced and inflammation is increased (Martinez-Vicente et al., 2010). In addition, their research in a mouse model of AD showed increased LAP activity in microglia located around Aβ plaques. This further supports the critical role of LAP in reducing Aβ in AD (Martinez-Vicente et al., 2010). A study led by Zhang et al. has presented evidence that triggering LANDO leads to nerve cell death and cognitive decline. Conversely, in a mouse model of AD, blocking LANDO was found to lead to reduced nerve cell death and improved cognitive performance. This raises the possibility that LANDO may be a potential candidate for AD therapy (Wang J. et al., 2019). Microglia, the resident immune cells of the CNS, play a crucial role in the pathogenesis of age-related neurodegenerative diseases (NDDs). Emerging evidence suggests that microglia coordinate the inflammatory responses in the CNS and contribute to the progression of NDDs (Lin et al., 2022). Microglial autophagy is involved in the regulation of neuronal homeostasis and the process of neuroinflammation. Dysregulation of microglial autophagy has been implicated in the pathogenesis of NDDs, including AD, PD, and HD (Navone et al., 2015; Lin et al., 2022). These findings suggest that the regulation of microglia autophagy and mitophagy may be a potential therapeutic target for the treatment of NDDs.
Gene therapy strategies such as overexpression of autophagy-related proteins, including Beclin-1, stimulate autophagy to reduce AD pathology in animal models (Pickford et al., 2008). In addition, small molecules capable of activating the autophagy function of Beclin-1 show protective effects in AD models (Seo et al., 2014). Thus, these early promising results demonstrate the potential of genetic and pharmacological approaches to enhance autophagy in AD therapy.
However, translating autophagy modulation to actual patients presents several challenges. The complex and dynamic nature of autophagy is closely intertwined with numerous cellular pathways, which may lead to physiological perturbations following non-selective autophagy induction (Ariosa et al., 2021; Manea and Ray, 2021). Unexpected cytotoxic or tumorigenic effects remain a concern with long-term use, as autophagy can oscillate between promoting cell survival and death (Zaarour et al., 2021).
Pharmacological agents are also not without problems—poor target specificity and minimal permeability of the blood–brain barrier, coupled with systemic side effects during long-term use, nullify their efficacy (Sitta, 2018; Zaarour et al., 2021). Therefore, the successful development of autophagy-based treatment strategies for neurodegeneration will require improvements in central nervous system (CNS) delivery, optimization of treatment duration and timing, and enhancement of cell and tissue specificity (Ajoolabady et al., 2021).
While preliminary results seem promising, most studies have examined short-term treatments in younger transgenic mouse models of AD (Oblak et al., 2021). The resulting long-term effects on cognition and pathology in older animal models, which better reflect AD progression, are areas that require further investigation (Munoz-Moreno et al., 2020). Future research should also examine the effects on neuroinflammation and synapse loss, which are key aspects of AD pathogenesis.
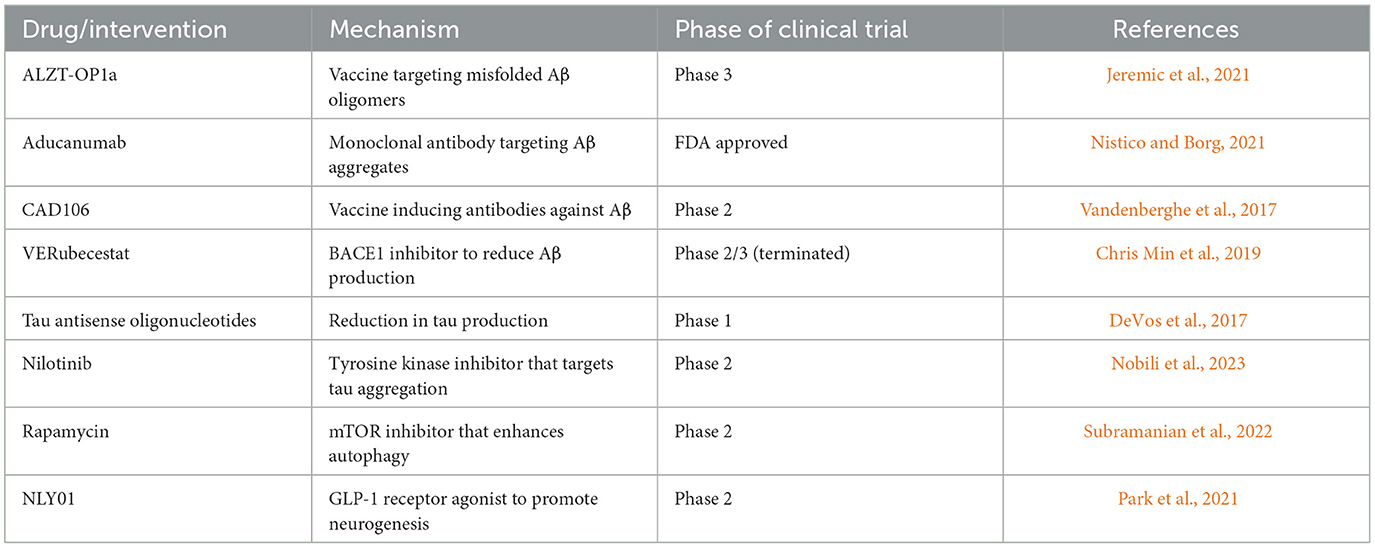
In summary, interventions aimed at restoring autophagy have been shown to ameliorate AD pathology and symptoms at the preclinical stage. However, successful translation to the clinic requires careful consideration of a variety of factors: unintended effects, disease stage, duration of treatment, and age-related changes in autophagy. Only then can autophagy be safely and effectively modulated for AD therapy. Table 3 illustrates a selection of ongoing clinical trials and drugs targeting proteostasis pathways for the treatment of AD.
9. Future directions and research gaps
A number of novel methodologies offer potential advances in our understanding of autophagy and proteostasis in AD. For example, the advent of super-resolution fluorescence microscopy has been instrumental in the study of autophagic structures at the nanoscale, allowing for a more comprehensive understanding of autophagosome formation, cargo sequestration, and lysosome fusion (Karanasios, 2019; Zheng et al., 2021). In addition, innovative live cell imaging methods provide the ability to monitor fluorescently tagged autophagy proteins together with selective autophagy substrates (Perez-Leal et al., 2021). Equipped with high-content screening platforms, it is becoming increasingly possible to identify novel compounds capable of modulating autophagy by allowing phenotype-based screening in neuronal models (Suenaga et al., 2021). Using the sophisticated techniques of single-cell transcriptomic profiling, the cell state-specific roles of autophagy genes in AD models can be more clearly delineated (Hu et al., 2017). These state-of-the-art methods are enabling an unprecedented level of understanding of autophagic mechanisms in the context of AD pathogenesis (Mijaljica, 2010).
Despite these advances, there are significant gaps in our knowledge of autophagy and proteostasis in AD. Determining the specific molecular pathways that link Aβ and tau pathologies to altered neuronal autophagy remains a critical task (Zhao et al., 2020). It is also unclear which aggregated forms exert the greatest toxicity and whether their accumulation is a cause or a symptom of autophagy dysfunction. The role of autophagy in brain cells such as microglia, which have been implicated in AD, remains largely unexplored (Pomilio et al., 2020). Understanding shifts in proteostasis networks during the early prodromal stages, before plaques and tangles accumulate, is critical. The question of why certain neuronal populations are more vulnerable than others also remains unanswered. The paucity of longitudinal patient studies monitoring cognitive decline along with cerebrospinal fluid (CSF) and imaging markers of autophagy calls for urgent attention (Mufson et al., 2016). Sophisticated investigations of these critical issues are expected to provide key insights into the pathogenesis of AD.
The understanding that diminished autophagy plays a role in AD progression has raised the potential of targeting autophagy with therapies. Drugs that induce the onset of autophagy have shown encouraging signs in AD models (Jogalekar et al., 2021; Kaushik et al., 2021). In addition, manipulation of late-stage autophagy may also prove beneficial. By controlling lysosomal dysfunction, proteostatic capacity can be significantly enhanced in AD models (Xu et al., 2022). While gene therapies aimed at increasing autophagy have shown efficacy in preclinical studies, most strategies aimed at modulating autophagy have yet to reach the clinic (Long and McWilliams, 2020). Predicting patient response, managing unanticipated side effects, and ensuring adequate CNS delivery of autophagy-modulating agents remain challenges. However, the identification of autophagy-related biomarkers may provide potential patient segmentation for targeted therapies (Pani and Keefe, 2019). Ultimately, a comprehensive approach to proteostasis therapies that simultaneously enhances autophagy, lysosomal function, and proteasomal activity may provide the most effective treatment for AD (Njomen and Tepe, 2019). Overall, modulation of neuronal autophagy and proteostasis represents a promising avenue for the development of individualized AD therapeutics.
10. Conclusion
Disturbances in proteostasis and autophagy are major contributors to AD. These perturbations result in the accumulation of misfolded proteins, including amyloid-beta and tau, due to an imbalance in protein production and clearance. The critical degradation function of autophagy is compromized, leading to the accumulation of toxic protein aggregates. Research has shown that molecular chaperones and the ubiquitin–proteasome system play an important role in managing proteostasis and influencing the aforementioned pathologies. Several autophagy pathways are critical for clearance of protein aggregates and dysfunctional mitochondria in AD. Impaired proteostasis and autophagy lead to oxidative stress, synaptic dysfunction, and neuronal death, accelerating AD neurodegeneration. A deeper understanding of these intricate cellular activities will be instrumental in identifying new drug targets and developing effective disease-modifying treatments. Efforts to repair proteostasis and enhance autophagy are promising therapeutic strategies for AD that require continued exploration to potentially move findings from the research stage to practical application. Ultimately, substantial evidence suggests that proteostasis networks and autophagy are critical in the development of AD and are promising areas for future treatments.
Author contributions
AN: Visualization, Writing—review & editing. HB: Visualization, Writing—original draft. FK-K: Supervision, Writing—review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agius, M. L. (2016). General concepts of neurodegeneration as specificity of mitochondrial pathobiology of neurons. J. Neurol. Neurophysiol. 7, 354. doi: 10.4172/2155-9562.1000354
Ajoolabady, A., Aslkhodapasandhokmabad, H., Henninger, N., Demillard, L. J., Nikanfar, M., Nourazarian, A., et al. (2021). Targeting autophagy in neurodegenerative diseases: from molecular mechanisms to clinical therapeutics. Clin. Exp. Pharmacol. Physiol. 48, 943–953. doi: 10.1111/1440-1681.13500
Al Mamun, A., Uddin, M. S., Kabir, M. T., Khanum, S., Sarwar, M. S., Mathew, B., et al. (2020). Exploring the promise of targeting ubiquitin-proteasome system to combat Alzheimer's disease. Neurotox Res. 38, 8–17. doi: 10.1007/s12640-020-00185-1
Alzheimer's Association (2023). 2023 Alzheimer's disease facts and figures. Alzheimers Dement. 19, 1598–695. doi: 10.1002/alz.13016
Anglada-Huguet, M., Rodrigues, S., Hochgrafe, K., Mandelkow, E., and Mandelkow, E. M. (2021). Inhibition of Tau aggregation with BSc3094 reduces Tau and decreases cognitive deficits in rTg4510 mice. Alzheimers Dement 7, e12170. doi: 10.1002/trc2.12170
Ariosa, A. R., Lahiri, V., Lei, Y., Yang, Y., Yin, Z., Zhang, Z., et al. (2021). A perspective on the role of autophagy in cancer. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166262. doi: 10.1016/j.bbadis.2021.166262
Boehi, F., Manetsch, P., and Hottiger, M. O. (2021). Interplay between ADP-ribosyltransferases and essential cell signaling pathways controls cellular responses. Cell Discov. 7, 104. doi: 10.1038/s41421-021-00323-9
Carosi, J. M., Hein, L. K., van den Hurk, M., Adams, R., Milky, B., Singh, S., et al. (2021). Retromer regulates the lysosomal clearance of MAPT/tau. Autophagy. 17, 2217–2237. doi: 10.1080/15548627.2020.1821545
Cheng, J., North, B. J., Zhang, T., Dai, X., Tao, K., Guo, J., et al. (2018). The emerging roles of protein homeostasis-governing pathways in Alzheimer's disease. Aging Cell. 17, e12801. doi: 10.1111/acel.12801
Chinnathambi, S., and Gorantla, N. V. (2021). Implications of valosin-containing protein in promoting autophagy to prevent Tau aggregation. Neuroscience. 476, 125–134. doi: 10.1016/j.neuroscience.2021.09.003
Chipurupalli, S., Samavedam, U., and Robinson, N. (2021). Crosstalk between ER stress, autophagy and inflammation. Front. Med. 8, 758311. doi: 10.3389/fmed.2021.758311
Chris Min, K., Dockendorf, M. F., Palcza, J., Tseng, J., Ma, L., Stone, J. A., et al. (2019). Pharmacokinetics and pharmacodynamics of the BACE1 inhibitor verubecestat (MK-8931) in healthy Japanese adults: a randomized, placebo-controlled study. Clin. Pharmacol. Ther. 105, 1234–1243. doi: 10.1002/cpt.1258
Ciccone, L., Shi, C., di Lorenzo, D., Van Baelen, A. C., and Tonali, N. (2020). The positive side of the Alzheimer's disease amyloid cross-interactions: the case of the abeta 1-42 peptide with Tau, TTR, CysC, and ApoA1. Molecules. 25, 439. doi: 10.3390/molecules25102439
Condon, K. J., and Sabatini, D. M. (2019). Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 132, jcs.222570. doi: 10.1242/jcs.222570
Cortes Sanchon, A., Santhosh Kumar, H., Mantovani, M., Osinnii, I., Mateos, J. M., Kaech, A., et al. (2021). ER-misfolded proteins become sequestered with mitochondria and impair mitochondrial function. Commun. Biol. 4, 1350. doi: 10.1038/s42003-021-02873-w
Dang, T. T., and Back, S. H. (2021). Translation inhibitors activate autophagy master regulators TFEB and TFE3. Int. J. Mol. Sci. 22, 12083. doi: 10.3390/ijms222112083
DeVos, S. L., Miller, R. L., Schoch, K. M., Holmes, B. B., Kebodeaux, C. S., Wegener, A. J., et al. (2017). Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 9, eaag0481. doi: 10.1126/scitranslmed.aag0481
Eisenberg, T., Abdellatif, M., Ljubojević-Holzer, S., and Sedej, S. (2022). Chapter 7 - Effects of Physiologic Inputs on Autophagy. 2Edn, eds A. Beverly and A. D. Rothermel. Elsevier.
Feng, Q., Luo, Y., Zhang, X. N., Yang, X. F., Hong, X. Y., Sun, D. S., et al. (2020). MAPT/Tau accumulation represses autophagy flux by disrupting IST1-regulated ESCRT-III complex formation: a vicious cycle in Alzheimer neurodegeneration. Autophagy. 16, 641–658. doi: 10.1080/15548627.2019.1633862
Ferreira-Marques, M., Carvalho, A., Cavadas, C., and Aveleira, C. A. (2021). PI3K/AKT/MTOR and ERK1/2-MAPK signaling pathways are involved in autophagy stimulation induced by caloric restriction or caloric restriction mimetics in cortical neurons. Aging. 13, 7872–7882. doi: 10.18632/aging.202805
Festa, B. P., Barbosa, A. D., Rob, M., and Rubinsztein, D. C. (2021). The pleiotropic roles of autophagy in Alzheimer's disease: from pathophysiology to therapy. Curr. Opin. Pharmacol. 60, 149–157. doi: 10.1016/j.coph.2021.07.011
Fhu, C. W., and Ali, A. (2021). Dysregulation of the ubiquitin proteasome system in human malignancies: a window for therapeutic intervention. Cancers. 13, 1513. doi: 10.3390/cancers13071513
Frumkin, A., Dror, S., Pokrzywa, W., Bar-Lavan, Y., Karady, I., Hoppe, T., et al. (2014). Challenging muscle homeostasis uncovers novel chaperone interactions in Caenorhabditis elegans. Front. Mol. Biosci. 1, 21. doi: 10.3389/fmolb.2014.00021
Gadhave, K., Kumar, D., Uversky, V. N., and Giri, R. (2020). A multitude of signaling pathways associated with Alzheimer's disease and their roles in AD pathogenesis and therapy. Med. Res. Rev. 41, 2689–2745. doi: 10.1002/med.21719
Gcwensa, N. Z., Russell, D. L., Cowell, R. M., and Volpicelli-Daley, L. A. (2021). Molecular mechanisms underlying synaptic and axon degeneration in Parkinson's disease. Front. Cell Neurosci. 15, 626128. doi: 10.3389/fncel.2021.626128
Gelinas, R., Dontaine, J., Horman, S., Beauloye, C., Bultot, L., Bertrand, L., et al. (2018). AMP-activated protein kinase and O-GlcNAcylation, two partners tightly connected to regulate key cellular processes. Front. Endocrinol. 9, e00519. doi: 10.3389/fendo.2018.00519
Gyparaki, M. T., Arab, A., Sorokina, E. M., Santiago-Ruiz, A. N., Bohrer, C. H., Xiao, J., et al. (2021). Tau forms oligomeric complexes on microtubules that are distinct from tau aggregates. Proc. Natl. Acad. Sci. U. S. A. 118, e2021461118. doi: 10.1073/pnas.2021461118
Hampel, H., Hardy, J., Blennow, K., Chen, C., Perry, G., Kim, S. H., et al. (2021). The amyloid-beta pathway in Alzheimer's disease. Mol. Psychiatry. 26, 5481–5503. doi: 10.1038/s41380-021-01249-0
Hohn, A., Tramutola, A., and Cascella, R. (2020). Proteostasis failure in neurodegenerative diseases: focus on oxidative stress. Oxid. Med. Cell Longev. 2020, 5497046. doi: 10.1155/2020/5497046
Hooper, K. M., Jacquin, E., Li, T., Goodwin, J. M., Brumell, J. H., Durgan, J., et al. (2021). V-ATPase is a universal regulator of LC3 associated phagocytosis and non-canonical autophagy. bioRxiv. doi: 10.1101/2021.05.20.444917
Hu, B., Zhou, Y., Sun, D., Yang, Y., Liu, Y., Li, X., et al. (2020). PROTACs: new method to degrade transcription regulating proteins. Eur. J. Med. Chem. 207, 112698. doi: 10.1016/j.ejmech.2020.112698
Hu, Y., Huang, Y., Yi, Y., Wang, H., Liu, B., Yu, J., et al. (2017). Single-cell RNA sequencing highlights transcription activity of autophagy-related genes during hematopoietic stem cell formation in mouse embryos. Autophagy. 13, 770–771. doi: 10.1080/15548627.2016.1278093
Hughes, W. E., Beyer, A. M., and Gutterman, D. D. (2020). Vascular autophagy in health and disease. Basic Res. Cardiol. 115, 41. doi: 10.1007/s00395-020-0802-6
Hyttinen, J., Blasiak, J., Tavi, P., and Kaarniranta, K. (2021). Therapeutic potential of PGC-1alpha in age-related macular degeneration (AMD) - the involvement of mitochondrial quality control, autophagy, and antioxidant response. Expert Opin Ther. Targets. 25, 773–785. doi: 10.1080/14728222.2021.1991913
Izumi, M. (2019). Heat shock proteins support refolding and shredding of misfolded proteins. Plant Physiol. 180, 1777–1778. doi: 10.1104/pp.19.00711
Jeremic, D., Jimenez-Diaz, L., and Navarro-Lopez, J. D. (2021). Past, present and future of therapeutic strategies against amyloid-beta peptides in Alzheimer's disease: a systematic review. Ageing Res. Rev. 72, 101496. doi: 10.1016/j.arr.2021.101496
Jogalekar, M. P., Veerabathini, A., and Gangadaran, P. (2021). Recent developments in autophagy-targeted therapies in cancer. Exp. Biol. Med. 246, 207–212. doi: 10.1177/1535370220966545
Jones, C. L., and Tepe, J. J. (2019). Proteasome activation to combat proteotoxicity. Molecules. 24, 2841. doi: 10.3390/molecules24152841
Karanasios, E. (2019). Correlative live-cell imaging and super-resolution microscopy of autophagy. Methods Mol. Biol. 1880, 231–242. doi: 10.1007/978-1-4939-8873-0_15
Kaushik, S., and Cuervo, A. M. (2018). The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381. doi: 10.1038/s41580-018-0001-6
Kaushik, S., Tasset, I., Arias, E., Pampliega, O., Wong, E., Martinez-Vicente, M., et al. (2021). Autophagy and the hallmarks of aging. Ageing Res. Rev. 72, 101468. doi: 10.1016/j.arr.2021.101468
Ke, P. Y. (2017). Horning cell self-digestion: autophagy wins the 2016. Nobel prize in physiology or medicine. Biomed. J. 40, 5–8. doi: 10.1016/j.bj.2017.03.003
Kocaturk, N. M., and Gozuacik, D. (2018). Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front. Cell Dev. Biol. 6, e00128. doi: 10.3389/fcell.2018.00128
Kong, K. E., Fischer, B., Meurer, M., Kats, I., Li, Z., Ruhle, F., et al. (2021). Timer-based proteomic profiling of the ubiquitin-proteasome system reveals a substrate receptor of the GID ubiquitin ligase. Mol Cell. 81, 2460–76.e11. doi: 10.1016/j.molcel.2021.04.018
Kroemer, G., and White, E. (2010). Autophagy for the avoidance of degenerative, inflammatory, infectious, and neoplastic disease. Curr. Opin. Cell Biol. 22, 121–123. doi: 10.1016/j.ceb.2010.02.003
Lee, J., Giordano, S., and Zhang, J. (2012). Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 441, 523–540. doi: 10.1042/BJ20111451
Lei, L., Wu, Z., and Winklhofer, K. F. (2021). Protein quality control by the proteasome and autophagy: a regulatory role of ubiquitin and liquid-liquid phase separation. Matrix Biol. 100–101, 9–22. doi: 10.1016/j.matbio.2020.11.003
Leveille, E., Ross, O. A., and Gan-Or, Z. (2021). Tau and MAPT genetics in tauopathies and synucleinopathies. Parkinsonism Relat. Disord. 90, 142–154. doi: 10.1016/j.parkreldis.2021.09.008
Li, J., Liu, W., Sun, W., Rao, X., Chen, X., Yu, L. A., et al. (2023). Study on autophagy related biomarkers in Alzheimer's disease based on bioinformatics. Cell Mol. Neurobiol. 43, 3693–3703. doi: 10.1007/s10571-023-01379-9
Lin, M., Yu, H., Xie, Q., Xu, Z., and Shang, P. (2022). Role of microglia autophagy and mitophagy in age-related neurodegenerative diseases. Front. Aging Neurosci. 14, 1100133. doi: 10.3389/fnagi.2022.1100133
Lin, T., Tjernberg, L. O., and Schedin-Weiss, S. (2021). Neuronal trafficking of the amyloid precursor protein-what do we really know? Biomedicines. 9, 801. doi: 10.3390/biomedicines9070801
Lipinski, M. M., Zheng, B., Lu, T., Yan, Z., Py, B. F., Ng, A., et al. (2010). Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 107, 14164–14169. doi: 10.1073/pnas.1009485107
Liu, J., and Li, L. (2019). Targeting autophagy for the treatment of Alzheimer's disease: challenges and opportunities. Front. Mol. Neurosci. 12, 203. doi: 10.3389/fnmol.2019.00203
Liu, L., Lauro, B. M., Wolfe, M. S., and Selkoe, D. J. (2021). Hydrophilic loop 1 of Presenilin-1 and the APP GxxxG transmembrane motif regulate gamma-secretase function in generating Alzheimer-causing Abeta peptides. J. Biol. Chem. 296, 100393. doi: 10.1016/j.jbc.2021.100393
Lloyd, G. M., Trejo-Lopez, J. A., Xia, Y., McFarland, K. N., Lincoln, S. J., Ertekin-Taner, N., et al. (2020). Prominent amyloid plaque pathology and cerebral amyloid angiopathy in APP V717I (London) carrier - phenotypic variability in autosomal dominant Alzheimer's disease. Acta Neuropathol. Commun. 8, 31. doi: 10.1186/s40478-020-0891-3
Long, M., and McWilliams, T. G. (2020). Monitoring autophagy in cancer: from bench to bedside. Semin. Cancer Biol. 66, 12–21. doi: 10.1016/j.semcancer.2019.05.016
Magne, J., and Green, D. R. (2022). LC3-associated endocytosis and the functions of Rubicon and ATG16L1. Sci. Adv. 8, eabo5600. doi: 10.1126/sciadv.abo5600
Manea, A. J., and Ray, S. K. (2021). Regulation of autophagy as a therapeutic option in glioblastoma. Apoptosis. 26, 574–599. doi: 10.1007/s10495-021-01691-z
Mani, S., Swargiary, G., Singh, M., Agarwal, S., Dey, A., Ojha, S., et al. (2021). Mitochondrial defects: an emerging theranostic avenue towards Alzheimer's associated dysregulations. Life Sci. 285, 119985. doi: 10.1016/j.lfs.2021.119985
Markaki, M., and Tavernarakis, N. (2020). Autophagy mechanisms and roles: recent advances and implications. FEBS J. 287, 5024–5026. doi: 10.1111/febs.15573
Martinez-Vicente, M., Talloczy, Z., Wong, E., Tang, G., Koga, H., Kaushik, S., et al. (2010). Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 13, 567–576. doi: 10.1038/nn.2528
Metur, S. P., and Klionsky, D. J. (2021). Autophagy under construction: insights from in vitro reconstitution of autophagosome nucleation. Autophagy. 17, 383–384. doi: 10.1080/15548627.2020.1835231
Mijaljica, D. (2010). Autophagy in 2020 and beyond: eating our way into a healthy future. Autophagy. 6, 194–196. doi: 10.4161/auto.6.1.10992
Mizushima, N. (2020). The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 63, 1–10. doi: 10.1016/j.ceb.2019.12.001
Montibeller, L., and de Belleroche, J. (2018). Amyotrophic lateral sclerosis (ALS) and Alzheimer's disease (AD) are characterised by differential activation of ER stress pathways: focus on UPR target genes. Cell Stress Chaperones. 23, 897–912. doi: 10.1007/s12192-018-0897-y
Morato, X., Pytel, V., Jofresa, S., Ruiz, A., and Boada, M. (2022). Symptomatic and disease-modifying therapy pipeline for Alzheimer's disease: towards a personalized polypharmacology patient-centered approach. Int. J. Mol. Sci. 23, 9305. doi: 10.3390/ijms23169305
Morimoto, R. I. (2020). Cell-nonautonomous regulation of proteostasis in aging and disease. Cold Spring Harb. Perspect Biol. 12, ea034074. doi: 10.1101/cshperspect.a034074
Morris, G., and Berk, M. (2016). The putative use of lithium in Alzheimer's disease. Curr. Alzheimer Res. 13, 853–861. doi: 10.2174/1567205013666160219113112
Mufson, E. J., Ikonomovic, M. D., Counts, S. E., Perez, S. E., Malek-Ahmadi, M., Scheff, S. W., et al. (2016). Molecular and cellular pathophysiology of preclinical Alzheimer's disease. Behav. Brain Res. 311, 54–69. doi: 10.1016/j.bbr.2016.05.030
Muller, L., Power Guerra, N., Stenzel, J., Ruhlmann, C., Lindner, T., Krause, B. J., et al. (2021). Long-term caloric restriction attenuates beta-amyloid neuropathology and is accompanied by autophagy in APPswe/PS1delta9 mice. Nutrients. 13, 985. doi: 10.3390/nu13030985
Munoz-Moreno, E., Tudela, R., Lopez-Gil, X., and Soria, G. (2020). Brain connectivity during Alzheimer's disease progression and its cognitive impact in a transgenic rat model. Netw. Neurosci. 4, 397–415. doi: 10.1162/netn_a_00126
Navone, F., Genevini, P., and Borgese, N. (2015). Autophagy and neurodegeneration: insights from a cultured cell model of ALS. Cells. 4, 354–386. doi: 10.3390/cells4030354
Nieto-Torres, J. L., and Hansen, M. (2021). Macroautophagy and aging: the impact of cellular recycling on health and longevity. Mol. Aspects Med. 82, 101020. doi: 10.1016/j.mam.2021.101020
Nistico, R., and Borg, J. J. (2021). Aducanumab for Alzheimer's disease: a regulatory perspective. Pharmacol. Res. 171, 105754. doi: 10.1016/j.phrs.2021.105754
Njomen, E., and Tepe, J. J. (2019). Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J. Med. Chem. 62, 6469–6481. doi: 10.1021/acs.jmedchem.9b00101
Nobili, A., D'Amelio, M., and Viscomi, M. T. (2023). Nilotinib: from animal-based studies to clinical investigation in Alzheimer's disease patients. Neural Regen. Res. 18, 803–804. doi: 10.4103/1673-5374.350700
Ntsapi, C. M., and Loos, B. (2021). Neurons die with heightened but functional macro- and chaperone mediated autophagy upon increased amyloid-ss induced toxicity with region-specific protection in prolonged intermittent fasting. Exp. Cell Res. 408, 112840. doi: 10.1016/j.yexcr.2021.112840
Oblak, A. L., Lin, P. B., Kotredes, K. P., Pandey, R. S., Garceau, D., Williams, H. M., et al. (2021). Comprehensive evaluation of the 5XFAD mouse model for preclinical testing applications: a MODEL-AD study. Front. Aging Neurosci. 13, 713726. doi: 10.3389/fnagi.2021.713726
Orr, M. E., and Oddo, S. (2013). Autophagic/lysosomal dysfunction in Alzheimer's disease. Alzheimers Res. Ther. 5, 53. doi: 10.1186/alzrt217
Pani, L., and Keefe, R. S. E. (2019). Approaches to attenuated psychosis syndrome treatments: a perspective on the regulatory issues. Schizophr. Res. Cogn. 18, 100155. doi: 10.1016/j.scog.2019.100155
Park, J. S., Kam, T. I., Lee, S., Park, H., Oh, Y., Kwon, S. H., et al. (2021). Blocking microglial activation of reactive astrocytes is neuroprotective in models of Alzheimer's disease. Acta Neuropathol. Commun. 9, 78. doi: 10.1186/s40478-021-01180-z
Patil, S. P., Maki, S., Khedkar, S. A., Rigby, A. C., and Chan, C. (2010). Withanolide A and asiatic acid modulate multiple targets associated with amyloid-beta precursor protein processing and amyloid-beta protein clearance. J. Nat. Prod. 73, 1196–1202. doi: 10.1021/np900633j
Perez-Leal, O., Nixon-Abell, J., Barrero, C. A., Gordon, J. C., Oesterling, J., Rico, M. C., et al. (2021). Multiplex gene tagging with CRISPR-Cas9 for live-cell microscopy and application to study the role of SARS-CoV-2 proteins in autophagy, mitochondrial dynamics, and cell growth. CRISPR J. 4, 854–871. doi: 10.1089/crispr.2021.0041
Pickford, F., Masliah, E., Britschgi, M., Lucin, K., Narasimhan, R., Jaeger, P. A., et al. (2008). The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J. Clin. Invest. 118, 2190–2199. doi: 10.1172/JCI33585
Pimenova, A. A., and Goate, A. M. (2020). Novel presenilin 1 and 2 double knock-out cell line for in vitro validation of PSEN1 and PSEN2 mutations. Neurobiol. Dis. 138, 104785. doi: 10.1016/j.nbd.2020.104785
Pomilio, C., Gorojod, R. M., Riudavets, M., Vinuesa, A., Presa, J., Gregosa, A., et al. (2020). Microglial autophagy is impaired by prolonged exposure to beta-amyloid peptides: evidence from experimental models and Alzheimer's disease patients. Geroscience. 42, 613–632. doi: 10.1007/s11357-020-00161-9
Powers, E. T., and Gierasch, L. M. (2021). The proteome folding problem and cellular proteostasis. J. Mol. Biol. 433, 167197. doi: 10.1016/j.jmb.2021.167197
Qi, L. F., Liu, S., Liu, Y. C., Li, P., and Xu, X. (2021). Ganoderic acid A promotes amyloid-beta clearance (in vitro) and ameliorates cognitive deficiency in Alzheimer's disease (mouse model) through autophagy induced by activating Axl. Int. J. Mol. Sci. 22, 5559. doi: 10.3390/ijms22115559
Qin, C., Bai, L., Li, Y., and Wang, K. (2022). The functional mechanism of bone marrow-derived mesenchymal stem cells in the treatment of animal models with Alzheimer's disease: crosstalk between autophagy and apoptosis. Stem Cell Res. Ther. 13, 90. doi: 10.1186/s13287-022-02765-8
Ramaian Santhaseela, A., and Jayavelu, T. (2021). Does mTORC1 inhibit autophagy at dual stages? A possible role of mTORC1 in late-stage autophagy inhibition in addition to its known early-stage autophagy inhibition. Bioessays. 43, e2000187. doi: 10.1002/bies.202000187
Runwal, G., Stamatakou, E., Siddiqi, F. H., Puri, C., Zhu, Y., Rubinsztein, D. C., et al. (2019). LC3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 9, 10147. doi: 10.1038/s41598-019-46657-z
Ruz, C., Alcantud, J. L., Vives Montero, F., Duran, R., and Bandres-Ciga, S. (2020). Proteotoxicity and neurodegenerative diseases. Int. J. Mol. Sci. 21, 646. doi: 10.3390/ijms21165646
Saibil, H. R. (2021). The PDB and protein homeostasis: from chaperones to degradation and disaggregase machines. J. Biol. Chem. 296, 100744. doi: 10.1016/j.jbc.2021.100744
Sarkar, A., and Nazir, A. (2022). Carrying excess baggage can slowdown life: protein clearance machineries that go awry during aging and the relevance of maintaining them. Mol. Neurobiol. 59, 821–840. doi: 10.1007/s12035-021-02640-2
Sayed, F. A., Kodama, L., Fan, L., Carling, G. K., Udeochu, J. C., Le, D., et al. (2021). AD-linked R47H-TREM2 mutation induces disease-enhancing microglial states via AKT hyperactivation. Sci. Transl. Med. 13, eabe3947. doi: 10.1126/scitranslmed.abe3947
Schaefer, L., and Dikic, I. (2021). Autophagy: instructions from the extracellular matrix. Matrix Biol. 100–101, 1–8. doi: 10.1016/j.matbio.2021.06.002
Schuck, S. (2020). Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci. 133, e246322. doi: 10.1242/jcs.246322
Sebastian, R. M., and Shoulders, M. D. (2020). Chemical biology framework to illuminate proteostasis. Ann. Rev. Biochem. 89, 529–555. doi: 10.1146/annurev-biochem-013118-111552
Senos Demarco, R., and Jones, D. L. (2020). EGFR signaling promotes basal autophagy for lipid homeostasis and somatic stem cell maintenance in the Drosophila testis. Autophagy. 16, 1145–1147. doi: 10.1080/15548627.2020.1739450
Seo, J., Giusti-Rodriguez, P., Zhou, Y., Rudenko, A., Cho, S., Ota, K. T., et al. (2014). Activity-dependent p25 generation regulates synaptic plasticity and Abeta-induced cognitive impairment. Cell. 157, 486–498. doi: 10.1016/j.cell.2014.01.065
Shimobayashi, M., Oppliger, W., Moes, S., Jeno, P., and Hall, M. N. (2013). TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell. 24, 870–881. doi: 10.1091/mbc.e12-10-0753
Sitta, R. (2018). Novel drug targeting and delivery techniques: avenues for the advancement of neuropharmacology. Brain Circ. 4, 79–80. doi: 10.4103/bc.bc_10_18
Steiner, A., Schlepckow, K., Brunner, B., Steiner, H., Haass, C., Hagn, F., et al. (2020). gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics. EMBO J. 39, e104247. doi: 10.15252/embj.2019104247
Subramanian, A., Tamilanban, T., Alsayari, A., Ramachawolran, G., Wong, L. S., Sekar, M., et al. (2022). Trilateral association of autophagy, mTOR and Alzheimer's disease: potential pathway in the development for Alzheimer's disease therapy. Front. Pharmacol. 13, 1094351. doi: 10.3389/fphar.2022.1094351
Suenaga, H., Kagaya, N., Kawada, M., Tatsuda, D., Sato, T., Shin-Ya, K., et al. (2021). Phenotypic screening system using three-dimensional (3D) culture models for natural product screening. J. Antibiot. 74, 660–666. doi: 10.1038/s41429-021-00457-8
Tambini, M. D., Yao, W., and D'Adamio, L. (2019). Facilitation of glutamate, but not GABA, release in Familial Alzheimer's APP mutant Knock-in rats with increased beta-cleavage of APP. Aging Cell. 18, e13033. doi: 10.1111/acel.13033
Tooze, S. A., Hannan, L. A., Marks, M. S., Stevens, T. H., and Schroer, T. A. (2017). Fundamental mechanisms deliver the Nobel Prize to Ohsumi. Traffic. 18, 93–95. doi: 10.1111/tra.12460
Uddin, M. N., Elahi, M., Shimonaka, S., Kakuta, S., Ishiguro, K., Motoi, Y., et al. (2021). Strain-specific clearance of seed-dependent tau aggregation by lithium-induced autophagy. Biochem. Biophys. Res. Commun. 543, 65–71. doi: 10.1016/j.bbrc.2020.12.113
Vandenberghe, R., Riviere, M. E., Caputo, A., Sovago, J., Maguire, R. P., Farlow, M., et al. (2017). Active abeta immunotherapy CAD106 in Alzheimer's disease: a phase 2b study. Alzheimers Dement. 3, 10–22. doi: 10.1016/j.trci.2016.12.003
Venkatramani, A., and Panda, D. (2019). Regulation of neuronal microtubule dynamics by tau: implications for tauopathies. Int. J. Biol. Macromol. 133, 473–483. doi: 10.1016/j.ijbiomac.2019.04.120
Vujic, N., Bradic, I., Goeritzer, M., Kuentzel, K. B., Rainer, S., Kratky, D., et al. (2021). ATG7 is dispensable for LC3-PE conjugation in thioglycolate-elicited mouse peritoneal macrophages. Autophagy. 17, 3402–3407. doi: 10.1080/15548627.2021.1874132
Wang, D., Huang, X., Yan, L., Zhou, L., Yan, C., Wu, J., et al. (2021). The structure biology of Tau and Clue for aggregation inhibitor design. Protein J. 40, 656–668. doi: 10.1007/s10930-021-10017-6
Wang, G., Xue, Y., Wang, Y., Dong, F., Shen, M., Zong, R., et al. (2019). The role of autophagy in the pathogenesis of exposure keratitis. J. Cell Mol. Med. 23, 4217–4228. doi: 10.1111/jcmm.14310
Wang, J., Zhou, P., Wang, X., Yu, Y., Zhu, G., Zheng, L., et al. (2019). Rab25 promotes erlotinib resistance by activating the beta1 integrin/AKT/beta-catenin pathway in NSCLC. Cell Prolif. 52, e12592. doi: 10.1111/cpr.12592
Wang, L., Klionsky, D. J., and Shen, H.-. M. (2022). The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 24, 186–203. doi: 10.1038/s41580-022-00529-z
Wang, Y. Y., Sun, Y. P., Luo, Y. M., Peng, D. H., Li, X., Yang, B. Y., et al. (2021). Biomarkers for the clinical diagnosis of Alzheimer's disease: metabolomics analysis of brain tissue and blood. Front. Pharmacol. 12, 700587. doi: 10.3389/fphar.2021.700587
Wang, Z., Wang, Q., Li, S., Li, X. J., Yang, W., He, D., et al. (2022). Microglial autophagy in Alzheimer's disease and Parkinson's disease. Front. Aging Neurosci. 14, 1065183. doi: 10.3389/fnagi.2022.1065183
Wu, S., Wei, Y., Li, J., Bai, Y., Yin, P., Wang, S., et al. (2021). SIRT5 represses neurotrophic pathways and abeta production in Alzheimer's disease by targeting autophagy. ACS Chem. Neurosci. 12, 4428–4437. doi: 10.1021/acschemneuro.1c00468
Xu, S., Yang, P., Qian, K., Li, Y., Guo, Q., Wang, P., et al. (2022). Modulating autophagic flux via ROS-responsive targeted micelles to restore neuronal proteostasis in Alzheimer's disease. Bioact. Mater. 11, 300–316. doi: 10.1016/j.bioactmat.2021.09.017
Yang, M., Wang, Y., Liang, G., Xu, Z., Chu, C. T., Wei, H., et al. (2019). Alzheimer's disease presenilin-1 mutation sensitizes neurons to impaired autophagy flux and propofol neurotoxicity: role of calcium dysregulation. J. Alzheimers Dis. 67, 137–147. doi: 10.3233/JAD-180858
Yang, Y., and Zhang, L. (2020). The effects of caloric restriction and its mimetics in Alzheimer's disease through autophagy pathways. Food Funct. 11, 1211–1224. doi: 10.1039/C9FO02611H
Zaarour, R. F., Azakir, B., Hajam, E. Y., Nawafleh, H., Zeinelabdin, N. A., Engelsen, A. S. T., et al. (2021). Role of hypoxia-mediated autophagy in tumor cell death and survival. Cancers. 13, 533. doi: 10.3390/cancers13030533
Zhang, Z., Yang, X., Song, Y. Q., and Tu, J. (2021). Autophagy in Alzheimer's disease pathogenesis: therapeutic potential and future perspectives. Ageing Res. Rev. 72, 101464. doi: 10.1016/j.arr.2021.101464
Zhao, Y., Zhang, Y., Zhang, J., Zhang, X., and Yang, G. (2020). Molecular mechanism of autophagy: its role in the therapy of Alzheimer's disease. Curr. Neuropharmacol. 18, 720–739. doi: 10.2174/1570159X18666200114163636
Zheng, Q., Huang, T., Zhang, L., Zhou, Y., Luo, H., Xu, H., et al. (2016). Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front. Aging Neurosci. 8, 303. doi: 10.3389/fnagi.2016.00303
Zheng, Y., Zhang, X., and Chen, Z. (2021). Monitoring autophagy by optical microscopy. Adv. Exp. Med. Biol. 1208, 117–130. doi: 10.1007/978-981-16-2830-6_8
Keywords: Alzheimer's disease, proteostasis, autophagy, neurodegeneration, oxidative stress
Citation: Barmaki H, Nourazarian A and Khaki-Khatibi F (2023) Proteostasis and neurodegeneration: a closer look at autophagy in Alzheimer's disease. Front. Aging Neurosci. 15:1281338. doi: 10.3389/fnagi.2023.1281338
Received: 23 August 2023; Accepted: 11 October 2023;
Published: 02 November 2023.
Edited by:
Shibin Cheng, Brown University, United StatesReviewed by:
Vassiliki Nikoletopoulou, Foundation for Research and Technology Hellas (FORTH), GreeceAriadna Recasens, Monash University, Australia
Copyright © 2023 Barmaki, Nourazarian and Khaki-Khatibi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alireza Nourazarian, bm9vcmF6YXJpYW5fYUBraG95dW1zLmFjLmly; Fatemeh Khaki-Khatibi, ZmF0ZW1laGtoYWtpa2hhdGliaUB5YWhvby5jb20=
 Haleh Barmaki1
Haleh Barmaki1 Alireza Nourazarian
Alireza Nourazarian Fatemeh Khaki-Khatibi
Fatemeh Khaki-Khatibi