
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Aging Neurosci., 13 December 2022
Sec. Cellular and Molecular Mechanisms of Brain-aging
Volume 14 - 2022 | https://doi.org/10.3389/fnagi.2022.1048333
 Nusrat Begum1
Nusrat Begum1 Aniket Mandhare1
Aniket Mandhare1 Kamatham Pushpa Tryphena1
Kamatham Pushpa Tryphena1 Saurabh Srivastava2*
Saurabh Srivastava2* Mohd Farooq Shaikh3*
Mohd Farooq Shaikh3* Shashi Bala Singh1
Shashi Bala Singh1 Dharmendra Kumar Khatri1*
Dharmendra Kumar Khatri1*Gut-brain axis is a dynamic, complex, and bidirectional communication network between the gut and brain. Changes in the microbiota-gut-brain axis are responsible for developing various metabolic, neurodegenerative, and neuropsychiatric disorders. According to clinical and preclinical findings, the gut microbiota is a significant regulator of the gut-brain axis. In addition to interacting with intestinal cells and the enteric nervous system, it has been discovered that microbes in the gut can modify the central nervous system through metabolic and neuroendocrine pathways. The metabolites of the gut microbiome can modulate a number of diseases by inducing epigenetic alteration through DNA methylation, histone modification, and non-coding RNA-associated gene silencing. Short-chain fatty acids, especially butyrate, are well-known histone deacetylases inhibitors. Similarly, other microbial metabolites such as folate, choline, and trimethylamine-N-oxide also regulate epigenetics mechanisms. Furthermore, various studies have revealed the potential role of microbiome dysbiosis and epigenetics in the pathophysiology of depression. Hence, in this review, we have highlighted the role of gut dysbiosis in epigenetic regulation, causal interaction between host epigenetic modification and the gut microbiome in depression and suggest microbiome and epigenome as a possible target for diagnosis, prevention, and treatment of depression.
Microbiota study has advanced dramatically over the past 20 years, and it is now becoming more evident how these microscopic inhabitants affect our daily lives in many ways. The microbiota plays a significant role in determining human health and disease and controlling host physiology. Several international projects such as Human Microbiome Project, Metagenomics of the Human Intestinal Tract, International Human Microbiome Consortium have characterized the human microbiota (Bäckhed et al., 2012; Huttenhower et al., 2012) which includes various microbiome such as gut microbiome, oral microbiome, vaginal microbiome, skin and placental microbiome. The gut microbiota has abundance of two bacterial phyla, the gram negative Bacteroidetes and the gram positive Firmicutes; Actinobacteria, Fusobacteria, and Verrucomicrobia levels are comparatively lower and varies remarkably among individuals (Arumugam et al., 2011; Bäckhed et al., 2012). The vaginal microbiome consists of over 200 phyla of which Firmicutes, Bacteroides, Actinobacteria and Fusobacteria are the predominant phyla (Romero et al., 2014). Lactobacillus sp. plays a major role in maintaining the acidic pH of vagina by secretion of lactic acid and hydrogen peroxide, failure to do so results in bacterial vaginosis, an ecological disorder of vaginal microbiota (Kenyon et al., 2013). The oral microbiome comprises of bacteria, fungi, viruses, protozoa, and archaea. It includes more than 20 bacterial phyla spread over more than 300 genera (Zhou et al., 2013; Jiang et al., 2014). Skin microbiome comprises more than 100 microbial phylotypes, both harmless or beneficial for host, which differ in their abundance and diversity depending upon race, skin color, and geographic location (Rosenthal et al., 2011; Ladizinski et al., 2014). The placental microbiome is composed of non-pathogenic commensal bacteria derived from phylum Firmicutes, Proteobacteria, Bacteroidetes, and Fusobacteria (Aagaard et al., 2014). The population of placental microbiota differ in preterm (gram negative Burkholderia) and normal delivery (gram positive Paenibacillus) (Groer et al., 2014).
Various environmental factors influence the composition of gut microbiota. They include macro environmental factors like socio economic, chemical, built environment and micro environmental factors like smoking, alcohol consumption, dietary habits (Ahn and Hayes, 2021). Many studies provide evidence that chemical substances like arsenic, triclocarban, triclosan upon being metabolized by gut microbiota in turn cause gut dysbiosis (Rook et al., 2014; Halden, 2016). The association between gut microbiota and built environment (infrastructures built by humans) is an emerging hypothesis and yet to be explored further (Ahn and Hayes, 2021). Low socioeconomic status, psychosocial stress, sedentary lifestyle also influences the gut microbiota as evidenced in studies conducted in some countries (Lin et al., 2013). Similarly, studies found that smokers had decreased diversity of gut microbiome. The studies demonstrate that Phyla Proteobacteria, Bacteroidetes and genera Clostridium, Bacteroides, Prevotella were increased and Actinobacteria, Firmicutes phyla were decreased (Savin et al., 2018). Chronic Alcohol consumption was found to alter the gut microbiota resulting in decreased Bacteroidetes and increased Proteobacteria (Mutlu et al., 2012). Finally, dietary habits like consumption of high fat, sugar, protein, and fiber intake also affect the gut microbiota composition. High fiber diet was proved to have beneficial effect on gut microbiome by speeding up the proliferation of microbiota as well as increase the diversity (Donohoe et al., 2014). A study conducted in Africa showed that children exposed to antibiotics and indoor cooking fires had decreased diversity and dysbiosis of gut microbiota (Nel Van Zyl et al., 2021). Food additives, organic pesticides, heavy metals also cause gut dysbiosis (Jin et al., 2017). Recent studies have emphasized the role of the microbiota in average intestinal growth and function, including digestion and nutrition intake, metabolism, tissue formation, and immunity (Hooper et al., 2012). Furthermore, alterations in the microbiota’s composition or abundance have been linked to several chronic human illnesses, including local diseases like inflammatory bowel disease (IBD) (Nagao-Kitamoto et al., 2016), metabolic diseases like obesity and diabetes (Napolitano and Covasa, 2020), cancer (Vivarelli et al., 2019) and neuropsychiatric disorders like autism (Iglesias–vázquez et al., 2020), schizophrenia (Szeligowski et al., 2020) or depression (Liu et al., 2021; Figure 1).
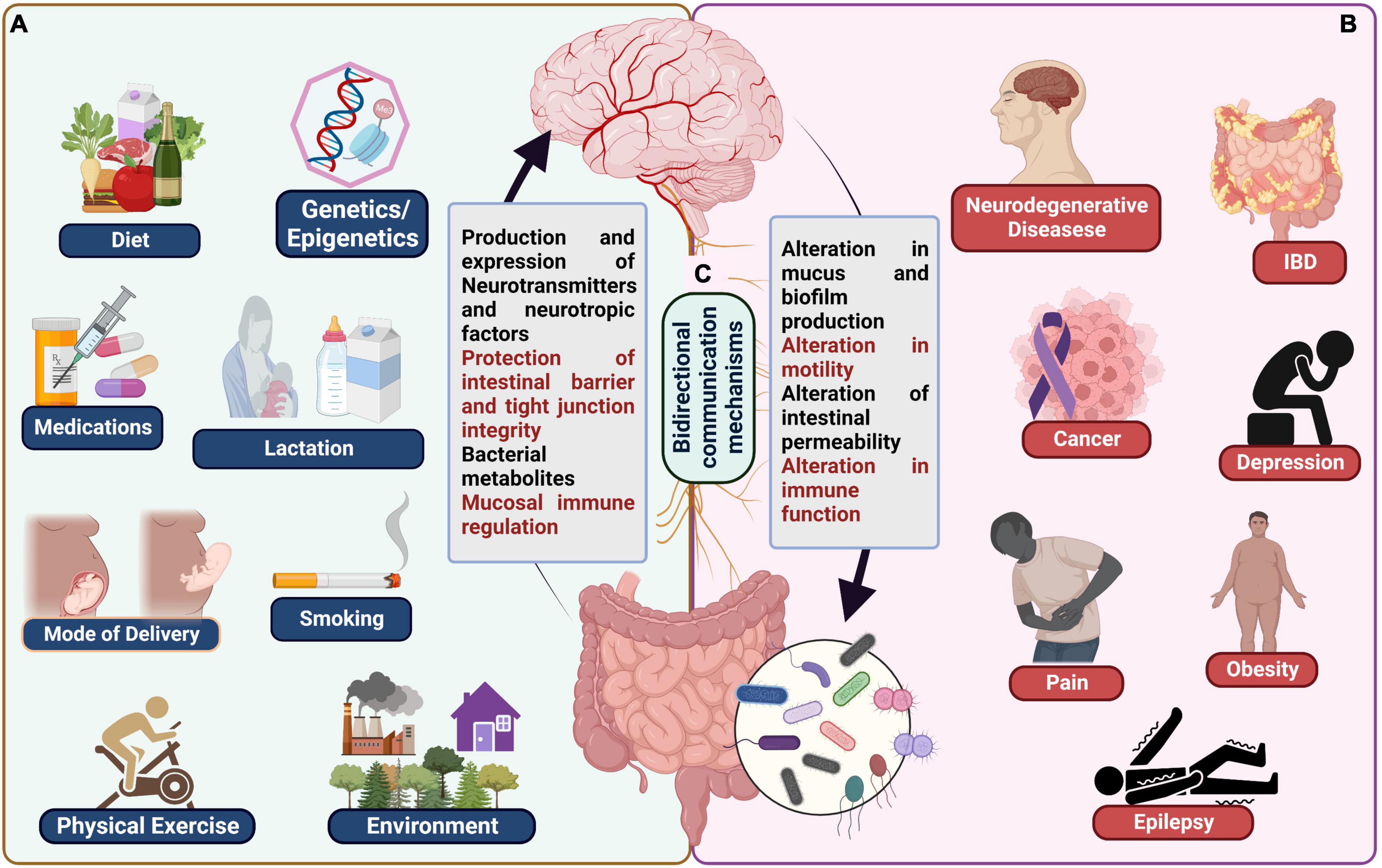
Figure 1. (A) Examples of typical influences on the microbiota gut-brain axis including genetics/epigenetics, medications, mode of delivery, lactation, smoking, physical exercise, and environment. (B) Various diseases known to be affected by gut brain axis dysbiosis, including neurodegenerative diseases, cancer, IBD, depression, obesity, pain, and epilepsy. (C) Bidirectional communication mechanisms between the gut and the brain.
Depression ranks second in the global disease burden ranking (Smith, 2014). Its heritability is only 37% (Sullivan et al., 2000) which is far less than other psychiatric diseases, e.g., Schizophrenia and bipolar disorders which account for 70–80% (Kendler, 1983). Previous research revealed an approximately 50% variability rate of depression in monozygotic twins, which indicates the involvement of various other non-genetic factors in the pathogenesis of depression (Fraga et al., 2005). Identification of gene loci using human genome-wide association studies failed to give reproducible results (Bosker et al., 2011). The factors involved in resilience and susceptibility to depression include variability penetrant, genetic differences, and environmental factors (Sun et al., 2013). Environmental factors such as lifestyle, diet, physical exercise, and stress can affect gut microbiome composition and epigenetic alterations. Various studies have provided insight into how the metabolites produced by the gut microbiome act as an epigenetic regulator by modifying epigenetic mechanisms such as DNA methylation (Kovacheva et al., 2007; Kok et al., 2015), histone modifications (Soliman et al., 2011; Krautkramer et al., 2016; Wang et al., 2019), or non-coding RNA associated silencing (Liang et al., 2015; Paul et al., 2015; Virtue et al., 2019). Despite the tremendous research, there are still significant gaps in our knowledge of how epigenetic mechanisms contribute to depression. Understanding of the pathological mechanisms of depression will advance with the closure of knowledge gaps on the multifactorial interaction between epigenetics, gut microbiome, and their antidepressant effects, which may also help in the development of more sophisticated pharmacological approaches. Thus, in this review, we have tried to establish a connection between gut dysbiosis, and epigenetics to suggest them as a possible target for diagnosis and treatment of depression.
The gut-brain axis (GBA) is a complex bidirectional network between gut microbiota and the brain (Iannone et al., 2019). It consists of several immunological, endocrinological, and neural mediators. Gut microbiota is a collection of all microbial strains present in a gut. Microbiota development is host-specific and develops fully in the first 3 years of life. Microbiota composition varies from individual to individual and is generally based on environmental factors, mode of delivery, food intake, and disease condition (Burokas et al., 2015). Intestinal microbiota, central nervous system (CNS), and enteric nervous system (ENS) mediate the GBA (Ambrosini et al., 2019). More than 75% of gut microbiota comprises Bacteroidetes and Firmicutes species. Depending upon the bacterial strain in the intestine, this microbiota can regulate various brain functions, such as alteration of memory, increased stress response, and anxiety-like behavior (Evrensel et al., 2020). The composition of gut microbiota differs from individual to individual. Changes in the balance of common gut microbiota affect the production of fermentation products of bacteria such as short-chain fatty acids (SCFAs), acetic acid, propionic acid, and butyric acid, which play an essential role in CNS function and regulate intestinal adaptive response (de Vadder et al., 2014).
The effect of microbiota on physiology and disease can be clearly understood by utilizing germ-free (free of all microorganisms; obtained via c-section and raised in sterile isolators) and Specific Pathogen Free (SPF) (free of specific disease-causing pathogens capable of altering mouse health and research outcome) mice. Experimental studies in Germ-free (GF) mice showed that gut microbiota is required to develop the hypothalamus pituitary adrenal (HPA) axis. GF mice showed increased levels of adrenocorticotropic hormone (ACTH) and corticosterone when exposed to stress conditions compared to normal and SPF mice. Studies also showed decreased BDNF expression levels in the cortical and hypothalamic regions of the brain in GF mice compared to SPF mice (Sudo et al., 2004).
The gastrointestinal tract of human beings contains 100 trillion different types of micro-organisms, which include bacteria, viruses, and yeast (Shi et al., 2017; Rinninella et al., 2019). Nearly 1,000–1,500 bacterial species are included in the gut microbiota, but only 160 species are present in any individual (Shi et al., 2017; Rowland et al., 2018). This shows that the microbiota between two individuals is different (Shi et al., 2017). The dominant phyla of bacteria in the gut are Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, and Verrucomicrobia. Firmicutes and Bacteroidetes constitute 90% of the gut microbiota. Lactobacillus, Bacillus, Clostridium, Enterococcus, and Ruminococcus constitute 95% of the Firmicutes, whereas Bacteroides and Prevotella are dominant among Bacteroidetes (Rinninella et al., 2019). Initially it was thought that the colonization of microbiota takes place during delivery and after birth. But with the advent of several molecular techniques like fluorescent in situ hybridization, Polymerase chain reaction etc., it has been revealed that the fetus encounters the commensal as well as pathogenic bacteria through umbilical cord blood, amniotic fluid, placenta, and fetal membrane (Stinson et al., 2016). It has been demonstrated that this encounter is necessary for the development of gastrointestinal tract in the fetus through the use of GF animal models (Chowdhury et al., 2007; Le Hurou-Luron et al., 2010). In some studies, bacteria of the adult gastrointestinal tract like E. coli, Enterococcus sp. had been found in the placenta, meconium and amniotic fluid respectively (DiGiulio et al., 2008; Aagaard et al., 2014). Proteobacteria were predominantly found in the amniotic fluid (Doyle et al., 2017). Duration of pregnancy also effects the composition of the neonates gut microbiota. Clostridium sp., E. coli, Enterococcus, Streptococcus, Staphylococcus, Klebsiella were predominantly found in the gut of premature infants. Maternal health, dietary habits, antibiotic intake etc influence the fetal gut microbiota composition (Hill et al., 2017). But results from some other studies prove the opposite (Kim et al., 2009; Boro et al., 2014). Hence it is still doubtful to conclude about the prenatal colonization and composition of gut microbiota. More studies with appropriate experimental design coupled with advanced analyzing techniques in the future might provide clear insights about this controversy. According to Gosalbes et al. (2013), two types of meconium microbiota are present, a less diverse Enterobacteriaceae predominant microbiota and a highly diverse Firmicutes predominant microbiota (Tanaka and Nakayama, 2017). Extensive colonization starts at birth during the delivery and breastfeeding. Gestational age, mode of delivery, feeding method, sanitation, and antibiotic treatment are some factors that influence the colonization of the infant’s gut. The diversity in the gut microbiota of neonates is low. Proteobacteria and Actinobacteria are the predominant phyla in the gut microbiota of neonates. Initially, the facultative anaerobes colonized and pave the way for the colonization of strict anaerobes. Mother’s milk is another primary source of microbiota which includes streptococci and staphylococci (Rodríguez et al., 2015). The breastfed infant’s gut microbiota consists of a more significant number of Bifidobacterium species that live on the Human Milk Oligosaccharides (HMO). The diversity of the gut increases after weaning and commencement of solid food (Rodríguez et al., 2015; Milani et al., 2017). Bacteroidetes and Firmicutes (Clostridia) help break down complex carbohydrates that predominate the gut microbiota after the introduction of solid food (Derrien et al., 2019). The child’s oral bacteria also colonize the gut (Rodríguez et al., 2015). The diversity of microbiota progresses with age (Rodríguez et al., 2015; Rinninella et al., 2019). Initially, it was thought that, as the child reaches age three, the composition and diversity of the gut microbiota becomes identical to that of the adult (Cresci and Bawden, 2015; Rinninella et al., 2019). But many studies suggest that gut microbiota composition continues to develop even after 3 years of age (Derrien et al., 2019). Bifidobacterium is present at higher levels in children than in adults (Derrien et al., 2019). It remains almost constant until the person ages (Cresci and Bawden, 2015). Firmicutes, Bacteroidetes, and Actinobacteria dominate the adult gut microbiota. As age progresses, the microbiota composition is altered due to a decrease in dentition, reduced salivary function and digestion, and alteration in intestinal transit time (Cresci and Bawden, 2015). After 70 years of age, it has been observed that anaerobic bacteria like Bifidobacterium decreases whereas Proteobacteria and Clostridium increase in abundance (Rinninella et al., 2019). Other than bacteria, methanogens of the archeal order, like Methanobrevibacter smithii and Methanosphaera stadtmanae, are also present in the adult gut microbiota (Derrien et al., 2019). Fungal cells are also present in gut microbiota but in less number than the bacteria. Fungi, like Aspergillus species, tremellomycetes are more predominant in children than in adults (Derrien et al., 2019).
The gut microbiota composition varies between individuals and depends on the factors like enterotypes, body mass index, lifestyle, ethnicity, dietary and cultural habits, exercise frequency (Rinninella et al., 2019), genetics (Wu et al., 2011; Rodríguez et al., 2015; Shi et al., 2017), stress, antibiotic use (Cresci and Bawden, 2015), geography (Derrien et al., 2019). Enterotypes are the specific clusters of bacteria that characterize the individual’s gut microbiota. There are three enterotypes: enterotype I, in which Bacteroides is the dominant cluster, Prevotella in enterotype II, and Ruminococcus in enterotype III. Each enterotype generates energy from the fermentable substrates present in the colon in a specific way (Rinninella et al., 2019) and is closely linked to long-term dietary patterns (Wu et al., 2011). Consumption of a high-fiber diet is associated with increased microbial diversity and stability (Derrien et al., 2019).
There is a mutually beneficial symbiotic relationship between the host and the gut microbiota (Shi et al., 2017). The microbiome, which is the collection of the microbiota genomes, harbors different genes that encode different types of proteins that are not encoded by the human genome (Kho and Lal, 2018). Gut bacteria play an essential role in the digestion of food, absorption of nutrients, and production of metabolites like SCFAs, lipids, vitamins, bile acids, branched-chain amino acids, tryptophan, and indole derivatives (Rinninella et al., 2019; Agus et al., 2021;Table 1). Gut microbiota also maintains the integrity of the intestinal epithelium and prevents bacterial invasion and pathogenic colonization in the gut (Rinninella et al., 2019).
Short chain fatty acid are produce in colon by gut microbiota as carbon and energy source through fermentation of complex and indigestible carbohydrates (Hugenholtz et al., 2013). Upon microbial hydrolysis of carbohydrate, pyruvate (a key precursor for SCFA) is produced by glycolytic pathway for hexoses and pentoses (Hugenholtz et al., 2013). The pathway for butyrate and propionate production are substrate specific and more conserved whereas pathways for acetate production are commonly spread among various bacterial classes (Morrison and Preston, 2016). Bacteroidetes and Negativicutes uses succinate pathway whereas Lachnospiraceae uses propanediol pathway for propionate production (Louis and Flint, 2017). The butyrate production is mediated by enzymes butyrate kinase (Coprococcus comes and Coprococcus eutactus) and butyryl coA-acetate coA transferases (Faecalibacterium prausnitzii, Eubacterium rectale, and Eubacterium hallii) (Venegas et al., 2019). The acetate, butyrate and propionate contribute 60, 20, and 20% respectively in total SCFA (Cummings et al., 1987). SCFAs are essential in maintaining the intestinal epithelium’s integrity by regulating the tight junction proteins. They also preserve glucose homeostasis through Free Fatty Acid Receptors 2/3 (FFAR2/3). Butyrate and acetate are lipogenic, whereas propionate is gluconeogenic in nature. They also play a role in gut-hormone-derived signals. Reduction in lipolysis and adipogenesis is seen with increased SCFAs in circulation. They also play an essential role in regulating appetite by modulating neuronal activity and visceral reflexes. SCFAs, especially butyrate, play a vital role in the regulation of the immune system and inflammation by inhibiting the Nuclear Factor Kappa B (NF-kB) activation in macrophages and also deter Histone Deacetylases (HDAC) (Koh et al., 2016; Morrison and Preston, 2016). SCFAs also regulate the microglia’s maturation and function in the CNS (Koh et al., 2016; Kho and Lal, 2018).
The gut microbiota has a vital role in synthesizing vitamins. They synthesize vitK2, which reduces the risk of cardiovascular diseases. They also produce vitamins B5, and B12, which are essential for proper neurological functioning (Kho and Lal, 2018). The gut microbiota also synthesizes group B vitamins, thiamine, riboflavin, nicotinic acid, pantothenic acid, pyridoxine, biotin, folates, and cyanocobalamin (Rowland et al., 2018). Vitamin B5 and B12 are necessary for proper neurological functioning (Kho and Lal, 2018).
The gut microbiota also plays a vital role in the metabolism of bile salts by secreting bile salt hydrolases and converting unabsorbed primary bile salts to secondary bile salts and deconjugating them. Then they are partly reabsorbed, and the remaining are excreted by the host. These secondary bile acids act as ligands for the host’s Farsenoid X nuclear receptors (FXR) and have anti-microbial effects (Kho and Lal, 2018; Rowland et al., 2018).
The gut microbiota also metabolizes polyphenols that are present in fruits and vegetables into simpler forms that are easily absorbed (Rowland et al., 2018). Tryptophan derivatives and indole derivatives, are also produced by gut microbiota (Koh et al., 2016; Kho and Lal, 2018; Caspani et al., 2019). Gut microbiota also plays an essential role in the absorption of minerals like iodine, iron, zinc, selenium, and copper (Bargiel et al., 2021).
Some metabolites like trimethyl-N-oxide (TMO), a metabolite produced from choline-containing foods are associated with cardiovascular risk. Similarly, Imidazole Propionate, a histidine utilization product, is found to increase insulin resistance and is associated with type 2 diabetes (Agus et al., 2021).
The most obvious method by which the microbiota could affect the neurological system is by altering host neurotransmitters and/or associated pathways. In fact, it has been discovered that a variety of important neurotransmitters can be produced by gut microbiota. It’s interesting to note that some bacterial species in the gut can also produce enzymes to facilitate the synthesis of neurotransmitters or their precursors. Additionally, some gut bacteria can communicate with intestinal enteroendocrine cells (EEC) via their metabolites to control the production and release of neurotransmitters (Yano et al., 2015; Kaelberer et al., 2018, 2020). It was recently discovered that a subpopulation of intestinal EEC synthesizes glutamate and uses it to send quick signals to the brain through the vagus nerve (VN) (Kaelberer et al., 2018). “Neuropod cells” are EEC that synapses with vagal neurons; they express the vesicular glutamate transporter 1 (VGLUT1) gene and release glutamate to transmit sensory stimuli from gut sugars to the brain in milliseconds (Frost et al., 2014). Coculture technique used in a recent study to find the key growth factor for bacterial survival revealed that gram positive human gut bacteria from Ruminococcaceae family require Bacteroides fragilis for their growth (Strandwitz et al., 2019). It was later found that B. fragilis primarily produces GABA as a growth factor. Additionally, this co-culture system demonstrated that Bifidobacterium, Parabacteroides, and Eubacterium, also synthesize GABA. Since it cannot cross the BBB, GABA produced by gut microbes may instead act locally on the VN or ENS. The early 1900s saw the discovery of acetylcholine (Ach) in a study of ergot on wheat rye; however, it wasn’t until much later that it was realized that Bacillus acetylcholini, not ergot, was the true producer of this neurochemical (O’Donnell et al., 2020). Since then, numerous microbes, including B. subtilis, L. plantarum, E. coli, and S. aureus, have been discovered to produce Ach. The Ach levels are notably higher in B. subtilis than E. coli or S. aureus (Koussoulas et al., 2018). Staphylococcus has been found to produce dopamine in the human intestine by the enzyme staphylococcal aromatic amino acid decarboxylase (SadA) which allows it to take up the precursor L-3,4-dihydroxy-phenylalanine (L-DOPA) and convert it into dopamine (Luqman et al., 2018). In humans, more than 50% of dopamine is synthesize in gut (Eisenhofer et al., 1997). Mucosal blood flow, gastric secretion and motility are regulated by dopamine and its receptor in the GIT (Vaughan et al., 2000; Al-Jahmany et al., 2004). In the human body, enterochromaffin cells (ECC), primarily in the intestinal epithelium, synthesize over 90% of serotonin. The bacterial kynurenine production pathway regulates how ECC in the gut use the amino acid tryptophan from dietary protein as a substrate to produce serotonin (Bailey and Cryan, 2017). Spore-forming bacteria in the gut (mostly Clostridia) might encourage the biosynthesis of serotonin by raising the gene expression of the rate-limiting enzyme tryptophan hydroxylase 1 (TPH1) in colonic ECC (Yano et al., 2015). Staphylococci have also been found to produce serotonin by decarboxylating the precursor 5-hydroxytryptophan (5-HTP) into serotonin using the enzyme SadA (Luqman et al., 2018). Numerous studies have shown that, in the absence of microbial colonization (in GF mice), level of neurotransmitters (GABA, serotonin, and Ach) as well as their precursors (tryptophan and choline) changes in the feces and serum (Yano et al., 2015). Similarly, acquired deprivation of gut microbiota (through antibiotic treatment) also results in changes in level of neurotransmitter and their precursors in gut and blood (Gao et al., 2019). It’s interesting to note that variations in gut microbial diversity also affect the expression of neurotransmitter receptors in the brain (Sudo et al., 2004; Bravo et al., 2011; Neufeld et al., 2011).
Prebiotics are fermentable ingredients utilized by bacteria and modulate bacterial activity by promoting the growth of symbiotic bacteria and decreasing the growth of pathogenic bacteria. In comparison, probiotics are living strains of microorganisms. Generally, prebiotics are beneficial and consumed by microbial flora, and these prebiotics are found in onion, wheat, and leafy vegetables in the form of polyphenols or oligosaccharides. A prebiotics-rich diet such as dietary fibers, oligosaccharides, and inulin significantly alters the microbial composition and activity. Bimuno-galactooligosaccharide consumption as a prebiotic therapy showed a decrease in the salivary cortisol awakening response and an improvement of mood and behavior in healthy volunteers compared to placebo (Schmidt et al., 2015).
Numerous advantages substantiated by research have been associated with regular consumption of probiotic foods and supplements includes reduction in inflammation (Hilton et al., 1997; Lai et al., 2019), management of diarrhea and other digestive complications (Hilton et al., 1997; Salazar-Lindo et al., 2004; Lahtinen et al., 2011; Aggarwal et al., 2014; Eskesen et al., 2015; Lai et al., 2019) as well as wide spectrum of other conditions from autoimmune diseases (Pham et al., 2008; Takada et al., 2017; Parker et al., 2018; Nishida et al., 2019; Ansari et al., 2020; Chao et al., 2020), emotional imbalance to cancer (Liu et al., 2011; Bajramagic et al., 2019; Sasidharan et al., 2019). Lactic acid bacteria, namely Bifidobacterium and Lactobacillus strains, are the most prevalent bacterial species used in modern probiotic products (Spano et al., 2019). Other bacterial species, such as Faecalibacterium prausnitzii and Akkermansia muciniphila, have been found in recent studies to possibly have positive effects when used as probiotics (Kumari et al., 2021). Their use in probiotic products is increasing as well (Saarela, 2019). The oral supplementation of B. fragilis has improved the gut permeability and microbial composition in maternal immune activation mouse model of autism spectrum disorder (Hsiao et al., 2013). Similarly, improvement in memory and learning was observed in rat model of AD upon supplementation of probiotics (L. acidophilus, L. fermentum, B. lactis, and B. longum) in drinking water for 8 weeks (Azm et al., 2018). Probiotics cannot always be considered an alternative to medication, especially in situations of severe disorders, despite the fact that many of their clinical advantages have undergone thorough testing. Early maternal separation causes a decrease in the expression of Corticotropin-releasing hormone receptor 1 (CRH R1) in the hippocampal region due to dysregulation of the HPA axis (Nemoto et al., 2015). Probiotic therapy of Lactobacillus rhamnosus GG in rodent studies showed a significant increase in the expression of CRH R1 in the hippocampal region and restoration of HPA activity (McVey Neufeld et al., 2019).
Mother-to-infant transmission is the first and primary approach to inhabiting gut microbiota during birth. Fecal examination of vaginally delivered infants showed a significant increase in the population of Bifidobacterium species such as Bifidobacterium adolescentis, B. catenulatum, and B. longum when compared with cesarean-born infants, which means the mother’s intestine and vaginal delivery are one of the crucial factors which regulate growth and species present in gut microbiota (Makino et al., 2013). As the infant grows, a sufficient number of prebiotics are given through breastfeeding as it is required for normal growth of microbial flora and different strains of lactobacillus species (Haarman and Knol, 2006).
Dietary intake and lifestyle greatly influence gut microbiota composition. Numerous popular diets, such as the Western (Drasar et al., 1973; Reddy et al., 1975; Wu et al., 2011), Mediterranean (Lopez-Legarrea et al., 2014; De Filippis et al., 2016), vegetarian, vegan, and gluten-free diets (Sanz, 2010; Pisarello et al., 2015; Bonder et al., 2016), have been investigated for their capacity to modify gut microbiota. The western diet (high protein/animal fat) is proved to increase Bacteroides and Enterobacteria while reducing the diversity of Eubacterium and Bifidobacterium sp. The Mediterranean diet (high fiber and antioxidants, low in red meat, high monounsaturated fat) is believed to reduce inflammation, improve lipid profiles, and reduce the risk of obesity (Sandhu et al., 2017). In terms of microbes, these traits were linked to increase in Prevotella, Bifidobacterium, and Lactobacillus, and decrease in Clostridium (Clemente-Postigo et al., 2012; Queipo-Ortuño et al., 2012; Fava et al., 2013; Koloverou et al., 2016). When compared to an unrestricted control diet, vegan and vegetarian diets were found to dramatically decrease the levels of Bacteroides and Bifidobacterium sp. (Wu et al., 2016). Incompetent dietary consumption reduces microbial nutrient intake and decreases the population of beneficial microbial flora. As a compensatory mechanism, microbial flora starts consuming glycoprotein-rich protective mucosal barrier, which can further lead to the leaky wall and dysbiotic condition to initiate inflammation (Tabrizi et al., 2019). Studies found that an inulin or Oligofructose-rich diet is responsible for improving memory, mood and cognition enhancement (Smith et al., 2015).
The difference in metabolic phenotype was observed between lean and genetically obese (fa/fa) rats and (ob/ob) mice (Turnbaugh et al., 2006; Waldram et al., 2009; Palmnäs et al., 2020). They are also characterized by difference in abundance of Firmicutes, Bacteroidetes, and Actinobacteria (Turnbaugh and Gordon, 2009). Calorie restriction in obese individuals resulted in decrease in Firmicutes/Bacteroidetes ratio which were higher at baseline (Ley et al., 2006; Karlsson et al., 2013). In children, low Bifidobacterium abundance is generally linked to obesity (Abenavoli et al., 2019). Additionally, a gut microbiome dominated primarily by Firmicutes revealed altered methylation in gene promoters associated with obesity and cardiovascular disease (Kumar et al., 2014).
In comparison to controls, human microbiome studies on tobacco smokers revealed a lower relative abundance of Bacteroides, a higher relative abundance of Prevotella, and a lower Shannon diversity (Stewart et al., 2018). Following smoking cessation, decrease in Bacteroides along with changes in abundance of alphaproteobacteria and betaproteobacteria were observed (Nolan-Kenney et al., 2020). In fact, quitting smoking caused significant changes in the gut microbiome, with an increase in Firmicutes and Actinobacteria and a decrease in Bacteroidetes and Proteobacteria. It also results in increase in microbial diversity (Biedermann et al., 2013). Importantly, when provided similar meals to prevent nutritional impacts, different gut microbiota composition was found in smokers and non-smokers (Kobayashi and Fujiwara, 2013).
There is mounting evidence that exercise can influence the human gut microbiome by altering the composition and function of gut microbiota (Clarke et al., 2014; Bressa et al., 2017; Barton et al., 2018; Durk et al., 2019). Matsumoto et al. (2008) discovered that butyrate synthesis of bacteria, like Bifidobacteria, increased after 5 weeks of exercise training in rats. In contrast to sedentary controls, women who exercised at least three hours per week had higher concentrations of Faecalibacterium prausnitzii, Roseburia hominis, and A. muciniphila (Bressa et al., 2017). Professional rugby players gut microbiota exhibited a higher relative abundance of 40 different bacterial taxa, increase in alpha diversity as well as reduced abundance of Lactobacillus and Bacteroides sp. as compared to lean sedentary group (Clarke et al., 2014). There is currently conflicting data regarding the relationship between exercise and the gut microbiome. For instance, some research on rodents found that exercise decreased the Firmicutes to Bacteroidetes ratio (Mika et al., 2015; Denou et al., 2016), whereas others found that exercise raised the ratio (Kang et al., 2014; Lambert et al., 2015).
Gut-microbiota modulates brain functions by stimulating neuronal responses such as VN or secreting metabolites that directly control brain behavior. The gut microbiota communicates with the brain through the nervous (VN), endocrine (HPA axis), immune, and metabolic systems. The gut communicates with the brain via two pathways: Direct gut-brain communication is mediated through ANS and the spinal cord. Communication between gut and brain happens through ENS in the gut and ANS; and VN in the spinal cord.
Although the gut microbiota can interact through immunological and endocrine pathways, the VN signaling is the quickest and most direct route for the microbiota to influence the brain. The 10th cranial nerve, or VN, connects the viscera to the brain. It is a paired nerve made up of sensory and motor neurons (afferent and efferent, respectively). Vagal efferent transmit messages “down” from the brain to the gut through efferent fibers, which make up 10–20% of all fibers, whereas vagal afferents transmit signals “up” from the intestinal wall to the brain, making up 80–90% of all fibers (Tubbs et al., 2015). The activation and regulation of the HPA axis, which regulates the adaptive responses to stressors, is mediated by the vagal afferent pathways (Tsigos and Chrousos, 2002). EECs, 1% of intestinal epithelial cells, communicate with vagal afferents either directly by releasing serotonin that activates 5-HT3 receptors on these fibers or indirectly through the action of gut hormones that target the brain, such as cholecystokinin (CCK), glucagon-like peptide-1, and peptide YY, via vagal afferents that express receptors for these anorexigenic or orexigenic (ghrelin, orexin) hormone (Strader and Woods, 2005). In addition to cell-mediated sensing, VN has direct mechanisms for sensing microbial signals. For example, depending on the compound, SCFAs activate vagal afferent fibers through multiple ways. For example, butyrate directly affects afferent terminals, but the long fatty acid oleate acts on vagal afferents via a CCK-mediated mechanism (Lal et al., 2001). Additionally, TLR4 is also expressed on the vagal afferent fibers, which can detect bacterial products like Lipopolysaccharide (LPS) and stimulate the brain (Goehler et al., 1999). LPS acts on TLR4 and initiates an inflammatory response to release various pro-inflammatory cytokines and chemokines (Peña et al., 2014; Covington et al., 2015). The team of Mark Lyte used c-fos as a marker of neuronal activation to map brain circuits activated by oral administration of Campylobacter jejuni at subclinical concentrations in mice, impacting behavior and brain functions, highlighting the indirect stimulation of vagal afferent fibers by microbes (Gaykema et al., 2004). Both, the nucleus tractus solitarius (NTS), the first entrance point of vagal afferents in the brain, and the NTS’s widespread projections exhibited the signs of brain activation. Chronic administration of L. rhamnosus in rodent species showed increased GABAB receptor expression in the prefrontal cortex, which is crucial for anti-depressant activity. L. rhamnosus is also responsible for reducing stress-induced corticosterone levels indicating the influence of Lactobacillus on CNS has significant impact on physiological levels. The absence of neurochemical and behavioral benefits of L. rhamnosus in vagotomized mice identifies the vagus as a significant modulatory constitutive communication channel between the gut and the brain (Bravo et al., 2011).
The composition and overall biomass of the intestinal microbiota are modulated by different types of psychological stressors. Short term stressors can also have an influence on the microbiota, with social stress exposure lasting only 2 h having a considerable impact on the community profile and a reduction in the relative proportions of the major microbiota phyla (Galley et al., 2014). Neurons, immunological cells, and ECC secrete signaling molecules under the control of the brain, which may have direct impact on the microbiota. According to a number of studies, bacteria have binding sites for enteric neurotransmitters produced by the host and can affect how certain components of the microbiota operate, which can increase susceptibility to inflammatory and infection stimuli (Hughes and Sperandio, 2008). P. fluorescens has been found to have high affinity for the GABA system and binding characteristics that are similar to those of brain receptors (Guthrie and Nicholson-Guthrie, 1989). Escherichia coli O157:H7 has a receptor for host-derived epinephrine/norepinephrine that can be selectively inhibited by adrenergic antagonists (Clarke et al., 2006). Additionally, the brain plays a significant role in the regulation of gut functions like motility, secretion of acid, mucus, and bicarbonates, handling of intestinal fluids, and the mucosal immune response, all of which are crucial for maintaining the mucus layer and biofilm, where various bacterial species grow in a variety of different microhabitats and metabolic niches linked to the mucosa (Macfarlane and Dillon, 2007). The disruption of the normal mucosal environment caused by a dysregulation of GBA can therefore have an impact on the gut microbiota. The quantity and quality of mucus vary in response to stress (Rubio and Huang, 1992). Acoustic stress has an impact on dogs’ intestinal and gastric postprandial motility, prolonging the recovery of the migratory motor complex pattern and temporarily reducing gastric emptying (Gué et al., 1989). Through the central release of CRF, mental stress also increases the frequency of cecocolonic spike-burst activity (Gue et al., 1991).
By altering intestinal permeability, which allows bacterial antigens to pass through the epithelium and trigger an immune response in the mucosa, the brain might also have an impact on the composition and function of the microbiota. Acute stress increased colonic paracellular permeability by increasing interferon-γ and downregulating ZO-2 and occluding mRNA expression (Demaude et al., 2006). The brain might also regulate immune function via the ANS. The sympathetic branch controls the quantity, degranulation, and activity of mast cells, resulting in an imbalance in tryptase and histamine release in stress-related muscular failure (Santos et al., 1998). It is crucial to note that gastrointestinal changes brought on by stress promote the production of pathogenic bacteria. The expression of P. aeruginosa is induced by norepinephrine produced during surgery, which might cause gut sepsis (Alverdy et al., 2000). Additionally, norepinephrine may promote the overgrowth of both pathogenic (E. coli 0157:H7:3) and non-pathogenic E. coli isolates (Freestone et al., 2002, 2003), as well as increase the virulence of Campylobacter jejuni and other intestinal pathogens (Cogan et al., 2007).
The HPA axis is essential for the species’ survival. It is one of the critical non-neuronal communication routes in the microbiota-gut-brain axis (mGBA). Upon disturbance in homeostasis, there is a release of corticotrophin-releasing factor (CRF) and arginine vasopressin (AVP), a nonapeptide, from the hypothalamus into the hypophysial bloodstream, which stimulates adenohypophysis of pituitary gland thus trigger the release of ACTH into systemic blood circulation. In adrenal glands (also known as supra renal glands), the released ACTH binds to the receptors in zona fasciculata (adrenal cortex) and stimulates the steroidogenic pathway by facilitating the conversion of cholesterol esters into free cholesterol. Upon a series of enzymatic reactions, the cholesterol is converted into various steroidal end products, the glucocorticoids, i.e., cortisol in humans and corticosterone in rodents. These freshly synthesized glucocorticoids are rapidly released into blood circulation. The released glucocorticoids bind to the glucocorticoid receptor (GR) present in the cytoplasm, and the glucocorticoid-receptor complex translocate into the nucleus and binds to DNA. Thus, it acts as a transcription factor and results in target genes’ transcription, leading to protein synthesis changes (Fulford and Harbuz, 2005). The negative feedback regulation of the HPA axis is governed by two mechanisms, rapid and delayed feedback. Glucocorticoids exert the rapid feedback at the level of the hypothalamus by inhibiting the synthesis and release of ACTH. Delayed feedback at the level of adenohypophysis where glucocorticoids inhibit the mRNA expression of pro-opiomelanocortin (POMC), the ACTH precursor protein (Fulford and Harbuz, 2005).
Hypothalamus pituitary adrenal axis interacts with various neuronal and non-neuronal pathways of communication between the gut and the brain. It is found that VN stimulation results in a rise in plasma ACTH and corticosterone levels in rodents by increasing the CRF mRNA expression in the hypothalamus. Increased levels of cortisol induce an inflammatory cytokine-mediated surge in anxiety, bowel movement, and intestinal permeability while altering intestine’s microbiota (Wallace and Milev, 2017).
The relationship between the HPA axis and the gut microbiota has been explained by a number of different mechanisms. First, Increased cytokine release and the production of small bioactive molecules because of the gut microbiota dysbiosis can cause certain cytokines, such as interleukin (IL)-1, IL-6, and tumor necrosis factor (TNFα) to pass through the BBB and act as potent HPA axis activators (Turnbull and Rivier, 1995; Banks, 2005). Second, the release of LPS and peptidoglycan, components of bacterial cell wall, can also activate the HPA axis (Arentsen et al., 2016). Third, the ClpB protein (mimics α-melanotrophin MSH) produced by E. coli stimulate the release of POMC thus promote ACTH synthesis (Breton et al., 2016). GF animal studies suggested that gut microbiota plays a significant role in the development and modulation of the HPA axis. Experimental studies in GF mice showed increased HPA axis activity and higher level of corticosterone when exposed to mild restraint stress as compared to SPF mice (Sudo et al., 2004). Brain derived neurotropic factor (BDNF) expression level was also decreased in the cortex and hippocampal region of GF mice compared with SPF mice (Sudo et al., 2004). This experiment demonstrates that vegetation of gut microbiota composition in early life is critical for the appropriate development of the HPA axis and stress response. Bacterial metabolites such as LPS can act on TLR4, which then initiates cytokine release and is responsible for stimulating the release of hypothalamic CRH. LPS can also directly stimulates adrenal glands to secret cortisol in humans and corticosterone in animals (Zacharowski et al., 2006). Several cytokines such as TNF-α and IL-12, are responsible for elevated corticosterone levels in animals.
Patients with different phases of mood and psychotic disorders can show disruption of the HPA axis. The precise mechanisms driving these findings are still unknown. Various studies have demonstrated a link between the composition of the gut microbiota and the HPA axis activity. Different stresses may affect intestinal integrity and abundance of Bacteroides, Lactobacilli, and Clostridium in animal models (Qu et al., 2021). Probiotics based on Lactobacillus and Bifidobacterium have also been shown to ameliorate depression- and anxiety-like symptoms, enhance learning, and restore stress-induced HPA axis dysfunction (Desbonnet et al., 2010). Furthermore, there is evidence that Lactobacillus farciminis supplementation in rats may dampen the HPA axis response to partial restraint stress by decreasing gut permeability (Ait-Belgnaoui et al., 2012). SCFAs have also been demonstrated to reduce the expression of genes that code for proteins involved in HPA axis (van de Wouw et al., 2018). Besides, maternal separation in animals creates an early life stress event, leading to an altered gut-brain axis and increases plasma corticosterone levels in the maternally separated animals. The maternally separated animals showed a higher immunological response and increased HPA activity (O’Mahony et al., 2009).
Depression is a mood-related disorder characterized by the depletion of monoamines such as serotonin, nor-adrenaline, and dopamine in the midbrain and nuclei of the brainstem. Several factors responsible for the cause of such disease include genetic factors, environmental factors, hyperactivity of the HPA axis, poor diet, low-grade inflammation, gut microbiota alteration, vitamin D deficiency, psychosocial stressors, etc. (Berk et al., 2013; Jokela et al., 2016). The proposed hypothesis suggests that increased CRH is secreted by neuronal cells of the paraventricular nucleus of the hypothalamus in patients suffering from depression (Raadsheer et al., 1994). CRH stimulates the pituitary gland to secrete ACTH, which further activates the release of corticosteroids from adrenal glands (Barden, 2004). An impaired feedback mechanism is also found in depressive patients who cannot control the excessive release of CRH and ACTH. Hypercortisolemia is generally seen in these patients, and further leads to a decrease in the sensitivity of GR (Liang et al., 2018). To overcome the stress response, the levels of ACTH and corticosterone are elevated in microbiota deficient mice compared to SPF mice (Sudo et al., 2004). Anhedonia and anxiety-like response are observed after pooled FMT from depressed patients than FMT from healthy rats (Kelly et al., 2016). All these results suggest the direct relationship between gut microbiota and the HPA axis.
Various studies conducted so far suggest the involvement of gut microbiota in the modulation of depression-like behavior, including pre/probiotic treatment, GF mice, antibiotic-induced gut microbiota alteration, FMT, and deliberate microbial contamination of GIT (Bravo et al., 2011; Messaoudi et al., 2011; Cryan and Dinan, 2012; Desbonnet et al., 2015; Wong et al., 2016; Gur et al., 2017). In recent studies, certain species of gut-microbiota such as Prevotella (type 2), Bacteroides (type 1), and Proteobacteria were also found to be increased and caused dysbiotic conditions in patients suffering from depression (Liu et al., 2016; Evrensel et al., 2020). These micro-organisms are responsible for the dysbiotic states, cause a leaky gut, and translocation of bacterial metabolites into systemic circulation. This further activates pro-inflammatory cytokinin such as IL-1, IL-1β, IL-6, and TNF-α, which promotes the activation of microglia and astrocytes (Song and Wang, 2011). Cytokines such as IL-1 are responsible for activating neurons in the paraventricular nucleus, releasing CRH, and promoting a chronic increase in blood corticosteroid level (Song et al., 2009). Patients suffering from depressive disorder were found to have decreased number of protective microbial species like Bifidobacterium and Lactobacillus in their intestinal flora, which may be a key factor for dysbiotic conditions and initiation of the inflammatory process. Probiotic treatment of these strains was effective in disease conditions (Aizawa et al., 2016; Evrensel et al., 2020). The probiotic therapy of L. Rhamnosus showed improvement in mood and behavior by decreasing the elevated level of corticosterone in a stress-induced depressive model of rodents (Bravo et al., 2011). The pharmacological anti-depressant-like effect of (R)-Ketamine, a dissociative hallucinogen, NMDA receptor antagonist, and as an antidepressant; is altered because of the dysbiotic gut microbiome in the chronic social defeat stress model of mice (Qu et al., 2017; Yang et al., 2017). Huang et al. (2019) revealed the role of phylum Actinobacteria and the class Coriobacteriia in the anti-depressant effect of ketamine. Furthermore, FMT from alcoholic individuals induced depression-like behavior, spontaneous alcohol preference as well as decreased BDNF, α-1 subunit of GABA A receptor (α-GABAA R) in medial PFC; and glutamate receptor 1 (mGluR1) and protein kinase C ε in NAc in C57BL/6J mice (Zhao et al., 2020). Contrarily, FMT from healthy individuals decreased alcohol-induced depressive behavior in mice (Xu et al., 2018). Another study on FMT from anhedonia-susceptible rats revealed exaggerated pain and depression-like behavior in antibiotic-treated pseudo-GF mice. In contrast, transplantation from resilient rats exhibits significant improvement in pain and depressive-like behavior (Yang et al., 2019). This signifies that gut microbiome, dysbiotic condition, and inflammation can be the underline cause of depression. Specific strains of prebiotic or probiotic therapy of gut microbiota can be the key factors in the treatment of depression.
For millions of years, the gut microbiota has coevolved and works in symbiotic association with humans. These prokaryotic microscopic members of the ‘human holobiont’ play a key role in maintaining normal physiology and homeostasis. Aberration in gut microbiome configuration and function is linked to being a causative factor for the development of various neurological diseases such as Alzheimer’s disease, Parkinson’s disease, depression, etc. These micro-organisms follow multiple complex molecular mechanisms for maintenance of normal homeostasis such as vagal nerve signaling (Berk et al., 2013) neuroendocrine signaling (Jokela et al., 2016) metabolic signaling (Bravo et al., 2011), immune system modification (Cryan and Dinan, 2012), and epigenetics (Abe-Higuchi et al., 2016; Higuchi et al., 2016). Epigenetics has a crucial role in regulating host physiology via alteration in gut microbiome metabolic activity, which depends on environment and diet. The metabolites produced by gut microbiota act as cofactor and substrate for various enzyme reactions; for example, cofactors for the activity of enzyme acetylases and methylases, which regulate histone modification and DNA methylation; comes from the gut microbiome (Figure 2)(Cai et al., 2015; Ledford, 2015; Abe-Higuchi et al., 2016). Regulation of epigenetics is a dynamic process and is subject to changes in exercise, nutrition, and microbiota composition (Libert et al., 2011).
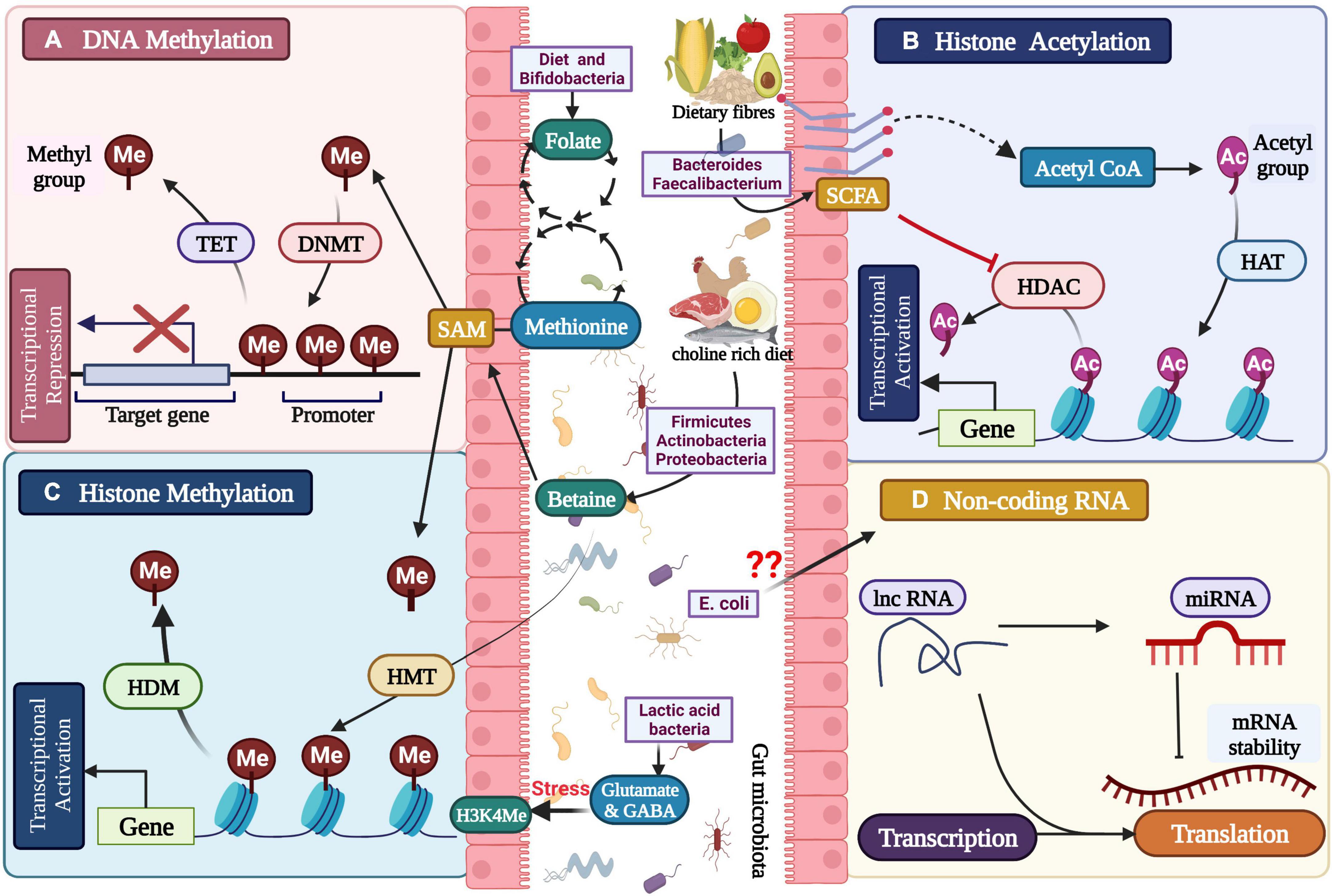
Figure 2. Gut microbial metabolites regulating the epigenetic mechanisms: (A) DNA methylation, (B) histone modification, (C) histone methylation, and (D) non-coding RNA associated gene silencing.
The literal meaning of epigenetics is “in addition to genetics.” It involves studying gene expression changes in the chromosome instead the DNA sequence. These changes are heritable as well as stable. Changes in Chromosomal superstructure and chemical modification of nitrogenous bases are mainly regulated by epigenetics without directly altering the DNA sequence. Various molecular mechanisms can lead to epigenetics, but the main pathways include DNA methylation and acetylation, histone modification, and RNA-associated silencing.
Methylation of DNA occurs covalently to the pyrimidine ring of cytosine, which results in a change in structure of major grooves. The methyl group is extracted from S-adenyl methionine (SAM). The enzymes DNA Methyltransferases (DNMTs) catalyze DNA methylation by transferring the methyl group to carbon-5 of cytosine and forming 5-methyl cytosine. There are three major DNMTs in mammals viz, DNMT1, DNMT3a, and DNMT 3b. DNMT3L is catalytically inactive, but can increase the activity of Dnmt3a/b to 15-fold upon binding with them. DNMT3a and DNMT3b are de novo DNMT because they establish new methylation patterns for unmodified DNA (Gowher et al., 2005). Cytosine is the most common site for methylation, followed by guanine nucleotide and CpG (5′-C-phosphate-G-3′) rich sites. The CpG-rich region in the DNA is called CpG Island. These DNMTs are extremely sensitive to the availability of nutrients, which can be influenced by the metabolic activities of the gut microbial species. Various metabolites such as cyanocobalamin, choline, folate, and betaine play an essential role in synthesizing 6-methyltetrahydrofolate, which acts as a methyl group donor for SAM that is directly involved in the DNA methylation process (Covington et al., 2009; Hunter et al., 2009). These metabolites are regulated by specific folate-producing gut microbiota such as Bifidobacteria and Lactobacillus (Otsuki et al., 2008; Uchida et al., 2011). DNA methylation plays a vital role in the inactivation of the X chromosome (Bosker et al., 2011; Hooper et al., 2012; Sun et al., 2013), regulation of tissue-specific gene expression, silencing of retroviral elements and genomic imprinting (Sullivan et al., 2000).
Pieces of evidence are available about methylation of various genes in depression. Increased CpG methylation in the BDNF gene’s promoter region has been linked to a reduction in the amount of BDNF produced by neurons (Martinowich et al., 2003). In mouse model of depression, epigenetic modifications result in alteration of chromatin structure of BDNF (Tsankova et al., 2006). Additionally, majority of depressed population exhibits increased DNA methylation on BDNF gene (Fuchikami et al., 2011). These results suggested that BDNF epigenetic alterations may be crucial to pathophysiology of depression and to its potential therapeutic use. Majority of the studies conducted so far, to establish the correlation between depression and DNA methylation of BDNF, revealed hypermethylation of BDNF gene in depressed patients compared to healthy individuals (Fuchikami et al., 2011; Kang et al., 2015). Contrarily, one study in pregnant women have associated depressive symptoms with reduced methylation of BDNF promotor in offspring (Braithwaite et al., 2015).
In humans SLC6A4, serotonin transporter, paly key role in development and function of brain region (Booij et al., 2013). DNA methylation has received a lot of attention recently in studies, and this focus may help to explain how the regulation of SLC6A4 gene expression affects changes in emotional behavior. Lower SLC6A4 mRNA levels were linked to higher methylation in human lymphoblast cell lines (Philibert et al., 2007), suggesting that SLC6A4 promoter methylation may be linked to gene expression. Additionally, it was discovered that SLC6A4 promoter methylation, both full and partial, dramatically decreased SLC6A4 expression levels (Olsson et al., 2010). Numerous studies have revealed association between SLC6A4 DNA methylation and depression (Philibert et al., 2007; Devlin et al., 2010; Olsson et al., 2010; Zhao et al., 2013). The majority of them demonstrated an increase in SLC6A4 DNA methylation in depressive disorders. Three studies, however, discovered that there was no variation in the DNA methylation of SLC6A4 between depressive patients and healthy controls (Olsson et al., 2010; Bayles et al., 2013; Chagnon et al., 2015). On the other hand, depression disorders are also linked to DNA methylation in the serotonin receptor (5-HTR) family (Perez-Cornago et al., 2014; Weder et al., 2014; Perroud et al., 2016). According to research by Perez-Cornago et al. (2014), a decrease in depression symptoms was associated with an increase in the DNA methylation of the 5-HTR2A gene. In contrast, Perroud et al. (2016) discovered that in bipolar disorder patients who attempted suicide, the level of 5-HTR3A methylation was higher at CpG2 III and CpG4 III and lower at CpG1 I and CpG5 III. Additionally, other genes related to the metabolism of 5-HT may be crucial in the development of depression. Three experiments with MAO-A, which catalyzes the oxidative deamination of 5-HT, revealed variable DNA methylation levels. Among 43 CpG sites, Katharina Domschke discovered only two hypomethylation sites (Domschke et al., 2014). Melas PA observed that the DNA methylation level of the MAO-A gene exon 1 rose in depressed individuals (Melas et al., 2013). In contrast, Melas PA and Forsell found it decreased in female patients (Melas and Forsell, 2015). In conclusion, numerous investigations have found a link between SLC6A4 methylation and depression. To further understand the involvement of epigenetic mechanisms of SLC6A4 methylation in the etiology of depression, more research in the field of epigenetics is required.
The GR, which is encoded by the protein-coding gene NR3C1 (Nuclear Receptor Subfamily 3, Group C, Member 1), is activated by cortisol (Francke and Foellmer, 1989). GR (ligand activated transcription factor) is necessary for the HPA axis to operate properly. Individuals with GR anomalies in their brains were found to be connected to bipolar disorder and schizophrenia. It has been suggested that the genetic variants in the GR (NR3C1) gene may be the mechanism causing HPA axis malfunction and GR abnormalities in psychiatric disorders (Turner et al., 2010). In particular, it has been shown that NR3C1 methylation contributes significantly to our understanding of both depression and dysregulation of the HPA axis. In three investigations, it was discovered that new-born of depressive mothers had higher NR3C1 DNA methylation levels (Oberlander et al., 2008; Conradt et al., 2013; Murgatroyd et al., 2015). However, Braithwaite et al. (2015) found that this rise exclusively affected male new-born. In addition, female depressive individuals with early parental deaths had hypermethylation. These findings imply that depression can have a gender-specific impact on the DNA methylation of the NR3C1 gene. Comparing NR3C1 methylation to controls in more recent research, however, did not reveal any correlation between the two conditions (Alt et al., 2010; Kim et al., 2016). Contradictory results were found in other research comparing depression to controls, with either increased (Melas et al., 2013) or decreased (Na et al., 2014) level of DNA methylation. According to some studies, the DNA methylation of FK506 binding protein (FKBP5), a co-chaperone of the GR, increased in depressed patients (Weder et al., 2014; Höhne et al., 2015) indicating a connection between depression and the DNA methylation in the GR system. Undoubtedly, there is conflicting and muddled evidence supporting the relationship between NR3C1 methylation and depression, the reason being small sample size, various tissue types, low effect size, different ethnicities etc. Therefore, prior to conducting the investigations, future research should take these factors into account.
Several physiological and behavioral functions, including the operation of the HPA axis, are regulated by the neuropeptide oxytocin, which is released by the paraventricular nucleus of the hypothalamus. Recent research suggests that the polymorphisms of the oxytocin receptor (OXTR) affect the neurocardiac response to the HPA function (Norman et al., 2012), suggesting that OXTR may have regulatory effects on the pathomechanisms of depression. There is evidence of OXTR CpG hypermethylation in depressed women (Bell et al., 2015; Chagnon et al., 2015; Reiner et al., 2015). However, DNA hypomethylation was observed in depressed African American cohort (Smearman et al., 2016). Contrarily, Kimmel et al. (2016) did not find any discernible genetic effects.
Histones are the core proteins which give the backbone to chromatin. DNA wraps around histones to form a nucleosome. The nucleosome consists of pair of histones (H2A, H2B, H3, and H4) at the core, around which DNA is tightly bound. The histone modification types include acetylation, methylation, phosphorylation, biotinylation, citrullination, SUMOylation, ubiquitination, and proline isomerization. Among all the mentioned modifications, methylation, acetylation, and deacetylation are the most common (Kendler, 1983; Sun et al., 2013). The N-terminal of H3 usually undergoes histone modification via methylation or acetylation of lysine and arginine residue, whereas phosphorylation occurs at serine and threonine residues (Tsankova et al., 2007). Methylation involves the addition of methyl groups to the histone proteins by histone methyl transferases (HMTs). The methylation of histone can lead to transcription activation [methylation of H3 lysine 4 (H3K4)] or transcription inactivation [methylation of H3 lysine 9 (H3K9) and H3K27] depending upon the specific residue (Peña et al., 2014; Yamagata et al., 2017). Acetylation of histones results in neutralization of N-terminal, leading to a reduction in affinity for DNA and loosening of chromatin conformation, thus leads to transcription activation (Fraga et al., 2005). In histone acetylation, the acetyl group from acetyl CoA is transferred to terminal lysine residue by Histone acetyltransferase (HATs). Various metabolites from the gut microbiome, such as SCFA, are proved to regulate the histone acetylation process (Hobara et al., 2010; Sarkar et al., 2014). Histone deacetylases (HDACs) remove the acetyl group from the terminal lysine residue, which favors chromatin compaction and results in transcription inactivation (Iga et al., 2007). Total of 13 HDACs are found in humans and are mainly classified into four classes: Class I includes HDACs 1, 2, 3, and 8 (most similar to yeast transcription regulator RPD3); Class II contains HDACs 4, 5, 7, and 9; Class IIb consist of HDACs 6 and 10. The class II HDACs are similar to yeast deacetylase, HDA1.; Class III includes Sirt1 to Sirt7, which deacetylates histone as well as non-histone proteins, and Class IV comprised of a single member, HDAC 11, which does not share any similarity with RPD3 and HDA1 (Xu et al., 2018; Zhao et al., 2020). HDACs are found to be overexpressed in various neurological and inflammatory disorders. The SCFAs produced by the gut microbiome are involved in HDAC inhibition; butyrate is a specific HDAC Class I and II inhibitor (Higuchi et al., 2016).
Gram-positive, anaerobic bacteria belonging to the genera Coprococcus and Faecalibacterium digest dietary fibers to produce SCFAs. One of the most prevalent genera of gut microbes, Faecalibacteria have significant immunological roles as well as therapeutic importance for a number of disorders, including depression (Jiang et al., 2015). SCFAs can bind to and activate G protein-coupled receptors GPR43/41 (FFAR2/3), as well as the less prevalent GPR109a and CPR164 (also known as OR51E1 and HCAR2, respectively). Since these receptors are widely expressed in many different human organs, such as EEC, adipocytes, immune cells, and neurons, it is possible that SCFAs can change behavior either directly by stimulating neuronal pathways or indirectly by activating neuroendocrine and immunological systems (Stilling et al., 2016). With regard to depression, lactate, one of the SCFA, has both preventive and reversible effects, and these effects are carried out on HDACs by distinct epigenetic mechanisms (Karnib et al., 2019). The social avoidance and anxiety behaviors that resulted from the 10-day social defeat task were prevented in control mice by chronic lactate administration before the challenge. The class I HDAC2/3 level and activity were increased in lactate treated mice. After the onset of depression, the effect of lactate was not mediated by HDAC2/3 but rather by a decrease in HDAC5 levels (Karnib et al., 2019). A lack of butyrate, acetate, and propionate was observed in MDD patients (Chen et al., 2015; Jiang et al., 2015; Zheng et al., 2016; Skonieczna-żydecka et al., 2018), and a high number of butyrate-producing bacteria, such as Faecalibacterium and Coprococcus sp., was found in subjects with greater quality of life indicators (Valles-Colomer et al., 2019), supporting SCFA involvement with the etiology of depression.
Non-coding RNA-associated silencing is a new epigenetic mechanism. Non-coding RNA (ncRNA) are nothing but RNA that is not transcribed into protein. They are classified into two types: housekeeping ncRNA and regulatory ncRNA. Based on the size, regulatory ncRNA are further divided into short chain ncRNA and long ncRNA (lncRNA). The short chain ncRNA includes small interfering RNA (siRNA), microRNA (miRNA) and piwi RNA (piRNA) (Desbonnet et al., 2015; Gur et al., 2017). Several studies have indicated the role of siRNA in gene silencing through DNA methylation and histone modification (Messaoudi et al., 2011; Wong et al., 2016; Qu et al., 2017). The direct role of miRNA in epigenetics has not been reported in mammalian cells. However, few studies suggest the change in whole DNA or chromatin state by miRNA by inhibiting the activity of chromatin remodeling enzymes (Yang et al., 2017; Huang et al., 2019). Several miRNAs were found to be dysregulated in depression. MiR-124, miR-139a-5p, miR-221, miR-218, miR-17-5p, miR-146a, miR-132, miR-425-3p, miR-184 were some of the miRNAs identified in various biological samples of depression patients as well as in preclinical models (Shi et al., 2021). Several miRNAs derived from the host exosomes play an important role in maintaining the gut microbiota and the host physiology. The importance of miRNAs in the maintenance of gastro-intestinal functions is also widely recognized (Zhao et al., 2021). But the relation between the gut dysbiosis induced miRNA dysregulation in depression has not been established. Future studies aimed at exploring this association might provide novel insights into pathogenesis as well as treatment of depression.
The epigenetic modifications play a key role in anti-depressant response as well as in the pathophysiology of depression (Tsankova et al., 2007; Sun et al., 2013; Peña et al., 2014; Yamagata et al., 2017). The levels of acetylated histones in the limbic region were affected by chronic stress (Covington et al., 2009; Hunter et al., 2009). Six weeks of chronic stress results in depressive behavior in mice by increasing the HDAC2 function in the ventral striatum (Uchida et al., 2011), indicating HDAC2 involvement in stress vulnerability, thus favoring the anti-depressant potential of HDAC2 inhibitors. Glial-derived neurotrophic factor (GDNF) level gets lowered in the ventral striatum in response to chronic stress, which is prevented by knockdown of HDAC2, suggesting that GDNF is a primary target of HDAC2. There is evidence of high HDAC2 expression and low GDNF expression in depressed patients (Otsuki et al., 2008; Hobara et al., 2010).
Similarly, HDAC4 and HDAC5 are also overexpressed in depression (Iga et al., 2007; Otsuki et al., 2008; Sarkar et al., 2014). A previous study has reported that hippocampal administration of HDAC4/5 inhibitors prevented chronic stress-induced depressive behavior (Higuchi et al., 2016), which can be considered a new treatment strategy for depression. Following chronic stress, SIRT1, a class III HDAC shows reduced activity in the dentate gyrus. Increased depressive behavior is observed after the blockade of hippocampal SIRT1 activity either with drugs or through a genetic approach. Contrarily, during chronic stress, hippocampal SIRT1 activation results in the blockade of depressive behavior and abnormal dendritic structures and increased phosphorylation of extracellular signal-regulated protein kinases 1 and 2 (ERK1/2). ERK2 overexpression in the hippocampus has an anti-depressant effect, whereas lower expression is associated with increased depressive symptoms, indicating the critical role of SIRT1 in regulating depression (Abe-Higuchi et al., 2016). SIRT1 exhibit a significant genome-wide association with MDD (Cai et al., 2015; Ledford, 2015). Single nucleotide polymorphism (SNP; rs12413112) in SIRT1 is associated with MDD (Libert et al., 2011). There are number of drugs with the epigenetic mechanism of action beneficial to counteract depressive symptoms and are listed in Table 2.
As discussed earlier, epigenetic modifications bring about changes in gene expression because of changes in the external or internal environment without altering the DNA sequence. These changes are stable for the long term, even if the exposure to that causative factor is for a short period. Microbiota is also one of the environmental factors that can alter host epigenome via GBA modification and results in visible behavioral or phenotypic changes. Though these changes are long-lasting but are not permanent and hence can be reversed at a later stage by various methods such as restoration of gut microbiome through pre and probiotic supplementation, FMT, and lifestyle modification approaches like proper sleep cycle, healthy eating habits, physical exercise, yoga, and meditation. All the above approaches are beneficial in diabetes, obesity, neurodegenerative diseases, and depression.
The microbial count of humans is 10 times higher than own human cells. The human gut comprises three to four million unique genes, which is 100–150 times more than our genome (Qin et al., 2010). These genes are responsible for the production of various proteins and metabolites of the microbiome. These metabolites directly or indirectly affect the host genome by modulating the host epigenome (Table 3).
Gut microbiota, for example, Firmicutes, produces SCFA by utilizing dietary fibers. These SCFA acts as HDAC inhibitors, thus promoting gene expression by blocking the removal of an acetyl group from histones (Zhu et al., 2010). The global histone acetylation (H3 and H4) and methylation (H3) along with chromatin modification were observed in a recent study in which C57BL/6 mice were supplemented with a mixture of SCFA (67.5 mM acetate, 40 mM propionate, and 25.9 mM butyrate) through drinking water (Louwies et al., 2020). Similarly, DNA and histone methylation are also regulated by gut microbiota by utilizing dietary methionine for the synthesis of SAM using the L-methionine S-adenosyl transferase (MAT) enzyme (Miro-Blanch and Yanes, 2019). SAM acts as a methyl donor for the transfer of methyl group to the 5th cytosine residue of the CpG Island by DNMTs and thus favors transcriptional repression. Ten eleven translocation (TET) enzyme functions for transcriptional activation through DNA methylation (Kumar et al., 2018). Folate, a methyl donor essential for the synthesis of SAM, is produced by Bifidobacterium sp. and Lactobacillus plantarum. Thus, gut dysbiosis can influence SAM levels, ultimately altering DNA and histone methylation state (Rossi et al., 2011).
Vagus nerve stimulation affects the hippocampal, cortical, and blood epigenetic transcriptomes in male Sprague Dawley rats and epigenetically modifies genes associated with neural plasticity and stress-response signaling (Sanders et al., 2019). Microbiota can cause an altered chromatin state, which might result in host immune maturation (Seeley et al., 2018). SCFAs are critical mediators of altered chromatin states. They regulate DNMTs and HDAC, thus regulating the expression of the MHC gene, which is involved in immune response (Ting and Trowsdale, 2002). According to a comparative NMR-based metabolome investigation using mouse models (Romano et al., 2017), there is a positive correlation between the luminal concentrations of gut metabolites and the number of Treg cells in the colon. Butyrate is a well-known anti-inflammatory molecule among SCFAs that promotes the differentiation of colonic Treg cells (Furusawa et al., 2013). Specific metabolite produced by gut microbiota acts as a cofactor for epigenetic reaction. In addition to being a necessary nutrient for the brain’s healthy development, choline also has a role in the production of SAM, a crucial methyl donor for the methylation of DNA and histones. Choline metabolizing bacteria competes with the host for dietary choline, thus resulting in low plasma and hepatic choline levels. Mothers with choline deficiency deliver fetus having hippocampal global DNA methylation and cholinergic neurons with altered function (Romano et al., 2017).
The majority of studies so far has been on the bacteria that dwell in our gut, although various species have diverse homes and functions throughout the body. Finding the correct data from the microbiome, which is necessary to understand how it affects our health, is one of the major hurdles in microbiome science. Before this cross-kingdom communication can be used to treat neurological illnesses, it is important to better understand the intricacy of the microbiota and its biochemical interaction with the host. This intricacy may be reflected in the conflicting results found in various investigations. The complex interplay exists between the metabolic pathways as well as bacterial and human metabolism. For instance, the abundance of SCFAs and bile acids in the gut is intrinsically linked to the production of intestinal neurotransmitters, and inflammatory molecules like nitrate encourage the metabolism of choline by choline-utilizing bacteria. This evidence suggests that the psychotropic effect of a particular metabolite may be tightly dependent on the presence of other metabolites. It is therefore difficult to determine how much of the observed impact on depressive behavior can be attributed to gut microbial metabolism. De Cremoux emphasized in a recent study from Seventure that it’s critical to remember that some microorganisms in the human microbiome are found in lesser numbers. The essential participants in a given condition could be concealed among less frequent species, despite the fact that it is easier to gather data on the most prevalent strains. Studies on the microbiome generate enormous volumes of data that are highly variable. Numerous studies in this area result in contradicting findings due to this variability.
Similarly, there are still significant gaps in our knowledge of how epigenetic mechanisms contribute to depression. The majority of studies describing epigenetic alterations in patients with depression have concentrated on DNA methylation and less research has been conducted on histone modifications and non-coding RNAs. The majority of research has used peripheral tissue, while those that have used post-mortem brain have typically used brain homogenate and smaller cohort size. These factors have made it challenging to confirm depression-related epigenetic changes across cohorts. The challenge has only been exacerbated by additional factors that are already part and parcel of psychiatric research, such as the polygenic architecture of depression, retrospective recollection of environmental exposures and symptom-based diagnoses.
Understanding of the pathological mechanisms of depression will advance with the closure of knowledge gaps on the multifactorial interaction between epigenetics, gut microbiome and their antidepressant effects, which may also help in the development of more sophisticated pharmacological approaches. Future study is critical to fully comprehend the gut flora, as shown by the aforementioned discoveries and the recent spike in media interest in gut health. Effective and widely available therapies for anxiety and depression would be helpful to millions of individuals worldwide since these conditions are becoming more prevalent globally.
The gut-brain axis is a dynamic, complex, and bidirectional communication network between the gut and brain. Since gut microbiota is a crucial regulator of the gut-brain axis, it is often referred to as microbiota-gut-brain axis. Some metabolites produced by the microbiome (SCFA, bile acids, tryptophan metabolites, vitamins, etc.) can cross BBB and exert a direct and indirect effect on the brain. Various preclinical and clinical studies revealed gut dysbiosis as one of the etiopathological factors in numerous metabolic, psychiatric, and neurodegenerative diseases. Gut metabolites can regulate the host genome by modifying host epigenome through various epigenetic mechanisms such as DNA methylation or acetylation, histone modification, RNA associated silencing, etc. butyrate, one of the SCFAs, is a well-known HDAC inhibitor, choline acts as methyl donor for DNA methylation, amino acids produced by gut bacteria acts as a precursor for the synthesis of neurotransmitters such as GABA, glutamate, epinephrine, nor-epinephrine, and dopamine. Depression is a multifactorial psychiatric disorder, and a plethora of studies has proved the separate involvement of gut microbiota as well as epigenetics as a key player in its etiopathogenesis. Despite the fact that gut microbiome is essential for modulating the host’s epigenetic processes, much research is required to understand the underlying molecular mechanisms and their biological effects on the host. For example, which bacterial species exhibit symbiotic relationships, how they interact with one another, and how their metabolites contribute to epigenetics as well as depression? Therefore, further research on particular gut microbial metabolites that can alters the host epigenome will provide new insight into human health and diseases. Additionally, studies examining the role of gut microbiome in depression and its impact on health conditions are becoming more prevalent. This may pave the way for the discovery of novel therapeutic mechanisms that could aid in the treatment and prevent depression by restoring the altered intestinal microbiome to a healthy state.
NB, AM, and KT drafted the manuscript. NB revised the manuscript critically for important intellectual content and made figures. SS, MS, SBS, and DK has made significant contributions to the development of the article’s concept and design, as well as to its peer review and editing. All authors read and approved the final manuscript.
The authors would like to acknowledge National Institute of Pharmaceutical Education and Research (NIPER) – Hyderabad and the Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Government of India for their support. All the figures in the manuscript are created at biorender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., and Versalovic, J. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6:237ra65. doi: 10.1126/SCITRANSLMED.3008599
Abe-Higuchi, N., Uchida, S., Yamagata, H., Higuchi, F., Hobara, T., Hara, K., et al. (2016). Hippocampal Sirtuin 1 signaling mediates depression-like behavior. Biol. Psychiatry 80, 815–826. doi: 10.1016/J.BIOPSYCH.2016.01.009
Abenavoli, L., Scarpellini, E., Colica, C., Boccuto, L., Salehi, B., Sharifi-Rad, J., et al. (2019). Gut microbiota and obesity: A role for probiotics. Nutrients 11:2690. doi: 10.3390/NU11112690
Aggarwal, S., Upadhyay, A., Shah, D., Teotia, N., Agarwal, A., and Jaiswal, V. (2014). Lactobacillus GG for treatment of acute childhood diarrhoea: An open labelled, randomized controlled trial. Indian J. Med. Res. 139, 379–385.
Agus, A., Clément, K., and Sokol, H. (2021). Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 70, 1174–1182. doi: 10.1136/GUTJNL-2020-323071
Ahn, J., and Hayes, R. B. (2021). Environmental influences on the human microbiome and implications for noncommunicable disease. Annu. Rev. Public Health 42:277. doi: 10.1146/ANNUREV-PUBLHEALTH-012420-105020
Ait-Belgnaoui, A., Durand, H., Cartier, C., Chaumaz, G., Eutamene, H., Ferrier, L., et al. (2012). Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 37, 1885–1895. doi: 10.1016/J.PSYNEUEN.2012.03.024
Aizawa, E., Tsuji, H., Asahara, T., Takahashi, T., Teraishi, T., Yoshida, S., et al. (2016). Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord 202, 254–257. doi: 10.1016/J.JAD.2016.05.038
Al-Jahmany, A. A., Schultheiss, G., and Diener, M. (2004). Effects of dopamine on ion transport across the rat distal colon. Pflugers Arch. 448, 605–612. doi: 10.1007/S00424-004-1299-9
Alt, S. R., Turner, J. D., Klok, M. D., Meijer, O. C., Lakke, E. A. J. F., DeRijk, R. H., et al. (2010). Differential expression of glucocorticoid receptor transcripts in major depressive disorder is not epigenetically programmed. Psychoneuroendocrinology 35, 544–556. doi: 10.1016/J.PSYNEUEN.2009.09.001
Alverdy, J., Holbrook, C., Rocha, F., Seiden, L., Wu, R. L., Musch, M., et al. (2000). Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: Evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann. Surg. 232, 480–489. doi: 10.1097/00000658-200010000-00003
Ambrosini, Y. M., Borcherding, D., Kanthasamy, A., Kim, H. J., Willette, A. A., Jergens, A., et al. (2019). The gut-brain axis in neurodegenerative diseases and relevance of the canine model: A review. Front. Aging Neurosci. 11:130. doi: 10.3389/FNAGI.2019.00130/FULL
Ansari, F., Pourjafar, H., Tabrizi, A., and Homayouni, A. (2020). The effects of probiotics and prebiotics on mental disorders: A review on depression, anxiety, alzheimer, and autism spectrum disorders. Curr. Pharm. Biotechnol. 21, 555–565. doi: 10.2174/1389201021666200107113812
Arentsen, T., Qian, Y., Gkotzis, S., Femenia, T., Wang, T., Udekwu, K., et al. (2016). The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol. Psychiatry 2, 257–266. doi: 10.1038/mp.2016.182
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/NATURE09944
Azm, S. A. N., Djazayeri, A., Safa, M., Azami, K., Ahmadvand, B., Sabbaghziarani, F., et al. (2018). Lactobacilli and bifidobacteria ameliorate memory and learning deficits and oxidative stress in β-amyloid (1–42) injected rats. Appl. Physiol. Nutr. Metab. 43, 718–726. doi: 10.1139/apnm-2017-0648
Bäckhed, F., Fraser, C. M., Ringel, Y., Sanders, M. E., Sartor, R. B., Sherman, P. M., et al. (2012). Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe 12, 611–622. doi: 10.1016/J.CHOM.2012.10.012
Bailey, M. T., and Cryan, J. F. (2017). The microbiome as a key regulator of brain, behavior and immunity: Commentary on the 2017 named series. Brain Behav. Immun. 66, 18–22. doi: 10.1016/J.BBI.2017.08.017
Bajramagic, S., Hodzic, E., Mulabdic, A., and Holjan, S. (2019). Usage of probiotics and its clinical significance at surgically treated patients sufferig from colorectal carcinoma. Med. Arch. 73, 316–320. doi: 10.5455/medarh.2019.73.316-320
Banks, W. (2005). Blood-brain barrier transport of cytokines: A mechanism for neuropathology. Curr. Pharm. Des. 11, 973–984. doi: 10.2174/1381612053381684
Barden, N. (2004). Implication of the hypothalamic-pituitary-adrenal axis in the physiopathology of depression. J. Psychiatry Neurosci. 29, 185–193.
Bargiel, P., Szczuko, M., Stachowska, L., Prowans, P., Czapla, N., Markowska, M., et al. (2021). Microbiome metabolites and thyroid dysfunction. J. Clin. Med. 10:3609. doi: 10.3390/JCM10163609
Barton, W., Penney, N. C., Cronin, O., Garcia-Perez, I., Molloy, M. G., Holmes, E., et al. (2018). The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 67, 625–633. doi: 10.1136/GUTJNL-2016-313627
Bayles, R., Baker, E. K., Jowett, J. B. M., Barton, D., and Esler, M. (2013). Methylation of the SLC6a2 gene promoter in major depression and panic disorder. PLoS One 8:83223. doi: 10.1371/journal.pone.0083223
Bell, A. F., Carter, C. S., Steer, C. D., Golding, J., Davis, J. M., Steffen, A. D., et al. (2015). Interaction between oxytocin receptor DNA methylation and genotype is associated with risk of postpartum depression in women without depression in pregnancy. Front. Genet. 6:243. doi: 10.3389/FGENE.2015.00243
Berk, M., Williams, L. J., Jacka, F. N., O’Neil, A., Pasco, J. A., Moylan, S., et al. (2013). So depression is an inflammatory disease, but where does the inflammation come from? BMC Med. 11:200. doi: 10.1186/1741-7015-11-200
Biedermann, L., Zeitz, J., Mwinyi, J., Sutter-Minder, E., Rehman, A., Ott, S. J., et al. (2013). Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 8:e59260. doi: 10.1371/JOURNAL.PONE.0059260
Bonder, M. J., Tigchelaar, E. F., Cai, X., Trynka, G., Cenit, M. C., Hrdlickova, B., et al. (2016). The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 8, 1–11. doi: 10.1186/S13073-016-0295-Y/TABLES/4
Booij, L., Wang, D., Lévesque, M. L., Tremblay, R. E., and Szyf, M. (2013). Looking beyond the DNA sequence: The relevance of DNA methylation processes for the stress-diathesis model of depression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120251. doi: 10.1098/RSTB.2012.0251
Boro, P., Kumaresan, A., Singh, A. K., Gupta, D., Kumar, S., Manimaran, A., et al. (2014). Expression of short chain fatty acid receptors and pro-inflammatory cytokines in utero-placental tissues is altered in cows developing retention of fetal membranes. Placenta 35, 455–460. doi: 10.1016/J.PLACENTA.2014.04.009
Bosker, F. J., Hartman, C. A., Nolte, I. M., Prins, B. P., Terpstra, P., Posthuma, D., et al. (2011). Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol. Psychiatry 16, 516–532. doi: 10.1038/MP.2010.38
Braithwaite, E. C., Kundakovic, M., Ramchandani, P. G., and Champagne, F. (2015). Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics 10, 408–417. doi: 10.1080/15592294.2015.1039221
Bravo, J. A., Forsythe, P., Chew, M. v, Escaravage, E., Savignac, H. M., Dinan, T. G., et al. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 108, 16050–16055. doi: 10.1073/PNAS.1102999108
Bressa, C., Bailén-Andrino, M., Pérez-Santiago, J., González-Soltero, R., Pérez, M., Montalvo-Lominchar, M. G., et al. (2017). Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One 12:e0171352. doi: 10.1371/JOURNAL.PONE.0171352
Breton, J., Tennoune, N., Lucas, N., Francois, M., Legrand, R., Jacquemot, J., et al. (2016). Gut commensal E. coli proteins activate host satiety pathways following nutrient-induced bacterial growth. Cell Metab. 23, 324–334. doi: 10.1016/J.CMET.2015.10.017
Burokas, A., Moloney, R. D., Dinan, T. G., and Cryan, J. F. (2015). Microbiota regulation of the mammalian gut-brain axis. Adv. Appl. Microbiol. 91, 1–62. doi: 10.1016/BS.AAMBS.2015.02.001
Cai, N., Bigdeli, T. B., Kretzschmar, W., Lei, Y., Liang, J., Song, L., et al. (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591. doi: 10.1038/NATURE14659
Caspani, G., Kennedy, S., Foster, J. A., and Swann, J. (2019). Gut microbial metabolites in depression: Understanding the biochemical mechanisms. Microbial. Cell 6, 454–481. doi: 10.15698/MIC2019.10.693
Chagnon, Y. C., Potvin, O., Hudon, C., and Préville, M. (2015). DNA methylation and single nucleotide variants in the brain-derived neurotrophic factor (BDNF) and oxytocin receptor (OXTR) genes are associated with anxiety/depression in older women. Front. Genet. 6:230. doi: 10.3389/FGENE.2015.00230
Chao, L., Liu, C., Sutthawongwadee, S., Li, Y., Lv, W., Chen, W., et al. (2020). Effects of probiotics on depressive or anxiety variables in healthy participants under stress conditions or with a depressive or anxiety diagnosis: A meta-analysis of randomized controlled trials. Front. Neurol. 11:421. doi: 10.3389/FNEUR.2020.00421/BIBTEX
Chen, J.-J., Zhou, C.-J., Liu, Z., Fu, Y.-Y., Zheng, P., Yang, D.-Y., et al. (2015). Divergent urinary metabolic phenotypes between major depressive disorder and bipolar disorder identified by a combined GC-MS and NMR spectroscopic metabonomic approach. J. Proteome Res. 14, 3382–3389. doi: 10.1021/acs.jproteome.5b00434
Chowdhury, S. R., King, D. E., Willing, B. P., Band, M. R., Beever, J. E., Lane, A. B., et al. (2007). Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics 8:215. doi: 10.1186/1471-2164-8-215
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: A bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/PNAS.0604343103
Clarke, S. F., Murphy, E. F., O’Sullivan, O., Lucey, A. J., Humphreys, M., Hogan, A., et al. (2014). Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920. doi: 10.1136/GUTJNL-2013-306541
Clemente-Postigo, M., Queipo-Ortuño, M. I., Murri, M., Boto-Ordoñez, M., Perez-Martinez, P., Andres-Lacueva, C., et al. (2012). Endotoxin increase after fat overload is related to postprandial hypertriglyceridemia in morbidly obese patients. J. Lipid Res. 53, 973–978. doi: 10.1194/JLR.P020909
Cogan, T. A., Thomas, A. O., Rees, L. E. N., Taylor, A. H., Jepson, M. A., Williams, P. H., et al. (2007). Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 56, 1060–1065. doi: 10.1136/GUT.2006.114926
Conradt, E., Lester, B. M., Appleton, A. A., Armstrong, D. A., and Marsit, C. J. (2013). The roles of DNA methylation of NR3C1 and 11β-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics 8, 1321–1329. doi: 10.4161/epi.26634
Covington, H. E., Maze, I., LaPlant, Q. C., Vialou, V. F., Ohnishi, Y. N., Berton, O., et al. (2009). Antidepressant actions of histone deacetylase inhibitors. J. Neurosci. 29, 11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009
Covington, H. E., Maze, I., Vialou, V., and Nestler, E. J. (2015). Antidepressant action of HDAC inhibition in the prefrontal cortex. Neuroscience 298, 329–335. doi: 10.1016/J.NEUROSCIENCE.2015.04.030
Cresci, G. A., and Bawden, E. (2015). Gut microbiome: What we do and don’t know. Nutr. Clin. Pract. 30, 734–746. doi: 10.1177/0884533615609899
Cryan, J. F., and Dinan, T. G. (2012). Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. doi: 10.1038/NRN3346
Cummings, J. H., Pomare, E. W., Branch, H. W. J., Naylor, E., and Macfarlane, G. T. (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227. doi: 10.1136/gut.28.10.1221
De Filippis, F., Pellegrini, N., Vannini, L., Jeffery, I. B., La Storia, A., Laghi, L., et al. (2016). High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65, 1812–1821. doi: 10.1136/GUTJNL-2015-309957
de Vadder, F., Kovatcheva-Datchary, P., Goncalves, D., Vinera, J., Zitoun, C., Duchampt, A., et al. (2014). Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96. doi: 10.1016/J.CELL.2013.12.016
Demaude, J., Salvador-Cartier, C., Fioramonti, J., Ferrier, L., and Bueno, L. (2006). Phenotypic changes in colonocytes following acute stress or activation of mast cells in mice: Implications for delayed epithelial barrier dysfunction. Gut 55, 655–661. doi: 10.1136/GUT.2005.078675
Denou, E., Marcinko, K., Surette, M. G., Steinberg, G. R., and Schertzer, J. D. (2016). High-intensity exercise training increases the diversity and metabolic capacity of the mouse distal gut microbiota during diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 310, E982–E993. doi: 10.1152/AJPENDO.00537.2015
Derrien, M., Alvarez, A. S., and de Vos, W. M. (2019). The gut microbiota in the first decade of life. Trends Microbiol. 27, 997–1010. doi: 10.1016/J.TIM.2019.08.001
Desbonnet, L., Clarke, G., Traplin, A., O’Sullivan, O., Crispie, F., Moloney, R. D., et al. (2015). Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav. Immun. 48, 165–173. doi: 10.1016/J.BBI.2015.04.004
Desbonnet, L., Garrett, L., Clarke, G., Kiely, B., Cryan, J. F., and Dinan, T. G. (2010). Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 170, 1179–1188. doi: 10.1016/J.NEUROSCIENCE.2010.08.005
Devlin, A. M., Brain, U., Austin, J., and Oberlander, T. F. (2010). Prenatal exposure to maternal depressed mood and the MTHFR C677T variant affect SLC6A4 methylation in infants at birth. PLoS One 5:e12201. doi: 10.1371/journal.pone.0012201
DiGiulio, D. B., Romero, R., Amogan, H. P., Kusanovic, J. P., Bik, E. M., Gotsch, F., et al. (2008). Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: A molecular and culture-based investigation. PLoS One 3:e3056. doi: 10.1371/JOURNAL.PONE.0003056
Domschke, K., Tidow, N., Schwarte, K., Ziegler, C., Lesch, K. P., Deckert, J., et al. (2014). Pharmacoepigenetics of depression: No major influence of MAO-A DNA methylation on treatment response. J.Neural Transm. 1, 99–108. doi: 10.1007/S00702-014-1227-X
Donohoe, D. R., Holley, D., Collins, L. B., Montgomery, S. A., Whitmore, A. C., Hillhouse, A., et al. (2014). A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate–dependent manner. Cancer Discov. 4:1387. doi: 10.1158/2159-8290.CD-14-0501
Doyle, R. M., Harris, K., Kamiza, S., Harjunmaa, U., Ashorn, U., Nkhoma, M., et al. (2017). Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS One 12:e0180167. doi: 10.1371/JOURNAL.PONE.0180167
Drasar, B. S., Crowther, J. S., Goddard, P., Hawksworth, G., Hill, M. J., Peach, S., et al. (1973). The relation between diet and the gut microflora in man. Proc. Nutr. Soc. 32, 49–52. doi: 10.1079/PNS19730014
Durk, R. P., Castillo, E., Márquez-Magaña, L., Grosicki, G. J., Bolter, N. D., Matthew Lee, C., et al. (2019). Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int. J. Sport. Nutr. Exerc. Metab. 29, 249–253. doi: 10.1123/IJSNEM.2018-0024
Eisenhofer, G., Aneman, Å, Friberg, P., Hooper, D., Fåndriks, L., Lonroth, H., et al. (1997). Substantial production of dopamine in the human gastrointestinal tract. J. Clin. Endocrinol. Metab. 82, 3864–3871. doi: 10.1210/JCEM.82.11.4339
Erburu, M., Muoz-Cobo, I., Diaz-Perdigon, T., Mellini, P., Suzuki, T., Puerta, E., et al. (2017). SIRT2 inhibition modulate glutamate and serotonin systems in the prefrontal cortex and induces antidepressant-like action. Neuropharmacology 117, 195–208. doi: 10.1016/J.NEUROPHARM.2017.01.033
Eskesen, D., Jespersen, L., Michelsen, B., Whorwell, P. J., Müller-Lissner, S., and Morberg, C. M. (2015). Effect of the probiotic strain Bifidobacterium animalis subsp. lactis, BB-12®, on defecation frequency in healthy subjects with low defecation frequency and abdominal. Br. J. Nutr. 114, 1638–1646. doi: 10.1017/S0007114515003347
Evrensel, A., Ünsalver, B. Ö, and Ceylan, M. E. (2020). Neuroinflammation, gut-brain axis and depression. Psychiatry Investig. 17, 2–8. doi: 10.30773/PI.2019.08.09
Fava, F., Gitau, R., Griffin, B. A., Gibson, G. R., Tuohy, K. M., and Lovegrove, J. A. (2013). The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short-chain fatty acid excretion in a metabolic syndrome “at-risk” population. Int. J. Obes. (Lond) 37, 216–223. doi: 10.1038/IJO.2012.33
Fraga, M. F., Ballestar, E., Paz, M. F., Ropero, S., Setien, F., Ballestar, M. L., et al. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U.S.A. 102, 10604–10609. doi: 10.1073/PNAS.0500398102
Francke, U., and Foellmer, B. E. (1989). The glucocorticoid receptor gene is in 5q31-q32. Genomics 4, 610–612. doi: 10.1016/0888-7543(89)90287-5
Freestone, P. P. E., Haigh, R. D., Williams, P. H., and Lyte, M. (2003). Involvement of enterobactin in norepinephrine-mediated iron supply from transferrin to enterohaemorrhagic Escherichia coli. FEMS Microbiol. Lett. 222, 39–43. doi: 10.1016/S0378-1097(03)00243-X
Freestone, P. P., Williams, P. H., Haigh, R. D., Maggs, A. F., Neal, C. P., and Lyte, M. (2002). Growth stimulation of intestinal commensal Escherichia coli by catecholamines: A possible contributory factor in trauma-induced sepsis. Shock 18, 465–470. doi: 10.1097/00024382-200211000-00014
Frost, G., Sleeth, M. L., Sahuri-Arisoylu, M., Lizarbe, B., Cerdan, S., Brody, L., et al. (2014). The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat. Commun. 5:3611. doi: 10.1038/NCOMMS4611
Fuchikami, M., Morinobu, S., Segawa, M., Okamoto, Y., Yamawaki, S., Ozaki, N., et al. (2011). DNA methylation profiles of the Brain-Derived Neurotrophic Factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS One 6:e23881. doi: 10.1371/JOURNAL.PONE.0023881
Fulford, A. J., and Harbuz, M. S. (2005). Chapter 1.3 An introduction to the HPA axis. Techniq. Behav. Neural Sci. 15, 43–65. doi: 10.1016/S0921-0709(05)80006-9
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/NATURE12721
Galley, J. D., Nelson, M. C., Yu, Z., Dowd, S. E., Walter, J., Kumar, P. S., et al. (2014). Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 14:189–201. doi: 10.1186/1471-2180-14-189
Gao, K., Pi, Y., Mu, C. L., Farzi, A., Liu, Z., and Zhu, W. Y. (2019). Increasing carbohydrate availability in the hindgut promotes hypothalamic neurotransmitter synthesis: Aromatic amino acids linking the microbiota-brain axis. J. Neurochem. 149, 641–659. doi: 10.1111/JNC.14709
Gaykema, R. P. A., Goehler, L. E., and Lyte, M. (2004). Brain response to cecal infection with Campylobacter jejuni: Analysis with Fos immunohistochemistry. Brain Behav. Immun. 18, 238–245. doi: 10.1016/j.bbi.2003.08.002
Goehler, L. E., Gaykema, R. P. A., Nguyen, K. T., Lee, J. E., Tilders, F. J. H., Maier, S. F., et al. (1999). Interleukin-1beta in immune cells of the abdominal vagus nerve: A link between the immune and nervous systems? J. Neurosci. 19, 2799–2806. doi: 10.1523/JNEUROSCI.19-07-02799.1999
Gosalbes, M. J., Llop, S., Vallés, Y., Moya, A., Ballester, F., and Francino, M. P. (2013). Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43, 198–211. doi: 10.1111/CEA.12063
Gowher, H., Liebert, K., Hermann, A., Xu, G., and Jeltsch, A. (2005). Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 280, 13341–13348. doi: 10.1074/JBC.M413412200
Groer, M. W., Luciano, A. A., Dishaw, L. J., Ashmeade, T. L., Miller, E., and Gilbert, J. A. (2014). Development of the preterm infant gut microbiome: A research priority. Microbiome 2:38. doi: 10.1186/2049-2618-2-38
Gue, M., Junien, J. L., and Bueno, L. (1991). Conditioned emotional response in rats enhances colonic motility through the central release of corticotropin-releasing factor. Gastroenterology 100, 964–970. doi: 10.1016/0016-5085(91)90270-U
Gué, M., Peeters, T., Depoortere, I., Vantrappen, G., and Buéno, L. (1989). Stress-induced changes in gastric emptying, postprandial motility, and plasma gut hormone levels in dogs. Gastroenterology 97, 1101–1107. doi: 10.1016/0016-5085(89)91678-8
Gur, T. L., Shay, L., Palkar, A. V., Fisher, S., Varaljay, V. A., Dowd, S., et al. (2017). Prenatal stress affects placental cytokines and neurotrophins, commensal microbes, and anxiety-like behavior in adult female offspring. Brain Behav. Immun. 64, 50–58. doi: 10.1016/J.BBI.2016.12.021
Guthrie, G. D., and Nicholson-Guthrie, C. S. (1989). gamma-Aminobutyric acid uptake by a bacterial system with neurotransmitter binding characteristics. Proc. Natl. Acad. Sci. U.S.A. 86, 7378–7381. doi: 10.1073/PNAS.86.19.7378
Haarman, M., and Knol, J. (2006). Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 72, 2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006
Halden, R. U. (2016). Lessons learned from probing for impacts of triclosan and triclocarban on human microbiomes. mSphere 1, e89–e16. doi: 10.1128/MSPHERE.00089-16
Han, A., Sung, Y. B., Chung, S. Y., and Kwon, M. S. (2014). Possible additional antidepressant-like mechanism of sodium butyrate: Targeting the hippocampus. Neuropharmacology 81, 292–302. doi: 10.1016/J.NEUROPHARM.2014.02.017
Higuchi, F., Uchida, S., Yamagata, H., Abe-Higuchi, N., Hobara, T., Hara, K., et al. (2016). Hippocampal microRNA-124 enhances chronic stress resilience in mice. J. Neurosci. 36, 7253–7267. doi: 10.1523/JNEUROSCI.0319-16.2016
Hill, C. J., Lynch, D. B., Murphy, K., Ulaszewska, M., Jeffery, I. B., O’Shea, C. A., et al. (2017). Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 5, 1–18. doi: 10.1186/S40168-016-0213-Y/FIGURES/8
Hilton, E., Kolakowski, P., Singer, C., and Smith, M. (1997). Efficacy of lactobacillus GG as a diarrheal preventive in travelers. J. Travel Med. 4, 41–43. doi: 10.1111/J.1708-8305.1997.TB00772.X
Hobara, T., Uchida, S., Otsuki, K., Matsubara, T., Funato, H., Matsuo, K., et al. (2010). Altered gene expression of histone deacetylases in mood disorder patients. J. Psychiatr. Res. 44, 263–270. doi: 10.1016/J.JPSYCHIRES.2009.08.015
Höhne, N., Poidinger, M., Merz, F., Pfister, H., Brückl, T., Zimmermann, P., et al. (2015). FKBP5 genotype-dependent DNA methylation and mrna regulation after psychosocial stress in remitted depression and healthy controls. International J. Neuropsychopharmacol. 18, 1–9. doi: 10.1093/IJNP/PYU087
Hooper, L., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/SCIENCE.1223490
Hsiao, E. Y., McBride, S. W., Hsien, S., Sharon, G., Hyde, E. R., McCue, T., et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463. doi: 10.1016/j.cell.2013.11.024
Huang, N., Hua, D., Zhan, G., Li, S., Zhu, B., Jiang, R., et al. (2019). Role of Actinobacteria and Coriobacteriia in the antidepressant effects of ketamine in an inflammation model of depression. Pharmacol. Biochem. Behav. 176, 93–100. doi: 10.1016/J.PBB.2018.12.001
Hugenholtz, F., Mullaney, J. A., Kleerebezem, M., Smidt, H., and Rosendale, D. I. (2013). Modulation of the microbial fermentation in the gut by fermentable carbohydrates. Amsterdam: Elsevier.
Hughes, D. T., and Sperandio, V. (2008). Inter-kingdom signalling: Communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. doi: 10.1038/NRMICRO1836
Hunter, R. G., McCarthy, K. J., Milne, T. A., Pfaff, D. W., and McEwen, B. S. (2009). Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc. Natl. Acad. Sci. U.S.A. 106, 20912–20917. doi: 10.1073/PNAS.0911143106
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., Chinwalla, A. T., et al. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/NATURE11234
Iannone, L. F., Preda, A., Blottière, H. M., Clarke, G., Albani, D., Belcastro, V., et al. (2019). Microbiota-gut brain axis involvement in neuropsychiatric disorders. Exp. Rev. Neurother 19, 1037–1050. doi: 10.1080/14737175.2019.1638763
Iga, J. I., Ueno, S. I., Yamauchi, K., Numata, S., Kinouchi, S., Tayoshi-Shibuya, S., et al. (2007). Altered HDAC5 and CREB mRNA expressions in the peripheral leukocytes of major depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 31, 628–632. doi: 10.1016/J.PNPBP.2006.12.014
Iglesias–vázquez, L., Van Ginkel Riba, G., Arija, V., and Canals, J. (2020). Composition of gut microbiota in children with autism spectrum disorder: A systematic review and meta-analysis. Nutrients 12:792. doi: 10.3390/NU12030792
Jiang, H., Ling, Z., Zhang, Y., Mao, H., Ma, Z., Yin, Y., et al. (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. doi: 10.1016/J.BBI.2015.03.016
Jiang, W., Ling, Z., Lin, X., Chen, Y., Zhang, J., Yu, J., et al. (2014). Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb. Ecol. 67, 962–969. doi: 10.1007/s00248-014-0372-y
Jin, Y., Wu, S., Zeng, Z., and Fu, Z. (2017). Effects of environmental pollutants on gut microbiota. Environ. Pollut. 222, 1–9. doi: 10.1016/J.ENVPOL.2016.11.045
Jokela, M., Virtanen, M., David Batty, G., and Kivimaki, M. (2016). Inflammation and specific symptoms of depression. JAMA Psychiatry 73, 87–88. doi: 10.1001/JAMAPSYCHIATRY.2015.1977
Kaelberer, M. M., Buchanan, K. L., Klein, M. E., Barth, B. B., Montoya, M. M., Shen, X., et al. (2018). A gut-brain neural circuit for nutrient sensory transduction. Science 361:eaat5236. doi: 10.1126/SCIENCE.AAT5236
Kaelberer, M. M., Rupprecht, L. E., Liu, W. W., Weng, P., and Bohorquez, D. V. (2020). Neuropod cells: The emerging biology of gut-brain sensory transduction. Annu. Rev. Neurosci. 43, 337–353. doi: 10.1146/ANNUREV-NEURO-091619-022657
Kang, H. J., Kim, J. M., Kim, S. Y., Kim, S. W., Shin, I. S., Kim, H. R., et al. (2015). A longitudinal study of BDNF promoter methylation and depression in breast cancer. Psychiatry Investig. 12, 523–531. doi: 10.4306/pi.2015.12.4.523
Kang, S. S., Jeraldo, P. R., Kurti, A., Miller, M. E. B., Cook, M. D., Whitlock, K., et al. (2014). Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol. Neurodegener. 9, 1–12. doi: 10.1186/1750-1326-9-36/FIGURES/6
Karlsson, F., Tremaroli, V., Nielsen, J., and Bäckhed, F. (2013). Assessing the human gut microbiota in metabolic diseases. Diabetes 62, 3341–3349. doi: 10.2337/DB13-0844
Karnib, N., El-Ghandour, R., El Hayek, L., Nasrallah, P., Khalifeh, M., Barmo, N., et al. (2019). Lactate is an antidepressant that mediates resilience to stress by modulating the hippocampal levels and activity of histone deacetylases. Neuropsychopharmacology 44, 1152–1162. doi: 10.1038/s41386-019-0313-z
Kelly, J. R., Borre, Y., O’ Brien, C., Patterson, E., El Aidy, S., Deane, J., et al. (2016). Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 82, 109–118. doi: 10.1016/J.JPSYCHIRES.2016.07.019
Kendler, K. S. (1983). Overview: A current perspective on twin studies of schizophrenia. Am. J. Psychiatry 140, 1413–1425. doi: 10.1176/AJP.140.11.1413
Kenyon, C., Colebunders, R., and Crucitti, T. (2013). The global epidemiology of bacterial vaginosis: A systematic review. Am. J. Obstet. Gynecol. 209, 505–523. doi: 10.1016/J.AJOG.2013.05.006
Kho, Z. Y., and Lal, S. K. (2018). The human gut microbiome - A potential controller of wellness and disease. Front. Microbiol. 9:1835. doi: 10.3389/FMICB.2018.01835
Kim, D., Kubzansky, L. D., Baccarelli, A., Sparrow, D., Spiro, A., Tarantini, L., et al. (2016). Psychological factors and DNA methylation of genes related to immune/inflammatory system markers: The VA Normative Aging Study. BMJ Open 6:e009790. doi: 10.1136/BMJOPEN-2015-009790
Kim, M. J., Romero, R., Gervasi, M. T., Kim, J. S., Yoo, W., Lee, D. C., et al. (2009). Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab. Investig. 89, 924–936. doi: 10.1038/labinvest.2009.49
Kimmel, M., Clive, M., Gispen, F., Guintivano, J., Brown, T., Cox, O., et al. (2016). Oxytocin receptor DNA methylation in postpartum depression. Psychoneuroendocrinology 69, 150–160. doi: 10.1016/j.psyneuen.2016.04.008
Kobayashi, T., and Fujiwara, K. (2013). Identification of Heavy Smokers through Their Intestinal Microbiota by Data Mining Analysis. Biosci. Microbiota Food Health 32, 77–80. doi: 10.12938/BMFH.32.77
Koh, A., de Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/J.CELL.2016.05.041
Kok, D. E. G., Dhonukshe-Rutten, R. A. M., Lute, C., Heil, S. G., Uitterlinden, A. G., van der Velde, N., et al. (2015). The effects of long-term daily folic acid and vitamin B12 supplementation on genomewide DNA methylation in elderly subjects. Clin. Epigenetics 7:121. doi: 10.1186/S13148-015-0154-5
Koloverou, E., Panagiotakos, D. B., Pitsavos, C., Chrysohoou, C., Georgousopoulou, E. N., Grekas, A., et al. (2016). Adherence to Mediterranean diet and 10-year incidence (2002-2012) of diabetes: Correlations with inflammatory and oxidative stress biomarkers in the ATTICA cohort study. Diabetes Metab. Res. Rev. 32, 73–81. doi: 10.1002/DMRR.2672
Koussoulas, K., Swaminathan, M., Fung, C., Bornstein, J. C., and Foong, J. P. P. (2018). Neurally released GABA acts via GABAC receptors to modulate Ca2+ transients evoked by trains of synaptic inputs, but not responses evoked by single stimuli, in myenteric neurons of mouse ileum. Front. Physiol. 9:97. doi: 10.3389/FPHYS.2018.00097
Kovacheva, V. P., Mellott, T. J., Davison, J. M., Wagner, N., Lopez-Coviella, I., Schnitzler, A. C., et al. (2007). Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J. Biol. Chem. 282, 31777–31788. doi: 10.1074/JBC.M705539200
Krautkramer, K. A., Kreznar, J. H., Romano, K. A., Vivas, E. I., Barrett-Wilt, G. A., Rabaglia, M. E., et al. (2016). Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992. doi: 10.1016/J.MOLCEL.2016.10.025
Kumar, H., Lund, R., Laiho, A., Lundelin, K., Ley, R. E., Isolauri, E., et al. (2014). Gut microbiota as an epigenetic regulator: Pilot study based on whole-genome methylation analysis. mBio 5, e2113–e2114. doi: 10.1128/MBIO.02113-14
Kumar, S., Chinnusamy, V., and Mohapatra, T. (2018). Epigenetics of modified DNA bases: 5-Methylcytosine and beyond. Front. Genet. 9:640. doi: 10.3389/FGENE.2018.00640
Kumari, M., Singh, P., Nataraj, B., Kokkiligadda, A., and Naithani, H. (2021). Fostering next-generation probiotics in human gut by targeted dietary modulation: An emerging perspective. Food Res. Int. 150:110716. doi: 10.1016/j.foodres.2021.110716
Ladizinski, B., Mclean, R., Lee, K. C., Elpern, D. J., and Eron, L. (2014). The human skin microbiome. Int. J. Dermatol. 53, 1177–1179. doi: 10.1111/IJD.12609
Lahtinen, S. J., Forssten, S., Aakko, J., Granlund, L., Rautonen, N., Salminen, S., et al. (2011). Probiotic cheese containing Lactobacillus rhamnosus HN001 and Lactobacillus acidophilus NCFM® modifies subpopulations of fecal lactobacilli and Clostridium difficile in the elderly. Age (Omaha) 34, 133–143. doi: 10.1007/S11357-011-9208-6
Lai, H. H., Chiu, C. H., Kong, M. S., Chang, C. J., and Chen, C. C. (2019). Probiotic Lactobacillus casei: Effective for managing childhood diarrhea by altering gut microbiota and attenuating fecal inflammatory markers. Nutrients 11:1150. doi: 10.3390/NU11051150
Lal, S., Kirkup, A. J., Brunsden, A. M., Thompson, D. G., and Grundy, D. (2001). Vagal afferent responses to fatty acids of different chain length in the rat. Am. J. Physiol. Gastrointest Liver Physiol. 281, G907–G915. doi: 10.1152/AJPGI.2001.281.4.G907
Lambert, J. E., Myslicki, J. P., Bomhof, M. R., Belke, D. D., Shearer, J., and Reimer, R. A. (2015). Exercise training modifies gut microbiota in normal and diabetic mice. Appl. Physiol. Nutr. Metab. 40, 749–752. doi: 10.1139/APNM-2014-0452
Laplant, Q., Vialou, V., Covington, H. E., Dumitriu, D., Feng, J., Warren, B. L., et al. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137–1143. doi: 10.1038/NN.2619
Le Hurou-Luron, I., Blat, S., and Boudry, G. (2010). Breast- v. formula-feeding: Impacts on the digestive tract and immediate and long-term health effects. Nutr. Res. Rev. 23, 23–36. doi: 10.1017/S0954422410000065
Ledford, H. (2015). First robust genetic links to depression emerge. Nature 523, 268–269. doi: 10.1038/523268A
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022A
Liang, L., Ai, L., Qian, J., Fang, J. Y., and Xu, J. (2015). Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 5:11763. doi: 10.1038/SREP11763
Liang, S., Wu, X., Hu, X., Wang, T., and Jin, F. (2018). Recognizing depression from the microbiota–gut–brain axis. Int. J. Mol. Sci. 19:1592. doi: 10.3390/IJMS19061592
Libert, S., Pointer, K., Bell, E. L., Das, A., Cohen, D. E., Asara, J. M., et al. (2011). SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell 147, 1459–1472. doi: 10.1016/J.CELL.2011.10.054
Lin, A., Bik, E. M., Costello, E. K., Dethlefsen, L., Haque, R., Relman, D. A., et al. (2013). Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One 8:e53838. doi: 10.1371/JOURNAL.PONE.0053838
Liu, Y., Wang, H., Gui, S., Zeng, B., Pu, J., Zheng, P., et al. (2021). Proteomics analysis of the gut–brain axis in a gut microbiota-dysbiosis model of depression. Transl. Psychiatry 11:568. doi: 10.1038/S41398-021-01689-W
Liu, Y., Zhang, L., Wang, X., Wang, Z., Zhang, J., Jiang, R., et al. (2016). Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin. Gastroenterol. Hepatol. 14, 1602.e–1611.e. doi: 10.1016/J.CGH.2016.05.033
Liu, Z., Qin, H., Yang, Z., Xia, Y., Liu, W., Yang, J., et al. (2011). Randomised clinical trial: The effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery–a. Wiley Online Library 33, 50–63. doi: 10.1111/j.1365-2036.2010.04492.x
Lopez-Legarrea, P., Fuller, N. R., Zulet, M. A., Martinez, J. A., and Caterson, I. D. (2014). The influence of Mediterranean, carbohydrate and high protein diets on gut microbiota composition in the treatment of obesity and associated inflammatory state. Asia Pac. J. Clin. Nutr. 23, 360–368. doi: 10.6133/APJCN.2014.23.3.16
Louis, P., and Flint, H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41. doi: 10.1111/1462-2920.13589
Louwies, T., Johnson, A. C., Orock, A., Yuan, T., and Greenwood-Van Meerveld, B. (2020). The microbiota-gut-brain axis: An emerging role for the epigenome. Exp. Biol. Med. 245, 138–145. doi: 10.1177/1535370219891690
Luqman, A., Nega, M., Nguyen, M. T., Ebner, P., and Götz, F. (2018). SadA-Expressing staphylococci in the human gut show increased cell adherence and internalization. Cell Rep. 22, 535–545. doi: 10.1016/J.CELREP.2017.12.058
Macfarlane, S., and Dillon, J. F. (2007). Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102, 1187–1196. doi: 10.1111/J.1365-2672.2007.03287.X
Makino, H., Kushiro, A., Ishikawa, E., Kubota, H., Gawad, A., Sakai, T., et al. (2013). Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One 8:e78331. doi: 10.1371/JOURNAL.PONE.0078331
Martinowich, K., Hattori, D., Wu, H., Fouse, S., He, F., Hu, Y., et al. (2003). DNA methylation-related chromatin remodeling in activity-dependent bdnf gene regulation. Science 302, 890–893. doi: 10.1126/SCIENCE.1090842/SUPPL_FILE/MARTINOWICH.SOM.REV.PDF
Matsumoto, M., Inoue, R., Tsukahara, T., Ushida, K., Chiji, H., Matsubara, N., et al. (2008). Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 72, 572–576. doi: 10.1271/BBB.70474
McVey Neufeld, K. A., O’Mahony, S. M., Hoban, A. E., Waworuntu, R. V., Berg, B. M., Dinan, T. G., et al. (2019). Neurobehavioural effects of Lactobacillus rhamnosus GG alone and in combination with prebiotics polydextrose and galactooligosaccharide in male rats exposed to early-life stress. Nutr. Neurosci. 22, 425–434. doi: 10.1080/1028415X.2017.1397875
Melas, P. A., Wei, Y., Wong, C. C. Y., Sjöholm, L. K., Åberg, E., Mill, J., et al. (2013). Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int. J. Neuropsychopharmacol. 16, 1513–1528. doi: 10.1017/S1461145713000102
Melas, P., and Forsell, Y. (2015). Hypomethylation of MAOA× s first exon region in depression: A replication study. Psychiatry Res. 226, 389–391. doi: 10.1016/j.psychres.2015.01.003
Messaoudi, M., Lalonde, R., Violle, N., Javelot, H., Desor, D., Nejdi, A., et al. (2011). Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br. J. Nutr. 105, 755–764. doi: 10.1017/S0007114510004319
Meylan, E. M., Halfon, O., Magistretti, P. J., and Cardinaux, J. R. (2016). The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: Possible relevance for treatment-resistant depression. Neuropharmacology 107, 111–121. doi: 10.1016/J.NEUROPHARM.2016.03.012
Mika, A., van Treuren, W., González, A., Herrera, J. J., Knight, R., and Fleshner, M. (2015). Exercise is more effective at altering gut microbial composition and producing stable changes in lean mass in juvenile versus adult male F344 rats. PLoS One 10:e0125889. doi: 10.1371/JOURNAL.PONE.0125889
Milani, C., Duranti, S., Bottacini, F., Casey, E., Turroni, F., Mahony, J., et al. (2017). The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 81, e36–e17. doi: 10.1128/MMBR.00036-17
Miro-Blanch, J., and Yanes, O. (2019). Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front. Genet. 10:638. doi: 10.3389/FGENE.2019.00638/XML/NLM
Morrison, D. J., and Preston, T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200. doi: 10.1080/19490976.2015.1134082
Muñoz-Cobo, I., Belloch, F. B., Díaz-Perdigón, T., Puerta, E., and Tordera, R. M. (2017). SIRT2 inhibition reverses anhedonia in the VGLUT1+/- depression model. Behav. Brain Res. 335, 128–131. doi: 10.1016/J.BBR.2017.07.045
Murgatroyd, C., Quinn, J. P., Sharp, H. M., Pickles, A., and Hill, J. (2015). Effects of prenatal and postnatal depression, and maternal stroking, at the glucocorticoid receptor gene. Transl. Psychiatry 5:e560. doi: 10.1038/TP.2014.140
Mutlu, E. A., Gillevet, P. M., Rangwala, H., Sikaroodi, M., Naqvi, A., Engen, P. A., et al. (2012). Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest Liver Physiol. 302:G966. doi: 10.1152/AJPGI.00380.2011
Na, K. S., Chang, H. S., Won, E., Han, K. M., Choi, S., Tae, W. S., et al. (2014). Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One 9:e85425. doi: 10.1371/JOURNAL.PONE.0085425
Nagao-Kitamoto, H., Shreiner, A. B., Gillilland, M. G., Kitamoto, S., Ishii, C., Hirayama, A., et al. (2016). Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. CMGH 2, 468–481. doi: 10.1016/J.JCMGH.2016.02.003
Napolitano, M., and Covasa, M. (2020). Microbiota transplant in the treatment of obesity and diabetes: Current and future perspectives. Front. Microbiol. 11:590370. doi: 10.3389/FMICB.2020.590370
Nel Van Zyl, K., Whitelaw, A. C., Hesseling, A. C., Seddon, J. A., Demers, A. M., and Newton-Foot, M. (2021). Association between clinical and environmental factors and the gut microbiota profiles in young South African children. Sci. Rep. 1, 1–13. doi: 10.1038/s41598-021-95409-5
Nemoto, T., Kakinuma, Y., and Shibasaki, T. (2015). Impaired miR449a-induced downregulation of Crhr1 expression in low-birth-weight rats. J. Endocrinol. 224, 195–203. doi: 10.1530/JOE-14-0537
Neufeld, K. M., Kang, N., Bienenstock, J., and Foster, J. A. (2011). Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 23, 255–264. doi: 10.1111/J.1365-2982.2010.01620.X
Nishida, K., Sawada, D., Kuwano, Y., Tanaka, H., and Rokutan, K. (2019). Health benefits of lactobacillus gasseri CP2305 tablets in young adults exposed to chronic stress: A randomized, double-blind, placebo-controlled study. Nutrients 11:1859. doi: 10.3390/NU11081859
Nolan-Kenney, R., Wu, F., Hu, J., Yang, L., Kelly, D., Li, H., et al. (2020). The association between smoking and gut microbiome in Bangladesh. Nicotine Tob. Res. 22, 1339–1346. doi: 10.1093/NTR/NTZ220
Norman, G. J., Hawkley, L., Luhmann, M., Ball, A. B., Cole, S. W., Berntson, G. G., et al. (2012). Variation in the oxytocin receptor gene influences neurocardiac reactivity to social stress and HPA function: A population based study. Horm. Behav. 61, 134–139. doi: 10.1016/J.YHBEH.2011.11.006
O’Donnell, M. P., Fox, B. W., Chao, P. H., Schroeder, F. C., and Sengupta, P. (2020). A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature 583, 415–420. doi: 10.1038/S41586-020-2395-5
O’Mahony, S. M., Marchesi, J. R., Scully, P., Codling, C., Ceolho, A. M., Quigley, E. M. M., et al. (2009). Early life stress alters behavior, immunity, and microbiota in rats: Implications for irritable bowel syndrome and psychiatric illnesses. Biol. Psychiatry 65, 263–267. doi: 10.1016/J.BIOPSYCH.2008.06.026
Oberlander, T. F., Weinberg, J., Papsdorf, M., Grunau, R., Misri, S., and Devlin, A. M. (2008). Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 3, 97–106. doi: 10.4161/EPI.3.2.6034
Olsson, C., Foley, D., Parkinson-Bates, M., Byrnes, G., McKenzie, M., Patton, G., et al. (2010). Prospects for epigenetic research within cohort studies of psychological disorder: A pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol. Psychol. 83, 159–165. doi: 10.1016/j.biopsycho.2009.12.003
Otsuki, K., Uchida, S., Watanuki, T., Wakabayashi, Y., Fujimoto, M., Matsubara, T., et al. (2008). Altered expression of neurotrophic factors in patients with major depression. J. Psychiatr. Res. 42, 1145–1153. doi: 10.1016/J.JPSYCHIRES.2008.01.010
Palmnäs, M., Brunius, C., Shi, L., Rostgaard-Hansen, A., Torres, N. E., González-Domínguez, R., et al. (2020). Perspective: Metabotyping-a potential personalized nutrition strategy for precision prevention of cardiometabolic disease. Adv. Nutr. 11, 524–532. doi: 10.1093/ADVANCES/NMZ121
Parker, E. A., Roy, T., D’Adamo, C. R., and Wieland, L. S. (2018). Probiotics and gastrointestinal conditions: An overview of evidence from the cochrane collaboration. Nutrition 45, 125.e–134.e. doi: 10.1016/J.NUT.2017.06.024
Paul, B., Barnes, S., Demark-Wahnefried, W., Morrow, C., Salvador, C., Skibola, C., et al. (2015). Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 7:112. doi: 10.1186/S13148-015-0144-7
Peña, C. J., Bagot, R. C., Labonté, B., and Nestler, E. J. (2014). Epigenetic signaling in psychiatric disorders. J. Mol. Biol. 426, 3389–3412. doi: 10.1016/J.JMB.2014.03.016
Perez-Cornago, A., Mansego, M. L., Zulet, M. A., and Martinez, J. A. (2014). DNA hypermethylation of the serotonin receptor type-2A gene is associated with a worse response to a weight loss intervention in subjects with metabolic syndrome. Nutrients 6, 2387–2403. doi: 10.3390/nu6062387
Perroud, N., Zewdie, S., Stenz, L., Adouan, W., Bavamian, S., Prada, P., et al. (2016). Methylation of serotonin receptor 3A in ADHD, Borderline personality, and bipolar disorders: Link with severity of the disorders and childhood maltreatment. Depress Anxiety 33, 45–55. doi: 10.1002/DA.22406
Pham, M., Lemberg, D., and Day, A. S. (2008). Probiotics: Sorting the evidence from the myths. Med. J. Austr. 188, 304–308.
Philibert, R., Madan, A., Andersen, A., Cadoret, R., Packer, H., and Sandhu, H. (2007). Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 144, 101–105. doi: 10.1002/AJMG.B.30414
Pisarello, M. L. J., Vintiñi, E. O., González, S. N., Pagani, F., and Medina, M. S. (2015). Decrease in lactobacilli in the intestinal microbiota of celiac children with a gluten-free diet, and selection of potentially probiotic strains. Can. J. Microbiol. 61, 32–37. doi: 10.1139/CJM-2014-0472
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/NATURE08821
Qu, Q., Li, H., Bai, L., Zhang, S., Sun, J., Lv, W., et al. (2021). Effects of heat stress on gut microbiome in rats. Indian J. Microbiol. 61, 338–347. doi: 10.1007/S12088-021-00948-0
Qu, Y., Yang, C., Ren, Q., Ma, M., Dong, C., and Hashimoto, K. (2017). Comparison of (R)-ketamine and lanicemine on depression-like phenotype and abnormal composition of gut microbiota in a social defeat stress model. Sci. Rep. 7:15725. doi: 10.1038/S41598-017-16060-7
Queipo-Ortuño, M. I., Boto-Ordóñez, M., Murri, M., Gomez-Zumaquero, J. M., Clemente-Postigo, M., Estruch, R., et al. (2012). Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 95, 1323–1334. doi: 10.3945/AJCN.111.027847
Raadsheer, F., Hoogendijk, W., Stam, F., Tilders, F., and Swaab, D. (1994). Increased numbers of corticotropin-releasing hormone expressing neurons in the hypothalamic paraventricular nucleus of depressed patients. Neuroendocrinology 60, 436–444.
Reddy, B. S., Weisburger, J. H., and Wynder, E. L. (1975). Effects of high risk and low risk diets for colon carcinogenesis on fecal microflora and steroids in man. J. Nutr. 105, 878–884. doi: 10.1093/JN/105.7.878
Reiner, I., van IJzendoorn, M. H., Bakermans-Kranenburg, M. J., Bleich, S., Beutel, M., and Frieling, H. (2015). Methylation of the oxytocin receptor gene in clinically depressed patients compared to controls: The role of OXTR rs53576 genotype. J. Psychiatr. Res. 65, 9–15. doi: 10.1016/J.JPSYCHIRES.2015.03.012
Rinninella, E., Raoul, P., Cintoni, M., Franceschi, F., Miggiano, G. A. D., Gasbarrini, A., et al. (2019). What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7:14. doi: 10.3390/MICROORGANISMS7010014
Rodríguez, J. M., Murphy, K., Stanton, C., Ross, R. P., Kober, O. I., Juge, N., et al. (2015). The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol. Health Dis. 26:26050. doi: 10.3402/MEHD.V26.26050
Romano, K. A., Martinez-del Campo, A., Kasahara, K., Chittim, C. L., Vivas, E. I., Amador-Noguez, D., et al. (2017). Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 22, 279.e–290.e. doi: 10.1016/J.CHOM.2017.07.021
Romero, R., Hassan, S. S., Gajer, P., Tarca, A. L., Fadrosh, D. W., Nikita, L., et al. (2014). The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2, 2045–2051. doi: 10.1186/2049-2618-2-4
Rook, G. A. W., Raison, C. L., and Lowry, C. A. (2014). Microbial ‘old friends’, immunoregulation and socioeconomic status. Clin. Exp. Immunol. 177:1. doi: 10.1111/CEI.12269
Rosenthal, M., Goldberg, D., Aiello, A., Larson, E., and Foxman, B. (2011). Skin microbiota: Microbial community structure and its potential association with health and disease. Infect. Genet. Evol. 11, 839–848. doi: 10.1016/J.MEEGID.2011.03.022
Rossi, M., Amaretti, A., and Raimondi, S. (2011). Folate production by probiotic bacteria. Nutrients 3, 118–134. doi: 10.3390/NU3010118
Rowland, I., Gibson, G., Heinken, A., Scott, K., Swann, J., Thiele, I., et al. (2018). Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24. doi: 10.1007/S00394-017-1445-8
Rubio, C., and Huang, C. B. (1992). Quantification of the sulphomucin-producing cell population of the colonic mucosa during protracted stress in rats. Vivo 6, 81–84.
Saarela, M. H. (2019). Safety aspects of next generation probiotics. Curr. Opin. Food Sci. 30, 8–13. doi: 10.1016/J.COFS.2018.09.001
Salazar-Lindo, E., Miranda-Langschwager, P., Campos-Sanchez, M., Chea-Woo, E., and Sack, R. B. (2004). Lactobacillus casei strain GG in the treatment of infants with acute watery diarrhea: A randomized, double-blind, placebo controlled clinical trial [ISRCTN67363048]. BMC Pediatr. 4:18. doi: 10.1186/1471-2431-4-18/FIGURES/1
Sales, A. J., and Joca, S. R. L. (2015). Effects of DNA methylation inhibitors and conventional antidepressants on mice behaviour and brain DNA methylation levels. Acta Neuropsychiatr. 28, 11–22. doi: 10.1017/NEU.2015.40
Sanders, T. H., Weiss, J., Hogewood, L., Chen, L., Paton, C., McMahan, R. L., et al. (2019). Cognition-enhancing vagus nerve stimulation alters the epigenetic landscape. J. Neurosci. 39, 3454–3469. doi: 10.1523/JNEUROSCI.2407-18.2019
Sandhu, K. V., Sherwin, E., Schellekens, H., Stanton, C., Dinan, T. G., and Cryan, J. F. (2017). Feeding the microbiota-gut-brain axis: Diet, microbiome, and neuropsychiatry. Transl. Res. 179, 223–244. doi: 10.1016/J.TRSL.2016.10.002
Santos, J., Saperas, E., Nogueiras, C., Mourelle, M., Antolin, M., Cadahia, A., et al. (1998). Release of mast cell mediators into the jejunum by cold pain stress in humans. Gastroenterology 114, 640–648. doi: 10.1016/S0016-5085(98)70577-3
Sanz, Y. (2010). Effects of a gluten-free diet on gut microbiota and immune function in healthy adult humans. Gut Microbes 1, 135–137. doi: 10.4161/GMIC.1.3.11868
Sarkar, A., Chachra, P., Kennedy, P., Pena, C. J., Desouza, L. A., Nestler, E. J., et al. (2014). Hippocampal HDAC4 contributes to postnatal fluoxetine-evoked depression-like behavior. Neuropsychopharmacology 39, 2221–2232. doi: 10.1038/NPP.2014.73
Sasidharan, B. K., Ramadass, B., Viswanathan, P., Samuel, P., Gowri, M., Pugazhendhi, S., et al. (2019). A phase 2 randomized controlled trial of oral resistant starch supplements in the prevention of acute radiation proctitis in patients treated for cervical cancer. J. Cancer Res. Ther. 15:1383. doi: 10.4103/JCRT.JCRT_152_19
Savin, Z., Kivity, S., Yonath, H., and Yehuda, S. (2018). Smoking and the intestinal microbiome. Arch. Microbiol. 200, 677–684. doi: 10.1007/S00203-018-1506-2
Schmidt, K., Cowen, P. J., Harmer, C. J., Tzortzis, G., Errington, S., and Burnet, P. W. J. (2015). Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology (Berl) 232, 1793–1801. doi: 10.1007/S00213-014-3810-0
Schroeder, F. A., Lewis, M. C., Fass, D. M., Wagner, F. F., Zhang, Y. L., Hennig, K. M., et al. (2013). A Selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS One 8:e71323. doi: 10.1371/JOURNAL.PONE.0071323
Schroeder, F. A., Lin, C. L., Crusio, W. E., and Akbarian, S. (2007). Antidepressant-like effects of the histone deacetylase inhibitor. Sodium Butyrate, in the Mouse. Biol. Psychiatry 62, 55–64. doi: 10.1016/J.BIOPSYCH.2006.06.036
Seeley, J. J., Baker, R. G., Mohamed, G., Bruns, T., Hayden, M. S., Deshmukh, S. D., et al. (2018). Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature 559, 114–119. doi: 10.1038/S41586-018-0253-5
Shi, N., Li, N., Duan, X., and Niu, H. (2017). Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 4:14. doi: 10.1186/S40779-017-0122-9
Shi, Y., Wang, Q., Song, R., Kong, Y., and Zhang, Z. (2021). Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 71:103569. doi: 10.1016/J.EBIOM.2021.103569
Skonieczna-żydecka, K., Grochans, E., Maciejewska, D., Szkup, M., Schneider-Matyka, D., Jurczak, A., et al. (2018). Faecal short chain fatty acids profile is changed in polish depressive women. Nutrients 10:1939. doi: 10.3390/NU10121939
Smearman, E. L., Almli, L. M., Conneely, K. N., Brody, G. H., Sales, J. M., Bradley, B., et al. (2016). Oxytocin receptor genetic and epigenetic variations: Association with child abuse and adult psychiatric symptoms. Child Dev. 87, 122–134. doi: 10.1111/cdev.12493
Smith, A. P., Sutherl, D., and Hewlett, P. (2015). An investigation of the acute effects of oligofructose-enriched inulin on subjective wellbeing, mood and cognitive performance. Nutrients 7, 8887–8896. doi: 10.3390/NU7115441
Soliman, M. L., Health, E., Rosenberger, T. A., and Soliman, M. L. (2011). Acetate supplementation increases brain histone acetylation and inhibits histone deacetylase activity and expression. Springer 352, 173–180. doi: 10.1007/s11010-011-0751-3
Song, C., and Wang, H. (2011). Cytokines mediated inflammation and decreased neurogenesis in animal models of depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 760–768. doi: 10.1016/J.PNPBP.2010.06.020
Song, C., Xiang, Y. Z., and Manku, M. (2009). Increased phospholipase A2 activity and inflammatory response but decreased nerve growth factor expression in the olfactory bulbectomized rat model of depression: Effects of chronic ethyl-eicosapentaenoate treatment. J. Neurosci. 29, 14–22. doi: 10.1523/JNEUROSCI.3569-08.2009
Spano, G., Kumar, M., Zhang, B., Huo, G., Luo, G., Li, B., et al. (2019). Major traditional probiotics: Comparative genomic analyses and roles in gut microbiome of eight cohorts. Front. Microbiol. 10:712. doi: 10.3389/fmicb.2019.00712
Stewart, C. J., Auchtung, T. A., Ajami, N. J., Velasquez, K., Smith, D. P., de La Garza, R., et al. (2018). Effects of tobacco smoke and electronic cigarette vapor exposure on the oral and gut microbiota in humans: A pilot study. PeerJ 6:e4693. doi: 10.7717/PEERJ.4693
Stilling, R. M., van de Wouw, M., Clarke, G., Stanton, C., Dinan, T. G., and Cryan, J. F. (2016). The neuropharmacology of butyrate: The bread and butter of the microbiota-gut-brain axis? Neurochem. Int. 99, 110–132. doi: 10.1016/J.NEUINT.2016.06.011
Stinson, L. F., Payne, M. S., and Keelan, J. A. (2016). Planting the seed: Origins, composition, and postnatal health significance of the fetal gastrointestinal microbiota. Crit. Rev. Microbiol. 43, 352–369. doi: 10.1080/1040841X.2016.1211088
Strader, A. D., and Woods, S. C. (2005). Gastrointestinal hormones and food intake. Gastroenterology 128, 175–191. doi: 10.1053/j.gastro.2004.10.043
Strandwitz, P., Kim, K. H., Terekhova, D., Liu, J. K., Sharma, A., Levering, J., et al. (2019). GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 4, 396–403. doi: 10.1038/S41564-018-0307-3
Sudo, N., Chida, Y., Aiba, Y., Sonoda, J., Oyama, N., Yu, X. N., et al. (2004). Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 558, 263–275. doi: 10.1113/JPHYSIOL.2004.063388
Sullivan, P. F., Neale, M. C., and Kendler, K. S. (2000). Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 157, 1552–1562. doi: 10.1176/APPI.AJP.157.10.1552
Sun, H., Kennedy, P. J., and Nestler, E. J. (2013). Epigenetics of the depressed brain: Role of histone acetylation and methylation. Neuropsychopharmacology 38, 124–137. doi: 10.1038/NPP.2012.73
Szeligowski, T., Yun, A. L., Lennox, B. R., and Burnet, P. W. J. (2020). The gut microbiome and schizophrenia: The current state of the field and clinical applications. Front. Psychiatry 11:156. doi: 10.3389/FPSYT.2020.00156
Tabrizi, A., Khalili, L., Homayouni-Rad, A., Pourjafar, H., Dehghan, P., and Ansari, F. (2019). Prebiotics, as promising functional food to patients with psychological disorders: A review on mood disorders, sleep, and cognition. NeuroQuantology 17, 1–9. doi: 10.14704/NQ.2019.17.6.2189
Takada, M., Nishida, K., Gondo, Y., Kikuchi-Hayakawa, H., Ishikawa, H., Suda, K., et al. (2017). Beneficial effects of Lactobacillus casei strain Shirota on academic stress-induced sleep disturbance in healthy adults: A double-blind, randomised, placebo-controlled. Benef Microbes 8, 153–162. doi: 10.3920/BM2016.0150
Tanaka, M., and Nakayama, J. (2017). Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 66, 515–522. doi: 10.1016/J.ALIT.2017.07.010
Ting, J. P. Y., and Trowsdale, J. (2002). Genetic control of MHC class II expression. Cell 109, S21–S33. doi: 10.1016/S0092-8674(02)00696-7
Tsankova, N., Berton, O., Renthal, W., Kumar, A., Neve, R., and Nestler, E. (2006). Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 9, 519–525. doi: 10.1038/nn1659
Tsankova, N., Renthal, W., Kumar, A., and Nestler, E. J. (2007). Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 8, 355–367. doi: 10.1038/NRN2132
Tsigos, C., and Chrousos, G. P. (2002). Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J. Psychosom Res. 53, 865–871. doi: 10.1016/S0022-3999(02)00429-4
Tubbs, R. S., Rizk, E., Shoja, M. M., Loukas, M., Barbaro, N., and Spinner, R. J. (2015). Nerves and nerve injuries. Cambridge, MA: Academic Press. doi: 10.1016/C2012-0-06700-2
Turnbaugh, P. J., and Gordon, J. I. (2009). The core gut microbiome, energy balance and obesity. J. Physiol. 587, 4153–4158. doi: 10.1113/JPHYSIOL.2009.174136
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/NATURE05414
Turnbull, A. V., and Rivier, C. (1995). Regulation of the HPA Axis by Cytokines. Brain Behav. Immun. 9, 253–275. doi: 10.1006/BRBI.1995.1026
Turner, J. D., Alt, S. R., Cao, L., Vernocchi, S., Trifonova, S., Battello, N., et al. (2010). Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem. Pharmacol. 80, 1860–1868. doi: 10.1016/J.BCP.2010.06.037
Uchida, S., Hara, K., Kobayashi, A., Otsuki, K., Yamagata, H., Hobara, T., et al. (2011). Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron 69, 359–372. doi: 10.1016/J.NEURON.2010.12.023
Valles-Colomer, M., Falony, G., Darzi, Y., Tigchelaar, E. F., Wang, J., Tito, R. Y., et al. (2019). The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 4, 623–632. doi: 10.1038/s41564-018-0337-x
van de Wouw, M., Boehme, M., Lyte, J. M., Wiley, N., Strain, C., O’Sullivan, O., et al. (2018). Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain–gut axis alterations. J. Physiol. 596, 4923–4944. doi: 10.1113/JP276431
Vaughan, C. J., Aherne, A. M., Lane, E., Power, O., Carey, R. M., and O’Connell, D. P. (2000). Identification and regional distribution of the dopamine D(1A) receptor in the gastrointestinal tract. Am. J. Physiol. Regul. Integr. Comp. Physiol. 279, R599–R609. doi: 10.1152/AJPREGU.2000.279.2.R599
Venegas, D. P., de La Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/FIMMU.2019.00277/FULL
Virtue, A. T., McCright, S. J., Wright, J. M., Jimenez, M. T., Mowel, W. K., Kotzin, J. J., et al. (2019). The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 11:eaav1892. doi: 10.1126/SCITRANSLMED.AAV1892
Vivarelli, S., Salemi, R., Candido, S., Falzone, L., Santagati, M., Stefani, S., et al. (2019). Gut microbiota and cancer: From pathogenesis to therapy. Cancers (Basel) 11:38. doi: 10.3390/CANCERS11010038
Waldram, A., Holmes, E., Wang, Y., Rantalainen, M., Wilson, I. D., Tuohy, K. M., et al. (2009). Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res. 8, 2361–2375. doi: 10.1021/PR8009885
Wallace, C. J. K., and Milev, R. (2017). The effects of probiotics on depressive symptoms in humans: A systematic review. Ann. Gen. Psychiatry 16:14. doi: 10.1186/S12991-017-0138-2
Wang, X., Wang, W., Wang, L., Yu, C., Zhang, G., Zhu, H., et al. (2019). Lentinan modulates intestinal microbiota and enhances barrier integrity in a piglet model challenged with lipopolysaccharide. Food Funct. 10, 479–489. doi: 10.1039/C8FO02438C
Weder, N., Zhang, H., Jensen, K., Yang, B. Z., Simen, A., Jackowski, A., et al. (2014). Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J. Am. Acad. Child Adolesc. Psychiatry 53, 417.e–424.e. doi: 10.1016/J.JAAC.2013.12.025
Wong, M. L., Inserra, A., Lewis, M. D., Mastronardi, C. A., Leong, L., Choo, J., et al. (2016). Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol. Psychiatry 21, 797–805. doi: 10.1038/MP.2016.46
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/SCIENCE.1208344
Wu, G. D., Compher, C., Chen, E. Z., Smith, S. A., Shah, R. D., Bittinger, K., et al. (2016). Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 65, 63–72. doi: 10.1136/GUTJNL-2014-308209
Xu, Z., Liu, Z., Dong, X., Hu, T., Wang, L., Li, J., et al. (2018). Fecal microbiota transplantation from healthy donors reduced alcohol-induced anxiety and depression in an animal model of chronic alcohol exposure. Chin. J. Physiol. 61, 360–371. doi: 10.4077/CJP.2018.BAH633
Yamagata, H., Uchida, S., Matsuo, K., Harada, K., Kobayashi, A., Nakashima, M., et al. (2017). Identification of commonly altered genes between in major depressive disorder and a mouse model of depression. Sci. Rep. 73044. doi: 10.1038/S41598-017-03291-X
Yang, C., Fang, X., Zhan, G., Huang, N., Li, S., Bi, J., et al. (2019). Key role of gut microbiota in anhedonia-like phenotype in rodents with neuropathic pain. Transl. Psychiatry 9:57. doi: 10.1038/S41398-019-0379-8
Yang, C., Qu, Y., Fujita, Y., Ren, Q., Ma, M., Dong, C., et al. (2017). Possible role of the gut microbiota–brain axis in the antidepressant effects of (R)-ketamine in a social defeat stress model. Transl. Psychiatry 7:1294. doi: 10.1038/s41398-017-0031-4
Yano, J. M., Yu, K., Donaldson, G. P., Shastri, G. G., Ann, P., Ma, L., et al. (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276. doi: 10.1016/J.CELL.2015.02.047
Zacharowski, K., Zacharowski, P. A., Koch, A., Baban, A., Tran, N., Berkels, R., et al. (2006). Toll-like receptor 4 plays a crucial role in the immune-adrenal response to systemic inflammatory response syndrome. Proc. Natl. Acad. Sci. U.S.A. 103, 6392–6397. doi: 10.1073/PNAS.0601527103
Zhao, J., Goldberg, J., Bremner, J. D., and Vaccarino, V. (2013). Association between promoter methylation of serotonin transporter gene and depressive symptoms: A monozygotic twin study. Psychosom Med. 75, 523–529. doi: 10.1097/PSY.0B013E3182924CF4
Zhao, W., Hu, Y., Li, C., Li, N., Zhu, S., Tan, X., et al. (2020). Transplantation of fecal microbiota from patients with alcoholism induces anxiety/depression behaviors and decreases brain mGluR1/PKC ε levels in mouse. Biofactors 46, 38–54. doi: 10.1002/BIOF.1567
Zhao, Y., Zeng, Y., Zeng, D., Wang, H., Zhou, M., Sun, N., et al. (2021). Probiotics and MicroRNA: Their Roles in the Host–Microbe Interactions. Front. Microbiol. 11:3363. doi: 10.3389/FMICB.2020.604462/BIBTEX
Zheng, P., Zeng, B., Zhou, C., Liu, M., Fang, Z., Xu, X., et al. (2016). Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 6, 786–796. doi: 10.1038/mp.2016.44
Zhou, Y., Gao, H., Mihindukulasuriya, K. A., Rosa, P. S. L., Wylie, K. M., Vishnivetskaya, T., et al. (2013). Biogeography of the ecosystems of the healthy human body. Genome Biol. 14, R1–R1. doi: 10.1186/GB-2013-14-1-R1
Keywords: gut-brain axis (GBA), microbiota, epigenetic, depression, microbiome, short-chain fatty acid (SCFA), DNA methylation, histone modifications
Citation: Begum N, Mandhare A, Tryphena KP, Srivastava S, Shaikh MF, Singh SB and Khatri DK (2022) Epigenetics in depression and gut-brain axis: A molecular crosstalk. Front. Aging Neurosci. 14:1048333. doi: 10.3389/fnagi.2022.1048333
Received: 19 September 2022; Accepted: 23 November 2022;
Published: 13 December 2022.
Edited by:
Shampa Ghosh, National Institute of Nutrition, IndiaReviewed by:
Pradeep Mishra, Institute for Stem Cell Science and Regenerative Medicine (inStem), IndiaCopyright © 2022 Begum, Mandhare, Tryphena, Srivastava, Shaikh, Singh and Khatri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saurabh Srivastava, c2F1cmFiaF9iaXRzcGlsYW5pQHlhaG9vLmNvLmlu; Mohd Farooq Shaikh, ZmFyb29xLnNoYWlraEBtb25hc2guZWR1; Dharmendra Kumar Khatri, ZGtraGF0cmkxMEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.