
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Aging Neurosci., 15 March 2021
Sec. Alzheimer's Disease and Related Dementias
Volume 13 - 2021 | https://doi.org/10.3389/fnagi.2021.641080
This article is part of the Research TopicNon-coding RNA in Alzheimer’s Pathology and Diagnosis.View all 7 articles
 Mohammad Samadian1†
Mohammad Samadian1† Mahdi Gholipour2†
Mahdi Gholipour2† Mohammadreza Hajiesmaeili1†
Mohammadreza Hajiesmaeili1† Mohammad Taheri3*†
Mohammad Taheri3*† Soudeh Ghafouri-Fard4*†
Soudeh Ghafouri-Fard4*†Alzheimer's disease (AD) is an irrevocable neurodegenerative condition characterized by the presence of senile plaques comprising amassed β-amyloid peptides (Aβ) and neurofibrillary tangles mainly comprising extremely phosphorylated Tau proteins. Recent studies have emphasized the role of microRNAs (miRNAs) in the development of AD. A number of miRNAs, namely, miR-200a-3p, miR-195, miR-338-5p, miR-34a-5p, miR-125b-5p, miR-132, miR-384, miR-339-5p, miR-135b, miR-425-5p, and miR-339-5p, have been shown to participate in the development of AD through interacting with BACE1. Other miRNAs might affect the inflammatory responses in the course of AD. Aberrant expression of several miRNAs in the plasma samples of AD subjects has been shown to have the aptitude for differentiation of AD subjects from healthy subjects. Finally, a number of AD-modifying agents affect miRNA profile in cell cultures or animal models. We have performed a comprehensive search and summarized the obtained data about the function of miRNAs in AD in the current review article.
Alzheimer's disease (AD) is an irrevocable neurodegenerative condition with a progressive course, and it is the chief reason for dementia in the elderly (Prince et al., 2013). AD is characterized by pervasive cognitive defects and memory deficits, leading to the dependence of the majority of AD patients on others for their routine activities. From a pathological point of view, AD is defined by the presence of senile plaques comprising amassed β-amyloid peptides (Aβ) and neurofibrillary tangles mainly comprising extremely phosphorylated Tau proteins (Ballard et al., 2011). The most accepted hypotheses for the development of AD are based on these two main pathological events [i.e., Aβ accumulation and Tau accumulation (Wang et al., 2019a)]. The amyloid precursor protein is converted to Aβ through consecutive enzymatic reactions catalyzed by β-secretase (BACE1) and γ-secretase (containing presenilin 1 and presenilin 2) (Querfurth and LaFerla, 2010). Recent studies have emphasized the role of microRNAs (miRNAs) in the development of AD (Wang et al., 2019a). These ~22 nucleotide transcripts post-transcriptionally regulate the expression of several target genes through binding with 3' UTR and destructing the target transcript or reducing its translation (O'Brien et al., 2018). Sequence complementarity mainly regulates the miRNA/mRNA interactions leading to the ability of one miRNA to target several genes and the possible regulation of one gene by multiple miRNAs. Therefore, miRNAs are potential means for investigating multifactorial disorders such as AD (Iqbal and Grundke-Iqbal, 2010). A leading investigation in this regard has examined the number of brain-associated miRNAs expressed in the human hippocampus specimens obtained from fetal, adult, and AD patients, revealing misregulation of certain miRNAs in the AD brain and their possible contribution to the pathological processes of this disorder (Lukiw, 2007). Dysregulation of other miRNAs has also been verified in multiple studies, and the underlying mechanisms of their contribution in AD have been identified in some cases. We have performed a comprehensive search and summarized the obtained data about the function of miRNAs in AD in this review article.
Dysregulation of miRNAs has been demonstrated in human AD subjects or animal models of AD. Moreover, several researchers have induced or suppressed the expression of some miRNAs in the cell/animal models of AD to appraise their function in the progression of AD. In a rat model of AD caused by the administration of Aβ25−35 into the brain, downregulation of SOX6 and over-expression of miR-129-5p have shortened the dormant escape period and enhanced the time of crossing platforms, repairing the pathological damage, blocking neuronal apoptosis, and decreasing inflammation. Based on the protective effects of miR-129-5p against nerve damage and inflammation, miR-129-5p has been suggested as a candidate for therapeutic options against AD, as it acts to suppress SOX6 (Zeng et al., 2019). Expression of miR-200a-3p has been shown to be repressed in animal and cell models of AD. miR-200a-3p can suppress cell apoptosis, inactivate Bax/caspase-3 axis, and decrease Aβ1−42 and tau phosphorylation in cell experiments. Mechanistically, these effects are mediated through the modulation of translocation of BACE1 and PRKACB. Taken together, the neuroprotective impact of miR-200a-3p is accomplished by inhibition of BACE1 expression and subsequent suppression of Aβ production as well as reduction of PKA expression and Tau phosphorylation (Wang et al., 2019b). miR-455-3p has been shown to bind with 3' UTR of APP gene to decrease its expression and reduce expression of mitochondrial fission proteins (Kumar et al., 2019). Mutant APP cells that show expression of miR-455-3p exhibit upregulation of synaptic genes. Over-expression of miR-455-3p in mutant APP cells reduces the number of mitochondria and increases the size of the mitochondria. Taken together, miR-455-3p controls APP processing and protects against mutant APP-associated mitochondrial dysfunction and synaptic anomalies in AD (Kumar et al., 2019). Expression of miR-455-3p has been shown to be increased in postmortem brain samples, fibroblasts, and plasma samples of patients with AD compared with controls (Kumar et al., 2017; Kumar and Reddy, 2018). As a primary event, expression of miR-409-5p has been decreased in an APP/PS1 double transgenic mice model of AD. Over-expression experiments have shown that this miRNA has a harmful impact on neurite outgrowth, reduces neuron survival, and quickens the progression of Aβ1−42-associated pathologic events (Guo et al., 2019). In line with the observed downregulation of miR-409-5p in APP/PS1 AD model, Aβ1−42 peptide has been shown to downregulate miR-409-5p levels. A luciferase study has shown that Plek is a target of miR-409-5p (Guo et al., 2019). Ectopic expression of miR-409-5p has induced neurotoxic effects and interferes with neuron survival and differentiation, while Plek upregulation could partly protect the neurite outgrowth from these toxic effects. Taken together, reduction of miR-409-5p expression in the early stages of AD might be a self-protective response to lessen the synaptic injury induced by Aβ (Guo et al., 2019). miR-132 is another downregulated miRNA in AD. Experiments in a rat model of AD have shown upregulation of AChE, iNOS, ROS, MDA, MAPK1, and p-MAPK1 and downregulation of SOD, GSH-Px, and miR-132. Over-expression of miR-132 has reversed these markers demonstrating the role of this miRNA in the suppression of hippocampal iNOS expression and oxidative stress through reduction of MAPK1 levels (Deng et al., 2020). However, expression of this miRNA has been demonstrated to be reduced in neurally-originated plasma exosomes of AD subjects (Cha et al., 2019). Table 1 shows the summary of studies that reported decreased levels of miRNAs in AD.
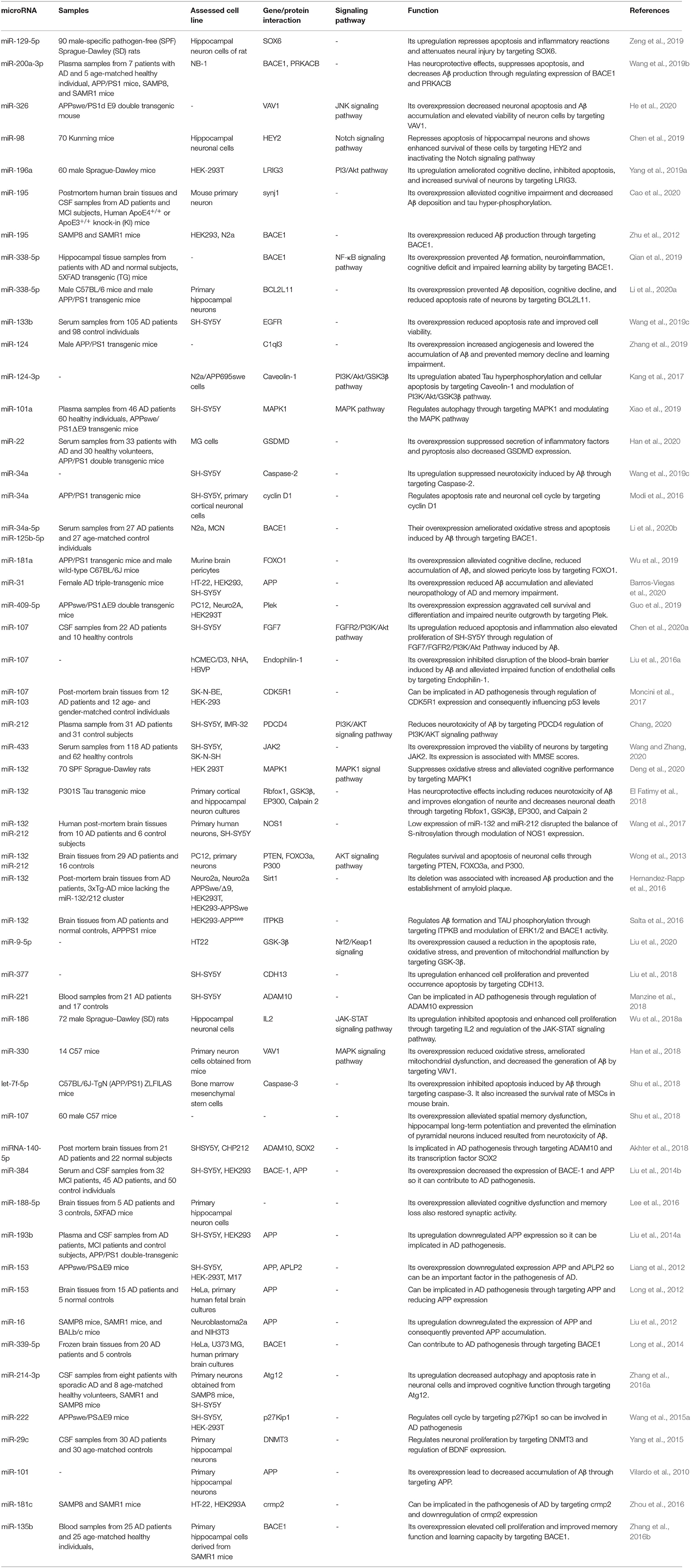
Table 1. Downregulated miRNAs in AD subject, animal models of AD, and related cell lines and their functions in progression of AD.
Although several studies have reported downregulation of miR-132 in AD (Wong et al., 2013; El Fatimy et al., 2018; Cha et al., 2019; Deng et al., 2020), Liu et al. have reported high levels of miR-132 in patients with mild cognitive impairment and AD vs. normal individuals. They have shown the impact of miR-132 upregulation in the induction of apoptosis in neurons through increasing Bax/Bcl-2 ratio (Liu and Zhang, 2019). Moreover, they have reported that miR-132 increases Tau phosphorylation and expression levels of Rb, Histone H1, and CDK-5. Collectively, they have suggested that miR-132 participates in AD by controlling cell apoptosis and the GTDC-1/CDK-5/Tau phosphorylation axis (Liu and Zhang, 2019). In addition to GTDC-1, miR-132 is also known to regulate the expression of synaptic proteins via complement C1q (Xu et al., 2019). Similarly, expression of miR-132 has been shown to be decreased in AD-derived plasma exosomes (Cha et al., 2019). miR-128 has also been over-expressed in the brain samples of AD patients (Liu et al., 2019). Experiments in AD mice have demonstrated parallel upregulation of miR-128 and downregulation of PPARγ in the cerebral cortex. The interaction between these two transcripts has been validated through functional assays. miR-128 silencing has suppressed AD-like features, amyloid plaque creation, Aβ production, and inflammation in AD mice through upregulating PPARγ (Liu et al., 2019). miR-425-5p is another upregulated miRNA in patients with AD and the cellular model of AD. Upregulation of miR-425-5p has induced cell apoptosis, stimulated expression of GSK-3β, and enhanced tau phosphorylation through targeting HSPB8 (Yuan et al., 2020). miR-146a is also upregulated in AD and participates in the pathogenesis of this condition via targeting Lrp2 and inhibiting the Akt signaling pathway, modulating ROCK1 expression and decreasing Tau phosphorylation, and influencing inflammatory responses via modulation of IRAK-1 (Cui et al., 2010; Wang et al., 2016). Insulin and liver X receptor (LXR) activators have been shown to increase the miR-7-1 levels. Expression of this miRNA has changed within the brains of diet-induced obese animals as well as AD patients, which is in parallel with the downregulation of its target genes IRS-2 and IDE. Upregulation of miR-7 has enhanced extracellular Aβ levels in neurons and interfered with the eradication of Aβ by microglia. Collectively, insulin can act via the HNRNPK-miR-7 cascade to post-transcriptionally affect metabolic pathways in AD (Fernández-de Frutos et al., 2019). Table 2 lists upregulated miRNAs in AD.
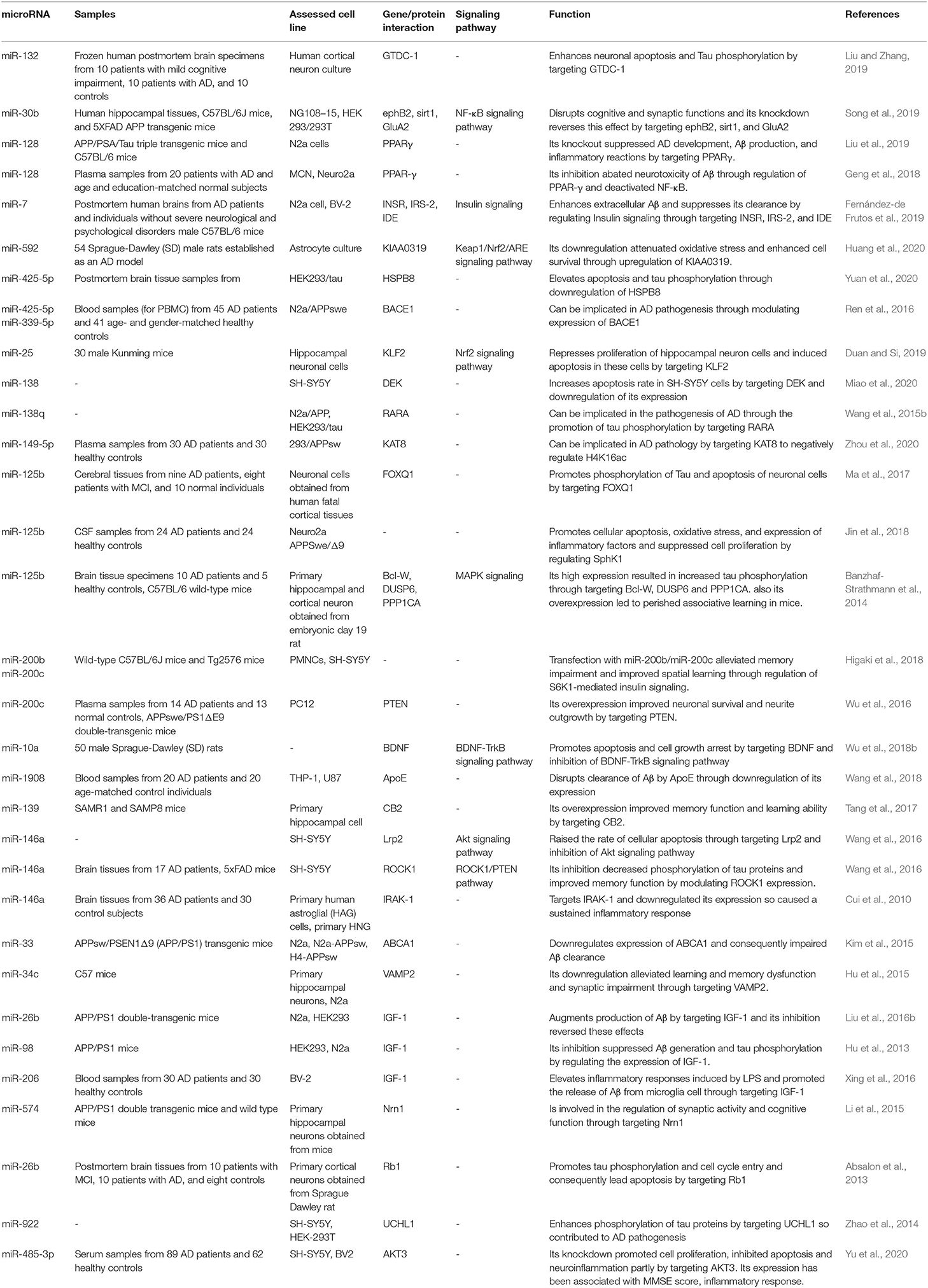
Table 2. Upregulated miRNAs in AD subject, animal models of AD or related cell lines and their functions in progression of AD.
Figure 1 demonstrates the function of a number of miRNAs in the pathogenesis of AD.
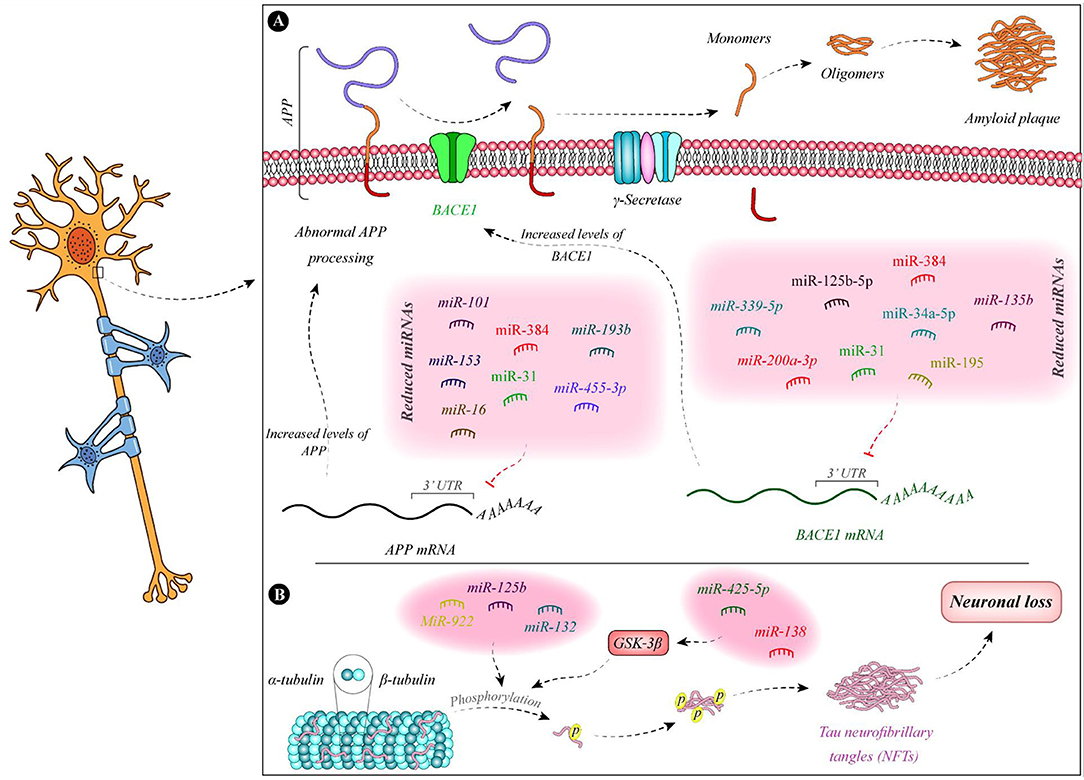
Figure 1. Summary of the function of miRNAs in the pathogenesis of AD. (A) Expressions of miR-135b (Zhang et al., 2016b), miR-195 (Zhu et al., 2012), miR-34a-5p (Liang et al., 2020), miR-384 (Liu et al., 2014b), miR-125b-5p (Liang et al., 2020), miR-31 (Barros-Viegas et al., 2020), miR-200a-3p (Pan et al., 2019), and miR-339-5p (Long et al., 2014) are decreased in patients with Alzheimer's disease. These miRNAs bind with the 3' UTR of BACE1 and decrease its expression. Therefore, the downregulation of these miRNAs leads to the upregulation of BACE1. In addition, expression levels of some APP-binding miRNAs namely miR-101 (Vilardo et al., 2010), miR-153 (Liang et al., 2012), miR-16 (Liu et al., 2012), miR-384 (Liu et al., 2014b), miR-31 (Barros-Viegas et al., 2020), miR-193b (Liu et al., 2014a), and miR-455-3p (Kumar et al., 2019) is decreased in patients with Alzheimer's disease. (B) Tau phosphorylation leads to defects in microtubules and induction of neurofibrillary tangles which result in neuron death. miR-138 and miR-425-5p are increased in Alzheimer's disease. These miRNAs regulate the expression of GSK-3β and enhance Tau phosphorylation (Wang et al., 2015b; Yuan et al., 2020). In addition, downregulation of miR-132 and upregulation of miR-125b and miR-922 leads to Tau hyperphosphorylation (Zhao et al., 2014; Salta et al., 2016; Ma et al., 2017).
The prognostic role of miRNAs in AD has been assessed in a single study. Xie et al. have evaluated serum levels of miR-206, miR-132, BDNF, and SIRT1 in a cohort of patients with amnestic mild cognitive impairment at baseline and after 5-year follow-up. Their results have shown higher levels of miR-206 in patients who converted to AD both at the baseline and after 5-year follow-up. However, miR-132 levels have been statistically similar between the conversion and non-conversion groups at both time points. Based on the Kaplan-Meier analysis, AD conversion has been correlated with over-expression of miR-206. In addition, multivariate Cox regression analysis has shown the suitability of serum miR-206 and its target BDNF as indicators of AD conversion (Xie et al., 2017). The diagnostic role of several miRNAs has been appraised in AD. For instance, downregulation of miR-433 and miR-133b in serum samples could precisely differentiate between AD subjects and normal persons (Yang et al., 2019b; Wang and Zhang, 2020). Moreover, the expression profile of the former miRNA in the cerebrospinal fluid (CSF) has appropriate diagnostic power for distinguishing AD patients (Wang and Zhang, 2020). The most astonishing results have been obtained for miR-34c. Expression of miR-34c has been increased in both cellular and plasma constituents of blood specimens of AD patients. The area under the receiver operating characteristic curve has been estimated to be 0.99. Moreover, expression of miR-34c has been inversely correlated with mental performance, as described by the mini-mental state examination. miR-34c has also been shown to affect the expression of numerous genes being involved in neuron survival and oxidative processes (Bhatnagar et al., 2014). Expression levels of miR-132 and miR-212 in neural-derived extracellular vesicles have been demonstrated to differentiate patients with AD from healthy subjects, yet their aptitude in identifying both AD and mild cognitive impairment as different from a healthy status has not been suitable (Cha et al., 2019). Table 3 summarizes the outlines of various studies that have reported on the diagnostic value of miRNAs in AD.
Boscher et al. have screened a larger cohort of early-onset AD (EOAD) patients who did not have autosomal dominant mutations for the presence of genetic polymorphisms. They have recognized 86 copy number variants (CNVs) in miRNA-coding genes, 31 of them being only present in EOAD cases. Duplication of the MIR138-2 locus has been one of these CNVs. Based on the role of miR-138 in Aβ production and tau phosphorylation, this CNV might be implicated in the risk of EOAD (Boscher et al., 2019). Functionally, miR-138 upregulation enhances Aβ synthesis and tau phosphorylation through modulation of GSK-3β and FERMT2 (Boscher et al., 2019). Other studies have demonstrated the role of rs2910164 of pri-miR-146a, rs57095329 of miR-146a, and rs2291418 of miR-1229 precursor in conferring risk of AD (Table 4). Zhang et al. have scanned the coding region of pri-miR-146a in AD patients. Among the four single nucleotide polymorphisms (SNPs) located in this genomic region, rs2910164 has been identified as a risk locus for AD as the C allele of this SNP has enhanced risk of AD.
Notably, this variant has been shown to reduce the expression of mature miR-146a-5p, releasing TLR2 from its inhibitory effects. Moreover, cell line studies have shown the impact of the C allele on upregulation of expression of TNF-α after induction with β-amyloid. Therefore, this SNP might predispose patients to AD by disturbing the production of mature miRNA and influencing the activity and expression level of TLR2 (Zhang et al., 2015). Cui et al. have analyzed the genotype and allele frequencies of rs2910464 and rs57095329 of miR-146a and have reported that the AA genotype of the former SNP increases susceptibility to AD and results in cognitive reduction in the affected individuals. Contrary to the previously mentioned study by Zhang et al., the risk genotype has been associated with higher levels of miR-146a in the PBMCs of control subjects and has exerted more robust effects on IL-6 and IL-1β synthesis following stimulation with LPS (Cui et al., 2014). Finally, in a genome-wide association study, Ghanbari et al. have detected an association between rs2291418 in the miR-1229 precursor and risk of AD. The risk allele of this SNP has been shown to increase the expression of miR-1229-3p, thus decreasing the expression of SORL1, an AD-associated gene. In addition, among more than 42,000 variants in miRNA-binding regions, 10 variants in the 3' UTR of nine genes have been associated with this disorder; among them has been rs6857, which enhances the miR-320e-mediated modulation of PVRL2 expression (Ghanbari et al., 2016).
Osthole, the active component of the fruits of the genus Cnidium moonnieri (L.) Cussion has been shown to affect the AD course via modulation of miRNAs expression. Lin et al. have shown miR-101a-3p as the main affected miRNA by osthole. APP has been identified as the target of miR-101a-3p. Osthole has enhanced the learning and memory aptitude in an animal model of AD, and it has inhibited APP levels by promoting the expression of miR-101a-3p (Lin et al., 2019). Other studies have verified the effects of Osthole on the expression of miR-9 (Li et al., 2016, 2017). Functionally, osthole enhances the viability of neurons, decreases apoptosis of these cells, and reverses the decline of synaptic proteins in APP-expressing cells by affecting miR-9 expression and consequently decreasing CAMKK2 and p-AMPKα levels (Li et al., 2016). Additionally, osthole has pro-survival effects in APP-expressing neural stem cells through suppression of the Notch pathway (Li et al., 2017). Moreover, Berberine has been shown to enhance proliferation and attenuate neuron apoptosis via regulation of miR-188/NOS1 molecular cascade (Chen et al., 2020b). Treatment of Aβ-treated murine microglia and neuroblastoma cells with this substance or upregulation of miR-188 in these cells has accelerated cell proliferation and suppressed caspase-3 activity and apoptosis (Chen et al., 2020b). Finally, exmedetomidine has been demonstrated to accomplish neuroprotective effects and enhance cognitive function in an animal model of AD by modulating the miR-129/YAP1/JAG1 cascade (Sun et al., 2020). Table 5 shows the effects of different AD-modifying compounds on the expression of miRNAs.
Numerous studies have demonstrated abnormal expression of miRNAs in AD subjects or cell/animal models of AD. However, each miRNA has been the subject of expression and functional assays in few independent studies. miR-146 has been among the miRNAs most assessed in the context of AD, as its expression levels, functions, and polymorphisms have been assessed in association with AD. miR-9 is another AD-associated miRNA whose expression has been altered following treatment of APP-expressing cells with anti-AD substances. In some cases, altered expression of a certain miRNA is regarded as a part of a self-protective process. For instance, the reduction of miR-409-5p expression in the early stages of AD might be associated with lower Aβ-induced synaptic injury (Guo et al., 2019). Similarly, upregulation of miR-200b and miR-200c has protective effects against AD-associated neurotoxicity (Higaki et al., 2018). However, in most cases, an aberrant miRNA signature directly participates in the pathogenesis of AD. miRNAs partake in the pathobiology of AD through various mechanisms, including the regulation of BACE1 activity. miR-200a-3p, miR-195, miR-338-5p, miR-34a-5p, miR-125b-5p, miR-132, miR-384, miR-339-5p, miR-135b, miR-425-5p, and miR-339-5p are among the miRNAs whose interaction with BACE1 has been verified in different investigation. Other miRNAs, such as miR-129-5p, miR-22, and miR-206, might affect the inflammatory responses in the course of AD. Moreover, a number of miRNAs, such as miR-326, miR-338-5p, miR-124-3p, miR-34a, miR-326, and miR-98, modulate apoptotic pathways in neurons, thereby affecting the AD course via this route. Tau phosphorylation can be modulated by some miRNAs, namely, miR-200a-3p, miR-326, miR-124-3p, miR-146a, miR-425-5p, and miR-132. Expression of miR-132 has been assessed by several investigations with most of them reporting its downregulation in AD (Wong et al., 2013; El Fatimy et al., 2018; Cha et al., 2019; Deng et al., 2020). Yet, Liu et al. have reported over-expression of miR-132 in patients with mild cognitive impairment and AD vs. normal individuals (Liu and Zhang, 2019).
Abnormal levels of miRNAs in serum or CSF samples have been shown to distinguish AD subjects from normal subjects, indicating their suitability as disease biomarkers. However, these studies have not been validated in independent cohorts. miR-103, miR-126, miR-93, miR-29, miR-424, and miR-181 are among AD-associated miRNAs with biomarker potential whose application as disease biomarkers has been validated in other disorders (So et al., 2020).
Animal studies have shown promising results regarding the impact of miRNA modifications on the course of AD. However, based on the unavailability of brain tissue for therapeutic interventions in human subjects, identification of appropriate transport mechanisms for delivery of anta-/ago-miRs to this tissue is an important issue.
The anti-AD effects of Osthole, Tiaoxin Recipe, Berberine, and Dexmedetomidine have been shown to be exerted through modulation of a number miRNAs, such as miR-101a-3p, miR-9, miR-34a, miR-188, and miR-129, emphasizing further the impact of miRNAs in the progression of AD. However, these results should be verified in human subjects as well.
Few studies have shown the association between miRNA CNVs/ SNPs and the risk of AD. However, these results have not been verified in different ethnic groups. Re-assessment of the results of genome-wide association studies with a focus on non-coding regions might lead to the identification of further risk loci for this multifactorial condition.
Finally, a limitation of several functional investigations in this field is that they have assessed miRNA functions in cell lines such as HEK293 and SH-SY5Y, which are not true models of AD.
MT and SG-F wrote the draft and revised it. MS, MH, and MG collected the data, designed the tables, and figures. All authors contributed to the article approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AD, Alzheimer's disease; microRNA, miRNA; Aβ, β-amyloid peptides; BACE1, β-secretase; EOAD, early-onset Alzheimer's disease; CNV, copy number variant; SNP, single nucleotide polymorphism; AChE, Acetylcholinesterase; iNOS, Inducible nitric oxide synthase; ROS, reactive oxygen species; MDA, Malondialdehyde; MAPK, mitogen-activated protein kinase; SOD, Superoxide dismutase; GSH-Px, glutathione peroxidase.
Absalon, S., Kochanek, D. M., Raghavan, V., and Krichevsky, A. M. (2013). MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 33, 14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013
Akhter, R., Shao, Y., Shaw, M., Formica, S., Khrestian, M., Leverenz, J. B., et al. (2018). Regulation of ADAM10 by miR-140-5p and potential relevance for Alzheimer's disease. Neurobiol. Aging. 63, 110–119. doi: 10.1016/j.neurobiolaging.2017.11.007
Ballard, C., Gauthier, S., Corbett, A., Brayne, C., Aarsland, D., and Jones, E. (2011). Alzheimer's disease. Lancet 377, 1019–1031. doi: 10.1016/S0140-6736(10)61349-9
Banzhaf-Strathmann, J., Benito, E., May, S., Arzberger, T., Tahirovic, S., Kretzschmar, H., et al. (2014). Micro RNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J. 33, 1667–1680. doi: 10.15252/embj.201387576
Barros-Viegas, A. T., Carmona, V., Ferreiro, E., Guedes, J., Cardoso, A. M., Cunha, P., et al. (2020). MiRNA-31 improves cognition and abolishes amyloid-β pathology by targeting APP and BACE1 in an animal model of Alzheimer's disease. Mol. Therapy Nucl. Acids 19, 1219–1236. doi: 10.1016/j.omtn.2020.01.010
Bhatnagar, S., Chertkow, H., Schipper, H. M., Shetty, V., Yuan, Z., Jones, T., et al. (2014). Increased microRNA-34c abundance in Alzheimer's disease circulating blood plasma. Front. Mol. Neurosci. 7:2. doi: 10.3389/fnmol.2014.00002
Boscher, E., Husson, T., Quenez, O., Laquerrière, A., Marguet, F., Cassinari, K., et al. (2019). Copy number variants in miR-138 as a potential risk factor for early-onset Alzheimer's disease. J. Alzheimer's Dis. 68, 1243–1255. doi: 10.3233/JAD-180940
Cao, J., Huang, M., Guo, L., Zhu, L., Hou, J., Zhang, L., et al. (2020). MicroRNA-195 rescues ApoE4-induced cognitive deficits and lysosomal defects in Alzheimer's disease pathogenesis. Mol. Psychiatry. 2020, 1–15. doi: 10.1038/s41380-020-0824-3
Cha, D. J., Mengel, D., Mustapic, M., Liu, W., Selkoe, D. J., Kapogiannis, D., et al. (2019). miR-212 and miR-132 are downregulated in neurally derived plasma exosomes of Alzheimer's patients. Front. Neurosci. 13:1208. doi: 10.3389/fnins.2019.01208
Chang, Y. W. Q. (2020). MicroRNA miR-212 regulates PDCD4 to attenuate Ab25–35-induced neurotoxicity via PI3K/AKT signaling pathway in Alzheimer's disease. Biotechnol. Lett. 42, 1789–1797. doi: 10.1007/s10529-020-02915-z
Chen, F. Z., Zhao, Y., and Chen, H. Z. (2019). MicroRNA-98 reduces amyloid β-protein production and improves oxidative stress and mitochondrial dysfunction through the Notch signaling pathway via HEY2 in Alzheimer's disease mice. Int. J. Mol. Med. 43, 91–102. doi: 10.3892/ijmm.2018.3957
Chen, M., Li, L., Liu, C., and Song, L. (2020b). Berberine attenuates Aβ-induced neuronal damage through regulating miR-188/NOS1 in Alzheimer's disease. Mol. Cell. Biochem. 474, 285–294. doi: 10.1007/s11010-020-03852-1
Chen, W., Wu, L., Hu, Y., Jiang, L., Liang, N., Chen, J., et al. (2020a). MicroRNA-107 ameliorates damage in a cell model of Alzheimer's disease by mediating the FGF7/FGFR2/PI3K/Akt pathway. J. Mol. Neurosci. 70, 1589–1597. doi: 10.1007/s12031-020-01600-0
Cui, J. G., Li, Y. Y., Zhao, Y., Bhattacharjee, S., and Lukiw, W. J. (2010). Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-κB in stressed human astroglial cells and in Alzheimer disease. J. Biol. Chem. 285, 38951–38960. doi: 10.1074/jbc.M110.178848
Cui, L., Li, Y., Ma, G., Wang, Y., Cai, Y., Liu, S., et al. (2014). A functional polymorphism in the promoter region of microRNA-146a is associated with the risk of Alzheimer disease and the rate of cognitive decline in patients. PLoS ONE 9:e89019. doi: 10.1371/journal.pone.0089019
Deng, Y., Zhang, J., Sun, X., Ma, G., Luo, G., Miao, Z., et al. (2020). miR-132 improves the cognitive function of rats with Alzheimer's disease by inhibiting the MAPK1 signal pathway. Exp. Therapeutic Med. 20:1. doi: 10.3892/etm.2020.9288
Denk, J., Oberhauser, F., Kornhuber, J., Wiltfang, J., Fassbender, K., Schroeter, M. L., et al. (2018). Specific serum and CSF microRNA profiles distinguish sporadic behavioural variant of frontotemporal dementia compared with Alzheimer patients and cognitively healthy controls. PLoS ONE 13:e0197329. doi: 10.1371/journal.pone.0197329
Duan, Q., and Si, E. (2019). MicroRNA-25 aggravates Aβ1-42-induced hippocampal neuron injury in Alzheimer's disease by downregulating KLF2 via the Nrf2 signaling pathway in a mouse model. J. Cell. Biochem. 120, 15891–15905. doi: 10.1002/jcb.28861
El Fatimy, R., Li, S., Chen, Z., Mushannen, T., Gongala, S., Wei, Z., et al. (2018). MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol. 136, 537–555. doi: 10.1007/s00401-018-1880-5
Fernández-de Frutos, M., Galán-Chilet, I., Goedeke, L., Kim, B., Pardo-Marqués, V., Pérez-García, A., et al. (2019). MicroRNA 7 impairs insulin signaling and regulates Aβ levels through posttranscriptional regulation of the insulin receptor substrate 2, insulin receptor, insulin-degrading enzyme, and liver X receptor pathway. Mol. Cell. Biol. 39, e00170–e00119. doi: 10.1128/MCB.00170-19
Gámez-Valero, A., Campdelacreu, J., Vilas, D., Ispierto, L., Reñé R, Álvarez, R., et al. (2019). Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer's disease and dementia with Lewy bodies. Transl. Neurodegeneration 8:31. doi: 10.1186/s40035-019-0169-5
Geng, L., Zhang, T., Liu, W., and Chen, Y. (2018). Inhibition of miR-128 abates Aβ-mediated cytotoxicity by targeting PPAR-γ via NF-κB inactivation in primary mouse cortical neurons and Neuro2a cells. Yonsei Med. J. 59, 1096–1106. doi: 10.3349/ymj.2018.59.9.1096
Ghanbari, M., Ikram, M. A., De Looper, H. W., Hofman, A., Erkeland, S. J., Franco, O. H., et al. (2016). Genome-wide identification of microRNA-related variants associated with risk of Alzheimer's disease. Sci. Rep. 6:28387. doi: 10.1038/srep28387
Guo, J., Cai, Y., Ye, X., Ma, N., Wang, Y., Yu, B., et al. (2019). MiR-409-5p as a regulator of neurite growth is down regulated in APP/PS1 murine model of Alzheimer's disease. Front. Neurosci. 13:1264. doi: 10.3389/fnins.2019.01264
Guo, R., Fan, G., Zhang, J., Wu, C., Du, Y., Ye, H., et al. (2017). A 9-microRNA signature in serum serves as a noninvasive biomarker in early diagnosis of Alzheimer's disease. J. Alzheimer's Dis. 60, 1365–1377. doi: 10.3233/JAD-170343
Han, C., Guo, L., Yang, Y., Guan, Q., Shen, H., Sheng, Y., et al. (2020). Mechanism of microRNA-22 in regulating neuroinflammation in Alzheimer's disease. Brain Behav. 2020:e01627. doi: 10.1002/brb3.1627
Han, L., Zhou, Y., Zhang, R., Wu, K., Lu, Y., Li, Y., et al. (2018). MicroRNA Let-7f-5p promotes bone marrow mesenchymal stem cells survival by targeting caspase-3 in alzheimer disease model. Front. Neurosci. 12:333. doi: 10.3389/fnins.2018.00333
Hara, N., Kikuchi, M., Miyashita, A., Hatsuta, H., Saito, Y., Kasuga, K., et al. (2017). Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer's disease. Acta Neuropathol. Commun. 5:10. doi: 10.1186/s40478-017-0414-z
He, B., Chen, W., Zeng, J., Tong, W., and Zheng, P. (2020). MicroRNA-326 decreases tau phosphorylation and neuron apoptosis through inhibition of the JNK signaling pathway by targeting VAV1 in Alzheimer's disease. J. Cell. Physiol. 235, 480–493. doi: 10.1002/jcp.28988
Hernandez-Rapp, J., Rainone, S., Goupil, C., Dorval, V., Smith, P. Y., Saint-Pierre, M., et al. (2016). microRNA-132/212 deficiency enhances Aβ production and senile plaque deposition in Alzheimer's disease triple transgenic mice. Sci. Rep. 6:30953. doi: 10.1038/srep30953
Higaki, S., Muramatsu, M., Matsuda, A., Matsumoto, K., Satoh, J-i, Michikawa, M., et al. (2018). Defensive effect of microRNA-200b/c against amyloid-beta peptide-induced toxicity in Alzheimer's disease models. PLoS ONE 13:e0196929. doi: 10.1371/journal.pone.0196929
Hu, S., Wang, H., Chen, K., Cheng, P., Gao, S., Liu, J., et al. (2015). MicroRNA-34c downregulation ameliorates amyloid-β-induced synaptic failure and memory deficits by targeting VAMP2. J. Alzheimer's Dis. 48, 673–686. doi: 10.3233/JAD-150432
Hu, Y-K., Wang, X., Li, L., Du, Y-H., Ye, H-T., and Li, C-Y. (2013). MicroRNA-98 induces an Alzheimer's disease-like disturbance by targeting insulin-like growth factor 1. Neurosci. Bulletin 29, 745–751. doi: 10.1007/s12264-013-1348-5
Huang, W., Wu, X., Xue, Y., Zhou, Y., Xiang, H., Yang, W., et al. (2020). MicroRNA-3614 regulates inflammatory response via targeting TRAF6-mediated MAPKs and NF-κB signaling in the epicardial adipose tissue with coronary artery disease. Int. J. Cardiol. 2020:45. doi: 10.1016/j.ijcard.2020.09.045
Iqbal, K., and Grundke-Iqbal, I. (2010). Alzheimer's disease, a multifactorial disorder seeking multitherapies. Alzheimers Dement. 6, 420–424. doi: 10.1016/j.jalz.2010.04.006
Jin, Y., Tu, Q., and Liu, M. (2018). MicroRNA-125b regulates Alzheimer's disease through SphK1 regulation. Mol. Med. Rep. 18, 2373–2380. doi: 10.3892/mmr.2018.9156
Kang, Q., Xiang, Y., Li, D., Liang, J., Zhang, X., Zhou, F., et al. (2017). MiR-124-3p attenuates hyperphosphorylation of Tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3β pathway in N2a/APP695swe cells. Oncotarget 8:24314. doi: 10.18632/oncotarget.15149
Kim, J., Yoon, H., Horie, T., Burchett, J. M., Restivo, J. L., Rotllan, N., et al. (2015). microRNA-33 regulates ApoE lipidation and amyloid-β metabolism in the brain. J. Neurosci. 35, 14717–14726. doi: 10.1523/JNEUROSCI.2053-15.2015
Kumar, S., Reddy, A. P., Yin, X., and Reddy, P. H. (2019). Novel MicroRNA-455-3p and its protective effects against abnormal APP processing and amyloid beta toxicity in Alzheimer's disease. Biochim. Biophys. Acta 1865, 2428–2440. doi: 10.1016/j.bbadis.2019.06.006
Kumar, S., and Reddy, P. H. (2018). MicroRNA-455-3p as a potential biomarker for Alzheimer's disease: an update. Front. Aging Neurosci. 10:41. doi: 10.3389/fnagi.2018.00041
Kumar, S., Vijayan, M., and Reddy, P. H. (2017). MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer's disease. Hum. Mol. Genet. 26, 3808–3822. doi: 10.1093/hmg/ddx267
Lee, K., Kim, H., An, K., Kwon, O-B, Park, S., Cha, J. H., et al. (2016). Replenishment of microRNA-188-5p restores the synaptic and cognitive deficits in 5XFAD mouse model of Alzheimer's disease. Sci Rep. 6, 1–14. doi: 10.1038/srep34433
Li, F., Wei, G., Bai, Y., Li, Y., Huang, F., Lin, J., et al. (2015). MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin. Brain Res. 1627, 177–188. doi: 10.1016/j.brainres.2015.09.022
Li, J., Li, D., Zhou, H., Wu, G., He, Z., Liao, W., et al. (2020a). MicroRNA-338-5p alleviates neuronal apoptosis via directly targeting BCL2L11 in APP/PS1 mice. Aging 12:10.18632. doi: 10.18632/aging.104005
Li, P., Xu, Y., Wang, B., Huang, J., and Li, Q. (2020b). miR-34a-5p and miR-125b-5p attenuate Aβ-induced neurotoxicity through targeting BACE1. J. Neurol. Sci. 2020:116793. doi: 10.1016/j.jns.2020.116793
Li, S., Yan, Y., Jiao, Y., Gao, Z., Xia, Y., Kong, L., et al. (2016). Neuroprotective effect of osthole on neuron synapses in an Alzheimer's disease cell model via upregulation of MicroRNA-9. J. Mol. Neurosci. 60, 71–81. doi: 10.1007/s12031-016-0793-9
Li, S-H., Gao, P., Wang, L-T., Yan, Y-H., Xia, Y., Song, J., et al. (2017). Osthole stimulated neural stem cells differentiation into neurons in an Alzheimer's disease cell model via upregulation of microRNA-9 and rescued the functional impairment of hippocampal neurons in APP/PS1 transgenic mice. Front. Neurosci. 11:340. doi: 10.3389/fnins.2017.00340
Liang, C., Zhu, H., Xu, Y., Huang, L., Ma, C., Deng, W., et al. (2012). MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 1455, 103–113. doi: 10.1016/j.brainres.2011.10.051
Liang, X., Wang, L., Wang, M., Liu, Z., Liu, X., Zhang, B., et al. (2020). MicroRNA-124 inhibits macrophage cell apoptosis via targeting p38/MAPK signaling pathway in atherosclerosis development. Aging 12:13005. doi: 10.18632/aging.103387
Lin, Y., Liang, X., Yao, Y., Xiao, H., Shi, Y., and Yang, J. (2019). Osthole attenuates APP-induced Alzheimer's disease through upregulating miRNA-101a-3p. Life Sci. 225, 117–131. doi: 10.1016/j.lfs.2019.04.004
Liu, C-G., Song, J., Zhang, Y-Q., and Wang, P-C. (2014a). MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer's disease. Mol. Med. Rep. 10, 2395–2400. doi: 10.3892/mmr.2014.2484
Liu, C-G., Wang, J-L., Li, L., and Wang, P-C. (2014b). MicroRNA-384 regulates both amyloid precursor protein and β-secretase expression and is a potential biomarker for Alzheimer's disease. Int. J. Mol. Med. 34, 160–166. doi: 10.3892/ijmm.2014.1780
Liu, D. Y., and Zhang, L. (2019). MicroRNA-132 promotes neurons cell apoptosis and activates Tau phosphorylation by targeting GTDC-1 in Alzheimer's disease. Eur. Rev. Med. Pharmacol. Sci. 23, 8523–32. doi: 10.26355/eurrev_201910_19166
Liu, F., Zhang, Z., Chen, W., Gu, H., and Yan, Q. (2018). Regulatory mechanism of microRNA-377 on CDH13 expression in the cell model of Alzheimer's disease. Eur. Rev. Med. Pharmacol. Sci. 22, 2801–8. doi: 10.26355/eurrev_201805_14979
Liu, H., Chu, W., Gong, L., Gao, X., and Wang, W. (2016b). MicroRNA-26b is upregulated in a double transgenic mouse model of Alzheimer's disease and promotes the expression of amyloid-β by targeting insulin-like growth factor 1. Mol. Med. Rep. 13, 2809–2814. doi: 10.3892/mmr.2016.4860
Liu, J., Zuo, X., Han, J., Dai, Q., Xu, H., Liu, Y., et al. (2020). MiR-9-5p inhibits mitochondrial damage and oxidative stress in AD cell models by targeting GSK-3β. Biosci. Biotechnol. Biochem. 84, 2273–2280. doi: 10.1080/09168451.2020.1797469
Liu, W., Cai, H., Lin, M., Zhu, L., Gao, L., Zhong, R., et al. (2016a). MicroRNA-107 prevents amyloid-beta induced blood-brain barrier disruption and endothelial cell dysfunction by targeting Endophilin-1. Exp. Cell Res. 343, 248–257. doi: 10.1016/j.yexcr.2016.03.026
Liu, W., Liu, C., Zhu, J., Shu, P., Yin, B., Gong, Y., et al. (2012). MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer's-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 33, 522–534. doi: 10.1016/j.neurobiolaging.2010.04.034
Liu, Y., Zhang, Y., Liu, P., Bai, H., Li, X., Xiao, J., et al. (2019). MicroRNA-128 knockout inhibits the development of Alzheimer's disease by targeting PPARγ in mouse models. Eur. J. Pharmacol. 843, 134–144. doi: 10.1016/j.ejphar.2018.11.004
Long, J. M., Ray, B., and Lahiri, D. K. (2012). MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 287, 31298–31310. doi: 10.1074/jbc.M112.366336
Long, J. M., Ray, B., and Lahiri, D. K. (2014). MicroRNA-339-5p downregulates protein expression of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J. Biol. Chem. 289, 5184–5198. doi: 10.1074/jbc.M113.518241
Lukiw, W. J. (2007). Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport 18, 297–300. doi: 10.1097/WNR.0b013e3280148e8b
Ma, X., Liu, L., and Meng, J. (2017). MicroRNA-125b promotes neurons cell apoptosis and Tau phosphorylation in Alzheimer's disease. Neurosci. Lett.661, 57–62. doi: 10.1016/j.neulet.2017.09.043
Madadi, S., Saidijam, M., Yavari, B., and Soleimani, M. (2020). Downregulation of serum miR-106b: a potential biomarker for Alzheimer disease. Archiv. Physiol. Biochem. 2020, 1–5. doi: 10.1080/13813455.2020.1734842
Manzine, P. R., Pelucchi, S., Horst, M. A., Vale, F. A., Pavarini, S. C., Audano, M., et al. (2018). microRNA 221 Targets ADAM10 mRNA and is Downregulated in Alzheimer's Disease. J. Alzheimer's Dis. 61, 113–123. doi: 10.3233/JAD-170592
McKeever, P. M., Schneider, R., Taghdiri, F., Weichert, A., Multani, N., Brown, R. A., et al. (2018). MicroRNA expression levels are altered in the cerebrospinal fluid of patients with young-onset Alzheimer's disease. Mol. Neurobiol. 55, 8826–8841. doi: 10.1007/s12035-018-1032-x
Miao, J., Jing, J., Shao, Y., and Sun, H. (2020). MicroRNA-138 promotes neuroblastoma SH-SY5Y cell apoptosis by directly targeting DEK in Alzheimer's disease cell model. BMC Neurosci. 21, 1–8. doi: 10.1186/s12868-020-00579-z
Modi, P. K., Jaiswal, S., and Sharma, P. (2016). Regulation of neuronal cell cycle and apoptosis by microRNA 34a. Mol. Cell. Biol. 36, 84–94. doi: 10.1128/MCB.00589-15
Moncini, S., Lunghi, M., Valmadre, A., Grasso, M., Del Vescovo, V., Riva, P., et al. (2017). The miR-15/107 family of microRNA genes regulates CDK5R1/p35 with implications for Alzheimer's disease pathogenesis. Mol. Neurobiol. 54, 4329–4342. doi: 10.1007/s12035-016-0002-4
Müller, M., Jäkel, L., Bruinsma, I. B., Claassen, J. A., Kuiperij, H. B., and Verbeek, M. M. (2016). MicroRNA-29a is a candidate biomarker for Alzheimer's disease in cell-free cerebrospinal fluid. Mol. Neurobiol. 53, 2894–2899. doi: 10.1007/s12035-015-9156-8
Nagaraj, S., Laskowska-Kaszub, K., Debski, K. J., Wojsiat, J., Dabrowski, M., Gabryelewicz, T., et al. (2017). Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer's disease patients from non-demented subjects. Oncotarget 8:16122. doi: 10.18632/oncotarget.15109
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9:402. doi: 10.3389/fendo.2018.00402
Pan, J., Zhang, D., Zhang, J., Qin, P., and Wang, J. (2019). LncRNA RMRP silence curbs neonatal neuroblastoma progression by regulating microRNA-206/tachykinin-1 receptor axis via inactivating extracellular signal-regulated kinases. Cancer Biol. Therapy 20, 653–665. doi: 10.1080/15384047.2018.1550568
Prince, M., Bryce, R., Albanese, E., Wimo, A., Ribeiro, W., and Ferri, C. P. (2013). The global prevalence of dementia: a systematic review and metaanalysis. Alzheimer's Dement. 9, 63–75 e2. doi: 10.1016/j.jalz.2012.11.007
Qian, Q., Zhang, J., He, F-P, Bao, W-X, Zheng, T-T, Zhou, D-M, et al. (2019). Downregulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer's disease. FASEB J. 33, 4404–44017. doi: 10.1096/fj.201801846R
Querfurth, H. W., and LaFerla, F. M. (2010). Alzheimer's disease. N. Engl. J. Med. 362, 329–344. doi: 10.1056/NEJMra0909142
Ren, R-J., Zhang, Y-F., Dammer, E. B., Zhou, Y., Wang, L-l., Liu, X-H., et al. (2016). Peripheral blood MicroRNA expression profiles in Alzheimer's disease: screening, validation, association with clinical phenotype and implications for molecular mechanism. Mol. Neurobiol. 53, 5772–5781. doi: 10.1007/s12035-015-9484-8
Salta, E., Sierksma, A., Vanden Eynden, E., and De Strooper, B. (2016). miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer's brain. EMBO Mol. Med. 8, 1005–1018. doi: 10.15252/emmm.201606520
Shu, B., Zhang, X., Du, G., Fu, Q., and Huang, L. (2018). MicroRNA-107 prevents amyloid-β-induced neurotoxicity and memory impairment in mice. Int. J. Mol. Med. 41, 1665–1672. doi: 10.3892/ijmm.2017.3339
So, J. B. Y., Kapoor, R., Zhu, F., Koh, C., Zhou, L., Zou, R., et al. (2020). Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2020:322065. doi: 10.1136/gutjnl-2020-322065
Song, Y., Hu, M., Zhang, J., Teng, Z-q, and Chen, C. (2019). A novel mechanism of synaptic and cognitive impairments mediated via microRNA-30b in Alzheimer's disease. EBioMedicine 39, 409–421. doi: 10.1016/j.ebiom.2018.11.059
Sun, W., Zhao, J., and Li, C. (2020). Dexmedetomidine provides protection against hippocampal neuron apoptosis and cognitive impairment in mice with Alzheimer's disease by mediating the miR-129/YAP1/JAG1 axis. Mol. Neurobiol. 57, 5044–5055. doi: 10.1007/s12035-020-02069-z
Tan, L., Yu, J-T., Tan, M-S., Liu, Q-Y., Wang, H-F., Zhang, W., et al. (2014). Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J. Alzheimer's Dis. 40, 1017–1027. doi: 10.3233/JAD-132144
Tang, Y., Bao, J., Su, J., and Huang, W. (2017). MicroRNA-139 modulates Alzheimer's-associated pathogenesis in SAMP8 mice by targeting cannabinoid receptor type 2. Genet. Mol. Res. 16:19166. doi: 10.4238/gmr16019166
Vilardo, E., Barbato, C., Ciotti, M., Cogoni, C., and Ruberti, F. (2010). MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 285, 18344–18351. doi: 10.1074/jbc.M110.112664
Wang, G., Huang, Y., Wang, L-L, Zhang, Y-F, Xu, J., Zhou, Y., et al. (2016). MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer's disease. Sci. Rep. 6:26697. doi: 10.1038/srep26697
Wang, J., Chen, C., and Zhang, Y. (2020). An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer's disease. J. Clin. Lab. Anal. 34:e23006. doi: 10.1002/jcla.23006
Wang, L., Liu, J., Wang, Q., Jiang, H., Zeng, L., Li, Z., et al. (2019b). MicroRNA-200a-3p mediates neuroprotection in Alzheimer-related deficits and attenuates amyloid-beta overproduction and tau hyperphosphorylation via coregulating BACE1 and PRKACB. Front. Pharmacol. 10:806. doi: 10.3389/fphar.2019.00806
Wang, M., Qin, L., and Tang, B. (2019a). MicroRNAs in Alzheimer's disease. Front. Genet. 10:153. doi: 10.3389/fgene.2019.00153
Wang, R., and Zhang, J. (2020). Clinical significance of miR-433 in the diagnosis of Alzheimer's disease and its effect on Aβ-induced neurotoxicity by regulating JAK2. Exp. Gerontol. 141:111080. doi: 10.1016/j.exger.2020.111080
Wang, X., Tan, L., Lu, Y., Peng, J., Zhu, Y., Zhang, Y., et al. (2015b). MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 589, 726–729. doi: 10.1016/j.febslet.2015.02.001
Wang, X., Xu, Y., Zhu, H., Ma, C., Dai, X., and Qin, C. (2015a). Downregulated microRNA-222 is correlated with increased p27Kip1 expression in a double transgenic mouse model of Alzheimer's disease. Mol. Med. Rep. 12, 7687–7692. doi: 10.3892/mmr.2015.4339
Wang, Y., Luo, X., Liu, Y., Han, G., and Sun, D. (2019c). Long noncoding RNA RMRP promotes proliferation and invasion via targeting miR-1-3p in non–small-cell lung cancer. J. Cell. Biochem. 120, 15170–15181. doi: 10.1002/jcb.28779
Wang, Y., Veremeyko, T., Wong, AH-K, El Fatimy, R., Wei, Z., Cai, W., et al. (2017). Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer's disease. Neurobiol. Aging 51, 156–166. doi: 10.1016/j.neurobiolaging.2016.12.015
Wang, Z., Qin, W., Wei, C., Tang, Y., Zhao, L., Jin, H., et al. (2018). The microRNA-1908 up-regulation in the peripheral blood cells impairs amyloid clearance by targeting ApoE. Int. J. Geriatric Psychiatry 33, 980–986. doi: 10.1002/gps.4881
Wong, H-KA., Veremeyko, T., Patel, N., Lemere, C. A., Walsh, D. M., Esau, C., et al. (2013). De-repression of FOXO3a death axis by microRNA-132 and-212 causes neuronal apoptosis in Alzheimer's disease. Hum. Mol. Genet. 22, 3077–3092. doi: 10.1093/hmg/ddt164
Wu, B. W., Wu, M. S., and Guo, J. D. (2018b). Effects of microRNA-10a on synapse remodeling in hippocampal neurons and neuronal cell proliferation and apoptosis through the BDNF-TrkB signaling pathway in a rat model of Alzheimer's disease. J. Cell. Physiol. 233, 5281–5292. doi: 10.1002/jcp.26328
Wu, D. M., Wen, X., Wang, Y. J., Han, X. R., Wang, S., Shen, M., et al. (2018a). Effect of microRNA-186 on oxidative stress injury of neuron by targeting interleukin 2 through the janus kinase-signal transducer and activator of transcription pathway in a rat model of Alzheimer's disease. J. Cell. Physiol. 233, 9488–9502. doi: 10.1002/jcp.26843
Wu, Q., Ye, X., Xiong, Y., Zhu, H., Miao, J., Zhang, W., et al. (2016). The protective role of microRNA-200c in Alzheimer's disease pathologies is induced by beta amyloid-triggered endoplasmic reticulum stress. Front. Mol. Neurosci. 9:140. doi: 10.3389/fnmol.2016.00140
Wu, Q., Yuan, X., Bai, J., Han, R., Li, Z., Zhang, H., et al. (2019). MicroRNA-181a protects against pericyte apoptosis via directly targeting FOXO1: implication for ameliorated cognitive deficits in APP/PS1 mice. Aging 11:6120. doi: 10.18632/aging.102171
Xiao, X., Gu, Y., Wang, G., and Chen, S. (2019). c-Myc, RMRP, and miR-34a-5p form a positive-feedback loop to regulate cell proliferation and apoptosis in multiple myeloma. Int. J. Biol. Macromol. 122, 526–537. doi: 10.1016/j.ijbiomac.2018.10.207
Xie, B., Liu, Z., Jiang, L., Liu, W., Song, M., Zhang, Q., et al. (2017). Increased serum miR-206 level predicts conversion from amnestic mild cognitive impairment to Alzheimer's disease: a 5-year follow-up study. J. Alzheimer's Dis. 55, 509–520. doi: 10.3233/JAD-160468
Xing, H., Guo, S., Zhang, Y., Zheng, Z., and Wang, H. (2016). Upregulation of microRNA-206 enhances lipopolysaccharide-induced inflammation and release of amyloid-β by targeting insulin-like growth factor 1 in microglia. Mol. Med. Rep. 14, 1357–1364. doi: 10.3892/mmr.2016.5369
Xu, N., Li, A-D, Ji, L-L, Ye, Y., Wang, Z-Y, and Tong, L. (2019). miR-132 regulates the expression of synaptic proteins in APP/PS1 transgenic mice through C1q. Eur. J. Histochem. 63:3008. doi: 10.4081/ejh.2019.3008
Yang, G., Song, Y., Zhou, X., Deng, Y., Liu, T., Weng, G., et al. (2015). DNA methyltransferase 3, a target of microRNA-29c, contributes to neuronal proliferation by regulating the expression of brain-derived neurotrophic factor. Mol. Med. Rep. 12, 1435–1442. doi: 10.3892/mmr.2015.3531
Yang, K., Feng, S., Ren, J., and Zhou, W. (2019a). Upregulation of microRNA-196a improves cognitive impairment and alleviates neuronal damage in hippocampus tissues of Alzheimer's disease through downregulating LRIG3 expression. J. Cell. Biochem. 120, 17811–17821. doi: 10.1002/jcb.29047
Yang, Q., Zhao, Q., and Yin, Y. (2019b). miR-133b is a potential diagnostic biomarker for Alzheimer's disease and has a neuroprotective role. Exp. Therapeutic Med. 18, 2711–2718. doi: 10.3892/etm.2019.7855
Yilmaz, S. G., Erdal, M. E., Özge, A. A., and Sungur, M. A. (2016). Can peripheral MicroRNA expression data serve as epigenomic (upstream) biomarkers of Alzheimer's disease? Omics 20, 456–461. doi: 10.1089/omi.2016.0099
Yu, L., Li, H., Liu, W., Zhang, L., Tian, Q., Li, H., et al. (2020). MiR-485-3p serves as a biomarker and therapeutic target of Alzheimer's disease via regulating neuronal cell viability and neuroinflammation by targeting AKT3. Mol. Genet. Genom. Med. 2020:e1548. doi: 10.1002/mgg3.1548
Yuan, J., Wu, Y., Li, L., and Liu, C. (2020). MicroRNA-425-5p promotes tau phosphorylation and cell apoptosis in Alzheimer's disease by targeting heat shock protein B8. J. Neural Transmission. 2020, 1–8. doi: 10.1007/s00702-019-02134-5
Zeng, Z., Liu, Y., Zheng, W., Liu, L., Yin, H., Zhang, S., et al. (2019). MicroRNA-129-5p alleviates nerve injury and inflammatory response of Alzheimer's disease via downregulating SOX6. Cell Cycle 18, 3095–3110. doi: 10.1080/15384101.2019.1669388
Zhang, B., Wang, A., Xia, C., Lin, Q., and Chen, C. (2015). A single nucleotide polymorphism in primary-microRNA-146a reduces the expression of mature microRNA-146a in patients with Alzheimer's disease and is associated with the pathogenesis of Alzheimer's disease. Mol. Med. Rep. 12, 4037–4042. doi: 10.3892/mmr.2015.3968
Zhang, S. Y., Huang, S. H., Gao, S. X., Wang, Y. B., Jin, P., and Lu, F. J. (2019). Upregulation of lncRNA RMRP promotes the activation of cardiac fibroblasts by regulating miR-613. Mol. Med. Rep. 20, 3849–3857. doi: 10.3892/mmr.2019.10634
Zhang, Y., Li, Q., Liu, C., Gao, S., Ping, H., Wang, J., et al. (2016a). MiR-214-3p attenuates cognition defects via the inhibition of autophagy in SAMP8 mouse model of sporadic Alzheimer's disease. Neurotoxicology 56, 139–149. doi: 10.1016/j.neuro.2016.07.004
Zhang, Y., Xing, H., Guo, S., Zheng, Z., Wang, H., and Xu, D. (2016b). MicroRNA-135b has a neuroprotective role via targeting of β-site APP-cleaving enzyme 1. Exp. Therapeut. Med. 12, 809–814. doi: 10.3892/etm.2016.3366
Zhao, X., Kang, J., Svetnik, V., Warden, D., Wilcock, G., David Smith, A., et al. (2020). A machine learning approach to identify a circulating MicroRNA signature for Alzheimer disease. J. Appl. Lab. Med. 5, 15–28. doi: 10.1373/jalm.2019.029595
Zhao, Z-B., Wu, L., Xiong, R., Wang, L-L, Zhang, B., Wang, C., et al. (2014). MicroRNA-922 promotes tau phosphorylation by downregulating ubiquitin carboxy-terminal hydrolase L1 (UCHL1) expression in the pathogenesis of Alzheimer's disease. Neuroscience 275, 232–237. doi: 10.1016/j.neuroscience.2014.06.013
Zhou, H., Zhang, R., Lu, K., Yu, W., Xie, B., Cui, D., et al. (2016). Deregulation of miRNA-181c potentially contributes to the pathogenesis of AD by targeting collapsin response mediator protein 2 in mice. J. Neurol. Sci. 367, 3–10. doi: 10.1016/j.jns.2016.05.038
Zhou, Y., Chen, Z., Chen, A., Ma, J., Qian, J., and Ge, J. (2020). Elevated serum miR-133a predicts patients at risk of periprocedural myocardial injury after elective percutaneous coronary intervention. Cardiol. J. 2020:34. doi: 10.5603/CJ.a2020.0034
Keywords: Alzheimer's disease, miRNA, marker, expression, polymorphism
Citation: Samadian M, Gholipour M, Hajiesmaeili M, Taheri M and Ghafouri-Fard S (2021) The Eminent Role of microRNAs in the Pathogenesis of Alzheimer's Disease. Front. Aging Neurosci. 13:641080. doi: 10.3389/fnagi.2021.641080
Received: 13 December 2020; Accepted: 19 February 2021;
Published: 15 March 2021.
Edited by:
Ramesh Kandimalla, Texas Tech University Health Sciences Center, United StatesReviewed by:
Rezvan Noroozi, Jagiellonian University, PolandCopyright © 2021 Samadian, Gholipour, Hajiesmaeili, Taheri and Ghafouri-Fard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==; Soudeh Ghafouri-Fard, cy5naGFmb3VyaWZhcmRAc2JtdS5hYy5pcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.