
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci., 25 February 2021
Sec. Alzheimer's Disease and Related Dementias
Volume 13 - 2021 | https://doi.org/10.3389/fnagi.2021.638922
This article is part of the Research TopicImmunological Mechanisms, Biomarkers and Immunotherapies of Alzheimer's DiseaseView all 10 articles
 Jagan A. Pillai1,2,3*
Jagan A. Pillai1,2,3* Gurkan Bebek4,5
Gurkan Bebek4,5 Maria Khrestian6,7
Maria Khrestian6,7 James Bena8
James Bena8 Cornelia C. Bergmann7
Cornelia C. Bergmann7 William S. Bush9
William S. Bush9 James B. Leverenz1,2,3
James B. Leverenz1,2,3 Lynn M. Bekris6,7 for the Alzheimer's Disease Neuroimaging Initiative†
Lynn M. Bekris6,7 for the Alzheimer's Disease Neuroimaging Initiative†Tumor necrosis factor receptor 2 (TNFR2) promotes neuronal survival downstream. This longitudinal study evaluated whether the TNFRSF1B gene encoding TNFR2 and levels of its soluble form (sTNFR2) affect Alzheimer disease (AD) biomarkers and clinical outcomes. Data analyzed included 188 patients in the Alzheimer's Disease Neuroimaging Initiative (ADNI) who had mild cognitive impairment (MCI) and AD dementia. Further, a replication study was performed in 48 patients with MCI with positive AD biomarkers who were treated at a memory clinic. Cerebrospinal fluid (CSF) sTNFR2 levels along with two related TNFRSF1B gene single nucleotide polymorphisms (SNPs) rs976881 and rs1061622 were assessed. General linear models were used to evaluate the effect of CSF sTNFR2 levels and each SNP in relationship to CSF t-tau and p-tau, cognitive domains, MRI brain measures, and longitudinal cognitive changes after adjustments were made for covariates such as APOE ε4 status. In the ADNI cohort, a significant interaction between rs976881 and CSF sTNFR2 modulates CSF t-tau and p-tau levels; hippocampal and whole brain volumes; and Digit Span Forwards subtest scores. In the replication cohort, a significant interaction between rs976881 and CSF sTNFR2 modulates CSF p-tau. A significant interaction between rs976881 and CSF sTNFR2 also impacts Clinical Dementia Rating Sum of Boxes scores over 12 months in the ADNI cohort. The interaction between TNFRSF1B variant rs976881 and CSF sTNFR2 levels was noted to modulate multiple AD-associated severity markers and cognitive domains. This interaction impacts resilience-related clinical outcomes in AD and lends support to sTNFR2 as a promising candidate for therapeutic targeting to improve clinical outcomes of interest.
Alzheimer disease (AD) is paradigmatically characterized by the presence of amyloid β plaques and tau neurofibrillary tangles in the brain. However, the cognitive and behavioral phenotypes of AD and their related CSF and MRI biomarker signatures can vary among patients, and molecular factors beyond amyloid β and tau likely play a role in this heterogeneity (van der Vlies et al., 2009; Whitwell et al., 2012; Dubois et al., 2014; Risacher et al., 2017; Pillai et al., 2019a). Multiple studies have evaluated APOE ε4 and other genes that may be related to the various MRI and clinical endophenotypes of AD (Hohman et al., 2014; Saykin et al., 2015; Louwersheimer et al., 2016; Therriault et al., 2020). Elucidating the interplay between genetic factors and these clinical phenotypes is crucial to developing precision medicine interventions and understanding cognitive “resilience,” a term used to describe better-than-expected cognitive performance in relation to the degree of AD pathology (Negash et al., 2013; Arenaza-Urquijo and Vemuri, 2018).
Accumulating evidence supports that inflammation-related changes may play a role in AD (Perry et al., 2010; Wyss-Coray and Rogers, 2012). We recently reported that key inflammatory analytes in the CSF related to rapid cognitive decline are pro-inflammatory among patients with the mild cognitive impairment (MCI) stage of AD (MCI-AD) (Pillai et al., 2020), whereas a cell-protective analyte profile is noted in the CSF correlating to neurodegeneration markers (Pillai et al., 2019b). Among the analytes that showed consistently high correlation with CSF t-tau, p-tau, and neuron-specific enolase was CSF soluble tumor necrosis factor receptor 2 (sTNFR2) (Pillai et al., 2019b). TNFR2 activation has been shown to promote downstream antiapoptotic responses and protect against oxidative stress-induced neuronal death and neurodegeneration (Fischer et al., 2011; Dong et al., 2016). sTNFR2 results from alternative splicing/shedding from membrane bound TNFR2 and, upregulation of the TNFR2 receptor is thought to be reflected in elevated sTNFR2 levels during inflammation (DeBerge et al., 2015). The TNFRSF1B gene (also known as TNFR2) and its variants have previously been linked with inflammatory responses, inflammatory disease outcomes, and rate of cancer progression (Fairfax et al., 2011; Steenholdt et al., 2012; Singhal et al., 2016), but the role of these gene variants in AD has yet to be elucidated.
We therefore sought to determine whether sTNFR2 and related TNFRSF1B gene variants affect the relationship between the biomarkers of tau pathology, MRI measures of disease severity, and cognitive outcomes. Our main hypothesis was that key TNFRSF1B gene variants and associated CSF sTNFR2 levels would modify AD biomarker levels of t-tau and p-tau, MRI brain volumes, and cognitive outcomes. After performing this analysis for patients enrolled in the Alzheimer's Disease Neuroimaging Initiative (ADNI), we evaluated the reliability of the findings by assessing the same factors in a group of patients with MCI-AD who were treated at a memory clinic.
For the ADNI study, all patient consent was obtained according to the Declaration of Helsinki, and the study was approved by local Institutional Review Boards. For the replication memory clinic cohort, all patients provided written informed consent, and CSF, plasma, and DNA samples were collected for the Lou Ruvo Center for Brain Health Aging and Neurodegeneration Biobank following approval by the Cleveland Clinic Institutional Review Board.
A multistage study design was used to collect and replicate findings in two independent cohorts (the ADNI cohort and the replication memory clinic cohort) for which data regarding TNFRSF1B variants, CSF sTNFR2, and CSF AD biomarkers and cognitive variables were available. The CSF data used in this analysis were cleaned and quality controlled according to methodology described previously1.
ADNI is a longitudinal multicenter study designed to develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of AD. The eligibility criteria for ADNI-1 (the first phase of the project) are described in the ADNI-1 protocol (available at http://adni.loni.usc.edu/methods/documents/). Briefly, eligible participants were aged 55 to 90 years, spoke either English or Spanish, and had an informant who was able to provide an independent evaluation of functioning. Eligible participants had also completed at least 6 years of education (or had a work history sufficient to exclude intellectual disability).
Our final analysis cohort consisted of 188 patients from ADNI-1 for whom CSF sTNFR2 levels and genetic status related to the TNFRSF1B SNPs of interest were available. In addition CSF Aβ42 levels were available for 177 and t-tau and p-tau levels for 182 of these patients. Details regarding the Elecsys method used to measure ADNI AD biomarkers are provided elsewhere (Shaw et al., 2019).
Levels of sTNFR2 were measured in ADNI CSF samples using the RBM Discovery Multi-Analyte Profiling (MAP) v.1.0 panel (Myriad Genetics, Salt Lake City, Utah), which uses a Luminex platform (Myriad Genetics). Data obtained from “Biomarkers Consortium CSF Proteomics Project RBM Multiplex Data and Primer.”
ADNI-1 patients were genotyped using Illumina's10 Human610-Quad BeadChip (San Diego, California), and intensity data were processed with GenomeStudio v2009.1 (Illumina). For this study, we analyzed two TNFRSF1B SNPs: rs976881 and rs1061622, which were found to be in linkage equilibrium in all populations (R2 = 0.022 and D′ = 0.50; chi square = 108.34; p < 0.0001) according to LDlink analysis (Machiela and Chanock, 2015). These two SNPs are located within or in close proximity to the TNFRSF1B gene in the chr1p36 intronic 5' region (rs976881) and a missense coding variant in exon 6 (rs1061622). These SNPs were chosen based on their association with TNFRSF1B-related clinical outcomes and peripheral sTNFR2 levels (Glossop et al., 2005; Vistoropsky et al., 2010; Medrano et al., 2014; Cohen-Woods et al., 2018). Reference, heterozygous, and alternate alleles were T/T, T/C, and C/C, respectively, for rs976881, and G/G, G/T, and T/T, respectively, for rs 1061622. The reference and alternate allele notations were determined per the ADNI dataset based on calls from the International HapMap Project phase 3 data (Biffi et al., 2010). APOE ε4 genotyping data were obtained from the ADNIMERGE table.
In ADNI-1 patients with MCI or dementia, results from 1.5T MRI scans were performed at baseline, and these results (taken from the ADNIMERGE primary table) were used for this analysis (Jack et al., 2008). The whole brain, ventricular volume, and hippocampal volume were the primary dependent variables, with intracranial volume used as a covariate in statistical models. These data are available as part of the ADNIMERGE package (downloaded on May 6, 2020).
Baseline neurocognitive scores for ADNI-1 patients with MCI or dementia were analyzed. All major cognitive domains included in ADNI were evaluated, including the Logical Memory delayed recall score to assess verbal episodic memory (Wechsler Memory Scale, Logical Memory subtest) (Wechsler, 1981). Attention was assessed using the Digit Span subtest (Digit Span Forward and Digit Span Backward), and executive functioning was quantified using the Trail Making Test Part B, both from the Wechsler Adult Intelligence Scale (Wechsler, 1981). Object naming was assessed using the Boston Naming Test (Kaplan et al., 1983), and verbal fluency was assessed using a category fluency test (animals) (Strauss et al., 2006). The Mini Mental State Examination (MMSE) (Folstein et al., 1975) and Clinical Dementia Rating Sum of Boxes (CDR-SB) score (Morris, 1993) were used to assess global cognition, and change in CDR-SB score over 12 months was used to assess longitudinal change in cognition.
A cross-sectional replication cohort consisting of 48 patients in the MCI stage of AD (MCI-AD) were recruited from a specialized memory clinic at Cleveland Clinic (Lou Ruvo Center for Brain Health, Cleveland site). Recruitment details have been described previously (Pillai et al., 2019b).
The diagnosis of MCI-AD in these patients was confirmed by the presence of CSF Aβ42 and p-tau levels consistent with a diagnosis of AD as the primary etiology; the diagnosis was also based on consensus evaluation from two neurologists using published criteria (Albert et al., 2011). A commercially available test (ADmark Alzheimer's Evaluation, Athena Diagnostics, Marlborough, Massachusetts) was used to measure CSF levels of Aβ42, t-tau, and p-tau. All patients met the CSF cutoffs of ≤ 530 pg/mL for Aβ42 and ≥60 pg/mL for p-tau, which are consistent with amyloid-positive status on Amyvid (florbetapir F18 injection) at our center. APOE status was determined with blood samples (10 ng per patient) dispensed into 96-well plates for TaqMan (Applied Biosystems, Waltham, Massachusetts) allelic discrimination assays for the detection of SNPs associated with APOE alleles (rs429358, rs7412). PCR was performed using a 9700 Gene Amp PCR system (Applied Biosystems) and an end-point read in a 7500 Real-Time PCR system (Applied Biosystems) (Bekris et al., 2008).
CSF levels of sTNFR2 were assessed as described previously (Pillai et al., 2019b). In brief, CSF was collected and analyzed by an independent laboratory via the RBM HumanMAP v.2.0, which is a subset of the RBM DiscoveryMAP v.1.0 used in the ADNI cohort; these platforms have the same quality control and thresholding process and are comparable. The lowest detectable dose of sTNFR2 was 0.0017 ng/mL. Samples of CSF were frozen within 15 min after collection and were processed (at −70°C in dry ice) and maintained at −80°C (in a non-frost-free refrigerator). The samples were shipped frozen in a Styrofoam container with sufficient dry ice to maintain the temperature at < -70°C for at least 48 h. Samples therefore underwent a single freeze-thaw cycle before analysis.
Genomic DNA was extracted from peripheral whole blood using standard protocols. Two TNFRSF1B SNPs (rs976881 and rs1061622) were characterized. TNFRSF1B variant allelic discrimination was performed using the 7500 Real Time PCR System and TaqMan SNP Genotyping Assays. The genotypes were determined based on sample clustering using the autocaller function of the Genotyping Application within Thermo Fisher Connect software.
MMSE and Montreal Cognitive Assessment (MoCA) scores (Nasreddine et al., 2005) obtained <6 months before AD biomarker testing were available for all patients. In addition baseline CDR-SB scores (Morris, 1993) were available to characterize patients' cognitive and functional deficits.
Participants in ADNI-1 who had either MCI or AD dementia at baseline were merged into one group to ensure adequate power of analysis. The normality of biomarkers was evaluated using graphical methods and the Shapiro-Wilk test. A log (base 2) transformation allowed Pearson correlations to be fit; the sTNFR2 and AD biomarker levels described are therefore dimensionless. Along with estimates of correlation, 95% CIs and p-values were calculated. ANOVA models were used to estimate the mean differences between CSF sTNFR2 and t-tau and p-tau levels among reference, alternate, and heterozygous alleles of the TNFRSF1B SNPs. Multivariate general linear models were used to test the effect of CSF sTNFR2 levels and each of the two TNFRSF1B SNPs on CSF t-tau and p-tau, MRI brain measures, and cognitive domains after adjustments were made for covariates such as age, sex, years of education, APOE ε4 status, and CSF Aβ42/p-tau ratio. Baseline MMSE and t-tau were also included as covariates in evaluations of cognitive domains as dependent variables, and baseline intracranial volumes and t-tau were included as covariates in evaluations of MRI measures as dependent variables.
First, we tested the effect of sTNFR2 levels on CSF t-tau and p-tau, MRI volumetric measures, and cognitive variables. Second, we tested the effect of each TNFRSF1B variant of interest on CSF t-tau and p-tau, MRI volumetric measures, and cognitive variables. Third, we tested whether the interaction effects between sTNFR2 and each TNFRSF1B variant and between sTNFR2 and t-tau were present for CSF t-tau and p-tau, MRI volumetric measures, and cognitive variables. Levene's test of equality of error variances, lack-of-fit F-test, and residual plots were used to assess model linearity and fit.
Model 1, evaluated the dependent variables CSF t-tau and p-tau. The main effects were CSF sTNFR2, TNFRSF1B SNP, and interaction effect between sTNFR2 and TNFRSF1B SNP. Covariates for this model included age, sex, CSF Aβ42, and APOEε4 status.
Model 2, evaluated the dependent variables hippocampal volume, ventricular volume, and whole brain volume. The main effects were CSF sTNFR2, TNFRSF1B SNP, CSF t-tau, and interaction effects between sTNFR2 and TNFRSF1B SNP and between sTNFR2 and t-tau. Covariates for this model included age, sex, years of education, CSF Aβ42/p-tau ratio, APOE ε4 status, and baseline intracranial volume.
Model 3, evaluated the dependent variables Logical Memory delayed recall score, Digit Span subtest score, Trail Making Test Part B score, category fluency test (animals), and Boston Naming Test score. The main effects were CSF sTNFR2, TNFRSF1B SNP, CSF t-tau, and interaction effects between sTNFR2 and TNFRSF1B SNP and between sTNFR2 and t-tau. The covariates for this model included age, sex, years of education, CSF Aβ42/p-tau ratio, APOE ε4 status, and baseline MMSE score.
Model 4, CDR-SB scores were log transformed because Levene's test demonstrated that the variances were significantly different for CDR-SB scores for the TNFRSF1B SNPs. A univariate general linear model was used to evaluate the interaction effect for each TNFRSF1B SNP and CSF sTNFR2 on change in CDR-SB score over 1 year in the ADNI cohort. CDR-SB score at 1 year was the dependent variable; the main effects were disease state (MCI or AD dementia), CSF t-tau, and interaction effect between sTNFR2 and TNFRSF1B SNP. The covariates included age, sex, years of education, APOE ε4 status, baseline CDR-SB score, and CSF Aβ42/p-tau.
Paralleling Model 1, here the dependent variables were again CSF t-tau and p-tau. The main effects were CSF sTNFR2, TNFRSF1B SNP, and interaction effect between sTNFR2 and TNFRSF1B SNP. Covariates for this model included age, sex, CSF Aβ42, and APOE ε4 status.
Sensitivity analysis with and without covariates were conducted. Proportion of variance associated with one or more main effects was calculated using eta squared. All tests were two-tailed, and statistical significance was set at p < 0.05. For model 3 (with more than three dependent variables in the outcome), the Benjamini-Hochberg false discovery rate was used to assess significance. IBM SPSS Statistics for Windows, Version 22.0 (Armonk, New York) and RStudio Team (2020) RStudio (Version 1.2.5042) were used for all analyses.
ADNI data are available on request at loni.ADNI.org.
Our analysis included a total of 188 ADNI-1 participants (59 with AD dementia; 129 with MCI) (Table 1).There were no differences in sTNFR2 or in CSF Aβ42, t-tau, or p-tau biomarker levels within the allelic subgroups of rs976881 or rs1061622.
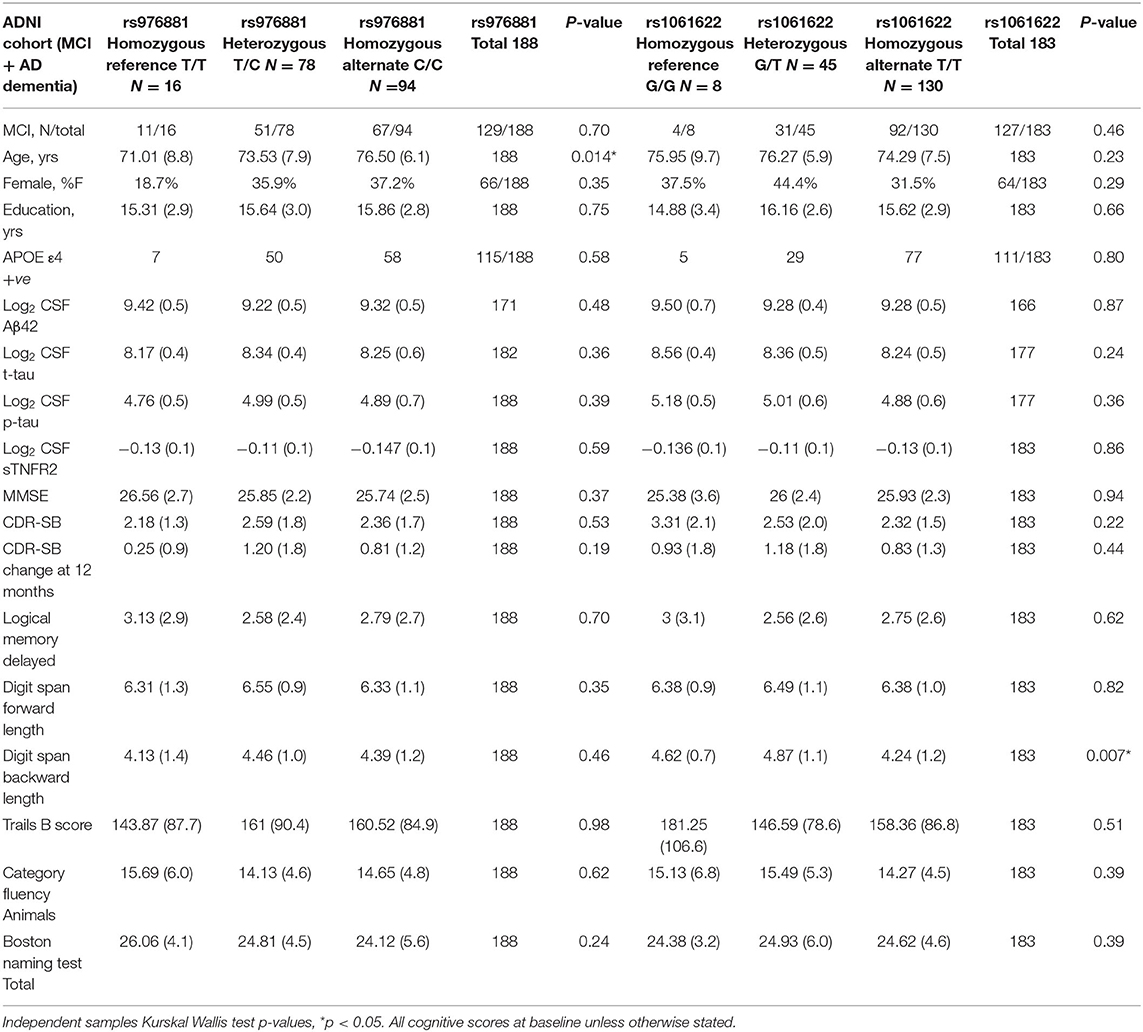
Table 1. Demographics of ADNI cohort with patients divided into allelic subgroups, Mean (Std dev), percent of total and counts are presented.
A total of 48 participants with MCI-AD were included in the replication memory clinic cohort (Table 2). There were again no differences in sTNFR2 or in CSF t-tau, or p-tau biomarker levels within the allelic subgroups of rs976881 and rs1061622. The differences between ADNI and replication cohort is noted in Supplementary Table 1. Both cohorts had similar baseline CDR-SB scores, years of education and were predominantly White and a slightly higher frequency of men. The replication cohort had a lower age, higher frequency of APOE ε4 carriers and a slightly lower baseline MMSE.
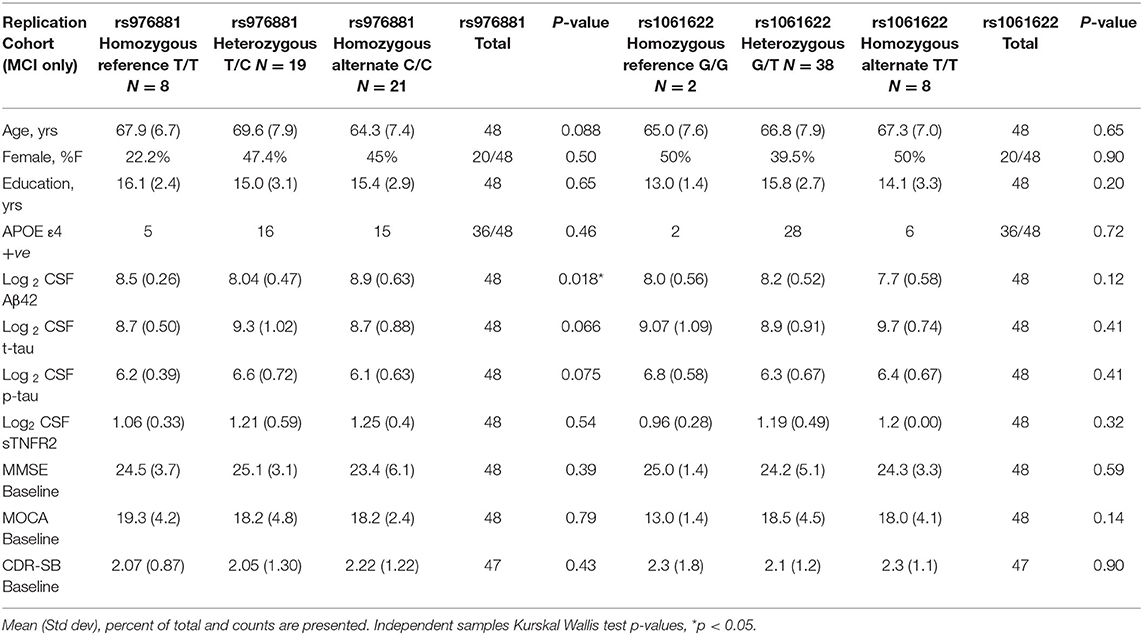
Table 2. Demographics of replication memory clinic cohort with patients divided into allelic subgroups.
A significant interaction effect between rs976881 and CSF sTNFR2 was associated with CSF t-tau levels, but this association with CSF t-tau levels was not seen for the interaction effect between rs1061622 and CSF sTNFR2 (Table 3 and Figure 1A). The effect sizes (by partial eta squared values) were 0.046 for CSF t-tau and 0.044 for CSF p-tau (i.e., 4.6% of all variance in CSF t-tau and 4.4% of all variance in CSF p-tau were attributable to the interaction effect between rs976881 and CSF sTNFR2). The T/T group was noted to have a shallower slope and poorer fit (R2 = 0.1) than C/C (R2 = 0.38), suggesting that sTNFR2 levels are not as closely related to t-tau levels in T/T as in C/C. The direct main effects of sTNFR2 on CSF t-tau and p-tau are shown in Supplementary Table 2.
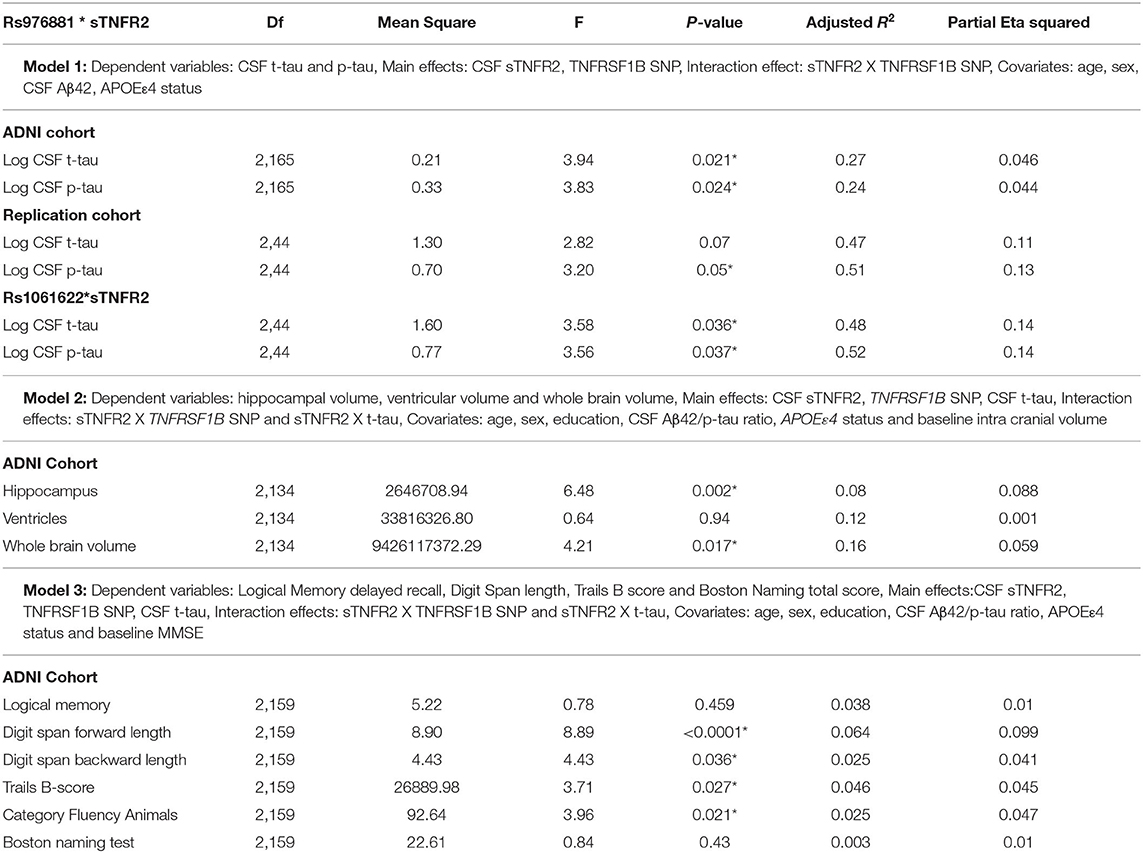
Table 3. Key results of the general linear models: Results from the ADNI and replication memory clinic cohorts, *p ≤ 0.05.
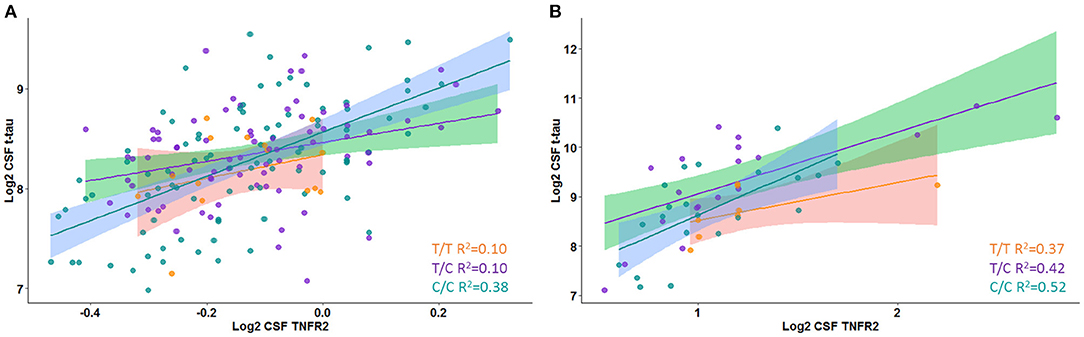
Figure 1. Interaction between rs976881 and soluble tumor necrosis factor receptor 2 (sTNFR2) and the effect on CSF t-tau in Alzheimer's Disease Neuroimaging Initiative (ADNI) and replication cohorts. Regression lines, 95% CI and R2 values shown for the alleles T/T, T/C, and C/C. (A) ADNI cohort. (B) Replication cohort.
A significant interaction effect between rs976881 and CSF sTNFR2 was associated with hippocampal volume and whole brain volume but not with ventricular volume (Table 3). 8.8% of all variance in hippocampal volume and 5.9% of all variance in whole brain volume were attributable to the interaction effect between rs976881 and CSF sTNFR2.
The MRI measures were not affected by rs1061622, nor by the interaction effect between sTNFR2 and CSF t-tau. CSF t-tau did have a significant direct main effect on ventricular volume and whole brain volume but not on hippocampal volume (Supplementary Table 2).
In Model 3, a significant interaction effect between rs976881 and CSF sTNFR2 was associated with the Digit Span subtest score, the Trail Making Test Part B score, and the category fluency test (animals) score (Table 3). 9.9% of all variance for the Digit Span Forward score and 4.7% of all variance for the category fluency (animals) score were attributable to the interaction effect between rs976881 and CSF sTNFR2. There was no significant interaction effect between sTNFR2 and CSF t-tau, nor was there a significant main effect of CSF t-tau on these cognitive measures (Figure 2A). As shown in Figure 2B, the T/T group had a steeper slope and a larger regression coefficient for the Digit Span Forward score (R2 = 0.33), suggesting that lower sTNFR2 levels (but not CSF t-tau levels) relate to poorer Digit Span Forward scores for this group, unlike for the C/C group (R2 = 0.004). The interaction effect between rs1061622 and CSF sTNFR2 had no effect on these cognitive measures. The Digit Span Forward score was significant even after false discovery rate correction (false discovery rate–corrected p-values are provided in the Supplementary Material).
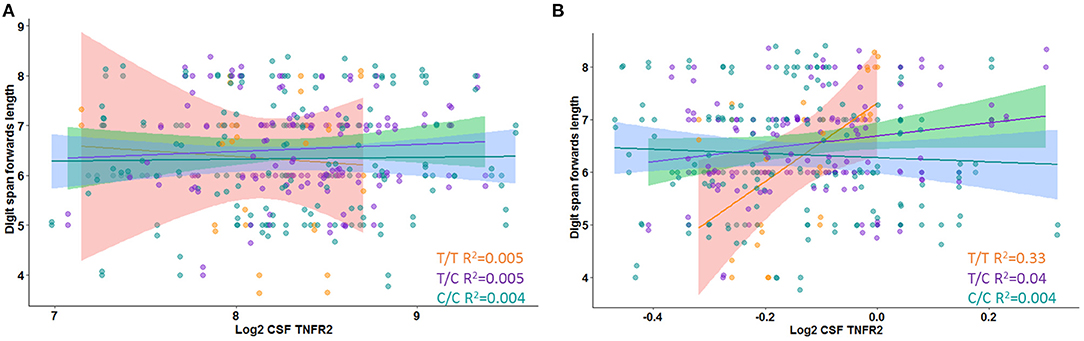
Figure 2. Interaction between rs976881 and soluble tumor necrosis factor receptor 2 (sTNFR2) and between rs976881 and CSF t-tau and the effect on Digit Span Forward score. (A) CSF t-tau (B) CSF sTNFR2. Regression lines, 95% CI and R2 values shown for the alleles T/T, T/C, and C/C. Alzheimer's Disease Neuroimaging Initiative (ADNI) data.
A significant interaction effect between rs976881 and sTNFR2 was associated with change in CDR-SB scores over 1 year (df 3, 175; mean square = 0.21; F = 2.83; adjusted R2 = 0.36). A total of 4.6% of all variance in CDR-SB change over 1 year was attributable to the interaction effect between rs976881 and CSF sTNFR2 (Figure 3A). As shown in Figure 3B, the T/T group had a smaller change (shallower slope) in CDR-SB change over 1 year than the C/C group (R2 = 0.52 vs 0.70). There was no significant effect of rs1061622 on CDR-SB change over 1 year (df 3, 170; F = 2.32; adjusted R2 = 0.35).
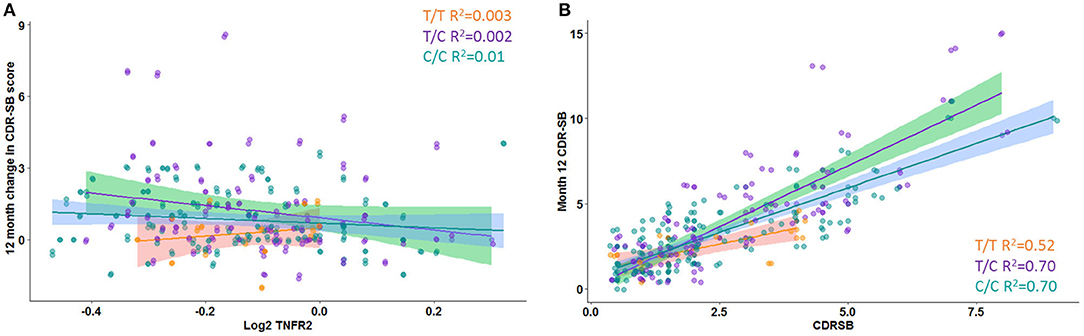
Figure 3. Change in Clinical Dementia Rating Sum of Boxes (CDR-SB) score over time. (A) Interaction between rs976881 and sTNFR2 level and the effect on change in CDR-SB score over 1 year. (B) Slope of change in score over 1 year. Regression lines, 95% CI and R2 values shown for the alleles T/T, T/C, and C/C. Alzheimer's Disease Neuroimaging Initiative (ADNI) data.
A significant interaction effect between rs976881 and CSF sTNFR2 was associated with CSF p-tau levels (Table 3). Eleven percentage of all variance in CSF t-tau and 13% of all variance in CSF p-tau were attributable to the interaction effect between rs976881 and CSF sTNFR2. The homozygous reference (T/T) group had a shallower slope and a smaller regression coefficient (R2 = 0.37) than the homozygous alternate allele (C/C) (R2 = 0.42) (Figure 1B), suggesting that sTNFR2 levels are not as closely related to t-tau levels in the T/T replication memory clinic group as they are in the ADNI cohort. Pairwise comparisons for the rs976881 alleles demonstrated significant differences between the T/T and T/C groups in CSF t-tau levels (mean difference = −0.58; 95% CI: 0.074 to 1.09) and CSF p-tau levels (mean difference = −0.39; 95% CI: 0.050–0.747). Supplementary Table 2 provides details regarding the direct main effects of significance.
A significant interaction effect between rs1061622 and CSF sTNFR2 was associated with CSF t-tau and p-tau levels. Fourteen percentage of all variance in CSF t-tau and 14% of all variance in CSF p-tau were attributable to the interaction effect between rs1061622 and CSF sTNFR2 (Table 3). Pairwise comparisons for the rs1061622 alleles demonstrated significant differences between the T/T and T/G groups in CSF t-tau levels (mean difference = −0.69; 95% CI: −0.079 to −1.29) and CSF p-tau levels (mean difference = −0.49; 95% CI: −0.067 to −0.91).
Immune-related genes and pathways have been found to play a role in AD pathophysiology (Fischer et al., 2011; Kunkle et al., 2019). Previous studies have assessed how these genetic immune and factors could affect AD biomarkers (Hohman et al., 2014; Louwersheimer et al., 2016; Deming et al., 2019) and clinical outcomes (Pillai et al., 2020), but little research has addressed their potential contribution to resilience. The inflammatory analyte TNF has been found to play a role in synaptic plasticity and modulating responses to neural injury and neurodegeneration (Beattie et al., 2002; Ellwardt et al., 2018), suggesting that it may also be involved in cognitive resilience (Pape et al., 2019). The current study demonstrated that the interaction between TNF pathway–related sTNFR2 CSF levels and TNFRSF1B gene variants contributed to significant differences in AD-associated severity markers such as CSF t-tau and p-tau, MRI measures of interest, and cognitive scores in the ADNI cohort. In a replication memory clinic cohort, the interaction between CSF sTNFR2 and rs976881 was also associated with differences in CSF p-tau biomarkers, with a significantly larger effect size than was seen in the ADNI cohort. In the ADNI cohort, rs976881 T/T allele (homozygous reference) carriers demonstrated slower rates of CDR-SB score changes in relation to corresponding sTNFR2 levels. Additionally, statistical interaction effects demonstrated that the strength of a linear relationship between sTNFR2 and (1) CSF t-tau and p-tau, (2) MRI whole brain and hippocampal volumes, and (3) Digit Forward test score varied for the rs976881 T/T allele compared to the C/C and T/C alleles. Studies in animal models have demonstrated that TNFR2 plays a protective role against neurodegeneration in the CNS (Marchetti et al., 2004; Fischer et al., 2011; Dong et al., 2016). Taken together, these findings support this genotypic variant is a potential marker of resilience in AD.
sTNFR2 is expressed primarily in immune and endothelial cells. TNFR2 receptor signaling is involved in pro-survival signaling pathways, which activate cellular inhibitors of apoptosis and the NF-κB pathway (Kanehisa et al., 2016). Subsequent downstream activation of PKB/Akt promotes cell survival and proliferation (Fischer et al., 2020). sTNFR2 has previously been found to be highly correlated with CSF t-tau, p-tau, and neuron-specific enolase in MCI-AD (Pillai et al., 2019b), and a similar profile with TNFRSF1B activation was noted in a parallel brain transcriptome analysis (Pillai et al., 2019b). It is possible, therefore, that sTNFR2 and TNFRSF1B SNPs play a modulating role in regard to clinical outcomes in AD, rather than serving as an AD risk gene that have been the focus of prior genome-wide association studies in AD. This is consistent with prior reports of gene variants related to resilience in AD which suggest that genetic architecture of resilience appears to be distinct from that of clinical AD (Dumitrescu et al., 2020).
Research has shown that rs976881 and rs1061622 are associated with sTNFR2 levels in the periphery, with the rs976881 reference allele (T/T) related to higher sTNFR2 levels than the alternate allele (C/C) (Vistoropsky et al., 2010; Cohen-Woods et al., 2018). In the replication memory clinic cohort in this study, patients with the rs976881 reference allele T/T had lower CSF t-tau and p-tau levels than patients with T/C (given their corresponding CSF sTNFR2 levels). Previous research has also found that rs976881 T/T is less responsive than T/C to anti-TNFα maintenance therapy (infliximab) in patients with Crohn's disease (Steenholdt et al., 2012). It is therefore likely that among minor allele (T/T) carriers with Crohn's disease, higher levels of TNFR2 counteract TNF-α, making infliximab less effective. In our study, the effect of rs976881 may also be tied to its effect on sTNFR2 levels, with the rs976881 minor allele (T/T) demonstrating a more robust sTNFR2 response than C/C and T/C alleles to CSF t-tau and p-tau levels. Studies have also demonstrated that the minor allele of rs1061622 is also related to sTNFR2 levels and response to anti-TNFα maintenance therapy in patients with Crohn's disease (Medrano et al., 2014). In our study, the significant interaction observed between rs1061622 and sTNFR2 and its effect on CSF t-tau and p-tau in the replication cohort but not in the ADNI cohort suggests differences in the nature of participants recruited, as discussed in further detail later.
In the ADNI cohort, a significant interaction between rs976881 and CSF sTNFR2 was strongest with regards to Digit Span Forwards subtest, but was also noted in Trail Making Part B test, and category fluency test (animals) scores. These measures assess overlapping but distinct cognitive skills, including working memory, complex attention, and speed of information processing, and can be categorized as contributing to domains of attention/executive functioning and processing speed. In this cohort, rs976881 status was also related to the change in CDR-SB score over 1 year. This is consistent with our previous finding that better initial performance on the Digit Span subtest and related working memory task is associated with a slower rate of functional decline on the CDR-SB test over time in patients with AD (Pillai et al., 2014). Additionally, higher levels of pro-cell survival and inflammation resolution chemokines have been found in the human temporal cortex and entorhinal cortex at autopsy among human brains resilient to AD pathology (Barroeta-Espar et al., 2019). These results are reflected in our study, which demonstrated that a significant interaction between rs976881 and CSF sTNFR2 modulated favorable outcomes in three measures related to AD severity: CSF biomarkers of neurodegeneration and tau (CSF p-tau and t-tau), MRI measures (hippocampal and whole brain volumes), and cognitive measures (Digit Span Forward score and CDR-SB score).
Strengths/limitations: The current study demonstrated directional replication by consistently noting a similar directional interaction effect of sTNFR2 and rs976881 on CSF p-tau and t-tau in both the ADNI and replication cohorts. Additionally, the effect of rs976881 on MRI and cognitive outcomes demonstrated in this study is consistent with the known relationship between these biomarkers (CSF p-tau, t-tau, hippocampal volume, and whole brain volumes) and longitudinal cognitive outcomes in the ADNI population. In the ADNI cohort for this study, 4.6% of the variance in CSF t-tau levels was explained by rs976881 and sTNFR2, whereas 11% of the variance in CSF t-tau was attributed to the same variables in the replication cohort. There were some differences between the cohorts, however. First, the positive correlation between the sTNFR2 and neurodegeneration biomarkers are consistent within each cohort but the replication cohort had 2.2 times higher mean levels of p-tau than ADNI (Pillai et al., 2019b). Mean sTNFR2 levels correlating with neurodegeneration biomarkers were also 2.4 times higher in the replication cohort than ADNI. This is likely due to different participant characteristics for the two cohorts; for instance, the replication cohort was a sample of memory clinic patients with a faster rate of disease progression than patients in the ADNI cohort (Pillai et al., 2020). Second, the replication cohort included participants at the MCI stage of AD, whereas the ADNI cohort included patients with MCI and AD dementia. As the replication cohort was not harmonized with the ADNI cohort for all of the neurocognitive variables and MRI volumetric measures, there could not be a close corroboration between the replication cohort and the ADNI cohort for the multiple variables of interest. These results are likely generalizable to MCI and AD dementia subjects with positive AD biomarkers but the fact that patients in both cohorts were predominantly Caucasian and had a higher level of education suggests the need for replication of these results among other racial and ethnic cohorts with a diverse education and socioeconomic backgrounds.
Within the ADNI cohort, MRI hippocampal volumes were noted to have a significant interaction effect between rs976881 and sTNFR2, while logical memory delayed recall scores did not meet significance. One possible reason for this is that the predominance of amnestic patients with MCI despite likely multiple etiologies of amnestic complaints in ADNI cohort, limits our ability to discriminate based on logical memory delayed recall scores alone given normative limits (Nettiksimmons et al., 2014). Additional corroboration of these results in a cohort with recruitment goals different from those of ADNI is therefore warranted. The lack of neuropathologic confirmation of diagnosis also limits our understanding regarding the potential role of mixed pathology on clinical outcomes.
The study is an evaluation of the association between CSF sTNFR level and TNFRSF1B SNPS on clinical outcomes, and does not address the mechanistic relationship between them on neurodegeneration. This study further does not provide a comprehensive survey of TNFRSF1B SNPs in AD, as we did not analyze other TNFRSF1B SNPs reported to be related to sTNFR2 levels (e.g., rs590368). These other SNPs were not included because of the smaller number of participants within ADNI who had these genetic variants and who also had measured levels of sTNFR2 available. Using multiple SNPs may allow researchers to define a stronger TNFRSF1B haplotype with regard to sTNFR2 levels, and some variables may become more or less salient considering additional SNP interactions. Independent replication using the same biomarkers used in ADNI would allow for further clarification. Type II errors have to be considered for this study, given the small number of patients within some of the allele groups; smaller effect sizes could therefore have been missed.
The rs976881 T/T reference genotype has been related to higher levels of sTNFR2 previously and activation of TNFR2 signaling has been posited as a promising strategy for AD therapy (Ortí-Casañ et al., 2019). We have now demonstrated that interaction between TNFRSF1B gene variant rs976881 and CSF sTNFR2 affects CSF and MRI biomarkers of neurodegeneration, cognitive profiles, and rate of functional decline over 1 year. These results provide important information regarding the molecular characterization of AD phenotypes and suggest that this genotypic variant could be used as a marker of resilience in AD. Independent confirmation of these results in other cohorts with mutidomain AD biomarkers is warranted.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://adni.loni.usc.edu/, genetic-data.
The studies involving human participants were reviewed and approved by Cleveland Clinic Institutional Review Board ADNI Individual Site Institutional Review Board. The ethics committee waived the requirement of written informed consent for participation.
JP: obtained funding for research project, organization of research project, execution of research project, design the statistical analysis, execution of statistical analysis, and writing the first draft of manuscript preparation. GB: execution of research project, review and critique of statistical analysis, and review and critique of manuscript preparation. MK: execution of research project and review and critique of manuscript preparation. JB: design the statistical analysis and review and critique of manuscript preparation. CB, WB, and JL: review and critique of statistical analysis and review and critique of manuscript preparation. LB: organization of research project, review and critique of statistical analysis, and review and critique of manuscript preparation. All authors contributed to the article and approved the submitted version.
The Funding was provided by 2018-AARG-592251 Alzheimer's Association, NIA K23AG055685, 1P30 AG062428-01, Keep Memory Alive Foundation and the Jane and Lee Seidman Fund. Research reported in this publication was supported by the National Institute On Aging of the National Institutes of Health under Award Number K23AG055685. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JP was a program participant of the Research Education Component of the Cleveland Alzheimer's Disease Research Center supported by NIA P30 AG062428. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI was funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
JP receives research funding from the National Institutes of Health, Alzheimer's Association, and Keep Memory Alive Foundation. CB receives research funding from the National Institutes of Health. WB receives research funding from the National Institutes of Health. JL has received consulting fees from Acadia, Aptnyx, Biogen, Eisai, GE Healthcare, Sanofi, and Takeda and grant support from the Alzheimer's Association, Alzheimer's Drug Discovery Foundation, Biogen, Department of Defense, GE Healthcare, Genzyme/Sanofi, Lewy Body Dementia Association, Michael J Fox Foundation, and National Institute of Health (NIA, NINDS).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the patients and families who took part in the replication cohort at Cleveland Clinic Lou Ruvo Center for Brain Health and Megan Griffiths and Dennis Lal for editorial support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2021.638922/full#supplementary-material
1. ^Biomarkers Consortium ADNI CSF Targeted Proteomics Project - Data Primer Version 28 Dec 2011. Available online at: http://adni.loni.usc.edu/methods/ (accessed December 1, 2018).
Albert, M. S., DeKosky, S. T., Dickson, D., Dubois, B., Feldman, H. H., Fox, N. C., et al. (2011). The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 7, 270–279. doi: 10.1016/j.jalz.2011.03.008
Arenaza-Urquijo, E. M., and Vemuri, P. (2018). Resistance vs resilience to Alzheimer disease: clarifying terminology for preclinical studies. Neurology 90, 695–703. doi: 10.1212/WNL.0000000000005303
Barroeta-Espar, I., Weinstock, L. D., Perez-Nievas, B. G., Meltzer, A. C., Siao Tick Chong, M., Amaral, A. C., et al. (2019). Distinct cytokine profiles in human brains resilient to Alzheimer's pathology. Neurobiol. Dis. 121, 327–337. doi: 10.1016/j.nbd.2018.10.009
Beattie, E. C., Stellwagen, D., Morishita, W., Bresnahan, J. C., Ha, B. K., Von Zastrow, M., et al. (2002). Control of synaptic strength by glial TNFalpha. Science 295, 2282–2285. doi: 10.1126/science.1067859
Bekris, L. M., Millard, S. P., Galloway, N. M., Vuletic, S., Albers, J. J., Li, G., et al. (2008). Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J. Alzheimers Dis. 13, 255–266. doi: 10.3233/JAD-2008-13303
Biffi, A., Anderson, C. D., Desikan, R. S., Sabuncu, M., Cortellini, L., and Schmansky, N. (2010) Genetic variation neuroimaging measures in Alzheimer disease. Arch. Neurol. 67, 677–685. doi: 10.1001/archneurol.2010.108.
Cohen-Woods, S., Fisher, H. L., Ahmetspahic, D., Douroudis, K., Stacey, D., Hosang, G. M., et al. (2018). Interaction between childhood maltreatment on immunogenetic risk in depression: discovery and replication in clinical case-control samples. Brain Behav. Immun. 67, 203–210. doi: 10.1016/j.bbi.2017.08.023
DeBerge, M. P., Ely, K. H., Wright, P. F., Thorp, E. B., and Enelow, R. I. (2015). Shedding of TNF receptor 2 by effector CD8+ T cells by ADAM17 is important for regulating TNF-α availability during influenza infection. J. Leukoc. Biol. 98, 423–434. doi: 10.1189/jlb.3A0914-432RR
Deming, Y., Filipello, F., Cignarella, F., Cantoni, C., Hsu, S., Mikesell, R., et al. (2019). The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer's disease risk. Sci. Transl. Med. 11:eaau2291. doi: 10.1126/scitranslmed.aau2291
Dong, Y., Fischer, R., Naudé, P. J, Maier, O., Nyakas, C., Duffey, M., et al. (2016). Essential protective role of tumor necrosis factor receptor 2 in neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 113, 12304–12309. doi: 10.1073/pnas.1605195113
Dubois, B., Feldman, H. H., Jacova, C., Hampel, H., Molinuevo, J. L., Blennow, K., et al. (2014). Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol. 13, 614–629. doi: 10.1016/S1474-4422(14)70090-0
Dumitrescu, L., Mahoney, E. R., Mukherjee, S., Lee, M. L., Bush, W. S., and Engelman, C. D. (2020) Genetic variants functional pathways associated with resilience to Alzheimer's disease. Brain 143, 2561–2575. doi: 10.1093/brain/awaa209.
Ellwardt, E., Pramanik, G., Luchtman, D., Novkovic, T., Jubal, E. R., Vogt, J., et al. (2018). Maladaptive cortical hyperactivity upon recovery from experimental autoimmune encephalomyelitis. Nat. Neurosci. 21, 1392–1403. doi: 10.1038/s41593-018-0193-2
Fairfax, B. P., Davenport, E. E., Makino, S., Hill, A. V., Vannberg, F. O., and Knight, J. C. (2011). A common haplotype of the TNF receptor 2 gene modulates endotoxin tolerance. J. Immunol. 186, 3058–3065. doi: 10.4049/jimmunol.1001791
Fischer, R., Kontermann, R. E., and Pfizenmaier, K. (2020). Selective targeting of TNF receptors as a novel therapeutic approach. Front. Cell. Dev. Biol. 8:401. doi: 10.3389/fcell.2020.00401
Fischer, R., Maier, O., Siegemund, M., Wajant, H., Scheurich, P., and Pfizenmaier, K. (2011). A TNF receptor 2 selective agonist rescues human neurons from oxidative stress-induced cell death. PLoS ONE 6:e27621. doi: 10.1371/journal.pone.0027621
Folstein, M. F., Folstein, S. E., and McHugh, P. R. (1975). “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 12, 189–198. doi: 10.1016/0022-3956(75)90026-6
Glossop, J. R., Dawes, P. T., Nixon, N. B., and Mattey, D. L. (2005). Polymorphism in the tumour necrosis factor receptor II gene is associated with circulating levels of soluble tumour necrosis factor receptors in rheumatoid arthritis. Arthritis Res. Ther. 7:R1227–R1234. doi: 10.1186/ar1816
Hohman, T. J., Koran, M. E., and Thornton-Wells, T. A. Alzheimer's Disease Neuroimaging Initiative. (2014). Genetic modification of the relationship between phosphorylated tau and neurodegeneration. Alzheimers Dement. 10, 637–645.e1. doi: 10.1016/j.jalz.2013.12.022
Jack, C. R. Jr., Bernstein, M. A., Fox, N. C., Thompson, P., Alexander, G., Harvey, D., et al. (2008). The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27, 685–691. doi: 10.1002/jmri.21049
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kaplan, E., Goodglass, H., and Weintraub, S. (1983). Boston Naming Test. Philadelphia, PA: Lea and Febiger.
Kunkle, B. W., Grenier-Boley, B., Sims, R., Bis, J. C., Damotte, V., and Naj, A. C. (2019) Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci implicates Aβ, tau, immunity lipid processing. Nat. Genet. 51, 414–430. doi: 10.1038/s41588-019-0358-2.
Louwersheimer, E., Wolfsgruber, S., Espinosa, A., Lacour, A., Heilmann-Heimbach, S., Alegret, M., et al. (2016). Alzheimer's disease risk variants modulate endophenotypes in mild cognitive impairment. Alzheimers Dement. 12, 872–881. doi: 10.1016/j.jalz.2016.01.006
Machiela, M. J., and Chanock, S. J. (2015). LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. doi: 10.1093/bioinformatics/btv402
Marchetti, L., Klein, M., Schlett, K., Pfizenmaier, K., and Eisel, U. L. (2004). Tumor necrosis factor (TNF)-mediated neuroprotection against glutamate-induced excitotoxicity is enhanced by N-methyl-D-aspartate receptor activation. Essential role of a TNF receptor 2-mediated phosphatidylinositol 3-kinase-dependent NF-kappa B pathway. J. Biol. Chem. 279, 32869–32881. doi: 10.1074/jbc.M311766200
Medrano, L. M., Taxonera, C., Márquez, A., Barreiro-de Acosta, M., Gómez-García, M., González-Artacho, C., et al. (2014). Role of TNFRSF1B polymorphisms in the response of Crohn's disease patients to infliximab. Hum. Immunol. 75, 71–75. doi: 10.1016/j.humimm.2013.09.017
Morris, J. C. (1993). The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43, 2412–2414. doi: 10.1212/WNL.43.11.2412-a
Nasreddine, Z. S., Phillips, N. A., Bédirian, V., Charbonneau, S., Whitehead, V., Collin, I., et al. (2005). The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J. Am. Geriatr. Soc. 53, 695–699. doi: 10.1111/j.1532-5415.2005.53221.x
Negash, S., Wilson, R. S., Leurgans, S. E., Wolk, D. A., Schneider, J. A., Buchman, A. S., et al. (2013). Resilient brain aging: characterization of discordance between Alzheimer's disease pathology and cognition. Curr. Alzheimer Res. 10, 844–851. doi: 10.2174/15672050113109990157
Nettiksimmons, J., DeCarli, C., Landau, S., and Beckett, L. Alzheimer's Disease Neuroimaging Initiative. (2014). Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimers Dement. 10, 511–521.e1. doi: 10.1016/j.jalz.2013.09.003
Ortí-Casañ, N., Wu, Y., Naudé, P. J. W, De Deyn, P. P., Zuhorn, I. S., and Eisel, U. L. M. (2019). Targeting TNFR2 as a novel therapeutic strategy for Alzheimer's disease. Front. Neurosci. 13:49. doi: 10.3389/fnins.2019.00049
Pape, K., Tamouza, R., Leboyer, M., and Zipp, F. (2019). Immunoneuropsychiatry - novel perspectives on brain disorders. Nat. Rev. Neurol. 15, 317–328. doi: 10.1038/s41582-019-0174-4
Perry, V. H., Nicoll, J. A., and Holmes, C. (2010). Microglia in neurodegenerative disease. Nat. Rev. Neurol. 6, 193–201. doi: 10.1038/nrneurol.2010.17
Pillai, J. A., Bena, J., Bebek, G., Bekris, L. M., Bonner-Jackson, A., Kou, L., et al. (2020). Inflammatory pathway analytes predicting rapid cognitive decline in MCI stage of Alzheimer's disease. Ann. Clin. Transl. Neurol. 7, 1225–1239. doi: 10.1002/acn3.51109
Pillai, J. A., Bonner-Jackson, A., Bekris, L. M., Safar, J., Bena, J., and Leverenz, J. B. (2019a). Highly elevated cerebrospinal fluid total tau level reflects higher likelihood of non-amnestic subtype of Alzheimer's Disease. J. Alzheimers Dis. 70, 1051–1058. doi: 10.3233/JAD-190519
Pillai, J. A., Bonner-Jackson, A., Walker, E., Mourany, L., and Cummings, J. L. (2014). Higher working memory predicts slower functional decline in autopsy-confirmed Alzheimer's disease. Dement. Geriatr. Cogn. Disord. 38, 224–233. doi: 10.1159/000362715
Pillai, J. A., Maxwell, S., Bena, J., Bekris, L. M., Rao, S. M., Chance, M., et al. (2019b). Key inflammatory pathway activations in the MCI stage of Alzheimer's disease. Ann. Clin. Transl. Neurol. 6, 1248–1262. doi: 10.1002/acn3.50827
Risacher, S. L., Anderson, W. H., Charil, A., Castelluccio, P. F., Shcherbinin, S., et al. (2017). Alzheimer disease brain atrophy subtypes are associated with cognition and rate of decline. Neurology 89, 2176–2186. doi: 10.1212/WNL.0000000000004670
RStudio Team. (2020). RStudio: Integrated Development for R. RStudio. Boston, MA: PBC. Available online at: http://www.rstudio.com/
Saykin, A. J., Shen, L., Yao, X., Kim, S., Nho, K., Risacher, S. L., et al. (2015). Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimers Dement. 11, 792–814. doi: 10.1016/j.jalz.2015.05.009
Shaw, L. M., Hansson, O., Manuilova, E., Masters, C. L., Doecke, J. D., Li, Q. X., et al. (2019). Method comparison study of the Elecsys® β-Amyloid (1-42) CSF assay versus comparator assays and LC-MS/MS. Clin. Biochem. 72, 7–14. doi: 10.1016/j.clinbiochem.2019.05.006
Singhal, P., Sharma, U., Hussain, S., Nag, A., and Bharadwaj, M. (2016). Identification of genetic variants in TNF receptor 2 which are associated with the development of cervical carcinoma. Biomarkers 21, 665–672. doi: 10.3109/1354750X.2016.1172109
Steenholdt, C., Enevold, C., Ainsworth, M. A., Brynskov, J., Thomsen, O. Ø., and Bendtzen, K. (2012). Genetic polymorphisms of tumour necrosis factor receptor superfamily 1b and fas ligand are associated with clinical efficacy and/or acute severe infusion reactions to infliximab in Crohn's disease. Aliment. Pharmacol. Ther. 36, 650–659. doi: 10.1111/apt.12010
Strauss, E., Sherman, E. M. S., and Spreen, O. (2006). A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary. 3rd Edn. New York, NY: Oxford University Press.
Therriault, J., Benedet, A. L., Pascoal, T. A., Mathotaarachchi, S., Savard, M., Chamoun, M., et al. (2020). APOEε4 potentiates the relationship between amyloid-β and tau pathologies. Mol. Psychiatry. doi: 10.1038/s41380-020-0688-6. [Epub ahead of print].
van der Vlies, A. E., Verwey, N. A., Bouwman, F. H., Blankenstein, M. A., Klein, M., Scheltens, P., et al. (2009). CSF biomarkers in relationship to cognitive profiles in Alzheimer disease. Neurology 72, 1056–1061. doi: 10.1212/01.wnl.0000345014.48839.71
Vistoropsky, Y., Ermakov, S., Toliat, M. R., Trofimov, S., Altmüller, J., Malkin, I., et al. (2010). Genetic determinants of circulating levels of tumor necrosis factor receptor II and their association with TNF-RII gene polymorphisms. Cytokine 51, 28–34. doi: 10.1016/j.cyto.2010.04.013
Wechsler, D. (1981). Wechsler Adult Intelligence Scale – Revised. San Antonio, TX: The Psychological Corporation.
Whitwell, J. L., Dickson, D. W., Murray, M. E., Weigand, S. D., Tosakulwong, N., Senjem, M. L., et al. (2012). Neuroimaging correlates of pathologically defined subtypes of Alzheimer's disease: a case-control study. Lancet Neurol. 11, 868–877. doi: 10.1016/S1474-4422(12)70200-4
Keywords: Alzheimer disease, neuroinflammation, TNFRSF1B, resilience, executive function, MCI, digit span
Citation: Pillai JA, Bebek G, Khrestian M, Bena J, Bergmann CC, Bush WS, Leverenz JB and Bekris LM (2021) TNFRSF1B Gene Variants and Related Soluble TNFR2 Levels Impact Resilience in Alzheimer's Disease. Front. Aging Neurosci. 13:638922. doi: 10.3389/fnagi.2021.638922
Received: 07 December 2020; Accepted: 01 February 2021;
Published: 25 February 2021.
Edited by:
Yang Xiang, Western Theater General Hospital, ChinaReviewed by:
Atsushi Iwata, Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology (TMGH-IG), JapanCopyright © 2021 Pillai, Bebek, Khrestian, Bena, Bergmann, Bush, Leverenz and Bekris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jagan A. Pillai, cGlsbGFpakBjY2Yub3Jn
†Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.