- 1Department of Neurology, Xuanwu Hospital of Capital Medical University, Beijing, China
- 2Department of Neurology, Zhejiang Taizhou Municipal Hospital, Taizhou, Zhejiang, China
- 3Department of Neurobiology, Xuanwu Hospital of Capital Medical University, Beijing, China
- 4Center of Alzheimer's Disease, Beijing Institute for Brain Disorders, Beijing, China
- 5National Clinical Research Center for Geriatric Disorders, Beijing, China
Objective: This study assessed the methylation of peripheral NCAPH2 in individuals with subjective cognitive decline (SCD), identified its correlation with the hippocampal volume, and explored whether the correlation is influenced by apolipoprotein E ε4 (APOE ε4) status.
Methods: Cognitively normal controls (NCs, n = 56), individuals with SCD (n = 81), and patients with objective cognitive impairment (OCI, n = 51) were included from the Sino Longitudinal Study on Cognitive Decline (NCT03370744). All participants completed neuropsychological assessments, blood tests, and structural MRI. NCAPH2 methylation was compared according to the diagnostic and APOE ε4 status. Partial correlation analysis was conducted to assess the correlations between the hippocampal volume, cognitive tests, and the NCAPH2 methylation levels.
Results: Individuals with SCD and patients with OCI showed significantly lower levels of NCAPH2 methylation than NCs, which were independent of the APOE ε4 status. The NCAPH2 methylation levels and the hippocampal volumes were positively correlated in the SCD APOE ε4 non-carriers but not in the OCI group. No association was found between the NCAPH2 methylation levels and the cognitive function.
Conclusion: Abnormal changes in blood NCAPH2 methylation were found to occur in SCD, indicating its potential to be used as a useful peripheral biomarker in the early stage of Alzheimer's disease screening.
Introduction
Alzheimer's disease (AD) is a progressive and highly debilitating neurodegenerative disorder, accounting for about two-thirds of 50 million people with dementia worldwide (Lane et al., 2018). There is still no therapy to treat the underlying cause of the disease or slow down its progression. The pathogenesis of AD is complex and may involve genetic and environmental factors. A growing body of evidence points to the epigenetic contribution of AD, and both global and gene-specific changes in DNA methylation have been observed in the affected postmortem brain regions (Bakulski et al., 2012; De Jager et al., 2014; Cronin et al., 2017). The search for peripheral blood epigenetic biomarkers of AD is of particular interest because of the unavailability of brain DNA samples until postmortem and the inability to collect longitudinal brain samples to track disease diagnosis (Bakulski et al., 2016).
Recently, two large-scale epigenome-wide association studies (EWAS) have reported the first replicable and robust association of brain methylation and AD pathology in independent cohorts (De Jager et al., 2014; Lunnon et al., 2014). Given the stable changes in DNA methylation levels in older asymptomatic individuals with amyloid pathology, the altered DNA methylation in the brain has been identified as an early feature of preclinical AD. NCAPH2 is a subunit of the condensin-2 complex involved in chromosome condensation during mitosis and meiosis (Martin et al., 2016). Peripheral blood DNA methylation levels in the NCAPH2/LMF2 promoter region were significantly decreased in patients with AD and those with amnestic mild cognitive impairment (aMCI) (Kobayashi et al., 2016); therefore, these are considered to be a potentially useful biomarker for the diagnosis of AD. However, while the hypomethylation of NCAPH2 has been reported in the Japanese population, its presence in the Chinese population remains unclear. Furthermore, whether NCAPH2 methylation changes across the AD spectrum is still unknown. To date, there have been relatively few studies devoted to the determination of NCAPH2 methylation patterns in peripheral blood in the early stages of AD.
Subjective cognitive decline (SCD) is characterized by subjective self-perception of worsening cognitive capacity but without any impairment observed in objective evaluations (Jessen et al., 2014; Molinuevo et al., 2017). It has been suggested that SCD precedes MCI and is the preclinical stage of AD (Rabin et al., 2017). Several studies have shown that SCD may be associated with risk factors for dementia, such as the apolipoprotein E (APOE) ε4 allele and the neuropathology of AD (Perrotin et al., 2015; Risacher et al., 2017; Vogel et al., 2017). Thus, recognizing the early characterization of DNA methylation patterns in SCD may help to better understand the role of methylation in the early stages of AD.
Recent research has demonstrated a robust decrease in global DNA methylation in the hippocampus of patients with AD, and this was significantly correlated with amyloid plaque load (Chouliaras et al., 2013). Moreover, a large multisite EWAS revealed DNA methylation in the superior temporal gyrus-mediated associations between blood DNA methylation and the hippocampal volume (Jia et al., 2019). These findings suggest that DNA methylation and the relationship with the hippocampal volume are involved in the pathophysiology of AD. However, whether DNA methylation changes in the blood are correlated with the hippocampal volume in the early stage of AD remain unknown.
Thus, the objectives of this study were (1) to identify cross-sectional differences in peripheral blood NCAPH2 methylation among patients with SCD and objective cognitive impairment (OCI) relative to normal control (NC) participants and (2) to identify the correlation between the hippocampal volume, the cognitive function, and the NCAPH2 methylation levels and their interaction with the APOE ε4 status.
Methods
Participants
Altogether, 56 NCs, 81 participants with SCD, and 51 participants with OCI (34 MCI and 17 AD) from the Sino Longitudinal Study on Cognitive Decline (SILCODE) project were included in the present research. The SILCODE project is a large, multicenter-based longitudinal observational study in China (ClinicalTrials.gov, NCT 03370744) that is based in Xuanwu Hospital, Capital Medical University, China, and aims to construct a high-precision multimodal model for the ultra-early diagnosis of AD. The study was approved by the institutional review board at Xuanwu Hospital in Capital Medical University, and written informed consent was obtained from all participants.
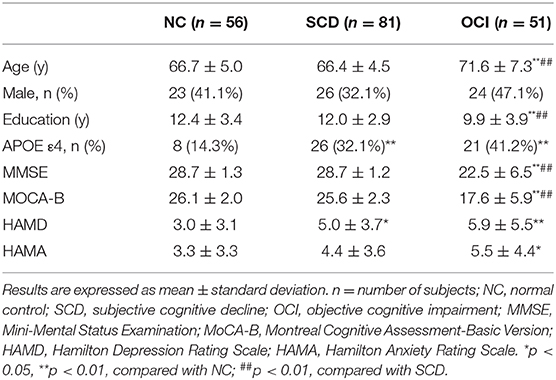
All 188 individuals fulfilled the inclusion criteria of the SILCODE project (Li et al., 2019). Briefly, SCD is defined using the following criteria (Jessen et al., 2014): (1) self-experienced persistent decline in memory rather than other domains of cognition within the last 5 years; (2) concerns related to SCD and a feeling of worsened performance when compared to others of the same age group; and (3) performance on standardized cognitive tests within age-, gender-, and education-adjusted norms and failure to meet the criteria for MCI or dementia. MCI was diagnosed using the Jak–Bondi approach (Bondi et al., 2014). Impairment in a cognitive domain was defined as having at least two tests >1.0 SD below the age-adjusted normative means or having impaired scores in each of the three sampled cognitive domains (memory, language, and speed/executive functioning). The diagnosis of AD was determined by the published diagnostic criteria (McKhann et al., 1984; Dubois et al., 2007). Individuals with no cognitive complaints and normal performance on the standardized neuropsychological tests were included as controls. The exclusion criteria contained brain trauma or disorder, including clinical stroke, severe psychiatric, and/or severe somatic disease, that may account for symptoms, intellectual disability or other developmental disorders, and other systemic diseases. All participants were assessed using a standardized clinical evaluation protocol comprised of the Mini-Mental State Examination (MMSE), Montreal Cognitive Assessment Basic Version (MoCA-B), Hamilton Depression Scale (HAMD), and Hamilton Anxiety Scale (HAMA).
Peripheral NCAPH2 Methylation Measurements
Venous blood samples were collected early in the morning from 6:00 a.m. to 8:00 a.m. Genomic DNA was prepared from leukocytes using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), with sodium bisulfite treatment performed as described previously (Ghodsi et al., 2020). Briefly, for the bisulfite reaction (in which cytosine is converted to uracil and 5-methylcytosine remains non-reactive), genomic DNA was initially denatured with 0.3 M NaOH. Sodium metabisulfite solution (pH 5.0) and hydroquinone were then added at final concentrations of 3.0 M and 0.5 mM, respectively. Reaction mixtures were incubated under mineral oil in the dark at 50°C for 16 h. Denatured DNA was purified with Wizard DNA Purification Resin (Promega, Fitchburg, WI, USA), and the reaction terminated by treatment with 0.3 M NaOH at 37°C for 15 min, followed by ethanol precipitation. For bisulfite pyrosequencing analysis, 50 ng of bisulfite-treated genomic DNA was amplified in a DNA Engine Opticon 2 system (MJ Research, Waltham, MA, USA) using a Taq DNA Polymerase kit, hot-start (Takara Bio, Otsu Shiga, Japan). The sequences of the PCR amplification primers, as well as the sequencing primer for NCAPH2, were as follows: forward: 5′-GTATTTTTTTGGGAGGGAATAGTAAAATG-3′, reverse: 5′-CCACCTCCCAATTCTTAATAAAA-3′, sequencing: 5′-AGTAAAATGGAGTTAGAATTAGTG-3′, with an amplicon of 187 bp. The reverse primer contained biotin at the 5′ position. The amplification conditions for NCAPH2 were as follows: 1 cycle of 94°C for 2 min; 50 cycles of 94°C for 20 s, 61°C for 20 s, and 72°C for 20 s; and 1 cycle of 72°C for 5 min. For the pyrosequencing reaction, single-stranded DNA templates were immobilized on Streptavidin Sepharose High Performance beads (GE Healthcare, Uppsala, Sweden) using the PSQ Vacuum Prep Tool and Vacuum Prep Worktable (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Reactions were incubated at 80°C for 2 min and allowed to anneal with the sequencing primers (0.4 mM) at room temperature. Pyrosequencing was performed using PyroMark Gold Reagents (Qiagen) on a PyroMark Vaccum Workstation (Qiagen), according to the manufacturer's instructions.
APOE Genotyping
Apolipoprotein E was amplified with the following primers: 5′-ACGCGGGCACGGCTGTCCAAGG-3′ (forward) and 5′-GGCGCTCGCGGATGGCGCTGA-3′ (reverse), using the following conditions: 1 cycle of 98°C for 10 s, 35 cycles of 72°C for 5 s, and 1 cycle of 72°C for 5 min. PCR was performed in a final volume of 30 μl, containing 10 pmol of forward and reverse primers and 50 ng of genomic DNA template, using PrimeSTAR HS DNA Polymerase with GC Buffer (Takara Bio), according to the manufacturer's instructions. Then, APOE was genotyped using the standard Sanger sequencing method (Sangon, Shanghai, China).
MRI Acquisition
All individuals were scanned on an integrated simultaneous 3.0 T TOF PET/MR (Signa PET/MR, GE Healthcare, WI, USA) at Xuanwu Hospital, Capital Medical University, China. The 3D BRAVO T1-weighted sequence was obtained with the following parameters: repetition time (TR)/echo time (TE) = 6.9 ms/2.98 ms, flip angle (FA) = 12°, inversion time (TI) = 450 ms, field of view = 256 × 256 mm2, matrix = 256 × 256, slices = 192, slice thickness = 1 mm, no interslice gap, and voxel size = 1 × 1 × 1 mm3.
MRI Processing
Briefly, the entire workflow was as follows: (1) spatially adaptive non-local means denoising, (2) rough inhomogeneity correction, (3) an aligned image into the Montreal Neurological Institute-Hospital (MNI) space, (4) inhomogeneity correction, (5) intensity normalization, (6) non-local intracranial cavity extraction, and (7) subcortical nucleus segmentation. Steps from 1 to 7 were implemented in the volBrain pipeline (http://volbrain.upv.es). The left and right hippocampal volumes were presented as relative values (%), which were measured in relation to the total intracranial volume (Manjon and Coupe, 2016).
Statistical Analysis
Demographic data and neuropsychological tests were compared by the ANOVA and the chi-squared test for continuous and categorical variables. To compare the NCAPH2 methylation levels and the hippocampal volumes, a one-way analysis of covariance (ANCOVA) was conducted with age, gender, and years of education as covariates. Bonferroni's multiple comparison tests were conducted for post-hoc comparison. We also performed group comparisons among the NC, SCD, MCI, and AD groups. A two-way ANCOVA was used to assess the interactions between the diagnostic status and the APOE ε4 allele on NCAPH2 methylation controlling for age, gender, and years of education. Then, several partial correlation analyses were conducted with age, gender, and years of education as covariates. First, we assessed the correlations between the hippocampal volumes (left and right side) and the NCAPH2 methylation levels in the SCD group and further in the SCD APOE ε4 non-carriers and carriers separately to examine whether the correlation differed according to the APOE ε4 status. We also assessed the correlation between the NCAPH2 methylation levels and the hippocampal volumes in NC and OCI groups. Second, we evaluated the correlation between the NCAPH2 methylation levels and the cognitive scores (MMSE and MoCA-B) in all individuals and subgroups (APOE ε4 non-carriers and carriers groups). All p-values were calculated using two-sided tests, and p < 0.05 was considered statistically significant. All analyses were performed using SPSS Statistics (version 24.0, IBM).
Results
Demographic Characteristics and Cognitive Function
Table 1 summarizes the demographic and neuropsychological scores for each group. All groups were statistically comparable in terms of sex distribution. The one-way ANOVA showed group differences in age (F = 15.79, p < 0.001) and years of education (F = 8.88, p < 0.001). The OCI group was significantly older and less educated than the control and SCD groups. There were significant differences in the APOE ε4 prevalence among groups (χ2 = 9.88, p = 0.007); patients with SCD and OCI had higher proportions of APOE ε4 carriers (32.1 and 41.2%, respectively) when compared to the controls (14.3%). Patients with OCI showed lower MMSE and MoCA-B scores than controls and individuals with SCD (MMSE: F = 53.85, p < 0.001; MoCA-B: F = 92.62, p < 0.001). The SCD and OCI groups scored significantly higher on the HAMD than the controls (F = 6.62, p = 0.002). Similarly, the HAMA score was higher among the patients with OCI than among the controls (F = 4.06, p = 0.019), but there was no difference between the SCD and control groups. No significant differences were found in the HAMD or HAMA scores between patients with SCD and patients with OCI (all p > 0.1).
Group Differences in the NCAPH2 Methylation Levels
One-way ANCOVA revealed that the NCAPH2 methylation levels were significantly different among the three groups (F = 6.43, p = 0.002) after controlling for age, gender, and years of education. Figure 1 shows that patients with SCD and patients with OCI had significantly lower NCAPH2 methylation levels than the controls (both p < 0.001; Bonferroni corrected), but there was no difference between patients with SCD and patients with OCI (p > 0.1; Bonferroni corrected). Further comparison among the four groups showed that the NCAPH2 methylation levels were lower in patients with MCI and patients with AD than the controls, but there was no difference among patients with SCD, patients with MCI, and patients with AD (Supplementary Figure 1).
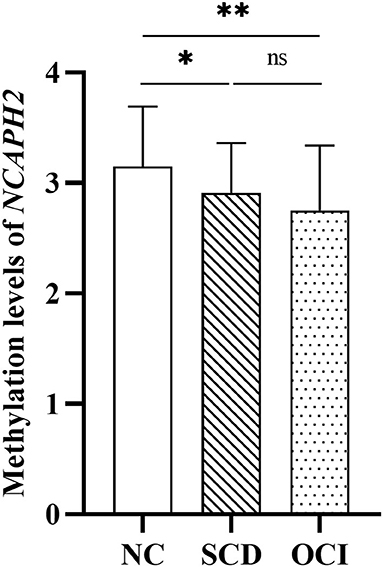
Figure 1. Group differences in the NCAPH2 methylation levels among the three groups. The analysis was adjusted with age, gender, and years of education. Asterisks indicate post-hoc comparisons with respect to controls (Bonferroni corrected). The error bars indicated SDs. *p < 0.05, **p < 0.01; ns, not significant. NC, normal control; SCD, subjective cognitive decline; OCI, objective cognitive impairment.
The NCAPH2 methylation levels were significantly low in the APOE ε4 carriers than in the APOE ε4 non-carriers (Supplementary Table 1 and Supplementary Figure 2). Furthermore, no significant additive interactions were observed in NCAPH2 methylation between the diagnostic groups and the APOE ε4 allele (F = 0.41, p = 0.665).
Group Differences in the Hippocampal Volume
Figure 2 shows the significant differences in the volume proportion of the bilateral hippocampus among the three groups (right hippocampus: F = 27.95, p < 0.001; left hippocampus: F = 25.61, p < 0.001). Compared with the controls and SCD groups, patients with OCI exhibited significant volume loss in the right and left hippocampus (all p < 0.001; Bonferroni corrected). However, there were no significant differences between the SCD and control groups (p > 0.05; Bonferroni corrected).
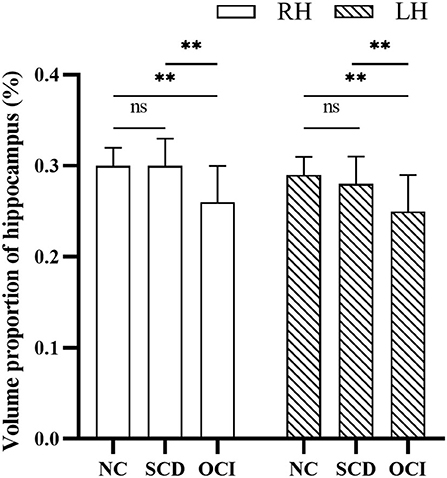
Figure 2. Group differences in the volume proportion of the right and left hippocampus. Results were corrected for multiple comparisons (Bonferroni corrected). **p < 0.01; ns, not significant. NC, normal control; SCD, subjective cognitive decline; OCI, objective cognitive impairment; LH, left hippocampus; RH, right hippocampus.
Relationship Between the NCAPH2 Methylation Levels and the Hippocampal Volume
In the SCD group, there was a significant positive correlation between the NCAPH2 methylation levels and the volume proportion of the left hippocampus (r = 0.245, p = 0.027); however, the positive correlation in the right hippocampus was not significant (r = 0197, p = 0.078). Moreover, the positive correlation between the NCAPH2 methylation levels and the volume proportion of the hippocampus was significant in the APOE ε4 non-carriers in the SCD group (left/right: r = 0.347/0.279, p = 0.009/0.039). Nevertheless, there were no significant associations in the APOE ε4 carriers in the SCD group (left/right: r = −0.017/−0.026, p = 0.935/0.900) (Figure 3).
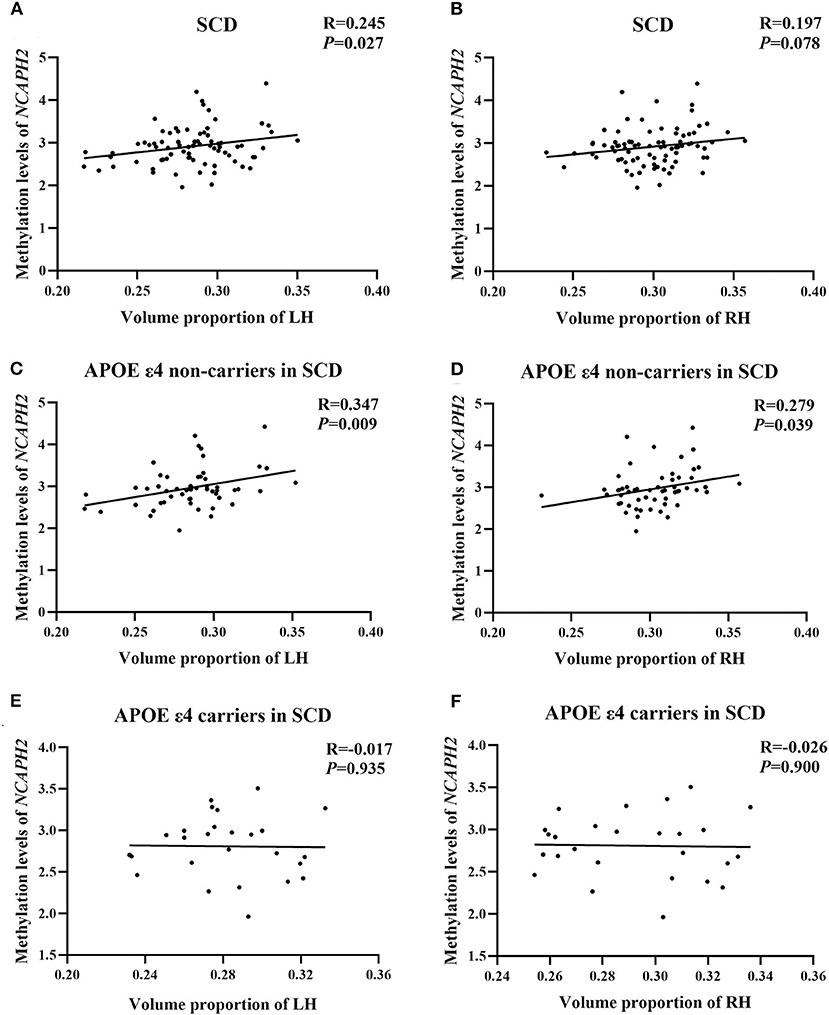
Figure 3. The relationship between the NCAPH2 methylation levels and the volume proportion of hippocampus in participants with SCD. (A) The NCAPH2 methylation levels were significantly associated with the volume proportion of the LH in participants with SCD; (B) The positive correlation was not significant in the RH; Furthermore, the NCAPH2 methylation levels were significantly associated with the volume proportion of both the LH (C) and the RH (D) in the APOE ε4 non-carriers. There were no significant associations in the APOE ε4 carriers in the LH (E) or the RH (F). SCD, subjective cognitive decline; LH, left hippocampus; RH, right hippocampus.
The correlations between the NCAPH2 methylation levels and the volume proportion of the hippocampus in NC and OCI groups were not significant (all p > 0.1, Supplementary Table 2).
Relationship Between the NCAPH2 Methylation Levels and the Cognitive Scores
There was no correlation between the NCAPH2 methylation levels and the cognitive scores in all individuals (MMSE: r = 0.077, p = 0.304; MoCA-B: r = 0.126, p = 0.095, Supplementary Figures 3, 4). Further, subgroup analysis showed no significant correlation in both APOE ε4 carriers and non-carriers groups (all p > 0.1, Supplementary Table 3).
Discussion
In the present study, we found that NCAPH2 methylation was decreased in the peripheral blood of patients with SCD and patients with OCI compared to the controls. Notably, NCAPH2 hypomethylation was significantly positively associated with the hippocampal volume in patients with SCD, especially in the APOE ε4 non-carriers. No association was found between the NCAPH2 methylation levels and the cognitive function.
It has been reported that DNA methylation modifications of several genes are fundamental to the development of AD, which has been considered relevant to AD progression. However, empirical evidence is hard to come by, and the results thus far have been conflicting and controversial. One study has confirmed that higher peripheral methylation levels of brain-derived neurotrophic factor, a member of the neurotrophin family, are associated with a significant AD conversion propensity for patients with MCI (Xie et al., 2017). Other β-amyloid precursor protein (APP)-related genes, such as BACE1, were hypomethylated in the peripheral blood of patients with AD (Marques et al., 2012). Nevertheless, one large case-control study found that there was no difference in the blood DNA methylation levels of PSEN1 or BACE1, both codes for proteins directly involved in the APP cleavage, between patients with AD and the controls, leading to the formation of the β-amyloid (Aβ) (Tannorella et al., 2015). The different methods of methylation analysis and small samples of the varying population may account for divergent findings (Fransquet et al., 2018). The decreased NCAPH2 methylation in patients with OCI found in our study was consistent with the hypomethylation of the NCAPH2/LMF2 promoter region reported in a previous Japanese study (Kobayashi et al., 2016). Moreover, the altered methylation pattern was also been found in individuals with SCD, and no difference among patients with SCD, patients with MCI, and patients with AD was observed. Our results indicated that the NCAPH2 methylation levels decreased during the SCD stage of the disease and reached a plateau at the cognitive impairment stage, suggesting that altered blood NCAPH2 methylation might be an early feature of AD pathology.
The hippocampus, a brain area critical for learning and memory, is vulnerable to damage at the early stages of AD. In addition, altered neurogenesis in the hippocampus has been suggested as an early critical event in AD due to its relevance for neural plasticity and network maintenance (Mu and Gage, 2011). DNA methylation during neurogenesis has been shown to be responsive to many extrinsic signals, both under normal conditions and during the development of the disease. Our study found that there was a relationship between the NCAPH2 methylation levels and the hippocampal volume in SCD individuals, but not in patients with MCI and patients with AD. This is not consistent with observations from the previous research, showing a correlation between NCAPH2/LMF2 methylation and hippocampal atrophy in AD (Shinagawa et al., 2016). A possible explanation for this difference in outcome could be that NCAPH2 methylation has reached a plateau phase in the early stage of AD, leading to little meaningful variance in methylation levels between patients with cognitive impairment. Thus, a dissociation between NCAPH2 hypomethylation and the markers of neurodegeneration, including hippocampal atrophy and cognitive impairment, was observed in the late stage of AD. Besides, we observed a significant positive association between the NCAPH2 methylation levels and the left hippocampal volume rather than the right hippocampus in the SCD group. A previous study of community-dwelling Chinese people showed asymmetry patterns in the hippocampus in SCD, and the correlation between the volume of the right hippocampus and the cognitive function was shown (Yue et al., 2018). Another study reported that SCD with a smaller left hippocampal volume was associated with greater depressive symptomatology (Buckley et al., 2016). Further work is needed to provide a better understanding of the different correlations with hippocampal asymmetry, for instance, in the context of brain function and compensation. Previous studies have shown that epigenetic regulation may be involved in defective T-cell function in NCAPH2 mutant mice (Gosling et al., 2008). Recent work has identified numerous extravascular CD8+ T-cells in the perivascular space of blood vessels with cerebral amyloid angiopathy in the hippocampi of patients with AD. Furthermore, there was a negative correlation in aMCI and AD between T-cells and cognition (Gate et al., 2020). We hypothesize that dysregulation of NCAPH2 methylation may lead to an abnormal immune response and finally to hippocampal atrophy. Therefore, a large sample and longitudinal analyses are required to investigate the role of NCAPH2 methylation, and the correlation with the hippocampus contributes to the pathologic process of AD.
Among the several genes that are considered as risk factors for late-onset AD, the APOE ε4 confers the strongest risk. APOE ε4 isoforms have been shown to affect the disease pathogenesis by regulating Aβ aggregation (Ramanan et al., 2013, 2015) and impairing Aβ clearance in the brain (Castellano et al., 2011). However, it is neither necessary nor sufficient for incident AD; thus, it is of great interest to identify AD risk factors for the APOE ε4-non-carriers population. In this study, we found a positive correlation between NCAPH2 hypomethylation and smaller hippocampal volume in the APOE ε4 non-carriers of the SCD group but not in the APOE ε4 carriers. Previous studies found that the APOE ε4 carriers have greater hippocampal atrophy than the non-carriers in patients with AD, cognitively normal elderly, and healthy young adults (O'Dwyer et al., 2012; Chang et al., 2019; Dong et al., 2019). This finding further strengthens the idea that AD pathology has multiple factors and suggests that hippocampal atrophy is partly due to DNA methylation in the early stage of the disease. It seems that NCAPH2 methylation is a useful peripheral biomarker to be used in combination with the analysis of genetic risk alleles to identify the disease pathogenesis, especially in the SCD APOE ε4-non-carriers risk population (Di Francesco et al., 2015).
This study has several limitations. First, the sample size in our study was relatively small. The lack of amyloid biomarkers for the underlying pathology of patients with SCD, patients with MCI, and patients with AD is another limitation of the current study. Further research, especially a follow-up large sample study with biomarkers of AD-type pathology in the preclinical disease stage, is needed to confirm this issue.
Taken together, our results indicated the NCAPH2 methylation patterns in peripheral blood of individuals with SCD. Moreover, our data revealed the relationship between the NCAPH2 methylation levels and the hippocampal volume in the APOE ε4-non-carriers of SCD. This information would suggest that changes in blood methylation may be an early indicator of individuals at risk for dementia as well as potential targets for intervention in the early stage of the disease.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YH, Y-NC, and X-NW provided data and designed the study. YC and T-RL assembled and analyzed the data, consulted literature, and drafted the manuscript. S-WH reviewed the manuscript. YH and Y-NC critically revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2016YFC1306300) and the National Natural Science Foundation of China (61633018 and 82020108013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Data collection and sharing for this study was funded by the Sino Longitudinal Study on Cognitive Decline (SILCODE) project (ClinicalTrials.gov, NCT 03370744). Colleagues of Xuanwu Hospital, Capital Medical University who have made contributions to build the SILCODE cohort are Weina Zhao, Ziqi Wang, Yuxia Li, Bin Mu, Xing Zhao, Jun Wang, Mingyan Zhao, Liu Yang, Xuanyu Li, Yu Sun, Guanqun Chen, Qin Yang, Can Sheng, Jiachen Li, Wenying Du, Xiaoqi Wang, Li Lin, Yang Zhan, Yi Liu, Lianghui Ni, and Xuan Jia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2021.632382/full#supplementary-material
References
Bakulski, K. M., Dolinoy, D. C., Sartor, M. A., Paulson, H. L., Konen, J. R., Lieberman, A. P., et al. (2012). Genome-wide DNA methylation differences between late-onset Alzheimer's disease and cognitively normal controls in human frontal cortex. J. Alzheimer's Dis. 29, 571–588. doi: 10.3233/jad-2012-111223
Bakulski, K. M., Halladay, A., Hu, V. W., Mill, J., and Fallin, M. D. (2016). Epigenetic research in neuropsychiatric disorders: the “tissue issue”. Curr. Behav. Neurosci. Rep. 3, 264–274. doi: 10.1007/s40473-016-0083-4
Bondi, M. W., Edmonds, E. C., Jak, A. J., Clark, L. R., Delano-Wood, L., McDonald, C. R., et al. (2014). Neuropsychological criteria for mild cognitive impairment improves diagnostic precision, biomarker associations, and progression rates. J. Alzheimers Dis. 42, 275–289. doi: 10.3233/JAD-140276
Buckley, R. F., Maruff, P., Ames, D., Bourgeat, P., Martins, R. N., Masters, C. L., et al. (2016). Subjective memory decline predicts greater rates of clinical progression in preclinical Alzheimer's disease. Alzheimers Dement 12, 796–804. doi: 10.1016/j.jalz.2015.12.013
Castellano, J. M., Kim, J., Stewart, F. R., Jiang, H., DeMattos, R. B., Patterson, B. W., et al. (2011). Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 3:89ra57. doi: 10.1126/scitranslmed.3002156
Chang, Y. T., Kazui, H., Ikeda, M., Huang, C. W., Huang, S. H., Hsu, S. W., et al. (2019). Genetic interaction of APOE and FGF1 is associated with memory impairment and hippocampal atrophy in Alzheimer's disease. Aging Dis. 10, 510–519. doi: 10.14336/AD.2018.0606
Chouliaras, L., Mastroeni, D., Delvaux, E., Grover, A., Kenis, G., Hof, P. R., et al. (2013). Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol. Aging 34, 2091–2099. doi: 10.1016/j.neurobiolaging.2013.02.021
Cronin, P., McCarthy, M. J., Lim, A. S. P., Salmon, D. P., Galasko, D., Masliah, E., et al. (2017). Circadian alterations during early stages of Alzheimer's disease are associated with aberrant cycles of DNA methylation in BMAL1. Alzheimers Dement 13, 689–700. doi: 10.1016/j.jalz.2016.10.003
De Jager, P. L., Srivastava, G., Lunnon, K., Burgess, J., Schalkwyk, L. C., Yu, L., et al. (2014). Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 17, 1156–1163. doi: 10.1038/nn.3786
Di Francesco, A., Arosio, B., Falconi, A., Micioni Di Bonaventura, M. V., Karimi, M., Mari, D., et al. (2015). Global changes in DNA methylation in Alzheimer's disease peripheral blood mononuclear cells. Brain Behav. Immunity 45, 139–144. doi: 10.1016/j.bbi.2014.11.002
Dong, Q., Zhang, W., Wu, J., Li, B., Schron, E. H., McMahon, T., et al. (2019). Applying surface-based hippocampal morphometry to study APOE-E4 allele dose effects in cognitively unimpaired subjects. Neuroimage Clin. 22:101744. doi: 10.1016/j.nicl.2019.101744
Dubois, B., Feldman, H. H., Jacova, C., Dekosky, S. T., Barberger-Gateau, P., Cummings, J., et al. (2007). Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 6, 734–746. doi: 10.1016/S1474-4422(07)70178-3
Fransquet, P. D., Lacaze, P., Saffery, R., McNeil, J., Woods, R., and Ryan, J. (2018). Blood DNA methylation as a potential biomarker of dementia: a systematic review. Alzheimer's Dement. 14, 81–103. doi: 10.1016/j.jalz.2017.10.002
Gate, D., Saligrama, N., Leventhal, O., Yang, A. C., Unger, M. S., Middeldorp, J., et al. (2020). Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer's disease. Nature 577, 399–404. doi: 10.1038/s41586-019-1895-7
Ghodsi, M., Shahmohammadi, M., Modarressi, M. H., and Karami, F. (2020). Investigation of promoter methylation of MCPH1 gene in circulating cell-free DNA of brain tumor patients. Exp. Brain Res. 238, 1903–1909. doi: 10.1007/s00221-020-05848-1
Gosling, K. M., Goodnow, C. C., Verma, N. K., and Fahrer, A. M. (2008). Defective T-cell function leading to reduced antibody production in akleisin-βmutant mouse. Immunology 125, 208–217. doi: 10.1111/j.1365-2567.2008.02831.x
Jessen, F., Amariglio, R. E., van Boxtel, M., Breteler, M., Ceccaldi, M., Chetelat, G., et al. (2014). A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer's disease. Alzheimers Dement 10, 844–852. doi: 10.1016/j.jalz.2014.01.001
Jia, T., Chu, C., Liu, Y., van Dongen, J., Papastergios, E., Armstrong, N. J., et al. (2019). Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes: findings from the ENIGMA Epigenetics Working Group. Mol. Psychiatry. doi: 10.1038/s41380-019-0605-z. [Epub ahead of print].
Kobayashi, N., Shinagawa, S., Nagata, T., Shimada, K., Shibata, N., Ohnuma, T., et al. (2016). Development of biomarkers based on DNA methylation in the NCAPH2/LMF2 promoter region for diagnosis of Alzheimer's disease and amnesic mild cognitive impairment. PLoS ONE 11:e0146449. doi: 10.1371/journal.pone.0146449
Lane, C. A., Hardy, J., and Schott, J. M. (2018). Alzheimer's disease. Europ. J. Neurol. 25, 59–70. doi: 10.1111/ene.13439
Li, X., Wang, X., Su, L., Hu, X., and Han, Y. (2019). Sino Longitudinal Study on Cognitive Decline (SILCODE): protocol for a Chinese longitudinal observational study to develop risk prediction models of conversion to mild cognitive impairment in individuals with subjective cognitive decline. BMJ Open 9:e028188. doi: 10.1136/bmjopen-2018-028188
Lunnon, K., Smith, R., Hannon, E., De Jager, P. L., Srivastava, G., Volta, M., et al. (2014). Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer's disease. Nat. Neurosci. 17, 1164–1170. doi: 10.1038/nn.3782
Manjon, J. V., and Coupe, P. (2016). volBrain: an online MRI brain volumetry system. Front. Neuroinform 10:30. doi: 10.3389/fninf.2016.00030
Marques, S. C. F., Lemos, R., Ferreiro, E., Martins, M., de Mendonça, A., Santana, I., et al. (2012). Epigenetic regulation of BACE1 in Alzheimer's disease patients and in transgenic mice. Neuroscience 220, 256–266. doi: 10.1016/j.neuroscience.2012.06.029
Martin, C.-A., Murray, J. E., Carroll, P., Leitch, A., Mackenzie, K. J., Halachev, M., et al. (2016). Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 30, 2158–2172. doi: 10.1101/gad.286351.116
McKhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D., and Stadlan, E. M. (1984). Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on Alzheimer's disease. Neurology 34, 939–944. doi: 10.1212/wnl.34.7.939
Molinuevo, J. L., Rabin, L. A., Amariglio, R., Buckley, R., Dubois, B., Ellis, K. A., et al. (2017). Implementation of subjective cognitive decline criteria in research studies. Alzheimer's Dement. 13, 296–311. doi: 10.1016/j.jalz.2016.09.012
Mu, Y., and Gage, F. H. (2011). Adult hippocampal neurogenesis and its role in Alzheimer's disease. Mol. Neurod. 6:85. doi: 10.1186/1750-1326-6-85
O'Dwyer, L., Lamberton, F., Matura, S., Tanner, C., Scheibe, M., Miller, J., et al. (2012). Reduced hippocampal volume in healthy young ApoE4 carriers: an MRI study. PLoS ONE 7:e48895. doi: 10.1371/journal.pone.0048895
Perrotin, A., de Flores, R., Lamberton, F., Poisnel, G., La Joie, R., de la Sayette, V., et al. (2015). Hippocampal subfield volumetry and 3d surface mapping in subjective cognitive decline. J. Alzheimer's Dis. 48, S141–S150. doi: 10.3233/jad-150087
Rabin, L. A., Smart, C. M., and Amariglio, R. E. (2017). Subjective cognitive decline in preclinical Alzheimer's disease. Annu. Rev. Clin. Psychol. 13, 369–396. doi: 10.1146/annurev-clinpsy-032816-045136
Ramanan, V. K., Risacher, S. L., Nho, K., Kim, S., Shen, L., McDonald, B. C., et al. (2015). GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer's disease implicates microglial activation gene IL1RAP. Brain 138(Pt 10), 3076–3088. doi: 10.1093/brain/awv231
Ramanan, V. K., Risacher, S. L., Nho, K., Kim, S., Swaminathan, S., Shen, L., et al. (2013). APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol. Psychiatry 19, 351–357. doi: 10.1038/mp.2013.19
Risacher, S. L., Tallman, E. F., West, J. D., Yoder, K. K., Hutchins, G. D., Fletcher, J. W., et al. (2017). Olfactory identification in subjective cognitive decline and mild cognitive impairment: association with tau but not amyloid positron emission tomography. Alzheimers Dement (Amst) 9, 57–66. doi: 10.1016/j.dadm.2017.09.001
Shinagawa, S., Kobayashi, N., Nagata, T., Kusaka, A., Yamada, H., Kondo, K., et al. (2016). DNA methylation in the NCAPH2/LMF2 promoter region is associated with hippocampal atrophy in Alzheimer's disease and amnesic mild cognitive impairment patients. Neurosci. Lett. 629, 33–37. doi: 10.1016/j.neulet.2016.06.055
Tannorella, P., Stoccoro, A., Tognoni, G., Petrozzi, L., Salluzzo, M. G., Ragalmuto, A., et al. (2015). Methylation analysis of multiple genes in blood DNA of Alzheimer's disease and healthy individuals. Neurosci. Lett. 600, 143–147. doi: 10.1016/j.neulet.2015.06.009
Vogel, J. W., Varga Dolezalova, M., La Joie, R., Marks, S. M., Schwimmer, H. D., Landau, S. M., et al. (2017). Subjective cognitive decline and beta-amyloid burden predict cognitive change in healthy elderly. Neurology 89, 2002–2009. doi: 10.1212/WNL.0000000000004627
Xie, B., Xu, Y., Liu, Z., Liu, W., Jiang, L., Zhang, R., et al. (2017). Elevation of Peripheral BDNF promoter methylation predicts conversion from amnestic mild cognitive impairment to Alzheimer's disease: a 5-year longitudinal study. J. Alzheimer's Dis. 56, 391–401. doi: 10.3233/jad-160954
Keywords: Alzheimer's disease, NCAPH2, methylation, subjective cognitive decline, SCD
Citation: Chen Y, Li T-R, Hao S-W, Wang X-N, Cai Y-N and Han Y (2021) Blood NCAPH2 Methylation Is Associated With Hippocampal Volume in Subjective Cognitive Decline With Apolipoprotein E ε4 Non-carriers. Front. Aging Neurosci. 13:632382. doi: 10.3389/fnagi.2021.632382
Received: 23 November 2020; Accepted: 06 January 2021;
Published: 02 February 2021.
Edited by:
Boon-Seng Wong, Singapore Institute of Technology, SingaporeReviewed by:
Nobuyuki Kobayashi, Jikei University School of Medicine, JapanBing Zhang, Nanjing Drum Tower Hospital, China
Copyright © 2021 Chen, Li, Hao, Wang, Cai and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Ni Wang, d3huYW5uYUAxNjMuY29t; Yan-Ning Cai, Y2FpeWFubmluZ0B4d2guY2NtdS5lZHUuY24=; Ying Han, aGFueWluZ0B4d2guY2NtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Ying Chen1,2†
Ying Chen1,2† Tao-Ran Li
Tao-Ran Li Xiao-Ni Wang
Xiao-Ni Wang Ying Han
Ying Han