
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Aging Neurosci. , 11 April 2019
Sec. Alzheimer's Disease and Related Dementias
Volume 11 - 2019 | https://doi.org/10.3389/fnagi.2019.00084
 Xinyu Fang1
Xinyu Fang1 Wei Tang2
Wei Tang2 Fuyin Yang2
Fuyin Yang2 Weihong Lu1
Weihong Lu1 Jun Cai1
Jun Cai1 Jianliang Ni3
Jianliang Ni3 Jiangtao Zhang3
Jiangtao Zhang3 Wenxin Tang4
Wenxin Tang4 Tao Li5
Tao Li5 Deng-Feng Zhang6
Deng-Feng Zhang6 Chen Zhang1*
Chen Zhang1*There is ample evidence suggesting that calcium/calmodulin-dependent protein kinase II alpha (CaMK2A) may play an important role in the pathophysiology of Alzheimer’s disease (AD). This genetic study aimed to investigate whether CaMK2A confers susceptibility to the development of AD in the Han Chinese population. A total of seven single nucleotide polymorphisms (SNPs) within CaMK2A were screened in two independent cohorts from southwestern China (333 AD patients and 334 controls) and eastern China (382 AD patients and 426 controls) to discern the potential association between this gene and AD. In addition, a cross-platform normalized expression resource was used to investigate whether CaMK2A is differentially expressed in the brain between individuals with AD and the controls. In addition, expression quantitative trait loci (eQTL) analysis was used to explore the differences in CaMK2A expression in the brain among different genotypes. The cross-platform normalized data showed significant differences in CaMK2A expression in the hippocampus, entorhinal cortex and temporal cortex between the AD patients and the control subjects (|log FC| > 0.1, P < 0.05); however, only the differences in the hippocampus and temporal cortex remained after the multiple comparisons correction [false discovery rate (FDR)-corrected, P < 0.05]. The frequency of the rs4958445 genotype was significantly different between the AD subjects and the controls from southwestern China (P = 0.013, P = 0.034 after FDR correction). When the two samples were combined, rs4958445 still showed a significant association with AD (P = 0.044). Haplotype analysis indicated that the T-A-C-A-T-C-C and T-G-C-A-T-C-C haplotypes in the southwestern cohort and the T-G-C-G-C-T-C haplotype in the eastern cohort, consisting of rs10051644, rs6869634, rs3797617, rs3756577, rs4958445, rs10515639 and rs6881743, showed a significant association with AD (P = 0.037, P = 0.026 and P = 0.045, respectively). Furthermore, the brain eQTL analysis revealed a significant association between the rs4958445 polymorphism and CaMK2A expression in the inferior olivary nucleus (P = 0.029). Our results suggest an important role for CaMK2A in the pathophysiology of AD in the Han Chinese population, especially the southwestern population.
Alzheimer’s disease (AD), which is the most common neurodegenerative disorder, is primarily characterized by progressive cognitive impairment in the elderly (Zhang D. F. et al., 2016; Xing et al., 2017). Epidemiological studies have demonstrated that AD will become more prevalent by the middle of this century and therefore represents a major global public health problem (Frozza et al., 2018). Although the underlying cause of AD is unclear in most cases, numerous genetic alterations have been identified as being associated with the risk of AD (Wang et al., 2016a,b; Zhang D. F. et al., 2016; Li G. D. et al., 2017; Tang et al., 2017), and signaling pathways involved in memory loss in AD are also under intense investigation (Vázquez-Higuera et al., 2011; Berridge, 2016).
It has been well established that altered synaptic Ca2+ signaling may be involved in the pathogenesis of AD (Jang and Chung, 2016). Calcium/calmodulin-dependent protein kinase II (CaMK2) is a multifunctional protein kinase that is highly expressed in the central nervous system (CNS) and is activated by the binding of Ca2+/calmodulin (Lisman et al., 2012). Early literature reported that CaMK2 plays a crucial role in gene expression, memory processing, learning and neuroplasticity in the CNS (Irvine et al., 2006; Lisman et al., 2012; Wang D. et al., 2018). The role of CaMK2A is especially important, as this subunit is the most abundant subunit of CaMK2 in the brain (Hanson and Schulman, 1992). There is accumulating evidence indicating that CaMK2A is required for long-term potential (LTP), which is the long-lasting form of synaptic plasticity and is required for memory formation (Irvine et al., 2006). Preclinical studies demonstrated that CaMK2A knock-in mutant mice have impaired memory and LTP (Giese et al., 1998; Yamagata et al., 2009). Moreover, postmortem analyses also confirmed that CaMK2A is dysregulated in the hippocampus in AD, and CaMK2A-expressing neurons are selectively lost at synaptic locations in the hippocampus of AD patients (Wang et al., 2005; Reese et al., 2011). These results suggest that aberrant synaptic CaMK2A activity may play an important role in the pathogenesis of AD.
The gene encoding CaMK2A is located at human chromosome 5q32. A recent study reported that the CaMK2A rs3756577 and rs3822606 polymorphisms confer susceptibility to AD in the Spanish population (Bufill et al., 2015), whereas earlier literature did not support the association of CaMK2A with AD in this population (Vázquez-Higuera et al., 2011). To the best of our knowledge, no genetic study to date has addressed the association of CaMK2A with AD in the Chinese Han population.
In the present study, we attempted to explore the potential association between common variants of CaMK2A and the genetic risk of AD in two independent Han Chinese samples from eastern and southwestern China. First, a public database was used to determine whether CaMK2A is differentially expressed in the brain between AD patients and healthy controls. Second, we completely genotyped seven CaMK2A single nucleotide polymorphisms (SNPs) in our samples. Third, brain expression quantitative trait loci (eQTL) analysis was used to determine whether CaMK2A SNPs that are associated with AD risk are differentially expressed in the brain among individuals with different genotypes.
Two independent samples of Chinese participants were recruited into this study. One cohort from eastern China was composed of 382 unrelated sporadic AD patients and 426 cognitively healthy individuals who were recruited from the Shanghai Mental Health Center and Tongde Hospital of Zhejiang Province. The cohort from southwestern China was enrolled at the Mental Health Center of Southwest China Hospital; this cohort was composed of 333 unrelated sporadic AD patients and 334 cognitively healthy controls. All patients were required to meet the following conditions: (1) diagnosed with AD according to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) and the National Institute of Neurological and Communicative Disorders and Stroke/AD and Related Disorders Association (NINCDSADRDA; McKhann et al., 1984); (2) Han Chinese and over 65 years old; (3) defined as sporadic, i.e., their family history did not mention any first-degree relative with AD; and (4) Mini Mental State Examination (MMSE) scores for the following educational levels: illiterate <17, primary school education <20, middle school or above education <24. Patients with vascular dementia, Vitamin B12 deficiency, major depressive disorder, hypothyroidism or any other diseases that may cause cognitive decline were excluded. The healthy controls arose from the same base population and met the following criteria: (1) Han Chinese and over 65 years old; (2) confirmed as cognitively intact by MMSE screening; (3) no family history of AD; and (4) no neurodegenerative diseases, vascular diseases or systemic diseases. The patients and controls were independently diagnosed and screened by two psychiatrists who had worked in clinical practice for at least 5 years. Written informed consent was obtained from each participant prior to the performance of any procedure related to this study. In addition, the study protocol was approved by all the institutional review boards involved.
To investigate whether CaMK2A was differentially expressed in the brain between AD patients and healthy controls, cross-platform normalized expression data for four brain regions [entorhinal cortex (EC), hippocampus (HP), temporal cortex (TC), and frontal cortex (FC)] were analyzed. Cross-platform normalization is a method that combines all expression data from multiple microarray studies into a unified dataset. Because cross-platform normalization removes artifacts between different platforms (batch effects) and preserves “real” biological differences between experimental conditions, this method is regarded to have a better performance in robust biomarker detection compared with the meta-analysis method (Taminau et al., 2014). Genes with a log 2 fold change greater than 0.1 (|log FC| > 0.1) and a false discovery rate (FDR) smaller than 0.05 (FDR < 0.05) were defined as being differentially expressed in AD patients. The above-mentioned analyses are accessible at the Alzdata.org web server1, and detailed information can be found in a previous study (Xu et al., 2018).
A total of 7 SNPs within CaMK2A (rs10051644, rs6869634, rs3797617, rs3756577, rs4958445, rs10515639 and rs6881743) were selected based on the following criteria: (1) SNPs are capable of tagging more SNPs based on the linkage disequilibrium (LD) pattern of the respective genes according to the data from the HapMap CHB dataset2 and 1,000 Genomes3, and (2) all eligible SNPs should have a minor allele frequency (MAF) >10% according to the HapMap CHB2 and dbSNP datasets4. Detailed information on each SNP is shown in Supplementary Table S1. Genomic DNA of all participants was extracted from peripheral blood using the AxyPrep™ Blood Genomic DNA Miniprep Kit (Axygen, USA). All SNPs were genotyped using the SNaPshot assay, as described in our previous studies (Wang et al., 2012; Bi et al., 2014).
To further explore the differences in CaMK2A expression in various brain regions among individuals with different genotypes, we performed a eQTL analysis using the brain eQTL database5, which is a large exon-specific eQTL dataset covering 10 human brain regions [substantia nigra (SNIG), thalamus (THAL), inferior olivary nucleus (MEDU), putamen (PUTM), hippocampus (HIPP), temporal cortex (TCTX), intralobular white matter (WHMT), frontal cortex (FCTX), occipital cortex (OCTX), and cerebellar cortex (CRBL)]. More detailed information about this database can be found in the original literature (Ramasamy et al., 2014).
Hardy–Weinberg equilibrium (HWE) and allele and genotype frequency analyses were performed using SHEsis6. SNPs with a P value less than 0.001 were considered as a departure from the HWE (Wang et al., 2016b). In addition, we used Haploview 4.2 to perform a pairwise LD estimation and haplotype analysis. A strict FDR of 0.05 was applied as the multiple comparison correction to reduce the Type 1 error rate. All values were two-tailed, and we considered a P-value < 0.05 or the P-value threshold defined by FDR to be significant. All statistical analyses were conducted in the two independent cohorts separately, as well as in the combined Han Chinese population. Power calculations performed with Quanto 1.2.3 (Gauderman, 2002) were used to calculate the statistical power of the case-control sample size, assuming the prevalence of AD to be 5.56% in the Han Chinese population aged more than 65 years (Huang et al., 2019) and the OR to be 1.4. As the MAF of the selected SNPs fluctuated from 12% to 49%, the power of the sample ranged from 87.86% to 99.98%.
Differential expression analysis from the cross-platform normalized data showed that there were significant differences in CaMK2A expression in the HP, EC and TC between the AD patients and the control subjects (|log FC| > 0.1, P < 0.05); however, only the differences in the HP and TC remained following the multiple comparisons correction (FDR-corrected P = 0.034, P = 0.017, respectively). We failed to detect a difference in CaMK2A expression in the FC between the two groups (Figure 1).
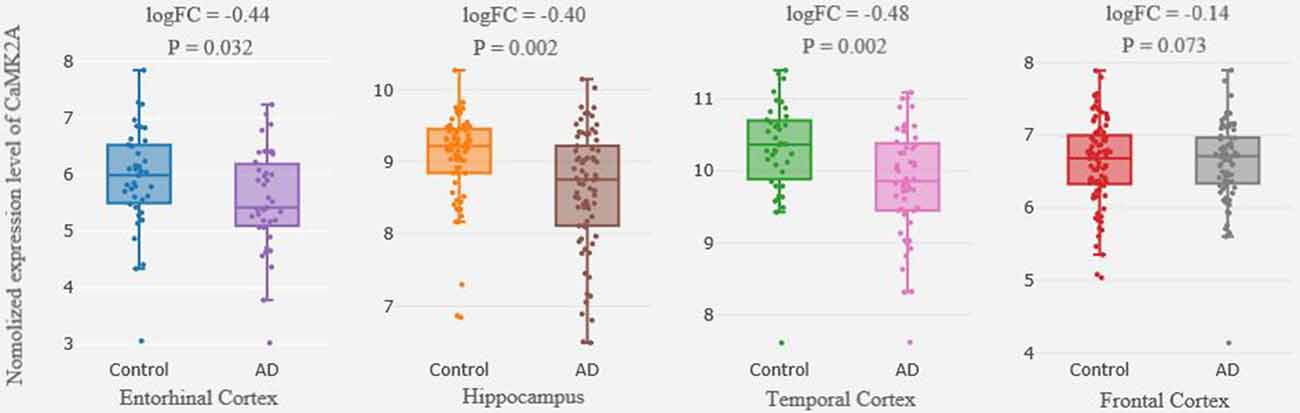
Figure 1. Differential expression of CaMK2A in brain between patients with AD and healthy controls. Each bar represents the average level of CaMK2A expression. Error bars represent the standard deviation of the mean value. Abbreviations: CaMK2A, calcium/calmodulin dependent protein kinase II alpha; AD, Alzheimer’s disease.
In total, seven CaMK2A SNPs were analyzed in the current study, and the genotyping call rate of all individuals was 99.5%. No deviation from HWE was observed in any SNP analyzed. The genotype frequency of the rs4958445 polymorphism was shown to be significantly different between the AD and control subjects from southwestern China before (P = 0.013) and after the multiple comparisons correction (adjusted P = 0.034), whereas this difference was not detected in the cohort from eastern China (P = 0.403). When the two cohorts were combined, the results remained significant (P = 0.044) and were almost significant even after the multiple comparisons correction (adjusted P = 0.084). However, we failed to discover any positive association between other CaMK2A SNPs and AD in either the separate cohorts or the combined Chinese Han population. In addition, the allele frequencies of all the seven SNPs showed no significant difference between patients with AD and the control subjects (Table 1). The LD structures of the 7 SNPs were similar between the AD patients and controls in the separate cohorts and the combined cohort (Figure 2). Haplotype analysis indicated that the T-A-C-A-T-C-C haplotype and T-G-C-A-T-C-C haplotype in the southwestern cohort, consisting of rs10051644, rs6869634, rs3797617, rs3756577, rs4958445, rs10515639 and rs6881743, showed a significant association with AD (P = 0.037, P = 0.026, separately). In addition, our results also showed a significant association between the rs10051644T-rs6869634G-rs3797617C-rs3756577G-rs4958445C-rs10515639T-rs6881743C haplotype and AD in the eastern China sample (P = 0.045). However, no significant difference was detected for any haplotype in the combined Chinese Han population (see Table 2).
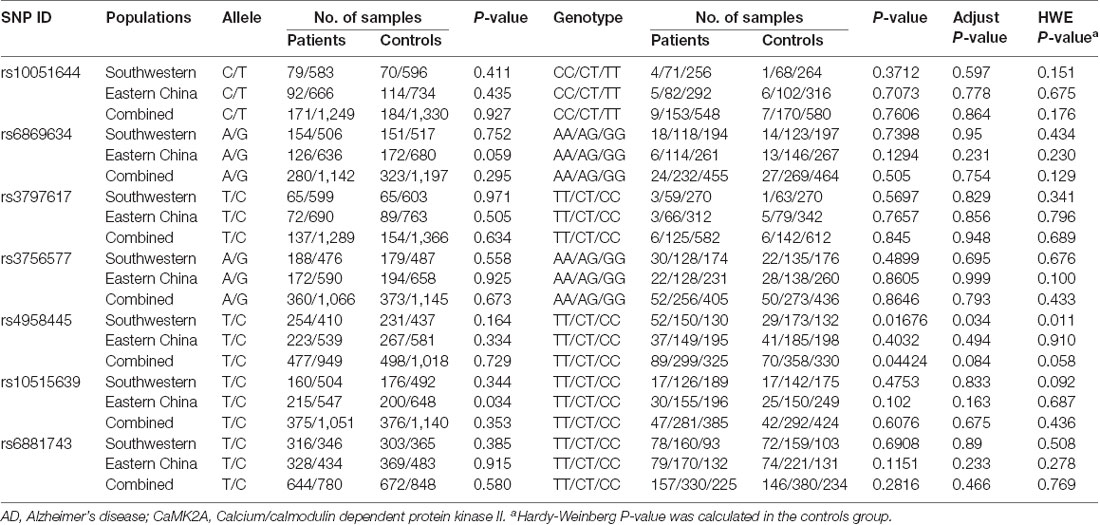
Table 1. Comparison of allele and genotype frequencies of the selected single nucleotide polymorphisms (SNPs) within CaMK2A between AD and healthy controls.
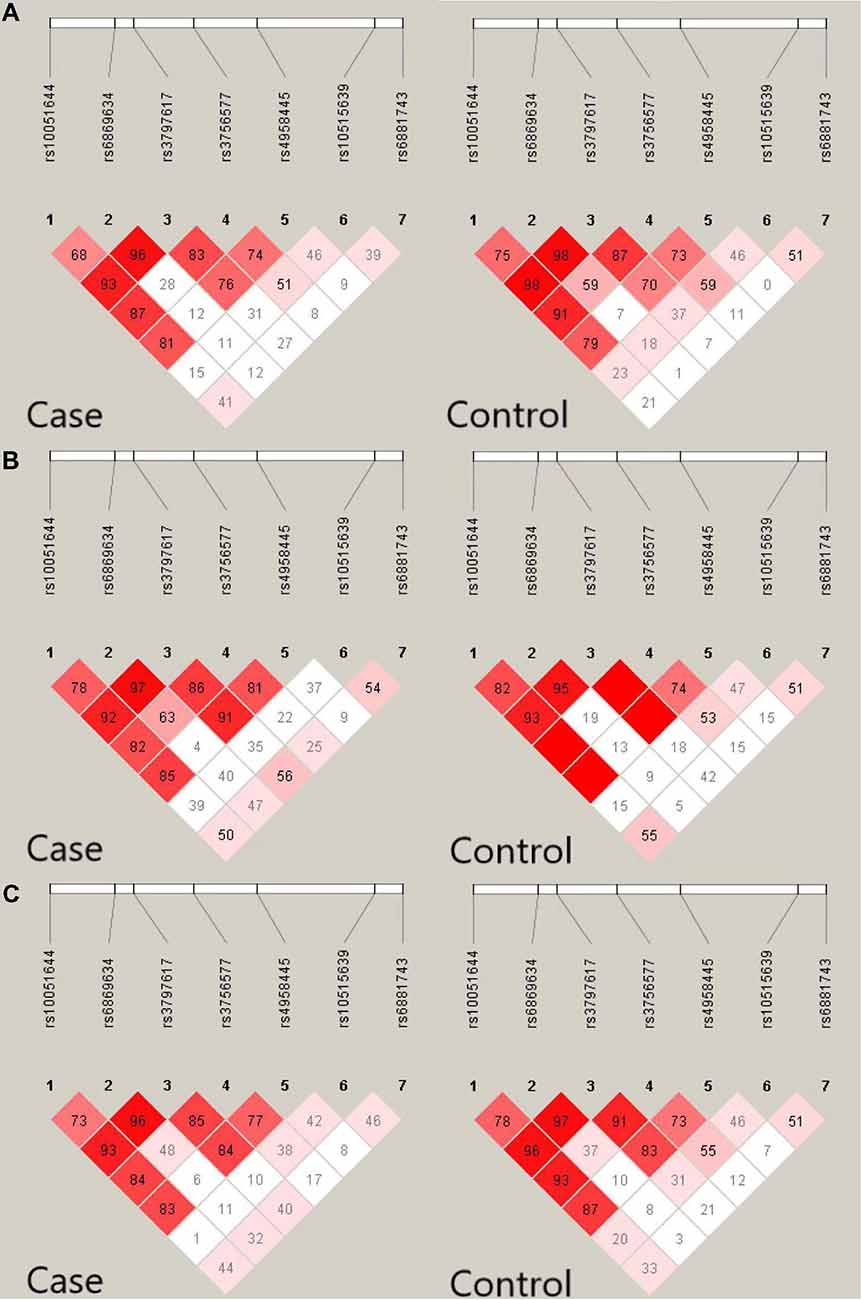
Figure 2. Linkage disequilibrium (LD) pattern of seven single nucleotide polymorphisms (SNPs) in the CaMK2A gene in AD patients and controls. Value in each square refers to r2 × 100. (A) LD pattern of seven SNPS in the CaMK2A gene in AD and controls from southwestern China. (B) LD pattern of seven SNPS in the CaMK2A gene in AD patients and controls from eastern China. (C) LD pattern of seven SNPS in the CaMK2A gene in AD and controls in the combined populations from eastern China and southwestern China.
To explore the role of various polymorphisms in the brain expression of CaMK2A, eQTL analysis was performed. Our results showed a significant association between the rs4958445 polymorphism and CaMK2A expression in the MEDU (inferior olivary nucleus, P = 0.029), and there was a clear trend toward significance in the OCTX (occipital cortex, P = 0.052). These results all emphasized that the expression of the AA genotype was higher than that of the A/G or GG genotype (Figure 3).
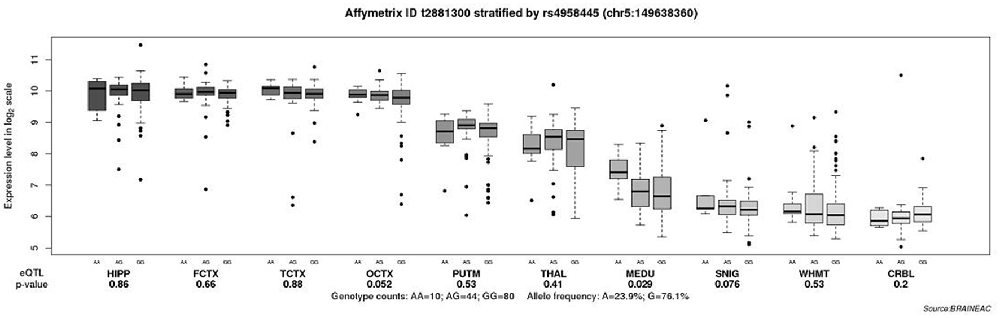
Figure 3. Association of rs5958445 with CaMK2A expression level in 10 brain regions (Affymetrix ID t2881300). Data were extracted from the BRAINEAC database (http://peana-od.inf.um.es:8080/UKBECv12/). Abbreviations: BRAINEAC, The Brain eQTL Almanac; CaMK2A, calcium/calmodulin dependent protein kinase II alpha; eQTL, expression quantitative trait locus; SNIG, substantia nigra; THAL, thalamus; MEDU, inferior olivary nucleus; PUTM, putamen; HIPP, hippocampus; TCTX, temporal cortex; WHMT, intralobular white matter; FCTX, frontal cortex; OCTX, occipital cortex; CRBL, cerebellar cortex.
Ample evidence supports that genetic susceptibility may contribute to the development of AD (Gao et al., 2019). Numerous studies have indicated that the major pathological characteristics of AD include the abnormal gene expression of amyloid beta (Aβ) and tau protein in the brain (Hung et al., 2016; Li J. et al., 2017; Wang C. et al., 2018). Furthermore, it has been reported that CaMK2A, which has a potential link with Aβ production, has an aberrant expression in the brains of patients with AD (Ghosh and Giese, 2015). On this premise, we attempted to investigate the role of CaMK2A in AD. Our differential expression analysis confirmed that there are significant differences in CaMK2A expression in the brains of AD patients compared with control subjects, especially in the HP and TC. It has been well documented that the HP and TC are significantly associated with cognitive deficits, such as learning and memory (Ezzyat et al., 2018), and substantial research has also revealed abnormalities of the HP and TC in AD (Ye et al., 2019; Savioz et al., 2016). The finding that CaMK2A expression is altered in the HP and TC in AD implies that CaMK2A may be involved in the pathogenesis of AD.
To the best of our knowledge, this is the first study to investigate the association between CaMK2A and AD in the Chinese Han population. Herein, we genotyped seven CaMK2A SNPs in two independent Han Chinese cohorts from southwestern and eastern China. Our results showed that the genotype frequency of one CaMK2A SNP, rs4958445, is significantly associated with AD risk in populations from southwestern China and in the combined sample, and this association persisted in the southwestern cohort following the multiple comparisons correction. However, we did not detect any differences in the genotype frequency of SNPs within the eastern cohort. This disparity may be attributed to the different genetic backgrounds of the two cohorts (Wang J. et al., 2016).
To date, numerous preclinical studies have suggested a function of CaMK2A in spatial learning and memory (Matsuo et al., 2009; Moriguchi et al., 2014), which is a proposed functional endophenotype of AD (Nogueira et al., 2018). In addition, associations between several variants of CaMK2A and AD have been investigated in other ethnic populations. Bufill et al. (2015) screened seven SNPs of CaMK2A (rs2241695, rs3756577, rs3822606, rs13357922 and others) in the Spanish population and determined that the TT genotype at rs3756577 and the GG genotype at rs3822606 are underrepresented in Spanish AD patients. However, the link between rs3756577 and AD risk was not confirmed in our current study, and similar conflicting result has been reported by previous studies, indicating that the real functional variants may be associated with different SNPs in different populations (Wang et al., 2016b).
It has been widely accepted that haplotypes can be more specific risk markers compared with single alleles; therefore, the use of haplotypes can reduce false-positive associations in common psychiatric disorders (Zhang X. Y. et al., 2016). Since the seven markers analyzed in the present study were in the same haplotype block, we performed a seven-marker haplotype analysis, and our results showed that the T-A-C-A-T-C-C, T-G-C-A-T-C-C and T-A-C-G-T-C-T haplotype in the southwestern cohort and the T-G-C-G-C-T-C haplotype in the eastern cohort, which included rs10051644, rs6869634, rs3797617, rs3756577, rs4958445, rs10515639 and rs6881743, are significantly associated with AD risk. The fact that this association did not persist when the two cohorts were combined further suggests that people in different regions may have different genetic backgrounds. However, the results of the haplotype analysis in the independent sample all highlighted the relationship between CaMK2A and AD pathogenesis.
As is well known, numerous previous studies have suggested that there are structural and functional changes in certain brain regions in the case of AD, including the OCTX (Seidl et al., 2001; Kemppainen et al., 2006) and MEDU (Dowson, 1982). To detect the differential expression of SNP rs4958445 in the brain, eQTL analysis was performed. Our results indicate that the expression of rs4958445 in the MEDU and OCTX appears to be higher in individuals with the A/A genotype compared with those with the A/G or G/G genotype. In addition, the results of our present study indicate that the genotype frequency of SNP rs4958445 is significantly associated with the risk of AD in the Han Chinese population, and previous research revealed abnormalities in the MEDU and OCTX of AD patients. Taken together, our comprehensive analysis suggests that CaMK2A may regulate the structure and function of brain regions related to AD through altered gene expression, thereby leading to the pathogenesis of AD. However, the influence of CaMK2A on gene expression and its relationship with AD in the Han Chinese population should be verified in the future with a larger cohort.
The strength of the present study is that we combined our original data with previous relevant databases to perform a comprehensive analysis of the relationship between CaMK2A and AD for the first time. In addition, due to the regional differences in the Chinese Han population, we discussed the southwestern and eastern Chinese Han populations separately. However, several limitations of this study should be noted when interpreting the results. First, owing to the modest sample size, the conclusions that can be drawn from our data are limited. Second, only common variants of the CaMK2A were investigated; we did not analyze the rare variants of CaMK2A. Admittedly, this limitation precluded the detection of the active roles of such rare variants in the pathophysiology of AD. Third, although all patients and healthy controls were over 65 years old, we did not collect information about the age, sex and education level of the participants, which limited the information that was included in the association analyses. Therefore, future research involving larger samples of AD patients and focusing on a larger number of functional polymorphisms and their interactions, as well as other rare variants, may provide a more adequate understanding of the role of CaMK2A in AD. Such future studies may reveal the biomarkers of AD to facilitate early diagnosis and provide a more specific treatment of AD.
In summary, our case-control association study provides support that CaMK2A variants may play a major role in the pathophysiology of AD in the Chinese Han population. However, this finding remains preliminary due to the limited sample size and the fact that rare variants were not analyzed. Further independent validating studies and essential functional assays are warranted to confirm our observations and characterize the putative role of these genes in the pathophysiology of AD.
This study was carried out in accordance with the recommendations of Ethics Committee of Shanghai Mental Health Center with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Ethics Committee of Shanghai Mental Health Center.
XF, TL, D-FZ and CZ contributed to the overall design of the study. All authors got involved in the sample collection. XF and CZ undertook the statistical analysis, interpretation of data and wrote the manuscript. All authors contributed to the approval of the final manuscript.
This work was supported by the National Key Research and Development Program of China (2018YFC1314302), the National Natural Science Foundation of China (81771450, 81471358), the Shanghai Science and Technology Commission Foundation (14411969000), the Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (20152530), the Shanghai Municipal Commission of Health and Family Planning Foundation (201540029) and the Shanghai Municipal Commission of Health and Family Planning, Key Developing Disciplines (2015ZB0405).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are deeply grateful to all participants.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2019.00084/full#supplementary-material
Berridge, M. J. (2016). Vitamin D, reactive oxygen species and calcium signalling in ageing and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150434. doi: 10.1098/rstb.2015.0434
Bi, R., Zhao, L., Zhang, C., Lu, W., Feng, J. Q., Wang, Y., et al. (2014). No association of the LRRK2 genetic variants with Alzheimer’s disease in Han Chinese individuals. Neurobiol. Aging 35, 444.e5–444.e9. doi: 10.1016/j.neurobiolaging.2013.08.013
Bufill, E., Roura-poch, P., Sala-matavera, I., Antón, S., Lleó, A., Sánchez-saudinós, B., et al. (2015). Reelin signaling pathway genotypes and Alzheimer disease in a Spanish population. Alzheimer Dis. Assoc. Disord. 29, 169–172. doi: 10.1097/WAD.0000000000000002
Dowson, J. H. (1982). Neuronal lipofuscin accumulation in ageing and alzheimer dementia: a pathogenic mechanism? Br. J. Psychiatry 140, 142–148. doi: 10.1192/bjp.140.2.142
Ezzyat, Y., Wanda, P. A., Levy, D. F., Kadel, A., Aka, A., Pedisich, I., et al. (2018). Closed-loop stimulation of temporal cortex rescues functional networks and improves memory. Nat. Commun. 9:365. doi: 10.1038/s41467-017-02753-0
Frozza, R. L., Lourenco, M. V., and De Felice, F. G. (2018). Challenges for Alzheimer’s disease therapy: insights from novel mechanisms beyond memory defects. Front Neurosci. 12:37. doi: 10.3389/fnins.2018.00037
Gao, Y., Ren, R. J., Zhong, Z. L., Dammer, E., Zhao, Q. H., Shan, S., et al. (2019). Mutation profile of APP, PSEN1, and PSEN2 in Chinese familial Alzheimer’s disease. Neurobiol. Aging 31, 154–157. doi: 10.1016/j.neurobiolaging.2019.01.018
Gauderman, W. J. (2002). Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol. 155, 478–484.
Ghosh, A., and Giese, K. P. (2015). Calcium/calmodulin-dependent kinase II and Alzheimer’s disease. Mol. Brain 8:78. doi: 10.1186/s13041-015-0166-2
Giese, K. P., Fedorov, N. B., Filipkowski, R. K., and Silva, A. J. (1998). Autophosphorylation at Thr286 of the α calcium-calmodulin kinase II in LTP and learning. Science 279, 870–873. doi: 10.1126/science.279.5352.870
Hanson, P. I., and Schulman, H. (1992). Neuronal Ca2+/calmodulin-dependent protein kinases. Annu. Rev. Biochem. 61, 559–601. doi: 10.1146/annurev.biochem.61.1.559
Huang, Y. Q., Wang, Y., Wang, H., Liu, Z. R., Yu, X., Yan, J., et al. (2019). Prevalence of mental disorders in China: a cross-sectional epidemiological study. Lancet Psychiatry 6, 211–224. doi: 10.1016/S2215-0366(18)30511-X
Hung, A. S., Liang, Y., Chow, T. C., Tang, H. C., Wu, S. L., Wai, M. S., et al. (2016). Mutated tau, amyloid and neuroinflammation in Alzheimer disease-A brief review. Prog. Histochem. Cytochem. 51, 1–8. doi: 10.1016/j.proghi.2016.01.001
Irvine, E. E., von Hertzen, L. S., Plattner, F., and Giese, K. P. (2006). αCaMKII autophosphorylation: a fast track to memory. Trends Neurosci. 29, 459–465. doi: 10.1016/j.tins.2006.06.009
Jang, S. S., and Chung, H. J. (2016). Emerging link between Alzheimer’s disease and homeostatic synaptic plasticity. Neural Plast. 2016:7969272. doi: 10.1155/2016/7969272
Kemppainen, N. M., Aalto, S., Wilson, I. A., Någren, K., Helin, S., Brück, A., et al. (2006). Voxel-based analysis of PET amyloid ligand [11C]PIB uptake in Alzheimer disease. Neurology 67, 1575–1580. doi: 10.1212/01.wnl.0000240117.55680.0a
Li, G. D., Bi, R., Zhang, D. F., Xu, M., Luo, R., Wang, D., et al. (2017). Female-specific effect of the BDNF gene on Alzheimer’s disease. Neurobiol. Aging 53, 192.e11–192.e19. doi: 10.1016/j.neurobiolaging.2016.12.023
Li, J., Zhang, Q., Chen, F., Meng, X., Liu, W., Chen, D., et al. (2017). Genome-wide association and interaction studies of CSF T-tau/Aβ42 ratio in ADNI cohort. Neurobiol. Aging 57, 247.e1–247.e8. doi: 10.1016/j.neurobiolaging.2017.05.007
Lisman, J., Yasuda, R., and Raghavachari, S. (2012). Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182. doi: 10.1038/nrn3192
Matsuo, N., Yamasaki, N., Ohira, K., Takao, K., Toyama, K., Eguchi, M., et al. (2009). Neural activity changes underlying the working memory deficit in α-CaMKII heterozygous knockout mice. Front. Behav. Neurosci. 320:20. doi: 10.3389/neuro.08.020.2009
McKhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D., and Stadlan, E. M. (1984). Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of department of health and human services task force on Alzheimer’s disease. Neurology 34, 939–944. doi: 10.1212/wnl.34.7.939
Moriguchi, S., Tagashira, H., Sasaki, Y., Yeh, J. Z., Sakagami, H., Narahashi, T., et al. (2014). CaMKII activity is essential for improvement of memory-related behaviors by chronic rivastigmine treatment. J. Neurochem. 128, 927–937. doi: 10.1111/jnc.12510
Nogueira, J., Freitas, S., Duro, D., Tábuas-Pereira, M., Guerreiro, M., Almeida, J., et al. (2018). Alzheimer’s disease assessment scale—cognitive subscale (ADAS-Cog): normative data for the portuguese population. Acta Med. Port. 31, 94–100. doi: 10.20344/amp.8859
Ramasamy, A., Trabzuni, D., Guelfi, S., Varghese, V., Smith, C., Walker, R., et al. (2014). Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428. doi: 10.1038/nn.3801
Reese, L. C., Laezza, F., Woltjer, R., and Taglialatela, G. (2011). Dysregulated phosphorylation of Ca2+/calmodulin-dependent protein kinase II-α in the hippocampus of subjects with mild cognitive impairment and Alzheimer’s disease. J. Neurochem. 119, 791–804. doi: 10.1111/j.1471-4159.2011.07447.x
Savioz, A., Giannakopoulos, P., Herrmann, F. R., Klein, W. L., Kovari, E., Bouras, C., et al. (2016). Study of Aβ oligomers in the temporal cortex and cerebellum of patients with neuropathologically confirmed Alzheimer’s disease compared to aged controls. Neurodegener. Dis. 16, 398–406. doi: 10.1159/000446283
Seidl, R., Cairns, N., Singewald, N., Kaehler, S. T., and Lubec, G. (2001). Differences between GABA levels in Alzheimer’s disease and Down syndrome with Alzheimer-like neuropathology. Naunyn Schmiedebergs Arch. Pharmacol. 363, 139–145. doi: 10.1007/s002100000346
Taminau, J., Lazar, C., Meganck, S., and Nowé, A. (2014). Comparison of merging and meta-analysis as alternative approaches for integrative gene expression analysis. ISRN Bioinform. 2014:345106. doi: 10.1155/2014/345106
Tang, W., Yang, F., Lu, W., Ni, J., Zhang, J., Tang, W., et al. (2017). Association study of CREB1 and CBP genes with Alzheimer’s disease in Han Chinese. Asia Pac. Psychiatry. 9:e12274. doi: 10.1111/appy.12274
Vázquez-Higuera, J. L., Mateo, I., Sánchez-Juan, P., Rodríguez-Rodríguez, E., Pozueta, A., Calero, M., et al. (2011). Genetic variation in the tau kinases pathway may modify the risk and age at onset of Alzheimer’s disease. J. Alzheimers Dis. 27, 291–297. doi: 10.3233/jad-2011-110794
Wang, H. Z., Bi, R., Hu, Q. X., Xiang, Q., Zhang, C., Zhang, D. F., et al. (2016a). Validating GWAS-identified risk loci for Alzheimer’s disease in Han Chinese populations. Mol. Neurobiol. 53, 379–390. doi: 10.1007/s12035-014-9015-z
Wang, H. Z., Bi, R., Zhang, D. F., Li, G. D., Ma, X. H., Fang, Y., et al. (2016b). Neprilysin confers genetic susceptibility to Alzheimer’s disease in han chinese. Mol. Neurobiol. 53, 4883–4892. doi: 10.1007/s12035-015-9411-z
Wang, Y. J., Chen, G. H., Hu, X. Y., Lu, Y. P., Zhou, J. N., and Liu, R. Y. (2005). The expression of calcium/calmodulin-dependent protein kinase II-α in the hippocampus of patients with Alzheimer’s disease and its links with AD-related pathology. Brain Res. 1031, 101–108. doi: 10.1016/j.brainres.2004.10.061
Wang, D., Feng, J. Q., Li, Y. Y., Zhang, D. F., Li, X. A., Li, Q. W., et al. (2012). Genetic variants of the MRC1 gene and the IFNG gene are associated with leprosy in Han Chinese from Southwest China. Hum. Genet. 131, 1251–1260. doi: 10.1007/s00439-012-1153-7
Wang, C., Saar, V., Leung, K. L., Chen, L., Wong, G., Calero, M., et al. (2018). Human amyloid β peptide and tau co-expression impairs behavior and causes specific gene expression changes in Caenorhabditis elegans. Neurobiol. Dis. 109, 88–101. doi: 10.1016/j.nbd.2017.10.003
Wang, D., Wang, X., Liu, X., Jiang, L., Yang, G., Shi, X., et al. (2018). Inhibition of miR-219 alleviates arsenic-induced learning and memory impairments and synaptic damage through up-regulating CaMKII in the hippocampus. Neurochem. Res. 43, 948–958. doi: 10.1007/s11064-018-2500-4
Wang, J., Zhao, S., Shugart, Y. Y., Zhou, Z., Jin, C., Yuan, J., et al. (2016). No association between ZNF804A rs1344706 and schizophrenia in a case-control study of Han Chinese. Neurosci. Lett. 618, 14–18. doi: 10.1016/j.neulet.2016.02.048
Xing, H. Y., Li, B., Peng, D., Wang, C. Y., Wang, G. Y., Li, P., et al. (2017). A novel monoclonal antibody against the N-terminus of Aβ1–42 reduces plaques and improves cognition in a mouse model of Alzheimer’s disease. PLoS One 12:e0180076. doi: 10.1371/journal.pone.0180076
Xu, M., Zhang, D. F., Luo, R., Wu, Y., Zhou, H., Kong, L. L., et al. (2018). A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer’s disease. Alzheimers Dement. 14, 215–229. doi: 10.1016/j.jalz.2017.08.012
Yamagata, Y., Kobayashi, S., Umeda, T., Inoue, A., Sakagami, H., Fukaya, M., et al. (2009). Kinase-dead knock-in mouse reveals an essential role of kinase activity of Ca2+/calmodulin-dependent protein kinase IIα in dendritic spine enlargement, long-term potentiation and learning. J. Neurosci. 29, 7607–7618. doi: 10.1523/JNEUROSCI.0707-09.2009
Ye, C. F., Mori, S., Chan, P., and Ma, T. (2019). Connectome-wide network analysis of white matter connectivity in Alzheimer’s disease. Neuroimage Clin. 22:101690. doi: 10.1016/j.nicl.2019.101690
Zhang, X. Y., Chen, D. C., Tan, Y. L., Tan, S. P., Luo, X., Zuo, L., et al. (2016). BDNF polymorphisms are associated with schizophrenia onset and positive symptoms. Schizophr. Res. 170, 41–47. doi: 10.1016/j.schres.2015.11.009
Keywords: Alzheimer’s disease, CaMK2A, gene, haplotype, eQTL, China
Citation: Fang X, Tang W, Yang F, Lu W, Cai J, Ni J, Zhang J, Tang W, Li T, Zhang D-F and Zhang C (2019) A Comprehensive Analysis of the CaMK2A Gene and Susceptibility to Alzheimer’s Disease in the Han Chinese Population. Front. Aging Neurosci. 11:84. doi: 10.3389/fnagi.2019.00084
Received: 13 January 2019; Accepted: 26 March 2019;
Published: 11 April 2019.
Edited by:
Laura Morelli, Leloir Institute Foundation (FIL), ArgentinaReviewed by:
Magdolna Pákáski, University of Szeged, HungaryCopyright © 2019 Fang, Tang, Yang, Lu, Cai, Ni, Zhang, Tang, Li, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Zhang, emhhbmdjaGVuNjQ1QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.