
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Aging Neurosci. , 15 May 2018
Sec. Cellular and Molecular Mechanisms of Brain-aging
Volume 10 - 2018 | https://doi.org/10.3389/fnagi.2018.00139
Prion diseases are fatal neurological disorders affecting various mammalian species including humans. Lack of proper diagnostic tools and non-availability of therapeutic remedies are hindering the control strategies for prion diseases. MicroRNAs (miRNAs) are abundant endogenous short non-coding essential RNA molecules that negatively regulate the target genes after transcription. Several biological processes depend on miRNAs, and altered profiles of these miRNAs are potential biomarkers for various neurodegenerative diseases, including prion diseases. Autophagic flux degrades the misfolded prion proteins to reduce chronic endoplasmic reticulum stress and enhance cell survival. Recent evidence suggests that specific miRNAs target and regulate the autophagic mechanism, which is critical for alleviating cellular stress. miRNAs-mediated regulation of these specific proteins involved in the autophagy represents a new target with highly significant therapeutic prospects. Here, we will briefly describe the biology of miRNAs, the use of miRNAs as potential biomarkers with their credibility, the regulatory mechanism of miRNAs in major neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and prion diseases, degradation pathways for aggregated prion proteins, the role of autophagy in prion diseases. Finally, we will discuss the miRNAs-modulated autophagic flux in neurodegenerative diseases and employ them as potential therapeutic intervention strategy in prion diseases.
Prion diseases also known as transmissible spongiform encephalopathies (TSEs) are a family of neurodegenerative diseases affecting several mammalian species including humans. The most commonly observed prion diseases of animals include bovine spongiform encephalopathies (BSE) in cattle, scrapie disease in sheep and goats, chronic wasting disease in elk and wild deer. Human populations are equally affected with TSEs, including kuru, Creutzfeldt-Jakob disease (CJD), Gerstmann–Straussler–Scheinker syndrome (GSS) and Fatal Familial Insomnia (FFI) (Nakagaki et al., 2013; Puig et al., 2016). The most critical step in prion propagation is the conversion of normal cellular prion protein (PrPc) into an abnormal and protease-resistant, misfolded and aggregated isoform termed PrPSc (Prusiner, 1998). The misfolded PrPSc is abundantly rich in beta-sheet conformation as compared to its predecessor PrPc, which is alpha-helical in structure (Pan et al., 1993; Mukherjee and Soto, 2011; Yunsheng et al., 2015; Song and Kim, 2017). Initial neuroinflammation leads to severe neurodegeneration in prion diseases due to accumulation and aggregation of PrPSc in the brains of affected individuals (Moreno et al., 2013; Shah et al., 2017a,b,c). The exact molecular mechanisms leading to neuronal inflammation and death are still unclear, and hence there is no cure available for this fatal neurodegenerative disorder (Moreno et al., 2013).
A cell is a fundamental unit of all living organisms, and it runs several biochemical and physiological reactions with high precision and accuracy. There are several regulatory components and pathways of these reactions, which tightly regulate cellular homeostasis. Researchers focus on cellular biomarkers for the study of these highly complicated cellular reactions. These biomarkers are not only critical for providing key information about the standard cellular functions, but they also provide information regarding the pathogenic conditions arising from the endogenous or exogenous stimulus. Modern molecular techniques such as microRNA (miRNA) array-analysis, Northern-analysis, quantitative RT-PCR analysis, and RNA-sequencing-based analyses have uncovered a small group of inducible, pathogenic miRNAs, which are significantly upregulated in the degenerating central nervous system of affected individuals. These miRNAs are involved in the dysregulated expression of disease-relevant messenger RNA (mRNA) targets (Lukiw et al., 2011; Yaghmoor et al., 2014; Zhao et al., 2015a,b; Bhattacharjee et al., 2016; Clement et al., 2016).
The miRNAs are crucial regulators at the post-transcriptional level of genes, and they take an active part in various normal biological and numerous pathological processes involved in these neurodegenerative diseases (Szafranski et al., 2015). miRNAs constitute of 21–23 nucleotides in length. At post-transcriptional level, some important genes are expressed under the influence of these miRNAs (Lee et al., 1993). Pathogenesis of neurodegenerative diseases is a very complex process, and more than 2000 miRNAs have been identified in humans. Large numbers of these miRNAs are directly involved in the disease progression, which gives them the unique role of potential biomarkers in many diseases including Alzheimer’s disease (AD), Parkinson’s disease (PD) and prion diseases (Lee et al., 1993; Cissell et al., 2008; Liu et al., 2008). Several brain pathologists have reported variation in the profile of miRNAs. Some important genes are targeted by these miRNAs in various neurodegenerative disorders (Eacker et al., 2009; Provost, 2010; Sonntag, 2010). Alteration in the synaptic structural plasticity is the earliest hallmark of all major neurodegenerative diseases. Boese et al. (2016) recently demonstrated that during preclinical stage of prion disease, miRNAs enriched in synaptoneurosomes including miRNA-124a-3p, miRNA-136-5p and miRNA-376a-3p were elevated. At the terminal stage of prion infection miRNA-146a-5p, miRNA-142-3p, miRNA-143-3p, miRNA-145a-5p, miRNA-451a, miRNA-let-7b, miRNA-320, and miRNA-150-5p were elevated. During clinical stage of prion disease, all members of the miRNA-200 family (such as miRNA-200a-3p, miRNA-200b-3p, miRNA-200c-3p), miRNA-141-3p, miRNA-429-3p, and 182 cluster miRNAs (miRNA-182-5p and miRNA-183-5p) were downregulated (Boese et al., 2016). Similarly, an important inflammation related miRNA, miRNA-146a expression was upregulated in CJD and GSS patients (Lukiw et al., 2011). Bellingham et al. (2012) demonstrated that those exosomes, released by the prion-infected neuronal cells, had increased levels of let-7b, let-7i, miRNA-128a, miRNA-21, miRNA-222, miRNA-29b, miRNA-342-3p and miRNA-424 levels with decreased miRNA-146a levels compared to control exosomes in non-infected individuals.
Autophagy plays an essential role in cellular housekeeping processes via the degradation, recycling, and removal of old and damaged dysfunctional organelles and protein aggregates (Yoshimori, 2004). Impairments of autophagy/mitophagy, autophagy-lysosomal pathway and ubiquitin-proteasome system (UPS) has been studied extensively in prion diseases (Cortes et al., 2013; Choi et al., 2014; Mckinnon et al., 2016; Khan et al., 2017). Recently many research groups focused on the role of miRNAs in regulation of autophagy in several neurodegenerative diseases (Cheng et al., 2016; Shi et al., 2016; Li et al., 2017; Sun L. et al., 2017; Wang Y. et al., 2017; Yang et al., 2017). Kim et al. (2016) demonstrated that miRNA-27a and miRNA-27b plays a crucial role in the regulation of autophagy for clearing damaged mitochondria via PTEN-induced putative kinase 1 (PINK1) gene. Similarly, Cheng et al. (2016) demonstrated that miRNA-181a sensitizes neuroblastoma cells to apoptosis by suppressing Parkin-mediated mitophagy. miRNA-299-5p plays a crucial role in AD pathogenesis by regulating apoptosis through autophagy in N2a and SH-SY5Y cells and a mouse model of the AD (Zhang et al., 2016).
In this review article, we will overview our current understanding and knowledge about miRNAs-mediated regulatory roles in major neurodegenerative diseases. Furthermore, we will describe the role played by miRNAs in the regulation of autophagy in prion diseases and the therapeutic potentials of miRNAs in treating individuals afflicted with prion diseases.
It was until the end of 20th century when the biological impacts of the small RNA molecules named as miRNAs by the research group of Prof. Victor Ambros opened up an entirely new avenue for molecular research (Lee et al., 1993). Since then, miRNAs are being considered and used as vital tools for the regulation of gene expression in many diseases (Lee et al., 1993). Molecular studies about their origin, biological synthesis, and activation mechanism have substantially advanced our knowledge about miRNAs. The biogenesis of miRNAs is a complex four-step process including transcription, nuclear processing, nuclear export and cytoplasmic processing. The transcription of miRNA genes usually occurs under the influence of RNA polymerase-2 enzyme found in the nucleus of eukaryotic cells. The RNA polymerase-2 enzyme binds with a promoter found near the DNA sequence; encoding would result in the hairpin loop of pre-miRNA (Lee et al., 2002; Reinhart et al., 2002; Gregory et al., 2004). The Animal miRNAs are initially transcribed as part of one arm of a ∼80 nucleotide RNA stem-loop that ultimately forms part of a several hundred nucleotide-long miRNA precursor known as primary miRNA (pri-miRNA). The presence of a stem-loop precursor in the 3′ UTR region will result in a transcript that may serve as pri-miRNA and a mRNA (Lee et al., 2002). There are one to six miRNA precursors in a single pri-miRNA. There are70 nucleotides each in these hairpin loop structures. Each hairpin is flanked by sequences necessary for efficient processing. The double-stranded RNA structure present in the hairpins of a pri-miRNA is recognizable by a nuclear protein called DiGeorge Syndrome Critical Region 8 (DGCR8 or “Pasha” in invertebrates). The DGCR8 associates with the enzyme Drosha, a protein that cuts RNA, to make a Microprocessor complex. The hairpins are liberated from pri-miRNAs by cleavage with RNA about 11 nucleotides from the base of the hairpin. The product that has 3′ hydroxyl and 5′ phosphate groups is termed as pre-miRNA. As much as 16% of the pre-miRNAs might be altered through nuclear RNA editing technique to alter downstream processing in the cytoplasm (Filipowicz et al., 2008; Huntzinger and Izaurralde, 2011). In next step, the pre-miRNA hairpins are exported from the nucleus in a process called nuclear export, which occurs under the influence of nucleocytoplasmic shuttler protein called exportin-5. The RNase-3 enzyme Drosha plays a vital role in the recognition of pre-miRNA hairpin via exportin-5. The transport to cytoplasm occurs under the influence of GTP bound to Ran protein. In the final step, the cytoplasmic processing is initiated, where the pre-miRNA hairpin is cleaved by RNase III enzyme Dicer. The Dicer complex interacts with the 5′ and 3′ end of the hairpin, and it cuts away the loop joining the 3′ and 5′ arms, making an imperfect miRNA:miRNA duplex of about 22 nucleotides in length. Although either of these strands may potentially act like a mature functional miRNA, only one of the strands is usually presented into the RNA-induced silencing complex (RISC), where the miRNA and its mRNA target interact (Lee et al., 2004). The RISC-bound miRNAs target its mRNA under the influence of the Argonaute (AGO) family proteins known as (Argonaute 1–Argonaute 4) (Figure 1) (Bartel, 2009). During the developmental stage of organisms, the specific cellular and tissues type have different levels of miRNAs. Apart from the endogenous stimulus, the exogenous environmental stimuli also play an important role in changing the levels of miRNAs (Aumiller and Forstemann, 2008). Abnormal miRNAs levels play a key role in the pathogenesis of neurodegenerative diseases. It might be possible that alterations in the biogenesis of miRNAs may provide important clues about the cellular and molecular mechanisms in these diseases; and it could be possible to generate novel targets based on miRNAs-mediated therapeutic interventions.

FIGURE 1. Schematic diagram of miRNA synthesis and biology. (1) In the first step, the transcription of pri-miRNAs occurs via MIRNA genes with the help of an RNA-polymerase-2 enzyme. (2) Pri-miRNAs size ranges from 70 to 100 nucleotides, and they fold into a hairpin-loop structure that consists of paired bases, which is composed of various mismatches and bulges. (3) The RNase III enzyme Drosha and its cofactor DGCR8/Pasha plays a crucial role in the excision of hairpin-loop structure to form pre-miRNAs. Ran-GTP-dependent exportin-5 protein helps in the transport of pre-miRNAs from the nucleus to the cytoplasm via nuclear pore complexes found in the nuclear membrane. (4) The pre-miRNAs is matured via RNase III, Dicer, in the cytoplasm. Dicer complex with transactivation responsive RNA binding protein (TRBP), protein activator of the interferon-induced protein kinase (PACT) cuts the pre-miRNAs and generates 21–23 nucleotide-long RNAs, which are mature miRNAs and ready to function. (5) Upon unwinding of the double helix, the mature miRNAs are joined to a ribonucleoprotein complex, known as “RNA-induced silencing complex” (RISC). The RISC complexes retain one of the strands as mature miRNA and eliminate the other strand. (6) Under the influence of Argonaute (AGO) family proteins, the mature miRNA targets the specific mRNA within the RISC. (7) Exact and partial complementation result in mRNA degradation and translation inhibition, respectively.
The biomarker can be defined as a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological responses to therapeutic intervention (Basak et al., 2016). The neurodegenerative disorders are highly heterogeneous in nature, and this heterogeneity necessitates the need for accurate diagnostic and prognostic biomarkers for these diseases. The molecular heterogeneity hampers the successful development of accurate and robust biomarkers for these diseases onsets and progressions. However, the recent findings that miRNAs are present in biofluids such as blood, cerebrospinal fluid, serum, and plasma (Gilad et al., 2008), as stable molecules has led to the development of recent studies based on miRNAs as potential candidates for biomarkers in several neurodegenerative diseases. To determine the potential of miRNAs as biomarkers for neurodegenerative diseases including prion diseases, we must examine the problems before implementing their routine use. A miRNAs should possess some of the basic characteristics of the ideal biomarker involving consideration of clinical, analytical, and practical criteria for implementation in these diseases. Some basic points or parameters before selection of miRNAs as biomarkers are:
(a) The optimal clinical sensitivity and specificity are important for selecting an ideal biomarker. The miRNA selected should provide high sensitivity in particular disease condition. In addition, an ideal miRNA expression profile must be demonstrated to be tissue-specific, however, this does not necessarily mean that translation is based on disease specificity as well.
(b) Detection and quantification of target miRNAs should be robust, rapid, simple, accurate, reproducible and inexpensive. Currently there are many issues regarding the use of miRNAs in neurodegenerative diseases as there is low correlation of results obtained from different platforms or even from the same platform using reagents from different companies. So the starting point should be standardization of these assays followed by normalization of data to get accurate results.
(c) The levels of biomarker should be proportional to the degree of severity of disease. This has been shown in various neurodegenerative diseases including AD, PD, and prion diseases, where the severity of disease has been shown to correlate with the levels of specific plasma or blood miRNAs.
(d) Minimal invasive procedures must be identified for obtaining the samples. The circulating miRNAs would merely require a blood sample, making it highly convenient for the patient, but in case of biopsy or as in all neurodegenerative diseases the brain tissue is affected, will make this process more difficult. The alternative could be obtaining postmortem brain tissues for sampling.
(e) The final and most important factor to remember is the pre-analytical variability of different samples. At present there are no guidelines for the collection, preparation, and extraction of samples for miRNA analysis. Different specimen types, such as brain tissue, cerebrospinal fluid, whole blood, plasma, or serum can introduce a variety of profound effects on miRNA concentrations. Biological variance is an important variable that may affect the clinical utility of miRNAs.
Before employing miRNAs as essential biomarkers for neurodegenerative diseases, we must understand the fact of species and tissue specificity. Roux et al. (2012) demonstrated that the expression of miRNAs was mostly tissue-dependent and the embryonic and nervous tissues shown little change in miRNAs expression with age. They further confirmed that the age of appearance was the primary factor affecting miRNAs expression (Roux et al., 2012). Rodent models are an important tool for molecular studies of neurodegenerative diseases. Human brain tissues and mouse models shown similarities in many aspects of transcriptome analysis but still, there are divergent points as well (Miller et al., 2010). Molecular studies related to miRNAs needs further molecular investigations in rodent models as most of the studies for miRNAs profiling were carried out on primate blood, serum, plasma, CSF or postmortem brain tissue from humans. In-depth studies on rodent models will surely show the extent of divergence amongst humans and rodents.
miRNAs are molecular biomarkers easily identifiable by using basic molecular identification techniques, such as genomics and proteomics. Genomics analysis includes northern blots for isolation of a set of RNA sequences. These RNA sequences can be analyzed by gene expression techniques or surveyed by using SAGE. The DNA microarray technology can be used to determine the frequency of each gene. The microarray and northern blots are usually consistent in data collection however at times; northern blots can detect small changes in gene expression that microarrays cannot detect (Taniguchi et al., 2001). On the other hand, microarrays are very popular for detecting a large number of genes as compared to northern blot (Taniguchi et al., 2001). Sample degradation by RNases from environmental contamination is a significant limitation in northern blot. Another limitation is the danger of chemicals used in northern blots such as formaldehyde, radioactive material, ethidium bromide, DEPC, and UV light are all harmful to the user. The polymerase chain reaction (PCR) technique is used to create many copies of the sequences for user-friendly approach. PCR is easy to use, a highly sensitive technique that can give results quickly but the major limitations are prior information about target sequence required, and DNA contamination will result in false positive results (Schochetman et al., 1988; Zhou et al., 1991). There are several proteomic techniques also available for identification of biomarkers such as 2D-PAGE, LC-MS, SELDI-TOF or MALDI-TOF, antibody array and tissue microarray. The tissue microarrays are combined with immunohistochemistry for cohort studies. Beside genomic and proteomic techniques there are other techniques used for the identification of biomarkers such as metabolomics, lipidomics, glycomics, and secretomics. Recently imaging techniques such as magnetic resonance imaging (MRI), computed tomography (CT), optical coherence tomography (OCT), positron emission tomography (PET) and near-infrared spectroscopy has also been used for the identification of biomarkers. MRI is generally safe technique, but scientists are advised not to overuse it, as it can cause injury due to foreign bodies in the patient’s tissue. MRI is preferred over CT, as it does not use any ionizing radiation. Although PET has gained the attention of many researchers working on AD and PD due to accurate glucose reactivity images, still there are specific limitations such as high costs of cyclotrons needed to produce short-lived radionuclides for PET.
It is evident from above discussion that not all biomarkers should be used as potential biomarkers for specific diseases and its outcome, and these biomarkers are difficult to validate. The biomarkers used for measuring the success of a therapeutic intervention must be directly related to the drug used.
Neuroinflammatory and neurodegenerative events in major neurodegenerative diseases such as PD, AD, and prion diseases are complex processes related to a combination of changes on genetic, molecular and environment basis (Bossy-Wetzel et al., 2004). Neurodegeneration is still poorly understood in all protein misfolding diseases, but recent advances in neuroscience unraveled numerous pathways that play a crucial role in the pathogenesis of these diseases. With the advancement of latest molecular technologies and the involvement of miRNAs in stem cell differentiation, neuritogenesis, synaptoplasticity, etc. has broadened our understanding of the miRNAs biogenesis and using them from potential biomarkers to therapeutic intervention strategies are all underway (Kim J. et al., 2007; Manakov et al., 2009; Zhou et al., 2009; Gao et al., 2010; Bruno et al., 2011; Olde Loohuis et al., 2012) (Table 1).
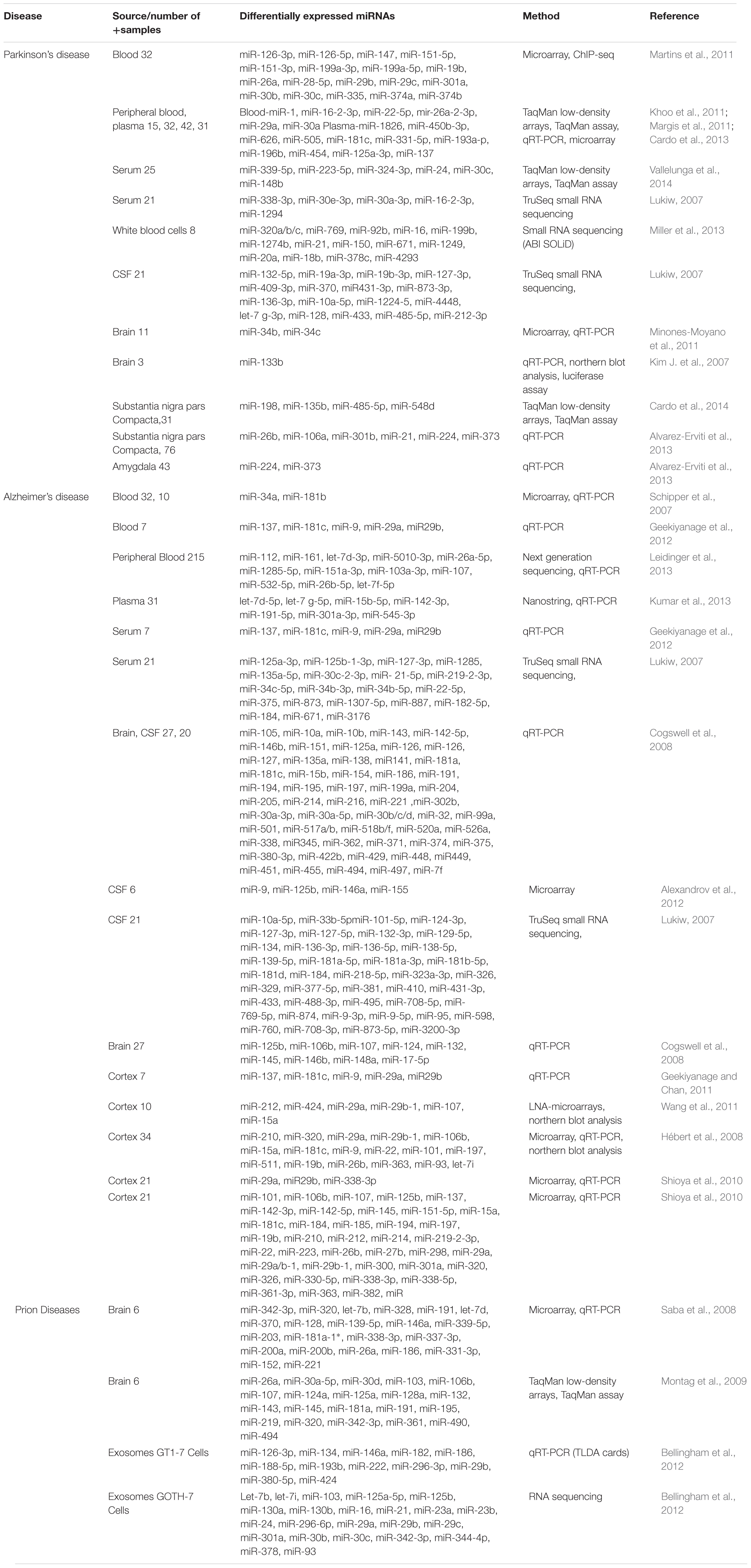
TABLE 1. Showing a list of important biomarkers identified in Parkinson’s, Alzheimer’s, and prion Diseases (adapted from Basak et al., 2016 with minor modifications).
Parkinson’s disease equally affects both younger, and older human populations across the world and an estimated 50,000 Americans are diagnosed each year with PD. PD is a chronic motor neurodegenerative disease with incidence rate almost as high as AD. There are several genes responsible for inherited PD, but five widely accepted genes responsible for late-onset are a-synuclein (SNCA), and leucine-rich repeat kinase 2 (LRRK2), whereas, for early onset, Parkin (PARK2), oncogene DJ1, and PTEN-induced PINK1 are important (Coppede, 2012). LRRK2 is downregulated by miR-205, and pathogenic mutations in the LRRK2 disrupts the critical signaling mechanism of let-7 and miRNA-184, which causes the deregulation of transcription factor E2F1/DP and cell survival is hampered (Gehrke et al., 2010; Cho et al., 2014). Minones-Moyano et al. (2011) demonstrated that miRNA-34b/c regulate DJ-1/PARK7 and Parkin protein expression in SHSY-5Y cell model and in the brains of patients afflicted with PD. Several research groups demonstrated that miRNA-548d, miRNA-224, miRNA-373, miRNA-198, miRNA-106a, miRNA-26b, and miRNA-301b show altered expression in PD patients (Alvarez-Erviti et al., 2013; Burgos et al., 2014; Cardo et al., 2014). The midbrain samples showed miR-133b downregulated in PD patients. Dopaminergic neurons are enriched with Pixt3, which is a crucial transcription factor and miRNA-33b directly increases the expression of Pixt3 (Goodall et al., 2013). Alpha-synuclein (SNCA) plays a vital role in the pathogenesis of PD. In addition, miR-7 and miR-153 target the 3.0 UTR of SNCA, bind directly to SNCA mRNA to downregulate its expression and are enriched in the brains of PD patients (Doxakis, 2010). A polymorphism in the expression of miRNA-433 increases the Fibroblast Growth Factor 20 (FGF20) gene expression. FGF20 ultimately upregulates the downstream target SNCA (Itoh and Ohta, 2013). Li et al. (2014) demonstrated that miRNA-320a plays an essential role in SNCA aggregation during PD pathogenesis.
Alzheimer’s disease is one the primary neurodegenerative disease affecting 44 million people around the world, and statistics reveal that only one-in-four people affected with AD have been diagnosed (Basak et al., 2016). Several studies demonstrated that miRNA-148a, miRNA-17-5p, miRNA-137, miRNA-181c, miRNA-101, miRNA-184, miRNA-15a, miRNA-185, and miRNA-210 are few of those miRNAs that are expressed in AD (Lukiw, 2007; Cogswell et al., 2008; Hébert et al., 2008; Geekiyanage and Chan, 2011; Wang et al., 2011). Tauopathies involved in the pathogenesis of AD have many key proteins, which are regulated by miRNAs. Dickson et al. (2013) demonstrated that endogenous tau aggregates were repressed by miRNA-34a in M17D cell model of the AD. Similarly, many researchers found that the amyloid precursor protein (APP) expression is also influenced by miRNA-101, miRNA-16, miRNA-106a, and miRNA-644 (Patel et al., 2008; Delay et al., 2011; Long and Lahiri, 2011; Liu et al., 2012). Furthermore, the amyloid protein fragment amyloid beta (Aβ), is also influenced by miRNA-24, miRNA-186, miRNA-455, miRNA-146a, and miRNA-98 (Delay et al., 2011; Li Y.Y. et al., 2011; Hu et al., 2013). The levels of Aβ in AD patients have been speculated to be regulated by miRNA-137, miRNA-146a, and miRNA-181c (Delay et al., 2011; Geekiyanage and Chan, 2011; Li Y.Y. et al., 2011; Hu et al., 2013). Interestingly, there are many miRNAs, such as miRNA-107, miRNA-29a-1/b-1, miRNa-9 and miRNA-124, which regulate the APP-processing enzyme b-secretase enzyme I (BACE1) in AD patients (Bossy-Wetzel et al., 2004; Wang et al., 2010; Fang et al., 2012). Some miRNAs have a substantial influence on age-related disorders such as AD. The anti-aging factor sirtuin-1 (SIRT1) plays a vital role in enhancing the neuronal survival and alleviating neurodegeneration in AD and ALS models. The SIRT1 expression is also regulated by miRNA-34a (Kim D. et al., 2007; Li X. et al., 2011; Smit-McBride et al., 2014). The demented aging brain showed an inverse relationship between SIRT1 and miRNA-34a. Similarly, it has been demonstrated that miRNA-144 can possibly bind to the 3UTR of programmed cell death protein 4 (PDCD4), which shows that miRNA-144 might be crucial in apoptotic mechanisms during AD development (Persengiev et al., 2011). Recently Wang Y. et al. (2017) showed that miRNA-132/212 impaired S-nitrosylation and induced tau phosphorylation via NOS1 pathway. Similarly, Lau et al. (2013) studied 90 samples of human brain affected with the AD, and found that miRNA-132-3p was strongly downregulated. In addition, the transcription factor FOXO1a was the target of miRNA-132-3p, which plays a crucial role in tau network (Lau et al., 2013). Song and Kim (2017) showed that miRNA-142-5p plays an important role in the pathogenesis of AD, as it is upregulated in the brains of AD patients and inhibition of miRNA-142-5p resulted in the rescue of synaptic dysfunction in Aβ42-treated SH-SY5Y cells. Zhang et al. (2017) demonstrated that miRNA-200a-3p promotes Aβ-induced neuronal apoptosis via downregulation of SIRT1 in a transgenic mouse model of the AD and PC12 cells. Kumar et al. (2017) demonstrated that upregulation of miRNA-455-3p, miRNA-4668-5p, miRNA-3613-3p, and miRNA-4674, while downregulation of miRNA-6722 in 10 AD positive brain samples.
While the misfolded protein diseases often feature complex interactions between aggregates of multiple proteins, all misfolded proteins share a common structural feature, known as the amyloid folds (Andreoletti et al., 2000, 2002). Prion disease pathogenesis is complex, and the involvement of miRNAs deregulation in prion disease pathogenesis is still unclear. It is demonstrated that during preclinical stage of prion disease, miRNAs abundant in synaptoneurosomes including miRNA-124a-3p, miRNA-136-5p and miRNA-376a-3p are increased. While in terminal stage of prion infection miRNA-146a-5p, miRNA-142-3p, miRNA-143-3p, miRNA-145a-5p, miRNA-451a, miRNA-let-7b, miRNA-320, and miRNA-150-5p are significantly elevated in the brains of prion-infected animals. miRNA-124 and miRNA-126 are also upregulated in prion, AD and PD patients (Lukiw, 2007; Cogswell et al., 2008; Martins et al., 2011; Bellingham et al., 2012). During clinical phase of prion disease all the members of the miRNA-200 family (miRNA-200a-3p, miRNA-200b-3p, and miRNA-200c-3p), miRNA-141-3p, and miRNA-429-3p and the 182 cluster (miRNA-182-5p and miRNA-183-5p) are downregulated (Boese et al., 2016). Similarly, miRNA-146a expression is upregulated in CJD and GSS patients (Lukiw et al., 2011). Recently, miRNA-200 has been found upregulated in AD models (Zhang et al., 2017). Bellingham et al. (2012) demonstrated that let-7b, let-7i, miRNA-128a, miRNA-21, miRNA-222, miRNA-29b, miRNA-342-3p, and miRNA-424 levels were upregulated, while miRNA-146a levels were downregulated in prion-infected individual. Different miRNAs profiles were observed in the brains of BSE- infected cynomolgus macaques (Montag et al., 2009). Through miRNA-microarray and qRT-PCR analysis, two miRNAs miRNA-342-3p and miRNA-494 were significantly upregulated in the brains of BSE-infected macaques compared to non-infected animals. In addition, the miRNA miRNA-342-3p was also upregulated in the brains of human type 1 and type 2 sporadic CJD patients. Similarly, miRNA-342-3p was also upregulated in scrapie-infected mice brains (Saba et al., 2008; Montag et al., 2009). These studies reveal that miRNA-342-3p might be a potential biomarker in the terminal stage of prion diseases. No validation of this hypothesis exists, due to the lack of knowledge about the target mRNAs for miRNA-342-3p. Although, the TargetScan algorithm shows some important genes targeted by miRNA-342-3p, such as ataxin 7, tau tubulin kinase 2 and huntingtin interacting protein 1 (HIP1). It is worthwhile to note, that these genes are primarily involved in the protein misfolding disorders. Further confirmation of the significance of miRNA-342-3p in prion diseases is required due to the small number of experimental animals and human brains available for the above-mentioned study (two sCJD and one healthy control) (Saba et al., 2008). Similarly, miR-342-3p and miR-21-5p are upregulated in the brains of sheep naturally infected with prion disease (Sanz Rubio et al., 2017).
Neuroinflammation and neurodegeneration are the hallmarks of prion diseases, and miRNA-146a plays a crucial role in activating the innate immune response and microglial activation during the course of prion diseases (Saba et al., 2012). Saba et al. (2012) showed that miRNA-146a is overexpressed during prion infection and microglial lineage cell lines, such as BV2 are directly modulated via TLR2 or TLR4 receptors. miRNA-146a mimic decreased the expression of interleukin-1 beta (IL-1β) and interleukin-18 (IL-18), while increased the expression of interleukin-10 (IL-10) (Saba et al., 2012). The role of miRNA-146a in the regulation of inflammatory responses during prion diseases and its modulatory effect on nuclear factor-kappa and the Janus kinase (JAK) signal transducer and activator of transcription (JAK-STAT) signaling makes it a new and attractive therapeutic target for prion diseases (Figure 3) (Saba et al., 2012). Similarly, overexpressed miRNA-146 is observed in prion and AD patients (Cogswell et al., 2008; Bellingham et al., 2012). During the preclinical phase of prion disease, a cluster of genes and miRNAs are dysregulated, such as miRNA-132-3p, miRNA-124a-3p, miRNA-16-5p, miRNA-26a-5p, miRNa-29a-3p, and miRNA-140-5p, and they follow associated patterns of expression (Majer et al., 2012). miRNA-125 is upregulated in prion, AD, and PD models (Cogswell et al., 2008; Bellingham et al., 2012; Cardo et al., 2013). Recently, Gao et al. (2017) worked on miRNA expression profile of three prion strains, 139A, ME7, and S15. They found that the most significantly upregulated and downregulated miRNAs in scrapie-infected mice were, mmu-miRNA-3473e (9.48 log2) and mmu-miRNA-141-5p (-7.33 log2) in the 139A group, mmu-miRNA-3473e (13.18 log2) and mmu-miRNA-200a-5p (-7.08 log2) in the ME7 group, and mmu miRNA-3473e (14.15 log2) and mmu-miRNA-183-3p (-10.15 log2) in the S15 group (Gao et al., 2017). Recently, Burak et al. (2018) demonstrated that miRNA-16 localized within the CA1 region of the hippocampus was upregulated in the early stage of prion infection. The novel targets of miRNA-16 were APP, BCL2, MAPK, and ERK. They showed that miRNA-16 plays a crucial role in neurite growth and branching (Burak et al., 2018). Similarly, Miller et al. (2013) also showed the upregulation of miRNA-16 in PD patients. Llorens et al. (2018) demonstrated the miRNA signature in frontal cortex (FC) and cerebellum (CB) of sporadic CJD patients. In sCJD FC, miRNAs 29b-3p, 342-3p, 146a-5p, 154-5p, 195-5p, 26a-5p, 16-5p, 449a, 142-3p, let7i-5p, and 135a-5p were upregulated, while miRNAs 124-3p, 331-3p, 877-5p, and 125a-5p were downregulated compared to controls. miRNAs 378a-3p and 5701, which expression was only altered in CB did not present changes in the FC. In CB, miRNAs 146a-5p, 154-5p, 26a-5p, 378a-3p, 449a, 142-3p, let7i-3p, and 5701 were upregulated, and miRNAs 124-3p and 877-5p were downregulated in sCJD. The rest of miRNAs, which expression was only altered in FC did not present changes in the CB (Llorens et al., 2018).
The essential cellular quality control pathways for prion protein degradation are ubiquitin-proteasomal system (UPS) and autophagy-lysosomal pathways (Goold et al., 2015; Shah et al., 2017b). UPS involves the attachment of ubiquitin chains, which are then recognized by proteasome for subsequent degradation. The substrate proteins are ubiquitinated in a three-step mechanism: firstly, an enzyme (E1) which activates the ubiquitin covalently binds with ubiquitin moiety in an ATP-dependent manner, secondly; already formed ubiquitin moiety is then exposed to another enzyme called E2 ubiquitin conjugation enzyme. In the third and final step, a third enzyme called E3 ubiquitin ligase controls the reaction based on the specificity of the substrate. These ubiquitination steps are run in several rounds to prolong the ubiquitin chain, and it will result in polyubiquitination of the target substrate protein. There are various modes of ubiquitination, dependent on the lysine residue that is used for connecting with different ubiquitin moieties. The proteasomal degradations are carried out via lysine-48 (K48). The substrates recognized for ubiquitination through K48 are targeted by the proteasome ubiquitination system, and they are eventually unfolded and degraded. While the ubiquitin used for all this process is cleaved off and recycled (Kang et al., 2004; Goold et al., 2015).
Autophagy is a bulk degradation process that starts with the development of a crescent shape double membrane known as phagophore, which then engulfs the damaged organelle or cargo, which is supposed to be degraded to form autophagosome. The autophagosome gets mature and fused with a lysosome to form autolysosome. The acidic lysosomal proteases degrade the cargo within the autolysosome (Figure 2) (Goold et al., 2015). Ubiquitin molecules also tag the substrate recognized for degradation by autophagy. Substrates are labeled by the ubiquitin and like UPS, which is dependent on the lysine residue; here the linkage is via lysine-63 (K63), and ultimately the autophagy initiation signals are started. The autophagy adaptor proteins recognize the polyubiquitin signals which mediate and simultaneous bind the ubiquitinated cargo with the autophagic machinery. Various adaptor proteins were recognized in different species including yeast and in mammalian cells. All these adapter proteins are characterized by the ubiquitin-binding domain. The best example of which is SQSTM1/p62 protein. The p62 protein posses a domain called ubiquitin-associated domain (UBA). UBA is involved in ubiquitin binding and an LC3-interacting region (LIR), which attaches with autophagosomes. Several researchers demonstrated that p62 protein plays a crucial role in the degradation of misfolded aggregated proteins and mitophagy (Ding et al., 2010; Geisler et al., 2010; Tung et al., 2010). Many scientists used p62 as a marker for staining the protein aggregates in various proteinopathies, including neurodegenerative diseases. This further elaborates the role of selective autophagy in the pathogenesis of misfolded protein aggregation diseases (Zatloukal et al., 2002) Neurodegenerative diseases like prion, AD, PD and Huntington’s disease are characterized by the deposition of intra and extracellular protein aggregation, which cannot be degraded by proteasomes. Thus, activation of autophagy is seen as an alternative therapeutic strategy for protein aggregation diseases.
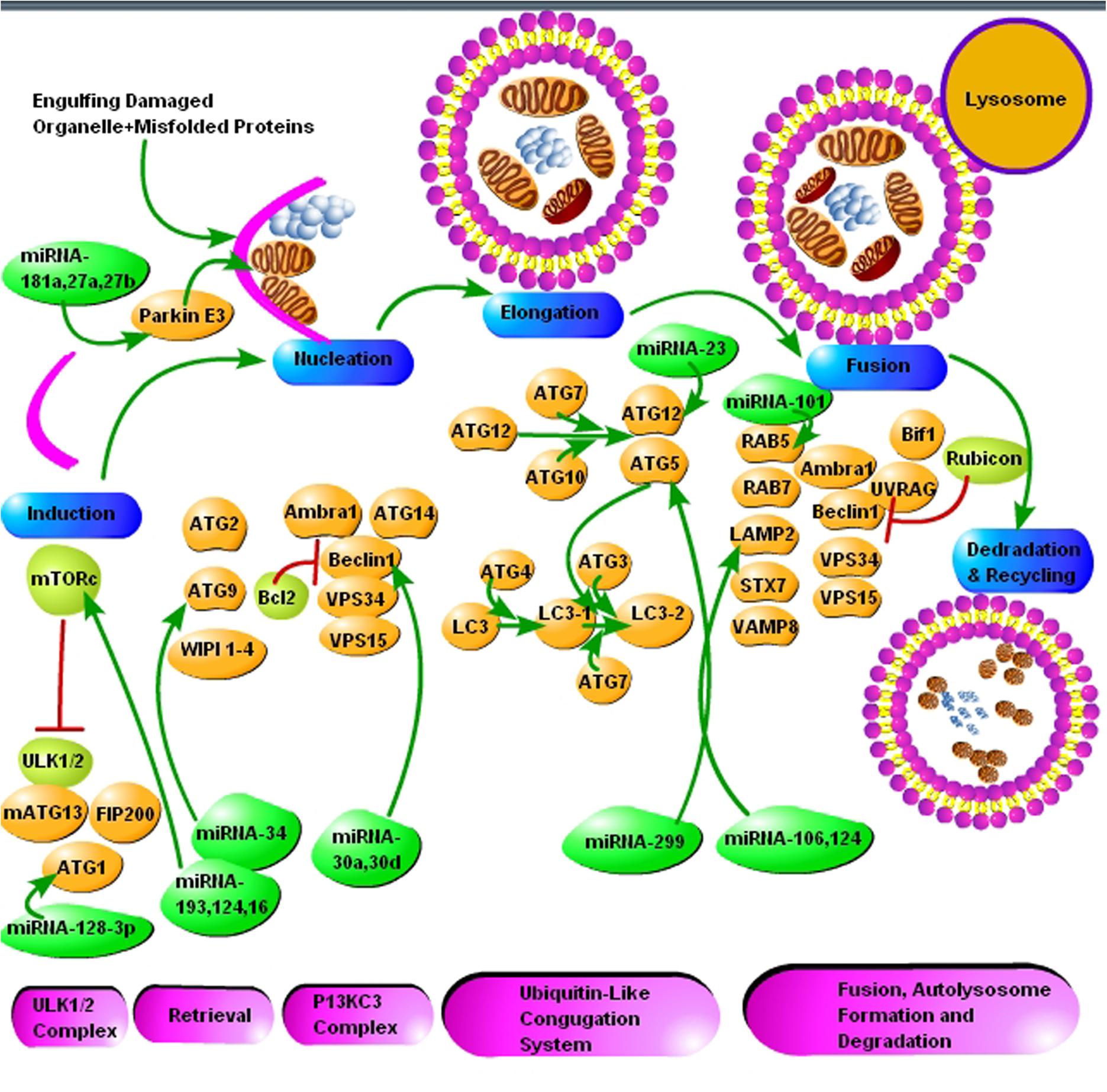
FIGURE 2. The schematic presentation of autophagy pathway and core autophagy-related proteins targeted by various miRNAs. The figure shows some significant miRNAs (dark green color) and their respective targets (light green and yellow color) for the regulation of autophagic mechanisms in Alzheimer’s and Parkinson’s disease. The figure shows that insufficient research has been conducted in neurodegenerative diseases for the regulation of autophagy via miRNAs and further molecular research is encouraged in all major neurodegenerative diseases including prion diseases.
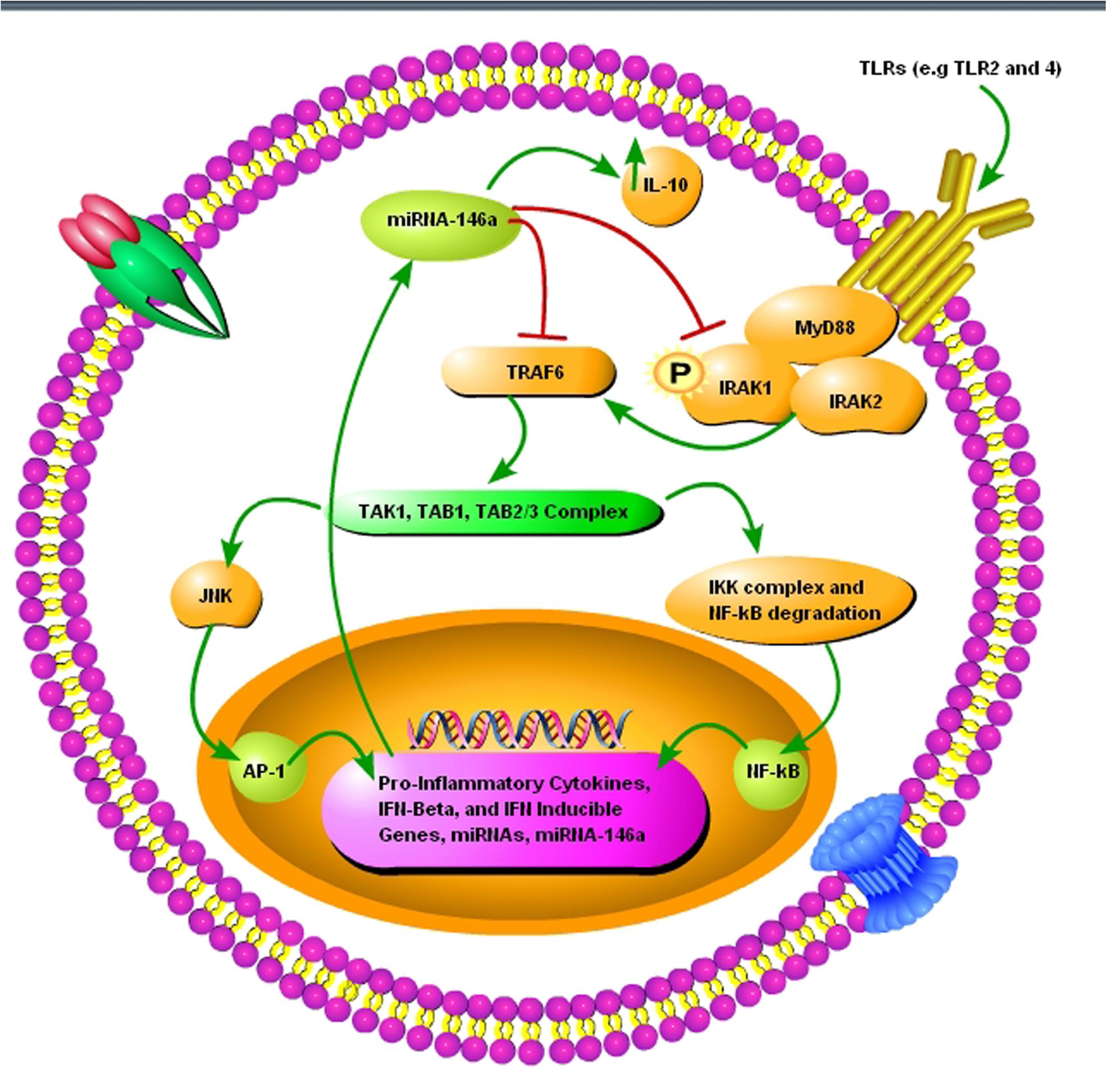
FIGURE 3. A schematic diagram showing the importance of pro-inflammatory miRNA-146a during prion infection. (1) The toll-like receptors TRL2 and TLR4 are activated upon misfolded prion protein formation. (2) TNF receptor-associated factor 6 (TRAF6) and interleukin-1 receptor-associated kinase 1 (IRAK1), interleukin-1 receptor-associated kinase 2 (IRAK2), myeloid differentiation primary response gene 88 (MyD88) complexes are modulated in response to activated TLR2 and TLR4 receptors. (3) TRAF6 activates the c-Jun N-terminal kinases (JNK) and IκB kinase (IKK) complex via TGF-beta activated kinase 1 (TAK1), TAK1-binding protein 1 (TAB1), and (TAB2/3) complex. (4) NF-κB degradation and nuclear translocation plus activator protein 1 (AP-1) activation via JNK result in upregulation of proinflammatory cytokines and several miRNAs including miRNA-146a. (5) Upregulated miRNA-146a inhibits IRAK1 and TRAF6 signaling to increase interleukin-10 (IL-10) secretion and reduce inflammation.
Autophagy is a primary cellular housekeeping process for the degradation and recycling of cellular components that maintains energy homeostasis via degradation of aggregated proteins within the cell (Clarke, 1990; Yoshimori, 2004). The induction of autophagic flux plays an essential role in the degradation of misfolded prion proteins (Yao et al., 2013). Any cellular modification leading to the disruption in autophagic mechanisms via the use of pharmacological interference or siRNA gene silencing dramatically reduce the efficiency of compound-induced autophagy, and ultimately the cellular levels of PrPSc remained high (Heiseke et al., 2010). Recent molecular techniques such as small molecules for the induction of autophagy have also shown the protective mechanism of autophagy in neurodegenerative diseases. Heiseke et al. (2009) demonstrated that lithium-induced clearance of PrPSc within the prion-infected cells was solely dependent on increased autophagy via mTOR-independent manner (Yao et al., 2013). Recently Moon et al. (2016b) demonstrated that plant extract hinokitiol successfully alleviated the prion-peptide induced toxicity via restoration of autophagic flux in primary neurons. Similarly, ginsenoside-rg3 attenuated prion peptide-induced neurotoxicity via autophagic flux through modulation of mitochondrial dysfunction (Moon et al., 2016a).
Mice treated with autophagy inducer rapamycin showed a prolonged incubation period in comparison with the vehicle-treated control mice (Cortes et al., 2012). Similarly, autophagy was protective against de novo formation of PrPSc in a yeast model of prion diseases, and increased autophagic flux resulted in the degradation of aggregated PrPSc proteins (Speldewinde et al., 2015). In another study, rapamycin reduced the quantity of misfolded PrPSc in a mTOR-dependent manner. Rapamycin treatment significantly improved mitochondrial dysfunction, neuroinflammation, and neurodegeneration in a mouse (Johnson et al., 2013). Recently Wang H. et al. (2017) demonstrated that overexpression of Polo-like kinase 3 (PLK3) played an important role in the degradation of PrPSc proteins. Some researchers proposed that beneficial effects of rapamycin are related to the indirect inhibition of protein synthesis, ultimately maintaining the ATP levels within the infected cells (Zheng et al., 2016). Modern molecular techniques such as small molecules can stimulate autophagy in mTOR-dependent and mTOR-independent manners. This shows the importance of small molecules for the possible treatment of patients with prion diseases by targeting mTOR-dependent and mTOR-independent pathways simultaneously. More detailed in vivo studies showing the prospects of small molecules as a treatment for prion diseases are necessary.
The miRNAs contribute to the development of neurons, neuritogenesis and synapse growth. miRNAs are also involved in the CNS ischemia, spinal cord injury, and traumatic brain injury (Bellingham et al., 2012). The prion gene-associated miRNAs play a vital role in neuronal apoptosis, differentiation, development, synaptic integrity, and neurogenesis (Shapshak, 2013). Studies involving prion-infected mouse brain showed that various gene promoters are related to miRNAs. Some important gene promoters identified were E2F-1 (cell cycle re-entry and neurodegeneration), MAZ (inflammatory response transcription factor), PAX6 (neurogenesis), KROX (transcription factor), and early growth response 1 (EGR1). Additionally, human studies involving the brains of individuals affected with prion diseases showed the effect on miRNAs on N-methyl-D-aspartic acid (NMDA) receptors and glutamate receptors (ionotropic, N-methyl-D-aspartate 2A, GRIN2A) (Majer et al., 2012; Saba et al., 2012).
Perturbation in the clearance of misfolded proteins by autophagic flux and UPS is the hallmark of all prion disorders. The UPS pathway is comprised of over 30 genes that are involved in the dysregulated miRNAs and been identified in prion diseases (Saba et al., 2008). Some of these important genes are involved in ubiquitin, ubiquitin-conjugating enzymes, and the ubiquitin protein ligase NEDD4. Unfolded-protein response genes are also the primary targets, such as valosin-containing protein, VCP (CDC48/p97). VCP plays an essential role in the shuttling of ubiquitinated proteins from the endoplasmic reticulum (ER) to the proteasome system, and the transcription factor X-box binding protein 1, XBP1, activated upon the accumulation of misfolded proteins in the ER (Saba et al., 2008).
Neuroinflammation is the hallmark of all protein misfolding diseases including prion diseases, and one genuinely noteworthy finding is that a considerable upregulation of inducible, pro-inflammatory pathogenic miRNAs such as miRNA-34a and/or miRNA-146a are collectively shared by AD and prion diseases (Hill and Lukiw, 2015; Zhao et al., 2015a,b). As these pro-inflammatory miRNAs are also crucial for plaque formation during AD and prion diseases, it could be involved in the regulation of autophagic mechanism during prion diseases. The detection of those miRNAs involved in the pathogenesis of disease process and the following interacting proteins provide several important junctures that might be targeted in devising treatment strategies and possible cures for prion diseases.
Protein aggregation or accumulation is a biological phenomenon commonly observed in neurodegenerative disorders known as amyloidosis, including ALS, AD, PD, and prion disease. The clearance of these aggregated proteins is necessary for cell survival and functioning. The aggregated proteins are mainly cleared through the UPS (Yedidia et al., 2001), but severe ER stress and a significant amount of aggregated proteins lead to dysfunction of the UPS (Jellinger, 2010). The failure of the UPS leads to triggering another clearance mechanism known as autophagy (Figure 2) (Lynch-Day et al., 2012). Defective UPS and autophagy are linked to the pathogenesis of AD, PD, and prion diseases (Kristiansen et al., 2007; Schapira and Gegg, 2011).
Autophagy does not degrade miRNAs directly, but it influences the proteins targeted by miRNAs and hence miRNAs-mediated repression of appropriate target genes is hampered. Gibbings et al. (2012) demonstrated that autophagy selectively degrades miRNA-processing enzyme, Dicer1 and the main miRNA effector protein, Ago2, by selective autophagy receptor NDP52, ultimately regulating miRNAs biogenesis. This shows that NDP52 and autophagy plays a crucial role in the homeostasis and activity of miRNAs (Gibbings et al., 2012). Similarly, it is demonstrated that miR-106a and miR-224 are upregulated in the brains of PD patients, ultimately leading to impaired chaperone-mediated autophagy (CMA) and α-synuclein accumulation. miR-106a and miR-224 cause a dose-dependent reduction in heat shock 70 kDa protein (hsc70) and lysosome-associated membrane protein 2 (LAMP-2A), respectively, to impair autophagic mechanism in SH-SY5Y cells (Alvarez-Erviti et al., 2013). Both hsc70 and LAMP-2A are active mediators of autophagy, and they play a crucial role in CMA (Majeski and Dice, 2004). Li et al. (2014) showed that miRNA-320a targeted the 3′ UTR of hsc70, decreased hsc70 expression in α-synuclein-overexpressed SH-SY5Y cells, and resulted in significant α-synuclein intracellular accumulation. miRNA-34 mutations extend the life of Caenorhabditis elegans by increasing autophagic flux. Furthermore, miR-34 represses autophagy by directly inhibiting the expression of the autophagy-related protein Atg9 in mammalian cells (Yang et al., 2013). Recently, Valera et al. (2017) demonstrated that overexpression of miRNA-101 in oligodendroglial cell cultures resulted in a significant increase of α-synuclein via disruption of autophagy. The predicted target of miRNA-101 was autophagy-related gene RAB5A. The stereotaxic injection with anti-miRNA-101 into the striatum of a mouse model of multiple system atrophy resulted in reduced oligodendroglial α-syn accumulation and enhanced autophagy (Valera et al., 2017). The role played by miRNAs in the regulation of autophagy is further supported by the work where essential autophagy-promoting protein Beclin 1(BECN1) was directly regulated by miRNA-30a. Furthermore, miRNA-206 regulated HDAC4, which is linked with autophagy (Williams et al., 2009; Zhu et al., 2009). Another miRNA known as miRNA-29a/b-1, downregulated in the brains of sporadic AD patients causes dysregulation of proteins like, APP or BACE1/β-secretase, resulting in an autophagy-dependent cell toxicity and death events (Hébert et al., 2008).
Li et al. (2017) demonstrated that miRNA-193b-3p plays a vital role in the pathogenesis of ALS. Downregulation of miRNA-193b-3p promoted autophagy and cell survival by targeting TSC1/mTOR signaling pathway in NSC-34 cell model of ALS. In contrast, upregulation of miRNA-193b-3p activated mTORC1 signaling leading to the inhibition of autophagy and promoted cell death (Li et al., 2017). Antagomirs of miRNA-134 rescued epileptic rats from lithium chloride-pilocarpine-induced status epilepticus via regulation of oxidative stress, mitochondrial dysfunction, and autophagy. Anti-134 treatment downregulated the expression of autophagy-associated proteins ATG 5, Beclin-1, and LC3-b, to enhance memory and survival in mouse model of epilepsy (Sun J. et al., 2017). miRNA-30d rescued rat astrocytes from oxygen and glucose deprivation by targeting Beclin 1 in cell injury under hypoxic condition (Zhao F. et al., 2017). miRNA-124 participates in neuronal protection against ischemic stroke by targeting P13K/Akt signaling pathway in PC12 cells (Wang C.M. et al., 2017; Wang Y. et al., 2017). Moreover, miRNA-124 is protective of dopaminergic neurons via regulation of AMPK/mTOR pathway in SH-SY5Y cell model of PD (Gong et al., 2016). Sun L. et al. (2017) showed that miRNA-23b improved cognitive impairment in a traumatic brain injury rat model by targeting ATG 12-mediated neuronal autophagy. miRNA-299-5p downregulation is observed in APPswe/PS1dE9 mice, cerebrospinal fluid of AD patients, N2a and SH-SY5Y cell models of the AD. It is demonstrated that miRNA-299-5p regulates autophagy by targeting ATG 5 directly (Zhang et al., 2016). The role of miRNA-16 in reversing autophagic and apoptotic changes during chronic stress are studied recently in a rat model of chronic stress. It is observed that miRNA-16 restores BDNF levels by downregulating p13-Akt-mTOR pathway (Yang et al., 2017). The involvement of miRNA-128-3p expression by directly targeting ATG 1 in cerebral ischemia rat model has been established recently (Shi et al., 2016). Cheng et al. (2016) found that miRNA-181a is a novel inhibitor of mitophagy and it directly targets Parkin E3 ubiquitin ligase. Similarly, miRNA-181a regulated apoptosis and autophagy in PD by inhibiting the two crucial p38 mitogen-activated protein kinase (MAPK)/c-Jun N-terminal kinases (JNK) signaling pathways (Liu et al., 2017). miRNA-27a and miRNA-27b play a vital role in the autophagic clearance of injured mitochondria by targeting PTEN-induced PINK1 in PD (Kim et al., 2016).
Kang et al. (2017) demonstrated that dual miRNA to cellular prion protein successfully inhibited the propagation of pathogenic prion in a mixed culture of neuronal and glial cells. Furthermore, this reduction in PrPSc was cell type-specific as no reduction was observed in reserve cells. Interestingly, a 20% decrease in PrPc levels led to more than 60% decreases in pathogenic misfolded form PrPSc (Kang et al., 2017). Although, this study did not focus on the role played by autophagy in the degradation of misfolded prions. However, it encourages the researchers that if an artificial dual miRNA and siRNA combination can reduce the pathogenic PrPSc levels substantially, then there might be prospects for finding the naturally present miRNAs that have an important role in targeting the autophagy-related proteins, and ultimate goal of PrPSc free cell can be achieved.
Our research group focused on the activity of repressor element-1 binding transcription factor (REST), or neuron-restrictive silencer factor (NRSF), in prion diseases (Song et al., 2016, 2017a,b). REST is an important monitor for the establishment of neuronal specificity, and it is highly expressed in neuronal tissues (Zhao Y. et al., 2017). Interestingly, REST is the target of many brain-derived miRNAs such as miR-124a, miR-9, and miR-132 (Wu and Xie, 2006). Lu et al. (2014) demonstrated that REST expression in the nucleus of neurons in the AD, frontotemporal dementia, and dementia with Lewy bodies patients was lower as compared to age-matched controls. REST is misplaced from the nucleus and can be found in autophagosomes together with pathological misfolded proteins (Lu et al., 2014). We found similar results with the PrP peptide 106–126-induced neurotoxicity in primary neurons and N2a cells after 24 h exposure (Song et al., 2016). Cho et al. (2014) demonstrated that REST regulates mTOR-signaling pathway in oral carcinoma. We also found REST mediated regulation of Akt-mTOR and Wnt-β-catenin pathway in 263K infected hamsters (Song et al., 2017a). We can speculate that REST-directed miRNA therapies in neurodegenerative diseases may be practical in regulating autophagic flux. The knowledge from these recent studies could play a crucial role in finding the therapeutic remedy toward autophagy regulation in prion-related neuropathologies as well.
As we know that upregulation or downregulation of a miRNA is dependent on the expression of a specific gene, so modern molecular techniques such as Clustered, Regularly Interspaced, Short Palindromic Repeats (CRISPER) -CRISPR-associated 9 (Cas9) can be used to control the expression levels of specific miRNAs. The CRISPER-Cas9 technology work by delivering the Cas9 nuclease complexed with a synthetic guided RNA (gRNA) into a cell, the cell’s genome can be cut at the desired location, allowing existing genes to be removed and/or new ones added (Hendel et al., 2015). The CRISPER-Cas9 is a gene editing technique, simply designed, with high efficiency at relatively low cost, and it has replaced other old techniques for gene manipulation. Recently, Chang et al. (2016) showed that CRISPER-Cas9 targeted miRNA-17, miRNA-200c and miRNA-141, repressed their activity in human colon cancer cell lines HCT116 and HT-29. Furthermore, in vivo targeting was effective for at least a month (Chang et al., 2016). However, off-target mutagenesis and effects of a single miRNA on various gene targets are the limitations to the use of this modern technology specifically in brain disorders like prion diseases (Hendel et al., 2015). Beside CRISPER-Cas9 there is another technique known as anti-miRNA oligonucleotides (AMOs), used to control cellular mechanics. These are synthetically designed molecules, used to neutralize the miRNAs function in cells for desired responses (Figure 4). By controlling miRNAs that regulate mRNAs expression in the cells, AMOs can be used for therapeutic treatment of certain neurodegenerative disorders. AMOs uses the steric-blocking mechanism for the regulation of miRNAs, and hybridization to target miRNAs (Lennox and Behlke, 2011). In order to determine the functionality of certain AMOs, the AMO/miRNA binding expression (transcript concentration) must be measured against the expressions of the isolated miRNA. The direct detection of differing levels of gene expression allows the relationship between AMOs and miRNAs. This can be detected through luciferase activity. AMOs are commonly used for the diseases involving miRNAs such as cancers, muscular diseases, autoimmune disorders, and viruses. Their efficacy in neurological disorders is still unclear, and there are some major constraints on to the delivery of AMO’s in the brain. Some of these constraints are the poor in vivo stability of AMO’s, Inappropriate biodistribution of AMO’s, disruption and saturation of all endogenous RNA machinery after AMO’s therapy and side effects of AMO’s related therapies. Hence, further molecular research is required for their therapeutic use in neurodegenerative disorders.
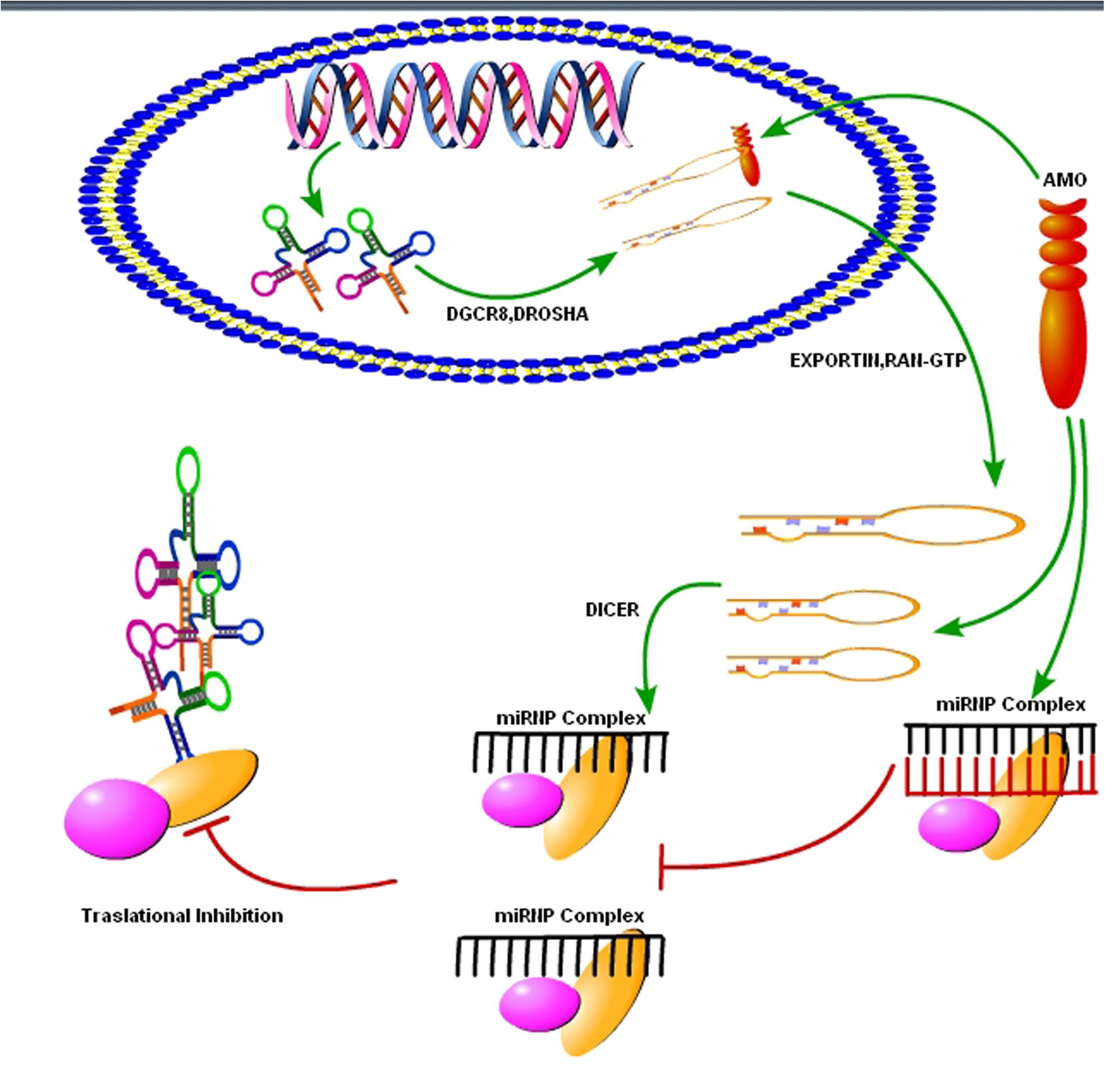
FIGURE 4. Use of synthetic oligonucleotides to interfere the activity of miRNAs. The Inhibition of the activity of miRNA may be achieved by using anti-miRNA oligonucleotides (AMOs) fully complementary to the pri-miRNA, the pre-miRNA or the mature miRNA (figure adapted from Weiler et al., 2006).
Many research groups focus on prion diseases since the discovery of a fatal prion disease in humans known as variant CJD (Somerville et al., 1997; Lezmi et al., 2003). Recently, molecular research increasing the lifespan in different animal models is a promising start toward finding the best possible therapeutic intervention strategy in prion diseases. Still, there is no available therapeutic strategy curing prion-afflicted individuals completely (Castilla et al., 2008). Recently, miRNAs-mediated molecular research in the field of neurodegenerative diseases, such as AD, PD, HD, and prion diseases has made significant progress. Autophagy plays a crucial role in protein misfolding neurodegenerative diseases. Autophagic flux is necessary for the degradation of misfolded/aggregated PrPSc in prion diseases (Speldewinde et al., 2015), and regulation of autophagic flux via miRNAs may prove beneficial in these diseases, as seen in other neurodegenerative diseases. The role of miRNAs in modulating autophagy is still unclear, and there are some research groups that found beneficial effects of reduced autophagy (Zhang et al., 2016), while on the other hand, some research groups found promotion of autophagic flux as a useful therapeutic strategy to reduce toxic effects of misfolded proteins (Kim et al., 2016). We propose a hypothesis based on previous data in the field of prion-related research, where several groups found autophagic flux as a useful therapeutic strategy toward degradation of misfolded prion proteins, and ultimately reducing the number of damaged organelles (Johnson et al., 2013; Speldewinde et al., 2015). This could be achieved through miRNAs-mediated regulation of autophagy. Some research groups already worked on miRNAs profile of prion disease-infected animals and individuals, but to date, none of the research group focused on miRNAs-mediated regulation of autophagy in prion diseases. Therefore, the starting point would be to identify those miRNAs, which regulate autophagy in prion diseases. Next step after identification of these miRNAs would be upregulation or downregulation of these miRNAs to regulate autophagic flux in prion diseases. Neuroinflammation and neurodegeneration are hallmarks of prion diseases, and discovery of miRNAs that are important for reducing neuroinflammation and neurodegeneration via modulating autophagy could make a breakthrough toward achieving the goal of therapeutic intervention in prion diseases.
SS wrote the manuscript. DZ conceived the idea for the study. TH and NS helped with figure preparation. LY critically reviewed the manuscript before final submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ALCC and handling Editor declared their shared affiliation.
This work was supported by Natural Science Foundation of China (Project No. 31472166), Ministry of Agriculture of China, 948 projects (2014-S9), and Chinese Universities Scientific Fund (Project No. 2017DY003).
Alexandrov, P. N., Dua, P., Hill, J. M., Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2012). microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 3, 365–373.
Alvarez-Erviti, L., Seow, Y., Schapira, A. H., Rodriguez-Oroz, M. C., Obeso, J. A., and Cooper, J. M. (2013). Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson’s disease. Cell Death Dis. 4:e545.
Andreoletti, O., Barthon, P., Marc, D., Sarradin, P., Grosclaude, J., Keulen, L. V., et al. (2000). Early accumulation of PrPSc in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J. Gen. Virol. 81, 3115–3126.
Andreoletti, O., Lacroux, C., Chabert, A., Monnereau, L., Tabouret, G., Lantier, F., et al. (2002). PrPSc accumulation in placentas of ewes exposed to natural scrapie: influence of foetal PrP genotype and effect on ewe-tolamb transmission. J. Gen. Virol. 83, 2607–2616.
Aumiller, V., and Forstemann, K. (2008). Roles of microRNAs beyond development–metabolism and neural plasticity. Biochim. Biophys. Acta 1779, 692–696. doi: 10.1016/j.bbagrm.2008.04.008
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Basak, I., Patil, K. S., Alves, G., Larsen, J. P., and Moller, S. G. (2016). microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell. Mol. Life Sci. 73, 811–827. doi: 10.1007/s00018-015-2093-x
Bellingham, S. A., Coleman, B. M., and Hill, A. F. (2012). Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 40, 10937–10949. doi: 10.1093/nar/gks832
Bhattacharjee, S., Zhao, Y., Dua, P., Rogaev, E. I., and Lukiw, W. J. (2016). microRNA-34a mediated down-regulation of the microglial-enriched triggering receptor and phagocytosis-sensor TREM2 in age-related macular degeneration. PLoS One 11:e0150211. doi: 10.1371/journal.pone.0150211
Boese, A. S., Saba, R., Campbell, K., Majer, A., Medina, S., Burton, L., et al. (2016). MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol. Cell. Neurosci. 71, 13–24. doi: 10.1016/j.mcn.2015.12.001
Bossy-Wetzel, E., Schwarzenbacher, R., and Lipton, S. A. (2004). Molecular pathways to neurodegeneration. Nat. Med. 10, S2–S9.
Bruno, I. G., Karam, R., Huang, L., Bhardwaj, A., Lou, C. H., Shum, E. Y., et al. (2011). Identification of microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell. 42, 500–510. doi: 10.1016/j.molcel.2011.04.018
Burak, K., Lamoureux, L., Boese, A., Majer, A., Saba, R., Niu, Y., et al. (2018). MicroRNA-16 targets mRNA involved in neurite extension and branching in hippocampal neurons during presymptomatic prion disease. Neurobiol. Dis. 112, 1–13. doi: 10.1016/j.nbd.2017.12.011
Burgos, K., Malenica, I., Metpally, R., Courtright, A., Rakela, B., Beach, T., et al. (2014). Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 9:e94839. doi: 10.1371/journal.pone.0094839
Cardo, L. F., Coto, E., de Mena, L., Ribacoba, R., Moris, G., Menéndez, M., et al. (2013). Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J. Neurol. 260, 1420–1422.
Cardo, L. F., Coto, E., Ribacoba, R., Menendez, M., Moris, G., Suarez, E., et al. (2014). MiRNA profile in the substantia nigra of Parkinson’s disease and healthy subjects. J. Mol. Neurosci. 54, 830–836. doi: 10.1007/s12031-014-0428-y
Castilla, J., Gonzalez-Romero, D., Saa, P., Morales, R., Castro, J. D., and Soto, C. (2008). Crossing the species barrier by PrPSc replication in vitro generates unique infectious prions. Cell 134, 757–768. doi: 10.1016/j.cell.2008.07.030
Chang, H., Yi, B., Ma, R., Zhang, X., Zhao, H., and Xi, Y. (2016). CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 6:22312. doi: 10.1038/srep22312
Cheng, M., Liu, L., Lao, Y., Liao, W., Liao, M., Luo, X., et al. (2016). MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 7, 42274–42287. doi: 10.18632/oncotarget.9786
Cho, H. J., Liu, G., Jin, S. M., Parisiadou, L., Xie, C., Yu, J., et al. (2014). MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 22, 608–620. doi: 10.1093/hmg/dds470
Choi, H.-S., Choi, Y.-G., Shin, H.-Y., Oh, J.-M., Park, J.-H., Kim, J. I., et al. (2014). Dysfunction of mitochondrial dynamics in the brains of scrapie-infected mice. Biochem. Biophys. Res. Commun. 448, 157–162. doi: 10.1016/j.bbrc.2014.04.069
Cissell, K. A., Rahimi, Y., Shrestha, S., Hunt, E. A., and Deo, S. K. (2008). Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal. Chem. 80, 2319–2325.
Clarke, P. G. (1990). Developmental cell death: morphological diversity and multiple mechanisms. Anat. Embryol. 181, 195–213.
Clement, C., Hill, J. M., Dua, P., Culicchia, F., and Lukiw, W. J. (2016). Analysis of RNA from Alzheimer’s disease post-mortem brain tissues. Mol. Neurobiol. 53, 1322–1328. doi: 10.1007/s12035-015-9105-6
Cogswell, J. P., Ward, J., Taylor, I. A., Waters, M., Shi, Y., Cannon, B., et al. (2008). Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 14, 27–41.
Cortes, C. J., Qin, K., Cook, J., Solanki, A., and Mastrianni, J. A. (2012). Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann–Straussler–Scheinker disease. J. Neurosci. 32, 12396–12405.
Cortes, C. J., Qin, K., Norstrom, E. M., Green, W. N., Bindokas, V. P., and Mastrianni, J. A. (2013). Early delivery of misfolded PrP from ER to lysosomes by autophagy. Int. J. Cell Biol. 560421, 1–18. doi: 10.1155/2013/560421
Delay, C., Calon, F., Mathews, P., and Hebert, S. S. (2011). Alzheimer-specific variants in the 30UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 6:70. doi: 10.1186/1750-1326-6-70
Dickson, J. R., Kruse, C., Montagna, D. R., Finsen, B., and Wolfe, M. S. (2013). Alternative polyadenylation and miR-34 family members regulate tau expression. J. Neurochem. 127, 739–749. doi: 10.1111/jnc.12437
Ding, W.-X., Ni, H. M., Li, M., Liao, Y., Chen, X., Stolz, D. B., et al. (2010). Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 285, 27879–27890. doi: 10.1074/jbc.M110.119537
Doxakis, E. (2010). Post-transcriptional regulation of alpha synuclein expression by mir-7 and mir-153. J. Biol. Chem. 285, 12726–12734.
Eacker, S. M., Dawson, T. M., and Dawson, V. L. (2009). Understanding microRNAs in neurodegeneration. Nat. Rev. Neurosci. 10, 837–841. doi: 10.1038/nrn2726
Fang, M., Wang, J., Zhang, X., Geng, Y., Hu, Z., Rudd, J. A., et al. (2012). The miR-124 regulates the expression of BACE1/p-secretase correlated with cell death in Alzheimer’s disease. Toxicol. Lett. 209, 94–105. doi: 10.1016/j.toxlet.2011.11.032
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114.
Gao, C., Wei, J., Zhang, B.-Y., Shi, Q., Chen, C., Wang, J., et al. (2017). MiRNA expression profiles in the brains of mice infected with scrapie agents 139A, ME7 and S15. Emerg. Microb. Infect. 5:e115. doi: 10.1038/emi.2016.120
Gao, J., Wang, W. Y., Mao, Y. W., Gräff, J., Guan, J. S., Pan, L., et al. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109. doi: 10.1038/nature09271
Geekiyanage, H., and Chan, C. (2011). MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J. Neurosci. 31, 14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011
Geekiyanage, H., Jicha, G. A., Nelson, P. T., and Chan, C. (2012). Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp. Neurol. 235, 491–496. doi: 10.1016/j.expneurol.2011.11.026
Gehrke, S., Imai, Y., Sokol, N., and Lu, B. (2010). Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466, 637–641. doi: 10.1038/nature09191
Geisler, S., Holmström, K. M., Skujat, D., Fiesel, F. C., Rothfuss, O. C., Kahle, P. J., et al. (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131. doi: 10.1038/ncb2012
Gibbings, D., Mostowy, S., Jay, F., Schwab, Y., Cossart, P., and Voinnet, O. (2012). Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 14, 1314–1321.
Gilad, S., Meiri, E., Yogev, Y., Benjamin, S., Lebanony, D., Yerushalmi, N., et al. (2008). Serum microRNAs are promising novel biomarkers. PLoS One 3:e3148. doi: 10.1371/journal.pone.0003148
Gong, X., Wang, H., Ye, Y., Shu, Y., Deng, Y., He, X., et al. (2016). miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am. J. Transl. Res. 8, 2127–2137.
Goodall, E. F., Heath, P. R., Bandmann, O., Kirby, J., and Shaw, P. J. (2013). Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front. Cell. Neurosci. 7:178. doi: 10.3389/fncel.2013.00178
Goold, R., Mckinnon, C., and Tabrizi, S. J. (2015). Prion degradation pathways: potential for therapeutic intervention. Mol. Cell. Neurosci. 66, 12–20.
Gregory, R. I., Yan, K. P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., et al. (2004). The microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240.
Hébert, S. S., Horré, K., Nicolaï, L., Papadopoulou, A. S., Mandemakers, W., Silahtaroglu, A. N., et al. (2008). Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/b-secretase expression. Proc. Natl. Acad. Sci. U.S.A. 105, 6415–6420. doi: 10.1073/pnas.0710263105
Heiseke, A., Aguib, Y., Riemer, C., Baier, M., and Schatzl, H. M. (2009). Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J. Neurochem. 109, 25–34. doi: 10.1111/j.1471-4159.2009.05906.x
Heiseke, A., Aguib, Y., and Schatzl, H. M. (2010). Autophagy, prion infection and their mutual interactions. Curr. Issues Mol. Biol. 12, 87–98.
Hendel, A., Bak, R. O., Clark, J. T., Kennedy, A. B., Ryan, D. E., Roy, S., et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989. doi: 10.1038/nbt.3290
Hill, J. M., and Lukiw, W. J. (2015). microRNA (miRNA)-mediated pathogenetic signaling in Alzheimer’s disease (AD). Neurochem. Res. 41, 96–100. doi: 10.1007/s11064-015-1734-7
Hu, N., Yu, J.-T., Tan, L., Wang, Y.-L., Sun, L., and Tan, L. (2013). Nutrition and the risk of Alzheimer’s disease. BioMed Res. Int. 2013:524820. doi: 10.1155/2013/524820
Huntzinger, E., and Izaurralde, E. (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 99–110. doi: 10.1038/nrg2936
Itoh, N., and Ohta, H. (2013). Roles of FGF20 in dopaminergic neurons and Parkinson’s disease. Front. Mol. Neurosci. 6:15. doi: 10.3389/fnmol.2013.00015
Jellinger, K. A. (2010). Basic mechanisms of neurodegeneration: a critical update. J. Cell Mol. Med. 14, 457–487. doi: 10.1111/j.1582-4934.2010.01010.x
Johnson, S. C., Yanos, M. E., Kayser, E.-B., Quintana, A., Sangesland, M., Castanza, A., et al. (2013). mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528. doi: 10.1126/science.1244360
Kang, S. C., Brown, D. R., Whiteman, M., Li, R., Pan, T., Perry, G., et al. (2004). Prion protein is ubiquitinated after developing protease resistance in the brains of scrapie-infected mice. J. Pathol. 203, 603–608. doi: 10.1002/path.1555
Kang, S.-G., Kim, C., Aiken, J., Yoo, H. S., and McKenzie, D. (2017). Dual MicroRNA to cellular prion protein inhibits propagation of pathogenic prion protein in cultured cells. Mol. Neurobiol. 55, 2384–2396. doi: 10.1007/s12035-017-0495-5
Khan, S. H., Zhao, D., Shah, S. Z., Hassan, M. F., Zhu, T., Song, Z., et al. (2017). Parkin overexpression ameliorates Prp106–126-induced neurotoxicity via enhanced autophagy in N2a cells. Cell. Mol. Neurobiol. 37, 717–728.
Khoo, S. K., Petillo, D., Kang, U. J., Resau, J. H., Berryhill, B., Linder, J., et al. (2011). Plasma-based circulating microRNA biomarkers for Parkinson’s disease. J. Parkinsons Dis. 2, 321–331.
Kim, D., Nguyen, M. D., Dobbin, M. M., Fischer, A., Sananbenesi, F., Rodgers, J. T., et al. (2007). SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 26, 3169–3179.
Kim, J., Inoue, K., Ishii, J., Vanti, W. B., Voronov, S. V., Murchison, E., et al. (2007). A microRNA feedback circuit in midbrain dopamine neurons. Science 317, 1220–1224.
Kim, J., Fiesel, F. C., Belmonte, K. C., Hudec, R., Wang, W.-X., Kim, C., et al. (2016). miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 11, 1–15. doi: 10.1186/s13024-016-0121-4
Kristiansen, M., Deriziotis, P., Dimchef, D. E., Jackson, G. S., Ova, H., Naumann, H., et al. (2007). Disease-associated prion protein oligomers inhibit the 26s proteasome. Mol. Cell 26, 175–188.
Kumar, P., Dezso, Z., MacKenzie, C., Oestreicher, J., Agoulnik, S., Byrne, M., et al. (2013). Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One 8:e69807. doi: 10.1371/journal.pone.0069807
Kumar, S., Vijayan, M., and Reddy, P.-H. (2017). MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum. Mol. Genet. 26, 3808–3822.
Lau, P., Bossers, K., Janky, R., Salta, E., Frigerio, C. S., Barbash, S., et al. (2013). Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 5, 1613–1634. doi: 10.1002/emmm.201201974
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.
Lee, Y., Jeon, K., Lee, J. T., Kim, S., and Kim, V. N. (2002). MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21, 4663–4670.
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060.
Leidinger, P., Backes, C., Deutscher, S., Schmitt, K., Mueller, S. C., Frese, K., et al. (2013). A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 14:R78. doi: 10.1186/gb-2013-14-7-r78
Lennox, K. A., and Behlke, M. A. (2011). Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 18, 1111–1120. doi: 10.1038/gt.2011.100
Lezmi, S., Benesik, A., Monks, E., Petit, T., and Barron, T. (2003). First case of feline spongiform encephalopathy in a captive cheetah born in France: PrPsc analysis in various tissues revealed unexpected targeting of kidney and adrenal gland. Histochem. Cell Biol. 119, 415–422.
Li, C., Chen, Y. P., Chen, X. P., Wei, Q. Q., Cao, B., and Shang, H. F. (2017). Downregulation of MicroRNA-193b-3p promotes autophagy and cell survival by targeting TSC1/mTOR signaling in NSC-34 cells. Front. Mol. Neurosci. 10:160. doi: 10.3389/fnmol.2017.00160
Li, G., Yang, H., Zhu, D., Huang, H., Liu, G., and Lun, P. (2014). Targeted suppression of chaperone-mediated autophagy by miR-320a promotes α-synuclein aggregation. Int. J. Mol. Sci. 15, 15845–15857. doi: 10.3390/ijms150915845
Li, X., Khanna, A., Li, N., and Wang, E. (2011). Circulatory miR34a as an RNA based, noninvasive biomarker for brain aging. Aging 3, 985–1002.
Li, Y. Y., Cui, J. G., Hill, J. M., Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2011). Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci. Lett. 487, 94–98.
Liu, C. G., Calin, G. A., Volinia, S., and Croce, C. M. (2008). MicroRNA expression profiling using microarrays. Nat. Protoc. 3, 563–578.
Liu, W., Liu, C., Zhu, J., Shu, P., Yin, B., Gong, Y., et al. (2012). MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 33, 522–534. doi: 10.1016/j.neurobiolaging.2010.04.034
Liu, Y., Song, Y., and Zhu, X. (2017). MicroRNA-181a regulates apoptosis and autophagy process in Parkinson’s disease by inhibiting p38 mitogen-activated protein kinase (MAPK)/c-Jun N-terminal kinases (JNK) signaling pathways. Med. Sci. Monit. 23, 1597–1606.
Llorens, F., Thüne, K., Martí, E., Kanata, E., Dafou, D., Díaz-Lucena, D., et al. (2018). Regional and subtype-dependent miRNA signatures in sporadic Creutzfeldt-Jakob disease are accompanied by alterations in miRNA silencing machinery and biogenesis. PLoS Pathog. 14:e1006802. doi: 10.1371/journal.ppat.1006802
Long, J. M., and Lahiri, D. K. (2011). MicroRNA-101 downregulates Alzheimer’s amyloid-b precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 404, 889–895.
Lu, T., Aron, L., Zullo, J., Pan, Y., Kim, H., Chen, Y., et al. (2014). REST and stress resistance in ageing and Alzheimer’s disease. Nature 507, 448–454. doi: 10.1038/nature13163
Lukiw, W. J. (2007). Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 18, 297–300. doi: 10.1097/WNR.0b013e3280148e8b
Lukiw, W. J., Dua, P., Pogue, A. I., Eicken, C., and Hill, J. M. (2011). Upregulation of microRNA-146a(miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker (GSS) syndrome. J. Toxicol. Environ. Health A 74, 1460–1468.
Lynch-Day, M. A., Mao, K., Wang, K., Zhao, M., and Klionsky, D. J. (2012). The role of autophagy in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2:a009357.
Majer, A., Medina, S. J., Niu, Y., Abrenica, B., Manguiat, K. J., Frost, K. L., et al. (2012). Early mechanisms of pathobiology are revealed by transcriptional temporal dynamics in hippocampal CA1Neurons of prion infected Mice. PLoS Pathog. 8:e1003002. doi: 10.1371/journal.ppat.1003002
Majeski, A. E., and Dice, J. F. (2004). Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36, 2435–2444.
Manakov, S. A., Grant, S. G., and Enright, A. J. (2009). Reciprocal regulation of microRNAs and mRNA profiles in neuronal development and synapse formation. BMC Genomics 10:419. doi: 10.1186/1471-2164-10-419
Margis, R., Margis, R., and Rieder, C. R. (2011). Identification of blood microRNAs associated to Parkinson’s disease. J. Biotechnol. 152, 96–101.
Martins, M., Rosa, A., Guedes, L. C., Fonseca, B. V., Gotovac, K., Violante, S., et al. (2011). Convergence of miRNA expression profiling, a-synuclein interaction and GWAS in Parkinson’s disease. PLoS One 6:e25443. doi: 10.1371/journal.pone.0025443
Mckinnon, C., Goold, R., Andre, R., Devoy, A., Ortega, Z., Moonga, J., et al. (2016). Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin–proteasome system. Acta Neuropathol. 131, 411–425. doi: 10.1007/s00401-015-1508-y
Miller, J. A., Horvath, S., and Geschwind, D. H. (2010). Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. 107, 12698–12703.
Miller, T. M., Pestronk, A., David, W., Rothstein, J., Simpson, E., Appel, S. H., et al. (2013). An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomized, first-in-man study. Lancet Neurol. 12, 435–442. doi: 10.1016/S1474-4422(13)70061-9
Minones-Moyano, E., Porta, S., Escaramıs, G., Rabionet, R., Iraola, S., Kagerbauer, B., et al. (2011). MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 20, 3067–3078.
Montag, J., Hitt, R., Opitz, L., Schulz-Schaeffer, W. J., Hunsmann, G., and Motzkus, D. (2009). Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Mol. Neurodegener. 4:36. doi: 10.1186/1750-1326-4-36
Moon, J.-H., Lee, J.-H., Lee, U. J., and Park, S.-Y. (2016a). Autophagy flux induced by ginsenoside-Rg3 attenuates human prion protein-mediated neurotoxicity and mitochondrial dysfunction. Oncotarget 7, 85697–85708. doi: 10.18632/oncotarget.13730
Moon, J.-H., Lee, J.-H., Lee, U. J., and Park, S.-Y. (2016b). Hinokitiol protects primary neuron cells against prion peptide-induced toxicity via autophagy flux regulated by hypoxia inducing factor-1. Oncotarget 7, 29944–29957. doi: 10.18632/oncotarget.8670
Moreno, J. A., Halliday, M., Molloy, C., Radford, H., Verity, N., Axten, J. M., et al. (2013). Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 5:206ra138. doi: 10.1126/scitranslmed.3006767
Mukherjee, A., and Soto, C. (2011). Role of calcineurin in neurodegeneration produced by misfolded proteins and endoplasmic reticulum stress. Curr. Opin. Cell Biol. 23, 223–230. doi: 10.1016/j.ceb.2010.12.006
Nakagaki, T., Satoh, K., Ishibashi, D., Fuse, T., Sano, K., Kamatari, Y. O., et al. (2013). FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy 9, 1386–1394. doi: 10.4161/auto.25381
Olde Loohuis, N. F., Kos, A., Martens, G. J., Van Bokhoven, H., Nadif Kasri, N., and Aschrafi, A. (2012). MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. 69, 89–102. doi: 10.1007/s00018-011-0788-1
Pan, K. M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., et al. (1993). Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966.
Patel, N., Hoang, D., Miller, N., Ansaloni, S., Huang, Q., Rogers, J. T., et al. (2008). MicroRNAs can regulate human APP levels. Mol. Neurodegener. 3:10. doi: 10.1186/1750-1326-3-10
Persengiev, S., Kondova, I., Otting, N., Koeppen, A. H., and Bontrop, R. E. (2011). Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol. Aging 32, e2317–e2327. doi: 10.1016/j.neurobiolaging.2010.03.014
Provost, P. (2010). MicroRNAs as a molecular basis for mental retardation. Alzheimer’s and prion diseases. Brain Res. 1338, 58–66. doi: 10.1016/j.brainres.2010.03.069
Puig, B., Altmeppen, H. C., Ulbrich, S., Linsenmeier, L., Krasemann, S., Chakroun, K., et al. (2016). Secretory pathway retention of mutant prion protein induces p38-MAPK activation and lethal disease in mice. Sci. Rep. 6:24970. doi: 10.1038/srep24970
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626.
Roux, J., Porta, M.-G., and Rechavi, M.-R. (2012). Comparative analysis of human and mouse expression data illuminates tissue-specific evolutionary patterns of miRNAs. Nucleic Acids Res. 40, 5890–5900. doi: 10.1093/nar/gks279
Saba, R., Goodman, C. D., Huzarewich, R. L., Robertson, C., and Booth, S. A. (2008). A miRNA signature of prion induced neurodegeneration. PLoS One 3:e3652. doi: 10.1371/journal.pone.0003652
Saba, R., Gushue, S., Huzarewich, R. L., Manguiat, K., Medina, S., Robertson, C., et al. (2012). MicroRNA 146a (miR-146a) is over-expressed during prion disease and modulates the innate immune response and the microglial activation state. PLoS One 7:e30832. doi: 10.1371/journal.pone.0030832
Sanz Rubio, D., López-Pérez,Ó., de Andrés Pablo,Á., Bolea, R., Osta, R., Badiola, J. J., et al. (2017). Increased circulating microRNAs miR-342-3p and miR-21-5p in natural sheep prion disease. J. Gen. Virol. 98, 305–310. doi: 10.1099/jgv.0.000685
Schapira, A. H., and Gegg, M. (2011). Mitochondrial contribution to Parkinson’s disease pathogenesis. Parkinsons Dis. 2011:159160.
Schipper, H. M., Maes, O. C., Chertkow, H. M., and Wang, E. (2007). microRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Biol. 1, 263–274.
Schochetman, G., Ou, C. Y., and Jones, W. K. (1988). Polymerase chain reaction. J. Infect. Dis. 158, 1154–1157.
Shah, S. Z. A., Hussain, T., Zhao, D., and Yang, L. (2017a). The role of unfolded protein response and mitogen-activated protein kinase signaling in neurodegenerative diseases with special focus on prion diseases. Front. Aging Neurosci. 9:120. doi: 10.3389/fnagi.2017.00120
Shah, S. Z. A., Zhao, D., Hussain, T., and Yang, L. (2017b). Role of the AMPK pathway in promoting autophagic flux via modulating mitochondrial dynamics in neurodegenerative diseases: insight into prion diseases. Ageing Res. Rev. 40, 51–63. doi: 10.1016/j.arr.2017.09.004
Shah, S. Z. A., Zhao, D., Taglialatela, G., Khan, S. H., Hussain, T., Dong, H., et al. (2017c). Early minocycline and late FK506 treatment improves survival and alleviates neuroinflammation, neurodegeneration, and behavioral deficits in prion-infected hamsters. Neurotherpeutics 14, 463–483. doi: 10.1007/s13311-016-0500-0
Shapshak, P. (2013). Molecule of the month: miRNA and Human Prion brain disease. Bioinformation 9, 659–660.
Shi, L., Liu, H., Zhang, M., Guo, Y., Song, C., Song, D., et al. (2016). MiR-128-3p activates autophagy in rat brain cells after focal cerebral ischemia reperfusion through targeting Atg1. Int. J. Clin. Exp. Med. 2016, 2486–2492.
Shioya, M., Obayashi, S., Tabunoki, H., Arima, K., Saito, Y., Ishida, T., et al. (2010). Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 36, 320–330. doi: 10.1111/j.1365-2990.2010.01076.x
Smit-McBride, Z., Forward, K. I., and Nguyen, A. T. (2014). Age-dependent increase in miR-34a expression in the posterior pole of the mouse eye. Mol. Vis. 20, 1569–1578.
Somerville, R. A., Birkett, C. R., Farquhar, C. F., Hunter, N., Goldmann, W., Dornan, J., et al. (1997). Immunodetection of PrPSc in spleens of some scrapie-infected sheep but not BSE-infected cows. J. Gen. Virol. 78, 2389–2396.
Song, J., and Kim, Y.-K. (2017). Identification of the role of miR-142-5p in Alzheimer’s disease by comparative bioinformatics and cellular analysis. Front. Mol. Neurosci. 10:227. doi: 10.3389/fnmol.2017.00227
Song, Z., Shah, S. Z. A., Yang, W., Dong, H., Yang, L., Zhou, X., et al. (2017a). Downregulation of the repressor element 1-silencing transcription factor (REST) Is Associated with Akt-mTOR and Wnt-b-catenin signaling in prion diseases models. Front. Mol. Neurosci. 10:128. doi: 10.3389/fnmol.2017.00128
Song, Z., Yang, W., Zhou, X., Yang, L., and Zhao, D. (2017b). Lithium alleviates neurotoxic prion peptide-induced synaptic damage and neuronal death partially by the upregulation of nuclear target REST and the restoration of Wnt signaling. Neuropharmacology 123, 332–348. doi: 10.1016/j.neuropharm.2017.05.021
Song, Z., Zhu, T., Zhou, X., Barrow, P., Yang, W., Cui, Y., et al. (2016). REST alleviates neurotoxic prion peptide-induced synaptic abnormalities, neurofibrillary degeneration and neuronal death partially via LRP6-mediated Wnt beta-catenin signaling. Oncotarget 7, 12035–12052. doi: 10.18632/oncotarget.7640
Sonntag, K. C. (2010). MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 1338, 48–57. doi: 10.1016/j.brainres.2010.03.106
Speldewinde, S. H., Doronina, V. A., and Grant, C. M. (2015). Autophagy protects against de novo formation of the [PSI+] prion in yeast. Mol. Biol. Cell 26, 4541–4551. doi: 10.1091/mbc.E15-08-0548
Sun, J., Gao, C., Meng, D., Xu, Y., Wang, X., Gu, C., et al. (2017). Antagomirs targeting MiroRNA-134 attenuates epilepsy in rats through regulation of oxidative stress. Mitochondrial Functions and Autophagy. Front. Pharmacol. 8:524. doi: 10.3389/fphar.2017.00524
Sun, L., Liu, A., Zhang, J., Ji, W., Li, Y., Yang, X., et al. (2017). miR-23b improves cognitive impairments in traumatic brain injury by targeting ATG12-mediated neuronal autophagy. Behav. Brain Res. 340, 126–136. doi: 10.1016/j.bbr.2016.09.020
Szafranski, K., Abraham, K. J., and Mekhail, K. (2015). Non-coding RNA in neural function, disease, and aging. Front. Genet. 6:87. doi: 10.3389/fgene.2015.00087
Taniguchi, M., Miura, K., Iwao, H., and Yamanaka, S. (2001). Quantitative assessment of DNA microarrays – comparison with northern blot analysis. Genomics 71, 34–39. doi: 10.1006/geno.2000.6427
Tung, Y. T., Hsu, W. M., Lee, H., Huang, W. P., and Liao, Y. F. (2010). The evolutionarily conserved interaction between LC3 and p62 selectively mediates autophagy-dependent degradation of mutant huntingtin. Cell. Mol. Neurobiol. 30, 795–806. doi: 10.1007/s10571-010-9507-y
Valera, E., Spencer, B., Mott, J., Trejo, M., Adame, A., Mante, M., et al. (2017). MicroRNA-101 modulates autophagy and oligodendroglial alpha-synuclein accumulation in multiple system atrophy. Front. Mol. Neurosci. 10:329. doi: 10.3389/fnmol.2017.00329
Vallelunga, A., Ragusa, M., Di Mauro, S., Iannitti, T., Pilleri, M., Biundo, R., et al. (2014). Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front. Cell. Neurosci. 8:156. doi: 10.3389/fncel.2014.00156
Wang, C. M., Wei, Z. J., Jiang, G. H., and Liu, H. J. (2017). Neuroprotective mechanisms of miR-124 activating PI3K/Akt signaling pathway in ischemic stroke. Exp. Ther. Med. 13, 3315–3318. doi: 10.3892/etm.2017.4424
Wang, H., Tian, C., Sun, J., Chen, L. N., Lv, Y., Yang, X.-D., et al. (2017). Overexpression of PLK3 mediates the degradation of abnormal prion proteins dependent on chaperone-mediated autophagy. Mol. Neurobiol. 54, 4401–4413. doi: 10.1007/s12035-016-9985-0
Wang, W. X., Huang, Q., Hu, Y., Stromberg, A. J., and Nelson, P. T. (2011). Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 121, 193–205.
Wang, W. X., Wilfred, B. R., and Madathil, S. K. (2010). miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am. J. Pathol. 177, 334–345. doi: 10.2353/ajpath.2010.091202
Wang, Y., Tatiana, V., Wong, A. H., El Fatimy, R., Wei, Z., Cai, W., et al. (2017). Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol. Aging 51, 156–166. doi: 10.1016/j.neurobiolaging.2016.12.015
Weiler, J., Hunziker, J., and Hall, J. (2006). Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 13, 496–502.
Williams, A. H., Valdez, G., Moresi, V., Qi, X., McAnally, J., Elliott, J. L., et al. (2009). MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554. doi: 10.1126/science.1181046
Wu, J., and Xie, X. (2006). Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 7:R85. doi: 10.1186/gb-2006-7-9-r85
Yaghmoor, F., Noorsaeed, A., Alsaggaf, S., Aljohani, W., Scholtzova, H., Boutajangout, A., et al. (2014). The role of TREM2 in Alzheimer’s and other neurological disorders. J. Alzheimers Dis. Parkinsonism 4, 160–165.
Yang, J., Chen, D., He, Y., Meléndez, A., Feng, Z., Hong, Q., et al. (2013). MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age 35, 11–22. doi: 10.1007/s11357-011-9324-3
Yang, Y., Hu, Z., Du, X., Davies, H., Huo, X., and Fang, M. (2017). miR-16 and fluoxetine both reverse autophagic and apoptotic change in chronic unpredictable mild stress model rats. Front. Neurosci. 11:428. doi: 10.3389/fnins.2017.00428
Yao, H., Zhao, D., Khan, S. H., and Yang, L. (2013). Role of autophagy in prion protein-induced neurodegenerative diseases. Acta Biochim. Biophys. Sin. 45, 494–502. doi: 10.1093/abbs/gmt022
Yedidia, Y., Horonchik, L., Tzaban, S., Yanai, A., and Taraboulos, A. (2001). Proteasome and ubiquitin are involved in the turn-over of the wild type prion protein. EMBO J. 20, 5383–5391.
Yoshimori, T. (2004). Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 313, 453–458.
Yunsheng, W., Zhao, D., Pan, B., Song, Z. Q., Shah, S. Z. A., Yin, X., et al. (2015). Death receptor 6 and caspase-6 regulate prion peptide-induced axonal degeneration in rat spinal neurons. J. Mol. Neurosci. 56, 966–976. doi: 10.1007/s12031-015-0562-1
Zatloukal, K., Stumptner, C., Fuchsbichler, A., Heid, H., Schnoelzer, M., Kenner, L., et al. (2002). p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160, 255–263.
Zhang, Q.-S., Liu, W., and Lu, G.-X. (2017). miR-200a-3p promotes b-Amyloid-induced neuronal apoptosis through down-regulation of SIRT1 in Alzheimer’s disease. J. Biosci. 42, 397–404.
Zhang, Y., Liu, C., Wang, J., Li, Q., Ping, H., Gao, S., et al. (2016). MiR-299-5p regulates apoptosis through autophagy in neurons and ameliorates cognitive capacity in APPswe/PS1dE9 mice. Sci. Rep. 6:24566. doi: 10.1038/srep24566
Zhao, F., Qu, Y., Wang, H., Huang, L., Zhu, J., Li, S., et al. (2017). The effect of miR-30d on apoptosis and autophagy in cultured astrocytes under oxygen-glucose deprivation. Brain Res. 1671, 67–76. doi: 10.1016/j.brainres.2017.06.011
Zhao, Y., Bhattacharjee, S., Dua, P., Alexandrov, P. N., and Lukiw, W. J. (2015a). microRNA-based biomarkers and the diagnosis of Alzheimer’s disease. Front. Neurol. 6:162. doi: 10.3389/fneur.2015.00162
Zhao, Y., Pogue, A. I., and Lukiw, W. J. (2015b). MicroRNA (miRNA) signaling in the human CNS in sporadic Alzheimer’s disease (AD)–novel and unique pathological features. Int. J. Mol. Sci. 16, 30105–30116. doi: 10.3390/ijms16122
Zhao, Y., Zhu, M., Yu, Y., Qiu, L., Zhang, Y., He, L., et al. (2017). Brain REST/NRSF is not only a silent repressor but also an active protector. Mol. Neurobiol. 54, 541–550.
Zheng, X., Boyer, L., Jin, M., Kim, Y., Fan, W., Bardy, C., et al. (2016). Alleviation of neuronal energy deficiency by mTOR inhibition as a treatment for 6 mitochondria-related neurodegeneration. eLife 5:e13378.
Zhou, R., Yuan, P., Wang, Y., Hunsberger, J. G., Elkahloun, A., Wei, Y., et al. (2009). Evidence of selective microRNAs and their effectors as common long-term targets for the action of mood stabilizers. Neuropsychopharmacology 34, 1395–1405. doi: 10.1038/npp.2008.131
Zhou, Y. H., Zhang, X. P., and Ebright, R. H. (1991). Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 19:6052. doi: 10.1093/nar/19.21.6052
Keywords: prion diseases, microRNAs (miRNAs), biological processes, biomarkers, autophagic flux, Alzheimer’s disease, Parkinson’s disease, therapeutic interventions
Citation: Shah SZA, Zhao D, Hussain T, Sabir N and Yang L (2018) Regulation of MicroRNAs-Mediated Autophagic Flux: A New Regulatory Avenue for Neurodegenerative Diseases With Focus on Prion Diseases. Front. Aging Neurosci. 10:139. doi: 10.3389/fnagi.2018.00139
Received: 05 December 2017; Accepted: 25 April 2018;
Published: 15 May 2018.
Edited by:
Catarina Oliveira, University of Coimbra, PortugalReviewed by:
Hermona Soreq, Hebrew University of Jerusalem, IsraelCopyright © 2018 Shah, Zhao, Hussain, Sabir and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lifeng Yang, eWFuZ2xmQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.