
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Adv. Opt. Technol., 13 March 2025
Sec. Optical Imaging
Volume 14 - 2025 | https://doi.org/10.3389/aot.2025.1546386
This article is part of the Research TopicDeep Learning Enhanced Computational Imaging: Leveraging AI for Advanced Image Reconstruction and AnalysisView all articles
The advent of X-ray Free Electron Lasers (XFELs) has opened unprecedented opportunities for advances in the physical, chemical, and biological sciences. With their state-of-the-art methodologies and ultrashort, and intense X-ray pulses, XFELs propel X-ray science into a new era, surpassing the capabilities of traditional light sources. Ultrafast X-ray scattering and imaging techniques leverage the coherence of these intense pulses to capture nanoscale structural dynamics with femtosecond spatial-temporal resolution. However, spatial and temporal resolutions remain limited by factors such as intrinsic fluctuations and jitters in the Self-Amplified Spontaneous Emission (SASE) mode, relatively low coherent scattering cross-sections, the need for high-performance, single-photon-sensitive detectors, effective sample delivery techniques, low parasitic X-ray instrumentation, and reliable data analysis methods. Furthermore, the high-throughput data flow from high-repetition rate XFEL facilities presents significant challenges. Therefore, more investigation is required to determine how Artificial Intelligence (AI) can support data science in this situation. In recent years, deep learning has made significant strides across various scientific disciplines. To illustrate its direct influence on ultrafast X-ray science, this article provides a comprehensive overview of deep learning applications in ultrafast X-ray scattering and imaging, covering both theoretical foundations and practical applications. It also discusses the current status, limitations, and future prospects, with an emphasis on its potential to drive advancements in fourth-generation synchrotron radiation, ultrafast electron diffraction, and attosecond X-ray studies.
The ability to capture ultrafast dynamics far-from-equilibrium with angstrom to nanometer spatial resolution has revolutionized the fundamental findings of physical, chemical, and biological sciences (Lindroth et al., 2019). X-ray free-electron lasers (XFELs) have turned the once-theoretical dream of capturing ultrafast phenomena at nanoscale resolutions into a practical reality (Pellegrini, 2016). As a groundbreaking light source, XFELs enable the creation of molecular movies, offering unprecedented insights into the fundamental interactions of matter at previously inaccessible temporal and spatial scales. These state-of-the-art facilities generate extremely intense, ultra-short X-ray pulses, allowing researchers to investigate electron and nuclear interactions on timescales as short as tens of femtoseconds. Since the operation of the world’s first hard X-ray free-electron laser at the Linac Coherent Light Source (LCLS) of SLAC National Accelerator Laboratory (Emma et al., 2010), XFELs have been utilized across nearly all fields of natural science. Applications span biological imaging, protein crystallography, femto-chemistry, condensed matter physics, and atomic, molecular, and optical (AMO) science (Bostedt et al., 2016).
In the early years, methods adapted from synchrotron sources and optical lasers—such as imaging, spectroscopy, scattering, and crystallography—were integrated into the XFEL community. Among these, lensless imaging techniques, particularly coherent diffraction imaging, have attracted significant attention due to their potential for achieving high-resolution images (Gaffney and Chapman, 2007; Miao, et al., 1998). When coupled with the ultrafast capabilities of XFELs, these methods have propelled imaging into an entirely new era (Chapman et al., 2006; Miao et al., 2015). A notable method is single-particle imaging (SPI) (Aquila et al., 2015). This technique relies on coherent diffraction and operates on the principle of “diffraction before destruction” (Neutze et al., 2000). SPI has emerged as a powerful approach for determining native structures, and it has been successfully applied to imaging a wide variety of targets, including biological viruses (Ekeberg et al., 2015; Seibert et al., 2011), bacteria (Kimura et al., 2014), organelles (Gallagher-Jones et al., 2014; Hantke et al., 2014), clusters (Bostedt et al., 2008; Gorkhover et al., 2016), aerosols (Bogan et al., 2010; Loh et al., 2012), self-assembled structures (Sun et al., 2018), and nanoparticles (Clark et al., 2015; Clark et al., 2013; Xu et al., 2014). The best spatial resolution achieved for reproducible objects is around 3 nm (Ayyer et al., 2021). In the field of time-resolved research, topics such as thermal and nonthermal melting (Ferguson et al., 2016; Ihm et al., 2019; Jung et al., 2021), as well as light-matter interactions (Gorkhover et al., 2012), have been particularly prevalent. Recent advances, including the establishment of high-repetition-rate XFEL facilities such as the European XFEL (Decking et al., 2020), LCLS-II/LCLS-II-HE (Zhou et al., 2021), and SHINE (Huang et al., 2021), have enabled the rapid collection of high-throughput diffraction data, greatly enhancing experimental efficiency. However, the high throughput data generated presents significant challenges, particularly in data processing. Moreover, processing time-resolved datasets to construct quasi-particle movies requires complex steps. These include pattern recognition, filtering, orientation determination, and phase retrieval. Traditional data-processing methods often struggle to handle these tasks efficiently.
In this context, machine learning (Baldi et al., 2002; Jordan and Mitchell, 2015), particularly deep learning (LeCun et al., 2015), has emerged as a powerful tool for addressing these challenges. Recent breakthroughs in artificial neural networks have sparked widespread interest across diverse sectors, including academia, industry, and beyond. Deep learning excels at identifying complex patterns in large datasets. Combined with increased computational power and the availability of big data, it has become a transformative tool in scientific research. In particular, deep learning has made significant strides in image processing, surpassing traditional methods in tasks such as object recognition and classification (Farabet et al., 2013; Krizhevsky et al., 2012; Simonyan and Zisserman, 2014; Ioffe and Szegedy, 2015; Tompson et al., 2014). This rapid evolution has led to a surge of interest and investment in deep learning, creating new opportunities for its application in diverse fields, including ultrafast scattering and imaging.
This overview provides a comprehensive outline of deep learning applications in ultrafast X-ray scattering and imaging. It explores both the theoretical foundations and practical implementations of deep learning techniques, focusing on their potential to revolutionize the analysis of XFEL data. The paper is organized into three sections: (1) A brief introduction to the core theory and key concepts of deep learning, including an overview of common models used in image processing and analysis; (2) A discussion of how deep learning can be applied to process and analyze XFEL data, outlining its advantages and challenges; and (3) An evaluation of the strengths and limitations of these methods, along with potential directions for future research. By addressing these aspects, the paper aims to demonstrate the significant impact of deep learning on ultrafast X-ray scattering and imaging, encouraging further exploration in this interdisciplinary area.
Machine learning focuses on developing computational techniques that learn patterns from data to build predictive models. These mathematical models are trained through parameter optimization using input-output examples, enabling them to generalize to new datasets. The training process involves iterative refinement guided by validation set performance, followed by final evaluation on test data simulating real-world applications. Three primary paradigms govern this field: “supervised learning” dominates current practice by training models on labeled datasets for pattern recognition tasks; “unsupervised learning” identifies inherent structures in unlabeled data through clustering and feature extraction; while “reinforcement learning” employs trial-and-error environmental interactions to optimize goal-oriented behaviors, exemplified by DeepMind’s AlphaGo systems (Silver et al., 2017).
Artificial neural networks (ANNs), inspired by biological neural systems, were introduced in the 1980s (Feldman, et al., 1988; Lippmann, 1987). These networks consist of interconnected nodes structured into layered architectures, including input, output, and variable hidden layers (Rosenblatt, 1958). Based on connection patterns, ANNs are categorized as “feedforward networks” (Figure 1 with unidirectional data flow for static input-output mapping, or “feedback networks” that use cyclic connections to achieve dynamic equilibria through iterative processing. Perceptron is a common feedforward network (Rosenblatt, 1958), while examples of feedback networks include the Hopfield net (Hopfield, 1982) and Kohonen self-organizing maps (Kangas, et al., 1990; Kohonen, 1990).
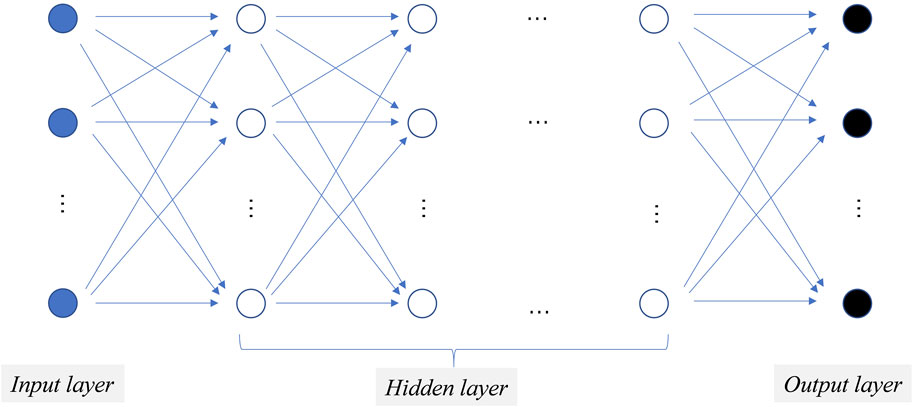
Figure 1. A typical three-layer feed-forward network architecture. Circular shapes represent neurons organized into layers, with a series of linearly arranged blue solid circles forming the input layer, blue hollow circles representing the hidden layer, and black solid circles representing the output layer.
The ANN employs a learning process to train the network by adjusting weights to desired values. Learning falls into two main categories: supervised learning and unsupervised learning. In supervised learning, a training set with input-output examples is provided to adjust weights and minimize output errors. The training set must be representative of the model for effective learning. Once trained, the network can be used when it produces desired outputs. Unsupervised learning, on the other hand, does not use target outputs and focuses on identifying patterns in input data alone. Different types of networks require specific learning processes.
While ANNs have long been valued for their ability to solve complex problems, their practical use was historically limited by computational costs and training challenges. Recent advancements in big data, GPU processing, and improved algorithms have elevated ANNs to a dominant position in machine learning. These developments have spurred rapid progress, influencing broader AI research. Today, ANNs excel at modeling nonlinear relationships, offering superior fault tolerance, speed, and scalability through parallel processing, outperforming traditional regression methods.
Deep learning has transformed artificial intelligence through multi-layered neural networks capable of autonomous pattern recognition (LeCun et al., 2015). While backpropagation (Hopfield, 1982) established foundational weight adjustment mechanisms, limitations like local optima and overfitting persisted. A breakthrough occurred with Hinton’s layer-wise greedy learning (Hinton et al., 2006), which combined unsupervised pre-training for feature extraction with supervised fine-tuning, leveraging big data to mitigate overfitting and improve convergence.
Critical technical advances enabled deeper architectures: “ReLU activation functions” (Nair and Hinton 2010) overcame vanishing gradients through non-saturating derivatives; “convolutional layers” (Krizhevsky et al., 2012; Lecun and Bengio, 1995) automated spatial feature learning, eliminating manual engineering. These innovations allowed networks to progressively abstract hierarchical representations through pooling and specialized layers, establishing end-to-end learning frameworks.
Convolutional Neural Networks (CNNs) are specialized architectures designed to efficiently process image data by preserving spatial relationships. Unlike traditional feedforward networks, which inefficiently connect all nodes across layers, CNNs employ structured layers (Figure 2): “convolutional layers” extract features using parameterized filters (O'Shea and Nash, 2015), reducing learnable weights through weight sharing; “activation layers” introduce non-linearity to feature maps (Klabjan and Harmon 2019); and “pooling layer” down-sample features by aggregating small grid regions into single outputs (Sun et al., 2017). Additionally, CNNs enhance performance through techniques like “dropout regularization”, which randomly deactivates neurons during training to prevent overfitting (Baldi and Sadowski, 2014), and “batch normalization”, which standardizes activation maps to accelerate convergence and reduce sensitivity to parameter initialization (Ioffe and Szegedy, 2015). Together, these components and techniques improve training efficiency, robustness, and overall model performance.
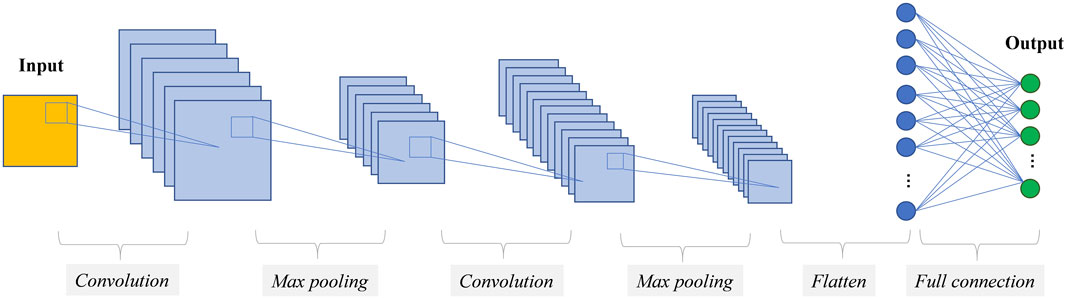
Figure 2. Illustration of the Convolutional Neural Network (CNN) architecture, with the blue-highlighted layers representing the feature extraction process. These convolutional and pooling layers automatically identify hierarchical patterns within the input data, which are subsequently processed by fully connected layers for prediction.
Modern CNN architectures combine these components in increasingly complex ways to optimize performance for specific image-oriented tasks. Popular CNN architectures include LeNet (Lecun, et al., 1998), AlexNet (Krizhevsky et al., 2012), ResNet (He et al., 2020), and VGG (Simonyan and Zisserman, 2014), each building on ideas and insights from previous architectures to push the state-of-the-art in image processing. Implementing CNNs is typically done using frameworks like TensorFlow, Keras, or Pytorch, which are built on NVIDIA’s CUDA platform and are actively developed in the machine learning research community.
The advancements discussed in the preceding sections have significantly enhanced signal, image, video, and audio processing, as elaborated earlier. While a comprehensive overview of all developments is beyond the scope of this document. In the following section, we will briefly touch upon select advanced network architectures that we consider to have had, or will have, a notable impact on the processing of ultrafast X-ray imaging data.
Autoencoders (Hinton and Salakhutdinov 2006, Vincent et al., 2008) are used for unsupervised learning and data compression. The main purpose of an autoencoder is to learn a compressed, lower-dimensional representation of the input data, and then reconstruct the original input data from this compressed representation. An autoencoder consists of an encoder network and a decoder network. The encoder network compresses the input data into a latent representation, while the decoder network reconstructs the input data from this compressed representation.
U-Net (Ronneberger et al., 2015) is a deep learning architecture designed for image segmentation tasks. It features a symmetric encoder-decoder structure with skip connections, making it particularly effective for high-resolution image segmentation. The U-Net architecture consists of an encoder that captures the contextual information of the input image and a decoder that generates the segmentation output. The skip connections between the encoder and decoder help preserve fine details and prevent information loss by connecting feature maps from different levels. U-Net has demonstrated remarkable performance in various image segmentation tasks, especially in scenarios with limited training data and the need for high-resolution segmentation results. Its simple yet efficient architecture has made U-Net a classic model in the field of image segmentation. Originally proposed in 2D, it has since been applied to 3D as well.
Generative adversarial networks (GANs) are a type of deep learning framework introduced by Ian Goodfellow (Goodfellow et al., 2014). GANs consist of two neural networks, the generator and the discriminator, trained simultaneously through a competitive process. The generator creates new data instances based on random noise input, aiming to produce data indistinguishable from real data. The discriminator evaluates generated data instances, distinguishing them from real data. During training, the generator and discriminator engage in a minimax game. The generator strives to produce realistic data to deceive the discriminator, while the discriminator attempts to distinguish between real and generated data. GANs have been used in various applications such as image generation (Karras et al., 2017; Radford et al., 2015), image-to-image translation (Isola et al., 2017; Zhu et al., 2017), and style transfer (Gatys et al., 2016; Johnson et al., 2016), showing success in generating high-quality data samples.
Google’s Inception Network, also known as GoogLeNet (Ioffe and Szegedy, 2015), is a deep convolutional neural network architecture developed by Google researchers in 2014. It is designed to achieve high accuracy in image classification tasks while being computationally efficient. The main highlight of the Inception Network is the introduction of the “inception block,” which allows for parallel computation of convolutions and pooling operations. By incorporating multiple parallel convolutional operations of different sizes (such as 1 × 1, 3 × 3, 5 × 5) and max pooling within the inception block, the network can capture features at various scales and resolutions simultaneously. In 2014, the Inception Network won the ImageNet Large Scale Visual Recognition Challenge (ILSVRC), demonstrating its effectiveness in image classification tasks. Since then, the Inception architecture has been widely adopted and adapted for various deep learning applications, including object detection, image segmentation, and more.
ResNets, or Residual Networks (He et al., 2016), were developed by researchers to enable training of very deep neural networks effectively. The key innovation of ResNet is the use of residual connections, or shortcut connections. It allows gradients to flow more easily through the network. By introducing residual blocks, ResNets address the issue of vanishing gradients in deep networks. The concept of residual learning in ResNets involves learning the residual mapping between input and output, rather than the direct mapping. This approach enables the training of very deep neural networks with hundreds or even thousands of layers. ResNets have been widely adopted in various deep learning tasks, such as image classification and object detection, and have achieved state-of-the-art performance on benchmark datasets.
YOLO (You Only Look Once) (Redmon et al., 2016) is a real-time object detection system proposed by Redmon et al., in 2016. It is known for its speed and accuracy in detecting objects in images or videos. YOLO adopts a single neural network that predicts bounding boxes and class probabilities directly from full images in one evaluation. This approach eliminates the need for multiple region proposals and extensive post-processing, making YOLO faster and more efficient than traditional object detection systems. The YOLO architecture divides the input image into a grid and predicts bounding boxes and class probabilities for each grid cell. YOLO has been widely used in various applications, including autonomous driving, surveillance, and image analysis. Its ability to provide real-time object detection with high accuracy has made YOLO a popular choice in the field of computer vision.
In ultra-fast imaging and scattering experiment setups, a succession of identical single particles will be intercepted by intense X-ray pulses to capture diffraction snapshots before each particle is destroyed. However, several challenges and complications often arise during this process. Firstly, the synchronization between particle injectors (DePonte et al., 2008; Kirian et al., 2015; Sierra et al., 2012) and X-ray pulses is often imperfect, leading to a mix of blank shots and successful exposures. Injected particles may also vary in composition, containing none, one, or multiple target particles, while contamination can introduce snapshots of other species. Fluctuations in beam intensity, incomplete particle exposure, and detection artifacts, such as saturation and charge bleeding, further complicate the data (Figure 3). These factors create large, heterogeneous datasets with blank frames, mixed-species snapshots, and noise, making it challenging to isolate single-particle hits. Snapshots of identical single particles are important for three-dimensional structure retrieval, but their scarcity severely restricts experimental efficiency.
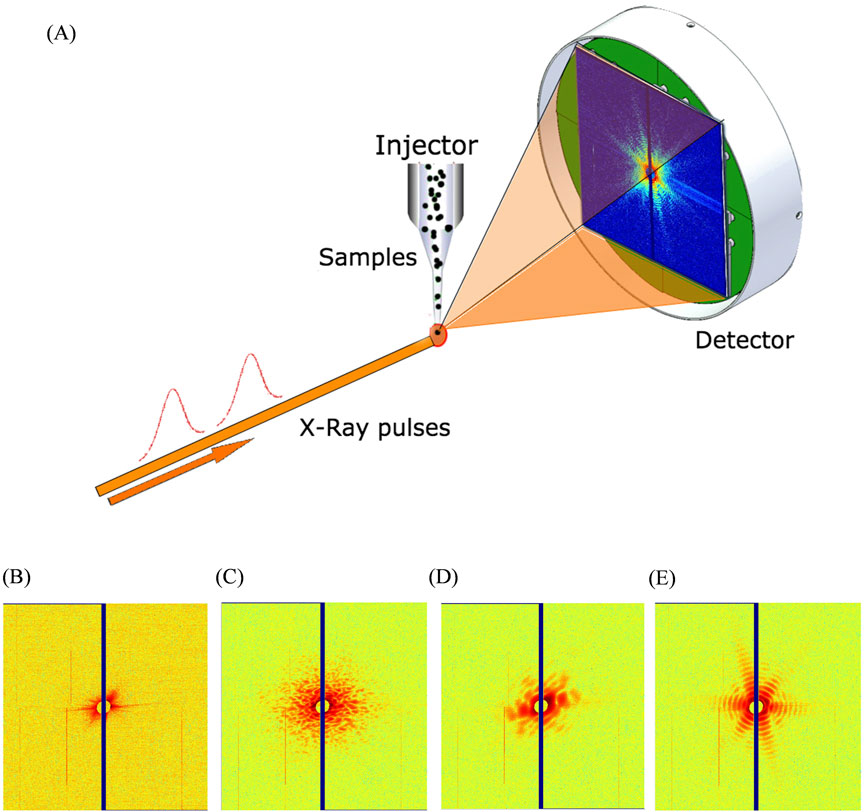
Figure 3. (A) SPI experiment setup. (B) Blank pattern. (C–D) Multiple particles. (E) Diffraction data of a single particle of an icosahedral virus. The first step in data analysis is to identify single-particle datasets within the massive dataset that can be used for reconstruction. Reproduced with permission from (Liu et al., 2019).
Initially, single-particle data classification was manually implemented. However, in many cases, especially with high repetition rate free-electron lasers, data volumes exceeding 106/h posed significant challenges for data classification. In 2011, C. Yoon et al. presented an unbiased, accurate, and computationally efficient method for classifying experimental X-ray diffraction snapshots without relying on templates, specific noise models, or user-directed learning (Yoon et al., 2011). The approach utilized spectral clustering, a kernel-based Principal Component Analysis (PCA) method (Ham et al., 2004), which leveraged nonlinear correlations in the dataset across various length scales to classify snapshots. The results demonstrated a 90% agreement with manual classification. In 2015,S. A. Bobkov et al. applied PCA and support vector machine (SVM) (Cortes and Vapnik, 1995) algorithms to the simulated and measured single particle imaging data sets (Bobkov et al., 2015). This approach relies on constructing a feature vector using parameters that reflect the underlying diffraction physics. These parameters capture the relationship between a particle’s real-space structure and its reciprocal-space intensity distribution. While both methods demonstrate accurate clustering of the simulated data, only the SVM algorithm allowed to classify different biological species in the experimental data set. In 2019, J. Liu et al. presented two supervised template-based learning methods for classifying SPI patterns (Liu et al., 2019). The Eigen-Image and LogLikelihood classifiers can quickly find the best-matched template for a single-molecule pattern within milliseconds and can be easily parallelized to match the XFEL repetition rate, allowing for on-site processing. These methods demonstrate stable performance on different types of synthetic data and were tested on a real mimivirus dataset (Arslan et al., 2011; Seibert, et al., 2011), achieving a classification accuracy of 0.9.
These custom-defined algorithms are developed with significant effort to approximate the specific features associated with an individual specimen so they can be effective to some extent. Nevertheless, when encountering diverse experimental conditions, these methods may struggle to generalize effectively. Researchers must carefully consider the advantages and disadvantages of utilizing generic machine learning classification methods as opposed to custom algorithms in order to determine the most appropriate approach for their research requirements. J. Zimmermann et al. presented the use of a deep neural network (Resnet 18) as a feature extractor for wide-angle diffraction images of helium nanodroplets (Figure 4), showcasing its effectiveness in sorting and classifying complex diffraction patterns (Zimmermann et al., 2019). The deep neural networks outperformed previous algorithms and provided valuable assistance in post-processing large amounts of experimental imaging data.
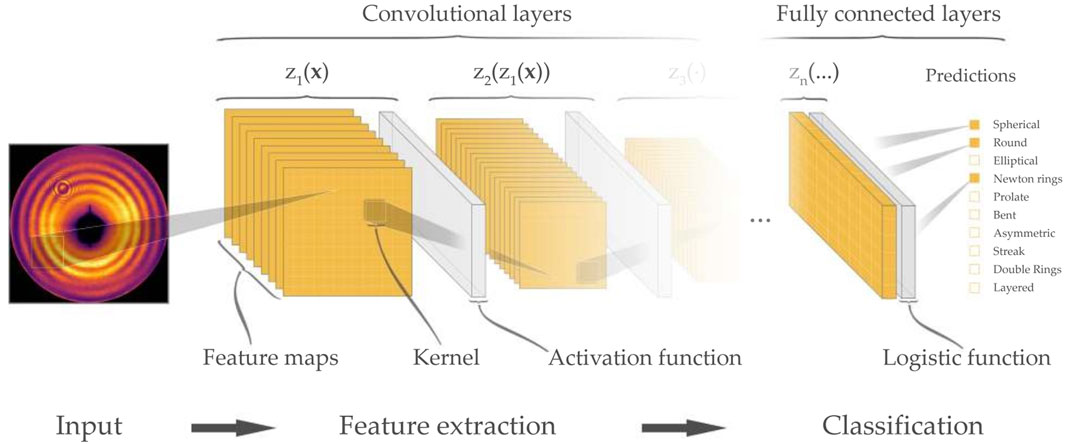
Figure 4. Schematic visualization of a convolutional neural network used for image classification. Reproduced with permission from (Zimmermann et al., 2019).
At the same time, two supervised algorithms based on the convolutional neural network (CNN) and graph cut (GC) framework were introduced to SPI data classification (Shi et al., 2019; Yin and Tai, 2017). The performance of these algorithms have been compared to the DM manifold embedding method (Giannakis et al., 2012; Giannakis et al., 2012) in PR772 virus particles (Coetzee et al., 1979). The results demonstrated that the common subset formed by the consensus of three methods might be less biased towards any particular conformational state. In 2021, Ignatenko et al. employed the fast object detector networks YOLOv2 and YOLOv3 to classify SPI data (Ignatenko et al., 2021). They compared the classification results of the two networks with different depth and architecture by applying them to the same SPI data with varied data representation. In the next year, they introduced two CNN configurations optimized for maximizing F1 score and high recall, respectively, and combines them with expectation-maximization (EM) selection and size filtering techniques (Assalauova et al., 2022). The introduction of CNNs streamlines the reconstruction pipeline (Figure 5), enables real-time pattern classification, and enhances control over experiment duration in SPI experiments.

Figure 5. CNN based SPI workflow. SPI workflow. Black arrows indicate the typical steps in SPI data analysis (Assalauova et al., 2020). Blue arrows show the implementation of CNN-based single-hit diffraction-pattern classification (Ignatenko et al., 2021). Red arrows show the modified workflow for CNN-based classification prior to the particle size filtering step. In the initial SPI data analysis workflow, Ivan et al. incorporated a CNN architecture for diffraction image classification. This method, in conjunction with various filtering techniques, resulted in a higher accuracy single-particle dataset, thereby improving the resolution of 3D reconstruction. Reproduced with permission from (Assalauova et al., 2022).
While supervised learning solutions based on artificial neural network (NN) models scale linearly, they necessitate a large number of labeled examples from data collected during beamtime and additional time for model training, thus preventing real-time classification. In 2023 C. Yoon et al. introduce SpeckleNN (Figure 6), a unified embedding model (Wang et al., 2023) for real-time speckle pattern classification with limited labeled examples that can scale linearly with dataset size. This method achieves few-shot classification on new, unseen samples. It performs robustly even with only a few dozen labels per category and can handle significant missing detector areas. Without the need for extensive manual labeling or even complete detector images, this method also showcases the great potential of deep learning in real-time high-throughput SPI experimental data classification. In the same year, J. Zimmermann introduced a Resnet50-D-based self-supervised contrastive projection learning method to find the semantic similarity in single-particle diffraction patterns (Zimmermann et al., 2023), achieving a dimensionality reduction producing semantically meaningful embeddings that align with physical intuition.
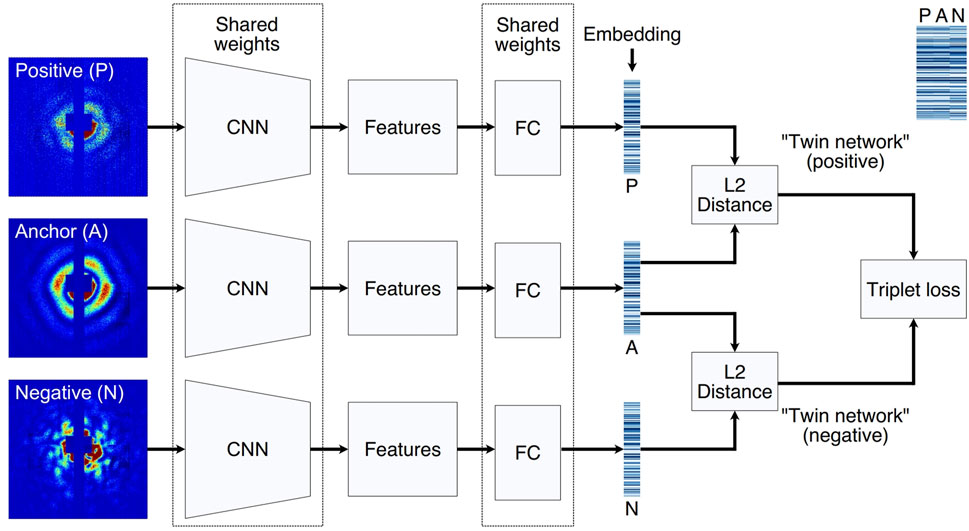
Figure 6. Triplet network architecture for model training. Three input examples (anchor, positive and negative) are propagated through the triplet NN simultaneously. Anchor and positive share the same label, thus forming a matching pair. In contrast, anchor and negative do not share the same label, thus forming an opposing pair. The three CNNs and FC layers share the same weights in the triplet network. After examples are embedded to a lowdimensional vector space, a triplet loss function is used to simultaneously maximize similarities between matching embeddings and minimize those between opposing embeddings. A side-by-side comparison of three embeddings in a triplet are annotated at the upper right corner. Reproduced with permission from (Wang et al., 2023).
Beyond single-particle classification, deep learning has found applications in other ultra-fast experimental data classifications. For example, in SAXS image classification, Convolutional Neural Networks (CNNs) and Convolutional Autoencoders have been employed (Wang et al., 2017). In defect classification, a 3D CNN was developed and trained to rapidly and accurately classify defects in nanocrystals of common face-centered cubic (fcc) transition metals (Lim et al., 2021). To further enhance defect identification efficiency, a novel data generation mechanism, termed “smart continual learning,” was introduced, surpassing previously published approaches (Yildiz et al., 2024). In femtosecond crystallography, three weakly supervised CNN models were utilized to identify SFX frames with crystal diffraction (Xie et al., 2023). Additionally, an unsupervised deep learning framework was developed for automated classification of relaxation dynamics in X-ray photon correlation spectroscopy. This approach requires no prior physical knowledge of the system, demonstrating its potential for analyzing experimental data (Horwath et al., 2024).
Single-particle imaging involves capturing the coherent X-ray diffraction intensity of particles in reciprocal space to determine their internal complex structure (Bielecki et al., 2020; Chapman and Nugent, 2010; Walmsley, 2015; Xiong et al., 2014). The phase information corresponding to this intensity is lost during measurement. To recover this missing phase information, iterative phase retrieval methods (Elser, 2003; Elser et al., 2007; Fienup, 1982; Gerchberg, 1972; Luke, 2005) alternate between detector and object spaces, applying constraints iteratively. Except for special cases of ill-posed structures, a unique reconstruction result can be expected in two or three dimensions, depending on the known symmetries of the Fourier transform.
However, these iterative phase retrieval methods typically require numerous iterations to converge to a confident solution, making them time-consuming and sensitive to factors like the initial guess of X-ray intensity phase, assumed support boundaries, and algorithm (Gao et al., 2021; Marchesini, 2007; Shechtman et al., 2015). Studies (Huang et al., 2010; Marchesini et al., 2003; Wang Z. et al., 2020) have highlighted the importance of correctly determining the support for convergence. While methods like “shrink-wrap” (Marchesini et al., 2003) support and adaptive support (Wang Z. et al., 2020) have been proposed, they still necessitate a substantial number of iterations to converge. Moreover, for structured nanoscale particles with complex phase-shifting domains, such as those in superconductors or catalysts, iterative methods struggle to provide high-confidence solutions due to their multi-center diffraction patterns. Consequently, the challenge of obtaining reliable unique solutions to the phase problem limits the effectiveness of iterative phase retrieval methods in single-particle imaging experiments.
Efforts have been made to overcome the issues caused by time-consuming and computationally expensive phase retrieval algorithms. In 2018, Cherukara et al. introduced CDI NN (Figure 7A), a pair of deep deconvolutional networks capable of predicting structure and phase in the real space of a 2D object from its far-field diffraction intensities (Cherukara et al., 2018). The network required two separate training stages, one for learning the mapping from 2D diffraction patterns to real-space structure and another for phase mapping. Trained CDI NN can quickly convert diffraction patterns into images within milliseconds on a standard desktop machine, enabling real-time imaging without the previous constraints of being time-consuming and computationally expensive. Building on this success, 2 years later, they proposed 3D-CDI-NN (Chan et al., 2020), a deep convolutional neural network and differential programming framework designed for 3D nanoscale X-ray imaging (Figure 7B). They enhanced the network architecture by introducing a novel design comprising one encoder and two decoders. In this updated network, the input consists of 3D intensity data, while the output includes both amplitude and phase information. This new architecture shifts the mapping pattern from a one-to-one mapping to a one-to-two mapping, allowing for the establishment of a comprehensive mapping relationship between the input data and the two output datasets through a single training process.
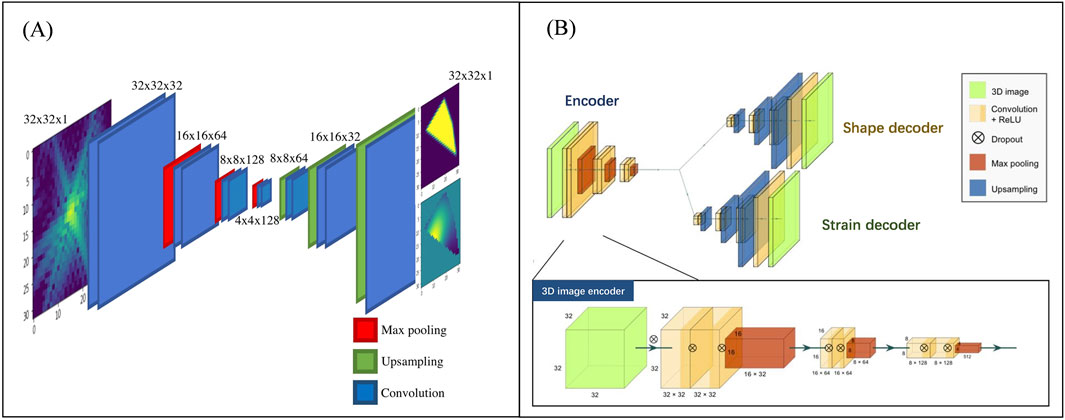
Figure 7. The network architecture used for phase recovery by Cherukara et al. (A) depicts the initial structure used for 2D data in 2018 (Cherukara et al., 2018) (B) shows the improved network structure in 2020 (Chan et al., 2020) Reproduced with permission from (Chan et al., 2020; Cherukara et al., 2018).
In 2021, I. Robinson et al. achieved the reconstruction of amplitude and phase information from the 2D diffraction modulus in reciprocal space with high accuracy and speed using a similar one2two-network architecture (Wu et al., 2020). Furthermore, in response to the challenge of achieving a distinct inversion of experimental data amidst noise, they proposed to overcome this limitation by integrating a similar network architecture to 3D intensity phase retrieval (Wu et al., 2020). Additionally, the model can refine predicted outcomes using transfer learning and learn missing phases in an image solely through minimizing a suitable ‘loss function.’ Demonstrations have shown significantly improved performance with experimental Bragg CDI data compared to traditional iterative phase retrieval algorithms.
However, such deep learning models require vast amounts of labeled data, which can only be obtained through simulation or performing computationally prohibitive phase retrieval on hundreds of or even thousands of experimental datasets. In 2021, Y. Yao introduced an unsupervised learning method named AutoPhaseNN (Yao et al., 2021) (Figure 8) to tackle the requirement for extensive labeled data. Using 3D nanoscale X-ray imaging (specifically Bragg Coherent Diffraction Imaging or BCDI) as an example, AutoPhaseNN is a deep learning-based approach that tackles the phase retrieval problem without requiring labeled data. By incorporating the physics of the imaging technique into the model during training, AutoPhaseNN can invert 3D BCDI data from reciprocal space to real space in a single step. Once trained, AutoPhaseNN is around one hundred times faster than traditional iterative methods while producing similar image quality.
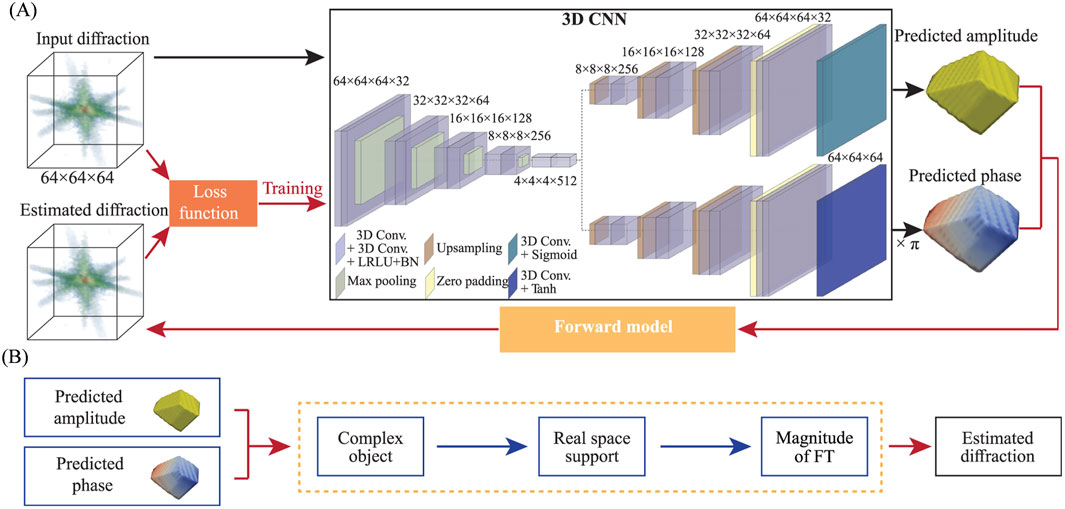
Figure 8. Diagram of the neural network structure of the AutoPhaseNN model. (A) The model consists of a 3D CNN and an X-ray scattering forward model. The 3D CNN utilizes one convolutional encoder and two deconvolutional decoders. (B) The X-ray scattering forward model includes numerical diffraction modeling and image shape constraints. It extracts amplitude and phase from the output of the 3D CNN to form a complex image. Subsequently, the estimated diffraction pattern is obtained by performing Fourier Transform on the estimate of the real-space image. The estimation in (B) is used to optimize the loss function in (A). Reproduced with permission from (Yao et al., 2021).
Later, Ian et al. further improved the network architecture (Yu et al., 2024). They noted that most previous studies used real-valued neural networks for phase retrieval problems, treating the amplitude and phase (or real and imaginary) information of a sample as two separate outputs with weak physical connections between them. Building on this observation, they introduced complex-valued operations in a CNN architecture (Figure 9) to better capture the relationship between phase and amplitude. The complex-valued neural network based approach outperforms the traditional real-valued neural network methods in both supervised and unsupervised learning manner.
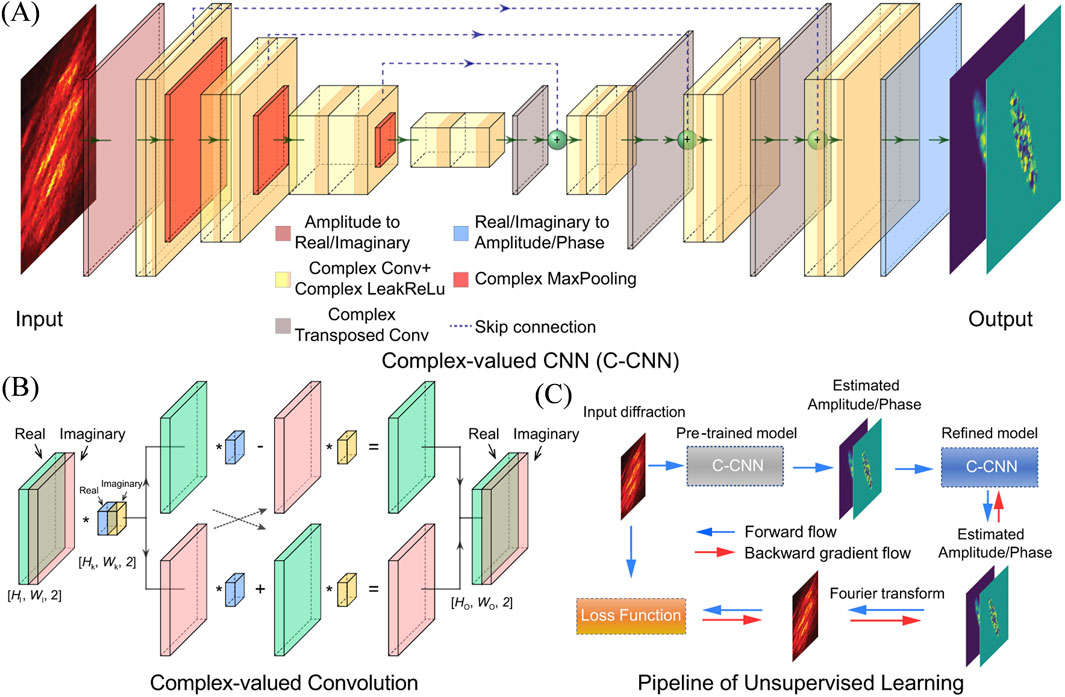
Figure 9. Schematic illustration of the complex-valued neural network for phase retrieval. (A) Architecture of the C-CNN model. (B) Convolutional operation for complex number. (C) Pipeline of the unsupervised learning for real experimental data. The output of the pre-trained model on synthetic data is employed as the starting point, and the training is continued with a Fourier transform constraint on experimental data. Reproduced with permission from (Yu et al., 2024).
Achieving high resolution for small biological structures, such as proteins, viruses, is challenging because it requires a low background and a high hit rate. Photons scattered from the sample delivery’s residual driving gas and optical instruments will add a background signal that is often comparable in strength to the signal originating from sample. The presence of experimental noise in largely fluctuating diffraction data poses practical challenges that impede consistent phase recovery.
Deep networks have also been applied in image denoising (Liang and Liu 2015; Xu et al., 2015), which is an important branch of image processing technologies. Deep learning has also been applied for image denoising in the field of ultrafast imaging. In 2021, C. Song et al. implemented a k-space CNN model (Figure 10) to address this issue (Lee et al., 2021). The network utilizes a U-Net architecture, where the decode layers have skipped connections with corresponding encode layers, and convolution operations are replaced by partial convolutions. Despite being a fully data-driven network, this model demonstrates excellent performance in improving image quality. The model effectively handles noise in coherent diffraction patterns, enhancing phase retrieval performance for single-pulse diffraction patterns obtained from XFEL experiments without requiring any prior object information.
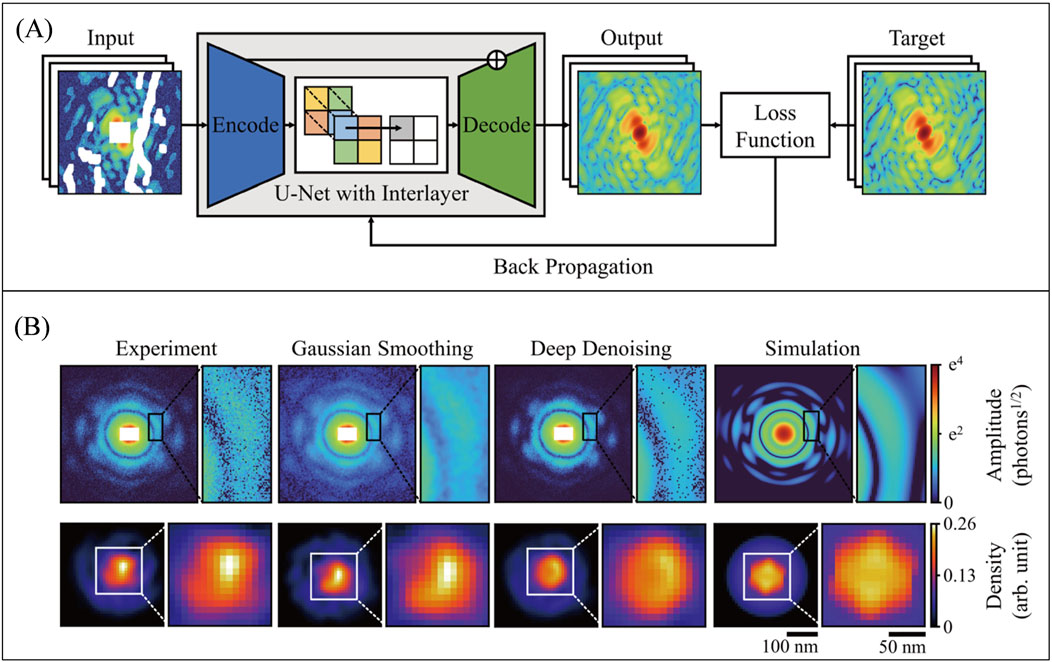
Figure 10. (A) The network structure for k-space denoising is based on a U-Net design, comprising an encoder and a decoder. Skipped connections exist between the encoder and decoder layers, and convolution operations are replaced by partial convolutions. (B) In the comparison of denoising results, it is evident that the deep learning denoising method, as opposed to Gaussian smoothing, preserves more details and closely resembles the true image. The reconstruction results also indicate an improvement in the reconstructed data after deep learning denoising compared to the original data. Reproduced with permission from (Lee et al., 2021).
Bellisario et al. also introduced U-Net network pipeline that aims to restore diffraction intensities to tackle the challenges posed by noise and masks (Bellisario et al., 2022). The method was compared with a low-pass filtering algorithm based on autocorrelation constraints. Results show a significant improvement in mean-squared error when masks were used, with demasking effective for masks smaller than half the central speckle size. This model demonstrates competitiveness in data processing and real-time restoration of diffraction intensities using deep learning. Preprocessing enhances orientation recovery reliability, especially for datasets with limited patterns, using the expansion-maximization-compression algorithm.
Additionally, regarding CDI phase retrieval, ambiguity is observed, leading to inconsistencies in the retrieved images under different initial conditions. Various guiding methods have been utilized to enhance the reliability of retrieval algorithms (Chen et al., 2007; Chou et al., 2003; Chou and Lee, 2002). To enhance robustness, the regularization-by-denoising framework and a convolutional neural network denoiser were leveraged to introduce prDeep (Metzler et al., 2018). Through simulations, prDeep exhibits noise resilience and can accommodate a variety of system models.
Moreover, the recommendation of utilizing free log-likelihood as an unbiased metric for CDI and the successful application of eigen-solution analysis through singular value decomposition (SVD) on datasets to reduce ambiguity have both been shown to be effective strategies (Favre-Nicolin et al., 2020). Later in 2024, A mixed-scale dense network architecture based on the Noise2Noise approach was proposed to denoise CDI images and mitigate ambiguity (Chu et al., 2024). This approach has the potential to enhance the quality of CDI reconstruction, providing benefits such as high-resolution output images from a trained network and rapid transformation from noisy to clear images.
For reproducible targets, small-angle X-ray scattering (SAXS) patterns can be used with conventional algorithms to reconstruct three-dimensional structures. A new algorithm based on a deep learning method for model reconstruction from SAXS data was presented (He et al., 2020) (Figure 11A). Later, Molodenskiy et al. proposed a method (Figure 11B) for primary SAXS data analysis. This approach predicts molecular weight and maximum intraparticle distance directly from experimental data with higher accuracy and better robustness against simulated experimental noise compared to existing methods (Molodenskiy et al., 2022).
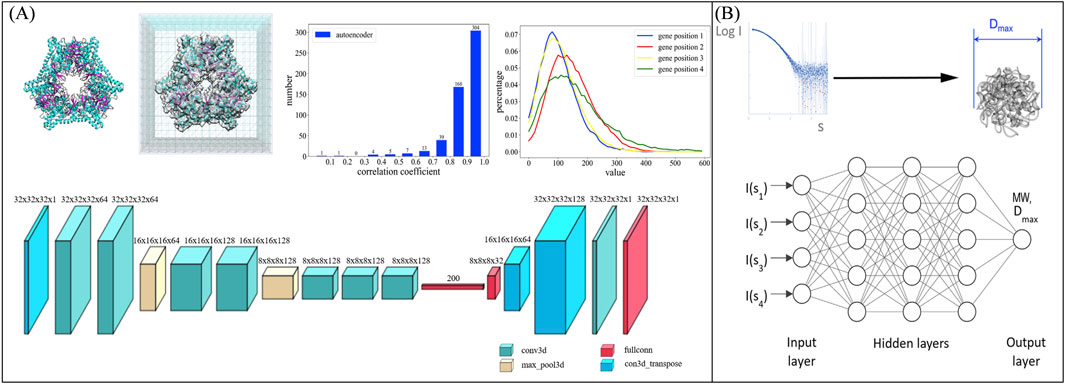
Figure 11. (A) Side shows a schematic diagram of the network structure for reconstructing low-resolution structures of samples from SAXS data (He et al., 2020), while (B) illustrates the network structure for extracting sample parameters from SAXS curves. Reproduced with permission from (Molodenskiy et al., 2022).
However, short-lived or non-reproducible objects lack of the multiple images required for three-dimensional reconstruction. Wide-angle scattering provides partial three-dimensional information (Barke et al., 2015; Langbehn et al., 2018; Rupp et al., 2017) but presents a more complex inversion challenge (Raines et al., 2010; Wang et al., 2011; Wei, 2011). The key obstacle is developing a rapid reconstruction method for effective single-shot structure characterization.
Wide-angle scattering differs from small-angle scattering in two main ways: the projection approximation is invalid due to the longitudinal wavevector component, and materials with non-unity refractive indices require consideration of multiple factors. Solving the full three-dimensional scattering problem using methods like FDTD, DDA, or MSFT allows for the description of wide-angle scattering patterns based on a nanoparticle’s geometry model (Gessner and Vilesov, 2019; Langbehn et al., 2018). Deriving the geometry from these patterns is highly complex due to the lack of a straightforward inversion method.
In 2020, T. Stielow et al. utilized the Resnet architecture (Figure 12A) with augmented theoretical scattering data to accurately and rapidly reconstruct wide-angle scattering images of individual icosahedral nanostructures (Stielow et al., 2020). Their results show that a network trained only on theoretical data can effectively analyze experimental scattering data. Image processing is completed in milliseconds, significantly faster than direct optimization methods. The next year, they developed a physics-informed deep neural network (Figure 12B) which can be used to reconstruct complete three-dimensional object models of uniform, convex particles on a voxel grid from single two-dimensional wide-angle scattering patterns (Stielow and Scheel, 2021). In classical supervised learning, the loss function is based on the binary cross entropy between the network prediction and the target entry for each data pair. In contrast, the loss function of the physics-associated network is computed in the scatter space, not the object space. This is done by simulating the scattering pattern of both the network prediction as well as the target object, and calculating their mean-squared difference (scatter loss). To enforce the binary nature of the object model, an additional regularization function (binary loss) is applied to the prediction.
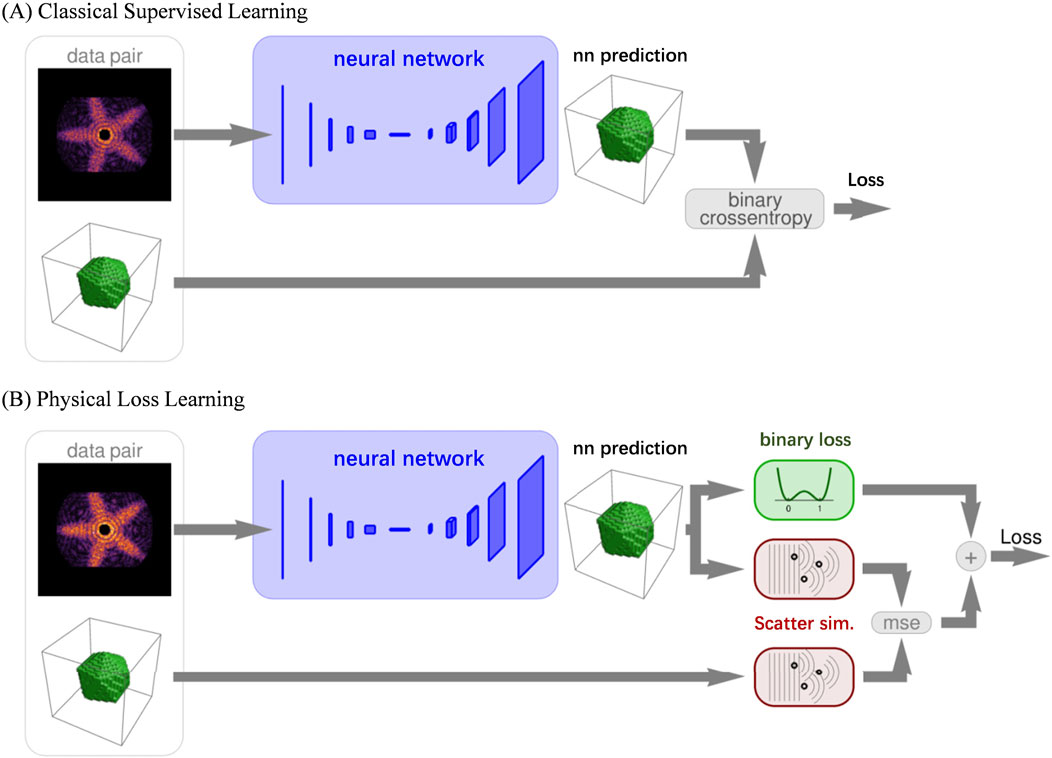
Figure 12. The network architecture for reconstructing sample information using WAXS. (A) Shows the network structure used for 2D reconstruction in 2020, utilizing ResNet as the network architecture and Crossentropy as the loss function. (B) Presents the enhanced network structure introduced in 2021, incorporating the physical process of sample diffraction as prior information to optimize the loss function. Reproduced with permission from (Stielow and Scheel, 2021). Copyright American Physical Society and SciPris.
In this review, we have summarized the current advancements and trends in the application of deep learning for ultrafast X-ray imaging. Specifically, we focused on its effectiveness in image classification, phase retrieval, and denoising, areas where deep learning algorithms have shown significant promise. The ability of deep learning to address the challenges posed by big data, leveraging AI-driven features, has significantly accelerated and improved the efficiency of scientific data processing. Moreover, the AI techniques employed require minimal prior domain-specific knowledge, making them highly adaptable. As a result, state-of-the-art architectures from fields such as computer vision can be seamlessly transferred and tailored to the ultrafast X-ray imaging domain. Nevertheless, there are still several aspects that need further refinement to fully harness the potential of deep learning for ultrafast X-ray imaging applications.
Deep neural networks require large datasets to perform effectively. This poses a significant challenge in fields like ultrafast X-ray imaging, particularly with high-repetition-rate free-electron lasers that produce millions of images per second. The large volume of high-throughput data makes data labeling both expensive and labor-intensive. To address this issue, several potential solutions are available.
By applying transformations, intensity adjustments, noise addition, and operations like cropping or padding to images, the dataset can be expanded. This method increases the diversity of the data, helping the model to learn more generalized features, thereby enhancing its robustness. However, in the context of experimental data, a neural network trained on fixed settings may struggle to adapt to different scenarios not explicitly represented in the training dataset.
Transfer learning involves pre-training a network on a data-rich task and then transferring the learned weights to a new task, leveraging similarities between domains (Figure 13). A common practice is to pre-train on large datasets such as ImageNet, as the features learned in the initial layers of the network are often transferable to other tasks. This not only provides a better starting point but also improves the network’s robustness, particularly when fine-tuning on smaller, domain-specific datasets. Well-known deep learning models, like ResNet and U-Net, are pre-trained on large datasets and then directly applied to ultrafast imaging data. These models, trained on general data, are adapted to the specific nature of ultrafast imaging data.
In some cases, combining simulated and real data can be effective. By initially training on simulated datasets and then fine-tuning with a small amount of real experimental data, the network can achieve good performance even with limited data. Pre-training the model on offline simulated data before applying it to real-time experimental data allows for real-time interpretation of ultrafast imaging data via deep learning techniques. However, the success of neural networks is highly dependent on the quality of the training data. Paired training sets typically contain fixed input-to-label mappings, which set an upper limit on the network’s learning capacity. If the distribution of the simulated training data differs significantly from real-world data, the network’s ability to generalize will be compromised.
The deep learning for phase recovery were divided into two class, “network-only” and “network-with-physics” strategies (Wang et al., 2024). The “network-only” category can be further classified into “data-driven” and “physics-driven” networks. In a “physics-driven network”, physical constraints are typically integrated into the loss function. In contrast, “network-with-physics” refers to the integration of network with physical processes, where each training iteration of the network corresponds to an iteration of the physical process itself. This classification framework is also applicable to the networks discussed in this paper.
As highlighted in the previous section, networks used for ultrafast imaging evolve from simple “data-driven networks” to more advanced “physics-driven networks”. Researchers are actively working to bridge the gap between the opaque nature of purely data-driven approaches and the underlying physical processes that govern the system.
However, these networks do not incorporate physical processes into network, and the training process lacks interpretation. The primary advantage of this approach lies in its simplicity and efficiency. Given the wide variety of deep learning architectures available for image processing tasks, users can easily design models by configuring the input and output layers and selecting or defining appropriate loss functions. During training, large datasets are processed, and millions of parameters are optimized to learn complex relationships within the data. It is challenging to understand the meaning of the features extracted by individual layers and the implications of internal parameters on the overall model performance. This lack of transparency complicates the identification of failure causes, hindering targeted optimization efforts. In such cases, the process may seem driven by chance, as it is difficult to predict success or failure based on physical principles before model construction, or to identify clear avenues for improvement post-failure.
Since light propagation can be described through physical models, the experimental process can be simulated numerically. This enables the use of a “network-with-physics” approach, where the training process is integrated with physical iterations. Several models already employ this approach for deep learning-based phase recovery (BaoShun and QiuSheng, 2022; Wang C. J. et al., 2020; Wu et al., 2022; Yang et al., 2022; Zhang et al., 2021). In these models, diffraction intensity is not the sole input; fixed or latent vectors are also incorporated. Each training iteration of the network is accompanied by a corresponding physical iteration. This integration of physical processes into the network training enhances interpretability and provides clearer insights into the model’s behavior, facilitating more informed optimization.
Although research on deep learning-based denoising methods in this field remains limited, existing studies have demonstrated that deep learning can improve the reconstruction of sample structures from noisy data. Even so, it is important to note that the effectiveness of deep neural networks in reconstructing “real images” from noisy or incomplete data has its limitations. As the signal-to-noise ratio drops and the amount of missing data grows, the difference between network-reconstructed images and the ground truth tends to increase. This highlights a fundamental limitation of even advanced deep learning techniques in recovering information that is not captured by the underlying hardware imaging system. The region highlighted in red in Figure 14 clearly illustrates this.
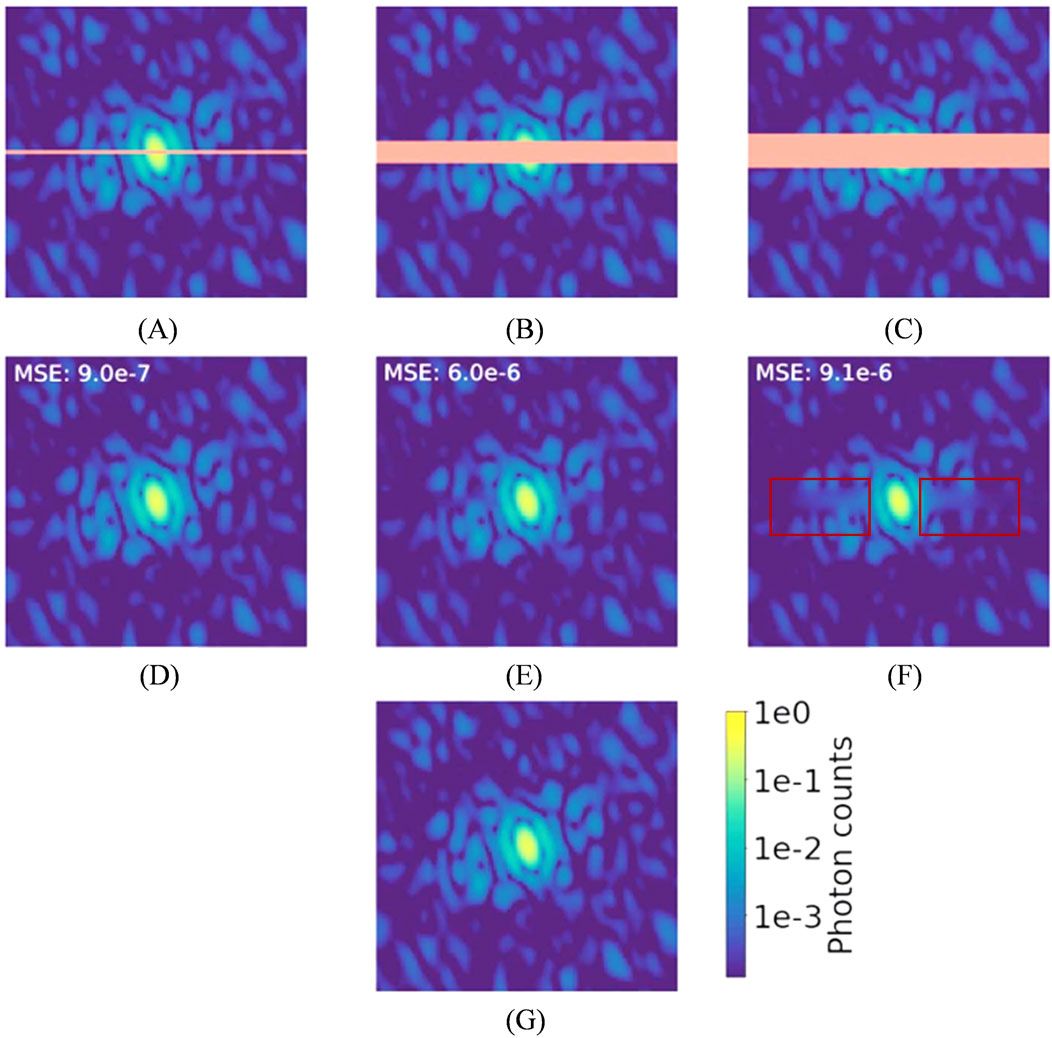
Figure 14. Diffraction patterns for human aurora A catalytic domain (PDB 4zs0; Kilchmann et al., 2016), logarithmic scale color map. Orange is used in the top row to highlight masked pixels. (A) A simulated diffraction pattern with a two pixel-wide mask. (B) A simulated diffraction pattern with a 10 pixel-wide mask. (C) A simulated diffraction pattern with a 15 pixelwide mask. (D–F) U-Net demasked output. (G) Simulated diffraction intensities without a mask. Reproduced with permission from (Bellisario et al., 2022).
Therefore, integrating data with hardware parameters and optimizing deep learning models based on diffraction or scattering physical models represents a promising direction for future research. The performance of neural networks in recovering missing data indicates that, despite their significant potential and efficiency, black-box models trained on large datasets lack the integration of physical knowledge. As a result, these models cannot guarantee the accuracy of their output results.
Given these challenges, it is equally crucial to develop physics-informed networks that integrate physical principles into the training process and network architecture, thereby providing greater physical interpretability. Furthermore, in practical scenarios, where the true values of data obscured by noise or missing information are unknown, there is a clear need to develop evaluation methods that assess the reliability of the network’s inference results.
Deep neural network architectures are computationally efficient, often outperforming many state-of-the-art algorithms. Their computational cost at inference time is typically lower than traditional methods. This runtime advantage comes at the expense of high computational costs during training, which can be time-consuming, even on GPU clusters. By optimizing problem domains and training setups, this efficiency can be leveraged to reduce runtime at the expense of extended training time.
The integration of deep learning into ultrafast X-ray scattering and imaging represents a transformative advancement, effectively addressing challenges in data processing, phase retrieval, denoising, and classification in XFEL experiments. Researchers have made substantial advances in high-throughput data processing, real-time pattern recognition, and nanoscale structural reconstruction by taking advantage of AI-driven approaches. These advancements enable the investigation of intricate physical phenomena with greater effectiveness and precision compared to traditional methods.
Notable challenges still persist. Deep learning models continue to rely heavily on big datasets, and the mismatch between simulated and experimental data frequently restricts their applicability in practical applications. Furthermore, the interpretability of many network topologies remains inadequate. This highlights the need for physics-informed designs that incorporate domain knowledge to enhance model reliability and robustness. Future research should prioritize creating data-efficient algorithms, integrating physical principles into AI models, and enhancing the interpretability of deep learning methods.
As the field evolves, the synergy between deep learning and ultrafast X-ray science holds immense potential to drive breakthroughs across disciplines such as biology, materials science, and chemistry. This progress underscores the critical importance of interdisciplinary collaboration, where expertise in AI, physics, and domain-specific knowledge converges to address some of the most complex challenges in ultrafast imaging and scattering.
MH: Writing–original draft, Writing–review and editing, Funding acquisition, Investigation. JF: Writing–review and editing. YT: Funding acquisition, Writing–review and editing. ZS: Writing–review and editing. HJ: Funding acquisition, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We would like to thank the National Natural Science Foundation of China (Grant No. 12105176) for its financial support. This research was also supported by the Strategic Priority Research Program of the Major State Basic Research Development Program of China (Grant No. 2022YFA1603703), and the National Natural Science Foundation of China (Grant No. 12335020) and Shanghai Action Plan for Science, Technology and Innovation (No. 24JD1402200).
We extend our appreciation to our colleagues for their constructive discussions and valuable suggestions, which have greatly enriched the quality of this work. We also gratefully acknowledge the support from the Shanghai High Repetition rate XFEL and Extreme Light Facility (SHINE) and the Shanghai Soft X-ray Free-Electron Laser User Facility.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/aot.2025.1546386/full#supplementary-material
Aquila, A., Barty, A., Bostedt, C., Boutet, S., Carini, G., dePonte, D., et al. (2015). The linac coherent light source single particle imaging road map. Struct. Dyn. 2 (4), 041701. doi:10.1063/1.4918726
Arslan, D., Legendre, M., Seltzer, V., Abergel, C., and Claverie, J. M. (2011). Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. 108 (42), 17486–17491. doi:10.1073/pnas.1110889108
Assalauova, D., Ignatenko, A., Isensee, F., Trofimova, D., and Vartanyants, I. A. (2022). Classification of diffraction patterns using a convolutional neural network in single-particle-imaging experiments performed at X-ray free-electron lasers. J. Appl. Crystallogr. 55 (3), 444–454. doi:10.1107/s1600576722002667
Assalauova, D., Kim, Y. Y., Bobkov, S., Khubbutdinov, R., Rose, M., Alvarez, R., et al. (2020). An advanced workflow for single particle imaging with the limited data at an X-ray free-electron laser. iUCrJ 7, 1102–1113. doi:10.1107/S2052252520012798
Ayyer, K., Xavier, P. L., Bielecki, J., Shen, Z., Daurer, B. J., Samanta, A. K., et al. (2021). 3D diffractive imaging of nanoparticle ensembles using an x-ray laser. Optica 8 (1), 15–23. doi:10.1364/OPTICA.410851
Baldi, P., and Sadowski, P. (2014). The dropout learning algorithm. Artif. Intell. 210, 78–122. doi:10.1016/j.artint.2014.02.004
BaoShun, S., and QiuSheng, L. (2022). DualPRNet: deep shrinkage dual frame network for deep unrolled phase retrieval. IEEE Signal Process. Lett. 29, 1177–1181. doi:10.1109/LSP.2022.3169695
Barke, I., Hartmann, H., Rupp, D., Flückiger, L., Sauppe, M., Adolph, M., et al. (2015). The 3D-architecture of individual free silver nanoparticles captured by X-ray scattering. Nat. Commun. 6 (1), 6187. doi:10.1038/ncomms7187
Bellisario, A., Maia, F. R. N. C., and Ekeberg, T. (2022). Noise reduction and mask removal neural network for X-ray single-particle imaging. J. Appl. Crystallogr. 55 (1), 122–132. doi:10.1107/s1600576721012371
Bielecki, J., Maia, F. R. N. C., and Mancuso, A. P. (2020). Perspectives on single particle imaging with x rays at the advent of high repetition rate x-ray free electron laser sources. Struct. Dyn. 7 (4), 040901. doi:10.1063/4.0000024
Bobkov, S. A., Teslyuk, A. B., Kurta, R. P., Gorobtsov, O. Y., Yefanov, O. M., Ilyin, V. A., et al. (2015). Sorting algorithms for single-particle imaging experiments at X-ray free-electron lasers. J. Synchrotron Radiat. 22 (6), 1345–1352. doi:10.1107/s1600577515017348
Bogan, M. J., Boutet, S., Chapman, H. N., Marchesini, S., Barty, A., Benner, W. H., et al. (2010). Aerosol imaging with a soft X-ray free electron laser. Aerosol Sci. Technol. 44 (3), i–vi. doi:10.1080/02786820903485800
Bostedt, C., Boutet, S., Fritz, D. M., Huang, Z., Lee, H. J., Lemke, H. T., et al. (2016). Linac coherent light source: the first five years. Rev. Mod. Phys. 88 (1), 015007. doi:10.1103/RevModPhys.88.015007
Bostedt, C., Thomas, H., Hoener, M., Eremina, E., Fennel, T., Meiwes-Broer, K. H., et al. (2008). Multistep ionization of argon clusters in intense femtosecond extreme ultraviolet pulses. Phys. Rev. Lett. 100 (13), 133401. doi:10.1103/PhysRevLett.100.133401
Chan, H., Nashed, Y. S. G., Kandel, S., Hruszkewycz, S., Sankaranarayanan, S., Harder, R. J., et al. (2020). Real-time 3D nanoscale coherent imaging via physics-aware deep learning. Phys. Comput. Sci. 16. doi:10.48550/arXiv.2006.09441
Chapman, H. N., Barty, A., Bogan, M. J., Boutet, S., Frank, M., Hau-Riege, S. P., et al. (2006). Femtosecond diffractive imaging with a soft-X-ray free-electron laser. Nat. Phys. 2 (12), 839–843. doi:10.1038/nphys461
Chapman, H. N., and Nugent, K. A. (2010). Coherent lensless X-ray imaging. Nat. Photonics 4 (12), 833–839. doi:10.1038/nphoton.2010.240
Chen, C. C., Miao, J., Wang, C. W., and Lee, T. K. (2007). Application of optimization technique to noncrystalline x-ray diffraction microscopy: guided hybrid input-output method. Phys. Rev. B. 76 (76), 064113. doi:10.1103/PhysRevB.76.064113
Cherukara, M. J., Nashed, Y. S. G., and Harder, R. J. (2018). Real-time coherent diffraction inversion using deep generative networks. Sci. Rep. 8 (1), 16520. doi:10.1038/s41598-018-34525-1
Chou, C. I., Han, R. S., Li, S. P., and Lee, T. K. (2003). Guided simulated annealing method for optimization problems. Phys. Rev. E 67 (6 Pt 2), 066704. doi:10.1103/PhysRevE.67.066704
Chou, C. I., and Lee, T. K. (2002). A guided simulated annealing method for crystallography. Acta Crystallogr. Sect. A Found. Crystallogr. 58 (1), 42–46. doi:10.1107/S0108767301015537
Chu, K.-C., Yeh, C.-H., Lin, J.-M., Chen, C.-Y., Cheng, C.-Y., Yeh, Y.-Q., et al. (2024). Using convolutional neural network denoising to reduce ambiguity in X-ray coherent diffraction imaging. J. Synchrotron Radiat. 31 (5), 1340–1345. doi:10.1107/s1600577524006519
Clark, J. N., Beitra, L., Xiong, G., Fritz, D. M., Lemke, H. T., Zhu, D., et al. (2015). Imaging transient melting of a nanocrystal using an X-ray laser. Proc. Natl. Acad. Sci. U. S. A. 112 (24), 7444–7448. doi:10.1073/pnas.1417678112
Clark, J. N., Beitra, L., Xiong, G., Higginbotham, A., Fritz, D. M., Lemke, H. T., et al. (2013). Ultrafast three-dimensional imaging of lattice dynamics in individual gold nanocrystals. Science 341 (6141), 56–59. doi:10.1126/science.1236034
Coetzee, J. N., Lecatsas, G., Coetzee, W. F., and Hedges, R. W. (1979). Properties of R Plasmid R772 and the corresponding pilus-specific phage PR772. J. General Microbiol. 110 (2), 263–273. doi:10.1099/00221287-110-2-263
Cortes, C., and Vapnik, V. (1995). Support-vector networks. Mach. Learn. 20 (3), 273–297. doi:10.1007/bf00994018
Decking, W., Abeghyan, S., Abramian, P., Abramsky, A., Aguirre, A., Albrecht, C., et al. (2020). A MHz-repetition-rate hard X-ray free-electron laser driven by a superconducting linear accelerator. Nat. Photonics 14 (6), 391–397. doi:10.1038/s41566-020-0607-z
DePonte, D. P., Weierstall, U., Schmidt, K., Warner, J., Starodub, D., Spence, J. C. H., et al. (2008). Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D Appl. Phys. 41 (19), 195505. doi:10.1088/0022-3727/41/19/195505
Ekeberg, T., Svenda, M., Abergel, C., Maia, F. R. N. C., Seltzer, V., Claverie, J.-M., et al. (2015). Three-dimensional reconstruction of the giant mimivirus particle with an x-ray free-electron laser. Phys. Rev. Lett. 114 (9), 098102. doi:10.1103/PhysRevLett.114.098102
Elser, V. (2003). Phase retrieval by iterated projections. J. Opt. Soc. Am. A 20 (1), 40–55. doi:10.1364/JOSAA.20.000040
Elser, V., Rankenburg, I. C., and Thibault, P. (2007). Searching with iterated maps. Proc. Natl. Acad. Sci. 104, 418–423. doi:10.1073/pnas.0606359104
Emma, P., Akre, R., Arthur, J., Bionta, R., Bostedt, C., Bozek, J., et al. (2010). First lasing and operation of an ångstrom-wavelength free-electron laser. Nat. Photonics 4, 641–647. doi:10.1038/nphoton.2010.176
Farabet, C., Couprie, C., Najman, L., and Lecun, Y. (2013). Learning hierarchical features for scene labeling. IEEE Trans. Pattern Anal. Mach. Intell. 35 (8), 1915–1929. doi:10.1109/tpami.2012.231
Favre-Nicolin, V., Leake, S., and Chushkin, Y. (2020). Free log-likelihood as an unbiased metric for coherent diffraction imaging. Sci. Rep. 10 (1), 2664. doi:10.1038/s41598-020-57561-2
Feldman, J. A., Fanty, M. A., and Goodard, N. H. (1988). Computing with structured neural networks. Computer 21 (3), 91–103. doi:10.1109/2.34
Ferguson, K. R., Bucher, M., Gorkhover, T., Boutet, S., Fukuzawa, H., Koglin, J. E., et al. (2016). Transient lattice contraction in the solid-to-plasma transition. Sci. Adv. 2 (1), e1500837. doi:10.1126/sciadv.1500837
Fienup, J. R. (1982). Phase retrieval algorithms: a comparison. Appl. Opt. 21 (15), 2758–2769. doi:10.1364/ao.21.002758
Gaffney, K. J., and Chapman, H. N. (2007). Imaging atomic structure and dynamics with ultrafast X-ray scattering. Science 316 (5830), 1444–1448. doi:10.1126/science.1135923
Gallagher-Jones, M., Bessho, Y., Kim, S., Park, J., Kim, S., Nam, D., et al. (2014). Macromolecular structures probed by combining single-shot free-electron laser diffraction with synchrotron coherent X-ray imaging. Nat. Commun. 5, 3798. doi:10.1038/ncomms4798
Gao, Y., Huang, X., Yan, H., and Williams, G. J. (2021). Bragg coherent diffraction imaging by simultaneous reconstruction of multiple diffraction peaks. Phys. Rev. B 103 (1), 014102. doi:10.1103/PhysRevB.103.014102
Gatys, L. A., Ecker, A. S., and Bethge, M. (2016). A neural algorithm of artistic style. J. Vis. 16, 326. doi:10.1167/16.12.326
Gerchberg, R. W. (1972). A practical algorithm for the determination of phase from image and diffraction plane pictures. Optik 35, 237–246.
Gessner, O., and Vilesov, A. F. (2019). Imaging quantum vortices in superfluid helium droplets. Annu. Rev. Phys. Chem. 70, 173–198. doi:10.1146/annurev-physchem-042018-052744
Giannakis, D., Schwander, P., and Ourmazd, A. (2012). The symmetries of image formation by scattering. I. Theoretical framework. Opt. Express 20 (12), 12799–12826. doi:10.1364/OE.20.012799
Goodfellow, I. J., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). “Generative adversarial nets,” in Proceedings of the Proceedings of the 27th International Conference on Neural Information Processing Systems 2, 2672–2680. doi:10.1145/3422622
Gorkhover, T., Adolph, M., Rupp, D., Schorb, S., Epp, S. W., Erk, B., et al. (2012). Nanoplasma dynamics of single large xenon clusters irradiated with superintense X-ray pulses from the linac coherent light source free-electron laser. Phys. Rev. Lett. 108 (24), 245005. doi:10.1103/PhysRevLett.108.245005
Gorkhover, T., Schorb, S., Coffee, R., Adolph, M., Foucar, L., Rupp, D., et al. (2016). Femtosecond and nanometre visualization of structural dynamics in superheated nanoparticles. Nat. Photonics 10, 93–97. doi:10.1038/nphoton.2015.264
Ham, J., Lee, D. D., Mika, S., and Schölkopf, B. (2004). “A kernel view of the dimensionality reduction of manifolds,” in Proceedings of the Proceedings of the twenty-first international conference on Machine learning, 47. doi:10.1145/1015330.1015417
Hantke, M. F., Hasse, D., Maia, F. R. N. C., Ekeberg, T., John, K., Svenda, M., et al. (2014). High-throughput imaging of heterogeneous cell organelles with an X-ray laser. Nat. Photonics 8, 943–949. doi:10.1038/nphoton.2014.270
He, H., Liu, C., and Liu, H. (2020). Model reconstruction from small-angle X-ray scattering data using deep learning methods. iScience 23 (3), 100906. doi:10.1016/j.isci.2020.100906
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770–778. doi:10.1109/CVPR.2016.90
Hinton, G. E., Osindero, S., and Teh, Y. (2006). A fast learning algorithm for deep belief nets. Neural Comput. 18 (7), 1527–1554. doi:10.1162/neco.2006.18.7.1527
Hinton, G. E., and Salakhutdinov, R. R. (2006). Reducing the dimensionality of data with neural networks. Science 313 (5786), 504–507. doi:10.1126/science.1127647
Hopfield, J. J. (1982). Neural networks and physical systems with emergent collective computational abilities. Proc. Natl. Acad. Sci. U. S. A. 79 (8), 2554–2558. doi:10.1073/pnas.79.8.2554
Horwath, J. P., Lin, X.-M., He, H., Zhang, Q., Dufresne, E. M., Chu, M., et al. (2024). AI-NERD: elucidation of relaxation dynamics beyond equilibrium through AI-informed X-ray photon correlation spectroscopy. Nat. Commun. 15 (1), 5945. doi:10.1038/s41467-024-49381-z
Huang, N., Deng, H., Liu, B., Wang, D., and Zhao, Z. (2021). Features and futures of X-ray free-electron lasers. Innovation 2 (2), 100097. doi:10.1016/j.xinn.2021.100097
Huang, X., Nelson, J., Steinbrener, J., Kirz, J., Turner, J. J., and Jacobsen, C. (2010). Incorrect support and missing center tolerances of phasing algorithms. Opt. Express 18 (25), 26441–26449. doi:10.1364/OE.18.026441
Ignatenko, A., Assalauova, D., Bobkov, S. A., Gelisio, L., Teslyuk, A. B., Ilyin, V. A., et al. (2021). Classification of diffraction patterns in single particle imaging experiments performed at X-ray free-electron lasers using a convolutional neural network. Mach. Learn. Sci. Technol. 2 (2), 025014. doi:10.1088/2632-2153/abd916
Ihm, Y., Cho, D. H., Sung, D., Nam, D., Jung, C., Sato, T., et al. (2019). Direct observation of picosecond melting and disintegration of metallic nanoparticles. Nat. Commun. 10 (1), 2411. doi:10.1038/s41467-019-10328-4
Ioffe, S., and Szegedy, C. (2015). “Batch normalization: accelerating deep network training by reducing internal covariate shift,” in Proceedings of the 32nd International Conference on International Conference on Machine Learning 37, 448–456. doi:10.5555/3045118.3045167
Isola, P., Zhu, J. Y., Zhou, T., and Efros, A. A. (2017). Image-to-Image translation with conditional adversarial networks. 2017 IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), 5967–5976. doi:10.1109/cvpr.2017.632
Johnson, J., Alahi, A., and Fei-Fei, L. (2016). Perceptual losses for real-time style transfer and super-resolution. Cham: Springer.
Jordan, M. I., and Mitchell, T. M. (2015). Machine learning: trends, perspectives, and prospects. Science 349 (6245), 255–260. doi:10.1126/science.aaa8415
Jung, C., Ihm, Y., Cho, D. H., Lee, H., Nam, D., Kim, S., et al. (2021). Inducing thermodynamically blocked atomic ordering via strongly driven nonequilibrium kinetics. Sci. Adv. 7 (52), eabj8552. doi:10.1126/sciadv.abj8552
Kangas, J. A., Kohonen, T. K., and Laaksonen, J. T. (1990). Variants of self-organizing maps. IEEE Trans. Neural Netw. 1 (1), 93–99. doi:10.1109/72.80208
Karras, T., Aila, T., Laine, S., and Lehtinen, J. (2017). Progressive growing of GANs for improved quality, stability, and variation.
Kilchmann, F., Marcaida, M. J., Kotak, S., Schick, T., Boss, S. D., Awale, W., et al. (2016). Discovery of a selective aurora a kinase inhibitor by virtual screening. J. Medici. Chemi. 59 (15), 7188–7211. doi:10.1021/acs.jmedchem.6b00709
Kimura, T., Joti, Y., Shibuya, A., Song, C., Kim, S., Tono, K., et al. (2014). Imaging live cell in micro-liquid enclosure by X-ray laser diffraction. Nat. Commun. 5, 3052. doi:10.1038/ncomms4052
Kirian, R. A., Awel, S., Eckerskorn, N., Fleckenstein, H., Wiedorn, M., Adriano, L., et al. (2015). Simple convergent-nozzle aerosol injector for single-particle diffractive imaging with X-ray free-electron lasers. Struct. Dyn. 2 (4), 041717. doi:10.1063/1.4922648
Klabjan, D., and Harmon, M. (2019). “Activation ensembles for deep neural networks,” in Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), 206–214. doi:10.1109/BigData47090.2019.9006069
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). ImageNet classification with deep convolutional neural networks. Communications of the ACM 60, 84–90. doi:10.1145/3065386
Langbehn, B., Sander, K., Ovcharenko, Y., Peltz, C., Clark, A., Coreno, M., et al. (2018). Three-dimensional shapes of spinning helium nanodroplets. Phys. Rev. Lett. 121 (25), 255301. doi:10.1103/PhysRevLett.121.255301
Lecun, Y., and Bengio, Y. (1995). Convolutional networks for images, speech, and time-series. Handb. Brain Theory and Neural Netw., 255–258. doi:10.5555/303568.303704
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521 (7553), 436–444. doi:10.1038/nature14539
Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998). Gradient-based learning applied to document recognition. Proc. IEEE 86 (11), 2278–2324. doi:10.1109/5.726791
Lee, S. Y., Cho, D. H., Jung, C., Sung, D., Nam, D., Kim, S., et al. (2021). Denoising low-intensity diffraction signals using k-space deep learning: applications to phase recovery. Phys. Rev. Res. 3 (4), 043066. doi:10.1103/PhysRevResearch.3.043066
Liang, J., and Liu, R. (2015). “Stacked denoising autoencoder and dropout together to prevent overfitting in deep neural network,” in Proceedings of the 2015 8th International Congress on Image and Signal Processing (CISP), 697–701. doi:10.1109/CISP.2015.7407967
Lim, B., Bellec, E., Dupraz, M., Leake, S., Resta, A., Coati, A., et al. (2021). A convolutional neural network for defect classification in Bragg coherent X-ray diffraction. npj Comput. Mater. 7 (1), 115. doi:10.1038/s41524-021-00583-9
Lindroth, E., Calegari, F., Young, L., Harmand, M., Dudovich, N., Berrah, N., et al. (2019). Challenges and opportunities in attosecond and XFEL science. Nat. Rev. Phys. 1 (2), 107–111. doi:10.1038/s42254-019-0023-9
Lippmann, R. (1987). An introduction to computing with neural nets. IEEE ASSP Mag. 4 (2), 4–22. doi:10.1109/MASSP.1987.1165576
Liu, J., van der Schot, G., and Engblom, S. (2019). Supervised classification methods for flash X-ray single particle diffraction imaging. Opt. Express 27 (4), 3884. doi:10.1364/oe.27.003884
Loh, N. D., Hampton, C. Y., Martin, A. V., Starodub, D., Sierra, R. G., Barty, A., et al. (2012). Fractal morphology, imaging and mass spectrometry of single aerosol particles in flight. Nature 486, 513–517. doi:10.1038/nature11222
Luke, D. R. (2005). Relaxed averaged alternating reflections for diffraction imaging. Inverse Probl. 21 (1), 37–50. doi:10.1088/0266-5611/21/1/004
Marchesini, S. (2007). Invited article: a [corrected] unified evaluation of iterative projection algorithms for phase retrieval. Rev. Sci. Instrum. 78 (1), 011301. doi:10.1063/1.2403783
Marchesini, S., He, H., Chapman, H. N., Hau-Riege, S. P., Noy, A., Howells, M. R., et al. (2003). X-ray image reconstruction from a diffraction pattern alone. Phys. Rev. B 68 (14), 140101. doi:10.1103/PhysRevB.68.140101
Metzler, C. A., Schniter, P., Veeraraghavan, A., and Baraniuk, R. G. (2018). prDeep: robust phase retrieval with a flexible deep network. Comput. Sci. Phys. Eng. doi:10.48550/arXiv.1803.00212
Miao, J., Ishikawa, T., Robinson, I. K., and Murnane, M. M. (2015). Beyond crystallography: diffractive imaging using coherent x-ray light sources. Science 348 (6234), 530–535. doi:10.1126/science.aaa1394
Miao, J., Sayre, D., and Chapman, H. N. (1998). Phase retrieval from the magnitude of the Fourier transforms of nonperiodic objects. J. Opt. Soc. Am. A 15, 1662–1669. doi:10.1364/JOSAA.15.001662
Molodenskiy, D. S., Svergun, D. I., and Kikhney, A. G. (2022). Artificial neural networks for solution scattering data analysis. Structure 30 (6), 900–908.e2. doi:10.1016/j.str.2022.03.011
Nair, V., and Hinton, G. E. (2010). “Rectified linear units improve restricted boltzmann machines,” in Proceedings of the Proceedings of the 27th International Conference on International Conference on Machine Learning, 807–814. doi:10.5555/3104322.3104425
Neutze, R., Wouts, R., van der Spoel, D., Weckert, E., and Hajdu, J. (2000). Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757. doi:10.1038/35021099
O'Shea, K., and Nash, R. (2015). An introduction to convolutional neural networks. Comput. Sci. doi:10.48550/arXiv.1511.08458
Pellegrini, C. (2016). X-ray free-electron lasers: from dreams to reality. Phys. Scr. T169, 014004. doi:10.1088/1402-4896/aa5281
Radford, A., Metz, L., and Chintala, S. (2015). Unsupervised representation learning with deep convolutional generative adversarial networks. doi:10.48550/arxiy1511.06434
Raines, K. S., Salha, S., Sandberg, R. L., Jiang, H., Rodríguez, J. A., Fahimian, B. P., et al. (2010). Three-dimensional structure determination from a single view. Nature 463 (7278), 214–217. doi:10.1038/nature08705
Redmon, J., Divvala, S., Girshick, R., and Farhadi, A. (2016). “You Only Look Once: Unified, Real-Time Object Detection,” in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 779–788. doi:10.1109/CVPR.2016.91
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-Net: Convolutional networks for biomedical image segmentation,” in Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 234–241. doi:10.1007/978-3-319-24574-4_28
Rosenblatt, F. (1958). The perceptron: a probabilistic model for information storage and organization in the brain. Psychol. Rev. 65 (6), 386–408. doi:10.1037/h0042519
Rupp, D., Monserud, N., Langbehn, B., Sauppe, M., Zimmermann, J., Ovcharenko, Y., et al. (2017). Coherent diffractive imaging of single helium nanodroplets with a high harmonic generation source. Nat. Commun. 8 (1), 493. doi:10.1038/s41467-017-00287-z
Seibert, M. M., Ekeberg, T., Maia, F. R. N. C., Svenda, M., Andreasson, J., Jonsson, O., et al. (2011). Single mimivirus particles intercepted and imaged with an X-ray laser. Nature 470 (7332), 78–81. doi:10.1038/nature09748
Shechtman, Y., Eldar, Y. C., Cohen, O., Chapman, H. N., Miao, J., and Segev, M. (2015). Phase retrieval with application to optical imaging: a contemporary overview. IEEE Signal Process. Mag. 32 (3), 87–109. doi:10.1109/MSP.2014.2352673
Shi, Y., Yin, K., Tai, X., DeMirci, H., Hosseinizadeh, A., Hogue, B. G., et al. (2019). Evaluation of the performance of classification algorithms for XFEL single-particle imaging data. IUCrJ 6 (2), 331–340. doi:10.1107/s2052252519001854
Sierra, R. G., Laksmono, H., Kern, J., Tran, R., Hattne, J., Alonso-Mori, R., et al. (2012). Nanoflow electrospinning serial femtosecond crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 68 (11), 1584–1587. doi:10.1107/S0907444912038152
Silver, D., Schrittwieser, J., Simonyan, K., Antonoglou, I., Huang, A., Guez, A., et al. (2017). Mastering the game of Go without human knowledge. Nature 550 (7676), 354–359. doi:10.1038/nature24270
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. Comput. Sci. doi:10.48550/arXiv.1409.1556
Stielow, T., and Scheel, S. (2021). Reconstruction of nanoscale particles from single-shot wide-angle free-electron-laser diffraction patterns with physics-informed neural networks. Phys. Rev. E 103 (5), 053312. doi:10.1103/PhysRevE.103.053312
Stielow, T., Schmidt, R., Peltz, C., Fennel, T., and Scheel, S. (2020). Fast reconstruction of single-shot wide-angle diffraction images through deep learning. Mach. Learn. Sci. Technol. 1 (4), 045007. doi:10.1088/2632-2153/abb213
Sun, M., Song, Z., Jiang, X., Pan, J., and Pang, Y. (2017). Learning pooling for convolutional neural network. Neurocomputing 224, 96–104. doi:10.1016/j.neucom.2016.10.049
Sun, Z., Fan, J., Li, H., Liu, H., Nam, D., Kim, C., et al. (2018). Necessary experimental conditions for single-shot diffraction imaging of DNA-based structures with X-ray free-electron lasers. ACS Nano 12 (8), 7509–7518. doi:10.1021/acsnano.8b01838
Tompson, J., Jain, A., Lecun, Y., and Bregler, C. (2014). Joint training of a convolutional network and a graphical model for human pose estimation. Eprint Arxiv, 1799–1807. doi:10.48550/arXiv.1406.2984
Vincent, P., Larochelle, H., Bengio, Y., and Manzagol, P.-A. (2008). “Extracting and composing robust features with denoising autoencoders,” in Proceedings of the Proceedings of the 25th international conference on Machine learning, 1096–1103. doi:10.1145/1390156.1390294
Walmsley, I. A. (2015). Quantum optics: science and technology in a new light. Science 348 (6234), 525–530. doi:10.1126/science.aab0097
Wang, C., Florin, E., Chang, H.-Y., Thayer, J., and Yoon, C. H. (2023). SpeckleNN: a unified embedding for real-time speckle pattern classification in X-ray single-particle imaging with limited labeled examples. IUCrJ 10 (5), 568–578. doi:10.1107/s2052252523006115
Wang, C. J., Wen, C. K., Tsai, S. H. L., and Jin, S. (2020b). Phase retrieval with learning unfolded expectation consistent signal recovery algorithm. IEEE Signal Process. Lett. 27 (99), 780–784. doi:10.1109/lsp.2020.2990767
Wang, G., Yu, H., Cong, W., and Katsevich, A. (2011). Non-uniqueness and instability of ‘ankylography. Nature 480 (7375), E2–E3. doi:10.1038/nature10635
Wang, K., Song, L., Wang, C., Ren, Z., Zhao, G., Dou, J., et al. (2024). On the use of deep learning for phase recovery. Light. and Appl. 13 (2), 4–235. doi:10.1038/s41377-023-01340-x
Wang, B., Yager, K., Yu, D., and Hoai, M. (2017). “X-Ray scattering image classification using deep learning,” in Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), 697–704. doi:10.1109/WACV.2017.83
Wang, Z., Gorobtsov, O., and Singer, A. (2020a). An algorithm for Bragg coherent x-ray diffractive imaging of highly strained nanocrystals. New J. Phys. 22 (1), 013021. doi:10.1088/1367-2630/ab61db
Wei, H. (2011). Fundamental limits of ‘ankylography’ due to dimensional deficiency. Nature 480 (7375), E1. doi:10.1038/nature10634
Wu, L., Juhas, P., Yoo, S., and Robinson, I. (2020). Complex imaging of phase domains by deep neural networks. IUCrJ 8 (1), 12–21. doi:10.1107/s2052252520013780
Wu, L., Yoo, S., Suzana, A. F., Assefa, T. A., Diao, J., Harder, R. J., et al. (2021). Three-dimensional coherent X-ray diffraction imaging via deep convolutional neural networks. npj Comput. Mater. 7 (1), 175. doi:10.1038/s41524-021-00644-z
Wu, X., Wu, Z., Shanmugavel, S. C., Yu, H. Z., and Zhu, Y. (2022). Physics-informed neural network for phase imaging based on transport of intensity equation. Opt. Express 30 (24), 43398–43416. doi:10.1364/OE.462844
Xie, J., Liu, J., Zhang, C., Chen, X., Huai, P., Zheng, J., et al. (2023). Weakly supervised learning for pattern classification in serial femtosecond crystallography. Opt. Express 31 (20), 32909. doi:10.1364/OE.492311
Xiong, G., Moutanabbir, O., Reiche, M., Harder, R., and Robinson, I. (2014). Coherent X-ray diffraction imaging and characterization of strain in silicon-on-insulator nanostructures. Adv. Mater. Deerf. Beach, Fla. 26 (46), 7747–7763. doi:10.1002/adma.201304511
Xu, Q., Zhang, C., and Zhang, L. (2015). “Denoising convolutional neural network,” in Proceedings of the 2015 IEEE International Conference on Information and Automation, 1184–1187. doi:10.1109/ICInfA.2015.7279466
Xu, R., Jiang, H., Song, C., Rodriguez, J. A., Huang, Z., Chen, C.-C., et al. (2014). Single-shot three-dimensional structure determination of nanocrystals with femtosecond X-ray free-electron laser pulses. Nat. Commun. 5, 4061. doi:10.1038/ncomms5061
Yang, Y., Qiusheng, L., Zhang, X., Zhang, D., and Zhang, H. (2022). HIONet: deep priors based deep unfolded network for phase retrieval. Digit. Signal Process. 132, 103797. doi:10.1016/j.dsp.2022.103797
Yao, Y., Chan, H., Sankaranarayanan, S., Balaprakash, P., Harder, R. J., and Cherukara, M. J. (2021). AutoPhaseNN: unsupervised physics-aware deep learning of 3D nanoscale coherent imaging. arXiv e-prints. doi:10.48550/arXiv.2109.14053
Yildiz, O., Raghavan, K., Chan, H., Cherukara, M. J., Balaprakash, P., Sankaranarayanan, S., et al. (2024). Automated defect identification in coherent diffraction imaging with smart continual learning. Neural Comput. Appl. 36 (35), 22335–22346. doi:10.1007/s00521-024-10415-8
Yin, K., and Tai, X. C. (2017). An effective region force for some variational models for learning and clustering. J. Sci. Comput. 74, 175–196. doi:10.1007/s10915-017-0429-4
Yoon, C. H., Schwander, P., Abergel, C., Andersson, I., Andreasson, J., Aquila, A., et al. (2011). Unsupervised classification of single-particle X-ray diffraction snapshots by spectral clustering. Opt. Express 19 (17), 16542–16549. doi:10.1364/oe.19.016542
Yu, X., Wu, L., Lin, Y., Diao, J., Liu, J., Hallmann, J., et al. (2024). Ultrafast Bragg coherent diffraction imaging of epitaxial thin films using deep complex-valued neural networks. npj Comput. Mater. 10 (1), 24. doi:10.1038/s41524-024-01208-7
Zhang, F., Liu, X., Guo, C., Lin, S., Jiang, J., and Ji, X. (2021). “Physics-based iterative projection complex neural network for phase retrieval in lensless microscopy imaging,”in 2021 IEEE/CVF conference on computer vision and pattern recognition (CVPR) (IEEE), 10518–10526. doi:10.1109/cvpr46437.2021.01038
Zhou, F., Adolphsen, C., Benwell, A., Brown, G., Dowell, D. H., Dunning, M., et al. (2021). Commissioning of the SLAC linac coherent light source II electron source. Phys. Rev. Accel. Beams 24 (7), 073401. doi:10.1103/PhysRevAccelBeams.24.073401
Zimmermann, J., Beguet, F., Guthruf, D., Langbehn, B., and Rupp, D. (2023). Finding the semantic similarity in single-particle diffraction images using self-supervised contrastive projection learning. npj Comput. Mater. 9 (1), 24. doi:10.1038/s41524-023-00966-0
Zimmermann, J., Langbehn, B., Cucini, R., Di Fraia, M., Finetti, P., LaForge, A. C., et al. (2019). Deep neural networks for classifying complex features in diffraction images. Phys. Rev. E 99 (6), 063309. doi:10.1103/PhysRevE.99.063309
Keywords: x-ray free electron Laser, coherent diffraction imaging, single particle imaging, deep Learning, machine learning
Citation: Hu M, Fan J, Tong Y, Sun Z and Jiang H (2025) Deep learning for ultrafast X-ray scattering and imaging with intense X-ray FEL pulses. Adv. Opt. Technol. 14:1546386. doi: 10.3389/aot.2025.1546386
Received: 16 December 2024; Accepted: 20 February 2025;
Published: 13 March 2025.
Edited by:
Bin Ji, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Lianghua Wen, Yibin University, ChinaCopyright © 2025 Hu, Fan, Tong, Sun and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaidong Jiang, amlhbmdoZEBzaGFuZ2hhaXRlY2guZWR1LmNu; Zhibin Sun, c3VuemhiQHNoYW5naGFpdGVjaC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.