
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Biosci. , 29 August 2016
Sec. Molecular Recognition
Volume 3 - 2016 | https://doi.org/10.3389/fmolb.2016.00044
This article is part of the Research Topic Modulating prokaryotic lifestyle by DNA-binding proteins View all 25 articles
In bacteria, active partition systems contribute to the faithful segregation of both chromosomes and low-copy-number plasmids. Each system depends on a site-specific DNA binding protein to recognize and assemble a partition complex at a centromere-like site, commonly called parS. Many plasmid, and all chromosomal centromere-binding proteins are dimeric helix-turn-helix DNA binding proteins, which are commonly named ParB. Although the overall sequence conservation among ParBs is not high, the proteins share similar domain and functional organization, and they assemble into similar higher-order complexes. In vivo, ParBs “spread,” that is, DNA binding extends away from the parS site into the surrounding non-specific DNA, a feature that reflects higher-order complex assembly. ParBs bridge and pair DNA at parS and non-specific DNA sites. ParB dimers interact with each other via flexible conformations of an N-terminal region. This review will focus on the properties of the HTH centromere-binding protein, in light of recent experimental evidence and models that are adding to our understanding of how these proteins assemble into large and dynamic partition complexes at and around their specific DNA sites.
In bacteria, the segregation, or partition, of low-copy-number plasmids and cellular chromosomes depends on the activity of site-specific DNA binding proteins to recognize one or more copies of a centromere-like DNA site. These “centromere-binding proteins” generally work in concert with an ATPase or GTPase, resulting in dynamic movement and positioning of plasmids or chromosomal domains during the cell cycle (reviewed in Wang et al., 2013; Baxter and Funnell, 2014; Bouet et al., 2014). Centromere-binding proteins fall into one of two structural classes, as helix-turn-helix (HTH), or ribbon-helix-helix site-specific DNA binding proteins. In bacteria, the proteins of all chromosomal, and many plasmid partition systems are members of the HTH class. They share similar properties in vivo and in vitro, including similar domain organization and DNA-binding properties, although there are also interesting differences. They all form large partition complexes in vivo that can be visualized as foci using fluorescence approaches. This review will focus on the properties of the HTH centromere-binding proteins, and in particular, how they assemble into large partition complexes. I will discuss the contribution of protein domains, the “spreading” phenomenon that has been reported as a general property of HTH centromere-binding proteins, and how flexibility in the protein allows multiple conformations and binding modes in complex assembly.
The components of partition systems are commonly named ParA (the partition ATPase), ParB (the centromere-binding protein), and parS (the centromere or partition site). Plasmids typically contain one parS located near the parA and parB genes, although some have multiple parS sites. Bacterial chromosomes contain several parS sites, which are primarily located in the chromosomal domain that contains the replication origin. Bacterial ParA and ParB are also called Soj and Spo0J, respectively, because their genes were first defined by roles in sporulation of Bacillus subtilis (Ireton et al., 1994). For simplicity, I will use the ParABS nomenclature, with specific names for some of the discussion to be consistent with published literature.
HTH ParBs share a similar domain organization although the primary sequence conservation is not high among members of this family. In general, the protein is divided into three regions: A central HTH DNA binding domain is flanked by a C-terminal dimer domain and an N-terminal region necessary for protein oligomerization (Figure 1A). Flexible linkers connect the domains, and flexibility in domain organization, orientation, and folding have been observed in biochemical experiments and crystal structures. The most highly conserved sequence among plasmid and chromosomal ParBs is a short arginine-rich motif in the N-terminus, which is often called an arginine patch (Yamaichi and Niki, 2000). ParA interactions are often specified by residues near the N-terminus of ParB (Radnedge et al., 1998; Figge et al., 2003; Leonard et al., 2005; Ah-Seng et al., 2009), although there are exceptions. For example, the ParA interactions for RK2 KorB and Pseudomonas aeruginosa ParB map to the center and dimer domain, respectively (Lukaszewicz et al., 2002; Bartosik et al., 2004). There are also added complexities to the general arrangement. For example, P1 ParB and its relatives contain an additional site-specific DNA binding activity within the dimer domain. These ParBs recognize both an inverted repeat and a second DNA motif in their parS sites, via their HTH domain and dimer domains, respectively (Schumacher and Funnell, 2005).
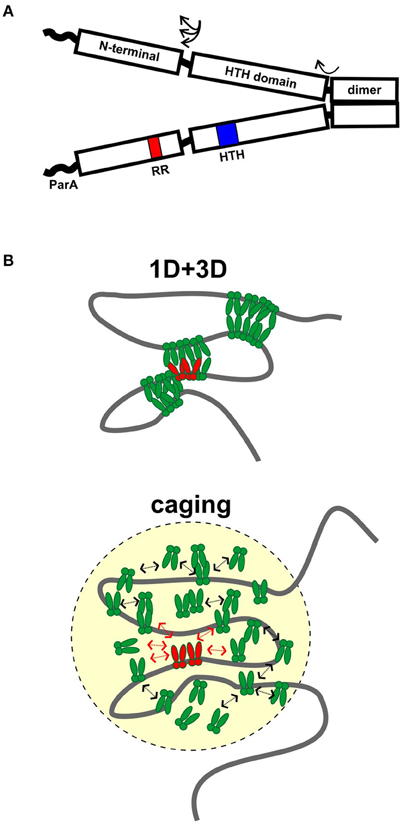
Figure 1. Assembly of ParB partition complexes. (A) Cartoon of the conserved domain structure of HTH ParBs, shown as a dimer. The black rectangles represent the regions of the protein for which there is some structural information: the C-terminal dimer domain, the HTH DNA binding domain, and the N-terminal domain. The three regions are connected by flexible linker sequences (arrows). The linker length here is represented as short, as in the HpSpo0J and P1 ParB published structures (Schumacher and Funnell, 2005; Chen et al., 2015), but may be longer in other ParBs. The wavy line represents the region that interacts with ParA in many, although not all, ParBs. The position of the HTH motif (blue) and the conserved arginine patch motif (RR, red) are indicated in one monomer. (B) Diagrams of 1D + 3D and caging models for higher-order ParB binding and partition complex assembly. ParB dimers bound to parS (in red) nucleate complex assembly and interact with other ParBs in green. Arrows in the caging architecture illustrate that dynamic associations maintain the cluster of ParB.
Partition complex assembly begins with the recognition of parS by a dimer of ParB, followed by loading of multiple ParB dimers to form a very large protein-DNA complex (Baxter and Funnell, 2014). These higher-order complexes are necessary as both the substrates and the activators of the mechanisms of partition. The number of ParB protein foci observed inside cells is usually lower than the number of parS sites, leading to the idea that inter and intra-molecular pairing of parS sites occurs in plasmids and chromosomal domains.
Spreading is an unusual feature for site-specific DNA binding proteins that is common to HTH ParBs, and it reflects how ParB assembles into higher-order complexes (Rodionov et al., 1999; Murray et al., 2006; Breier and Grossman, 2007; Sanchez et al., 2015). Measured by ChIP approaches, in vivo ParB binding extends beyond parS into the surrounding non-specific DNA, often many kb away from the site. Binding is maximal at parS, and diminishes non-linearly as a function of distance from parS. Spreading can be impeded by “roadblocks,” which are strong binding sites for other proteins (Rodionov et al., 1999; Murray et al., 2006). Spreading has also been inferred from the ability of some ParBs, especially when overexpressed, to silence expression of nearby genes (Lynch and Wang, 1995; Rodionov et al., 1999; Bartosik et al., 2004; Bingle et al., 2005; Kusiak et al., 2011). Silencing is likely a consequence of protein overexpression and is not necessary for partition (Rodionov and Yarmolinsky, 2004). Spreading ability is required however, as ParB mutants that do not spread are defective in partition (Rodionov et al., 1999; Autret et al., 2001; Breier and Grossman, 2007; Kusiak et al., 2011; Graham et al., 2014). In particular, the arginine patch in ParB is essential for spreading, focus formation, and partition activity in vivo.
The first and simplest model described spreading as lateral protein-protein association along the DNA as a one-dimensional filament (Rodionov et al., 1999). However some properties of ParBs lead investigators to question this idea. Studies with plasmid KorB and with B. subtilis Spo0J (BsSpo0J) argued that the intracellular concentration of ParB was insufficient to account for the amount of spreading observed in vivo if arranged as a one-dimensional filament (Bingle et al., 2005; Graham et al., 2014). It was also difficult to demonstrate biochemically that ParB binding to parS increased the affinity of ParB for adjacent non-specific DNA.
Two other models have emerged recently, based on sophisticated microscope and ChIP-seq technologies as well as computer modeling and traditional biochemistry (Broedersz et al., 2014; Graham et al., 2014; Sanchez et al., 2015). The first proposes that limited lateral ParB-ParB interactions (1D) in combination with inter and intra-molecular looping and bridging (3D) act to build large complexes and coalesce many ParB molecules into foci (Broedersz et al., 2014; Graham et al., 2014; Figure 1B). Computer modeling was used to argue that neither 1D nor 3D interactions alone could generate ParB foci; that the 1D + 3D arrangement allows focus formation because it creates a surface tension on the ParB cluster that counteracts the tendency for entropy to disperse the protein on DNA (Broedersz et al., 2014). Elegant in vitro TIRF microscopy experiments provided experimental support for the bridging activity: flow-stretched DNA was condensed by BsSpo0J in a manner that is most consistent with ParB bridging across loops within the same or across different DNA molecules (Graham et al., 2014). The experiments were however unable to demonstrate any sequence-specificity for parS, leading to the suggestion that experiments on flow-stretched DNA were not recapitulating an undefined aspect of critical nucleation properties of ParB bound to parS. For example, one factor missing from these experiments is DNA supercoiling, which affects chromosome compaction and may strongly influence the DNA binding properties of ParB in higher-order complexes.
Mutations in conserved arginine residues of the arginine patch motif eliminated bridging in the TIRF assay, consistent with the requirement for this motif for spreading and partition in vivo (Graham et al., 2014). Interestingly, BsSpo0J mutated at one residue in the arginine patch (G77S), which is unable to form foci or spread in vivo, was still able to bridge DNA, and with slightly higher stability than that of wild-type BsSpo0J. Therefore the bridging activity of ParB is necessary but not sufficient for complex formation. These observations lead to the suggestion that the G77S mutation may promote inappropriate bridging and/or alter the dynamics of bridging necessary for proper complex assembly in vivo. Modeling the spreading/bridging behavior also predicted that roadblocks would decrease the probability of loops forming in their vicinity, interfering with complex assembly beyond the roadblock (Broedersz et al., 2014).
In contrast, a second model proposes that a network of stochastic binding of ParB explains the clustering of ParB molecules around parS (Sanchez et al., 2015). In essence, the nucleation of ParB by parS creates, and maintains a very high localized concentration of ParB in a “cage” by many weak but dynamic interactions with itself (dimer-dimer interactions via the N-terminal domains), as well as with non-specific DNA around parS (Figure 1B). In caging, these interactions do not need to occur simultaneously or to bridge DNA (Figure 1B). Computer modeling of the patterns of ParB occupancy around parS measured by ChIP was used to argue that they are not consistent with either 1D lateral spreading or a combination of 1D spreading and 3D bridging. Biochemical examination showed no evidence that binding of one ParB could stabilize binding of an adjacent ParB. The caging model neither requires nor excludes bridging, although bridging interactions are intuitively attractive as part of the dynamic glue. The model does depend on other properties of the DNA chromosome, such as topology or organization by other nucleoid-binding proteins in vivo to help restrict the DNA within the cage. Roadblocks could alter local DNA organization and reduce the proximity of parS to the rest of the DNA in three-dimensional space; that is, place this DNA outside the cage.
Both models agree that ParB binding to parS must nucleate the formation of higher-order complexes to explain ParB clustering and foci in vivo (Broedersz et al., 2014; Sanchez et al., 2015). This is most simple to envision in plasmid systems such as P1, in which ParB's affinity for parS is at least 10,000-fold higher than that for non-specific DNA (Funnell, 1991). However this affinity difference is small for some ParBs (Broedersz et al., 2014; Taylor et al., 2015), and it was suggested that a conformational change is induced in ParB by parS-specific binding to effectively anchor the focus at the site. Both models agree that multiple ParB-ParB interaction interfaces must be involved in assembly of higher-order partition complexes, and that the dynamics of these interactions are critical for proper assembly and function in partition. How bridging activities detected in vitro contribute to ParB activity in vivo remains to be resolved. Further refinement of these models will depend on the ability to completely reconstitute the parS-dependent complex assembly in vitro, which will in turn depend on identifying the other factors necessary for caging or bridging, and on the nature of ParB-ParB and ParB-DNA interactions at the molecular level. The influence of ParA on complex architecture and dynamics has also yet to be defined (see below).
Site-specific DNA binding of ParBs to cognate parS sites has been examined directly and in detail in many different partition systems, but the parS-dependent formation of higher order, large partition complexes has been difficult to reconstitute in vitro. However, there are insights arising from structural biology of several plasmid and chromosomal ParBs, which are leading to a preliminary, albeit incomplete, picture of partition complex assembly.
Although there are no structures of a full length ParB, those of individual domains or combinations of domains, with and without parS DNA, have provided clues concerning the three dimensional organization of the protein with respect to DNA and to itself. There are structures of the HTH domains of three plasmid ParBs (P1 ParB, F SopB, and RP4 KorB) in complex with their specific DNA sites, and of their dimer domains (SopB and KorB dimer domains solved separately from the HTH; Delbrück et al., 2002; Khare et al., 2004; Schumacher and Funnell, 2005; Schumacher et al., 2010). Structures of two chromosomal ParB fragments, each containing the N-terminal region and adjacent HTH domain, have visualized the oligomerization interactions of the proteins. These ParBs are Thermus thermophilus Spo0J (TtSpo0J) and Helicobacter pylori Spo0J (HpSpo0J); structure of the latter was solved bound to parS (Leonard et al., 2004; Chen et al., 2015).
One of the first themes to highlight is that of flexibility. Taken together, the structures indicate that these three regions of the protein are connected by flexible linkers, and that their orientation with respect to each other can vary. The conformation of the N-terminus is particularly flexible (Chen et al., 2015).
Second, ParBs are bona-fide HTH site-specific DNA binding proteins, but with a twist. As expected, the HTH domains contact inverted repeat sequences within parS via helix insertion into the major groove of DNA. However, unexpected features emerged from the structures. First, residues outside of the recognition helix also contribute to specificity for parS in SopB, KorB, and HpSpo0J (Khare et al., 2004; Schumacher et al., 2010; Sanchez et al., 2013; Chen et al., 2015). Second, both the P1 ParB and HpSpo0J structures demonstrated bridging across parS sites mediated by the dimer and N-terminal domains, respectively (Schumacher and Funnell, 2005; Chen et al., 2015). Each monomer of a P1 ParB dimer interacts with a half-site on a different DNA molecule, effectively pairing two parS sites. HpSpo0J-parS bridging interactions are mediated by the N-terminal oligomerization regions of the protein (Figure 2). Four monomers of HpSpo0J (monomeric because it lacks the C-terminal dimer domain) interact with two parS oligos and with each other in a cross-bridge arrangement across the DNA molecules (molecules A–D in Figure 2). The monomers share a common HTH domain, but show different conformations in the extended N-terminal regions as well as different interactions with each other. There are two adjacent (AB and CD) and one transverse (AC) sets of protein-protein interactions, which are distinct. For example, the conserved arginine patch motifs are close to each other at the AC interface, but not at the AB or CD interfaces (Figure 2). In the structure, there is no BD interaction, leaving these surfaces available, perhaps for interactions with different conformations of ParB or with different partners.
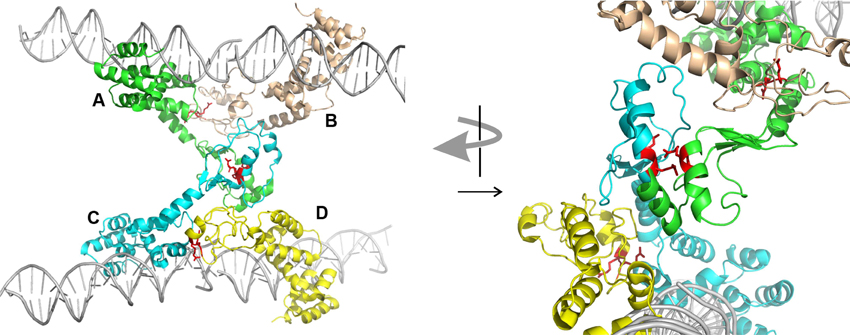
Figure 2. Structure of HpSpo0J monomers (lacking C-terminal dimer domain) bound to and across parS DNA (PDB 4UMK, Chen et al., 2015). Four monomers (A to D) make adjacent (AB and CD) and transverse (AC) interactions. The arginine patch motif is illustrated by two red arginines from each monomer. The arrangement on the left is rotated approximately 90° and magnified on the right to illustrate the environments of these arginines in the different interactions. The images were generated using the PyMOL Molecular Graphics System, Version 1.8.2.1 Schrödinger, LLC.
The overall fold of HpSpo0J is similar to that of TtSpo0J, except for a bend in the linker between the N-terminal and HTH domains (Leonard et al., 2004; Chen et al., 2015). It was suggested that the TtSpo0J structure may represent a closed conformation that opens up following DNA binding to the HpSpo0J architecture.
How do these structures inform us of higher-order complex assembly, particularly when ParB binds, bridges, and spreads on non-specific DNA adjacent to and away from parS? The simplest model is that the HTH is responsible for both specific and non-specific DNA interactions, which is supported by the observation that a triple substitution in the HTH domain of F SopB impairs both DNA binding activities (Ah-Seng et al., 2009). In this case the HpSpo0J-DNA structure may represent ParB-ParB and ParB-DNA interactions during spreading at any DNA site. The requirement for the N-terminus in higher-order complex formation in vivo is also consistent with this picture. The flexibility of and variation in the cross-monomer interactions and interfaces seen in the HpSpo0J structure make it an attractive model for the ability of ParB to make multiple and flexible interactions during complex assembly and maintenance. However, one recent study suggests that the specific and non-specific DNA binding activities may be distinct (Taylor et al., 2015). The specific and non-specific DNA binding activities of BsSpo0J showed different properties in vitro, including different abilities to protect the HTH domain from proteolysis. The results lead to the proposal that the N-terminus contains a DNA binding region that is distinct from the HTH. These observations may also reflect differences between plasmid and chromosomal ParBs. We must await identification of the protein-DNA contacts at non-specific sites before we can confirm the organization of ParB during higher-order partition complex assembly.
During partition, ParAs form patterns on the surface of the bacterial nucleoid due to dynamic interactions with ParB bound to parS (Hatano et al., 2007; Ringgaard et al., 2009; Hatano and Niki, 2010; Ah-Seng et al., 2013). The patterning is necessary for the segregation of plasmids and chromosomal domains, and the molecular mechanisms involved are still being defined. ParA is not necessary to form the large ParB-DNA complexes seen in vivo and in vitro, but ParA can influence or modulate these complexes. For example, the behavior of several parA and parB mutants supports a proposal that ParA is necessary to separate pairs or groups of plasmids during segregation (Fung et al., 2001; Ah-Seng et al., 2013). For the ParBs that interact with ParA via N-terminal regions that are adjacent to the flexible ParB-ParB oligomerization interface, one attractive idea is that ParA-ParB interactions at the N-terminus may influence the available conformations for ParB-ParB interactions next door. For example, specific interference with the transverse ParB-ParB interactions in the HpSpo0J structure might favor intramolecular associations over intermolecular ones (Figure 2).
Taken together, the biochemistry and structural biology of the N-terminal regions of ParBs imply that the folding and structures are flexible, dynamic, and fluid. Recent experiments using magnetic tweezers support the idea that the complexes are not highly ordered (Taylor et al., 2015). ParB-ParB (dimer-dimer) interactions via the N-terminal domain must contribute to higher-order complex assembly and function. It is attractive to consider that the flexibility of the N-terminus, in folding and conformation, is important for the dynamics and architecture of the large higher-order partition complex in vivo. This conformational flexibility resembles the properties of so-called “intrinsically disordered” proteins and domains whose unstructured properties allow proteins to sample and bind to multiple targets and in multiple ways (Wright and Dyson, 1999; Uversky, 2016). The same region in ParB could be involved, directly or indirectly, in different binding scenarios with different partners, including ParA, and potentially non-specific DNA. Why are flexibility, dynamics, and size important for these partition complexes? ParBs are engaging in multiple and constantly changing interactions during partition. Clustering creates a condensed, organized DNA substrate and provides a high density of ParBs available to ParA. Defining the molecular nature of these interactions continues to be an essential step toward the understanding of these intriguing DNA binding proteins.
The author confirms being the sole contributor of this work and approved it for publication.
This work was supported by Canadian Institutes of Health Research grant 133613.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ah-Seng, Y., Lopez, F., Pasta, F., Lane, D., and Bouet, J.-Y. (2009). Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein. J. Biol. Chem. 284, 30067–30075. doi: 10.1074/jbc.M109.044800
Ah-Seng, Y., Rech, J., Lane, D., and Bouet, J. Y. (2013). Defining the role of ATP hydrolysis in mitotic segregation of bacterial plasmids. PLoS Genet 9:e1003956. doi: 10.1371/journal.pgen.1003956
Autret, S., Nair, R., and Errington, J. (2001). Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41, 743–755. doi: 10.1046/j.1365-2958.2001.02551.x
Bartosik, A. A., Lasocki, K., Mierzejewska, J., Thomas, C. M., and Jagura-Burdzy, G. (2004). ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 186, 6983–6998. doi: 10.1128/jb.186.20.6983-6998.2004
Baxter, J. C., and Funnell, B. E. (2014). Plasmid partition mechanisms. Microbiol. Spectr. 2, 6. doi: 10.1128/microbiolspec.PLAS-0023-2014
Bingle, L. E. H., Macartney, D. P., Fantozzi, A., Manzoor, S. E., and Thomas, C. M. (2005). Flexibility in repression and cooperativity by KorB of broad host range IncP-1 plasmid RK2. J. Mol. Biol. 349, 302–316. doi: 10.1016/j.jmb.2005.03.062
Bouet, J. Y., Stouf, M., Lebailly, E., and Cornet, F. (2014). Mechanisms for chromosome segregation. Curr. Opin. Microbiol. 22, 60–65. doi: 10.1016/j.mib.2014.09.013
Breier, A. M., and Grossman, A. D. (2007). Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64, 703–718. doi: 10.1111/j.1365-2958.2007.05690.x
Broedersz, C. P., Wang, X. D., Meir, Y., Loparo, J. J., Rudner, D. Z., and Wingreen, N. S. (2014). Condensation and localization of the partitioning protein ParB on the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 111, 8809–8814. doi: 10.1073/pnas.1402529111
Chen, B.-W., Lin, M.-H., Chu, C.-H., Hsu, C.-E., and Sun, Y.-J. (2015). Insights into ParB spreading from the complex structure of Spo0J and parS. Proc. Natl. Acad. Sci. U.S.A. 112, 6613–6618. doi: 10.1073/pnas.1421927112
Delbrück, H., Ziegelin, G., Lanka, E., and Heinemann, U. (2002). An Src Homology 3-like domain is responsible for dimerization of the repressor protein KorB encoded by the promiscuous IncP plasmid RP4. J. Biol. Chem. 277, 4191–4198. doi: 10.1074/jbc.M110103200
Figge, R. M., Easter, J., and Gober, J. W. (2003). Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol. Microbiol. 47, 1225–1237. doi: 10.1046/j.1365-2958.2003.03367.x
Fung, E., Bouet, J.-Y., and Funnell, B. E. (2001). Probing the ATP-binding site of P1 ParA: partition and repression have different requirements for ATP binding and hydrolysis. EMBO J. 20, 4901–4911. doi: 10.1093/emboj/20.17.4901
Funnell, B. E. (1991). The P1 partition complex at parS: the influence of Escherichia coli integration host factor and of substrate topology. J. Biol. Chem. 266, 14328–14337.
Graham, T. G. W., Wang, X., Song, D., Etson, C. M., van Oijen, A. M., Rudner, D. Z., et al. (2014). ParB spreading requires DNA bridging. Genes Dev. 28, 1228–1238. doi: 10.1101/gad.242206.114
Hatano, T., and Niki, H. (2010). Partitioning of P1 plasmids by gradual distribution of the ATPase ParA. Mol. Microbiol. 78, 1182–1198. doi: 10.1111/j.1365-2958.2010.07398.x
Hatano, T., Yamaichi, Y., and Niki, H. (2007). Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 64, 1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x
Ireton, K., Gunther, N. W., and Grossman, A. D. (1994). spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176, 5320–5329.
Khare, D., Ziegelin, G., Lanka, E., and Heinemann, U. (2004). Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 11, 656–663. doi: 10.1038/nsmb773
Kusiak, M., Gapczyńska, A., Płochocka, D., Thomas, C. M., and Jagura-Burdzy, G. (2011). Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J. Bacteriol. 193, 3342–3355. doi: 10.1128/JB.00328-11
Leonard, T. A., Butler, P. J. G., and Löwe, J. (2004). Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 53, 419–432. doi: 10.1111/j.1365-2958.2004.04133.x
Leonard, T. A., Butler, P. J., and Löwe, J. (2005). Bacterial chromosome segregation: structure and DNA binding of the Soj dimer – a conserved biological switch. EMBO J. 24, 270–282. doi: 10.1038/sj.emboj.7600530
Lukaszewicz, M., Kostelidou, K., Bartosik, A. A., Cooke, G. D., Thomas, C. M., and JaguraBurdzy, G. (2002). Functional dissection of the ParB homologue (KorB) from IncP-1 plasmid RK2. Nucl. Acids Res. 30, 1046–1055. doi: 10.1093/nar/30.4.1046
Lynch, A. S., and Wang, J. C. (1995). SopB protein-meditated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl. Acad. Sci. U.S.A. 92, 1896–1900.
Murray, H., Ferreira, H., and Errington, J. (2006). The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 61, 1352–1361. doi: 10.1111/j.1365-2958.2006.05316.x
Radnedge, L., Youngren, B., Davis, M., and Austin, S. (1998). Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 17, 6076–6085.
Ringgaard, S., van Zon, J., Howard, M., and Gerdes, K. (2009). Movement and equipositioning of plasmids by ParA filament disassembly. Proc. Natl. Acad. Sci. U.S.A. 106, 19369–19374. doi: 10.1073/pnas.0908347106
Rodionov, O., Lobocka, M., and Yarmolinsky, M. (1999). Silencing of genes flanking the P1 plasmid centromere. Science 283, 546–549.
Rodionov, O., and Yarmolinsky, M. (2004). Plasmid partitioning and the spreading of P1 partition protein ParB. Mol. Microbiol. 52, 1215–1223. doi: 10.1111/j.1365-2958.2004.04055.x
Sanchez, A., Cattoni, D. I., Walter, J.-C., Rech, J., Parmeggiani, A., Nollmann, M., et al. (2015). Stochastic self-assembly of ParB proteins builds the bacterial DNA segregation apparatus. Cell Syst. 1, 163–173. doi: 10.1016/j.cels.2015.07.013
Sanchez, A., Rech, J., Gasc, C., and Bouet, J. Y. (2013). Insight into centromere-binding properties of ParB proteins: a secondary binding motif is essential for bacterial genome maintenance. Nucl. Acids Res. 41, 3094–3103. doi: 10.1093/nar/gkt018
Schumacher, M. A., and Funnell, B. E. (2005). ParB-DNA structures reveal DNA-binding mechanism of partition complex formation. Nature 438, 516–519. doi: 10.1038/nature04149
Schumacher, M. A., Piro, K. M., and Xu, W. J. (2010). Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes. Nucl. Acids Res. 38, 4514–4526. doi: 10.1093/nar/gkq161
Taylor, J. A., Pastrana, C. L., Butterer, A., Pernstich, C., Gwynn, E. J., Sobott, F., et al. (2015). Specific and non-specific interactions of ParB with DNA: implications for chromosome segregation. Nucl. Acids Res. 43, 719–731. doi: 10.1093/nar/gku1295
Uversky, V. N. (2016). Dancing protein clouds: the strange biology and chaotic physics of intrinsically disordered proteins. J. Biol. Chem. 291, 6681–6688. doi: 10.1074/jbc.R115.685859
Wang, X. D., Llopis, P. M., and Rudner, D. Z. (2013). Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14, 191–203. doi: 10.1038/nrg3375
Wright, P. E., and Dyson, H. J. (1999). Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331. doi: 10.1006/jmbi.1999.3110
Keywords: chromosome dynamics, segregation, ParABS, DNA-binding, bridging
Citation: Funnell BE (2016) ParB Partition Proteins: Complex Formation and Spreading at Bacterial and Plasmid Centromeres. Front. Mol. Biosci. 3:44. doi: 10.3389/fmolb.2016.00044
Received: 27 June 2016; Accepted: 15 August 2016;
Published: 29 August 2016.
Edited by:
Manuel Espinosa, Spanish National Research Council - Centro de Investigaciones Biológicas, SpainReviewed by:
Christopher Morton Thomas, University of Birmingham, UKCopyright © 2016 Funnell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Funnell, Yi5mdW5uZWxsQHV0b3JvbnRvLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.