- 1Ministry of Agriculture Laboratory for Risk Assessment of Quality and Safety of Livestock and Poultry Products, Huazhong Agricultural University, Wuhan, China
- 2National Reference Laboratory of Veterinary Drug Residues (HZAU) and Ministry of Agriculture Key Laboratory for the Detection of Veterinary Drug Residues in Foods, Huazhong Agricultural University, Wuhan, China
The outbreak of antimicrobial resistance, together with the lack of newly developed antimicrobial drugs, represents an alarming signal for both human and animal healthcare worldwide. Selection of rational dosage regimens for traditional antimicrobial drugs based on pharmacokinetic/pharmacodynamic principles as well as development of novel antimicrobials targeting new bacterial targets or resistance mechanisms are key approaches in tackling AMR. In addition to the cellular level resistance (i.e., mutation and horizontal gene transfer of resistance determinants), the community level resistance (i.e., bilofilms and persisters) is also an issue causing antimicrobial therapy difficulties. Therefore, anti-resistance and antibiofilm strategies have currently become research hotspot to combat antimicrobial resistance. Although metallic nanoparticles can both kill bacteria and inhibit biofilm formation, the toxicity is still a big challenge for their clinical applications. In conclusion, rational use of the existing antimicrobials and combinational use of new strategies fighting against antimicrobial resistance are powerful warranties to preserve potent antimicrobial drugs for both humans and animals.
Introduction
Antimicrobial resistance (AMR) is one of the ultimate fears to the health of humans and animals worldwide. Use of antimicrobial drugs in humans or animals results in the emergence and dissemination of resistant bacteria, and overuse or abuse of them makes this situation worse. Thus, it is important to simultaneously preserve effective antimicrobials as long as possible as well as continue to employ them for the service of human and animal health (Chang et al., 2015).
The dissemination of AMR has not been paralleled by the development of novel antimicrobials. This is due to that the process of drug discovery and clinical trials of new antimicrobials takes a long time, and only a few new agents have recently been approved for use. These situations prompt the efforts to develop alternatives to traditional antimicrobials, as described in our previous review (Cheng et al., 2014). However, some of the alternatives are only used for the prevention of bacterial infections (e.g., vaccines); some show indirect effect against pathogens (e.g., immunomodulators, feed enzymes); some are of complex composition (e.g., probiotics, plant extracts), thus the effects vary greatly; and the antimicrobial peptides, such as one of the bacteriocins, lantibiotics, have been reported causing bacterial resistance (Draper et al., 2015).
In this review, we briefly focus on old and novel antimicrobial agents in tackling AMR. The AMR occurs on two levels, the cellular level resistance (mutation and horizontal gene transfer (HGT) of resistance determinants) and the community level resistance (biofilm and persister cells) (Penesyan et al., 2015). The studies reviewed suggest that only rational use of existing old antimicrobial drugs and combinational use of anti-resistance or antibiofilm strategies with antimicrobials as well as continuing development of new antimicrobial agents could fight against AMR.
Mechanisms of AMR
AMR includes two levels of resistance, the cellular level resistance and the community level resistance (Penesyan et al., 2015; Table 1). The development of cellular resistance occurs due to endogenous gene mutations as well as via HGT of resistance determinants from other microorganisms. Also, a group of bacteria can be tolerant to the environmental stress that individual cells cannot, which is called the community level resistance. Such tolerance can cause an increased resistance to antimicrobials (Penesyan et al., 2015). For example, the resistance obtained by microorganisms in biofilm can be up to 1000 times higher than that gained by their planktonic counterparts, which impairs the treatment of biofilm-associated infections in clinical treatment (Lebeaux et al., 2014b; Penesyan et al., 2015). The main mechanism currently proposed to explain such tolerance is the presence of persister cells. The persisters can escape the lethal action of antimicrobials by entering a physiological state in which the antimicrobials do not kill them, a phenomenon known as bacterial persistence (Maisonneuve and Gerdes, 2014). Moreover, the cellular and community levels of resistance can be synergistic, thereby greatly enhancing the overall AMR of the microbial community (Penesyan et al., 2015).

Table 1. Mechanisms of AMR (derived from Penesyan et al., 2015).
Revisit to Old Antimicrobials
As AMR to commonly used antimicrobial drugs increases, older antimicrobials are being “revived” and attracting attention. These old antimicrobials represent a new alternative for the control of AMR (Pulcini et al., 2012). Cassir et al. (2014) have provided a collection of microbiological and clinical data on potentially useful older antimicrobials for the successful treatment of multidrug-resistant (MDR) Gram-negative bacterial infections (e.g., polymyxins, fosfomycin, mecillinam, temocillin, and nitrofurantoin), MDR Gram-positive infections (e.g., trimethoprim-sulfamethoxazole, tetracyclines, chloramphenicol, clindamycin, pristinamycin, rifampicin, and fusidic acid), and MDR tuberculosis (e.g., clofazimine, amoxicilline-clavulanate, trimethoprim-sulfamethoxazole, and minocycline). Since older antimicrobials have rarely been subjected to present drug development procedures, they are less considered in practice guidelines. Therefore, their efficacy and safety must be revaluated to optimize therapy.
A number of antimicrobials discovered decades ago that have unique chemical scaffolds and/or utilize novel modes of action to interact with bacterial targets, such as ribosome (Arenz and Wilson, 2016). For example, dityromycin, a cyclic decapeptide antibiotic produced by Streptomyces sp., can uniquely bind to ribosomal protein S12 in the small ribosomal subunit, a mode of action different than any other known translational inhibitor (Bulkley et al., 2014). In many cases, these “forgotten” compounds display cytotoxicity against eukaryotic cells and thus were abandoned (Arenz and Wilson, 2016). However, recent structure-function analysis gives us better understanding of the similarities and differences between bacterial targets and their eukaryotic counterparts, thereby guiding the future development of more specific and less toxic inhibitors. With the increased understanding of AMR mechanisms, revisiting the known antimicrobials are helpful to the exploration of the next generation of antimicrobial drugs.
Procedures for registration of antimicrobials drugs have improved significantly. Both EU (EMA, 2013) and US authorities (FDA, 2010) have published numerous guidance documents, and addressed the increasing need for antimicrobials that are active against MDR bacteria. The guidance documents include recommendations for dosage regimens based on pharmacokinetic (PK)/pharmacodynamic (PD) relationships. PK/PD provides a universal framework for exposure-response relationships, and the responses include efficacy, toxicity, and emergence of resistance (Muller et al., 2015). Exposure-response relationships also provide a means to translate experimental and preclinical data into the clinical settings, including setting clinical breakpoints, as extensively described by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (Mouton et al., 2012). To determine the optimal dose, several key features of the exposure-response relationship need to be determined, including MIC distribution of the interested microorganisms, the PK profiles for a variety of doses and patient populations, and the exposure-response relationship and PD target (Muller et al., 2015).
There are knowledge gaps for those revived antimicrobials in the areas of PK profiling in patients, as well as PD targets derived from preclinical and clinical studies (Muller et al., 2015). Although the regulatory requirements for new antimicrobial agents have become more and more rigorous, updates of the product information for old antimicrobials are either missing or insufficient, which would pose significant risks of potential harm to the patients. In addition, there is no motivation for companies to develop antimicrobials when the cost and time of drug approval is far beyond commercial interests, even if there is an obvious medical need.
In summary, redevelopment of an old antimicrobials leads to an improved understanding of its chemistry, PK/PD as well as optimizing its clinical use in different patient populations. Optimization of antimicrobial therapy in terms of PK/PD is essential to improve therapeutic efficacy but minimize the toxicity and the risk of resistance development during treatment (Mouton et al., 2011). As old antimicrobials are rarely included in surveillance programs, the evaluation of the risks of drug resistance is lacking. The prescription of old antimicrobials needs to be regulated by professional antimicrobial stewardships. Besides, as public health concern, cost effectiveness should be integrated in further comparisons between old and currently used antimicrobials.
Development of New Antimicrobials
The current antimicrobials, mainly derived from natural sources, inhibit cellular processes such as cell wall biosynthesis, DNA replication, and protein synthesis. With the worldwide emergence of AMR, there is renewed interest in the investigation of alternative essential cellular processes, including bacterial central metabolic pathways, as the drug targets for the next generation of antimicrobials (Murima et al., 2014). For examples, bedaquiline is an antitubercular drug targeting the F0F1 ATP synthase (Andries et al., 2005). Like bedaquiline, Q203, an optimized imidazopyridine amide compound, selectively inhibits the respiratory cytochrome bc1 complex in mycobacteria regardless of architectural conservation of the bc1 complex in many species (Pethe et al., 2013). The inhibition of the bacterial divisome, mainly by targeting the central cell division mediator FtsZ, has been accepted as a promising strategy for antimicrobial attack by either interfering with the natural dynamics and functions of FtsZ during the cell cycle or activating a bacterial protease to degrade FtsZ, thus causing bacterial death in a suicidal manner (Sass and Brötz-Oesterhelt, 2013). The mode of action of alkyl gallate is a combination of direct targeting of FtsZ and permeabilization of bacterial membranes, which is a promising hit for the further development of antibacterials (Król et al., 2015). Recent efforts have also been devoted to developing drugs that interrupt the assimilation of iron by bacteria, a process that is vital to cellular homeostasis (Foley and Simeonov, 2012). The unique asymmetric outer membrane in Gram-negative bacteria, which acts as a permeability barrier that protects the cell from external stresses such as the presence of antimicrobials, has become an attractive drug target. A novel β-hairpin macrocyclic peptide, JB-95, exhibits an ability to selectively disrupt the outer membrane through interactions with selected β-barrel outer membrane proteins including BamA and LptD, but not the inner membrane of E. coli (Urfer et al., 2016). Furthermore, the bacterial protein secretion pathway is a target for eliminating or disarming pathogens. Targeting the Sec-pathway for novel antimicrobial agents focuses on two key components: SecA, the ATP-driven motor protein responsible for driving preproteins across the cytoplasmic membrane and the Type I signal peptidase which is responsible for the removal of the signal peptide to allow the release of the mature protein from the membrane (Rao et al., 2014).
Targeting resistance, usually used in combination with the traditional antimicrobials, is another strategy to fight against AMR. Among the four general resistance mechanisms, which include target alteration (Hooper and Jacoby, 2015), drug inactivation (Shaw et al., 1993; Bush and Fisher, 2011), decreased permeability (Nikaido, 2003) and increased efflux (Sun et al., 2014; Li et al., 2015), drug extrusion by multidrug efflux pumps serves as an important mechanism of MDR. Efflux pumps also have physiological functions in response to various of environmental and physiological signals (Sun et al., 2014). Recent studies have tried to reverse the resistance phenotype conferred by efflux pump activation (Opperman and Nguyen, 2015; Venter et al., 2015). It was observed that the addition of efflux pump inhibitors partially restored drug susceptibility in vitro and in vivo in the anti-Mycobacterium tuberculosis therapy (Pule et al., 2016). The class of zinc-dependent hydrolases, metallo-β-lactamase, can confer bacteria with extended spectrum β-lactam resistance. The design of compounds with the β-lactam core scaffold is an attractive approach to the development of metallo-β-lactamase inhibitors. Some promising inhibitors, including cephalosporin derivatives, penicillin derivatives, carbapenem derivatives, cyclobutanone β-lactam analogs, thiol derivatives, succinic acid derivatives, mercaptoacetic acid thioester derivatives, pyrrole-, pyridine- and triazole-containing compounds, and DNA aptamer, have been thoroughly reviewed by King and Strynadka (King and Strynadka, 2013). Targeting the resistance mechanism itself by a vaccine is an interesting but hitherto unexplored approach (Henriques-Normark and Normark, 2014). Vaccination directed against the resistance mechanism can be possible when resistance is mediated by an enzyme whose activity can be inhibited by neutralizing antibodies.
Except for the above inhibitors targeting resistance, drugs in already-known classes such as new β-lactams, quinolones, aminoglycosides, and tetracyclines have been designed to escape from many of the known resistance mechanisms. BAL30072, a siderophore monosulfactam similar to aztreonam, exhibits antibacterial activity against most species of aerobic Gram-negative bacteria (Page et al., 2010). It is stable toward metallo-β-lactamases and is a poor substrate for many serine carbapenemases. Several new quinolones, such as avarofloxacin, delafloxacin, finafloxacin and the desfluoroquinolone nemonoxacin, which show enhanced activity against fluoroquinolone-resistant Gram-positive bacteria including MRSA are in clinical development (Page and Bush, 2014). A modified aminoglycoside, plazomicin, has been demonstrated activity against both Gram-negative and Gram-positive bacterial pathogens (Zhanel et al., 2012). Modified tetracyclines, such as tigecycline, omadacycline and eravacycline are of interest for their activity against many MDR Enterobacteriaceae and Acinetobacter spp., including isolates expressing tetracycline-specific efflux and ribosomal protection proteins (Sutcliffe, 2011).
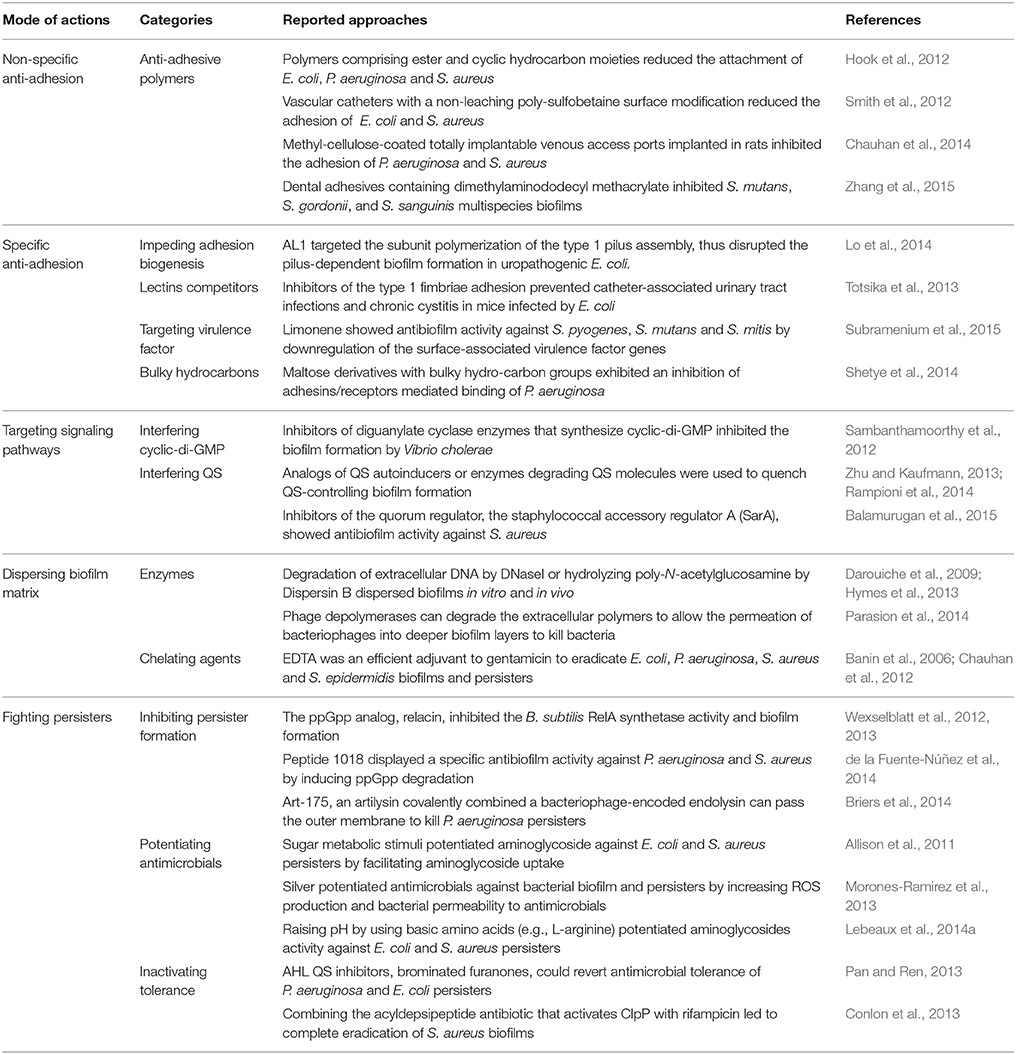
Approaches to Combat Biofilms
The approaches to combat biofilms are extensively reviewed by Beloin et al. (2014) (Table 2). During the biofilm development, the bacteria initially adhere to a surface that ultimately leads to colonization and infection by pathogenic bacteria. Therefore, reducing adhesion is a strategy to prevent biofilm formation and related infections (Veerachamy et al., 2014). Recently, non-specific inhibition of adhesion vs. targeting specific adhesions has been developed to reduce bacterial adhesion. Quorum sensing (QS), which controls many important physiological processes such as biofilm development, is a widespread intercellular form of communicating and cooperative activities of bacteria at the population level, and it depends on the production, secretion, and detection of small diffusible autoinducers, such as acyl-homoserine lactones, auto-inducing peptides and autoinducer 2 (Zhang and Li, 2016). Cyclic di-GMP is a second messenger that acts to regulate a wide range of functions including developmental transitions, adhesion and biofilm formation (Caly et al., 2015). Targeting these signaling pathways is also a strategy to prevent biofilm development. Moreover, using enzymes or chelating agents can hydrolyze biofilm components or destabilize biofilm matrix. On the other hand, persister cells have recently been subjected to an intensive research in order to limit biofilm-associated antimicrobial tolerance. The formation of persister cells depends on the ubiquitous bacterial regulatory nucleotides tetra and penta-guanosine phosphate (ppGpp) that activate inhibitors of cell growth (Germain et al., 2015). Therefore, interfering ppGpp could inhibit the formation of persister cells.
The deep research on the mechanism of biofilm formation leads to the emergence of numerous promising antibiofilm approaches. However, the conversion of experimental data into clinical settings is time-consuming and to some extent unsatisfactory. Non-biocidal anti-adhesive or anti-virulence strategies face the diversity of bacterial phenotypes and may only be active against a subpopulation of bacteria encountered in clinical practice, therefore limiting their overall efficacy. In vitro biofilms are probably structurally different from in vivo biofilms (Lebeaux et al., 2013). Currently, due to the diversity of the in vivo conditions leading to the complexity of clinical biofilm situations, the diversity of persister phenotypes is unknown. Most of the studies use rodent models, but these in vivo models may not properly replicate real clinical state. Furthermore, as for clinical trials, rigorous statistical analysis is compulsory in order to avoid any false positive results. Most importantly, molecules identified in vitro should be validated using in vivo models not only for the antibiofilm activity but also non-toxicity.
Metallic Nanoparticles (NPs)
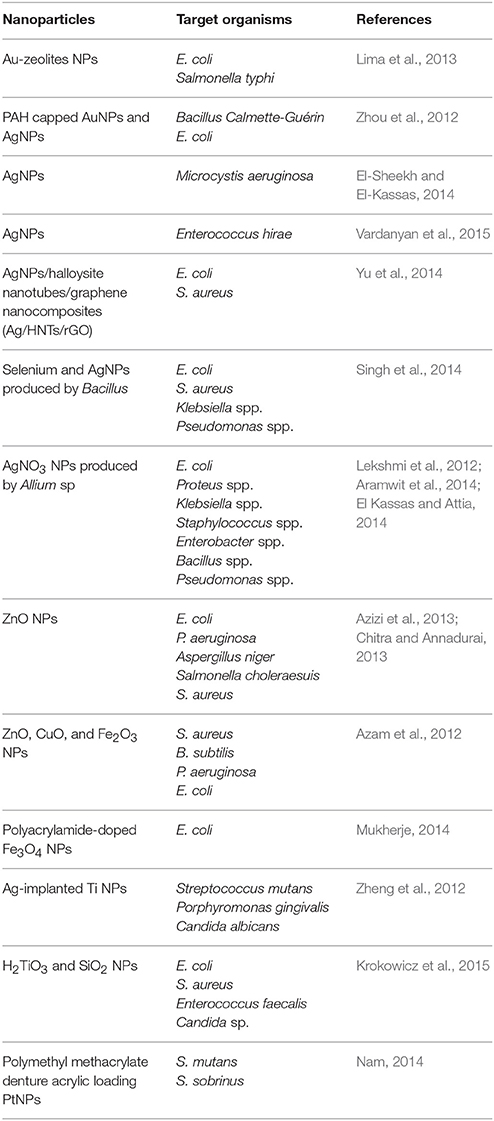
Physiochemical properties of nanomolecules as antimicrobial agents are widely used in human and veterinary medicine. Metallic NPs are of great interest for use as potential antimicrobial agents because of their unique optical, electronic, and magnetic properties (Kandi and Kandi, 2015). The electrostatic interaction of NPs with negatively charged bacterial surfaces draws the particles to the bacteria and promotes their penetration into the membrane, causing membrane disruption, bacterial flocculation, and a reduction in viability. The generation of reactive oxygen species (ROS) is also a mechanism of antibacterial activity of NPs (Thekkae Padil and Černík, 2013). Further actions of NPs as antimicrobial agents include disrupting DNA during the replication and cell division of microorganisms, compromising the bacterial membrane integrity via physical interactions with the microbial cell, and releasing toxic metal ions and causing lysis of cells (Franci et al., 2015). Recently, the silicon dioxide NPs (Si-NP) were engineered to target the signaling molecules (i.e., acylhomoserine lactones) used for QS in order to halt bacterial communication (Miller et al., 2015). The bactericidal activity of NPs depends on size, stability and concentration in the growth medium (Tillotson and Theriault, 2013). The applications of nanomolecules in medicine have recently been evaluated in reports highlighting the in vitro antimicrobial activities of NPs (Table 3), and also the possible potential adverse effects of nanomolecules on human health and the environment (Kandi and Kandi, 2015).
As various metallic NPs and their oxides have already been used as potent antimicrobial agents, silver or ionic form is most toxic for microorganisms when compared to other metals (Seil and Webster, 2012). This makes silver of particularly interests. Silver NPs (AgNPs) probably have multiple mechanisms of antibacterial action (Markowska et al., 2013). For example, (1) AgNPs can affect bacterial membrane permeability by attaching to the cell membrane surface and modifying the cell potential; (2) AgNPs can causes oxidative damage by production of ROS (Kim et al., 2007; Xu et al., 2012); (3) AgNPs can interact with the SH-groups of bacterial membrane proteins and intracellular proteins, and also can interact with the phosphate residues in DNA, thus to interfere with protein synthesis and function and cell division (Durán et al., 2016). However, due to the current lack of knowledge, the exact basis for the activity of AgNPs is still uncharacterized.
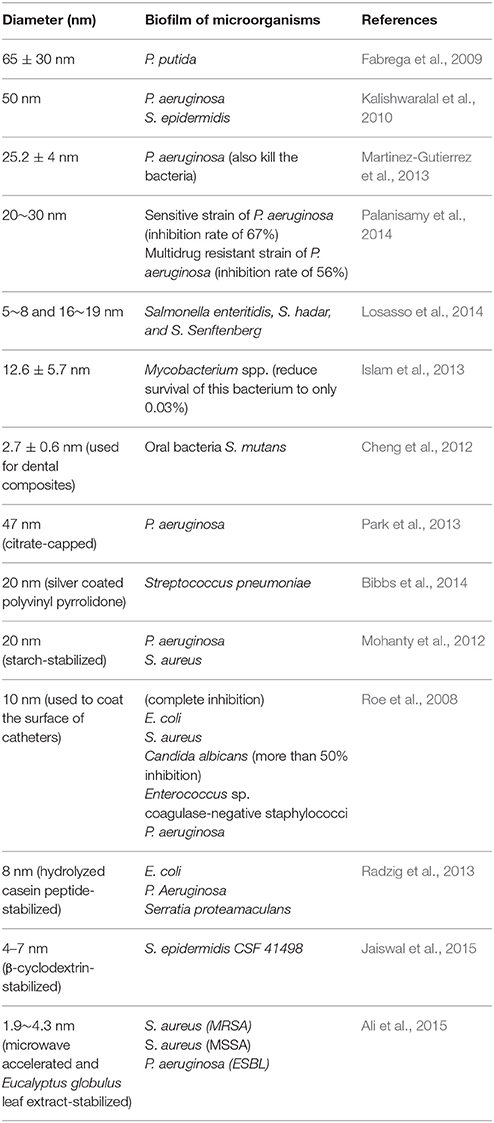
The antibiofilm activity of AgNPs has also been demonstrated in a number of studies (Table 4). Most of the AgNPs are within the range of 1~100 nm. Although smaller AgNPs may have greater biological activity, it is important to note that differences in the chemical and physical properties of AgNP may cause variation in its antimicrobial and antibiofilm efficacy (Markowska et al., 2013). Because of the relatively low stability of colloidal solutions, some researchers propose the usage of stabilized AgNPs (Table 4). AgNPs can also enhance the antibacterial and antibiofilm activity of conventional antimicrobials. There are reports describing synergistic activity between AgNPs and, e.g., ampicillin, kanamycin, streptomycin or vancomycin against E. coli and P. aeruginosa (Wolska et al., 2012). Some AgNPs have been subjected to clinical trials (Franci et al., 2015).
Although metallic NPs have great potential in the future to become antimicrobial agents, the local and systemic toxic complications and the deleterious effect they have on beneficial bacteria in humans and animals may be a cause for concern (Zhang et al., 2010; Prabhu and Poulose, 2012). NPs have the ability to spread throughout the body, cross the blood-brain barrier, cause haemolysis, and may result in degradation products which have toxic effects and influence blood coagulation pathways (Kandi and Kandi, 2015). Most studies have not conclusively evaluated the exact mechanism by which nanomolecules contribute to toxic complications, and many of the interactions of the AgNPs with the human or animal body are still poorly understood (dos Santos et al., 2013). It has been observed that the larger size of NP, the greater is the risk of adverse health effects (De Jong and Borm, 2008). Research is necessary to clearly understand the interaction of nanomolecules with living cells, the extent of their distribution in the body, and their specific organ toxicity.
Concluding Remarks
The paradox of antimicrobial drugs is that through their use, they not only inhibit an infection but also select for the emergence and spread of AMR, directly counteracting their long-term efficacy. Considerable inappropriate use of both prophylactic and therapeutic antimicrobials in human and veterinary medicine highlights an urgent need for antibiotic stewardship initiatives. At the present time, rational use of existing antibiotics based on PK/PD dosage-regimen is a key strategy in tackling AMR, thus to preserve potent antimicrobials for both humans and animals. At the same time, we should never stop discovering novel inhibitors with new bacterial targets and digging the treasure box of “old” and “forgotten” antimicrobials. Compounds showing profound anti-resistance and antibiofilm effects are in research hotspot, but they still have limitations. Combining existing antimicrobials with compounds that inhibit their specific resistance mechanisms would be a good choice. Although metallic NPs can both kill bacteria and inhibit biofilm formation, the toxicity is still a big challenge for their clinical applications (dos Santos et al., 2013). With single-drug therapy, there is always a selective advantage to resistance; specific combinations of drugs can inhibit bacterial growth while disfavoring resistance to the individual components. These approaches can be used to invert the selective advantage of resistant bacteria competing with their sensitive cousins, or even drive a resistant bacterial population back toward drug sensitivity (Baym et al., 2016). Besides, screening and developing multiple-target inhibitors as “resistance-resistant” antimicrobials can reduce the effects of target mutation (Oldfield and Feng, 2014). The natural products have also been a prolific and unsurpassed source for new lead antimicrobial compounds, but target identification and validation has remained a major bottleneck (Farha and Brown, 2016). Functional genomics techniques are proved to be indispensible for in vitro target authentication and elucidating mechanism of action of novel antimicrobials (Khan and Khan, 2016). Since most of the new strategies described in this review are only at the early basic experimental stage, their potential for clinical applications requires more extensive investigations.
Author Contributions
GC contributed to the design of the review and wrote the review. MD, SA, HH, and XW revised the review. ZY contributed to the conception of the review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Basic Research (973) Program of China (No. 2013CB127201), the National Natural Science Foundation of China (No. 31502115) and the National Program for Risk Assessment of Quality and Safety of Livestock and Poultry Products (GJFP2016008).
Abbreviations
AMR, antimicrobial resistance; HGT, horizontal gene transfer; MDR, multidrug-resistant; MIC, minimum inhibitory concentration; NPs, nanoparticles; PK, pharmacokinetic; PD, pharmacodynamic; QS, quorum sensing; ROS, reactive oxygen species.
References
Ali, K., Ahmed, B., Dwivedi, S., Saquib, Q., Al-Khedhairy, A. A., and Musarrat, J. (2015). Microwave accelerated green synthesis of stable silver nanoparticles with eucalyptus globulus leaf extract and their antibacterial and antibiofilm activity on clinical isolates. PLoS ONE 10:e0131178. doi: 10.1371/journal.pone.0131178
Allison, K. R., Brynildsen, M. P., and Collins, J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. doi: 10.1038/nature10069
Andries, K., Verhasselt, P., Guillemont, J., Göhlmann, H. W., Neefs, J. M., Winkler, H., et al. (2005). A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227. doi: 10.1126/science.1106753
Aramwit, P., Bang, N., Ratanavaraporn, J., and Ekgasit, S. (2014). Green synthesis of silk sericin-capped silver nanoparticles and their potent anti-bacterial activity. Nanoscale Res. Lett. 9:79. doi: 10.1186/1556-276X-9-79
Arenz, S., and Wilson, D. N. (2016). Blast from the past: reassessing forgotten translation inhibitors, antibiotic selectivity, and resistance mechanisms to aid drug development. Mol. Cell 61, 3–14. doi: 10.1016/j.molcel.2015.10.019
Azam, A., Ahmed, A. S., Oves, M., Khan, M. S., Habib, S. S., and Memic, A. (2012). Antimicrobial activity of metal oxide nanoparticles against Gram-positive and Gram-negative bacteria: a comparative study. Int. J. Nanomedicine 7, 6003–6009. doi: 10.2147/IJN.S35347
Azizi, S., Ahmad, M., Mahdavi, M., and Abdolmohammadi, S. (2013). Preparation, characterization, and antimicrobial activities of ZnO nanoparticles/cellulose nanocrystal nanocomposites. Bioresources 8, 1841–1851. doi: 10.15376/biores.8.2.1841-1851
Balamurugan, P., Hema, M., Kaur, G., Sridharan, V., Prabu, P. C., Sumana, M. N., et al. (2015). Development of a biofilm inhibitor molecule against multidrug resistant Staphylococcus aureus associated with gestational urinary tract infections. Front. Microbiol. 6:832. doi: 10.3389/fmicb.2015.00832
Banin, E., Brady, K. M., and Greenberg, E. P. (2006). Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72, 2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006
Baym, M., Stone, L. K., and Kishony, R. (2016). Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351:aad3292. doi: 10.1126/science.aad3292
Beloin, C., Renard, S., Ghigo, J. M., and Lebeaux, D. (2014). Novel approaches to combat bacterial biofilms. Curr. Opin. Pharmacol. 18, 61–68. doi: 10.1016/j.coph.2014.09.005
Bernier, S. P., Lebeaux, D., DeFrancesco, A. S., Valomon, A., Soubigou, G., Coppée, J. Y., et al. (2013). Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 9:e1003144. doi: 10.1371/journal.pgen.1003144
Bibbs, R. K., Harris, R. D., Peoples, V. A., Barnett, C., Singh, S. R., Dennis, V. A., et al. (2014). Silver polyvinyl pyrrolidone nanoparticles exhibit a capsular polysaccharide influenced bactericidal effect against Streptococcus pneumoniae. Front. Microbiol. 5:665. doi: 10.3389/fmicb.2014.00665
Blázquez, J. (2003). Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 37, 1201–1209. doi: 10.1086/378810
Briers, Y., Walmagh, M., Grymonprez, B., Biebl, M., Pirnay, J. P., Defraine, V., et al. (2014). Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58, 3774–3784. doi: 10.1128/AAC.02668-14
Bulkley, D., Brandi, L., Polikanov, Y. S., Fabbretti, A., O'Connor, M., Gualerzi, C. O., et al. (2014). The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation. Cell Rep. 6, 357–365. doi: 10.1016/j.celrep.2013.12.024
Bush, K., and Fisher, J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478. doi: 10.1146/annurev-micro-090110-102911
Caly, D. L., Bellini, D., Walsh, M. A., Dow, J. M., and Ryan, R. P. (2015). Targeting cyclic di-GMP signalling: a strategy to control biofilm formation? Curr. Pharm. Des. 21, 12–24. doi: 10.2174/1381612820666140905124701
Cassir, N., Rolain, J. M., and Brouqui, P. (2014). A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front. Microbiol. 5:551. doi: 10.3389/fmicb.2014.00551
Chang, Q., Wang, W., Regev-Yochay, G., Lipsitch, M., and Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: how worried should we be? Evol. Appl. 8, 240–247. doi: 10.1111/eva.12185
Chauhan, A., Bernardin, A., Mussard, W., Kriegel, I., Estève, M., Ghigo, J. M., et al. (2014). Preventing biofilm formation and associated occlusion by biomimetic glycocalyxlike polymer in central venous catheters. J. Infect. Dis. 210, 1347–1356. doi: 10.1093/infdis/jiu249
Chauhan, A., Lebeaux, D., Ghigo, J. M., and Beloin, C. (2012). Full and broad-spectrum in vivo eradication of catheter-associated biofilms using gentamicin-EDTA antibiotic lock therapy. Antimicrob. Agents Chemother. 56, 6310–6318. doi: 10.1128/AAC.01606-12
Cheng, G., Hao, H., Xie, S., Wang, X., Dai, M., Huang, L., et al. (2014). Antibiotic alternatives: the substitution of antibiotics in animal husbandry? Front. Microbiol. 5:217. doi: 10.3389/fmicb.2014.00217
Cheng, L., Weir, M. D., Xu, H. H., Antonucci, J. M., Kraigsley, A. M., Lin, N. J., et al. (2012). Antibacterial amorphous calcium phosphate nanocomposites with a quaternary ammonium dimethacrylate and silver nanoparticles. Dent. Mater. 28, 561–572. doi: 10.1016/j.dental.2012.01.005
Chiang, W. C., Nilsson, M., Jensen, P. Ø., Høiby, N., Nielsen, T. E., Givskov, M., et al. (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361. doi: 10.1128/AAC.00001-13
Chitra, K., and Annadurai, G. (2013). Antimicrobial activity of wet chemically engineered spherical shaped ZnO nanoparticles on food borne pathogen. Int. Food Res. J. 20, 59–64.
Conibear, T. C., Collins, S. L., and Webb, J. S. (2009). Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS ONE 4:e6289. doi: 10.1371/journal.pone.0006289
Conlon, B. P., Nakayasu, E. S., Fleck, L. E., LaFleur, M. D., Isabella, V. M., Coleman, K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370. doi: 10.1038/nature12790
Darouiche, R. O., Mansouri, M. D., Gawande, P. V., and Madhyastha, S. (2009). Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J. Antimicrob. Chemother. 64, 88–93. doi: 10.1093/jac/dkp158
D'Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. (2006). Sampling the antibiotic resistome. Science 311, 374–377. doi: 10.1126/science.1120800
De Jong, W. H., and Borm, P. J. (2008). Drug delivery and nanoparticles:applications and hazards. Int. J. Nanomedicine 3, 133–149. doi: 10.2147/IJN.S596
de la Fuente-Núñez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. doi: 10.1371/journal.ppat.1004152
Domingues, S., Harms, K., Fricke, W. F., Johnsen, P. J., da Silva, G. J., and Nielsen, K. M. (2012). Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 8:e1002837. doi: 10.1371/journal.ppat.1002837
dos Santos, C. A., Seckler, M. M., Ingle, A. P., Gupta, I., Galdiero, S., Galdiero, M., et al. (2013). Silver nanoparticles: therapeutical uses, toxicity, and safety issues. J. Pharm. Sci. 103, 1931–1944. doi: 10.1002/jps.24001
Draper, L. A., Cotter, P. D., Hill, C., and Ross, R. P. (2015). Lantibiotic resistance. Microbiol. Mol. Biol. Rev. 79, 171–191. doi: 10.1128/MMBR.00051-14
Durán, N., Durán, M., de Jesus, M. B., Seabra, A. B., Fávaro, W. J., and Nakazato, G. (2016). Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine 12, 789–799. doi: 10.1016/j.nano.2015.11.016
El Kassas, H. Y., and Attia, A. A. (2014). Bactericidal application and cytotoxic activity of biosynthesized silver nanoparticles with an extract of the red seaweed Pterocladiella capillacea on the HepG2 cell line. Asian Pac. J. Cancer Prev. 15, 1299–1306. doi: 10.7314/APJCP.2014.15.3.1299
El-Sheekh, M. M., and El-Kassas, H. Y. (2014). Application of biosynthesized silver nanoparticles against a cancer promoter cyanobacterium, Microcystis aeruginosa. Asian Pac. J. Cancer Prev. 15, 6773–6779. doi: 10.7314/APJCP.2014.15.16.6773
EMA (2013). New EMA Guidance on Development of Antibacterials to Help in the Fight against Multidrug-Resistant Pathogens. European Medicines Agency. Available online at: http://www.ema.europa.eu/ema/index.jsp?curl=pages/news_and_events/news/2013/11/news_detail_001944.jsp&mid=WC0b01ac058004d5c1
Fabrega, J., Renshaw, J. C., and Lead, J. R. (2009). Interactions of silver nanoparticles with Pseudomonas putida biofilms. Environ. Sci. Technol. 43, 9004–9009. doi: 10.1021/es901706j
Farha, M. A., and Brown, E. D. (2016). Strategies for target identification of antimicrobial natural products. Nat. Prod. Rep. doi: 10.1039/C5NP00127G. [Epub ahead of print].
FDA (2010). US Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER). Guidance for industry antibacterial drug products: use of noninferiority trials to support approval. Available online at: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm070951.pdf
Foley, T. L., and Simeonov, A. (2012). Targeting iron assimilation to develop new antibacterials. Expert Opin. Drug Discov. 7, 831–847. doi: 10.1517/17460441.2012.708335
Franci, G., Falanga, A., Galdiero, S., Palomba, L., Rai, M., Morelli, G., et al. (2015). Silver nanoparticles as potential antibacterial agents. Molecules 20, 8856–8874. doi: 10.3390/molecules20058856
Germain, E., Roghanian, M., Gerdes, K., and Maisonneuve, E. (2015). Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 112, 5171–5176. doi: 10.1073/pnas.1423536112
Gillings, M. R., Holley, M. P., and Stokes, H. W. (2009). Evidence for dynamic exchange of qac gene cassettes between class 1 integrons and other integrons in freshwater biofilms. FEMS Microbiol. Lett. 296, 282–288. doi: 10.1111/j.1574-6968.2009.01646.x
Gillings, M. R., and Stokes, H. W. (2012). Are humans increasing bacterial evolvability? Trends Ecol. Evol. 27, 346–352. doi: 10.1016/j.tree.2012.02.006
Henriques-Normark, B., and Normark, S. (2014). Bacterial vaccines and antibiotic resistance. Ups. J. Med. Sci. 119, 205–208. doi: 10.3109/03009734.2014.903324
Hook, A. L., Chang, C. Y., Yang, J., Luckett, J., Cockayne, A., Atkinson, S., et al. (2012). Combinatorial discovery of polymers resistant to bacterial attachment. Nat. Biotechnol. 30, 868–875. doi: 10.1038/nbt.2316
Hooper, D. C., and Jacoby, G. A. (2015). Mechanisms of drug resistance: quinolone resistance. Ann. N.Y. Acad. Sci. 1354, 12–31. doi: 10.1111/nyas.12830
Hymes, S. R., Randis, T. M., Sun, T. Y., and Ratner, A. J. (2013). DNase inhibits Gardnerella vaginalis biofilms in vitro and in vivo. J. Infect. Dis. 207, 1491–1497. doi: 10.1093/infdis/jit047
Islam, M. S., Larimer, C., Ojha, A., and Nettleship, I. (2013). Antimycobacterial efficacy of silver nanoparticles as deposited on porous membrane filters. Mater. Sci. Eng. 33, 4575–4581. doi: 10.1016/j.msec.2013.07.013
Jaiswal, S., Bhattacharya, K., McHale, P., and Duffy, B. (2015). Dual effects of beta-cyclodextrin-stabilised silver nanoparticles: enhanced biofilm inhibition and reduced cytotoxicity. J. Mater. Sci. 26:5367. doi: 10.1007/s10856-014-5367-1
Kalishwaralal, K., BarathManiKanth, S., Pandian, S. R., Deepak, V., and Gurunathan, S. (2010). Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surfaces 79, 340–344. doi: 10.1016/j.colsurfb.2010.04.014
Kandi, V., and Kandi, S. (2015). Antimicrobial properties of nanomolecules: potential candidates as antibiotics in the era of multi-drug resistance. Epidemiol. Health 37:e2015020. doi: 10.4178/epih/e2015020
Khan, S. N., and Khan, A. U. (2016). Breaking the Spell: Combating Multidrug Resistant ‘Superbugs’. Front. Microbiol. 7:174. doi: 10.3389/fmicb.2016.00174
Khan, W., Bernier, S. P., Kuchma, S. L., Hammond, J. H., Hasan, F., and O'Toole, G. A. (2010). Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int. Microbiol. 13, 207–212. doi: 10.2436/20.1501.01.127
Kim, J. S., Kuk, E., Yu, K. N., Kim, J. H., Park, S. J., Lee, H. J., et al. (2007). Antimicrobial effects of silver nanoparticles. Nanomedicine 3, 95–101. doi: 10.1016/j.nano.2006.12.001
King, D. T., and Strynadka, N. C. (2013). Targeting metallo-beta-lactamase enzymes in antibiotic resistance. Future Med. Chem. 5, 1243–1263. doi: 10.4155/fmc.13.55
Krokowicz, L., Tomczak, H., Bobkiewicz, A., Mackiewicz, J., Marciniak, R., Drews, M., et al. (2015). In vitro studies of antibacterial and antifungal wound dressings comprising H2TiO3 and SiO2 nanoparticles. Pol. J. Microbiol. 64, 137–142.
Król, E., de Sousa Borges, A., da Silva, I., Polaquini, C. R., Regasini, L. O., Ferreira, H., et al. (2015). Antibacterial activity of alkyl gallates is a combination of direct targeting of FtsZ and permeabilization of bacterial membranes. Front. Microbiol. 6:390. doi: 10.3389/fmicb.2015.00390
Lebeaux, D., Chauhan, A., Létoffé, S., Fischer, F., de Reuse, H., Beloin, C., et al. (2014a). pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J. Infect. Dis. 210, 1357–1366. doi: 10.1093/infdis/jiu286
Lebeaux, D., Chauhan, A., Rendueles, O., and Beloin, C. (2013). From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356. doi: 10.3390/pathogens2020288
Lebeaux, D., Ghigo, J. M., and Beloin, C. (2014b). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543. doi: 10.1128/MMBR.00013-14
Lekshmi, N. C., Sumi, S. B., Viveka, S., Jeeva, S., and Brindha, J. R. (2012). Antibacterial activity of nanoparticles from Allium sp. J. Microbiol. Biotechnol. Res. 2, 115–119.
Lewis, K. (2008). Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322, 107–131. doi: 10.1007/978-3-540-75418-3_6
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Li, X. Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Lima, E., Guerra, R., Lara, V., and Guzmán, A. (2013). Gold nanoparticles as efficient antimicrobial agents for Escherichia coli and Salmonella typhi. Chem. Cent. J. 7:11. doi: 10.1186/1752-153X-7-11
Lo, A. W., Van de Water, K., Gane, P. J., Chan, A. W., Steadman, D., Stevens, K., et al. (2014). Suppression of type 1 pilus assembly in uropathogenic Escherichia coli by chemical inhibition of subunit polymerization. J. Antimicrob. Chemother. 69, 1017–1026. doi: 10.1093/jac/dkt467
Losasso, C., Belluco, S., Cibin, V., Zavagnin, P., Micetic, I., Gallocchio, F., et al. (2014). Antibacterial activity of silver nanoparticles: sensitivity of different Salmonella serovars. Front. Microbiol. 5:227. doi: 10.3389/fmicb.2014.00227
Maisonneuve, E., and Gerdes, K. (2014). Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548. doi: 10.1016/j.cell.2014.02.050
Markowska, K., Grudniak, A. M., and Wolska, K. I. (2013). Silver nanoparticles as an alternative strategy against bacterial biofilms. Acta Biochim. Pol. 60, 523–530.
Martinez-Gutierrez, F., Boegli, L., Agostinho, A., Sánchez, E. M., Bach, H., Ruiz, F., et al. (2013). Anti-biofilm activity of silver nanoparticles against different microorganisms. Biofouling 29, 651–660. doi: 10.1080/08927014.2013.794225
Miller, K. P., Wang, L., Chen, Y. P., Pellechia, P. J., Benicewicz, B. C., and Decho, A. W. (2015). Engineering nanoparticles to silence bacterial communication. Front. Microbiol. 6:189. doi: 10.3389/fmicb.2015.00189
Mohanty, S., Mishra, S., Jena, P., Jacob, B., Sarkar, B., and Sonawane, A. (2012). An investigation on the antibacterial, cytotoxic, and antibiofilm efficacy of starch-stabilized silver nanoparticles. Nanomedicine 8, 916–924. doi: 10.1016/j.nano.2011.11.007
Morita, Y., Tomida, J., and Kawamura, Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422. doi: 10.3389/fmicb.2013.00422
Morones-Ramirez, J. R., Winkler, J. A., Spina, C. S., and Collins, J. J. (2013). Silver enhances antibiotic activity against gram-negative bacteria. Sci. Transl. Med. 5, 190ra181. doi: 10.1126/scitranslmed.3006276
Mouton, J. W., Ambrose, P. G., Canton, R., Drusano, G. L., Harbarth, S., MacGowan, A., et al. (2011). Conserving antibiotics for the future: new ways to use old and new drugs from a pharmacokinetic and pharmacodynamic perspective. Drug Resist. Updat. 14, 107–117. doi: 10.1016/j.drup.2011.02.005
Mouton, J. W., Brown, D. F., Apfalter, P., Cantón, R., Giske, C. G., Ivanova, M., et al. (2012). The role of pharmacokinetics/pharmacodynamics in setting clinical MIC breakpoints: the EUCAST approach. Clin. Microbiol. Infect. 18, E37–E45. doi: 10.1111/j.1469-0691.2011.03752.x
Mukherje, M. (2014). In vitro antimicrobial activity of polyacrylamide doped magnetic iron oxide nanoparticles. Int. J. Mater. Mech. Manuf. 2, 64–66. doi: 10.7763/ijmmm.2014.v2.101
Mulcahy, H., Charron-Mazenod, L., and Lewenza, S. (2008). Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4:e1000213. doi: 10.1371/journal.ppat.1000213
Muller, A. E., Theuretzbacher, U., and Mouton, J. W. (2015). Use of old antibiotics now and in the future from a pharmacokinetic/pharmacodynamic perspective. Clin. Microbiol. Infect. 21, 881–885. doi: 10.1016/j.cmi.2015.06.007
Murima, P., McKinney, J. D., and Pethe, K. (2014). Targeting bacterial central metabolism for drug development. Chem. Biol. 21, 1423–1432. doi: 10.1016/j.chembiol.2014.08.020
Nam, K. Y. (2014). Characterization and bacterial anti-adherent effect on modified PMMA denture acrylic resin containing platinum nanoparticles. J. Adv. Prosthodont. 6, 207–214. doi: 10.4047/jap.2014.6.3.207
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. doi: 10.1126/science.1211037
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. doi: 10.1128/MMBR.67.4.593-656.2003
Oldfield, E., and Feng, X. (2014). Resistance-resistant antibiotics. Trends Pharmacol. Sci. 35, 664–674. doi: 10.1016/j.tips.2014.10.007
Opperman, T. J., and Nguyen, S. T. (2015). Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 6:421. doi: 10.3389/fmicb.2015.00421
Page, M. G., and Bush, K. (2014). Discovery and development of new antibacterial agents targeting Gram-negative bacteria in the era of pandrug resistance: is the future promising? Curr. Opin. Pharmacol. 18, 91–97. doi: 10.1016/j.coph.2014.09.008
Page, M. G., Dantier, C., and Desarbre, E. (2010). In vitro properties of BAL3(0072). a novel siderophore sulfactam with activity against multiresistant gram-negative bacilli. Antimicrob. Agents Chemother. 54, 2291–2302. doi: 10.1128/AAC.01525-09
Palanisamy, N. K., Ferina, N., Amirulhusni, A. N., Mohd-Zain, Z., Hussaini, J., Ping, L. J., et al. (2014). Antibiofilm properties of chemically synthesized silver nanoparticles found against Pseudomonas aeruginosa. J. Nanobiotechnology 12:2. doi: 10.1186/1477-3155-12-2
Pan, J., and Ren, D. (2013). Structural effects on persister control by brominated furanones. Bioorg. Med. Chem. Lett. 23, 6559–6562. doi: 10.1016/j.bmcl.2013.10.070
Parasion, S., Kwiatek, M., Gryko, R., Mizak, L., and Malm, A. (2014). Bacteriophages as an alternative strategy for fighting biofilm development. Pol. J. Microbiol. 63, 137–145.
Park, H. J., Park, S., Roh, J., Kim, S., Choi, K., Yi, J., et al. (2013). Biofilm-inactivating activity of silver nanoparticles: a comparison with silver ions. J. Ind. Eng. Chem. 19, 614–619. doi: 10.1016/j.jiec.2012.09.013
Penesyan, A., Gillings, M., and Paulsen, I. T. (2015). Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20, 5286–5298. doi: 10.3390/molecules20045286
Pethe, K., Bifani, P., Jang, J., Kang, S., Park, S., Ahn, S., et al. (2013). Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 19, 1157–1160. doi: 10.1038/nm.3262
Prabhu, S., and Poulose, E. K. (2012). Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int. Nano Lett. 2:32. doi: 10.1186/2228-5326-2-32
Pulcini, C., Bush, K., Craig, W. A., Frimodt-Møller, N., Grayson, M. L., Mouton, J. W., et al. (2012). Forgotten antibiotics: an inventory in Europe, the United States, Canada, and Australia. Clin. Infect. Dis. 54, 268–274. doi: 10.1093/cid/cir838
Pule, C. M., Sampson, S. L., Warren, R. M., Black, P. A., van Helden, P. D., Victor, T. C., et al. (2016). Efflux pump inhibitors: targeting mycobacterial efflux systems to enhance TB therapy. J. Antimicrob. Chemother. 71, 17–26. doi: 10.1093/jac/dkv316
Radzig, M. A., Nadtochenko, V. A., Koksharova, O. A., Kiwi, J., Lipasova, V. A., and Khmel, I. A. (2013). Antibacterial effects of silver nanoparticles on gram-negative bacteria: influence on the growth and biofilms formation, mechanisms of action. Colloids Surfaces 102, 300–306. doi: 10.1016/j.colsurfb.2012.07.039
Rampioni, G., Leoni, L., and Williams, P. (2014). The art of antibacterial warfare: deception through interference with quorum sensing-mediated communication. Bioorg. Chem. 55, 60–68. doi: 10.1016/j.bioorg.2014.04.005
Rao, C. V. S., De Waelheyns, E., Economou, A., and Anné, J. (2014). Antibiotic targeting of the bacterial secretory pathway. Biochim. Biophy. Acta 1843, 1762–1783. doi: 10.1016/j.bbamcr.2014.02.004
Roe, D., Karandikar, B., Bonn-Savage, N., Gibbins, B., and Roullet, J. B. (2008). Antimicrobial surface functionalization of plastic catheters by silver nanoparticles. J. Antimicrob. Chemother. 61, 869–876. doi: 10.1093/jac/dkn034
Sambanthamoorthy, K., Sloup, R. E., Parashar, V., Smith, J. M., Kim, E. E., Semmelhack, M. F., et al. (2012). Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob. Agents Chemother. 56, 5202–5211. doi: 10.1128/AAC.01396-12
Sass, P., and Brötz-Oesterhelt, H. (2013). Bacterial cell division as a target for new antibiotics. Curr. Opin. Microbiol. 16, 522–530. doi: 10.1016/j.mib.2013.07.006
Seil, J. T., and Webster, T. J. (2012). Antimicrobial applications of nanotechnology: methods and literature. Int. J. Nanomedicine 7, 2767–2781. doi: 10.2147/IJN.S24805
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163.
Shetye, G. S., Singh, N., Jia, C., Nguyen, C. D., Wang, G., and Luk, Y. Y. (2014). Specific maltose derivatives modulate the swarming motility of nonswarming mutant and inhibit bacterial adhesion and biofilm formation by Pseudomonas aeruginosa. Chembiochem 15, 1514–1523. doi: 10.1002/cbic.201402093
Singh, N., Saha, P., Rajkumar, K., and Abraham, J. (2014). Biosynthesis of silver and selenium nanoparticles by Bacillus sp. JAPSK2 and evaluation of antimicrobial activity. Der. Pharm. Lett. 6, 175–181.
Smith, R. S., Zhang, Z., Bouchard, M., Li, J., Lapp, H. S., Brotske, G. R., et al. (2012). Vascular catheters with a nonleaching poly-sulfobetaine surface modification reduce thrombus formation and microbial attachment. Sci. Transl. Med. 4, 153ra132. doi: 10.1126/scitranslmed.3004120
Subramenium, G. A., Vijayakumar, K., and Pandian, S. K. (2015). Limonene inhibits streptococcal biofilm formation by targeting surface-associated virulence factors. J. Med. Microbiol. 64, 879–890. doi: 10.1099/jmm.0.000105
Sun, J., Deng, Z., and Yan, A. (2014). Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. doi: 10.1016/j.bbrc.2014.05.090
Sutcliffe, J. A. (2011). Antibiotics in development targeting protein synthesis. Ann. N.Y. Acad. Sci. 1241, 122–152. doi: 10.1111/j.1749-6632.2011.06323.x
Thekkae Padil, V. V., and Černík, M. (2013). Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application. Int. J. Nanomedicine 8, 889–898. doi: 10.2147/IJN.S40599
Tillotson, G. S., and Theriault, N. (2013). New and alternative approaches to tackling antibiotic resistance. F1000Prime Rep. 5:51. doi: 10.12703/P5-51
Totsika, M., Kostakioti, M., Hannan, T. J., Upton, M., Beatson, S. A., Janetka, J. W., et al. (2013). A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli ST131. J. Infect. Dis. 208, 921–928. doi: 10.1093/infdis/jit245
Townsend, J. P., Nielsen, K. M., Fisher, D. S., and Hartl, D. L. (2003). Horizontal acquisition of divergent chromosomal DNA in bacteria: effects of mutator phenotypes. Genetics 164, 13–21.
Urfer, M., Bogdanovic, J., Lo Monte, F., Moehle, K., Zerbe, K., Omasits, U., et al. (2016). A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J. Biol. Chem. 291, 1921–1932. doi: 10.1074/jbc.M115.691725
Vardanyan, Z., Gevorkyan, V., Ananyan, M., Vardapetyan, H., and Trchounian, A. (2015). Effects of various heavy metal nanoparticles on Enterococcus hirae and Escherichia coli growth and proton-coupled membrane transport. J. Nanobiotechnol. 13, 69. doi: 10.1186/s12951-015-0131-3
Veerachamy, S., Yarlagadda, T., Manivasagam, G., and Yarlagadda, P. K. (2014). Bacterial adherence and biofilm formation on medical implants: a review. Proc. Inst. Mech. Eng. 228, 1083–1099. doi: 10.1177/0954411914556137
Venter, H., Mowla, R., Ohene-Agyei, T., and Ma, S. (2015). RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6:377. doi: 10.3389/fmicb.2015.00377
Wexselblatt, E., Kaspy, I., Glaser, G., Katzhendler, J., and Yavin, E. (2013). Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur. J. Med. Chem. 70, 497–504. doi: 10.1016/j.ejmech.2013.10.036
Wexselblatt, E., Oppenheimer-Shaanan, Y., Kaspy, I., London, N., Schueler-Furman, O., Yavin, E., et al. (2012). Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 8:e1002925. doi: 10.1371/journal.ppat.1002925
Wolska, K. I., Grzes, K., and Kurek, A. (2012). Synergy between novel antimicrobials and conventional antibiotics or bacteriocins. Pol. J. Microbiol. 61, 95–104.
Xu, H., Qu, F., Xu, H., Lai, W., Andrew Wang, Y., Aguilar, Z. P., et al. (2012). Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7. Biometals 25, 45–53. doi: 10.1007/s10534-011-9482-x
Yu, L., Zhang, Y., Zhang, B., and Liu, J. (2014). Enhanced antibacterial activity of silver nanoparticles/halloysite nanotubes/graphene nanocomposites with sandwich-like structure. Sci. Rep. 4, 45–51. doi: 10.1038/srep04551
Zhanel, G. G., Lawson, C. D., Zelenitsky, S., Findlay, B., Schweizer, F., Adam, H., et al. (2012). Comparison of the next-generation aminoglycoside plazomicin to gentamicin, tobramycin and amikacin. Expert Rev. Anti Infect. Ther. 10, 459–473. doi: 10.1586/eri.12.25
Zhang, K., Wang, S., Zhou, X., Xu, H. H., Weir, M. D., Ge, Y., et al. (2015). Effect of antibacterial dental adhesive on multispecies biofilms formation. J. Dent. Res. 94, 622–629. doi: 10.1177/0022034515571416
Zhang, L., and Mah, T. F. (2008). Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190, 4447–4452. doi: 10.1128/JB.01655-07
Zhang, L., Pornpattananangku, D., Hu, C. M., and Huang, C. M. (2010). Development of nanoparticles for antimicrobial drug delivery. Curr. Med. Chem. 17, 585–594. doi: 10.2174/092986710790416290
Zhang, W., and Li, C. (2016). Exploiting quorum sensing interfering strategies in gram-negative bacteria for the enhancement of environmental applications. Front. Microbiol. 6:1535. doi: 10.3389/fmicb.2015.01535
Zheng, Y., Li, J., Liu, X., and Sun, J. (2012). Antimicrobial and osteogenic effect of Ag-implanted titanium with a nanostructured surface. Int. J. Nanomedicine 7, 875–884. doi: 10.2147/IJN.S28450
Zhou, Y., Kong, Y., Kundu, S., Cirillo, J. D., and Liang, H. (2012). Antibacterial activities of gold and silver nanoparticles against Escherichia coli and Bacillus Calmette-Guerin. J. Nanobiotechnology 10:19. doi: 10.1186/1477-3155-10-19
Keywords: antimicrobial resistance, biofilm, persisters, antimicrobial drug, nanoparticles
Citation: Cheng G, Dai M, Ahemd S, Hao H, Wang X and Yuan Z (2016) Antimicrobial Drugs in Fighting against Antimicrobial Resistance. Front. Microbiol. 7:470. doi: 10.3389/fmicb.2016.00470
Received: 22 December 2015; Accepted: 21 March 2016;
Published: 08 April 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Santi M. Mandal, Vidyasagar University, IndiaXian-Zhi Li, Health Canada, Canada
William Charles Nierman, J. Craig Venter Institute, USA
Copyright © 2016 Cheng, Dai, Ahmed, Hao, Wang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zonghui Yuan, eXVhbjU4MDJAbWFpbC5oemF1LmVkdS5jbg==
 Guyue Cheng
Guyue Cheng Menghong Dai1
Menghong Dai1 Haihong Hao
Haihong Hao Xu Wang
Xu Wang