- 1Key Laboratory of Zoonosis of Ministry of Agriculture, College of Veterinary Medicine, China Agricultural University, Beijing, China
- 2Department of Biomedical and Diagnostic Sciences, College of Veterinary Medicine, The University of Tennessee, Knoxville, TN, USA
Development of highly effective, broad-spectrum antiviral agents is the major objective shared by the fields of virology and pharmaceutics. Antiviral drug development has focused on targeting viral entry and replication, as well as modulating cellular defense system. High throughput screening of molecules, genetic engineering of peptides, and functional screening of agents have identified promising candidates for development of optimal broad-spectrum antiviral agents to intervene in viral infection and control viral epidemics. This review discusses current knowledge, prospective applications, opportunities, and challenges in the development of broad-spectrum antiviral agents.
The epidemic viral diseases of severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), Western/Eastern equine encephalitis (WEE/EEE), and Ebola, as well as pandemic influenza A (H1N1) occurred in 2003, 2012, 2013, 2014, and 2009, respectively. The emerging SARS, H1N1, and MERS, as well as re-emerging WEE/EEE and EBOVes are lethal and transmissible through travelers. They have been identified as high priority biodefense pathogens by the United States. The high virulence of these viruses and the absence of effective therapies have posed an ongoing threat to the public health. The conventional one-bug-one-drug paradigm is insufficient to address the challenge of emerging and re-emerging viral pathogens, and few drugs are currently available to a prompt control of epidemic viral diseases (Chan et al., 2013; Raveh et al., 2013; Carossino et al., 2014). Thus, it is imperative to develop a broad-spectrum class of antiviral agents. Current strategies for the development of broad-spectrum antiviral agents focus on two aspects of targeting viral infectivity and modulating host defense system, as discussed in this review.
Agents Target on Viral Infectivity
Current strategies of controlling viral infectivity focus on identification of agents capable of intervening in the essential steps for viral infection, including viral attachment, fusion/endocytosis, replication, assembly and budding in addition to drugs targeting viral envelope, as detailed in the following subsections, and in Table 1 and Figure 1.
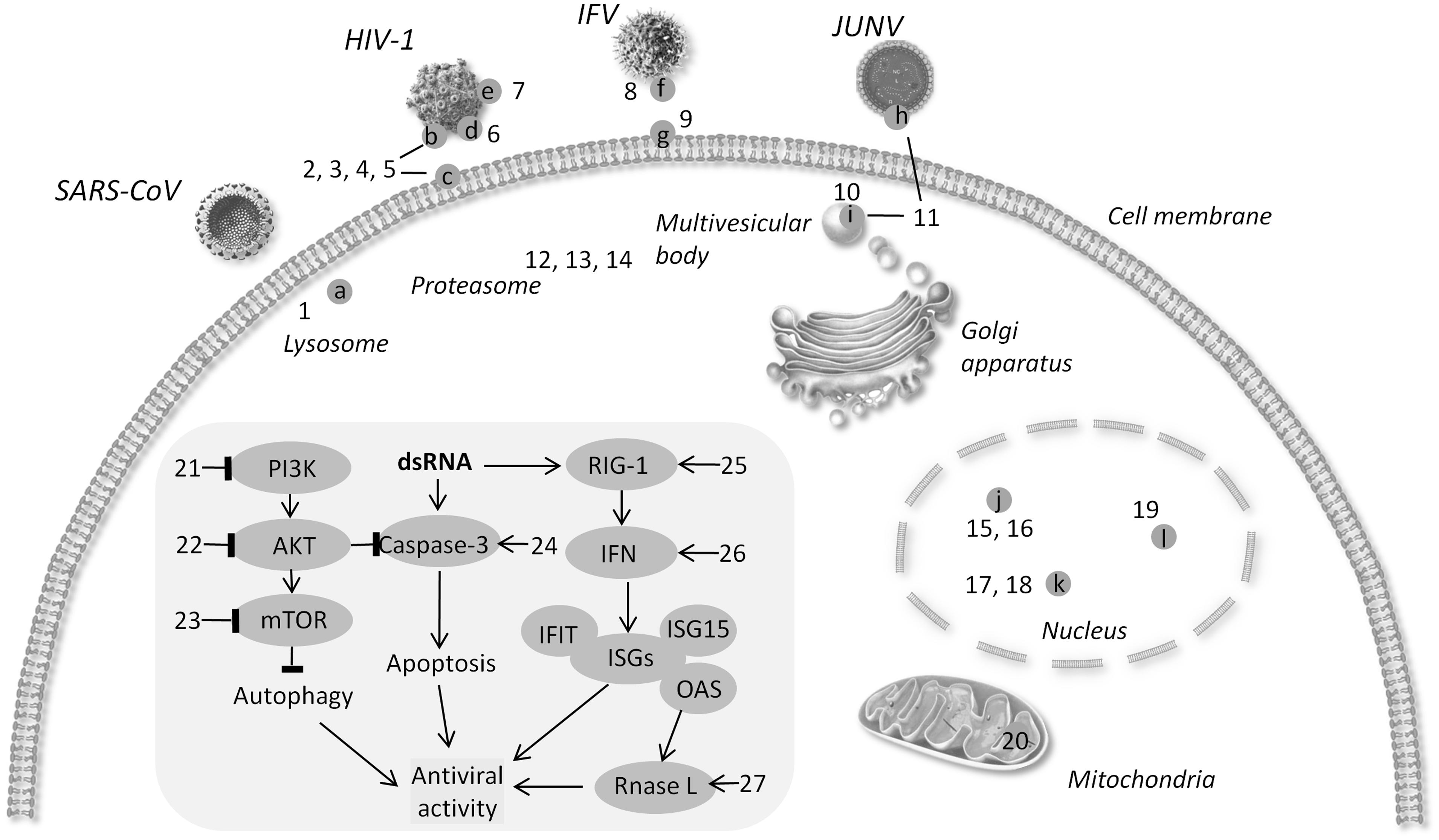
FIGURE 1. Schematic diagram of targets for broad-spectrum antiviral agents. (a) Cathepsin L (b) gP120 (c) CD4 (d) six-helix bundle (e). N-glycan terminal mannose residues (f) hemagglutinin (g) sialic acid (h) protein Z (i) TSG101 (j) viral RNA polymerase (k) DHODH l. XPO1 → activate - l block. (1) 5705213 (2) PVP-coated nano-silver (3) HP-OVA (4) CV-N 5. eCD4Ig. (6) p20 (7) Griffithsin (8) CR6261 (9) DAS181 (10) FGI-104 (11) Compound 0013 (12) VL-01 (13) Bortezomib (14) MG132 (15) JMN3-003 (16) BCX4430. (17) Compound A3 (18) Cmp1 (19) KPT-335 (20) Antimycin A (21) BEZ-235. (22) Akt-IV (23) Rapamycin (24) DRACOs (25) 5′pppRNA (26) GSK983. (27) Compounds C1 and C2.
Viral Attachment
Viral attachment to host cells is a specific interaction between viral surface proteins and host cell surface receptors and is a critical step for most viral entry into cells. Thus, blocking the interaction between viral particles and host cells is an important task being seriously considered in development of broad-spectrum antiviral agents to control viral infection, such as IFV and HIV-1.
IFV-A infects host cells through interaction of its surface antigen haemagglutinin with host cell surface receptor complex containing sialic acid (Nicholls et al., 2008). The highly conserved epitope at membrane-proximal stem of hemagglutinin has been successfully targeted by the specific monoclonal antibody CR6261 to inhibit IFV-A infection via blocking the conformational changes of haemagglutinins during viral attachment to host cells. Pre- or post-treatment of mice with CR6261 effectively inhibit infection with various serotypes of IFV and protect animals from IFV infection, demonstrating CR6261′s potential for a broad-spectrum antiviral agent (Friesen et al., 2010). On the other hand, the sialic acid of host cell surface receptor is also a recognized target for development of broad-spectrum antiviral agents. Sialidase hydrolyzes cell surface sialic acid and prevents viral attachment to cells. DAS181, a sialidase fusion protein consisting of a sialidase enzymatic domain and an amphiregulin glycosaminoglycan binding sequence to respiratory epithelium, has been shown to efficiently protect mice from infection of various serotypes of IFV, such as A/PR/8/34 (H1N1) and A/Victoria/3/75 (H3N2), as well as PIV. Treatment of IFV-infected animals with DAS181 also prevents pneumonia, indicating that DAS181 treatment prevents animals from secondary bacterial infection (Hedlund et al., 2010). Thus, CR6261 and DAS181 are candidates for developing broad-spectrum antiviral agents to control infection by IFV variants.
The HIV-1 particle carries only one envelope glycoprotein precursor gp160, which can be cleaved by host proteases, such as Furin, into gp120 and gp41; the resulting gp120 binds to the cell surface receptor CD4 and coreceptors (CCR5, CXCR4) for viral attachment and entry into cells (Wilen et al., 2012). Three reagents silver, ovalbumin (OVA), and lectin have been considered for development of broad-spectrum antiviral agents; they are all cost-efficient materials for antiviral agent development. Silver is a common expedient for cooking procedures and preserving water from contamination. Coating nano-silver with polyvinylpyrrolidone (PVP) has been shown to competitively bind to gp120, resulting in blocking viral attachment to the host cell receptor CD4 and preventing viral infection (Galdiero et al., 2011). In addition, PVP-coated nano-silver exhibits antiviral ability to directly inhibit the infectivity of viral particles in a cell-independent manner (Lara et al., 2010). However, what other metal nanoparticles may interfere with the infectivity of viral particles for developing broad-spectrum antiviral agents remains to be investigated. Similarly, OVA shows anti-viral ability, and 3-hydroxy phthalic anhydride-modified OVA (HP-OVA) exhibits a broad-spectrum antiviral activity by interfering with gp120 binding to CD4 (Li et al., 2010). Many natural lectins, such as Concanavalin A and Lentil lectin isolated from plants, have been shown to be cytotoxic, to non-specifically activate T cells, and to agglutinate red blood cells; these concerns have limited the use of lectins in antiviral drug development (Barton et al., 2014). Mori’s group revealed that the 12 kDa red algae lectin Griffithsin (GRFT) binds N-glycan terminal mannose residues of gp120, blocks viral binding to CD4-expressing cells, and inhibits HIV-1 entry into cells in a glycosylation-dependent manner (Mori et al., 2005). GRFT has been shown to possess a broad-spectrum antiviral activity, and it is potentially safe in systemic treatment of HIV-1 infection without inducing pro-inflammatory cytokines (Mori et al., 2005; Barton et al., 2014). Cyanovirin-N (CV-N), a natural lectin isolated from the cyanobacterium Nostoc ellipsosporum, has been shown its high-affinity to interact with the N-linked high-mannose oligosaccharides of HIV gp120 and block the ability of gp120 to mediate membrane fusion (Botos and Wlodawer, 2003). However, the associated cytotoxicity and immunogenicity have hindered the development of CV-N to be a viable therapeutic agent. CV-N also inhibits EOBV and HCV infection effectively by interacting with viral envelope glycoproteins at the early stages of viral entry (Barrientos et al., 2004; Helle et al., 2006). Recently, (GGGGS)3, which is a flexible and hydrophilic linker, was used to conjugate with CV-N to produce LCV-N; then PEG was used to modify the N-terminus of LCV-N to produce PEGylated LCV-N (PEG-LCV-N); and PEG-LCV-N is effective to control IFN infection in vivo (Wu et al., 2015). Accordingly, CV-N and its derivatives, such as PEG-LCV-N, should be further developed and used as broad-spectrum antiviral agents. Thus, PVP-coated nano-silver particles, HP-OVA, GRFT, and CV-N should be further studied for developing broad-spectrum antiviral agents to control HIV-1 infection.
Broad-spectrum neutralizing antibodies (bNAbs) are being developed to target HIV gp120 for inhibition of HIV infection. However, all the tested bNAbs were able to neutralize 10–50% of HIV-1 isolates (Su and Moog, 2014). Recent development of antiviral agents recognizes that two most conserved epitopes of HIV-1 gp120 are CD4- and CCR5-binding sites (Gardner et al., 2015). Fusion of CD4-Ig, which is a recombinant fusion protein consisting of CD4 and Fc region, with a small CCR5-mimetic sulfopeptide resulted in an effective gp120-targeted agent eCD4-Ig. eCD4-Ig has been shown to effectively neutralize 100% of a panel of HIV-1, HIV-2, and simian immunodeficiency viruses, including bNAbs-resistant viral isolates (Gardner et al., 2015). Accordingly, eCD4-Ig should be further developed as an antiviral agent to control HIV infection.
Membrane Fusion and Endocytosis
Severe acute respiratory syndrome coronavirus, EBOV, HeV, and NiV are highly pathogenic to a broad range of hosts including human and various species of animals. The entry of these viruses into host cells is dependent on cathepsin L (CatL), which is a cellular lysosomal protease required for processing gS-glycoprotein cleavage, to generate fusion of viral envelope with cellular membrane after viral attachment (Pager and Dutch, 2005; Simmons et al., 2005; Pager et al., 2006; Kaletsky et al., 2007). High throughput approaches have identified 50 molecules from 5000-molecule libraries by their ability to inhibit CatL and block viral entry into cells. The small molecule 5705213 and its derivative 7402683 have been shown to efficiently inhibit CatL-mediated cleavage of gS-glycoprotein and effectively block SARS-CoV entry into cells (Elshabrawy et al., 2014). The molecule 5705213 also blocks the entry of EBOV, HeV, and NiV into cells via inhibition of glycoprotein cleavage (Elshabrawy et al., 2014). Accordingly, identification of small molecules to inhibit CatL is a promising approach of developing broad-spectrum antiviral agents to interfere with virus-cell membrane fusion, endocytosis, and viral entry into cells.
Binding the gp120 of HIV-1 to cellular receptors triggers a conformational change of the subunit gp41 for the fusion of viral envelope and cellular membrane (Wilen et al., 2012). The gp41 is a transmembrane protein, and the core of gp41 consists of three units of heptad repeat 1 (HR1) and three units of HR2; HR2 units surround the HR1 units in an anti-parallel manner to form a six-helix bundle structure. Conformational changes of the six-helix bundle activate its function to mediate the fusion of viral and cellular membranes together (Colman and Lawrence, 2003). The HR2-mimicking peptide Enfuvirtide (Fuzeon, a currently, clinically approved peptide fusion inhibitor) can bind HR1 and interfere with conformational changes, thereby blocking the formation of active six-helix bundle structure (Eckert and Kim, 2001). Although Enfuvirtide is effective in control of wild-type HIV-1 and variants, Enfuvirtide treatment results in resistant HIV-1 variants, which are also resistant to reverse transcriptase inhibitors (such as Zidovudine) and protease inhibitors (such as Ritonavir; Eckert and Kim, 2001). Thus, development of next-generation inhibitors of HRs is urgent. Recently, the P20, homologous to human troponin I type 3 interacting kinase (TNNI3K)-like protein, is identified by screening a DNA library of human bone marrow cells through yeast two-hybridization method (Zhu et al., 2010). The P20 does not share any homology with Enfuvirtide, but it binds to six-helix bundle and inhibit infection by various HIV-1 strains. It is reported that P20 variants with a common motif (WGRLEGRRT) exhibit potent activity to inhibit HIV-1 infection (Zhu et al., 2010); apparently, this common motif can be used as a lead to develop optimal inhibitors for controlling HIV-1 infection.
Similar to HIV-1, a six-helix bundle structure is reported to mediate the entry of class I enveloped viruses into host cells, including coronavirus and paramyxovirus (Liu et al., 2004; Wang et al., 2005); the six-helix bundle structure appears to be an optimal target for developing broad-spectrum antiviral agents. To achieve this end, the HR2 sequences of the paramyxovirus NDV and the coronavirus IBV were systemically analyzed. The primary structure of each helix consists of heptad repeats containing seven commonly residues (a–b–c–d–e–f–g)n. Replacement of the non-conservative sites b, c, f, and g with the charged residues glutamic acid or lysine followed by screening revealed a novel peptide AU (VNKKIEEIDKKIEELNKKLEELEKKLEEVNKK) that is able to completely inhibit single or double infection of cells with NDV and IBV in vitro and in vivo (Wang et al., 2011). Furthermore, tagging a cholesterol unit to the N-terminus of AU, resulting in C-AU, not only prolongs the half-life of AU in vivo but also increases its potency to control viral infection. For example, intramuscular injection of chicken with C-AU one day prior to virus inoculation and then every 3 days for three times after viral inoculation resulted in a 70% protection of animals from NDV infection (Li et al., 2013). This in vivo study demonstrated feasibility of using antiviral peptides in clinical application to control viral infection.
Viral Replication
RNA polymerase is essential for the replication of numerous RNA viruses (te Velthuis, 2014). BCX4430, a novel nucleoside analog, is an inhibitor specific for viral RNA polymerase and exhibits broad-spectrum antiviral activity against viruses including filoviruses, flaviviruses, arenaviruses, paramyxoviruses, coronaviruses, and bunyaviruses (Warren et al., 2014). BCX4430 treatment completely protected golden hamsters from death caused by infection with YFV (flavivirus), and the treatment also improved symptoms including weight loss and viremia. BCX4430 also completely protected cynomolgus macaques from infection with Marburg virus, a hemorrhagic fever virus of the Filoviridae family, even it administered 48 h after viral infection (Julander et al., 2014; Warren et al., 2014). Similarly, JMN3-003 can block the replication of various viruses (paramyxoviruses, orthomyxoviruses, etc.) via specific inhibition of viral RNA polymerases; interestingly, JMN3-003 treatment does not affect mRNA synthesis or protein translation of host cells (Krumm et al., 2011). Both BCX4430 and JMN3-003 are able to specifically target viral RNA polymerases without causing side effects to hosts. In addition, resistant viral variants were not detectable in animals after treatment with BCX4430 or JMN3-003 (Krumm et al., 2011; Warren et al., 2014). The pyrazine derivative favipiravir (T-705) targets viral RNA-dependent RNA polymerases, and T705 treatment inhibits almost all RNA viruses without affecting DNA or RNA synthesis of host cells. Clinically, T-705 was reportedly effective in controlling EBOV infection (Oestereich et al., 2014; De Clercq, 2015). In addition, T-705 treatment of IFV-A-infected cells did not result in detectable resistant variants (Mendenhall et al., 2011). Accordingly, agents, targeting viral RNA polymerases, are optimal candidates of broad-spectrum antiviral agents to be fully developed for the control of RNA viruses.
Dihydroorotate dehydrogenase (DHODH) dehydrogenizes dihydroorotate to orotic acid that is the key step of biosynthesis of de novo pyrimidine to generate uracil essential for replication of both viral RNA and DNA (Evans and Guy, 2004). Compound A3, which was identified by high-throughput screening of 61,600 small-molecular-weight compounds, is able to target DHODH, interrupt the biosynthesis of pyrimidine, and inhibit replication of viruses including negative strand RNA viruses (such as IFV-A and B), positive strand RNA viruses (such as hepatitis C virus, HCV and DENV), and DNA viruses (such as VACV and adenovirus; Hoffmann et al., 2011). Compound A3 treatment also causes damages to viral genomic RNA and reduces viral infectivity (Ortiz-Riano et al., 2014). Co-treatment with Ribavirin enhances the therapeutic efficacy of A3 in control of LCMV (Ortiz-Riano et al., 2014). Ribavirin is a broad-spectrum antiviral agent and is approved for treatment of RSV and HCV infection (Naik and Tyagi, 2012). Ribavirin, a guanosine analog, has been used to inhibit synthesis of viral RNAs (Parker, 2005). Ribavirin also inhibits RNA polymerase of HCV to interfere with viral replication and induces formation of GTP analog RTP to cause translational error of viral proteins (Cannon et al., 2009; Shah et al., 2010). In addition, Ribavirin is known to inhibit replication of RNA and DNA viruses via distinctive mechanisms and replication of the same virus via distinct mechanisms in different cell types, depending on intracellular Ribavirin metabolism (Shah et al., 2010). Interestingly, treatment with Ribavirin alone showed a modest or no effect on viral replication in HCV-infected patients; however, combination with IFN dramatically improved long-term antiviral response in patients (Dixit and Perelson, 2006). Another DHODH inhibitor, compound 1 (Cmp1), was identified by the silico fragment-based drug design (FBDD); Cmp1 inhibits both human and murine DHODH, in vitro, in a dose-dependent manner (Konteatis, 2010). Cmp1, at low micro- to nano-molar levels, is highly effective in inhibiting replication of human and murine cytomegaloviruses (HCMV and MCMV) as well as HAdV-2 (Marschall et al., 2013). An in vivo study showed that treatment of mice with Cmp1 at 30 mg/kg/day for 7 days resulted in a significant suppression of MCMV replication in various organs, including salivary gland, lung, liver, and spleen (Marschall et al., 2013).
Antimycin A, isolated from Streptomyces kaviengensis, displays a potent activity to control equine encephalitis viruses (EEV) infection; treatment with antimycin A increased survival rates of EEV-infected mice and reduced viral replication (Raveh et al., 2013). Antimycin A is also able to control infection by various RNA viruses, including members of the Togaviridae, Flaviviridae, Bunyaviridae, Picornaviridae, and Paramyxoviridae families (Raveh et al., 2013). Interestingly, antimycin A inhibits cellular mitochondrial electron transport chain and de novo pyrimidine synthesis, indicating that host mitochondrial electron transport is a viable target for identifying broad-spectrum antiviral agents. The commercialized antiprotozoal agent Nitazoxanide has been recently identified as a first-in-class broad-spectrum antiviral agent to inhibit the replication of various RNA and DNA viruses, including HIV, IFV, DENV, HCV, HBV, and JEV (Rossignol, 2014). Nitazoxanide is shown to mobilize Ca2+, resulting in chronic sub-lethal stress of ER and disruption of viral protein glycosylation and trafficking (Ashiru et al., 2014).
Based on phylogenetic analysis, positive-sense RNA viruses picornaviruses, caliciviruses, and coronaviruses are recognized as members of picornavirus-like supercluster. These viruses possess 3C or 3C-like proteases (3Cpro or 3CLpro, respectively), which contain a typical chymotrypsin-like fold and a catalytic triad (or dyad) with a Cys residue as a nucleophile (Xia and Kang, 2011). 3Cpro and 3CLpro act as key enzymes for proteolytic processing of viral polyproteins for virus replication (Wang and Liang, 2010). To target 3Cpro and 3CLpro of supercluster members, the aldehyde GC373, the α-ketoamide GC375, and the bisulfite adduct GC376, which share a common dipeptidyl residue, are identified with the ability to inhibit the enzymatic activity of 3Cpro or 3CLpro via covalent binding to a nucleophilic Cys residue in the conserved catalytic site of 3Cpro and 3CLpro (Kim et al., 2012, 2013). GC373, GC375, and GC376 are being developed as broad-spectrum antiviral agents to control viral infection by picornavirus-like supercluster members.
Viral Assembly and Budding
Formation of viral ribonucleoprotein (vRNP) complexes, consisting of viral RNA, RNA polymerase, and nucleoprotein, is essential for the process in replication of negative-sense single-stranded RNA viruses, such as IFV-A. After replication of IFV genomic RNA in the nucleus, the vRNP complexes translocate into cytoplasm for assembly of viral nucleocapsid through a series of nuclear export pathways, such as the nuclear export receptor chromosome region maintenance 1 (CRM1)-mediated pathway (Mao and Yang, 2013). Leptomycin B (LMB) is a highly specific inhibitor of CRM1, and LMB treatment blocks the export of vRNP from nucleus to cytoplasm, thereby suppressing the assembly of IFV-B particles (Dauber et al., 2009). LMB has also been shown to inhibit replications of DNA viruses, such as HSV-1 (Park et al., 2013). Although LMB treatment was effective, its toxicity was detectable in animals (Perwitasari et al., 2014). Thus, LMB needs to be further studied and modified to improve its therapeutic value in control of viral infection.
The human CRM1 homolog exportin 1 (XPO1) mediates transport of leucine-rich nuclear export signal-dependent cellular protein and RNAs for cell growth and differentiation (Askjaer et al., 1998). The nuclear export of vRNPs complexes is mediated by the interaction of XPO1 with viral nuclear export protein of IFV-A NS2 (Neumann et al., 2000). KPT-335 (also Verdinexor) is a small molecule antagonist of XPO1 (Perwitasari et al., 2014). In vitro studies showed that KPT-335 is able to selectively and effectively inhibit replication of various IFV A and B strains, including pandemic H1N1, highly pathogenic AIV H5N1, and the recently emerged H7N9. In vivo studies showed that prophylactic and therapeutic administration of KPT-335 protected mice from IFV A/California/04/09 or IFV A/Philippines/2/82-X79, and KPT-335 treatment reduced production of viral particles and proinflammatory cytokines in lung with non-significant toxicity (Perwitasari et al., 2014). Thus, KPT-335 appears to be a promising broad-spectrum antiviral agent.
It has been recently reported that enveloped viruses, such as EBOV, utilize the cellular endosomal sorting complexes required for transport (ESCRT) pathway to release their progenies from infected cells; the ESCRT pathway is also the major escaping route for enveloped viruses (Votteler and Schubert, 2008; Bucci, 2013; Snyder et al., 2013; Dolnik et al., 2014). The tumor susceptibility gene 101 product TSG101 is a subunit of ESCRT-I (Luyet et al., 2008). The phenolic compound FGI-104 targets TSG101 and inhibits viral budding of EBOV; treatment with FGI-104 at 10 mg/kg/day for 7 days protected mice from EBOV-caused death. FGI-104 treatment also intervenes in HCV, HIV-1, IFV-A, and HSV-1 infection via inhibition of TSG101 (Kinch et al., 2009). Recently, Lu’s group reported that TSG101 serves as a target of the Z protein PTAP-L domain of JUNV, which causes Argentine hemorrhagic fever, to facilitate viral budding (Lu et al., 2014). Using the structure of TSG101-PTAP interaction site as a target for silico screening of competitive binding molecules, a novel compound 0013 was identified by its ability to block the interaction of protein Z with TSG101 (Lu et al., 2014); the 0013 inhibited budding of JUNV particles. It is known that PTAP L-domain-containing proteins are generally required for separation of RNA viral particles from host cells (Chen and Lamb, 2008). Studies showed that the 0013 is effective in inhibiting PTAP-dependent budding of arenavirus, filovirus, and retrovirus (Lu et al., 2014). Thus, compound 0013 can be used as a lead compound for further development of broad-spectrum antiviral agents to control viral budding.
Viral Envelope
Cells are able to promptly repair damages, ranging from 0.2 to 10 μm, of cell membrane by cytoplasmic endomembrane; however, viral particles are incapable of repairing damaged viral envelope (McMahon and Gallop, 2005). LJ001 is a type II photosensitizer derived from aromatic methyl diene rhodanine (Wolf et al., 2010). Treatment of cells with LJ001 results in generating singlet oxygen (1O2) in the membrane bilayer, and 1O2-mediated lipid oxidation permanently changes biophysical properties of viral envelope, resulting in interruption of virus-cell fusion; in contrast, LJ001 treatment does not result in a permanent damage of cell membrane due to the ability of cells to repair lipid biosynthesis (Wolf et al., 2010). Thus, LJ001 treatment is able to irreversibly damage the viral envelope bilayer structure, but not host cell membrane, to protect cells from fusion with enveloped viruses, including bunyaviruses, filoviruses, poxviruses, arenaviruses, paramyxoviruses, flaviviruses, IFV-A, and HIV-1 in vitro (Wolf et al., 2010). Using LJ001 as a lead compound, a new class of membrane-targeted photosensitizers (i.e., oxazolidine-2,4-dithione derivatives: JL118 and JL122) has been developed to improve solubility and bioavailability for in vivo application. In vitro studies showed that JL118 and JL122 are able to increase 1O2 level 100-fold than LJ001 can do in cells. In vivo studies showed that JL118 and JL122 treatment significantly delayed the death of RVFV (bunyaviruses)-infected mice (Vigant et al., 2013).
Recently, mastoparan, which is a natural host defense peptide (NHP) isolated from the invertebrate wasp, has been shown to enter into viral envelop and interact with the lipid components (Albiol Matanic and Castilla, 2004). The mastoparan-derived peptide MP7-NH2 is able to alter the structure of viral envelope and directly inactivate the infectivity of enveloped viruses, including rhabdoviruses, flaviviruses, herpesviruses, and poxviruses, but not IFV-A (Sample et al., 2013). Possibly, IFV-A envelope is less organized/ordered than the envelope of viruses, such as VSV of rhabdoviruses, contributing to IFV resistance to MP7-NH2 (Doms et al., 1988; Mukhopadhyay et al., 2005; Harris et al., 2006). Pre-exposure of VSV particles to MP7-NH2 significantly reduced the infectivity of VSV in vivo (Sample et al., 2013). Thus, LJ001 and MP7-NH2 should be recognized as lead compounds for further development of broad-spectrum agents to control viral infection by targeting viral envelope.
The indole derivative Arbidol has been shown to block the interaction between cell membrane and HCV glycoproteins to inhibit viral entry and fusion into cells (Boriskin et al., 2008). Arbidol also inhibits viral replication of HCV by altering cytoplasmic proteins, which are essential for intracellular trafficking (e.g., clathrin coat components, elements of the cytoskeleton) and viral replication (e.g., membranous web), and by hindering membrane rearrangements involved in viral budding. Arbidol has been clinically shown to inhibit infection by enveloped viruses, such as IFV-A and coronavirus, as well as non-enveloped reovirus; thus, Arbidol is considered as a broad-spectrum antiviral agent worthy of further development (Blaising et al., 2014). Arbidol has been clinically used to treat viral infections in Russia since 2005; however, it has not been approved for use in Western countries (Blaising et al., 2014).
Summary for the First Section
In general, antiviral agents, such as DAS181 and C-AU, which target viral attachment or virus-cell membrane fusion/endocytosis are specific to viruses; however, their treatments often result in drug-resistant viral variants, and their specificity to viruses limits their potential for development of broad-spectrum antiviral agents. Agents, such as BCX4430 and JMN3-003, inhibit viral RNA polymerases and are non-cytotoxic; their ability to block replication of various RNA viruses without resulting in drug-resistant viral variants positions their candidacy to be developed into optimal broad-spectrum agents to inhibit RNA viruses. Compound A3, which interrupts biosynthesis of pyrimidine in cells, appears to be a broad-spectrum antiviral agent to control both RNA and DNA viruses; A3 treatment does not result in drug-resistant viral variants. KPT-335, which blocks viral assembly in cytoplasm, appears to be another promising broad-spectrum antiviral agent to control RNA and DNA viruses. FGI-104, which targets the cellular exporting ESCRT system, inhibits viral budding without resulting in drug-resistant viral variants. Although compound A3, KPT-335, and FGI-104, which target on cellular machinery, are able to act as broad-spectrum antiviral agents, their potential side effects on patients need to be addressed. Whether combination of these agents will be optimal to control various viruses remains to be determined.
Agents Based on Host Defense
Host cellular defense is the first hurdle against viral infection. However, viruses are able to utilize cellular machineries for viral replication. Growing knowledge has helped identify broad-spectrum antiviral agents by targeting the cellular machineries of defense, programmed cell death, and metabolism, as presented in the following subsections, and in Table 2 and Figure 1.
Interferon (IFNs)
Type I IFNs [alpha and beta interferons (IFN-α/β)] are the first-line cytokines for host’s antiviral defense via immune system and rapid induction of cellular modulators to inhibit viral replication and spread (Burke et al., 2014). Viral infection yields highly conserved pathogen-associated molecular patterns (PAMPs), such as double-stranded RNA (dsRNA). Viral RNA contains short hairpin dsRNA with triphosphorylated 5′ end (5′pppRNA) that preferentially activates host’s pattern recognition receptors (PRRs), such as retinoic acid inducible gene-I (RIG-I) and related receptors (Ng and Gommerman, 2013). 5′pppRNA-activated RIG-I interacts with mitochondrial antiviral-signaling protein (MAVS) to result in activation of transcription factors, such as IFN regulatory factor-3/7 (IRF-3/7), through the IKK-related kinases TBK1 and IKK𝜀, to induce IFN gene expression (Schlee et al., 2009; Wilkins and Gale, 2010). An analog of 5′pppRNA, derived from the 5′ and 3′ untranslated regions of VSV genome (Schlee et al., 2009), has been shown to induce IFN expression and multiple innate antiviral responses, including IRF3/7, to control infections with IFV-A, DENV, VSV, HCV, and VACV (Goulet et al., 2013; Olagnier et al., 2014). Mice, treated with 5′pppRNA, fully recovered (100% survival) from a lethal infection with H1N1 A/PR/8/34 IFV; 5′pppRNA treatment completely inhibited viral replication in the lung (Goulet et al., 2013).
Secretion of type I IFNs from virus-infected cells may trigger antiviral activity in not only infected cells but also surrounding uninfected cells in an autocrine- or a paracrine-dependent manner. IFNs bind to cell surface IFN receptor (IFNAR) to activate the Janus kinase signal transducer and activator of transcription (JAK-STAT) pathway, leading to upregulation of hundreds of IFN-stimulated gene (ISG) products, such as ISG15, ITIT, IFITM and 2–5′ oligoadenylate synthetase (2-5A OAS; Burke et al., 2014). In general, ISGs have been shown to exhibit a wide range of antiviral abilities, including the ability to degrade viral nucleic acids, to inhibit viral gene expression, and to serve as PRRs to amplify IFN signals and elevate IFN expression (Bisbal et al., 2001; Wilkins and Gale, 2010).
ISG15 may act as an ubiquitin cross-reactive protein (Morales and Lenschow, 2013). ISG15 is able to inhibit ubiquitination of HIV-1 Gag protein and the interaction of Gag with TSG101, which are important for releasing HIV-1 particles from cells (Pincetic and Leis, 2009). ISG15 may block the ESCRT pathway (see Viral Assembly and Budding) and disrupt cell release of viral particles, such as HIV-1, EBOV, and ASLV (Morales and Lenschow, 2013). ISG15 is also involved in ISGylation of viral proteins that contributes to IFN-mediated inhibition of virus replication. HERC5 is an IFN-induced E3 ubiquitin ligase for ISG15 to mediate ISGylation of the IFV-A protein NS1 that directly inhibits viral replication (Tang et al., 2010; Zhao et al., 2010). ISG15 is also able to help other ISGs, such as IRF3. ISGylation of IRF3 inhibits ubiquitination and degradation of IRF3 (Lu et al., 2006). Stabilized IRF3 forms complex with CREB binding protein (CBP) that translocates into the nucleus and activates the transcription of IFNs and other ISGs genes (Prinarakis et al., 2008). IRF3 has been shown to play a critical role in the innate immune response against DNA and RNA viruses. Overexpression of ISG15 resulted in increased antiviral activity in vitro and in vivo, and knockdown of ISG15 resulted in increased replication of VSV, SeV, and NDV in cells (Shi et al., 2010) that further demonstrated the ISG15 antiviral ability.
IFIT (Interferon induced proteins with tetratricopeptide repeats) and IFITM (IFN-induced transmembrane protein) are two families, involved in IFN activation, with distinctive genetics and functions. Both IFIT and IFITM show broad-spectrum antiviral activity to interfere with viral replication, transmission, and virulence (Diamond and Farzan, 2013). Four human family members (IFIT1, IFIT2, IFIT3, and IFIT5) and three rodent family members (Ifit1, Ifit2, Ifit3) are identified. IFIT inhibits viral infections through multiple mechanisms; IFIT1 and IFIT2 suppress the translation initiation (Hui et al., 2003), IFIT1 binds uncapped or incompletely capped viral RNA (Pichlmair et al., 2011) and sequesters viral protein (e.g., HPV E1) or RNA in the cytoplasm (Terenzi et al., 2008). On the other hand, human IFITM family composes of four members IFITM1, IFITM2, IFITM3, and IFITM5; and murine IFITM composes of Ifitm1, Ifitm2, Ifitm3, and Ifitm5. IFITM proteins traverse cell membrane and are abundant in endosomes and lysosomes, and they can prevent enveloped viruses from crossing endosomal or lysosomal membranes and penetrating into the cytoplasm (Diamond and Farzan, 2013). IFITM members also interfere with the membrane fusion and endocytosis of viral particles to effectively inhibit viral infection by enveloped viruses; such as IFV, EBOV, SARS-CoV and DENV (Brass et al., 2009; Huang et al., 2011). It is shown that IFITM3 is able to limit IFV-A to the hemagglutinin attachment phase of viral replication, contributing to control of IFV-A infection in vivo (Brass et al., 2009; Feeley et al., 2011). However, the detailed mechanisms for IFITM members in differential inhibition of enveloped viruses and whether IFITM may interfere with replication of non-enveloped viruses remain to be studied.
The 2-5A OAS/RNase L pathway has been detected in various types of mammalian cells (Bisbal et al., 2001). Upon activation by viral dsRNA, 2-5A OAS synthesizes 2–5′ oligoadenylates (2-5A) from ATPs. The resulting 2-5A binds to the inactive monomeric form of RNase L to result in dimerization and activation of RNase L, and activated RNase L mediates degradation of viral RNA, contributing to IFN antiviral activity (Chakrabarti et al., 2011; Silverman and Weiss, 2014). Thus, activators of RNase L are considered for developing antiviral agents. However, 2-5A is labile and cytotoxic; it may not be an optimal antiviral agent candidate (Gale and Sen, 2009). High throughput screening of the chemical library ChemBridge DIVERset of 30,000 small molecules with the technique of fluorescence resonance energy transfer (FRET) has identified seven compounds effective in activating RNase L at micromolar concentrations (Thakur et al., 2007). Two lead compounds, C1 and C2, are able to enter into cells and induce RNase L activity without showing cytotoxicity. Both C1 and C2 show broad-spectrum antiviral activity against RNA viruses, including picornaviruses, rhabdoviruses, paramyxoviruses, and retroviruses (Thakur et al., 2007). In addition, GSK983, a novel tetrahydrocarbazole, has been shown to induce significant increases of seven ISGs, including ISG15, ISG56, OAS1, OAS2, OASL, IL6, and TRAIL. GSK983 treatment resulted in growth inhibition of cells infected by HPV, adenovirus Ad-5, or EBV, possibly via induction of ISGs (Harvey et al., 2009). However, the exact mechanism for GSK983 antiviral activity still needs a clarification.
Programmed Cell Death
Apoptosis and autophagy are two major forms of programmed cell death and play essential roles in not only homeostasis but also survival in multicellular organisms (Lockshin and Zakeri, 2007; He and Klionsky, 2009). Apoptosis and autophagy are distinguished on the basis of cell morphological changes (Hacker, 2000; Lockshin and Zakeri, 2007). Apoptosis is characterized by activation of proteolytic caspases and formation of apoptotic bodies that mediate degradation of cellular organelles. Autophagy is characterized by the formation of autophagosomes to sequestrate and transport cytoplasmic materials, including damaged proteins and organelles, to lysosomes for degradation; it is regarded as “self-eating” catabolism.
In response to viral infection, induction of apoptosis may limit viral production and spread into adjacent cells, while simultaneously promoting host innate and inflammatory responses (Taylor et al., 2008; Upton and Chan, 2014). However, viruses, such as HSV-1 and EBOV, are able to escape from apoptotic control or reverse the process to facilitate viral replication, respectively (Martins et al., 2012; Fuentes-Gonzalez et al., 2013). Infection of viruses, such as paramyxoviruses and orthomyxoviruses, may result in activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway to block apoptosis for the benefit of viral replications (Sun et al., 2008; Liu et al., 2010; Diehl and Schaal, 2013). Activated Akt phosphorylates downstream transcription factors, such as FoxO1; FoxO1 promotes degradation of IRF3 in suppression of cellular antiviral response (see Programmed Cell Death; Lei et al., 2013). The PI3K/Akt pathway inhibitor Akt-IV was identified by high throughput screening of inhibitors of FoxO1 (Ramaswamy et al., 1999; Sun et al., 2008). Akt-IV has been shown to inhibit replication of the paramyxoviruses PIV5 and RSV. Interestingly, although the PI3k/Akt pathway is not involved in the replication of VSV, Akt-IV is able to inhibit VSV replication (Sun et al., 2008; Dunn et al., 2009). It has also been shown that the PI3K inhibitor BEZ-235 is effective in inhibiting the production of LCMV and JUNV viral particles without causing cell toxicity (Urata et al., 2012).
The dsRNA-activated caspase oligomerizer (DRACO) is a new antiviral agent and is currently under preclinical studies. DRACO consists of three components: a dsRNA recognition domain derived from protein kinase R (PKR) for activating the defensive IFN pathway, apoptotic protease activating factor-1 (Apaf-1) for inducing apoptotic caspase pathways, and a transduction peptide derived from HIV-1 TAT protein for entering into cells (Rider et al., 2011). Thus, the DRACO approach induces both IFN and apoptosis pathways for controlling virus-infected cells. Studies showed that DRACO is able to induce death of cells infected by either enveloped or non-enveloped viruses with undetectable toxicity to uninfected cells (Rider et al., 2011). DRACO has been reportedly to effectively control infections by various viruses, including orthomyxovirus, flavivirus, picornavirus, arenavirus, bunyavirus, reovirus, and adenovirus (Rider et al., 2011). Animal studies showed that administration of DRACO from 1 day prior to viral infection through 3 days post to viral infection significantly reduced production of viral particles and morbidity of animals intranasally infected with IFV H1N1 A/PR/8/34 (Rider et al., 2011). DRACO appears to be an optimal broad-spectrum antiviral agent for further development.
Autophagy is a basic cellular mechanism for degradation of unnecessary or dysfunctional cellular components through the actions of lysosomes to recycle cellular components (He and Klionsky, 2009). Autophagy is reportedly involved in both pro-survival and pro-death pathways depending on individual cellular events (Nikoletopoulou et al., 2013). Autophagy has been recognized to play roles in controlling intracellular pathogens and fueling innate and adaptive immune responses to viral infections (Liang et al., 1998; Nakashima et al., 2006; He and Klionsky, 2009). Viruses, such as HSV-1 ICP34.5, can block the formation of autolysosomes by targeting the autophagic protein Beclin-1 to inhibit autophagy and allow viral replication (Orvedahl et al., 2007). The non-structural protein 2C of foot-and-mouth disease virus (FMDV) can interact with Beclin-1 for facilitating viral replication (Gladue et al., 2012). Viruses, such as flaviviruses, orthomyxoviruses, and coronaviruses, may also utilize autophagic machinery to facilitate viral replication (Prentice et al., 2004; Jackson et al., 2005; Zhou et al., 2009; Heaton and Randall, 2011; Dumit and Dengjel, 2012). Thus, whether autophagy is an optimal machinery targeted for developing broad-spectrum antiviral agents remains to be addressed. Treatment with the autophagy inhibitor Baicalin, a natural product isolated from Scutellariaradix, resulted in suppressing expression of autophagic Atg5–Atg12 complex and autophagosomal marker LC3-II in cells (Zhu et al., 2015). Baicalin treatment has been shown to suppress viral replication of the flavivirus DENV in vitro (Moghaddam et al., 2014). Baicalin treatment also attenuated IFV-A-induced autophagy in cells and effectively increased survival rates of mice infected with IFV H3N2 or H1N1 (Nayak et al., 2014; Zhu et al., 2015).
Mammalian target of rapamycin (mTOR) has been shown to play roles in regulation of cell growth and autophagy in response to changes of nutritional supplies, growth factors, and stresses (Jung et al., 2010). The mTOR is a serine/threonine kinase and mediates signals to block autophagy via phosphorylation of Atg13 and ULK1/2. Treatment of cells with the mTOR inhibitor Rapamycin leads to dephosphorylation of ULK1/2 and Atg13, resulting in induction of autophagy (Jung et al., 2010). HIV-1 infection may result in induction of mTOR for protein synthesis that downregulates autophagy in lymphocytes and dendritic cells, contributing to its blockage of host’s immune responses (Zhou and Spector, 2008; Gougeon and Piacentini, 2009; van der Vlist et al., 2011; Rehman et al., 2012). Treatment with Rapamycin blocked HIV-1 infection of T cells in vitro (Roy et al., 2002; Oswald-Richter et al., 2004) and inhibited HIV-1 infection of human leucocytes in SCID mice (Nicoletti et al., 2009; Nicoletti et al., 2011). Combination of Rapamycin with the viral entry inhibitor Vicriviroc synergistically enhanced suppression of HIV-1 infection in vitro (Heredia et al., 2008). Rapamycin treatment has also been shown to reduce replication of avian leukemia virus (ALV; Liu et al., 2013). These results suggested that Rapamycin is a potential candidate for anti-HIV drug development. However, how to adequately use agents to suppress or activate autophagy for control of viral infection still needs to be addressed.
Ubiquitin-Proteasome System (UPS)
Ubiquitination and ubiquitination-like modifications (e.g., SUMOylation) are widely involved in stability, localization, and recycling of cellular proteins, as well as interactions of proteins with their substrates (Hershko, 1983; Hochstrasser, 2009). Ubiquitination and ubiquitination-like modifications play roles in regulation of cellular proteins for nuclear transportation, proteolysis, translation, autophagy, and antiviral responses. Ubiquitination is a post-translational modification via attachment of ubiquitin (UB) to a substrate protein. UB is a small regulatory protein (MW 8.5 kDa) and found in almost all eukaryotic cells. Ubiquitination is initially induced by interaction of the UB-activating enzyme E1 to activate UB, followed by interaction with the UB-conjugating enzyme E2 and the UB ligase E3 to form an isopeptide bond between the carboxyl terminus of UB and the 𝜀-amino group of lysine residues on a targeted protein (Pickart, 2001). Subsequently, the resulting UB-tagged protein binds to proteasome for degradation and recycling. Accordingly, assembly of ubiquitination-involved components and proteasomes together constructs the UB-proteasome system (UPS; Coux et al., 1996).
Ubiquitination and ubiquitination-like modifications are known to play important roles in regulation of viral components, such as proteasomal degradation of HBV X protein (Kim et al., 2008), HCV core protein (Shirakura et al., 2007), and HAV 3C proteases (Schlax et al., 2007). It is logical to enhance the host cell defense ability, via modulation of ubiquitin system, to control viral infection. However, it has been reported that viruses are able to utilize UPS to help viral replication (Isaacson and Ploegh, 2009). For example, HIV-1 and IFV-A and B utilize UPS to modify their viral proteins HIV-Gag and IFV-M1, respectively, to facilitate viral replication. Ubiquitination of HIV-Gag, which is a major structural protein, enhances its binding to TSG101 (subunit of ESCRT-I, see Viral Assembly and Budding) for assembly of viral particles and completion of viral budding and release (Pornillos et al., 2002; Klinger and Schubert, 2005). SUMOylation of IFV-A-M1 plays an important role in modulating assembly of viral particles (Pal et al., 2011; Rossman and Lamb, 2011; Wu et al., 2011). Viruses are also able to utilize UPS to subvert or interfere with host innate defense system (such as APOBEC3G and NF-kB). The HIV-1 proteins Vif, Nef, Vpu, and Vpr, are essential for viral replication, and they are important for evading host immune system (Strebel, 2013). For example, the unique zinc-finger motif of Vif recruits E3 ubiquitin ligase to form a complex of CBF-β-CUL5-ELOB-ELOC, which ubiquitinates the human antiviral protein APOBEC3G for degradation (Huthoff and Towers, 2008; Guo et al., 2014). NF-kB is sequestered by IkB in the cytoplasm; viral infection, such as HIV-1 and EBV, may result in the activation of NF-kB through UPS-dependent degradation of IkB, leading to up-regulation of a variety of antiviral genes (Magnani et al., 2000; Mingyan et al., 2009; Sun and Cesarman, 2011). However, IFV-A is able to utilize NF-kB-dependent signaling pathways, via degradation of IkB to activate NF-kB, for viral replication (Ludwig and Planz, 2008). It has been reported that the proteasome inhibitors MG-132 (Carbobenzoxy-L-leucyl-L-leucyl-L-leucinal, peptide aldehyde), Lactacystin, and Bortezomib are able to block NF-kB activation, via inhibition of IkB degradation (Omura et al., 1991; Palombella et al., 1994; Hideshima et al., 2009). MG132 treatment inhibits IFV-A replication (Widjaja et al., 2010). Treatment of cells with Bortezomib (PS-341), at non-cytotoxic levels, significantly reduces IFV-A and VSV replication (Dudek et al., 2010). The proteasome inhibitor VL-01 is able reduce viral replication of three IFV strains, including A/Puerto Rico/8/34 (H1N1), A/Regensburg/D6/09 (H1N1v) and A/Mallard/Bavaria/1/2006 (H5N1). Treatment of IFV-infected mice with VL-01 resulted in reducing viral replication in the lung and increasing mice survival without any side effects (Haasbach et al., 2011).
Paramyxoviruses (SeV and RSV) may induce ubiquitination of STATs to evade host IFN response [see Interferon (IFNs)]; the SeV C protein and the RSV NS protein are able to induce ubiquitination of STAT1 and STAT2, respectively, for degradation (Garcin et al., 2002; Elliott et al., 2007). Treatment of SeV- or RSV-infected cells with MG132 reduced viral replication in a dose-dependent manner (Watanabe et al., 2005; Lupfer and Pastey, 2010). More than ten families of viruses utilize UPS to complete their replication for a successful infection (Calistri et al., 2014). Treatment with MG-132 or Lactacystin effectively blocks maturation, budding, and infectivity of HIV-1 particles (Schubert et al., 2000; Klinger and Schubert, 2005; Votteler and Schubert, 2008). Proteasome inhibitors have also been shown to interfere with replication of herpesviruses (Delboy et al., 2008), coronaviruses (Yu and Lai, 2005), and rotaviruses (López et al., 2011). Thus, it is promising to develop optimal broad-spectrum non-cytotoxic antiviral agents by inhibiting UPS.
Native Barrier
Mucus plays an important role in protection of epithelial cells from invading pathogens in the respiratory, gastrointestinal, urogenital, visual, and auditory systems of animals (Thornton and Sheehan, 2004; Martinez et al., 2015). Mucins, the major components of mucus, are a family of high molecular weight glycoproteins (up to hundreds of kDa) produced by epithelial tissues in both vertebrates and invertebrates. Mucins are able to form a gel-like barrier to protect cells from viral attachment (Lieleg et al., 2012). Porcine mucins, isolated from gastric mucous epithelium, has been shown to form a shield to trap viral particles, such as HPV and IFV IFV-A with sizes of ∼50 and∼100 nm, respectively (Buck et al., 2005), as well as HIV (∼120 nm) and HSV (∼180 nm; Lai et al., 2010; Lieleg et al., 2012). Porcine gastric mucins may be used as antiviral agents for future biomedical applications.
Summary for the Second Section
More than 200 cellular functional proteins are directly and indirectly involved in replication of HIV-1 (Brass et al., 2008; Dziuba et al., 2012). Understanding the roles of cellular proteins, and associated pathways, in viral replication has helped design new strategies for developing broad-spectrum antiviral agents, such as the PI3K inhibitor Akt-IV and BEZ-235, the mTOR inhibitor Rapamycin, and the proteasome inhibitors MG132 and VL-01. Eventually, each agent has its own specificity to intervene in replication of targeted viruses. However, challenges of drug toxicity to hosts and generation of resistant viral progenies remain to be addressed.
Conclusion and Perspectives
Current development of broad-spectrum antiviral agents targeting viral infectivity and modulating host defense system has substantially advanced the fields of virology and pharmaceutics and significantly contributed to the health care of human and animals. However, there are concerns of viral resistance associated with agents targeting viral components and non-specific side effects associated with agents targeting cellular machineries. Accordingly, how to reduce viral resistance and increase drug specificity are current challenges to be addressed. Whether combined uses of agents to target both viral components and cellular machineries may improve antiviral efficacy, reduce viral resistance, and minimize toxicity in the control of viral infection and epidemic viral diseases needs to be clarified.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Beijing Natural Science Foundation of China, Grant No. 6142013, and the program of Education Ministry of China for New Century Excellent Talents in University (NCET-11-0486/2015QC085), as well as the University of Tennessee, Center of Excellence in Livestock Diseases and Human Health (H-CW).
Abbreviations
AIV, avian influenza virus; ASLV, avian sarcoma leucosis virus; DENV, dengue virus; EBOV, Ebola virus; EBV, Epstein-Barr virus; HAdV-2, human adenovirus type 2; HBV, hepatitis B virus; HCMV, human cytomegalo virus; HCV, hepatitis C virus; HeV, Hendra virus; HIV, human immunodeficiency virus; HPV, human papillomavirus; HSV-1, herpes simplex virus 1; IBV, infectious bronchitis virus; IFV-A, influenza virus Type A; IFV-B, influenza virus Type B; JEV, Japanese encephalitis virus; JUNV, Junin arenavirus; LCMV, lymphocytic choriomeningitis virus; LFV, Lassa fever virus; MCMV, murine cytomegalo virus; NDV, Newcastle disease virus; NiV, Nipah virus; PIV, parainfluenza virus; RSV, respiratory syncytial virus; RVFV, Rift Valley fever virus; SARS-CoV, severe acute respiratory syndrome coronavirus; SeV, sendai virus; VACV, vaccinia virus; VSV, vesicular stomatitis virus; WEEV, equine encephalitis virus; YFV, yellow fever virus.
References
Albiol Matanic, V., and Castilla, V. (2004). Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int. J. Antimicrob. Agents 23, 382–389. doi: 10.1016/j.ijantimicag.2003.07.022
Ashiru, O., Howe, J. D., and Butters, T. D. (2014). Nitazoxanide, an antiviral thiazolide, depletes ATP-sensitive intracellular Ca(2+) stores. Virology 462–463, 135–148. doi: 10.1016/j.virol.2014.05.015
Askjaer, P., Jensen, T. H., Nilsson, J., Englmeier, L., and Kjems, J. (1998). The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273, 33414–33422. doi: 10.1074/jbc.273.50.33414
Barrientos, L. G., Lasala, F., Otero, J. R., Sanchez, A., and Delgado, R. (2004). In vitro evaluation of cyanovirin-N antiviral activity, by use of lentiviral vectors pseudotyped with filovirus envelope glycoproteins. J. Infect. Dis. 189, 1440–1443. doi: 10.1086/382658
Barton, C., Kouokam, J. C., Lasnik, A. B., Foreman, O., Cambon, A., Brock, G., et al. (2014). Activity of and effect of subcutaneous treatment with the broad-spectrum antiviral lectin griffithsin in two laboratory rodent models. Antimicrob. Agents Chemother. 58, 120–127. doi: 10.1128/aac.01407-13
Bisbal, C., Salehzada, T., Silhol, M., Martinand, C., Le Roy, F., and Lebleu, B. (2001). The 2-5A/RNase L pathway and inhibition by RNase L inhibitor (RLI). Methods Mol. Biol. 160, 183–198. doi: 10.1385/1-59259-233-3:183
Blaising, J., Polyak, S. J., and Pecheur, E. I. (2014). Arbidol as a broad-spectrum antiviral: an update. Antiviral. Res. 107, 84–94. doi: 10.1016/j.antiviral.2014.04.006
Boriskin, Y. S., Leneva, I. A., Pecheur, E. I., and Polyak, S. J. (2008). Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 15, 997–1005. doi: 10.2174/092986708784049658
Botos, I., and Wlodawer, A. (2003). Cyanovirin-N: a sugar-binding antiviral protein with a new twist. Cell. Mol. Life Sci. 60, 277–287. doi: 10.1007/s000180300023
Brass, A. L., Dykxhoorn, D. M., Benita, Y., Yan, N., Engelman, A., Xavier, R. J., et al. (2008). Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926. doi: 10.1126/science.1152725
Brass, A. L., Huang, I. C., Benita, Y., John, S. P., Krishnan, M. N., Feeley, E. M., et al. (2009). The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254. doi: 10.1016/j.cell.2009.12.017
Bucci, M. (2013). Cell biology: viruses get an ESCRT. Nat. Chem. Biol. 9, 408–408. doi: 10.1038/nchembio.1280
Buck, C. B., Thompson, C. D., Pang, Y. Y., Lowy, D. R., and Schiller, J. T. (2005). Maturation of papillomavirus capsids. J. Virol. 79, 2839–2846. doi: 10.1128/jvi.79.5.2839-2846.2005
Burke, J. D., Platanias, L. C., and Fish, E. N. (2014). Beta interferon regulation of glucose metabolism is PI3K/Akt dependent and important for antiviral activity against coxsackievirus B3. J. Virol. 88, 3485–3495. doi: 10.1128/jvi.02649-13
Calistri, A., Munegato, D., Carli, I., Parolin, C., and Palu, G. (2014). The ubiquitin-conjugating system: multiple roles in viral replication and infection. Cells 3, 386–417. doi: 10.3390/cells3020386
Cannon, N. A., Donlin, M. J., Mayes, L. M., Lyra, A. C., Di Bisceglie, A. M., and Tavis, J. E. (2009). Evidence for action of ribavirin through the hepatitis C virus RNA polymerase. J. Viral. Hepat. 16, 595–604. doi: 10.1111/j.1365-2893.2009.01109.x
Carossino, M., Thiry, E., De La Grandiere, A., and Barrandeguy, M. E. (2014). Novel vaccination approaches against equine alphavirus encephalitides. Vaccine 32, 311–319. doi: 10.1016/j.vaccine.2013.11.071
Chakrabarti, A., Jha, B. K., and Silverman, R. H. (2011). New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 31, 49–57. doi: 10.1089/jir.2010.0120
Chan, J. F. W., Chan, K.-H., Kao, R. Y. T., To, K. K. W., Zheng, B.-J., Li, C. P. Y., et al. (2013). Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 67, 606–616. doi: 10.1016/j.jinf.2013.09.029
Chen, B. J., and Lamb, R. A. (2008). Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372, 221–232. doi: 10.1016/j.virol.2007.11.008
Colman, P. M., and Lawrence, M. C. (2003). The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell. Biol. 4, 309–319. doi: 10.1038/nrm1076
Coux, O., Tanaka, K., and Goldberg, A. L. (1996). Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847. doi: 10.1146/annurev.bi.65.070196.004101
Dauber, B., Martinez-Sobrido, L., Schneider, J., Hai, R., Waibler, Z., Kalinke, U., et al. (2009). Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 5:e1000473. doi: 10.1371/journal.ppat.1000473
De Clercq, E. (2015). Ebola virus (EBOV) infection: therapeutic strategies. Biochem. Pharmacol. 93, 1–10. doi: 10.1016/j.bcp.2014.11.008
Delboy, M. G., Roller, D. G., and Nicola, A. V. (2008). Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J. Virol. 82, 3381–3390. doi: 10.1128/jvi.02296-07
Diamond, M. S., and Farzan, M. (2013). The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57. doi: 10.1038/nri3344
Diehl, N., and Schaal, H. (2013). Make yourself at home: viral hijacking of the PI3K/Akt signaling pathway. Viruses 5, 3192–3212. doi: 10.3390/v5123192
Dixit, N. M., and Perelson, A. S. (2006). The metabolism, pharmacokinetics and mechanisms of antiviral activity of ribavirin against hepatitis C virus. Cell. Mol. Life Sci. 63, 832–842. doi: 10.1007/s00018-005-5455-y
Dolnik, O., Kolesnikova, L., Welsch, S., Strecker, T., Schudt, G., and Becker, S. (2014). Interaction with Tsg101 is necessary for the efficient transport and release of nucleocapsids in marburg virus-infected cells. PLoS Pathog. 10:e1004463. doi: 10.1371/journal.ppat.1004463
Doms, R. W., Ruusala, A., Machamer, C., Helenius, J., Helenius, A., and Rose, J. K. (1988). Differential effects of mutations in three domains on folding, quaternary structure, and intracellular transport of vesicular stomatitis virus G protein. J. Cell. Biol. 107, 89–99. doi: 10.1083/jcb.107.1.89
Dudek, S. E., Luig, C., Pauli, E. K., Schubert, U., and Ludwig, S. (2010). The clinically approved proteasome inhibitor PS-341 efficiently blocks influenza A virus and vesicular stomatitis virus propagation by establishing an antiviral state. J. Virol. 84, 9439–9451. doi: 10.1128/jvi.00533-10
Dumit, V. I., and Dengjel, J. (2012). Autophagosomal protein dynamics and influenza virus infection. Front. Immunol. 3:43. doi: 10.3389/fimmu.2012.00043
Dunn, E. F., Fearns, R., and Connor, J. H. (2009). Akt inhibitor Akt-IV blocks virus replication through an Akt-independent mechanism. J. Virol. 83, 11665–11672. doi: 10.1128/jvi.01092-09
Dziuba, N., Ferguson, M. R., O’Brien, W. A., Sanchez, A., Prussia, A. J., Mcdonald, N. J., et al. (2012). Identification of cellular proteins required for replication of human immunodeficiency virus type 1. AIDS. Res. Hum. Retroviruses 28, 1329–1339. doi: 10.1089/aid.2011.0358
Eckert, D. M., and Kim, P. S. (2001). Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810. doi: 10.1146/annurev.biochem.70.1.777
Elliott, J., Lynch, O. T., Suessmuth, Y., Qian, P., Boyd, C. R., Burrows, J. F., et al. (2007). Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J. Virol. 81, 3428–3436. doi: 10.1128/jvi.02303-06
Elshabrawy, H. A., Fan, J., Haddad, C. S., Ratia, K., Broder, C. C., Caffrey, M., et al. (2014). Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay. J. Virol. 88, 4353–4365. doi: 10.1128/jvi.03050-13
Evans, D. R., and Guy, H. I. (2004). Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J. Biol. Chem. 279, 33035–33038. doi: 10.1074/jbc.R400007200
Feeley, E. M., Sims, J. S., John, S. P., Chin, C. R., Pertel, T., Chen, L. M., et al. (2011). IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7:e1002337. doi: 10.1371/journal.ppat.1002337
Friesen, R. H., Koudstaal, W., Koldijk, M. H., Weverling, G. J., Brakenhoff, J. P., Lenting, P. J., et al. (2010). New class of monoclonal antibodies against severe influenza: prophylactic and therapeutic efficacy in ferrets. PLoS ONE 5:e9106. doi: 10.1371/journal.pone.0009106
Fuentes-Gonzalez, A. M., Contreras-Paredes, A., Manzo-Merino, J., and Lizano, M. (2013). The modulation of apoptosis by oncogenic viruses. Virol. J. 10, 182. doi: 10.1186/1743-422x-10-182
Galdiero, S., Falanga, A., Vitiello, M., Cantisani, M., Marra, V., and Galdiero, M. (2011). Silver nanoparticles as potential antiviral agents. Molecules 16, 8894–8918. doi: 10.3390/molecules16108894
Gale, M. J., and Sen, G. C. (2009). Viral evasion of the interferon system. J. Interferon Cytokine Res. 29, 475–476. doi: 10.1089/jir.2009.0078
Garcin, D., Marq, J. B., Strahle, L., Le Mercier, P., and Kolakofsky, D. (2002). All four Sendai Virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295, 256–265. doi: 10.1006/viro.2001.1342
Gardner, M. R., Kattenhorn, L. M., Kondur, H. R., von Schaewen, M., Dorfman, T., Chiang, J. J., et al. (2015). AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519, 87–91. doi: 10.1038/nature14264
Gladue, D. P., O’donnell, V., and Baker-Branstetter, R. (2012). Foot-and-mouth disease virus nonstructural protein 2C interacts with Beclin1, modulating virus replication. J. Virol. 86, 12080–12090. doi: 10.1128/JVI.01610-12
Gougeon, M. L., and Piacentini, M. (2009). New insights on the role of apoptosis and autophagy in HIV pathogenesis. Apoptosis 14, 501–508. doi: 10.1007/s10495-009-0314-1
Goulet, M. L., Olagnier, D., Xu, Z., Paz, S., Belgnaoui, S. M., Lafferty, E. I., et al. (2013). Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 9:e1003298. doi: 10.1371/journal.ppat.1003298
Guo, Y., Dong, L., Qiu, X., Wang, Y., Zhang, B., Liu, H., et al. (2014). Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233. doi: 10.1038/nature12884
Haasbach, E., Pauli, E. K., Spranger, R., Mitzner, D., Schubert, U., Kircheis, R., et al. (2011). Antiviral activity of the proteasome inhibitor VL-01 against influenza A viruses. Antiviral. Res. 91, 304–313. doi: 10.1016/j.antiviral.2011.07.006
Hacker, G. (2000). The morphology of apoptosis. Cell. Tissue. Res. 301, 5–17. doi: 10.1007/s004410000193
Harris, A., Cardone, G., Winkler, D. C., Heymann, J. B., Brecher, M., White, J. M., et al. (2006). Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 103, 19123–19127. doi: 10.1073/pnas.0607614103
Harvey, R., Brown, K., Zhang, Q., Gartland, M., Walton, L., Talarico, C., et al. (2009). GSK983: a novel compound with broad-spectrum antiviral activity. Antiviral. Res. 82, 1–11. doi: 10.1016/j.antiviral.2008.12.015
He, C., and Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. doi: 10.1146/annurev-genet-102808-114910
Heaton, N. S., and Randall, G. (2011). Dengue virus and autophagy. Viruses 3, 1332–1341. doi: 10.3390/v3081332
Hedlund, M., Aschenbrenner, L. M., Jensen, K., Larson, J. L., and Fang, F. (2010). Sialidase-based anti-influenza virus therapy protects against secondary pneumococcal infection. J. Infect. Dis. 201, 1007–1015. doi: 10.1086/651170
Helle, F., Wychowski, C., Vu-Dac, N., Gustafson, K. R., Voisset, C., and Dubuisson, J. (2006). Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J. Biol. Chem. 281, 25177–251783. doi: 10.1074/jbc.M602431200
Heredia, A., Latinovic, O., Gallo, R. C., Melikyan, G., Reitz, M., Le, N., et al. (2008). Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc. Natl. Acad. Sci. U.S.A. 105, 20476–20481. doi: 10.1073/pnas.0810843106
Hershko, A. (1983). Ubiquitin: roles in protein modification and breakdown. Cell 34, 11–12. doi: 10.1016/0092-8674(83)90131-9
Hideshima, T., Ikeda, H., Chauhan, D., Okawa, Y., Raje, N., Podar, K., et al. (2009). Bortezomib induces canonical nuclear factor-kappaB activation in multiple myeloma cells. Blood 114, 1046–1052. doi: 10.1182/blood-2009-01-199604
Hochstrasser, M. (2009). Origin and function of ubiquitin-like proteins. Nature 458, 422–429. doi: 10.1038/nature07958
Hoffmann, H. H., Kunz, A., Simon, V. A., Palese, P., and Shaw, M. L. (2011). Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 5777–5782. doi: 10.1073/pnas.1101143108
Huang, I. C., Bailey, C. C., Weyer, J. L., Radoshitzky, S. R., Becker, M. M., Chiang, J. J., et al. (2011). Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7:e1001258. doi: 10.1371/journal.ppat.1001258
Hui, D. J., Bhasker, C. R., Merrick, W. C., and Sen, G. C. (2003). Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2. GTP. Met-tRNAi. J. Biol. Chem. 278, 39477–39482. doi: 10.1074/jbc.M305038200
Huthoff, H., and Towers, G. J. (2008). Restriction of retroviral replication by APOBEC3G/F and TRIM5alpha. Trends Microbiol. 16, 612–619. doi: 10.1016/j.tim.2008.08.013
Isaacson, M. K., and Ploegh, H. L. (2009). Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell. Host. Microbe. 5, 559–570. doi: 10.1016/j.chom.2009.05.012
Jackson, W. T., Giddings, T. J., Taylor, M. P., Mulinyawe, S., Rabinovitch, M., Kopito, R. R., et al. (2005). Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. doi: 10.1371/journal.pbio.0030156
Julander, J. G., Bantia, S., Taubenheim, B. R., Minning, D. M., Kotian, P., Morrey, J. D., et al. (2014). BCX4430, a novel nucleoside analog, effectively treats yellow fever in a hamster model. Antimicrob. Agents Chemother. 58, 6607–6614. doi: 10.1128/AAC.03368-14
Jung, C. H., Ro, S. H., Cao, J., Otto, N. M., and Kim, D. H. (2010). mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295. doi: 10.1016/j.febslet.2010.01.017
Kaletsky, R. L., Simmons, G., and Bates, P. (2007). Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81, 13378–13384. doi: 10.1128/jvi.01170-07
Kim, J. H., Sohn, S. Y., Benedict, Y. T., and Ahn, B. Y. (2008). Ubiquitin-dependent and -independent proteasomal degradation of hepatitis B virus X protein. Biochem. Biophys. Res. Commun. 366, 1036–1042. doi: 10.1016/j.bbrc.2007.12.070
Kim, Y., Lovell, S., Tiew, K. C., Mandadapu, S. R., Alliston, K. R., Battaile, K. P., et al. (2012). Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 86, 11754–11762. doi: 10.1128/JVI.01348-12
Kim, Y., Mandadapu, S. R., Groutas, W. C., and Chang, K. O. (2013). Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease. Antiviral. Res. 97, 161–168. doi: 10.1016/j.antiviral.2012.11.005
Kinch, M. S., Yunus, A. S., Lear, C., Mao, H., Chen, H., Fesseha, Z., et al. (2009). FGI-104: a broad-spectrum small molecule inhibitor of viral infection. Am. J. Transl. Res. 1, 87–98. doi: 10.1016/j.antiviral.2009.02.082
Klinger, P. P., and Schubert, U. (2005). The ubiquitin-proteasome system in HIV replication: potential targets for antiretroviral therapy. Expert Rev. Anti. Infect. Ther. 3, 61–79. doi: 10.1586/14787210.3.1.61
Konteatis, Z. D. (2010). In silico fragment-based drug design. Expert Opin. Drug Discov. 5, 1047–1065. doi: 10.1517/17460441.2010.523697
Krumm, S. A., Ndungu, J. M., Yoon, J. J., Dochow, M., Sun, A., Natchus, M., et al. (2011). Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS ONE 6:e20069. doi: 10.1371/journal.pone.0020069
Lai, S. K., Wang, Y. Y., Hida, K., Cone, R., and Hanes, J. (2010). Nanoparticles reveal that human cervicovaginal mucus is riddled with pores larger than viruses. Proc. Natl. Acad. Sci. U.S.A. 107, 598–603. doi: 10.1073/pnas.0911748107
Lara, H. H., Ixtepan-Turrent, L., Garza-Trevino, E. N., and Rodriguez-Padilla, C. (2010). PVP-coated silver nanoparticles block the transmission of cell-free and cell-associated HIV-1 in human cervical culture. J. Nanobiotechnol. 8, 15. doi: 10.1186/1477-3155-8-15
Lei, C. Q., Zhang, Y., Xia, T., Jiang, L. Q., Zhong, B., and Shu, H. B. (2013). FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J. Biol. Chem. 288, 12596–12604. doi: 10.1074/jbc.M112.444794
Li, C. G., Tang, W., Chi, X. J., Dong, Z. M., Wang, X. X., and Wang, X. J. (2013). A cholesterol tag at the N terminus of the relatively broad-spectrum fusion inhibitory peptide targets an earlier stage of fusion glycoprotein activation and increases the peptide’s antiviral potency in vivo. J. Virol. 87, 9223–9232. doi: 10.1128/jvi.01153-13
Li, L., He, L., Tan, S., Guo, X., Lu, H., Qi, Z., et al. (2010). 3-hydroxyphthalic anhydride-modified chicken ovalbumin exhibits potent and broad anti-HIV-1 activity: a potential microbicide for preventing sexual transmission of HIV-1. Antimicrob. Agents Chemother. 54, 1700–1711. doi: 10.1128/aac.01046-09
Liang, X. H., Kleeman, L. K., Jiang, H. H., Gordon, G., Goldman, J. E., Berry, G., et al. (1998). Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72, 8586–8596.
Lieleg, O., Lieleg, C., Bloom, J., Buck, C. B., and Ribbeck, K. (2012). Mucin biopolymers as broad-spectrum antiviral agents. Biomacromolecules 13, 1724–1732. doi: 10.1021/bm3001292
Liu, H., Cao, W., Li, Y., Feng, M., Wu, X., Yu, K., et al. (2013). Subgroup J avian leukosis virus infection inhibits autophagy in DF-1 cells. Virol. J. 10, 196. doi: 10.1186/1743-422x-10-196
Liu, H., Golebiewski, L., Dow, E. C., Krug, R. M., Javier, R. T., and Rice, A. P. (2010). The ESEV PDZ-binding motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis by directly targeting Scribble. J. Virol. 84, 11164–11174. doi: 10.1128/jvi.01278-10
Liu, S., Xiao, G., Chen, Y., He, Y., Niu, J., Escalante, C. R., et al. (2004). Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363, 938–947. doi: 10.1016/s0140-6736(04)15788-7
Lockshin, R. A., and Zakeri, Z. (2007). Cell death in health and disease. J. Cell. Mol. Med. 11, 1214–1224. doi: 10.1111/j.1582-4934.2007.00150.x
López, T., Silva-Ayala, D., Lopez, S., and Arias, C. F. (2011). Replication of the rotavirus genome requires an active ubiquitin-proteasome system. J. Virol. 85, 11964–11971. doi: 10.1128/jvi.05286-11
Lu, G., Reinert, J. T., Pitha-Rowe, I., Okumura, A., Kellum, M., Knobeloch, K. P., et al. (2006). ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell. Mol. Biol. (Noisy-le-grand) 52, 29–41.
Lu, J., Han, Z., Liu, Y., Liu, W., Lee, M. S., Olson, M. A., et al. (2014). A host-oriented inhibitor of Junin Argentine hemorrhagic fever virus egress. J. Virol. 88, 4736–4743. doi: 10.1128/jvi.03757-13
Ludwig, S., and Planz, O. (2008). Influenza viruses and the NF-kappaB signaling pathway - towards a novel concept of antiviral therapy. Biol. Chem. 389, 1307–1312. doi: 10.1515/bc.2008.148
Lupfer, C., and Pastey, M. K. (2010). Decreased replication of human respiratory syncytial virus treated with the proteasome inhibitor MG-132. Virus Res. 149, 36–41. doi: 10.1016/j.virusres.2009.12.010
Luyet, P. P., Falguieres, T., Pons, V., Pattnaik, A. K., and Gruenberg, J. (2008). The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA. Traffic 9, 2279–2290. doi: 10.1111/j.1600-0854.2008.00820.x
Magnani, M., Crinelli, R., Bianchi, M., and Antonelli, A. (2000). The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-kB (NF-kB). Curr. Drug Targets 1, 387–399. doi: 10.2174/1389450003349056
Mao, L., and Yang, Y. (2013). Targeting the nuclear transport machinery by rational drug design. Curr. Pharm. Des. 19, 2318–2325. doi: 10.2174/1381612811319120018
Marschall, M., Niemann, I., Kosulin, K., Bootz, A., Wagner, S., Dobner, T., et al. (2013). Assessment of drug candidates for broad-spectrum antiviral therapy targeting cellular pyrimidine biosynthesis. Antiviral. Res. 100, 640–648. doi: 10.1016/j.antiviral.2013.10.003
Martinez, J. P., Sasse, F., Bronstrup, M., Diez, J., and Meyerhans, A. (2015). Antiviral drug discovery: broad-spectrum drugs from nature. Nat. Prod. Rep. 32, 29–48. doi: 10.1039/c4np00085d
Martins, S. T., Silveira, G. F., Alves, L. R., Duarte, D. S. C., and Bordignon, J. (2012). Dendritic cell apoptosis and the pathogenesis of dengue. Viruses 4, 2736–2753. doi: 10.3390/v4112736
McMahon, H. T., and Gallop, J. L. (2005). Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596. doi: 10.1038/nature04396
Mendenhall, M., Russell, A., Smee, D. F., Hall, J. O., Skirpstunas, R., Furuta, Y., et al. (2011). Effective oral favipiravir (T-705) therapy initiated after the onset of clinical disease in a model of arenavirus hemorrhagic Fever. PLoS Negl. Trop. Dis. 5:e1342. doi: 10.1371/journal.pntd.0001342
Mingyan, Y., Xinyong, L., and De Clercq, E. (2009). NF-kappaB: the inducible factors of HIV-1 transcription and their inhibitors. Mini. Rev. Med. Chem. 9, 60–69. doi: 10.2174/138955709787001677
Moghaddam, E., Teoh, B. T., Sam, S. S., Lani, R., Hassandarvish, P., Chik, Z., et al. (2014). Baicalin, a metabolite of baicalein with antiviral activity against dengue virus. Sci. Rep. 4, 5452. doi: 10.1038/srep05452
Morales, D. J., and Lenschow, D. J. (2013). The antiviral activities of ISG15. J. Mol. Biol. 425, 4995–5008. doi: 10.1016/j.jmb.2013.09.041
Mori, T., O’keefe, B. R., Sowder, R. N., Bringans, S., Gardella, R., Berg, S., et al. (2005). Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 280, 9345–9353. doi: 10.1074/jbc.M411122200
Mukhopadhyay, S., Kuhn, R. J., and Rossmann, M. G. (2005). A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3, 13–22. doi: 10.1038/nrmicro1067
Nakashima, A., Tanaka, N., Tamai, K., Kyuuma, M., Ishikawa, Y., Sato, H., et al. (2006). Survival of parvovirus B19-infected cells by cellular autophagy. Virology 349, 254–263. doi: 10.1016/j.virol.2006.03.029
Nayak, M. K., Agrawal, A. S., Bose, S., Naskar, S., Bhowmick, R., Chakrabarti, S., et al. (2014). Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 69, 1298–1310. doi: 10.1093/jac/dkt534
Neumann, G., Hughes, M. T., and Kawaoka, Y. (2000). Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19, 6751–6758. doi: 10.1093/emboj/19.24.6751
Ng, D., and Gommerman, J. L. (2013). The regulation of immune responses by DC derived type I IFN. Front. Immunol. 4:94. doi: 10.3389/fimmu.2013.00094
Nicholls, J. M., Chan, R. W., Russell, R. J., Air, G. M., and Peiris, J. S. (2008). Evolving complexities of influenza virus and its receptors. Trends Microbiol. 16, 149–157. doi: 10.1016/j.tim.2008.01.008
Nicoletti, F., Fagone, P., Meroni, P., Mccubrey, J., and Bendtzen, K. (2011). mTOR as a multifunctional therapeutic target in HIV infection. Drug Discov. Today 16, 715–721. doi: 10.1016/j.drudis.2011.05.008
Nicoletti, F., Lapenta, C., Donati, S., Spada, M., Ranazzi, A., Cacopardo, B., et al. (2009). Inhibition of human immunodeficiency virus (HIV-1) infection in human peripheral blood leucocytes-SCID reconstituted mice by rapamycin. Clin. Exp. Immunol. 155, 28–34. doi: 10.1111/j.1365-2249.2008.03780.x
Nikoletopoulou, V., Markaki, M., Palikaras, K., and Tavernarakis, N. (2013). Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta. 1833, 3448–3459. doi: 10.1016/j.bbamcr
Naik, G. S, and Tyagi, M. G. (2012). A pharmacological profile of ribavirin and monitoring of its plasma concentration in chronic hepatitis C infection. J. Clin. Exp. Hepatol. 2, 42–54. doi: 10.1016/S0973-6883(12)60090-5
Oestereich, L., Ludtke, A., Wurr, S., Rieger, T., Munoz-Fontela, C., and Gunther, S. (2014). Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral. Res. 105, 17–21. doi: 10.1016/j.antiviral.2014.02.014
Olagnier, D., Scholte, F. E., Chiang, C., Albulescu, I. C., Nichols, C., He, Z., et al. (2014). Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J. Virol. 88, 4180–4194. doi: 10.1128/jvi.03114-13
Omura, S., Fujimoto, T., Otoguro, K., Matsuzaki, K., Moriguchi, R., Tanaka, H., et al. (1991). Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J. Antibiot. (Tokyo) 44, 113–116. doi: 10.7164/antibiotics.44.113
Ortiz-Riano, E., Ngo, N., Devito, S., Eggink, D., Munger, J., Shaw, M. L., et al. (2014). Inhibition of arenavirus by A3, a pyrimidine biosynthesis inhibitor. J. Virol. 88, 878–889. doi: 10.1128/jvi.02275-13
Orvedahl, A., Alexander, D., Tallóczy, Z. (2007). HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell. Host. Microbe. 1, 23–35. doi: 10.1016/j.chom.2006.12.001
Oswald-Richter, K., Grill, S. M., Leelawong, M., and Unutmaz, D. (2004). HIV infection of primary human T cells is determined by tunable thresholds of T cell activation. Eur. J. Immunol. 34, 1705–1714. doi: 10.1002/eji.200424892
Pager, C. T., Craft, W. J., Patch, J., and Dutch, R. E. (2006). A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346, 251–257. doi: 10.1016/j.virol.2006.01.007
Pager, C. T., and Dutch, R. E. (2005). Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79, 12714–12720. doi: 10.1128/jvi.79.20.12714-12720.2005
Pal, S., Santos, A., Rosas, J. M., Ortiz-Guzman, J., and Rosas-Acosta, G. (2011). Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 158, 12–27. doi: 10.1016/j.virusres.2011.02.017
Palombella, V. J., Rando, O. J., Goldberg, A. L., and Maniatis, T. (1994). The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell 78, 773–785. doi: 10.1016/S0092-8674(94)90482-0
Park, D., Lengyel, J., and Rice, S. A. (2013). Role of immediate early protein ICP27 in the differential sensitivity of herpes simplex viruses 1 and 2 to leptomycin B. J. Virol. 87, 8940–8951. doi: 10.1128/jvi.00633-13
Parker, W. B. (2005). Metabolism and antiviral activity of ribavirin. Virus Res. 107, 165–171. doi: 10.1016/j.virusres.2004.11.006
Perwitasari, O., Johnson, S., Yan, X., Howerth, E., Shacham, S., Landesman, Y., et al. (2014). Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo. J. Virol. 88, 10228–10243. doi: 10.1128/JVI.01774-14
Pichlmair, A., Lassnig, C., Eberle, C. A., Gorna, M. W., Baumann, C. L., Burkard, T. R., et al. (2011). IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat. Immunol. 12, 624–630. doi: 10.1038/ni.2048
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Pincetic, A., and Leis, J. (2009). The Mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009, 6239691–6239699. doi: 10.1155/2009/623969
Pornillos, O., Alam, S. L., Davis, D. R., and Sundquist, W. I. (2002). Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9, 812–817. doi: 10.1038/nsb856
Prentice, E., Jerome, W. G., Yoshimori, T., Mizushima, N., and Denison, M. R. (2004). Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279, 10136–10141. doi: 10.1074/jbc.M306124200
Prinarakis, E., Chantzoura, E., Thanos, D., and Spyrou, G. (2008). S-glutathionylation of IRF3 regulates IRF3-CBP interaction and activation of the IFN beta pathway. EMBO J. 27, 865–875. doi: 10.1038/emboj.2008.28
Ramaswamy, S., Nakamura, N., Vazquez, F., Batt, D. B., Perera, S., Roberts, T. M., et al. (1999). Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. U.S.A. 96, 2110–2115. doi: 10.1073/pnas.96.5.2110
Raveh, A., Delekta, P. C., Dobry, C. J., Peng, W., Schultz, P. J., Blakely, P. K., et al. (2013). Discovery of potent broad spectrum antivirals derived from marine actinobacteria. PLoS ONE 8:e82318. doi: 10.1371/journal.pone.0082318
Rehman, S., Husain, M., Yadav, A., Kasinath, B. S., Malhotra, A., and Singhal, P. C. (2012). HIV-1 promotes renal tubular epithelial cell protein synthesis: role of mTOR pathway. PLoS ONE 7:e30071. doi: 10.1371/journal.pone.0030071
Rider, T. H., Zook, C. E., Boettcher, T. L., Wick, S. T., Pancoast, J. S., and Zusman, B. D. (2011). Broad-spectrum antiviral therapeutics. PLoS ONE 6:e22572. doi: 10.1371/journal.pone.0022572
Rossignol, J. F. (2014). Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral. Res. 110, 94–103. doi: 10.1016/j.antiviral.2014.07.014
Rossman, J. S., and Lamb, R. A. (2011). Influenza virus assembly and budding. Virology 411, 229–236. doi: 10.1016/j.virol.2010.12.003
Roy, J., Paquette, J. S., Fortin, J. F., and Tremblay, M. J. (2002). The immunosuppressant rapamycin represses human immunodeficiency virus type 1 replication. Antimicrob. Agents Chemother. 46, 3447–3455. doi: 10.1128/AAC.46.11.3447-3455.2002
Sample, C. J., Hudak, K. E., Barefoot, B. E., Koci, M. D., Wanyonyi, M. S., Abraham, S., et al. (2013). A mastoparan-derived peptide has broad-spectrum antiviral activity against enveloped viruses. Peptides 48, 96–105. doi: 10.1016/j.peptides.2013.07.014
Schlax, P. E., Zhang, J., Lewis, E., Planchart, A., and Lawson, T. G. (2007). Degradation of the encephalomyocarditis virus and hepatitis A virus 3C proteases by the ubiquitin/26S proteasome system in vivo. Virology 360, 350–363. doi: 10.1016/j.virol.2006.10.043
Schlee, M., Roth, A., Hornung, V., Hagmann, C. A., Wimmenauer, V., Barchet, W., et al. (2009). Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31, 25–34. doi: 10.1016/j.immuni.2009.05.008
Schubert, U., Ott, D. E., Chertova, E. N., Welker, R., Tessmer, U., Princiotta, M. F., et al. (2000). Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. U.S.A. 97, 13057–13062. doi: 10.1073/pnas.97.24.13057
Shah, N. R., Sunderland, A., and Grdzelishvili, V. Z. (2010). Cell type mediated resistance of vesicular stomatitis virus and Sendai virus to ribavirin. PLoS ONE 5:e11265. doi: 10.1371/journal.pone.0011265
Shi, H. X., Yang, K., Liu, X., Liu, X. Y., Wei, B., Shan, Y. F., et al. (2010). Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 30, 2424–2436. doi: 10.1128/mcb.01466-09
Shirakura, M., Murakami, K., Ichimura, T., Suzuki, R., Shimoji, T., Fukuda, K., et al. (2007). E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 81, 1174–1185. doi: 10.1128/JVI.01684-06
Silverman, R. H., and Weiss, S. R. (2014). Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J. Interferon Cytokine Res. 34, 455–463. doi: 10.1089/jir.2014.0007
Simmons, G., Gosalia, D. N., Rennekamp, A. J., Reeves, J. D., Diamond, S. L., and Bates, P. (2005). Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881. doi: 10.1073/pnas.0505577102
Snyder, J. C., Samson, R. Y., Brumfield, S. K., Bell, S. D., and Young, M. J. (2013). Functional interplay between a virus and the ESCRT machinery in archaea. Proc. Natl. Acad. Sci. U.S.A. 110, 10783–10787. doi: 10.1073/pnas.1301605110
Strebel, K. (2013). HIV accessory proteins versus host restriction factors. Curr. Opin. Virol. 3, 692–699. doi: 10.1016/j.coviro.2013.08.004
Su, B., and Moog, C. (2014). Which Antibody Functions are Important for an HIV Vaccine? Front. Immunol. 5:289. doi: 10.3389/fimmu.2014.00289
Sun, M., Fuentes, S. M., Timani, K., Sun, D., Murphy, C., Lin, Y., et al. (2008). Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J. Virol. 82, 105–114. doi: 10.1128/jvi.01520-07
Sun, S. C., and Cesarman, E. (2011). NF-kappaB as a target for oncogenic viruses. Curr. Top. Microbiol. Immunol. 349, 197–244. doi: 10.1007/82_2010_108
Tang, Y., Zhong, G., Zhu, L., Liu, X., Shan, Y., Feng, H., et al. (2010). Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J. Immunol. 184, 5777–5790. doi: 10.4049/jimmunol.0903588
Taylor, R. C., Cullen, S. P., and Martin, S. J. (2008). Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell. Biol. 9, 231–241. doi: 10.1038/nrm2312
Terenzi, F., Saikia, P., and Sen, G. C. (2008). Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 27, 3311–3321. doi: 10.1038/emboj.2008.241
te Velthuis, A. J. (2014). Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 71, 4403–4420. doi: 10.1007/s00018-014-1695-z
Thakur, C. S., Jha, B. K., Dong, B., Das, G. J., Silverman, K. M., Mao, H., et al. (2007). Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc. Natl. Acad. Sci. U.S.A. 104, 9585–9590. doi: 10.1073/pnas.0700590104
Thornton, D. J., and Sheehan, J. K. (2004). From mucins to mucus: toward a more coherent understanding of this essential barrier. Proc. Am. Thorac. Soc. 1, 54–61. doi: 10.1513/pats.2306016
Upton, J. W., and Chan, F. K. (2014). Staying alive: cell death in antiviral immunity. Mol. Cell. 54, 273–280. doi: 10.1016/j.molcel.2014.01.027
Urata, S., Ngo, N., and De La Torre, J. C. (2012). The PI3K/Akt pathway contributes to arenavirus budding. J. Virol. 86, 4578–4585. doi: 10.1128/jvi.06604-11
van der Vlist, M., Van Der Aar, A. M., Gringhuis, S. I., and Geijtenbeek, T. B. (2011). Innate signaling in HIV-1 infection of dendritic cells. Curr. Opin. HIV. AIDS. 6, 348–352. doi: 10.1097/COH.0b013e328349a2d1
Vigant, F., Lee, J., Hollmann, A., Tanner, L. B., Akyol, A. Z., Yun, T., et al. (2013). A mechanistic paradigm for broad-spectrum antivirals that target virus-cell fusion. PLoS Pathog. 9:e1003297. doi: 10.1371/journal.ppat.1003297
Votteler, J., and Schubert, U. (2008). Ubiquitin ligases as therapeutic targets in HIV-1 infection. Expert Opin. Ther. Targets 12, 131–143. doi: 10.1517/14728222.12.2.131
Wang, H. M., and Liang, P. H. (2010). Picornaviral 3C protease inhibitors and the dual 3C protease/ coronaviral 3C-like protease inhibitors. Expert Opin. Ther. Pat. 20, 59–71. doi: 10.1517/13543770903460323
Wang, X. J., Bai, Y. D., Zhang, G. Z., Zhao, J. X., Wang, M., and Gao, G. F. (2005). Structure and function study of paramyxovirus fusion protein heptad repeat peptides. Arch. Biochem. Biophys. 436, 316–322. doi: 10.1016/j.abb.2005.02.004
Wang, X. J., Li, C. G., Chi, X. J., and Wang, M. (2011). Characterisation and evaluation of antiviral recombinant peptides based on the heptad repeat regions of NDV and IBV fusion glycoproteins. Virology 416, 65–74. doi: 10.1016/j.virol.2011.05.001
Warren, T. K., Wells, J., Panchal, R. G., Stuthman, K. S., Garza, N. L., Van Tongeren, S. A., et al. (2014). Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 508, 402–405. doi: 10.1038/nature13027
Watanabe, H., Tanaka, Y., Shimazu, Y., Sugahara, F., Kuwayama, M., Hiramatsu, A., et al. (2005). Cell-specific inhibition of paramyxovirus maturation by proteasome inhibitors. Microbiol. Immunol. 49, 835–844. doi: 10.1111/j.1348-0421.2005.tb03672.x
Widjaja, I., De Vries, E., Tscherne, D. M., Garcia-Sastre, A., Rottier, P. J., and De Haan, C. A. (2010). Inhibition of the ubiquitin-proteasome system affects influenza A virus infection at a postfusion step. J. Virol. 84, 9625–9631. doi: 10.1128/jvi.01048-10
Wilen, C. B., Tilton, J. C., and Doms, R. W. (2012). Molecular mechanisms of HIV entry. Adv. Exp. Med. Biol. 726, 223–242. doi: 10.1007/978-1-4614-0980-9-10
Wilkins, C., and Gale, M. J. (2010). Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22, 41–47. doi: 10.1016/j.coi.2009.12.003
Wolf, M. C., Freiberg, A. N., Zhang, T., Akyol-Ataman, Z., Grock, A., Hong, P. W., et al. (2010). A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. U.S.A. 107, 3157–3162. doi: 10.1073/pnas.0909587107
Wu, C., Chen, W., Chen, J., Han, B., Peng, Z., Ge, F., et al. (2015). Preparation of monoPEGylated Cyanovirin-N’s derivative and its anti-influenza A virus bioactivity in vitro and in vivo. J. Biochem. doi: 10.1093/jb/mvv013 [Epub ahead of print].
Wu, C. Y., Jeng, K. S., and Lai, M. M. (2011). The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J. Virol. 85, 6618–6628. doi: 10.1128/jvi.02401-10
Xia, B., and Kang, X. (2011). Activation and maturation of SARS-CoV main protease. Protein. Cell. 2, 282–290. doi: 10.1007/s13238-011-1034-1
Yu, G. Y., and Lai, M. M. (2005). The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J. Virol. 79, 644–648. doi: 10.1128/jvi.79.1.644-648.2005
Zhao, C., Hsiang, T. Y., Kuo, R. L., and Krug, R. M. (2010). ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 107, 2253–2258. doi: 10.1073/pnas.0909144107
Zhou, D., and Spector, S. A. (2008). Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 22, 695–699. doi: 10.1097/QAD.0b013e3282f4a836
Zhou, Z., Jiang, X., Liu, D., Fan, Z., Hu, X., Yan, J., et al. (2009). Autophagy is involved in influenza A virus replication. Autophagy 5, 321–328. doi: 10.4161/auto.5.3.7406
Zhu, H. Y., Han, L., Shi, X. L., Wang, B. L., Huang, H., Wang, X., et al. (2015). Baicalin inhibits autophagy induced by influenza A virus H3N2. Antiviral. Res. 113, 62–70. doi: 10.1016/j.antiviral.2014.11.003
Keywords: broad-spectrum, antiviral agent, viral entry, replication, cellular defense
Citation: Zhu J-D, Meng W, Wang X-J and Wang H-CR (2015) Broad-spectrum antiviral agents. Front. Microbiol. 6:517. doi: 10.3389/fmicb.2015.00517
Received: 19 March 2015; Accepted: 09 May 2015;
Published online: 22 May 2015
Edited by:
Slobodan Paessler, University of Texas Medical Branch, USAReviewed by:
Chris Sullivan, University of Texas at Austin, USABin Su, INSERM U941, Université Paris Diderot – Paris 7, France
Copyright © 2015 Zhu, Meng, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Jia Wang, Key Laboratory of Zoonosis of Ministry of Agriculture, College of Veterinary Medicine, China Agricultural University, Beijing 100193, China,d2FuZ3hqQGNhdS5lZHUuY24=; Hwa-Chain R. Wang, Department of Biomedical and Diagnostic Sciences, College of Veterinary Medicine, The University of Tennessee, Knoxville, TN 37996, USA,aGNyd2FuZ0B1dGsuZWR1
 Jun-Da Zhu
Jun-Da Zhu Wen Meng1
Wen Meng1 Xiao-Jia Wang
Xiao-Jia Wang Hwa-Chain R. Wang
Hwa-Chain R. Wang