- 1Multidisciplinary Research Unit (MRU), Assam Medical College and Hospital (AMCH), Dibrugarh, Assam, India
- 2Department of Pediatrics, Assam Medical College and Hospital (AMCH), Dibrugarh, Assam, India
- 3Regional Viral Research and Diagnostic Laboratory (Regional VRDL), ICMR-RMRCNE, Dibrugarh, Assam, India
Purpose: The present study explored the molecular characterization of human Adenovirus (HAdV) and its strains among hospitalized SARI cases in the pediatric unit of a tertiary care hospital in North-East India.
Methods: Nasal and throat swabs were collected from 70 patients of Pediatric Unit, of a tertiary hospital in NE India from April 2023-October 2023. The samples were screened for the presence of HAdV using an adenovirus-specific Real-Time PCR Kit. For molecular characterization, Next Generation Sequencing (NGS) was performed by targeting the hexon gene of HAdV followed by post-sequencing analysis.
Results: Overall, 18.57% (13/70) of samples were positive for HAdV. In context of the severity of illness, 3/13 adenovirus-positive patients (23.07%) died after hospitalization, had severe pneumonia among which two were of less than one year of age. Molecular characterization using NGS indicated that 4/13 individuals were infected with HAdV-B type 3 and 5/13 patients were infected with HAdV type 7. Notably, 4/7 cases of severe pneumonia were under five years of age and associated with HAdV type 7 infection. The ratio of non-synonymous to synonymous mutation (dN/dS) was comparatively low in HAdV type 7 positive samples (dN/dS=0.31). No non-synonymous mutation was observed in HAdV-B type 3 positive samples. The higher neutrophil percentage among the death cases suggested an acute immune response.
Conclusion: The study demonstrated HAdV type 7 and HAdV-B type 3 as strains associated with pediatric SARI cases from April 2023-October 2023. Further, HAdV type 7 infection was primarily linked with lower respiratory tract infections mainly severe pneumonia.
1 Introduction
Severe acute respiratory infections (SARI) are a leading cause of increased mortality rates among children worldwide, particularly in low-resource setting (1). Approximately, 33% of deaths in pediatric cases of less than five years of age in developing countries are SARI-associated (2, 3). Viruses are the major pathogens attributed to Acute Respiratory Infection (ARI) including influenza viruses A and B, respiratory syncytial viruses, parainfluenza viruses, human adenovirus (HAdV), human rhinovirus, human metapneumovirus, and human coronaviruses, etc. (4). HAdV associated ARI is highly contagious and mainly characterized by rapid transmission through respiratory secretions and direct contact. HAdV is a ubiquitous, non-enveloped, double-stranded DNA virus of ~ 34 to 36 kb that belongs to the Adenoviridae family genus Mastadenovirus (5, 6). Entry of HAdV into the human body is mediated via receptor-mediated endocytosis using the coxsackievirus-adenovirus receptor (CAR) followed by the synthesis of viral proteins and viral replication (7). In addition, receptors including CD80/CD86, CD46, desmoglein 2 (DSG2) and sialic acid are also associated with the infection caused by HAdV B or HAdV D (8).
Overall, the prevalence rate of respiratory HAdV has been reported to vary between ∼3 and 24% (9–11). HAdV have been classified into seven species (A-G) consisting of approximately 103 HAdV types of which HAdV-1 to HAdV-51 were serotypes and HAdV-52 to HAdV-103 were genotypes (12–14). Among the HAdV types, HAdV species-B types 3, 4, 7, 14, 55, 21, and 11 were reported to be commonly associated with severe ARI outbreaks (15, 16). The severity of adenoviral infection mainly depends upon the patient’s age, surrounding environment, and immune tolerance of the patients. The unavailability of specific antiviral drugs for adenovirus till date further enhanced the risk of the severity and transmissibility of the disease.
India has witnessed several HAdV-mediated outbreaks in the past years including a recent outbreak in West Bengal India (17–20). In addition, a multicentric surveillance study from India documented 3.1% and 2.9% prevalence rates for HAdV in ARI and SARI cases respectively (21).
Considering that adenovirus is one of the important respiratory pathogens, routine systematic surveillance is essentially required for early diagnosis to avoid major complications. However, till date, no molecular epidemiological studies related to HAdV species types associated with severe acute respiratory infections (SARI) have been reported from North East India. The present study explored the molecular characterization of HAdV and its strains, among hospitalized SARI cases in the pediatric unit of a tertiary care hospital, in North-East India from April 2023- October 2023.
2 Materials and methods
2.1 Study site and design
The investigation was carried out in the Multidisciplinary Research Unit (MRU) of a tertiary care hospital (Assam Medical College & Hospital) and Indian Council of Medical Research-Regional Medical Research Centre (ICMR-RMRC) Northeast Region located in Dibrugarh, Assam. This study was performed from April 2023 to October 2023. The cases were enrolled in the study after obtaining written and informed consent. A predesigned proforma was used for the collection of relevant demographic and clinical information from each of the participants. The study was approved by the Institutional Ethics Committee (IEC) and conducted in accordance with the ethical guidelines of both the participating sites.
2.2 Sample collection
Nasal and throat swabs were collected from seventy clinically suspected cases from the Department of Pediatrics of the participating hospital from April 2023 to October 2023. The samples were collected in a sterile corning tube containing 2ml of phosphate buffer saline (PBS, pH 7.2). PBS is generally considered to be a suitable collection medium for real time PCR based amplification. Viral Transport Medium (VTM) was not used for collection of samples as the samples were not supposed to be processed for culture. Further, the collected samples were transported to the designated laboratory for molecular screening and characterization of the causative organism. As per the clinical case definition in the study, Acute Respiratory Illness (ARI) includes both upper and lower respiratory tract infections with the symptoms like short and rapid breathing, fever, cough, runny nose or nasal congestion, sore throat etc. (22, 23). ARI is a broad term used to describe infections affecting the respiratory system, including the upper (e.g., common cold) and lower (e.g., bronchitis, pneumonia) airways. It can range from mild to severe and is often caused by viruses or bacteria. Pneumonia is a type of lower respiratory infection characterized by inflammation of the air sacs (alveoli) in one or both lungs, which may fill with fluid or pus, leading to symptoms like cough, fever, and difficulty breathing. Pneumonia symptoms include -history of cough or breathlessness, inability to feed, raised respiratory rate, lower chest wall indrawing, fever, and tachycardia (24). Severe pneumonia shows the symptoms like central cyanosis, severe difficulty breathing, difficulty breastfeeding/drinking, vomiting, lethargy, unconsciousness, and convulsions (22, 24).
2.3 Laboratory investigation
2.3.1 Nucleic acid extraction and real-time PCR
The genomic DNA was extracted from nasopharyngeal & throat swab samples in PBS within 24 hours using a QIAamp DNA Mini extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The extracted DNA was then processed for screening of the samples for adenovirus followed by viral load estimation using the Helini Adenovirus Real-Time PCR (RT PCR) Kit according to the manufacturer’s protocol (Helini Biomolecules, India).
2.3.2 Targeted sequencing using next generation sequencing approach
The targeted sequencing was performed using Next Generation Sequencing Platform (NGS) in the Illumina MiSeq machine. The sequencing was run using the COVIDSeq test kit manufactured by Illumina, CA, USA. This kit can be used to sequence the other pathogens also apart from SARS-CoV-2. However, the targeted primers used for the complete sequencing of the Hexon gene (approximately 2.8 kb) were designed by the authors (as presented in Supplementary Table S1). The hexon gene was selected for differentiating strains of adenovirus by sequencing as the hexon protein also helps to classify adenoviruses into their 51 serotypes. All other reagents and protocols have been used as per the manufacturer’s instructions of COVID Seq Test kit. The sequencing reaction was executed in the MiSeq platform with a 2 × 75 paired-end v3 Flow Cell. Analysis of NGS data was performed employing the Linux Command Line Interface (CLI).
2.3.3 Post-sequencing data analysis
The raw fastq files were analyzed for quality and quantity using the fastp (v. 0.20.0) tool (25). The reads were aligned against multiple the Human Adenovirus types including type 7 and Human adenovirus B1 NCBI Reference sequences (accession numbers: AC_000018.1 and NC_011203.1). Alignment was performed using a bowtie2 aligner (v. 2.5.2) (26). Before generating the consensus sequences, the mapped ‘binary alignment files’ were sorted and indexed by using samtools (v. 1.19) (27). The consensus sequences thus generated were further utilized for downstream analysis.
2.3.4 Sequence alignment, mutational analysis, and phylogenetic analysis
To investigate the adenovirus species responsible for SARI, evolutionary analysis was performed to determine the phylogenetic correlation of the adenovirus-positive samples. The target sequences of the adenovirus were compared with closely related complete reference sequences and the nearest available sequences in NCBI’s GenBank. Basic Local Alignment Search Tool (BLAST) was used for the alignment search on the NCBI website for a selection of the nearest available sequences. Further, the obtained sequences were analyzed for existing amino acid and protein mutations in comparison to reference sequences of adenovirus Type 7 (AC_000018.1) and adenovirus Type 3 (MK847517.1 and NC_011203.1). In addition, the sequences were also examined for non-synonymous (dN) and synonymous (dS) mutations and the dN/dS ratio was calculated in MEGA X software by using the Nei–Gojobori model (28).
For evolutionary analysis, the ClustalW alignment program was used in MEGAX software. The best nucleotide model was determined using MEGA X’s model test and a phylogenetic tree was constructed by employing maximum likelihood (29). The process of building the evolutionary tree comprised of general time reversal, gamma distribution, and invariants for calculating rate differences among sites. A total of 500 bootstrap replications have been implemented to enhance the strength of the tree. A total of 23 nucleotide sequences have been included for evolutionary analysis (eight nucleotide sequences from the present study along with 15 complete reference sequences of different adenovirus types).
3 Results
3.1 Demographic and clinical characteristics of HAdV-positive pediatric cases
The present study collected 70 samples from pediatric ARI cases of which 18.57% (13/70) of samples were found to be positive for HAdV. Among the positive subjects, the mean age was found to be 3.4 years (SD± 3.7) ranging from 5 months to 10 years. A total of 76.9% of the adenovirus-positive cases were males (10/13) and 23.07% were females (03/13). The demographic data of all the enrolled cases has been presented in Table 1. The monthly prevalence rates of the HAdV infection were calculated as 12.5% (2/16), 42.85% (3/7), 14.28% (2/14), 25% (5/20), 14.28% (1/7), 0% (0/3), 0% (0/3) respectively from April 2023 to October 2023. While comparing the age of the positive cases, it was observed that the majority of HAdV infections (10/13, 76.92%) were associated with children under the age of 5 years. The median duration of illness of the enrolled subjects was found to be 5 days (SD± 3.8).
In the context of severity of illness, it was noted that 7/13 (53.8%) cases were diagnosed with severe pneumonia followed by 1/13 cases (7.6%) with acute encephalitis syndrome respectively. It is important to note that 3 out of 13 adenovirus-positive patients (23.1%) who died after hospitalization had severe pneumonia, with two of them being less than one year old. In terms of symptoms, it was observed that all the adenovirus-positive cases were associated with fever (100%, 13/13). In addition, primely associated symptoms included cough (76.9%, 10/13), breathlessness (53.8%, 7/13), Sore throat (46.2%, 6/13) and Seizure (46.2%, 6/13) respectively. Interestingly, three HAdV positive cases had shown pink eye symptom. While speculating the percentage of neutrophils among the HAdV positive cases it was noted that the neutrophil percentage of the three death cases was above 70%. Notably, the neutrophil to lymphocyte (NLR) ratio of all the three death cases were observed to be 12.8, 3.3 and 4.5 respectively suggesting altered host immune response.
The HAdV viral load (copies/ml) ranged from 6.86 × 102 to 1.46 × 108 copies/ml in HAdV-positive samples. However, the estimated viral load was higher among the three death cases with viral loads of 1.18 × 108, 1.46 × 108 and 4.85× 105 copies/ml respectively.
3.2 Adenovirus typing and phylogenetic analysis
Molecular characterization of HAdV of complete hexon gene (~2800 bp) using NGS revealed that among the positive cases, 4/13 were infected with HAdV-B type 3 and 5/13 were infected with HAdV type 7. Average sequencing depth of all the HAdV -positive samples were found to be 6850.2 ± 746.4 (Mean ± SD). Phylogenetic analysis indicated that the type 3 adenovirus-positive samples showed 100% resemblance with the complete reference sequence of Human adenovirus B1 (NC_011203.1) and clustered separately showing closest resemblance with Human Mastadenovirus type B (accession number: MK847517.1) strain collected in 2009 from Shanghai, China and one recent complete adenovirus sequence collected from Hannover, Germany in 2023 (accession number: OR853835) as presented in Figure 1. Additionally, all the five-adenovirus type 7 positive sequences showed 99.9% resemblance with the complete reference sequence of type 7 Adenovirus collected from USA (accession number: AC_000018.1) along with one recent adenovirus type 7 strain isolated from Marburg, Germany in 2023 (accession number: OR487156.1).
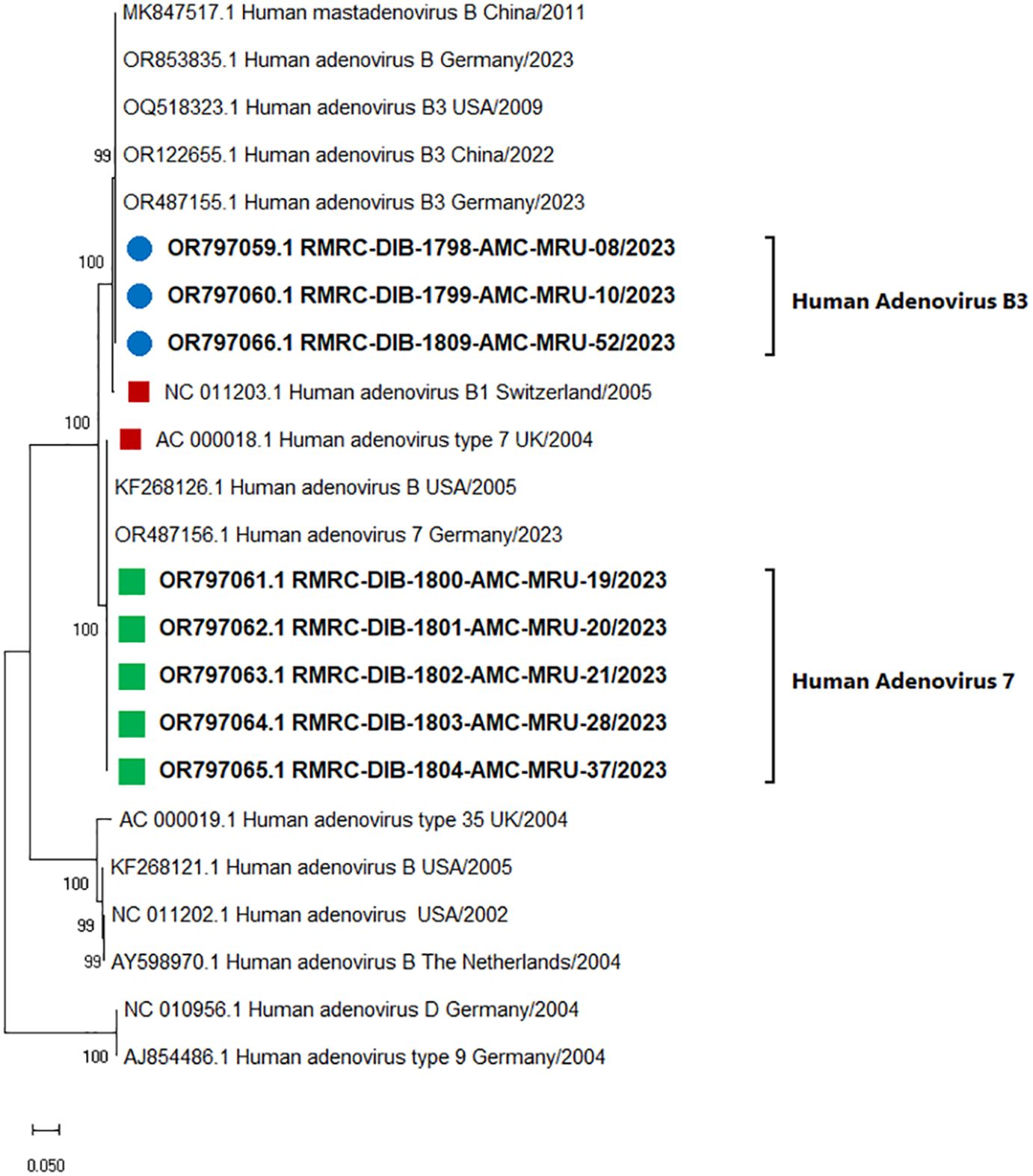
Figure 1. Evolutionary analysis for human adenovirus types of the present study. The evolutionary analysis was performed using the Maximum Likelihood method and General Time Reversible model (23). A total of 23 nucleotide sequences have been included for evolutionary analysis (eight nucleotide sequences from the present study covering the hexon gene ~ 2.8 kb along with 15 complete reference sequences of adenovirus types). A 500-bootstrap iterations was performed to make the tree robust. Evolutionary analyses were conducted in MEGA X. A discrete Gamma distribution was used to model evolutionary rate differences among sites. The rate variation model allowed for some sites to be evolutionarily invariable. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
While comparing the clinical data with the NGS results it was observed that 4/7 severe pneumonia cases were infected with HAdV type 7 and 2/7 infected with adenovirus B type 3. Of the HAdV type 3 infected pneumonia cases, two subjects died during hospitalization.
3.3 Mutational analysis
Mutational analysis of HAdV-B type 3 and HAdV type 7 positive samples has been performed and the synonymous and non-synonymous substitutions for the hexon gene of HAdV were examined. No non-synonymous substitutions (dN) were observed in the hexon gene of HAdVB type 3 positive samples. In contrast, HAdVB type 7 positive samples showed one non-synonymous mutation. The ratio of non-synonymous mutation to synonymous mutation (dN/dS) was comparatively low in the hexon gene of HAdV type 7 positive samples (dN/dS=0.31). Mutational analysis of adenovirus type 7 positive samples showed that there was a substitution from asparagine to threonine (N589T) in all the five isolates of 7 positive samples (Figure 2).
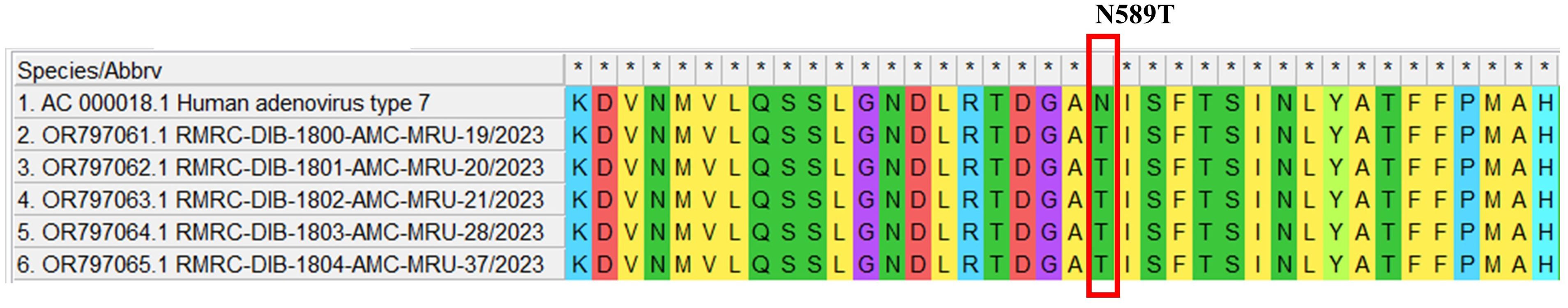
Figure 2. Mutational analysis of HAdV type7 amino acid sequences in the study subjects. The symbol * indicates an amino acid match in the table. If the same amino acid is present in all sequences, it is marked with *.
3.4 GenBank accession numbers of submitted sequences
The newly obtained sequences of HAdV-B type 3 and HAdV type 7 have been submitted to NCBI GenBank (GenBank accession numbers: OR797059.1-OR797066.1).
4 Discussion
Severe Acute Respiratory Infections (SARI) caused by human adenovirus and other respiratory viruses constitute a marked global health concern with significant morbidity and mortality worldwide (1). Adenovirus-associated SARI outbreaks have been reported throughout the world and the comprehensive influence on patient health reflects the necessity for early and rapid detection, and effective control strategies. Despite being one of the commonly associated pathogens of ARI infection, improper and misdiagnosis, self-medication, and delay in seeking medical care have become the triggering factors for the increased mortality and lack of true epidemiological information on adenovirus-associated SARI (16). The present study identified adenovirus-mediated SARI infection in the pediatric cases of the study site from April 2023 to October 2023. Our data on molecular diagnosis and sequencing-based characterization demonstrates the causative agent of the pediatric cases of SARI in the present study to be from HAdV type 7 (38.46%) and HAdV-B type 3 (30.76%). The association of HAdVB7 in the studied SARI samples was in accordance with the earlier findings of Zadheidar S et al., 2022 where authors also obtained 75% association of HAdV B-7 with SARI cases (30). Similarly, our finding on the association of HAdVB-3 in the three samples of SARI was concomitant with the findings of Xu et al., 2022 who reported HAdVB-3 as the most prevalent strain related to SARI infection in Huzhou, China during 2017-2019 (14). Further, it was inferred from the phylogenetic analysis that adenovirus-positive samples from the present study showed 100% resemblance with the complete reference sequences of (hexon gene of approximately 2800bp size, position: 18666 - 21470) HAdVB type 3 (accession numbers: NC_011203.1, MK847517.1and OR853835) and HAdV type 7 (accession numbers: AC_000018.1and OR487156.1) including the recently circulating strains of both the types from Germany and USA in 2023 respectively. Our data on the circulation of HAdV type 7 and HAdV-B type 3 during the study period was further supported by a recently published whole genome sequencing-based study from West Bengal India in which it was documented that HAdV type 7 and HAdV-B type 3 were the mainly circulating strains of adenovirus during 2022-23 in West Bengal, India (31).
Although adenovirus-mediated respiratory infections can be observed globally throughout the year, the seasonal distinctness in the trend of its positivity rates cannot be neglected (32–34). A previously published study has shown the presence of HAdV throughout the year without any seasonal preferences (35). Contradictorily, the prevalence of adenovirus infection in summer was also reported previously (36). Of note, the month-wise distribution of data from the present study among the collected samples showed that all the adenovirus-positive samples have been collected from April 2023 to August 2023 and the positivity rate was nil after August 2023 suggesting the possibility of seasonal variation in adenovirus circulation. Our findings further highlighted the point that human adenovirus infection gradually declined by autumn which is consistent with similar studies documented from Northern and Southern China where the highest prevalence of the virus was observed during summer (37, 38).
It is pertinent to mention here that, adenovirus associated respiratory infections are generally evidenced by having a broad spectrum of symptoms starting from asymptomatic to severe depending on the age of the host, immunotolerance status of the patient, and the type of the associated adenoviral strain (39, 40). Notably, HAdV species B, C, and E are commonly involved in respiratory disease outbreaks of which serotypes, 7 and 3 are suggested to be the major contributors in the occurrence of various adenoviral outbreaks (10, 41, 42). The molecular characterization data obtained from the present study indicated HAdV type 7 and HAdV B type 3 to be associated with SARI infections of under five years children. This inference from our study was well supported by the previously published reports which reported that both HAdV-B type 3 and type 7 are predominantly associated with lower respiratory infections among children under five years of age (43).
It is worthy to note that 4/7 cases of severe pneumonia were of those under five years of age and associated with HAdV type 7 infection. Our data was in agreement with a previously published study that documented HAdV type 7 to be commonly associated with severe infections among children under five years (34). Additionally, as per a report published earlier, HAdV type 3 and type 7 were the most common adenoviral strains responsible for causing pneumonia in China from 1958 to 1990 (44). This finding is in agreement with the comparative analysis between clinical profile and molecular profile of the patients from the present study as our data showed that 66.66% of the severe pneumonia cases were infected with HAdV type 7. Mutational analysis of adenovirus type 7 positive samples showed that there was a substitution of asparagine to threonine (N589T) in all the five type 7 positive samples compared to the reference sequence (Figure 2). To the best of our knowledge this is a novel mutation reported in our study. Further, to assess the epitope binding sites, these amino acid sequences has been analyzed in Immune Epitope Database (IEDB) but no epitopic regions have been detected spanning the novel mutation. Interestingly, no non-synonymous substitutions (dN) was observed in the HAdVB type 3 positive samples while only one non-synonymous mutation was found in HAdVB type 7 positive samples.
High percentage of neutrophils and the neutrophil to lymphocyte ratio among the HAdV positive death cases of the present study suggested altered immune response with severe clinical outcome including pneumonia as documented previously (45). Since the adenovirus-positive cases from the present study also possessed Influenza-like illness (ILI) symptoms, those samples were evaluated for the presence of influenza A, Influenza B and SARS-CoV-2. Three out of thirteen HAdV-positive samples were also found to be positive for influenza A. All three cases were cured without any further complications. To the best of our knowledge, this is the first documented report of HAdV-associated respiratory infections among the pediatric population, in North Eastern India. However, this study had several limitations. Firstly, detection for other viral or bacterial pathogens was not performed apart from the 13 positive cases. Additionally, NGS-based molecular characterization focused primarily on the complete hexon gene rather than the entire viral genome. Moreover, the small sample size limited the ability to draw definitive conclusions, and a larger sample pool is necessary to gain a clearer understanding of adenoviral epidemiology and disease pathogenesis.
The study highlighted the importance of screening for Adenovirus, which can lead to high mortality in pediatric SARI cases. In conclusion, it identified HAdV type 7 and HAdV-B type 3 as the strains responsible for the severe acute respiratory infections among pediatric cases from April to October 2023. The findings also emphasized that HAdV types 3 and 7 are linked to more severe lower respiratory tract infections, such as severe pneumonia.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR797059.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797060.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797061.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797062.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797063.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797064.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797065.1 https://www.ncbi.nlm.nih.gov/genbank/, OR797066.1.
Ethics statement
The studies involving humans were approved by Institutional Ethics Committee (IEC), ICMR-RMRCNE, Dibrugarh Assam, Institutional Ethics Committee (IEC), AMCH, Dibrugarh Assam. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
RNa: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – review & editing. GC: Formal analysis, Investigation, Methodology, Writing – review & editing. AG: Investigation, Supervision, Writing – review & editing. NS: Investigation, Writing – original draft, Writing – review & editing. NB: Investigation, Writing – original draft. AS: Formal analysis, Investigation, Writing – original draft. RNe: Investigation, Writing – original draft. MD: Investigation, Writing – original draft. AJ: Formal analysis, Writing – review & editing. BB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The financial support for conducting the laboratory investigations is obtained under the scheme Regional VRDL, ICMR-RMRCNE and MRU, AMCH which are funded by the Department of Health Research (DHR), Government of India.
Acknowledgments
The authors sincerely acknowledge the Department of Health Research (DHR), Govt. of India for funding the Multidisciplinary Research Unit (MRU), Assam Medical College and Hospital (AMCH), and Regional Viral Research and Diagnostic Laboratory (Regional VRDL), ICMR-RMRCNE, Dibrugarh where the present study was carried out. Authors sincerely acknowledge the Department of pediatrics, AMCH for the kind support towards collection of samples and clinical data. In addition, authors would also like to acknowledge the German Epidemic Preparedness Team (SEEG), Germany for providing the Illumina MiSeq NGS system and the required consumables to carry out the molecular characterization of the adenovirus positive samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2024.1462907/full#supplementary-material
References
1. Ogunbayo AE, Mogotsi MT, Sondlane H, Nkwadipo KR, Sabiu S, Nyaga MM. Pathogen profile of children hospitalised with severe acute respiratory infections during COVID-19 pandemic in the free state province, South Africa. Int J Environ Res Public Health. (2022) 19:10418. doi: 10.3390/ijerph191610418
2. Balasubramani K, Prasad KA, Kodali NK, Abdul Rasheed NK, Chellappan S, Sarma DK, et al. Spatial epidemiology of acute respiratory infections in children under 5 years and associated risk factors in India: District-level analysis of health, household, and environmental datasets. Front Public Health. (2022) 10:906248. doi: 10.3389/fpubh.2022.906248
3. Um S, Vang D, Pin P, Chau D. Trends and determinants of Acute Respiratory Infection symptoms among Under-five children in Cambodia: Analysis of 2000 to 2014 Cambodia Demographic and Health Surveys. PloS Global Public Health. (2023) 3:e0001440. doi: 10.1371/journal.pgph.0001440
4. Liu T, Li Z, Zhang S, Song S, Julong W, Lin Y, et al. Viral Etiology of acute respiratory tract infections in hospitalized children and adults in Shandong Province, China. Virol J. (2015) 12:1–7. doi: 10.1186/s12985-015-0388-z
5. Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, Hess M, et al. ICTV virus taxonomy profile: Adenoviridae 2022. J Gen Virol. (2022) 103:001721. doi: 10.1099/jgv.0.001721
6. Wang X, Wang D, Umar S, Qin S, Ling Q, Gray GC, et al. Molecular typing of human adenoviruses among hospitalized patients with respiratory tract infections in a tertiary Hospital in Guangzhou, China between 2017 and 2019. BMC Infect Dis. (2021) 21:748. doi: 10.1186/s12879-021-06412-0
7. Crenshaw BJ, Jones LB, Bell CR, Kumar S, Matthews QL. Perspective on adenoviruses: epidemiology, pathogenicity, and gene therapy. Biomedicines. (2019) 7:61. doi: 10.3390/biomedicines7030061
8. Yao J, Atasheva S, Wagner N, Di Paolo NC, Stewart PL, Shayakhmetov DM. Targeted, safe, and efficient gene delivery to human hematopoietic stem and progenitor cells in vivo using the engineered AVID adenovirus vector platform. Mol Ther. (2024) 32:103–23. doi: 10.1016/j.ymthe.2023.10.023
9. Abbas KZ, Lombos E, Duvvuri VR, Olsha R, Higgins RR, Gubbay JB. Temporal changes in respiratory adenovirus serotypes circulating in the greater Toronto area, Ontario, during December 2008 to April 2010. Virol J. (2013) 10:1–7. doi: 10.1186/1743-422X-10-15
10. Barnadas C, Schmidt DJ, Fischer TK, Fonager J. Molecular epidemiology of human adenovirus infections in Denmark, 2011–2016. J Clin Virol. (2018) 104:16–22. doi: 10.1016/j.jcv.2018.04.012
11. Bastug A, Altas AB, Koc BT, Bayrakdar F, Korukluoglu G, Bodur H, et al. Molecular characterization of human adenoviruses associated with respiratory infection in Turkey. Apmis. (2021) 129:23–31. doi: 10.1111/apm.13088
12. Xu L, Liu J, Liu C, Duan Y, Zhu Y, Xu B, et al. Case-control study of the epidemiological and clinical features of human adenovirus 55 and human adenovirus 7 infection in children with acute lower respiratory tract infections in Beijing, China, 2008-2013. BMC Infect Dis. (2018) 18:634. doi: 10.1186/s12879-018-3520-z
13. Zhao SH, Wan CS, Ke CW, Seto J, Dehghan S, Zou LR. Re-emergent human adenovirus genome type 7d caused an acute respiratory disease outbreak in southern China after a twenty-one year absence. Sci Rep. (2014) 4:7365. doi: 10.1038/srep07365
14. Xu D, Chen L, Wu X, Ji L. Molecular typing and epidemiology profiles of human adenovirus infection among hospitalized patients with severe acute respiratory infection in Huzhou, China. PloS One. (2022) 17:e0265987. doi: 10.1371/journal.pone.0265987
15. Wen S, Lin Z, Zhang Y, Lv F, Li H, Zhang X, et al. The epidemiology, molecular, and clinical of human adenoviruses in children hospitalized with acute respiratory infections. Front Microbiol. (2021) 12:629971. doi: 10.3389/fmicb.2021.629971
16. Lynch JP III, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. In: Seminars in respiratory and critical care medicine, vol. 37. Thieme Medical Publishers (2016). p. 586–602.
17. Mishra P, Nayak L, Das RR, Dwibedi B, Singh A. Viral agents causing acute respiratory infections in children under five: a study from Eastern India. Int J Pediatr. (2016) 2016(1):7235482. doi: 10.1155/2016/7235482
18. Malhotra B, Swamy MA, Reddy PJ, Gupta ML. Viruses causing severe acute respiratory infections (SARI) in children≤ 5 years of age at a tertiary care hospital in Rajasthan, India. Indian J Med Res. (2016) 144:877. doi: 10.4103/ijmr.IJMR_22_15
19. Bhardwaj S, Choudhary ML, Jadhav S, Vipat V, Ghuge R, Salvi S, et al. A retrospective analysis of respiratory virus transmission before and during the COVID-19 pandemic in Pune the western region of India. Front Public Health. (2022) 10:936634. doi: 10.3389/fpubh.2022.936634
20. Saha R, Majumdar A, Chaudhuri RD, Chatterjee A, Lo M, Dutta S, et al. Molecular epidemiology of circulating human adenoviruses among acute respiratory infection patients seeking healthcare facilities in West Bengal, India. Virology. (2023) 588:109912. doi: 10.1016/j.virol.2023.109912
21. Chadha M, Prabhakaran AO, Choudhary ML, Biswas D, Koul P, Kaveri K, et al. Multisite surveillance for influenza and other respiratory viruses in India: 2016–2018. PloS Global Public Health. (2022) 2:e0001001. doi: 10.1371/journal.pgph.0001001
22. Simoes EAF, Cherian T, Chow J, Shahid-Salles SA, Laxminarayan R, John TJ, et al. Acute Respiratory Infections in Children. In: Jamison DT, Breman JG, Measham AR, et al, editors. Disease Control Priorities in Developing Countries, 2nd edition. Oxford University Press, New York (2006). Available at: https://www.ncbi.nlm.nih.gov/books/NBK11786/Co-. Washington (DC): The International Bank for Reconstruction and Development/The World BankChapter 25.
23. Mir F, Ariff S, Bhura M, Chanar S, Nathwani AA, Jawwad M, et al. Risk factors for acute respiratory infections in children between 0 and 23 months of age in a peri-urban district in Pakistan: A matched case–control study. Front Pedia. (2022) 9:704545. doi: 10.3389/fped.2021.704545
24. Scott JA, Wonodi C, Moïsi JC, Deloria-Knoll M, DeLuca AN, Karron RA, et al. The definition of pneumonia, the assessment of severity, and clinical standardization in the Pneumonia Etiology Research for Child Health study. Clin Infect Dis. (2012) 54:S109–16. doi: 10.1093/cid/cir1065
25. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. (2018) 34:i884–90. doi: 10.1093/bioinformatics/bty560
26. Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. (2012) 9:357–9. doi: 10.1038/nmeth.1923
27. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
28. Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. (1986) 3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410
29. Tamura K, Stecher G, Kumar S. MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
30. Zadheidar S, Yavarian J, Heydarifard Z, Nejati A, Sadeghi K, Ghavami N, et al. Molecular epidemiology of human adenoviruses in children with and without respiratory symptoms: Preliminary findings from a case-control study. BMC Pedia. (2022) 22:1–7. doi: 10.1186/s12887-022-03625-3
31. Chatterjee A, Bhattacharjee U, Gupta R, Debnath A, Majumdar A, Saha R, et al. Genomic expedition: deciphering human adenovirus strains from the 2023 outbreak in West Bengal, India: insights into viral evolution and molecular epidemiology. Viruses. (2024) 16:159. doi: 10.3390/v16010159
32. Wang C, Liu J, Mi Y, Chen J, Bi J, Chen Y. Clinical features and epidemiological analysis of respiratory human adenovirus infection in hospitalized children: a cross-sectional study in Zhejiang. Virol J. (2021) 18:1–1. doi: 10.1186/s12985-021-01705-x
33. Sandkovsky U, Vargas L, Florescu DF. Adenovirus: current epidemiology and emerging approaches to prevention and treatment. Curr Infect Dis Rep. (2014) 16:1–8. doi: 10.1007/s11908-014-0416-y
34. Yu Z, Zeng Z, Zhang J, Pan Y, Chen M, Guo Y, et al. Fatal community-acquired pneumonia in children caused by re-emergent human adenovirus 7d associated with higher severity of illness and fatality rate. Sci Rep. (2016) 6:37216. doi: 10.1038/srep37216
35. Price RH, Graham C, Ramalingam S. Association between viral seasonality and meteorological factors. Sci Rep. (2019) 9:929. doi: 10.1038/s41598-018-37481-y
36. Pscheidt VM, Gregianini TS, Martins LG, Veiga AB. Epidemiology of human adenovirus associated with respiratory infection in southern Brazil. Rev Med Virol. (2021) 31:e2189. doi: 10.1002/rmv.2189
37. Yao LH, Wang C, Wei TL, Wang H, Ma FL, Zheng LS. Human adenovirus among hospitalized children with respiratory tract infections in Beijing, China, 2017–2018. Virol J. (2019) 16:1–8. doi: 10.1186/s12985-019-1185-x
38. Chen Y, Liu F, Wang C, Zhao M, Deng L, Zhong J, et al. Molecular identification and epidemiological features of human adenoviruses associated with acute respiratory infections in hospitalized children in Southern China, 2012-2013. PloS One. (2016) 11:e0155412. doi: 10.1371/journal.pone.0155412
39. Shafiei-Jandaghi NZ, Yavarian J, Malekshahi SS, Naseri M, Shadab A, Ghavami N, et al. Identification of adenovirus species in Iranian pediatric population with severe acute respiratory infections. Future Virol. (2019) 14:577–83. doi: 10.2217/fvl-2019-0074
40. Akhil C, Suresha PG, Sabeena S, Hindol M, Arunkumar G. Genotyping of human adenoviruses circulating in Southwest India. Virusdisease. (2016) 27:266–70. doi: 10.1007/s13337-016-0337-5
41. Xie L, Zhang B, Xiao N, Zhang F, Zhao X, Liu Q, et al. Epidemiology of human adenovirus infection in children hospitalized with lower respiratory tract infections in Hunan, China. J Med Virol. (2019) 91:392–400. doi: 10.1002/jmv.25333
42. Callaway Z, Kim SH, Kim JY, Kim DW, Kim CK. Adenovirus infection with serious pulmonary sequelae in Korean children. Clin Respir J. (2011) 5:92–8. doi: 10.1111/j.1752-699X.2010.00204.x
43. Fu Y, Tang Z, Ye Z, Mo S, Tian X, Ni K, et al. Human adenovirus type 7 infection causes a more severe disease than type 3. BMC Infect Dis. (2019) 19:1–1. doi: 10.1186/s12879-018-3651-2
44. Li QG, Zheng QJ, Liu YH, Wadell G. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J Med Virol. (1996) 49:170–7. doi: 10.1002/(ISSN)1096-9071
Keywords: severe acute respiratory infections, SARI, pediatric, adenovirus, HAdV, pneumonia, ARI, Assam
Citation: Nath R, Choudhury G, Gogoi A, Sarmah N, Bhattacharya N, Siddique AI, Neog R, Dutta M, Jakharia A and Borkakoty B (2024) Molecular characterization of human adenovirus associated with pediatric severe acute respiratory infections in a tertiary care hospital in North East India. Front. Virol. 4:1462907. doi: 10.3389/fviro.2024.1462907
Received: 10 July 2024; Accepted: 27 September 2024;
Published: 21 October 2024.
Edited by:
Susan Prockop, Boston Children’s Hospital and Harvard Medical School, United StatesReviewed by:
Carmen Baur Vieira, Fluminense Federal University, BrazilYeon Joo Lee, Memorial Sloan Kettering Cancer Center, United States
Copyright © 2024 Nath, Choudhury, Gogoi, Sarmah, Bhattacharya, Siddique, Neog, Dutta, Jakharia and Borkakoty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biswajyoti Borkakoty, Ymlzd2Fib3JrYWtvdHlAZ21haWwuY29t
 Reema Nath1
Reema Nath1 Neelanjana Sarmah
Neelanjana Sarmah Rahul Neog
Rahul Neog Biswajyoti Borkakoty
Biswajyoti Borkakoty