
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol., 22 January 2024
Sec. Viral Disease Investigation
Volume 4 - 2024 | https://doi.org/10.3389/fviro.2024.1335752
This article is part of the Research TopicEnterovirus Surveillance in Europe and BeyondView all 6 articles
 Tina Uršič1*
Tina Uršič1* Monika Jevšnik Virant1
Monika Jevšnik Virant1 Rok Kogoj1
Rok Kogoj1 Uros Krivec2,3
Uros Krivec2,3 Joanna Prusnik2
Joanna Prusnik2 Minca Mramor4
Minca Mramor4 Sara Lovšin4
Sara Lovšin4 Miroslav Petrovec1
Miroslav Petrovec1Introduction: Enterovirus D68 (EV-D68) belongs to the Picornaviridae family, genus Enterovirus. It is mostly known as a respiratory virus causing upper and lower respiratory tract infections, but it is also rarely associated with a variety of central nervous system complications, with acute flaccid myelitis being reported most frequently. This study assesses the incidence, seasonality, clinical presentation, and molecular epidemiology of the EV-D68 strain in EV-positive children hospitalized between 2014 and 2022 at the largest pediatric medical center in Slovenia.
Methods: EV-D68 was detected using specific qRT-PCR, whereas partial VP1 sequences were obtained with Sanger sequencing, and further analyzed using the software CLC Main Workbench version 7 and MEGA version X.
Results: EV-D68 was detected in 154 out of 1,145 (13.4%) EV-positive children. In the two epidemic years, 2014 and 2016, EV-D68 was most frequently detected in the summer and early autumn, peaking in September. The median age of EV-D68–infected children was 3 years (IQR 1–3 years), with a female: male ratio of 1:1.17. Rhinorrhea was present in 74.0% of children, respiratory distress in 82.5%, and hypoxemia requiring supplemental oxygen in 44.1%. Out of 154 patients, 80.0% were hospitalized, with a median stay of 2 days (IQR 1–3 days). Lower respiratory tract infection was observed in 89.0% of EV-D68–positive patients, with bronchitis and bronchiolitis being most frequently diagnosed. No central nervous system manifestations of EV-D68 infection were observed in the study cohort. Phylogenetic analysis of partial VP1 sequences of EV-D68 revealed close similarity to the EV-D68 variants that were circulating in other European countries in these years.
Discussion: Slovenia faced two EV-D68 epidemics in 2014 and 2016; however, after 2016 only nine more cases were detected until the end of the study period. Based on the results of this study, EV-D68 was a frequent cause of lower respiratory tract infection among EV-positive patients. However, none of the patients we studied needed ICU treatment, and none developed acute flaccid paralysis. Our results indicate that EV-D68 is not present constantly, so additional monitoring studies should be conducted in the future to better understand the implications of this EV type in human disease.
Enterovirus-D68 (EV-D68) is a non-enveloped, positive-sense ssRNA virus that belongs to the Picornaviridae family, genus Enterovirus. It was first isolated in 1962 from children with pneumonia and bronchiolitis (1). Like rhinoviruses, EV-D68 is transmitted via the respiratory route, with a seasonal pattern, and infections peak in late summer and early autumn in countries with temperate climates (2). Enterovirus-D68 infections usually result in mild upper respiratory symptoms and rarely in severe infections that may lead to hospitalization, intensive care unit (ICU) treatment, or even fatal outcomes (3). Children are at increased risk for severe EV-D68 infections, but publications show that immunocompromised adults are also at high risk (4–6). Large EV-D68 outbreaks with severe clinical presentation in the United States and Europe in 2014 gained attention. In the following years, the research trend and interest in EV-D68 increased significantly because it was found that the virus can cause severe lower respiratory tract infection (LRTI), which can lead to ICU admission or fatality (2, 7). Moreover, systemic dissemination and various central nervous system complications (CNS) such as acute flaccid myelitis (AFM) can occur, which clinically overlaps largely with acute flaccid paralysis (AFP) caused by poliovirus (3).
Phylogenetic analyses of EV-D68 sequences from individual outbreaks in various years revealed genetic differences among circulating strains in the United States, Europe, and Asia, indicating the presence of multiple clades (A, B, and C). These clades can be divided further into subclades A1, A2/D (which can be subdivided to D1 and D2) and B1–B3. Some subclades are more prevalent than others (8–11), with different clusters of B3 subclades being detected most frequently in recent years (12–16). Previous studies have shown that the ability to invade and replicate in the CNS is not a clade-specific feature (17–19). In Slovenia, to date no data exists on the circulation of predominant types of enteroviruses, neither in the general pediatric population nor among hospitalized pediatric patients as enteroviruses are not typed in routine diagnostics. Due to the increasing interest in enteroviruses in recent years and known disease outbreaks caused by EV-D68 in Europe and elsewhere, we decided to examine more closely the population of pediatric patients with confirmed enterovirus infection over a longer period. This 9-year retrospective study assesses the incidence, seasonality, clinical presentation, and molecular epidemiology of EV-D68–positive children (<17 years of age), who were admitted (hospitalized) or examined (non-hospitalized) due to respiratory infection or other clinical indications between 2014 and 2022 at the Department of Infectious Diseases and Children’s Hospital, which are both part of Slovenia’s largest hospital, the Ljubljana University Medical Centre.
All pediatric patients who are examined and/or hospitalized due to respiratory tract infection receive complete molecular respiratory virus panel testing (15 viruses), and in winter epidemic peaks of FluA/B and RSV, a smaller panel containing 4 viruses (FluA/B, RSV, SARS-CoV-2) is also available if so chosen by the clinician. Clinicians may also order additional respiratory virus panel testing in asymptomatic cases when hospitalization of the children are due to other reasons which could influence disease prognosis following positive respiratory virus identification (chemotherapy implementation, worsening of underlying neurologic diseases, etc.). This testing algorithm stayed the same through-out the nine-year study period, however, in 2021 the short FluA/B & RSV panel was exchanged for SARS-CoV-2 FluA/B & RSV when introduced to the market. When looking at the two years before the emergence of SARS-CoV-2 (Jan 2018- Mar 2020), 60.8% of all the children examined at the pediatric clinic were tested with respiratory virus panel. In contrast, after the SARS-CoV-2 emergence (between Apr 2020 and Dec 2022) the percentage of pediatric samples sent for respiratory virus panel diagnostics increased to 70.4%. This percentage is calculated for only children (< 17 years) and the result cannot be extrapolated for other hospitalized patients in other age groups.
In our 9-year retrospective study, all children under 17 years of age with detected EV-D68 in respiratory samples, between January 2014 and December 2022, who were admitted (hospitalized) or examined (non-hospitalized) at the Department of Infectious Diseases and Children’s Hospital, both part of Slovenia’s largest hospital, the Ljubljana University Medical Centre, were included. Children for whom the clinicians concluded after examination that hospitalization was not necessary, due to the mild course of the respiratory infection, were discharged. AFM or AFP symptoms were not required for inclusion.
We retrospectively collected demographic and clinical data for all included patients. All signs and symptoms of infection were recorded and obtained on examination after arrival at the hospital.
Nucleic acids (NA) were isolated and eluted in 100 µl from 200 µl of respiratory samples—nasopharyngeal swab (NPS), throat swabs, bronchoalveolar lavages (BAL), and tracheal aspirates (AT)—using a MagNA Pure Compact Instrument following the manufacturer’s instructions (Roche Applied Science, Mannheim, Germany) at the time of collection and stored at −20°C until the time of this study. Routine diagnostics of respiratory viruses included the detection of influenza virus A/B (FluA/B), human respiratory syncytial virus (HRSV), human mastadenoviruses (HAdV), human metapneumovirus (HMPV), parainfluenza viruses 1–4 (PIV 1–4), human coronaviruses (HCoV-NL63, HCoV-HKU1, HCoV-OC43, and HCoV-229E), human bocavirus 1 (HBoV1), human rhinovirus (HRV), and enterovirus (EV). Two different diagnostic platforms were used: in-house qRT-PCR between 2014 and 2018 and the AusDiagnostics 16-well respiratory panel (AusDiagnostics Mascot, Australia) between 2018 and 2022, as described previously (20, 21).
The specific EV-D68 qRT-PCR was performed with the addition of 5 µl of extracted NA in a total reaction volume of 20 µl using TaqMan™ Fast Virus 1-Step Master Mix following the manufacturer’s instructions (Thermo Fisher Scientific, Baltics UAB, Vilnius, Lithuania). The primers and probe were used as described previously (22) with a slight modification of the WashU 1 protocol. Sequences from EV-D68 isolates available in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 22 December 2023) were aligned including recent years 2021 and 2022) and the primers were corrected in a way that all aligned sequences covered clades and subclades which should be detected by the modified primers; (primer EV-D68-L1 CACTGAACCAGAAGAAGCTA, primer EV-D68 R1 CCAAAGCTGCTCTACTGAGAAA, probe EV–D68 P1 FAM-TCGCACAGTGATAAATCARCAYGG-BBQ and thermocycling condition conditions were 50°C for 10 min, followed by 95°C for 2 min, and then 40 cycles of 95°C for 15 s and 60°C 30 s.
A 575 bp VP1 gene region of EV-D68qRT-pcr positive samples was amplified by using SuperScript® III One-Step RT-PCR platinum® Taq HiFi kit (Invitrogen by Thermo Fisher Scientific, Carlsbad, USA), with in-house developed primers and PCR conditions shown in Table 1.
The PCR products were detected with 1.2% agarose gel electrophoresis and then enzymatically purified using Fast AP + Exonuclease I (Thermo Fischer Scientific, Waltham, MA, USA) and sequenced. Sanger sequencing was performed using BigDye terminator chemistry and the v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions on an ABI-3500 genetic analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
Nucleotide sequences were assembled and analyzed using CLC Main Workbench software version 7. Consensus sequences were generated from forward and reverse trace files using the Assemble Sequences tool in CLC Main Workbench with default parameters (Minimum aligned read length 50, Vote conflicts, and Create full contigs, including trace data). Each sequence pair was manually trimmed on both ends to exclude single-strand ends and further inspected for conflicts throughout the double-stranded region. Conflicts were corrected where applicable or re-sequenced if needed.
Trimmed sequences (534 bp) were aligned and compared with reference sequences (using Basic Local Alignment Search Tool (BLAST) available at the NCBI website. We also used the Enterovirus Genotyping Tool for enterovirus sequence type identification (23). Reference sequences were obtained from GenBank (acc. no.: OP321143, OP572095, OP572088, OP321146, OQ160431, MK419065, MK491182, MN896983, MN896982, MN896985, MN896986, MN812202, OM831204, OK085437, OK085464, MN809623, MN245441, MK121717, MH674156, ON006421, OM831187, OM831202, OP267497, MN726800, MN935870, MN896984, LR743440, MN896976, MN896977, LR743439, MN894268, MN896974, MN896981, LT745912, MT791927, LN681320, AB601883, LR743438, LR743441, NC_038308; originating from countries: USA, Australia, Japan, France, Germany, Denmark, Netherlands and Sweden). The sequences were aligned in MEGA-X (software v.10.1.7) using the Clustal W Algorithm with default options (Gap Opening penalty 15, Gap Extension penalty 6.66 for both pairwise and multiple alignments), DNA Weight Matrix set to IUB nucleotide codes, transition Weight set to 0.50, without Negative Matrix and Delay Divergent cut off set to 30%. After further trimming to achieve uniform sequence length a Phylogenetic tree was constructed using the Neighbor-joining Statistical Method with 500 Bootstrap replications. Then the Best-Fit substitution model algorithm revealed, Tamura 3-parameter model with Gamma Distributed Rates among sites and the Nearest-Neighbor-Interchange ML Heuristic Method without Branch Swap Filter to be the most suitable for phylogenetic tree construction.
Pearson’s chi-squared test was used to compare categorical variables between hospitalized and non-hospitalized patients.
Over the 9 years (2014–2022), 17,163 respiratory samples from 12,675 children were received for molecular detection of respiratory viruses. A total of 1,145 (6.7%) respiratory samples (97.3% were NPSs, 1.7% bronchoalveolar lavages and tracheal aspirates, and 1.0% throat swabs), collected from 1,028 (8.1%) of the children, were positive for enterovirus (EV). All 1,145 enterovirus-positive samples were analyzed using EV-D68–specific qRT-PCR with EV-D68 detected in 154 children from 154 NPS. Only one NPS was obtained from each EV-D68 positive patient. The majority of EV-D68–positive children were detected in two seemingly epidemic years, 2014 and 2016. EV-D68 was detected in NPSs of 91 (59.1%) children in 2014, and NPSs of 54 (35.1%) children in 2016, and the percentage of EV-D68 positive patients that were hospitalized was nearly the same between both epidemic years; 78% (71/91) versus 79.6% (43/54). After 2016, EV-D68 was detected in only nine (5.8%) children, in 2018 in three (1.9%) children, in 2019 in four (2.6%) children, and in 2022 in two (1.3%) children. No cases of EV-D68 among hospitalized and non-hospitalized children were detected in 2015, 2017, 2020, and 2021, as shown in Figure 1.
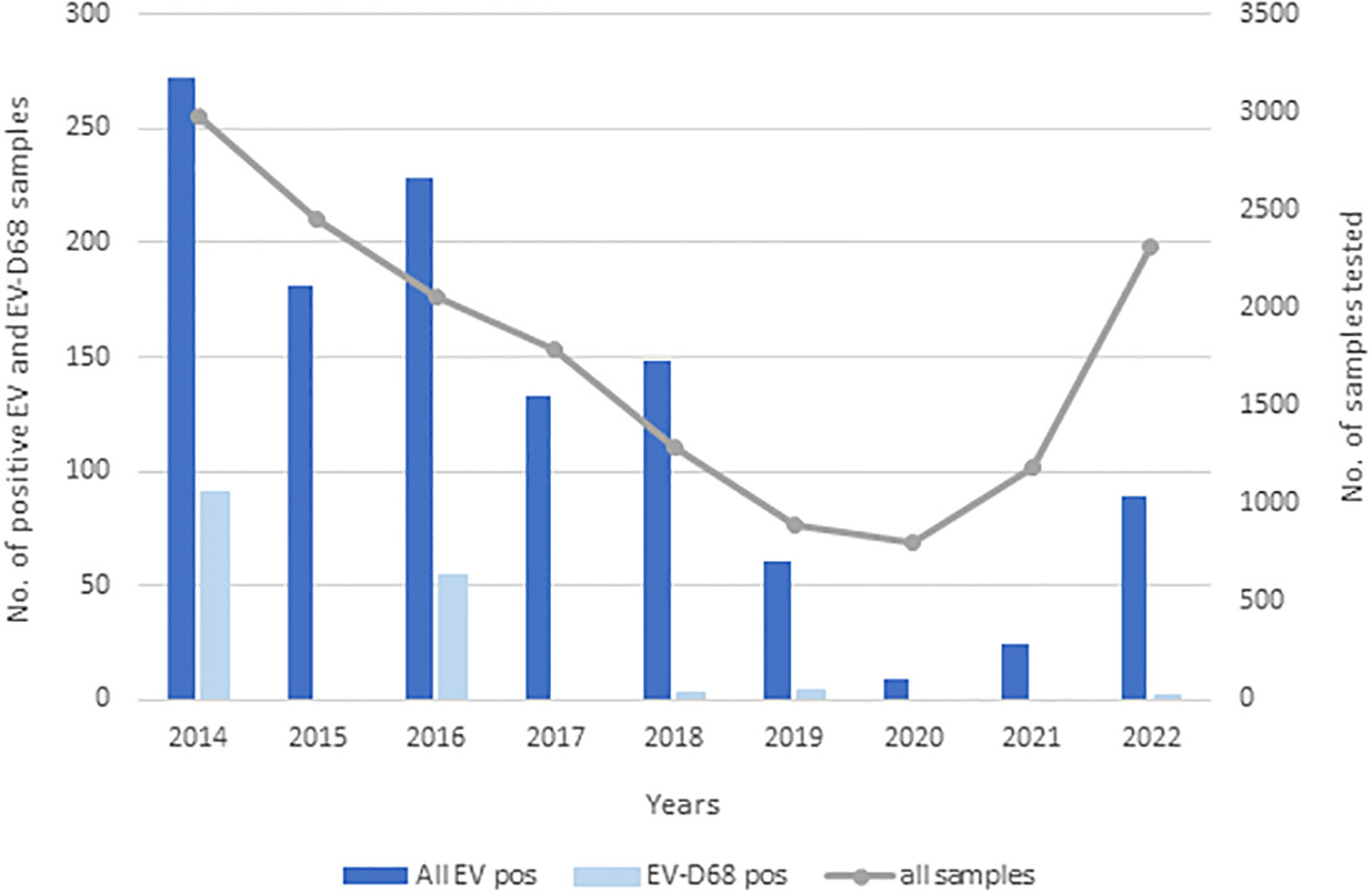
Figure 1 Incidence of enteroviruses and EV-D68 between 2014 and 2022. Dark blue bars represent all EVs detected, light blue EV-D68 detected, and the gray line all samples that were sent for diagnostics of respiratory viruses.
Because infections caused by enteroviruses in Slovenia are seasonally distributed with traditionally observed incidence peaks in summer and autumn, a similar analysis was also performed for the EV-D68 cases detected. The data revealed a similar pattern, with the greatest number of infections in September in both epidemic years, as shown in Figures 2A, B. In these two years, EV-D68 represented 33.5% (91/272) and 23.7% (54/228) of all enterovirus infections, respectively.
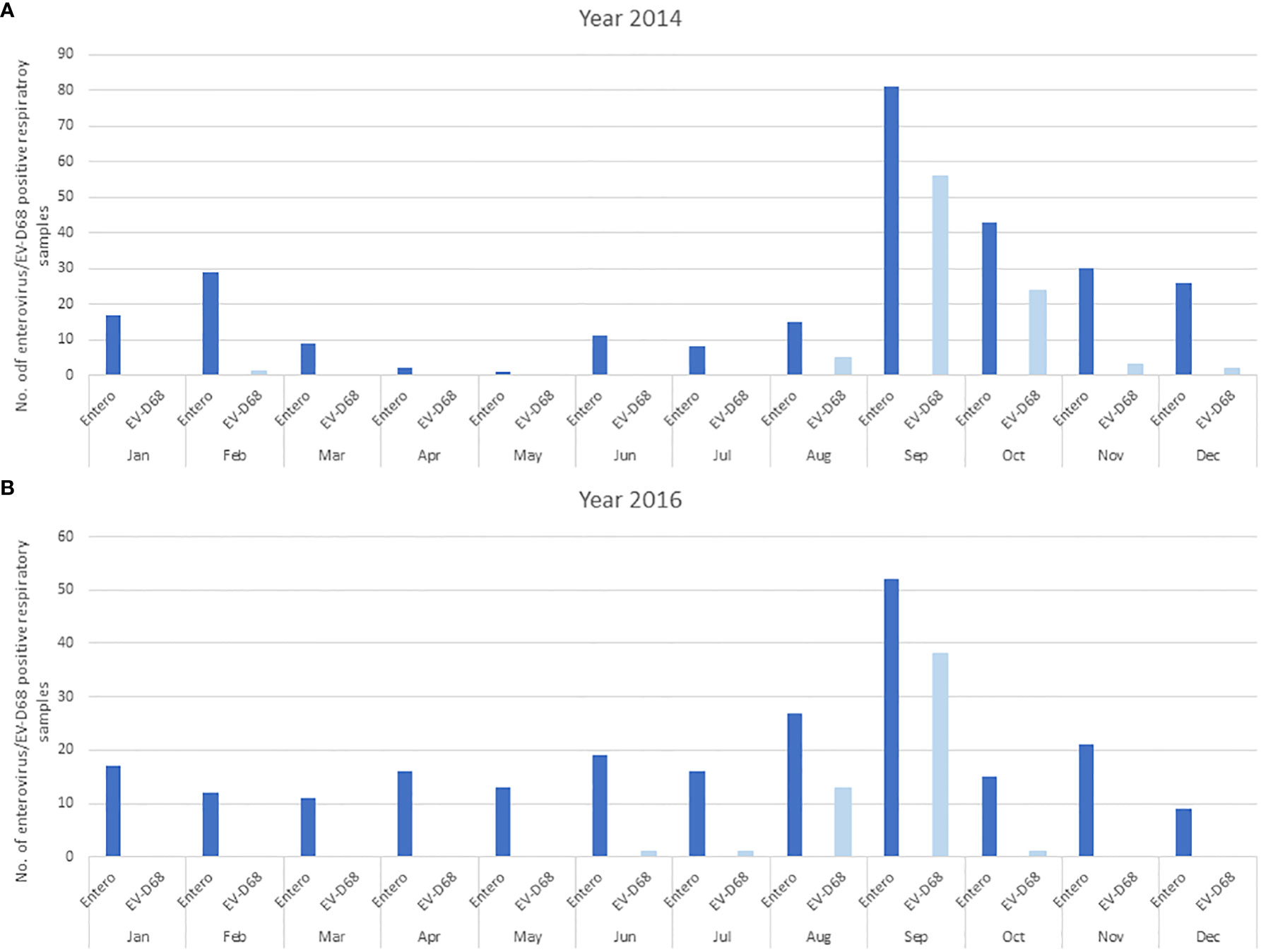
Figure 2 The distribution of enterovirus and EV-D68 infections in hospitalized and non-hospitalized children is presented monthly in the two epidemic years 2014 (A) and 2016 (B). The dark blue bars represent all EVs detected, and light blue EV-D68 detected.
In the 9-year span from 2014 to 2022, 154 children with EV-D68 infection were confirmed. Most patients were young children, with a median age of 3 years and an interquartile range of 1 to 4 years. Out of 154 children, 123 (79.9%) required hospitalization, and 31 (21.1%) were solely examined as outpatients (non-hospitalized) and did not need additional diagnostics, treatment, or hospitalization due to a mild disease course.
Dyspnea was the most frequent clinical finding, occurring in 82.5% of 154 EV-D68–positive cases. LRTI was diagnosed in 89.0% of children. Bronchitis and bronchiolitis were the most common diagnoses, accounting for 33.1% and 24.0% of the patients, respectively. Almost half of the patients (44.2%) required oxygen therapy. All children were treated with low or high oxygen therapy, and no child received any higher mode of respiratory support, but due to the retrospective design of the study further stratification was not available in the collected data set. None of the children in our cohort needed intensive care treatment and none developed AFM. Upon admission to the hospital, laboratory tests revealed low levels of inflammatory markers, a mean CRP value of 32.9 mg/L, and a mean leukocyte count of 14.2 × 109/L. The average stay in the hospital was 2.6 days.
Altogether, 149 children (96.8%) were included due to respiratory conditions. An additional five patients with EV-D68 detection were not primarily hospitalized due to respiratory infection; one patient due to the start of chemotherapy treatment, one due to acute atopic dermatitis, and two, due to urinary tract infection. However, four patients also had an upper respiratory tract infection (URTI) at admission. One of the five patients was primarily hospitalized due to a relapse of acute disseminated encephalomyelitis (ADEM) but also had positive EV-D68 NPS, however, the patient had no respiratory symptoms (according to the child’s clinical records).
Out of 154 children, 58 had an underlying condition or chronic disease that could influence the course of the EV-D68 infection, with allergies and asthma/recurrent wheezing reported in 30 children (19.5%) and 17 children (11.0%), respectively. More detailed demographic and clinical characteristics (at admission) of EV–D68–infected hospitalized and non-hospitalized children are shown in Table 2.
In 13 out of 154 EV–D68–positive children (8.4%), other viruses or bacteria were co-detected. In six out of 13 children (3.9%), another virus was co-detected; HAdV in three children, HRV in two, and HBoV1 in one. A bacterial co-infection was clinically suspected in seven children (4.5%). Of these, three cases were confirmed microbiologically in separate tests, as bacteria are not detected in the routine respiratory virus panel. Mycoplasma pneumoniae was detected in one child, and Bordetella pertussis in one child from additional pharyngeal swabs using molecular diagnostics (using PCR). In one child, Streptococcus pyogenes was detected by bacterial culture. The other four patients had to be treated with antibiotics due to clinical and biochemical laboratory parameters that indicated a bacterial co-infection but without microbiologic investigation. The mean hospital stay of children with co-detection was 2.9 days. In children with co-detections, rhinorrhea was assessed in 11 children (84.6%) and dyspnea in seven (53.8%), and only two children (15.4%) required oxygen therapy. Nine children had LRTI, and five of them were diagnosed with pneumonia.
There were no gender or age differences between hospitalized and non-hospitalized pediatric patients with EV-D68 infection. There were also no differences in body temperature, CRP levels, leukocyte count, and the presence of rhinorrhea. However, dyspnea occurred more frequently in hospitalized patients and the difference was statistically significant (chi-square; p<0.0001). In addition, hospitalized patients were significantly more likely to be diagnosed with LRTI, and non-hospitalized patients were significantly more likely to be diagnosed with URTI (chi-square; p<0.0001), Table 2.
Among the 154 EV–D68–positive patients, we randomly selected samples of 12 patients from each epidemic year (2014 and 2016) and all nine samples in which EV-D68 was detected after 2016. Suitable sequence quality was obtained from all 33 samples; 28 were obtained from hospitalized and 5 from non-hospitalized pediatric patients. The phylogenetic analysis on the one hand revealed high similarity or even 100% sequence identity with other European or American EV-D68 isolates obtained in comparative years, but at the same time, it revealed the presence of many variants within the same year, as shown in detail in Figure 3. After sequence quality trims, 534 bp partial VP1 EV-D68 gene sequences were obtained and deposited in GenBank under accession Nos.: OR365002–OR365034. Circulation of different EV-D68 strains belonging to several clades throughout the study period was observed. Out of 12 partial VP1 sequences from 2014, nine belonged to subclade B2 and three sequences to subclade B1. All 12 partial VP1 sequences from 2016 belonged to subclade B3, as did all the sequences from 2018, 2019, and 2022. Interestingly, there was one patient from 2014 for whom at position 490 in the sequenced VP1 gene both adenine (A) and guanine (G) were detected, showing a single nucleotide polymorphism (SNP); as seen in the both sequencing traces with CLC Main Workbench 7. For this particular sample, sequencing was later repeated three independent times and the same results were obtained, suggesting the presence of two subpopulations of the virus.

Figure 3 Phylogenetic tree of EV-D68 VP1 partial sequences (534 bp long) from hospitalized and non-hospitalized children in Slovenia. A partial polyprotein region was inferred by using the Neighbor-joining method under the Tamura–Nei substitution model. Evolutionary analyses were conducted in MEGA X. The sequences obtained from different years (reference no. OR365002–OR365034) were aligned with the sequences circulating in the same years with accession numbers that were selected from GenBank (OP321143, OP572095, OP572088, OP321146, OQ160431, MK419065, MK491182, MN896983, MN896982, MN896985, MN896986, MN812202, OM831204, OK085437, OK085464, MN809623, MN245441, MK121717, MH674156, ON006421, OM831187, OM831202, OP267497, MN726800, MN935870, MN896984, LR743440, MN896976, MN896977, LR743439, MN894268, MN896974, MN896981, LT745912, MT791927, LN681320, AB601883, LR743438, LR743441, NC_038308; encompassing countries: USA, Australia, Japan, France, Germany, Denmark, Netherlands and Sweden). Numbers along branches are bootstrap values. subclades were defined by using Enterovirus Genotyping Tool (23).
Since its discovery, EV-D68 has mostly been described sporadically in isolated cases or small breakouts of respiratory illnesses, with overall mild clinical presentation. Interest in EV-D68 infections arose in 2014 when large outbreaks of severe EV-D68 respiratory infections were detected in the United States, Canada, and Europe (6, 12, 24–26), and due to its potential neurotropism with a significant increase in AFM cases reported compared to non-outbreak years (27–31).
Before this study, no data were available on EV-D68 circulation for our region. We now show that high rates of EV-D68 cases among enterovirus infections in hospitalized and non-hospitalized children were present in 2014 and 2016. After 2016 and until the end of this study, only sporadic cases remained. No EV-D68 infections were seen in hospitalized pediatric patients in 2015 and 2017, in agreement with its biennial circulation pattern already observed previously (2, 12, 32, 33). Similarly, its disruption in 2020 due to the SARS-CoV-2 epidemic and then the upsurge of EV-D68 infections in some European countries in 2021 after easing COVID-19 preventive measurements (21) also seem to agree with our data. However, a more in-depth analysis in Slovenia interestingly shows that EV-D68’s biennial circulation pattern, with the highest peaks in 2014 (59.1%) and in 2016 (35.1%), broke down after 2017, with only sporadic cases in 2018, 2019, and 2022. Namely, instead of a clear biennial circulation, the disruption of this typical pattern in Slovenia appears to have happened much earlier than the emergence of SARS-CoV-2, as would have been expected.
Unlike in some European countries (2, 12), only three cases of EV-D68 were seen in 2018 and four in 2019, and no evidence of EV-D68 circulation in hospitalized and non-hospitalized Slovenian pediatric patients was observed in 2020 and 2021. Furthermore, in 2022 EV-D68 infection was seen in only two hospitalized pediatric patients from the central Slovenian region. We can only speculate on the reason for this observation, but most probably it is connected to both SARS-CoV-2 non-pharmaceutical interventions and the strong laboratory focus on solely SARS-CoV-2 diagnostics. Although in 2018 AusDiagnostics platform was introduced into routine diagnostics of respiratory viruses this decrease does not seem to be linked to the platform change. Namely, during the evaluation process, we found AusDiagnostics 16-well respiratory panel to be better in distinguishing between enteroviruses and rhinoviruses. In fact, many rhinoviruses detected with the in-house assay turned out to be enteroviruses, which was also confirmed with sequencing. Nevertheless, the number of tests for enterovirus detection and the consequent number of positives are almost one-fourth and one-tenth of the level in pre–COVID-19 years, respectively. In fact, in 2022, when the number of tests and positives rose to a similar extent as in 2019, EV-D68 cases were again observed. Follow-up studies are needed to better explain whether the disappearance was due to COVID-19 or to other EV-D68–related factors, like mutations that could lead to decreased virus fitness as well as decreased pathogenicity.
In a study of EV-D68 reemergence in Europe after the COVID-19 lockdown in 2021, coinfections with other respiratory viruses were reported, but the study does not report possible bacterial coinfections (12). In the present study, co-detections with other respiratory viruses and bacteria were infrequently detected; only in 8.4% (3.9% with virus and 4.5% with bacteria) of EV-D68–positive children. A similar result among hospitalized children was previously published in a study from France (34).
In our study, LRTI was identified in 89.0% of studied EV-D68–positive pediatric patients, with the difference being statistically significant between hospitalized and non-hospitalized patients (94.3% vs. 67.7%; chi-square, p<0.0001). All the patients studied exhibited symptoms of respiratory infection except one, who was hospitalized due to ADEM. Therefore, we believe this was the only patient in which the EV-D68 was detected as an innocent bystander and not a cause of infection. In all other 153 cases, EV-D68 was the cause of respiratory infection, whether detected as a sole pathogen or in co-detection with another virus or bacteria.
Furthermore, dyspnea was observed in 82.5% of the children in our study; with the difference being statistically significant between hospitalized and non-hospitalized patients (91.9% vs. 45.2%; chi-square, p<0.0001), and 44.2% of them required oxygen therapy. Overall, 37.7% of pediatric patients had some kind of preexisting (underlying) condition, with asthma being assessed most frequently, which is also a recognized risk factor for severe clinical presentations (35–37) in EV-D68-positive patients. However, there was no difference in frequency of underlying diseases between hospitalized and non-hospitalized patients (38.5% vs. 35.5%; chi-square, p=0.6896), as well as no difference in the clinical presentation of EV-D68 infection between the patients with underlying diseases and without. We also have to mention the high percentage of children needing hospitalization in our study, which is probably due to the fact that all children referred to our tertiary clinic are seen beforehand by a pediatrician at the primary level, and only those that are assessed as needing additional diagnostics or hospitalization are then referred to our center.
We acknowledge the significance of EV-D68 in individuals with asthma and the associated apprehensions regarding EV-D68 infection, as outlined in current literature (38, 39). However, the retrospective chart review methodology in data collection presents inherent limitations in information gathering. Clinical insights from our study indicate a notably low prevalence of physician-diagnosed asthma within our study group. As a result, the utilization of bronchodilators or inhaled steroids was infrequent, and their impact on overall results was not clinically significant. In contrast to American studies from 2014, in which 59% of patients studied with EV-D68 infection required ICU admission, none of our pediatric patients required ICU treatment and none developed AFM (11). Similarly, a Spanish study on EV-D68 in hospitalized children between 2014 and 2021 showed a high proportion of children with LRTI. However, in contrast to our findings, almost 12% of the children they studied required ICU admission, and one patient developed AFM (2). Previous studies suggested that the ability to invade and replicate in the CNS is not a clade-specific feature; nevertheless, some studies have shown clade B3 isolates to cause CNS complications (18, 19, 40). Although clade B3 dominated in our study in 2016 and the majority of our children were hospitalized (79.9%), none of them required ICU treatment and none developed AFM. Moreover, during the study period there were no suspected AFM patients.
Three different clades of EV-D68 (A–C) have been described, and subclades A1, A2/D, B1, B2, and B3 cocirculate variably (8, 14). In our study, subclade B2 predominated over the subclade B1 in 2014, the results being in line with the findings of a French study (16, 34). In the following seasons, subclade B3 completely predominated, which was also seen in other European countries (2, 34). Some phylogenetic analyses showed a global rise in subclades B3 and D1 between 2009 and 2010, but in 2018 D1 reemerged (9, 34). After 2018, EV-D68 was detected in only nine hospitalized pediatric patients and no circulation of subclade D1 was seen in our study. One reason could lie in a very low number of EV-D68 cases detected, but another could be due to the nature of the study focusing on the pediatric population. Moreover, the children included in our study were from the central part of Slovenia and, even though Slovenia is a small country, children from three other pediatric hospitals that cover other geographical areas were not included. The inclusion of the adult population (especially immunocompromised) might result in a larger number of EV-D68–positive patients and thus also a greater variety in cocirculating EV-D68 variants and subclades, as observed by others (9, 34). In Slovenia, Sanger sequencing of enteroviruses from clinical samples (enterovirus typing) is still only used for research purposes and is not part of routine diagnostics. Therefore, we believe that the introduction of efficient and standardized laboratory characterization of circulating EV-D68 strains for a better understanding of EV-D68 and the disease it causes is an important step toward continuous global surveillance activities.
We acknowledge some study limitations. One is the retrospective nature of clinical data assessment of EV-D68–positive hospitalized patients, which may not be of the same quality if they were studied prospectively. Another limitation of the study is not testing the samples from children with suspected neurological disease for EV-D68. The study does not include adults and the elderly. Last but not least, we are aware of the phylogenetic analysis limitation derived from partial VP1 sequencing only.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR365002–OR365034.
The studies involving humans were approved by National Medical Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
TU: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. MV: Conceptualization, Data curation, Formal analysis, Investigation, Writing – review & editing. RK: Formal analysis, Investigation, Supervision, Validation, Writing – original draft, Writing – review & editing. UK: Conceptualization, Supervision, Validation, Visualization, Writing – review & editing. JP: Data curation, Formal analysis, Writing – review & editing. MM: Conceptualization, Data curation, Formal analysis, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. SL: Conceptualization, Data curation, Formal analysis, Writing – review & editing. MP: Conceptualization, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Slovenian Research and Innovation Agency (Research Program P3-0083). This study was approved by the National Medical Ethics Committee of Slovenia (no. 0120-56/2023/3).
The authors express their gratitude to Viktorija Mlinšek for her excellent laboratory work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Schieble JH, Fox VL, Lennette EH. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol (1967) 85(2):297–310. doi: 10.1093/oxfordjournals.aje.a120693
2. Andres C, Vila J, Creus-Costa A, Pinana M, Gonzalez-Sanchez A, Esperalba J, et al. Enterovirus D68 in hospitalized children, barcelona, Spain, 2014-2021. Emerg Infect Dis (2022) 28(7):1327–31. doi: 10.3201/eid2807.220264
3. Sooksawasdi Na Ayudhya S, Laksono BM, van Riel D. The pathogenesis and virulence of enterovirus-D68 infection. Virulence (2021) 12(1):2060–72. doi: 10.1080/21505594.2021.1960106
4. Waghmare A, Pergam SA, Jerome KR, Englund JA, Boeckh M, Kuypers J. Clinical disease due to enterovirus D68 in adult hematologic Malignancy patients and hematopoietic cell transplant recipients. Blood (2015) 125(11):1724–9. doi: 10.1182/blood-2014-12-616516
5. Giombini E, Rueca M, Barberi W, Iori AP, Castilletti C, Scognamiglio P, et al. Enterovirus D68-associated acute flaccid myelitis in immunocompromised woman, Italy. Emerg Infect Dis (2017) 23(10):1690–3. doi: 10.3201/eid2310.170792
6. Poelman R, Schuffenecker I, Van Leer-Buter C, Josset L, Niesters HG, Lina B, et al. European surveillance for enterovirus D68 during the emerging North-American outbreak in 2014. J Clin Virol (2015) 71:1–9. doi: 10.1016/j.jcv.2015.07.296
7. Schuster JE, Miller JO, Selvarangan R, Weddle G, Thompson MT, Hassan F, et al. Severe enterovirus 68 respiratory illness in children requiring intensive care management. J Clin Virol (2015) 70:77–82. doi: 10.1016/j.jcv.2015.07.298
8. Tokarz R, Firth C, Madhi SA, Howie SRC, Wu W, Sall AA, et al. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol (2012) 93(Pt 9):1952–8. doi: 10.1099/vir.0.043935-0
9. Bal A, Sabatier M, Wirth T, Coste-Burel M, Lazrek M, Stefic K, et al. Emergence of enterovirus D68 clade D1, France, August to November 2018. Euro Surveill (2019) 24(3). doi: 10.2807/1560-7917.ES.2019.24.3.1800699
10. Pellegrinelli L, Giardina F, Lunghi G, Uceda Renteria SC, Greco L, Fratini A, et al. Emergence of divergent enterovirus (EV) D68 sub-clade D1 strains, northern Italy, September to October 2018. Euro Surveill (2019) 24(7). doi: 10.2807/1560-7917.ES.2018.24.7.1900090
11. Greninger AL, Naccache SN, Messacar K, Clayton A, Yu G, Somasekar S, et al. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect Dis (2015) 15(6):671–82. doi: 10.1016/S1473-3099(15)70093-9
12. Benschop KS, Albert J, Anton A, Andres C, Aranzamendi M, Armannsdottir B, et al. Re-emergence of enterovirus D68 in Europe after easing the COVID-19 lockdown, September 2021. Euro Surveill (2021) 26(45). doi: 10.2807/1560-7917.ES.2021.26.45.2100998
13. Shah MM, Perez A, Lively JY, Avadhanula V, Boom JA, Chappell J, et al. Enterovirus D68-Associated Acute Respiratory Illness horizontal line New Vaccine Surveillance Network, United States, July-November 2018-2020. MMWR Morb Mortal Wkly Rep (2021) 70(47):1623–8. doi: 10.15585/mmwr.mm7047a1
14. Midgley SE, Benschop K, Dyrdak R, Mirand A, Bailly JL, Bierbaum S, et al. Co-circulation of multiple enterovirus D68 subclades, including a novel B3 cluster, across Europe in a season of expected low prevalence, 2019/20. Euro Surveill (2020) 25(2). doi: 10.2807/1560-7917.ES.2020.25.2.1900749
15. Fall A, Gallagher N, Morris CP, Norton JM, Pekosz A, Klein E, et al. Circulation of enterovirus D68 during period of increased influenza-like illness, maryland, USA, 2021. Emerg Infect Dis (2022) 28(7):1525–7. doi: 10.3201/eid2807.212603
16. Fall A, Han L, Abdullah O, Norton JM, Eldesouki RE, Forman M, et al. An increase in enterovirus D68 circulation and viral evolution during a period of increased influenza like illness, The Johns Hopkins Health System, USA, 2022. J Clin Virol (2023) 160:105379. doi: 10.1016/j.jcv.2023.105379
17. Hixon AM, Clarke P, Tyler KL. Evaluating treatment efficacy in a mouse model of enterovirus D68-associated paralytic myelitis. J Infect Dis (2017) 216(10):1245–53. doi: 10.1093/infdis/jix468
18. Rosenfeld AB, Warren AL, Racaniello VR. Neurotropism of enterovirus D68 isolates is independent of sialic acid and is not a recently acquired phenotype. mBio (2019) 10(5). doi: 10.1128/mBio.02370-19
19. Sooksawasdi Na Ayudhya S, Meijer A, Bauer L, Oude Munnink B, Embregts C, Leijten L, et al. Enhanced enterovirus D68 replication in neuroblastoma cells is associated with a cell culture-adaptive amino acid substitution in VP1. mSphere (2020) 5(6). doi: 10.1128/mSphere.00941-20
20. Ursic T, Miksic NG, Lusa L, Strle F, Petrovec M. Viral respiratory infections in a nursing home: a six-month prospective study. BMC Infect Dis (2016) 16(1):637. doi: 10.1186/s12879-016-1962-8
21. Jevsnik Virant M, Ursic T, Kogoj R, Korva M, Petrovec M, Avsic-Zupanc T. Evaluation of two broadly used commercial methods for detection of respiratory viruses with a recently added new target for detection of SARS-coV-2. Viruses (2022) 14(7). doi: 10.3390/v14071530
22. Wylie KM, Wylie TN, Orvedahl A, Buller RS, Herter BN, Magrini V, et al. Genome sequence of enterovirus D68 from St. Louis, Missouri, USA. Emerg Infect Dis (2015) 21(1):184–6. doi: 10.3201/eid2101.141605
23. Kroneman A, Vennema H, Deforche K, v d Avoort H, Penaranda S, Oberste MS, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol (2011) 51(2):121–5. doi: 10.1016/j.jcv.2011.03.006
24. Meijer A, Benschop KS, Donker GA, van der Avoort HG. Continued seasonal circulation of enterovirus D68 in the Netherlands, 2011-2014. Euro Surveill (2014) 19(42). doi: 10.2807/1560-7917.es2014.19.42.20935
25. Rahamat-Langendoen J, Riezebos-Brilman A, Borger R, van der Heide R, Brandenburg A, Scholvinck E, et al. Upsurge of human enterovirus 68 infections in patients with severe respiratory tract infections. J Clin Virol (2011) 52(2):103–6. doi: 10.1016/j.jcv.2011.06.019
26. Midgley CM, Jackson MA, Selvarangan R, Turabelidze G, Obringer E, Johnson D, et al. Severe respiratory illness associated with enterovirus D68 - Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep (2014) 63(36):798–9. doi: 10.1111/ajt.13035
27. Lang M, Mirand A, Savy N, Henquell C, Maridet S, Perignon R, et al. flaccid paralysis following enterovirus D68 associated pneumonia, France, 2014. Euro Surveill (2014) 19(44). doi: 10.2807/1560-7917.es2014.19.44.20952
28. Messacar K, Schreiner TL, Maloney JA, Wallace A, Ludke J, Oberste MS, et al. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet (2015) 385(9978):1662–71. doi: 10.1016/S0140-6736(14)62457-0
29. Pfeiffer HC, Bragstad K, Skram MK, Dahl H, Knudsen PK, Chawla MS, et al. Two cases of acute severe flaccid myelitis associated with enterovirus D68 infection in children, Norway, autumn 2014. Euro Surveill (2015) 20(10):21062. doi: 10.2807/1560-7917.es2015.20.10.21062
30. Esposito S, Lunghi G, Zampiero A, Tagliabue C, Orlandi A, Torresani E, et al. Enterovirus-D68 in the cerebrospinal fluid of two children with aseptic meningitis. Pediatr Infect Dis J (2016) 35(5):589–91. doi: 10.1097/INF.0000000000001085
31. Aliabadi N, Messacar K, Pastula DM, Robinson CC, Leshem E, Sejvar JJ, et al. Enterovirus D68 infection in children with acute flaccid myelitis, colorado, USA, 2014. Emerg Infect Dis (2016) 22(8):1387–94. doi: 10.3201/eid2208.151949
32. Kramer R, Sabatier M, Wirth T, Pichon M, Lina B, Schuffenecker I, et al. Molecular diversity and biennial circulation of enterovirus D68: a systematic screening study in Lyon, France, 2010 to 2016. Euro Surveill (2018) 23(37). doi: 10.2807/1560-7917.ES.2018.23.37.1700711
33. Messacar K, Pretty K, Reno S, Dominguez SR. Continued biennial circulation of enterovirus D68 in Colorado. J Clin Virol (2019) 113:24–6. doi: 10.1016/j.jcv.2019.01.008
34. Duval M, Mirand A, Lesens O, Bay JO, Caillaud D, Gallot D, et al. Retrospective study of the upsurge of enterovirus D68 clade D1 among adults (2014-2018). Viruses (2021) 13(8). doi: 10.3390/v13081607
35. Itagaki T, Aoki Y, Matoba Y, Tanaka S, Ikeda T, Mizuta K, et al. Clinical characteristics of children infected with enterovirus D68 in an outpatient clinic and the association with bronchial asthma. Infect Dis (Lond) (2018) 50(4):303–12. doi: 10.1080/23744235.2017.1400176
36. Korematsu S, Nagashima K, Sato Y, Nagao M, Hasegawa S, Nakamura H, et al. "Spike" in acute asthma exacerbations during enterovirus D68 epidemic in Japan: A nation-wide survey. Allergol Int (2018) 67(1):55–60. doi: 10.1016/j.alit.2017.04.003
37. Foster CB, Coelho R, Brown PM, Wadhwa A, Dossul A, Gonzalez BE, et al. A comparison of hospitalized children with enterovirus D68 to those with rhinovirus. Pediatr Pulmonol (2017) 52(6):827–32. doi: 10.1002/ppul.23661
38. Midgley CM, Watson JT, Nix WA, Curns AT, Rogers SL, Brown BA, et al. Severe respiratory illness associated with a nationwide outbreak of enterovirus D68 in the USA (2014): a descriptive epidemiological investigation. Lancet Respir Med (2015) 3(11):879–87. doi: 10.1016/S2213-2600(15)00335-5
39. Moyer K, Wang H, Salamon D, Leber A, Mejias A. Enterovirus D68 in hospitalized children: sequence variation, viral loads and clinical outcomes. PloS One (2016) 11(11):e0167111. doi: 10.1371/journal.pone.0167111
Keywords: Enterovirus D68, epidemic, seasonality, PCR detection, clinical presentation, sequencing, phylogenetic analysis
Citation: Uršič T, Jevšnik Virant M, Kogoj R, Krivec U, Prusnik J, Mramor M, Lovšin S and Petrovec M (2024) Enterovirus D68 circulation between 2014 and 2022 in Slovenian children. Front. Virol. 4:1335752. doi: 10.3389/fviro.2024.1335752
Received: 09 November 2023; Accepted: 10 January 2024;
Published: 22 January 2024.
Edited by:
Kimberley Benschop, National Institute for Public Health and the Environment, NetherlandsReviewed by:
Hai Nguyen-Tran, University of Colorado, United StatesCopyright © 2024 Uršič, Jevšnik Virant, Kogoj, Krivec, Prusnik, Mramor, Lovšin and Petrovec. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tina Uršič, dGluYS51cnNpY0BtZi51bmktbGouc2k=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.