- 1Department of Infection Biology, Faculty of Medicine, University of Tsukuba, Tsukuba, Japan
- 2Graduate School of Comprehensive Human Sciences, University of Tsukuba, Tsukuba, Japan
- 3Transborder Medical Research Center, University of Tsukuba, Tsukuba, Japan
- 4Microbiology Research Center for Sustainability, University of Tsukuba, Tsukuba, Japan
Introduction: The influenza virus genome consists of single-stranded RNAs and forms viral ribonucleoprotein (RNP) complexes. After viral genome replication in the nucleus, the viral RNP interacts with viral protein M1. The M1-viral RNP complex is exported to the cytoplasm via the CRM1-dependent pathway using NS2/NEP as an export adaptor protein. NEP is a 14 kDa protein and diffusely localizes in the nucleus and cytoplasm. Upon binding to the NLS motif of M1, NEP inhibits the nuclear accumulation of M1 and promotes the nuclear export of M1-viral RNP complex. However, the detail mechanism by which NEP binds to M1 only in the nucleus remains unclear.
Methods: To visualize the interaction of NEP with M1 in the formation of vRNP export complexes, we performed in situ proximity ligation assays. The close proximity of N-terminal and C-terminal domains of NEP was tested by split Renilla luciferase complementation assays in which the N-terminal and C-terminal fragments of Renilla luciferase were fused to the N-terminus and C-terminus of NEP, respectively.
Results and discussion: We found that the intramolecular interaction of NEP inhibits the interaction of NEP with M1. The intramolecular interaction of NEP was mediated through the interaction of the N-terminal NES motif with the M1-binding domain at the C-terminus. By adding leptomycin B, a potent inhibitor of CRM1, the interaction of NEP with M1 was impaired. These results suggest that CRM1 disrupts the intramolecular interaction of NEP by recognizing the NES motif at the N-terminus of NEP, thereby promoting the interaction of NEP with M1. We also found that NEP mutant deficient in the intramolecular interaction was co-localized with M1 at the plasma membrane and did not show nuclear localization with M1. Based on these results, we propose that the intramolecular interaction of NEP regulated by CRM1 ensures the unidirectional transport of M1.
1 Introduction
The nuclear export process is typically mediated by nuclear export receptors and Ran GTPase cycle (1). CRM1/XPO1 is one of the nuclear export receptors responsible for the nuclear export of many different cargo proteins by recognizing the nuclear export signal (NES) consisting of conserved hydrophobic residues. Some RNA cargos, including tRNA and miRNA, are directly recognized by export receptors for their RNA secondary structures, but other RNA species require adaptor proteins harboring NES with RNA binding activity for recruiting export receptors (2). CRM1 is involved in the nuclear export of ribosomal RNA and small nuclear (sn)RNA through adaptor proteins, such NMD3 and PHAX (3–5). The nuclear export machinery is recruited to the RNA cargos after RNA processing and maturation as a quality control mechanism for RNAs.
Influenza A virus (IAV) has single-stranded RNA genomes of negative polarity. The replication of virus genome takes place in the nucleus. The replicated virus genome forms a viral ribonucleoprotein complexes (vRNP) with viral RNA polymerases and nucleoprotein (NP). The progeny vRNP interacts with viral matrix protein M1 and the M1-vRNP complexes are exported from the nucleus by the CRM1-dependent pathway through NS2/NEP adaptor protein (6–8). NEP is a monomeric 14-kDa protein consisting of N-terminal domain (1-53 a.a. position) and C-terminal domain (54-112 a.a. position). The N-terminal domain is highly mobile and is in an exposed state (9), and contains a cluster of hydrophobic residues between amino acid positions 12 and 21 that function as NES for CRM1 binding (10–12). The C-terminal domain of NEP forms a relatively rigid conformation consisting of two α-helices (helix C1 and C2) that assemble an antiparallel amphiphilic hairpin structure (13). The hydrophilic surface of the amphiphilic hairpin interacts with the basic nuclear localization signal (NLS) of M1 through the exposed Trp78 surrounded by a cluster of glutamic acid residues in the C-terminal domain of NEP (13).
Although the mechanism of apical transport of M1 remains unclear (14–18), M1 protein mainly accumulates beneath the plasma membrane and functions as a scaffold of viral particles budding from the plasma membrane. A part of the M1 protein shuttles into the nucleus in an NLS-dependent manner (19), and terminates the viral genome replication to destine the newly-replicated vRNP to the nuclear export (20). Most M1 proteins localize to the plasma membrane, however, NEP is not recruited to the plasma membrane and localizes diffusely in the nucleus and the cytoplasm due to its low molecular weight (6), indicating that the recognition of M1 by NEP is restricted in the nucleus possibly for efficient nuclear export of vRNP. However, the detailed mechanism of the viral RNP export complex formation remains unclear.
Here, we found that NEP interacts with the M1 in a CRM1-dependent manner. The N-terminal domain of NEP was in close intramolecular proximity to the C-terminal domain. The intramolecular interaction is mediated by the N-terminal NES motif and the hydrophobic surface of the C-terminal M1-binding domain. NEP harboring I76T/L106T mutation in the M1-binding domain, which does not form the intramolecular interaction, is co-localized with M1 at the plasma membrane, without nuclear localization of M1 as well as NEP, indicating that the intramolecular interaction of NEP inhibits the interaction with M1 prior to the nuclear import, and that CRM1 releases this inhibition by recognizing the NES motif of NEP in the nucleus. Our results demonstrate the molecular mechanism of NEP recruitment to the M1-vRNP complexes regulated by CRM1 to ensure the unidirectional transport of M1.
2 Methods
2.1 Biological materials
A rabbit polyclonal antibody against M1 and a rat polyclonal antibody against NEP were prepared as previously described (21). A rabbit monoclonal antibody against β-actin (CST; 8547) was purchased. HeLa cells present in this study were obtained from ATCC. HeLa cells and HeLa-GFP-emerin cells (22) were grown in Dulbecco’s minimal essential medium (DMEM) containing 10% bovine fetal calf serum and incubated at 37°C in 5% CO2. pPol-I-M and pPol-I-NS were kindly gifted by Dr. Fumitaka Momose (Kitasato University). pCAGGS-PB1, pCAGGS-PB2, pCAGGS-PA, and pCAGGS-NP were prepared as previously reported (23). For the construction of plasmid expressing N-terminal or C-terminal Renilla luciferase-fused NEP, FLAG-tagged NEP cDNA, which was amplified with primers, 5’-CTGCTAGCGCCGCCACCATGGACTACAAGGACGACGATGACAAGATGGACCCAAACACTGTGTC-3’ and 5’-CCGGAATTCTTAAATAAGCTGAAACGAGAAAG-3’, was cloned into pcDNA3.1-NR, pcDNA3.1-CR, or pcDNA3.1-NR-CR plasmids (24). The NES motif of NEP was mutated by the PCR-based site-directed mutagenesis using primers, 5’-GGACGTCAAAAACGCAGTCGGAGTCC-3’ and 5’- TCTGCTGTGTGTCCTGGAAGAGAAGG-3’. For the construction of plasmid expressing delNES mutant harboring additional NES motif at the C-terminus (exoNES), NEP cDNA was amplified with primers, 5’-CCAAGGATCCCATATGGACCCAAACACTGTGTCAAGC-3’ and 5’-CCAAGGATCCAATAAGCTGAAACGAGAAAGTTC-3’. Influenza A/Puerto Rico/8/34 (A/PR/8/34) virus was grown at 35.5°C for 48 h in allantoic sacs of 11-days-old embryonated eggs, and then the infected allantoic fluids were collected and stored at -80°C until use.
2.2 Intracellular localization of viral proteins and viral genome
Indirect immunofluorescence assays and fluorescence in situ hybridization (FISH) assays were carried out as previously described (25). Briefly, cells were fixed with 4% PFA for 10 min. After being washed with PBS, cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min. After incubation in PBS containing 1% bovine serum albumin for 30 min, the coverslips were incubated with each antibody for 1 h and then further incubated with Alexa Fluor 488- and 568-conjugated secondary antibodies, respectively (Life Technologies). FISH assays were performed after indirect immunofluorescence assays using an RNA probe complementary to the segment 8 virus genome. PLA was carried out using Duolink In Situ PLA kit (Olink Bioscience) according to the manufacturer’s protocol. The number of the PLA signals was measured with reconstructed 3D images from z-stack series using IMARIS software (Carl Zeiss). Images were acquired by a confocal laser scanning microscopy (LSM700; Carl Zeiss) using ×63 Apochromat objective (NA = 1.4).
2.3 Split Renilla luciferase complementation assay
HeLa cells were transfected with the pcDNA3.1 plasmid encoding N-terminal (NR; 1-229 a.a.) and/or C-terminal (CR; 230-311 a.a.) fragment of Renilla luciferase fused to NEP. At 24 h post-transfection, cells were collected and lysed in a cell lysis buffer for Renilla luciferase (Promega) by three freezing-thawing cycles. Renilla luciferase activity was measured according to the manufacturer’s instruction (Promega). Relative luminescence intensity was measured for 10 sec with a Mini Lumat (Berthold Technologies).
3 Results
3.1 CRM1 promotes the interaction of NEP with M1
To visualize the interaction of NEP with M1 in the formation of vRNP export complexes, we performed in situ proximity ligation assays (PLA) with rat polyclonal anti-NEP and rabbit polyclonal anti-M1 antibodies in IAV-infected HeLa cells expressing GFP-emerin, a marker of the nuclear envelope. In the antibody-based PLA system, when a pair of DNA primer-conjugated secondary antibodies is in close proximity, the primers hybridize with a connector oligo and serve as a template for rolling circle amplification, which can generate DNA targets for a fluorescently labeled DNA probe (Figure 1A and Supplementary Figure 1). Strong punctate PLA signals (red) were observed in the nucleus and the cytoplasm of infected cells (Figure 1B). The number of PLA signals in the nucleus and cytoplasm were counted from the reconstructed z-stack images, respectively. We found that the number of PLA signals increased in both the nucleus and cytoplasm along with the progression of infection, but at 11 h post-infection, most PLA signals were observed in the cytoplasm (Figure 1C). We next examined the close proximity between NEP and M1 in the presence of leptomycin B (LMB), a potent inhibitor of the interaction between CRM1 and its cargo protein through NES motif (Figure 2). At 5 h post-infection, before the nuclear export of vRNP, infected HeLa cells were treated with 10 nM LMB for 2 h and subjected to indirect immunofluorescence assays (Figure 2A) and in situ PLA with anti-NEP and anti-M1 antibodies (Figures 2B, C). The intracellular localization of NEP was unchanged by the addition of LMB, possibly due to the passive diffusion of NEP in the nucleus and cytoplasm (Figure 2A, green). In contrast, M1 was mainly localized to the cytoplasm and cell periphery in the absence of LMB, but the level of M1 protein increased in the nucleus by LMB treatment (Figure 2A, red). The nuclear accumulation of M1 may be due to the inhibition of NEP-mediated nuclear export of M1, which has an NLS motif and is actively imported to the nucleus. In contrast, the localization pattern of NEP was not changed by LMB treatment, possibly due to passive diffusion of NEP by the low molecular weight (Figure 2A).
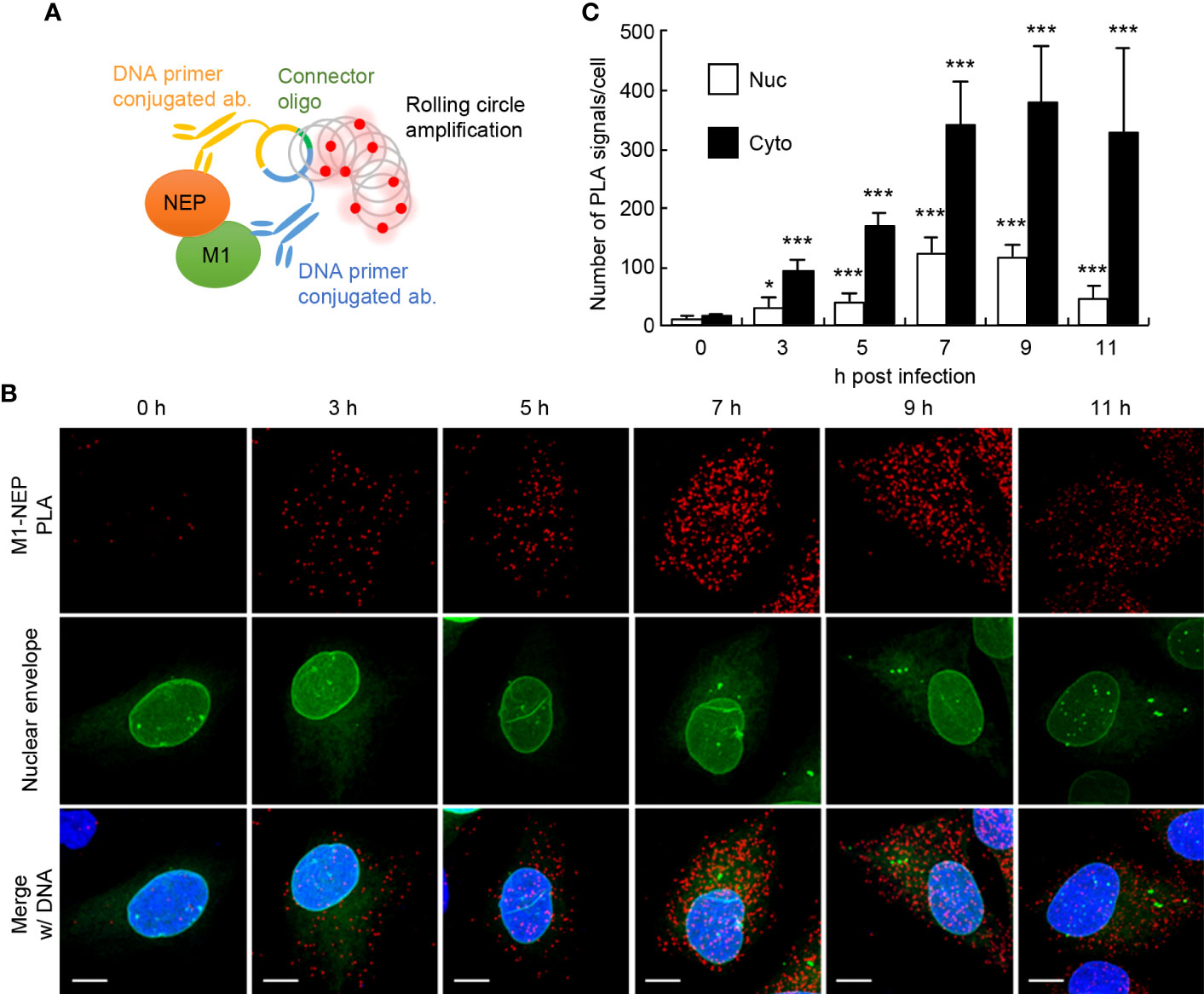
Figure 1 The close proximity between NEP and M1 proteins. HeLa cells constitutively expressing GFP-emerin as a marker of the nuclear envelope were infected with influenza virus at MOI of 10. At 0, 3, 5, 7, 9, and 11 h post-infection, the cells were subjected to in situ PLA with rat anti-NEP and rabbit anti-M1 antibodies (red). Nuclei were stained with DAPI (blue). (A) A schematic diagram the in situ PLA used to analyze the association between M1 and NEP. (B) Representative images from the three independent experiments at 0, 3, 5, 7, 9, and 11 h post-infection are shown. Scale bar, 10 μm. (C) The number of PLA signals in the nucleus and cytoplasm was counted using IMARIS software (n = 4-6 cells/group). Results are means ± SDs. *P < 0.05, ***P < 0.001; one-way ANOVA with Tukey’s test against uninfected cells.
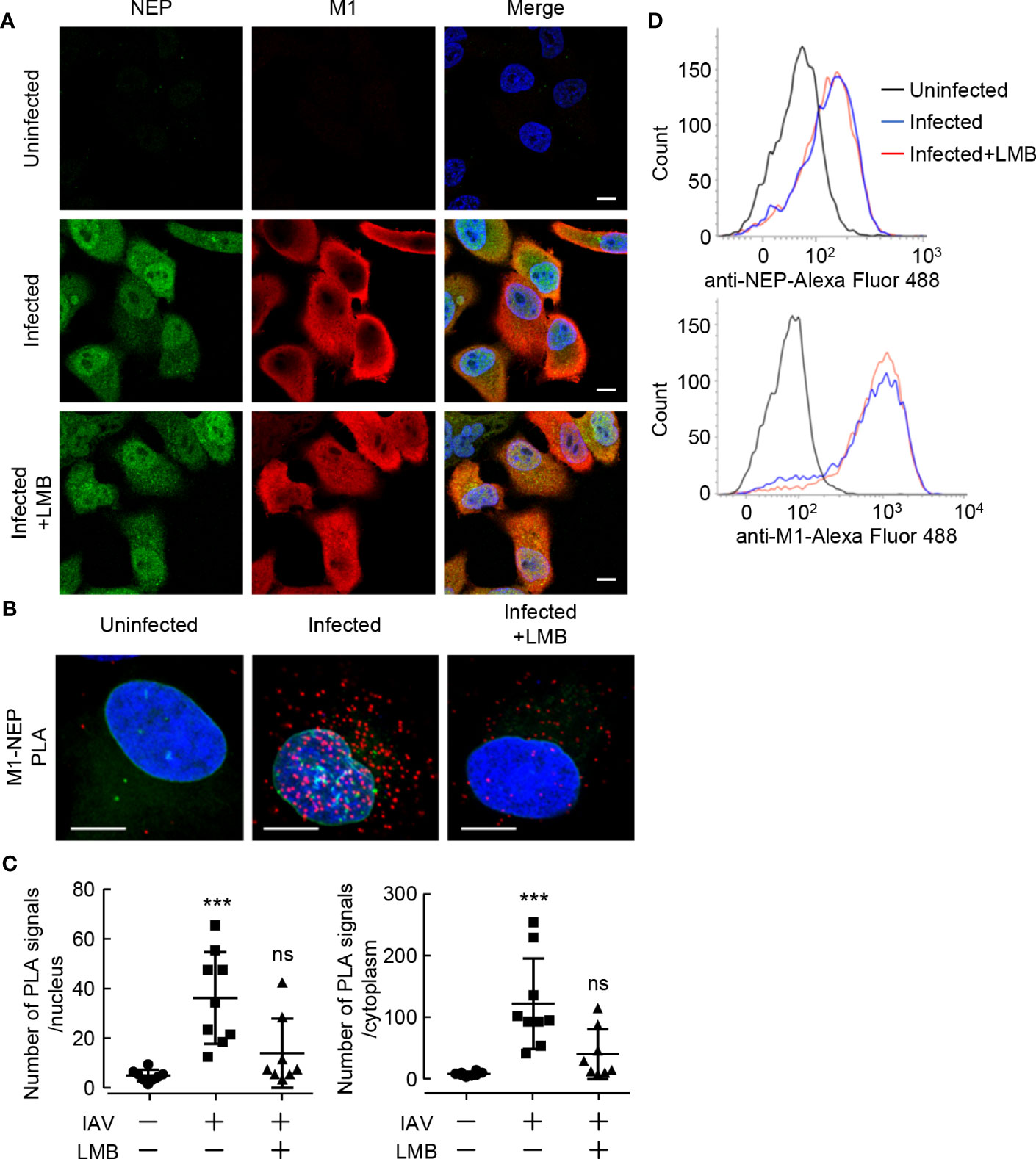
Figure 2 Leptomycin B inhibits the interaction between NEP and M1. (A) HeLa cells were infected with influenza virus at MOI of 10. At 5 h post-infection, the infected cells were treated with 10 nM LMB for 2 h. The infected cells were subjected to indirect immunofluorescence assays with anti-NEP (green) and anti-M1 antibodies (red). Nuclei were stained with DAPI (blue). Scale bar, 10 μm. (B, C) HeLa-GFP-emerin cells were infected with influenza virus at MOI of 10. At 5 h post-infection, the infected cells were treated with 10 nM LMB for 2 h. The infected cells were subjected to in situ PLA with anti-NEP and anti-M1 antibodies (red). Nuclei were stained with DAPI (blue). Scale bar, 10 μm. The numbers of PLA signals in the nucleus and cytoplasm were counted using IMARIS software (n = 8-9 cells/group), respectively. ***P < 0.001, ns, not significant; Kruskal-Wallis test against uninfected cells. (D) The expression levels of NEP and M1 were examined by FACS with anti-NEP and anti-M1 antibodies in the presence or absence of LMB.
LMB treatment significantly reduced the number of PLA signals not only in the cytoplasm but also in the nucleus (Figures 2B and C), although M1 accumulated in the nucleus (Figure 2A). This suggests that NEP requires the interaction with CRM1 via the NES motif of NEP to assemble the vRNP export complexes (NEP-M1-vRNP). The expression levels of NEP and M1 were comparable with LMB treatment for 2 h (Figure 2D).
3.2 The NES motif of NEP inhibits the interaction of NEP with M1
NEP has a NES motif consisting of multiple hydrophobic residues at amino acid positions 12 to 21 in the N-terminal mobile domain (10, 11). We next examined the interaction of M1 with NEP harboring point mutations in the NES motif (delNES mutant). The hydrophobic residues critical for NES function were mutated to hydrophilic residues to prevent the interaction of CRM1 with NEP (Figure 3A). A mutant virus defective in NES function cannot be recovered using the reverse genetics system because the NEP-mediated nuclear export of vRNP is an essential process for virus production. Thus, cells were transfected with pPol I plasmids, containing M and NS segments (pPol-I-M and pPol-I-NS), and expression plasmids for PB1, PB2, PA, and NP (Figure 3B). In this system, M and NS segments are replicated by viral polymerases and NP, then M1/M2 and NS1/NEP genes are expressed from the reconstructed viral replicons. Therefore, as in infected cells at 11 h post-infection, the synthesized virus genome was successfully exported to the cytoplasm and localized as punctate structures in the recycling endosomes at 24 h post-transfection (26) (Figure 3B; WT). PLA signals were observed at the periphery of endosomal puncta, although clear colocalization was not found (Figure 3B; enlarged panel). This may be due to the fact that PLA signal is produced by the rolling-circle amplification of short DNA primers conjugated with secondary antibodies, indicating that the pattern of PLA signals is possibly different from the exact localization of the target proteins. The delNES mutation was introduced in NEP ORF without any amino acid changes in NS1 ORF. The virus genome was found in the nucleus in cells transfected with delNES mutant (Figures 3B, C), indicating that NEP is responsible for the nuclear export of virus genome in an NES motif-dependent manner. Unexpectedly, PLA signals between delNES mutant and M1 were mainly observed in the cytoplasm and increased to about 1.6-fold those of WT (Figures 3B, D). Further, this was not complemented by the insertion of an exogenous NES (N-ILLRMSKMQL-C) at the C-terminus of the delNES mutant (Figures 3B–D). These findings indicate that the endogenous NES motif of NEP intrinsically inhibits the interaction with M1 possibly in the cytoplasm.
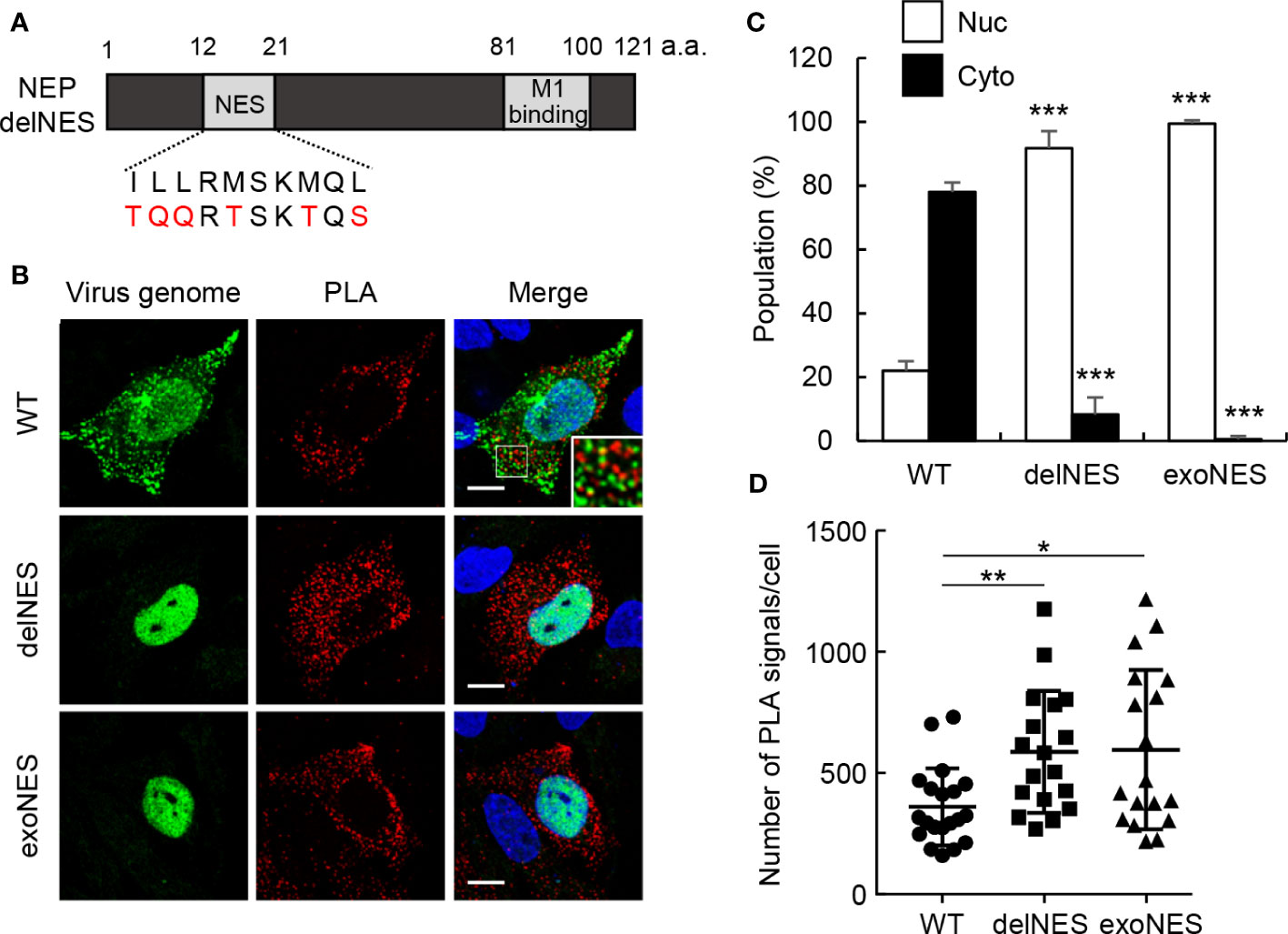
Figure 3 The NES motif of NEP inhibits the interaction of NEP with M1. (A) The schematic diagram of NEP shows the NES motif and M1-binding domain. The NES motif and the mutated sequence are indicated. (B–D) HeLa cells were transfected with pPol I plasmids, containing M and NS segments (pPol-I-M and pPol-I-NS), and expression plasmids for PB1, PB2, PA, and NP. At 24 h post-transfection, the cells were subjected to in situ PLA (red) and FISH assays (green). Nuclei were stained with DAPI (blue). Scale bar, 10 μm. The populations of cells in which the virus genome was localized only in the nucleus (Nuc) and cells in which the virus genome was localized not only in the nucleus but also in the cytoplasm (Cyto) are shown. The number of PLA signals in each cell was counted using IMARIS software [n = 18-20 cells/group; panel (D)]. Results are means ± SDs obtained from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with Tukey’s test against WT for panel (C) Kruskal-Wallis test against WT for panel (D).
3.3 The intramolecular interaction of NEP is mediated by the NES motif and M1-binding domain
The M1 binding domain is located at the C-terminal domain of NEP and consists of the antiparallel hairpin structure with hydrophobic and hydrophilic interfaces (13). The endogenous NES motif may have additional functions other than nuclear export, because the exogenously added NES motif did not result in the functional nuclear export of the virus genome (Figure 3). Thus, we hypothesized that the NES motif in the N-terminal domain associates with the M1 binding domain intramolecularly to inhibit the interaction of NEP with M1. To address this possibility, we next performed split Renilla luciferase complementation assays to examine the close proximity of N-terminal and C-terminal domains of NEP (27). The N-terminal (NR; 1-229 a.a.) and C-terminal (CR; 230-311 a.a.) fragments of Renilla luciferase were fused to the N-terminus and C-terminus of NEP, respectively, with a linker sequence (GGGSGGGS) (Figure 4A). At 24 h post-transfection, the cell lysates were prepared and subjected to the luciferase assays. Although the luciferase activity was not detected in the cell lysates transfected with NR-NEP or NEP-CR, the complementation of luciferase activity was observed by expressing NEP harboring NR and CR (NR-NEP-CR) (Figure 4A). Co-transfection of NR-NEP and NEP-CR did not show the complemented luciferase activity, suggesting that the close association of N- and C-termini of NEP is mediated by the intramolecular interaction rather than the intermolecular interaction. We also found that the level of luciferase complementation in the lysates expressing the NES-mutated NEP with NR and CR (NR-delNES-CR) was reduced to about 10% of that of WT NEP (Figure 4A).
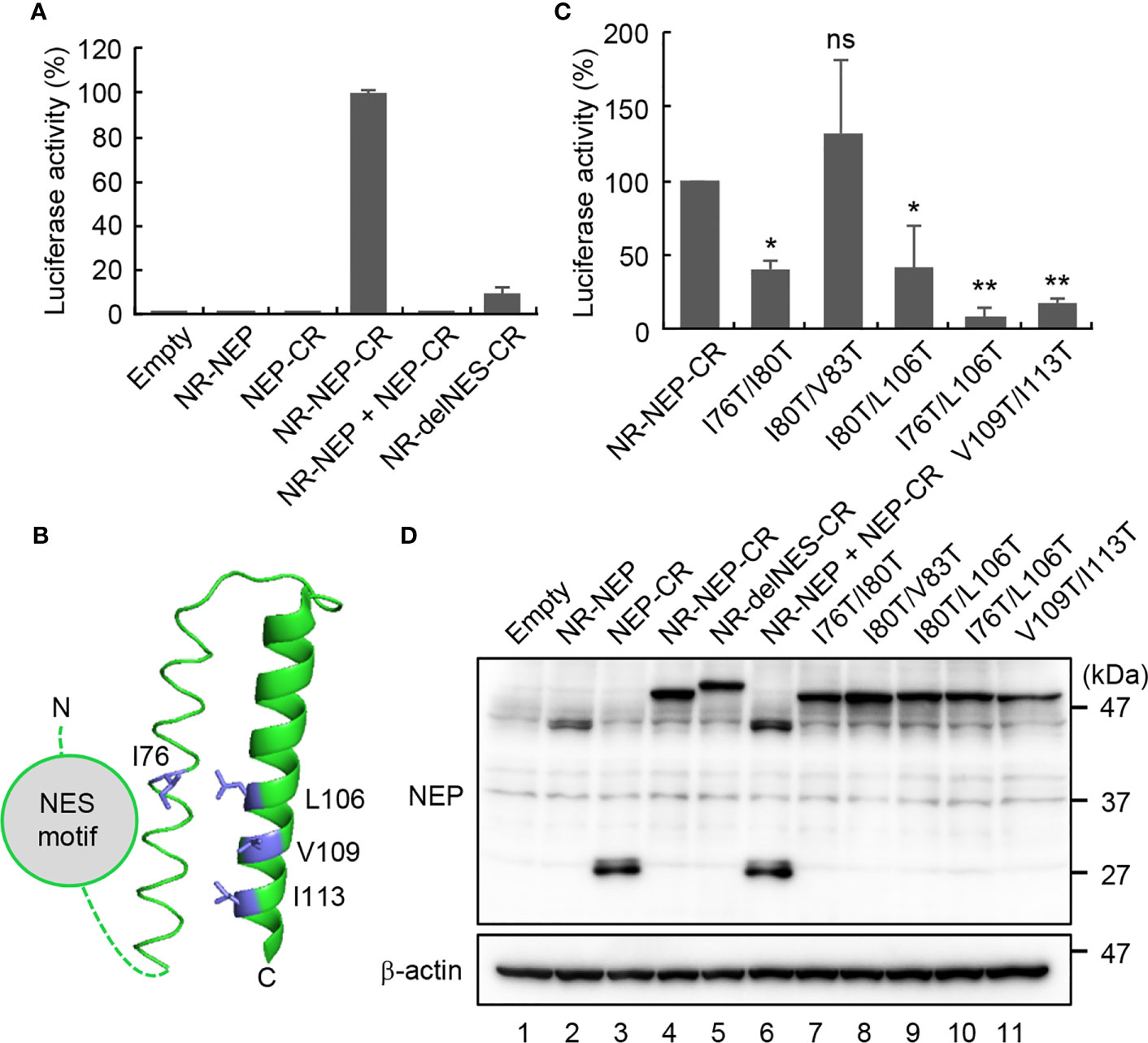
Figure 4 Intramolecular interaction of NEP mediated by the NES motif and M1-binding domain. HeLa cells were transfected with indicated plasmids expressing N-terminal (NR; 1-229 a.a.) and C-terminal (CR; 230-311 a.a.) fragments of Renilla luciferase fused to NEP WT (A), delNES mutant (A), or point mutants in the M1-binding domain (C), respectively. At 24 h post-transfection, the lysates were subjected to the luciferase assay. Results are means ± SDs obtained from three independent experiments. *P < 0.05, **P < 0.01, ns, not significant; one-way ANOVA with Tukey’s test against WT. (B) shows a ribbon diagram of the M1-binding domain (PDB 1PD3). The cell lysates were also analyzed by SDS-PAGE followed by western blotting assays with anti-FLAG and anti-β-actin antibodies (D).
The hydrophobic surface of the amphiphilic hairpin structure in the M1-binding domain is thought to be exposed to solvent (13). Thus, it is possible that the hydrophobic NES motif in the N-terminal domain associates intramolecularly with the M1-binding domain through the hydrophobic interaction (Figure 4B). To address this possibility, we examined the split Renilla luciferase complementation assays with NEP proteins mutated at the hydrophobic residues (I76, I80, V83, L106, V109, and I113) aligned to the exposed surface of the antiparallel amphiphilic hairpin structure (Figure 4C). I76T/I80T and I80T/L106T partially impaired the complemented luciferase activity, while I80T/V83T did not. Furthermore, I76T/L106T and V109T/I113T, reduced the intramolecular interaction to less than 20% of WT NEP. Notably, the expression levels of mutated NEP proteins were unchanged compared to WT NEP (Figure 4D). To elucidate the biological significance of the intramolecular interaction of NEP, we next examined the intracellular localization of NEP harboring I76T/L106T mutations. HeLa cells were transfected with pPol-I-M and either WT or I76T/L106T-mutated pPol-I-NS, and expression plasmids for PB1, PB2, PA, and NP (Figure 5). WT NEP was diffusively localized in the nucleus and the cytoplasm (Figure 5A, upper panels and Figure 5B). Since M1 was also localized in the nucleus, but it was predominantly observed in the cytoplasm (Figure 5A). In contrast, the I76T/L106T mutant was localized to the cell periphery with M1 (Figure 5A, middle panels and Figure 5B), and M1 was not observed in the nucleus. Although a milder phenotype than I76T/L106T, similar results were obtained for V109T/I113T mutant (Figure 5A, lower panels and Figure 5B). We also examined the close proximity of M1 with either I76T/L106T or V109T/I113T mutant by in situ PLA (Figures 5C, D). Although the number of PLA signals between M1 and NEP mutants defective in the intramolecular interaction tended to be reduced compared to NEP WT, a significant amount of NEP mutants was still associated with M1 (Figure 5D).
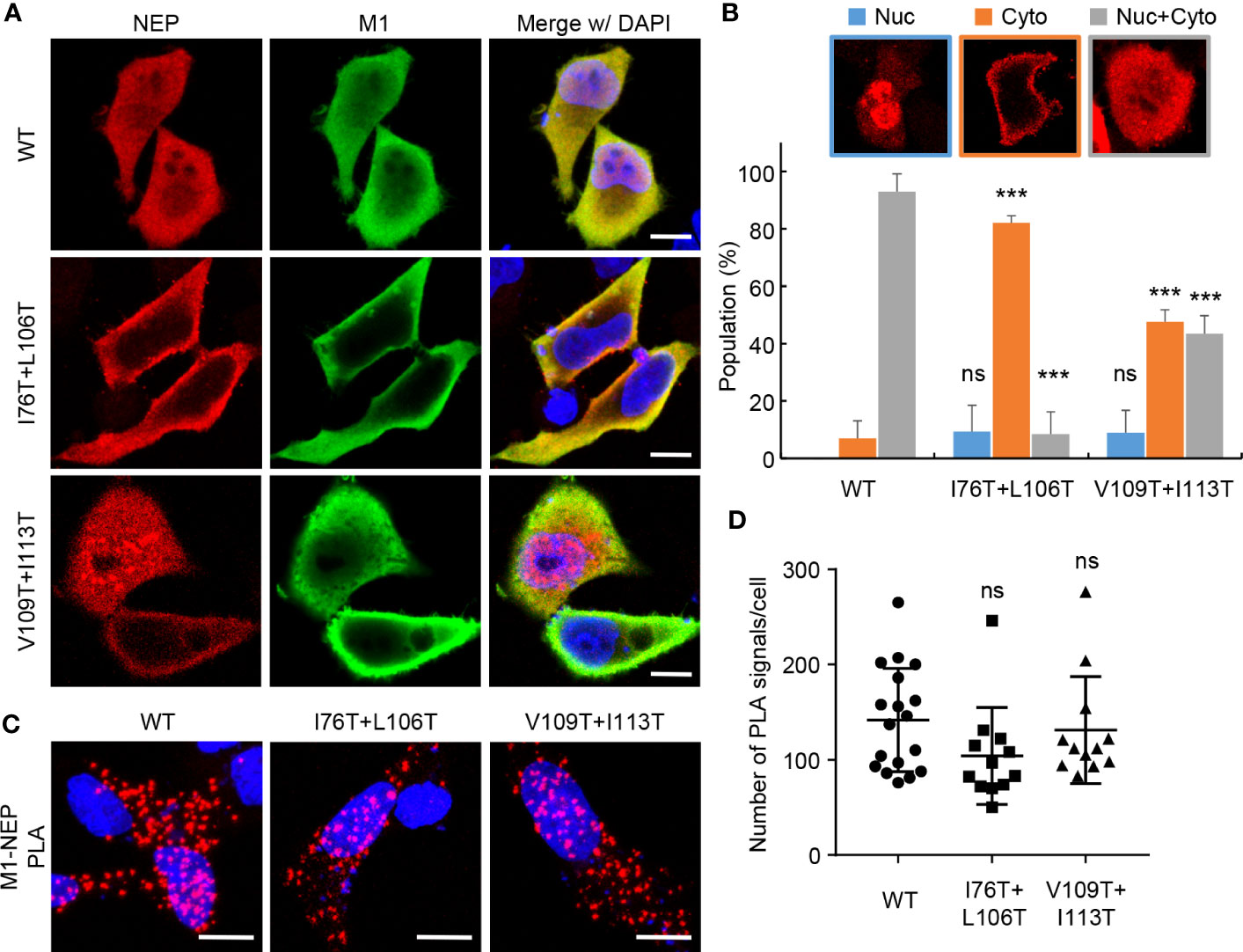
Figure 5 Intracellular localization of NEP mutants defective in the intramolecular interaction. HeLa cells were transfected with pPol I-M and either pPol-I-NS-WT, pPol-I-NS-I76T/L106T, or pPol-I-NS-V109T/I113T, and expression plasmids for PB1, PB2, PA, and NP. At 24 h post-transfection, the cells were subjected to indirect immunofluorescence assays with anti-NEP (red) and anti-M1 (green) antibodies (A). The ratio of cells in which NEP was observed in the nucleus, cytoplasm and both are analyzed using IMARIS software (n = 25 cells/group), respectively (B). The transfected cells were subjected to in situ PLA with anti-NEP and anti-M1 antibodies (red) (C). The number of PLA signals in each cell was counted using IMARIS software (D). Nuclei were stained with DAPI (blue). Scale bar, 10 μm. Results are means ± SDs obtained from three independent experiments. ns, not significant, ***P < 0.001; one-way ANOVA with Tukey’s test against WT.
4 Discussion
NEP functions as an export adaptor protein by interacting with vRNP through M1 (28). It is proposed that NEP binds to the NLS motif of M1, thereby inhibiting re-import of M1 to ensure the unidirectional transport of vRNP from the nucleus to the cytoplasm (29). Because of its low molecular weight, NEP is diffusively observed in the nucleus and cytoplasm, but for efficient export of vRNP, the interaction of NEP with M1 needs to be restricted to the nucleus. Here, we found that NEP forms the intramolecular interaction between its globular N-terminal domain and C-terminal amphiphilic hairpin domain in an NES motif-dependent manner (Figure 4). It is also observed that the point mutations of I76T/L106T or V109T/I113T in the C-terminal domain disrupted the intramolecular interaction of NEP (Figure 4), and the expression of mutants defective in the intramolecular interaction resulted in the co-localization of NEP with M1 at the plasma membrane, without nuclear localization of M1 and NEP (Figure 5). Thus, by lacking intramolecular interaction, NEP may interact with M1 in the cytoplasm and inhibit the NLS-dependent nuclear accumulation of M1 (Figure 6). Based on these findings, it is possible that the intramolecular interaction of NEP is required to repress the interaction of NEP with M1 in the cytoplasm, and that CRM1 releases this inhibition by recognizing the NES motif of NEP, enabling NEP to interact with M1 only in the nucleus to ensure the unidirectional transport of M1-vRNP complex (Figure 6). However, CRM1 regulates the nuclear export of a large number of cellular proteins. Although there are no reports on inhibitory factors of the NEP-M1 interaction, a major limitation of this study is that an unknown CRM1 cargo, which accumulates in the nucleus upon LMB treatment, may interfere with the formation of NEP-M1-vRNP complex. Notably, mutant viruses with either I76T/L106T or V109T/I113T could not be recovered using the reverse genetics system. These amino acid residues are highly conserved among seasonal and avian influenza A viruses and conserve the hydrophobic character compared to influenza B virus (Supplementary Figure 2). It is likely that the spatiotemporal regulation of NEP for vRNP export, regulated by the intramolecular interaction of NEP, is critical in virus production among influenza viruses. The C-terminal M1-binding domain consists of helices C1 (amino acids 64-85) and C2 (amino acids 94-115) that assemble an antiparallel hairpin structure mainly composed of hydrophobic residues over the entire length (13). The antiparallel hairpin structure is further stabilized by polar residues such as the hydrogen bonding between R84 and Q96, and the salt bridges between R55, E74, and E110 (13). The surface of helical hairpin is amphiphilic, and the hydrophilic face is proposed to interact with the NLS domain of M1. In this study, we found that the amino acid residues (I76, L106, V109, and I113) in the hydrophobic face of helices C1/C2 were responsible for the intramolecular interaction with the N-terminal NES motif of NEP (Figure 4). We also observed that leptomycin B inhibits the interaction between NEP and M1 (Figure 2B), suggesting that the interaction of CRM1 with the N-terminal NES domain of NEP is crucial for the efficient binding of NEP with M1. Further, the delNES mutant increased the PLA signal between NEP and M1 (Figure 3), indicating that the NES domain intrinsically inhibits the interaction between NEP and M1. Thus, it is possible that NEP interacts with M1 by releasing the C-terminal helical hairpin from the N-terminal NES domain through a competitive interaction with CRM1. It is reported that W78 residue in the hydrophilic face protrudes from helices C1/C2, which has been proposed to be responsible for the interaction with M1 (13). It remains unclear how the intramolecular interaction between the N-terminal NES motif and the hydrophobic face of helical hairpin changes the structure of the hydrophilic face. Further structural analysis is required to reveal the mechanistic insight of NEP-M1 complex formation.
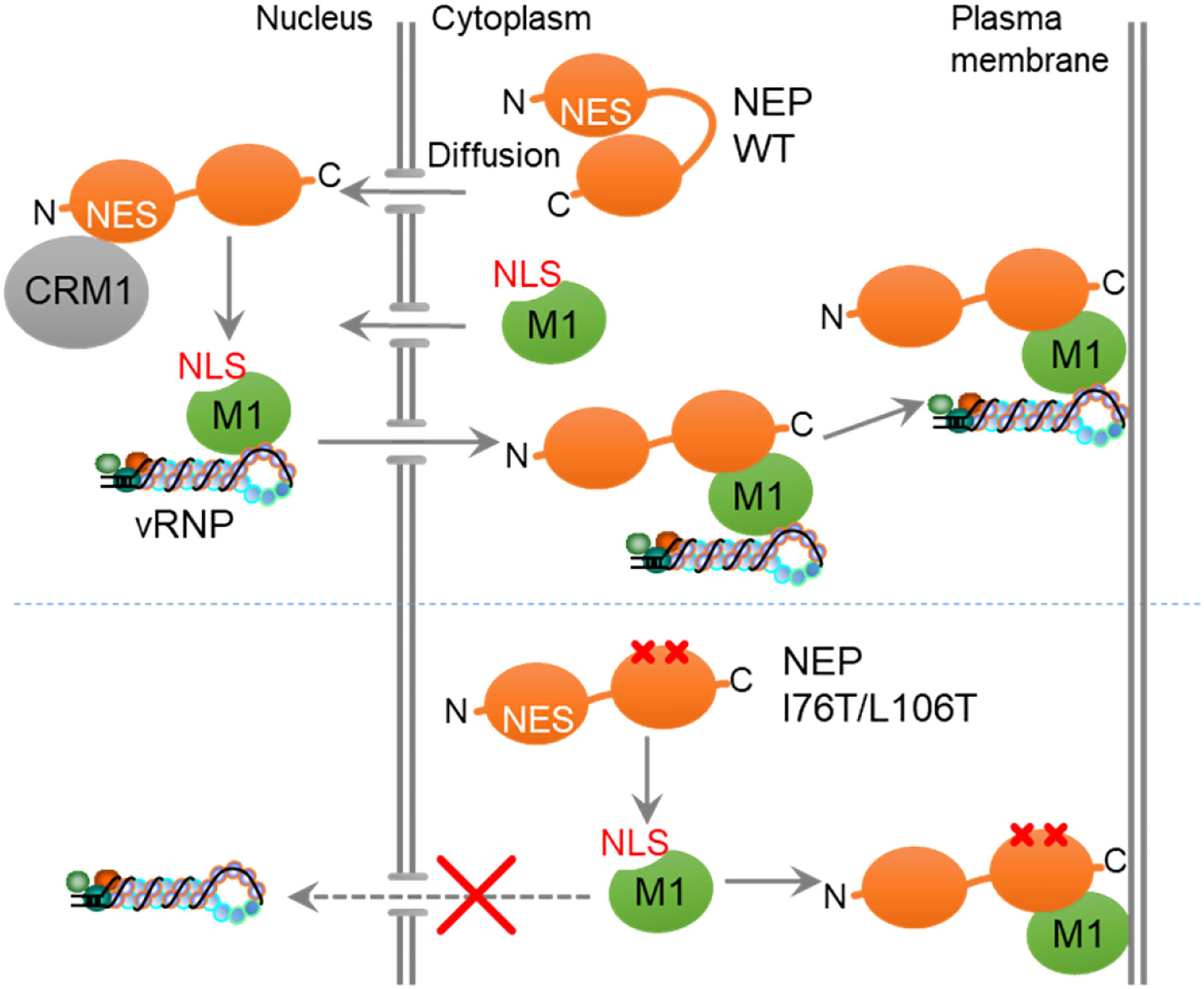
Figure 6 A proposed model. The intramolecular interaction of NEP inhibits the interaction with M1, and CRM1 releases this inhibition by recognizing the NES motif of NEP, enabling NEP to interact with M1 in the nucleus to ensure the unidirectional transport of M1.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
AK conceived and designed the experiments; MH, SY, MA, TK, and AK performed the experiments and analyzed the data; MH, SY, MA, TK, and AK contributed reagents/materials/analysis tools; MH and AK wrote the manuscript.
Funding
This research was supported in part by grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (16H05192, 19H03475, and 22H02874 to AK); the Research Program on Emerging and Re-emerging Infectious Diseases (JP20fk0108076h0003. JP22fk0108638h0001 to AK), the Japan Program for Infectious Diseases Research and Infrastructure (JP20wm0325019h0001 to AK), and AMED-CREST (JP22gm1610008h0001) from the Japan Agency for Medical Research and Development; COI-NEXT (JPMJPF2017 to AK) and CREST (JPMJCR20H6) from the Japan Science and Technology Agency; and the Takeda Science Foundation (to AK).
Acknowledgments
We thank T. Mayers (Medical English Communications Center, University of Tsukuba) for critical review of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author AK declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1232906/full#supplementary-material
References
1. Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: from importin to exportin. Cell (1997) 90(6):967–70. doi: 10.1016/s0092-8674(00)80361-x
2. Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol (2007) 8(10):761–73. doi: 10.1038/nrm2255
3. Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol (2001) 21(10):3405–15. doi: 10.1128/MCB.21.10.3405-3415.2001
4. Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol (2000) 151(5):1057–66. doi: 10.1083/jcb.151.5.1057
5. Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell (2000) 101(2):187–98. doi: 10.1016/S0092-8674(00)80829-6
6. Watanabe K, Takizawa N, Katoh M, Hoshida K, Kobayashi N, Nagata K. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res (2001) 77(1):31–42. doi: 10.1016/s0168-1702(01)00263-5
7. Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, et al. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J Virol (2001) 75(1):408–19. doi: 10.1128/JVI.75.1.408-419.2001
8. Ma K, Roy AM, Whittaker GR. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology (2001) 282(2):215–20. doi: 10.1006/viro.2001.0833
9. Lommer BS, Luo M. Structural plasticity in influenza virus protein NS2 (NEP). J Biol Chem (2002) 277(9):7108–17. doi: 10.1074/jbc.M109045200
10. O'Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J (1998) 17(1):288–96. doi: 10.1093/emboj/17.1.288
11. Iwatsuki-Horimoto K, Horimoto T, Fujii Y, Kawaoka Y. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J Virol (2004) 78(18):10149–55. doi: 10.1128/JVI.78.18.10149-10155.2004
12. Neumann G, Hughes MT, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J (2000) 19(24):6751–8. doi: 10.1093/emboj/19.24.6751
13. Akarsu H, Burmeister WP, Petosa C, Petit I, Muller CW, Ruigrok RW, et al. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J (2003) 22(18):4646–55. doi: 10.1093/emboj/cdg449
14. Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res (2009) 143(2):147–61. doi: 10.1016/j.virusres.2009.05.010
15. Wang D, Harmon A, Jin J, Francis DH, Christopher-Hennings J, Nelson E, et al. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J Virol (2010) 84(9):4673–81. doi: 10.1128/JVI.02306-09
16. Ruigrok RW, Barge A, Durrer P, Brunner J, Ma K, Whittaker GR. Membrane interaction of influenza virus M1 protein. Virology (2000) 267(2):289–98. doi: 10.1006/viro.1999.0134
17. Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol (2000) 74(10):4634–44. doi: 10.1128/jvi.74.10.4634-4644.2000
18. McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol (2006) 80(16):8178–89. doi: 10.1128/JVI.00627-06
19. Ye Z, Robinson D, Wagner RR. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J Virol (1995) 69(3):1964–70. doi: 10.1128/JVI.69.3.1964-1970.1995
20. Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol (1996) 70(1):241–7. doi: 10.1128/JVI.70.1.241-247.1996
21. Kawaguchi A, Hirohama M, Harada Y, Osari S, Nagata K. Influenza virus induces cholesterol-enriched endocytic recycling compartments for budozone formation via cell cycle-independent centrosome maturation. PloS Pathog (2015) 11(11):e1005284. doi: 10.1371/journal.ppat.1005284
22. Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem (2003) 75(7):1584–9. doi: 10.1021/ac020731c
23. Paterson D, Fodor E. Emerging roles for the influenza A virus nuclear export protein (NEP). PloS Pathog (2012) 8(12):e1003019. doi: 10.1371/journal.ppat.1003019
24. Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS Lett (2011) 585(1):41–6. doi: 10.1016/j.febslet.2010.11.017
25. Takizawa N, Watanabe K, Nouno K, Kobayashi N, Nagata K. Association of functional influenza viral proteins and RNAs with nuclear chromatin and sub-chromatin structure. Microbes Infect (2006) 8(3):823–33. doi: 10.1016/j.micinf.2005.10.005
26. Kawaguchi A, Asaka MN, Matsumoto K, Nagata K. Centrosome maturation requires YB-1 to regulate dynamic instability of microtubules for nucleus reassembly. Sci Rep (2015) 5:8768. doi: 10.1038/srep08768
27. Kawaguchi A, Naito T, Nagata K. Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes. J Virol (2005) 79(2):732–44. doi: 10.1128/JVI.79.2.732-744.2005
28. Ueshima S, Nagata K, Okuwaki M. Internal associations of the acidic region of upstream binding factor control its nucleolar localization. Mol Cell Biol (2017) 37(22):e00218-17. doi: 10.1128/MCB.00218-17
Keywords: CRM1, nuclear export, NEP, intramolecular interaction, influenza A virus
Citation: Hirohama M, Yamashita S, Asaka MN, Kuroki T and Kawaguchi A (2023) Intramolecular interaction of NEP regulated by CRM1 ensures the unidirectional transport of M1 for the nuclear export of influenza viral ribonucleoprotein. Front. Virol. 3:1232906. doi: 10.3389/fviro.2023.1232906
Received: 01 June 2023; Accepted: 08 August 2023;
Published: 17 August 2023.
Edited by:
Seiya Yamayoshi, The University of Tokyo, JapanReviewed by:
I-Hsuan Wang, Academia Sinica, TaiwanMasahiro Oka, National Institutes of Biomedical Innovation, Health and Nutrition, Japan
Copyright © 2023 Hirohama, Yamashita, Asaka, Kuroki and Kawaguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Atsushi Kawaguchi, YXRzLWthd2FndWNoaUBtZC50c3VrdWJhLmFjLmpw
 Mikako Hirohama1
Mikako Hirohama1 Takahiro Kuroki
Takahiro Kuroki Atsushi Kawaguchi
Atsushi Kawaguchi