
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Virol. , 12 October 2023
Sec. Emerging and Reemerging Viruses
Volume 3 - 2023 | https://doi.org/10.3389/fviro.2023.1208853
Viruses are one of the major restraining factors in pepper cultivation globally. Among different viruses, pepper mild mottle virus (PMMoV) is one of the most detrimental plant viruses infecting Capsicum spp. belonging to the genus Tobamovirus and Virgaviridae family. It has a monopartite positive-sense single-stranded RNA genome of 6.35 kb size. On an average, PMMoV results in 15%–40% losses in capsicum fruit yield. However, the incidence of PMMoV can reach as high as 95%, leading to substantial yield losses ranging from 75% to 95%. The virus is transmitted via contact, soil, and seeds rather than via insect vectors. PMMoV, because of its seed-borne nature, now occurs worldwide. PMMoV mainly infects Capsicum spp. under natural conditions; however, it can experimentally be transmitted to other plants species belonging to the families Solanaceae, Cucurbitaceae, Labiatae, Chenopodiaceae, and Plantaginaceae. The resistance to tobamoviruses in capsicum is conferred by L locus. Mutations in the coat protein of PMMoV are responsible for the emergence of L-mediated resistance-breaking pathotypes. The highly contagious nature of the virus, seed transmission behavior, and the emergence of virulence complicate its management through a single approach. Therefore, efforts are directed towards providing a more practical and efficient integrated management solution using the RNA interference approach; exploitation of the L gene for resistance breeding; and the inhibitory potential of natural products, systemic resistance-inducing antagonistic bacteria, and chemically synthesized silver nanoparticles. Markers linked to L alleles have been observed to accelerate capsicum breeding programs through marker-assisted selection. In this study, an attempt has been made to compile the recent developments in PMMoV biology, pathogenic variability, genomic organization, and management strategies.
According to the United Nations, the global human population has surpassed 8.0 billion and is anticipated to escalate to 9.7 billion by the 2050s. To sustain this growing population, we must augment our global food production. Moreover, to guarantee nutritional security, we must produce a varied range of nourishing and nutrient-rich foods. Currently, micronutrient deficiency is the dominant cause of impaired human health in many developing nations (1). Vegetables are crucial to ensuring food and nutritional security as they are a rich source of vitamins, minerals, and dietary fibers (2). Among different vegetables, capsicum (Capsicum spp.), also known as bell pepper, is an exceptional source of vitamins C, A, and E; carotenoids; and other essential nutrients such as fiber, potassium, and folate (3). Globally, the total capsicum production was 752,000 tons in 2017, with 4.1 billion dollars in total market revenue (2018) (https://www.researchandmarkets.com/reports/4701016/world-pepper-market-analysis-forecast). Its versatility and accessibility render it an essential constituent of a balanced and nutritious diet. Globally, capsicum is widely cultivated in varied climates spanning tropical to temperate regions, under both open and protected cultivation (1).
Capsicum production faces numerous constraints; of these, diseases and insect pests are the most significant (4–6). A wide range of phytopathogens, including fungi (such as Colletotrichum truncatum, Phytopthora capsici, and Leveillula taurica), bacteria (Xanthomonas and Ralstonia), viruses (such as tobamoviruses, tospoviruses, potyviruses, and cucumoviruses), nematodes, and insects, damage capsicum plants, resulting in extensive yield losses (6). Of all the categories of phytopathogens, viruses are the most destructive biotic agents in capsicum in particular (7, 8). At present, 68 virus species have been reported in capsicum. Among these, 45 species belonging to eight genera, viz., Alfamovirus, Begomovirus, Cucumovirus, Potyvirus, Polerovirus, Potexvirus, Tobamovirus, and Tospovirus, cause considerable economic loss in capsicum production worldwide (7). Among all, tobamoviruses (family Virgaviridae) cause severe losses to the crop and are one of the most studied plant viruses (7, 9).
In capsicum, tobamoviruses are one of the most damaging viruses, particularly in protected cultivation. Tobacco mosaic virus (TMV), pepper mild mottle virus (PMMoV), tomato mosaic virus (ToMV), tobacco mild green mosaic virus (TMGMV), bell pepper mottle virus (BPeMV), paprika mild mottle virus (PaMMV), and Obuda pepper virus (OBPV) are the most widespread tobamoviruses infecting capsicum worldwide (7). PMMoV was initially recognized in the United States as a latent strain of TMV and named as South Carolina mild mottling strain of TMV. It causes mottling in pepper plants but is not capable of inducing systemic infection in tomato (10). However, in 1984, PMMoV was identified as a distinct virus from TMV (11). PMMoV imposes a global threat to capsicum production as its incidence can reach as high as 95%, resulting in yield losses of 75% to 95% (12–15).
PMMoV causes mild to severe symptoms in infected plants, including leaf mosaic, distortion, and stunting, in addition to fruit deformities and reduced yields. The extent of losses depends on the crop stage at which plants become infected. Farmers and researchers face a significant challenge in controlling the spread of PMMoV due to the virus’s ability to form highly stable viral particles. These particles can persist and remain infectious in a variety of environments, including irrigation and natural water sources, soil, compost, and plant debris (16–18). This makes it difficult to eliminate the virus and prevent its transmission among crops. To develop effective management strategies, a thorough understanding of PMMoV, its geographical occurrence, diversity, and existent sources of host resistance are prerequisites.
The focus of our current review revolves around PMMoV, which is accountable for inflicting significant economic losses for capsicum cultivators worldwide (15, 19). The current review provides a comprehensive analysis of the PMMoV pathogen, encompassing its geographical spread, its pathogenic and genomic organization, its variation among isolates, management strategies, genetic resources currently available for future breeding endeavors, and novel strategies for PMMoV management.
PMMoV was reported as a distinct virus for the first time in Sicily, Italy (11). Since then, the virus has been reported in different countries where capsicum is widely cultivated, indicating its worldwide occurrence (11, 20–31) (Figure 1). The virus is highly stable and remains infective even in irrigation/river/seawater, soil, compost, plant debris, etc. (16, 17). In India, PMMoV was first time reported in Himachal Pradesh in capsicum plants grown under protected cultivation (27). In addition to Himachal Pradesh, PMMoV has also been reported in Maharashtra, Karnataka, and Northeast India (32–34).
The disease symptoms on capsicum appear mostly on the leaves and fruits (35). The symptoms caused by PMMoV on pepper plants are mild leaf chlorosis and noticeable growth reduction in case of natural infection, which subsequently result in deformed, mottled leaves with chlorotic appearance (36). Stunting is very severe in plants infected at early growth stages (37) (Figures 2A–C), with young infected plants exhibiting more pronounced symptoms than those infected at later stages (38). The symptoms on fruits include distortion, such as a lumpy appearance, and reduced fruit size (39). Other symptoms caused by PMMoV include mosaic on leaves and fruits, mottling, puckering of leaves, vein thickening, stunting, leaf upward cupping, and fruit deformations (Figure 2) (14, 19, 20, 40). Fruits infected with the virus are reduced in size and show mottling, changes in color, and necrosis, which ultimately result in significant yield losses in both the greenhouse and the field (19) (Figures 2D–F). The fruits infected with PMMoV have significantly lower total antioxidant status (TAS) levels and vitamin C levels, though their levels of total oxidant status (TOS), total phenol (TP), free phenol (FP), and conjugated phenol are higher than those of healthy fruits (41). At times, plants infected with PMMoV demonstrate very mild symptoms that can easily be overlooked (14).
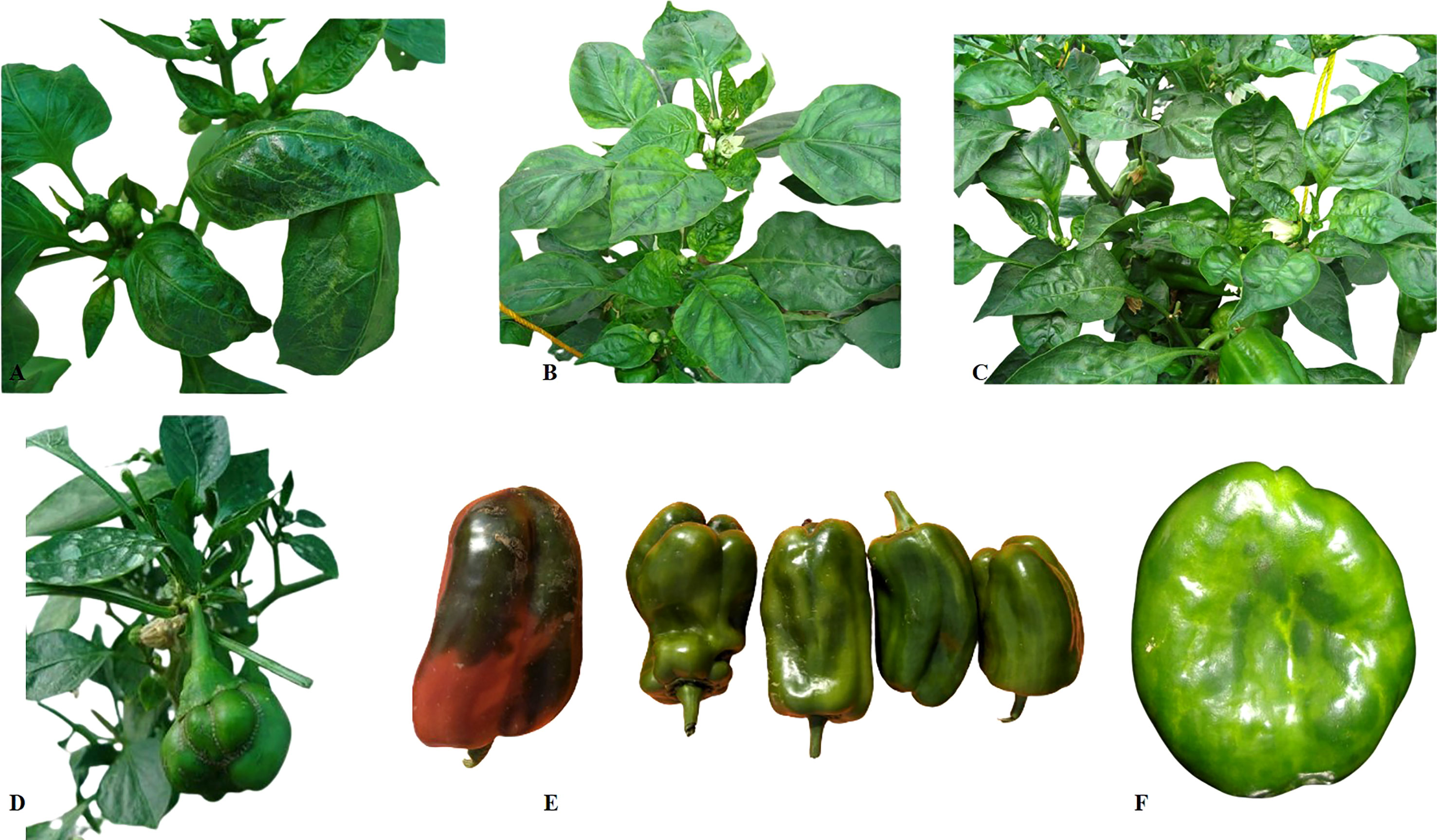
Figure 2 Symptoms of PMMoV observed on capsicum plants and fruits. (A–C) represent mottling and puckering in the young leaves, vein banding, and mosaic on leaves along with downward cupping of the leaves; (E–F) represent the effect of PMMoV infection on fruit size and quality.
The magnitude of yield losses due to PMMoV varies depending on the stage of crop infection. On an average, PMMoV can result in 15%–40% losses in capsicum fruit yield (42). In cases where the incidence of PMMoV reaches up to 80%, significant losses ranging from 50% to 100% have been reported (13, 43). In Himachal Pradesh, India, where the crop is largely grown under protected cultivation, the pathogen may result in 100% disease incidence, which causes yield losses of up to 90%. Yield reduction of up to 86.51% is observed in plants raised from infected seeds (44). Furthermore, plants infected at the 3–4 leaf stage exhibit a yield reduction of 78.38%, whereas those infected at the initiation of flowering and fruiting exhibit yield reductions of 65.35% and 40.33%, respectively (44).
Although the Capsicum spp. are the main host of PMMoV, the virus may infect approximately another 24 plant species belonging to the family Solanaceae (tobacco, eggplant) and also other species in the Cucurbitaceae, Labiatae, Chenopodiaceae, and Plantaginaceae families (11). Capsicum annuum (bell pepper), Capsicum baccatum (pepper), and Capsicum frutescens (chili) are reported to be major hosts of the virus (45, 46). The Nicotiana species (N. glutinosa, N. clevelandii, N. benthamiana, N. occidentalis, and N. tabacum), Chenopodium amaranticolor, and Datura stramonium are local lesion hosts of PMMoV. In addition to these species, PMMoV has also been detected in Dracaena braunii, Paris polyphylla var. yunnanensis, Physalis angulata (cut leaf ground cherry), Achyranthes aspera, Trachelospermum asiaticum, Rorippa palustris, Hydrangea macrophylla, and Leonurus sibiricus (47–50). Until 2009, tomato was considered to be a non-host for PMMoV (51); however, recent studies from the USA and China have shown that it may infect tomato plants naturally (48, 52).
PMMoV transmits by means of seeds, soil, and contact (53). Similar to other tobamoviruses, insect transmission is not observed in the case of PMMoV. The mechanical transmission of PMMoV occurs through contact, and standard agricultural practices such as pinching and pruning that entail equipment usage can exacerbate its dissemination. Since PMMoV can be transmitted through contaminated seeds and virus-infected soils, it has the potential to spread during the planting process. Many studies have reported on the seed-borne nature of PMMoV (10, 54–56). PMMoV resides on the outer coat of seeds but is very rarely found in the embryo and endosperm, which means that its transmission occurs non-embryonically (57). The virus transmits via seed coats to seedlings during transplanting, or during other cultural operations during by mechanical contamination. Abrasions or wounds on the seeds and plant parts during transplanting open the door for viral entry into the host. This serves as a primary infection source. A seed tobamoviruses transmission rate of between 0% and 65.3% was reported for the Capsicum species (10, 40, 54, 56, 58). In contrast to the high rate of seed transmission observed by other researchers, an average seed transmission rate of 10%–30% was recorded for the ‘California Wonder’, ‘Marconi’, ‘Yolo Wonder’, and ‘Anaheim’ pepper varieties (59). In our experiment, average seed transmission rates of 55.56%, 40.64%, and 28.96% were recorded for ‘California Wonder’, ‘Doux des Landes’, and ‘Yolo Wonder’, respectively, using a direct RT-PCR assay of their seeds (56). The rate of seed transmission decreased with an increase in seed storage duration at room temperature (40, 58, 60). Even a very low proportion of infected seeds can cause an epidemic in a field or polyhouse (56). PMMoV absorbed by the soil, plant debris (leaves, stems, and roots), greenhouse structures, humus, and working tools can remain stable for extended periods, serving as an inoculum (12, 61).
Belonging to the genus Tobamovirus, the PMMoV genome comprises a positive-sense single-stranded RNA (+ve ssRNA) that encodes four open reading frames (ORFs) corresponding to four proteins (62) (Figure 3). The first ORF that begins from the 70th position and terminates with an amber codon at nucleotide 3423 encodes a 126-kDa protein. Associated with replication, the 126-kDa protein comprises a methyltransferase, an intervening region, and helicase-like domains (63). It appears that the replication protein of tobamoviruses, including PMMoV, possesses a dual role, serving as both a viral replication factor and an RNA-silencing suppressor (64). The second ORF starts at the same position (i.e., the 70th base) and through an amber codon behavior terminates at nucleotide 4908 encodes a 183-kDa protein. In addition to methyltransferase, the second ORF is an intervening region and helicase-like domains, encoding an additional RNA polymerase domain (62).
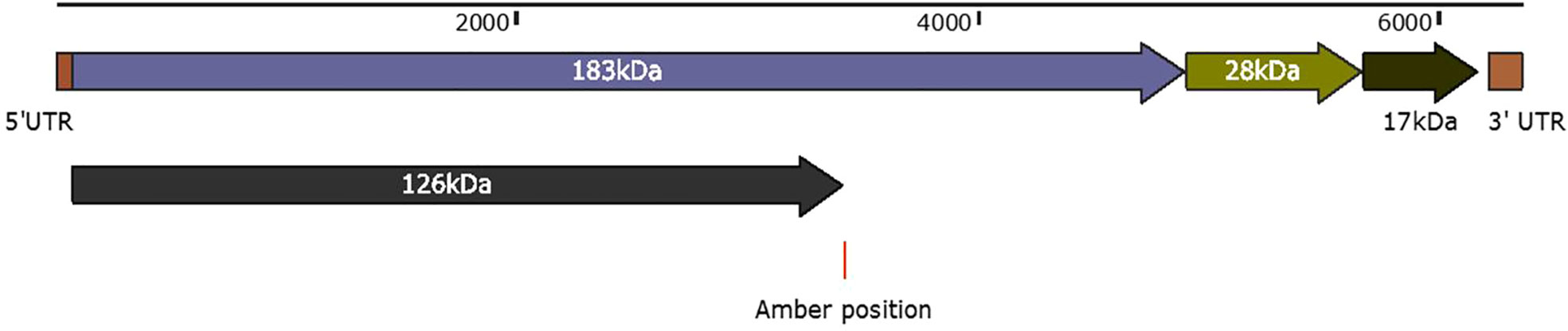
Figure 3 Schematic representation of PMMoV genome organization. The first ORF that begins from position 70 and terminates with an amber codon at nucleotide 3423 encodes a 126-kDa protein. Associated with replication, the 126-kDa protein comprises a methyltransferase, an intervening region, and helicase-like domains. The second ORF starts at the same positions, and through an amber codon behavior, terminates at nucleotide 4908 to encode a 180-kDa protein. The second ORF in addition to methyltransferase, an intervening region and helicase-like domains, encodes an RNA polymerase domain. The third ORF begins at nucleotide 4909 and ends at 5682 nucleotide position, encodes a 28-kDa protein that is responsible for the cell-to-cell movement of the virus. The fourth ORF begins at nucleotide 5685 and ends at nucleotide 6158 to encode a 17-kDa capsid/coat protein. The 5′ upstream and 3′ downstream comprise 69 nucleotides and 198 nucleotides.
The third ORF (4909–5682 nt position) encodes a 28-kDa protein that is responsible for the cell-to-cell movement of the virus (65). The fourth (5685 to 6158 nt position) encodes a 17-kDa capsid/coat protein (65). The role of PMMoV coat protein (CP) as an elicitor in breaking L-mediated resistance has been much highlighted. A single amino acid mutation in CP can result in symptom variation and varied sub-cellular localization of PMMoV in host plants (66). The 5′ upstream and 3′ downstream of PMMoV genome, sequenced using rapid amplification of cDNA ends-polymerase chain reaction (RACE-PCR), showed that the region upstream to first ORF, i.e., 5′ UTR, consists of 69 nucleotides (1–69 nt), whereas the region downstream to fourth ORF, i.e., 3′ UTR, comprises 199 nucleotides (6159–6356 nt) (14, 65). The 5′ leader sequence consists of a 69-nucleotide-long region that is guanosine free and differs from the corresponding genomic region of TMV and ToMV. The 3′ non-coding region of the PMMoV genome is 199 nucleotides long, with a potential secondary structure similar to other tobamoviruses (67–70). The 3′ non-coding region (NCR) has two subregions: one comprises 3–5 consecutive pseudoknots followed by another subregion spanning the last 106 nucleotides; this forms the tRNA-like structure which can be charged with histidine (67, 69). This tRNA-like structure is likely to functionally substitute for the poly(A)-rich region (71). The nucleotide sequence of the 5′ and 3′ non-coding regions of PMMoV was first determined in 1989 (70). Two years later the first entire genomic RNA of Spanish isolate of PMMoV (PMMoV-S) was determined (72). PMMoV-S was classified as a resistance-breaking isolate because of its ability to infect pepper plants that are typically resistant to TMV and ToMV (72). Currently, there are about 50 full genome sequences of PMMoV isolated from Capsicum spp., Nicotiana spp., and other grass hosts available in the NCBI database (Table 1). From India, four full genomes (MN496154, MN734123, MN496153, and KJ631123) are available in the NCBI database. The first complete nucleotide sequence of isolate PMMoV-HP1 collected from Himachal Pradesh was determined in 2015 (14). Based on a pairwise homology index deduced from CLUSTALW software, it was observed that the Indian isolate PMMoV-HP1 is almost identical (i.e., with more than 99% similarity) to isolates from Japan (AB000709), Spain (NC_003630), China (MG515725), Venezuela (KU312319), and Slovenia (MN267898). However, PMMoV-HP1 is least identical to isolates from the Netherlands (MT385868) followed by those from Germany (OP357934), with 89.11% and 93.93% similarity, respectively (Supplementary Table 1).
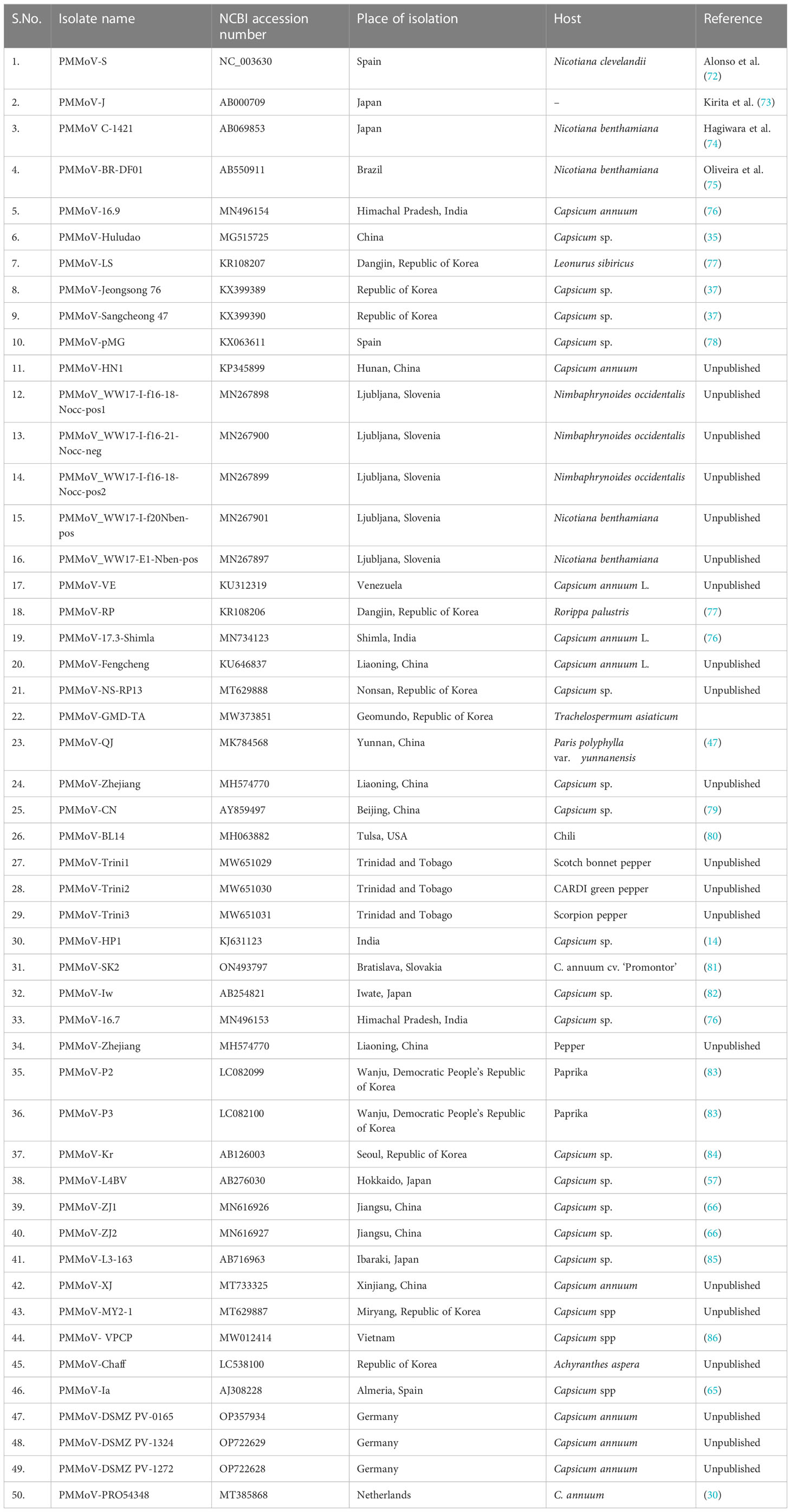
Table 1 Complete genome sequences of PMMoV available in the National Center for Biotechnology Information database.
Various techniques have been employed for the identification and detection of PMMoV utilizing serological, molecular, and high-throughput sequencing (HTS) methods. The identification of PMMoV predominantly entails the synergistic employment of electron microscopy, DAS-ELISA, and PCR (14, 75, 87). The timeline of diagnostic methods for PMMoV since 1984 is depicted in Figure 4. Electron microscopy is the preeminent technique employed by virologists to ascertain the association of tobamoviruses with corresponding plant samples by means of visualizing rod-shaped rigid virion particles (11, 24, 87). DAS-ELISA, a widely employed serological assay for high-throughput screening, is frequently utilized to detect PMMoV infection in plant tissues, including seeds, and in soil (14, 38, 43, 56, 61, 79, 88). In addition to DAS-ELISA, the utilization of non-precoated indirect ELISA (Id-ELISA) has also proven valuable for the detection of PMMoV in infected soil (61, 89). Indirect ELISA allows for the simultaneous detection of multiple serologically related viruses using a single polyclonal antiserum. By employing Id-ELISA and DAS-ELISA prior to planting in the field, it becomes possible to assess the risk of PMMoV, facilitating effective risk management strategies (61, 90). In a few initial studies, tissue blot immunobinding assay (TBIA) and dot blot immunobinding assay (DBIA) using PMMoV-specific polyclonal antibodies were used for PMMoV detection in plant tissues (87, 91, 92). In a recent development, monoclonal antibodies (MAbs) were developed for PMMoV detection using hybridoma technology (93). Utilizing these MAbs, the triple antibody sandwich ELISA (TAS-ELISA) procedure was developed for PMMoV detection in plant sap, with sensitivity up to 1:5120 dilution (93).
RT-PCR and its variants are DNA-based diagnostic methods that have been used to detect PMMoV since 2000. These methods typically involve the use of primers targeting the coat protein (CP) or RNA-dependent RNA polymerase (RdRp) gene (14, 94, 95). Two variants of PCR, namely immunocapture RT-PCR (IC-RT-PCR) and multiplex-PCR (m-PCR), have been developed for the detection of various economically important viruses, including PMMoV (93, 96). A novel single-tube multiplex IC-RT-PCR assay was developed, enabling the simultaneous detection of PMMoV and TMGMV (96). This assay demonstrated a significant advancement, with a 1,000-fold increase in sensitivity compared with ELISA, and also reduced time and cost requirements in comparison with traditional RT-PCR methods (96). A straightforward m-PCR method was developed to detect six different capsicum viruses, namely PMMoV, capsicum chlorosis orthotospovirus (CaCV), chili leaf curl virus (ChiLCV), chili veinal mottle virus (ChiVMV), cucumber mosaic virus (CMV), and large cardamom chirke virus (LCCV) (34). Real-time quantitative PCR (RT-qPCR) is being used for the detection and quantification of PMMoV from water, fecal, and plant samples (66, 97). Some of the PMMoV-specific primers which have been previously used for specific PMMoV detection, along with their target gene and amplification product size, are shown in Table 2. In a recent investigation, 12 viruses and 2 viroids, including PMMoV, were identified in pepper using metatranscriptomics (86). Commercially available rapid immunological assays, such as ImmunoStrip® (Agdia, USA), have emerged as convenient and efficient tools for detecting PMMoV in leaf tissue or seeds. These assays do not rely on laboratory facilities, making them field-applicable and highly accessible. Consequently, they are frequently employed by researchers for diagnosis purposes (100–102).
The tobamoviruses have pathotypes that are determined by their ability to infect Capsicum spp. possessing different alleles, namely L1, L2, L3, and L4 at the L locus (103–105). Resistance corresponding to the pathotypes 0 (P0), P1, P12 and P123 is conferred by L1-, L2-, L3-, and L4-resistant genes, respectively. The L1 gene in capsicum cultivars ‘Bruinsma Wonder’ and ‘Verbeterde glas’ is capable of localization of P0 strain of TMV. Similarly, the L2 gene in C. frutescens cv. ‘Tabasco’ localize both P0 and P1 strains of TMV; L3 localizes the P0, P1, and P12 of TMV strains in C. chinense PIs and the L4 gene is responsible for the localization of the P0, P1, P12, and P123 strains of TMV in C. chacoense accessions PI260429 and SA185 (103–105). The P0 pathotype is incapable of infecting plants that contain the L1 gene. The P1 pathotype consists of viruses capable of infecting plants that carry the L1 gene which are unable to infect Capsicum spp. plants possessing the L2 gene. Similarly, the L2 gene confers resistance to the P1 pathotype, the L3 gene confers resistance to the P0, P1, and P12 pathotypes, and the L4 gene confers resistance to the P0, P1, P12, and P123 pathotypes (106, 107). However, the P1234 pathotype is capable of overcoming the resistance conferred by the L4 gene. PMMoV has three pathotypes, viz., P12, P123, and P1234. As a consequence of the extensive cultivation of capsicum cultivars with the L4 gene that are resistant to the P123 pathotype, the pathogen has adapted and exhibited L4 gene-mediated resistance-breaking abilities, which has resulted in the emergence of a pathotype, namely P1234 (108).
Different capsicum differential cultivars harboring different L alleles have been identified and are being used by researchers for PMMoV pathotype characterization (https://worldseed.org/wp-content/uploads/2020/04/Pepper_tobamo_virus_february2020final.pdf) (24, 30). C. annuum cvs. ‘Sivria-RY’/’Yolo wonder’, C. annuum cvs. ‘Sivria’/’Doux des Landes’, C. frutescense cv ‘Tabasco’, C. chinense line PI-159236, and C. chacoense line PI-260429 harbour L1, L+, L2, L3, and L4, respectively. According to various reports, mutations occurring in the CP of PMMoV have been observed to be effective in overcoming the L-mediated resistance in pepper plants (82, 106–110). Most of the field isolates of PMMoV belong to the P12 pathotype; however, others have reported the occurrence of PMMoV belonging to the P123 and P1234 pathotypes infecting pepper under field conditions (82, 107–110). As suggested by different studies, it is likely that mutations occurring in the CP of PMMoV can trigger the breakdown of resistance conferred by the L3 and L4 genes, leading to the evolution of the P123 and P1234 pathotypes (78).
A single amino acid mutation of asparagine to methionine at the 138th position (N138M) of the PMMoV-I isolate was sufficient to induce a hypersensitive response (HR) and localization of the virus in C. chinense plants with the L3 gene (106). This mutation (N138M) is observed in all PMMoV isolates characterized as P123 pathotypes (18, 78, 106, 111). The mutations in PMMoV CP at the 7th position from alanine to serine (A7S), and the 81st position from serine to alanine (S81A) had L3-mediated resistance-inducing and -breaking abilities, respectively (112). The synergistic effect of two mutations in CP at the 43rd position from threonine to lysine (T43K)—the 50th position from aspartic acid to glycine (D50G) in the case of isolate PMMoV-Ij (110)—and the 13th position from leucine to phenylalanine (L13F)—the 66th position from glycine to valine (G66V) in the case of PMMoV-Is (82)—has been shown to break L3-mediated resistance. PMMoV CP mutants analysis containing one or both of these amino acid changes has shown that both mutations are responsible for efficiently inducing necrosis and hence overcoming L3-mediated resistance (82). Two mutations at the 46th and 85th positions in PMMoV CP, of glutamine to arginine (Q46R) and glycine to lysine (G85K), are likely to be responsible for eliciting L4-mediated resistance breaking (57). Two mutations were observed in the CP of PMMoV isolate, which were capable of overcoming L4-mediated resistance at the 47th and 87th positions of leucine to glutamine (L47Q) and alanine to glycine (A87G), respectively (108).
The differentiation of PMMoV pathotypes is time consuming and often difficult. However, few attempts have been made in the direction of differential detection of PMMoV pathotypes. The antisera and RNA hybridization methods available for the detection of PMMoV are insufficient to resolve the pathotypes. Therefore, sequence analysis is of particular importance in resolving the pathotypes. For instance, EcoRI restriction digestion of 836 bp amplicons of PMMoV-S genome (P12) and PMMoV-Ia (P123) genome obtained using primer pair P12/3 and P12/3A revealed two fragments of 260 and 570 bp, respectively, from the genome of PMMoV-Ia, whereas only a single fragment was obtained for PMMoV-S (65). The available genomic data of PMMoV were mined for identifying the informative sites in PMMoV genome for its pathotype determination (18). The findings indicate that the nucleotide composition at positions 552, 565, 639, 666, 708, 5921, 5975, and 6002 can serve as a reliable marker for distinguishing three PMMoV pathotypes, viz., P12, P123, and P1234 (18). PMMoV isolates with TGGTC nucleotides at positions 565, 639, 708, 5975, and 6002 were all characterized as P12, whereas those with the GATTT combination belonged to the P123 pathotype. Isolates with GGTAC nucleotides at these positions can be identified as the P12 pathotype if it has ACT or ACC nucleotide combination at additional informative positions, viz., 552, 666, and 5921, whereas isolates with GTA combinations belong to the P123 pathotype. The pathotypes of 10 PMMoV sequences (nine full genome sequences and one partial genome sequence) were appropriately predicted using the given model (18).
Phytoviruses are widespread in fruits and vegetables worldwide. Tobamoviruses have an extensive host range, infecting over than 150 plant species, which include vegetables such as tomato, capsicum, turnip, and cucumber (113). Therefore, humans have been exposed to plant viruses for a long time (114). It is believed that plant viruses cannot infect humans as plant and animal viruses differ significantly in the mechanisms they employ to enter and propagate within their host cells. Animal viruses rely on specific interactions with host cell receptors for viral entry by endocytosis or fusion; on the other hand, phytoviruses enter plant cells via injuries caused by insects, infected seeds, agricultural practices, or by transmission directly into the phloem by whiteflies, nematodes, mites, or fungi, and this process does not involve specific molecular interactions. Subsequently, plant viruses propagate from cell to cell through plasmodesmata, which require a movement protein that is exclusively encoded by plant virus genomes. These critical differences have led to a paradigm that plant viruses are safe for human beings. This paradigm regarding the nature of phytoviruses, which has persisted for a long time, has recently been challenged by metagenomic studies of the human gut microbiome and human fecal samples (115–118). The metagenomics analysis of human feces to identify human enteric or other viruses has surprisingly discovered a large and diverse community of plant RNA viruses, predominated by PMMoV (115). It was hypothesized that capsicum-based food consumption could be the source of fecal-borne PMMoV. In a separate study, PMMoV was found in the stools of 7.2% of adult subjects (sample size: 304) and it was observed that PMMoV-positive people were more likely to experience fever and abdominal pain (119). In addition, the detection of anti-PMMoV IgM antibodies in PMMoV positive individuals indicates the non-neutral existence of PMMoV in human gut and their association with clinical symptoms such as fever, abdominal pains, and skin itching (119). This was the first evidence of association of any plant virus with such clinical symptoms. In a few other studies, PMMoV was detected in high abundance in the gut of children less than 1 year old in clinical samples of acute encephalitis/encephalopathy patients; however, further exhaustive investigations are required to confirm whether PMMoV has some pathogenic association with clinical symptoms in humans or is just a neutral member of the human gut microbiome (116–118).
It would not be inaccurate to assert that PMMoV exhibits a ubiquitous presence, as it has been identified in various mediums, including plant tissues such as leaves and seeds, soil, human fecal matter, and water (14, 56, 97, 119). The presence of PMMoV has been observed frequently in different types of water sources, ranging from surface water to drinking water, and streams that are dominated by wastewater effluent, indicating fecal contamination (97, 120–122). PMMoV is considered highly suitable as an indicator of fecal contamination in water sources because of its great abundance and persistence in environmental samples alongside human enteric viruses without any seasonal fluctuation (16, 123). PMMoV has been demonstrated to be effective for assessing water quality and monitoring the effectiveness of water reclamation plants. Due to its high concentration levels in wastewater (in contrast to other enteric viruses), it can be used to measure how much virus copy number is removed from wastewater through large-scale wastewater treatment facilities. This illustrates the potential of PMMoV, which is otherwise an agricultural menace, as an effective tool in water-quality assessment and monitoring water reclamation facilities (123). However, a more comprehensive investigation into PMMoV behavior in aquatic environments and its correlation with other viruses that pose public health risks is necessary to gain a better understanding of its effectiveness as an indicator of water quality (124).
Managing plant virus diseases is an exceptionally challenging task as recovery is nearly impossible once plants have been infected. Currently, PMMoV management options involve the integration of several approaches important for field sanitation: the use of healthy seeds and planting material; disinfection of hands, agricultural tools and implements to prevent mechanical transmission; timely detection and eradication of infected plants including main and other weed hosts; cultivation of disease resistant varieties; and improved cultural practices. In recent years research for eco-friendly control measures has also been accelerated.
Host resistance is one of the most safe, economical, and environmentally friendly methods for the management of plant diseases in general and plant viruses in particular. The resistance to tobamoviruses in capsicum is conferred by the L locus, which consists of a single gene with different alleles encoding an R protein with varying recognition spectra (125). The initial documentation of resistance in pepper against tobamoviruses occurred with the discovery of three alleles that govern varying levels of resistance against TMV (126). These alleles were L, Li, and L+, which were responsible for the complete localization of TMV, substandard localization of TMV, and mottling caused by TMV, respectively (126). L alleles exhibit monogenic with incomplete dominant inheritance. A new allele designated as L3 was identified in C. chinense (103). A new allelic series was proposed as L3>L2>L1>L+ (103). Later, another new allele L4 was reported in C. chacoense PI260429 (105). L4 has a broader resistance spectrum than other resistance alleles. A new tobamovirus resistance gene, L1a, in sweet pepper was also identified later (91). Both L1a homozygote and heterozygote plants showed resistance against the P0 pathotype of tobamoviruses when incubated at 24°C–30°C. Another tobamovirus resistance gene that differed from the L gene is Hk in C. annuum L. cultivar ‘Nanbu Ohnaba’, which confers temperature-dependent resistance (127). The L locus is mapped on chromosome 11 of C. annuum L. at 4.0 cM from TG36 marker (random fragment length polymorphism) in the sub-telomeric region (128, 129). The L locus in capsicum shares synteny with the I2 gene in tomato, which confers resistance to vascular wilt pathogen (Fusarium spp.) (130–133). The entire leucine rich repeat (LRR) domain and the C terminal of L protein is essential for tobamovirus resistance and resistance spectrum determination (125).
In pepper breeding programs, L gene (a partially dominant gene) has been utilized to confer broad resistance to tobamoviruses, including PMMoV. Initially workers screened capsicum germplasm including wild relatives for PMMoV resistance under field conditions. Twenty-eight open pollinated progenies of C. chacoense 'PI 260429' and C. annuum genotypes (L+L+)/C. chinense (L3), including its F1 hybrid and backcross progeny, have been evaluated for PMMoV-P123 resistance and 11 out of 28 lines were resistant (134). C. baccatum (PI 439381-L3) and C. chinense (PI 159236, L3 gene) displayed resistance against PMMoV, as these accessions remained free from infection when planted in PMMoV-infested soil (135). In addition, C. baccatum (PI 439381-L3) showed systemic necrosis after artificial inoculation, indicating that this line has partial resistance which might protect a plant from the PMMoV infection through soil-borne infection under field conditions (135). Between 2012 and 2017, we conducted a screening of 290 capsicum germplasm varieties to evaluate their resistance to PMMoV P12 using bioassay and identified two accessions, viz., PI-159236 and PI-260429, which exhibited resistance (76, 136).
To accelerate the capsicum breeding program for PMMoV resistance, various workers have identified markers linked to L3 and L4 resistant alleles in capsicum (Table 3). Predominantly these markers are employed for selecting the accession lines with L3 or L4 alleles through marker-assisted selection (MAS) (36, 140, 141). Among 10 paprika cultivars, five cultivars, viz., ‘Easy’, ‘Magnipico’, ‘Scirocco’, ‘Orange glory F1’, and ‘Special F1’, were identified as resistant to PMMoV P12 and P123 pathotypes through bioassay and genetic markers (SCAR and Cleaved amplified polymorphic sequence markers) linked to L locus (142). This study revealed that ‘Magnipico’ and ‘Easy’ were homozygous for L4; ‘Scirocco’ and ‘Orange glory F1’ had L4L3; and ‘Special F1’ had L4L1; all these genotypes produced necrotic spots when challenge inoculated with PMMoV pathotypes P12 and P123 (142). Two SCAR markers were applied (143), viz., PMFR11269 (140) and L4SC340 (138), for molecular selection of pepper lines with L resistance alleles and identified three resistant lines derived from the cross of two pepper cultivars, ‘Brill’ and ‘Brilliant’, with the L3 gene.
The studies focusing on the development of PMMoV-resistant commercial capsicum varieties with desirable agronomic traits are very limited. In Japan, during the period between 1985 and 2006, four capsicum varieties, viz., ‘Tosahime R’, ‘Tosajishi-beauty’, ‘Tosajishi-slim’, and ‘Tosa-P-Red’, with PMMoV P12 resistance were developed by Kochi Prefectural Agricultural Research Center, Japan, using a conventional breeding approach (144–147). A new cultivar, ‘Murasaki L4 Daisuke’, for rootstock purposes in grafting, was developed by incorporating resistance to PMMoV (P123) along with bacterial wilt (R. solanacearum) (148). The technique of gene pyramiding was employed to develop a superior sweet Charleston pepper line using the 'Y-CAR' variety as the recurrent parent which exhibited desirable agronomic characteristics (149). This line was, however, found to be vulnerable to three viruses, viz., potato virus Y (PVY), tomato spotted wilt virus (TSWV), and PMMoV. To confer resistance against these viruses, the researchers introduced multiple genes, namely Pvr4, Tsw, and L4, into the ‘Y-CAR’ line. As a result of this gene-pyramiding strategy, the new pepper line exhibited resistance to all three viruses, thereby improving its overall suitability for cultivation. Cultivars, viz., ‘LET-1’, a sweet Charleston type; ‘ENT-1’, a white Hungarian type, and ‘RAZ-1’, a sweet lamuyo type, were used as resistance sources for TSWV, PMMoV, and PVY, respectively. The pathotype used for biological assays of PMMoV was P123. The markers AP-7/AP-8 (CGTACTGTGGCTCAAAACTC/ATTCGCACCGTTTAGCCCGT), linked to the L4 locus (137), were used for marker-assisted selection of PMMoV-resistant seedlings after phenotypic screening. Similarly, markers linked to the Pvr4 locus for PVY resistance and the Tsw locus for TSWV resistance were used for marker-assisted selection of PVY and TSWV resistance. The gene pyramiding strategy followed is presented in Figure 5 (149).
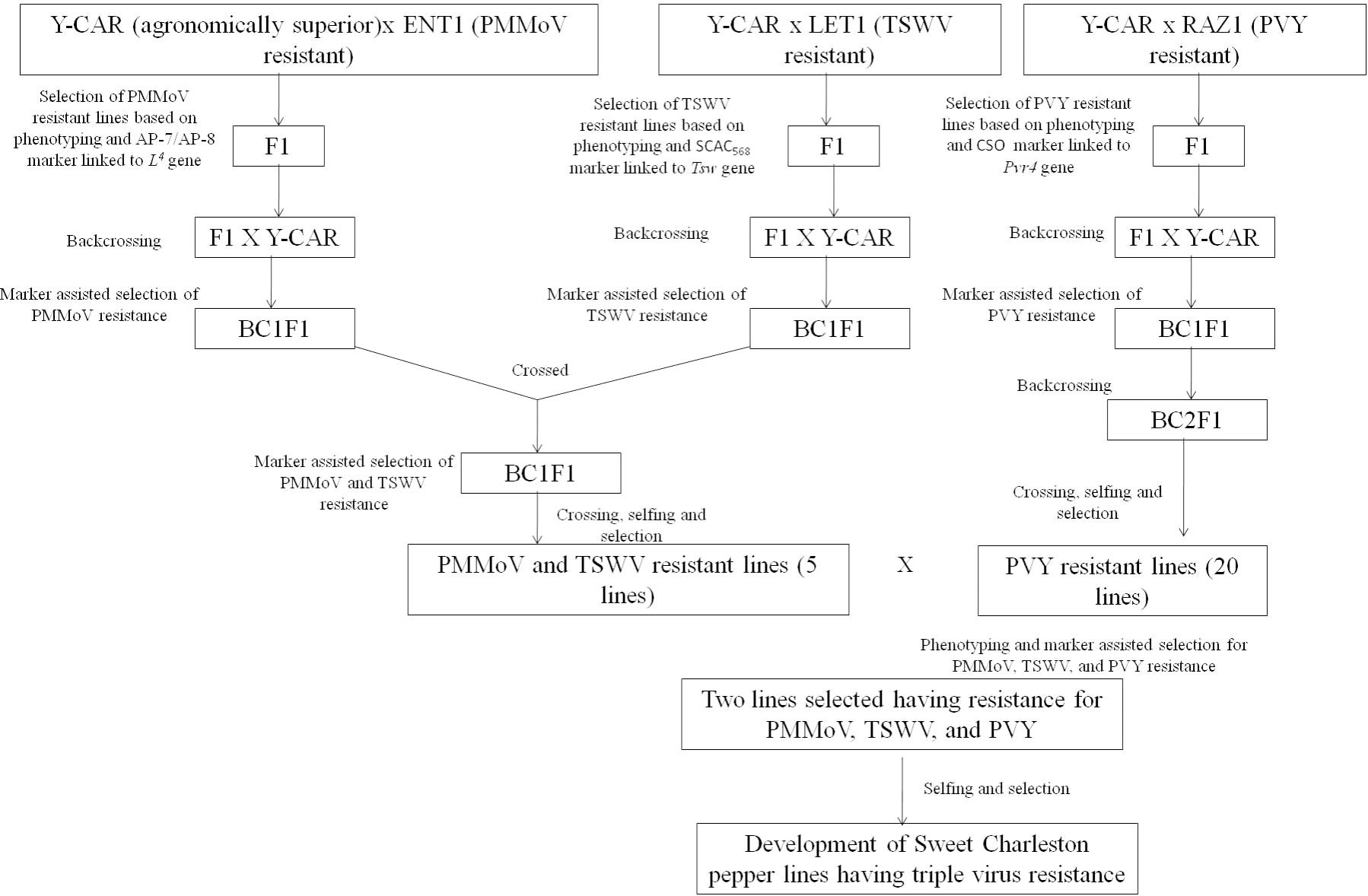
Figure 5 The gene pyramiding strategy followed by Ozkaynak et al. (2014) to develop a new pepper line exhibiting resistance to TSWV, PMMoV, and PVY.
The use of L3- and L4-mediated resistance is the most feasible approach for PMMoV management, but there are still concerns about the emergence of PMMoV strains that would be able to overcome these resistance genes (57, 108). In this regard, cross-protection using an attenuated strain offers an effective alternative approach. The protection of plants from a deadly virus using an attenuated or mild strain of the same virus is one method of crop protection, and the phenomenon is called cross-protection or interference (150). Tobacco plants infected with the yellow strain of TMV were resistant to the severe TMV strain; this phenomenon is referred to as “cross-protection” (150). The successful field application of any attenuated strain for cross-protection depends on the ability of the attenuated strain to induce no or very mild symptoms, a high viability rate in their host plants, a high level of interference with the virulent strains, its genetic stability, and restricted non-intentional spread (151, 152). In tobamoviruses, several attenuated strains have been reported to be used as biological control of viruses, such as ToMV in tomato, PMMoV in pepper, and CGMMV in muskmelon (153). In Japan, an attenuated strain of PMMoV, C-1421, was obtained by pre-heating the pepper stem pieces inoculated with PMMoV at 25°C for 4 days, followed by treatment at 35°C for 16 days (154). The attenuated isolate C-1421 either did not produce symptoms or produced very mild mottling on the pepper seedlings of various cultivars, and provided protection against mechanical challenge inoculation with the severe PMMoV strain. Based on the complete genome sequence of PMMoV-C1421, it was revealed that amino acid substitution of alanine in place of valine at the 649th position (V649A) in 126-kDa replicase protein caused symptom suppression and restricted the accumulation of virion particles in pepper plants (74). Two other attenuated strains, one isolated from the field (TPO-2-19) and the other obtained from heat treatment (Pa-18), were evaluated for their potential to cross-protect pepper plants in the field from the severe PMMoV strain (62). These attenuated strains resulted in a 20%–30% increase in the crop yield of capsicum plants planted in heavily contaminated fields (62). These previously developed attenuated strains (C-1421, TPO-2-19, and Pa-18) were successfully applied for controlling PMMoV in plants without any L resistance gene; however, later they largely remain unused, as these strains produced symptoms on capsicum plants at higher temperatures and could not infect the capsicum cultivars containing the L3 gene (62, 71). Another attenuated strain named TPa18ch was prepared using parent PMMoV-attenuated strains, viz., TPO-2-19 and Pa-18, by inducing amino acid changes at positions 549, 556, 649, and 760 in 126-/183-kDa proteins (mutations at these positions were held responsible for symptom reduction in TPO-2-19 and Pa-18). The major objective of this study was to develop an improved attenuated strain from the P123 pathotype for the effective control of PMMoV in capsicum plants that possess the L3 resistance gene. Pre-inoculation with this attenuated strain followed by challenge inoculation with PMMoV-J isolate (severe strain) resulted in no symptoms in capsicum cv. ‘New tosahikari’ or ‘Miogi ‘(possess L3 gene); however, the TPa18ch strain required more time to accumulate in host cells to exhibit the cross-protection abilities (62). In the Ministry of Agriculture, Forestry and Fisheries, Japan, an attenuated strain, PMMoV-L3-163 (P123), is commercially registered as “Green Pepper PM” for the biological management of PMMoV in the field (71, 85). The plants treated with the attenuated strain PMMoV-L3-163 remained completely free from PMMoV infection until 55 days after treatment; however, > 5% of treated plants produced mild symptoms after 55 days, whereas in control plants the PMMoV incidence was 71%–76% (71). Under field conditions, to achieve significant protection from the wild strain of PMMoV, 50%–75% prior infection with attenuated strain is needed (71, 155).
Post-transcriptional gene silencing is a component of plants’ viral defense mechanisms. The target gene is silenced by the introduction of double-stranded RNA (dsRNA) molecules that have sequence homology with the target viral genome (156). RNA silencing/RNA interference (RNAi) technology has been successfully employed in developing resistance against different economically important plant viruses (Table 4). In PMMoV, dsRNA-mediated resistance has been demonstrated by different workers (169–172). Complete and delayed resistance was achieved in N. benthamiana plants which were transformed with the 54kDa-encoding region of the PMMoV replicase gene, although this resistance was independent of transcript level (173). Inhibition of virus infection was also achieved in the case of co-inoculation of in vitro-transcribed dsRNA molecules homologous to target the virus sequence and target the virus via Agrobacterium-mediated transient expression of dsRNA in Nicotiana spp. However, the in vitro synthesis of transcribed dsRNA molecules is expensive and thus not feasible for large-scale applications (171). In this regard, an in vitro expression system for producing a large number of dsRNA molecules derived from PMMoV replicase gene (IR) in E. coli strain HT115 (DE3) deficient for RNase III was developed (171). N. benthamiana plants inoculated with the total nucleic acid extracted from bacteria expressing 997 bp of PMMoV replicase gene remained free from any symptoms even 10 weeks post PMMoV inoculation. A similar observation on interference with PMMoV infection upon exogenous application of bacterial crude extract expressing PMMoV-sequence in N. benthamiana plants was made.
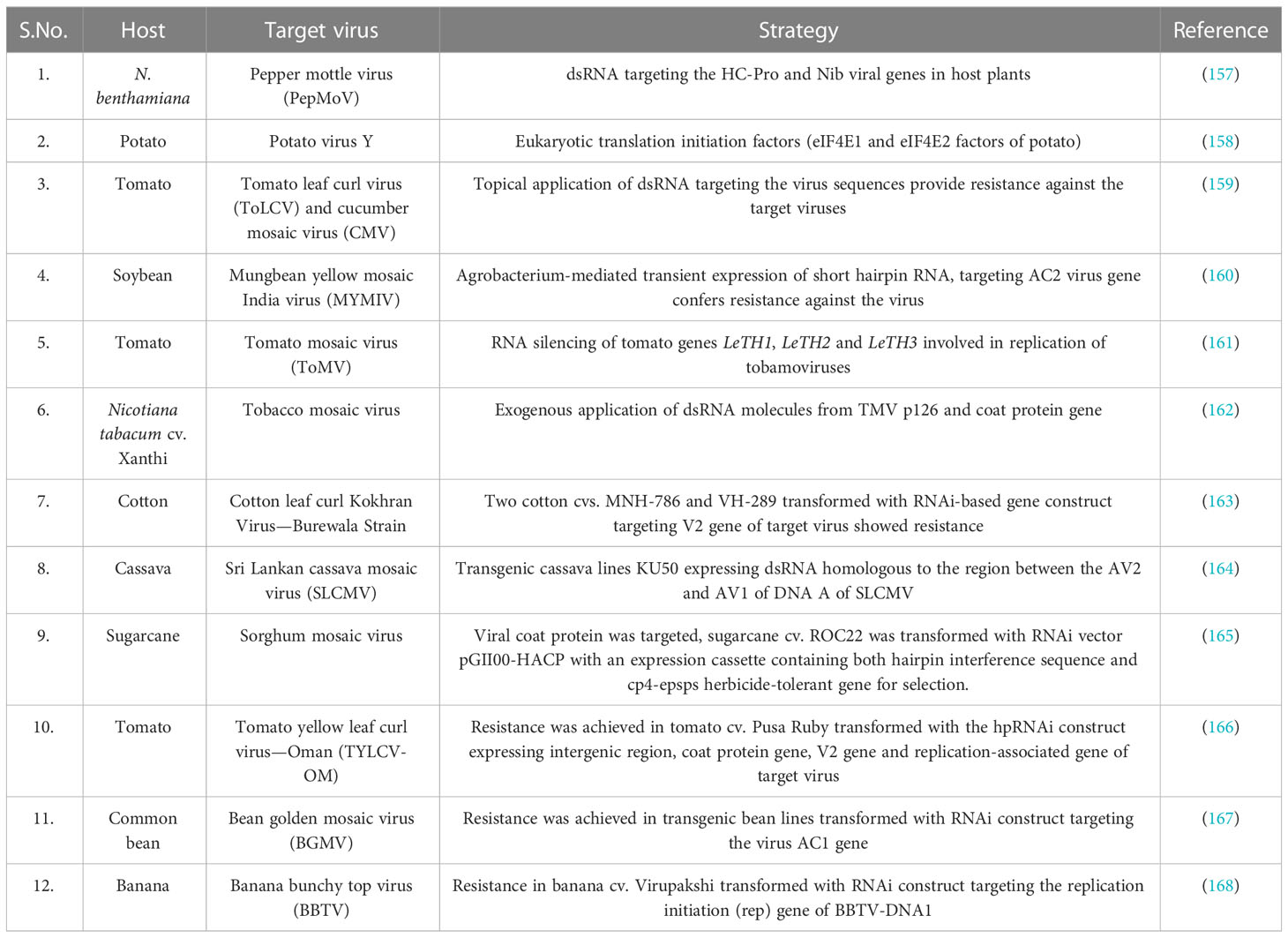
Table 4 Application of RNA interference technology in inducing resistance in crop plants against economically important plant viruses.
Although the successful targeting of economically important plant viruses has primarily relied on genetically modified methods, these approaches present drawbacks such as time and cost intensiveness and challenges related to regulation and public acceptance. To overcome these limitations and address public concerns, alternative strategies involving the exogenous application of naked dsRNA have been proven effective in triggering the RNAi pathway against pathogenic viruses. However, a notable drawback of this approach is its limited virus protection window, typically lasting only 5–7 days following application. To address this issue, the use of layered double hydroxide (LDH) clay nanosheets as a carrier for dsRNA was investigated (172). It was demonstrated that loading dsRNA onto LDH clay nanosheets, which was termed as “Bioclay”, resulted in prolonged stability of the dsRNA and was even detectable after 30 days of topical application. Double-stranded RNA (dsRNA) is released in a sustainable manner from the bioclay once LDH degrades, which occurs as a result of exposure to atmospheric CO2 and water (172). To determine the extent of protection provided by BioClay loaded with PMMoV dsRNA (PMMoVIR54-BioClay), both PMMoVIR54-BioClay and naked PMMoVIR54-dsRNA were sprayed on N. tabacum cv. ‘Xanthi’ leaves and subsequently challenged with PMMoV on the 5th and 20th days post spraying. PMMoVIR54-BioClay provided extended and systemic protection against PMMoV in N. tabacum, thus greatly suppressing the impact of PMMoV. The capacity of BioClay to revolutionize plant protection by overcoming the challenges encountered by genetically modified crops is noteworthy (172, 174).
The antiviral activity of chitosan (C) and phosphate-linked chitosan (PC) at concentrations from 0.01% to 0.1% against PMMoV and other viruses such as cucumber mosaic virus (CMV) in Nicotiana glutinosa plants has been explored. Counting the total number of lesions of N. glutinosa leaves showed that 0.1% concentration of C or PC caused 40%–75% suppression of PMMoV symptoms. The evaluation of viral load by DAS-ELISA assay showed that it reduced 40% of CMV accumulation, which likely upregulates the NPR1, PR-1, LOX, PAL, CRF3, SRC2, and ERF4 expression in chili pepper plants (175). The treatment with silver nanoparticles (AgNPs) at 200, 300, and 400 µg/L after 24h of viral inoculation reduces the leaf deformation and mosaic symptoms (176). Two antiviral compounds, viz., murrayafoline-A (1) and isomahanine isolated from leaves of Glycosmis stenocarpa, a shrub from Rutaceae family have inhibitory properties against PMMoV (177). Cellulose and commercial cellulases have PMMoV-inhibitory properties (178, 179). When soil that has been contaminated with PMMoV-infected capsicum plant roots is supplemented with cellulose, PMMoV becomes inactivated due to an elevation in the activity of antiviral microorganisms present in the soil (178). Commercially available cellulases derived from Trichoderma reesei, T. viride, and Aspergillus niger also possess antiviral properties and the potential to inhibit PMMoV infection in N. glutinosa and pepper plants (179). Nevertheless, the mechanism through which these cellulases inhibit PMMoV infection is still to be fully elucidated.
Vanisulfane, a novel antiviral agent with the chemical name 2,2-(((4-((4-chlorobenzyl)oxy)-3-methoxyphenyl)methylene) bis-(2-hydroxyethyl) dithioacetal, has been designated as a plant vaccine or plant immunity booster (180). This is due to the fact that vanisulfane has the ability to mitigate the activity of defense-related enzymes such as phenylalanine ammonia lyase (PAL), catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD), in addition to other enzymes that play a role in sugar and starch metabolism, photosynthesis, oxidative phosphorylation, and the MAPK signaling pathway in capsicum (181, 182). In addition to its potent ability to inactivate PMMoV, vanisulfane has also been found to exhibit a strong affinity towards PMMoV coat protein. Thus, it has been suggested that vanisulfane could potentially serve as an anti-PMMoV drug, specifically targeting the PMMoV CP (181, 183). With the aim of developing an antiviral drug, 34 novel chalcone derivatives comprised of 1,2,4-oxadiazole moiety was synthesized (184), and evaluated their antiviral abilities (TMV, PMMoV, and TSWV) (184). Few of these compounds exhibited excellent curative and protective activity against PMMoV (up to 56.5% and 71.8%, respectively) and thus can be considered as potential candidates in the development of antiviral drugs (184).
Recently, the utilization of beneficial microorganisms, such as plant growth-promoting rhizobacteria (PGPR), has emerged as a promising solution for protecting crops against virus damage (185). Extracts from different bacteria and plant origins have been explored for their antiviral activity. For example, the culture supernatant of Pseudomonas oleovorans strain KBPF-004 (ATCC 8062) was found to reduce the viral load of PMMoV and cucumber green mottle mosaic virus (CGMMV). The reduction of viral load is linked to the remodeling of the 126-kDa protein and localization of the movement protein of PMMoV, which abolished the interaction of the movement protein with the microtubule network and even reduced the transmission of viruses (186). The application of cell-free supernatants of Pseudomonas spp. (Pseudomonas putida and P. fluorescens) and Bacillus spp. (Bacillus licheniformis and B. amyloliquifaciens), and Serratia marcescens resulted in a significant (51%–66%) decrease in viral accumulation in capsicum plants. The gas chromatography-mass spectrometry (GC-MS) analysis identified 24 different compounds including alkanes, alcohols, ketones, and aromatic compounds in the bacterial supernatants (101). The antiviral activity of different rhizobacteria has also been explored in the case of other tobamoviruses. For example, Stenotrophomonas maltophilia HW2, a PGPR isolated from healthy cucumber plants, has displayed a biocontrol efficiency of 52.61% against CGMMV and also delayed its replication in the host plant post inoculation by 3 days (185). Consortia containing B. thuringiensis, P. aeruginosa, B. subtilis, B. macerans, and B. cereus have significantly suppressed the symptoms caused by tobamoviruses on tomato plants (187).
There is a growing need to develop environmentally compatible methods for virus management fields. In this context, various plant extracts have been extensively studied to identify compounds with controlling or inhibitory effects against plant pathogens (188). Botanicals have antiviral factors which include alkaloids, furocoumarins, terpenoids, lignins, or ribosome-inactivating proteins, and can thus induce systemic resistance in plants against viruses and inhibit their replication in host plants (188). The extracts of several plants, viz., Mirabilis jalapa, Eucalyptus, Camellia sinensis, Glycyrrhiza glabra L., Phyllanthus sp., Thuja sp., Musa acuminate Colla, and Musa balbisiana Colla, have antiviral properties against tobamoviruses (189–192). The extract of Populus nigra promoted the plant growth and fruit quality of PMMoV-infected C. annuum under greenhouse conditions (193). In addition, the structurally modified whey proteins with quercetin and onion extract also reduced the viral load of PMMoV and the disease severity 14 days post-inoculation in pepper plants (194). The treatment with modified proteins enhanced plant growth and boosted the total antioxidant status and vitamin C level compared with the control. Furthermore, it also upregulated the expression of defense-related genes, including PR4, PR9, TIN1, and PIN2 (194).
Apart from a serious agricultural problem, PMMoV has great potential as a tool for monitoring water quality. PMMoV has been constantly detected from human feces and is a predominant member of the human gut microbiome. However, extensive investigations are required to confirm whether PMMoV causes any pathological disorders in humans or is just a passive bystander. In plants, managing viruses is a difficult task, especially when dealing with soil and seed-transmitted viral pathogens such as PMMoV. To manage PMMoV in the field, it is critical to develop sensitive and reliable methods for the differential detection of its pathotypes. Knowing the PMMoV pathotypes beforehand is crucial to selecting appropriate cultivars for cultivation to avoid exerting selection pressure on the virus population in the field. Consequently, prior diagnosis and preventing transmission through real-time and point-of-use (PoU) diagnostics which allows in situ rapid PMMoV detection from plants, seeds, and soil even by an untrained person is the most practical solution. The implementation and utilization of cutting-edge PoU diagnostic tools in the field of plant virology have been slower than their adoption in animal virology. In recent studies, isothermal amplification methods such as recombinase polymerase amplification (RPA) assay and loop-mediated isothermal amplification (LAMP) has emerged as a rapid, straightforward, highly sensitive, and cost-effective method for detecting DNA and RNA plant viruses. These methods can easily integrate with lateral flow immunoassay and thus be adapted for PoU detection. Such PoU diagnostic tools for PMMoV detection from plants, seed soil, and water are essential.
The utilization of host resistance is considered the most sustainable approach for managing plant virus diseases. It is vital to undertake the task of mapping the L3 and L4 genes and identify the genetic markers that are closely linked to them to expedite the molecular selection of capsicum germplasm with L3 and L4 genes through MAS. The PMMoV pathotypes P1234 can overcome the L4 gene, which is known to have the broadest resistance spectrum among all resistance alleles against tobamoviruses in capsicum germplasm. Therefore, apart from identifying the existing resistant sources, we must also direct our future research toward the use of modern genetic engineering tools for developing resistance against PMMoV. CRISPR (clustered regularly interspaced short palindromic repeats)/CRISPR-associated (Cas) systems are very effective and convenient tools to edit endogenous and exogenous DNA or RNA sequences across a wide range of organisms. With the discovery of RNA targeting and editing CRISPR-associated proteins, the application of CRISPR-Cas systems has been expanded from DNA to RNA genomes. Generally, CRISPR/Cas technology can be utilized to either target or manipulate the viral genome which can inhibit the viral replication and infection cycle of the virus in its hosts, or it can be utilized to edit the host factors which are indispensable for completing the viral replication cycle in their host plants. After conducting an extensive search, we did not find any research studies focused on using CRISPR-Cas technology for the detection or management of PMMoV. The use of CRISPR technology for PMMoV management also requires a comprehensive understanding of PMMoV–capsicum interaction at the molecular level using a next-generation sequencing approach. It will provide baseline information for devising efficient, sustainable, and novel management strategies for PMMoV management. Nevertheless, the advancement of RNAi-based biopesticides is gaining momentum as a targeted and specific approach for plant virus management. Provided a suitable delivery mechanism, topically applied target virus-specific dsRNA can be deemed as a secure and efficacious approach in the management of PMMoV through RNA interference. However, CRISPR-Cas, an efficient genome editing tool, has many advantages over the RNAi-based management approach. CRISPR-Cas has been applied for generating resistance in plants against masterviruses, begomoviruses, caulimoviruses, cucumovirus, badnaviruses, potyviruses, etc. (195).
Nanotechnology has emerged as a promising approach to plant virus management. A number of studies have been published in the last decade investigating the antiviral role of engineered nanoparticles (NPs) and their application in plant virus management (196). Nanomaterials with inherent antiviral properties, such as metal nanoparticles or nanocomposites, have shown promise for plant virus management (197, 198). Another application of nanotechnology in plant virus management is in the area of virus detection (198, 199). Nanoparticles offer versatility in their applications and can be combined with various probes to detect viral nucleic acids or proteins, upon binding. In the realm of virus detection, nanoscale biosensors utilizing nanomaterials such as graphene oxide, silica, carbon nanotubes, quantum dots, gold, silver, zinc oxide, and magnetic nanoparticles have been developed to specifically target and identify human pathogenic viruses, and their application in plant virus detection is limited but expanding. Although nanotechnology shows great promise in phytoviruses management, there are notable challenges that can impede its implementation. It is crucial to allocate substantial efforts toward comprehending the mechanism of interaction between NPs and plant viruses so that sustainable management strategies with optimal efficacy can be designed.
NK, VS, and PS contributed to the conceptualization of the manuscript. NK, VS, and PP drafted the original manuscript. VS and NK created the figures. PS organized the funding acquisition. All authors contributed to the article and approved the submitted version.
The work was funded by Indian Council of Agricultural Research, New Delhi (ICAR 017-17 NSP-Seeds), and UGC, Govt of India (award letter no.43-3/2014-SR).
We acknowledge support from ICAR and UGC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1208853/full#supplementary-material
1. Olatunji TL, Afolayan AJ. The suitability of chili pepper (Capsicum annuum L.) for alleviating human micronutrient dietary deficiencies: A review. Food Sci Nutr (2018) 6(8):2239–51. doi: 10.1002/fsn3.790
2. Schreinemachers P, Simmons EB, Wopereis MC. Tapping the economic and nutritional power of vegetables. Glob Food Sec (2018) 16:36–45. doi: 10.1016/j.gfs.2017.09.005
3. Sun T, Powers JR, Tang J. Evaluation of the antioxidant activity of asparagus, broccoli and their juices. Food Chem (2007) 105(1):101–6. doi: 10.1016/j.foodchem.2007.03.048
4. Adigun JA. Evaluation of some pre-emergence herbicide mixtures for weed control in chilli pepper (Capsicum frutescens L) in Northern Guinea Savanna of Nigeria. Agric Environ (2001) 1(1):27–33.
5. Anaya-Esparza LM, Mora ZV, Vázquez-Paulino O, Ascencio F, Villarruel-López A. Bell peppers (Capsicum annum L.) losses and wastes: Source for food and pharmaceutical applications. Mol (2021) 26(17):5341. doi: 10.3390/molecules26175341
6. Parisi M, Alioto D, Tripodi P. Overview of biotic stresses in pepper (Capsicum spp.): Sources of genetic resistance, molecular breeding and genomics. Int J Mol Sci (2020) 21(7):2587. doi: 10.3390/ijms21072587
7. Kenyon L, Kumar S, Tsai WS, Hughes JA. Virus Diseases of Peppers (Capsicum spp.) and their Control'. Adv Virus Res (2014) 90:297–343. doi: 10.1016/B978-0-12-801246-8.00006-8
8. Waweru BW, Miano DW, Kilalo DC, Rukundo P, Kimenju JW. Detection and distribution of viruses infecting hot pepper (Capsicum spp.) in Rwanda. J Plant Pathol (2021) 103:573–85. doi: 10.1007/s42161-021-00811-7
9. Ilyas R, Rohde MJ, Richert-Pöggeler KR, Ziebell H. To be seen or not to be seen: latent infection by Tobamoviruses. Plants (2022) 11(16):2166. doi: 10.3390/plants11162166
10. McKinney HH. Two strains of Tobacco-mosaic virus, one of which is seed-borne in an etch-immune pungent Pepper. Plant Dis Rep (1952) 36(5):184–7.
11. Wetter C, Conti M, Altschuh D, Tabillion R, Van Regenmortel MH. Pepper mild mottle virus, a tobamovirus infecting pepper cultivars in Sicily. Phytopathology (1984) 74(4):405–10. doi: 10.1094/Phyto-74-405
12. Lamb EM, Adkins SE, Shuler KD, Roberts PD. Pepper mild mottle virus. Univ Florida IFAS Extension Bulletin. (2001) 808–9.
13. Guldur ME, Çaglar BK. Outbreaks of Pepper mild mottle virus in greenhouses in Sanliurfa, Turkey. J Plant Pathol (2006) 88(3):339.
14. Rialch N, Sharma V, Sharma A, Sharma PN. Characterization and complete nucleotide sequencing of pepper mild mottle virus infecting bell pepper in India. Phytoparasitica (2015) 43:327–37. doi: 10.1007/s12600-015-0453-6
15. Ochar K, Ko HC, Woo HJ, Hahn BS, Hur O. Pepper mild mottle virus: an infectious pathogen in pepper production and a potential indicator of domestic water quality. Viruses (2023) 15(2):282. doi: 10.3390/v15020282
16. Rosario K, Symonds EM, Sinigalliano C, Stewart J, Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl Environ Microbiol (2009) 75(22):7261–7. doi: 10.1128/AEM.00410-09
17. Hamza IA, Jurzik L, Überla K, Wilhelm M. Methods to detect infectious human enteric viruses in environmental water samples. Int J Hyg Environ Health (2011) 214(6):424–36. doi: 10.1016/j.ijheh.2011.07.01
18. Deri BB, Pajic V, Dudic D. Revealing new information from existing genomic data for pepper mild mottle virus pathotype determination. Crop Prot (2018) 107:93–103. doi: 10.1016/j.cropro.2018.01.017
19. Petrov N. Effect of pepper mild mottle virus infection on pepper and tomato plants. Sci Technol (2014) 4(6):61–4.
20. Alonso E, Garcia Luque I, Avila-Rincon MJ, Wicke B, Serra MT, Díaz-Ruiz JR. A tobamovirus causing heavy losses in protected pepper crops in Spain. J Phytopathol (1989) 25(1):67–76. doi: 10.1111/j.1439-0434.1989.tb01057.x
22. Beczner L, Rochon DM, Hamilton RI. Characterization of an isolate of pepper mild mottle tobamovirus occurring in Canada. Can J Plant Pathol (1997) 19(1):83–8. doi: 10.1080/07060669709500579
23. Adkins S, Lamb EM, Roberts PD, Gooch MD, Breman L, Shuler KD. Identification of Pepper mild mottle virus in commercial bell pepper in Florida. Plant Dis (2001) 85(6):679. doi: 10.1094/PDIS.2001.85.6.679D
24. Svoboda J, Cervena G, Rodova J, Jokes M. First report of Pepper mild mottle virus in pepper seeds produced in the Czech Republic–Short communication. Plant Prot Sci (2006) 42(1):34. doi: 10.17221/2694-PPS
25. Herrera-Vásquez JA, Córdoba-Sellés MC, Cebrián MC, Alfaro-Fernández A, Jordá C. First report of Pepper mild mottle virus and Tobacco mild green mosaic virus infecting pepper in Panama. Plant Pathol (2009) 58(4):786. doi: 10.1111/j.1365-3059.2009.02030.x
26. Lima MF, Inoue-Nagata AK, Reifschneider FJB, Souza KRR, Ulhoa AB, Ferraz RM. Detection, occurrence and natural incidence of Pepper mild mottle virus (PMMoV) in hot peppers in Brazil. Acta Hortic (2011) 917:269–73. doi: 10.17660/ActaHortic.2011.917.37
27. Sharma PN, Patiyal K. Status of viruses infecting sweet pepper under polyhouse cultivation in Himachal Pradesh. Plant Dis Res (2011) 26(2):185.
28. Ahmad A, Tiberini A, Ashfaq M, Tomassoli L. First report of Pepper mild mottle virus infecting chilli pepper in Pakistan. New Dis Rep (2015) 32(31):2044–0588. doi: 10.5197/j.2044-0588.2015.032.031
29. Li XD, An MN, Wu YH. First report of pepper mild mottle virus in Northeast China. Plant Dis (2016) 100(2):541. doi: 10.1094/PDIS-06-15-0722-PDN
30. Berendsen SM, Schravesande WE. Complete genome sequence of a novel genotype of pepper mild mottle virus infecting pepper in Chile. Microbiol Resour Announc (2020) 9(47):e01183–20. doi: 10.1128/MRA.01183-20
31. Vélez-Olmedo JB, Fribourg CE, Melo FL, Nagata T, de Oliveira AS, Resende RO. Pepper mild mottle virus isolates from Peru induce severe symptoms in susceptible pepper plants and belong to the P 1, 2 pathotype. Trop Plant Pathol (2021) 46:381–5. doi: 10.1007/s40858-020-00402-4
32. Verma R, Tripathi S, Gorane T, Naik AA, Nikam TD, Ade AB, et al. First report of zucchini yellow mosaic virus in bell pepper, capsicum annum, in India. Plant Dis (2019) 103(10):2700. doi: 10.1094/PDIS-04-19-0740-PDN
33. Channakeshava C, Patil MS, Moger NB, Mantur SM, Prashanthi SK. Incidence of viral diseases on capsicum (sweet pepper) under protected conditions in Karnataka. Int J Chem Stud (2019) 7(6):1548–51.
34. Devi OP, Sharma SK, Sanatombi K, Devi KS, Pathaw N, Roy SS, et al. A simplified multiplex PCR assay for simultaneous detection of six viruses infecting diverse chilli species in India and its application in field diagnosis. Pathogens (2022) 12(1):6. doi: 10.3390/pathogens12010006
35. Yu M, Zhou T, Wu Y, An M. Complete Genome Sequence of a Pepper mild mottle virus Isolate from Northeast China. Genome Announc (2018) 6(9):e01500–17. doi: 10.1128/genomeA.01500-17
36. Kim JS, Lee SH, Choi HS, Kim MK, Kwak HR, Kim JS, et al. Characteristics of plant virus infections on crop samples submitted from agricultural places. Res Plant Dis (2012) 18(4):277–89. doi: 10.5423/RPD.2012.18.4.277
37. Han SH, Park JS, Han JY, Gong JS, Park CH, Kim JK, et al. New Korean isolates of Pepper mild mottle virus (PMMoV) differ in symptom severity and subcellular localization of the 126 kDa protein. Virus Genes (2017) 53(3):434–45. doi: 10.1007/s11262-017-1432-4
38. Sevik MA. Occurrence of pepper mild mottle virus in greenhousegrown pepper (Capsicum annuum L.) in the West Mediterranean region of Turkey. Afr J Biotechnol (2011) 10(25):4976–9.
39. Brunt AA, Crabtree K, Dallwitz AJ, Gibbs AJ, Watson L. Viruses of plants: Descriptions and lists from the vide database. Cambridge: Cab International, University Press (1996) p. 947–9.
40. Nagai Y. Control of mosaic disease of tomato and sweet pepper caused by tobacco mosaic virus. Spec Bull Chiba Agric Exp Station (1981) 9:1–09.
41. Dikilitas M, Guldur ME, Deryaoglu A, Ozcan ER. Antioxidant and oxidant levels of pepper (Capsicum annuum cv.'Charlee') infected with pepper mild mottle virus. Notulae Botanicae Horti Agrobotanici Cluj-Napoca (2011) 39(2):58–63. doi: 10.15835/nbha3925881
42. Kim MS, Kim MJ, Hong JS, Choi JK, Ryu KH. Patterns in disease progress and the influence of single and multiple viral infections on pepper (Capsicum annuum L.) growth. Eur J Plant Pathol (2010) 127:53–61. doi: 10.1007/s10658-009-9570-8
43. Martínez-Ochoa N, Langston DB, Mullis SW, Flanders JT. First Report of Pepper mild mottle virus in Jalapeno pepper in Georgia. Plant Health Prog (2003) 4(1):26. doi: 10.1094/PHP-2003-1203-01-BR
44. Patel PB, Sharma PN. Investigation on Seed Borne Nature of Pepper mild mottle virus and Assessment of Yield Losses in Capsicum. Palampur (HP: Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya (2019).
45. Yoon JY, Her NH, Cho IS, Chung BN, Choi SK. First report of a resistance-breaking strain of Tomato spotted wilt orthotospovirus infecting Capsicum annuum carrying the Tsw resistance gene in South Korea. Plant Dis (2021) 105(8):2259. doi: 10.1094/PDIS-09-20-1952-PDN
46. Gong MX, Zhao H, Wu X, Wang M, Zhao ZJ, He Z, et al. First report of Capsicum frutescens endornavirus 1 infecting pepper in China. J Plant Pathol (2022) 104(2):809–10. doi: 10.1007/s42161-021-00982-3
47. Wen GS, Yang LY, Anane RF, Chen ZL, Yang YH, Chen L, et al. First report of pepper mild mottle virus in Paris polyphylla var. yunnanensis China. Plant Dis (2019) 103(12):3289. doi: 10.1094/PDIS-03-19-0445-PDN
48. Zhou WP, Li YY, Li F, Tan GL. First report of natural infection of tomato by pepper mild mottle virus in China. J Plant Pathol (2021) 103:363. doi: 10.1007/s42161-020-00688-y
49. Kim SW, Jeong Y, Yang KY, Jeong RD. First report of natural infection of Dracaena braunii by pepper mild mottle virus in Korea. J Plant Pathol (2022) 104:1579. doi: 10.1007/s42161-022-01205-z
50. Kim SW, Lee GE, Lee HJ, Kim HJ, Yang KY, Jeong RD. First report of natural infection of Hydrangea macrophylla by pepper mild mottle virus in Korea. J Plant Pathol (2023) 2:1. doi: 10.1007/s42161-023-01394-1
51. Ishibashi K, Naito S, Meshi T, Ishikawa M. An inhibitory interaction between viral and cellular proteins unde1rlies the resistance of tomato to nonadapted tobamoviruses. Proc Natl Acad Sci (2009) 106(21):8778–83. doi: 10.1073/pnas.0809105106
52. Li Y, Tan G, Lan P, Zhang A, Liu Y, Li R, et al. Detection of tobamoviruses by RT-PCR using a novel pair of degenerate primers. J Virol Methods (2018) 259:122–8. doi: 10.1016/j.jviromet.2018.06.012
53. Lewandowski DJ, Dawson W. Genus tobamovirus. In: Virus taxonomy: Seventh report of the International Committee for the Taxonomy of Viruses. USA: Academic Press (1999). p. 889–94. doi: 10.1006/rwvi.1999.0282
54. Tosic M, Sutic D, Pesic Z. Transmission of tobacco mosaic virus through pepper (Capsicum annuum L.) seed. Phytopathologische Z (1980) 97(1):10–3. doi: 10.1111/j.1439-0434.1980.tb04595.x
55. Paludan N. Virus attack in Danish cultures of sweet pepper (Capsicum annuum L.) specially concerning Tobacco mosaic virus. Acta Hortic (1982) 127:65–78. doi: 10.17660/ActaHortic.1983.127.4
56. Patel P, Kumari N, Sharma PN. RT-PCR based detection of Pepper mild mottle virus from capsicum seeds and seed transmission assay. Virus Dis (2023) 34(1):50–5. doi: 10.1007/s13337-023-00807-0
57. Genda Y, Kanda A, Hamada H, Sato K, Ohnishi J, Tsuda S. Two Amino Acid Substitutions in the Coat Protein of Pepper mild mottle virus Are Responsible for Overcoming the L4 Gene-Mediated Resistance in Capsicum spp. Phytopathology (2007) 97(7):787–93. doi: 10.1094/phyto-97-7-0787
58. Demski JW. Tobacco mosaic virus in seedborne in pimiento peppers. Plant Dis (1981) 65(9):723–4. doi: 10.1094/PD-65-723
59. Alwabli AS, Khattab EA, Farag AG. Biological, serological and molecular characterization of pepper mild mottle virus isolated from weast region of kingdom of Saudi Arabia. Res J Infect Dis (2017) 5(1):1–11. doi: 10.7243/2052-5958-5-1
61. Ikegashira Y, Ohki T, Ichiki UT, Higashi T, Hagiwara K, Omura T, et al. An immunological system for the detection of Pepper mild mottle virus in soil from green pepper fields. Plant Dis (2004) 88(6):650–6. doi: 10.1094/PDIS.2004.88.6.650
62. Ichiki TU, Nagaoka EN, Hagiwara K, Uchikawa K, Tsuda S, Omura T. Integration of mutations responsible for the attenuated phenotype of Pepper mild mottle virus strains results in a symptomless cross-protecting strain. Arch Virol (2005) 150(10):2009–20. doi: 10.1007/s00705-005-0551-8
63. Palukaitis P, Zaitlin M. Tobacco mosaic virus infectivity and replication. Plant Viruses: Rod-Shaped Plant Viruses (1986) 2:105–31. doi: 10.1007/978-1-4684-7026-0_5
64. Tsuda S, Kubota K, Kanda A, Ohki T, Meshi T. Pathogenicity of Pepper mild mottle virus is controlled by the RNA silencing suppression activity of its replication protein but not the viral accumulation. Phytopathology (2007) 97(4):412–20. doi: 10.1094/PHYTO-97-4-0412
65. Velasco L, Janssen D, Ruiz-Garcia L, Segundo E, Cuadrado IM. The complete nucleotide sequence and development of a diferential detection assay for a pepper mild mottle virus (PMMoV) isolate that overcomes L3 resistance in pepper. J Virol Methods (2002) 106(1):135–40. doi: 10.1016/S0166-0934(02)00144-1
66. Han K, Zheng H, Ji M, Cui W, Hu S, Peng J, et al. A single amino acid in coat protein of Pepper mild mottle virus determines its subcellular localization and the chlorosis symptom on leaves of pepper. J Gen Virol (2020) 101(5):565–70. doi: 10.1099/jgv.0.001398
67. Rietveld K, Linschooten K, Pleij CW, Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J (1984) 3(11):2613–9. doi: 10.1002/j.1460-2075.1984.tb02182.x
68. Van Belkum A, Abrahams JP, Pleij CW, Bosch L. Five pseudoknots are present at the 204 nucleotides long 3'noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res (1985) 13(21):7673–86. doi: 10.1093/nar/13.21.7673
69. García-Arenal F. Sequence and structure at the genome 3′ end of the U2-strain of tobacco mosaic virus, a histidine-accepting tobamovirus. Virology (1988) 167(1):201–6. doi: 10.1016/0042-6822(88)90070-0
70. Avila-Rincon MJ, Ferrero ML, Alonso E, Garcí-Luque I, Díaz-Ruíz* JR. Nucleotide sequences of 5′ and 3′ non-coding regions of pepper mild mottle virus strain S RNA. J Gen Virol (1989) 70(11):3025–31. doi: 10.1099/0022-1317-70-11-3025
71. Ogai R, Kanda-Hojo A, Tsuda S. An attenuated isolate of Pepper mild mottle virus for cross protection of cultivated green pepper (Capsicum annuum L.) carrying the L3 resistance gene. Crop Prot (2013) 54:29–34. doi: 10.1016/j.cropro.2013.07.008
72. Alonso E, García-Luque I, de la Cruz A, Wicke B, Avila-Rincon MJ, Serra MT, et al. Nucleotide sequence of the genomic RNA of pepper mild mottle virus, a resistance-breaking tobamovirus in pepper. J Gen Virol (1991) 72(12):2875–84. doi: 10.1099/0022-1317-72-12-2875
73. Kirita M, Akutsu K, Watanabe Y, Tsuda S. Nucleotide sequence of the Japanese isolate of pepper mild mottle tobamovirus (TMV-P) RNA. Japanese J Phytopathol (1997) 63(5):373–6. doi: 10.3186/jjphytopath.63.373
74. Hagiwara K, Ichiki TU, Ogawa Y, Omura T, Tsuda S. A single amino acid substitution in 126-kDa protein of Pepper mild mottle virus associates with symptom attenuation in pepper; the complete nucleotide sequence of an attenuated strain, C-1421. Arch Virol (2002) 147:833–40. doi: 10.1007/s007050200030
75. Oliveira LM, Inoue-Nagata AK, Nagata T. Complete genome nucleotide sequence of Pepper mild mottle virus isolated in the Federal District, Brazil. Trop Plant Pathol (2010) 35:373–6. doi: 10.1590/S1982-56762010000600006
76. Kumari N, Sharma V, Patel P, Sharma PN. Heterologous expression of pepper mild mottle virus coat protein encoding region and its application in immuno-diagnostics. Virus Dis (2020) 3):323–32. doi: 10.1007/s13337-020-00597-9
77. Kwon SJ, Choi GS, Yoon JY, Seo JK, Choi HS. Identification of Leonurus sibiricus as a weed reservoir for three pepper-infecting viruses. Plant Pathol J (2006) 32(1):65. doi: 10.5423/PPJ.NT.07.2015.0138
78. Moreno-Pérez MG, García-Luque I, Fraile A, García-Arenal F. Mutations that determine resistance breaking in a plant RNA virus have pleiotropic effects on its fitness that depend on the host environment and on the type, single or mixed, of infection. J Virol (2016) 90(20):9128–37. doi: 10.1128/JVI.00737-16
79. Wang X, Liu F, Zhou G, Li XH, Li Z. Detection and molecular characterization of Pepper mild mottle virus in China. J Phytopathol (2006) 154(11-12):755–7. doi: 10.1111/j.1439-0434.2006.01186.x
80. Secrist SJ, Ali GS. First complete genome sequence of Pepper mild mottle virus from chili pepper in the United States. Genome Announc (2018) 6(18):10–128. doi: 10.1128/genomea.00331-18
81. Mihalik D, Hancinsky R, Kaňukova Š, Mrkvova M, Kraic J. Elicitation of Hyoscyamine Production in Datura stramonium L. Plants Using Tobamoviruses. Plants (2022) 11(23):3319. doi: 10.3390/plants11233319
82. Hamada H, Tomita R, Iwadate Y, Kobayashi K, Munemura I, Takeuchi S, et al. Cooperative effect of two amino acid mutations in the coat protein of Pepper mild mottle virus overcomes L 3-mediated resistance in Capsicum plants. Virus Genes (2007) 34:205–14. doi: 10.1007/s11262-006-0049-9
83. Choi SK, Choi GS, Kwon SJ, Yoon JY. Complete nucleotide sequences and genome organization of two pepper mild mottle virus isolates from Capsicum annuum in South Korea. Genome Announc (2016) 4(3):10–128. doi: 10.1128/genomea.00411-16
84. Yoon JY, Hong JS, Kim MJ, Ha JH, Choi GS, Choi JK, et al. Molecular characterization and infectious cDNA clone of a Korean isolate of pepper mild mottle virus from pepper. Plant Pathol J (2005) 21(4):361–8. doi: 10.5423/PPJ.2005.21.4.361
85. Ogai R, Kanda A, Kubota K, Tsuda S. Characterization and field assessment of L3-163, an attenuated strain of Pepper mild mottle virus. In XV Int Congress Virology IUMS (2011). The Unlimited World of Microbes, Japan. Final Program VI-PO23-7.
86. Choi H, Jo Y, Cho WK, Yu J, Tran PT, Salaipeth L, et al. Identification of viruses and viroids infecting tomato and pepper plants in Vietnam by metatranscriptomics. Int J Mol Sci (2020) 21(20):7565. doi: 10.3390/ijms21207565
87. Ali I, Ali A. First report of Pepper mild mottle virus in peppers in Oklahoma. Plant Dis (2015) 99(5):736. doi: 10.1094/PDIS-11-14-1144-PDN
88. Li XD, An MN, Wu YH. First report of pepper mild mottle virus in Northeast China. Plant Dis (2016) 100(2):541. doi: 10.1094/PDIS-06-15-0722-PDN
89. Takeuchi S, Hikichi Y, Kawada Y, Okuno T. Detection of tobamoviruses from soils by non-precoated indirect ELISA. J Gen Plant Pathol (2000) 66:153–8. doi: 10.1007/PL00012938
90. Ogawa T, Haga Y, Tsuda S. PMMoV-disease risk assessment using a DAS-ELISA of soil from green pepper fields. Japanese J Phytopathol (2012) 78(1):18–21. doi: 10.3186/jjphytopath.78.18
91. Sawada H, Takeuchi S, Hamada H, Kiba A, Matsumoto M, Hikichi Y. A new tobamovirus-resistance gene, L1a, of sweet pepper (Capsicum annuum L.). J Jpn Soc Hortic Sci (2004) 73(6):552–7. doi: 10.2503/jjshs.73.552
92. Damayanti TA, Kurniawati F. Pepper mild mottle virus infection in cayenne and sweet pepper in Indonesia. Australas Plant Dis Notes (2022) 17(1):4. doi: 10.1007/s13314-022-00451-5
93. Phatsaman T, Hongprayoon R, Wasee S. Monoclonal antibody-based diagnostic assays for pepper mild mottle virus. J Plant Pathol (2020) 102:327–33. doi: 10.1007/s42161-019-00421-4
94. Mnari-Hattab M, Ezzaier K. Biological, serological, and molecular characterization of Pepper mild mottle virus (PMMoV) in Tunisia. Tunis J Plant Prot (2006) 1:1–2.
95. Peng J, Shi B, Zheng H, Lu Y, Lin L, Jiang T, et al. Detection of pepper mild mottle virus in pepper sauce in China. Arch Virol (2015) 160:2079–82. doi: 10.1007/s00705-015-2454-7
96. Kim JH, Choi GS, Kim JS, Lee SH, Choi JK, Ryu KR. Development of single-tube multiplex immunocapture RT-PCR assay for simultaneous detection of two pepper tobamoviruses. Plant Pathol J (2006) 22(2):164–7. doi: 10.5423/PPJ.2006.22.2.164
97. Haramoto E, Kitajima M, Kishida N, Konno Y, Katayama H, Asami M, et al. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl Environ Microbiol (2013) 79(23):7413–8. doi: 10.1128/AEM.02354-13
98. Çaglar JY, Fidan JS, Elbeaino MJ. Detection and Molecular Characterization of Pepper Mild Mottle Virus from Turkey. J Phytopathol (2013) 1(6):434–8. doi: 10.1111/jph.12068
99. Choi GS, Kim JH, Lee DH, Kim JS, Ryu KH. Occurrence and distribution of viruses infecting pepper in Korea. Plant Pathol J (2005) 21(3):258–61. doi: 10.5423/PPJ.2005.21.3.258
100. Stommel JR, Dumm JM, Hammond J. Effect of ozone on inactivation of purified Pepper mild mottle virus and contaminated pepper seed. PhytoFront (2021) 1(2):85–93. doi: 10.1094/PHYTOFR-09-20-0020-R
101. Gangireddygari VS, Cho IS, Choi S, Yoon JY. Inhibitory effects of pepper mild mottle virus infection by supernatants of five bacterial cultures in capsicum annuum L. Plant Pathol J (2022) 38(6):646–55. doi: 10.5423/PPJ.OA.08.2022.0110
102. Groth-Helms D, Juszczak S, Adkins S. First report of Chili pepper mild mottle virus in calibrachoa in the United States. New Dis Rep (2022) 46(1):e12120. doi: 10.1002/ndr2.12120
103. Boukema IW. Allelism of genes controlling resistance to TMV in Capsicum. Euphytica (1980) 29:433–9. doi: 10.1007/BF00025143
104. Boukema IW. Resistance to TMV in Capsicum chacoense Hunz. is governed by allele of the L-locus. Capsicum Newslett (1982) 3:47–8.
105. Boukema IW. Resistance to TMV in Capsicum chacoense Hunz is governed by an allele of the L-locus. Capsicum Newslett (1984) 3:47–8.
106. Berzal-Herranz A, de la Cruz A, Tenllado F, Diaz-Ruiz JR, López L, Sanz AI, et al. The capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology (1995) 209:498–505. doi: 10.1006/viro.1995.1282
107. Gilardi P, Garcia-Luque I, Serra MT. The coat protein of tobamovirus acts as elicitor of both L 2 and L 4 gene-mediated resistance in Capsicum. J Gen Virol (2004) 85(7):2077–85. doi: 10.1099/vir.0.80017-0
108. Antignus Y, Lachman O, Pearlsman M, Maslenin L, Rosner A. A new pathotype of Pepper mild mottle virus (PMMoV) overcomes the L 4 resistance genotype of pepper cultivars. Plant Dis (2008) 92(7):1033–7. doi: 10.1094/PDIS-92-7-1033
109. De La Cruz A, Lopez L, Tenllado F, Díaz-Ruíz JR, Sanz AI, Vaquero C, et al. The coat protein is required for the elicitation of the Capsicum L2 gene-mediated resistance against the tobamoviruses. Mol Plant-Microbe Interact (1997) 10(1):107–13. doi: 10.1094/MPMI.1997.10.1.107
110. Tsuda S, Kirita M, Watanabe Y. Characterization of a pepper mild mottle tobamovirus strain capable of overcoming the L3 gene-mediated resistance, distinct from the resistance-breaking Italian isolate. Mol Plant-Microbe Interact (1998) 11(4):327–31. doi: 10.1094/MPMI.1998.11.4.327
111. Choi GS, Choi SK, Cho JD, Cho IS. A pathotype of pepper mild mottle virus causing necrotic spot symptoms in paprika fruit. Res Plant Dis (2013) 19(2):124–7. doi: 10.5423/rpd.2013.19.2.124
112. Hamada H, Takeuchi S, Kiba A, Tsuda S, Hikichi Y, Okuno T. Amino acid changes in Pepper mild mottle virus coat protein that affect L 3 gene-mediated resistance in pepper. J Gen Plant Pathol (2002) 68:155–62. doi: 10.1007/PL00013069
113. Smith E, Dombrovsky A. Aspects in Tobamovirus management in intensive agriculture. InPlant diseases-current threats and management trends. Intech Open (2019) 31–41. doi: 10.5772/intechopen.87101
114. Balique F, Lecoq H, Raoult D, Colson P. Can plant viruses cross the kingdom border and be pathogenic to humans? Viruses (2015) 7(4):2074–98. doi: 10.3390/v7042074
115. Aggarwala V, Liang G, Bushman FD. Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mobile DNA (2017) 8(1):1–10. doi: 10.1186/s13100-017-0095-y
116. Kawada J, Okuno Y, Torii Y, Okada R, Hayano S, Ando S, et al. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Sci Rep (2016) 6:33452. doi: 10.1038/srep33452
117. Taboada B, Moran P, Serrano-Vazquez A, Isa P, Rojas-Velázquez L, Perez-Juarez H, et al. The gut virome of healthy children during the first year of life is diverse and dynamic. PloS One (2021) 16(4):e0240958. doi: 10.1371/journal.pone.0240958
118. Aguado-García Y, Taboada B, Morán P, Rivera-Gutiérrez X, Serrano-Vázquez A, Iša P, et al. Tobamoviruses can be frequently present in the oropharynx and gut of infants during their first year of life. Sci Rep (2020) 10(1):1–1. doi: 10.1038/s41598-020-70684-w
119. Colson P, Richet H, Desnues C, Balique F, Moal V, Grob JJ, et al. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PloS One (2010) 5(4):e10041. doi: 10.1371/journal.pone.0010041
120. Rosiles-González G, Ávila-Torres G, Moreno-Valenzuela OA, Acosta-González G, Leal-Bautista RM, Grimaldo-Hernández CD, et al. Occurrence of pepper mild mottle virus (PMMoV) in groundwater from a karst aquifer system in the yucatan peninsula, Mexico. Food Environ Virol (2017) 9(4):487–97. doi: 10.1007/s12560-017-9309-1
121. Sassi HP, Tuttle KD, Betancourt WQ, Kitajima M, Gerba CP. Persistence of viruses by qPCR downstream of three effluent-dominated rivers in the western United States. Food Environ Virol (2018) 10(3):297–304. doi: 10.1007/s12560-018-9343-7
122. Canh VD, Torii S, Furumai H, Katayama H. Application of capsid integrity (RT-) qPCR to assessing occurrence of intact viruses in surface water and tap water in Japan. Water Res (2021) 189:116674. doi: 10.1016/j.watres.2020.116674
123. Symonds EM, Rosario K. Breitbart M Pepper mild mottle virus: Agricultural menace turned effective tool for microbial water quality monitoring and assessing (waste) water treatment technologies. PloS Pathog (2019) 15(4):e1007639. doi: 10.1371/journal.ppat.1007639
124. Kitajima M, Sassi HP, Torrey JR. Pepper mild mottle virus as a water quality indicator. NPJ Clean Water (2018) 1(1):19. doi: 10.1038/s41545-018-0019-5
125. Tomita R, Sekine KT, Mizumoto H, Sakamoto M, Murai J, Kiba A, et al. Genetic basis for the hierarchical interaction between Tobamovirus spp. and L resistance gene alleles from different pepper species. Mol Plant-Microbe Interact (2011) 24(1):108–17. doi: 10.1094/mpmi-06-10-0127
126. Holmes FO. Inheritance of resistance to tobacco-mosaic disease in the pepper. Phytopathology (1937) 27:637–42.
127. Sawada H, Takeuchi S, Matsumoto K, Hamada H, Kiba A, Matsumoto M, et al. A new Tobamovirus-resistance gene, Hk, in Capsicum annuum. J Jpn Soc Hortic Sci (2005) 74(4):289–94. doi: 10.2503/jjshs.74.289
128. Lefebvre V, Pflieger S, Thabuis A, Caranta C, Blattes A, Chauvet JC, et al. Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome (2002) 45(5):839–54. doi: 10.1139/g02-053
129. Tomita R, Murai J, Miura Y, Ishihara H, Liu S, Kubotera Y, et al. Fine mapping and DNA fiber FISH analysis locates the tobamovirus resistance gene L 3 of Capsicum chinense in a 400-kb region of R-like genes cluster embedded in highly repetitive sequences. Theor Appl Genet (2008) 117:1107–18. doi: 10.1007/s00122-008-0848-6
130. Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, et al. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell (1997) 9(4):521–32. doi: 10.1105/tpc.9.4.521
131. Livingstone KD, Lackney VK, Blauth JR, Van Wijk RI, Jahn MK. Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics (1999) 152(3):1183–202. doi: 10.1093/genetics/152.3.1183
132. Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, et al. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell (1998) 10(6):1055–68. doi: 10.1105/tpc.10.6.1055
133. Grube RC, Radwanski ER, Jahn M. Comparative genetics of disease resistance within the Solanaceae. Genetics (2000) 155(2):873–87. doi: 10.1093/genetics/155.2.873
134. Marte M, Castagnoli F, Saccardo F. Screening peppers for resistance to pepper mild mottle virus (tobamoviruses). Screening peppers for resistance to pepper mild mottle virus (tobamoviruses). Capsicum Newslett (1992) (Special issue), 144–9.
135. Suzuki K, Kuroda T, Miura Y, Murai J. Screening and field trials of virus resistant sources in Capsicum spp. Plant Dis (2003) 87(7):779–83. doi: 10.1094/PDIS.2003.87.7.779
136. Kumari N, Sharma PN. Molecular characterization of PMMoV and evaluation of resistance in capsicum. Palampur (HP: Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya (2014).
137. Matsunaga H, Saito T, Hirai M, Nunome T, Yoshida T. DNA markers linked to Pepper mild mottle virus (PMMoV) resistant locus (L4) in Capsicum. J Jpn Soc Hortic Sci (2003) 72(3):218–20. doi: 10.2503/jjshs.72.218
138. Kim HJ, Han JH, Yoo JH, Cho HJ, Kim BD. Development of a sequence characteristic amplified region marker linked to the L 4 locus conferring broad spectrum resistance to tobamoviruses in pepper plants. Mol Cells (2008) 25(2):205–10.
139. Yang HB, Liu WY, Kang WH, Jahn M, Kang BC. Development of SNP markers linked to the L locus in Capsicum spp. by a comparative genetic analysis. Molecular Breeding (2009) 24:433–46. doi: 10.5423/PPJ.2005.21.3.258
140. Sugita T, Yamaguchi K, Sugimura Y, Nagata R, Yuji K, Kinoshita T, et al. Development of SCAR markers linked to L3 gene in Capsicum. Breed Sci (2004) 54(2):111–5. doi: 10.1270/jsbbs.54.111
141. Barka GD, Lee J. Molecular marker development and gene cloning for diverse disease resistance in pepper (Capsicum annuum L.): Current status and prospects. Plant Breed Biotechnol (2020) 8(2):89–113. doi: 10.9787/PBB.2020.8.2.89
142. Choi GS, Choi SK, Cho IS, Kwon SJ. Resistance screening to pepper mild mottle virus pathotypes in paprika cultivars. Res Plant Dis (2014) 20(4):299–302. doi: 10.5423/rpd.2014.20.4.29
143. Hudcovicová M, Korbelová E, Šliková S, Klčová L, Mihálik D, Kraic J. Molecular selection of tomato and pepper breeding lines possessing resistance alleles against tobamoviruses. Agriculture (2015) 61(1):33–7. doi: 10.1515/agri-2015-0008
144. Matsumoto M, Nitta M, Sawada H, Nomachi A. 'Tosahime R', a PMMoV [pepper mild mottle tobamovirus] resistant sweet pepper [Capsicum annuum]. Bull Kochi Agric Res Center (Japan) (1999) 8:47–52.
145. Sawada H, Nomachi A, Matsumoto M. Breeding of the new PMMoV (P1, 2) resistant sweet pepper [Capsicum annuum] cultivar 'Tosajishi-beauty'. Bull Kochi Agric Res Center (Japan) (2002).
146. Sawada H, Yoshimoto E, Matsumoto M. Breeding of the PMMoV (P1, 2) resistant sweet pepper [Capsicum annuum] cultivar'Tosajishi-slim'. Bull Kochi Agric Res Center (Japan) (2008).
147. Hosomi Y, Sawada H, Nitta M, Kataoka A, Matsumoto M. Breeding of the PMMoV (P1, 2) resistant sweet pepper cultivar 'Tosa-P-Red'. Bull Kochi Agric Res Center (Japan) (2009).
148. Genda Y, Momma N, Ishikawa KE, Ogawa H, Kimura M, Nunomura OS, et al. Breeding and characteristics of a pepper (Capsicum annuum L.) rootstock cultivar 'Murasaki L4 Daisuke' with violet hypocotyl and resistance to bacterial wilt and PMMoV. Hort Res (2017) 16(2):203–10. doi: 10.2503/hrj.16.203
149. Özkaynak E, Devran Z, Kahveci E, Doğanlar S, Başköylü B, Doğan F, et al. Pyramiding multiple genes for resistance to PVY, TSWV and PMMoV in pepper using molecular markers. Europ J Hort Sci (2014) 79(4):233–9.
150. McKinney H. Mosaic diseases in the Canary Islands, West Africa and Gibraltar. J Agric Res (1929) 39:577–8.
151. Lecoq H. “Control of plant virus diseases by cross protection”. In: Hadidi A, Khetarpal RK, Koganezawa H, editors. Plant virus disease control. St. Paul: APS Press (1998). p. 33e40.
152. Nakazono-Nagaoka E, Omura T, Uehara-Ichiki T. A single amino acid substitution in the 126-kDa protein of pepper mild mottle virus controls replication and systemic movement into upper non-inoculated leaves of bell pepper plants. Arch Virol (2011) 156(5):897–901. doi: 10.1007/s00705-011-0919-x
153. Slavokhotova AA, Istomina EA, Andreeva EN, Korostyleva TV, Pukhalskij VA, Shijan AN, et al. An attenuated strain of cucumber green mottle mosaic virus as a biological control agent against pathogenic viral strains. Am J Plant Sci (2016) 7(5):724–32. doi: 10.4236/ajps.2016.75066
154. NAGAI Y. Production of C-1421, an attenuated mutant of pepper strain of tobacco mosaic virus. Japanese J Phytopathol (1987) 53(2):168–74. doi: 10.3186/jjphytopath.53.168
155. Tsuda S. Development of plant virus symptoms and attenuated virus strains to control viral disease. J Gen Plant Pathol (2020) 86(6):538–42. doi: 10.1007/s10327-020-00951-1
156. Tenllado F, Llave C, Diaz-Ruiz JR. RNA interference as a new biotechnological tool for the control of virus diseases in plants. Virus Res (2004) 102(1):85–96. doi: 10.1016/j.virusres.2004.01.019
157. Yoon J, Fang M, Lee D, Park M, Kim KH, Shin C. Double-stranded RNA confers resistance to pepper mottle virus in Nicotiana benthamiana. Appl Biol Chem (2021) 64(1):1–8. doi: 10.1186/s13765-020-00581-3
158. Miroshnichenko D, Timerbaev V, Okuneva A, Klementyeva A, Sidorova T, Pushin A, et al. Enhancement of resistance to PVY in intragenic marker-free potato plants by RNAi-mediated silencing of eIF4E translation initiation factors. Plant Cell Tissue Organ Culture (2020) 140(3):691–705. doi: 10.1007/s11240-019-01746-9
159. Namgial T, Kaldis A, Chakraborty S, Voloudakis A. Topical application of double-stranded RNA molecules containing sequences of Tomato leaf curl virus and Cucumber mosaic virus confers protection against the cognate viruses. Physiol Mol Plant Pathology (2019) 108:101432. doi: 10.1016/j.pmpp.2019.101432
160. Ramesh SV, Shivakumar M, Praveen S, Chouhan BS, Chand S. Expression of short hairpin RNA (shRNA) targeting AC2 gene of Mungbean yellow mosaic India virus (MYMIV) reduces the viral titre in soybean. 3 Biotech (2019) 9:1–9. doi: 10.1007/s13205-019-1865-7
161. Ali ME, Ishii Y, Taniguchi JI, Waliullah S, Kobayashi K, Yaeno T, et al. Conferring virus resistance in tomato by independent RNA silencing of three tomato homologs of Arabidopsis TOM1. Arch Virol (2018) 163:1357–62. doi: 10.1007/s00705-018-3747-4
162. Konakalla NC, Kaldis A, Berbati M, Masarapu H, Voloudakis AE. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta (2016) 244:961–9. doi: 10.1007/s00425-016-2567-6
163. Yasmeen A, Kiani S, Butt A, Rao AQ, Akram F, Ahmad A, et al. Amplicon-based RNA interference targeting V2 gene of cotton leaf curl Kokhran Virus-Burewala strain can provide resistance in transgenic cotton plants. Mol Biotechnol (2016) 58:807–20. doi: 10.1007/s12033-016-9980-8
164. Ntui VO, Kong K, Khan RS, Igawa T, Janavi GJ, Rabindran R, et al. Resistance to Sri Lankan Cassava Mosaic Virus (SLCMV) in Genetically Engineered Cassava cv. KU50 through RNA Silencing. PloS One (2015) 10(4):e0120551. doi: 10.1371/journal.pone.0120551
165. Guo J, Gao S, Lin Q, Wang H, Que Y, Xu L. Transgenic sugarcane resistant to Sorghum mosaic virus based on coat protein gene silencing by RNA interference. BioMed Res Int (2015) 2015:861907. doi: 10.1155/2015/861907
166. Mansoor S, Saeed M, Amin I, Briddon RW, Al-Sadi AM. RNA interference-based resistance in transgenic tomato plants against Tomato yellow leaf curl virus-Oman (TYLCV-OM) and its associated betasatellite. Virol J (2015) 12(1):1–2. doi: 10.1186/s12985-015-0263-y
167. Bonfim K, Faria JC, Nogueira EO, Mendes ÉA, Aragão FJ. RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant-Microbe Interact (2007) 20(6):717–26. doi: 10.1094/MPMI-20-6-0717
168. Elayabalan S, Kalaiponmani K, Subramaniam S, Selvarajan R, Panchanathan R, Muthuvelayoutham R, et al. Development of Agrobacterium-mediated transformation of highly valued hill banana cultivar Virupakshi (AAB) for resistance to BBTV disease. World J Microbiol Biotechnol (2013) 29:589–96. doi: 10.1007/s11274-012-1214-z
169. Tenllado F, Diaz-Ruiz JR. Complete resistance to pepper mild mottle tobamovirus mediated by viral replicase sequences partially depends on transgene homozygosity and is based on a gene silencing mechanism. Transgenic Res (1999) . 8:83–93. doi: 10.1023/A:1008910400047
170. Zhao Y, Yang X, Zhou G, Zhang T. Engineering plant virus resistance: from RNA silencing to genome editing strategies. Plant Biotechnol J (2020) 18(2):328–36. doi: 10.1111/pbi.13278
171. Tenllado F, Martínez-García B, Vargas M, Díaz-Ruíz JR. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol (2003) 3:1–1. doi: 10.1186/1472-6750-3-3
172. Mitter N, Worrall EA, Robinson KE, Li P, Jain RG, Taochy C, et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants (2017) 3(2):1–10. doi: 10.1038/nplants.2016.207
173. Tenllado F, Garcia-Luque I, Serra MT, Diaz-Ruiz JR. Nicotiana benthamiana plants transformed with the 54-kDa region of the pepper mild mottle tobamovirus replicase gene exhibit two types of resistance responses against vital infection. Virology (1995) 211(1):170–83. doi: 10.1006/viro.1995.1398
174. Roy AS, Kesavan Pillai S, Ray SS. Layered double hydroxides for sustainable agriculture and environment: an overview. ACS omega (2022) 7(24):20428–40. doi: 10.1021/acsomega.2c01405
175. Gangireddygari VSR, Chung BN, Cho IS, Yoon JY. Inhibitory effect of chitosan and phosphate cross-linked chitosan against cucumber mosaic virus and pepper mild mottle virus. Plant Pathol J (2021) 37:632–40. doi: 10.5423/PPJ.OA.10.2021.0155
176. Elbeshehy EKF, Hassan WM, Baeshen AA. Controlling pepper mild mottle virus (PMMoV) infection in pepper seedlings by use of chemically synthetic silver nanoparticles. Molecules (2022) 28(1):139. doi: 10.3390/molecules28010139
177. Kim JH, Yoon JY, Kwon SJ, Cho IS, Nguyen MC, Choi SK, et al. Inhibitory components from Glycosmis stenocarpa on pepper mild mottle virus. J Microbiol Biotechnol (2016) 26(12):2138–40. doi: 10.4014/jmb.1607.07034
178. Oka N, Ohki T, Honda Y, Matsumoto H, Nishio T. Inactivation of Pepper mild mottle virus in soil amended with cellulose. Jpn J Soil Sci Plant Nutr (2004) 75:673–7.
179. Oka N, Ohki T, Honda Y, Nagaoka K, Takenaka M. Inhibition of Pepper mild mottle virus with Commercial Cellulases. J Phytopathol (2008) 156(2):65–7. doi: 10.1111/j.1439-0434.2007.01316.x
180. Zhang J, Zhao L, Zhu C, Wu Z, Zhang G, Gan X, et al. Facile synthesis of novel vanillin derivatives incorporating a bis (2-hydroxyethyl) dithhioacetal moiety as antiviral agents. J Agric Food Chem (2017) 65(23):4582–8. doi: 10.1021/acs.jafc.7b01035
181. Shi J, He H, Hu D, Song B. Defense mechanism of capsicum annuum l. infected with pepper mild mottle virus induced by vanisulfane. J Agric Food Chem (2022) 70(12):3618–32. doi: 10.1021/acs.jafc.2c00659
182. Meng X, Wang N, Long X, Hu D. Degradation of a novel pesticide antiviral agent vanisulfane in aqueous solution: kinetics, identification of photolysis products, and pathway. ACS omega (2020) 5(38):24881–9. doi: 10.1021/acsomega.0c03661
183. Shi J, He H, Liu Z, Hu D. Pepper mild mottle virus coat protein as a novel target to screen antiviral drugs. J Agric Food Chem (2022) 70(27):8233–42. doi: 10.1021/acs.jafc.2c02667
184. Luo L, Liu D, Lan S and Gan X. Design, synthesis, and biological activity of novel chalcone derivatives containing an 1,2,4- oxadiazole moiety. Front Chem (2022) 10:943062. doi: 10.3389/fchem.2022.943062
185. Li H, Huang W, Xu L, Zhou X, Liu H, Cheng Z. Stenotrophomonas maltophilia HW2 enhanced cucumber resistance against cucumber green mottle mosaic virus. J Plant Biol (2016) 59:488–95. doi: 10.1007/s12374-016-0246-6
186. Kim NG, Seo EY, Han SH. Pseudomonas oleovorans Strain KBPF-004 Culture Supernatants Reduced Seed Transmission of Cucumber green mottle mosaic virus and Pepper mild mottle virus, and Remodeled Aggregation of 126 kDa and Subcellular Localization of Movement Protein of Pepper mild'. Plant Pathol J (2017) 33:393–401. doi: 10.5423/PPJ.OA.03.2017.0047
187. Adedire OM, Aduramigba-Modupe AO, Olaoye SO, Ogundipe WF, Farinu AO. The effect of rhizosphere bacterial consortium on the manifestation of tobamoviral infection symptoms on tomato. Int J Plant Soil Sci (2018) 25(2):1–8. doi: 10.9734/IJPSS/2018/44637
188. Khosroshahi TS, Canto T, Rakhshandehroo F, Salehi Jouzani G. Molecular characterization and transient expression in plants of a Mirabilis jalapa antiviral protein (MAP), and its use in functional studies. Eur J Plant Pathol (2022) 1:1–8. doi: 10.1007/s10658-021-02412-7
189. Hamdi RF, Owaid ZM, Omer OA, Kafi FN, Luhemus HH. Biological control on Tomato Mosaic Virus (ToMV) by using some plant extracts. Systematic Rev Pharm (2020) 1:11(12).
190. Nurviani N, Somowiyarjo S, Sulandari S, Subandiyah S. The inhibition of Tobamovirus by using the extract of banana flower. Jurnal Perlindungan Tanaman Indonesia (2018) 22(2):181–5. doi: 10.22146/jpti.31595
191. Deepthi KN, Madhusudhan AC, Uday Shankar H, Kumar B, Prakash HS, Shetty HS. Effect of plant extracts and acetone precipitated proteins from six medicinal plants against tobamovirus infection. Int J Virol (2007) 3(2):80–7.
192. Parizipour MG, Shahriari AG. Investigation of antiviral potential of licorice (Glycyrrhiza glabra L.) crude extract against Tobacco mosaic virus. J Anim. Plant Sci (2020) 30(1):107–14.
193. Gharib HA, Mandour AM. Effect of Populus nigra spring and autumn leaves extract on Capsicum annuum infected with pepper mild mottle virus. Sci Rep (2022) 12:22194. doi: 10.1038/s41598-022-24786-2
194. Elsharkawy MM, Al-Askar AA, Abdelkhalek A, Behiry SI, Kamran M, Ali M. Suppression of pepper mild mottle virus (PMMoV) by modified whey proteins. Life (2022) 12(8):1165. doi: 10.3390/life12081165
195. Khan ZA, Kumar R, Dasgupta I. CRISPR/cas-mediated resistance against viruses in plants. Int J Mol Sci (2022) 23(4):2303. doi: 10.3390/ijms23042303
196. Vargas-Hernandez M, Macias-Bobadilla I, Guevara-Gonzalez RG, Rico-Garcia E, Ocampo-Velazquez RV, Avila-Juarez L, et al. Nanoparticles as potential antivirals in agriculture. Agriculture (2020) 10(10):444. doi: 10.3390/agriculture10100444
197. Worrall EA, Hamid A, Mody KT, Mitter N, Pappu HR. Nanotechnology for plant disease management. Agronomy (2018) 8(12):285. doi: 10.3390/agronomy8120285
198. Khiyami MA, Almoammar H, Awad YM, Alghuthaymi MA, Abd-Elsalam KA. Plant pathogen nanodiagnostic techniques: forthcoming changes? Biotechnol Biotechnol Equip (2014) 28(5):775–85. doi: 10.1080/13102818.2014.960739
Keywords: capsicum, Tobamovirus, PMMoV, resistance, L gene, integrated management
Citation: Kumari N, Sharma V, Patel P and Sharma PN (2023) Pepper mild mottle virus: a formidable foe of capsicum production—a review. Front. Virol. 3:1208853. doi: 10.3389/fviro.2023.1208853
Received: 19 April 2023; Accepted: 25 July 2023;
Published: 12 October 2023.
Edited by:
Ashish Srivastava, Amity University, IndiaReviewed by:
Amit Kumar Kesharwani, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2023 Kumari, Sharma, Patel and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nidhi Kumari, bmlkaGlfcmlhbGNoQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.