
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol. , 29 August 2023
Sec. Translational Virology
Volume 3 - 2023 | https://doi.org/10.3389/fviro.2023.1163385
 Neale Harrison1†
Neale Harrison1† Lauren Richardson1†
Lauren Richardson1† Chiara Pallini1
Chiara Pallini1 Ines Morano1
Ines Morano1 Elizabeth Jinks1
Elizabeth Jinks1 Jamie Cowley1
Jamie Cowley1 Hujo Chan1
Hujo Chan1 Harriet J. Hill2
Harriet J. Hill2 Aekkachai Tuekprakhon2
Aekkachai Tuekprakhon2 Zhi Li1
Zhi Li1 Cristina Matas de las Heras1
Cristina Matas de las Heras1 Ana Teodosio1
Ana Teodosio1 Andrea S. Lavado1
Andrea S. Lavado1 Robert Moring1
Robert Moring1 Ayesha Ashraf1
Ayesha Ashraf1 Timothy R. Dafforn3
Timothy R. Dafforn3 Dimitris K. Grammatopoulos4,5
Dimitris K. Grammatopoulos4,5 John Gordon1
John Gordon1 Catherine A. Brady1
Catherine A. Brady1 Lawrence S. Young4
Lawrence S. Young4 Nicholas M. Barnes1,6‡
Nicholas M. Barnes1,6‡ Zania Stamataki2‡
Zania Stamataki2‡ Omar S. Qureshi1*‡
Omar S. Qureshi1*‡Introduction: The engagement of the SARS-CoV-2 spike protein with ACE2 is a critical step for viral entry to human cells, and, therefore, blocking this interaction is a major determinant of the efficacy of monoclonal antibody therapeutics and vaccine elicited serum antibodies. The emergence of SARS-CoV-2 variants has necessitated the development of adaptable assays that can be applied to assess the effectiveness of antibody-based therapeutics.
Methods: Through the testing of a range of recombinant spike proteins, we have developed a cell-based, ACE2/spike protein interaction assay that characterises monoclonal anti-spike protein antibodies and neutralising antibodies in donor serum. The assay uses high-content imaging to quantify cell-bound spike protein fluorescence.
Results: Using spike proteins from the original “Wuhan” SARS-CoV-2 strain and the Delta and Omicron variants, we identified differential blocking activity of three monoclonal antibodies directed against the spike receptor-binding domain. Importantly, biological activity in the spike interaction assay translated to efficacy in a SARS-CoV-2 infection assay.
Discussion: The spike protein interaction assay can be used to monitor anti-spike antibodies against the major known SARS-CoV-2 variants and is readily adaptable for quantification of the impact of antibodies against new and emerging SARS-CoV-2 variants.
The ongoing SARS-CoV-2 pandemic necessitates the development of tools that can support the advance of therapeutic antibodies that are efficacious against both current and future viral variants (1). Quantifying the activity of neutralising antibodies, whether they are recombinant monoclonals or elicited by vaccination, is therefore of considerable importance for drug development and monitoring effective immunity (2, 3). Emerging SARS-CoV-2 variants may exhibit escape or reduced neutralisation by current monoclonal antibodies, or from antibodies generated by vaccination to target earlier viral strains (4), and knowledge of these limitations informs health and political strategies in the face of societal challenges arising from the pandemic.
The SARS-CoV-2 spike protein engagement with the host ACE2 receptor is a key step in viral entry (5) and is therefore a major target for therapeutic interventions (6–9). It also serves as a relevant mechanism for studying the efficacy of neutralising antibodies in serum (10, 11). Functional virus neutralisation experiments require biosafety level 3 facilities, which limit the accessibility of these assays in many settings, and also require the propagation of strains prior to infection studies, which carries an inherent risk. Several other techniques have been developed to simplify and reduce this infectious risk, including the use of pseudotyped viruses and the surrogate neutralisation assay (12, 13). Reduced risk procedures using pseudoviruses still require the packaging and expression of viral particles (13). We therefore sought to develop a reductionist cell-based model, suitable for high-throughput screening, that quantifies the inhibition of SARS-CoV-2 spike protein interactions with mammalian cells and is suitable for assisting in therapeutic antibody development and immune monitoring.
See Table 1 for a list of recombinant spike proteins used in this study. Hoechst-33342 was obtained from ThermoFisher Scientific (H21492), His tag® antibody [HIS.H8] was obtained from Abcam (ab18184), Rho1D4 antibody was obtained from Cube Biotech (40020), and goat anti-Mouse IgG H&L (Alexa Fluor® 488) was obtained from ThermoFisher Scientific (A-11001). See Table 2 for a list of antibodies against spike proteins.
A549 lung carcinoma cells, or A549 cells expressing human ACE2, were obtained from Invivogen (a549 or a549-hace2, respectively). Cells were cultured in F-12K Ham Nutrient Mixture media (ThermoFisher Scientific; 21127022) and supplemented with 10% fetal bovine serum (Sigma-Aldrich; F9665) and 1% penicillin-streptomycin (ThermoFisher Scientific; 15140122) in a tissue culture incubator at 37°C (5% CO2) using standard cell culture practices.
Cells were seeded in 96-well plates overnight prior to incubation with recombinant spike proteins. Recombinant spike proteins (4.0 μg/mL, unless otherwise indicated) were pre-incubated with anti-His detection antibody (8.0 μg/mL), and blocking antibodies or plasma were pre-incubated for 1 hour prior to the addition to cells for 1 hour at 37°C. This was followed by preparation for confocal microscopy.
Media was removed from cells and wells washed with PBS (ThermoFisher Scientific; 14190144). Cells were then fixed with 4% paraformaldehyde for 10 minutes before being washed twice in PBS, once in permeabilisation buffer (ThermoFisher Scientific; 00–8333–56), prior to blocking (1% goat serum in permeabilisation buffer). Cells were then incubated with Alexa Fluor 488-conjugated anti-mouse IgG (1:500) and Hoechst (1:1,000) prepared in permeabilisation buffer before being washed three times with permeabilisation buffer, twice in PBS, followed by imaging in PBS. Confocal microscopy was carried out using a Yokogawa CQ1 spinning-disc microscope using a × 40 objective and appropriate excitation/emission settings for Hoechst and Alexa Fluor 488. Z-stack images were acquired and displayed as maximum intensity projections. Image analysis was carried out using Yokogawa image analysis software.
Levels of IgG, IgA, and IgM antibodies in plasma samples were measured by protein ELISA using the IgG/A/M Sars-COV-2 ELISA kit, following the manufacturer’s instructions (Binding Site; MK654).
Vero cells were cultured in DMEM, supplemented with 10% foetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, 10 µg/mL streptomycin, and 1% non-essential amino acids (cDMEM), and then seeded into 96-well plates. SARS-CoV-2-England 2 (Wuhan strain) at 106 IU/mL (GSAID Accession ID EPI_ISL_407073) was kindly gifted to us by Christine Bruce, Public Health England. Antibodies were pre-incubated with virus for 1 hour prior to being added to the Vero cells. After a 48-hour incubation at 37°C, cells were fixed with ice-cold methanol (5 minutes), washed with PBS, and stained with rabbit anti-SARS-CoV-2 spike protein, subunit 1 antibody (CR3022, The Native Antigen Company), detected by Alexa Fluor 555-conjugated goat anti-rabbit IgG secondary antibody (Invitrogen, ThermoFisher Scientific). Cell nuclei were stained with Hoechst 33342 (ThermoFisher Scientific). Cells were washed with PBS and imaged and analysed using a ThermoFisher Scientific CellInsight CX5 High-Content Screening (HCS) platform. Infected cells and cell viability were detected by measuring perinuclear fluorescence above a set threshold that was determined by positive (untreated) and negative (uninfected) controls. Automated quantification algorithms were developed with assistance from Dr Henri Huppert, ThermoFisher Scientific, UK.
Data are presented as mean + SEM or mean ± SD from at least three independent experimental setups unless otherwise indicated. Curve-fitting data analysis was conducted using KaleidaGraph (version 3.5). All statistical analysis was carried out using GraphPad Prism software (version 9.2). Data were tested for normality using the Shapiro–Wilk test and normally distributed data were analysed using one-way ANOVA and post hoc Tukey tests (unless specified), with corrected p-values where multiple comparisons were carried out. A Kruskal–Wallis test was used for data that were not considered to be normally distributed.
All samples were obtained with informed consent and with approval from the appropriate research ethics committee (REC Reference 20/WA/0216). The human studies reported in the manuscript abide by the Declaration of Helsinki principles.
Initial experiments investigated the interaction of a recombinant His-tagged SARS-CoV-2 spike protein (parental strain, Wuhan 1) to lung epithelial A549 cell lines, either wild-type (A549-WT) or over-expressing ACE2 (A549-ACE2). The His-tagged spike protein was detected using a mouse IgG anti-His antibody, followed by an AF488-conjugated anti-mouse IgG secondary antibody, and visualised using high-content imaging (Figure 1A), quantifying the intensity of spike protein labelling per cell, which encompasses both surface-bound spike protein and that which binds and is subsequently internalised (Figure 1C). Although the labelling of wild-type A549 cells with spike protein was close to background levels, clear punctate labelling of spike protein was evident on A549 cells over-expressing ACE2. Next, we tested a spike protein within a detergent micelle (Alpha), which likely contains the trimeric spike in its native configuration, and which also exhibited clear labelling as detected by an anti-RhoD1A4 antibody and an AF488-conjugated anti-mouse IgG secondary antibody (Figures 1B, C).
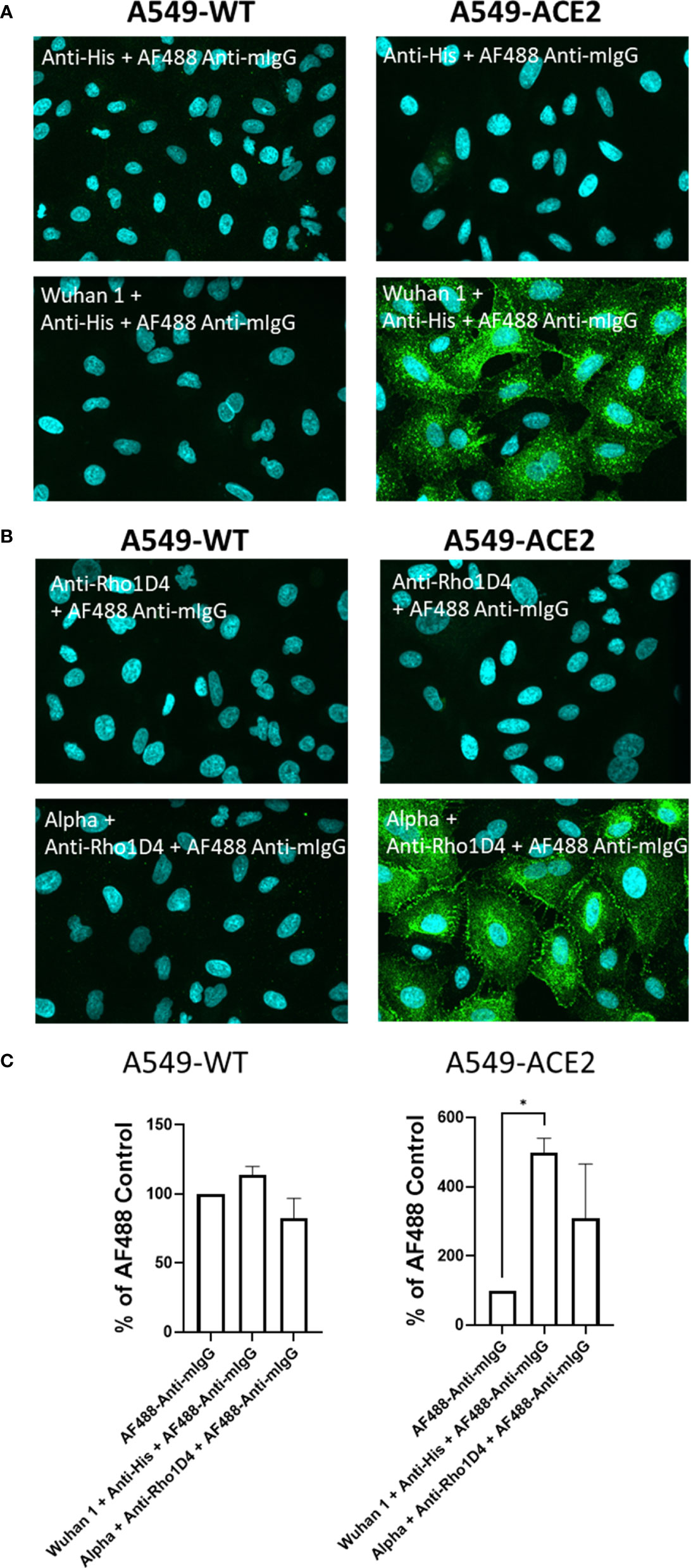
Figure 1 Labelling of ACE2 expressing A549 cells by recombinant spike proteins. (A) Representative confocal images showing labelling of wild type (A549-WT) and ACE2-overexpressing (A549-ACE2) cells by recombinant spike proteins (“Wuhan 1”) and anti-His + AF488-conjugated anti-mIgG detection antibodies. (B) Representative confocal images showing labelling of A549 cells (A549-WT) and ACE2-overexpressing (A549-ACE2) cells by recombinant spike proteins (“Alpha”) and anti-Rho1D4 + AF488-conjugated anti-mIgG detection antibodies (green; nuclei in cyan). (C) Quantification of spike protein labelling intensity from images acquired by high content confocal microscopy for cells labelled in (A, B). A mixed-effects one-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each spike protein to AF488 Anti-mIgG control. *P<0.05. Data expressed as mean + SD from three independent experiments.
Subsequent evaluation studied a range of recombinant spike proteins, including full-length wild types 1–6, D614G 1–2, Alpha, and Delta; RBD of Alpha and Delta, and Spike S2 wild type (listed in Table 1) to A549-WT and A549-ACE2 cells to assess their activity in a cell-based interaction assay. In accordance with the previous experiment, the labelling of A549-WT was generally low (Figure 2A). In A549-ACE2 cells, detection of the full-length spike proteins from different preparations (or from different suppliers, Table 1) gave comparable fluorescent intensities, except for Wuhan 3 (from Acrobio Biosystems), which showed relatively high intensities, and Wuhan 4, which showed the lowest intensities. In contrast, Alpha RBD with one amino acid mutation at position 501 from asparagine to tyrosine (N501Y), showed the highest intensity (Figure 2B). A comparison of a delta spike protein (Delta) vs. a delta spike protein RBD (Delta RBD) revealed that for an equivalent molar concentration, the full-length spike protein exhibited greater binding (Figure 2C). These data confirmed that both full-length spike and RBD allowed the clear labelling of A549-ACE2 cells.
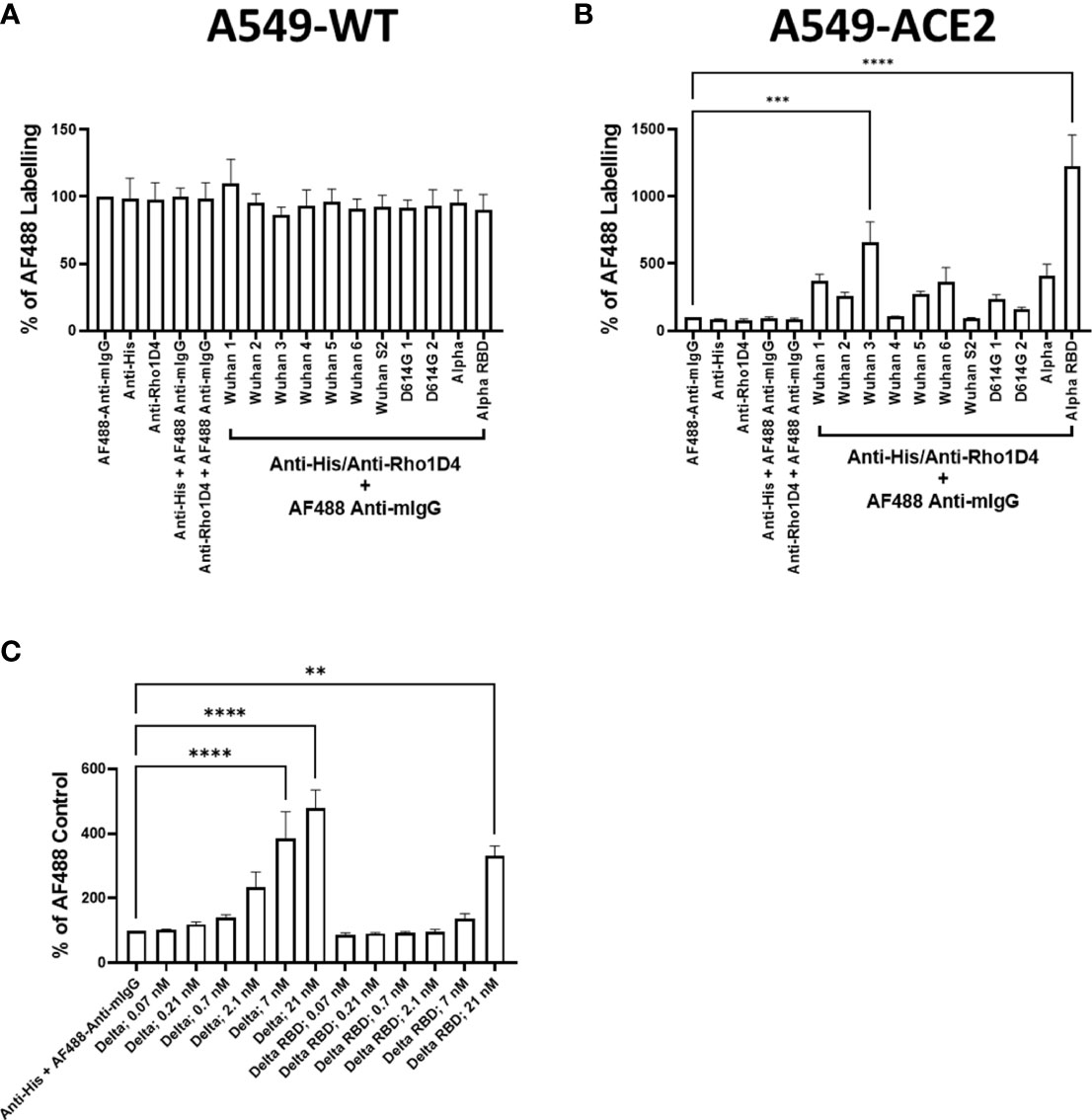
Figure 2 Comparison of recombinant spike proteins. Labelling of A549-WT (A) and A549-ACE2 (B) cells by recombinant spike proteins (listed in Table 1). Spike proteins were detected using anti-His + AF488-conjugated anti-mouse IgG antibodies, except for Alpha, which was detected using anti-Rho1D4 + AF488-conjugated anti-mouse IgG antibodies, followed by visualisation and quantification of spike protein labelling intensity by high-content confocal microscopy. (C) Labelling of A549-ACE2 cells by full-length Delta and Delta RBD. Non-parametric Kruskal–Wallis test was carried out for comparison of each spike protein to AF488 Anti-mIgG control (A), and a one-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each spike protein to AF488 Anti-mIgG control (B). One-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each concentration of spike L or spike L RBD to Anti-His + AF488 Anti-mIgG control (C). **P<0.01, ***P<0.001, ****P<0.0001. Data expressed as mean + SEM from three independent experiments.
To test the utility of this assay for monoclonal antibody screening, the impact of a range of commercially available anti-spike antibodies (Table 2) on the labelling of ACE2-A549 cells by Wuhan 1 spike (Figure 3A) and Alpha spike (Figure 3B) were investigated. Antibodies anti-RBD1, anti-RBD7, and anti-RBD8 consistently reduced the labelling of both spikes, and three of these antibodies were selected for further confirmatory studies. Titration of anti-RBD1, anti-RBD7, and anti-RBD8 revealed that there was a concentration-dependent blockade of Wuhan 1 spike (Figure 4A) and Wuhan 3 spike (Figure 4B) labelling, and enabled the quantification of IC50 values for each of the antibodies (Figure 4; Table 3).
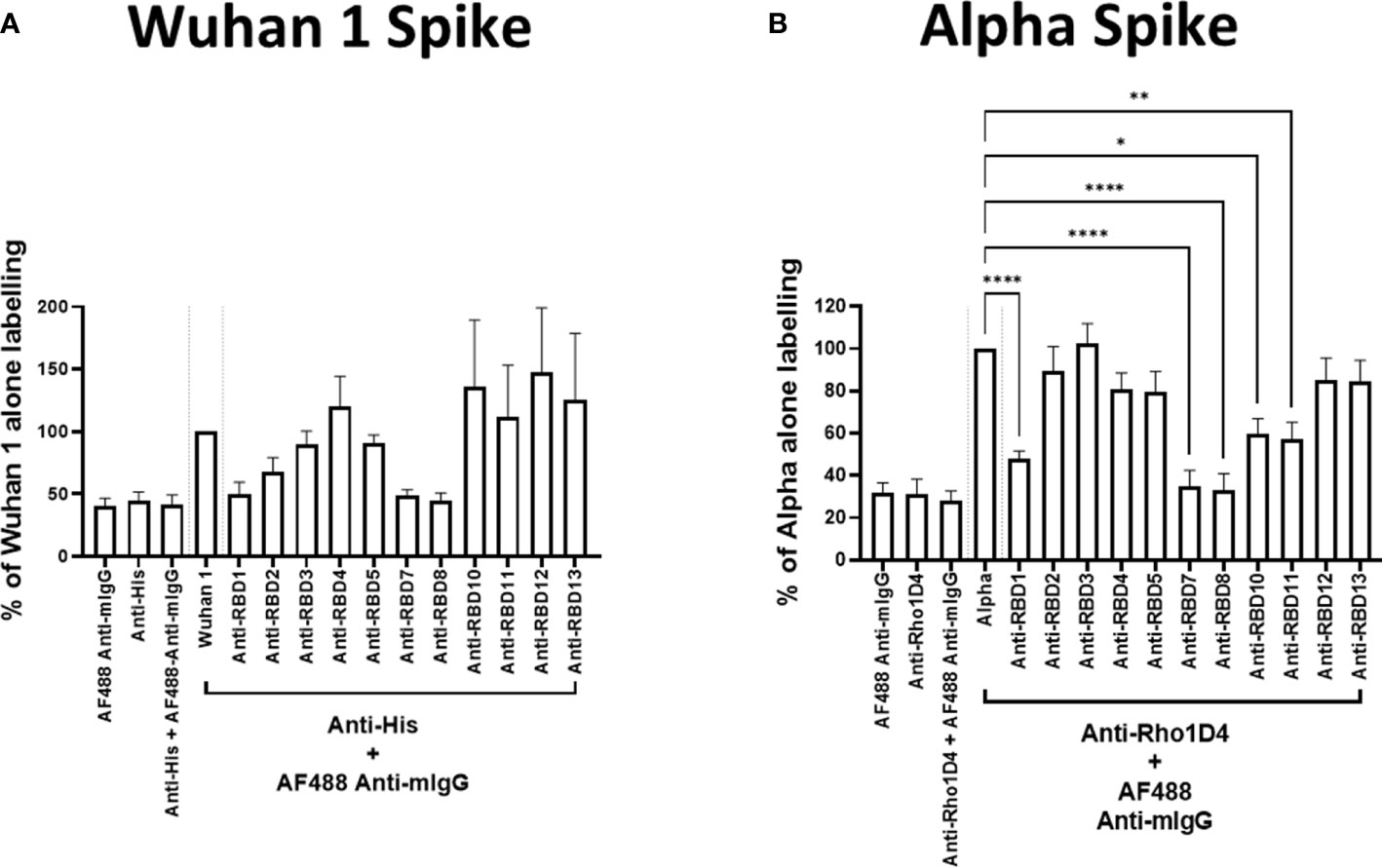
Figure 3 Antibody blockade of spike-protein binding. Labelling of A549-ACE2 cells by recombinant spike proteins Wuhan 1 (A) or Alpha (B) that had been pre-incubated in the absence or presence of anti-spike antibodies (as listed in Table 2) at 10 μg/mL, followed by detection using anti-His or anti-Rho1D4 + AF488-conjugated anti-mouse IgG antibodies, and visualisation and quantification of spike protein labelling intensity by high-content confocal microscopy. One-way ANOVA with Holm–Šídák post hoc multiple comparison test was carried out for comparison of each concentration of RBD antibody to Wuhan 1 (A) or Alpha alone (B). *P<0.05, **P<0.01, ****P<0.0001. Data expressed as mean + SEM from at least three independent experiments.
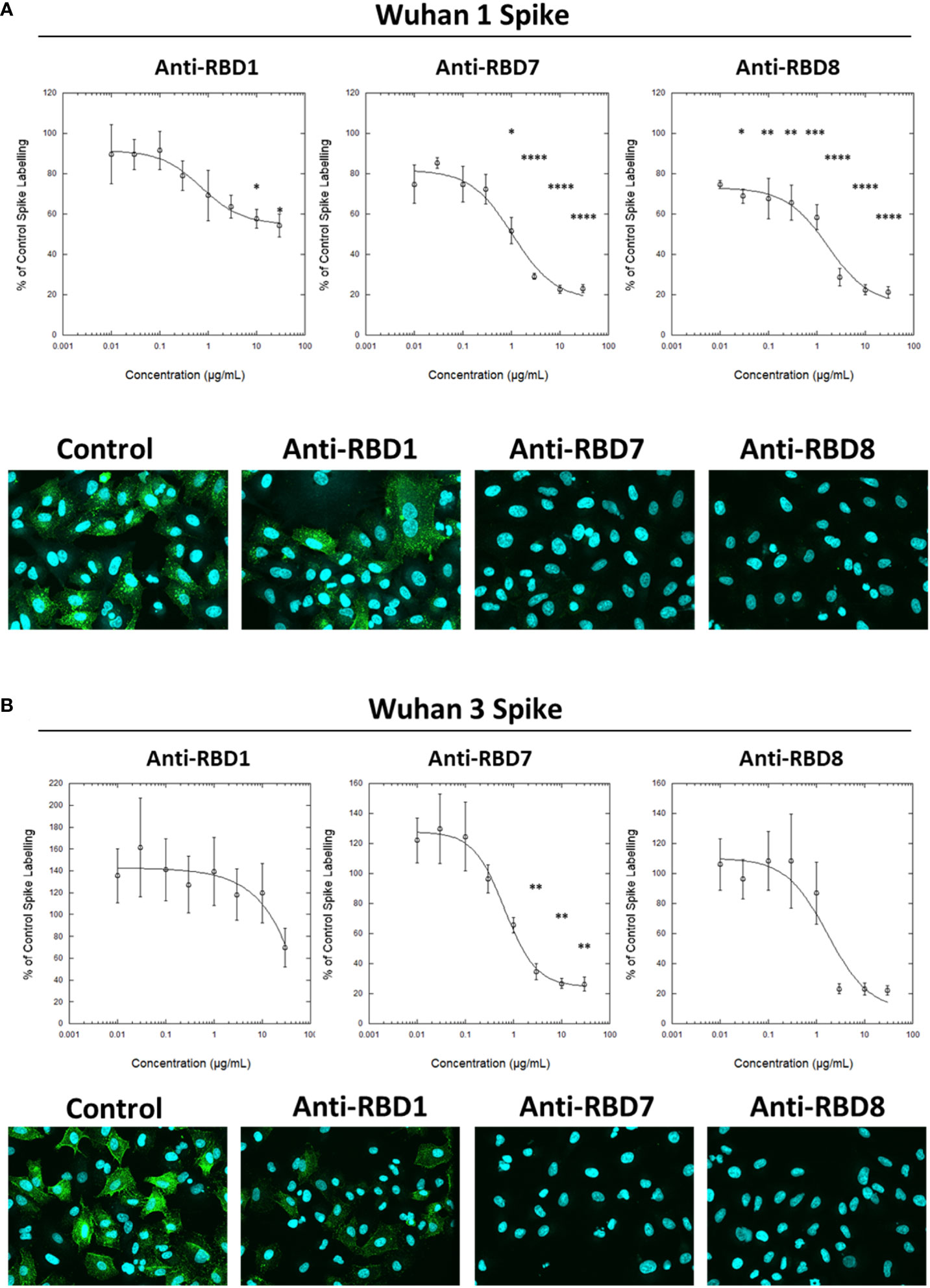
Figure 4 Characterisation of spike blocking antibodies. Labelling of A549-ACE2 cells by recombinant spike protein Wuhan 1 (A) or Wuhan 3 (B) that had been pre-incubated in the absence or presence of the indicated concentrations of antibodies anti-RBD1, anti-RBD7, or anti-RBD8, followed by detection using anti-His + AF488-conjugated anti-mouse IgG antibodies, and visualisation and quantification of spike protein labelling intensity by confocal microscopy. Plots show data expressed as mean ± SEM of three independent experiments. One-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each concentration of anti-RBD antibody to their relative vehicle control. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Solid line represents non-linear regression using a 4-parameter logistic equation. Representative confocal images show labelling of cells under control conditions or with spike protein pre-incubated with 10 μg/mL anti-RBD1, anti-RBD7, or anti-RBD8.
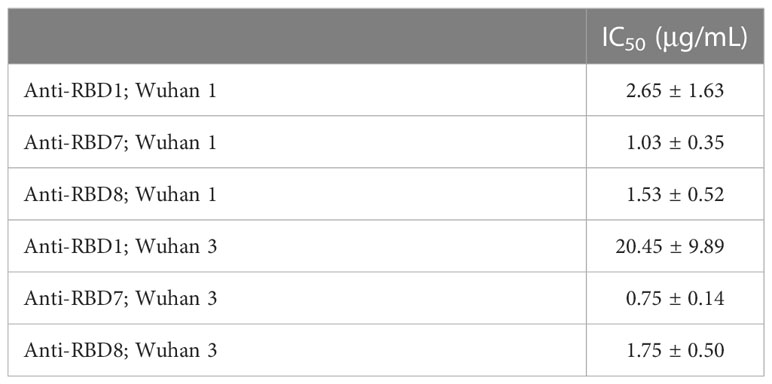
Table 3 IC50 values ± SEM from four independent experiments; Anti-RBD1, Anti-RBD7 and Anti-RBD8; Wuhan 1 and Wuhan 3.
To assess the translational relevance of the assay, the abilities of anti-RBD1, anti-RBD7, and anti-RBD8 to neutralise a live SARS-CoV-2 (wild-type virus, Wuhan Hu-1) infection of Vero cells was investigated. The result was in accordance with the spike-labelling results for Wuhan 1 spike (Figure 4A), in which anti-RBD1 exhibited the lowest potency in neutralising viral infection, whereas both anti-RBD7 and anti-RBD8 effectively prevented viral infection, with anti-RBD8 exhibiting greater potency than in the spike-binding assay (Figure 5; Table 4).
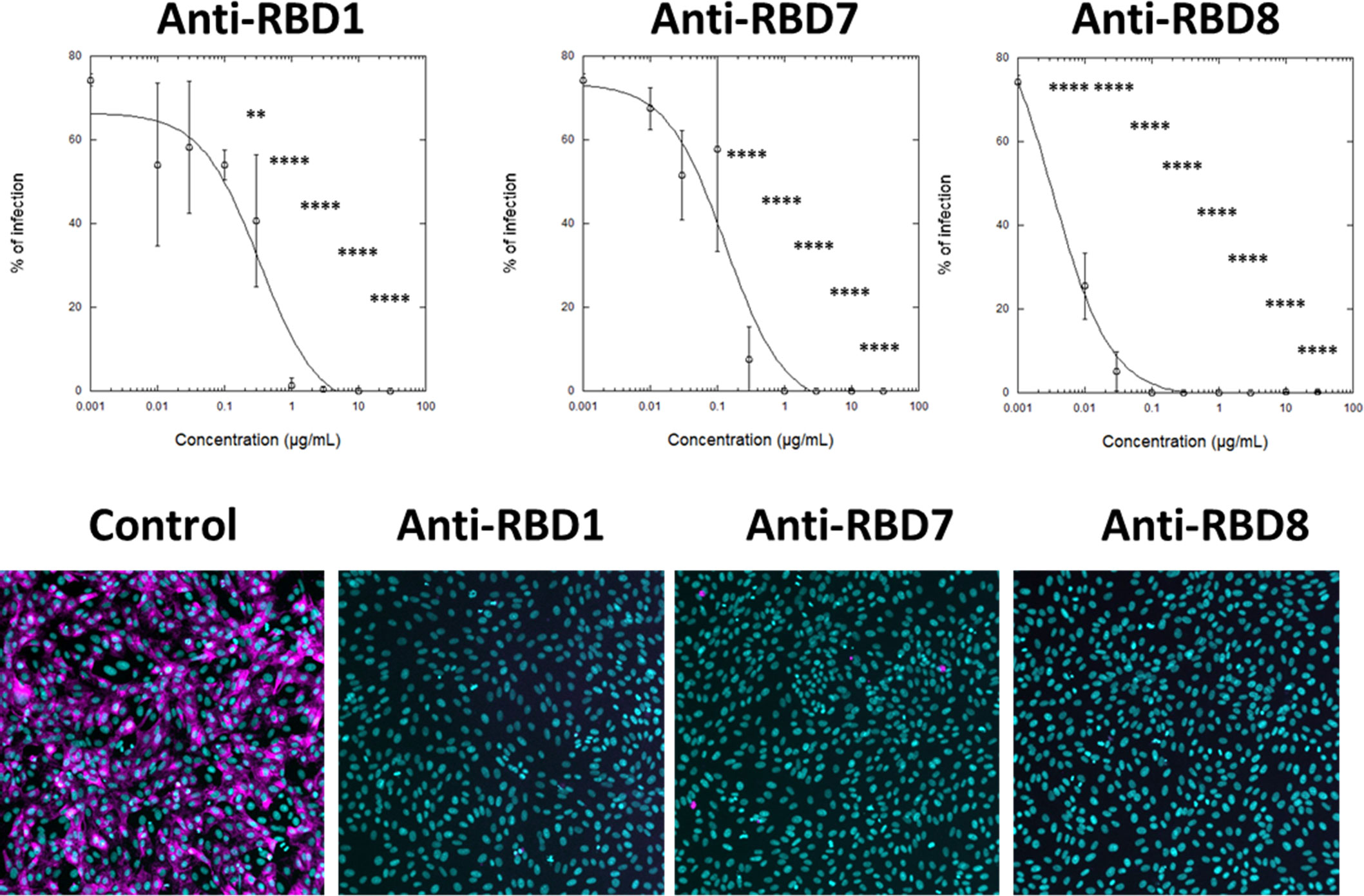
Figure 5 Impact of spike blocking antibodies on SARS-CoV-2 infection of Vero cells. Vero cells were infected with 3.3 × 103 IU/mL of hCOV-19/England/2/2020 virus isolate in the absence or presence of the indicated concentrations of anti-RBD1, anti-RBD7, or anti-RBD8. Infection rates were assessed at 24 h by staining Vero cells for viral spike protein (magenta) and counterstaining nuclei with Hoechst (cyan). Plots show data expressed as mean ± SD of replicate wells. One-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each concentration of RBD antibody to their relative vehicle control. **P<0.01, ****P<0.0001. Solid line represents non-linear regression using a 4-parameter logistic equation. Representative confocal images show labelling of cells under control conditions or with spike protein pre-incubated with 10 μg/mL anti-RBD1, anti-RBD7, or anti-RBD8.
To expand the potential utility of the spike interaction assay, we first determined the total IgG/A/M against SARS-CoV-2 spike protein of five donor plasma samples by ELISA (Figure 6A). A value of ≥ 1.0 was considered positive in this assay, and the plasma samples containing levels of anti-spike IgG/A/M >3 exhibited concentration-dependent spike-blocking activity, whereas those with levels <3 did not (Figure 6B).
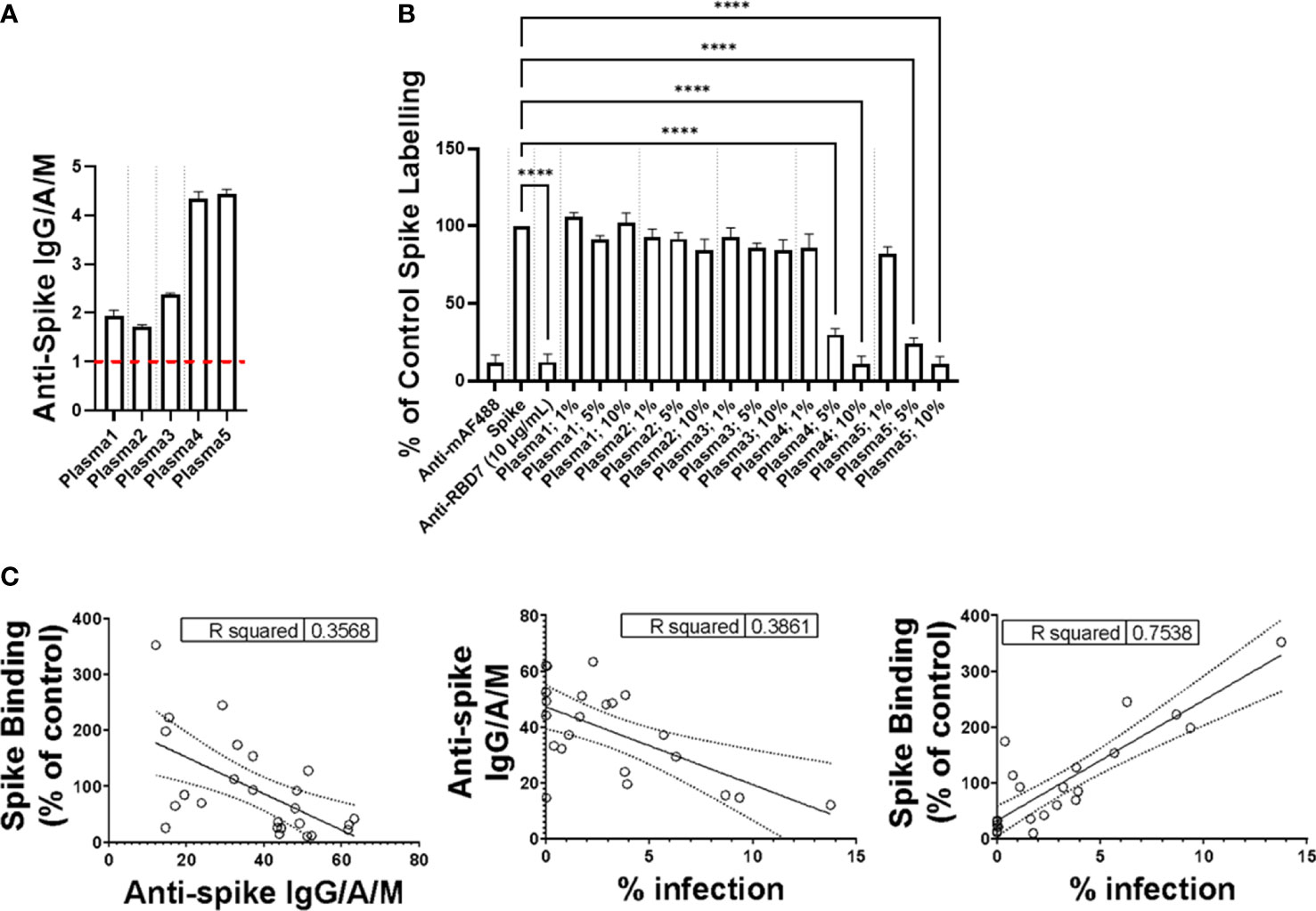
Figure 6 Impact of plasma on spike protein binding. (A) Plasma levels of anti-Spike IgG/M/A quantified by ELISA. Data expressed are mean + SD from three replicate wells. Dashed red line indicates threshold for positive for SARS-CoV-2 antibodies. (B) Labelling of A549-ACE2 by Wuhan 3 pre-incubated in the absence or presence of plasma at 1%, 5%, or 10%, or RBD7 (10 μg/mL), followed by detection using anti-His + AF488-conjugated anti-mouse IgG antibodies, and visualisation and quantification of spike protein labelling intensity by confocal microscopy. Data expressed as mean + SEM from three independent experiments. One-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each plasma at 1%, 5%, or 10% to untreated Wuhan 3 control. ****P<0.0001. (C) Correlation of plasma anti-Spike IgG/M/A quantified by ELISA, spike binding (as described in “B”) and the viral infection neutralisation assay carried out in A549-ACE2 cells. Data points represent mean of triplicate wells.
To further assess the relationship between spike interaction blockade, levels of anti-spike antibodies, and viral neutralisation, we screened the activity of donor plasma in the spike-binding assay, the anti-spike IgG/A/M ELISA, and an in vitro viral infection assay (Figure 6C), and analysed the correlation between the three assays. Although there was a correlation between all three assays, the correlation was strongest (R2 >0.75) between the spike-binding assays and the viral infection assay. This emphasises the fact that the spike interaction blockade assay provides information about the relevant functional activity of anti-spike antibodies. Taken together, these data suggest that the spike-binding assay is suitable for both monoclonal-antibody characterisation and monitoring neutralising antibody activity in plasma.
During this study, the emergence of the SARS-CoV-2 variants “Delta” and “Omicron BA.1” highlighted the need to rapidly assess the activity of potential therapeutics against the variant spike proteins. The activity of anti-RBD7, anti-RBD8, and anti-RBD14 was compared with the Delta and Omicron-BA.1 spike protein-labelling of A549-ACE2 cells. Although anti-RBD7, anti-RBD8, and anti-RBD14 effectively reduced labelling by the Delta spike proteins (Figure 7A; Table 5; with IC50 values of 0.26, 0.65, and 1.56 μg/mL, respectively), only anti-RBD14 displayed a comparable efficacy against the Omicron spike proteins (Figure 7B; Table 5; IC50 of 0.96 μg/mL). As there was variation in the IC50 values when averaged across independent experiments, we also ranked the antibodies by potency in each of the independent experiments (fitted to a four-parameter logistic equation, without normalisation to control staining, constraining the Hill coefficient to −1, and setting the minimum response to labelling in the absence of spike protein). For the Omicron variant, where the antibodies exhibited quite distinct blocking activities, the ranking of antibody IC50s (anti-RBD14 < anti-RBD8 < anti-RBD7, Table 6) was consistent throughout the experiments that were conducted. For the Delta variant, where the blocking activity was more comparable, it was not possible to distinguish between the relatively equipotent anti-RBD7 and anti-RBD14, although anti-RBD8 was less potent across all independent repeats (ranking of IC50s: anti-RBD7 ≈ anti-RBD14 < anti-RBD8, Table 6). Therefore, this assay, at least for the above antibodies, shows consistent relative potencies across independent experiments.
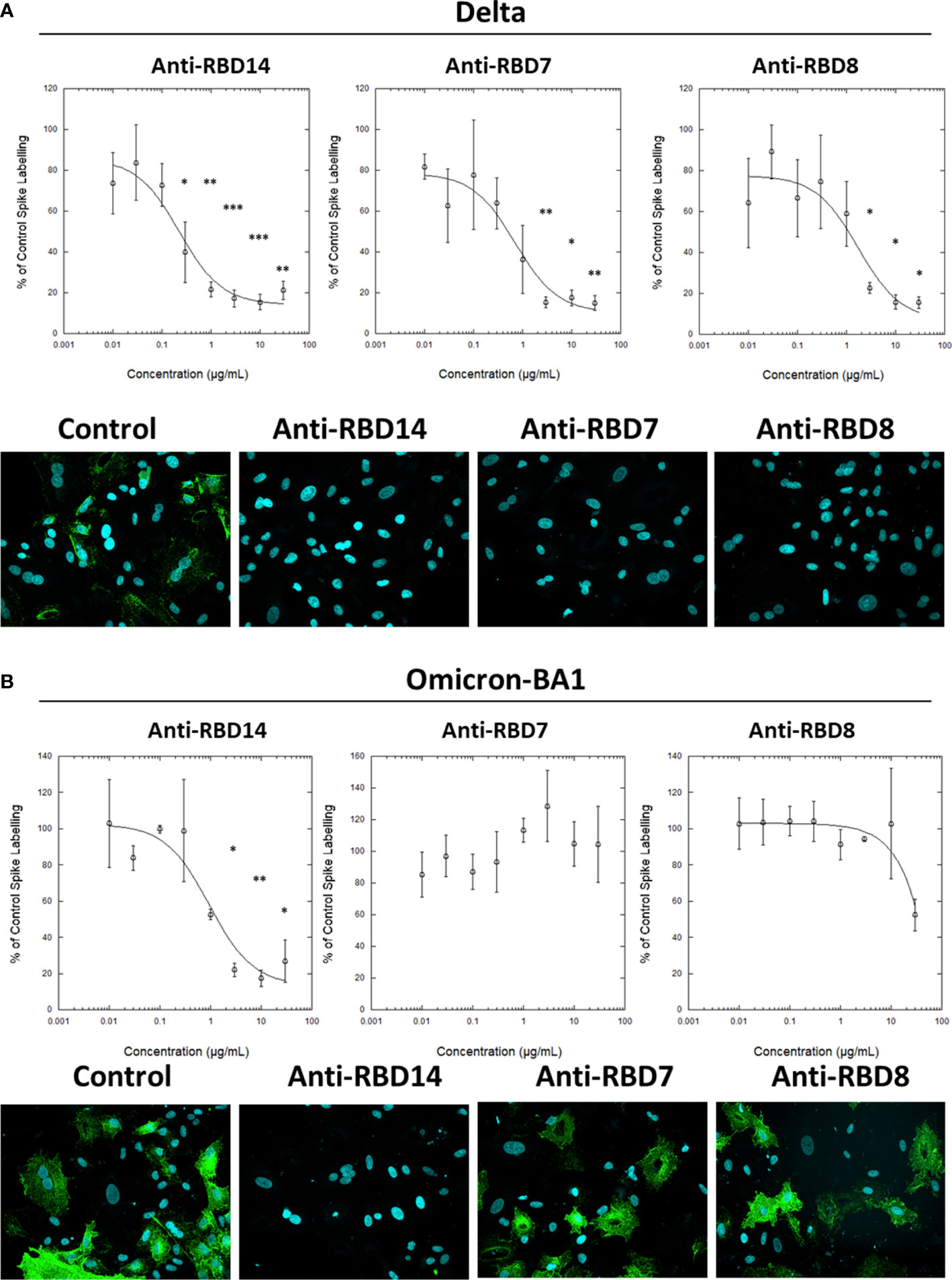
Figure 7 Characterisation of spike blocking antibodies against variants Delta and Omicron. Labelling of A549-ACE2 cells by recombinant Delta spike protein (A) and Omicron BA.1 spike protein (B) pre-incubated in the absence or presence of the indicated concentrations of antibodies anti-RBD14, anti-RBD7, or anti-RBD8, followed by detection using anti-His + AF488-conjugated anti-mouse IgG antibodies, and visualisation and quantification of spike protein labelling intensity by confocal microscopy. Plots show data expressed as mean ± SEM of three independent experiments. One-way ANOVA with Tukey’s post hoc multiple comparison test was carried out for comparison of each concentration of RBD antibody to their relative vehicle control. *P<0.05, **P<0.01, ***P<0.001. Solid line represents non-linear regression using a 4-parameter logistic equation. Representative confocal images show labelling of cells under control conditions (in the absence of anti-spike antibody) or with spike protein pre-incubated with 10 μg/mL anti-RBD14, anti-RBD7, or anti-RBD8.
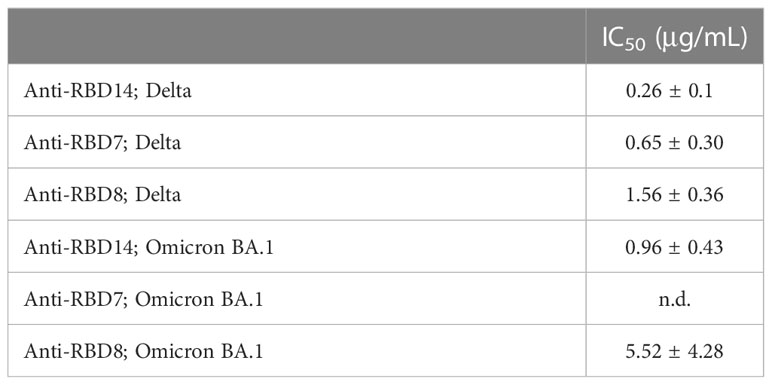
Table 5 IC50 values ± SEM from three independent experiments; Anti-RBD14, Anti-RBD7 and Anti-RBD8; Delta and Omicron BA.1.
In summary, we have developed a high-content imaging assay to quantify SARS-CoV-2 spike protein interaction with ACE2-expressing target cells. ACE2 expression in A549 cells was sufficient to detect various recombinant spike proteins from the Wuhan strain and from the variants of concern: Alpha, Delta, and Omicron. We demonstrated that some commercially available monoclonal antibodies were able to neutralise recombinant spike labelling in a manner comparable to replicating virus neutralisation.
It was previously reported that a single mutation of N501Y in the Alpha RBD increases the affinity towards ACE2 receptor, even when compared with the full-length Alpha spike protein (14, 15). Using our system, we found that the Alpha RBD (N501Y mutation) displayed the highest spike intensity (although this could also be accounted for by the different format of the spike protein). Interestingly, this was not the case between the full-length spike and RBD of Delta. This might be attributed to the two mutations in the Delta RBD (L452R and T478K) that showed only a modest increase of affinity towards ACE2 (16). Overall, these results indicated that our platform was in accordance with previous data.
Characterisation of spike (RBD)-specific antibodies is important to monitor immunological memory (17), and there is evidence that repertoires of SARS-CoV-2 epitopes targeted by antibodies vary according to severity of COVID-19 (18). Although ELISA-based assays are important to characterise antibody repertoires by assessing recognition of immobilised spike proteins, the spike-interaction bioassay offers the advantage of a functional assay that assesses the ability to neutralise receptor interactions without the need for high-containment facilities. Nevertheless, cell-based assays, while potentially providing additional information, may still be relatively time consuming and require cell culture facilities. An ELISA-based surrogate neutralisation assay has been developed (12), which offers an accessible method for assessing neutralising antibodies. Other assays, such as cPASS™ (19) and MSD V-PLEX (20), also utilise recombinant human ACE2, and, although they can identify blocking antibodies and have benefits of convenience, do not utilise ACE2 expressed in its native cellular environment and do not enable quantification at the single-cell level. As with all studies defining pharmacological potency, when ligands (in the present case antibodies) display similar potency (e.g., anti-RBD14 and anti-RBD7 antibodies against the Delta variant), establishing a consistent rank order of potency is challenging. However, when there are clear potency differences in blocking activity, such as for the Omicron variant, the rank order of potency for the antibodies was consistent across independent experiments, further emphasising the utility of the assay.
As this assay system uses a “His tag” for detection, which is frequently used for the purification, on initial expression, of a recombinant protein, His-tagged recombinant viral variant spike proteins are often rapidly available and accessible, enabling the assay to be quickly adapted to test the activity of therapeutics as new strains of SARS-CoV-2 emerge.
Across viral variants, although antibody affinity for spike protein may remain approximately constant, the affinity of spike protein for ACE2 may increase significantly and be associated with reduced antibody neutralisation (21). Measuring functional antibody blockade of the spike protein/ACE2 interaction is therefore likely to become increasingly important, and we envisage that this cell-based, spike protein interaction assay would complement existing biochemical and pseudovirus assays in the development of therapeutics and may be adapted for other cell types to investigate the potentially ACE2-independent binding of spike protein.
The raw data supporting the conclusions of this article will be made available by the authors on reasonable request.
The studies involving human participants were reviewed and approved by Wales REC 6 (REC Reference 20/WA/0216). The patients/participants provided their written informed consent to participate in this study.
NH (conceptualisation, methodology, investigation, resources, writing, formal analysis, visualisation, supervision), LR (methodology, investigation, writing, formal analysis, visualisation), CP (methodology, investigation, writing), IM (methodology, investigation, writing), EJ (methodology, investigation, writing), JC (methodology, investigation, writing), HC (methodology, investigation, writing, formal analysis), HH (methodology, investigation, formal analysis, writing), ZL (methodology, investigation, writing), CH (methodology, investigation, writing), ATe (methodology, investigation, writing), AL (methodology, investigation, writing), RM (writing, formal analysis), ATu (methodology, investigation, writing), AA (methodology, investigation, writing), TD (conceptualisation, methodology, writing, funding acquisition), DG (conceptualisation, methodology, writing, funding acquisition), JG (conceptualisation, methodology, writing, funding acquisition), CB (conceptualisation, methodology, writing, funding acquisition), LY (conceptualisation, methodology, writing, funding acquisition), NB (conceptualisation, methodology, writing, funding acquisition), ZS (conceptualisation, methodology, writing, funding acquisition), and OQ (conceptualisation, methodology, funding acquisition, resources, writing, formal analysis, visualisation, supervision). RM performed and advised on statistical testing, interpretation of data and final approval of manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Innovate UK [84361] and Celentyx Ltd. ATu, HJH and ZS are funded by a Medical Research Foundation intermediate career fellowship to ZS (UKRI, Grant number MRF-169-0001-F-STAM-C0826).
NH, LR, CP, IM, EJ, JC, HC, CM, ATe, AL, ZL, JG, CB, NB, and OS are current or former employees of Celentyx Ltd and/or hold stock or stock options in Celentyx Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors ZL and DG declared that they were editorial board members of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Robinson PC, Liew DFL, Tanner HL, Grainger JR, Dwek RA, Reisler RB, et al. COVID-19 therapeutics: challenges and directions for the future. Proc Natl Acad Sci (2022) 119(15):e2119893119. doi: 10.1073/pnas.2119893119
2. Hwang Y-C, Lu R-M, Su S-C, Chiang P-Y, Ko S-H, Ke F-Y, et al. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J Biomed Sci (2022) 29(1). doi: 10.1186/s12929-021-00784-w
3. Galipeau Y, Greig M, Liu G, Driedger M, Langlois M-A. Humoral responses and serological assays in SARS-CoV-2 infections. Front Immunol (2020) 11. doi: 10.3389/fimmu.2020.610688
4. Ramesh S, Govindarajulu M, Parise RS, Neel L, Shankar T, Patel S, et al. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines (2021) 9:1195. doi: 10.3390/vaccines9101195
5. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell (2020) 181:271–80. doi: 10.1016/j.cell.2020.02.052
6. Wang B, Zhao J, Liu S, Feng J, Luo Y, He X, et al. ACE2 decoy receptor generated by high-throughput saturation mutagenesis efficiently neutralizes SARS-CoV-2 and its prevalent variants. Emerging Microbes Infections (2022) 11(1):1488–99. doi: 10.1080/22221751.2022.2079426
7. Shah M, Ung Moon S, Hyun Kim J, Thanh Thao T, Goo Woo H. SARS-CoV-2 pan-variant inhibitory peptides deter S1-ACE2 interaction and neutralize delta and omicron pseudoviruses. Comput Struct Biotechnol J (2022) 20:2042–56. doi: 10.1016/j.csbj.2022.04.030
8. Zhang L, Narayanan KK, Cooper L, Chan KK, Devlin CA, Aguhob A, et al. An engineered ACE2 decoy receptor can be administered by inhalation and potently targets the BA.1 and BA.2 omicron variants of SARS-CoV-2. bioRxiv (2022). doi: 10.1101/2022.03.28.486075
9. Shah M, Woo HG. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front Immunol (2022) 12. doi: 10.3389/fimmu.2021.830527
10. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the sars-cov-2 spike glycoprotein. Cell (2020) 181:281–92. doi: 10.1016/j.cell.2020.02.058
11. Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, et al. SARS-CoV-2 omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell (2022) 185(3):467–84. doi: 10.1016/j.cell.2021.12.046
12. Abe KT, Zhijie L, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight (2020) 5(19):e142362. doi: 10.1172/jci.insight.142362
13. Tandon R, Mitra D, Sharma P, McCandless MG, Stray SJ, Bates JT, et al. Effective screening of SARS-CoV-2 neutralizing antibodies in patient serum using lentivirus particles pseudotyped with SARS-CoV-2 spike glycoprotein. Sci Rep (2020) 10:19076. doi: 10.1038/s41598-020-76135-w
14. Ali F, Kasry A, Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med Drug Discovery (2021) 10:100086.
15. Supasa P, Zhou D, Dejnirattisai W, Liu C, Mentzer AJ, Ginn HM, et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell (2021) 184(8):2201–11.
16. Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell (2021) 184(16):4220–36.
17. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science (2021) 371(6529):eabf4063. doi: 10.1126/science.abf4063
18. Gregory DJ, Vannier A, Duey AH, Roady TJ, Dzeng RK, Pavlovic MN, et al. Repertoires of SARS-CoV-2 epitopes targeted by antibodies vary according to severity of COVID-19. Virulence (2022) 13:890–902. doi: 10.1080/21505594.2022.2073025
19. Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol (2020) 38(9):1073–8. doi: 10.1038/s41587-020-0631-z
20. Johnson M, Wagstaffe HR, Gilmour KC, Mai AL, Lewis J, Hunt A, et al. Evaluation of a novel multiplexed assay for determining IgG levels and functional activity to SARS-CoV-2. J Clin Biol (2020) 130:104572. doi: 10.1016/j.jcv.2020.104572
Keywords: SARS-CoV-2, ACE2, spike protein, antibody discovery, virology, variant, antiviral
Citation: Harrison N, Richardson L, Pallini C, Morano I, Jinks E, Cowley J, Chan H, Hill HJ, Tuekprakhon A, Li Z, Matas de las Heras C, Teodosio A, Lavado AS, Moring R, Ashraf A, Dafforn TR, Grammatopoulos DK, Gordon J, Brady CA, Young LS, Barnes NM, Stamataki Z and Qureshi OS (2023) A cell-based, SARS-CoV-2 spike protein interaction assay to inform the neutralising capacity of recombinant and patient sera antibodies. Front. Virol. 3:1163385. doi: 10.3389/fviro.2023.1163385
Received: 10 February 2023; Accepted: 21 June 2023;
Published: 29 August 2023.
Edited by:
Matloob Husain, University of Otago, New ZealandReviewed by:
Joe Grove, MRC-University of Glasgow Centre For Virus Research (MRC), United KingdomCopyright © 2023 Harrison, Richardson, Pallini, Morano, Jinks, Cowley, Chan, Hill, Tuekprakhon, Li, Matas de las Heras, Teodosio, Lavado, Moring, Ashraf, Dafforn, Grammatopoulos, Gordon, Brady, Young, Barnes, Stamataki and Qureshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Omar Qureshi, b21hci5xdXJlc2hpQGNlbGVudHl4LmNvbQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.