- 1Department of Clinical Virology, Institute of Liver and Biliary Sciences, New Delhi, India
- 2Genome Sequencing Laboratory, Institute of Liver and Biliary Sciences, New Delhi, India
- 3Department of Pathology, Institute of Liver and Biliary Sciences, New Delhi, India
Despite the three years spent navigating the COVID-19 pandemic, scientists are still having to react to the disease due to the constant evolution of novel variants/subvariants. Over the last few months, a global plummet in COVID-19 cases has suggested we are transitioning towards endemic COVID-19. However, the new omicron offshoots (XBB variants) are driving a new surge of cases around the world. A few preliminary research findings suggest that the XBB.1.5 subvariant is more immune-evasive and displays higher binding to ACE2 human receptor than its other related omicron subvariants in circulation. In this first-of-its-kind report, we discuss a few XBB.1.5 cases and its clinical characteristics reported in Delhi State, North India.
Introduction
Since the start of the Coronavirus (COVID-19) pandemic in December 2019, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been continuously evolving and challenging all spheres of human life. The emergence of new COVID-19 variants and their sub-lineages is still on the increase due to the immune pressure conferred by the combined effects of vaccination and natural infection (1, 2). Among the menagerie of immune-evasive variants, B.1.1.529 (Omicron) is one such variant which is highly transmissible (3). Since January 2022, it has been the predominant variant in circulation globally and driven the emergence of more than 680 sub-lineages (https://cov-lineages.org/lineage_list.html), resulting in vaccine breakthrough and reinfections in many countries (4). Based on current available evidence, the World Health Organization (WHO) has termed BQ.1.1 and XBB* (XBB and its sub-lineages, including XBB.1.5) as “Omicron subvariants under monitoring” (5), as no dramatic increase in hospitalizations and severity has been observed yet. The emergence of new subvariants, especially Omicron sub-lineages and the evolution of recombinants between two or more sub-lineages (XBB*, X signifies that this subvariant is formed because of recombination), raises a concern for public health as an accumulation of mutation constellation within the viral genome substantially boosts viral transmissibility and host immune-evasiveness (6, 7).
XBB.1.5 is a recombinant of its two predecessor sub-lineages, BA.2.10.1 and BA.2.75, and was first reported in the US (collection date: 26th Oct, 2022, accessed at https://gisaid.org/hcov19-variants/, (8). Scientists believe that XBB.1.5 increases the susceptibility to reinfections by virtue of an escalated transmissibility and more enhanced immune dodging properties than other omicron subvariants (6, 9). The prevalence of the XBB.1.5 subvariant is on the rise globally, with a steep increase from 2% (early December) to 50% (early January) initially observed in some parts of the US (10). The mutation of F486P at the receptor-binding domain of the spike glycoprotein makes the XBB.1.5 different from its predecessors, XBB and XBB.1. This amino acid position is prone to mutation in different lineages e.g., F (Phenylalanine) is changed to V (Valine) in BQ.1.1 and to S (Serine) in XBB.1 and to P (Proline) in XBB.1.5. (See Supplementary Figure 1). F486P has been previously linked to increased ACE-2 receptor binding as compared to F486S (11). In a recent published article, Yue et al. suggests an increased ability to bind to the human ACE2 receptor as one of the underlying factors for higher transmissibility of the XBB.1.5 subvariant (7).
As of 6th April 2023, there have been approximately 141,100 cases of XBB.1.5* (now diversified into 26 further sub-lineages of XBB1.5.x) from 98 countries across the globe, with approximately 58% sequences from the US (https://outbreak.info/). The first submitted case belonged to the US with a collection date of 26th Oct, 2022. As per the last update on the CDC COVID data tracker, on 1st Apr, 2023, XBB.1.5 accounted for 90% of total cases. As shown in Supplementary Figure 2, the prevalence of XBB* sub-lineages started rising after September 2022 and is gaining ground slowly. Interestingly, between the time frame we submitted this paper (1st week Feb, 2023) and revised it (Aril 2nd week, 2023), the Omicron subvariant XBB.1.16 has begun to replace the current circulating subvariants in India (Figure 1).
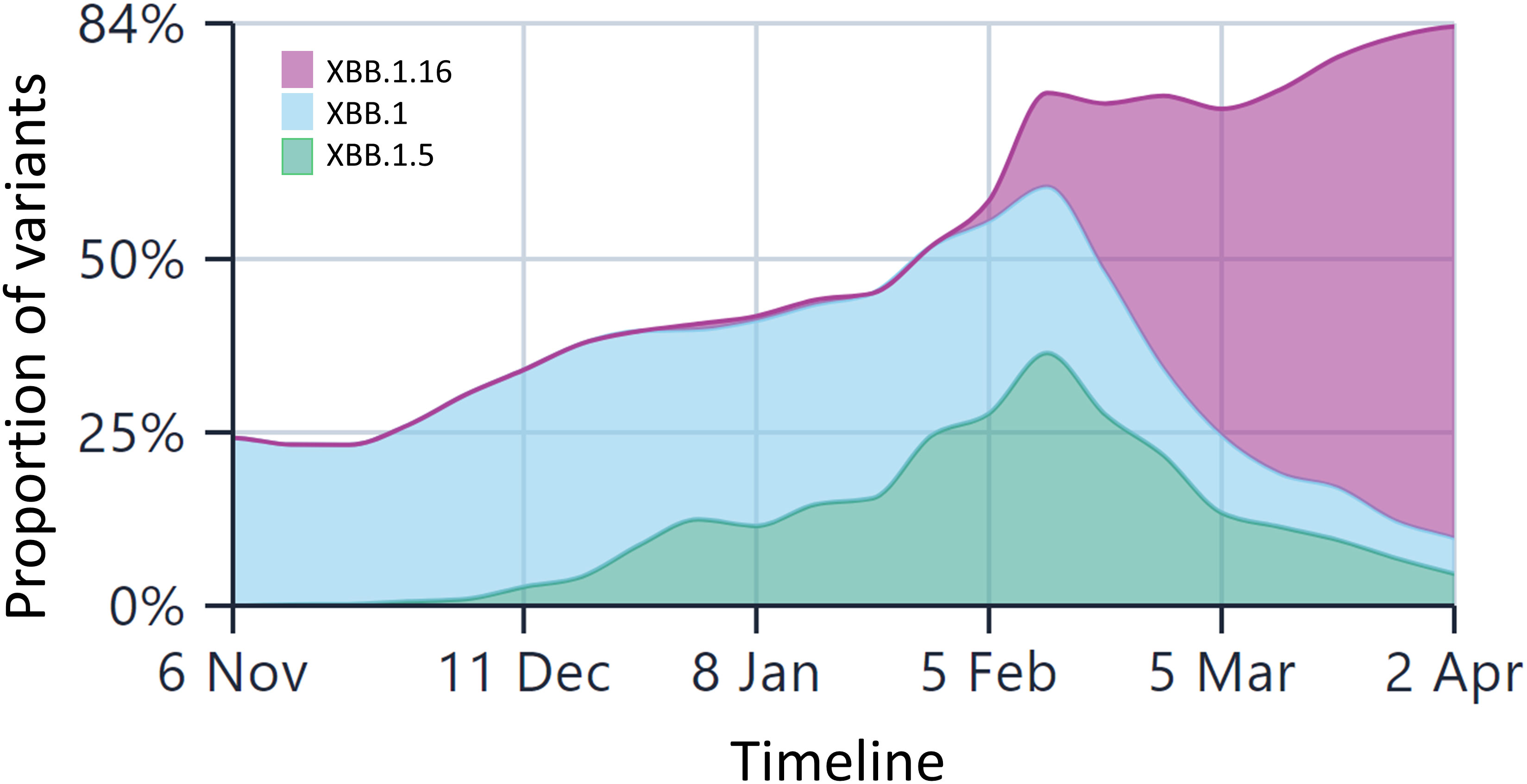
Figure 1 The lineage prevalence in India over the past 6 months (Comparison shown only for XBB.1.5 and XBB.1.16 and other Omicron lineages). The x-axis is time and y-axis is the relative frequencies of the lineages. The less frequent lineages are not depicted in the figure for the sake of comparison between these three lineages only. Currently, XBB.1.16 is the most predominant lineage in India, surpassing other Omicron lineages by a huge margin. The data are taken from the public repository, https://covglobe.org. The time period is from the 1st week November, 2022 till the last week of March, 2023.
The UK Health Security Agency (UKHSA), the European Centre for Disease Prevention and Control (ECDC), and the Centers for Disease Control and Prevention have anticipated that, being a master immune escape subvariant, XBB.1.5* may increase in overall incidence in the coming months (after January 2023), however, until now, no major hospitalization and severity has been associated with this variant.
Our experience with XBB.1.5
While we were writing this manuscript in January 2023, limited data about the XBB.1.5 subvariant was available. As this variant was gradually out-competing its other immune-dodging counterparts, we report the first few cases of the XBB.1.5* subvariant from North India (Delhi state). This study is the first of its kind to describe clinical characteristics of subvariant XBB.1.5 cases in an Indian population. As a part of Delhi Government’s and INSACOG’s (Indian SARS-CoV-2 Genomics Consortium) genome sequencing laboratory, our center has been instrumental in tracing and reporting SARS-CoV-2 variants in the Delhi population (12, 13). The data from India (accessed https://outbreak.info/ on 24th April 2023, (14) suggest that a total of 375 cases of XBB.1.5* have been confirmed and documented. The first case was detected in Gujarat, with a collection date of 8th Nov, 2022. The other states where it was found initially were Maharashtra (7 cases), New Delhi (5 cases), Gujarat (5 cases), Karnataka (2 cases), Tamil Nadu (2 cases), Telangana (2 cases), and Kerala (1 case).
Till date (6th April, 2023), we have detected 65 cases of XBB.1.5 (as uploaded on GISAID) (Figure 2). In our genomic surveillance data, phylogeny revealed clustering of XBB.1.5 sequences together compared to other sequences of XBB sub-lineage (Figure 2). The basic demographic and clinical details of the people infected with the XBB.1.5 subvariant is described briefly in this manuscript. Overall, the median age of the cases was 43 years (IQR: 30.5-57.5) and mean Ct value of real-time RT PCR was ~20.8 ± 3.8. The male:female ratio was 0.8:1. All the cases presented with mild or asymptomatic SARS-CoV-2 infection and recovered fully. No severe hospitalization was seen. Among these, five (8%) cases had a recent history of international travel. All the cases were fully vaccinated (two doses of vaccine). All cases belonged to different geographical locations and were not related to each other. As per the information available to us, in only one case was COVID-19 transmission to other family members (sequencing details are not available for them) observed.
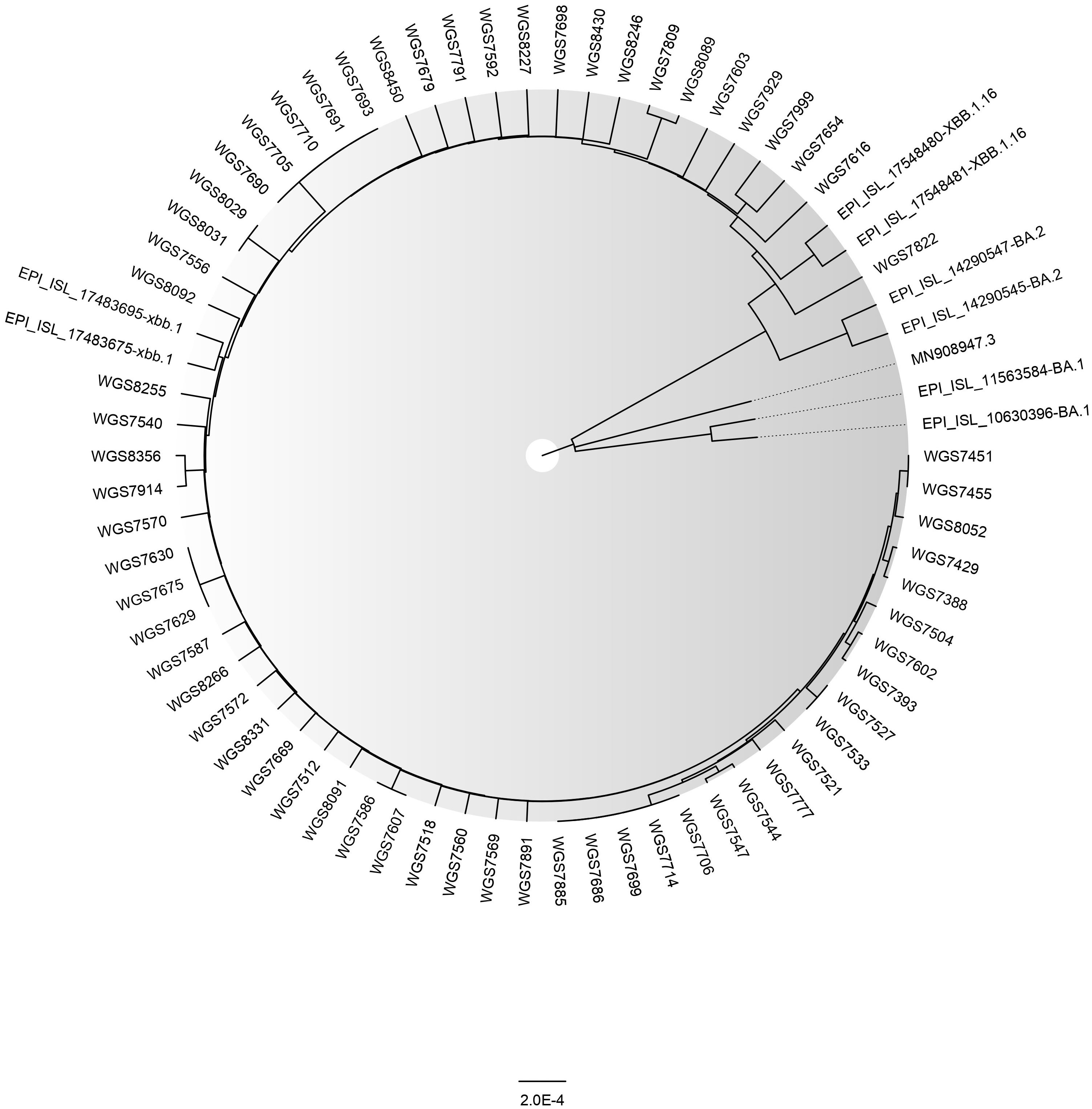
Figure 2 The Phylogenetic tree of a sample set of 65 SARS-CoV-2 XBB.1.5 sequences (GISAID IDs of corresponding samples are provided in the Supplementary Material) along with BA.1, BA.2, XBB.1, and XBB.1.16 reported from ILBS hospital, New Delhi. The reference strain’s ID is MN908947.3. The tree was constructed in MEGA (v. 11.0.13) using the Neighbor Joining method and model Maximum Composite Likelihood and graphically viewed in FigTree (v. 1.4.4).
Discussion and conclusion
Our initial data of 65 cases suggest that XBB.1.5 causes mild infection. We observed no cluster formation, nor did these cases add to an increase in positivity of cases in their particular geographical area. Although this is an initial observation from samples sequenced in state-level genome sequencing labs (covering Delhi population mainly), the data collected from our center and the constant low numbers of COVID-19 cases across the country indicate less likelihood of a surge in cases of XBB.1.5. In addition, we did not observe any association of XBB.1.5 with severity. Nevertheless, whether India will see a surge in cases or not is still under review and we need to closely monitor the changing landscape of the circulating variants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author contributions
JS and AB: Writing original Draft and data analysis, VS and PG: Data curation, analysis, and editing manuscript, RA and CB: Data collection and manuscript editing, and EG: Conceptualization, Supervision, review, and editing the manuscript. All authors contributed to the article and approved the submitted version
Funding
This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Govt. Of India (Grant numbers: BT/PR7403/MED/29/657/2012 and RAD-22017/28/2020-KGD-DBT) and the Government of National Capital Territory of Delhi, India (Grant number: F.8 (343)/ILBS/VAR/SEQ/P-I/2021/21705/CD-000654524/215).
Acknowledgments
We acknowledge the Government of NCT, Delhi for facilitating the Whole-Genome Sequencing Laboratory for COVID-19 at our institute. We also extend our gratitude to the National Liver Disease Biobank, ILBS, New Delhi for providing Next Generation Sequencing platforms. We also thank NCDC, New Delhi and INSACOG for their unconditional support. We thank ILBS technical and administrative staff for their help and support in project implementation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1158703/full#supplementary-material
References
1. Ahmad L. Implication of SARS-CoV-2 immune escape spike variants on secondary and vaccine breakthrough infections. Front Immunol (2021) 12(November):1–7. doi: 10.3389/fimmu.2021.742167
2. Padhi AK, Tripathi T. Can SARS-CoV-2 accumulate mutations in the s-protein to increase pathogenicity? ACS Pharmacol Transl Sci (2020) 3(5):1023–6. doi: 10.1021/acsptsci.0c00113
3. Rahimi F, Talebi Bezmin Abadi A. Omicron: a highly transmissible SARS-CoV-2 variant. Gene Rep (2022) 27:101549. doi: 10.1016/j.genrep.2022.101549
4. Dhawan M, Saied AA, Mitra S, Alhumaydhi FA, Emran Tb, Wilairatana P. Omicron variant (B.1.1.529) and its sublineages: what do we know so far amid the emergence of recombinant variants of SARS-CoV-2? Biomedicine Pharmacother (2022), 154. doi: 10.1016/j.biopha.2022.113522
5. World Health Organization. COVID-19 weekly epidemiological update.World health organisation (2022). Available at: https://www.who.int/news/item/27-10-2022-tag-ve-statement-on-omicron-sublineages-bq.1-and-xbb.
6. Davis-Gardner ME, Lai L, Wali B, Samaha H, Solis D, Lee M, et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. New Engl J Med (2023) 388(2):183–5. doi: 10.1056/NEJMc2214293
7. Yue C, Song W, Wang L, Jian F, Chen X, Gao F, et al. Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. bioRxiv (2023) 18(3):278–80. doi: 10.1101/2023.01.03.522427
8. Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall (2017) 1(1):33–46. doi: 10.1002/gch2.1018
9. Tamura T, Ito J, Keiya U, Zahradnik J, Kida I, Nasser H, et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two omicron subvariants. bioRxiv (2023), 14(1):2800. doi: 10.1038/s41467-023-38435-3
10. Callaway E. Is coronavirus variant XBB.1.5 a global threat? Nature (2023) 613:222–3. doi: 10.1038/d41586-023-00014-3
11. Starr TN, Greaney AJ, Stewart CM, Walls AC, Hannon WW, Veesler D, et al. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 omicron BA.1 and BA.2 receptor-binding domains. PloS Pathog (2022) 18(11). doi: 10.1371/journal.ppat.1010951
12. Garg R, Gautam P, Suroliya V, Agarwal R, Bhugra A, Kaur US, et al. Evidence of early community transmission of omicron (B1.1.529) in Delhi- a city with very high seropositivity and past-exposure. Travel Med Infect Dis (2022), 46. doi: 10.1016/j.tmaid.2022.102276
13. Gautam P, Paul D, Suroliya V, Garg R, Agarwal R, Das S, et al. SARS-CoV-2 lineage tracking, and evolving trends seen during three consecutive peaks of infection in Delhi, India: a clinico-genomic study. Microbiol Spectr (2022) 10(2):e0272921. doi: 10.1128/spectrum.02729-21
Keywords: COVID-19, XBB.1.5, Omicron, XBB*, immune-evasive, BQ.1.1, India
Citation: Samal J, Bhugra A, Suroliya V, Gautam P, Agarwal R, Bihari C and Gupta E (2023) The emergence of the Omicron XBB.1.5 variant in India: a brief report on clinical presentation of a few cases. Front. Virol. 3:1158703. doi: 10.3389/fviro.2023.1158703
Received: 04 February 2023; Accepted: 19 May 2023;
Published: 02 June 2023.
Edited by:
John-Sebastian Eden, The University of Sydney, AustraliaReviewed by:
Sandeep Swargam, Central University of Himachal Pradesh, IndiaAditya Kumar Padhi, Indian Institute of Technology (BHU), India
Copyright © 2023 Samal, Bhugra, Suroliya, Gautam, Agarwal, Bihari and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ekta Gupta, ZWd1cHRhQGlsYnMuaW4=, ZWt0YWdhdXJpc2hhQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
 Jasmine Samal
Jasmine Samal Arjun Bhugra
Arjun Bhugra Varun Suroliya
Varun Suroliya Pramod Gautam
Pramod Gautam Reshu Agarwal
Reshu Agarwal Chhagan Bihari
Chhagan Bihari Ekta Gupta
Ekta Gupta