
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol., 01 March 2023
Sec. Modeling of Viral Replication and Pathogenesis
Volume 3 - 2023 | https://doi.org/10.3389/fviro.2023.1114827
 Juliette Lewis1
Juliette Lewis1 Shijun Zhan1
Shijun Zhan1 Allison C. Vilander1
Allison C. Vilander1 Anna C. Fagre1
Anna C. Fagre1 Tawfik A. Aboellail1
Tawfik A. Aboellail1 Hippokratis Kiaris2
Hippokratis Kiaris2 Tony Schountz1*
Tony Schountz1*Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus that causes coronavirus disease-19 (COVID-19), emerged in late 2019 in Wuhan, China and its rapid global spread has resulted in millions of deaths. An important public health consideration is the potential for SARS-CoV-2 to establish endemicity in secondary animal reservoirs outside of Asia or acquire adaptations that result in new variants with the ability to evade the immune response and reinfect the human population. Previous work has shown that North American deer mice (Peromyscus maniculatus) are susceptible and can transmit SARS-CoV-2 to naïve conspecifics, indicating its potential to serve as a wildlife reservoir for SARS-CoV-2 in North America. In this study, we report experimental SARS-CoV-2 susceptibility of two additional subspecies of the North American deer mouse and two additional deer mouse species, with infectious virus and viral RNA present in oral swabs and lung tissue of infected deer mice and neutralizing antibodies present at 15 days post-challenge. Moreover, some of one species, the California mouse (P. californicus) developed clinical disease, including one that required humane euthanasia. California mice often develop spontaneous liver disease, which may serve as a comorbidity for SARS-CoV-2 severity. The results of this study suggest broad susceptibility of rodents in the genus Peromyscus and further emphasize the potential of SARS-CoV-2 to infect a wide array of North American rodents.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus that causes coronavirus disease-19 (COVID-19), emerged in late 2019 in Wuhan, China with rapid global spread resulting in millions of deaths (1). Molecular evidence suggests the zoonotic origin of SARS-CoV-2 were horseshoe bats of the genus Rhinolophus, which carry many SARS-related coronaviruses (2). COVID-19 remains an ongoing pandemic with hundreds of millions of cases and substantial global economic impacts.
The long-term pandemic status of COVID-19 is partially facilitated by the periodic emergence of new variants of SARS-CoV-2 that overcome immunity from vaccination or prior infection. The emergence of the omicron variant of SARS-CoV-2 in November 2021 sparked a new wave of infections globally due to its increased ability to infect convalescent and vaccinated individuals (3). The rapid accumulation of spike protein mutations that led to the omicron variant has led to the hypothesis that its progenitor may have acquired mutations during adaptation to a non-human host, perhaps rodents (4, 5). The potential for new variants to emerge because of adaptation to non-human hosts must be considered, and therefore identification and characterization of prospective non-human hosts is necessary to facilitate preparedness.
Several mammalian species have been shown to be susceptible to SARS-CoV-2, including tree shrews, Syrian hamsters, ferrets, cats, Egyptian fruit bats, mink, deer mice, and several species of non-human primates (6–13). Of note was the human-to-mink transmission of SARS-CoV-2 on mink farms in several countries, which resulted in localized human outbreaks of a mink-adapted SARS-CoV-2 variant (8). This exemplifies the need to examine animals living in close proximity to humans for their potential to act as hosts to SARS-CoV-2.
In our previous work, we determined that one subspecies of North American deer mice (Peromyscus maniculatus nebrascensis) is susceptible to experimental infection with SARS-CoV-2 and capable of transmission to naïve conspecifics for at least two passages. Another group reported similar findings in another subspecies of deer mouse (P. m. rufinus) (7). Deer mice are common peridomestic rodents that occupy a wide geographical range in North America (14, 15) and are the principal reservoir hosts of Sin Nombre orthohantavirus (16) and Borrelia burgdorferi, the causative agent of Lyme disease (17). Given their prominence in North American ecosystems, including on and around mink farms, it is feasible that SARS-CoV-2 could be introduced into wild deer mouse populations.
In this work, we expand the evidence of peromyscine rodent susceptibility to infection with a human isolate of SARS-CoV-2 to include two additional subspecies of the North American deer mouse group, including the Sonora white-footed mouse (P. m. sonoriensis) and the prairie deer mouse (P. m. bairdii). We also demonstrate susceptibility of two additional deer mouse species, the Oldfield mouse (P. polionotus subgriseus) and the California mouse (P. californicus), suggesting that rodents in genus Peromyscus are broadly susceptible to SARS-CoV-2. The examined species also expand the geographical range in which susceptible deer mice exist, increasing the risk of zooanthroponotic transmission of SARS-COV-2 from humans to wild rodents.
SARS-CoV-2 (isolate 2019-nCoV/USA-WA1, NR52281) was obtained from BEI Resources. Virus was passaged twice on Vero E6 cells (ATCC CRL-1586) in 2% FBS-DMEM containing 10,000 IU/ml penicillin and streptomycin at 37°C under 5% CO2 to generate stock virus used in these experiments. Virus was stored at −80°C. All work with infectious virus was performed at BSL-3 with approval from the Colorado State University Institutional Biosafety Committee.
All rodent work was approved by the CSU Institutional Animal Care and Use Committee (protocol 993). Deer mice were intranasally inoculated under inhalation of isoflurane anesthesia with 1x104 (P. bairdii), 1.5x104 (P. sonoriensis), or 2×104 (P. polionotus and P. californicus) TCID50 SARS-CoV-2 (dose based on appropriate inoculation volume for body size). On days 3, 6 and 15 post-challenge, all animals were orally swabbed and weighed, and three inoculated mice of each species were euthanized by inhalation anesthesia of 3% isoflurane to effect, and thoracotomy followed by cardiac exsanguination. Oral swabs and weights were taken for remaining mice on day 10 post-challenge. Control rodents were euthanized on day 0 using procedure described above. For all euthanized animals, necropsies were performed and samples (blood, lung) were collected for viral RNA detection (by RT-qPCR), virus isolation, and serology. Remaining whole carcasses were fixed in 10% neutral buffered formalin for histopathology and immunohistochemistry.
Eighteen California mice were intranasally inoculated under inhalation isoflurane anesthesia with 2×104 TCID50 SARS-CoV-2 and four were euthanized as controls on either day 3 or 10 post-challenge. All mice were orally swabbed, weighed, and observed for clinical signs daily. Necropsies were performed on 6 mice euthanized on days 3, 6, and 10 post-challenge, and tissues (blood, lung) were collected for viral RNA detection (by RT-qPCR), virus isolation, and serology. Remaining whole carcass was fixed in 10% neutral buffered formalin for histopathology and immunohistochemistry
Swabs were stored in 200 μL of viral transport media (VTM) at -80° C until processing and analysis. Swabs were vortexed for 10 seconds followed by centrifugation at 1,500 rcf for 10 minutes to pellet debris. Resulting supernatant was used in downstream assays. Tissues were stored in 500 μL of VTM and either flash frozen in LN2 and stored at -80° C until processing and analysis (multi-species experiment) or immediately processed following collection and stored at -80° C until analysis (California mouse experiment). Processing of tissue samples included homogenization on a TissueLyser II (Qiagen) at 50 Hz for 5 minutes followed by centrifugation at 10,000 rcf to pellet debris. Resulting supernatants were used in downstream analysis including infectious virus isolation and RNA extraction for viral RNA quantification. Blood was collected in a serum separator tube, allowed to clot for 20 to 30 minutes, and then centrifuged at 20,000 rcf for 90 seconds. Sera were collected and stored at -80° C until analysis.
Supernatants from swabs or lung homogenates were plated in triplicate on 96-well plates seeded with Vero E6 cells in a 10-fold dilution series from 100 to 10-3. Samples that were above the limit of detection were re-plated in triplicate on 96-well plates seeded with Vero E6 cells in a 10-fold dilution series from 10-2 to 10-5. All plates were scored four days post-infection and titers were calculated using the Reed-Meunch method (18).
RNA was extracted from swab or tissue supernatant using the Mag-Bind Viral DNA/RNA 96 Kit (Omega Biotek) on the KingFisher FLEX System (ThermoFisher Scientific) according to manufacturer’s instructions. Reverse transcriptase quantitative PCR (RT-qPCR) was run using the GoTaq RT-qPCR System (Promega) with the IDT E Assay First Line Screening primers and probe (Cat #10006804) on a LightCycler 96 System (Roche Diagnostics, Cat# 05815916001).
Serum neutralization was performed starting at a 1:10 dilution with a 2-fold dilution series. An equal volume of SARS-CoV-2 containing 100 TCID50s was added (final serum dilution of 1:20) and incubated for 1 hr at 37°C. The mixture was plated on Vero E6 cells and scored for cytopathic effect after four days. The titer was the reciprocal of the greatest dilution that conferred 100% protection.
Recombinant SARS-CoV-2 nucleocapsid antigen in PBS was coated onto 96-well plates overnight at 4°C. Plates were washed 3x with PBS-0.5% TWEEN-20 and 3x with PBS between each step, then blocked with 0.5% gelatin in PBS for 30 min. Serum samples were diluted 2-fold in 0.5% BSA-PBS starting 1:100 and incubated for 2 hours at room temperature. Detection antibody, anti-Peromyscus leucopus IgG H&L-HRP (SeraCare), was diluted 1:1000 and incubated for 1 hour, followed by ABTS substrate. Plates were read at 405 nm. Samples were considered positive if the OD was 0.200 above the mean of the negative control serum samples (uninfected deer mice).
After samples were collected for molecular analysis, the entire carcass was fixed in 10% neutral buffered formalin. Representative samples from the lungs, liver, heart, and the entire head (sagittal section) were collected and routinely processed for histopathology. Tissues were labeled with hematoxylin and eosin (H&E) for histologic analysis. Anti-SARS-CoV-2 immunohistochemistry was performed on embedded lung tissue from a representative subset of lung tissues at days 3 and 4 post-challenge, selected based on virus detection by RT PCR, using rabbit anit-SARS-CoV-2 nucleocapsid antibody (NP1189, ProSci) as a primary antibody with hematoxylin counterstain.
To investigate the susceptibility of different deer mouse species and subspecies, nine young, adult deer mice from each species or subspecies and of both sexes were intranasally challenged with SARS-CoV-2. Three unchallenged controls from each species group were euthanized and necropsied on day zero of the experiment. Three challenged mice from each species group were euthanized and necropsied on days 3, 6, and 15 post-challenge with one additional oral swab taken at day 10. Infectious virus was detected in 1/9 oral swabs from each species group on day 3 post-challenge, and viral RNA was detected in most lung samples from mice euthanized on days 3 and 6 post-challenge. Viral RNA was detected in oral swabs of all challenged mice on day 3, and infectious virus was detected in oral swabs from 3/4 species on day 3 post-challenge (prairie deer mouse, California mouse, and Oldfield mouse). Three of the four species (prairie deer mouse, Sonora white-footed mouse, and Oldfield mouse) had detectable neutralizing antibodies on day 15 post-challenge, with an average titer of 87 (SD ± 39). California mouse samples for this time point were compromised by mold contamination. A summary of virus detection and serology can be found in Table 1, with exact values available in the Supplementary Materials.
No clinical signs, including weight loss (Figure 1), were observed in the prairie deer mouse, Sonora white-footed mouse, or Oldfield mouse at any timepoint in the study. On day 3 post challenge, two California mice (1 female, 1 male) were observed to have ruffled fur, a hunched posture, and lethargic behavior. When the mice were re-examined on day 4, the male appeared to have improved whereas the female was moribund and humanely euthanized for necropsy following the same methods for scheduled necropsies. This mouse had hepatomegaly and prominent hepatic reticular pattern. It was suspected that the mouse was suffering from hepatic lipidosis (or in humans, non-alcoholic fatty liver disease (NAFLD)). Three other California mice had a grossly prominent hepatic reticular pattern and hepatomegaly upon necropsy, including the male that displayed clinical signs on day 3. California mice lost significantly more weight 3 days post-challenge than any other group. To further investigate California mice as a model for SARS-CoV-2 infection, additional California mice were infected with SARS-CoV-2. Serial necropsies were performed on days 3, 6, and 10 post-challenge. No clinical signs were observed in these mice for the duration of the study.
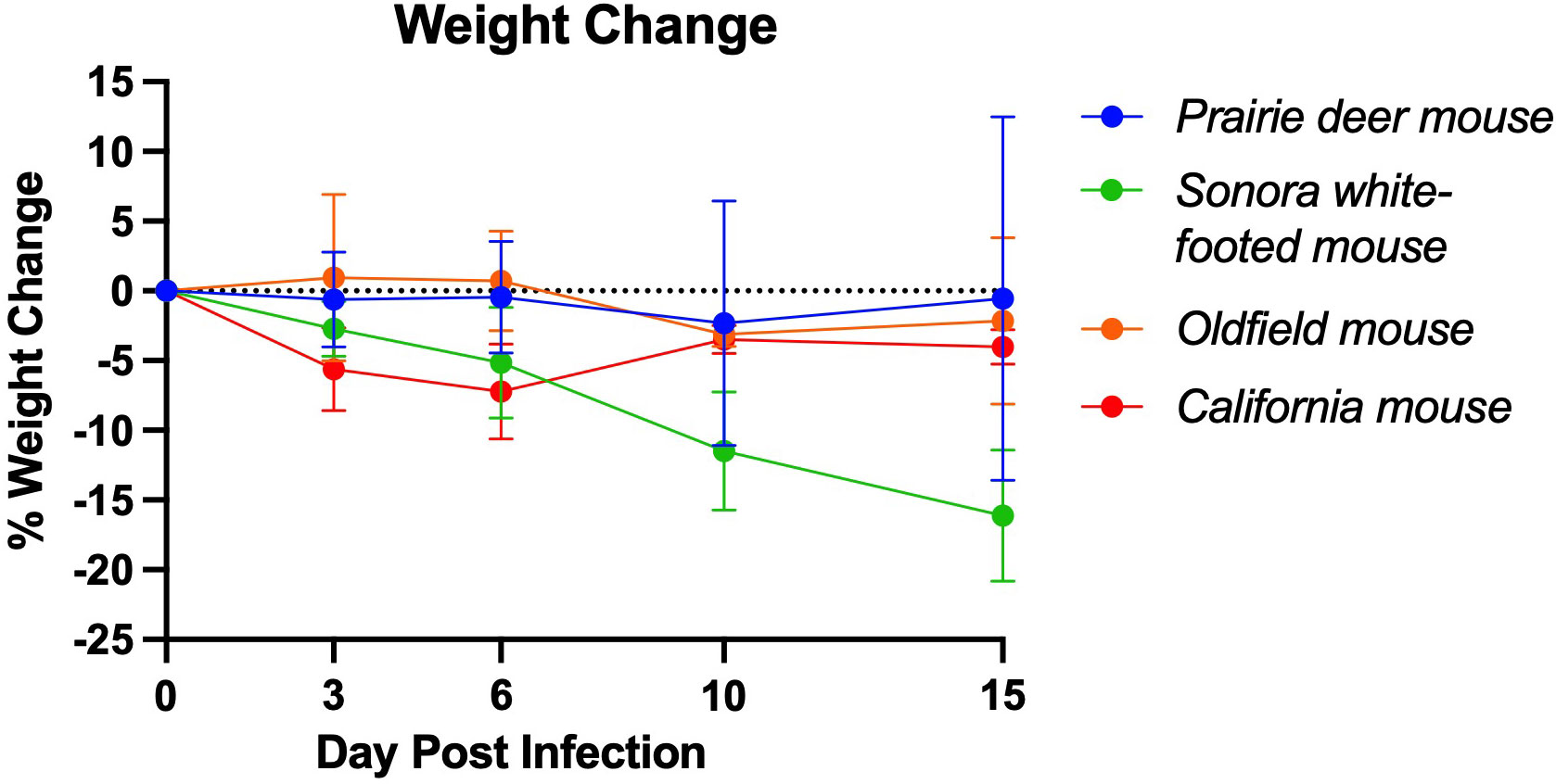
Figure 1 Mean weight change of deer mouse species during SARS-CoV-2 infection. On day 3 post-challenge, the California mice had significantly greater weight loss than the prairie deer mouse (p=0.004), Sonora white-footed mouse (p=0.027), and Oldfield mouse (p=0.010). At 6 days post-challenge, the California mice had significantly greater weight loss than the prairie deer mice (p=0.016) and the Oldfield mice (P=0.005). Points represent mean weight change of the group. Full list of p-values as calculated by unpaired t-test available in Supplementary Table 1. For each group, n=9 on day 3, n=6 on day 6, n=3 on day 15 except group D, for which n=2 on day 15.
We further developed California mice as an animal model of SARS-CoV-2 infection following our observation of clinical disease in two mice during our initial investigation. Our follow-up study indicated that the mice became infected, with infectious virus detectable in oral swabs on days 1, 2, and 3 post-challenge (average 3.16 x 102 ± 2.93 TCID50/mL, Figures 2A, B) and in all day 3 lungs (average 5.60 x 104 ± 5.39 x 104 TCID50/mL, Figures 2C, D). Viral RNA was detectable through day 10 post-challenge in both lungs and oral swabs. Serum IgG was detectable in all mice at 10 days post-challenge (geometric mean titer 400, 95%CI [180,887], Figure 2E), and neutralizing antibody was detectable in 5 out of 6 mice (geometric mean titer 92, 95% CI [45,189], Figure 2F). The SARS-CoV-2 challenged group showed significantly greater weight loss than the uninfected controls at days 1 and 3 post-challenge (Figure 3). At day 10 the control mice had a significant decrease in weight compared to infected controls. The cause of the weight loss is unknown. As the infected mice had all cleared detectible infectious virus by day 10, the weight loss in the control group at this time point is not significant to the conclusions of this study.
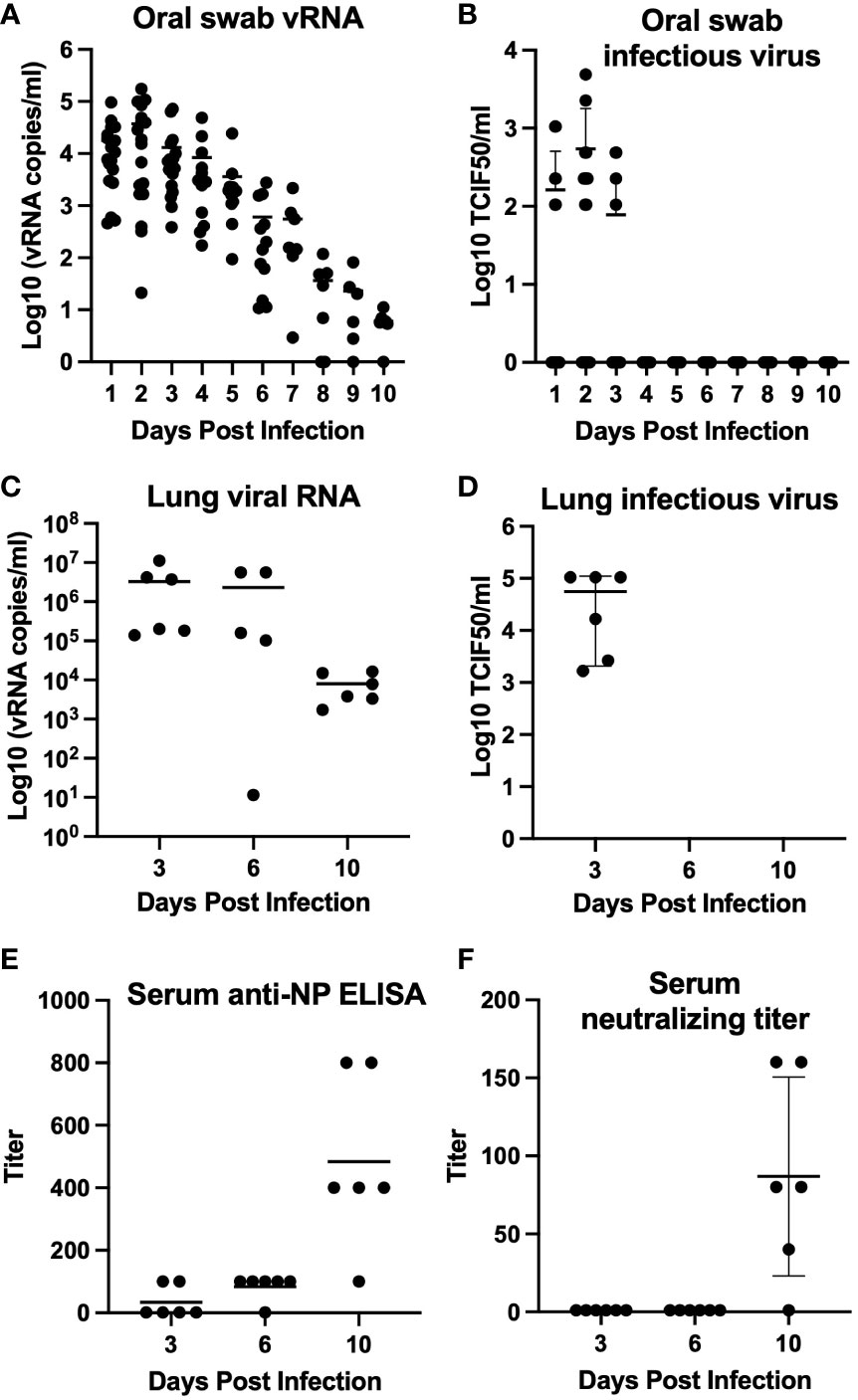
Figure 2 Mean virus detection and serology for California mice (P. californicus insignis). Oral swabs collected daily (days 0–10 post-challenge) (n=18 for days 0-3, n=12 for days 4-6, n=6 for days 7-10) were tested for viral RNA (A) and infectious virus (B). Lung homogenate collected from mice euthanized on days 3 (n=6), 6 (n=6), and 10 (n=6) post-challenge were also tested for viral RNA (C) and infectious virus (D). Serum collected from mice on days 3 (n=6), 6 (n=6), and 10 (n=6) post-challenge was evaluated for quantification of serum IgG determined by indirect ELISA (E) and serum neutralizing antibody titer determined by serum neutralization assay (F).
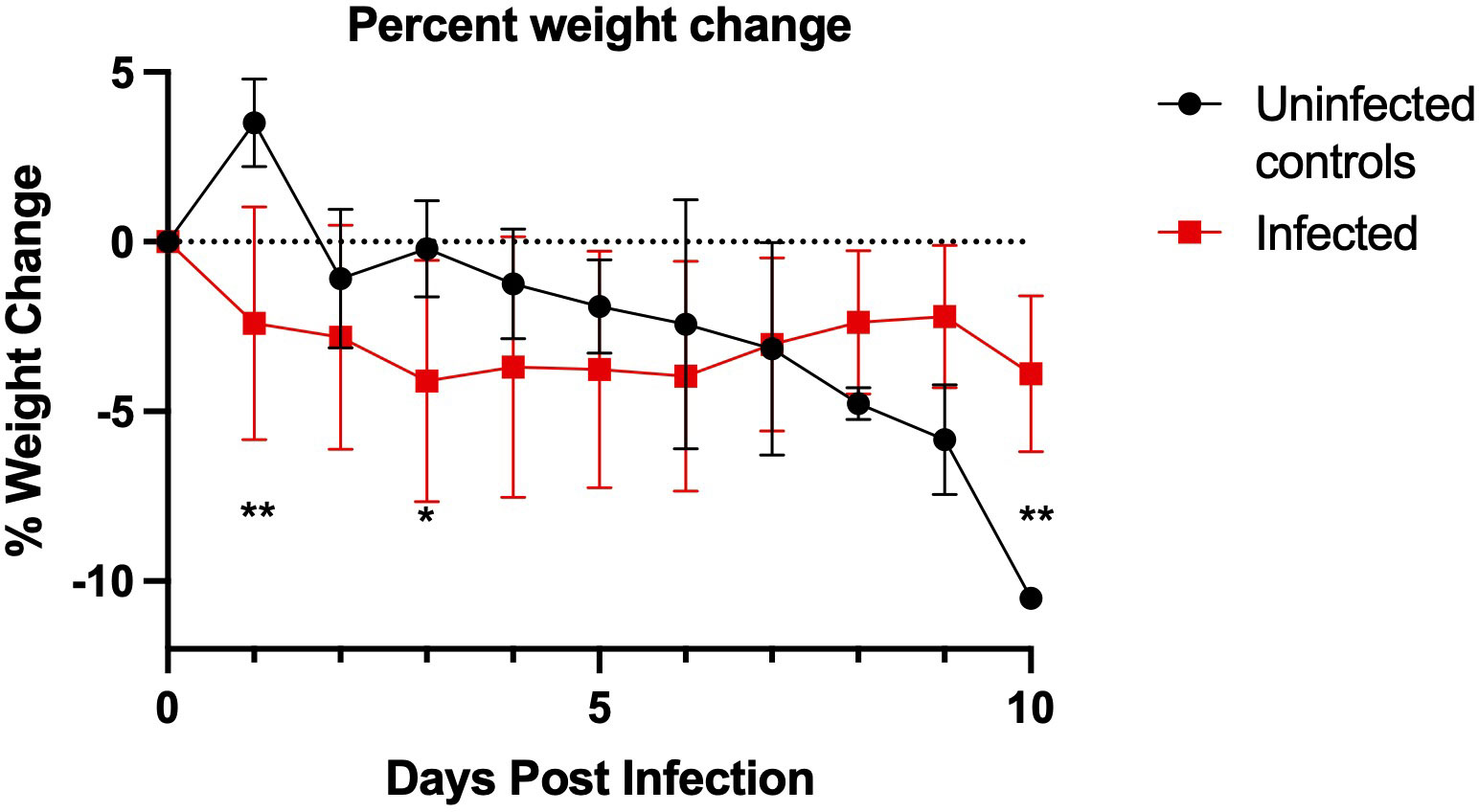
Figure 3 California mouse (P. californicus insignis) weight change during SARS-CoV-2 infection. SARS-CoV-2-challenged California mice had significantly greater weight loss on day 1 post-challenge (**p=0.003) and day 3 post-challenge (*p=0.047) as compared to the uninfected controls. The uninfected control group had significantly more weight loss than the challenged group at day 10 post-challenge (P=0.008). P-values calculated by unpaired t-test. Points represent mean weight change of the group. For the infected group, n=18 for days 0-3, n=12 for days 4-6, and n=6 for days 7-10. For the uninfected control group, n=4 for days 0-3 and n=2 for days 4-10.
Lung, liver, spleen, nasal skin and mucosa, and heart from all mice were evaluated for pathology. The most severe inflammation was observed in the lungs and nasal passages in all species. The nasal skin and mucosa, heart, and liver did not show histologic differences in any of the groups. Lung was the most affected tissue and infected mice showed variable severity of interstitial lymphohistiocytic and fibrinous pneumonia with edema, reactive vascular endothelium with neutrophilic margination and perivasculitis, and rarely minimal to mild bronchial epithelial hyperplasia (Figures 4A, B). Necrosuppurative rhinitis was the second most common finding in infected mice (Figure 4C). Other observed pathology included mild lymphohistiocytic myocarditis, microvascular hepatic lipidosis, necrotizing and lymphohistiocytic dermatitis of the muzzle and lymphoplasmacytic perineuritis. Lesions in all species, when present, were most severe at day 3 post-challenge versus days 6, 10, and 15. Pneumonia was present in all groups but was minimal to mild in the Sonora white-footed mouse. Immunohistochemistry for anti-SARS-CoV-2 nucleocapsid had minimal rare positive labeling of bronchiolar epithelium in mice with virus detectible by RT-PCR (Figure 4D).
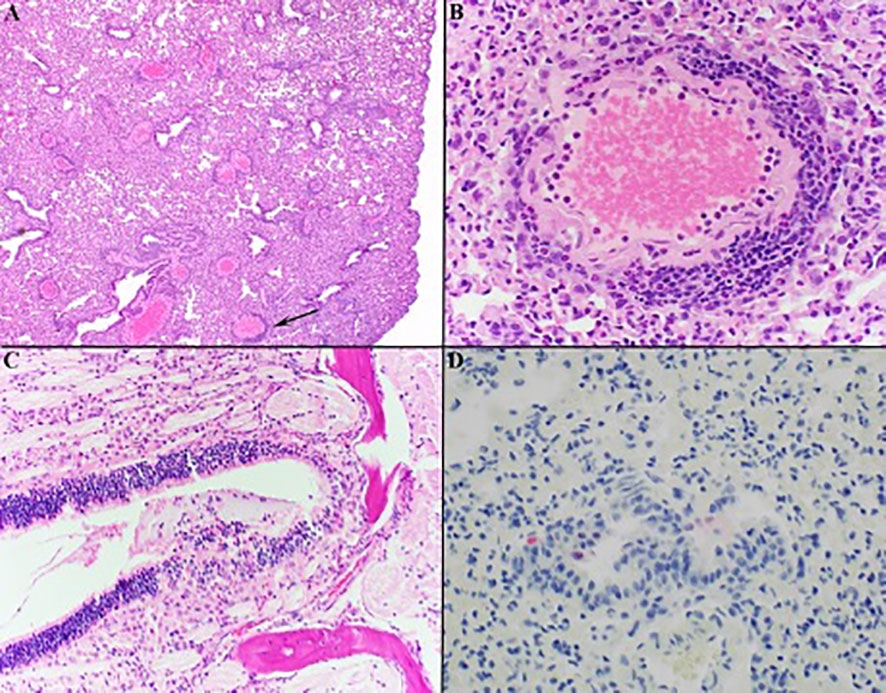
Figure 4 Histology of representative lesions in SARS-CoV-2 infected Peromyscus spp. Interstitial lymphohistiocytic pneumonia (A) Oldfield mouse, 100x) was the most common finding with multifocal vessels with segmental reactive endothelium, neutrophilic margination, and perivasculitis (indicated by black arrow in (A) magnified in (B) 400x). Necrosuppurative rhinitis was the second most common finding (C) California mouse, 200x). Anti-SARS-CoV-2 IHC shows rare positive labeling (red) of bronchial epithelial cells within the lungs (D) Sonora white-footed mouse, 400x). Hematoxylin counterstain.
In six of the twelve California mice, there was hepatic lipidosis that ranged from mild to severe. The mouse that became moribund during the study demonstrated the most severe hepatic lipidosis histologically (Figure 5A). Lymphohistiocytic pneumonia in this mouse was minimal to mild but there was increased bronchial epithelial hyperplasia compared to other animals and this animal had the strongest anti-SARS-CoV-2 IHC labeling of the hyperplastic epithelium and scattered histiocytes (Figures 5B, C).
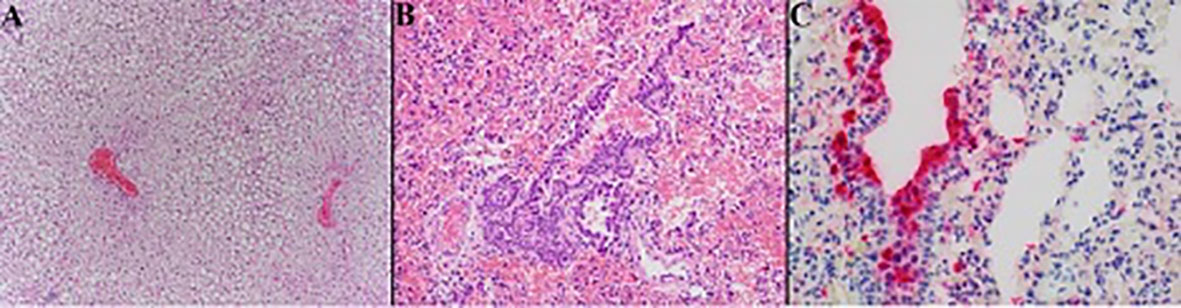
Figure 5 Photomicrographs of California mouse liver and pulmonary lesions. A single California mouse became moribund and at necropsy was found to have hepatomegaly. Histopathology showed severe diffuse distention of hepatocytes by lipid vacuoles consistent with hepatic lipidosis (A) 200x). The lungs of this animal exhibited only minimal to mild lymphohistiocytic pneumonia with moderate hyperplasia of the bronchiolar epithelium [(B), 200x] but there was marked anti-SARS-CoV-2 positive labeling (red) of hyperplastic bronchial epithelium and histiocytes [(C), 400x]. Hematoxylin counterstain.
The North American genus Peromyscus, collectively known as deer mice, contains more than 50 species, and has been studied extensively for their potential use as animal models and for their ability to transmit medically significant pathogens (19). Many peromyscine rodents live in peri-domestic habitats, encountering humans through shared habitats as well as during outdoor activities (e.g., camping, field research). The experiments described here, along with previous work, confirm that 3 species of deer mice (the North American deer mouse, the Oldfield mouse, and the California mouse) and 4 subspecies of the North American deer mouse are susceptible to experimental SARS-CoV-2 infection with a human isolate of the virus (6, 7). There is some uncertainty as to the phylogenetic classification of the subspecies of deer mice used in this work; it has been recently proposed that the North American deer mouse (P. maniculatus) should be reclassified as the eastern deer mouse (P. maniculatus) and the western deer mouse (P. sonoriensis) (20). This taxonomic change would increase the number of confirmed susceptible deer mouse species to four.
Experimental challenge resulted in asymptomatic infection in all species but the California mouse, of which two individuals displayed clinical signs and one required euthanasia. Interestingly, the moribund California mouse that was euthanized early had significant immunostaining for SARS-CoV-2 nucleocapsid in the lungs but minimal histological signs of pneumonia, whereas asymptomatic animals in this study had more signs of pneumonia and very little immunostaining. This could indicate that immunosuppression or immunocompromise played a role in the infection outcome for that animal. For most animals, infection lasted through day 10 post-challenge, and by day 15 neutralizing antibodies were present in all species. The mostly asymptomatic and short-lived nature of SARS-CoV-2 infection in the observed species warrants investigation into their potential use as animal models for asymptomatic infection. The weight loss in some uninfected California mice was striking; however, this species is monogamous and we have observed loss of appetite after disruption of breeding pairs, particularly males, in the University of South Carolina breeding colony. The separation of these animals after arrival at CSU may have contributed to weight loss. Deer mice can live up to 8 years in captivity (21), making them more attractive as small animal models for immune durability in vaccine and reinfection studies than Syrian hamsters and human ACE2-transgenic mice, which only live about 2 years.
Future studies of SARS-CoV-2 in these species of deer mice should explore susceptibility to more current strains of SARS-CoV-2, because the Washington strain used in this study in 2020 has since been displaced by omicron variants in humans. Additionally, although previous work has shown that two subspecies of North American deer mouse mouse are capable of sustained conspecific transmission, this should be confirmed in the species and subspecies used in this study (6, 7).
The California mouse has been studied as a model for several obesity-related conditions including type II diabetes mellitus and NAFLD (22, 23). California mice spontaneously develop these conditions when consuming a high-fat diet. In this study, we determined that California mice are experimentally susceptible to SARS-CoV-2, with evidence of infectious virus in the lungs, oral shedding, and seroconversion. Both California mice that exhibited clinical signs in this study had hepatic lipidosis as determined by blinded pathological review, indicating that obesity-related conditions may have similar consequences in California mice as in humans during SARS-CoV-2 infection. NAFLD has been shown to predict COVID-19 progression and severity and COVID-19-related liver injury in human patients (24, 25). Therefore, the California mouse may be a suitable comorbidity model for NAFLD or other obesity related conditions and SARS-CoV-2. A larger study using California mice that have been fed a high-fat diet could address questions about liver dysfunction and SARS-CoV-2 disease, along with metabolic testing of the animals to confirm the conditions.
Peromyscine rodents and Syrian hamsters belong to the family Cricetidae and they each develop similar disease upon SARS-CoV-2 infection. However, California mice are the first small animal model to develop disease following SARS-CoV-2 infection, other than transgenic mice that express human ACE2. Considering their long lifespans, their outbred nature, availability of annotated genomes for several species, ready availability, and easy laboratory management, peromyscine rodents may serve as a superior to existing small animal models used in some SARS-CoV-2 studies.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Colorado State University Institutional Animal Care and Use Committee.
JL designed and performed experiments; SZ performed experiments; AV and TA performed histopathology; AF performed animal infections; HK designed experiments; TS supervised work, performed bench work. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Institute of Allergy and Infectious Disease (R01 AI140442, TS), the National Science Foundation (2033260).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1114827/full#supplementary-material
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
2. Hu B, Zeng L-P, Yang X-L, Ge X-Y, Zhang W, Li B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PloS Pathog (2017) 13:e1006698. doi: 10.1371/journal.ppat.1006698
3. Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, et al. Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. bioRxiv (2021). doi: 10.1101/2021.12.03.471045:2021.12.03.471045
4. Sun Y, Lin W, Dong W, Xu J. Origin and evolutionary analysis of the SARS-CoV-2 omicron variant. J biosafety Biosecur (2022) 4:33–7. doi: 10.1016/j.jobb.2021.12.001
5. Wei C, Shan K-J, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS-CoV-2 omicron variant. J Genet Genomics (2021). doi: 10.1016/j.jgg.2021.12.003:S1673-8527(21)00373-8
6. Fagre A, Lewis J, Eckley M, Zhan S, Rocha SM, Sexton NR, et al. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: Implications for spillback to new world rodents. PloS Pathog (2021) 17:e1009585. doi: 10.1371/journal.ppat.1009585
7. Griffin BD, Chan M, Tailor N, Mendoza EJ, Leung A, Warner BM, et al. SARS-CoV-2 infection and transmission in the north American deer mouse. Nat Commun (2021) 12:3612. doi: 10.1038/s41467-021-23848-9
8. Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science (2021) 371:172–7. doi: 10.1126/science.abe5901
9. Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe (2020) 1:e218–25. doi: 10.1016/S2666-5247(20)30089-6
10. Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science (2020) 368:1016–20. doi: 10.1126/science.abb7015
11. Sia SF, Yan L-M, Chin AWH, Fung K, Choy K-T, Wong AYL, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature (2020) 583:834–8. doi: 10.1038/s41586-020-2342-5
12. Zhao Y, Wang J, Kuang D, Xu J, Yang M, Ma C, et al. Susceptibility of tree shrew to SARS-CoV-2 infection. Sci Rep (2020) 10:16007. doi: 10.1038/s41598-020-72563-w
13. Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther (2020) 5:157. doi: 10.1038/s41392-020-00269-6
15. Lawlor TE, Hall ER. The mammals of north America. J Mammalogy (1982) 63:718–9. doi: 10.2307/1380296
16. Schmaljohn C, Hjelle B. Hantaviruses: A global disease problem. Emerg Infect Dis (1997) 3:95–104. doi: 10.3201/eid0302.970202
17. Bosler EM, Ormiston BG, Coleman JL, Hanrahan JP, Benach JL. Prevalence of the Lyme disease spirochete in populations of white-tailed deer and white-footed mice. Yale J Biol Med (1984) 57:651–9.
18. Reed LJ, Muench H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. Am J Epidemiol (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
19. Kirkland GL, Layne JN. Advances in the study of peromyscus (Rodentia). Lubbock, Texas Tech University Press (1989).
20. Greenbaum I, Honeycutt R, Chirhart S. Taxonomy and phylogenetics of the peromyscus maniculatus species group. Lubbock, Texas: Museum of Texas Tech University. (2019) 71:559–75.
21. Sacher GA, Hart RW. Longevity, aging, and comparative cellular and molecular biology of the house mouse, mus musculus, and the white-footed mouse, peromyscus leucopus. Birth Defects Orig Artic Ser; (United States) (1978) 14 71–96.
22. Krugner-Higby L, Caldwell S, Coyle K, Bush E, Atkinson R, Joers V. The effects of diet composition on body fat and hepatic steatosis in an animal (Peromyscus californicus) model of the metabolic syndrome. Comp Med (2011) 61:31–8.
23. Krugner-Higby L, Shadoan M, Carlson C, Gendron A, Cofta P, Marler C, et al. Type 2 diabetes mellitus, hyperlipidemia, and extremity lesions in California mice (Peromyscus californicus) fed commercial mouse diets. Comp Med (2000) 50:412–8.
24. Ji D, Qin E, Xu J, Zhang D, Cheng G, Wang Y, et al. Non-alcoholic fatty liver diseases in patients with COVID-19: A retrospective study. J Hepatol (2020) 73:451–3. doi: 10.1016/j.jhep.2020.03.044
Keywords: SARS-CoV-2, Peromyscus, cricetid, zoonosis, deer mouse
Citation: Lewis J, Zhan S, Vilander AC, Fagre AC, Aboellail TA, Kiaris H and Schountz T (2023) SARS-CoV-2 infects multiple species of North American deer mice and causes clinical disease in the California mouse. Front. Virol. 3:1114827. doi: 10.3389/fviro.2023.1114827
Received: 02 December 2022; Accepted: 14 February 2023;
Published: 01 March 2023.
Edited by:
Takaaki Koma, Tokushima University, JapanReviewed by:
Mark Parcells, University of Delaware, United StatesCopyright © 2023 Lewis, Zhan, Vilander, Fagre, Aboellail, Kiaris and Schountz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tony Schountz, dG9ueS5zY2hvdW50ekBjb2xvc3RhdGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.