- 1Department of Microbiology, Graduate School of Medicine, Tokushima University, Tokushima, Japan
- 2Division of Interdisciplinary Researches for Medicine and Photonics, Institute of Post−LED Photonics, Tokushima University, Tokushima, Japan
- 3Division of Next-generation Photonics, Institute of Post−LED Photonics, Tokushima University, Tokushima, Japan
- 4Department of Immunology and Parasitology, Graduate School of Medicine, Tokushima University, Tokushima, Japan
- 5Department of Microbiology, Kansai Medical University, Hirakata, Japan
Deep ultraviolet light (UV) is useful for the disinfection of microorganisms, including bacteria and viruses. Although genome damage by UV has been widely accepted, the adverse effects of UV on the activity and/or function of viral proteins including the envelope components are poorly documented. Worthy of note, the observed unfavorable UV-effects for viruses are only insufficiently analyzed in association with the reduction in viral infectivity. In this study, we aimed to clarify which component of virions affected by UV significantly correlates with the loss of viral infectivity using HIV-1 as a model for single-stranded RNA enveloped viruses. Using our UV irradiation apparatus at three wavelengths (265, 280, and 300 nm), we first quantitatively determined the UV power density and irradiation period of each wavelength required for a reduction in infectivity. A heat-treated sample as a control drastically reduced the virion-associated reverse transcriptase (RT) activity and Gag-p24 level. The UV-irradiated samples at the three wavelengths, completely lacking viral infectivity, showed p24 levels similar to those without irradiation. While the virion-associated RT activity was gradually decreased in a wavelength and power density dependent manner, this reduction did not explain the loss of viral infectivity by UV. Remarkably, virological assays revealed that the entry efficiency of the UV-irradiated virus samples at the three wavelengths is comparable to those without irradiation. Importantly, this result shows that, even the virions exposed to UV of various wavelengths at the lethal level, still maintain the function of their envelope composed of a host lipid bilayer and viral proteins. In sharp contrast, UV-induced genome damage shown by semiquantitative RT-PCR correlated well with the reduction in viral infectivity, indicating that it is a major determinant for virus inactivation by UV. The degree of damage was found to be distinct among the regions analyzed. This was probably due to the different nucleotide sequences in those genomic regions amplified by PCR. Our data clearly demonstrate a principal mechanism for viral inactivation by UV and provide information contributing to the improvement of UV-based disinfection technology for microorganisms.
Introduction
The disinfection of pathogenic microorganisms in various environments is vital for public health. It is critically important not only in this particular COVID-19 pandemic period but also in other periods of time as a general and highly effective anti-infectious disease strategy. Of the several commonly used disinfectants, including heat and alcohol, deep ultraviolet light (UV) is a time-saving and efficacious means to disinfect/inactivate microorganisms such as bacteria and viruses (1–3). We and others have shown that UV is effective to inactivate a variety of viruses including SARS-CoV-2 (4–8). UV irradiation to inactivate microorganisms can be applied to various environments, such as public spaces and water, by using different kinds of light sources with distinct energy or by combining it with photosensitizer agents.
Based on the wavelength, UV is categorized into UV-A (315-400 nm), UV-B (280-315 nm), and UV-C (200-280 nm). UV-A inactivates microorganisms in the presence of photochemical agents such as titanium dioxide and amotosalen HCl. This is probably due to the induction of oxidative stress through the production of reactive oxygen species (9–11). UV-B and UV-C, close to the adsorption peak of nucleic acids at 265 nm, can be absorbed by viral genomes (DNA or RNA). It is well established that UV damages RNA through photochemical modification of nucleotides such as the formation of pyrimidine dimers, and through crosslinks of RNA-RNA and RNA-protein (12–18; for review, see 19, 20). UV-C at lower wavelengths of 254 and 222 nm also exhibits antimicrobial activities (21, 22; for review, see 19, 20). UV-C also has been reported to degrade the major outer viral protein capsid of bacteriophage MS2 (non-enveloped RNA virus), feline calicivirus (non-enveloped RNA virus), and mouse norovirus (non-enveloped RNA virus) by oxidation, whereas proteins of human adenovirus (non-enveloped DNA virus), Tulane virus (non-enveloped RNA virus), and rotavirus (non-enveloped RNA virus) are not significantly affected by UV-C irradiation (23–27). For SARS-CoV-2 (enveloped RNA virus), while the virion morphology exposed to UV appears to be unchanged, whether UV affects viral proteins S and N has not been concluded yet (28, 29). Moreover, the envelope integrity of hepatitis C virus (enveloped RNA virus) is not affected by UV irradiation, whereas UV can act on lipids constituting the envelope of herpes simplex virus (enveloped DNA virus) (30, 31). Studies so far have extensively investigated the effects of UV on apparent changes, integrity loss, or reduction in levels of virion components. However, even if viral proteins and the envelope are not apparently damaged by UV, it is possible that UV can affect the activity and function of virion components essential for viral replication. In this regard, targets directly related to the loss of viral infectivity by UV need to be determined.
We have reported a quantitative evaluation of SARS-CoV-2 inactivation by UV at three wavelengths (265, 280, and 300 nm) by calculating the dose of UV energy irradiated to the virus itself through the culture media (8). In our previous study, we definitely showed the length of the UV irradiation periods, which wavelength, and what doses of UV are required for SARS-CoV-2 inactivation. Our unique and useful UV irradiation apparatuses enable us to quantitatively evaluate the decrease in viral infectivity. As described above, UV irradiation can affect proteins, lipids, and nucleic acids. UV-irradiation in large quantities, of course, induces the loss of viral infectivity and also adversely affects proteins, lipids, and nucleic acids. For enveloped viruses like HIV-1 and SARS-CoV-2, if UV detrimentally and decisively affects lipids and Envelope (Env) proteins, major components of viral envelope, the entry process would be hampered. Our purpose in this study is to clarify which component of virions affected by UV significantly correlates with the loss of viral infectivity, and this would provide an important aspect of virus inactivation. To this end, we utilized HIV-1 as a model of single-stranded RNA enveloped viruses. HIV-1 has measurable reverse transcriptase (RT) and p24 proteins within virions, and various experimental systems to precisely analyze each viral replication step have been well-established for HIV-1 (Figure 1). After irradiation to cell-free virions, we can investigate the direct effect of UV on components within virions including envelope, enzymatic activity, and RNA genome (Figure 1). Through virological analyses, we demonstrate that UV can have an effect on the enzymatic activity of a viral protein but not the entry process at all, and that the loss of viral infectivity principally results from UV-induced genome damage.
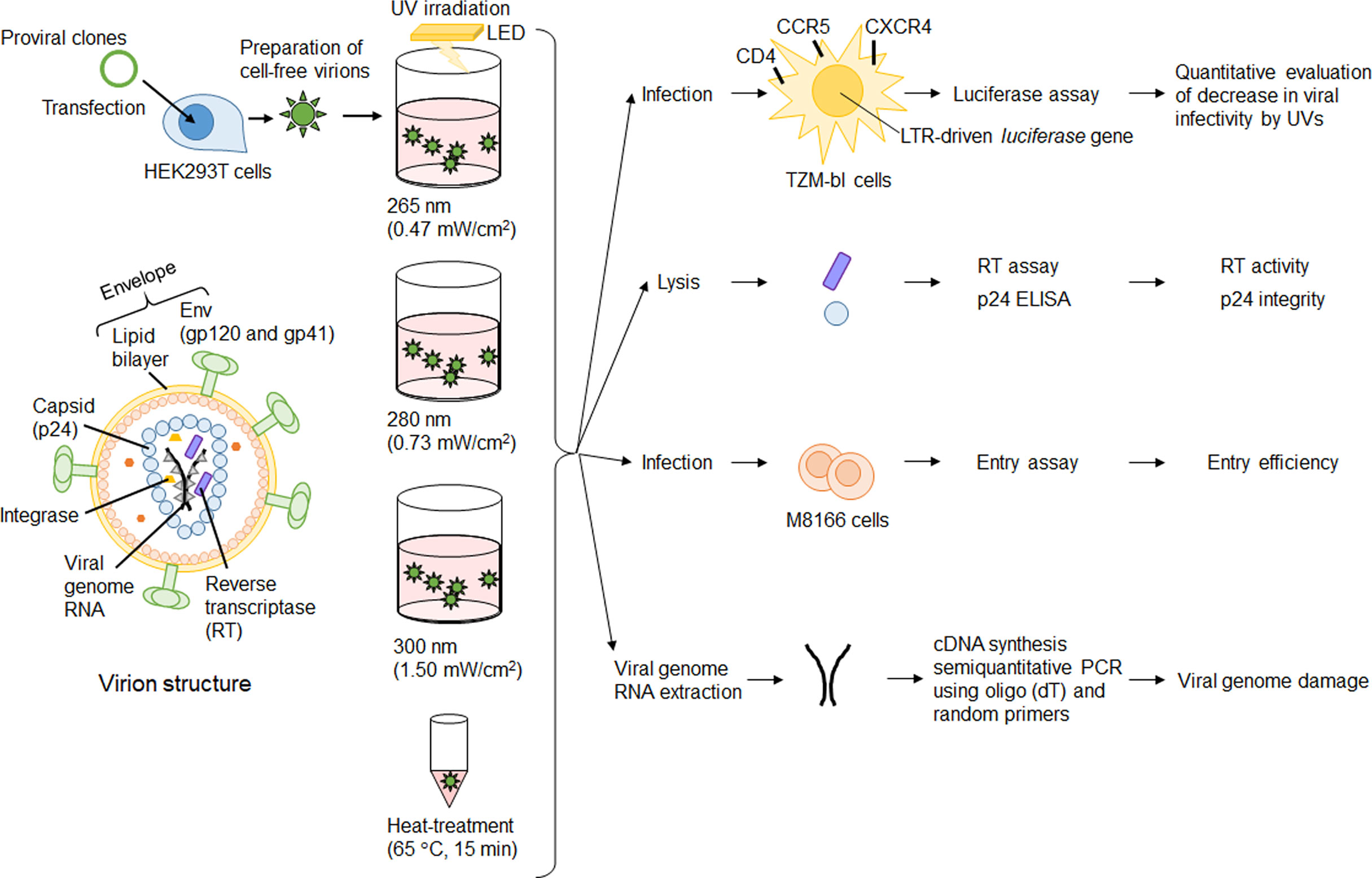
Figure 1 Experimental outline. Major virion components are depicted on the left. Cell-free virions were prepared from 293T cells transfected with proviral clones. Cell-free virions were treated with UV or heat, and then used for various assays. Details of assay procedures were described in the Materials and Methods section.
Materials and Methods
Cells
Monolayer cell lines HEK293T (ATCC CRL-1573) and HeLa-derived reporter TZM-bl (32) were cultured and maintained in Eagle’s minimal essential medium containing 10% heat-inactivated fetal bovine serum (FBS) as previously described (33). A lymphocyte line M8166 was cultured and maintained in RPMI 1640 medium containing 10% heat-inactivated FBS.
Virus preparations
HIV-1 proviral clones pNL4-3 (34) and pNL-Kp (an env-deficient clone) (35) were used in this study. Proviral clones were transfected into HEK293T cells by the calcium phosphate co-precipitation method as previously described (34, 36). The virus amounts were determined by virion-associated RT assays as previously described (36, 37). For quantification of the Gag-CA (p24) level, an HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix Corporation) was used according to the manufacturer’s protocol. When necessary, HIV-1 virus stocks were treated with Recombinant DNase I (RNase-free) (TAKARA BIO INC.) to remove the contamination of transfected plasmid DNA. Heat-treated virus stocks were prepared by incubating at 65°C for 15 min.
UV irradiation
UV irradiation apparatuses with three different wavelengths (265 nm, 280 nm, and 300 nm) that we made previously (8), were used in this study. Our irradiation apparatuses carry UV-light emitting diode (LED). The distance from the UV-LED chip to the virus inoculum was about 30 mm. The UV light from the UV-LED was collimated by a reflector and irradiated to a well. The UV-LED irradiation area was set large enough to obtain uniform irradiation. These apparatuses enable us to estimate doses irradiated to the virus itself by calculating the transmittances through the culture medium (8). To ensure repeated and reproducible irradiation by UV-LED light, a virus solution was placed in a defined well of a 96-well plate as a chamber, and set directly under the UV-LED upon irradiation. Prior to UV irradiation, the virus stocks were appropriately diluted with PBS containing 2% FBS (104 RT units/100 μL). The diluted samples were exposed to UV with different irradiation power densities at each wavelength for appropriate time periods (8).
Virus infectivity assay
TZM-bl cells (5 × 103/well) were seeded onto a 96-well plate and cultured overnight. The UV-irradiated virus samples were inoculated into the cells. Virus samples without any treatment and with heat-treatment were used as controls. On day 2 post-infection, cell lysates were prepared for luciferase assays (Promega) as previously described (33). HIV-1 infectivity was calculated as a relative light unit (RLU) of the UV- or heat-treated virus samples relative to that without any treatments.
Entry assay
Virus stocks were prepared from transfected HEK293T cells, diluted (105 RT units/100 μL in PBS containing 2% FBS), and then subjected to UV irradiation. Heat-treated virus stocks were also diluted in the same manner. Entry assays were carried out similarly to as described previously (38–40). Briefly, 200 μL of each sample was inoculated into M8166 cells (2 × 106), and the cells were incubated for 2 h at 4°C, extensively washed, and collected as viral binding fractions. To measure the substantial level of the virus entry, following incubation at 4°C as above, cells were trypsinized for 5 min at 37°C, extensively washed, and incubated for 2h at 37°C to collect the viral entry fractions. Cells collected as binding and entry fractions were lysed for p24 ELISA. The entry efficiency of each sample was calculated as the p24 level of the entry fraction/p24 level of the binding fraction. NL-Kp (delta Env) and heat-treated virus were used as controls.
Semiquantitative RT-PCR analysis
To assess the genome damage by UV, semiquantitative RT-PCR analysis was performed similarly to as described previously (33, 41). Briefly, DNase I-treated virus stocks were diluted and subjected to UV irradiation as above. Heat-treated virus samples were used as controls. The viral genome was isolated from the samples using a QIAamp Viral RNA Mini Kit (Qiagen GmbH), and reverse transcribed by using mixed primers (oligo (dT) and random hexamer) and the SuperScript III first-strand synthesis system (Life Technologies Corporation). The primer pairs used for semiquantitative RT-PCR analysis were as follows: primer set A (forward) (GGCCTGAAAATCCATACAAT) and (reverse) (TCTAAAAGGCTCTAAGATTTTTGTCAT); primer set B (forward) (CTTGGCACTAGCAGCATTA) and (reverse) (CTGGATGCTTCCAGGGCTCT); primer set C (forward) (GGAATAACATGACCTGGATG) and (reverse) (CGCAGATCGTCCCAGATAAGTG); primer set D (forward) (GAGGATTGTGGAACTTCT) and (reverse) (CTAGGTCTCGAGATACTGCTC). To gain PCR products amplified within a linear range, the cycle number of PCR reactions was carefully determined using a control sample without UV irradiation (33, 41). Negative controls for semiquantitative RT-PCR were prepared by reverse transcription of the UV-irradiated or heat-treated samples without any primers. PCR amplicons were separated on 2% Metaphor agarose gel (Lonza Ltd.), and stained with ethidium bromide. The signal intensities of the PCR amplicons were quantified by using the Amersham Imager 600 instrument (GE Healthcare UK Ltd.). For viral RNA samples without irradiation and with heat-treatment, the signal intensities of the PCR products were calculated by subtracting the signal intensity of each negative control from that of each sample.
Results
Experimental outline
To examine the direct effect of UV on components of HIV-1 virions, cell-free virions were exposed to UV using our irradiation apparatuses (Figure 1). First, we quantitatively evaluated energy doses of UVs with three wavelengths (265 nm, 280 nm, and 300 nm) required for the reduction in viral infectivity. UVs irradiation conditions, which give an around half reduction in viral infectivity or the complete loss of viral infectivity, were determined. Then, we examined the effect of UVs on viral components/function (virion-associated RT activity and p24 integrity, and entry ability) under the irradiation conditions we determined. Finally, UV-induced genome damage was investigated by semiquantitative RT-PCR using cDNA synthesized from viral RNA isolated from cell-free virions exposed to UV under the determined conditions.
Quantitative evaluation of HIV-1 inactivation by UV irradiation
To study the viral components responsible for the reduction in infectivity by UV, we used HIV-1 in which a number of experimental systems to analyse its replication step have been established. We first quantitatively evaluated a dose-dependent reduction in viral infectivity using our UV-LED irradiation apparatuses with major wavelengths (265, 280, and 300 nm). The reasons why we chose these three wavelengths in this study are as follows: 265 nm is an absorption peak wavelength of RNAs, at which the inactivation effect must be maximum in the UV-C region. 280 nm is an absorption peak wavelength of proteins, and RNAs also have moderate absorption at 280 nm. Furthermore, an LED at 280 nm has excellent characteristics of power and lifetime in commercial products. 300 nm is far from the absorption peak of RNAs and proteins, which can be used as a control. Viruses were prepared from HEK293T cells transfected with a proviral clone. The infectivity of viruses exposed to UV was measured using a HeLa-derived TZM-bl cell line. TZM-bl cells express HIV receptor CD4 and coreceptors CCR5/CXCR4, and carry a long terminal repeat-driven luciferase gene (32). Upon infection, HIV-coded Tat protein trans-activates luciferase expression in TZM-bl cells, and thus infectivity can be measured as luciferase activity. A heat-treated virus, which has a different inactivation process from UV, was used as a control (29). The power densities of UV at three wavelengths were determined by preliminary experiments to give a similar level of luciferase activity for the same irradiation period (0.47 mW/cm2 for 265 nm, 0.73 mW/cm2 for 280 nm, and 1.50 mW/cm2 for 300 nm). As shown in the left panel of Figure 2, for samples irradiated with UV for 20 sec at each wavelength, the virus infectivity was decreased to 20~40% relative to that without irradiation. A longer irradiation period was associated with a further reduction in viral infectivity. Infection was not detected for viruses exposed to UV at three wavelengths for 160 sec or treated with heat. Consistent with our previous result for SARS-CoV-2 inactivation (8), viral infectivity was similarly decreased by the UV energy dose for 265 nm and 280 nm, whereas a higher dose was required for the complete loss of viral infectivity at 300 nm (Figure 2, right panel).
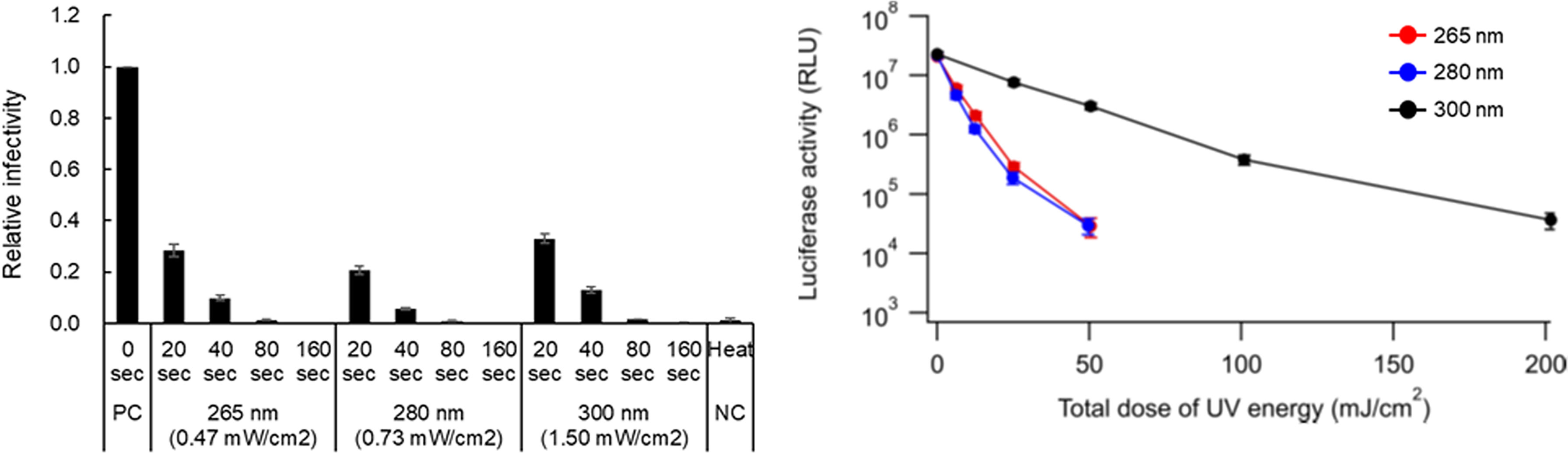
Figure 2 Effect of UV-irradiation with various wavelengths on HIV-1 infectivity. Virus stocks prepared from transfected HEK293T cells were diluted with PBS containing 2% FBS to make 104 RT units/100 µL of virus samples. The diluted samples were exposed to UV at three wavelengths with the indicated power densities for the indicated time periods. Heat-treated virus stocks were also diluted in the same manner as the UV-irradiated samples. The samples were inoculated into TZM-bl cells, and on day 2 post-infection, the cells were lysed and subjected to luciferase assay. (Left panel) Viral infectivity is presented as relative RLU of UV-irradiated and heat-treated samples to that of the sample without any treatment. (Right panel) The inactivation efficacy at various total doses of UV energy at three wavelengths is shown. Mean values with standard errors (SE) are shown (n=3). PC, positive control; NC, negative control.
UV irradiation can reduce the enzymatic activity of HIV-1 reverse transcriptase (RT) but this reduction does not explain the loss of infectivity
The HIV-1 virion core is formed by Gag-p24 proteins which play important roles in viral replication, and also contains two copies of single-stranded RNA genome and two enzymes, RT and integrase, essential for the early replication phase (42) (Figure 1). The inhibitory effects of UV on the activity/function of RT or Gag-p24, if any, are very likely to impair HIV-1 replication. To examine how UV affects the RT activity and p24 level in virions, viruses prepared from transfected HEK293T cells were exposed to UV for 20 and 160 sec at a fixed power density of each wavelength, which resulted in the reduction to 20~40% and the complete loss of infectivity, respectively (Figure 2). We first monitored the Gag-p24 levels of virus samples as shown in Figure 3. A heat-treated virus showed a significant decrease in the Gag-p24 level. The UV-irradiated samples tested exhibited a similar Gag-p24 level to that without irradiation, although the Gag-p24 levels were marginally decreased for samples exposed to UV at 280 nm and 300 nm for 160 sec (Figure 3). On the one hand, RT activity was undectable in heat-treated viruses as expected, and a longer UV irradiation time at three wavelengths resulted in a decline in the RT activity, particularly for 160 sec irradiations of UV at 280 nm and 300 nm (Figure 3). The RT activity was moderately reduced to around 60% compared to that without irradiation. The results here suggest that there may be effects of UV on viral proteins in a wavelength- and dose-dependent manner. It has been reported that some HIV-1 RT mutants with about half activity relative to a wild-type virus can moderately replicate in lymphocyte cells (primary cells and MT-2) (43). Taken together, these results indicated that while UV can induce a decrease in enzymatic activity and/or denaturation of viral proteins, these changes in the viral proteins do not directly lead to the complete loss of viral infectivity.
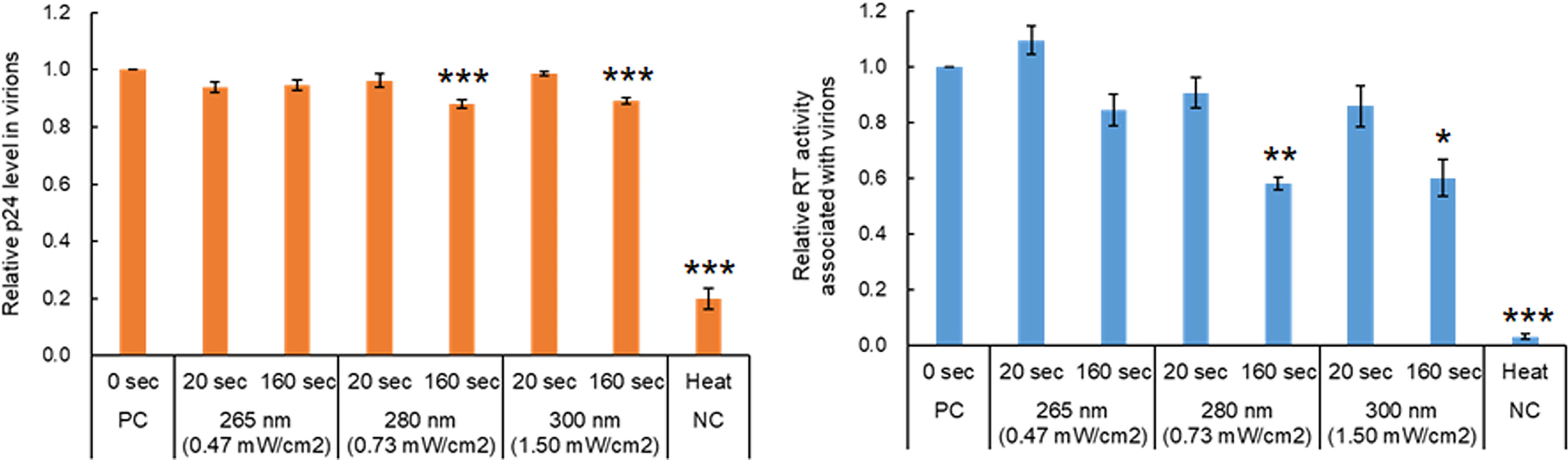
Figure 3 Effect of UV-irradiation with various wavelengths on HIV-1 proteins in virions. Viruses were prepared from HEK293T cells transfected with proviral clones. Virus stocks with and without heat-treatment were diluted as described in Figure 1. The diluted samples were exposed to UV at three wavelengths for the indicated time periods, and then were subjected to p24 ELISA and RT assays. The relative p24 level (left) and RT activity (right) in each sample to those in the sample without any treatment are presented. Mean values with SE are shown (n=4). Significance relative to the sample without any treatment as calculated by Welch’s t test is shown (*P < 0.05; **P < 0.02, ***P < 0.01). PC, positive control; NC, negative control.
UV irradiation does not affect the HIV-1 entry process
Since an HIV-1 virion has an envelope composed of a host lipid bilayer and viral envelope Env proteins, the viral entry process is expected to be hampered if UV adversely affects these components. It has been reported that no effect of UV irradiation on virus binding to cells is observed for the non-enveloped Tulane virus and rotavirus, and for the enveloped influenza virus (27, 44). Functional HIV-1 Env is a trimer of a heterodimer consisting of Env-gp120 and Env-gp41, which are produced by cleavage of precursor Env-gp160 with furin or furin-like protease within the Golgi apparatus (42). HIV-1 Env-gp160 has been shown to have the CD4 binding capability but not the entry ability (45, 46). Thus, it is necessary to examine the entry potential as the function of Env. To determine whether viral Env exposed to UV is functional, entry assays were performed. Since the Gag-p24 level was not significantly changed after UV irradiation (Figure 3), the entry efficiency was calculated by measuring the amounts of virus-binding and entry using Gag-p24 ELISA. Env-deleted or heat-treated virus samples were used as negative controls. Virus samples were exposed to UV at three wavelengths with a fixed power density for 160 sec, which resulted in the complete loss of infectivity (Figure 2). As shown in Figure 4, heat-treated viruses were binding-incompetent and thus the entry efficiency was not detected. Entry efficiency was dramatically reduced for Env-deficient virus samples. Of note, viruses exposed to UV still displayed a similar level of entry efficiency to that without irradiation, that is, viral Env is fully functional even after UV irradiation at a lethal level. Importantly, the results here indicate that the complete loss of HIV-1 infectivity is not explained by the impairment of the Env function via the adverse effect of UV on Env proteins and/or lipid bilayers.
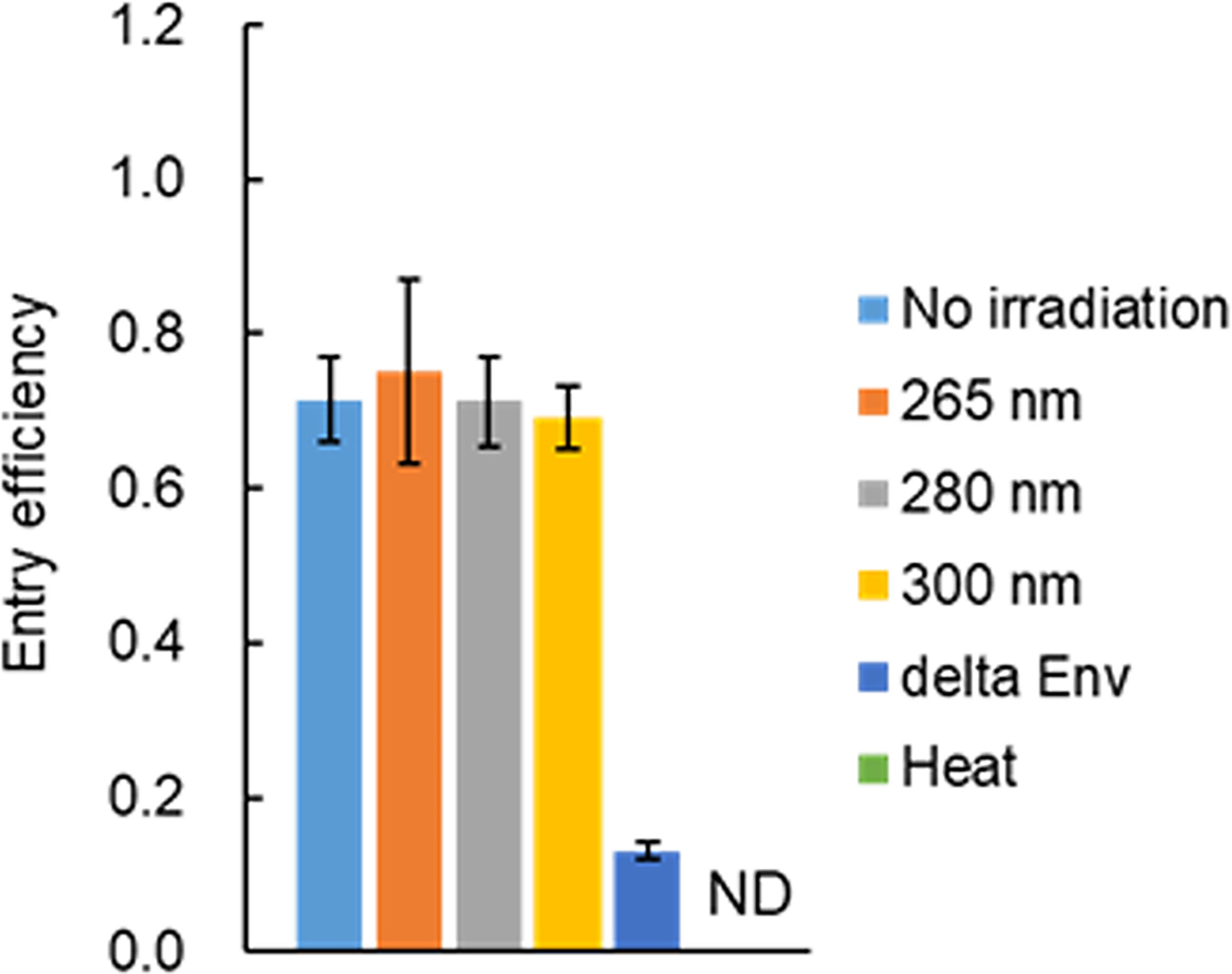
Figure 4 Effect of UV-irradiation on the viral entry process. Viruses were prepared from HEK293T cells transfected with proviral clones pNL4-3 (WT) or pNL-Kp (delta Env). Virus stocks were diluted with PBS containing 2% FBS, and exposed to UV at three wavelengths with various power densities (0.47 mW/cm2 for 265 nm, 0.73 mW/cm2 for 280 nm, 1.50 mW/cm2 for 300 nm) for 160 sec. Heat-treated WT virus stocks were also prepared and diluted in the same manner as UV-irradiated samples. The diluted UV- or heat-treated samples were used for the entry assay. The assays were carried out as described in “Materials and Methods” (Entry assay). The entry efficiency of each sample, calculated as the p24 level of entry fraction/p24 level of the binding fraction, is presented. Mean values with SE are shown (n=3). The control heat-treated virus sample did not bind to cells and thus the entry efficiency was not determined. ND, not determined.
Viral genome is damaged by UV irradiation, leading to the loss of viral infectivity
The results in this study so far indicated that while UV can moderately affect the HIV-1 RT activity, the viral core and envelope still remain to be functionally normal after sufficient UV irradiation to cause the complete loss of infectivity (Figures 1–4). It is well known that UV damages RNA including viral genome through photochemical modification of nucleotides such as the formation of pyrimidine dimers, and through crosslinks of RNA-RNA and RNA-protein (12–18; for review, see 19, 20). For analysis of genome damage by UV, it has been reported that conventional RT-qPCR, which amplifies 128 bp of PCR products in the N gene region of the SARS-CoV-2 genome, failed to detect viral genome damage (28). This was because the length of PCR products was too short to detect genome damage (28). Thus, to investigate the correlation between the reduction in viral infectivity and genome damage, semiquantitative RT-PCR analysis was carried out. Primer sets used for the analysis were designed to amplify four different regions (~0.4 kb in length), and enable us to examine whether the difference in positions and/or sequences of the genome affects UV-induced damage (Figure 5 and Supplementary Figure 1). Viruses were exposed to UV with a fixed power density for 20 and 160 sec, which resulted in a 60-80% reduction and complete loss of infectivity, respectively (Figure 2). A heat-treated virus was used as a control. Consistent with a previous report (29), heat treatment at a lethal level did not strongly induce genome damage (Figure 5 and Supplementary Figure 1). A longer UV-irradiation at three wavelengths was associated with a lower signal intensity of PCR products, suggesting that the genome damage is linked to the reduction in infectivity. This is probably due to inefficient cDNA synthesis using UV-damaged RNA genome (modification of nucleotides and crosslinking) as a template. It is well known that RNA quality affects the efficiency and quality of cDNA synthesized by RT, and that subsequent PCR reaction is also affected. This inferiority of cDNA synthesized from UV-damaged RNA may be responsible for the inhibition/failure of the subsequent PCR amplification. All virus samples exposed to UV at 300 nm showed relatively higher signal intensities compared to those irradiated UV at 265 and 280 nm, implicating weaker damage by UV irradiation at 300 nm and the difference in the efficacy of genome damage by UV wavelengths. This result may explain why a higher level of energy dose of UV at 300 nm is required for HIV-1 inactivation. In conclusion, the results demonstrated that UV-induced genome damage is primarily responsible for the loss of infectivity.
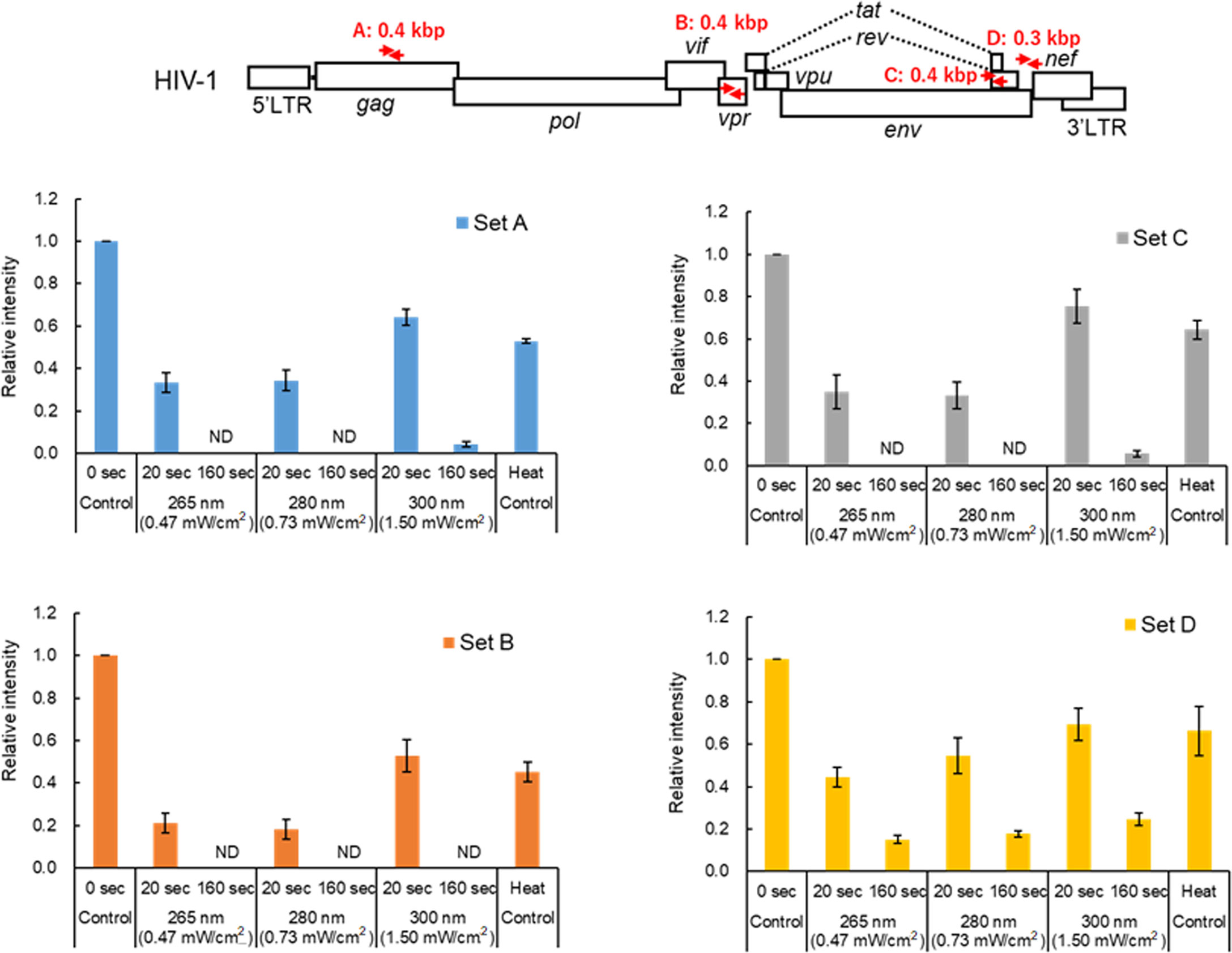
Figure 5 Effect of UV-irradiation on the HIV-1 genome. Virus stocks were prepared from HEK293T cells transfected with pNL4-3, and virus samples were appropriately prepared as indicated. The viral genome isolated was subjected to semiquantitative RT-PCR analysis using specific primer sets. The primer sets used in the analysis and the sizes of the RT-PCR products are shown in red arrows and letters in the schematic HIV-1 genome at the top. For details of the primers, see “Materials and Methods” (Semiquantitative RT-PCR analysis). Signal intensities of the semiquantitative RT-PCR products were quantitated. The relative intensity is presented as a signal intensity of each sample relative to that of the sample without UV irradiation. Mean values with SE are shown (n=3). ND, not detected.
Discussion
For virus inactivation by UV, its effects on viral components including genome damage and protein degradation have been reported (for review, see 19, 20). However, whether adverse effects of UV on the activity and function of viral components can lead to the loss of viral infectivity have been poorly documented. To address this issue, we investigated major targets for virus inactivation by UV using HIV-1 as a model for single-stranded RNA enveloped viruses. The reduction in HIV-1 infectivity was quantitatively evaluated using our UV-LED irradiation apparatuses with three wavelengths (265, 280, and 300 nm). The power densities of these apparatuses were fixed to give around 60-80% reduction and an undetectable level of infectivity by irradiating UV to viruses for 20 and 160 sec, respectively. Under these conditions, the Gag-p24 level was not significantly affected, whereas the RT activity was gradually decreased in virus samples exposed to UV, especially at 280 and 300 nm. This attenuated RT activity did not explain the complete loss of infectivity, since it has been shown to be still able to support viral replication in cells (43). Unexpectedly, our entry assays revealed that even for virions exposed to UV at a lethal level, the entry process normally proceeded, indicating no appreciable effect of UV on the function of viral Env composed of proteins and lipids. Unlike these viral components, viral genome damage by UV irradiation was obviously correlated to the loss of infectivity. In addition, the requirement of a higher energy dose of UV at 300 nm for viral inactivation may be related to its weaker damage to the viral genome compared to those by 265 and 280 nm. In terms of the HIV-1 life cycle, UV irradiation to cell-free virions can directly damage both tRNA primer and viral genome RNA within the virions. Such damaged RNA would not be a good template/primer for cDNA synthesis with HIV-1 RT and tRNA primer in infected cells. Thus, the loss of infectivity for UV-irradiated virions is highly likely to result from inhibition of the reverse transcription process through UV-induced damage to RNA such as viral genome and tRNA. In conclusion, we clearly demonstrate here that viral genome damage is a major target for UV-induced complete loss of HIV-1 infectivity.
Although UV can attack viral proteins and lipids, which are components of the envelope, our entry assay revealed that UV does not affect the HIV-1 entry into cells. A bacteriophage MS2 exposed to UV has been shown to have attachment ability but reduced genome penetration ability to enter into bacteria (47). This may be due to the difference in methods used to analyze the entry process. In our assays, viral entry was assessed by measuring the Gag-p24 level which is not affected by UV, whereas for MS2, genome penetration assays as monitored by quantitative PCR were performed. Alternatively, it may be due to the difference in the entry process between membrane fusion for HIV-1 and genome penetration for bacteriophages. The degree of damage to viral proteins may vary with the light sensitivity of the protein itself and with the wavelength and energy dose of UV. Indeed, the HIV-1 RT activity tended to decrease in virions exposed UV, particularly at 280 and 300 nm, which have higher energy doses relative to that of 265 nm. This implies that UV can adversely affect the enzymatic activity of viral proteins in a wavelength- and energy dose-dependent manner, although it is not directly associated with the loss of infectivity.
The degree of HIV-1 genome damage by UV correlated well with the loss of infectivity. In our semiquantitative assays (Figure 5), PCR products for primer set D were still detected in the virus samples exposed to UV at the three wavelengths for 160 sec. The efficiency in PCR amplification was different between primer sets C and D, which were designed in a close position on the viral genome. This suggests that the degree of genome damage may be different by sequences of amplified regions of the genome. In this regard, the reason why we designed primers to adjacent positions on the viral genome is that the ratio of contiguous pyrimidine nucleotides (UU) in the two regions is different (8.2% for set C and 4.3% for set D). Considering the formation of pyrimidine dimers by UV, a higher rate of UU sequence may lead to a reduction in amounts of PCR products. Indeed, the ratio of UU sequences within set D region (4.3%) is lower than those within sets A (7.4%) and B (5.7%). There are some reports on the prediction of UV susceptibility based on the nucleotide sequences (20). The establishment of such prediction methods would contribute to the development of more efficient and effective inactivation techniques by UV. Further studies are required to examine whether a higher ratio of UU in the sequences is associated with a higher rate of genome damage. The genome damage by UV has been reported for various viruses (for review, see 19, 20). We were interested in the genome damage in SARS-CoV-2, since we have reported the quantitative evaluation of SARS-CoV-2 inactivation by UV at three wavelengths (8). In the previous study, we showed that undetectable SARS-CoV-2 infectivity as determined by plaque assay was achieved by distinct UV energy doses at different wavelengths (3 mJ/cm2 for 265 nm, 5 mJ/cm2 for 280 nm, and 30 mJ/cm2 for 300 nm). These UV energy doses required for SARS-CoV-2 inactivation were around 7 to 10-fold lower than those for HIV-1 (50 mJ/cm2 for 265 and 280 nm and 200 mJ/cm2 for 300 nm) (Figure 2). The data do not necessarily imply that SARS-CoV-2 is more susceptible to UV irradiation compared to HIV-1. Although the sensitivity of the two distinct assay methods using different viruses/cells is absolutely not comparable, the difference in the UV energy doses that are required for HIV-1 and SARS-CoV-2 inactivation can be accounted for by the sensitivity of the infectivity assays used (luciferase expression vs plaque formation, respectively). The genome damage in SARS-CoV-2 was investigated in our laboratory by semiquantitative RT-PCR analysis using samples exposed to UV under each condition which inactivates SARS-CoV-2 and HIV-1 (Supplementary Figure 2). Two regions, orf1b and nucleocapsid, of the SARS-CoV-2 genome were targeted for PCR amplification. All the PCR products amplified were still detected in virus samples exposed to UV at energy doses which lead to the loss of SARS-CoV-2 infectivity (3 mJ/cm2 for 265 nm, 5 mJ/cm2 for 280 nm, and 30 mJ/cm2 for 300 nm), although the signal intensities of the PCR products were decreased compared to those without irradiation. As expected, the PCR products were undetectable or only slightly detectable for virus samples irradiated with UV at lethal levels of energy doses for HIV-1 (50 mJ/cm2 for 265 and 280 nm, and 200 mJ/cm2 for 300 nm). Again, these results underscore our conclusion that genome damage is a major determinant for virus inactivation by UV, because virions that are exposed to UV at a lethal level for HIV-1 still retain the Gag-p24 level and entry ability into cells (Figures 3 and 4). For RT-PCR analysis, RT and PCR enzymes available for experiments are powerful tools to amplify nucleic acids. In terms of virus inactivation by UV, we may need to prudently consider whether severe genome damage to the extent that cannot be amplified by RT-PCR is necessary for virus disinfection/inactivation. Avoiding an overdose of UV to inactivate viruses would lead to reducing the effect of UV on the environment and the waste of light sources and electricity.
In this study, we showed that HIV-1 is inactivated even without apparent destruction of proteins and lipids composing virions by UV irradiation. To determine the relationship between virus inactivation and energy dose of UV, we have properly quantified the actual dose of UV irradiated to virus itself by measuring values for irradiated power densities minus those for the light absorbance by the culture media (this study and 8). However, the efficacy of UV irradiation on virus inactivation can be affected by environments/conditions surrounding virions, e.g. virions in air, droplets, or blood. Further studies are required to solve this issue by considering the effect of environmental factors on virus inactivation by UV. UV is a highly efficacious means to disinfect/inactivate pathogenic microorganisms, such as HIV-1 and SARS-CoV-2, by causing genome damage. Actually, there are commercially available blood photochemical treatment systems with UV to reduce pathogens in blood (48). Our apparatuses and data here would be useful to determine the energy dose of UV required for disinfection/inactivation of pathogens. The future issue for UV-based inactivation would include what kind of light to use and in what situations to use it, considering the wavelength and power, the light sources, the cost performance, and the effects on the human body. It is necessary to consider how UV can be effectively and efficiently used for public spaces and clinical sites.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
TM and MN contributed to the experimental design. TK, ND, and MN performed the experiments. AS, KN, TY, and TM contributed to the UV irradiation apparatuses. AA, TM, and MN wrote the initial drafts of the manuscript. TK, ND, TY, KY, AA, TM, and MN performed the data analysis. All the authors contributed to editing of the manuscript.
Funding
This work was supported in part by grants as follows: The Japan Agency for Medical Research and Development (AMED) Research Program on HIV/AIDS (21443907 and 22580694); Medical Research Grants from the Takeda Science Foundation; The project on the Promotion of Regional Industries and Universities, Cabinet Office, Japan (The Plan for Industry Promotion and Young People’s Job Creation by the Creation and Application of Next-Generation Photonics by Tokushima Prefecture); Tokushima Prefecture Industry-Academia-Government Collaboration Research and Development Expenses Subsidy for Countermeasures against the Novel Coronavirus, etc. from the Tokushima Prefectural Government, Japan; JST Adaptable and Seamless Technology transfer Program through Target-driven R&D (A-STEP) from Japan Science and Technology Agency (JST), Japan (Grant Number: JPMJTM20Y8).
Acknowledgments
We thank Yayoi Shono for experimental assistance. We also thank Kazuko Yoshida and Kyoko Inui for editorial assistance. We appreciate the Support Center for Advanced Medical Sciences, Institute of Biomedical Sciences, Tokushima University Graduate School for the experimental facilities and technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.994842/full#supplementary-material
References
1. Kampf G. Efficacy of ethanol against viruses in hand disinfection. J Hosp. Infect (2018) 98:331–8. doi: 10.1016/j.jhin.2017.08.025
2. Pastorino B, Touret F, Gilles M, de Lamballerie X, Charrel RN. Heat inactivation of different types of SARS-CoV-2 samples: what protocols for biosafety, molecular detection and serological diagnostics? Viruses (2020) 12:735. doi: 10.3390/v12070735
3. Viana Martins CP, Xavier CSF, Cobrado L. Disinfection methods against SARS-CoV-2: a systematic review. J Hosp. Infect (2022) 119:84–117. doi: 10.1016/j.jhin.2021.07.014
4. Duan SM, Zhao XS, Wen RF, Huang JJ, Pi GH, Zhang SX, et al. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed Environ Sci (2003) 16:246–55.
5. Eickmann M, Gravemann U, Handke W, Tolksdorf F, Reichenberg S, Müller TH, et al. Inactivation of three emerging viruses - severe acute respiratory syndrome coronavirus, Crimean-Congo haemorrhagic fever virus and nipah virus - in platelet concentrates by ultraviolet c light and in plasma by methylene blue plus visible light. Vox Sang. (2020) 115:146–51. doi: 10.1111/vox.12888
6. Heilingloh CS, Aufderhorst UW, Schipper L, Dittmer U, Witzke O, Yang D, et al. Susceptibility of SARS-CoV-2 to UV irradiation. Am J Infect Control. (2020) 48:1273–5. doi: 10.1016/j.ajic.2020.07.031
7. Inagaki H, Saito A, Sugiyama H, Okabayashi T, Fujimoto S. Rapid inactivation of SARS-CoV-2 with deep-UV LED irradiation. Emerg Microbes Infect (2020) 9:1744–7. doi: 10.1080/22221751.2020.1796529
8. Minamikawa T, Koma T, Suzuki A, Mizuno T, Nagamatsu K, Arimochi H, et al. Quantitative evaluation of SARS-CoV-2 inactivation using a deep ultraviolet light-emitting diode. Sci Rep (2021) 11:5070. doi: 10.1038/s41598-021-84592-0
9. Van Voorhis WC, Barrett LK, Eastman RT, Alfonso R, Dupuis K. Trypanosoma cruzi inactivation in human platelet concentrates and plasma by a psoralen (amotosalen HCl) and long-wavelength UV. Antimicrob Agents Chemother (2003) 47:475–9. doi: 10.1128/AAC.47.2.475-479.2003
10. Kim JY, Lee C, Cho M, Yoon J. Enhanced inactivation of e. coli and MS-2 phage by silver ions combined with UV-a and visible light irradiation. Water Res (2008) 42:356–62. doi: 10.1016/j.watres.2007.07.024
11. Lee JE, Ko G. Norovirus and MS2 inactivation kinetics of UV-a and UV-b with and without TiO2. Water Res (2013) 47:5607–13. doi: 10.1016/j.watres.2013.06.035
12. Pearson M, Johns HE. Suppression of hydrate and dimer formation in ultraviolet-irradiated poly (A plus U) relative to poly U. J Mol Biol (1966) 20:215–29. doi: 10.1016/0022-2836(66)90061-1
13. Miller N, Cerutti P. Structure of the photohydration products of cytidine and uridine. Proc Natl Acad Sci U. S. A. (1968) 59:34–8. doi: 10.1073/pnas.59.1.34
14. Small GD, Tao M, Gordon MP. Pyrimidine hydrates and dimers in ultraviolet-irradiated tobacco mosaic virus ribonucleic acid. J Mol Biol (1968) 38:75–87. doi: 10.1016/0022-2836(68)90129-0
15. Singer B. Chemical modification of viral ribonucleic acid. IX. the effect of ultraviolet irradiation on TMV-RNA and other polynucleotides. Virology (1971) 45:101–7. doi: 10.1016/0042-6822(71)90117-6
16. Zwieb C, Ross A, Rinke J, Meinke M, Brimacombe R. Evidence for RNA-RNA cross-link formation in escherichia coli ribosomes. Nucleic Acids Res (1978) 5:2705–20. doi: 10.1093/nar/5.8.2705
17. Wilms C, Noah JW, Zhong D, Wollenzien P. Exact determination of UV-induced crosslinks in 16S ribosomal RNA in 30S ribosomal subunits. RNA (1997) 3:602–12.
18. Wigginton KR, Menin L, Sigstam T, Gannon G, Cascella M, Hamidane HB, et al. UV Radiation induces genome-mediated, site-specific cleavage in viral proteins. Chembiochem (2012) 13:837–45. doi: 10.1002/cbic.201100601
19. Wurtmann EJ, Wolin SL. RNA Under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol (2009) 44:34–49. doi: 10.1080/10409230802594043
20. Hadi J, Dunowska M, Wu S, Brightwell G. Control measures for SARS-CoV-2: a review on light-based inactivation of single-stranded RNA viruses. Pathogens (2020) 9:737. doi: 10.3390/pathogens9090737
21. Storm N, McKay LGA, Downs SN, Johnson RI, Birru D, de Samber M, et al. Rapid and complete inactivation of SARS-CoV-2 by ultraviolet-c irradiation. Sci Rep (2020) 10:22421. doi: 10.1038/s41598-020-79600-8
22. Kitagawa H, Nomura T, Nazmul T, Kawano R, Omori K, Shigemoto N, et al. Effect of intermittent irradiation and fluence-response of 222 nm ultraviolet light on SARS-CoV-2 contamination. Photodiagnosis Photodyn Ther (2021) 33:102184. doi: 10.1016/j.pdpdt.2021.102184
23. Rule Wigginton K, Menin L, Montoya JP, Kohn T. Oxidation of virus proteins during UV(254) and singlet oxygen mediated inactivation. Environ Sci Technol (2010) 44:5437–43. doi: 10.1021/es100435a
24. Bosshard F, Armand F, Hamelin R, Kohn T. Mechanisms of human adenovirus inactivation by sunlight and UVC light as examined by quantitative PCR and quantitative proteomics. Appl Environ Microbiol (2013) 79:1325–32. doi: 10.1128/AEM.03457-12
25. Park D, Shahbaz HM, Kim SH, Lee M, Lee W, Oh JW, et al. Inactivation efficiency and mechanism of UV-TiO2 photocatalysis against murine norovirus using a solidified agar matrix. Int J Food Microbiol (2016) 238:256–64. doi: 10.1016/j.ijfoodmicro.2016.09.025
26. Tanaka T, Nogariya O, Shionoiri N, Maeda Y, Arakaki A. Integrated molecular analysis of the inactivation of a non-enveloped virus, feline calicivirus, by UV-c radiation. J Biosci Bioeng. (2018) 126:63–8. doi: 10.1016/j.jbiosc.2018.01.018
27. Araud E, Fuzawa M, Shisler JL, Li J, Nguyen TH. UV Inactivation of rotavirus and Tulane virus targets different components of the virions. Appl Environ Microbiol (2020) 86:e02436–19. doi: 10.1128/AEM.02436-19
28. Lo CW, Matsuura R, Iimura K, Wada S, Shinjo A, Benno Y, et al. UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins. Sci Rep (2021) 11:13804. doi: 10.1038/s41598-021-93231-7
29. Loveday EK, Hain KS, Kochetkova I, Hedges JF, Robison A, Snyder DT, et al. Effect of inactivation methods on SARS-CoV-2 virion protein and structure. Viruses (2021) 13:562. doi: 10.3390/v13040562
30. Pfaender S, Brinkmann J, Todt D, Riebesehl N, Steinmann J, Steinmann J, et al. Mechanisms of methods for hepatitis c virus inactivation. Appl Environ Microbiol (2015) 81:1616–21. doi: 10.1128/AEM.03580-14
31. Bui TKN, Mawatari K, Emoto T, Fukushima S, Shimohata T, Uebanso T, et al. UV-LED irradiation reduces the infectivity of herpes simplex virus type 1 by targeting different viral components depending on the peak wavelength. J Photochem Photobiol B (2022) 228:112410. doi: 10.1016/j.jphotobiol.2022.112410
32. Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol (2009) 83:8289–92. doi: 10.1128/JVI.00709-09
33. Nomaguchi M, Doi N, Sakai Y, Ode H, Iwatani Y, Ueno T, et al. Natural single-nucleotide variations in the HIV-1 genomic SA1prox region can alter viral replication ability by regulating vif expression levels. J Virol (2016) 90:4563–78. doi: 10.1128/JVI.02939-15
34. Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol (1986) 59:284–91. doi: 10.1128/JVI.59.2.284-291.1986
35. Sakai H, Shibata R, Miura T, Hayami M, Ogawa K, Kiyomasu T, et al. Complementation of the rev gene mutation among human and simian lentiviruses. J Virol (1990) 64:2202–7. doi: 10.1128/JVI.64.5.2202-2207.1990
36. Nomaguchi M, Yokoyama M, Kono K, Nakayama EE, Shioda T, Doi N, et al. Generation of rhesus macaque-tropic HIV-1 clones that are resistant to major anti-HIV-1 restriction factors. J Virol (2013) 87:11447–61. doi: 10.1128/JVI.01549-13
37. Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, et al. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol (1988) 62:139–47. doi: 10.1128/JVI.62.1.139-147.1988
38. von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol (1993) 67:4945–55. doi: 10.1128/JVI.67.8.4945-4955.1993
39. Adachi A, Kawamura M, Tokunaga K, Sakai H. “Methods for HIV/SIV gene analysis”. In: Adolph KW, editor. Viral genome methods. Boca Raton, FL: CRCPress (1996). p. 43–53.
40. Yokoyama M, Nomaguchi M, Doi N, Kanda T, Adachi A, Sato H. In silico analysis of HIV-1 env-gp120 reveals structural bases for viral adaptation in growth-restrictive cells. Front Microbiol (2016) 7:110. doi: 10.3389/fmicb.2016.00110
41. Jablonski JA, Caputi M. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J Virol (2009) 83:981–92. doi: 10.1128/JVI.01801-08
42. Freed EO, Martin MA. “Human immunodeficiency viruses: replication,”. In: Knipe DM, Howly PM, editors. Fields viology. Philadelphia, PL: Lippincott Williams and Wilkins, a Wolters Kluwer business (2013). p. 1502–60.
43. Chunduri H, Crumpacker C, Sharma PL. Reverse transcriptase mutation K65N confers a decreased replication capacity to HIV-1 in comparison to K65R due to a decreased RT processivity. Virology (2011) 414:34–41. doi: 10.1016/j.virol.2011.03.007
44. Nishisaka-Nonaka R, Mawatari K, Yamamoto T, Kojima M, Shimohata T, Uebanso T, et al. Irradiation by ultraviolet light-emitting diodes inactivates influenza a viruses by inhibiting replication and transcription of viral RNA in host cells. J Photochem Photobiol B (2018) 189:193–200. doi: 10.1016/j.jphotobiol.2018.10.017
45. Earl PL, Moss B, Doms RW. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol (1991) 65:2047–55. doi: 10.1128/JVI.65.4.2047-2055.1991
46. Fenouillet E, Jones IM. The glycosylation of human immunodeficiency virus type 1 transmembrane glycoprotein (gp41) is important for the efficient intracellular transport of the envelope precursor gp160. J Gen Virol (1995) 76:1509–14. doi: 10.1099/0022-1317-76-6-1509
47. Rattanakul S, Oguma K. Analysis of hydroxyl radicals and inactivation mechanisms of bacteriophage MS2 in response to a simultaneous application of UV and chlorine. Environ Sci Technol (2017) 51:455–62. doi: 10.1021/acs.est.6b03394
Keywords: HIV-1, UV, envelope, reverse transcriptase, p24, virus inactivation, inactivation mechanism
Citation: Koma T, Doi N, Suzuki A, Nagamatsu K, Yasui T, Yasutomo K, Adachi A, Minamikawa T and Nomaguchi M (2022) Major target for UV-induced complete loss of HIV-1 infectivity: A model study of single-stranded RNA enveloped viruses. Front. Virol. 2:994842. doi: 10.3389/fviro.2022.994842
Received: 15 July 2022; Accepted: 02 September 2022;
Published: 20 September 2022.
Edited by:
Marie-Claire Gauduin, Texas Biomedical Research Institute, United StatesReviewed by:
Baolin Liao, Guangzhou Medical University, ChinaAna Paula Moreira Franco Luiz, Oswaldo Cruz Foundation (Fiocruz), Brazil
Copyright © 2022 Koma, Doi, Suzuki, Nagamatsu, Yasui, Yasutomo, Adachi, Minamikawa and Nomaguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeo Minamikawa, minamikawa.takeo@tokushima-u.ac.jp; Masako Nomaguchi, nomaguchi@tokushima-u.ac.jp
†These authors have contributed equally to this work
 Takaaki Koma
Takaaki Koma Naoya Doi
Naoya Doi Akihiro Suzuki2
Akihiro Suzuki2 Koji Yasutomo
Koji Yasutomo Akio Adachi
Akio Adachi Takeo Minamikawa
Takeo Minamikawa Masako Nomaguchi
Masako Nomaguchi