- Department of Biosciences and Bioengineering, Indian Institute of Technology Roorkee, Roorkee, Uttarakhand, India
Arboviruses have the potential to spread quickly and cause a global health emergency. These are RNA viruses that use RNA-dependent RNA polymerase (RdRp) for their replication. RdRp lacks proofreading activity, leading to high error rates, low replicative fidelity, and more genetic variability. In addition, shorter generation time and faster evolutionary rate of these viruses lead to re-emergence and recurrence of arboviral infections due to the emergence of new variants and the development of antiviral resistance. During the replication inside the host cell through protein-protein interactions (PPIs), these viruses interact with several host factors and utilize the host cellular machinery for their benefit. Besides this, viruses employ several transmission strategies to combat host innate and adaptive immune responses by manipulating the signaling and metabolic pathways of the hosts. Hence, antiviral therapies targeting host-virus PPIs can provide an alternative broad-spectrum strategy against RNA viruses. The approach of targeting virus-specific proteins for developing antivirals is expected to solve the problem of antiviral drug resistance and combat emerging new variants of these viruses. This review focuses on host-virus PPIs of arboviral infections that directly affect the host immune signaling and metabolic pathways. Better understanding of these mechanisms will develop new therapeutic tools to treat viral infections.
Introduction
Various viral pathogens, including arboviruses, are re-emerging and causing worldwide infections in humans and animals (1). In 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its various variants emerged and caused an acute respiratory disease pandemic called 'Coronavirus disease 2019' (COVID-19). This pandemic has threatened human health and public safety (2). Most emerging and re-emerging viruses appear to be caused by a change in virus spread due to deforestation, environmental change, microbial adaptation/change, commerce, technology, ecological changes, agricultural development, failure of public health measures, and lack of public health infrastructure (2).
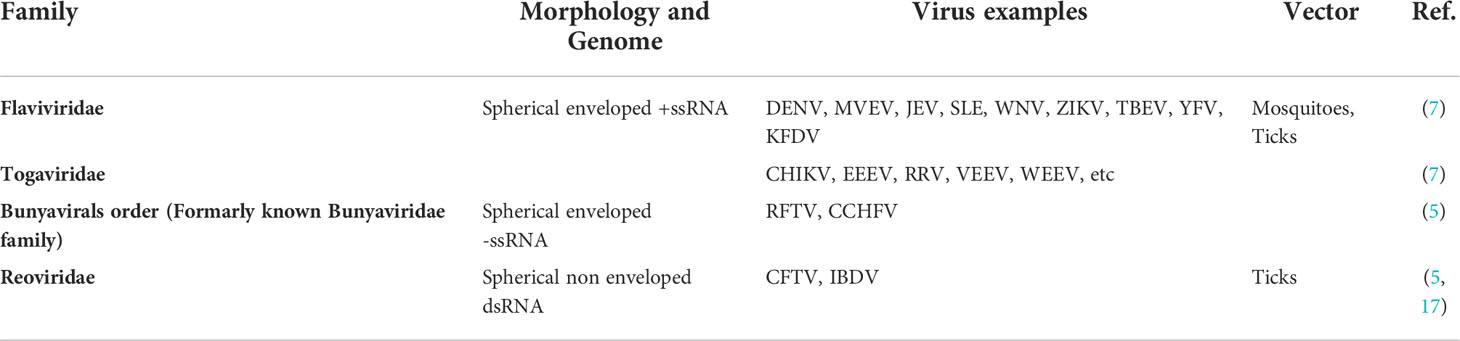
Arboviruses are a diverse group of arthropod-borne viruses that spread from infected to susceptible hosts by arthropod vectors such as mosquitoes, sandflies, ticks, or biting midges (1). Besides insect bites, viruses can also be transmitted through blood transfusion, pregnancy, and childbirth from mother to child (3, 4). Flaviviridae, Togaviridae, Bunyaviridae, and Reoviridae are the prominent families of economically important arboviruses that cause human infections (5–7).
Most infections caused by arboviruses are asymptomatic. Nevertheless, these viruses can show symptoms ranging from a mild flu-like illness to encephalitis, i.e., brain inflammation. Furthermore, the symptoms and clinical characteristics are distributed mainly into two subgroups such as neuroinvasive and non-neuroinvasive (8). Neuroinvasive arboviruses frequently cause meningitis or encephalitis. In addition, neuroinvasive arboviruses symptoms include the rapid onset of fever along with muscle pain, stiff neck, headache, weakness in the arms and legs, confusion or disorientation, and seizures. On the other hand, non-neuroinvasive arboviruses do not affect the nervous system, but they may cause fever, headache, joint pain, muscle aches, vomiting, diarrhea, upset stomach, nausea, and rash (8). To date, no specific treatment is commercially available against arboviruses.
The core of any viral infection is that protein-protein interactions (PPIs) are significant for the biological system (9, 10). Virus-host PPIs play a vital role in running the host's metabolic and signaling pathways and also help activate the host immune system to counter the viral infection. Detailed knowledge of interacting proteins is essential for understanding viral diseases and discovering potential drugs (11).
Therapeutic intervention against infectious viral diseases using small molecule inhibitors either directly acts on viral proteins or modulates the host immune system (12). Few potential host protein targets also act as receptors for the virus, facilitating viral attachment and entry inside the host cell (13–15). Moreover, host metabolic processes such as glycosylation, autophagy, actin polymerization, fatty acid biosynthesis, and host-mediated proteolytic cleavage are necessary for viral survival making them potential drug targets (13–16). Therapeutic strategies targeting both host and virus commonly aim to suppress viral infection to prevent the disease. This review focuses on host-virus PPIs of arboviral infections that directly affect host immune signaling and metabolic pathways.
General introduction about economically important arboviruses
Arboviruses are maintained in nature through biological transmission between susceptible vertebrate hosts and blood-sucking arthropods. These viruses consist variety of genome types such as positive single-stranded RNA (+ssRNA), negative-sense single-stranded RNA (-ssRNA) and double-stranded RNA (dsRNA), etc. Various families such as Flaviviridae, Togaviridae, Bunyaviridae, and Reoviridae are some crucial families of arboviruses (5–7) (Table 1).
Flaviviridae
Flaviviridae family includes spherical enveloped +ssRNA viruses. Flavivirus genus incorporates Dengue virus (DENV), Murray Valley encephalitis virus (MVEV), Japanese encephalitis virus (JEV), Saint Louis encephalitis virus (SLE), West Nile virus (WNV), Zika virus (ZIKV), Tick-borne encephalitis virus (TBEV), Yellow fever virus (YFV), Kyasanur Forest disease virus (KFDV), etc. (7). Phylogenetic analysis of the sequence alignment, generated for individual coding regions and the whole genome sequences of flavivirus species, revealed that the species belonging to the same vector group usually show more similarity than the species of different vector groups (18–20).
Most flaviviral infections remain asymptomatic, but mild to severe symptoms may occur in a few cases. Generally, mild flaviviral infection show symptoms such as fever, conjunctivitis, rash, malaise or headache, and muscle and joint pain. A safe, affordable, and effective vaccine can prevent only a handful of flaviviral infections caused by YFV, JEV, and KFDV (21–23). Overall, no specific antiviral drug treatment or efficacious licensed vaccines are available for flaviviral infection (24–27)
Dengue fever is an ancient disease caused by DENV. Aedes aegypti mosquitoes transmit dengue viruses that infect an estimated 400 million globally every year (28, 29). DENV has 4 serotypes, and the severe infection of DENV causes life-threatening illnesses such as dengue shock syndrome and dengue hemorrhagic fever (23). On the other hand, JEV is transmitted by Culex species mosquitoes and maintained in a cycle between mosquitoes and vertebrate hosts such as pigs and wading birds. As humans are not primary hosts, they do not harbor high virus titer concentrations in their blood to further pass it on to mosquitoes (22). JEV is endemic in various counties of Asia and the Western Pacific Region. It is estimated that around 68,000 clinical cases per year expose 3 billion people to risk (30). For JEV infection, severe symptoms include sudden onset of high fever, headache disorientation, coma, neck stiffness, spastic paralysis, and seizures. Simillarly, ZIKV is transmitted mainly by Aedes species mosquitoes. ZIKV infection during pregnancy can cause preterm birth and miscarriage (31). In addition, certain birth defects can occur, where infants born with microcephaly and other congenital malformations, also known as congenital Zika syndrome. Until now, around 86 countries and regions have reported evidence of mosquito-borne ZIKV infection, which can transmit to the additional areas where the Aedes species mosquitoes are found (31). Yellow fever is an acute viral hemorrhagic disease caused by YFV infection. It is transmitted by infected Aedes or Haemagogus mosquitoes. YFV is endemic in tropical areas of Africa and South America (32–34). Similarly, WNV, MVEV, and SLE, also causes various neurological disease and is transmitted by Culex species of mosquitoes. WNV, MVEV, and SLE were reported in multiple places worldwide, such as Africa, Europe, the Middle East, North America, and West Asia (35–37). TBEV spreads through the bite of an infected tick. In some cases, TBEV is transmitted through raw dairy product consumption from infected goats, sheep, or cows. It infects the central nervous system of the host. Every year approximately 10 to 12 thousand cases are reported from various regions stretching from Northern and Western Europe to Eastern and Northern Asia. Until now, no cure or vaccine against TBEV is available (38, 39). KFDV was first isolated in 1957 from the diseased monkey of Kyasanur Forest in Karnataka, India. KFDV is transmitted through tick bites and has caused hemorrhagic fever in some patients (26).
Togaviridae
This family consists of +ssRNA viruses. The Alphavirus genus incorporates numerous viruses such as Chikungunya virus (CHIKV), Eastern equine encephalitis virus (EEEV), Ross River virus (RRV), Venezuelan equine encephalitis virus (VEEV), Western equine encephalitis virus (WEEV), etc. (7). Among alphaviruses most of the protein coding region of the genome is highly conserved, with the variable regions occurring at the end of both protein coding and non coding sections of genome (40).
Chikungunya is a viral infection caused by the CHIKV and is transmitted to humans by infected Aedes mosquitoes. Most CHIKV infections remain asymptomatic, but in some cases, mild to severe symptoms such as fever, myalgia, headache, rash, and polyarthralgia can be seen (41). The disease was first reported in 1952 in Tanzania and is now endemic in various countries of Africa, and Asia, with some occasional outbreaks in Europe and America (41–43). Similarly, the Sindbis virus (SINV) causes Sindbis fever. This virus is similar to CHIKV and has caused a few outbreaks in South Africa and Northern Europe (44, 45). EEEV, RRV, VEEV, and WEEV are also some crucial viruses that have caused sporadic cases all around the world. Overall, no treatment or vaccine is available against these viruses (42, 46–49).
Bunyavirals
Bunyavirals, formerly known as the Bunyaviridae family, is an order which consists of various a family's such as Phenuiviridae, Nairoviridae, Hantaviridae, etc. These families incorporate the genome of three segments of -ssRNA. These viruses are enveloped, spherical RNA viruses (5). Bunyaviridae family consists of important arboviruses such as Rift Valley fever virus (RFTV) and Crimean-Congo he morrhagic fever virus (CCHFV). These viruses mainly infect animals, but hemorrhagic fever cases were also reported in humans. RFTV and CCHFV get transmitted via mosquitoes and ticks, respectively (5, 50, 51).
Reoviridae
The Reoviridae family consists of dsRNA viruses. It incorporates various viruses such as Rotavirus, Infectious bursal disease virus (IBDV), and Colorado tick fever virus (CFTV) (17). CTFV is a tick-borne rare viral disease from the western United States and Canada. The amino acid in different genera of reoviruses has a sequence identity of less than 30% in RNA-dependent RNA polymerase. Exceptions include Rotaviruses B with 22% identity to other rotaviruses, while Aquareovirus and Orthoreovirus, show an amino acid sequence identity of up to 42% (52, 53). CFTV has a relatively large genome as compared to other arboviruses. CFTV also has an icosahedral capsid structure. Further, as per literature, no vaccines or medicines are available against CFTV (6, 17).
Common viral targets and antiviral resistance
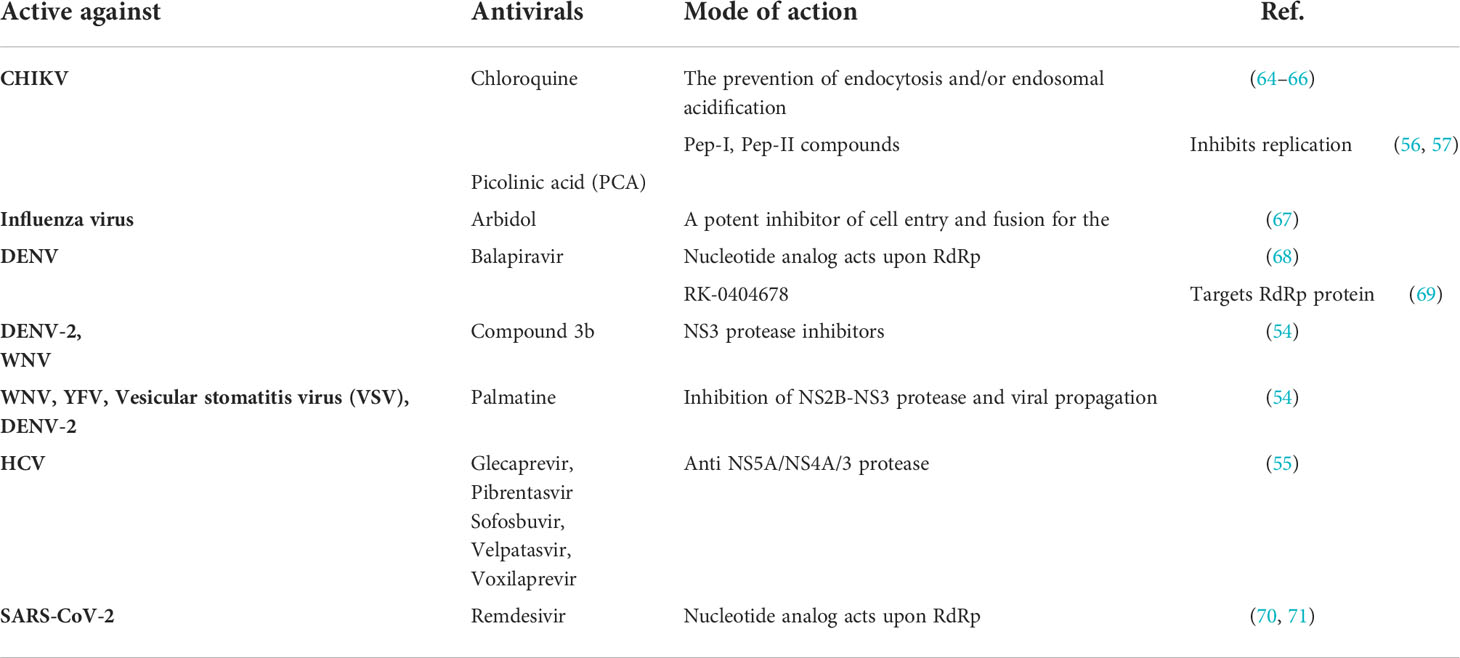
Ideal antiviral targets are those viral proteins that are essential for the completion of the viral life cycle. Additionally, it should have a lower rate of mutations, an important attribute in the case of RNA viruses like flaviviruses and alphaviruses. The most attractive molecular targets for the development of broad-spectrum antiviral compounds are viral protease or polymerase (54). In Addition to this, extensively studied antiviral targets also include viral surface proteins, which function as an anchor for the attachment and cell entry (55). Several studies are based on small molecule inhibitors targeting structural proteins such as envelop protein (E) of DENV, Spike protein (S) of SARS-CoV-2, and Capsid protein of CHIKV (11, 46–49, 56, 57). Other studies targeting various essential non-structural viral proteins (nsPs) such as non structural protein (nsP5) of DENV, RNA polymerase of ZIKV, Proteases of SARS-CoV-2, viral capping enzyme nsP1 of CHIKV, and two-component nsP2B-nsP3 protease of ZIKV, DENV, and WNV (28, 54–62).
Despite the enormous research, there is a dire need for antiviral therapies or broadly acting vaccines for the treatment of flaviviral and CHIKV infection (28, 54, 58). Only one vaccine is available against the DENV virus, i.e., Dengvaxia® licensed by the pharmaceutical company Sanofi Pasteur. This vaccine has limited use due to the high risk of severe dengue outbreak in vaccinated individuals. Antivirals used to treat viral infections generally act upon either viral or host proteins (61). The antivirals that directly act against the viral proteins are known as Direct-acting antivirals (DAA) (62) (Table 2). Using DAA against viral infection provides a promising approach, as DAA specifically acts upon viral protein and, therefore, generally shows low toxicity and a wide treatment window (72). For instance, Remdesivir, a nucleotide analog, acts upon RdRp of SARS-CoV-2 and inhibits viral replication (70, 71). Apart from that, Chloroquine is approved as an antimalarial drug and is a well-known DAA against CHIKV; it acts by inhibiting the endosomal acidification, thereby preventing the virus entry (64–66). Similarly, Arbidol is an antiviral drug and a potent inhibitor of cell entry and fusion for the influenza virus (67). It also acts in the early stage of CHIKV and blocks the virus attachment and entry (73). Some common bottlenecks of DAA are the high risk for the development of antiviral resistance, short window period of effectiveness, harmful side effects, and ineffectiveness against latent viruses (12, 42, 74). Mutations of the nsp5 protein of CHIKV cause resistance to favipiravir (T-107), a broadly acting antiviral (75). In flavivirus resistance to ciprofloxacin, is reported after seven passages with the drug (76). For CHIKV development of partial resistance is reported after fifteen passages against 4-OHT (4-Hydroxytamoxifin) (77). The RNA viruses have two mechanisms for drug resistance either through specific opposition to drug action, which results in decreased fitness in the absence of the drug, or a general increase in fitness, which is sometimes continued even after drug withdrawal (78–84). Antiviral therapy targeting multifunctional common nodes of interaction occurring between host and viral proteins have the potential to target a wide range of viral infection (12).
Host factors and their importance in the virus life cycle
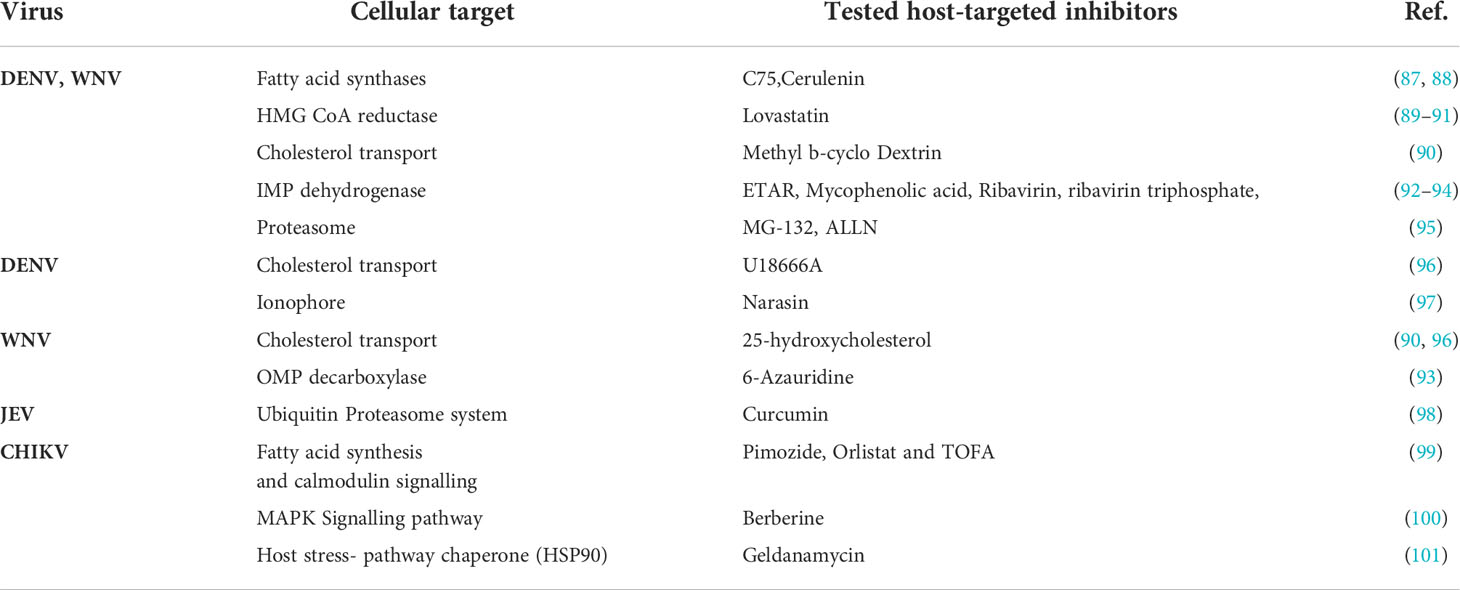
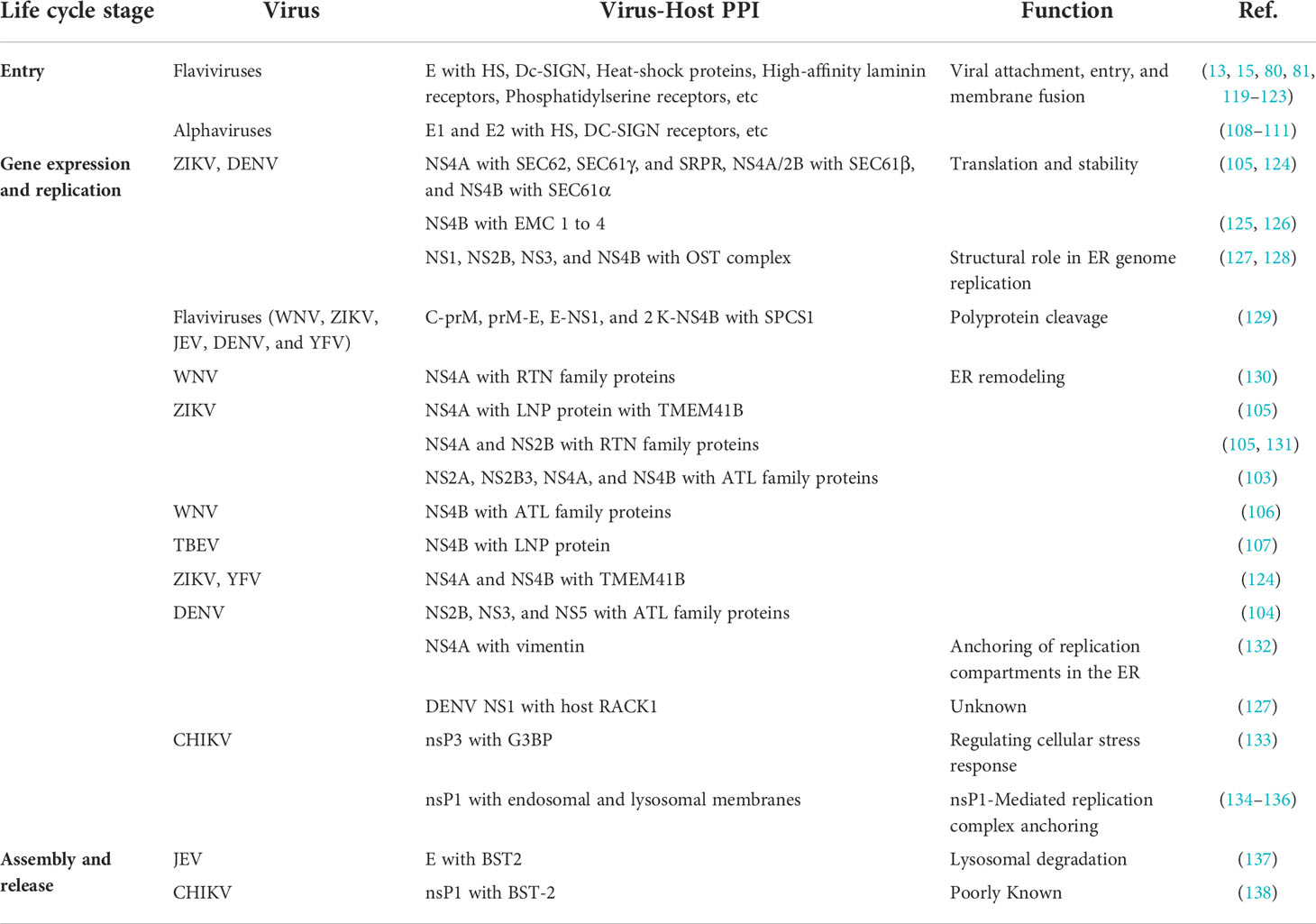
Proteins or RNA encoded by host genes that either positively or negatively impact an intruding virus are called host factors. The host factor that ablates viral survival or dissemination is called restriction or antiviral factors. On the contrary proviral factors or dependency factors promote viral growth and enable viral infection (72, 85). Viruses are gene-poor relative to their host, as the viral genome encodes hundreds of genes, whereas host cells typically encode thousands of genes (74). During viral infection, several host factors play a critical role in every facet of the viral life cycle (86) (Table 3). For instance, viruses utilize various biomolecules present on the host cell surface as an anchor for their attachment and entry inside the host cell. Flaviviruses such as DENV, WNV, JEV, YFV, and ZIKV binds through viral envelop protein (E) via heparan sulphate (HS), dendritic cell-intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN), heat-shock proteins (HSPs), and high-affinity laminin receptors, phosphatidylserine receptors (80, 81). In flavivirus, viral genome replication occurs in the host endoplasmic reticulum (ER) and uses ER-associated ribosome for translation (102). Also, in WNV, DENV, and ZIKV, many viral non-structural proteins interact with different host proteins of the reticulon (RTN) family, atlastin (ATL) family, and several transmembranes for ER remodeling and vesicle formation (103–107). Alphaviruses such as CHIKV, EEEV, RRV, VEEV, and WEEV utilize E1 and E2 with HS and DC-SIGN receptors to gain access to the surface and enter inside the host cells (82, 83). Similarly, the virus controls the eukaryotic initiation factors 4F (eIF4F) complex of a host, thereby suppressing the expression of the host gene, ensuring the uninterrupted synthesis of viral proteins in the host cell, and evasion of the host immune system (84) occurs in the endoplasmic reticulum and requires host viral RNA is initiated by nsP1 of the virus to guide the eukaryotic initiation factors 4E (eIF4E) for the identification and initiating translation and shielding the mRNA from cellular exonucleases (108–111). Another example of viral hijacking is the manipulation of host protein complexes to either delay or accelerate the host cell cycle to promote the replication of the viral genome (112, 113). In DENV, host cellular pathways such as autophagy, actin polymerization, and fatty acid biosynthesis are required for viral replication (114). Additionally, the formation and assembly of the mature infectious viral particle occurred in the endoplasmic reticulum and required the host glycoprotein-processing pathway (115).
Protein-protein interactions in the replication cycle of arboviruses
PPIs serve as the foundation of important cellular processes. During viral infections, the structural and functional characteristics of various signaling, metabolic and regulatory pathways of the host are controlled by host-virus PPIs (116). PPIs research has slowly and considerably evolved with technological and conceptual growth in diverse research areas over the past centuries (116). The basic level of PPIs investigation is crucially important because of their involvement in cellular processes like metabolism, signalling and their concerned diseases. Therefore, molecular interaction studies help in establishing molecular-level strategies to ascertain valuable therapeutics (117). Progression of in-vivo and in-vitro studies includes co-purification of protein complexes, TAP (tandem affinity purification), pull-down assays, proteomics, and Y2H (yeast 2 hybrid) system, etc., which are vital for identifying PPIs. Recent advancements in in silico methods like sequence and structure-based approaches, chromosome proximity, gene fusion, in silico 2 hybrid, phylogenetic tree, phylogenetic profile prediction, etc. makes it easy to predict and know about PPI-based multifaceted diseases and apply the knowledge towards the cure of these diseases (11). Viruses are sub-microscopic obligate intracellular microbes. They depend on host cells for their life cycle, which can be divided into three stages containing several steps, Entry (attachment, entry, uncoating), replication (gene expression and replication), and budding (assembly and release). At every step, viruses interact and manipulate host cellular machinery for its replication (118) (Table 4).
Virus entry
Virus attachment to target cells relies upon the interaction between viral surface proteins and cell membrane proteins. Flaviviruses structural protein consists of three proteins designated C (capsid), prM (precursor of membrane), and E (envelope). E protein consists of 3 homodimers and has an essential role in viral attachment, entry, and membrane fusion (139–141). Further, each E monomer is divided into 3 ectodomains (ED) EDI, EDII, and EDIII. These ectodomains are connected via flexible linkers. Further, two transmembrane (TM) domains, TM1 and TM2, has a crucial role in fusion (142, 143). HS is a type of glycosaminoglycan (GAG) that play a role of epithelial cell receptor. Several research articles have shown that flaviviruses interact with host cells by oppositely charged residue interactions between E and GAG (80, 119, 120). Furthermore, in DENV and WNV, DC-SIGN assists the virus in entry into Dendritic cells (15, 123). In addition to this, Fc receptors are also important in antibody dependant enhanced infection of monocytes with DENV (121). Above and beyond, various HSPs, high-affinity laminin receptors, phosphatidylserine receptors, etc., are also important in numerous Flavivirus entry mechanisms (13, 81, 122).
Alphaviruses are composed of five structural proteins, namely C, E3, E2, 6K, and E. E1 is a pH-dependent fusion protein, and E2 is a vital protein essential for receptor recognition. Hence, both are crucial proteins for viral attachment and entry into host cells (82). Alphaviruses utilize several cell surface molecules, including HS, DC-SIGN receptors, etc., to carry out an initial attachment to target cells (108–111). Many alphaviruses are internalized via clathrin-mediated endocytosis. Later, the clathrin-coated pits are transported inside the cell leading to the dissociation of clathrin molecules and virus delivery to endosomes (83). To mediate the fusion of the viral membrane with the endosomal membrane, at the low-pH environment of the endosomes, E1/E2 glycoproteins undergo conformational changes (83).
Gene expression and replication: -
In flaviviruses, several host pathways play an important role in the translation of the flaviviral genome into its functional proteins. All viruses initially need host ribosomes to translate its gene into viral polyproteins. However, Flavivirus replication remains confined to the ER and utilizes ER-associated ribosomes for its translation (102). Furthermore, numerous host proteases cleave polyproteins into singular proteins. These NS proteins (NS2A, NS2B, NS4A, and NS4B) comprise various TM domains which need to be properly inserted into the ER membrane to be functional (144–148). The signal-recognition particle (SRP), host SEC61 translocon, and ER membrane complex (EMC) were demonstrated to be crucial host factors for the stability and insertion of these proteins in flaviviruses. Additionally, signal peptidase complex (SPCS) and oligosaccharyltransferase (OST) complex were seen to be indispensable for the replication of several flaviviruses (128, 129, 149–151). The flaviviral NS protein also recruits and takes over the functions of other host protein families, including ATL, RTN, and the Lunapark (LNP) protein, Transmembrane Protein 41B (TMEM41B), and Vacuole membrane protein 1 (VMP1), Vimentin, Receptor for Activated C Kinase 1 (RACK1) protein, etc (127, 130, 152–154).
ZIKV and DENV have shown several PPIs with a host, such as NS4A with SEC62, SEC61γ, and SRPR, NS4A/2B with SEC61β, and NS4B with SEC61α (105, 124). Similarly, both the viruses have demonstrated PPI between NS4B and EMC family proteins (125, 126). Furthermore, SPS1 has shown polyprotein cleavage activity for C-prM, prM-E, E-NS1, and 2 K-NS4B (82). OST complex proteins which have an important role in ER genome replication, show PPI with NS1, NS2B, NS3, and NS4B of ZIKV and DENV (127, 128). In WNV-host PPI, NS4A interacted with RTN family proteins for ER remodeling. On the other hand, for ZIKV-host, NS4A and NS2B interacted with RTN family proteins (105, 130, 131). Additionally, various ATL, LNP, and TMEM41B family proteins have shown PPI with flavivirus NS proteins for ER remodeling (103–106, 124). More on this, DENV NS4A interacts with vimentin, which plays an important role in the anchoring of replication compartments in the ER (153). In addition, DENV NS1 interacts with host RACK1, although its function is unknown (127).
Alphaviral RNA replication is host-dependent and occurs in cytoplasmic vacuoles derived from endosomal and lysosomal membranes (134–136). Alphaviruses comprise four non-structural proteins (nsPs), specifically nsP1, nsP2, nsP3, and nsP4, which are indispensable in the life cycle of the viruses. The nsP1-mediates anchoring of replication complex on these membranes (132). The nsP1 also causes the capping of viral RNA to assist eukaryotic translational initiation factor 4E (eIF4E) in identification and initiating translation and shielding the mRNA from cellular exonucleases (155). Yet, nsP3's precise role in replication complexes is less clear; it interacts with Ras-GAP SH3 domain-binding protein (G3BP), regulating cellular stress response (133).
Assembly and release
Flavivirus assembly occurs in the ER. Then, the virion is further transferred for maturation and furin cleavage in the trans-Golgi-network (TGN), and at the end, the virus is released out of the cell (156). In the virion maturation process, prM protein on the outermost part gets cleaved by host proteases within the Golgi apparatus. An acidic environment in Golgi and secretory vesicles makes the cleavage site accessible (157–159). Bone marrow stromal cell antigen 2 (BST2), also called tetherin, restricts the replication of various viruses. In the case of JEV, BST2-E protein interactions lead to its lysosomal degradation, promoting virus replication (137).
Alphaviral subgenomic RNA is translated to produce structural proteins in the cytoplasm (160). Further, E3-E2-6K-E1/E2-E2-TK translocated into ER for post-translational modification of E2/E1 (160). Later, these proteins get mature through TGN and are deposited on the plasma membrane. Lastly, icosahedral nucleocapsids are formed by the interaction between capsid and genomic RNA, subsequently releasing mature progeny virions (160). Alphaviruses are also dependent on host machinery for the transfer of their E protein to the plasma membrane (161). nsP1 mediated down-regulation of BST-2 expression overcomes the BST-2 facilitated CHIKV and SFV virus release (138).
Host immune response in arbovirus infections
Host immune response plays a critical role during primary infection of arboviruses and the carrier (162). The first line of defense mechanism against any arbovirus infection is Skin. The skin possesses some specialized cells that provide safety against arthropod bite and the relevant agent (163). The Skin has three structural layers, including the uppermost epidermis, lowermost dermis, and the connecting air interface (164). Cells responsible for immunity like T and B cells, dermal macrophages, and dendritic cells exist in the epidermis, while the hypodermis has lesser numbers of immune cells in healthy skin (165). At the time of viral infection, host epidermal cells release cytokines and antimicrobial peptides; however, the inflammation and wounding process is continued by innate and adaptive immune cells (21, 166). Simultaneously neutrophils, monocytes, and other peripheral immune cells secret chemokines that help in the reduction of viral load (163). Additionally, the lymph nodes, the macrophages, and dendritic cells introduce antigens to B and T cells of adaptive immunity (163, 167). The B and T cells target the infected cells and produce virus-specific antibodies (165, 166). Innate and adaptive immunity work together to improve the healing from infection and tenderness at the bite site (168). Skin immune responses protect the host from arboviral infections, but evolution in arboviruses enables them to hijack protection mechanisms (169). Epidermal cells of the skin are prone to mosquito-borne DENV and WNV infection (170). Whereas the Langerhans cells (LCs) of the epidermis are more prone to infection by CHIKV, DENV, WNV, and TBEV (171). The migratory nature of LCs makes them move from epidermis to dermis and reach lymph nodes. It increases the viral load to the rest of the body parts and directly contributes to the viral-born infection of naive ticks feeding on infected cells (172). Salivary Gland Protein (SGP) secretion also plays a role in suppressing Skin Host Immune Responses (173). Even with evolutionary branching between mosquitoes and ticks, the SGP in both is functionally conserved concerning their interaction with the mammalian immune system (174). The SGP of ticks and mosquitoes suppresses T cell proliferation and inhibits and alters the release of key inflammatory cytokines at the time of viral infection (175). The progression of cytokine production during infection recruits monocytes from the blood. Monocytes inhibit viral propagation to PNS (Peripheral Nervous System) and CNS (Central Nervous System) (172, 176, 177).
The arboviral infection (SINV and WNV) induces a membrane attack complex (MAC), which activates the complement system that initiates the lysis of viral particles/virus-infected cells (178). The activation of the complement system also helps in producing antibodies that lead to the clearance of viral load (179). DENV and ZIKV mainly target monocytes and decreases vascular permeability through epithelial cell damage (180–183). Therefore, arboviruses assist their access to the remaining hosts by hijacking their dermal immune system, which is crucial for antiviral defense (184).
In the case of arthropods, the immune system protects the host from viral infection. Likewise, mosquitoes adopt an innate immune pathway (JAK-STAT and TOLL receptor) against viral infection (185) (Figure 1). Viral infection initiates a defense system by the formation of a complex consisting of transcription factors, protein kinases, and several regulatory molecules to keep balance on downstream gene expression (186). The regulatory genes are responsible for handling antimicrobial peptides (AMPs) and the effect of immune response on viruses. Mosquitoe express cytokine receptors and tyrosine kinase and together these two are responsible for further phosphorylation and release of transcription activator (STAT) (187). Self-phosphorylation of tyrosine kinase (JAK) leads to phosphorylation of cytosolic STAT (188). PIAS is a negative regulator of the Janus kinase/signal transducers and activators of the transcription (JAK-STAT) pathway (189) (Figure 1). STAT self-dimerize and translocate to the nucleus of the infected cell, which further activates effector genes and makes the immune system functional (190). The Toll pathway is well known for the production of innate immunity against pathogens (191). This immune path is initiated by the degradation of a cytokine ligand (Spätzle or Spz) that can bind to a transmembrane receptor toll-like receptor (Toll) (192). Activation of Toll pass on its signal and further activates MyD88 (adaptor proteins associated with Toll) (Figure 1) (193). CACTUS is a negative regulator of the Toll pathway (194). It gets phosphorylated and degraded (by proteasome) and leads to translocation of the transcription factor Relish 1 (Rel1) from the cytoplasm to the nucleus of the infected cell (195) (Figure 1).
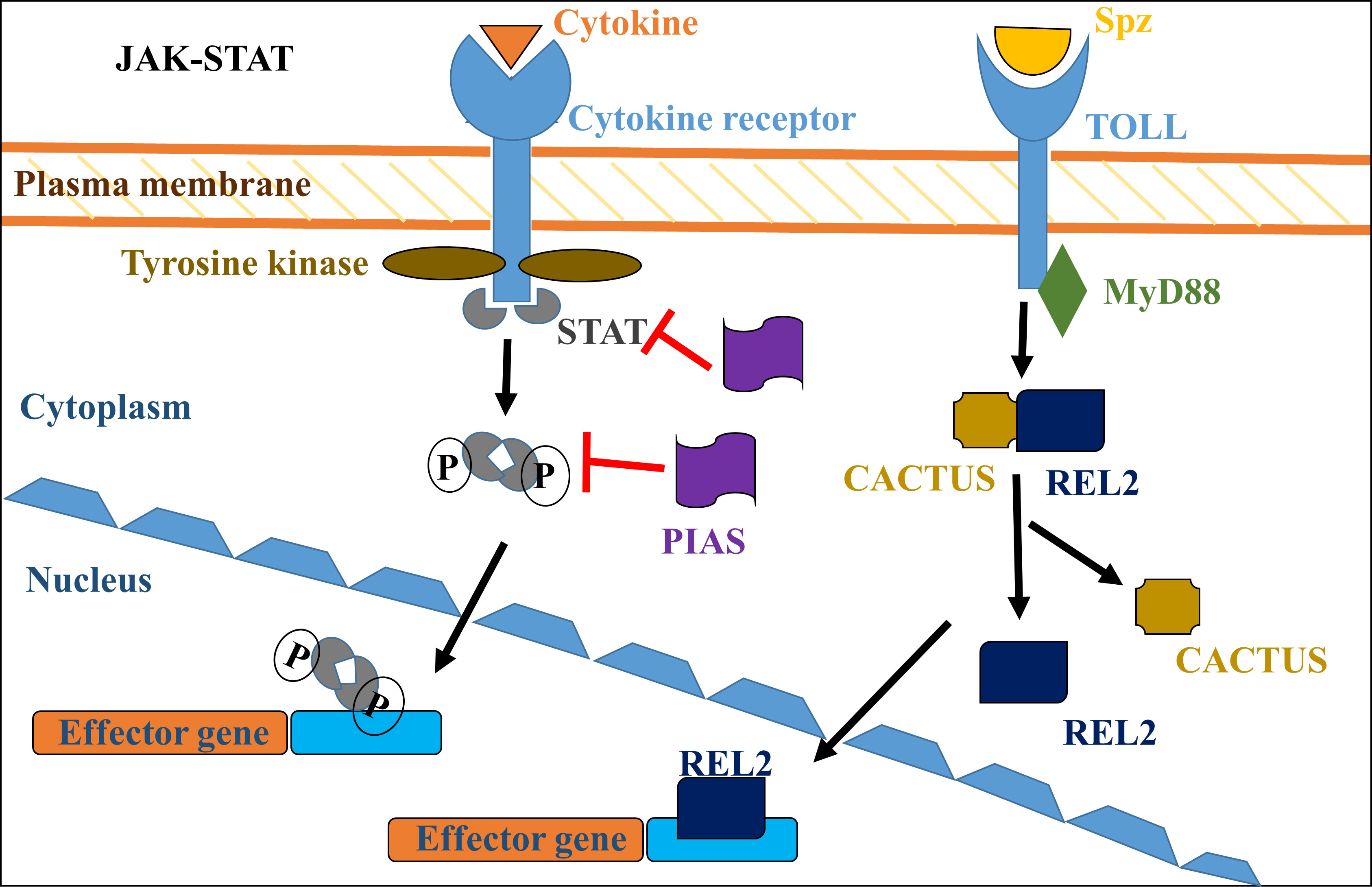
Figure 1 JAK-STAT and Toll-signaling pathway responsible for the activation of the immune system in mosquitoes. Activation of the signaling pathway initiates the formation of a multiprotein complex consisting of protein kinases, transcription factors, and the regulatory molecules to control the downstream genes that initiate and stimulate the immune system.
The parallel evolution of the viral infection strategies with the host immune system is responsible for viral evasion from host immune cells (196). Viral translational machinery is inhibited by interferon (IFN) as host antiviral responses (197), but due to evolution in viral infection strategies, IFN induction is hampered by the virus (198). The capability of many viral proteins for immune evasion makes them a potential target for antiviral therapies (9). The nsP2 of alphaviruses has a virulence feature mainly responsible for inhibiting transcription and translation in infected host cells (199). Likewise, non-strucrural proteins of the Flaviviridae family also help in immune escape for the viruses (200). WNV nsP1 acts via the Toll-like receptor 3 (TLR3) pathway, and DENV nsP2 based STimulator of INterferon Genes (STING) cleavage results in a decrease in Interferon β (IFNβ) level (201). It leads to the inhibition of antiviral molecules by viral proteins at ease. DENV nsP4 inhibits IFN-mediated phosphorylation of signal transducer and activator of transcription (STAT) (202). Therefore, viruses can malfunction the antiviral activity. Understanding Immune system evasion provides target protein for the development of potent antiviral drug molecules in the future (10).
Role of host-virus protein-protein interactions in the signaling pathway
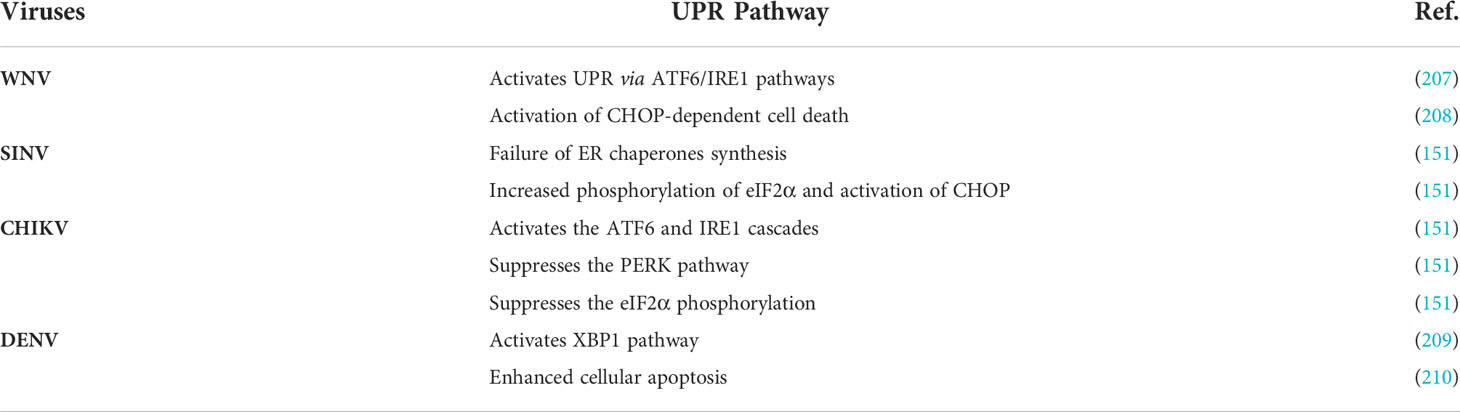
Arboviruses have a unique mode of life cycle as they require hosts (mostly humans, small mammals, and birds) for their replication and arthropods (commonly mosquitoes) as a vector for their transmission (203, 204). Similar to other viruses, arboviruses also induce host cell infection via cellular responses like unfolded protein response (UPR), apoptosis, and autophagy (205). In prolonged stress conditions, UPR and autophagy are both directly connected to programmed cell death (apoptosis) by Bcl2 family proteins and pro-apoptotic proteins like CHOP and Beclin-1 to maintain cellular homeostasis (205).
Unfolded protein response
Various kinds of cell stress like heat shock/UPR and programmed cell death play a crucial role in protecting other cells in different physiological conditions (206) (Table 5). However, stress response like autophagy is principally involved in the denaturation and recycling of proteins and cell organelles (211). The host cell endoplasmic reticulum (ER) is mainly responsible for the proper folding and maturation of proteins (205). During cell stress conditions, UPR stimulates ER membrane stress transducer, leading to activation of stress sensors like inositol-requiring enzyme I (IRE1), protein kinase RNA-activated-like ER kinase (PERK), and transcription factor6 (ATF6) (207). The mechanism of UPR regulation is quite different in similar viruses, such as flaviviruses that encourage the host protein UPR response, whereas WNV activates UPRs transcription and translation pathway (212–214). There are three pathways for UPR response. Among these three UPR pathways, proteasomal degradation of ATF and the PERK pathway play an essential role in the phosphorylation of eIF2α and induce the pro-apoptotic protein CHOP (215). This protein eventually leads to CHOP-dependent cell death to inhibit WNV infection (216). The ATF6 signaling pathway is critically responsible for innate immune response inhibition, which also helps in cell survival (217). The other arboviruses like SINV, CHIKV, and DENV also control the host's UPR system through different mechanisms (205). SINV induces premature cell death due to less control on UPR by inhibition of ER chaperones, increased eIF2α phosphorylation, and subsequently activation of CHOP protein (218). CHIKV suppresses eIF2α phosphorylation and PERK pathway and activates the IRE1-ATF6 cascades, whereas DENV triggers XBP1 signaling and enhances cellular apoptosis, leading to virus-induced cytotoxicity (171).
Apoptosis
Programmed cell death is termed apoptosis (219). Apoptosis regulates virus pathogenesis and mortality in viral infection (220). Apoptosis plays a vital role in the activation and initiation of the immune system, which is essential for arbovirus replication, infection, and pathogenesis (216, 221) (Table 6). In general, viral infection can use intrinsic and extrinsic apoptotic pathways. Flaviviruses include; DENV and WNV, which can delay apoptosis by activating several cell survival pathways (132). CHIKV uses external factors/ligands like tumor necrosis factor (TNF) as an extrinsic signal to activate caspases-8, which further triggers the activation of caspases-3, -6, and -7 (227). WNV and DENV both follow the intrinsic pathway of apoptosis via stimulation of P53 (228). The activation of P53 facilitates mitochondria-dependent apoptosis of cells (133). Viral infection initiates apoptosis by inducing dsRNA-dependent protein kinases (PKR) as it regulates eIF2a and activates effector caspases (229).
The apoptosis process induced by SINV is vital for its pathogenicity (230). A double-stranded RNA intermediate is formed into the host cell at the time of SINV entry. PKR helps in the identification of invaded viral particles (231). PKR kinase protein phosphorylates eukaryotic translation initiation factor 2A (eIF2A) to block cellular translation and inhibition of Mcl-1 (Bcl2 family anti-apoptotic protein) biosynthesis (232). PKR-mediated insulin receptor substrates (IRS1) phosphorylation of c-Jun N-terminal kinases (JNK) activates 14-3-3 (phosphorylation-dependent regulatory/effector proteins), which provides substrate access to kinases (Bad) (233). This process induces the interruption of the complex between anti-apoptotic proteins from the Bcl2 family, B-cell lymphoma-extra-large (Bcl-xl), Bak, and the release of Bad (234). Bax and Bak (pro-apoptotic proteins) are members of the Bcl-2 family and core regulators of the intrinsic pathway of apoptosis (234). Further, Bak replaces Bad and Bik (pro-apoptotic protein) from Mcl-1. The displacement of Bad by Bak oligomerization and release of cytochrome c induces apoptosis (235).
Apoptosis is essential for CHIKV replication as the virus elicits the apoptosis process to escape from the immune system of the host and can infect neighboring cells (236). CHIKV infection triggers apoptosis in two different manners: intrinsically, by helping in replication along with caspase-9 activation, and extrinsically, by activation of caspase-8 (mediators of apoptosis) (171). CHIKV uses external factors/ligands like tumor necrosis factor (TNF) as an extrinsic signal to activate caspases-8, which further triggers the activation of caspases-3, -6, and -7 (227). Eventually, both actuate caspase-3 and finally lead to cell death and expediting virus discharge and its spread (232). The histopathological studies of DENV-2 infection in the liver of mice provided an insight into that activation of the mitogen-activated protein kinase (MAPK) pathway blocks phosphatidylinositol 3 kinase (PI3K), which is an intracellular regulatory kinase (226). MAPK cascades are key signaling pathways that regulate a wide variety of cellular processes. Therefore, no impact was seen on DENV virus particle production. DENV infection also induces a cytopathic effect that triggers the process of apoptosis in infected cells (203). Similar to SINV, WNV also follows the analogous mechanism to uphold their pathogenesis using apoptosis (204). These viruses trigger signaling for cell survival through the PI3K-AKT signaling pathway (221). The PI3K-Akt pathway is an intracellular signaling pathway that promotes cell growth, angiogenesis, cell metabolism, proliferation, and cell survival, in response to extracellular signals (71). Anti-apoptotic activities have also been seen in some viruses, such as; DENV and WNV (207, 217). The regulation of cell cycle machinery by apoptosis and related signaling pathways are important for the determination of disease and identification of potential drug molecules for future arboviral infections (212).
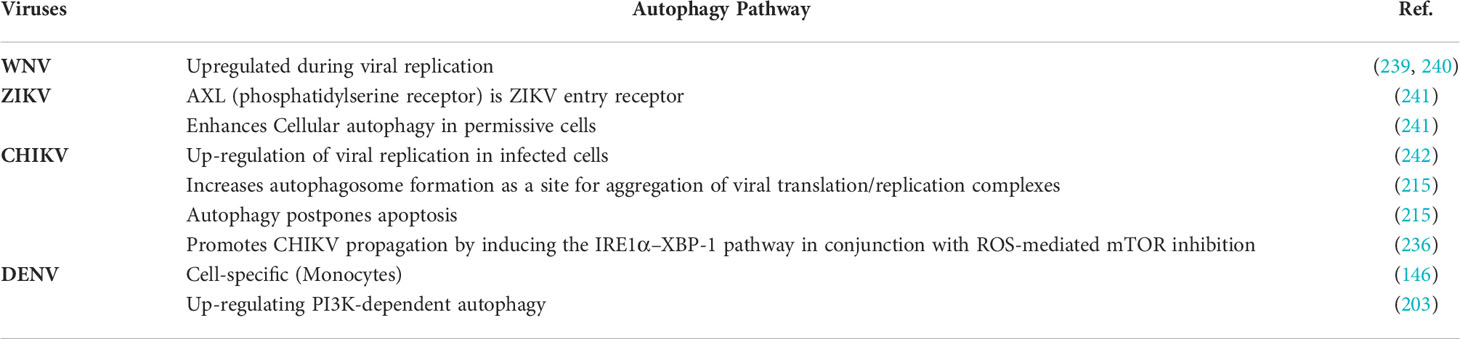
Autophagy
Autophagy is a process of eliminating damaged cells and facilitating the regeneration of new and healthy cells (237). In arbovirus infection, autophagy is an antiviral immunological cellular response that activates innate immune response with the assistance of pattern recognition receptor (PRR) based interferon production (238) (Table 7). It also shifted towards adaptive immunity when the virus delivers the antigen T-lymphocytes (212). Some studies on WNV infection showed that the replication of WNV is independent of autophagy; however, autophagy was induced (214). It is reported that autophagy can directly influence pro-viral mechanisms such as; entry, replication of the virus, lipid metabolism, and inhibiting innate immune responses, leading to cell death (213). In the case of DENV-2 infection, amphisomes (fusion vacuoles) play a vital role in the survival of the virus in host cells, while DENV-3 shows interaction with amphisomes as well as autophagolysosomes (202). In both fusion vacuoles, viral translation/replication occurs because these vacuoles are composed of viral RNA and proteins (243). CHIKV infection also facilitates autophagosome formation at the aggregation site of viral replication/transcription complex that helps in viral entry (171). Autophagy regulates the degradation of cytosolic triglycerides during lipid metabolism, and these triglycerides accumulate in the form of lipid droplets (244). At the time of DENV infection, the virus also uses these lipid droplets as an energy source for replication (114). Lysosomes convert lipid droplets to free fatty acids (FFAs) from triglycerides (114). β-oxidation of FFAs takes place in mitochondria, and viruses utilizes the released energy for viral replication (114). Therefore, the formation of amphisome is supposed to be advantageous for the virus to host cell access and reproduction (245). The autophagosome is also important for the replication of some viruses (216). Viruses follow the autophagy path to break lipid droplets into free fatty acids, and the generated energy is utilized as a source of energy/ATP for viral replication (246). Therefore, viruses use autophagy to disrupt the host immune response and eventually improve the replication rate and enhance their survival (247, 248).
Role of host-virus PPIs in the metabolic pathway
Understanding arbovirus infection and developing innovative ideas for their control is very important to develop effective therapeutics against them. We are aware that viruses lack metabolic enzymes and their machinery. They are dependent on the host for their reproduction and survival (249). Here, polyamines are essential in their life cycle from viral infection to viral replication (250, 251). Polyamines (putrescine, spermidine, and spermine) are positive, small amine units found in mammalian cells. They are helpful in virus survival and indulged in the immune system response of different organisms (250). Metabolism-related studies are essential to detect alterations in host physiology during infection. Earlier studies demonstrate that the evolutionarily conserved pathways, including glucose, fatty acid, protein, and nucleotide metabolism, are mainly manipulated by viral infection to increase their replication (252–254). Targeting metabolic and signaling pathways provides the central understanding of the host-virus interactions and further helps in biochemical research and therapeutics to treat viral infections (255). Togaviruses and flaviviruses depend on the β-oxidation of fatty acids or glycolysis process for energy production as they need it for their multiplication (256, 257). In contrast, DENV upregulates β-oxidation to fulfill energy requirements for other anabolic processes (173, 258). The formation of fatty acids and glycerophospholipids from acetyl-CoA as a byproduct of the β-oxidation process induces membrane architecture for viral replication (258, 259). Membrane-bound vesicles play an essential role in cell entry, replication, assembly, and exit of the virus (260). After arbovirus infection, the level of glycerophospholipids increases in mosquito cells and human host cells because their level is vital for viral replication and virus genome protection (260, 261). A decrease in the level of fatty acid formation leads to a decrease in DENV and WNV formation (114).
Regulation of host metabolism is essential for the survival and replication of arboviruses. Studies explain that host lipid metabolism plays a crucial role in the viral life cycle (262). Likewise, manipulating glucose metabolism using the intracellular Phosphatidylinositol 3-kinase (P13K) or AMP-activated protein kinase (AMPK) signaling pathway controls virus survival and replication (263). Virus replication is also activated in host cells through a proto-oncogene (Myc), sterol regulatory-element binding proteins (SREBPs), and the hypoxia-inducible factor 1-alpha (HIF-1α) ubiquitination signaling pathway (264). Inhibition of defined signaling pathways was found responsible for viral replication inhibition in vitro and in vivo (265). Detailed understanding of the immune responses, signaling pathways, host factors, and different phases of the viral life cycle are prerequisites for providing molecular insights for improving broad-spectrum antiviral therapy against repetitive viral infections (266).
Methods to identify host protein interaction:
Affinity purification
In this technique, the target protein is immobilized on the resin, and then cell lysate/extract is passed through the column, and the binding of proteins present in cell lysate to the target protein is detected by mass spectrometry (MS) by comparing the profile of protein retained in column with the target to the protein retained in the negative control sample (267). MS is used to determine the molecular weight of proteins by calculating the mass to-charge ratio (m/z) (268). There are three components of the mass spectrometer, an ion source, an ion detection system, and a mass analyzer. The overall process has three main steps for the analysis of proteins a) Ionization of proteins and transformation to gas-phase ions, b) separation of ions in the analyzer compartment based on m/z values, and c) finally separated ions were detected, and m/z values were measured for each species (268, 269). A combined proteomic study based on affinity purification-mass spectrometry (AP-MS) and RNAi screening to study virus-host interactions of DENV and ZIKV with humans and mosquitoes revealed important interactions (Figure 2) (105).
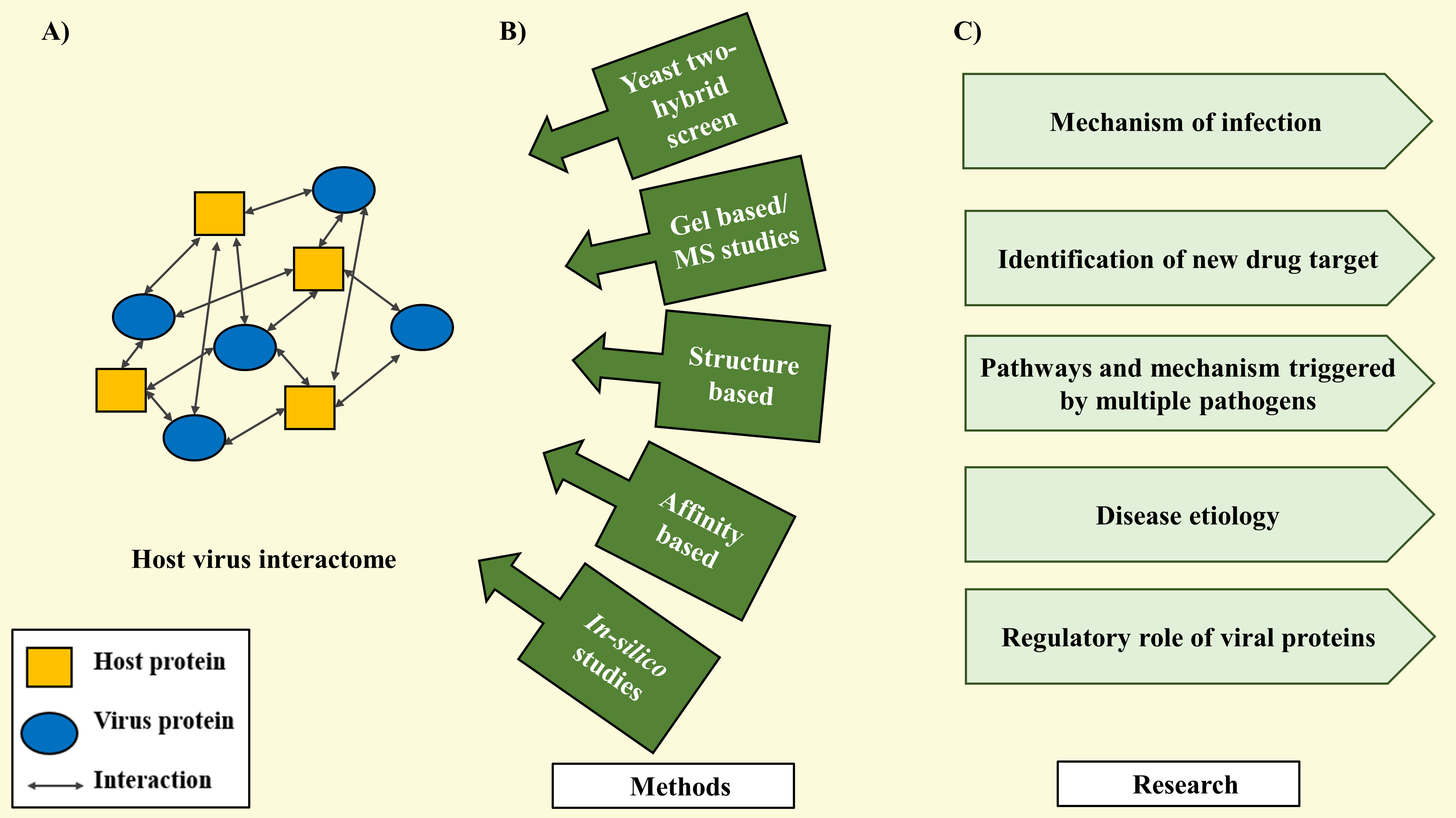
Figure 2 Methods to study host-virus interactome. (A) Host-virus interactome showing protein-protein interaction. (B) Different methods to study host-virus interactions. (C) Relevant information gained from studying host-virus interaction.
2D-polyacrylamide gel electrophoresis & (MS)
The effect of the viral infection on host cells can be understood by conducting various proteomic studies (Figure 2). A comparative 2D polyacrylamide gel electrophoresis is a study of whole cell lysate taken before and after infection and identification of detected protein through mass spectrometric studies. These studies can provide the degree of changes in the expression pattern of host proteins upon viral infection and also shed light on the regulatory role played by the viral protein. For instance, if the level of specific host proteins increases upon infection, this shows viral infection contributes to the enhanced production of mRNA, increased translation, and enhanced stability for certain proteins (270). Proteome-based studies coupled with mass spectroscopic studies were used for the detection of viral components inducing changes in host cellular proteome and identification of host-virus interactions (267). A study using 2D-polyacrylamide gel electrophoresis and mass spectrometry led to the identification of a total of 14 host proteins that were binding with the core protein of Hepatitis C virus (HCV) (271). Using a 2-dimensional electrophoretic approach followed by Liquid Chromatography-Mass Spectroscopy LC-MS/MS identified 15 differentially phosphorylated proteins present in host signaling pathways involved in secondary infection of DENV. Further, in this study, pyruvate kinase M2 (PKM2) was validated as a potential target, and it was shown that inhibitors of PKM2 were able to decrease infection and viral load (272).
GST pull-down assay
Pull-down assays are a common approach in which one of the interacting partners, i.e., purified protein, acts as bait and uses the second partner, i.e., cellular lysate or purified protein, as the prey (Figure 2). Among all the GST pulldowns in which bait proteins are expressed as a GST- tagged chimeric protein are very common. This is a low-cost, easy-to-use method for analysis of protein-protein interaction, in which one can adjust the concentrations of purified proteins for better sensitivity and can use different concentrations for the estimation of binding (273). The major drawbacks of this technique are Cloning, expression, and purification of interacting protein partners with GST tag is time-consuming, and the binding of GST tag can cause changes in the overall folding of proteins. The expression of proteins with GST tags can cause the accumulation of inclusion bodies which hinders the purification of active proteins. Moreover, the isolation of protein from their natural environment can lead to improper folding and eventually affects the interaction with other protein (274).
GST pull-down assay is successfully used for the analysis of the interactome of HIV-1 Tat protein and Hepatitis B virus HBx protein; for this, the researcher identified a protein involved in the metabolism of lipid and cholesterol known as apolipoprotein A-I (apoA-I) as a strong binder (248). The researcher further showed that increasing the apoA-I protein by transfection imparts a protective effect on mice infected with HBV, and this suggests apoA-I can be used as a possible target for the development of anti-HBV; therapeutics (275). For the Chikungunya virus multifunctional domain AUD (Alphavirus unique domain) of nsP3 protein is critical for the life cycle (276). Utilizing yeast two-hybrid system of human proteins interacting with CHIKV AUD was identified SNAP associated protein (SNAPIN)) and NEDD4-binding protein 2-like 1 (N4BP2L2). Further, their interaction was validated using GST pull-down assay, SNAPIN is involved in intracellular vesicle trafficking linked with autophagy, and N4BP2L2 has kinase activity. Still, its role in viral infection is unknown (277).
Yeast two-hybrid screen
This method is useful for screening protein libraries to find the binding of multiple proteins to a single protein of interest (278) (Figure 2). After the identification of Protein-Protein interaction, this assay can be further employed to map the interface of interacting proteins by making a set of chimeric bait and prey combinations from the fragments of interacting proteins (274). Shortcomings of this method are cloning of bait and prey is time-consuming; also, the transcription factor domain fusion to protein (bait) may induce structural changes that can alter the binding interface. Colocalized binding of bait and prey protein in the nucleus is necessary for the expression of the reporter gene, and this forced colocalization leads to the occurrence of false binding events (279). The yeast-two hybrid method is only suitable for soluble protein, and the interaction involving transmembrane or membrane protein is difficult to screen using this method. Analysis of the interactome of membrane-bound proteins is done using the split ubiquitin membrane yeast two-hybrid (MYTH or MbYTH) method. In this, one chimeric protein binds to N- the terminal region of a ubiquitin moiety (Nub), and another chimeric protein binds with the C-terminal region (Cub) linked to a reporter protein. The binding of two proteins will result in the joining of Nub and Cub regions and the formation of complete ubiquitin moiety that can bind with ubiquitin-specific proteases and will cleave the reporter protein from the protein-protein complexes that are interacting and producing positive results (258, 280). Proteome mapping of Flavivirus NS3 and NS5 proteins using a yeast two-hybrid screen identified 108 human proteins that were interacting either with nsP3 or nsP5 or with both proteins (278). Another study identified human proteins SNAPIN and N4BP2L2 interacting with the nsP3 protein of CHIKV (277).
Structure-based proteomics
Structural information provides knowledge about the biological function and physiological role of proteins. Structural proteomics studies are based on the 3D structure determination using cryo-EM, X-ray, or NMR and high throughput characterization of protein complexes (281) (Figure 2). Structural studies are important to interpret the multifunctional nature of proteins involved in viral infection and to understand how different protein interacts to form active complexes (282). Structural studies of DENV proteins revealed valuable information about conformational changes occurring in E protein and the unique fold present in prM protein (158, 283, 284). Another structural analysis of WNV and Dengue NS3 protease clarifies the mechanism of NS3pro activation and identifies the critical residue for substrate binding (285). Using structural proteomics approaches, thousands of protein structures have been determined and submitted to the protein data bank (PDB) (286). Determining the structure of viral and host protein-protein complexes are attractive targets as their structure reveals essential information relating to the molecular description of the interface and key residue of interaction (287).
Computational study of virus-host interactions
Computational prediction methods and strategies to map the protein-protein interaction are attractive, cost-effective, less laborious, and comparatively fast alternatives to experimental methods. Also, computational methods can be utilized to map the pairwise associations of proteins into a complete network relating to their different functional levels (288). The first published computational study was on 190 pathogen strains to identify the host-pathogen interaction, the data generated from experiments and databases had integrated to identify 10,477 human-pathogen interactions (289).
Large-scale computational studies were conducted on five viruses Influenza virus (H1N1), Human immunodeficiency virus-1 (HIV), Epstein-Barr virus (EBV), Human papillomavirus (HPV), and Hepatitis C virus (HCV) by integrating available knowledge about human – virus PPIs and human complexes, led to fundamental analysis and identification of complexes targeted by viruses (VTCs) (288). During the host cell cycle, a dynamic expression study for the subunits of host protein complexes targeted by viruses shows the manipulative tendency of viruses toward host protein complexes viruses (288). They also developed a web portal called VTcomplex (http://zzdlab.com/vtcomplex/index.php) which contains significant information about the virally-targeted human protein complexes in the form of text and graphics for easy understanding and a brief overview of complexes (288).
Database for virus-host interactions
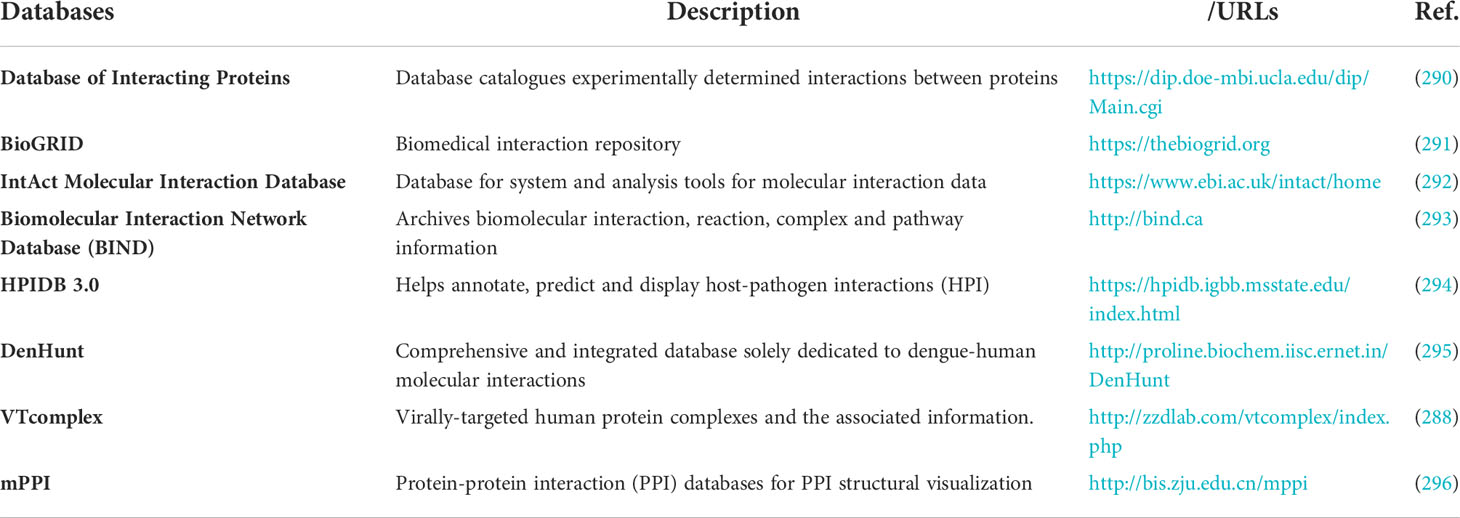
Detailed knowledge of the cellular interactions occurring between virus and host proteins is needed to understand the physiological changes and consequences of viral infection in the host. To make the information regarding PPIs readily accessible, researchers have curated the information in publically available databases (Table 8). Virus molecular interactions (VirusMINT) database collects virus-host (human) protein-protein interaction data from the available literature and integrates this data in an interaction network of proteins. VirusMINT presently has a collection of more than 5000 interactions with over 490 interactions exclusive for viral protein from different viral strains (297). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a specific database curated for PPIs of virus-virus and virus-host. The interactions involved are either physical (direct) or functional (indirect) associations. The sources of these interactions are genomic context predictions, high-throughput lab experiments, (conserved) co-expression, automated text mining, and previous knowledge. STRING currently has around 9,643,763 proteins from 2,031 organisms and a total of 1,380,838,440 interactions (298, 299). VirHostNet is a knowledge-based bioinformatics database with integrated information for virus-host, virus-virus, and host-host PPIs. This information system is committed to the biocuration of data from original literature, the integration of data, and the visualization of Virus/Host protein-protein interactions Networks based on graph theory (248).
Broad-spectrum antivirals targeting the host-virus protein-protein interaction
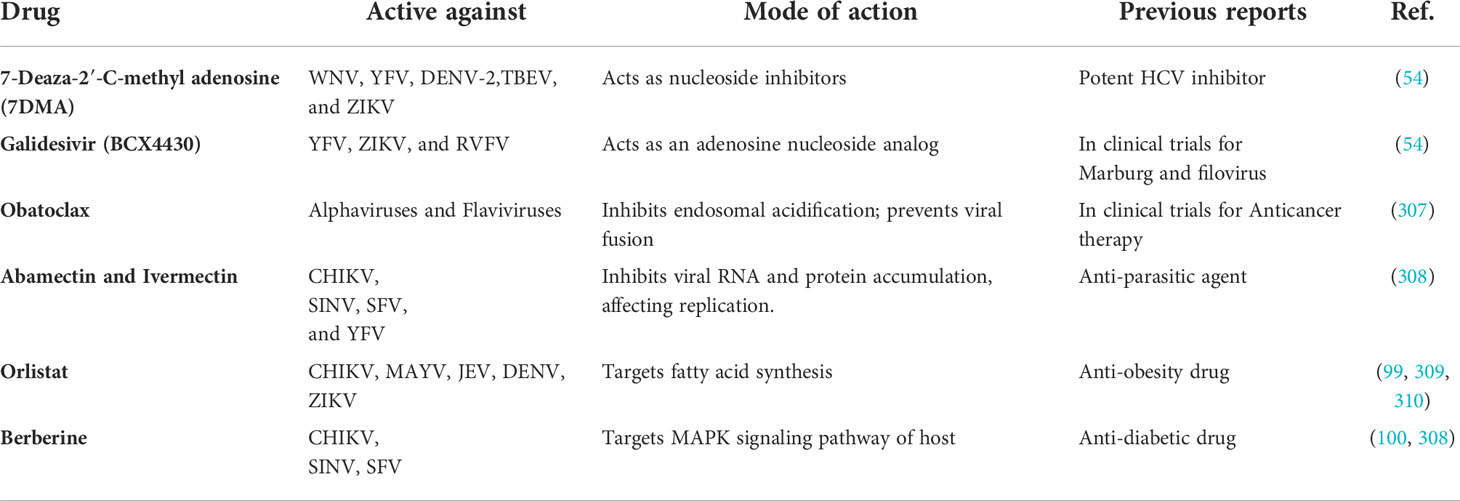
The trademark for RNA viruses is a heavy dependence on the host cell machinery for the replication of the viral genome, shorter generation time, and fast evolutionary time (300, 301). RNA viruses have high mutation rates, approximately 6 times higher than their cellular host (301). Spontaneous mutation per site per generation for DENV 2.64 × 10−5, E. coli 2 × 10−10, and yeast 3.3 × 10−10 (302–304). The rising number of viral emergence and onset over the last few years created the demand for novel, widely acting antivirals to reduce the burden of these infections on health and commerce. It is well-known that viruses exploit the host's translational assembly and protein folding machinery (149, 305). Developing antiviral compounds targeting pathways in host cells that facilitate viral replication is an appealing method of antiviral drug discovery, particularly with novel viruses. This approach would provide an attractive alternative for developing broadly acting antivirals with a lower risk of antiviral resistance (306) (Table 9).
Screening for antiviral through an image-based phenotypic technique and large-scale target identification led to the discovery of broad-spectrum antiviral having inhibitory activity against different RNA viruses like SARS-CoV-2, CCHFV, and Ebola virus (EBOV), target host proteostasis pathways and interfere with interactions of host cellular HSP70 complex and viral proteins (311). Obatoclax, an antagonist of the B-cell lymphoma 2 (Bcl-2) family of regulator protein that regulate cell death, has reached clinical trials as an anticancer therapy. It is reported to be active against flaviviruses, alphaviruses, and influenza viruses. It acts upon endosomal acidification and thus prevents viral membrane fusion and entry (307). In another study, bioactive compounds and FDA-approved drugs were screened using a CHIKV replicon cell line that identified Abamectin, Ivermectin, and Berberine as a potential inhibitors of CHIKV replication. Moreover, these compounds were also active against other alphaviruses such as SINV and SFV, and Abamectin and Ivermectin were also active against YFV.
Further studies on berberine reported that it acts on the MAPK (mitogen-activated protein kinase) signaling pathway during CHIKV infection (308). HSP-90, host-stress pathway chaperone, has been identified as a crucial factor for viral replication (312). HSP-90 is reported to interact with nsP3 and nsP4 proteins of CHIKV to help in viral replication. Also, during CHIKV infection Akt pathway is activated through the interaction of HSP90 via phosphorylation of mTOR to regulate the translation of viral mRNA I (101, 313). The inhibitors for HSP-90 have been well studied for anticancer therapy, and drugs acting against HSP 90 could decrease inflammation and viral titers upon CHIKV infection (313). The fusion of CHIKV and MAYV depends on the sphingolipid and cholesterol in the host cell membrane. Fatty acid synthase (FASN) and Stearoyl-CoA desaturase-1 (SCD1) were identified as conserved druggable host factors against Alphavirus. Inhibiting the enzyme Fatty acid synthase using cerulenin and orlistat and enzyme (SCD1) using CAY10566 decreased viral replication (99, 309). Moreover, orlistat also acts against other mosquito-transmitted viruses such as JEV, DENV, ZIKV, and CHIKV (310). An interesting study identified a small molecule inhibitor RK-0404678 of RdRp, i.e., NSP5 of dengue virus. The crystal structure revealed that RK-0404678 was able to bind at two different sites in the RdRp domains of DENV (69). The common structural features of these sites are cysteine residues. Moreover, these cysteine residues present in NS5 proteins are conserved across the DENV, JEV, WNV, and ZIKV, indicating that RK-0404678 could also bind to these flavivirus RdRps (69, 314, 315).
Conclusion
Arboviral diseases have a substantial impact on humans. The emergence of resistance towards antivirals is either due to the prolonged use or the error-prone replication cycle of RNA viruses. The use of therapeutic approaches targeting host factors interacting with viral proteins could be a promising alternative therapy. To perform replication, the arboviral genome requires host-virus PPI. Some of these PPIs seem to be broadly conserved among flaviviruses and alphaviruses. These viruses use various host proteins and physically interact with another cellular signaling, metabolomic pathways, and immune responses, which are critical to virus replication. Hence virus-host PPIs can be significant in the development of pathogenesis. The use of therapeutic approaches targeting host factors interacting with viral proteins could be a promising alternative therapy. PPI biology has great potential in identifying potential drug molecules to combat future viral infections.
Author contributions
MB, VS, PD, and ST reviewed the literature, critically analyzed it, and authored the article. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors duly thank Dr. Ravi Saini, Mr. Ankur Singh for critical reading and help in corrections in the manuscript. M.B. thankful to council of scientific and industrial research (CSIR), V.S. thankful to Indian Council of Medical Research (ICMR), P.D. thankful to Ministry of Human Resource Development (MHRD) Government of India for research fellowships.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Davis LE, Beckham JD, Tyler KL. North American encephalitic arboviruses. Neurol Clin (2008) 26(3):727.
2. Sabin NS, Calliope AS, Simpson SV, Arima H, Ito H, Nishimura T, et al. Implications of human activities for (re)emerging infectious diseases, including COVID-19. J Physiol Anthropol (2020) 39(1):1–12. doi: 10.1186/s40101-020-00239-5
3. Foy BD, Kobylinski KC, Foy JLC, Blitvich BJ, da Rosa AT, Haddow AD, et al. Probable non–vector-borne transmission of zika virus, Colorado, USA. Emerg Infect Dis (2011) 17(5):880.
4. McCarthy M. Zika virus was transmitted by sexual contact in Texas, health officials report. BMJ (2016) 352:i720.
5. Bunyavirales | viral hemorrhagic fevers (VHFs). CDC. Available at: https://www.cdc.gov/vhf/virus-families/bunyaviridae.html.
6. Virus | Colorado tick fever. CDC. Available at: https://www.cdc.gov/coloradotickfever/virus.html.
7. Schmaljohn AL, McClain D. Alphaviruses (Togaviridae) and flaviviruses (Flaviviridae) (1996). Available at: https://www.ncbi.nlm.nih.gov/books/NBK7633/.
8. Arboviral diseases, neuroinvasive and non-neuroinvasive 2015 case definition. CDC. Available at: https://ndc.services.cdc.gov/case-definitions/arboviral-diseases-neuroinvasive-and-non-neuroinvasive-2015/.
9. Shahfiza N, Osman H, Hock TT, Abdel-Hamid A-HZ. Metabolomics approach for multibiomarkers determination to investigate dengue virus infection in human patients. Acta Biochim Pol (2017) 64(2):215–219.
10. van de Leemput J, Han Z. Understanding individual SARS-CoV-2 proteins for targeted drug development against COVID-19. Mol Cell Biol (2021) 41(9):e00185–21.
11. Rao VS, Srinivas K, Sujini GN, Kumar GNS. Protein-protein interaction detection: Methods and analysis. Int J Proteomics (2014) 2014:1–12.
12. Rocha RF, Del Sarto JL, Marques RE, Costa VV, Teixeira MM. Host target-based approaches against arboviral diseases. Biol Chem (2018) 399(3):203–17.
13. Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe (2012) 12(4):544–557.
14. Miller JL, DeWet BJM, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, et al. The mannose receptor mediates dengue virus infection of macrophages. PloS Pathog (2008) 4(2):e17. doi: 10.1371/journal.ppat.0040017
15. Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med (2003) 197(7):823–829. doi: 10.1084/jem.20021840
16. Low JG, Sung C, Wijaya L, Wei Y, Rathore APS, Watanabe S, et al. Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect Dis (2014) 14(8):706–15.
17. Urbano P, Urbano FG. The reoviridae family. Comp Immunol Microbiol Infect Dis (1994) 17(3–4):151–61.
18. Schubert AM, Putonti C. Evolution of the sequence composition of flaviviruses. Infect Genet Evol (2010) 10(1):129–36.
19. Billoir F, De Chesse R, Tolou H, De Micco P, Gould EA, De Lamballerie X. Phylogeny of the genus flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J Gen Virol (2000) 81(Pt 3):781–90.
20. Cook S, Holmes EC. A multigene analysis of the phylogenetic relationships among the flaviviruses (Family: Flaviviridae) and the evolution of vector transmission. Arch Virol (2006) 151(2):309–25.
21. Coates M, Blanchard S, MacLeod AS. Innate antimicrobial immunity in the skin: A protective barrier against bacteria, viruses, and fungi. PloS Pathog (2018) 14(12):e1007353.
22. Japanese Encephalitis. Available at: https://www.who.int/news-room/fact-sheets/detail/japanese-encephalitis.
23. Dengue guidelines for diagnosis, treatment, prevention and control: new edition. Available at: https://apps.who.int/iris/handle/10665/44188.
24. About zika virus disease | zika virus. CDC. Available at: https://www.cdc.gov/zika/about/index.html.
25. Kyasanur forest disease. National Health Portal Of India. Available at: https://www.nhp.gov.in/disease/communicable-disease/kyasanur-forest-disease.
26. Rajaiah P. Kyasanur forest disease in India: innovative options for intervention. Hum Vaccin Immunother (2019) 15(10):2243.
27. Shearer FM, Moyes CL, Pigott DM, Brady OJ, Marinho F, Deshpande A, et al. Global yellow fever vaccination coverage from 1970 to 2016: an adjusted retrospective analysis. Lancet Infect Dis (2017) 17(11):1209.
28. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature (2013) 496(7446):504–7.
29. Simmons CP, Farrar JJ, van Vinh Chau N, Wills B, Ham Tu B, Chi Minh City H. Dengue. New England Journal of Medicine (2012) 366(15):1423–32. doi: 10.1056/nejmra1110265
30. Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, et al. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ (2011) 89(10):766.
31. Zika virus. Available at: https://www.who.int/news-room/fact-sheets/detail/zika-virus.
32. Quaresma JAS, Pagliari C, Medeiros DBA, Duarte MIS, Vasconcelos PFC. Immunity and immune response, pathology and pathologic changes: progress and challenges in the immunopathology of yellow fever. Rev Med Virol (2013) 23(5):305–18.
33. Yellow fever. Available at: https://www.who.int/news-room/fact-sheets/detail/yellow-fever.
34. Yellow fever. Available at: https://www.cdc.gov/yellowfever/index.html.
35. West Nile Virus. Available at: https://www.who.int/news-room/fact-sheets/detail/west-nile-virus.
36. Murray Valley encephalitis virus | disease directory. Travelers’ Health | CDC. Available at: https://wwwnc.cdc.gov/travel/diseases/murray-valley-encephalitis-virus.
37. St. Louis encephalitis | st. Louis encephalitis. CDC. Available at: https://www.cdc.gov/sle/index.html.
38. Tick-borne encephalitis. Available at: https://www.who.int/health-topics/tick-borne-encephalitis#tab=tab_1.
39. Tick-borne encephalitis (TBE). Tick-borne encephalitis. CDC. Available at: https://www.cdc.gov/tick-borne-encephalitis/.
40. Kutchko KM, Madden EA, Morrison C, Plante KS, Sanders W, Vincent HA, et al. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res (2018) 46(7):3657–70. https://academic.oup.com/nar/article/46/7/3657/4817396.
41. Chikungunya fact sheet. Available at: https://www.who.int/news-room/fact-sheets/detail/chikungunya.
42. Chikungunya virus. CDC. Available at: https://www.cdc.gov/chikungunya/index.html.
43. Robinson MC. An epidemic of virus disease in southern province, tanganyika territory, in 1952–1953. i. clinical features. Trans R Soc Trop Med Hyg (1955) 49(1):28–32.
44. Facts about sindbis fever. Available at: https://www.ecdc.europa.eu/en/sindbis-fever/facts.
45. Ling J, Smura T, Lundström JO, Pettersson JH-O, Sironen T, Vapalahti O, et al. Introduction and dispersal of sindbis virus from central Africa to Europe. J Virol (2019) 93(16):e00620–19. doi: 10.1128/JVI.00620-19
46. Crosby B, Crespo ME. Venezuelan Equine encephalitis. StatPearls (2021). Available at: https://www.ncbi.nlm.nih.gov/books/NBK559332/.
47. Eastern Equine encephalitis | Eastern equine encephalitis. CDC. Available at: https://www.cdc.gov/easternequineencephalitis/index.html.
48. Ross River virus disease | disease directory. Travelers’ Health | CDC. Available at: https://wwwnc.cdc.gov/travel/diseases/ross-river-virus-disease.
49. Simon LV, Coffey R, Fischer MA. Western Equine encephalitis. StatPearls (2022). Available at: https://www.ncbi.nlm.nih.gov/books/NBK470228/.
50. Leventhal SS, Wilson D, Feldmann H, Hawman DW. A look into bunyavirales genomes: Functions of non-structural (NS) proteins. Viruses (2021) 13(2):314.
51. Estrada DF, De Guzman RN. Structural characterization of the Crimean-Congo hemorrhagic fever virus gn tail provides insight into virus assembly. J Biol Chem (2011) 286(24):21678–86.
52. Quito-Avila D, Jelkmann W, Tzanetakis I, Research KK-V. Complete sequence and genetic characterization of raspberry latent virus, a novel member of the family reoviridae. Elsevier (2011). Available at: https://www.sciencedirect.com/science/article/pii/S0168170210004168.
53. Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard, et al. Reoviridae. In: Virus taxon. p. 541–637. Elsevier
54. Boldescu V, Behnam MAM, Vasilakis N, Klein CD. Broad-spectrum agents for flaviviral infections: dengue, zika and beyond. Nat Rev Drug Discovery (2017) 16(8):565–86.
55. Lim SP. Dengue drug discovery: Progress, challenges and outlook. Antiviral Res (2019) 163:156–78.
56. Singh H, Mudgal R, Narwal M, Kaur R, Singh VA, Malik A, et al. Chikungunya virus inhibition by peptidomimetic inhibitors targeting virus-specific cysteine protease. Biochimie (2018) 149:51–61.
57. Sharma R, Fatma B, Saha A, Bajpai S, Sistla S, Dash PK, et al. Inhibition of chikungunya virus by picolinate that targets viral capsid protein. Virology (2016) 498:265–76.
58. Aliota MT, Bassit L, Bradrick SS, Cox B, Garcia-Blanco MA, Gavegnano C, et al. Zika in the americas, year 2: What have we learned? what gaps remain? a report from the global virus network. Antiviral Res (2017) 144:223–46.
59. Thomas SJ, Yoon IK. A review of dengvaxia®: development to deployment. Hum Vaccin Immunother (2019) 15(10):2295–314. doi: 10.1080/21645515.2019.1658503
60. Liao W, Liu X, Yang Q, Liu H, Liang B, Jiang J, et al. Deguelin inhibits HCV replication through suppressing cellular autophagy via down regulation of Beclin1 expression in human hepatoma cells. Antiviral Res (2020) 174:104704.
61. Troost B, Smit JM. Recent advances in antiviral drug development towards dengue virus. Curr Opin Virol (2020) 43:9–21.
62. Low JG, Gatsinga R, Vasudevan SG, Sampath A. Dengue antiviral development: A continuing journey. Adv Exp Med Biol (2018) 1062:319–32. doi: 10.1007/978-981-10-8727-1_22
63. Zhou HY, Gao SQ, Gong YS, Lin T, Tong S, Xiong W, et al. Anti-HSV-1 effect of dihydromyricetin from ampelopsis grossedentata via the TLR9-dependent anti-inflammatory pathway. J Glob Antimicrob Resist (2020) 23:370–6.
64. Bernard E, Solignat M, Gay B, Chazal N, Higgs S, Devaux C, et al. Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PloS One (2010) 5(7):e11479.
65. Khan M, Santhosh SR, Tiwari M, Lakshmana Rao PV, Parida M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against chikungunya virus in vero cells. J Med Virol (2010) 82(5):817–24.
66. De Lamballerie X, Boisson V, Reynier JC, Enault S, Charrel RN, Flahault A, et al. On chikungunya acute infection and chloroquine treatment. J Vector Borne Zoonotic Dis (2008) 8(6):837–9. doi: 10.1089/vbz.2008.0049
67. Leneva IA, Russell RJ, Boriskin YS, Hay AJ. Characteristics of arbidol-resistant mutants of influenza virus: Implications for the mechanism of anti-influenza action of arbidol. Antiviral Res (2009) 81(2):132–40.
68. Nguyen NM, Tran CNB, Phung LK, Duong KTH, Huynh HLA, Farrar J, et al. A randomized, double-blind placebo controlled trial of balapiravir, a polymerase inhibitor, in adult dengue patients. J Infect Dis (2013) 207(9):1442–50.
69. Shimizu H, Saito A, Mikuni J, Nakayama EE, Koyama H, Honma T, et al. Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase. PloS Negl Trop Dis (2019) 13(11):e0007894.
70. Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem (2020) 295(20):6785–97.
71. Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science (2020) 368(6498):1499–504.
72. Krishnan MN, Garcia-Blanco MA. Targeting host factors to treat West Nile and dengue viral infections. Viruses (2014) 6:683–708.
73. Delogu I, Pastorino B, Baronti C, Nougairède A, Bonnet E, de Lamballerie X. In vitro antiviral activity of arbidol against chikungunya virus and characteristics of a selected resistant mutant. Antiviral Res (2011) 90(3):99–107.
74. Ahlquist P, Noueiry AO, Lee W-M, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol (2003) 77(15):8181–6.
75. Delang L, Guerrero NS, Tas A, Quérat G, Pastorino B, Froeyen M, et al. Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J Antimicrob Chemother (2014) 69(10):2770–84.
76. Scroggs SLP, Gass JT, Chinnasamy R, Widen SG, Azar SR, Rossi SL, et al. Evolution of resistance to fluoroquinolones by dengue virus serotype 4 provides insight into mechanism of action and consequences for viral fitness. Virology (2021) 552:94–106.
77. Mudgal R, Bharadwaj C, Dubey A, Choudhary S, Nagarajan P, Aggarwal M, et al. Selective estrogen receptor modulators limit alphavirus infection by targeting the viral capping enzyme nsP1. Antimicrob Agents Chemother (2022) 66(3):e01943–21. doi: 10.1128/aac.01943-21
78. Jensen SB, Fahnøe U, Pham LV, Serre SBN, Tang Q, Ghanem L, et al. Evolutionary pathways to persistence of highly fit and resistant hepatitis c virus protease inhibitor escape variants. Hepatology (2019) 70(3):771–87. doi: 10.1002/hep.30647
79. Quiñones-Mateu ME, Tadele M, Parera M, Mas A, Weber J, Rangel HR, et al. Insertions in the reverse transcriptase increase both drug resistance and viral fitness in a human immunodeficiency virus type 1 isolate harboring the multi-nucleoside reverse transcriptase inhibitor resistance 69 insertion complex mutation. J Virol (2002) 76(20):10546–52. doi: 10.1128/JVI.76.20.10546-10552.2002
80. Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med (1997) 3(8):866–71.
81. Das S, Laxminarayana SV, Chandra N, Ravi V, Desai A. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology (2009) 385(1):47–57.
82. Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure (2006) 14(1):63.
83. van Duijl-Richter MKS, Hoornweg TE, Rodenhuis-Zybert IA, Smit JM. Early events in chikungunya virus infection–from virus cell binding to membrane fusion. Viruses (2015) 7(7):3647.
84. Walsh D, Mohr I. Viral subversion of the host protein synthesis machinery. J Nat Rev Microbiol (2011) 9(12):860–875.
85. King CR, Mehle A. The later stages of viral infection: An undiscovered country of host dependency factors. PloS Pathog (2020) 16(8):e1008777.
86. Pérez-Vilaró G, Jungfleisch J, Saludes V, Scheller N, Giménez-Barcons M, Díez J. Host factors in viral life cycles. Math Model Nat Phenom (2012) 7(5):123–32.
87. Martín-Acebes MA, Blázquez AB, Jiménez de Oya N, Escribano-Romero E, Saiz JC. West Nile Virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PloS One (2011) 6(9):e24970.
88. Heaton NS, Perera R, Berger KL, Khadka S, LaCount DJ, Kuhn RJ, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A (2010) 107(40):17345–50.
89. Martínez-Gutierrez M, Castellanos JE, Gallego-Gómez JC. Statins reduce dengue virus production via decreased virion assembly. Intervirology (2011) 54(4):202–16.
90. Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe (2007) 2(4):229–39.
91. Rothwell C, LeBreton A, Young Ng C, Lim JYH, Liu W, Vasudevan S, et al. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology (2009) 389(1–2):8–19.
92. Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J Gen Virol (2006) 87(Pt 7):1947–52.
93. Morrey JD, Smee DF, Sidwell RW, Tseng C. Identification of active antiviral compounds against a new York isolate of West Nile virus. Antiviral Res (2002) 55(1):107–16.
94. Leyssen P, Balzarini J, De Clercq E, Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol (2005) 79(3):1943–7.
95. Fink J, Gu F, Ling L, Tolfvenstam T, Olfat F, Chin KC, et al. Host gene expression profiling of dengue virus infection in cell lines and patients. PloS Negl Trop Dis (2007) 1(2):e86.
96. Poh MK, Shui G, Xie X, Shi PY, Wenk MR, Gu F. U18666A, an intra-cellular cholesterol transport inhibitor, inhibits dengue virus entry and replication. Antiviral Res (2012) 93(1):191–8.
97. Low JSY, Wu KX, Chen KC, Ng MML, Chu JJH. Narasin, a novel antiviral compound that blocks dengue virus protein expression. Antivir Ther (2011) 16(8):1203–18.
98. Dutta K, Ghosh D, Basu A. Curcumin protects neuronal cells from japanese encephalitis virus-mediated cell death and also inhibits infective viral particle formation by dysregulation of ubiquitin-proteasome system. J Neuroimmune Pharmacol (2009) 4(3):328–37.
99. Karlas A, Berre S, Couderc T, Varjak M, Braun P, Meyer M, et al. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat Commun (2016) 7(1):1–14.
100. Varghese FS, Thaa B, Amrun SN, Simarmata D, Rausalu K, Nyman TA, et al. The antiviral alkaloid berberine reduces chikungunya virus-induced mitogen-activated protein kinase signaling. J Virol (2016) 90(21):9743–57.
101. Das I, Basantray I, Mamidi P, Nayak TK, Pratheek BM, Chattopadhyay S, et al. Heat shock protein 90 positively regulates chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PloS One (2014) 9(6):e100531.
102. Rothan HA, Kumar M. Role of endoplasmic reticulum-associated proteins in flavivirus replication and assembly complexes. Pathogens (2019) 8(3):148.
103. Monel B, Rajah MM, Hafirassou ML, Ahmed SS, Burlaud-Gaillard J, Zhu P-P, et al. Atlastin endoplasmic reticulum-shaping proteins facilitate zika virus replication. J Virol (2019) 93(23):e01047–19.
104. Neufeldt CJ, Cortese M, Scaturro P, Cerikan B, Wideman JG, Tabata K, et al. ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat Microbiol (2019) 4(12):2416.
105. Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell (2018) 175(7):1931.
106. Li M, Johnson JR, Truong B, Kim G, Weinbren N, Dittmar M, et al. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol (2019) 4(6):985.
107. Tran PTH, Asghar N, Johansson M, Melik W. Roles of the endogenous lunapark protein during flavivirus replication. Viruses (2021) 13(7):1198.
108. Bernard KA, Klimstra WB, Johnston RE. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology (2000) 276(1):93–103.
109. Byrnes AP, Griffin DE. Large-Plaque mutants of sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J Virol (2000) 74(2):644–51.
110. Gardner CL, Ebel GD, Ryman KD, Klimstra WB. Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc Natl Acad Sci U S A (2011) 108(38):16026–31.
111. Klimstra WB, Nangle EM, Smith MS, Yurochko AD, Ryman KD. DC-SIGN and l-SIGN can act as attachment receptors for alphaviruses and distinguish between mosquito cell- and mammalian cell-derived viruses. J Virol (2003) 77(22):12022.
112. Porcellini S, Gubinelli F, Alberici L, Piovani BM, Rizzardi GP, Bovolenta C. Chim3 confers survival advantage to CD4+ T cells upon HIV-1 infection by preventing HIV-1 DNA integration and HIV-1-induced G2 cell-cycle delay. Blood (2010) 115(20):4021–9.
113. Wang J, Reuschel EL, Shackelford JM, Jeang L, Shivers DK, Diehl JA, et al. HIV-1 vif promotes the G1- to s-phase cell-cycle transition. Blood (2011) 117(4):1260–9.
114. Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe (2010) 8(5):422–32.
115. Whitby K, Pierson TC, Geiss B, Lane K, Engle M, Zhou Y, et al. Castanospermine, a potent inhibitor of dengue virus infection in vitro and In vivo. J Virol (2005) 79(14):8698–706.
116. Braun P, Gingras AC. History of protein-protein interactions: From egg-white to complex networks. Proteomics (2012) 12(10):1478–98.
117. Zhang A, Hwang W-c, Chanda P, Shi L, Cho Y-r. Protein interaction networks: Computational analysis - aidong zhang - Google books. (2009) New York : Cambridge University Press
119. Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RWH, et al. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology (2002) 292(1):162–8.
120. Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virology (2003) 308(1):92–100.
121. Mady BJ, Kurane I, Erbe DV, Fanger MW, Ennis FA. Neuraminidase augments fcy receptor II-mediated antibody-dependent enhancement of dengue virus infection. J Gen Virol (1993) 74(5):839–44.
122. Thepparit C, Smith DR. Serotype-specific entry of dengue virus into liver cells: Identification of the 37-Kilodalton/67-Kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J Virol (2004) 78(22):12647.
123. Davis CW, Nguyen H-Y, Hanna SL, Sánchez MD, Doms RW, Pierson TC. West Nile Virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J Virol (2006) 80(3):1290–301.
124. Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, et al. An orthogonal proteomic survey uncovers novel zika virus host factors. Nature (2018) 561(7722):253–7.
125. Barrows NJ, Anglero-Rodriguez Y, Kim B, Jamison SF, Le Sommer C, McGee CE, et al. Dual roles for the ER membrane protein complex in flavivirus infection: viral entry and protein biogenesis. Sci Rep (2019) 9(1):1–16.
126. Lin DL, Inoue T, Chen Y-J, Chang A, Tsai B, Tai AW. The ER membrane protein complex promotes biogenesis of dengue and zika virus non-structural multi-pass transmembrane proteins to support infection in brief. Cell Rep (2019) 27(6):1666–74.
127. Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, Bonnet-Madin L, et al. A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell Rep (2017) 21(13):3900–13.
128. Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, et al. Genetic dissection of flaviviridae host factors through genome-scale CRISPR screens. Nature (2016) 535(7610):159–63.
129. Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature (2016) 535(7610):164.
130. Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM. The host protein reticulon 3.1A is utilized by flaviviruses to facilitate membrane remodelling. Cell Rep (2017) 21(6):1639–54.
131. Golubeva VA, Nepomuceno TC, de Gregoriis G, Mesquita RD, Li X, Dash S, et al. Network of interactions between ZIKA virus non-structural proteins and human host proteins. Cells (2020) 9(1):153.
132. Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. Biogenesis of the semliki forest virus RNA replication complex. J Virol (2001) 75(8):3873–84.
133. Fros JJ, Domeradzka NE, Baggen J, Geertsema C, Flipse J, Vlak JM, et al. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J Virol (2012) 86(19):10873–9.
134. Barton DJ, Sawicki SG, Sawicki DL. Solubilization and immunoprecipitation of alphavirus replication complexes. J Virol (1991) 65(3):1496–506.
135. Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol (1988) 107(6 Pt 1):2075–86.
136. Kuhn RJ, Niesters HGM, Hong Z, Strauss JH. Infectious RNA transcripts from Ross river virus cDNA clones and the construction and characterization of defined chimeras with sindbis virus. Virology (1991) 182(2):430–41.
137. Li M, Wang P, Zheng Z, Hu K, Zhang M, Guan X, et al. Japanese Encephalitis virus counteracts BST2 restriction via its envelope protein e. Virology (2017) 510:67–75.
138. Jones PH, Maric M, Madison MN, Maury W, Roller RJ, Okeoma CM. BST-2/tetherin-mediated restriction of chikungunya (CHIKV) VLP budding is counteracted by CHIKV non-structural protein 1 (nsP1). Virology (2013) 438(1):37–49.
139. Anderson R, King AD, Innis BL. Correlation of e protein binding with cell susceptibility to dengue 4 virus infection. J Gen Virol (1992) 73(Pt 8):2155–9.
140. Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J (2004) 23(4):728.
141. Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A (2003) 100(12):6986–91.
142. Fritz R, Blazevic J, Taucher C, Pangerl K, Heinz FX, Stiasny K. The unique transmembrane hairpin of flavivirus fusion protein e is essential for membrane fusion. J Virol (2011) 85(9):4377.
143. Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 a resolution. Nature (1995) 375(6529):291–8.
144. Li Y, Li Q, Wong YL, Liew LSY, Kang C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim Biophys Acta - Biomembr (2015) 1848(10):2244–52.
145. Li Y, Wong YL, Lee MY, Li Q, Wang QY, Lescar J, et al. Secondary structure and membrane topology of the full-length dengue virus NS4B in micelles. Angew Chem Int Ed Engl (2016) 55(39):12068–72.
146. Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem (2007) 282(12):8873–82.
147. Xie X, Gayen S, Kang C, Yuan Z, Shi P-Y. Membrane topology and function of dengue virus NS2A protein. J Virol (2013) 87(8):4609–22.
148. Kuhn: Fields virology: emerging viruses - Google scholar. Available at: https://scholar.google.com/scholar_lookup?title=Fields+Virology&author=R.J.+Kuhn&publication_year=2013&.
149. Heaton NS, Moshkina N, Fenouil R, Gardner TJ, Aguirre S, Shah PS, et al. Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity (2016) 44(1):46–58.
150. Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, et al. Discovery of insect and human dengue virus host factors. Nat (2009) 458(7241):1047–50. https://www.nature.com/articles/nature07967.
151. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature (2020) 583(7816):459–68.
152. Goyal U, Blackstone C. Untangling the web: Mechanisms underlying ER network formation. Biochim Biophys Acta - Mol Cell Res (2013) 1833(11):2492–8.
153. Teo CSH, Chu JJH. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J Virol (2014) 88(4):1897.
154. Wang S, Tukachinsky H, Romano FB, Rapoport TA. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. Elife (2016) 5(September):e18605.
155. Filipowicz W, Furuichi Y, Sierra JM, Muthukrishnan S, Shatkin AJ, Ochoa S. A protein binding the methylated 5’-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc Natl Acad Sci U S A (1976) 73(5):1559–63.
156. Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol (2005) 3(1):13–22.
157. Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol (1997) 71(11):8475–81.
158. Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science (2008) 319(5871):1834–7.
159. Zheng A, Yuan F, Kleinfelter LM, Kielian M. A toggle switch controls the low pH-triggered rearrangement and maturation of the dengue virus envelope proteins. Nat Commun (2014) 5(1):1–9.
160. Silva LA, Dermody TS. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest (2017) 127(3):737.
161. Knipe DM ed. R.J k. togaviridae. In: Fields virology, 6th ed. Philadelphia, PA, USA: Vo. Lippincott, Williams and Wilkins.
162. Melo CFOR, Delafiori J, Dabaja MZ, de Oliveira DN, Guerreiro TM, Colombo TE, et al. The role of lipids in the inception, maintenance and complications of dengue virus infection. Sci Rep (2018) 8(1):1–9.
163. Deckers J, Hammad H, Hoste E. Langerhans cells: Sensing the environment in health and disease. Front Immunol (2018) 9:93.
164. Briant L, Desprès P, Choumet V, Missé D. Role of skin immune cells on the host susceptibility to mosquito-borne viruses. Virology (2014) 464–465(1):26–32.
165. Geherin SA, Gómez D, Glabman RA, Ruthel G, Hamann A, Debes GF. IL-10 + innate-like b cells are part of the skin immune system and require α4β1 integrin to migrate between the peritoneum and inflamed skin. J Immunol (2016) 196(6):2514–25.
166. Tay SS, Roediger B, Tong PL, Tikoo S, Weninger W. The skin-resident immune network. Curr Dermatol Rep (2013) 3(1):13–22.
167. Romani N, Koide S, Crowley M, Witmer-Pack M, Livingstone AM, Fathman CG, et al. Presentation of exogenous protein antigens by dendritic cells to T cell clones. intact protein is presented best by immature, epidermal langerhans cells. J Exp Med (1989) 169(3):1169–78.
168. Duque GA, Descoteaux A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front Immunol (2014) 5.
169. Schneider CA, Calvo E, Peterson KE. Arboviruses: How saliva impacts the journey from vector to host. Int J Mol Sci (2021) 22(17):9173.
170. Surasombatpattana P, Hamel R, Patramool S, Luplertlop N, Thomas F, Desprès P, et al. Dengue virus replication in infected human keratinocytes leads to activation of antiviral innate immune responses. Infect Genet Evol (2011) 11(7):1664–73.
171. Krejbich-Trotot P, Denizot M, Hoarau J, Jaffar-Bandjee M, Das T, Gasque P. Chikungunya virus mobilizes the apoptotic machinery to invade host cell defenses. FASEB J (2011) 25(1):314–25.
172. Labuda M, Austyn JM, Zuffova E, Kozuch O, Fuchsberger N, Lysy J, et al. Importance of localized skin infection in tick-borne encephalitis virus transmission. Virology (1996) 219(2):357–66.
173. Francischetti TN, Mather JMC, Ribeiro IMBP, Conrads TD, Veenstra JF, Andersen A, et al. Prostaglandin E2 is a major inhibitor of dendritic cell maturation and function in ixodes scapularis saliva. Am Assoc Immnol (2007) 179(3):1497–1505.
174. Demarta-Gatsi C, Mécheri S, Paul R. Arthropod saliva and its role in pathogen transmission: Insect saliva. In: Skin arthropod vectors. p. 83–119. Academic press
175. Schneider BS, Soong L, Coffey LL, Stevenson HL, McGee CE, Higgs S. Aedes aegypti saliva alters leukocyte recruitment and cytokine signaling by antigen-presenting cells during West Nile virus infection. PloS One (2010) 5(7):e11704.
176. Krylova NV, Smolina TP, Leonova GN. Molecular mechanisms of interaction between human immune cells and far eastern tick-borne encephalitis virus strains. Viral Immunol (2015) 28(5):272–81.
177. Wei J, Kang X, Li Y, Wu X, Zhang Y, Yang Y. Pathogenicity of tick-borne encephalitis virus to monocytes. Wei Sheng Wu Xue Bao (2013) 53(11):1221–1225.
178. Mehlhop E, Whitby K, Oliphant T, Marri A, Engle M, Diamond MS. Complement activation is required for induction of a protective antibody response against West Nile virus infection. J Virol (2005) 79(12):7466–77.
179. Morrison TE, Heise MT. The host complement system and arbovirus pathogenesis. Curr Drug Targets (2008) 9(2):165–72.
180. Ayala-Nunez NV, Follain G, Delalande F, Hirschler A, Partiot E, Hale GL, et al. Zika virus enhances monocyte adhesion and transmigration favoring viral dissemination to neural cells. Nat Commun (2019) 10(1):1–16.
181. Foo SS, Chen W, Chan Y, Bowman JW, Chang LC, Choi Y, et al. Asian Zika virus strains target CD14 + blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nat Microbiol (2017) 2(11):1558–70.
182. Lum F-M, Lee D, Chua T-K, Tan JJL, Lee CYP, Liu X, et al. Zika virus infection preferentially counterbalances human peripheral monocyte and/or NK cell activity. Am Soc Microbiol (2018) 3(2):e00120–18.
183. Michlmayr D, Andrade P, Gonzalez K, Balmaseda A, Harris E. CD14 + CD16 + monocytes are the main target of zika virus infection in peripheral blood mononuclear cells in a paediatric study in Nicaragua. Nat Microbiol (2017) 2(11):1462–70.
184. Castillo JA, Naranjo JS, Rojas M, Castaño D, Velilla PA. Role of monocytes in the pathogenesis of dengue. Arch Immunol Ther Exp (Warsz) (2019) 67(1):27–40.
185. de Haan CAM, Rottier PJM. Molecular interactions in the assembly of coronaviruses. Adv Virus Res (2005) 64:165–230.
186. Xi Z, Ramirez JL, Dimopoulos G. The aedes aegypti toll pathway controls dengue virus infection. PloS Pathog (2008) 4(7):e1000098.
187. Lee WS, Webster JA, Madzokere ET, Stephenson EB, Herrero LJ. Mosquito antiviral defense mechanisms: A delicate balance between innate immunity and persistent viral infection. Parasites Vectors (2019) 12(1):1–12.
188. Dostert C, Jouanguy E, Irving P, Troxler L. The jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol (2005) 6(9):946–953.
189. Niu GJ, Xu JD, Yuan WJ, Sun JJ, Yang MC, He ZH, et al. Protein inhibitor of activated STAT (PIAS) negatively regulates the JAK/STAT pathway by inhibiting STAT phosphorylation and translocation. Front Immunol (2018) 9:2392.
190. Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci U S A (2009) 106(42):17841.
191. Duneau DF, Kondolf HC, Im JH, Ortiz GA, Chow C, Fox MA, et al. The toll pathway underlies host sexual dimorphism in resistance to both gram-negative and gram-positive bacteria in mated drosophila. BMC Biol (2017) 15(1):1–17.
192. Weber ANR, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, et al. Binding of the drosophila cytokine spätzle to toll is direct and establishes signaling. Nat Immunol (2003) 4(8):794–800.
194. Valanne S, Wang J-H, Rämet M. The drosophila toll signaling pathway. J Immunol (2011) 186(2):649–56.
195. Kim M, Hee Lee J, Young Lee S, Kim E, Chung J. Caspar, A suppressor of antibacterial immunity in drosophila. Natl Acad Sci (2006) 103(44):16358–16363.
196. Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun (2020) 12(1):4–20.
197. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol (2014) 14(1):36.
198. García-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe (2017) 22(2):176.
199. Bakar FA, Ng LFP. Nonstructural proteins of alphavirus–potential targets for drug development. Viruses (2018) 10(2):71.
200. Chen S, Yang C, Zhang W, Mahalingam S, Wang M, Cheng A. Flaviviridae virus nonstructural proteins 5 and 5A mediate viral immune evasion and are promising targets in drug development. Pharmacol Ther (2018) 190:1–14.
201. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PloS Pathog (2012) 8(10):e1002934.
202. Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, Simon V, et al. Dengue virus Co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PloS Pathog (2013) 9(3):e1003265.
203. Avila-Bonilla RG, Yocupicio-Monroy M, Marchat LA, De Nova-Ocampo MA, María Del Ángel R, Santiago Salas-Benito J. Analysis of the miRNA profile in C6/36 cells persistently infected with dengue virus type 2. Virus Res (2017) 232:139–51.
204. Lewis J, Wesselingh SL, Griffin DE, Hardwick JM. Alphavirus-induced apoptosis in mouse brains correlates with neurovirulence. J Virol (1996) 70(3):1828–35.
205. Mehrbod P, Ande SR, Alizadeh J, Rahimizadeh S, Shariati A, Malek H, et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence (2019) 10(1):376–413.
206. Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: Cell survival and cell death. Int J Cell Biol (2010).
207. Iranpour M, Moghadam AR, Yazdi M, Ande SR, Alizadeh J, Wiechec E, et al. Apoptosis, autophagy and unfolded protein response pathways in arbovirus replication and pathogenesis. Expert Rev Mol Med (2016) 18.
208. Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, DeFilippis V, et al. West Nile Virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol (2007) 81(20):10849–60.
209. Perera N, Miller JL, Zitzmann N. The role of the unfolded protein response in dengue virus pathogenesis. Cell Microbiol (2017) 19(5):e12734. doi: 10.1111/cmi.12734
210. Lin J-C, Lin S-C, Chen W-Y, Yen Y-T, Lai C-W, Tao M-H, et al. Dengue viral protease interaction with NF-κB inhibitor α/β results in endothelial cell apoptosis and hemorrhage development. J Immunol (2014) 193(3):1258–67.
211. Gladwyn-Ng I, Cordón-Barris L, Alfano C, Creppe C, Couderc T, Morelli G, et al. Stress-induced unfolded protein response contributes to zika virus-associated microcephaly. Nat Neurosci (2018) 21(1):63–73.
212. Romao S, Gannage M, Münz C. Checking the garbage bin for problems in the house, or how autophagy assists in antigen presentation to the immune system. Semin Cancer Biol (2013) 23(5):391–6.
213. Shi J, Luo H. Interplay between the cellular autophagy machinery and positive-stranded RNA viruses. Acta Biochim Biophys Sin (Shanghai) (2012) 44(5):375–84.
214. Vandergaast R, Fredericksen BL. West Nile Virus (WNV) replication is independent of autophagy in mammalian cells. PloS One (2012) 7(9):e45800.
215. Jackson WT, Giddings TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PloS Biol (2005) 3(5):0861–71.
217. Urbanowski MD, Hobman TC. The West Nile virus capsid protein blocks apoptosis through a phosphatidylinositol 3-kinase-dependent mechanism. J Virol (2013) 87(2):872–81.
218. Del Carmen Parquet M, Kumatori A, Hasebe F, Morita K, Igarashi A. West Nile Virus-induced bax-dependent apoptosis. FEBS Lett (2001) 500(1–2):17–24.
219. Xu X, Lai Y, Hua ZC. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep (2019) 39(1).
220. Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol (2003) 23(20):7198–209.
221. Wei L, Zhu S, Wang J, Liu J. Activation of the phosphatidylinositol 3-Kinase/Akt signaling pathway during porcine circovirus type 2 infection facilitates cell survival and viral replication. J Virol (2012) 86(24):13589.
222. Fujikura D, Ito M, Chiba S, Harada T, Perez F, Reed JC, et al. CLIPR-59 regulates TNF-α-induced apoptosis by controlling ubiquitination of RIP1. Cell Death Dis (2012) 3(2):e264.
223. Stollar V, Shenk TE, Stollar BD. Double-stranded RNA in hamster, chick, and mosquito cells infected with sindbis virus. Virology (1972) 47(1):122–32.
224. Domingo-Gil E, Toribio R, Nájera JL, Esteban M, Ventoso I. Diversity in viral anti-PKR mechanisms: a remarkable case of evolutionary convergence. PloS One (2011) 6(2):e16711.
225. Lee C-J, Liao C-L, Lin Y-L. Flavivirus activates phosphatidylinositol 3-kinase signaling to block caspase-dependent apoptotic cell death at the early stage of virus infection. J Virol (2005) 79(13):8388–99.
226. Sakinah S, Priya SP, Kumari S, Amira F, Poorani K, Alsaeedy H, et al. Impact of dengue virus (serotype DENV-2) infection on liver of BALB/c mice: A histopathological analysis. Tissue Cell (2017) 49(1):86–94.
227. Ghosh Roy S, Datan E, Sadigh B, Lockshin RA, Zakeri Z. Regulation of cell survival and death during flavivirus infections. World J Biol Chem (2014) 5(2):93.
228. Pan Y, Cheng A, Wang M, Yin Z, Jia R. The dual regulation of apoptosis by flaviviru. Front Microbiol (2021) 12:574.
229. Gamil AAA, Xu C, Mutoloki S, Evensen Ø. PKR activation favors infectious pancreatic necrosis virus replication in infected cells. Viruses (2016) 8(6):173.
230. Tan Z, Zhang W, Sun J, Fu Z, Ke X, Zheng C, et al. ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells. J Neuroinflammation (2018) 15(1):1–16.
231. Orvedahl A, MacPherson S, Sumpter R, Tallóczy Z, Zou Z, Levine B. Autophagy protects against sindbis virus infection of the central nervous system. Cell Host Microbe (2010) 7(2):115–27.
232. Akgul C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell Mo Life (2009) 66(8):1326–36.
233. Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-jun n-terminal kinases. Microbiol Mol Biol Rev (2006) 70(4):1061.
234. Shamas-Din A, Kale J, Leber B, Andrews DW. Mechanisms of action of bcl-2 family proteins. Cold Spring Harb Perspect Biol (2013) 5(4):1–21.
235. Venticinque L, Meruelo D. Sindbis viral vector induced apoptosis requires translational inhibition and signaling through mcl-1 and bak. Mol Cancer (2010) 9:1–16.
236. Joubert PE, Werneke SW, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L, et al. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J Exp Med (2012) 209(5):1029–47.
237. Alirezaei M, Kemball CC, Flynn CT, Wood MR, Whitton JL, Kiosses WB. Short-term fasting induces profound neuronal autophagy. Autophagy (2010) 6(6):702–10.
238. Yordy B, Iwasaki A. Autophagy in the control and pathogenesis of viral infection. Curr Opin Virol (2011) 1(3):196–203.
239. Beatman E, Oyer R, Shives KD, Hedman K, Brault AC, Tyler KL, et al. West Nile Virus growth is independent of autophagy activation. Virology (2012) 433(1):262–72.
240. Kobayashi S, Orba Y, Yamaguchi H, Takahashi K, Sasaki M, Hasebe R, et al. Autophagy inhibits viral genome replication and gene expression stages in West Nile virus infection. Virus Res (2014) 191(1):83–91.
241. Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, et al. Biology of zika virus infection in human skin cells. J Virol (2015) 89(17):8880–96.
242. Krejbich-Trotot P, Gay B, Li-Pat-Yuen G, Hoarau JJ, Jaffar-Bandjee MC, Briant L, et al. Chikungunya triggers an autophagic process which promotes viral replication. Virol J (2011) 8(1):1–10.
243. Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-Localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol (2009) 90(2):448–56.
244. Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol (2012).
245. Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol (2014) 6(8):a016972.
248. Navratil V, De Chassey B, Ne Meyniel L, Phane Delmotte S, Gautier C, André P, et al. VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res (2009) 37:661–8.
250. Huang M, Zhang W, Chen H, Zeng J. Targeting polyamine metabolism for control of human viral diseases. Infect Drug Resist (2020) 13:4335.
251. Bhutkar M, Ruchi R, Kothiala A, Mahajan S, Waghmode B, Kumar R, et al. Elucidation of the antiviral mechanism of natural therapeutic molecules herbacetin and caffeic acid phenethyl ester against chikungunya and dengue virus. bioRxiv (2022). doi: 10.1101/2022.05.31.494145v1
252. Thaker SK, Ch’ng J, Christofk HR. Viral hijacking of cellular metabolism. BMC Biol (2019) 17(1):59.
253. Girdhar K, Powis A, Raisingani A, Chrudinova M, Huang R, Tran T, et al. Viruses and metabolism: The effects of viral infections and viral insulins on host metabolism. Annu Rev Virol (2021) 8(1):373.
254. Fritsch SD, Weichhart T. Effects of interferons and viruses on metabolism. Front Immunol (2016) 7:630.
255. Goodacre N, Devkota P, Bae E, Wuchty S, Uetz P. Protein-protein interactions of human viruses. Semin Cell Dev Biol (2020) 99:31–9.
256. Byers NM, Fleshman AC, Perera R, Molins CR. Metabolomic insights into human arboviral infections: Dengue, chikungunya, and zika viruses. Viruses (2019) 11(3):225.
257. Findlay JS, Ulaeto D. Semliki forest virus and sindbis virus, but not vaccinia virus, require glycolysis for optimal replication. J Gen Virol (2015) 96(9):2693–6.
258. Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci U S A (1994) 91(22):10340–4.
259. Chatr-Aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, et al. VirusMINT: A viral protein interaction database. Nucleic Acids Res (2008) 37:669–73. www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions.
260. Sridharan A, Chen Q, Tang KF, Ooi EE, Hibberd ML, Chen J. Inhibition of megakaryocyte development in the bone marrow underlies dengue virus-induced thrombocytopenia in humanized mice. J Virol (2013) 87(21):11648–58.
261. Farfan-Morales CN, Cordero-Rivera CD, Reyes-Ruiz JM, Hurtado-Monzón AM, Osuna-Ramos JF, González-González AM, et al. Anti-flavivirus properties of lipid-lowering drugs. Front Physiol (2021) 12.
262. Zhang Z, He G, Filipowicz NA, Randall G, Belov GA, Kopek BG, et al. Host lipids in positive-strand RNA virus genome replication. Front Microbiol (2019) 10:286.
263. Brunton J, Steele S, Ziehr B, Moorman N, Kawula T. Feeding uninvited guests: MTOR and AMPK set the table for intracellular pathogens. PloS Pathog (2013) 9(10):e1003552.
264. Pant A, Dsouza L, Yang Z. Alteration in cellular signaling and metabolic reprogramming during viral infection. MBio (2021) 12(5):e00635-21.
265. Girdhar K, Powis A, Raisingani A, Chrudinová M, Huang R, Tran T, et al. Annual review of virology viruses and metabolism: The effects of viral infections and viral insulins on host metabolism. Annual review of virology (2021) 8(1):373. doi: 10.1146/annurev-virology-091919-
266. Mahajan S, Choudhary S, Kumar P, Tomar S. Antiviral strategies targeting host factors and mechanisms obliging +ssRNA viral pathogens. Bioorg Med Chem (2021) 46:116356.
268. Yates JR, Yates III JR. A century of mass spectrometry: from atoms to proteomes. Nat Methods (2011) 8(8):633–7.
269. Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem (2001) 70:437–73.
271. Kang SM, Shin MJ, Ki JH, Oh JW. Proteomic profiling of cellular proteins interacting with the hepatitis c virus core protein. Proteomics (2005) 5(8):2227–37. doi: 10.1002/pmic.200401093
272. Wongtrakul J, Thongtan T, Pannengpetch S, Wikan N, Kantamala D, Kumrapich B, et al. Phosphoproteomic analysis of dengue virus infected U937 cells and identification of pyruvate kinase M2 as a differentially phosphorylated phosphoprotein. Sci Rep (2020) 10(1):1–13.
273. Lapetina S, Gil-Henn H. A guide to simple, direct, and quantitative in vitro binding assays. J Biol Methods (2017) 4(1):62.
274. Ewing R, Sorokina O, Roux KJ, Nita-Lazar A, Gillen J. Experimental analysis of viral-host interactions. Front Physiol (2019) 10:425.
275. Zhang T, Xie N, He W, Liu R, Lei Y, Chen Y, et al. An integrated proteomics and bioinformatics analyses of hepatitis b virus X interacting proteins and identification of a novel interactor apoA-I. J Proteomics (2013) 84:92–105. doi: 10.1016/j.jprot.2013.03.028
276. Gao Y, Goonawardane N, Ward J, Tuplin A, Harris M. Multiple roles of the non-structural protein 3 (nsP3) alphavirus unique domain (AUD) during chikungunya virus genome replication and transcription. PloS Pathog (2019) 15(1):e1007239. doi: 10.1371/journal.ppat.1007239
277. Ghildiyal R, Gabrani R. Deciphering the human cellular interactors of alphavirus unique domain of chikungunya virus. Virus Res (2021) 295:198288.
278. Le Breton M, Meyniel-Schicklin L, Deloire A, Coutard B, Canard B, De Lamballerie X, et al. Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol (2011) 11:1–11.
279. Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Brief Funct Genomic Proteomic (2007) 6(4):302–12.
280. Stagljar I, Korostensky C, Johnsson N, Te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci U S A (1998) 95(9):5187–92.
281. Graves PR, Haystead TAJ. Molecular biologist’s guide to proteomics. Microbiol Mol Biol Rev (2002) 66(1):39–63. doi: 10.1128/MMBR.66.1.39-63.2002
282. Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol (2008) 11(4):369–77.
283. Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, et al. Conformational changes of the flavivirus e glycoprotein. Structure (2004) 12(9):1607–18.
284. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature (2004) 427(6972):313–9.
285. Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol (2006) 13(4):372–3.
286. Manjasetty BA, Turnbull AP, Panjikar S. The impact of structural proteomics on biotechnology. Biotechnol Genet Eng Rev (2010) 26(1):353–70.
287. Manjasetty BA, Büssow K, Panjikar S, Turnbull AP. Current methods in structural proteomics and its applications in biological sciences. 3 Biotech (2012) 2(2):89.
288. Yang S, Fu C, Lian X, Dong X, Zhang Z, Cristea IM. Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems (2019) 4(2):e00303–18.
289. Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PloS Pathog (2008) 4(2):e32.
290. Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res (2000) 28(1):289–91.
291. Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci (2021) 30(1):187.
292. del Toro N, Shrivastava A, Ragueneau E, Meldal B, Combe C, Barrera E, et al. The IntAct database: efficient access to fine-grained molecular interaction data. Nucleic Acids Res (2022) 50(D1):D648–53.
293. Bader GD, Donaldson I, Wolting C, Ouellette BFF, Pawson T, Hogue CWV. BIND–the biomolecular interaction network database. Nucleic Acids Res (2001) 29(1):242.
294. Ammari MG, Gresham CR, McCarthy FM, Nanduri B. HPIDB 2.0: a curated database for host-pathogen interactions. Database (Oxford) (2016) 2016.
295. Karyala P, Metri R, Bathula C, Yelamanchi SK, Sahoo L, Arjunan S, et al. DenHunt - a comprehensive database of the intricate network of dengue-human interactions. PloS Negl Trop Dis (2016) 10(9):e0004965.
296. Zhou Y, Chen H, Li S, Chen M. MPPI: A database extension to visualize structural interactome in a one-to-many manner. Database (2021) 2021.
297. Chatr-Aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, et al. VirusMINT: A viral protein interaction database. Nucleic Acids Res (2009) 37(SUPPL. 1):669–73.
298. Cook HV, Doncheva NT, Szklarczyk D, von Mering C, Jensen LJ. Viruses.STRING: A virus-host protein-protein interaction database. Viruses (2018) 10(10).
299. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res (2014) 43:447–52.
300. Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature (2012) 487(7408):486–490.
301. Holmes EC. The evolutionary genetics of emerging viruses. Annu Rev Ecol Evol Syst (2009) 40:353–72.
302. Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, et al. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. (2003) 20(10):1650–1658
303. Kondrashov FA, Kondrashov AS. Measurements of spontaneous rates of mutations in the recent past and the near future. J Philos Trans R Soc Lond B Biol Sci. (2010) 365(1544):1169–1176
304. Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A (2012) 109(41):E2774–E2783. doi: 10.1073/pnas.1210309109
305. Walsh D, Mathews MB, Mohr I. Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol (2013) 5(1):a012351.
306. Pérez-Pérez MJ, Delang L, Ng LFP, Priego EM. Chikungunya virus drug discovery: still a long way to go? Expert Opin Drug Discovery (2019) 14(9):855–66.
307. Varghese FS, Rausalu K, Hakanen M, Saul S, Kümmerer BM, Susi P, et al. Obatoclax inhibits alphavirus membrane fusion by neutralizing the acidic environment of endocytic compartments. Antimicrob Agents Chemother (2017) 61(3):e02227–16. doi: 10.1128/AAC.02227-16
308. Varghese FS, Kaukinen P, Gläsker S, Bespalov M, Hanski L, Wennerberg K, et al. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Res (2016) 126:117–24.
309. Bakhache W, Neyret A, McKellar J, Clop C, Bernard E, Weger-Lucarelli J, et al. Fatty acid synthase and stearoyl-CoA desaturase-1 are conserved druggable cofactors of old world alphavirus genome replication. Antiviral Res (2019) 172:172.
310. Hitakarun A, Khongwichit S, Wikan N, Roytrakul S, Yoksan S, Rajakam S, et al. Evaluation of the antiviral activity of orlistat (tetrahydrolipstatin) against dengue virus, Japanese encephalitis virus, zika virus and chikungunya virus. Sci Rep 10(1).
311. Tampere M, Pettke A, Salata C, Wallner O, Koolmeister T, Cazares-Körner A, et al. Novel broad-spectrum antiviral inhibitors targeting host factors essential for replication of pathogenic RNA viruses. Viruses (2020) 12:1423. www.mdpi.com/journal/viruses.
312. Wang Y, Jin F, Wang R, Li F, Wu Y, Kitazato K, et al. HSP90: a promising broad-spectrum antiviral drug target. Arch Virol (2017) 162(11):3269–82. doi: 10.1007/s00705-017-3511-1
313. Rathore APS, Haystead T, Das PK, Merits A, Ng ML, Vasudevan SG. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo.Antiviral Res (2014) 103(1):7–16.
314. Yap TL, Xu T, Chen Y-L, Malet H, Egloff M-P, Canard B, et al. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol (2007) 81(9):4753–65. doi: 10.1128/JVI.02283-06
Keywords: arboviruses, protein protein interaction (PPI), host factors, broad spectrum antivirals, host immune response
Citation: Bhutkar M, Singh V, Dhaka P and Tomar S (2022) Virus-host protein-protein interactions as molecular drug targets for arboviral infections. Front. Virol. 2:959586. doi: 10.3389/fviro.2022.959586
Received: 01 June 2022; Accepted: 18 July 2022;
Published: 04 August 2022.
Edited by:
Sujatha Sunil, International Centre for Genetic Engineering and Biotechnology, IndiaReviewed by:
Punit Kaur, All India Institute of Medical Sciences, IndiaAshok Varma, Research and Education in Cancer, India
Manisha Yadav, University of Delhi, India
Copyright © 2022 Bhutkar, Singh, Dhaka and Tomar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shailly Tomar, c2hhaWxseS50b21hckBidC5paXRyLmFjLmlu
 Mandar Bhutkar
Mandar Bhutkar Shailly Tomar
Shailly Tomar