- 1Office of Clinical Evaluation of Biological Products, Center for Drug Evaluation, Beijing, China
- 2Division of Hepatitis and Enterovirus Vaccines, Institute of Biological Products, National Institutes for Food and Drug Control, Beijing, China
- 3Center for Food and Drug Inspection, Beijing, China
- 4Division of Respiratory Virus Vaccines, Institute of Biological Products, National Institutes for Food and Drug Control, Beijing, China
Mumps, a disease caused by the mumps virus (MuV), has been spread widely across the world, especially among children and adolescents. Recent frequent local mumps outbreaks were reported worldwide, which may be caused by the decline in the neutralization ability of the existing attenuated live mumps vaccines against circulating MuV strains which were different from the genotype A or B vaccine strains. There is an urgent need to understand the genotypes of MuV strains currently circulated globally and in China. The gene sequences of MuV strains circulated globally were collected and phylogenetic trees were constructed using different strategies. The results showed that the MuV strains previously circulated globally were predominantly genotype G, while genotype F was predominantly circulated in China, followed by genotype G. The molecular evolution of genotype F MuV strains circulated in China is at a low genetic mutation rate, and the analysis of population dynamics pattern indicates that the incidence of genotype F mumps in China showed a rebound trend. These findings provide a basis for the selection or design of vaccine strains, and the decision of the evaluation strains for immunogenicity and protective efficacy, which laid the foundation for the research and development, as well as the application of next-generation MuV vaccines.
Introduction
Mumps is an acute respiratory infection caused by the mumps virus (MuV) that mainly spreads among children and adolescents. The main clinical symptoms include swelling of the parotid glands, pain, and fever. Mumps may be complicated by pancreatitis, orchitis, deafness, aseptic meningitis, and encephalitis (1, 2). Prior to the mumps vaccination program, mumps was highly prevalent worldwide, resulting in high morbidity and mortality (3). The first inactivated mumps vaccine was approved in the United States in 1948 and was used from 1950, but it was later discontinued due to its short-term immunity of low protective efficacy (4, 5). It was not until the 1960s that live attenuated mumps vaccines were approved for use in the United States and the Soviet Union (1, 6). The vaccine strains used in these two countries were the Jeryl Lynn and Leningrad-3 strains, respectively. Since then, several virus strains have been used worldwide for the production of mumps vaccines, with the Jeryl Lynn and Urabe Am9 strains being the most widely used (6, 7). The mumps-containing vaccines currently used in China include the monovalent live attenuated mumps vaccine and the measles-mumps-rubella (MMR) vaccine, with the virus strain S79 from Jeryl Lynn, which is geontype A (8). The total lot release number of mumps-containing vaccines were 18.5 million doses in China during 2015~2019. The mumps strains currently licensed for use in mumps-containing vaccines are all derived from strains that were prevalent in the mid-twentieth century, which are mostly genotype A or B. After the approval of the live attenuated mumps vaccine, the good immunogenicity and widespread vaccination have been effective in reducing the incidence of mumps (9).
The earliest report of MuV in China was a strain detected in specimens from 1995 to 1996, which were identified as genotype F (10). In the following two decades, genotype F was the predominant genotype in China (11, 12). Since the implementation of infectious disease surveillance in China in 2004, the average annual incidence of mumps has been 24/100,000 population (13). Although the MMR vaccine was introduced in the Expanded Program on Immunization (EPI) in 2008, local outbreaks among the vaccinated population have been reported each year in China (14). In recent years, several large mumps outbreaks have been reported in developed countries, including Canada, Australia, the United Kingdom, France, and the United States, where genotype G is the predominant strain (15–23). In 2016 and 2017, the number of mumps cases in the United States nearly tripled compared with the number of cases reported in the past five years (3).
Although there is only one serotype of MuV, there are 12 genotypes, and the antigenicity of MuV varies between genotypes. Decreased cross-neutralizing capacity of the current vaccine against the prevalent strains with different MuV genotypes has been suggested as a possible explanation for mumps outbreaks in highly vaccinated populations (24–26). Won et al. measured the cross-neutralization of sera immunized with the mumps vaccine against the MuV F, H, and I wild-type strains circulated in Republic of Korea from 1998 to 2016, and the results showed that the potency of the neutralizing antibody in the sera immunized with vaccine against genotypes F, H, and I was significantly lower than that against genotype A (27). A growing number of research indicates the urged need to develop next-generation mumps vaccines against the circulating genotypes (28, 29).
The molecular epidemiological data of viruses are the basis for the screening of strains for vaccine production and efficacy evaluation. With the increasing application of sequencing technology, more knowledge has been obtained on the different genotypes of MuV (7). The small hydrophobic (SH) gene has the highest number of sequence variations, making it commonly used for the standardized nomenclature of MuV (30). In 2012, the World Health Organization (WHO) updated and assigned standard names for the 12 MuV genotypes by the letters A to N (excluding E and M) based on the SH and haemagglutinin-neuraminidase (HN) sequences of the MuV genome. Recent findings suggested that the analysis of the SH gene sequences in combination with sequence data from the non-coding region (NCR) of the genome was a useful approach for improving genotyping resolution (31). In this study, gene sequences of MuV strains circulated globally and in China were collected and analyzed using bioinformatics methods. As the basis for the selection or design of vaccine strains, and the decision of the evaluation strains for immunogenicity and protective efficacy, the molecular epidemiological characteristics of MuV strains would lay the foundation for the research and development (R&D) of next-generation MuV vaccines.
Materials and Methods
Data Screening
The MuV gene sequences were searched and downloaded from the Genbank database (https://www.ncbi.nlm.nih.gov/genbank/). PhyloSuite_v1.2.1 software was used to extract a total of 12,601 sequences, of which 932 complete genome sequences were selected. After aligning the sequences using mafft-7.037, the online tool CD-HIT Suite (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit-est) was used to remove the redundant sequences (the cut-off of similarity was set as 99.7%) to obtain 111 complete genome sequences. SH, nucleocapsid protein and phosphoprotein (N-P), phosphoprotein and matrix protein (P-M), and matrix and F protein (M-F) genes were selected respectively, and concatenated using the PhyloSuite_v1.2.1 software. These procedures were performed in order to create a dataset of SH genes and the non-coding regions (NCR) fragments of MuV strains circulated worldwide. In this study, the NCR segment from Bodewes et al. was used as the reference sequence to confirm the length of the intercepted segment (a total of 1954 nt) (31).
The sequences of MuV strains circulated in China were searched and downloaded from the Genbank database. The data were screened to obtain 418 SH gene sequences that provided information on isolation time and geographical location. A phylogenetic tree based on the maximum likelihood (ML) estimation was constructed using iqtree-1.6.2 software. For the SH genes of the circulated genotype F strains, the online tool CD-HIT Suite was applied to remove the redundant sequences and build the SH gene dataset of genotype F MuV strains circulated in China, which contained 252 sequences.
Genotyping of MuV Strains Circulated Worldwide
The SH gene sequences of representative strains of a total of 12 genotypes, namely A, B, C, D, F, G, H, I, J, K, L, and N, from a study by Cheng et al. were used as reference sequences during genotyping (32). The reference sequences were combined with the SH gene (316 nt) dataset of the MuV strains circulated worldwide. Iqtree-1.6.2 software was used to construct the ML phylogenetic tree with bootstrap values set to 2000, while the nucleotide substitution model was determined as TVM+F+G4, which was calculated by this software. Then FigTree.v1.4.4 and Adobe Illustrator CC 2019 were used to visualize the ML phylogenetic tree. Based on the result from the SH gene phylogenetic tree, the NCR segment (1954 nt) dataset was used to construct the ML phylogenetic tree with bootstrap values set to 2000, while the nucleotide substitution model was determined as GTR+F+R3.
Genotyping of MuV Strains Circulated in Mainland China
Currently, there are relatively few complete genome data of MuV in China. Emphasis was placed on the SH gene sequences to study the genetic characteristics of mumps virus sequences in China. The gene sequences of representative strains of a total of 12 genotypes, namely A, B, C, D, F, G, H, I, J, K, L, and N, from a study by Cheng et al. were used as reference sequences during genotyping (32). The reference sequences were combined with almost complete SH genes (313 bp) of 418 MuV strains epidemic in China. Iqtree-1.6.2 software was used to construct the ML phylogenetic tree with the bootstrap value set to 2000, while the nucleotide substitution model was determined as K3Pu+F+G4.
Phylogenetic Tree Using Molecular Clocks for Genotype F Strains Circulated in China
A cluster-based Bayesian Markov chain Monte Carlo (MCMC) approach was used to construct the Maximum Clade Credibility (MCC) tree using BEAST.v1.10.4 based on the SH gene dataset of the genotype F MuV strains circulated in China, which contained 252 sequences. Breifly, jModeltest software was used to select GTR+G+I as the optimal nucleotide substitution model. Then the uncorrelated lognormal relaxed clock and the optimal a priori model (Bayesian Skyline) was selected through the Model comparison function in Tracer.v1.7.1. Province was used as a trait, the chain length was set as 200,000,000, sampling frequency was set as 20,000, and three duplicate chains were set up. LogCombiner v1.10.4 in the BEAST.v1.10.4 package was used to combine the log files and tree files separately. For the combined log file, the first 10% of aged samples were discarded, and the remaining samples were checked if there were sufficient effective sample size (ESS) values (> 200) for all estimated parameters to converge using Tracer. The TreeAnnotator v1.10.4 program in the BEAST.v1.10.4 package was then used to discard the first 10% of the combined tree to generate the final MCC tree, and the node height of the tree was set as the median. MEGA 7.0.20 software was used to calculate the intra- and inter-group evolutionary distances for the different lineages of genotype F strains.
Population Dynamics Study of Genotype F Strains Circulating in China
BEAST.v1.10.4 software was used based on the above optimal nucleotide substitution model GTR+G+I, the uncorrelated lognormal relaxed clock, and the optimal a priori model Bayesian Skyline. The chain length was set as 200,000,000, sampling frequency was set as 20,000, and three duplicate chains were set up. LogCombiner v1.10.4 in the BEAST.v1.10.4 package was used to combine the log files and tree files separately. For the combined log file, the first 10% of aged samples were discarded, and the remaining samples were checked if there were sufficient ESS values (> 200) for all estimated parameters to converge using Tracer. The Bayesian skyline plot (BSP) was made using the Tracer.v1.7.1 software. Mumps incidence data in 2008-2019 were obtained from the National Notifiable Diseases Reporting System (NNDRS) of China (http://www.nhc.gov.cn/jkj/s2907/new_list.shtml?tdsourcetag=s_pcqq_aiomsg).
As is shown in Figure 1, an analysis flow was depicted briefly.
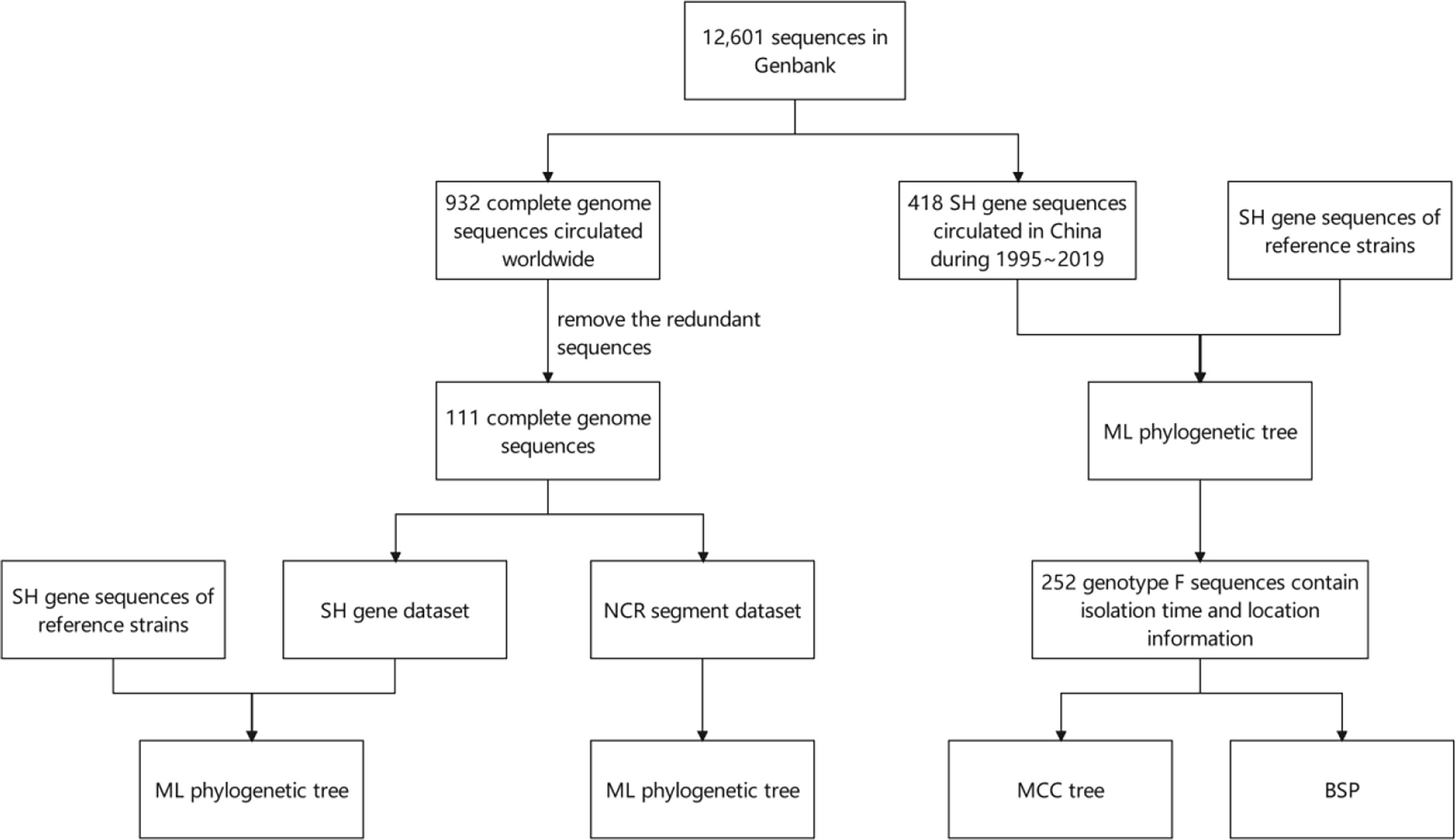
Figure 1 Schematic diagram of the analysis flow. SH, small hydrophobic; NCR, non-coding region; ML, maximum likelihood; MCC, Maximum Clade Credibility; BSP, Bayesian skyline plot.
Results
Genotyping of MuV Strains Circulated Worldwide
From the ML phylogenetic analysis of the SH gene of the 111 selected MuV strains collected worldwide since 1970 (Figure 2), there was a total of 12 genotypes circulated worldwide, namely genotypes A, B, C, D, F, G, H, I, J, K, L and N. The circulating MuV strains after 2010 included A, C, F, G, H, I, and K, with genotype G being the most prevalent. Among these, the genotype A strain (Genbank no. KX223397) was special in that it was obtained from a brain tissue specimen of a child who died of chronic encephalitis in 2014 and the strain belonged to the Jeryl Lynn 5 (JL5)-like strain of the MMR vaccine (33). The genotype G strain was widespread and has been reported in the Americas (USA and Canada), Europe (Netherlands), Australia (Australia and New Zealand), Asia (India and Japan), and Africa (Gabon) between 2010 and 2019. The genotype F strain was predominantly prevalent in China. In addition, it was also reported in Republic of Korea in 2012 (Genbank no. MN630056). In addition to G and F strains, genotype C strain had been reported in India/Canada/the Netherlands, genotype H in Republic of Korea and the United States, genotype I in South Korea, and genotype K in the United States and the Netherlands.
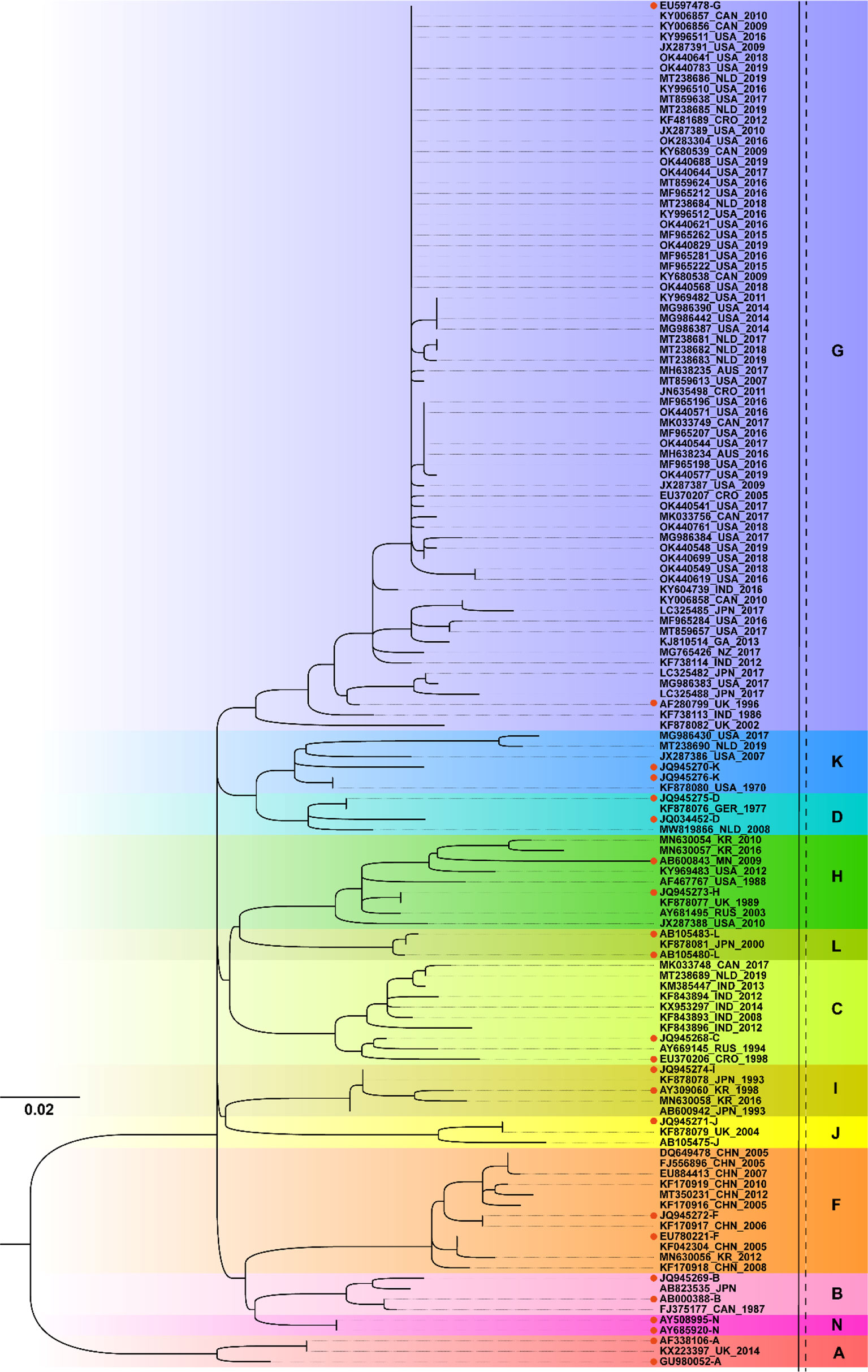
Figure 2 Phylogenetic analysis of mumps strains circulated worldwide based on 316 nucleotides of the SH gene, with reference strains of 12 genotypes (A, B, C, D, F, G, H, I, J, K, L, and N). The nucleotide substitution model used for the phylogenetic tree was TVM+F+G4, and the bootstrap value was set to 2000. • marks the reference sequences of the 12 genotypes; AUS, Australia; CAN, Canada; CHN, China; CRO, Croatia; GER, Germany; GA, Gabon; IND, India; JPN, Japan; KR, Republic of Korea; MN, Mongolia; NLD, Netherlands; NZ, New Zealand; RUS, Russia; UK, United Kingdom; USA, United States.
Figure 3 showed the ML phylogenetic tree constructed based on the NCR segment sequences. Similarly, the MuV strains circulated worldwide can be classified into a total of 11 genotypes, namely A, B, C, D, F, G, H, I, J, K, and L, based on the NCR gene. It was presumed that the complete genome sequence of the genotype N strains was not included in the database. The prevalent MuV strains after 2010 included genotypes A, C, F, G, H, I, and K, with genotype G being the most prevalent. The result was consistent with SH gene phylogenetic analysis. From the results obtained both with the SH and NCR sequences, the genotype G was the prevalent circulating MuV strains worldwide, while genotype F was the major strain in China.
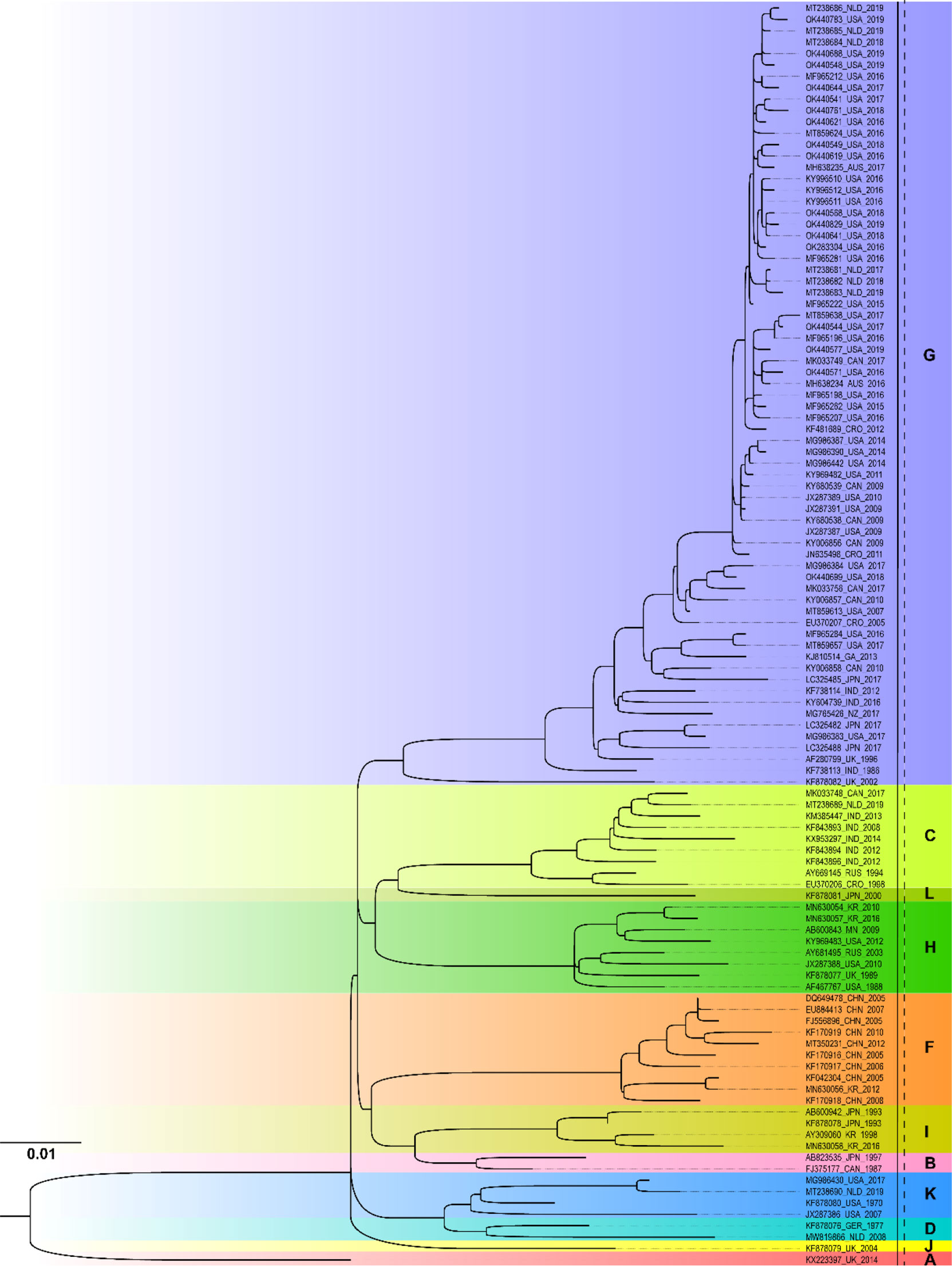
Figure 3 Phylogenetic analysis of mumps strains circulated worldwide based on 1954 nucleotides of the NCR sequence. The nucleotide substitution model used in this phylogenetic tree was GTR+F+R3, and the bootstrap value was set as 2000. AUS, Australia; CAN, Canada; CHN, China; CRO, Croatia; GER, Germany; GA, Gabon; IND, India; JPN, Japan; KR, Republic of Korea; MN, Mongolia NLD, Netherlands; NZ, New Zealand; RUS, Russia; UK, United Kingdom; USA, United States.
Genotyping of MuV Strains Circulated in China
To investigate the molecular epidemiological patterns during the spread history in China, 418 SH gene sequences of MuV reported in 22 different provinces/municipalities in China from 1995 to 2019 were obtained. An ML phylogenetic tree was constructed based on the SH gene, and the results were shown in Figure 4. The predominant strains circulated in China were genotype F, including 406 sequences from East China (Anhui, Jiangsu, Shandong, Shanghai, Zhejiang, and Jiangxi), North China (Beijing, Inner Mongolia, and Shanxi), South China (Guangdong and Guangxi), Central China (Henan, Hubei, and Hunan), Southwest China (Sichuan and Yunnan), Northeast China (Heilongjiang, Jilin, and Liaoning), and Northwest China (Gansu, Ningxia, and Shaanxi). In addition, genotype G was prevalent in Shaanxi, Liaoning, and Fujian in 2011, as well as in Gansu in 2013. A genotype K strain imported from Vietnam was reported in Guangxi in 2016 (34). These results indicated that, the genotypes of the main MuV strains currently prevalent in China are significantly different from those worldwide, however, there was also a risk of imported transmission.
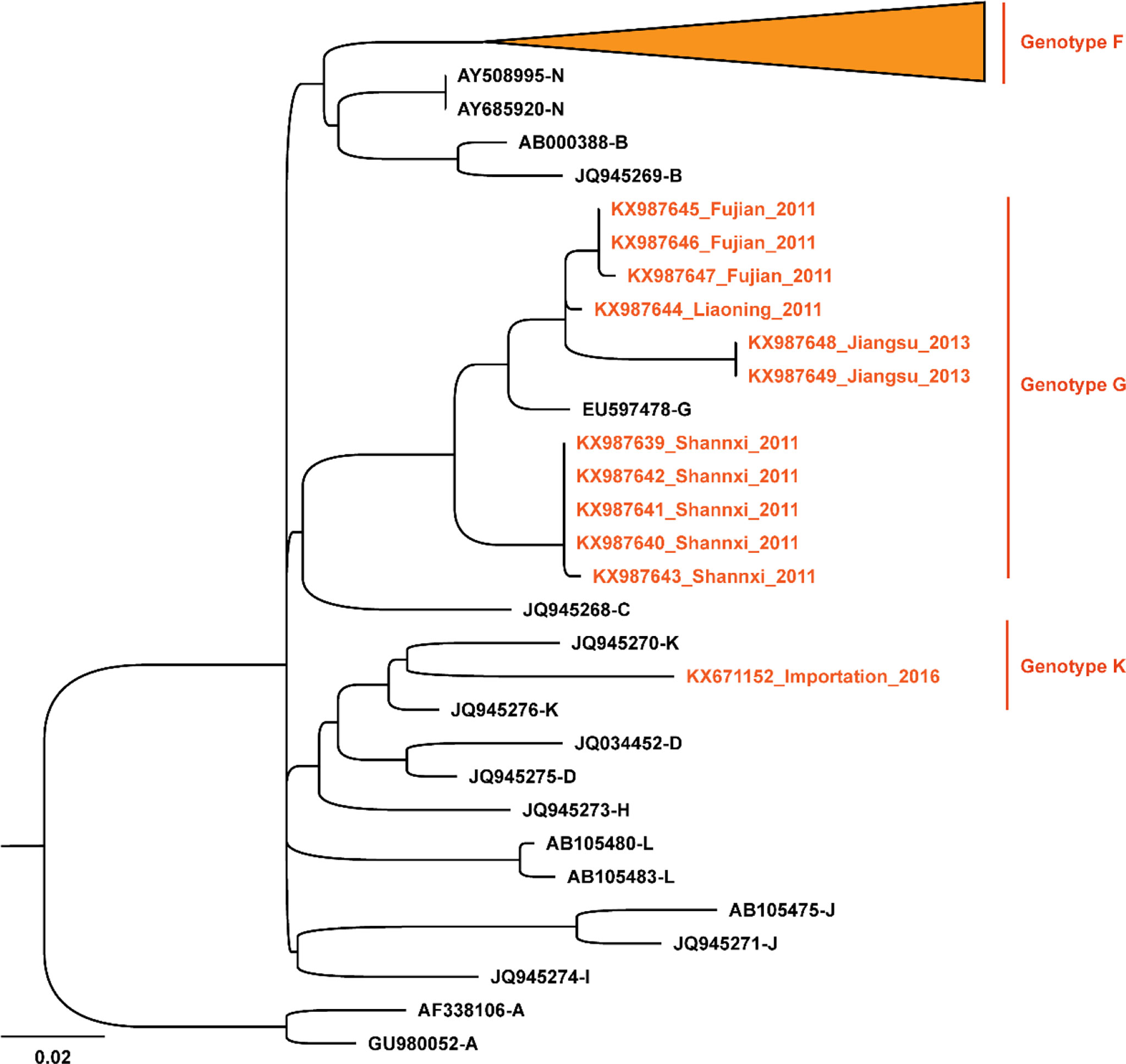
Figure 4 Phylogenetic analysis of mumps strains circulated in China based on 313 nucleotides of the SH gene, with reference strains of 12 genotypes (A, B, C, D, F, G, H, I, J, K, L, and N). The nucleotide substitution model used in this phylogenetic tree was K3Pu+F+G4, and the bootstrap value was set as 2000. The orange font and the sequences marked by the orange ▴ symbols are the strains circulated in China.
Evolutionary Origin of Genotype F Strains Circulated in China
To elucidate the evolution of MuV in China, the SH gene dataset of the genotype F strains circulating from 1995 to 2019 were analyzed. As shown in Figure 5, the average base substitution rate of the SH genes of genotype F MuV strains circulated in China was 2.02 × 10-3/site/year (95% highest posterior density (HPD) interval 1.68-2.40 × 10-3/site/year) between 1995 and 2019, and the most recent common ancestor (tMRCA) of genotype F MuV strains in China could be traced back to the year 1989.69 (95% HPD interval of 1979.47~1998.37). Using province as trait, the root node of the entire phylogenetic tree was Shanghai, with a posterior probability of 0.623. It might been speculated that the genotype F strains which persistently circulated in China originated in Shanghai around 1989, and then gradually spread to other regions.
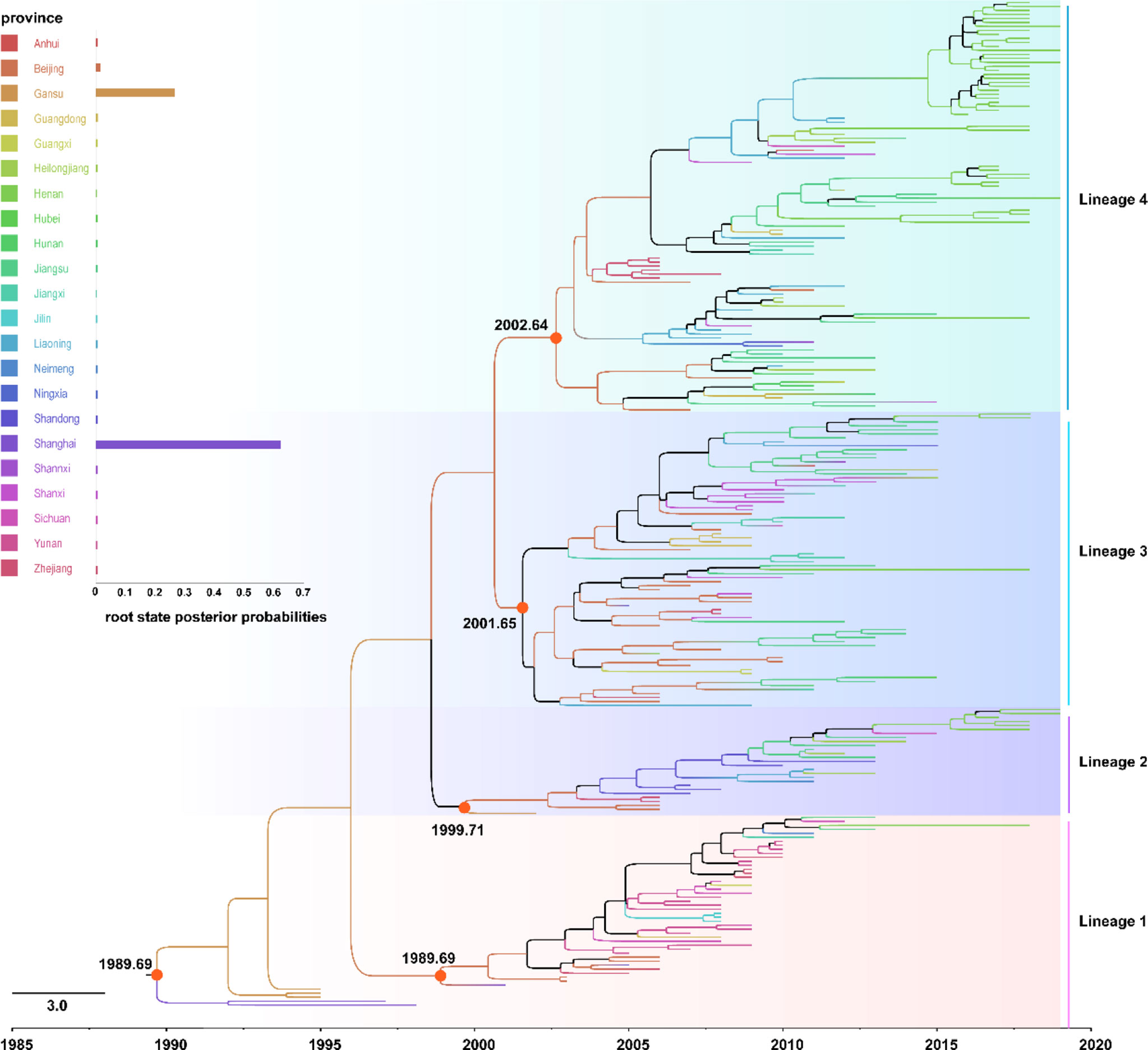
Figure 5 The Maximum Clade Credibility tree of genotype F strains circulated in China with geographic information based on 313 nucleotides of the SH gene.
The genotype F strains circulated in China were clustered into four lineages in the MCC tree, and namely lineages 1-4, respectively. The within group mean distance of the four lineages were calculated using MEGA7.0.20 software as 0.017, 0.019, 0.031, and 0.029, respectively, while the between group mean distance were between 0.036 and 0.049. The root nodes of the major circulated strains of lineages 2-4 and lineage 1 were between 1989.69 and 2002.64 years, with all of these still circulating after 2010. There were no significant differences in the timing and geographic location of transmission between the strains in different lineages. These results indicated that the genetic distance of genotype F MuV circulated in China was relatively close, and the strains of different lineages might be similar in biological manifestation, antigenicity and other characteristics.
Population Dynamic Patterns of Genotype F Strains in China
From the BSP result shown in Figure 6, there were two significant population expansions of genotype F MuV strains in China between 1995 and 2019. The population expansion event that started in 1995 experienced a rapid expansion period during 1995-2005 and a slow expansion period between 2005 and 2011, followed by population contraction period in 2014 -2015. After a second rapid expansion period in 2016, the population size stabilized from 2017 to 2019, and the effective population size (Ne) remained at a high level.
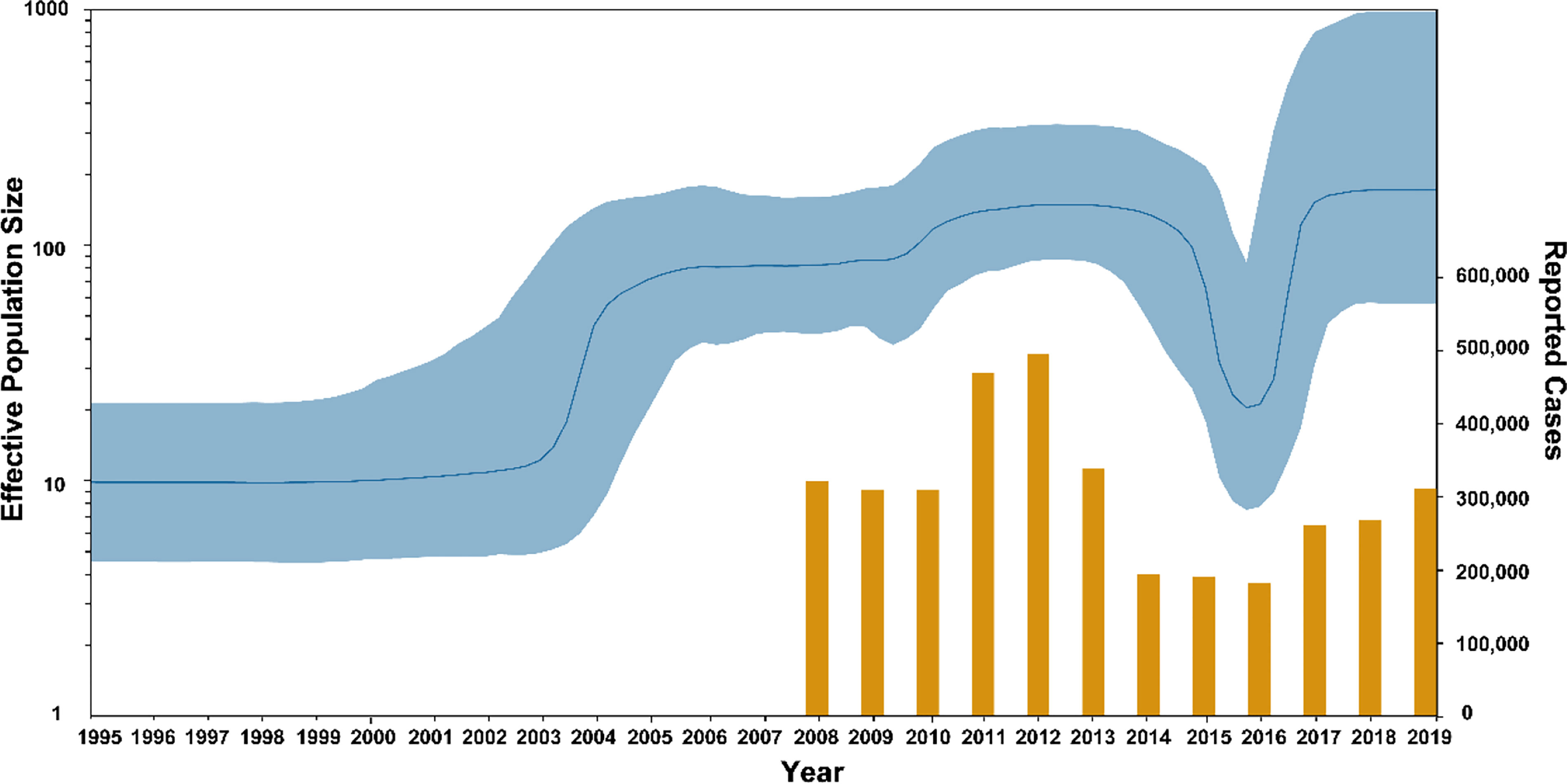
Figure 6 The population dynamics of MuV genotype F circulated in China. Yellow bars indicate the number of mumps cases reported by National Notifiable Diseases Reporting System.
Statistical analysis was performed on the number of mumps cases reported in the NNDRS of China from 2008 to 2019, and the average number were 293,988 cases/year (35). The results show that the number of annual cases peaked in 2011 and 2012 (454,400 and 479,500 cases), after which the number of cases declined and remained at a low level from 2014 to 2016, followed by a rebound trend in 2017-2019. The result was consistent with the trend in the BSP. The above results indicated that the incidence of mumps in China is currently on the rise, which means that strengthening mumps surveillance is necessary, along with research on the immunogenicity and effectiveness of the mumps vaccine.
Discussion
The mumps vaccine has significantly reduced the incidence of mumps in the past 60 years (9, 36). However, there have been frequent mumps outbreaks among populations with high rates of vaccination in recent years. Although studies have found that antibody responses induced by one or two doses of mumps vaccine in children provides protection for at least 10 years (37), as time passes, the protection provided by the vaccine decreases and leads to local outbreaks of mumps (21, 38). A third vaccine dose has been considered as a positive strategy to prevent recurrence (39), whereas the efficacy of the current vaccine against different genotypes of MuV has decline (24–26). Besides, the threshold potency of neutralizing antibodies required for the mumps vaccine to induce immunity remains unclear (40). These findings suggest that the cross-neutralizing and protective efficacy of mumps vaccine using genotype A or B strains against currently prevalent MuV genotypes should be evaluated, and that the R&D of a next-generation MuV vaccines should be promoted.
In recent years, genotype G strains have mainly circulated in Europe, the United States, Australia, and New Zealand, while genotype C strains have mainly circulated in India. Genotype F strains were prevalent mainly in China, in addition to being reported in ten other countries in North America, Europe, and Asia (41). In this study, strains circulated worldwide were analyzed based on the SH gene and NCR fragment sequences. It was found that the circulated strains reported after 2010 included genotypes C, F, G, H, I, and K. Among them, genotype G was more widely distributed, while genotype F strains were prevalent in China, which were significantly different from the genotypes of the current vaccine strains. The fact that multiple genotypes strains co-circulated worldwide makes the development and evaluation more difficult.
Xu et al. showed that a single dose of MuV had a cross-protective efficacy against genotype F MuV. However, the protection was limited, which might be due to insufficient doses of MuCV vaccine and genotype-specific differences in antigens between different MuV strains (42). A clinical study of an F-genotype live attenuated mumps vaccine found that this vaccine was safe, and its immunogenicity against homologous viruses was not inferior to previous A genotype live attenuated mumps vaccine (43). Although genotype F has been the dominant MuV circulated in China, there have been local mumps outbreaks caused by genotype G strains in recent years. In addition, imported genotype K MuV strains have also been reported (34). Therefore, a novel mumps vaccine induced broad-spectrum cross-protective antibody immunity against major genotypes such as F and G should be the target of development (14, 44).
The COVID-19 pandemic began in the end of 2019 and spread around the world. In the past two years, a variety of preventive and control measures have been taken in China, including wearing masks, restricting the flow of people in public spaces, and adopting dynamic zeroing measures for epidemic areas. These measures have had a significant impact on mumps, which is also caused by a respiratory pathogen. In the report from NNDRS in 2020 and 2021, the number of mumps cases reported in China fell to 129,120 and 97,300 cases, respectively, which were the lowest in the last decade or so. However, with the possible upcoming changes to COVID-19 pandemic, after the prevention and control measures are lifted, respiratory diseases such as mumps may still rebound and the dominant strains may change. We may learn from the COVID-19 prevention and control strategies and strengthen the epidemiological surveillance, especially the molecular epidemiological surveillance of mumps. In addition, new technologies and platforms have been widely used in the development of COVID-19 vaccines, which has accelerated vaccine development (45). The use of these technologies and platforms in the novel mumps vaccine in mumps vaccine should be promoted. All of these measures will contribute to the inhibition of MuV transmission as well as local outbreaks of mumps, and facilitate the R&D of novel mumps vaccines.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
LB and ZL designed and supervised the study. YS and JL co-drafted the article. YS, LB, and MCL did the literature and sequence data search, set up the database and did all statistical analysis and figures. LB, QW and ZL designed and supervised the study. All authors contributed to review and revision and approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Betáková T, Svetlíková D, Gocník M. Overview of Measles and Mumps Vaccine: Origin, Present, and Future of Vaccine Production. Acta Virol (2013) 57(2):91–6. doi: 10.4149/av_2013_02_91
2. Rubin S, Kennedy R, Poland G. Emerging Mumps Infection. Pediatr Infect Dis J (2016) 35(7):799–801. doi: 10.1097/INF.0000000000001182
3. Marlow MA, Marin M, Moore K, Patel M. CDC Guidance for Use of a Third Dose of MMR Vaccine During Mumps Outbreaks. J Public Health Manag Pract (2020) 26(2):109–15. doi: 10.1097/PHH.0000000000000962
4. Hviid A, Rubin S, Mühlemann K. Mumps. Lancet (2008) 371(9616):932–44. doi: 10.1016/S0140-6736(08)60419-5
5. World Health Organization. Weekly Epidemiological Record (2001). Available at: https://apps.who.int/iris/handle/10665/231692?show=full (Accessed 16 June, 2016).
6. Kowalzik F, Faber J, Knuf M. MMR and MMRV Vaccines. Vaccine (2018) 36(36):5402–7. doi: 10.1016/j.vaccine.2017.07.051
7. Jin L, Örvell C, Myers R, Rota PA, Nakayama T, Forcic D, et al. Genomic Diversity of Mumps Virus and Global Distribution of the 12 Genotypes. Rev Med Virol (2015) 25(2):85–101. doi: 10.1002/rmv.1819
8. National Medical Products Administration. Chinese Family Medicine Handbook (Vaccines and Immunizations) (2019). Available at: https://www.nmpa.gov.cn/xxgk/kpzhsh/kpzhshyp/20191105120001286.html (Accessed 05 November, 2019).
9. World Health Organization. Mumps Virus Nomenclature Update: 2012 = Nomenclature Des Virus Ourliens: Mise À Jour 2012. Weekly Epidemiol Rec = Relevé épidémiologique hebdomadaire (2012) 8 Available at: https://apps.who.int/iris/handle/10665/241922?locale-attribute=en&. (Accessed 1 June, 2012.
10. Wu L, Bai Z, Li Y, Rima BK, Afzal MA. Wild Type Mumps Viruses Circulating in China Establish a New Genotype. Vaccine (1998) 16(2):281–5. doi: 10.1016/s0264-410x(97)00173-4
11. Cui A, Zhu Z, Chen M, Zheng H, Liu L, Wang Y, et al. Epidemiologic and Genetic Characteristics of Mumps Viruses Isolated in China From 1995 to 2010. Infect Genet Evol (2014) 21:384–90. doi: 10.1016/j.meegid.2013.12.005
12. Cui A, Zhu Z, Zhang Y, Qiu Q, Mao N, Xu S, et al. Viral Surveillance and Genetic Characteristics of Mumps Viruses in China During 2001-2017. Chin J Vaccines Immunization. (2018) 24(3):249–55.
13. Su QR, Liu J, Ma C, Fan CX, Wen N, Luo HM, et al. Epidemic Profile of Mumps in China During 2004-2013. Zhonghua Yu Fang Yi Xue Za Zhi (2016) 50(7):611–4. doi: 10.3760/cma.j.issn.0253-9624.2016.07.009
14. Zengel J, Phan SI, Pickar A, Xu P, He B. Immunogenicity of Mumps Virus Vaccine Candidates Matching Circulating Genotypes in the United States and China. Vaccine (2017) 35(32):3988–94. doi: 10.1016/j.vaccine.2017.05.084
15. Peltola H, Kulkarni PS, Kapre SV, Paunio M, Jadhav SS, Dhere RM. Mumps Outbreaks in Canada and the United States: Time for New Thinking on Mumps Vaccines. Clin Infect Dis (2007) 45(4):459–66. doi: 10.1086/520028
16. Westphal DW, Eastwood A, Levy A, Davies J, Huppatz C, Gilles M, et al. A Protracted Mumps Outbreak in Western Australia Despite High Vaccine Coverage: A Population-Based Surveillance Study. Lancet Infect Dis (2019) 19(2):177–84. doi: 10.1016/S1473-3099(18)30498-5
17. Bag SK, Dey A, Wang H, Beard F. Australian Vaccine Preventable Disease Epidemiological Review Series: Mumps 2008-2012. Commun Dis Intell Q Rep (2015) 39(1):E10–18.
18. Vyse AJ, Gay NJ, Hesketh LM, Pebody R, Morgan-Capner P, Miller E. Interpreting Serological Surveys Using Mixture Models: The Seroepidemiology of Measles, Mumps and Rubella in England and Wales at the Beginning of the 21st Century. Epidemiol Infect (2006) 134(6):1303–12. doi: 10.1017/S0950268806006340
19. Aasheim ET, Inns T, Trindall A, Emmett L, Brown KE, Williams CJ, et al. Outbreak of Mumps in a School Setting, United Kingdo. Hum Vaccin Immunother (2014) 10(8):2446–9. doi: 10.4161/hv.29484
20. Coffinières E, Turbelin C, Riblier D, Aouba A, Levy-Bruhl D, Arena C, et al. Mumps: Burden of Disease in France. Vaccine (2012) 30(49):7013–8. doi: 10.1016/j.vaccine.2012.09.070
21. Vygen S, Fischer A, Meurice L, Mounchetrou Njoya I, Gregoris M, Ndiaye B, et al. Waning Immunity Against Mumps in Vaccinated Young Adults, France 2013. Euro Surveill (2016) 21(10):30156. doi: 10.2807/1560-7917.ES.2016.21.10.30156
22. Fields VS, Safi H, Waters C, Dillaha J, Capelle L, Riklon S, et al. Mumps in a Highly Vaccinated Marshallese Community in Arkansas, USA: An Outbreak Report. Lancet Infect Dis (2019) 19(2):185–92. doi: 10.1016/S1473-3099(18)30607-8
23. Dayan GH, Quinlisk MP, Parker AA, Barskey AE, Harris ML, Schwartz JM, et al. Recent Resurgence of Mumps in the United States. N Engl J Med (2008) 358(15):1580–9. doi: 10.1056/NEJMoa0706589
24. Örvell C, Tecle T, Johansson B, Saito H, Samuelson A. Antigenic Relationships Between Six Genotypes of the Small Hydrophobic Protein Gene of Mumps Virus. J Gen Virol (2002) 83(10):2489–96. doi: 10.1099/0022-1317-83-10-2489
25. Rubin S, Mauldin J, Chumakov K, Vanderzanden J, Iskow R, Carbone K. Serological and Phylogenetic Evidence of Monotypic Immune Responses to Different Mumps Virus Strains. Vaccine (2006) 24(14):2662–8. doi: 10.1016/j.vaccine.2005.10.050
26. Latner DR, Hickman CJ. Remembering Mumps. PloS Pathog (2015) 11(5):e1004791. doi: 10.1371/journal.ppat.1004791
27. Won H, Kim AR, Yoo JS, Chung GT, Kang HJ, Kim SJ, et al. Cross-Neutralization Between Vaccine and Circulating Wild-Type Mumps Viruses in Korea. Vaccine (2021) 39(13):1870–6. doi: 10.1016/j.vaccine.2021.01.039
28. Carr MJ, Moss E, Waters A, Dean J, Jin L, Coughlan S, et al. Molecular Epidemiological Evaluation of the Recent Resurgence in Mumps Virus Infections in Ireland. J Clin Microbiol (2010) 48(9):3288–94. doi: 10.1128/JCM.00434-10
29. Park SH. Resurgence of Mumps in Korea. Infect Chemother (2015) 47(1):1–11. doi: 10.3947/ic.2015.47.1.1
30. Jin L, Rima B, Brown D, Orvell C, Tecle T, Afzal M, et al. Proposal for Genetic Characterisation of Wild-Type Mumps Strains: Preliminary Standardisation of the Nomenclature. Arch Virol (2005) 150(9):1903–9. doi: 10.1007/s00705-005-0563-4
31. Bodewes R, van Rooijen K, Cremer J, Veldhuijzen IK, van Binnendijk R. Optimizing Molecular Surveillance of Mumps Genotype G Viruses. Infect Genet Evol (2019) 69:230–4. doi: 10.1016/j.meegid.2019.02.005
32. Cheng WY, Liu MT. Molecular Characteristics of Mumps Viruses Isolated in Taiwan From 2006 to 2016. Heliyon (2018) 4(2):e00518–8. doi: 10.1016/j.heliyon.2018.e00518
33. Morfopoulou S, Mee ET, Connaughton SM, Brown JR, Gilmour K, Chong WK, et al. Deep Sequencing Reveals Persistence of Cell-Associated Mumps Vaccine Virus in Chronic Encephalitis. Acta Neuropathol (2017) 133(1):139–47. doi: 10.1007/s00401-016-1629-y
34. Liu W, Deng L, Lin X, Wang X, Ma Y, Deng Q, et al. Importation of Mumps Virus Genotype K to China From Vietnam. Emerg Infect Dis (2018) 24(4):774–8. doi: 10.3201/eid2404.170591
35. National Health Commission of the People's Republic of China. Infectious Disease Prevention and Control (2022). Available at: http://www.nhc.gov.cn/jkj/s2907/new_list.shtml?tdsourcetag=s_pcqq_aiomsg (Accessed 15 March, 2022).
36. McLean HQ, Fiebelkorn AP, Temte JL, Wallace GS. Prevention of Measles, Rubella, Congenital Rubella Syndrome, and Mumps 2013: Summary Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep (2013) 62(Rr-04):1–34.
37. Carryn S, Feyssaguet M, Povey M, Di Paolo E. Long-Term Immunogenicity of Measles, Mumps and Rubella-Containing Vaccines in Healthy Young Children: A 10-Year Follow-Up. Vaccine (2019) 37(36):5323–31. doi: 10.1016/j.vaccine.2019.07.049
38. Cohen C, White JM, Savage EJ, Glynn JR, Choi Y, Andrews N, et al. Vaccine Effectiveness Estimates 2004-2005 Mumps Outbreak, England. Emerg Infect Dis (2007) 13(1):12–7. doi: 10.3201/eid1301.060649
39. Cardemil CV, Dahl RM, James L, Wannemuehler K, Gary HE, Shah M, et al. Effectiveness of a Third Dose of MMR Vaccine for Mumps Outbreak Control. N Engl J Med (2017) 377(10):947–56. doi: 10.1056/NEJMoa1703309
40. Almansour I. Mumps Vaccines: Current Challenges and Future Prospects. Front Microbiol (2020) 11:1999. doi: 10.3389/fmicb.2020.01999
41. Cui A, Zhu Z, Hu Y, Deng X, Sun Z, Zhang Y, et al. Mumps Epidemiology and Mumps Virus Genotypes Circulating in Mainland China During 2013-2015. PloS One (2017) 12(1):e0169561. doi: 10.1371/journal.pone.0169561
42. Cui A, Zhu Z, Mao N, Si Y, Ma Y, Hu Y, et al. Assessment of One-Dose Mumps-Containing Vaccine Effectiveness on Wild-Type Genotype F Mumps Viruses Circulating in Mainland China. Vaccine (2018) 36(38):5725–31. doi: 10.1016/j.vaccine.2018.08.028
43. Liang Y, Che Y, Yang B, Zhan F, Li H, Guan X, et al. Immunogenicity and Safety of an F-Genotype Attenuated Mumps Vaccine in Healthy 8- to 24-Month-Old Children. J Infect Dis (2018) 219(1):50–8. doi: 10.1093/infdis/jiy469
44. May M, Rieder CA, Rowe RJ. Emergent Lineages of Mumps Virus Suggest the Need for a Polyvalent Vaccine. Int J Infect Dis (2018) 66:1–4. doi: 10.1016/j.ijid.2017.09.024
45. World Health Organization. COVID-19 Vaccine Tracker and Landscape (2021). Available at: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (Accessed 15 March, 2022).
Keywords: mumps, mumps vaccines, phylogenetic trees, genotype F, population dynamic patterns
Citation: Su Y, Liu J, Liu M, Li M, Gao F, Li C, Liang Z, Wu X, Mao Q, Wang Q and Bian L (2022) Genotype F Mumps Viruses Continue to Circulate in China, From 1995 to 2019. Front.Virol. 2:901618. doi: 10.3389/fviro.2022.901618
Received: 22 March 2022; Accepted: 06 May 2022;
Published: 02 June 2022.
Edited by:
Shuofeng Yuan, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Lihua Song, Beijing University of Chemical Technology, ChinaLaura Bubba, University of Milan, Italy
Copyright © 2022 Su, Liu, Liu, Li, Gao, Li, Liang, Wu, Mao, Wang and Bian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Wang, cXdhbmcxODdAMTI2LmNvbQ==; Lianlian Bian, YmlhbmxpYW4yMDE1QDEyNi5jb20=
†These authors have contributed equally to this work
 Yao Su1,2†
Yao Su1,2† Jianyang Liu
Jianyang Liu Lianlian Bian
Lianlian Bian