- 1Doctorate Program in Human Genetics, Health Sciences Campus (CUCS), Guadalajara University, Guadalajara, Mexico
- 2Personalized Medicine National Laboratory (LAMPER), Medical and Pharmaceutical Biotechnology Department, Research Center in Technology and Design Assistance of Jalisco State (CIATEJ), National Council of Science and Technology (CONACYT), Guadalajara, Mexico
Long non-protein coding RNAs (lncRNAs, predicted to be up to 200,000 in the human genome) are nucleic acids of more than 200 nucleotides that not only play primordial roles in the regulation of chromatin states, but also are capable of decoying or scaffolding proteins (e.g., transcription factors, TFs; host and viral proteins), DNA (e.g., promoters and enhancers), and RNA (e.g., miRNAs and hnRNAs) in 3D conformations acting in a chaperonin-like fashion. Specifically, lncRNAs modulate gene expression during the regulatory layers of transcription, RNA processing (splicing and indirectly editing), translation, and post-translational modifications including phosphorylation, acetylation, and ubiquitination. Accumulated evidence indicates that lncRNAs regulate antiviral immune responses mainly by transcription of IFN regulatory factors 1 (IRF1) and 4 (IRF4), which contribute to type I interferon (IFNα and IFNβ) upregulation. Some of the most common TFs regulated by lncRNAs are TP53, CTCF, MYC, SOX2, EZH2 SFPQ, SUZ12, STAT1, STAT3, and NF-kappa B. In this review, the known functions of selected lncRNAs genes in HIV/AIDS (MALAT1, HEAL, NRON, TAR-gag, TP53COR1/lincRNA-p21, NEAT1, NKILA, LINC01426 [formerly Uc002yug.2], FAS-AS1, LINC00173 [formerly FLJ42957/NCRNA00173], GAS5, and HIV-encoded antisense lncRNA) and COVID-19 (EGOT, MALAT1, NEAT1, DANCR, HOTAIR, FENDRR, LINC1505, FALCOR, and HISLA) are discussed. Furthermore, MALAT1 is also involved in subsequent complications such as deep vein thrombosis (DVT) in COVID-19. In addition, after the increased understanding of the role of lncRNAs from Human Endogenous Retroviruses (HERVs, predicted to be at least 582 different with 725,763 repeats of them in the human genome) in cancer (TROJAN) and heart development (BANCR), transcripts of HERVs as lnc-EPAV and lnc-ALVA1-AS1 have recently drawn attention as host protective agents against viral infections. A deeper knowledge of host and viral lncRNAs interactions and their regulation will pave the way for the design of novel drugs inspired by host- and viral-encoded lncRNAs. These novel drugs have the potential to reduce the burden of HIV/AIDS and COVID-19 twofold: (1) by increasing their efficacy and (2) by minimizing the side effects of current drugs. We expect that lncRNA drugs will be able to modulate human and viral transcription in an unprecedented way but still effectively maintain homeostasis by deploying functionality below the pathogenic threshold.
Introduction
Non-coding RNAs (ncRNA) were initially classified as “evolutionary junk”, but their important role in the regulation of transcription (1), cell differentiation (2), development (3), metabolic reprograming (4), and many others has been demonstrated [reviewed in (5–7)]. Because they act at the RNA level and most of them are not transcribed, they were originally called “riboregulators” (8). They can be divided into two large classes: the small non-coding RNAs are commonly from ∽20 to 200 nucleotides (nt), while those longer than 200 nt and up to ∽100 kb and lacking open reading frames are long non-coding RNAs (lncRNA) (9–11). The non-coding elements comprise 80% of the human genome (10). It has been suggested that between ~21,488 and ~200,000 lncRNAs (12, 13) and a subset of these, the lincRNAs, sum up >14,279 (14, 15) in humans. RNA polymerase II is responsible for lncRNA transcription (8, 16), and its biogenesis is similar to mRNAs in the sense that they are polyadenylated, capped, and spliced (5). However, it is worth mentioning that lincRNAs are less efficiently spliced. This can be partially explained because lincRNAs have weaker splicing-related motifs and lower binding to U2 small nuclear RNA auxiliary factor 2 (U2AF2, formerly U2AF65) (17). Depending on their functions, lncRNAs can be classified as cis-acting (within 10 k upstream or downstream of the coding genes they regulate) or trans-acting [identified by expression levels, according to Pearson’s correlation coefficient |r| > 0.95 (18) reviewed in (5)]. LncRNA scaffolds, guide lncRNAs, lncRNA decoys, enhancer RNAs (eRNAs), promoter-associated lncRNAs (PALRs), precursor lncRNA [reviewed in (19)], and a heterogeneous subgroup are involved in X-chromosome inactivation, telomere regulation, and imprinting (reviewed in (6)). Depending on their position in the genes, lncRNAs can also be classified as exonic, intronic, or intergenic (20). An additional classification regarding evolution are the Transcribed Ultra-Conserved Regions lncRNAs (T-UCRs, ≥481) (21, 22). An alternative and comprehensive classification can be found at (23).
LncRNAs regulate innate immunity in host–virus interactions. During viral infection, cellular lncRNAs directly regulate viral genes and modify both viral replication and pathology through changes mediated by virus in transcriptome of the host (24–26). Also, several lncRNAs could upregulate IFN-I (IFNα and IFNβ) expression to induce an antiviral response (27–30), reviewed in (31). Although lncRNAs seem to not have protein-coding function, some of them actually encode functional small peptides (32–34) [reviewed in (35, 36)]. In this review, we will focus on lncRNAs. LncRNAs can regulate different gene modulation processes. They regulate gene expression during the regulatory layers of transcription (37), RNA processing as splicing and indirectly editing (38), and/or translation (39). LncRNAs can control post-translational modification of proteins such as phosphorylation (40), acetylation (41), and ubiquitination (42, 43). In brief, they can act as a decoy or scaffold of other molecules such as proteins (44–46), RNA,and DNA (47). Also, their subcellular location gives important clues about its function in different biological processes. In the nucleus, they guide transcription factors or complexes that modify chromatin. In the cytosol, they control stability of mRNA (48) or compete with endogenous mRNA for access to the protein expression machinery (23). In the cytoplasm, lncRNAs can bind directly to transcription factors (TFs) (49). Interestingly, several lncRNAs are located near genes that encode TFs as well (50). However, particularly long intergenic non-coding RNAs (lincRNAs) regulate both the expression of neighboring genes and distant genomic sequences (6). At the transcription level, lncRNAs regulate accessibility to chromatin in processes that involve RNA polymerase and histone modification enzymes. Furthermore, their ability to bypass chromatin-modifying complexes to impact the basal transcriptional machinery and gene promoters has been demonstrated (51). One classical example of lncRNA in a non-disease state is the X-inactivation specific transcript (XIST) in women that recruits the polycomb complex. XIST induces chromosome silencing of the sexual chromosome from which it was transcribed (52) [reviewed in (53)]. LncRNAs can also stabilize mRNAs and regulate their decay by diverse mechanisms such as (1) binding to target mRNAs, (2) sequestering miRNAs and/or ribosome-binding proteins [RBPs, these lncRNAs are also called competitive endogenous RNAs (ceRNAs), decoys, or sponges], and (3) acting as scaffolds to enhance interaction of RBP-mRNA or interacting with m6A machinery to modulate m6A levels of target mRNAs. This induces gene expression (54). In addition to the complexity of their regulation, lncRNAs have slicing variants that are expressed in a tissue- and/or cell-specific manner (54). In recent years, an increased number of academic groups and pharmaceutical companies became interested in the development of novel therapies based on lncRNA regulation. Currently, at least 61 lncRNA-related clinical trials are ongoing worldwide for cancer, cardiovascular disease, diabetes, and other conditions/diseases (https://www.clinicaltrials.gov/ and Figure 1).
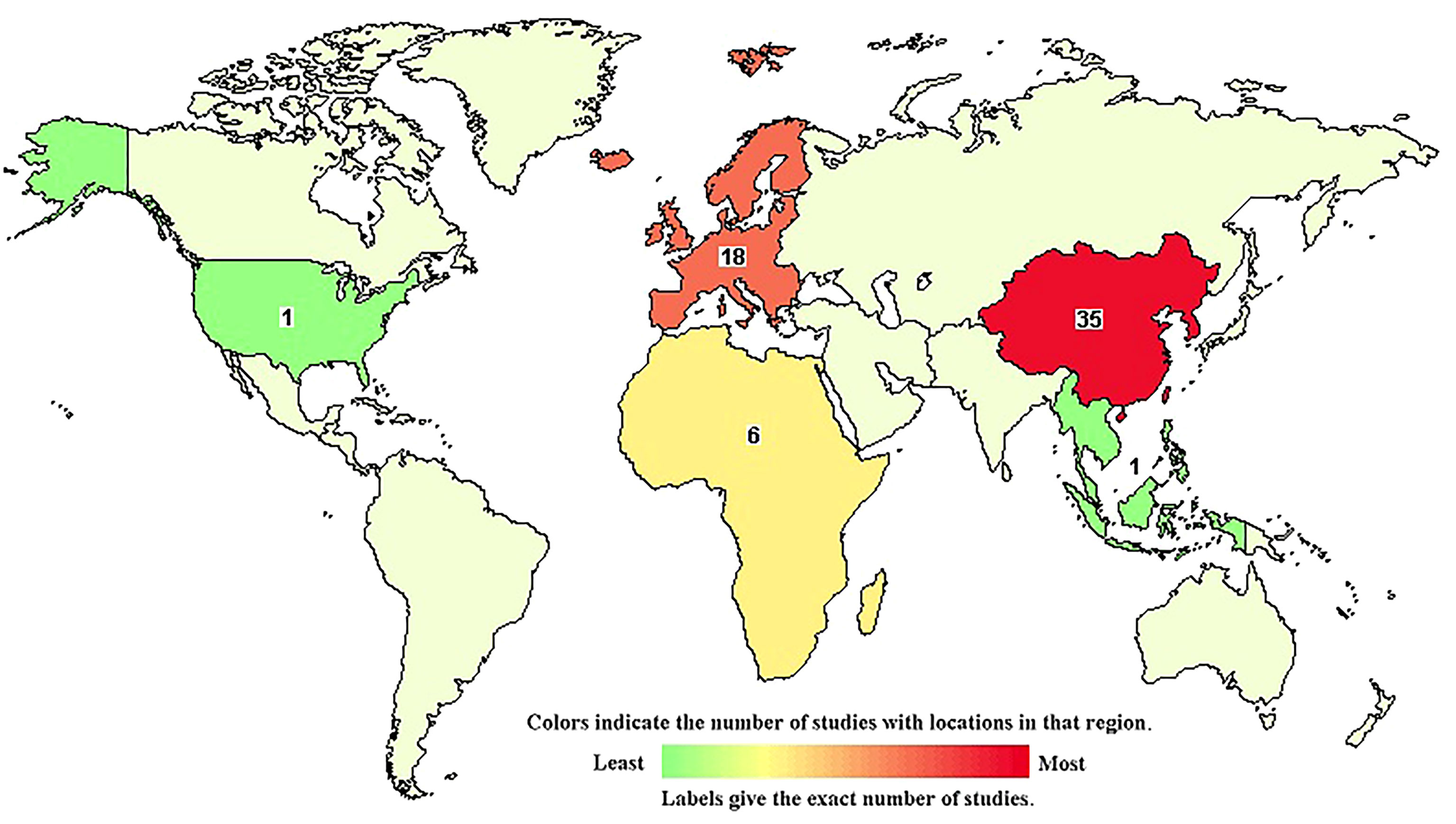
Figure 1 World map of ongoing lncRNAs clinical trials according to (www.clinicaltrials.gov, accessed on March 15th, 2022). Numbers indicate the current clinical trials per country/region where the active substance of the treatment arm contains a lncRNA. Currently, there are 61 registered clinical trials, but none of them are for HIV-AIDS or COVID-19. The color of the country/region refers to the relative frequency of registered clinical trials in comparison with the other countries/regions (see the key of the figure).
LncRNAs are also involved in pathological processes in viral infections and cancer. HIV is the causative agent of AIDS. There are two strains of HIV, HIV-1 and HIV-2 (55). In this review, we will focus on lncRNAs encoded in HIV-1 and SARS-CoV-2 genomes as well in lncRNAs of human endogenous retroviruses (HERVs). For data of lncRNAs in other viruses, see (10). With the emergence of SARS-CoV-2 in 2019, the importance and the role that non-coding RNAs play in the infection process have been highlighted and recently investigated (56).
LncRNAs of HERVs and other host lncRNAs appear as a potential novel strategy to combat viral infections. It is important to know the mechanisms of action of lncRNAs in the development and outcome of infections for the development of more effective therapeutic strategies based on these complex gene regulation layer. In order to develop an adequate infectious process, viruses have the ability to decrease or increase the expression of lncRNAs; these changes can generate antiviral or proviral functions and completely modify the development of a disease. HIV alters the expression of important cellular lncRNAs (57), but the participation of other viruses has been just recently studied (10). Ma et al. (2021), reported 242 differentially expressed (DE) lncRNAs in early infection with HIV and might show cis-dependent regulation of B and T lymphocyte-associated protein gene (BTLA), zeta chain-associated protein kinase gene (ZAP70) and growth factor receptor-bound protein 2-related adaptor protein gene (GRAP). Some lncRNAs were predicted to act as ceRNAs to regulate expression of fos protooncogene, AP1 transcription factor subunit (FOS), fosb protooncogene, AP1 transcription factor subunit (FOSB), V-jun avian sarcoma virus 17 oncogene homolog (JUN), transcription factor 7-like 2 (TCF7L2), and hypoxia-inducible factor 1, α-subunit (HIF1A) (58). LncRNAs are crucial for viral reproduction (production of new infectious virus particles), either repressing or activating HIV-transcription. After HIV or SARS-CoV-2 infection, ncRNAs, including miRNAs, small interfering RNAs (siRNAs), and lncRNAs, are modulated by the virus, in order to achieve the successful replication (nucleic acid synthesis) cycle. Attempts have been made to compare the different datasets obtained by research groups on expression changes during infection, but it represents a great challenge because the expression levels are highly sensitive to external stimuli and it is difficult to define the basal levels that normally exist in cells, because interindividual variation exists (59). It has been identified as a deregulation response of many lncRNAs in response to either an infection process or changes in cytokines such as IFN, which end up impacting the viral replication that may occur (60). In addition, about 50% of HIV-1-positive patients receive combined antiretroviral therapy (cART), which includes a cocktail of inhibitors that target viral processes including entry, reverse transcription, integration, and/or protease-mediated cleavage (61). HIV-1 latency is a state in which viral RNA is hard to detect by conventional/clinic techniques. HIV-1 latency is one of the major barriers to eradicate HIV/AIDS because the virus remains largely undetected by the surveillance of the immune system.
In this review, we summarize the role of lncRNAs in some important viral infections such as COVID-19 and HIV-1/AIDS as well as briefly underline the importance of lncRNAs transcribed from HERVs, their functions, mechanisms, and some of their nucleic acid- and protein-binding partners. To avoid confusion in this review, lncRNA genes are written in italics and the transcripts of lncRNAs are not.
Role of Selected lncRNAs in HIV/AIDS
Role of Metastasis-Associated Lung Adenocarcinoma Transcript 1
MALAT1 modulates epigenetic regulation of HIV-1 and alternative splicing of cellular precursor mRNAs (pre-mRNAs). Polycomb repressive complex 2 (PRC2) primarily trimethylates H3K27me3 to silence gene transcription. MALAT1 prevents the binding of the catalytic subunit of PRC2 named enhancer of zeste 2 (EZH2) with HIV 5´-long terminal repeat (LTR, Figure 2). This reduces the LTR methylation mediated by PRC2 and activates HIV-1 replication (62). Experiments on overexpression of MALAT1 in CD4+ T cells increase HIV-1 replication, suggesting a potential application in reactivation of latent HIV-1 (Table 1).
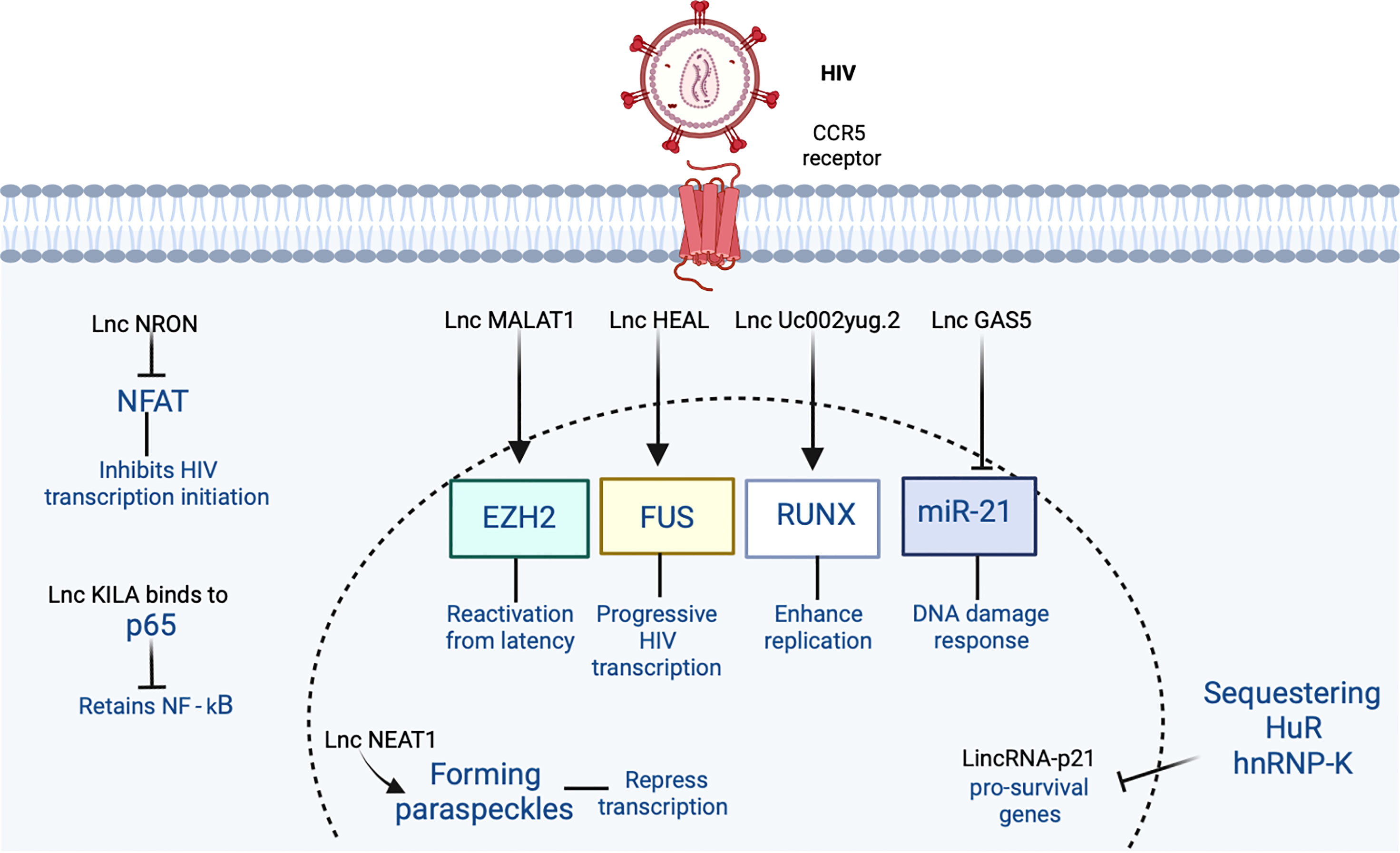
Figure 2 The participation of lncRNA in HIV-1 infection is shown. HIV-1 binds to the CCR5 receptor in the cell. In the figure, the lncRNAs that induce an increase or inhibition are shown with different indicators (↑ and ⊥ respectively). Figure was designed in Biorender by AVRR (https://biorender.com/).
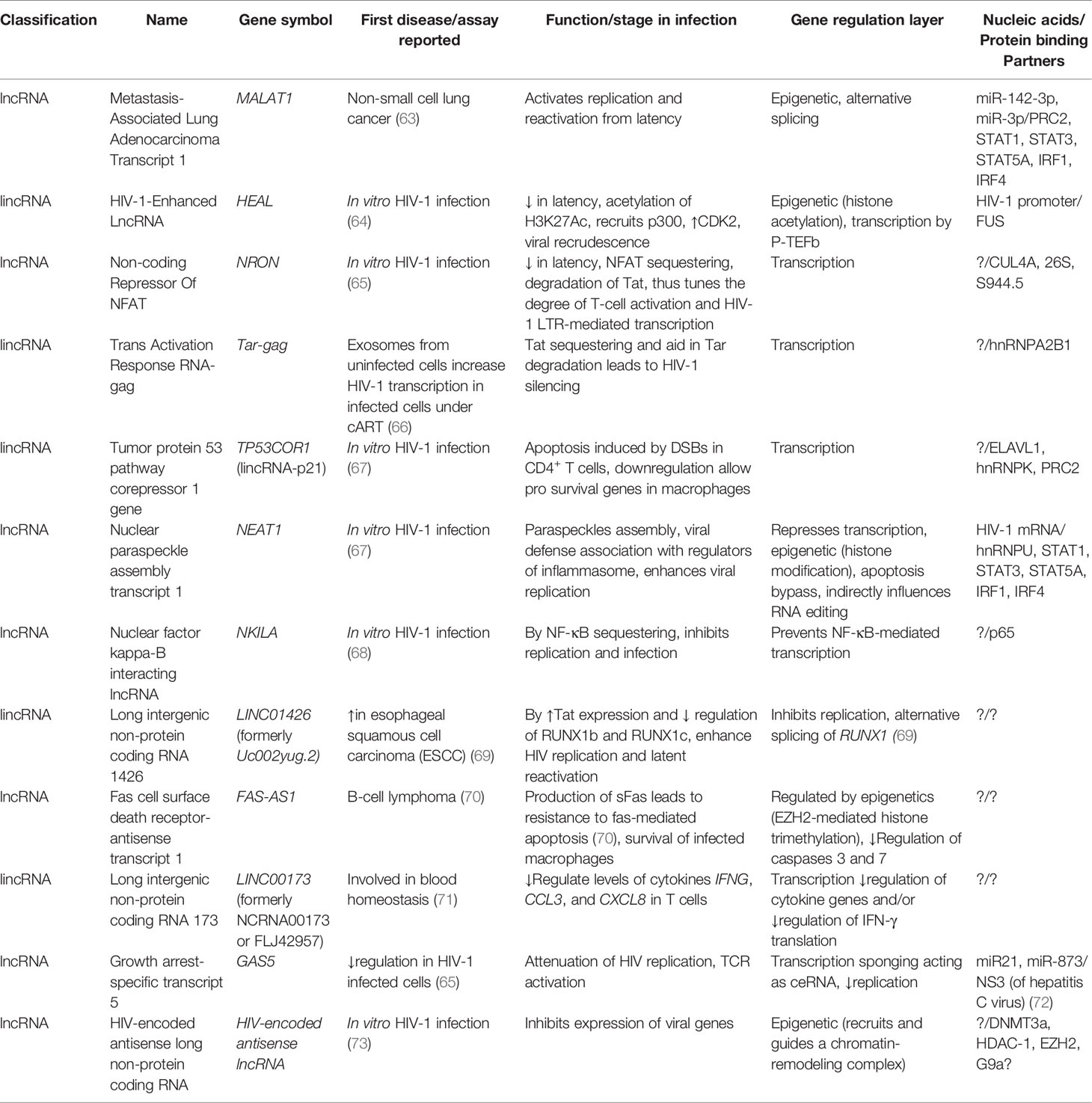
Table 1 Selected lncRNAs of HIV-1 including their first report, known function, and nucleic acid/protein-binding partners.
Role of HIV-1-Enhanced lncRNA
HEAL is a lincRNA with a function in epigenetic regulation of HIV transcription. Active HIV infection upregulates HEAL in monocyte-derived macrophages (MDMs), T lymphocytes, and microglia. Its expression is dependent of viral replication and independent of viral entry. HEAL expression is downregulated in HIV-1-latent CD4+T cells. HEAL targets the HIV promoter and upregulates viral transcription by histone acetylation and elongation by positive transcription elongation factor b (P-TEFb). Furthermore, HEAL binds to the transcription factor FUS (Figure 2), an RNA binding protein that together with HEAL recruits histone acetyltransferase p300 to the HIV-1 promoter. This also activates H3K27ac in the viral promoter and its progressive transcription. Blocking of HEAL could indirectly inhibit cyclin-dependent kinase 2 (CDK2) expression without altering its other relevant functions. Blocking HEAL also prevents viral recrudescence in both MDMs and T cells when azidothymidine (AZT) is discontinued (64). Downregulation of HEAL suggests a new epigenetic strategy to slow down HIV-1 infection (Table 1).
Role of the Non-Coding Repressor of Nuclear Factor of Activated T Cells
NRON serves as a scaffold and, together with ubiquitin ligase cullin A (CUL4A) and protease 26S, subunit 9 (S9p44.5) enzymes, sequesters the phosphorylated form of nuclear factor of activated T cell (NFAT) transcription factor in the cytoplasm. This inhibits HIV-1 transcription initiation (Figure 2). HIV-1 causes downregulation of NRON (74), so this mechanism increases gene expression and finally an increased efficacy of HIV-1 active replication in the cells (65). NRON expression in the HIV-1 cycle is complex. Firstly, early in the viral cycle, Nef inhibits NRON, thus activating viral transcription. Secondly, in the late stage of infection, the Vpu viral protein induces NRON expression to swap from viral transcription to viral assembly and budding (65). Thirdly, NRON can also regulate HIV latency in an NFAT-independent fashion by enhancing ubiquitin-proteasome-dependent degradation of the viral activator of transcription (Tat) (75) (Table 1). This suppresses viral transcription and facilitates HIV latency preventing transcriptional activation of proviruses. Knockdown of NRON could enhance viral production from primary CD4+ T lymphocytes latently infected with HIV-1. In HIV-infected individuals receiving combination antiretroviral therapy (cART), expression of endogenous NRON in resting CD4+T cells is inversely correlated with the expression level of intracellular viral RNA (75).
Role of Trans-Activation Response RNA-gag
This lncRNA was named TAR-gag because it contains the complete trans-activation response RNA (TAR) and U5 sequence of the 5-LTR and part of the gag gene (for an interaction network of gag, nc, ma, and rev viral proteins with human proteins, see Figure 3). Heterogeneous nuclear ribonucleoprotein A2/B1 protein (hnRNPA2B1) may package TAR and TAR-gag into exosomes by binding to their 5´-GGAG-3´ motif. For this reason, the authors suggest that true HIV-1 latency in vivo is very unlikely due to the high number of exosomes (108–1011) to which tissues and cells in fluids are constantly exposed (66) (Table 1). TAR-gag can also silence HIV-1 transcription and latency sequestering Tat and promote its degradation (for an interaction network of Tat, Vpr, and vif viral proteins with human proteins, see Figure 4). The TAR-gag mechanism acts as an “RNA machine” by binding to proteins that suppress transcription and forming an RNA–protein complex that regulates the gene expression of HIV-1 (61).
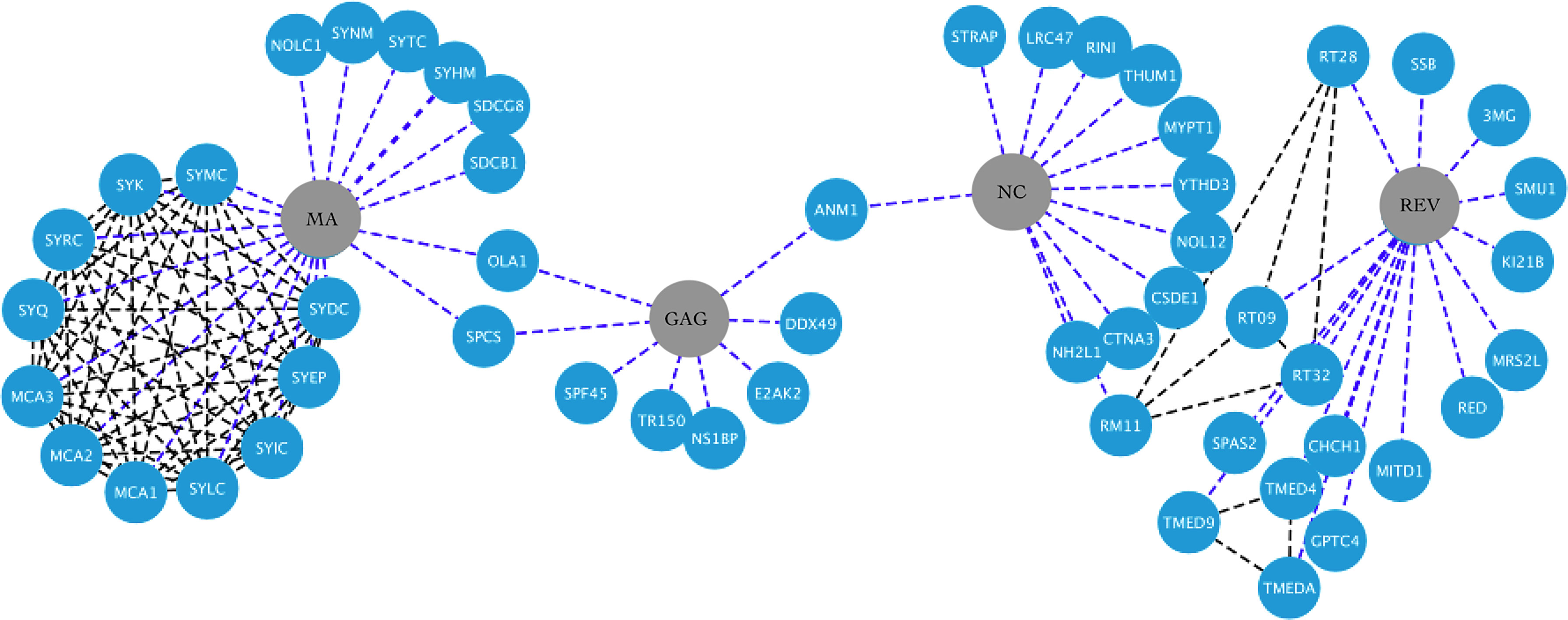
Figure 3 HIV-1-host protein interaction network. HIV-1 proteins Ma, Gag, Nc and Rev are shown in the central nodes in gray color, contacting each of them with the human proteins, which are nodes in blue color. Interactions between viral and human proteins are shown in the blue dotted lines, while interactions between the same human proteins are shown in the black dotted lines. Figure created in Cytoscape (https://cytoscape.org/download.html).
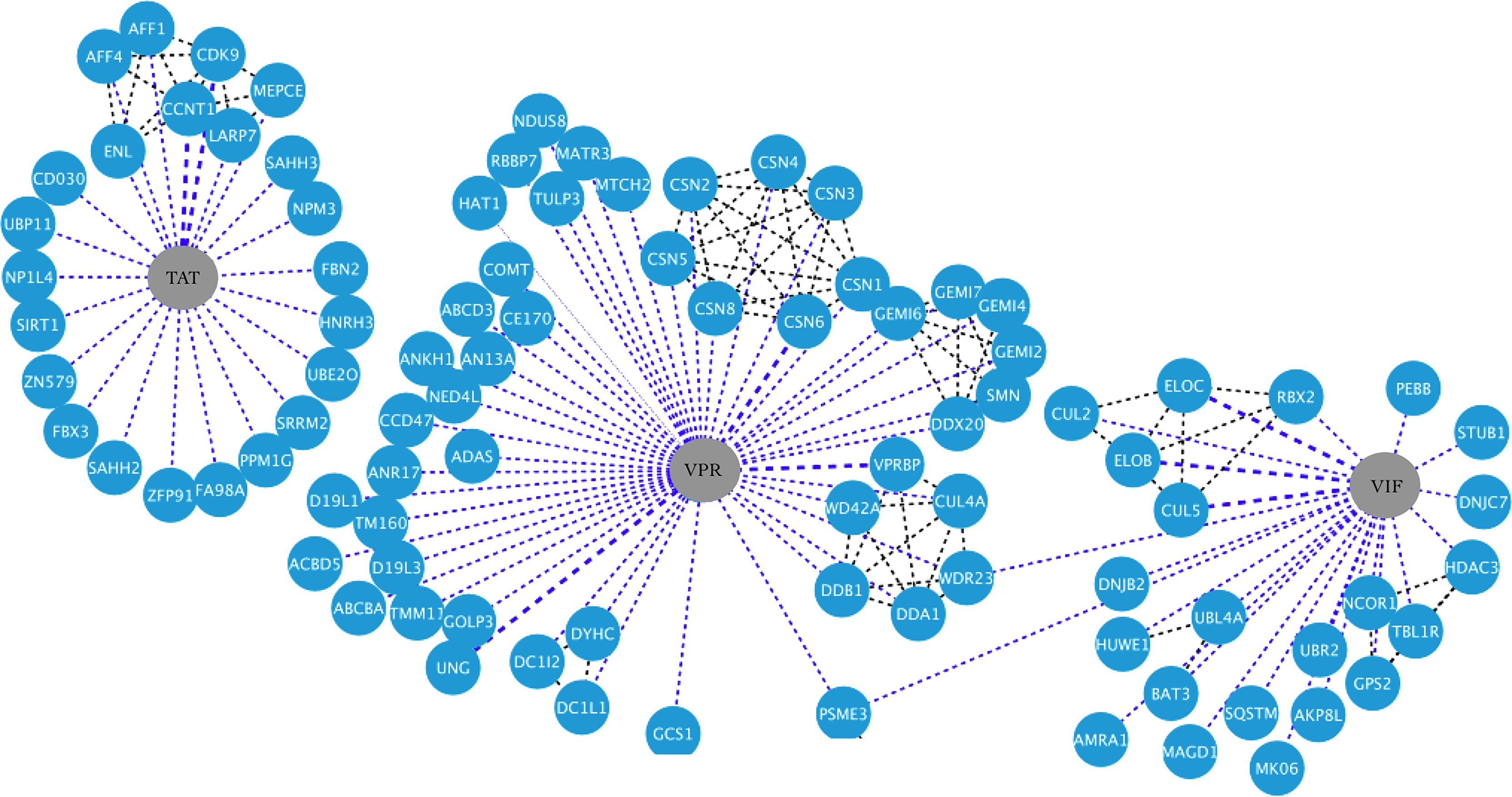
Figure 4 HIV-1-host protein interaction network. HIV-1 proteins Tat, Vpr, and Vif are shown in the central nodes in gray color, contacting each of them with the human proteins, which are nodes in blue color. Interactions between viral and human proteins are shown in the blue dotted lines, while interactions between the same human proteins are shown in the black dotted lines. Figure created in Cytoscape (https://cytoscape.org/download.html).
Role of Long Intergenic Non-Protein Coding RNA p21
LincRNA-p21 is encoded by the tumor protein p53 pathway corepressor 1 gene (TP53COR1, www.genenames.org). Apoptosis is caused by double-strand breaks (DSBs) such as those induced by retrovirus integration as in the case of HIV-1. Apoptosis is coordinated by lincRNA-p21, which is p53-regulated. LincRNA-p21 mediated its function by binding in a complex with one of its three protein partners ELAV-like RNA-binding protein 1 (ELAVL1, formerly called Hu-antigen R or HuR), heterogeneous nuclear ribonucleoprotein K (hnRNPK), or PRC2 (67) (Figure 2). ELAVL1 contains three ribonucleoprotein-type consensus motifs and bind to cis-acting AU-rich elements (AREs). A core element of 27 nucleotides contains AUUUA, AUUUUA, and AUUUUUA motifs (76). HIV enables lincRNA-p21 degradation by sequestering ELAVL1 in the nucleus. Also, HIV sequesters hnRNPK in the cytoplasm through the pro-survival MAP2K1/ERK2 pathway, thus inducing transcription of pro-survival genes in macrophages. This is in contrast to T cells, where maturing cells switch off the MAP2K1/ERK2 cascade (67). In brief, HIV-1 induces apoptosis in CD4+ T cells but mainly is non-cytopathic in macrophages leading to the long-term dissemination of the virus (Table 1).
Role of Nuclear Paraspeckle Assembly Transcript 1
Paraspeckles are non-membranous organelles located in the nucleus where many RNAs reside and RNA-binding proteins (RBPs) as well. Two of these RBPs are insulin-like growth factor-binding protein 7 (IGFBP7, formerly prostacyclin-stimulating factor or PSF) and non-Pou domain-containing octamer-binding protein (NONO, formerly p54 nuclear RNA-binding protein or p54NRB). The function of paraspeckle bodies is suggested to be gene regulation through nuclear retention of RNA for editing by the adenosine deaminase acting on RNA 1 (ADAR1) enzyme. NEAT1 controls RNA regulatory processes by forming a critical scaffold to assemble paraspeckles, substructures that are very important for HIV replication (77). NEAT1 is involved in different pathways as viral defense, innate, and inflammatory responses. NEAT1 has two isoforms, NEAT1_1 or MENε (3.7 kb) and NEAT1_2 or MENβ (23 kb). Upon HIV-1 infection in diverse cell lines such as Jurkat, MT4, THP1, and THP1 differentiated with phorbol-12-myristate-13-acetate (PMA), isoforms NEAT1_1 and NEAT1_2 were upregulated ~3- to 7-fold and ~6- to 11-fold, respectively (77). NEAT1 represses HIV transcription (58) (Figure 2) and this enhances HIV-1 replication in activated CD4+T cells in a post-transcriptional stage. Jurkat CD4+ T-cell lines with a knockout (KO) of the NEAT1 gene were more prone to the induction of apoptosis when compared with parental Jurkat cells (78). Quantitative PCR (qPCR) showed that NEAT1 is downregulated in the plasma of infected patients and this expression correlates with CD4+ T-cell count. Furthermore, in PBMCs, NEAT1 was upregulated in highly active antiretroviral therapy (HAART)-naïve patients (n = 31) and downregulated in patients receiving HAART (n = 28) (79). NEAT1 is further associated with regulator proteins of inflammasome and by its assembly with the immune-controlled ribonuclear complex (15). NEAT1 inhibits virus production through nuclear-to-cytoplasm export of Rev-dependent instability element (INS)-containing HIV-1 mRNAs. Thus, this evidence suggests that paraspeckles are indeed the depot for storing HIV-1 Rev-dependent INS-containing RNAs that are diverted away from splicing (77). In HeLa cells, NEAT1 KO can promote the export of HIV-1 mRNA from the nucleus towards the cytoplasm and enhance viral production (15). NEAT1 contributes to antiviral response in a similar way to MALAT1 when the former interacts with STAT1, STAT3, STAT5A, and interferon regulatory factors 1 and 4 (IRF1 and IRF4) TFs. NEAT1 also interacts with heterogeneous nuclear ribonucleoprotein U (hnRNPU) altering histone modification of target genes (10). HIV-1 takes advantage of NEAT1 downregulation by promoting infection and enhancing viral replication (79) (Table 1).
Role of Nuclear Factor Kappa-B-Interacting lncRNA
NKILA potently inhibits the replication and infection production of clones of HIV-1 with different co-receptor tropisms including R5X4-tropic 89.6, brain-derived Yu2, and macrophage-tropic AD8 strains. KILA binds to p65 and retains NF-κB in the cytoplasm (Figure 2). HIV-1 counterattacks downregulating NKILA by decreasing H3K27 acetylation in the NKILA promoter (68) (Table 1).
Role of Long Intergenic Non-Protein Coding RNA 1426
LINC01426 (formerly Uc002yug.2) is encoded by gene LINC01426 (www.genecards.org). LINC01426 enhances replication of HIV and latent HIV activation by increasing the expression of Tat protein and downregulating Runt-related transcription factor 1b and 1c (RUNX1b and RUNX1c). In primary CD4+T cells derived from HIV-1 patients successfully treated with HAART, the expression of LINC01426 was lower than in those HAART-naïve patients. Overexpression of LINC01426 in resting CD4+ T cells derived from individuals treated with HAART and after T-cell activation by phytohemagglutinin M (PHA-M) led to significant HIV reactivation (80) (Table 1).
Role of fas Cell Surface Death Receptor-Antisense Transcript 1
In macrophages, which are resistant to virus-induced apoptosis, FAS-AS1 [formerly SAF; for correct guidelines of lncRNA nomenclature according to the Human Genome Organization Gene Nomenclature Committee (HGNC), see (81, 82)] regulates apoptotic effector caspases. Downregulation of FAS-AS1 increases the caspase-3 and caspase-7 activities in HIV-infected MDMs and exclusively induces cell death in macrophages infected with HIV. In bronchoalveolar lavage (BAL), macrophages obtained from HAART-naïve individuals show increased FAS-AS1 expression. FAS-AS1 is useful in the survival of persistently HIV-1-infected macrophages. Also, FAS-AS1 is increased in HIV-positive lung alveolar macrophages from chronically infected patients (83) (Table 1).
Role of Long Intergenic Non-Protein Coding RNA 173
Two independent RNA-seq studies revealed that expression of LINC00173 (formerly NCRNA00173 or FLJ42957, https://www.genenames.org) is increased ≥2-fold during HIV-1 infection and the location of this lncRNA is mainly in the nucleus. There are two isoforms of this lncRNAs, lnc173 TSV1 and lnc173 TSV2, which are polyadenylated. These can be detected by qPCR (84). It is likely that LINC00173 has a role in the regulation of cytokine levels in T cells during immune responses associated with HIV-1 because Jurkat cells with KO for LINC00173 and exposed to PMA and ionomycin express higher mRNA levels of cytokine genes IFNG, CCL3, and CXCL8 than control cells, in contrast with IL2 and TNF genes, whose mRNA levels do not differ. Furthermore, in the same report, LINC00173 KO clones also produced increased protein levels of IFN-γ (84) (Table 1).
Role of Growth Arrest-Specific Transcript 5
Gas5 is downregulated in HIV-1-infected cells (65). GAS5 could act as ceRNA, because it suppresses miR-873 expression, which can promote replication of HIV. Thus GAS5 downregulates HIV replication (85). Furthermore, GAS5 regulates functions of CD4+ T cells through mir21-mediated signaling in people living with HIV (PLHIV, Figure 2). This regulates signaling molecules involved in the damage of DNA and cellular responses after T-cell receptor (TCR) activation (86) (Table 1).
Role of HIV-Encoded Antisense lncRNA
This antisense lncRNA is localized in the 5´LTR and replaces the components of endogenous cellular pathways. This lncRNA recruits and guides a chromatin-remodeling complex by binding to methyltransferase 3a (DNMT3a), histone deacetylase 1 (HDAC-1), and the polycomb protein enhancer of zeste homolog 2 (EZH2). The presence of these proteins at 5´LTR leads to epigenetic changes in histones associated with the viral promoter. This results in a downregulation of HIV-1 gene expression. Suppression of this lncRNA with small single-stranded antisense RNAs leads to the activation of viral gene expression DNA (73) (Table 1).
Other lncRNAs in HIV Infection
In monocyte-derived macrophages, lncRNA differentiation antagonizing non-protein coding RNA (DANCR) is downregulated by IFN-ε stimulation, and 21 interactions with mRNAs were predicted. This was in contrast to classic lncRNAs such as NEAT1, GAS5, and MALAT1, which showed no differential expression in HIV-infected MDMs (87). Another lncRNA is AK130181, which represses HIV transcription and induces latency through NF-κB signaling. Interestingly, lncRNA LOC102549805 (lncRNA-U1) modulates neurotoxicity of HIV Tat protein (88). Additional lncRNAs in HIV-1 infection were recently reviewed in (89).
Potential of lncRNAs in HIV/AIDS Diagnostics
Plasma levels of lincRNA chr12:57761837-57762303 and lincRNA:chr2:165509129-165519404 were better markers for HIV-1 infection whereas LincRNA chr5:87580664-87583451, XLOC_001148, and lincRNA chr10:128586385-128592960 were better markers for HIV-2 infection (90). The presence of NEAT1 in plasma has been suggested as a biomarker of HIV-1 infection (79).
Potential of lncRNAs in Prognosis of HIV/AIDS
Schynkel et al. (2020) analyzed the transcriptomic change of macrophages treated with three types of IFNs (IFN-α, IFN-γ, and IFN-ϵ) and 6 h post-infection with HIV-1. They found differential expression of 474 genes. From these, 112 (24%) were classified as non-coding; within these seven DE genes, they coincided between the four conditions of IFN finding five lncRNAs, namely, AC053503.1, NRIR, C8orf3, AL359551.1, and MIR3945HG, as well as two pseudogenes, namely, neutrophil cytosolic factor 1C pseudogene (NCF1C) and guanylate binding protein 1 pseudogene 1 (GBP1P1) (87).
In elite controllers of HIV, it has been suggested that in myeloid dendritic cells, the lnc MIR4435-2 host gene (MIR4435-2HG) enhances metabolic function through epigenetic mechanisms including H3K27 enrichment at an intronic enhancer of the regulatory associated protein of mammalian target of rapamycin [mTOR] (RPTOR) locus, which is the main component of the mTOR complex 1 (mTORC1) by means of a predicted triple helix of RNA–DNA. These changes shows features of trained innate immunity (91).
RUNX1 overlapping RNA (RUNXOR) is an lncRNA and an important regulator of myeloid-derived suppressor gene. RUNXOR and RUNX are upregulated in MDSCs that expand and accumulate in human PBMCs in people living with HIV (PLHIV). RUNXOR and RUNX are associated with expression of immunosuppressive molecules such as reactive oxygen species, arginase 1, signal transducer and activator of transcription 3 (STAT3), IL-6, and inducible nitric oxide (NO) synthase (92).
A total of 242 lncRNAs were DE in HAART-naïve people living with HIV (PLHIV) patients. Seventeen of those lncRNAs were predicted to regulate TCF7L2 and HIF1A genes, which are involved in HIV transcription. Twenty DE lncRNAs probably share miRNA response elements with JUN and FOS, which are associated with both HIV-1 replication and immune activation (58).
Chemokine, CC motif, receptor 5-antisense (CCR5-AS) is an lncRNA that regulates differential expression of CCR5, the major co-receptor of HIV. High expression of CCR5-AS sequesters the RALY heterogeneous nuclear ribonuclear protein (RALY) preventing its binding to CCR5 3´-UTR. This prevents the decay of CCR5 mRNA (93).
LncRNAs as Potential Therapy in HIV/AIDS
For the complete transcription of HIV-1, two stimuli are necessary. Firstly, the activation of T cells, and secondly, the translation of the viral protein Tat. With this rationale, one therapeutic approach is that peptides capable of competing for Tat binding and stimulating a viral ncRNA response have been applied. Peptide F07 #13 can induce the silencing of transcriptional genes by inducing viral ncRNA (e.g., TAR-gag and TAR) that act with transcriptional suppressor proteins cullin 4B (CUL4B), PRC2, and SIN3 transcription regulator family member A (SIN3A), ultimately leading to ubiquitination of Tat (61). Inhibition of lncRNAs that induce latency can be achieved by post-transcriptional degradation of the lncRNA with short hairpin RNA (shRNA), antisense oligonucleotide (ASO), or siRNA. Improved design of modified siRNA and ASOs has led to a higher efficacy and stability, while off-target effects are reduced. This can be attained by lncRNA transcription inhibition of by gene editing by CRISPR-Cas or promoter blockade (94, 95). As the function of lncRNAs is through formation of secondary structures with both proteins and nucleic acids, a diverse collection of RNA decoys, nanobodies, small molecules, and aptamers to induce steric hindrance are being evaluated in ongoing efforts (96). Most of the current approaches for therapy are focused on pathogenic lncRNA inhibition by delivery of lncRNA made of synthetic RNA. For lncRNA delivery, a feasible approach is to use adeno-associated virus, herpes virus, ncRNA sponges, chemical modified ncRNAs, non-viral vectors, computed tomography-guided injections, and endoscopic ultrasound (97).
The lack of sequence conservation across species is one of the main challenges of lncRNA research. One solution could be humanized mouse models. In addition, specific markers of HIV-1 latently infected cells remain to be identified. Induction of apoptosis in virus-infected cell only, is caused by downregulation of the lncRNA heterogeneous nuclear ribonucleoprotein U gene (HNRNPU, formerly SAF) with siRNA. This leaves unaffected the bystander cells (83). The effectiveness and safety of lncRNA-mediated therapy should be demonstrated in non-human primates as well as in clinical trials. Importantly, the elimination of reservoirs of latent HIV-1 remains the major hurdle to find a complete cure for HIV-1 infection. The approaches to trying to tackle this are killing infected cells, gene therapy, “shocking and kill” HIV-1 out of latency [reviewed in (97)], and/or HIV-1 silencing, e.g., through the histone chaperones, namely, chromatin assembly factor I, subunits A and B (CHAF1A and CHAF1B) (98). Nevertheless, none of these potential solutions are likely to eradicate HIV in the short term; a combination of them could lead to a feasible functional cure in the mid or long term.
Expression of lncRNAs in COVID-19 and Murine Models
The role of lncRNAs in both the initiation and progression of viral infection has been reported. In a mouse model, RNA sequencing of influenza A virus and SARS-CoV-2-infected lung tissues also demonstrated the key roles of lncRNAs in pathogenesis of respiratory virus by inducing interferon (IFN). In this model, the pathogenesis regulation is mediated by signal transducer and activation of transcription (STAT1). In COVID-19 patients, an increasing chemokine expression has been observed in reduced or absence of type I (IFNα and IFNβ) and III (IFNλ1, IFNλ2, IFNλ3, and IFNλ4) interferons (99). Thus, the lncRNA network of COVID-19 patients showed a significant downregulation of IFN-I response (15). Furthermore, in SARS-CoV-2 infection, 527 bronchoalveolar lavage fluid (BALF) and 239 protein-coding genes (PCGs in peripheral blood mononuclear cell-PBMC) could be influenced by 162 and 106 cis-acting lncRNAs, respectively. Main pathways involved are myeloid local site activation, immune system, iron homeostasis regulation, and degranulation of neutrophils in the tissue-specific regulation of host pyrin domain (PYD) and C-terminal caspase-recruitment domain (CARD)-containing protein (PYCARD) and IFNG gene (PBMC) as well as calpain 1 (CAPN1) gene mediated by MALAT1 and NEAT1 lncRNAs (99). Another report suggests that 195 and 155 lncRNAs are down- and upregulated, respectively (100). Furthermore, another group reported that, in the GEO database (GSE147507) in more than one cell line, the DE lncRNAs were LINC00312, LINC00473, LINC00605, HLA complex group 11 (HCG11), HIF1A-antisense 2 (HIF1A-AS2), eosinophil granule ontogeny (EGOT), erythrocyte membrane protein band 4.1-like 4A-antisense 1 (EPB41L4A-AS1), LINC00115, LINC00174, LINC00265, LINC00662, LINC00842, myocardial infarction-associated transcript (MIAT), NEAT1, MALAT1, maternally expressed 3 and 9 (MEG3 and MEG9), RNA component mitochondrial RNA processing endoribonuclease (RMRP), telomerase RNA component (TERC), and zinc finger protein 674–antisense 1 (ZNF674-AS1) and decreased expressions of TP3 target 1 (TP53TG1), LINC00488, prostate androgen-regulated transcript 1 (PART1), and LINC00857 (101). Also in a murine model, a report suggests that there are 5,329 different lncRNAs (15). In the binding of lncRNA and transcription factors, those more relevant were IRF1, STAT1, and MYC1. Notably, in lungs of COVID-19 patients, expression of NEAT1 was also increased significantly (101). Interestingly, expression of MALAT, NEAT1, and MIAT was common between SARS-CoV-2-infected cells and those infected with HIV. An increased expression of MALAT1 and abnormal expression of other lncRNAs was reported in BAL fluid of COVID-19 patients (15, 99, 101). Hox antisense intergenic RNA (HOTAIR), NEAT1, MALAT1, LINC00589 (formerly tumor-suppressor lncRNA on chromosome 8p12 or TSLNC8), and cardiac autophagy inhibitory factor (CAIF) lncRNAs may regulate IL-6 expression. In contrast, XIST, cyclin-dependent kinase inhibitor 2B-antisense RNA gene (CDKN2B-AS1, formerly antisense RNA in the INK4 locus or ANRIL), repulsive guidance molecule domain family, member B-antisense 1 (RGMB-AS1), NEAT1, MAS-related G-protein coupled receptor, member D (MRGPRD, formerly Gm499), and cytochrome C oxidase subunit II (MT-CO2, formerly Cox2) lncRNAs encoded in the mitochondria genome have been implicated in the formation of inflammasome (100). Another group reported 410 differentiated lncRNA among COVID-19 patients and controls (20). Different results, particularly the role of IFN-I in COVID-19 patients, must be due to the application of different methodologies, different tested time points, and non-comparable cell lines analyzed (101). The majority of these lncRNAs have been predicted by in silico approaches except NEAT1 (upregulation) and DANCR (downregulation). LncRNAs in cytokine storm are RAS gene associated with diabetes 51-antisense 1 (RAD51-AS1, formerly TODRA), non-coding RNA activated by DNA damage (NORAD, formerly LINC00657), and GAS5 (15).
Role of MALAT1
Regarding the possible complications of COVID-19 infection, MALAT1 ameliorates deep vein thrombosis (DVT). The mechanism is through inhibition of proliferation and migration of endothelial progenitor cells and finally thrombosis dissolution via the Wnt/β-catenin signaling pathway (102). In addition, MALAT1 reduces the epigenetic silencing of viral transcription by regulating interactions of promoter-enhancer upregulating viral transcription and infection. MALAT1 interacts with STAT1, STAT3, STAT5A, IRF1, and IRF4. MALAT1 is a negative regulator of type I IFN production (10) (Figure 5) and sequesters miR-142-3p and miR-146a-5p to repress their anti-inflammatory activities and its overexpression, thus promoting inflammation (10). Deletion of MALAT1 led to the activation of p53 and its target genes, which affect the normal progression of the cell cycle (15) (Table 2). In addition, in a mouse model of SARS-CoV-2 infection, Malat1 was downregulated by tubulin α1 (TubA1A), the 60S ribosomal protein (Rlp6), and the endoplasmic reticulum protein retention receptor 3 (Kdelr3) (15).
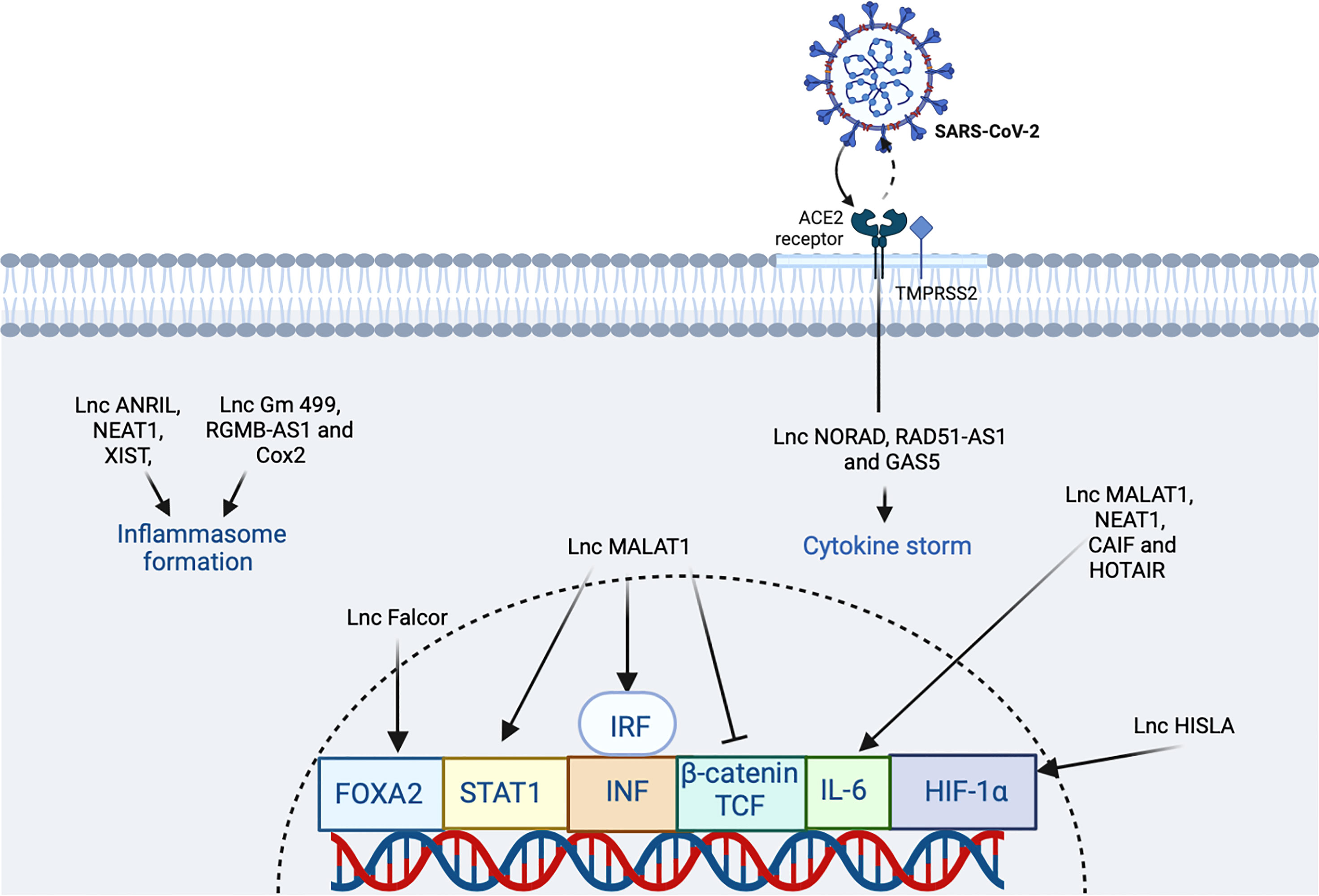
Figure 5 The participation of lncRNA in SARS-CoV2 infection. The virus binds the ACE2 receptor in the host cell. In the figure, the lncRNAs that induce an increase or inhibition are shown with different indicators (↑ and ⊥ respectively). The figure was designed in Biorender by AVRR (https://biorender.com/).
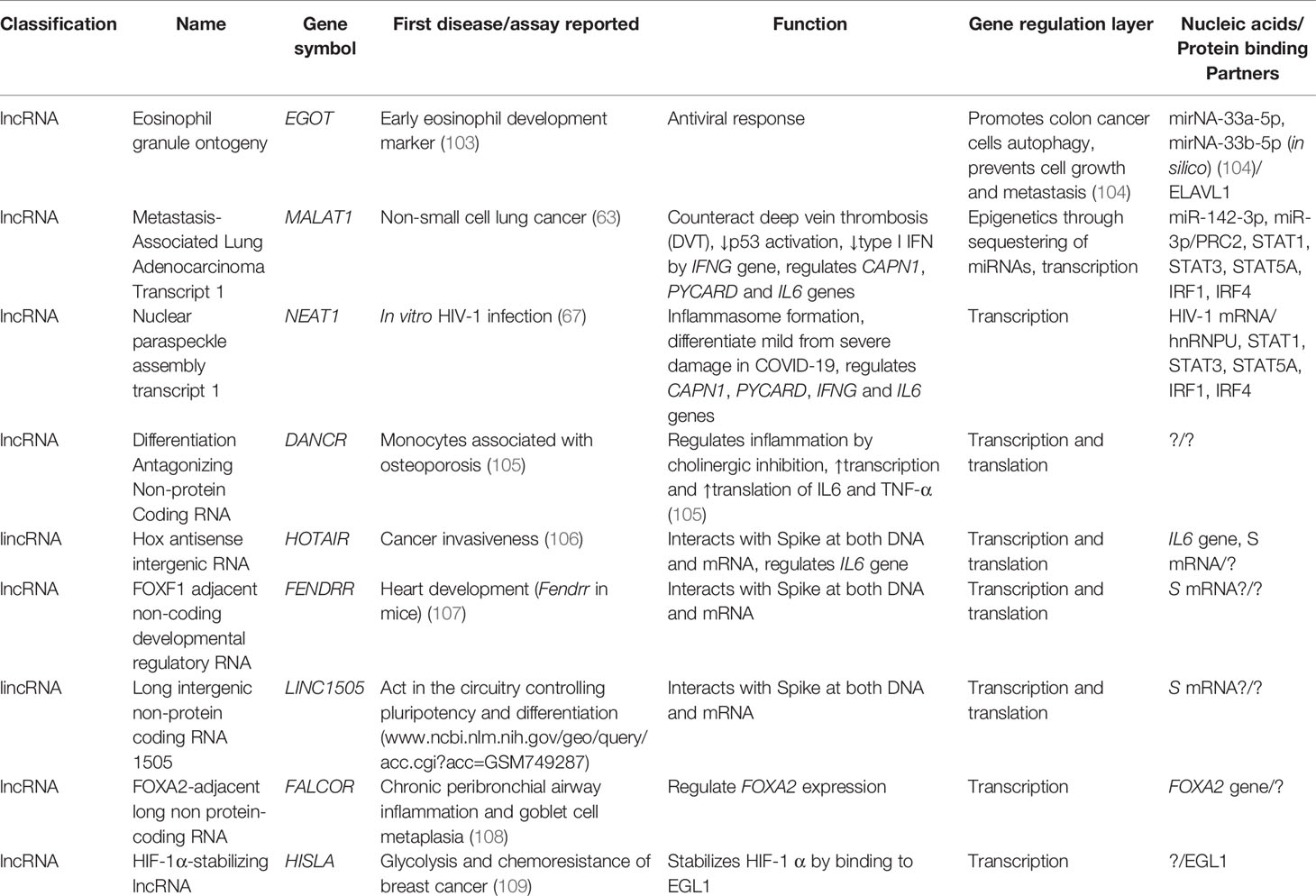
Table 2 Selected lncRNAs of SARS-CoV-2 including their first report, known function, and nucleic acid/protein-binding partners.
lncRNAs in Potential Prognosis in COVID-19
Also, in the case of autoimmunity in SARS-CoV-2-infected patients, some lncRNAs act through inflammatory pathways including TLR and NF-κB signaling (10). Approximately 15%–20% of COVID-19 patients develop severe pneumonia and acute respiratory distress syndrome (ARDS). LncRNA GATA5 is elevated and positively correlated with severe COVID-19 (11). In another study, vitamin D receptor gene (VDR) expression was lower in COVID-19 patients compared with non-infected controls. Differences were not found with Linc00511 or Linc00346 (110). Also, DANCR (in lungs) and NEAT1 lncRNAs targeting microRNAs were associated with genes that differentiate mild from severe damage in SARS-CoV-2 (111).
LncRNAs in COVID-19 Diagnosis
Genotyping of SARS-CoV-2 has been very useful in monitoring both viral evolution and transmission during the COVID-19 pandemic (112). However, in a more practical and affordable way, alternative strategies have been developed to demonstrate the presence of the virus in the samples of suspected individuals. The most common methods to identify the presence/absence of SARS-CoV-2 is reverse transcription (RT)-PCR (sensitivity = 82.2%; specificity = 100%) (113). Additional strategies include RT loop-mediated isothermal amplification (without the need for RNA extraction) (sensitivity = 87.5%; specificity = 100%) (114) and deep learning computer-aided diagnosis (CAD) based on digital chest x-ray images (overall detection accuracy = 96.31%; classification accuracy = 97.40%) (115). Transcript level combination of cytochrome p450, subfamily XXVIIB, polypeptide 1 (CYP27B1), VDR, small nucleolar RNA host gene 16 (SNHG16), structural maintenance of chromosomes 3 (SMC3, formerly chondroitin sulfate proteoglycan 6 or CSHG6), Linc00511, and Linc00346 could be a potential alternative method to COVID-19 diagnostics (sensitivity = 0.62; specificity = 0.81) (110).
LncRNAs as Potential Drug Targets in COVID-19
LncRNAs interactions with IFN in SARS-CoV-2 is an interesting research topic (15). The clotting alteration in COVID-19 may be related to inflammatory cytokines; thus, a better understanding of the cytokine regulation by lncRNAs will be helpful to understand this clinical phenotype and develop therapeutic targets to lncRNAs or molecules mimicking their function. Forty percent of known lncRNAs were found regulating different processes in the central nervous system (CNS) (15).
It has been suggested that lncRNAs can target and bind cytokine nucleotide sequences leading to downregulation of these cytokines. IFN-α is a common antiviral therapy in critically ill COVID-19 patients. Induction of MALAT1 expression by viral infection promotes activation of interferon regulatory factor 3 (IRF3) and production of type I IFN (Figure 5). Inhibition of the excessive inflammation may be a useful complement to COVID-19 therapy. Potential approaches in the clinic include oligonucleotide knockdown, RNAi knockdown, and viral gene therapy interfering with gene transcription, protein translation, and extranuclear messengers (15). Furthermore, lncRNAs are spatially correlated with TFs across the genome as, e.g., the case of FOXA2-adjacent long non protein-coding RNA (lncRNA Falcor) that can regulate forkhead box A2 (FOXA2) expression. Alteration of the regulatory feedback loop of Falcor-FoxA2 leads to altered cell migration and adhesion, thus causing goblet cell metaplasia and chronic peribronchial airway inflammation (15). Also, IL-6, transforming growth factor, beta 1 (TGF-β1, formerly TGF-β), nucleotide oligomerization domain-like receptor family, pyrin domain-containing 3 (NLRP3) inflammasome, and cholinergic signaling have been proposed as potential targets in COVID-19 (15). In the cholinergic inhibition, DANCR was found to potentially regulate inflammation. The identification of dysregulated or altered lncRNAs during SARS-CoV-2 infection could be a strategy for mitigating symptoms in patients and to limit viral genome amplification enhancing the immune response (15). The most relevant lncRNAs during COVID-19 seem to be HOTAIR, FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR, formerly FOXF1-AS1), LINC01505 (Table 2), and imprinted maternally expressed non-coding transcript (H19). Furthermore, FENDRR, HOTAIR, and LINC1505 potentially interact with the mRNA of spike protein (116). In macrophages, HIF-1α-stabilizing lncRNA (HISLA) regulates HIF-1α transcription factor by inhibition of its binding to EGL9 family hypoxia-inducible factor 1 protein (EGL1, formerly prolyl hydrolase domain-containing protein 2 or PHD2) (Table 2) (for an interaction network of SARS-CoV-2 proteins with human proteins, see Figure 6). This keeps continuous activation of an aerobic state in tumor cells. In the case of the treatment of COVID-19 using the strategy of an aptamer-siRNA chimera-mediated HISLA knockdown, this approach is promising but requires further investigation (15) (Figure 5).
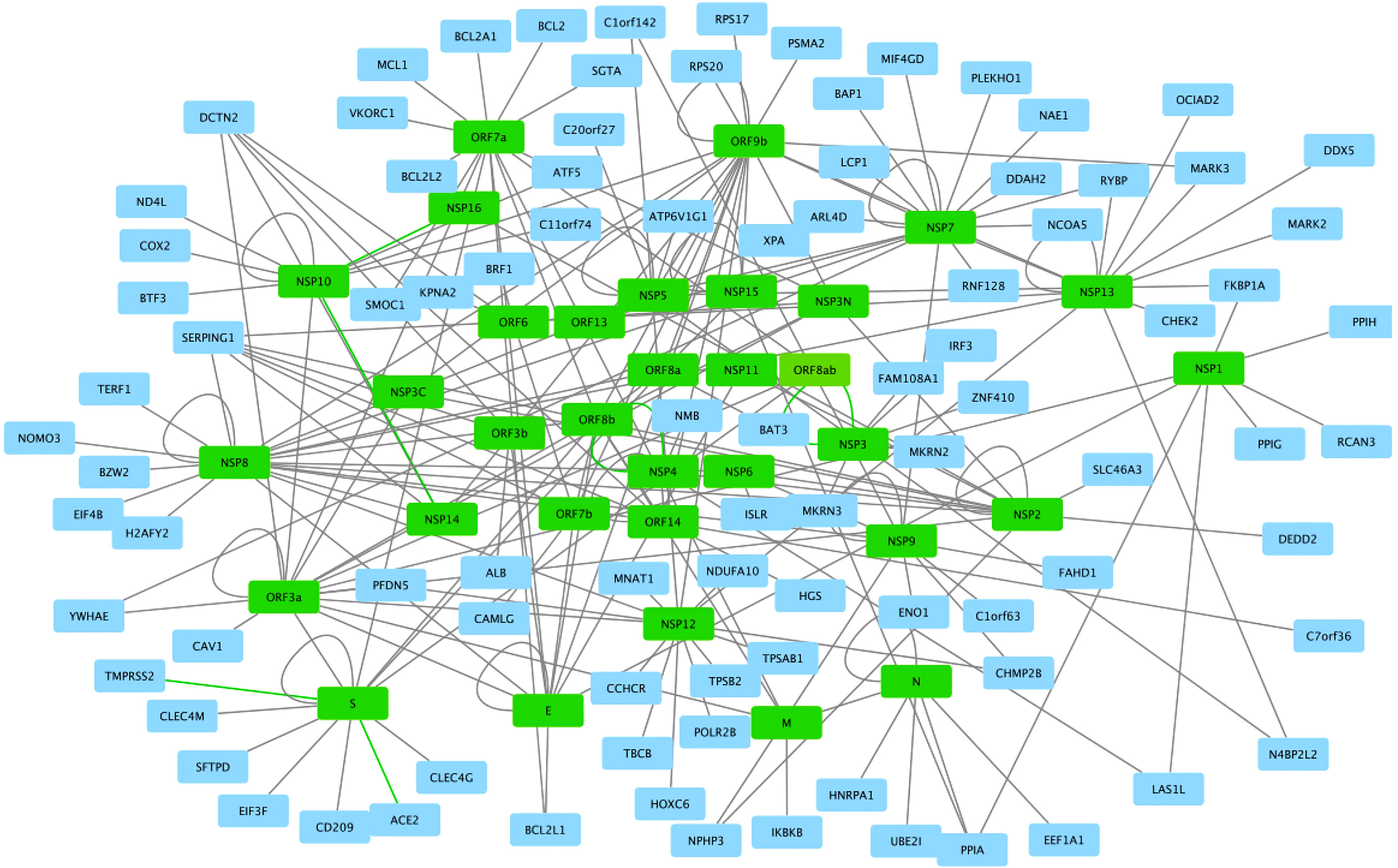
Figure 6 SARS-CoV2 -host protein interaction network. Viral proteins are shown in green boxes and human proteins are shown in blue boxes. Interactions between viral and human proteins are shown in the black lines. The figure was created in Cytoscape (https://cytoscape.org/download.html).
Host lncRNAs from Endogenous Retroviruses
Human endogenous retroviruses [HERVs, reviewed in (117)] are transposable elements comprising ∽10% of our genome and ranging from full-length proviruses to short gene fragments. HERVs are remnants of infectious exogenous retroviruses that became part of our DNA and now are inherited in a Mendelian manner. The HERV database [HERVd, http://herv.img.cas.cz (118)] contains 519,060 entities and 725,763 repeats of 582 different ERVs (accessed on March 3, 2022) that belong to 150 families. HERVs, working as Pathogen Associated Molecular Patterns (PAMPs), have become important parts of immune mechanisms to fight invading pathogens at least for dengue virus, herpes simplex viruses, influenza viruses, and HIV-1. Mechanisms used by ERVs are regulation of viral expression, enhancement of cellular sensing pathways, direct restriction of virion assembly, and blocking of entry receptors (117). Also, some ERVs are responsible for inserting proviruses in host genome facilitating infection and/or viral latency.
Host cells are armed with a variety of pattern recognition receptors (PRRs) that recognize nucleic acids from virus such as phospholipase A and acyltransferase 4 (PLAAT4, formerly retinoid acid-inducible gene 1or RIG-1) that recognizes 5´-triphosphate RNA, interferon induced with helicase C domain 1 (formerly melanoma differentiation-associated protein 5 or MDA5) that recognizes double-stranded RNAs (dsRNA), toll-like receptors 7 and 8 (TLR7 and TLR8) that recognize single-stranded RNA (ssRNA), toll-like receptor 3 (TLR3) that recognizes both dsRNA and poly I:C, toll-like receptor 9 that recognizes CpG DNA and DNA-immunoglobulin (Ig) complexes, and DNA-dependent activator of IRFs (DAI) that recognizes double-stranded DNA (dsDNA) [reviewed in (119)].
One classic example of ERV is a murine lncRNA called lncRNA ERV-derived positively regulating antiviral responses (lnc-Epav). This lncRNA boosts antiviral gene expression by increasing levels of RelA, a subunit of NF-κB transcription factor. The way that lnc-EPAV works in RELA promoter is competitively binding and displacing the splicing factor, proline- and glutamine-rich (SFPQ, an inhibitor of RELA gene) (47) (Table 3). Also, mice deficient in lnc-EPAV shows a reduced expression of type I interferons. In humans, the role of EPAV seems to be performed by interacting to MER9a2, LTR5A, and MLT2A1 HERVs. Results were concordant with a mouse model increasing levels of RELA in SFPQ knockdown human cells (47) (Figure 7).
Table 3 Selected lncRNAs of HERVs including their first report, known function, and nucleic acid/protein-binding partners.
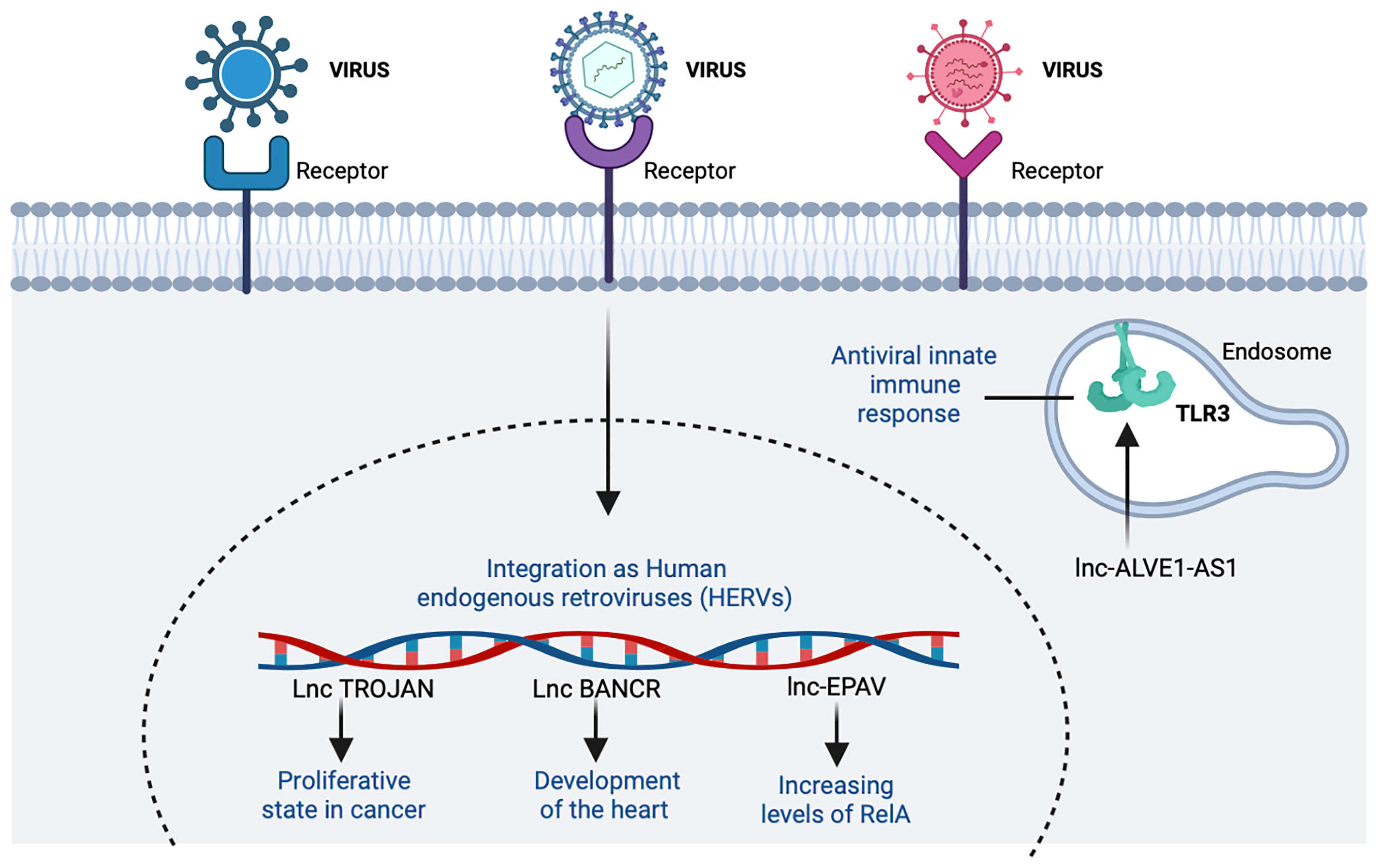
Figure 7 The participation of host lncRNAs from endogenous retroviruses (ERVs). ERVs are remnants of infectious exogenous retroviruses that became fixed in the DNA. In the figure, the lncRNAs that induce an increase are shown with the indicator (↑). Figure was designed in Biorender by AVRR (https://biorender.com/).
Another example of interaction with the immune response is the long non-coding endogenous avian leukosis virus on chromosome 1-antisense 1 (lnc-ALVE1-AS1), transcribed from ALVE1 gene, that can activate or bind to the Toll-like receptor 3 (TLR3) inducing an antiviral innate immune response (123) (Table 3 and Figure 7).
The lncRNAs related to HERVs have also been associated with functions in tissue growth, as in the case of V-raf murine sarcoma viral oncogene homolog B1 (BRAF)-regulated long non-coding RNA (BANCR), related to pro-migratory functions necessary in the development of the heart in humans and primates (121) (Table 3). This relationship with different tissues has also been analyzed with different types of cancer. In addition, lncRNA TROJAN is known to be involved in the proliferative state of breast cancer (BrCa) cells (Table 3). By reducing TROJAN expression, the proliferative potential in vitro is prevented, so this could reduce tumor volume and be a therapeutic target in the future (124). Even when these examples are in cancer, the similar cell cycle alterations with viral infection lead to hypothesize about a potential role of these HERVs in HIV/AIDS and/or COVID-19. Nevertheless, the importance of ERVs to antiviral immune response is becoming clear (47, 117); further basic and applied research is needed in this area.
Concluding Remarks
LncRNAs modulate both growth and viral replication as well as type I IFN expression mainly by stability regulation and interaction with mRNAs and proteins. The most relevant transcription factors that are regulated by lncRNAs are TP53, CTCF, MYC, SOX2, EZH2, SFPQ, and SUZ12 (125). In addition, lncRNAs seem to control the effector function of genes and peptides such as STAT1, STAT3, NF-κB, and IRFs and of many TFs, whose interactions remain to be discovered. The modulation of lncRNAs might regulate these transcription factors at both protein levels and at nucleotide motifs/structures. The key targeted potential therapies for SARS-Cov2 involve interfering gene transcription messengers and protein translation (15). Even when there are several questions to be solved, e.g., whether the cytokine storm activates ERVs in response to SARS-CoV-2 infection in COVID-19 (as is true for viral lncRNAs), a better understanding of how lncRNAs regulate gene expression in a cell/tissue specific manner by interactions with mRNAs, these new therapeutic targets and biomarkers hold promise for the treatment and risk stratification of patients with HIV/AIDS and COVID-19.
In addition, a promising area of research is to catalog and interpret the polymorphisms, mutations, and indels in lncRNAs (126, 127), as well as their epitranscriptomic modifications (128–130) with ethnic and cell/tissue specificity. As an example of this, our group reported that the regulatory SNP (rSNP) rs5743417, which is common in African and Afro-Americans, causes a 33% transcription downregulation in the constitutively expressed gene human β-defensin 1 (DEFB1) in the A549 pulmonary epithelial cell line (131). DEFB1 is a primordial gene of innate immunity involved in the defense against viruses [reviewed in (132)] including HIV-1 (133). In addition, hBD-1 peptide has other functions as well [reviewed in (134, 135)]. Interestingly, SNP rs5743417 overlaps with GS1-24F4.2, a lincRNA gene complementary to the X Kell blood-related 5 (XKR5) mRNA. GS1-24F4.2 has a testis-specific expression that could shed light on male infertility associated with COVID-19 (136). Results like these increase the difficulty of interpretation of genetic variants at different regulation layers but at the same time open new opportunities to dissect both molecular and clinical impact of polymorphisms, whose interpretation in the light of functional genomics remained elusive in the past decades. In general, the integral impact of rSNPs could be explained (a) by the fact that SNPs could alter transcription factor binding sites (TFBS) mainly in promoters or enhancers (137), thus causing an up- or downregulation (131); (b) by the fact that SNPs can create novel TFBS (138) and thus modify homeostatic gene expression pathways; (c) by altering the normal structural and/or sponging function of lncRNAs (54); and (d) probably by indirectly introducing variations in the epitranscriptome.
With no doubt, lncRNAs research is coming of age. These novel drugs inspired by host- and viral-encoded lncRNA have the potential to reduce the burden of HIV/AIDS and COVID-19 twofold: (1) by increasing their efficacy and (2) by minimizing the side effects by modulating gene transcription instead of the more traditional approaches such as complete silencing, knockout/editing of genes, non-selective DNA methylation, or blockade/neutralization of host peptides and proteins. We expect that a new generation of lncRNAs could modulate human and viral transcription in a more specific and unprecedented way but still maintaining homeostasis below the pathogenic threshold.
Author Contributions
AVRR: writing—original draft preparation and figures. EMP: writing and editing manuscript and tables. All authors contributed to the article and approved the submitted version.
Conflict of Interest
EP has a granted patent that protects a small molecule (and its synthesis and purification) that induces DEFB1 transcription. This synthetic molecule could be applied as therapy for viral diseases including but not limited to HIV/AIDS and COVID-19. This patent is property of CIATEJ.
The remaining author declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to the reviewers for their useful comments. AR is a Scholarship recipient of the Doctorate Program in Human Genetics, Guadalajara University/CIBO-IMSS, CONACYT. EP is a SNI-CONACYT Fellow since 2009.
References
1. Liu J, Ma L, Song C, Xing H, Cen S, Lin W. Anti-HIV Effects of Baculiferins Are Regulated by the Potential Target Protein DARS. ACS Chem Biol (2021) 16(8):1377–89. doi: 10.1021/acschembio.1c00148
2. Song C, Guo Y, Chen F, Liu W. LncRNA MALAT1 Promotes Osteogenic Differentiation Through the miR-217/AKT3 Axis: A Possible Strategy to Alleviate Osteoporosis. J Gene Med (2022):e3409. doi: 10.1002/jgm.3409
3. Amaral PP, Mattick JS. Noncoding RNA in Development. Mamm Genome (2008) 19(7-8):454–92. doi: 10.1007/s00335-008-9136-7
4. Zhang H, Yu L, Chen J, Liu L, Yang X, Cui H, et al. Role of Metabolic Reprogramming of Long non-Coding RNA in Clear Cell Renal Cell Carcinoma. J Cancer (2022) 13(2):691–705. doi: 10.7150/jca.62683
5. Gil N, Ulitsky I. Regulation of Gene Expression by Cis-Acting Long non-Coding RNAs. Nat Rev Genet (2020) 21(2):102–17. doi: 10.1038/s41576-019-0184-5
6. Esteller M. Non-Coding RNAs in Human Disease. Nat Rev Genet (2011) 12(12):861–74. doi: 10.1038/nrg3074
7. Mattick JS. The Genetic Signatures of Noncoding RNAs. PloS Genet (2009) 5(4):e1000459. doi: 10.1371/journal.pgen.1000459
8. Lottin S, Vercoutter-Edouart AS, Adriaenssens E, Czeszak X, Lemoine J, Roudbaraki M, et al. Thioredoxin Post-Transcriptional Regulation by H19 Provides a New Function to mRNA-Like non-Coding RNA. Oncogene (2002) 21(10):1625–31. doi: 10.1038/sj.onc.1205233
9. Beermann J, Piccoli MT, Viereck J, Thum T. Non-Coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev (2016) 96(4):1297–325. doi: 10.1152/physrev.00041.2015
10. Kesheh MM, Mahmoudvand S, Shokri S. Long Noncoding RNAs in Respiratory Viruses: A Review. Rev Med Virol (2021):e2275. doi: 10.1002/rmv.2275
11. Cheng J, Zhou X, Feng W, Jia M, Zhang X, An T, et al. Risk Stratification by Long non-Coding RNAs Profiling in COVID-19 Patients. J Cell Mol Med (2021) 25(10):4753–64. doi: 10.1111/jcmm.16444
12. Foulkes AS, Selvaggi C, Cao T, O'Reilly ME, Cynn E, Ma P, et al. Nonconserved Long Intergenic Noncoding RNAs Associate With Complex Cardiometabolic Disease Traits. Arterioscler Thromb Vasc Biol (2021) 41(1):501–11. doi: 10.1161/ATVBAHA.120.315045
13. Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, et al. LNCipedia: A Database for Annotated Human lncRNA Transcript Sequences and Structures. Nucleic Acids Res (2013) 41(Database issue):D246–51. doi: 10.1093/nar/gks915
14. More S, Zhu Z, Lin K, Huang C, Pushparaj S, Liang Y, et al. Long non-Coding RNA PSMB8-AS1 Regulates Influenza Virus Replication. RNA Biol (2019) 16(3):340–53. doi: 10.1080/15476286.2019.1572448
15. Yang Q, Lin F, Wang Y, Zeng M, Luo M. Long Noncoding RNAs as Emerging Regulators of COVID-19. Front Immunol (2021) 12:700184. doi: 10.3389/fimmu.2021.700184
16. Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, et al. Identification and Characterization of a Novel, Psoriasis Susceptibility-Related Noncoding RNA Gene, PRINS. J Biol Chem (2005) 280(25):24159–67. doi: 10.1074/jbc.M501704200
17. Mele M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL. Chromatin Environment, Transcriptional Regulation, and Splicing Distinguish lincRNAs and mRNAs. Genome Res (2017) 27(1):27–37. doi: 10.1101/gr.214205.116
18. Huang X, Sun W, Yan Z, Shi H, Yang Q, Wang P, et al. Integrative Analyses of Long Non-Coding RNA and mRNA Involved in Piglet Ileum Immune Response to Clostridium Perfringens Type C Infection. Front Cell Infect Microbiol (2019) 9:130. doi: 10.3389/fcimb.2019.00130
19. Napoli S. LncRNAs and Available Databases. Methods Mol Biol (2021) 2348:3–26. doi: 10.1007/978-1-0716-1581-2_1
20. Li CX, Chen J, Lv SK, Li JH, Li LL, Hu X. Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed mRNAs, miRNAs, and lncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients. Mediators Inflamm (2021) 2021:6635925. doi: 10.1155/2021/6635925
21. Siqueira E, Obiols-Guardia A, Jorge-Torres OC, Oliveira-Mateos C, Soler M, Ramesh-Kumar D, et al. Analysis of the circRNA and T-UCR Populations Identifies Convergent Pathways in Mouse and Human Models of Rett Syndrome. Mol Ther Nucleic Acids (2022) 27:621–44. doi: 10.1016/j.omtn.2021.12.030
22. Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved Elements in the Human Genome. Science (2004) 304(5675):1321–5. doi: 10.1126/science.1098119
23. Hull R, Mbita Z, Dlamini Z. Long non-Coding RNAs (LncRNAs), Viral Oncogenomics, and Aberrant Splicing Events: Therapeutics Implications. Am J Cancer Res (2021) 11(3):866–83. doi: 10.1126/science.1098119
24. Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, et al. The Role of MALAT1 Correlates With HPV in Cervical Cancer. Oncol Lett (2014) 7(6):2135–41. doi: 10.3892/ol.2014.1996
25. Wen J, Liu Y, Liu J, Liu L, Song C, Han J, et al. Expression Quantitative Trait Loci in Long non-Coding RNA ZNRD1-AS1 Influence Both HBV Infection and Hepatocellular Carcinoma Development. Mol Carcinog (2015) 54(11):1275–82. doi: 10.1002/mc.22200
26. Campbell M, Kung HJ, Izumiya Y. Long non-Coding RNA and Epigenetic Gene Regulation of KSHV. Viruses (2014) 6(11):4165–77. doi: 10.3390/v6114165
27. Joslyn RC, Forero A, Green R, Parker SE, Savan R. Long Noncoding RNA Signatures Induced by Toll-Like Receptor 7 and Type I Interferon Signaling in Activated Human Plasmacytoid Dendritic Cells. J Interferon Cytokine Res (2018) 38(9):388–405. doi: 10.1089/jir.2018.0086
28. Zhou Y, Li M, Xue Y, Li Z, Wen W, Liu X, et al. Interferon-Inducible Cytoplasmic Lnclrrc55-AS Promotes Antiviral Innate Responses by Strengthening IRF3 Phosphorylation. Cell Res (2019) 29(8):641–54. doi: 10.1038/s41422-019-0193-0
29. Liu W, Wang Z, Liu L, Yang Z, Liu S, Ma Z, et al. LncRNA Malat1 Inhibition of TDP43 Cleavage Suppresses IRF3-Initiated Antiviral Innate Immunity. Proc Natl Acad Sci U S A (2020) 117(38):23695–706. doi: 10.1073/pnas.2003932117
30. Xu J, Wang P, Li Z, Li Z, Han D, Wen M, et al. IRF3-Binding lncRNA-ISIR Strengthens Interferon Production in Viral Infection and Autoinflammation. Cell Rep (2021) 37(5):109926. doi: 10.1016/j.celrep.2021.109926
31. Ng CS, Kato H, Fujita T. Fueling Type I Interferonopathies: Regulation and Function of Type I Interferon Antiviral Responses. J Interferon Cytokine Res (2019) 39(7):383–92. doi: 10.1089/jir.2019.0037
32. Minati L, Firrito C, Del Piano A, Peretti A, Sidoli S, Peroni D, et al. One-Shot Analysis of Translated Mammalian lncRNAs With AHARIBO. Elife (2021) 10. doi: 10.7554/eLife.59303
33. Ji Z, Song R, Regev A, Struhl K. Many lncRNAs, 5'utrs, and Pseudogenes are Translated and Some are Likely to Express Functional Proteins. Elife (2015) 4:e08890. doi: 10.7554/eLife.08890
34. Prabakaran S, Hemberg M, Chauhan R, Winter D, Tweedie-Cullen RY, Dittrich C, et al. Quantitative Profiling of Peptides From RNAs Classified as Noncoding. Nat Commun (2014) 5:5429. doi: 10.1038/ncomms6429
35. Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of Noncoding RNAs and Cancer. Cancer Lett (2021) 497:89–99. doi: 10.1016/j.canlet.2020.10.002
36. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and circRNA. Mol Cancer (2020) 19(1):22. doi: 10.1186/s12943-020-1147-3
37. Wang Y, Feng F, Zheng P, Wang L, Wang Y, Lv Y, et al. Dysregulated lncRNA and mRNA may Promote the Progression of Ischemic Stroke via Immune and Inflammatory Pathways: Results From RNA Sequencing and Bioinformatics Analysis. Genes Genomics (2022) 44(1):97–108. doi: 10.1007/s13258-021-01173-1
38. Shi L, Magee P, Fassan M, Sahoo S, Leong HS, Lee D, et al. A KRAS-Responsive Long non-Coding RNA Controls microRNA Processing. Nat Commun (2021) 12(1):2038. doi: 10.1038/s41467-021-22337-3
39. Dong X, Chen X, Lu D, Diao D, Liu X, Mai S, et al. LncRNA Mir205hg Hinders HNRNPA0 Translation: Anti-Oncogenic Effects in Esophageal Carcinoma. Mol Oncol (2022) 16(3):795–812. doi: 10.1002/1878-0261.13142
40. Wu J, Huang Y, Zhang J, Xiang Z, Yang J. LncRNA CPhar Mediates Exercise-Induced Cardioprotection by Promoting eNOS Phosphorylation at Ser1177 via DDX17/PI3K/Akt Pathway After MI/RI. Int J Cardiol (2022) 350:16. doi: 10.1016/j.ijcard.2021.12.040
41. Ding H, Wang F, Shi X, Ma H, Du Y, Hou L, et al. LncRNA MALAT1 Induces the Dysfunction of Beta Cells via Reducing the Histone Acetylation of the PDX-1 Promoter in Type 1 Diabetes. Exp Mol Pathol (2020) 114:104432. doi: 10.1016/j.yexmp.2020.104432
42. Kong D, Hou Y, Li W, Ma X, Jiang J. LncRNA-ZXF1 Stabilizes P21 Expression in Endometrioid Endometrial Carcinoma by Inhibiting Ubiquitination-Mediated Degradation and Regulating the miR-378a-3p/PCDHA3 Axis. Mol Oncol (2022) 16(3):813–29. doi: 10.1002/1878-0261.12940
43. Zhu QQ, Lai MC, Chen TC, Wang X, Tian L, Li DL, et al. LncRNA SNHG15 Relieves Hyperglycemia-Induced Endothelial Dysfunction via Increased Ubiquitination of Thioredoxin-Interacting Protein. Lab Invest (2021) 101(9):1142–52. doi: 10.1038/s41374-021-00614-5
44. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The Long Noncoding RNA THRIL Regulates TNFalpha Expression Through its Interaction With hnRNPL. Proc Natl Acad Sci U S A (2014) 111(3):1002–7. doi: 10.1073/pnas.1313768111
45. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST Long ncRNA Controls Microbial Susceptibility and Epigenetic Activation of the Interferon-Gamma Locus. Cell (2013) 152(4):743–54. doi: 10.1016/j.cell.2013.01.015
46. Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R, et al. Cutting Edge: A Natural Antisense Transcript, AS-IL1alpha, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1alpha. J Immunol (2015) 195(4):1359–63. doi: 10.4049/jimmunol.1500264
47. Zhou B, Qi F, Wu F, Nie H, Song Y, Shao L, et al. Endogenous Retrovirus-Derived Long Noncoding RNA Enhances Innate Immune Responses via Derepressing RELA Expression. mBio (2019) 10(4). doi: 10.1128/mBio.00937-19
48. Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, et al. Natural Antisense Transcript Stabilizes Inducible Nitric Oxide Synthase Messenger RNA in Rat Hepatocytes. Hepatology (2008) 47(2):686–97. doi: 10.1002/hep.22036
49. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-Binding Long Noncoding RNA lnc-DC Controls Human Dendritic Cell Differentiation. Science (2014) 344(6181):310–3. doi: 10.1126/science.1251456
50. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin Signature Reveals Over a Thousand Highly Conserved Large non-Coding RNAs in Mammals. Nature (2009) 458(7235):223–7. doi: 10.1038/nature07672
51. Yang L, Froberg JE, Lee JT. Long Noncoding RNAs: Fresh Perspectives Into the RNA World. Trends Biochem Sci (2014) 39(1):35–43. doi: 10.1016/j.tibs.2013.10.002
52. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A Gene From the Region of the Human X Inactivation Centre is Expressed Exclusively From the Inactive X Chromosome. Nature (1991) 349(6304):38–44. doi: 10.1038/349038a0
53. Siniscalchi C, Di Palo A, Russo A, Potenza N. The lncRNAs at X Chromosome Inactivation Center: Not Just a Matter of Sex Dosage Compensation. Int J Mol Sci (2022) 23(2). doi: 10.3390/ijms23020611
54. Sebastian-delaCruz M, Gonzalez-Moro I, Olazagoitia-Garmendia A, Castellanos-Rubio A, Santin I. The Role of lncRNAs in Gene Expression Regulation Through mRNA Stabilization. Noncoding RNA (2021) 7(1). doi: 10.3390/ncrna7010003
55. Caffrey M. HIV Envelope: Challenges and Opportunities for Development of Entry Inhibitors. Trends Microbiol (2011) 19(4):191–7. doi: 10.1016/j.tim.2011.02.001
56. Henzinger H, Barth DA, Klec C, Pichler M. Non-Coding RNAs and SARS-Related Coronaviruses. Viruses (2020) 12(12). doi: 10.3390/v12121374
57. Carnero E, Barriocanal M, Prior C, Pablo Unfried J, Segura V, Guruceaga E, et al. Long Noncoding RNA EGOT Negatively Affects the Antiviral Response and Favors HCV Replication. EMBO Rep (2016) 17(7):1013–28. doi: 10.15252/embr.201541763
58. Ma L, Zhang H, Zhang Y, Li H, An M, Zhao B, et al. Integrated Analysis of lncRNA, miRNA and mRNA Profiles Reveals Potential lncRNA Functions During Early HIV Infection. J Transl Med (2021) 19(1):135. doi: 10.1186/s12967-021-02802-9
59. Lazar DC, Morris KV, Saayman SM. The Emerging Role of Long non-Coding RNAs in HIV Infection. Virus Res (2016) 212:114–26. doi: 10.1016/j.virusres.2015.07.023
60. Qiu L, Wang T, Tang Q, Li G, Wu P, Chen K. Long Non-Coding RNAs: Regulators of Viral Infection and the Interferon Antiviral Response. Front Microbiol (2018) 9:1621. doi: 10.3389/fmicb.2018.01621
61. Pinto DO, Scott TA, DeMarino C, Pleet ML, Vo TT, Saifuddin M, et al. Effect of Transcription Inhibition and Generation of Suppressive Viral non-Coding RNAs. Retrovirology (2019) 16(1):13. doi: 10.1186/s12977-019-0475-0
62. Qu D, Sun WW, Li L, Ma L, Sun L, Jin X, et al. Long Noncoding RNA MALAT1 Releases Epigenetic Silencing of HIV-1 Replication by Displacing the Polycomb Repressive Complex 2 From Binding to the LTR Promoter. Nucleic Acids Res (2019) 47(6):3013–27. doi: 10.1093/nar/gkz117
63. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a Novel Noncoding RNA, and Thymosin Beta4 Predict Metastasis and Survival in Early-Stage non-Small Cell Lung Cancer. Oncogene (2003) 22(39):8031–41. doi: 10.1038/sj.onc.1206928
64. Chao TC, Zhang Q, Li Z, Tiwari SK, Qin Y, Yau E, et al. The Long Noncoding RNA HEAL Regulates HIV-1 Replication Through Epigenetic Regulation of the HIV-1 Promoter. mBio (2019) 10(5). doi: 10.1128/mBio.02016-19
65. Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON Modulates HIV-1 Replication in a NFAT-Dependent Manner and is Differentially Regulated by Early and Late Viral Proteins. Sci Rep (2015) 5:8639. doi: 10.1038/srep08639
66. Barclay RA, Schwab A, DeMarino C, Akpamagbo Y, Lepene B, Kassaye S, et al. Exosomes From Uninfected Cells Activate Transcription of Latent HIV-1. J Biol Chem (2017) 292(28):11682–701. doi: 10.1074/jbc.M117.793521
67. Barichievy S, Naidoo J, Boulle M, Scholefield J, Parihar SP, Coussens AK, et al. Viral Apoptosis Evasion via the MAPK Pathway by Use of a Host Long Noncoding RNA. Front Cell Infect Microbiol (2018) 8:263. doi: 10.3389/fcimb.2018.00263
68. Wang H, Liu Y, Huan C, Yang J, Li Z, Zheng B, et al. NF-kappaB-Interacting Long Noncoding RNA Regulates HIV-1 Replication and Latency by Repressing NF-kappaB Signaling. J Virol (2020) 94(17). doi: 10.1128/JVI.01057-20
69. Wu H, Zheng J, Deng J, Zhang L, Li N, Li W, et al. LincRNA-Uc002yug.2 Involves in Alternative Splicing of RUNX1 and Serves as a Predictor for Esophageal Cancer and Prognosis. Oncogene (2015) 34(36):4723–34. doi: 10.1038/onc.2014.400
70. Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, et al. FAS-Antisense 1 lncRNA and Production of Soluble Versus Membrane Fas in B-Cell Lymphoma. Leukemia (2014) 28(12):2376–87. doi: 10.1038/leu.2014.126
71. Schwarzer A, Emmrich S, Schmidt F, Beck D, Ng M, Reimer C, et al. The non-Coding RNA Landscape of Human Hematopoiesis and Leukemia. Nat Commun (2017) 8(1):218. doi: 10.1038/s41467-017-00212-4
72. Qian X, Xu C, Zhao P, Qi Z. Long non-Coding RNA GAS5 Inhibited Hepatitis C Virus Replication by Binding Viral NS3 Protein. Virology (2016) 492:155–65. doi: 10.1016/j.virol.2016.02.020
73. Saayman S, Ackley A, Turner AW, Famiglietti M, Bosque A, Clemson M, et al. An HIV-Encoded Antisense Long Noncoding RNA Epigenetically Regulates Viral Transcription. Mol Ther (2014) 22(6):1164–75. doi: 10.1038/mt.2014.29
74. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT. Science (2005) 309(5740):1570–3. doi: 10.1126/science.1115901
75. Li J, Chen C, Ma X, Geng G, Liu B, Zhang Y, et al. Long Noncoding RNA NRON Contributes to HIV-1 Latency by Specifically Inducing Tat Protein Degradation. Nat Commun (2016) 7:11730. doi: 10.1038/ncomms11730
76. Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and Characterization of HuR, a Ubiquitously Expressed Elav-Like Protein. J Biol Chem (1996) 271(14):8144–51. doi: 10.1074/jbc.271.14.8144
77. Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 Long Noncoding RNA and Paraspeckle Bodies Modulate HIV-1 Posttranscriptional Expression. mBio (2013) 4(1):e00596-12. doi: 10.1128/mBio.00596-12
78. Liu H, Hu PW, Couturier J, Lewis DE, Rice AP. HIV-1 Replication in CD4(+) T Cells Exploits the Down-Regulation of Antiviral NEAT1 Long non-Coding RNAs Following T Cell Activation. Virology (2018) 522:193–8. doi: 10.1016/j.virol.2018.07.020
79. Jin C, Peng X, Xie T, Lu X, Liu F, Wu H, et al. Detection of the Long Noncoding RNAs Nuclear-Enriched Autosomal Transcript 1 (NEAT1) and Metastasis Associated Lung Adenocarcinoma Transcript 1 in the Peripheral Blood of HIV-1-Infected Patients. HIV Med (2016) 17(1):68–72. doi: 10.1111/hiv.12276
80. Huan C, Li Z, Ning S, Wang H, Yu XF, Zhang W. Long Noncoding RNA Uc002yug.2 Activates HIV-1 Latency Through Regulation of mRNA Levels of Various RUNX1 Isoforms and Increased Tat Expression. J Virol (2018) 92(9). doi: 10.1128/JVI.01844-17
81. Wright MW. A Short Guide to Long non-Coding RNA Gene Nomenclature. Hum Genomics (2014) 8:7. doi: 10.1186/1479-7364-8-7
82. Seal RL, Chen LL, Griffiths-Jones S, Lowe TM, Mathews MB, O'Reilly D, et al. A Guide to Naming Human non-Coding RNA Genes. EMBO J (2020) 39(6):e103777. doi: 10.15252/embj.2019103777
83. Boliar S, Gludish DW, Jambo KC, Kamng'ona R, Mvaya L, Mwandumba HC, et al. Inhibition of the lncRNA SAF Drives Activation of Apoptotic Effector Caspases in HIV-1-Infected Human Macrophages. Proc Natl Acad Sci U S A (2019) 116(15):7431–8. doi: 10.1073/pnas.1818662116
84. Postler TS, Pantry SN, Desrosiers RC, Ghosh S. Identification and Characterization of a Long non-Coding RNA Up-Regulated During HIV-1 Infection. Virology (2017) 511:30–9. doi: 10.1016/j.virol.2017.08.006
85. Chen L, Chen L, Zuo L, Gao Z, Shi Y, Yuan P, et al. Short Communication: Long Noncoding RNA GAS5 Inhibits HIV-1 Replication Through Interaction With miR-873. AIDS Res Hum Retroviruses (2018) 34(6):544–9. doi: 10.1089/aid.2017.0177
86. Nguyen LNT, Nguyen LN, Zhao J, Schank M, Dang X, Cao D, et al. Long Non-Coding RNA GAS5 Regulates T Cell Functions via Mir21-Mediated Signaling in People Living With HIV. Front Immunol (2021) 12:601298. doi: 10.3389/fimmu.2021.601298
87. Schynkel T, Szaniawski MA, Spivak AM, Bosque A, Planelles V, Vandekerckhove L, et al. Interferon-Mediated Long Non-Coding RNA Response in Macrophages in the Context of HIV. Int J Mol Sci (2020) 21(20). doi: 10.3390/ijms21207741
88. Torkzaban B, Natarajaseenivasan K, Mohseni Ahooyi T, Shekarabi M, Amini S, Langford TD, et al. The lncRNA LOC102549805 (U1) Modulates Neurotoxicity of HIV-1 Tat Protein. Cell Death Dis (2020) 11(10):835. doi: 10.1038/s41419-020-03033-4
89. Ghafouri-Fard S, Mahmud Hussen B, Abak A, Taheri M, Abdulmajid Ayatollahi S. Emerging Role of non-Coding RNAs in the Course of HIV Infection. Int Immunopharmacol (2022) 103:108460. doi: 10.1016/j.intimp.2021.108460
90. Biswas S, Haleyurgirisetty M, Ragupathy V, Wang X, Lee S, Hewlett I, et al. Differentially Expressed Host Long Intergenic Noncoding RNA and mRNA in HIV-1 and HIV-2 Infection. Sci Rep (2018) 8(1):2546. doi: 10.1038/s41598-018-20791-6
91. Hartana CA, Rassadkina Y, Gao C, Martin-Gayo E, Walker BD, Lichterfeld M, et al. Long Noncoding RNA MIR4435-2HG Enhances Metabolic Function of Myeloid Dendritic Cells From HIV-1 Elite Controllers. J Clin Invest (2021) 131(9). doi: 10.1172/JCI146136
92. Zhang J, Thakuri BKC, Zhao J, Nguyen LN, Nguyen LNT, Khanal S, et al. Long Noncoding RNA RUNXOR Promotes Myeloid-Derived Suppressor Cell Expansion and Functions via Enhancing Immunosuppressive Molecule Expressions During Latent HIV Infection. J Immunol (2021) 206(9):2052–60. doi: 10.4049/jimmunol.2001008
93. Kulkarni S, Lied A, Kulkarni V, Rucevic M, Martin MP, Walker-Sperling V, et al. CCR5AS lncRNA Variation Differentially Regulates CCR5, Influencing HIV Disease Outcome. Nat Immunol (2019) 20(7):824–34. doi: 10.1038/s41590-019-0406-1
94. Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, et al. Genome-Scale Deletion Screening of Human Long non-Coding RNAs Using a Paired-Guide RNA CRISPR-Cas9 Library. Nat Biotechnol (2016) 34(12):1279–86. doi: 10.1038/nbt.3715
95. Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-Based Genome-Scale Identification of Functional Long Noncoding RNA Loci in Human Cells. Science (2017) 355(6320). doi: 10.1126/science.aah7111
96. Gutschner T, Richtig G, Haemmerle M, Pichler M. From Biomarkers to Therapeutic Targets-the Promises and Perils of Long non-Coding RNAs in Cancer. Cancer Metastasis Rev (2018) 37(1):83–105. doi: 10.1007/s10555-017-9718-5
97. Boliar S, Russell DG. Lnc(ing)RNAs to the "Shock and Kill" Strategy for HIV-1 Cure. Mol Ther Nucleic Acids (2021) 23:1272–80. doi: 10.1016/j.omtn.2021.02.004
98. Geis FK, Sabo Y, Chen X, Li Y, Lu C, Goff SP. CHAF1A/B Mediate Silencing of Unintegrated HIV-1 DNAs Early in Infection. Proc Natl Acad Sci U S A (2022) 119(4). doi: 10.1073/pnas.2116735119
99. Moazzam-Jazi M, Lanjanian H, Maleknia S, Hedayati M, Daneshpour MS. Interplay Between SARS-CoV-2 and Human Long non-Coding RNAs. J Cell Mol Med (2021) 25(12):5823–7. doi: 10.1111/jcmm.16596
100. Serpeloni JM, Lima Neto QA, Lucio LC, Ramao A, Carvalho de Oliveira J, Gradia DF, et al. Genome Interaction of the Virus and the Host Genes and non-Coding RNAs in SARS-CoV-2 Infection. Immunobiology (2021) 226(5):152130. doi: 10.1016/j.imbio.2021.152130
101. Laha S, Saha C, Dutta S, Basu M, Chatterjee R, Ghosh S, et al. In Silico Analysis of Altered Expression of Long non-Coding RNA in SARS-CoV-2 Infected Cells and Their Possible Regulation by STAT1, STAT3 and Interferon Regulatory Factors. Heliyon (2021) 7(3):e06395. doi: 10.1016/j.heliyon.2021.e06395
102. Du B, Wang J, Zang S, Mao X, Du Y. Long non-Coding RNA MALAT1 Suppresses the Proliferation and Migration of Endothelial Progenitor Cells in Deep Vein Thrombosis by Regulating the Wnt/beta-Catenin Pathway. Exp Ther Med (2020) 20(4):3138–46. doi: 10.3892/etm.2020.9066
103. Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, et al. EGO, a Novel, Noncoding RNA Gene, Regulates Eosinophil Granule Protein Transcript Expression. Blood (2007) 109(12):5191–8. doi: 10.1182/blood-2006-06-027987
104. Liu Y, Zhang B, Cao WB, Wang HY, Niu L, Zhang GZ. Study on Clinical Significance of LncRNA EGOT Expression in Colon Cancer and Its Effect on Autophagy of Colon Cancer Cells. Cancer Manag Res (2020) 12:13501–12. doi: 10.2147/CMAR.S285254
105. Tong X, Gu PC, Xu SZ, Lin XJ. Long non-Coding RNA-DANCR in Human Circulating Monocytes: A Potential Biomarker Associated With Postmenopausal Osteoporosis. Biosci Biotechnol Biochem (2015) 79(5):732–7. doi: 10.1080/09168451.2014.998617
106. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
107. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The Tissue-Specific lncRNA Fendrr is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev Cell (2013) 24(2):206–14. doi: 10.1016/j.devcel.2012.12.012
108. Swarr DT, Herriges M, Li S, Morley M, Fernandes S, Sridharan A, et al. The Long Noncoding RNA Falcor Regulates Foxa2 Expression to Maintain Lung Epithelial Homeostasis and Promote Regeneration. Genes Dev (2019) 33(11-12):656–68. doi: 10.1101/gad.320523.118
109. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular Vesicle-Packaged HIF-1alpha-Stabilizing lncRNA From Tumour-Associated Macrophages Regulates Aerobic Glycolysis of Breast Cancer Cells. Nat Cell Biol (2019) 21(4):498–510. doi: 10.1038/s41556-019-0299-0
110. Taheri M, Rad LM, Hussen BM, Nicknafs F, Sayad A, Ghafouri-Fard S. Evaluation of Expression of VDR-Associated lncRNAs in COVID-19 Patients. BMC Infect Dis (2021) 21(1):588. doi: 10.1186/s12879-021-06248-8
111. Meydan C, Madrer N, Soreq H. The Neat Dance of COVID-19: NEAT1, DANCR, and Co-Modulated Cholinergic RNAs Link to Inflammation. Front Immunol (2020) 11:590870. doi: 10.3389/fimmu.2020.590870
112. Kubik S, Marques AC, Xing X, Silvery J, Bertelli C, De Maio F, et al. Recommendations for Accurate Genotyping of SARS-CoV-2 Using Amplicon-Based Sequencing of Clinical Samples. Clin Microbiol Infect (2021) 27(7):1036.e1–e8. doi: 10.1016/j.cmi.2021.03.029
113. Williams TC, Wastnedge E, McAllister G, Bhatia R, Cuschieri K, Kefala K, et al. Sensitivity of RT-PCR Testing of Upper Respiratory Tract Samples for SARS-CoV-2 in Hospitalised Patients: A Retrospective Cohort Study. Wellcome Open Res (2020) 5:254. doi: 10.12688/wellcomeopenres.16342.1
114. Anahtar MN, McGrath GEG, Rabe BA, Tanner NA, White BA, Lennerz JKM, et al. Clinical Assessment and Validation of a Rapid and Sensitive SARS-CoV-2 Test Using Reverse Transcription Loop-Mediated Isothermal Amplification Without the Need for RNA Extraction. Open Forum Infect Dis (2021) 8(2):ofaa631. doi: 10.1093/ofid/ofaa631
115. Al-Antari MA, Hua CH, Bang J, Lee S. "Fast Deep Learning Computer-Aided Diagnosis of COVID-19 Based on Digital Chest X-Ray Images". Appl Intell (Dordr) (2020), 1–18. doi: 10.21203/rs.3.rs-36353/v2
116. Natarelli L, Parca L, Mazza T, Weber C, Virgili F, Fratantonio D. MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2. Noncoding RNA (2021) 7(1):14. doi: 10.3390/ncrna7010014
117. Srinivasachar Badarinarayan S, Sauter D. Switching Sides: How Endogenous Retroviruses Protect Us From Viral Infections. J Virol (2021) 95(12). doi: 10.1128/JVI.02299-20
118. Paces J, Pavlicek A, Zika R, Kapitonov VV, Jurka J, Paces V. HERVd: The Human Endogenous RetroViruses Database: Update. Nucleic Acids Res (2004) 32(Database issue):D50. doi: 10.1093/nar/gkh075
119. Schlee M, Hartmann G. Discriminating Self From non-Self in Nucleic Acid Sensing. Nat Rev Immunol (2016) 16(9):566–80. doi: 10.1038/nri.2016.78
120. Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, et al. BRAFV600E Remodels the Melanocyte Transcriptome and Induces BANCR to Regulate Melanoma Cell Migration. Genome Res (2012) 22(6):1006–14. doi: 10.1101/gr.140061.112
121. Wilson KD, Ameen M, Guo H, Abilez OJ, Tian L, Mumbach MR, et al. Endogenous Retrovirus-Derived lncRNA BANCR Promotes Cardiomyocyte Migration in Humans and Non-Human Primates. Dev Cell (2020) 54(6):694–709.e9. doi: 10.1016/j.devcel.2020.07.006
122. Yu X, Zheng H, Tse G, Chan MT, Wu WK. Long non-Coding RNAs in Melanoma. Cell Prolif (2018) 51(4):e12457. doi: 10.1111/cpr.12457
123. Chen S, Hu X, Cui IH, Wu S, Dou C, Liu Y, et al. An Endogenous Retroviral Element Exerts an Antiviral Innate Immune Function via the Derived lncRNA lnc-ALVE1-As1. Antiviral Res (2019) 170:104571. doi: 10.1016/j.antiviral.2019.104571
124. Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ, et al. The Endogenous Retrovirus-Derived Long Noncoding RNA TROJAN Promotes Triple-Negative Breast Cancer Progression via ZMYND8 Degradation. Sci Adv (2019) 5(3):eaat9820. doi: 10.1126/sciadv.aat9820
125. Liu CJ, Gao C, Ma Z, Cong R, Zhang Q, Guo AY. Lncrinter: A Database of Experimentally Validated Long non-Coding RNA Interaction. J Genet Genomics (2017) 44(5):265–8. doi: 10.1016/j.jgg.2017.01.004
126. Xiang X, Chen L, He J, Ma G, Li Y. LncRNA GAS5 Rs145204276 Polymorphism Reduces Renal Cell Carcinoma Susceptibility in Southern Chinese Population. J Inflammation Res (2022) 15:1147–58. doi: 10.2147/JIR.S348628
127. Miao YR, Liu W, Zhang Q, Guo AY. Lncrnasnp2: An Updated Database of Functional SNPs and Mutations in Human and Mouse lncRNAs. Nucleic Acids Res (2018) 46(D1):D276–D80. doi: 10.1093/nar/gkx1004
128. Manfredonia I, Incarnato D. RNA Post-Transcriptional Modification Mapping Data Analysis Using RNA Framework. Methods Mol Biol (2021) 2298:3–13. doi: 10.1007/978-1-0716-1374-0_1
129. Liu Q, Lang X, Gregory RI. An Informatics Pipeline for Profiling and Annotating RNA Modifications. Methods Mol Biol (2021) 2298:15–27. doi: 10.1007/978-1-0716-1374-0_2
130. Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL, et al. RMBase V2.0: Deciphering the Map of RNA Modifications From Epitranscriptome Sequencing Data. Nucleic Acids Res (2018) 46(D1):D327–D34. doi: 10.1093/nar/gkx934
131. Cruz Diaz LA, Gutierrez Ortega A, Chavez Alvarez RDC, Velarde Felix JS, Prado Montes de Oca E. Regulatory SNP Rs5743417 Impairs Constitutive Expression of Human Beta-Defensin 1 and has High Frequency in Africans and Afro-Americans. Int J Immunogenet (2020) 47(4):332–41. doi: 10.1111/iji.12475
132. Ryan LK, Diamond G. Modulation of Human Beta-Defensin-1 Production by Viruses. Viruses (2017) 9(6). doi: 10.3390/v9060153
133. Sun L, Finnegan CM, Kish-Catalone T, Blumenthal R, Garzino-Demo P, La Terra Maggiore GM, et al. Human Beta-Defensins Suppress Human Immunodeficiency Virus Infection: Potential Role in Mucosal Protection. J Virol (2005) 79(22):14318–29. doi: 10.1128/JVI.79.22.14318-14329.2005
134. Alvarez AH, Martinez Velazquez M, Prado Montes de Oca E. Human Beta-Defensin 1 Update: Potential Clinical Applications of the Restless Warrior. Int J Biochem Cell Biol (2018) 104:133–7. doi: 10.1016/j.biocel.2018.09.007
135. Prado-Montes de Oca E. Human Beta-Defensin 1: A Restless Warrior Against Allergies, Infections and Cancer. Int J Biochem Cell Biol (2010) 42(6):800–4. doi: 10.1016/j.biocel.2010.01.021
136. Sabetian S, Castiglioni I, Jahromi BN, Mousavi P, Cava C. In Silico Identification of miRNA-lncRNA Interactions in Male Reproductive Disorder Associated With COVID-19 Infection. Cells (2021) 10(6). doi: 10.3390/cells10061480
137. Flores Saiffe Farias A, Jaime Herrera Lopez E, Moreno Vazquez CJ, Li W, Prado Montes de Oca E. Predicting Functional Regulatory SNPs in the Human Antimicrobial Peptide Genes DEFB1 and CAMP in Tuberculosis and HIV/AIDS. Comput Biol Chem (2015) 59:117–25. doi: 10.1016/j.compbiolchem.2015.09.002
138. Prado-Montes de Oca E, Velarde-Felix JS, Rios-Tostado JJ, Picos-Cardenas VJ, Figuera LE. SNP 668C (-44) Alters a NF-Kappab1 Putative Binding Site in non-Coding Strand of Human Beta-Defensin 1 (DEFB1) and is Associated With Lepromatous Leprosy. Infect Genet Evol (2009) 9(4):617–25. doi: 10.1016/j.meegid.2009.03.006
Keywords: long noncoding RNA, acquired immunodeficiency syndrome (AIDS), SARS-CoV-2, COVID-19, endogenous retroviruses (ERVs), HIV-1, drug development, gene regulation and expression
Citation: Ruiz Ramírez AV and Prado Montes de Oca E (2022) Therapeutic Potential of Long Non-Coding RNAs of HIV-1, SARS-CoV-2, and Endogenous Retroviruses. Front.Virol. 2:849349. doi: 10.3389/fviro.2022.849349
Received: 06 January 2022; Accepted: 21 March 2022;
Published: 29 April 2022.
Edited by:
Cybele Carina Garcia, University of Buenos Aires, ArgentinaReviewed by:
Dinesh Devadoss, Florida International University, United StatesPrashanth N. Suravajhala, Amrita Vishwa Vidyapeetham University, India
Copyright © 2022 Ruiz Ramírez and Prado Montes de Oca. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ernesto Prado Montes de Oca, ZXByYWRvQGNpYXRlai5teA==
 Andrea Virginia Ruiz Ramírez
Andrea Virginia Ruiz Ramírez Ernesto Prado Montes de Oca
Ernesto Prado Montes de Oca