
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Virol. , 14 December 2022
Sec. Fundamental Virology
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.1078454
This article is part of the Research Topic Post-transcriptional regulation of viral protein expression and function View all 5 articles
RNA metabolism in the eukaryotic cell includes the formation of ribonucleoprotein complexes (RNPs) that, depending on their protein components, have a different function. Cytoplasmic RNPs, such as stress granules (SGs) or P-bodies (PBs) are quite relevant during infections modulating viral and cellular RNA expression and as key players in the host cell antiviral response. RNA helicases are abundant components of RNPs and could have a significant effect on viral infection. This review focuses in the role that RNPs and RNA helicases have during coronavirus (CoVs) infection. CoVs are emerging highly pathogenic viruses with a large single-stranded RNA genome. During CoV infection, a complex network of RNA-protein interactions in different RNP structures is established. In general, RNA helicases and RNPs have an antiviral function, but there is limited knowledge on whether the viral protein interactions with cell components are mediators of this antiviral effect or are part of the CoV antiviral counteraction mechanism. Additional data is needed to elucidate the role of these RNA-protein interactions during CoV infection and their potential contribution to viral replication or pathogenesis.
Eukaryotic cell RNA is associated with proteins, forming ribonucleoprotein complexes (RNPs). The subcellular location of mRNAs in RNPs is a powerful mechanism for the spatial and temporal regulation of RNA processing events in the cell (1). In addition, by sharing components, different RNPs form a large regulatory network in cells.
The cytoplasm contains a number of RNPs that include specific mRNAs at various stages of post-transcriptional processing, including stress granules (SGs), processing bodies (PBs), neuronal bodies and exosome bodies (2). SGs and PBs are the most studied ones. Neuronal bodies are associated with the transport of translationally arrested mRNAs along the axon to dendrites (3, 4). Exosome bodies were proposed as sites for AU-rich element mediated mRNA decay containing exosome subunits (5). In the nucleus, the most studied RNPs are the Cajal bodies (CBs) (6), although the nucleus contains other RNPs such as nucleolus, nuclear speckles, nuclear stress bodies, histone locus bodies, paraspeckles and promyelocytic leukemia (PML) bodies (7–11). In general, these nuclear RNPs are sites of defined biochemical reactions, by concentrating reaction components in a confined space, and of gene activation or repression (12).
RNA helicases belong to an abundant protein family that is conserved from bacteria to humans, and are associated with all cellular processes involving RNA (13–16). RNA helicases are abundant components of cellular RNPs (17, 18). For instance, during gene expression, RNA helicases catalyze RNPs rearrangements starting with gene transcription and continuing with consecutive post-transcriptional steps, such as pre-mRNA splicing, mRNA export, translation and turnover (19–21).
Both cytoplasmic and nuclear RNPs have been involved in several disease conditions, including viral infections. This review is focused in the interplay between cytoplasmic RNPs, RNA helicases, and coronavirus (CoV) infections.
The most studied cytoplasmic RNPs are SGs and PBs. These structures are highly dynamic centers of mRNA sorting, storage and degradation, where the processes of splicing, non-sense mediated RNA decay (NMD), translation, turnover and RNA silencing intersect (Figure 1) (22–27). SGs are transient foci enriched in translation initiation factors and 40S ribosomal subunits, while PBs are enriched for RNA decay machinery. SGs and PBs each contain unique marker proteins although many proteins can be found in both SGs and PBs, such as eukaryotic translation initiation factor 4E (eIF4E), AU-rich RNA binding protein tristetraprolin (TTP), argonaute RISC catalytic component 2 (AGO2), apolipoprotein B mRNA-editing complex 3 (APOBEC3), poly(rC) binding protein 2 (PCBP2) and others (23, 28, 29). In fact, apart from having shared protein components, PBs and SGs dynamically exchange RNP cargo, often form co-aggregates and have been proposed to serve as nucleation sites for SGs formation (30, 31). SGs, PBs and other cytoplasmic foci are highly dynamic structures, although PBs are quite stable over the time (32). They are in a dynamic steady state with other RNPs, such as polysomes, in response to the translational state of the cell (23) (Figure 1).
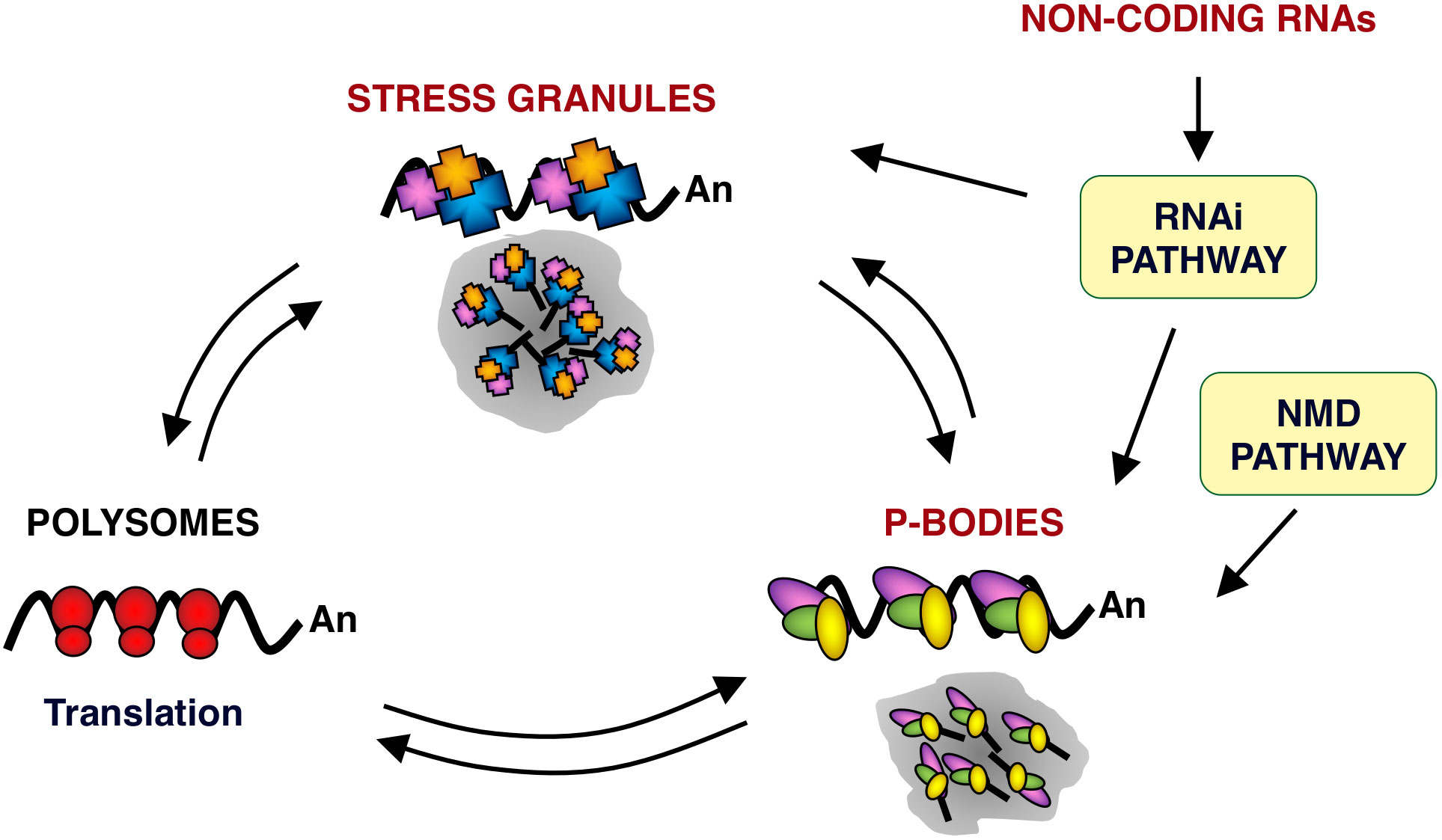
Figure 1 mRNA metabolism in eukaryotic cells. Schematic representation of mRNA locations in cells. mRNAs are found in three major RNPs: polysomes, PBs and SGs. PBs and SGs are large dynamic centers of mRNA sorting, storage and degradation. RNPs from and to polysomes and from and to SGs and PBs are in a dynamic equilibrium. Within PBs, RNPs could undergo further remodeling and define a path to follow, including their return to polysomes. In addition, PBs have been shown to interact and exchange components with SGs. PBs are also related to mRNA quality control mechanisms, such as NMD pathway. In silencing, non-coding RNAs function as the guide molecules that are incorporated into RNA-induced silencing complex (RISC) to control the translation and degradation of their target mRNAs within PBs and SGs. Arrows indicate the different equilibrium between RNPs. Ribosomes (red ovals), SG protein components (blue, orange and purple polygons) and PB protein components (yellow, green and purple ovals) are also represented.
SGs are 200-400 nm dynamic structures quickly formed when cells encounter external stresses and translation rates decline, and disperse when translation conditions are restored (33, 34). The most commonly described triggers of SG formation are oxidative, starvation, heat stresses and infection with different pathogens. These stimuli activate eukaryotic translation initiation factor 2 alpha (eIF2α) kinases: heme-regulated kinase (HRI), general control non-depressible 2 kinase, (GCN2), dsRNA-activated protein kinase R (PKR) and PKR-like endoplasmic reticulum kinase (PERK). The eIF2α phosphorylation blocks translation, forcing an accumulation of the stalled 43S and 48S ribosomal preinitiation complexes. Inhibition of the function of eukaryotic translation initiation factors eIF4G or eIF4A (DDX2A) is also linked to SG formation (35) and some mechanisms of SG formation can proceed in the absence of eIF2α phosphorylation (36–38). The components of SGs can be classified into three main groups: core components, such as stalled initiation complexes; RNA-binding proteins associated to silencing and transcript stability, such as the scaffold proteins T-cell-restricted intracellular antigen 1 (TIA-1) or TIA-1-related protein (TIAR), and RNA-binding proteins associated to mRNA metabolism (23, 39–42). The key function of SGs is to protect the mRNAs during cell stress, altering the composition of the RNPs in a reversible manner (43). Moreover, SGs are generally believed to have an antiviral activity upon viral infection and many viruses manipulate SGs to evade host responses (2, 44–47).
PBs act as temporary reservoirs for non-translated mRNAs which may further enter translation or be degraded. PBs contain components of the 5’ to 3’ decay machinery, NMD pathway and RNA-induced silencing machinery (48). This enrichment on mRNA decay machinery components led to propose that significant RNA decay occurs within PBs. However, this is controversial since some activators of mRNA decay pathways found in PBs are translational regulators also present in SGs (29, 49). PBs are constitutively present in the eukaryotic cells but increase in size and number when translational arrest occurs, coincident with SG formation, resulting in physical interactions among SG and PB structures (23, 30). It has been described that mRNA degradation occurs in PBs and depends on the existence of degradation enzymes and mRNA degradation intermediates (50, 51). Many of the PB components are not restricted to these foci and are also present in the soluble cytoplasm and nucleus, suggesting that the different processes might start before the mRNAs entry into PBs.
Virus infection activates cell stress responses at many levels that modulate RNA granules. In general, RNP granules can represent an obstacle for virus replication and also serve as sensors to mount the innate immune response. Therefore, viruses have evolved different mechanisms to control the assembly and functions of RNP granules and, in some cases the components of RNPs are co-opted into novel virus-specific structures required for virus replication (52).
Mammalian orthoreovirus (MRV), a dsRNA virus, induces SGs at an early stage of the infection, correlating with reduced translation of both cellular and viral mRNAs and increased phosphorylation of eIF2α (53). Poliovirus inhibits SGs assembly at the late stages of infection, by a mechanism involving cleavage of SG protein Ras GTPase-activating protein-binding protein 1 (G3BP1), which is mediated by the viral 3C protease (54). In addition, TIA-1-containing granules persisted even at late stage of poliovirus infection, but those RNPs are remnants of normal SGs that do not correlate with translational repression (55, 56). Unlike MRV and poliovirus, Influenza A virus (IAV) prevents SG formation at all stages of the infection, by a mechanism depending on NS1 protein expression, since SGs inhibit IAV replication (57).
As an example of viral co-opting of RNP components, RNAs from flaviviruses, such as West Nile virus and Dengue virus, bind TIA-1 and sequester many PB components near or within perinuclear viral replication centers while the number of PBs was decreased (58). Human immunodeficiency virus (HIV)-1 TAT protein interacts with the PB component DDX3 to facilitate viral mRNA translation (59), and HIV-1 TAT protein is antagonized by antiviral factors APOBEC3 and Moloney leukemia virus 10 (MOV10), that are PB constituents (60–63).
CoVs are single-stranded RNA viruses with large genomes of around 30 Kb, which life cycle occurs in the cytoplasm of the infected cell. CoV RNA synthesis, including replication and transcription, is a complex process involving host cell membrane rearrangements, viral and cellular proteins (64, 65). Regarding the role of cytoplasmic RNPs in CoV infection, it has been shown that mouse hepatitis virus (MHV) replication induces host translational shutoff and mRNA decay, with concomitant formation of SGs and PBs (66). Porcine transmissible gastroenteritis virus (TGEV) triggers SG formation and interferes with PB formation, correlating with viral replication and transcription (67). Severe and acute respiratory syndrome (SARS)-CoV, SARS-CoV-2 and avian infectious bronchitis virus (IBV) nucleocapsid (N) proteins have been found to interact with the SG component G3BP1, and this has been proposed as a mechanism to avoid SG formation (68–72). In addition, porcine epidemic diarrhea virus (PEDV) promotes G3BP1 cleavage by caspase-8 (73) and Middle East respiratory syndrome (MERS)-CoV 4a accessory protein interferes with SG formation by inhibiting the activation of PKR binding to dsRNA, thereby inhibiting the formation of SGs and ensuring viral protein translation and efficient virus replication (74, 75). It has been recently reported that SARS-CoV-2, HCoV-229E and HCoV-OC43 cause PB disassembly. In the case of SARS-CoV-2, this process may be mediated by N protein (76).
RNA helicases share conserved helicase domains, all containing ATP binding motifs, providing the basis for helicases classification (Figure 2). Most cellular RNA helicases belong to the SF2 superfamily and only a few of them, the up-frameshift suppressor 1(Upf1)-like helicases (i.e., UPF1, also known as RENT1, and MOV10 helicase), belong to SF1. In contrast, many viral RNA helicases, such as CoV nsp13, belong to SF1 superfamily (77). The SF1 and SF2 helicases contain seven to nine conserved motifs that constitute the helicase core. In addition, SF2 RNA helicases, generally referred to as DExD/H box RNA helicases, are divided in five different subgroups (Figure 2): DEAD box (for the conserved amino acid residues Asp-Glu-Ala-Asp), DEAH (for the conserved amino acid residues Asp-Glu-Ala-His)/RHA, Ski2 (Superkiller-like 2)-like, retinoic acid-inducible gene I (RIG-I)-like, and Viral DExH proteins, named after one of the consensus amino acid sequence motifs (16, 78–80). The DEAD box (DDX) helicases are the largest subgroup within SF2, most of them involved in RNA metabolism, from transcription to degradation and in establishment of larger RNA-protein complexes, such as ribosomes (81, 82).
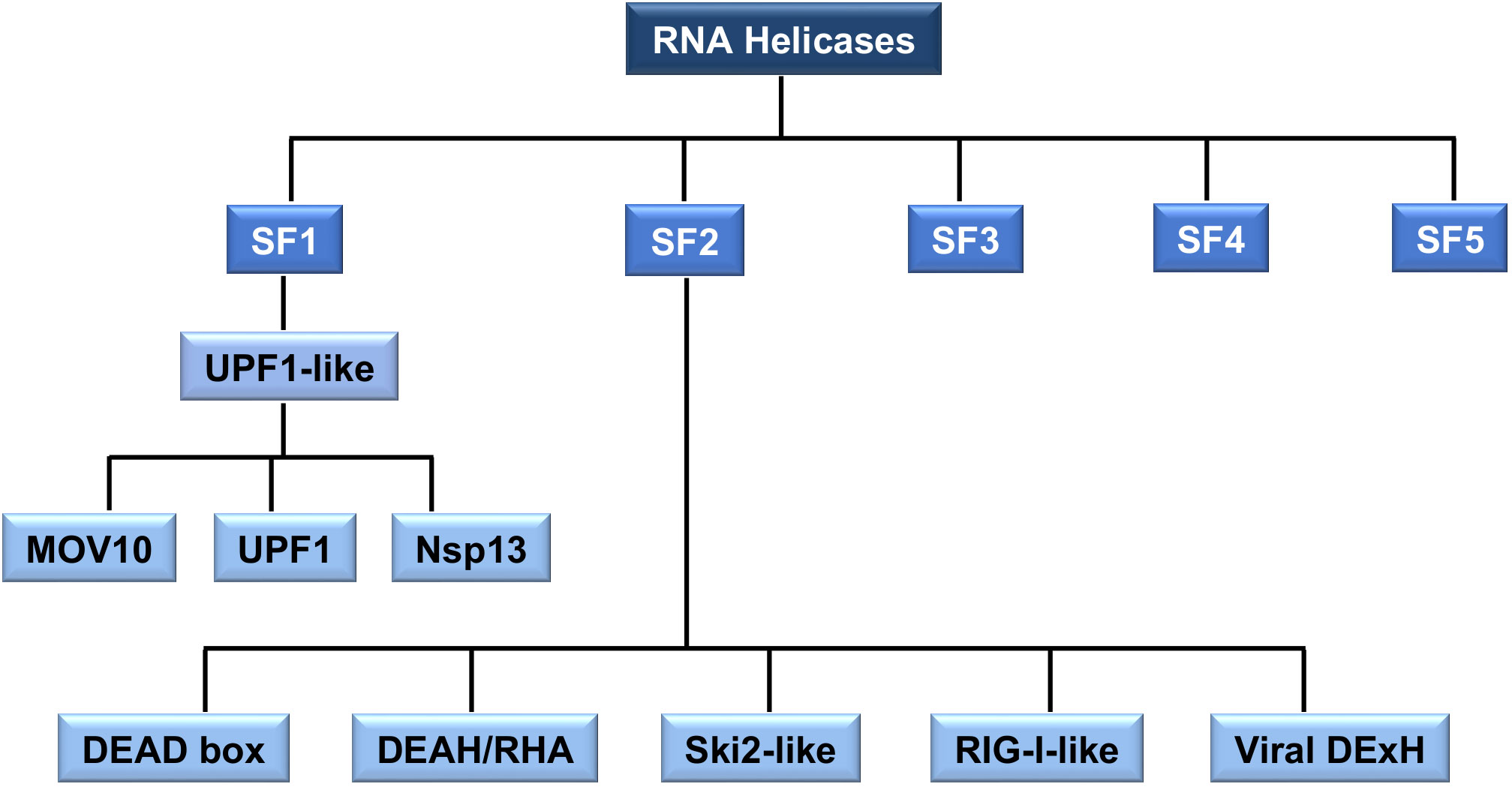
Figure 2 The classification of RNA helicases. All RNA helicases are classified into five superfamilies (SF) 1-5. SF1 and SF2 are sub-divided into distinct families and groups. SF1 is divided into Upf1-like family. SF2 is divided into five families. SF2 RNA helicase, DEAD box, DEAH/RHA, Ski2-like, RIG-I-like and Viral DExH, are collectively referred to as the DExD/H box of RNA helicases. The type of RNA helicases mentioned in this review, such as MOV10, UPF1, CoV helicase (nsp13), DEAD box and DEAH-box is indicated.
Cellular DDX RNA helicases play essential roles in a broad array of biological processes and serve multiple roles at the virus-host interface (83–86). Specifically, DDX RNA helicases are hijacked by CoVs and participate in essential DDX-mediated viral replication steps (87). During CoV infection, these RNA helicases interact with viral proteins, as described below (Table 1). In addition, many of them interact with viral RNA (88–90) (Table 1), raising the possibility that these RNA helicase-CoV protein interactions are mediated by RNA.
DDX1, interacts with the nsp14 protein both from SARS-CoV and IBV, suggesting that this interaction may be conserved for other CoVs. DDX1-nsp14 interaction contributes to efficient CoV RNA replication in cell cultures (91). DDX1 also interacts with the nsp14 protein from TGEV, inducing interferon (IFN) production (92) and suggesting that nsp14 would be the viral component sensed by DDX1-DDX21-DHX36 cytoplasmic sensor, described in dendritic cells (93). Supporting CoV interaction with the DDX1-DDX21-DHX36 complex, N proteins of SARS-CoV-2 and IBV interact with DDX21, although the consequences of this interaction are still unclear (71, 94). In addition, phosphorylation of MHV N protein allows the recruitment of DDX1 to the CoV replication transcription complex (RTC), increasing the synthesis of longer viral RNAs, suggesting that this is one of the viral strategies to support the transition from discontinuous to continuous transcription (95).
The interaction between DDX3X and N proteins from SARS-CoV, SARS-CoV-2, or IBV was described (71, 96). Since DDX3X is involved in the activation of the innate immune response at different levels (97–99), it was suggested that N protein can modulate the immune response by binding to DDX3X and inhibiting these antiviral pathways, as demonstrated for other viruses (100). SARS-CoV nsp13 interacts with DDX5 and inhibition of DDX5 results in the suppression of viral replication. A proviral function for DDX5 has been suggested, maybe acting as nsp13 co-activator during RNA synthesis (101). In addition, since DDX5 is involved in nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) response, the binding between nsp13 and DDX5 can also have the role of evading host inflammatory response (102). Interestingly, DDX5 interacts with other DEAD box helicases, such as DDX3X and its close homolog DDX17. The phosphorylation-dependent interaction between DDX5 and DDX3X was proposed as a combined mechanism of action for DEAD box helicases involved in RNP remodeling and splicing, by forming a complex that functions in shuttling RNP export from the nucleus to the cytoplasm (103, 104).
DDX6, a helicase located in SGs and PBs, is downregulated during PEDV infection to reduce endoplasmic reticulum (ER) stress, facilitating viral replication (105). The accumulation of DDX6 is decreased by HCoV-OC43 infection, while it does not change with HCoV-229E or SARS-CoV-2 infection (76). DDX6 interacts with RIG-I to increase antiviral signaling and IFN induction (106). However, the potential effect of DDX6 innate immunity modulation during PEDV infection was not analyzed as IFN incompetent Vero cells were used.
There is also recent evidence of other RNA helicases being involved in the immune response during SARS-CoV-2 infection. DHX16 has been described as a novel sensor for SARS-CoV-2 replication, triggering IFN response (107). The levels of DHX9 expression in effector TCD8+ cells have been correlated with better COVID-19 outcome (108).
MOV10 protein is a UPF1-like RNA helicase (109, 110). MOV10 helicase has more than 1060 interactors, according to BioGRID database (111), some of them linking MOV10 to RNA metabolism pathways such as the NMD pathway or the siRNA gene silencing pathway. In fact, depletion of MOV10 mRNA by using siRNAs interferes with RNAi activity (110, 112). In addition, MOV10 is an IFN-stimulated gene (ISG) (113) and MOV10 protein is involved in IFN induction after viral infection (114).
Both proviral and antiviral functions have been reported for MOV10. As a proviral factor, it is required for hepatitis delta virus (HDV) replication but not for the translation of its mRNA (115) and facilitates enterovirus replication (116). As an antiviral factor it inhibits: (i) HIV-1 and other retroviruses replication at multiple steps (63), (ii) nuclear import of influenza virus nucleoprotein (117), (iii) porcine reproductive and respiratory syndrome virus (PRRSV) replication by avoiding nucleocapsid protein trafficking to the nucleus (118), (iv) hepatitis C virus (HCV) and Dengue virus replication by a partly unknown mechanisms (119, 120), and (v) bunyavirus replication by blocking several nucleoprotein functions (121). In the case of hepatitis B virus (HBV), contradictory data has been reported for both, its proviral (122) and antiviral activities (123).
The role of MOV10 during CoV replication has recently been analyzed by our group. The interaction between endogenous MOV10 and N protein during infection was demonstrated for porcine TGEV virus, and both mild (HCoV-229E) and highly pathogenic CoVs (SARS-CoV, MERS-CoV and SARS-CoV-2) (124). This interaction was also reported by other authors, outside the infection context using N protein overexpression (71, 94, 125). During MERS-CoV infection, both MOV10 and N protein co-localized in cytoplasmic RNPs, not related with CoV RTCs, that may eventually contain also other cellular proteins, such as TIAR, AGO2 or UPF1 (124) (Figure 3). Functional analyses indicated that MOV10 has antiviral activity during MERS-CoV and SARS-CoV-2 infection, but not during HCoV-229E infection, suggesting a relationship with CoV pathogenesis that will be further explored (124). In agreement with other previously described MOV10 interactions with cellular proteins, MOV10 interaction with CoV N protein was RNA-dependent (124). Interestingly, MOV10 interaction with SARS-CoV-2 RNA was recently described (88, 90, 126–128) and it was conserved for other human CoVs (127). This issue opens the possibility of N protein having an active role in the MOV10 antiviral activity or of its non-specific recruitment to MOV10 RNPs, mediated by its binding to viral RNAs. In contrast with other viral infections, MOV10 helicase activity was required for antiviral function during MERS-CoV infection (124). The binding of MOV10 and CoV N protein was conserved independently of MOV10 antiviral activity, i.e., with a MOV10 mutant without helicase activity, or in HCoV-229E infection (124), suggesting that N binding to MOV10 is not related with MOV10 antiviral function. In addition, CoV N protein has been proposed to subvert SGs or counteract NMD pathway to facilitate viral replication (129, 130). Therefore, additional experimental evidence is needed to determine whether N protein may have an active role in the formation or function of MOV10 RNPs.
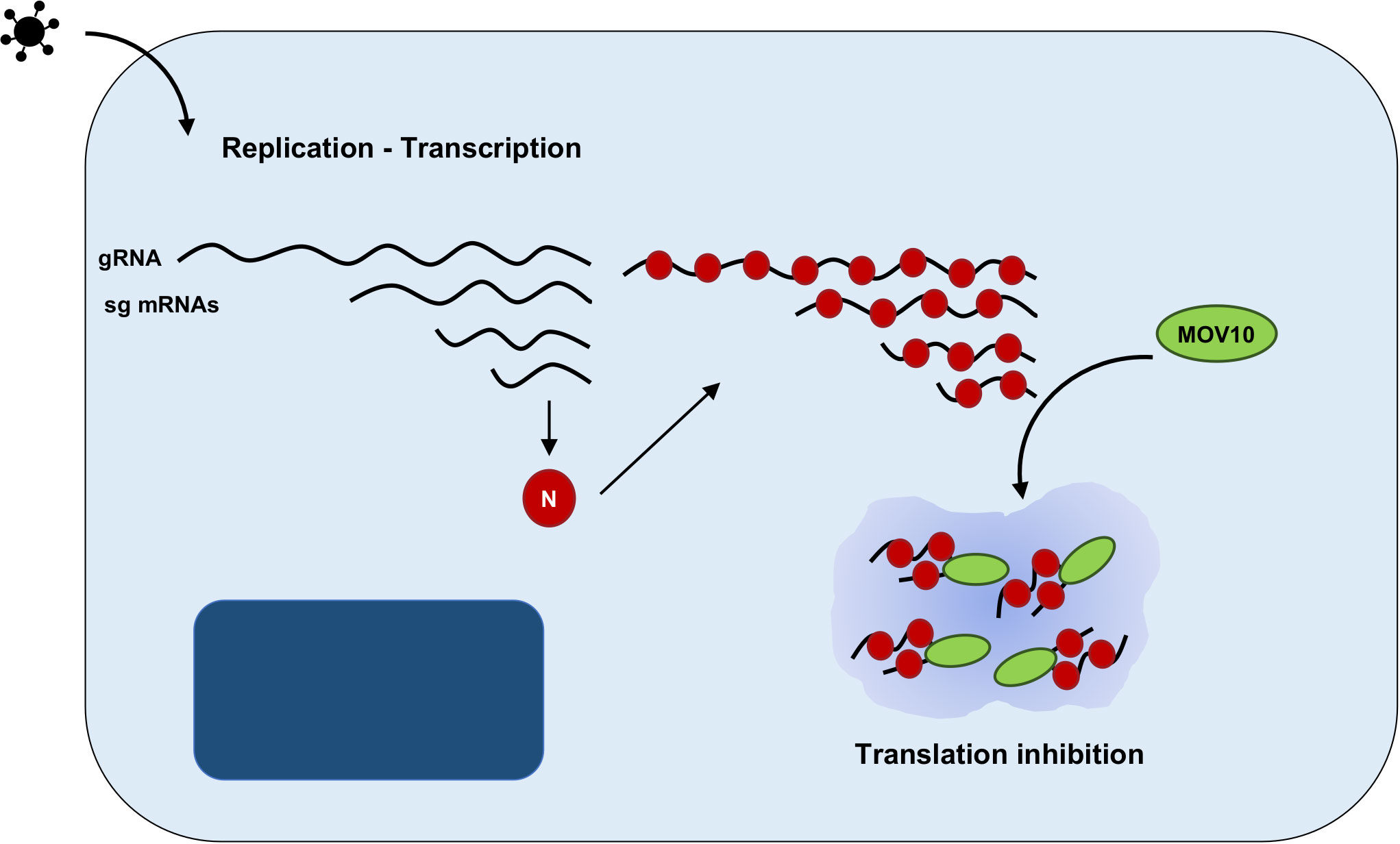
Figure 3 Proposed model of MOV10 antiviral function during MERS-CoV infection. Schematic representation of an infected cell, including nucleus (dark blue) and cytoplasm (light blue). CoV replication and transcription leads to the synthesis of a nested set of viral RNAs, both viral genome (gRNA) and subgenomic mRNAs (sgmRNAs). N protein is translated from the subgenomic RNA and binds to viral RNAs. The RNA-dependent interaction between N protein and MOV10 may lead to the accumulation of viral RNAs and N protein in cytoplasmic RNPs, inhibiting viral translation.
RNA-protein interactions leading to RNPs formation represent one of the mechanisms for post-transcriptional regulation of protein expression. Viruses interact with host cell RNPs to facilitate viral replication and to counteract antiviral responses. In the case of CoVs, cytoplasmic RNPs have, in general, an antiviral effect. Therefore, CoVs have developed mechanisms to antagonize RNPs formation or function.
The interaction between CoVs and cellular RNA helicases is a clear example of the complex network of interactions that may have different contributions to the outcome of the infection. Some RNA helicases, as DDX1, are recruited to CoVs RTCs to facilitate viral RNA synthesis, having a proviral function. On the other hand, RNA helicases have also an antiviral function mediated by their role in the innate immune response to the infection or by forming RNPs. These issues are frequently related, as RNPs components, both RNAs and proteins, may have a direct effect on the expression of innate immune factors.
There is limited knowledge on the interactions and functions of RNA helicases or RNPs in CoV infection, especially in the infection context. Recent omics approaches may help to decipher the composition and function of RNPs during CoV infection. However, some of the interactions occur between cellular proteins and essential viral factors, which cannot be modified to directly analyze the impact on infection. In addition, RNPs components often have a functional redundancy or are shared between different RNPs with different functions. Moreover, RNPs are dynamic and could be dependent on the cell type or infection stage. Therefore, it would be challenging to unravel these virus-host interaction networks and how they contribute to viral replication and pathogenesis.
LW, MG and SZ made the literature review, prepare the figures and tables, and wrote the manuscript. IS, SZ and LE assisted in the writing and revision of the manuscript. All authors contributed to the review article and approved the submitted version.
This work was supported by grants from the Government of Spain (PID2019-107001RB-I00 AEI/FEDER, UE; SEV 2017-0712 and PIE_INTRAMURAL_LINEA 1- 202020E079), the CSIC (PIE_INTRAMURAL-202020E043), the European Commission (ISOLDA_848166 H2020-SC1- 2019-Two-Stage-RTD, RIA; MANCO_101003651 H2020-SC1- PHE-CORONAVIRUS-2020 RIA), and the U.S. National Institutes of Health (NIH_2P01AI060699). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Marga Gonzalez (CNB-CSIC) for her technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-Binding proteins and post-transcriptional gene regulation. FEBS Lett (2008) 582:1977–86. doi: 10.1016/j.febslet.2008.03.004
2. White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol (2012) 20:175–83. doi: 10.1016/j.tim.2012.02.001
3. Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron (2001) 32:683–96. doi: 10.1016/s0896-6273(01)00508-6
4. Formicola N, Vijayakumar J, Besse F. Neuronal ribonucleoprotein granules: Dynamic sensors of localized signals. Traffic (2019) 20:639–49. doi: 10.1111/tra.12672
5. Lin WJ, Duffy A, Chen CY. Localization of AU-rich element-containing mRNA in cytoplasmic granules containing exosome subunits. J Biol Chem (2007) 282:19958–68. doi: 10.1074/jbc.M702281200
6. Stanek D. Cajal bodies and snRNPs - friends with benefits. RNA Biol (2017) 14:671–9. doi: 10.1080/15476286.2016.1231359
7. Bond CS, Fox AH. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol (2009) 186:637–44. doi: 10.1083/jcb.200906113
8. Sandqvist A, Sistonen L. Nuclear stress granules: The awakening of a sleeping beauty? J Cell Biol (2004) 164:15–7. doi: 10.1083/jcb.200311102
9. Ilik IA, Aktas T. Nuclear speckles: Dynamic hubs of gene expression regulation. FEBS J (2021) 289:7234–45. doi: 10.1111/febs.16117
10. Lallemand-Breitenbach V, de The H. PML nuclear bodies: From architecture to function. Curr Opin Cell Biol (2018) 52:154–61. doi: 10.1016/j.ceb.2018.03.011
11. Galganski L, Urbanek MO, Krzyzosiak WJ. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res (2017) 45:10350–68. doi: 10.1093/nar/gkx759
12. Sawyer IA, Dundr M. Nuclear bodies: Built to boost. J Cell Biol (2016) 213:509–11. doi: 10.1083/jcb.201605049
13. Silverman E, Edwalds-Gilbert G, Lin RJ. DEXD/H-box proteins and their partners: Helping RNA helicases unwind. Gene (2003) 312:1–16. doi: 10.1016/s0378-1119(03)00626-7
14. Rocak S, Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat Rev Mol Cell Biol (2004) 5:232–41. doi: 10.1038/nrm1335
15. Fuller-Pace FV. DEXD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res (2006) 34:4206–15. doi: 10.1093/nar/gkl460
16. Linder P. DEAD-box proteins: A family affair–active and passive players in RNP-remodeling. Nucleic Acids Res (2006) 34:4168–80. doi: 10.1093/nar/gkl468
17. Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell (2016) 164:487–98. doi: 10.1016/j.cell.2015.12.038
18. Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A. Dance with the devil: Stress granules and signaling in antiviral responses. Viruses (2020) 12:984. doi: 10.3390/v12090984
19. Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science (2005) 309:1514–8. doi: 10.1126/science.1111443
20. Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DEXH/D RNA helicase. Science (2001) 291:121–5. doi: 10.1126/science.291.5501.121
21. Ulvila J, Hultmark D, Ramet M. Rna silencing in the antiviral innate immune defence–role of DEAD-box RNA helicases. Scand J Immunol (2010) 71:146–58. doi: 10.1111/j.1365-3083.2009.02362.x
22. Anderson P, Kedersha N. Stress granules: The Tao of RNA triage. Trends Biochem Sci (2008) 33:141–50. doi: 10.1016/j.tibs.2007.12.003
23. Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol (2005) 169:871–84. doi: 10.1083/jcb.200502088
24. Buchan JR. mRNP granules. assembly, function, and connections with disease. RNA Biol (2014) 11:1019–30. doi: 10.4161/15476286.2014.972208
25. Erickson SL, Lykke-Andersen J. Cytoplasmic mRNP granules at a glance. J Cell Sci (2011) 124:293–7. doi: 10.1242/jcs.072140
26. Campos-Melo D, Hawley ZCE, Droppelmann CA, Strong MJ. The integral role of RNA in stress granule formation and function. Front Cell Dev Biol (2021) 9:621779. doi: 10.3389/fcell.2021.621779
27. Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol (2019) 11:a032813. doi: 10.1101/cshperspect.a032813
28. Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol (2007) 431:61–81. doi: 10.1016/S0076-6879(07)31005-7
29. Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell (2009) 36:932–41. doi: 10.1016/j.molcel.2009.11.020
30. Lloyd RE. Regulation of stress granules and p-bodies during RNA virus infection. Wiley Interdiscip Rev RNA (2013) 4:317–31. doi: 10.1002/wrna.1162
31. Stoecklin G, Kedersha N. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol (2013) 768:197–211. doi: 10.1007/978-1-4614-5107-5_12
32. Aizer A, Shav-Tal Y. Intracellular trafficking and dynamics of p bodies. Prion (2008) 2:131–4. doi: 10.4161/pri.2.4.7773
33. Moser JJ, Fritzler MJ. Relationship of other cytoplasmic ribonucleoprotein bodies (cRNPB) to GW/P bodies. Adv Exp Med Biol (2013) 768:213–42. doi: 10.1007/978-1-4614-5107-5_13
34. Reineke LC, Lloyd RE. Diversion of stress granules and p-bodies during viral infection. Virology (2013) 436:255–67. doi: 10.1016/j.virol.2012.11.017
35. Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, et al. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell (2006) 17:4212–9. doi: 10.1091/mbc.e06-04-0318
36. Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, et al. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine a. J Biol Chem (2006) 281:32870–8. doi: 10.1074/jbc.M606149200
37. Emara MM, Fujimura K, Sciaranghella D, Ivanova V, Ivanov P, Anderson P. Hydrogen peroxide induces stress granule formation independent of eIF2alpha phosphorylation. Biochem Biophys Res Commun (2012) 423:763–9. doi: 10.1016/j.bbrc.2012.06.033
38. Reineke LC, Dougherty JD, Pierre P, Lloyd RE. Large G3BP-induced granules trigger eIF2alpha phosphorylation. Mol Biol Cell (2012) 23:3499–510. doi: 10.1091/mbc.E12-05-0385
39. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-Binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol (1999) 147:1431–42. doi: 10.1083/jcb.147.7.1431
40. Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, et al. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol Biol Cell (2005) 16:405–20. doi: 10.1091/mbc.E04-06-0516
41. Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of argonaute protein reveals MicroRNA-dependent localization to stress granules. Proc Natl Acad Sci USA (2006) 103:18125–30. doi: 10.1073/pnas.0608845103
42. Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, et al. The rasgap-associated endoribonuclease G3BP assembles stress granules. J Cell Biol (2003) 160:823–31. doi: 10.1083/jcb.200212128
43. Layana C, Ferrero P, Rivera-Pomar R. Cytoplasmic ribonucleoprotein foci in eukaryotes: Hotspots of Bio(Chemical)Diversity. Comp Funct Genomics (2012) 2012:504292. doi: 10.1155/2012/504292
44. Prasad K, Alasmari AF, Ali N, Khan R, Alghamdi A, Kumar V. Insights into the SARS-CoV-2-Mediated alteration in the stress granule protein regulatory networks in humans. Pathogens (2021) 10:1459. doi: 10.3390/pathogens10111459
45. Gerassimovich YA, Miladinovski-Bangall SJ, Bridges KM, Boateng L, Ball LE, Valafar H, et al. Proximity-dependent biotinylation detects associations between SARS coronavirus nonstructural protein 1 and stress granule-associated proteins. J Biol Chem (2021) 297:101399. doi: 10.1016/j.jbc.2021.101399
46. Gao B, Gong X, Fang S, Weng W, Wang H, Chu H, et al. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease Nsp15 ensures efficient virus replication. PloS Pathog (2021) 17:e1008690. doi: 10.1371/journal.ppat.1008690
47. Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, et al. New world and old world alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PloS Pathog (2016) 12:e1005810. doi: 10.1371/journal.ppat.1005810
49. Arribere JA, Doudna JA, Gilbert WV. Reconsidering movement of eukaryotic mRNAs between polysomes and p bodies. Mol Cell (2011) 44:745–58. doi: 10.1016/j.molcel.2011.09.019
50. Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPss to mammalian processing bodies. RNA (2005) 11:717–27. doi: 10.1261/rna.2340405
51. Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science (2005) 310:486–9. doi: 10.1126/science.1115791
52. Tsai WC, Lloyd RE. Cytoplasmic RNA granules and viral infection. Annu Rev Virol (2014) 1:147–70. doi: 10.1146/annurev-virology-031413-085505
53. Qin Q, Hastings C, Miller CL. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J Virol (2009) 83:11090–101. doi: 10.1128/JVI.01239-09
54. White JP, Cardenas AM, Marissen WE, Lloyd RE. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe (2007) 2:295–305. doi: 10.1016/j.chom.2007.08.006
55. Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE. Stable formation of compositionally unique stress granules in virus-infected cells. J Virol (2010) 84:3654–65. doi: 10.1128/jvi.01320-09
56. White JP, Lloyd RE. Poliovirus unlinks Tia1 aggregation and mRNA stress granule formation. J Virol (2011) 85:12442–54. doi: 10.1128/jvi.05888-11
57. Khaperskyy DA, Hatchette TF, McCormick C. Influenza a virus inhibits cytoplasmic stress granule formation. FASEB J (2012) 26:1629–39. doi: 10.1096/fj.11-196915
58. Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci USA (2007) 104:9041–6. doi: 10.1073/pnas.0703348104
59. Yasuda-Inoue M, Kuroki M, Ariumi Y. DDX3 RNA helicase is required for HIV-1 tat function. Biochem Biophys Res Commun (2013) 441:607–11. doi: 10.1016/j.bbrc.2013.10.107
60. Izumi T, Burdick R, Shigemi M, Plisov S, Hu WS, Pathak VK. MOV10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol (2013) 87:11047–62. doi: 10.1128/JVI.02070-13
61. Abudu A, Wang X, Dang Y, Zhou T, Xiang SH, Zheng YH. Identification of molecular determinants from moloney leukemia virus 10 homolog (MOV10) protein for virion packaging and anti-HIV-1 activity. J Biol Chem (2012) 287:1220–8. doi: 10.1074/jbc.M111.309831
62. Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, et al. P body-associated protein MOV10 inhibits HIV-1 replication at multiple stages. J Virol (2010) 84:10241–53. doi: 10.1128/JVI.00585-10
63. Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, et al. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J Biol Chem (2010) 285:14346–55. doi: 10.1074/jbc.M110.109314
64. Enjuanes L, Almazan F, Sola I, Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol (2006) 60:211–30. doi: 10.1146/annurev.micro.60.080805.142157
65. Sola I, Almazán F, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol (2015) 2:265–88. doi: 10.1146/annurev-virology-100114-055218
66. Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol (2007) 9:2218–29. doi: 10.1111/j.1462-5822.2007.00951.x
67. Sola I, Galan C, Mateos-Gomez PA, Palacio L, Zuñiga S, Cruz JL, et al. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J Virol (2011) 85:5136–49. doi: 10.1128/JVI.00195-11
68. Nabeel-Shah S, Lee H, Ahmed N, Burke GL, Farhangmehr S, Ashraf K, et al. SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience (2022) 25:103562. doi: 10.1016/j.isci.2021.103562
69. Somasekharan SP, Gleave M. SARS-CoV-2 nucleocapsid protein interacts with immunoregulators and stress granules and phase separates to form liquid droplets. FEBS Lett (2021) 595:2872–96. doi: 10.1002/1873-3468.14229
70. Cai T, Yu Z, Wang Z, Liang C, Richard S. Arginine methylation of SARS-CoV-2 nucleocapsid protein regulates RNA binding, its ability to suppress stress granule formation, and viral replication. J Biol Chem (2021) 297:100821. doi: 10.1016/j.jbc.2021.100821
71. Emmott E, Munday D, Bickerton E, Britton P, Rodgers MA, Whitehouse A, et al. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J Virol (2013) 87:9486–500. doi: 10.1128/jvi.00321-13
72. Cascarina SM, Ross ED. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J (2020) 34:9832–42. doi: 10.1096/fj.202001351
73. Sun L, Chen H, Ming X, Bo Z, Shin HJ, Jung YS, et al. Porcine epidemic diarrhea virus infection induces caspase-8-Mediated G3BP1 cleavage and subverts stress granules to promote viral replication. J Virol (2021) 95:e02344–20. doi: 10.1128/JVI.02344-20
74. Rabouw HH, Langereis MA, Knaap RC, Dalebout TJ, Canton J, Sola I, et al. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PloS Pathog (2016) 12:e1005982. doi: 10.1371/journal.ppat.1005982
75. Nakagawa K, Narayanan K, Wada M, Makino S. Inhibition of stress granule formation by middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol (2018) 92:e00902–18. doi: 10.1128/JVI.00902-18
76. Kleer M, Mulloy RP, Robinson CA, Evseev D, Bui-Marinos MP, Castle EL, et al. Human coronaviruses disassemble processing bodies. PloS Pathog (2022) 18:e1010724. doi: 10.1371/journal.ppat.1010724
77. Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell (2007) 27:339–52. doi: 10.1016/j.molcel.2007.07.014
78. Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol (2002) 12:123–33. doi: 10.1016/s0959-440x(02)00298-1
79. de la Cruz J, Kressler D, Tollervey D, Linder P. DOB1p (MTR4p) is a putative ATP-dependent RNA helicase required for the 3' end formation of 5.8S rRNA in saccharomyces cerevisiae. EMBO J (1998) 17:1128–40. doi: 10.1093/emboj/17.4.1128
80. Tanner NK, Linder P. DEXD/H box RNA helicases: From generic motors to specific dissociation functions. Mol Cell (2001) 8:251–62. doi: 10.1016/s1097-2765(01)00329-x
81. Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol (2011) 12:505–16. doi: 10.1038/nrm3154
82. Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci (Landmark Ed) (2012) 17:2070–88. doi: 10.2741/4038
83. Fullam A, Schroder M. DEXD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta (2013) 1829:854–65. doi: 10.1016/j.bbagrm.2013.03.012
84. Oshiumi H, Kouwaki T, Seya T. Accessory factors of cytoplasmic viral RNA sensors required for antiviral innate immune response. Front Immunol (2016) 7:200. doi: 10.3389/fimmu.2016.00200
85. Stunnenberg M, Geijtenbeek TBH, Gringhuis SI. DDX3 in HIV-1 infection and sensing: A paradox. Cytokine Growth Factor Rev (2018) 40:32–9. doi: 10.1016/j.cytogfr.2018.03.001
86. Ariumi Y. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Front Genet (2014) 5:423. doi: 10.3389/fgene.2014.00423
87. Squeglia F, Romano M, Ruggiero A, Maga G, Berisio R. Host DDX helicases as possible SARS-CoV-2 proviral factors: A structural overview of their hijacking through multiple viral proteins. Front Chem (2020) 8:602162. doi: 10.3389/fchem.2020.602162
88. Schmidt N, Lareau CA, Keshishian H, Ganskih S, Schneider C, Hennig T, et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat Microbiol (2021) 6:339–53. doi: 10.1038/s41564-020-00846-z
89. Baggen J, Vanstreels E, Jansen S, Daelemans D. Cellular host factors for SARS-CoV-2 infection. Nat Microbiol (2021) 6:1219–32. doi: 10.1038/s41564-021-00958-0
90. Labeau A, Fery-Simonian L, Lefevre-Utile A, Pourcelot M, Bonnet-Madin L, Soumelis V, et al. Characterization and functional interrogation of the SARS-CoV-2 RNA interactome. Cell Rep (2022) 39:110744. doi: 10.1016/j.celrep.2022.110744
91. Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol (2010) 84:8571–83. doi: 10.1128/JVI.00392-10
92. Zhou Y, Wu W, Xie L, Wang D, Ke Q, Hou Z, et al. Cellular RNA helicase DDX1 is involved in transmissible gastroenteritis virus Nsp14-induced interferon-beta production. Front Immunol (2017) 8:940. doi: 10.3389/fimmu.2017.00940
93. Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity (2011) 34:866–78. doi: 10.1016/j.immuni.2011.03.027
94. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-Cov-2 protein interaction map reveals targets for drug repurposing. Nature (2020) 583:459–68. doi: 10.1038/s41586-020-2286-9
95. Wu CH, Chen PJ, Yeh SH. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe (2014) 16:462–72. doi: 10.1016/j.chom.2014.09.009
96. Ciccosanti F, Di Rienzo M, Romagnoli A, Colavita F, Refolo G, Castilletti C, et al. Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection. Antiviral Res (2021) 190:105064. doi: 10.1016/j.antiviral.2021.105064
97. Gu L, Fullam A, Brennan R, Schroder M. Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol Cell Biol (2013) 33:2004–15. doi: 10.1128/MCB.01603-12
98. Szappanos D, Tschismarov R, Perlot T, Westermayer S, Fischer K, Platanitis E, et al. The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity. PloS Pathog (2018) 14:e1007397. doi: 10.1371/journal.ppat.1007397
99. Kienes I, Bauer S, Gottschild C, Mirza N, Pfannstiel J, Schroder M, et al. DDX3X links NLRP11 to the regulation of type I interferon responses and NLRP3 inflammasome activation. Front Immunol (2021) 12:653883. doi: 10.3389/fimmu.2021.653883
100. Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J (2008) 27:2147–57. doi: 10.1038/emboj.2008.143
101. Chen JY, Chen WN, Poon KM, Zheng BJ, Lin X, Wang YX, et al. Interaction between SARS-CoV helicase and a multifunctional cellular protein (DDX5) revealed by yeast and mammalian cell two-hybrid systems. Arch Virol (2009) 154:507–12. doi: 10.1007/s00705-009-0323-y
102. Tanaka K, Tanaka T, Nakano T, Hozumi Y, Yanagida M, Araki Y, et al. Knockdown of DEAD-box RNA helicase DDX5 selectively attenuates serine 311 phosphorylation of NF-KappaB P65 subunit and expression level of anti-apoptotic factor bcl-2. Cell Signal (2020) 65:109428. doi: 10.1016/j.cellsig.2019.109428
103. Ogilvie VC, Wilson BJ, Nicol SM, Morrice NA, Saunders LR, Barber GN, et al. The highly related DEAD box RNA helicases P68 and P72 exist as heterodimers in cells. Nucleic Acids Res (2003) 31:1470–80. doi: 10.1093/nar/gkg236
104. Choi YJ, Lee SG. The DEAD-box RNA helicase DDX3 interacts with DDX5, Co-localizes with it in the cytoplasm during the G2/M phase of the cycle, and affects its shuttling during mRNP export. J Cell Biochem (2012) 113:985–96. doi: 10.1002/jcb.23428
105. Xu X, Wang J, Zhang Y, Yan Y, Liu Y, Shi X, et al. Inhibition of DDX6 enhances autophagy and alleviates endoplasmic reticulum stress in vero cells under PEDV infection. Vet Microbiol (2022) 266:109350. doi: 10.1016/j.vetmic.2022.109350
106. Nunez RD, Budt M, Saenger S, Paki K, Arnold U, Sadewasser A, et al. The RNA helicase DDX6 associates with RIG-I to augment induction of antiviral signaling. Int J Mol Sci (2018) 19:1877. doi: 10.3390/ijms19071877
107. Hage A, Bharaj P, van Tol S, Giraldo MI, Gonzalez-Orozco M, Valerdi KM, et al. The RNA helicase DHX16 recognizes specific viral RNA to trigger RIG-I-Dependent innate antiviral immunity. Cell Rep (2022) 38:110434. doi: 10.1016/j.celrep.2022.110434
108. Jiao A, Sun C, Wang X, Lei L, Liu H, Li W, et al. DEXD/H-box helicase 9 intrinsically controls CD8(+) T cell-mediated antiviral response through noncanonical mechanisms. Sci Adv (2022) 8:eabk2691. doi: 10.1126/sciadv.abk2691
109. Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol (2010) 20:313–24. doi: 10.1016/j.sbi.2010.03.011
110. Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, et al. Identification of novel argonaute-associated proteins. Curr Biol (2005) 15:2149–55. doi: 10.1016/j.cub.2005.10.048
111. Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res (2019) 47:D529–D41. doi: 10.1093/nar/gky1079
112. Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature (2007) 447:823–8. doi: 10.1038/nature05841
113. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature (2011) 472:481–5. doi: 10.1038/nature09907
114. Cuevas RA, Ghosh A, Wallerath C, Hornung V, Coyne CB, Sarkar SN. MOV10 provides antiviral activity against RNA viruses by enhancing RIG-I-MAVS-Independent IFN induction. J Immunol (2016) 196:3877–86. doi: 10.4049/jimmunol.1501359
115. Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol (2008) 15:714–21. doi: 10.1038/nsmb.1440
116. Wang H, Chang L, Wang X, Su A, Feng C, Fu Y, et al. MOV10 interacts with enterovirus 71 genomic 5'UTR and modulates viral replication. Biochem Biophys Res Commun (2016) 479:571–7. doi: 10.1016/j.bbrc.2016.09.112
117. Zhang J, Huang F, Tan L, Bai C, Chen B, Liu J, et al. Host protein moloney leukemia virus 10 (MOV10) acts as a restriction factor of influenza a virus by inhibiting the nuclear import of the viral nucleoprotein. J Virol (2016) 90:3966–80. doi: 10.1128/JVI.03137-15
118. Zhao K, Li LW, Zhang YJ, Jiang YF, Gao F, Li GX, et al. MOV10 inhibits replication of porcine reproductive and respiratory syndrome virus by retaining viral nucleocapsid protein in the cytoplasm of MARC-145cells. Biochem Biophys Res Commun (2018) 504:157–63. doi: 10.1016/j.bbrc.2018.08.148
119. Balinsky CA, Schmeisser H, Wells AI, Ganesan S, Jin T, Singh K, et al. IRAV (FLJ11286), an interferon-stimulated gene with antiviral activity against dengue virus, interacts with MOV10. J Virol (2017) 91:e01606–16. doi: 10.1128/JVI.01606-16
120. Liu D, Ndongwe TP, Puray-Chavez M, Casey MC, Izumi T, Pathak VK, et al. Effect of p-body component MOV10 on HCV virus production and infectivity. FASEB J (2020) 34:9433–49. doi: 10.1096/fj.201800641R
121. Mo Q, Xu Z, Deng F, Wang H, Ning YJ. Host restriction of emerging high-pathogenic bunyaviruses Via MOV10 by targeting viral nucleoprotein and blocking ribonucleoprotein assembly. PloS Pathog (2020) 16:e1009129. doi: 10.1371/journal.ppat.1009129
122. Ma YX, Li D, Fu LJ, Fu BQ, Chen SJ, Xu WZ, et al. The role of moloney leukemia virus 10 in hepatitis b virus expression in hepatoma cells. Virus Res (2015) 197:85–91. doi: 10.1016/j.virusres.2014.12.011
123. Puray-Chavez MN, Farghali MH, Yapo V, Huber AD, Liu D, Ndongwe TP, et al. Effects of moloney leukemia virus 10 protein on hepatitis b virus infection and viral replication. Viruses (2019) 11:651. doi: 10.3390/v11070651
124. Wang L, Sola I, Enjuanes L, Zuñiga S. MOV10 helicase interacts with coronavirus nucleocapsid protein and has antiviral activity. mBio (2021) 12:e01316–21. doi: 10.1128/mBio.01316-21
125. Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science (2020) 370:eabe9403. doi: 10.1126/science.abe9403
126. Sun L, Li P, Ju X, Rao J, Huang W, Ren L, et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell (2021) 184:1865–83. doi: 10.1016/j.cell.2021.02.008
127. Lee S, Lee YS, Choi Y, Son A, Park Y, Lee KM, et al. The SARS-CoV-2 RNA interactome. Mol Cell (2021) 81:2838–50.e6. doi: 10.1016/j.molcel.2021.04.022
128. Kamel W, Noerenberg M, Cerikan B, Chen H, Jarvelin AI, Kammoun M, et al. Global analysis of protein-RNA interactions in SARS-CoV-2-Infected cells reveals key regulators of infection. Mol Cell (2021) 81(13):2851–67.e7. doi: 10.1016/j.molcel.2021.05.023
129. Moosa MM, Banerjee PR. Subversion of host stress granules by coronaviruses: Potential roles of pi-rich disordered domains of viral nucleocapsids. J Med Virol (2020) 92:2891–3. doi: 10.1002/jmv.26195
Keywords: coronavirus, stress granules, P-bodies, RNA helicase, MOV10, virus-host interaction
Citation: Wang L, Guzmán M, Sola I, Enjuanes L and Zuñiga S (2022) Cytoplasmic ribonucleoprotein complexes, RNA helicases and coronavirus infection. Front. Virol. 2:1078454. doi: 10.3389/fviro.2022.1078454
Received: 24 October 2022; Accepted: 30 November 2022;
Published: 14 December 2022.
Edited by:
Atsushi Kawaguchi, University of Tsukuba, JapanReviewed by:
Koji Onomoto, Chiba University, JapanCopyright © 2022 Wang, Guzmán, Sola, Enjuanes and Zuñiga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis Enjuanes, bC5lbmp1YW5lc0BjbmIuY3NpYy5l; Sonia Zuñiga, c3p1bmlnYUBjbmIuY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.