
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Virol. , 02 November 2022
Sec. Virus and Host Immunity
Volume 2 - 2022 | https://doi.org/10.3389/fviro.2022.1049134
This article is part of the Research Topic Role of genetic and structural insights of neutralizing antibodies to guide viral vaccine design View all 5 articles
A correction has been applied to this article in:
Corrigendum: The influenza hemagglutinin stem antibody CR9114: Evidence for a narrow evolutionary path towards universal protection
 Anna L. Beukenhorst1,2†
Anna L. Beukenhorst1,2† Jacopo Frallicciardi1*†
Jacopo Frallicciardi1*† Clarissa M. Koch1
Clarissa M. Koch1 Jaco M. Klap1
Jaco M. Klap1 Angela Phillips3
Angela Phillips3 Michael M. Desai3
Michael M. Desai3 Kanin Wichapong4
Kanin Wichapong4 Gerry A. F. Nicolaes4
Gerry A. F. Nicolaes4 Wouter Koudstaal1
Wouter Koudstaal1 Galit Alter1,5
Galit Alter1,5 Jaap Goudsmit1,6
Jaap Goudsmit1,6Human monoclonal antibodies (hmAbs) that protect against all influenza A and B strains are considered the road to universal influenza vaccines. Based on publicly-available data, we analyze the mechanistic and structural basis of pan-influenza protection by CR9114, a hemagglutinin (HA) stem-reactive antibody that protects against influenza subtypes from groups A1, A2, and B. The mechanistic basis of CR9114’s universal protection is not limited to in vitro neutralization, as CR9114 also protects in vivo from strains that escape its neutralizing activity: some H2 strains and influenza B. Fusion inhibition, viral egress inhibition, and activation of Fc-mediated effector functions are key contributors to CR9114’s universal protection. A comparative analysis of paratopes – between CR9114 (pan-influenza protection) and structurally similar VH1-69 hmAb CR6261 (influenza A1 protection) – pinpoints the structural basis of pan-influenza protection. CR9114’s heterosubtypic binding is conferred by its ability to bind HA with multiple domains: three HCDR loops and FR3. In contrast to other VH1-69 hmAbs, CR9114 uses a long and polar side chain of tyrosine (Y) residues on its HCDR3 for crucial H-bonds with H3, H5, and B HA. The recognition of a highly conserved epitope by CR9114 results in a high genetic barrier for escape by influenza strains. The nested, hierarchical structure of the mutations between the germline ancestor and CR9114 demonstrates that it is the result of a narrow evolutionary pathway within the B cell population. This rare evolutionary pathway indicates an immuno-recessive epitope and limited opportunity for vaccines to induce a polyclonal CR9114-like response.
Influenza viruses cause epidemics and pose pandemic threats. Influenza A and B cause seasonal influenza epidemics in humans, resulting in significant morbidity [3-5 million deaths yearly (1)], mortality [290,000-650,000 deaths (1)], and economic burden [$10-$80 billion yearly in the US (2, 3)]. Elderly and immunocompromised individuals are at higher risk of severe influenza A infection, whereas children and adolescents are more susceptible to influenza B (4). New influenza strains or subtypes from animal reservoirs could cause a pandemic, as they can potentially evolve the ability for human-to-human transmission (5–7). Such influenza pandemics could result in vastly higher morbidity, mortality, and economic burden than seasonal epidemics or the 2020 SARS-CoV-2 pandemic (8, 9).
The marked genetic variation between and within influenza A and B impedes broad, pan-influenza protection. Influenza A and B viruses are phylogenetically diverse. They can be grouped based on their glycoproteins: hemagglutinin (HA), which binds sialic acid and enables cell fusion (10), and neuraminidase (NA), which enables egress through cleaving sialic acid (11). A wide variety of influenza subtypes can thus be formed, with nomenclature based on their HA (influenza A1, comprising H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18; influenza A2, comprising H3, H4, H7, H10, H14, and H15; B/Yamagata lineage; B/Victoria lineage) (12–14) and NA (group NA1, comprising N1, N4, N5, and N8; group NA2, comprising N2, N3, N6, N7, N9; influenza B NA). Further genetic variation is caused by the exchange of HA and NA genes [antigenic shift (15)] and random point mutations in viral epitopes [antigenic drift (16)].
Human monoclonal antibodies (hmAbs) protecting against all influenza strains are therefore of interest for universal influenza protection. However, human antibody responses after infection typically lack cross-reactivity for other influenza subtypes. Antibody responses after vaccination also mostly induce neutralizing antibodies against the strain that elicited them and closely related strains, and no pan-influenza vaccine currently exists (17–19). As a consequence, influenza vaccine efficacy is suboptimal: if the vaccine matches circulating strains, it is around 60% for adults under 65 and 40% for the elderly, whereas in the case of mismatch efficacy is around 45% for adults under 65 and only 13% for the elderly (20, 21).
To protect humans from influenza, various moments in the lifecycle of virions can be targeted: viral entry, cell fusion, and egress. When influenza virions reach the host’s respiratory epithelium, they bind sialic acid on host cell receptors, leading to viral entry. After internalization, the virion fuses with the endosomal vesicle. Cell fusion is dependent on the HA structural changes in response to endosomal acidification and consequent proteolytic activation (10). The virus’s RNA is released in the host cell cytoplasm and imported into the nucleoplasm, where viral transcription and replication occur (22). The newly synthesized viral surface proteins present themselves on the infected host cell membrane, and after cleavage of sialic acid by NA, new virions egress. NA cleavage also prevents virion agglutination, ensuring the efficient spread of newly produced virions in the host respiratory tract and, potentially, transmission (23).
HmAbs can interfere at one or more moments in this viral lifecycle. For pan-influenza protection, interference at certain moments is more effective than at others. To distinguish between mechanisms of protection, antibody activity can be assessed by in vitro assays. Assays that are statistically correlated with protection (‘correlates of protection’) can also be used to predict breadth and potency in vivo (24). Although receptor-binding inhibition is an effective strategy for protection by HA head-reactive antibodies, such protection is not broad: epitopes on the HA head are more susceptible to antigenic drift and shift and vary within influenza subtypes (25).
On the other hand, the HA-stem domain is relatively conserved within and across influenza groups A1, A2, and B HAs. Thus, HA-stem reactive antibodies show various degrees of breadth, from subtype-specific to heterosubtypic (26). Even within the realm of HA-stem reactive hmAbs, universal protection is rare. For instance, the hmAbs 2G02 (27) and FI6 (28) are highly cross-reactive but fail to interact with B HAs. The broadest HA-stem reactive hmAb is the VH1-69 antibody CR9114 (29): prophylactic systemic administration of CR9114 fully protects mice from mortality and weight loss when challenged with influenza A1, A2, B/Yamagata, and B/Victoria strains (29–31) (Figure 1A). CR9114 was recovered from a combinatorial display library constructed from human B cells of healthy recently vaccinated volunteers (29). CR9114 neutralizes in vitro most influenza viruses except for some H2N2 and H7, and influenza B strains (Figure 1B) (29, 32–38). Although these are not neutralized in vitro by CR9114, the hmAb still protects mice from lethal challenge with these viruses (29, 30). This demonstrates that HA neutralization is not the only mechanism by which CR9114 protects against influenza. Microneutralization assays specifically detect whether antibodies prevent infection of mammalian cells by (pseudo)virions. Even though CR9114 does not neutralize some H2N2, some H7, or influenza B in vitro, it still protects in vivo. This shows that in vitro neutralization of HA is an imperfect correlate of protection, as it is not always a predictor of protection (24).
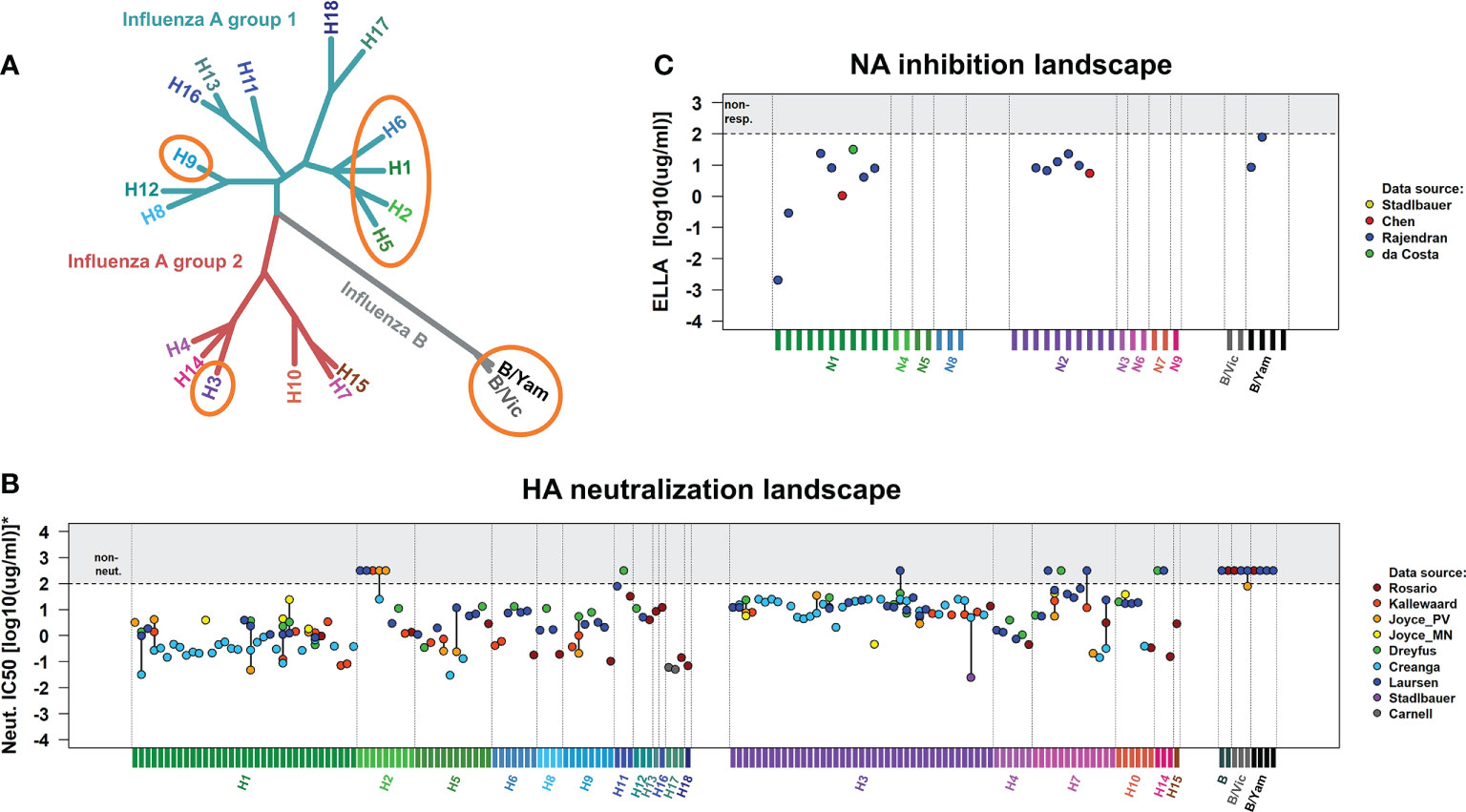
Figure 1 In vivo protection and in vitro neutralization of CR9114 against influenza virus groups A1, A2, and B. (A) Phylogenetic tree of influenza A and B viruses, indicating the subtypes for which CR9114’s prophylactic protection has been demonstrated through lethal challenges in the murine model (circled in orange). CR9114 has been shown to fully protect mice against challenges with lethal doses of influenza A H1N1, H3N2 (29), H3N1, H5N1, and H9N2 (31) in line with its ability to neutralize the same subtypes in vitro (B). Interestingly, CR9114 grants in vivo protection in mice challenged with lethal doses of influenza virus B (Florida/4/2006 and Malaysia/2506/2004 and Phuket/3037/2013) (29, 31) and H2 (A/Ann Arbor/6/1960 and A/swine/MO/4296424/06) (30), despite not showing any in vitro neutralizing activity against influenza B and H2 (B). (B) HA neutralization landscapes of CR9114 in influenza virus A1, A2, and B strains. Neutralization results (IC50) have been measured by microneutralization assays. IC50 values have been collected from various studies (29, 32–38), including all viral strains investigated in these studies. To allow for comparability across these studies, results have been normalized (scaled) to virus subtypes that were in common in most studies. The set of reference strains includes pH1N1 (A/California/4/2009, A/California/7/2009 and A/England/195/2009) and H3N2 (A/Hong Kong/1/1968, A/Hong Kong/8/1968, and A/Aichi/2/1968). Normalized IC50 results are presented in Supplementary Table 1. (C) NA inhibition landscapes of CR9114 in influenza virus A1, A2, and B strains. The results (IC50), collected from various works and here presented in Supplementary Table 2, have been measured by ELLA assay (39–41). CR9114 inhibits NA activity through steric hindrance by blocking access to HA-bound sialic acid (39). Version of the landscapes in (B, C) with the names of all the viral strains here investigated are presented in Figure S1.
Besides CR9114, another two broadly neutralizing antibodies (bnAb) cross-react with influenza groups A1, A2, and B: the camelid multi-domain Ab MD3606 (32) and the hmAb 1G01 (34). MD3606’s broad breadth of cross-reactivity rests on the four single single-domain antibodies of which it is composed: two against influenza A and two against influenza B. The efficacy and safety of camelid multi-domain antibodies in humans have yet to be determined (42). 1G01 targets a conserved epitope close to the NA’s active site and inhibits its catalytic activity. Thus, 1G01 mostly acts by preventing viral egress, whereas CR9114 interferes at more than one stage of the viral lifecycle.
CR9114’s protection hinges on its ability to interfere with the viral lifecycle (schematically depicted in Figure 2) at multiple steps. CR9114 does not prevent host receptor binding, as demonstrated by the absence of in vitro neutralizing activity in HA inhibition assays against influenza groups A1, A2, or B (29). This assay measures receptor-binding inhibition, typically demonstrated by head-reactive antibodies, but not stem-reactive antibodies such as CR9114 (43). Rather, CR9114 protects by preventing fusion of the viral envelope with the endocytic vesicular membrane. CR9114 blocks HA acidification-induced conformational changes associated with membrane fusion (Figure 2B) (10). In influenza A1 and A2, CR9114 hampers the transition from pre- to post-fusion conformation (29). On the other hand, CR9114 does not effectively prevent influenza B HA from transitioning to the post-fusion conformation and depends on other mechanisms of protection for this genus (29, 32).
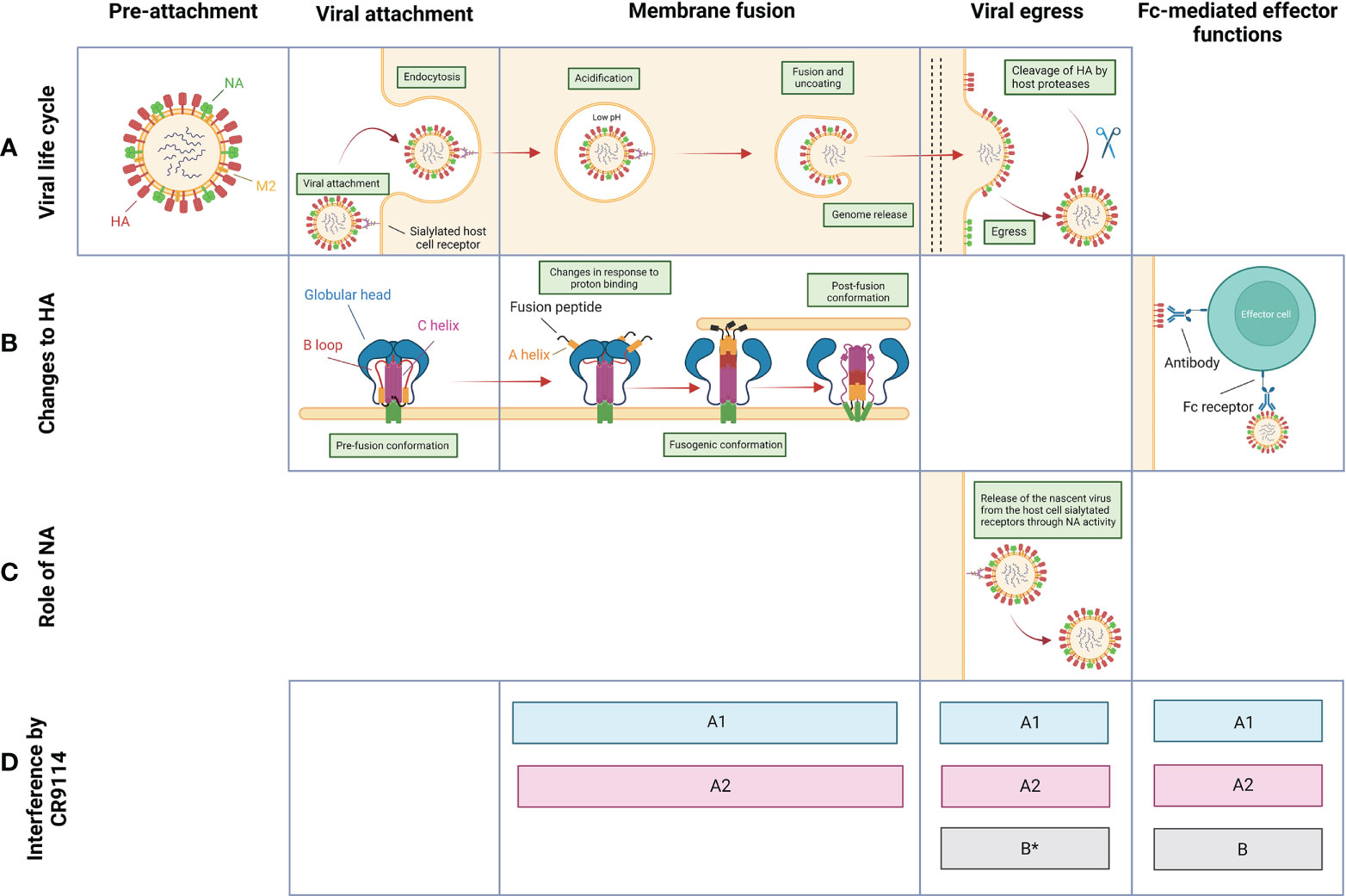
Figure 2 Overview of the proposed mechanisms of protection by CR9114 against influenza virus throughout the viral life cycle. (A) Schematic pictograms of the life cycle of influenza virus. Stages between membrane fusion/genome release and budding/viral egress are not depicted. (B) Structural changes to and function of HA through different stages of the virus life cycle. (C) Function of NA through different stages of the virus life cycle. (D) Stages at which CR9114 is known to interfere with the viral life cycle of influenza group A1 (light blue), A2 (red), and B (grey). *Contrasting data regarding the implication of CR9114 in preventing the viral egress of influenza B virions (32, 40).
CR9114 further provides protection by harnessing the host’s immune system through Fc-mediated effector functions. These include antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent phagocytosis by neutrophils and macrophages (44, 45), which result in viral clearance. Fc-mediated effector functions might represent the dominant mechanism of protection against viral strains that CR9114 does not neutralize. CR9114 recruits effector cells, as shown by ADCC reporter assays against strains of influenza virus groups A1, A2, and B (30, 32, 34). Specifically, CR9114 has been shown to activate FcγRIIIa in an ADCC reporter assay to H3 influenza virus (H3 A/Wisconsin/67/05 (32)). When mutated to decrease binding to FcγRs and knock down antibody effector function (LALA mutant), CR9114 fails to induce activation of both receptors FcRIV and FcRIII against H2 (30), indicating that the LALA mutations ablate FcR binding. This translates to less effective protection in mice treated with CR9114-LALA and then challenged with influenza virus H2 (30), confirming that CR9114 confers in vivo protection also through FcR-dependent mechanisms. Similarly, CR9114 has been shown to activate FcγRIIIa against H1N1 (A/California/07/09) and B (Florida/4/2006) (46). Inducing ADCC is only one of the possible Fc effector functions. Further work is needed to fully understand whether this bnAb can recruit other populations of immune cells. Of note, Fc-mediated effector functions triggered by antibodies can lead to inappropriate amplification of local inflammatory responses, also called ‘antibody-dependent enhancement (47). Lung pathology data collected in the work of Sutton et al (30) show that inflammation levels in the airways of H2-infected mice are lower when treated with CR9114 compared to untreated mice.
In later stages of the influenza life cycle, CR9114 can prevent viral egress (39, 40). CR9114 indirectly inhibits NA catalytic activity by steric hindrance, as measured by enzyme-linked lectin assay (ELLA) (Figure 1C) and confirmed by the NA-Star assay (34, 39–41). CR9114 binds the stalk region of HA and occupies the space on the side of the viral protein, blocking the access of NA to the sialic acids bound by HA (39, 40). This steric hindrance is dependent on the Fc tail of CR9114: an antibody-binding fragment of CR9114 without Fc tail has reduced NI activity (39, 40). Whether or not CR9114 can prevent the egress of influenza B virions is in doubt, due to contrasting data between different assays: the bnAb is able to neutralize B NA subtypes by steric hindrance in an ELLA assay (Figure 1C) but does not appear to prevent viral egress of influenza B (B/Malaysia/2506/04) virus in a viral egress inhibition assay (32).
To better understand the structural basis of pan-influenza protection, we compared sequences, three-dimensional structures, and interactions between HAs and CR9114 or CR6261 (Figure 3). These two antibodies differ in breadth: CR9114 is pan-influenza, whereas CR6261 protects only against subtypes from group A1. In other respects, the antibodies are very similar: they derive from similar germline sequences, share a high percent sequence identity (86.2%, for the heavy chain, Figure 3B) have similar 3D structures (RMSD of the heavy chain = 1.34 Å) and bind a similar epitope with their heavy chain only (Figure 3A) (29, 48). Both CR9114 and CR6261 use three heavy-chain complementarity determining regions (HCDR1-3) and one framework (FR3) for binding HA.
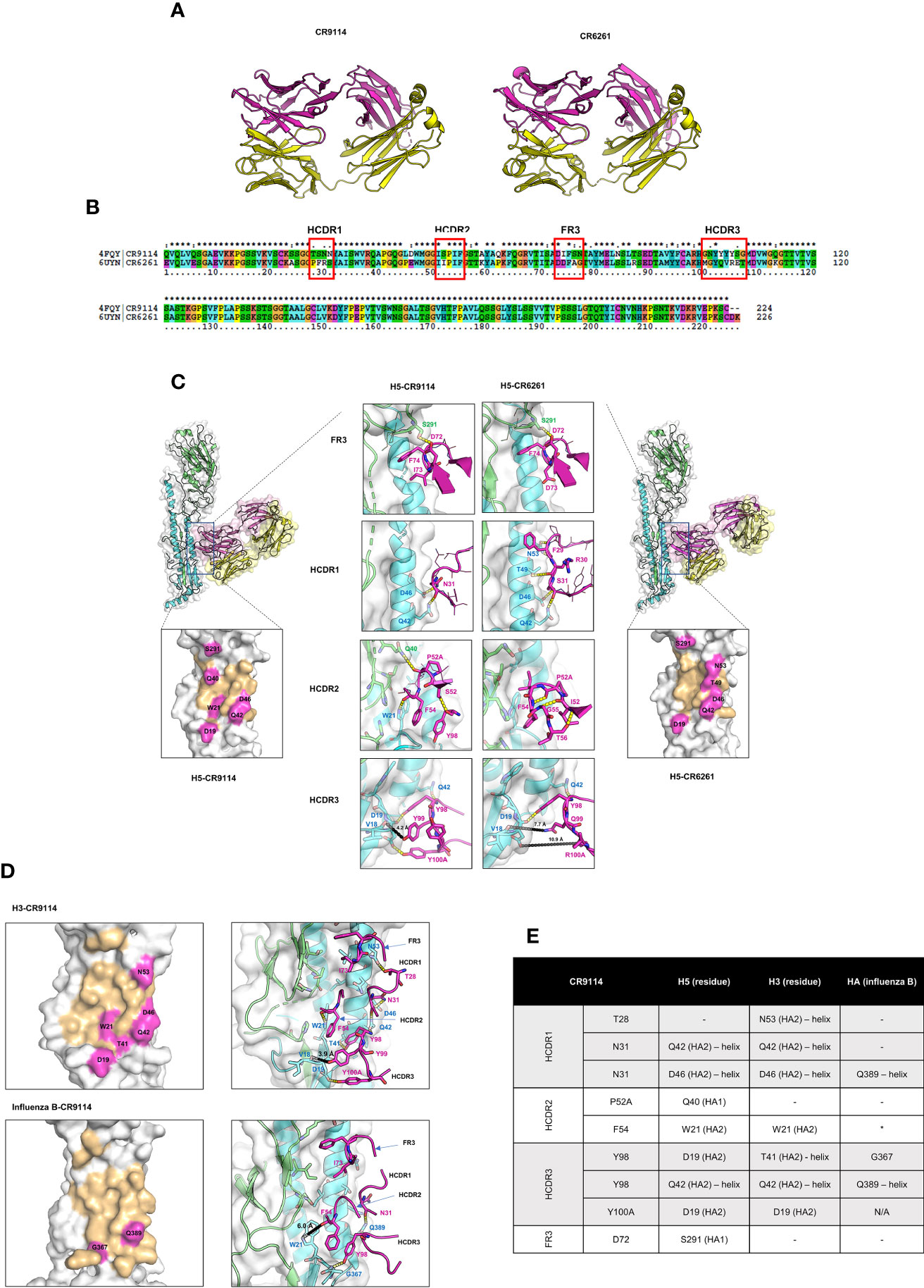
Figure 3 Comparison of 3D structures, sequence, binding, and interactions between CR9114 Fab and CR6261 Fab with different HA subtypes. (A) 3D structures of CR9114 and CR6261. The heavy and light chains of the antibodies are displayed in magenta and in yellow, respectively. (B) Sequence alignment between the variable part of the heavy chains of CR9114 and CR6261. (C) Comparison of the epitope mapping and the H-bond interactions between the H5-CR9114 complex (left panels) and the H5-CR6261 complex (right panels). (D) Epitope mapping (left panels) and H-bond interactions (right panels) of different H3- and B HA-CR9114 complexes. (E) Table summarizing the H-bond interactions between CR9114 and H3, H5, and HA of influenza B. In all epitope mappings (Figure 3C, D), binding epitopes of HAs with antibodies are shown in light orange, while hot spots of HAs, defined as residues of HAs that form H-bonds with the heavy chain of the antibodies as summarized in 3E (as indicated by residue name and ID), are shown in magenta. The same colour scheme is used in Figures 3C, D for H-bond interactions: the magenta cartoons/sticks/lines/labels represent residues from the heavy chain of antibodies; green and cyan cartoons and labels represent HA1 and HA2, respectively, of influenza H3, H5, and HA-influenza B; the molecular surface of all HAs is depicted in white; H-bonds are shown as yellow dashed lines; the different loops of antibodies are indicated by arrows. The comparative analysis reveals three main differences in the binding paratopes that make CR9114 a broad-spectrum antibody across HAs: (i) I73 in the FR3 loop does not form an internal H-bond, allowing CR9114 to flip into the hydrophobic groove of influenza H5 (C) and H3 (D); (ii) S52 in the HCDR2 establishes an internal H-bond with Y98 (HCDR3), inducing enlargement of the HCDR2 loop and blocking a larger space at the binding groove of HAs (C, D); (iii) the “quadruplet 98YYYY100A” in the HCDR3 establishes pivotal H-bonds with H3 (D) and H5 (C). (E) Table listing CR9114’s residues involved in interactions with the residues of H5, H3, or B HA. *The distance between Phe54 and Trp39 is 6Å. The following PDB IDs were exploited to investigate interactions between HAs and antibodies by using PyMol program (PDB ID: 4FQI (H5-CR9114 complex), PDB ID: 3GBM (H5-CR6261 complex), PDB ID: 6CNV (HA Influenza B-CR9114).
Both CR9114 and CR6261 derive from the VH1–69 germline. This is one of the most polymorphic loci within the human VH gene cluster (49–51) and is known for producing heterosubtypic protection against influenza A1. The VH1–69 gene encodes two hydrophobic residues in the heavy-chain complementarity determining regions (HCDR) 2 loop responsible for the conserved hydrophobic epitope recognition: (i) a canonical hydrophobic residue at position 53 (usually isoleucine) and (ii) phenylalanine at position 54 (Figure 3B). Some influenza VH1–69 antibodies also have a conserved tyrosine in the HCDR3 important for binding to the HA stem. The long HCDR3 loop helps access epitopes that are partially obscured, especially by surrounding glycans, or which require an extensive interacting surface (51).
Monoclonal antibody CR6261 was isolated from the immune repertoire of a healthy, vaccinated individual by using phage display selection on recombinant H5 HA (52). X-ray structural analysis revealed that CR6261 recognizes a highly conserved helical region in the membrane-proximal stem of HA1 and HA2: despite contacting HA1 residues in the stem region, the primary interactions are with the HA2 A helix (48). CR6261 can neutralize multiple influenza subtypes, including H1, H2, H5, H6, H8, and H9, and protects mice from lethal challenges with H1N1 and H5N1 (52). However, this anti-stem antibody’s neutralization function is limited to strains from group 1 of influenza A viruses, in contrast to the pan-influenza mAb CR9114 (48).
The comparative analysis shows that the several differences in the binding paratopes of CR9114 and CR6261 result in different binding profiles to HA. The I73 in the FR3 loop allows CR9114 to flip into the hydrophobic groove of influenza H5 (Figure 3C) and H3 (Figure 3D). I73 does not form an internal H-bond: its side chain can freely rotate to interact with HAs. At the same position, CR6261 has D73. Its side chain has a different conformation and flips out of the hydrophobic groove of H5 (Figure 3C). Within CR6261, R30—D73 form a stable and preferable internal H-bond, potentially explaining why that residue is not available for interacting with HAs. The R30-D73 H-bond induces intramolecular H-bond networks between D73(FR3)—R30 (HCDR1) —Y32 (HCDR1) and A33(HCDR1)—G97 (HCDR3) in CR6261 resulting in a larger loop conformation of HCRD1 of CR6261 as compared to CR9114 (Figure 3C). CR6261’s expanded HCDR1 loop brings F29 into the groove nearby for interacting with the hydrophobic pocket of H5 (Figure 3C), compensating for the lost interaction between D73 and HAs.
At HCDR2, one amino acid residue difference between CR9114 and CR6261 makes a major contribution to their dissimilar binding profiles: polar S52 (CR9114) vs. hydrophobic I52 (CR6261). In CR9114, S52 establishes an internal H-bond with Y98 (HCRD3) which can induce enlargement of the HCDR2 loop of CR9114. As a result, the HCDR2 loop of CR9114 can block a larger space at the binding groove of HAs, and several residues at this loop can interact with H3 HA (F54-W21(H3), Figure 3D) and H5 HA (F54-W21 and P52A-Q40, Figure 3C). CR6261’s I52 is hydrophobic and cannot form an H-bond with any residue. However, backbone atoms of I52-G55/T56 and P52A-F54 pair form H-bonds (Figure 3C), resulting in a small and rigid HCDR2 loop of CR6261. The small and rigid loop cannot properly bind to HAs and several essential interactions cannot be formed with H5 (Figure 3C).
The HCDR3 loop shows somewhat larger differences between the two antibodies 96GNYYYYSG100C (CR9114) vs 96MGYQVRET100C (CR6261). In CR9114’s HCDR3, the long and polar side chain of tyrosine (Y) residue promotes the “quadruplet 98YYYY100A”. It establishes crucial H-bonds with H5 HA (Y98-Q42, Y98/Y100A-D19, Figure 3C) and H3 HA (Y98-T41/Q42, Y100A-D19, Figure 3D). A short distance between Y99 (CR9114) to the backbone atom of V18 (H5 and H3; approximately 4 Å as shown in Figures 3C, D) can induce a strong charge-charge interaction between CR9114 and HAs. For CR6261, only Y98 can form an H-bond with a residue of the fusion peptide domain of H5 (Y98—D19, Figure 3C) and additional with Q42 (H5, Figure 3C) located at the helix. Longer distances between CR6261 residues and HA residues V18 and D19 result in very weak interactions: 7-8 Å for Q99–V18 and 11 Å for R100A-D19 (Figure 3C).
In summary, CR9114 represents a broad-spectrum antibody due to these key features: (i) I73 at the FR3 loop for binding with the hydrophobic groove of HAs, (ii) the flexibility of the HCDR2 loop which can induce a formation of H-bonds with HAs, and (iii), a quadruplet Y residue in the HCDR3 loop “ 98YYYY100A” which can establish essential interactions with HAs.
What led to the difference in the breadth of protection against influenza virus between CR9114 and CR6261? And why is CR9114’s signature broad protection so rare, even within the VH1-69 family? Our hypothesis is that the difference in the breadth of protection resides in the evolutionary paths that, from the germline, led to the somatic variants of CR6261 and CR9114. More specifically, in how the two bnAbs’ evolutionary histories diverged. Mutational pathways to CR6261 can simultaneously improve in affinity to multiple A1 strains. The evolution of breadth in this antibody could therefore have resulted from simultaneous exposure to different strains of influenza group A1. On the other hand, pathways to CR9114 are constrained to acquire affinity in a sequential manner – first to A1, then to A2, and finally to B – suggesting that sequential exposure to distinct antigens may be needed to produce bnAbs like CR9114 (53).
The divergence of CR9114’s and CR6261’s evolution is reflected in the number of mutations separating the germline from the somatic variants that are required for broad binding affinity (53): if only a few mutations are needed for CR9114 to improve the germline affinity for H1, more mutations are necessary to bind H3, and even more to bind B HA. Moreover, these mutations are organized into a nested, hierarchical structure: mutations beneficial for H1 are required for binding H3, and mutations required for H3 are required for binding influenza B HA. CR9114 has relatively few mutations (16) from its germline (shown in Figure 4), but almost all of them are required for binding influenza B, and most alternative evolutionary routes lead to mAbs with a more limited breadth. In contrast, many possible evolutionary intermediates leading to CR6261 have intermediate affinity for both H1 and H9.
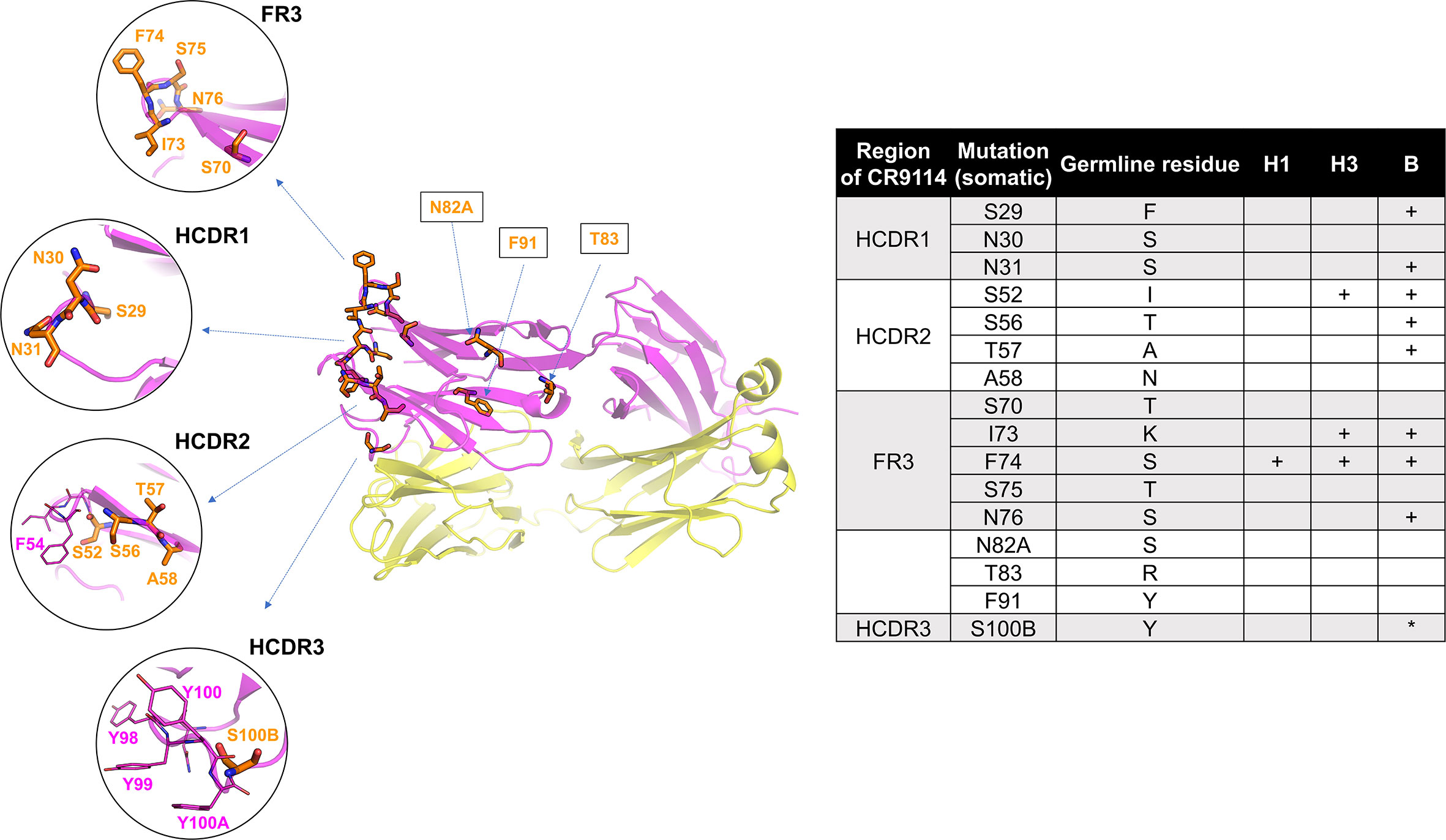
Figure 4 Structure of CR9114, indicating the 16 AAs that differ between the germline and the somatic variant. Mutated residues (53) (16 positions) are shown in orange sticks. Residues IDs as labelled in orange are converted to the somatic sequence as in the PDB structure (table). Mutated residues required (53) for gaining affinity for H1, H3, and B HA are indicated in the table (+ required mutation, *mutation improves binding). Some key residues, such as F54 (HCDR2) and a quadruplet “YYYY” in HCDR3, are additionally displayed in magenta sticks. The heavy and light chains are shown in magenta and yellow, respectively.
CR9114’s proven optimal binding affinity for the H1 subtypes corroborates the idea of a constricted path to broad breadth acquisition, that required the exposition to A1 subtypes as the first step(s). This also manifests itself in the inability of H1 subtypes to escape from binding to CR9114 and, thereby, in vitro neutralization and induced Fc-effector mediated functions (26). Even after 15 passages of selection pressure from CR9114, H1 mutants do not escape binding and neutralization by, nor Fc-mediated effector functions of CR9114. The resultant mutant is unfit, and not lethal even when administered to mice at very high dosages. The acquisition of binding affinity to the A2 group is subordinate to the A1 group. Thus, the genetic barrier to escape binding is smaller for H3 than H1. For H1, the relative resistance of all escape mutants to CR9114 is low (54), and CR9114 retains binding and in vivo efficacy to H1 escape mutants (26). For H3, the relative resistance of the vast majority of variants to CR9114 is low, but three mutants escape in vivo protection by CR9114 (54). These three mutants have a point mutation at location 45 that rarely occurs in human H3: their natural occurrence frequencies lie between 0.0001% and 0.01% (54).
Our structural analysis presented here is in accordance with the work on CR9114’s evolutionary path (53). Most of CR9114’s mutations from the germline sequence are mostly located throughout FW3 and the HCDRs, specifically at the sites forming hydrophobic contacts and hydrogen bonds with residues in the conserved HA stem epitope. Three specific mutations are required for binding to H3 (S52, I73, and F74) and eight specific mutations are required for binding to influenza B (table in Figure 4). Notably, many of these breadth-conferring mutations are absent in CR6261, particularly those in HCDR2 (29, 48).
Three specific mutations to the germline ancestor enable binding to H3 HA: I73 and F74 at the FR3, which form hydrophobic contacts with HA, and S52 which reorients the CDR2 loop. I73 and F74 bind at the hydrophobic groove of A1 HAs as previously described (and displayed in Figure 3), possibly due to removing steric hindrance. For example, K-lysine (germline ancestor) has a long side chain which may prevent the binding of the germline ancestor with HAs, whereas I73 (CR9114) has a short side chain that does not sterically hinder binding. In the case of S52 at HCDR2, the original I-isoleucine residue from the germline may induce similar effects as observed in CR6261: the formation of an internal H-bond with several residues at HCDR2 resulting in a rigid loop that does not bind to HAs (Figure 3C). S52, S56, and T57 at HCDR2 may contribute to the flexible HCDR2 loop in CR9114. S29 (CR9114) at HCDR1 doesn’t show direct interaction with HAs, but it may have removed adverse effects of the original F29 (germline ancestor), which is a larger amino acid and more likely to cause steric hindrance.
The quest for a universal influenza vaccine, inducing a polyclonal CR9114-like response, has not yet been fulfilled. CR9114 provides molecular insights into virtually universal antibody protection that may guide those on the pan-influenza quest. The protection that CR9114 provides in vivo exceeds its broad neutralization span, due to its ability to evoke Fc-effector mediated functions also against those strains that elude neutralization in vitro. This is rooted in CR9114’s binding affinity for a very broad range of viral hemagglutinin subtypes, covering influenza groups A1, A2, and B. The analysis of CR9114’s binding affinity to different HA subtypes reveals the history behind its evolution from the germline and the path to the acquisition of its heterosubtypic breadth. CR9114 is most likely the result of sequential exposure to the different viral subtypes, as demonstrated by the nested structure of the set of mutations that, from the germline, led to its broad breadth. The acquisition of affinity for all the considered viral subtypes happened progressively and through a rare, highly constrained evolutionary path. For a pan flu vaccine, it may be needed to find a way to shift from a recessive response to the CR9114 epitope to a dominant one. Alternatives to a pan-influenza vaccine, such as passive immunization using CR9114, could be considered for episodic prophylaxis.
AB and JF contributed equally to the writing of this article. AB, JF, CK, JK and JG designed this article. WK, GA, CK, and JG contributed to the article and provided critical remarks. KW and GAFN performed the structural analysis, JK analyzed the in vitro neutralization data for the landscape. AP and MMD contributed to the parts regarding the evolutionary pathway of pan-influenza protection. All authors contributed to the article and approved the submitted version.
We thank Carolyn Boudreau for the useful discussions.
AB, JF, CK, JK, WK, GA, and JG have disclosed that they are employees of Leyden Laboratories with stock options in this company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.1049134/full#supplementary-material
2. Molinari NAM, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, et al. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine (2007) 25:5086–96. doi: 10.1016/j.vaccine.2007.03.046
3. Putri WCWS, Muscatello DJ, Stockwell MS, Newall AT. Economic burden of seasonal influenza in the united states. Vaccine (2018) 36:3960–6. doi: 10.1016/j.vaccine.2018.05.057
4. Sharma L, Rebaza A, Cruz CSD. When ‘b’ becomes a’’: The emerging threat of influenza b virus. Eur Respir J (2019) 54:1901325. doi: 10.1183/13993003.01325-2019
5. Grange ZL, Goldstein T, Johnson CK, Anthony S, Gilardi K, Daszak P, et al. Ranking the risk of animal-to-human spillover for newly discovered viruses. PREDICT Consortium (2021) 118(15). doi: 10.1073/pnas.2002324118/-/DCSupplemental
6. Shi W, Gao GF. Emerging H5N8 avian influenza viruses: The global spread of H5N8 avian influenza viruses is a public health concern. Science (2021) 372:784–6. doi: 10.1126/science.abg6302
7. Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, De Wit E, Munster VJ, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science (2012) 336(6088)1534–41. doi: 10.1126/science.1213362
8. Morens DM, Taubenberger JK. The mother of all pandemics is 100 years old (and going strong)! Am J Public Health (2018) 108:1449–54. doi: 10.2105/AJPH.2018.304631
9. Krammer F. The human antibody response to influenza a virus infection and vaccination. Nat Rev Immunol (2019) 19:383–97. doi: 10.1038/s41577-019-0143-6
11. Wohlbold TJ, Krammer F. In the shadow of hemagglutinin: A growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses (2014) 6:2465–94. doi: 10.3390/v6062465
12. Wu NC, Wilson IA. Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol (2018) 25:115–21. doi: 10.1038/s41594-018-0025-9
13. Jang J, Bae SE. Comparative Co-evolution analysis between the HA and NA genes of influenza a virus. Virol Res Treat (2018) 9. doi: 10.1177/1178122X18788328
14. Corti D, Cameroni E, Guarino B, Kallewaard NL, Zhu Q, Lanzavecchia A. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol (2017) 24:60–9. doi: 10.1016/j.coviro.2017.03.002
15. Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature (1982) 296:115–21. doi: 10.1038/296115a0
16. Both GW, Sleigh MJ, Cox NJ, Kendal AP. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: Multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol (1983) 48:52–60. doi: 10.1128/jvi.48.1.52-60.1983
17. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol (2013) 87:6542–50. doi: 10.1128/JVI.00641-13
18. Nachbagauer R, Palese P. Is a universal influenza virus vaccine possible? Annu Rev Med (2020) 71:315–27. doi: 10.1146/annurev-med-120617-041310
19. Roozendaal R, Tolboom J, Roos A, Riahi S, Theeuwsen J, Bujny MV, et al. Transient humoral protection against H5N1 challenge after seasonal influenza vaccination of humans. PloS One (2014) 9:e103550. doi: 10.1371/journal.pone.0103550
20. Demicheli V, Jefferson T, Ferroni E, Rivetti A, Di Pietrantonj C. Vaccines for preventing influenza in healthy adults. Cochrane Database Syst Rev (2018) 2(2):CD001269. doi: 10.1002/14651858.CD001269.pub6
21. Rondy M, El Omeiri N, Thompson MG, Levêque A, Moren A, Sullivan SG. Effectiveness of influenza vaccines in preventing severe influenza illness among adults: A systematic review and meta-analysis of test-negative design case-control studies. J Infect (2017) 75:381–94. doi: 10.1016/j.jinf.2017.09.010
22. Dou D, Revol R, Östbye H, Wang H, Daniels R. Influenza a virus cell entry, replication, virion assembly and movement. Front Immunol (2018) 9:1581. doi: 10.3389/fimmu.2018.01581
23. Palese P, Tobita K, Ueda M, Compans RW. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology (1974) 61:397–410. doi: 10.1016/0042-6822(74)90276-1
24. Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol (2010) 17:1055–65. doi: 10.1128/CVI.00131-10
25. Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep (2018) 8:e03617–20. doi: 10.1038/s41598-018-28706-1
26. Kirkpatrick Roubidoux E, Carreño JM, Mcmahon M, Jiang K, Van Bakel H, Wilson P, et al. Mutations in the hemagglutinin stalk domain do not permit escape from a protective, stalk-based vaccine-induced immune response in the mouse model. Scientific Reports (2021) 8(1). doi: 10.1038/s41598-018-28706-1
27. Nachbagauer R, Shore D, Yang H, Johnson SK, Gabbard JD, Tompkins SM, et al. Broadly reactive human monoclonal antibodies elicited following pandemic H1N1 influenza virus exposure protect mice against highly pathogenic H5N1 challenge. J Virol (2018) 92:e00949–18. doi: 10.1128/JVI.00949-18
28. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza a hemagglutinins. Science (2011) 333:850–6. doi: 10.1126/science.1205669
29. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza b viruses. Science (2012) 337:1343–8. doi: 10.1126/science.1222908
30. Sutton TC, Lamirande EW, Bock KW, Moore IN, Koudstaal W, Rehman M, et al. In vitro neutralization is not predictive of prophylactic efficacy of broadly neutralizing monoclonal antibodies CR6261 and CR9114 against lethal H2 influenza virus challenge in mice. Science (2017) 366(6464)499–504. doi: 10.1128/JVI
31. Wang X, Zhou P, Wu M, Yang K, Guo J, Wang X, et al. Adenovirus delivery of encoded monoclonal antibody protects against different types of influenza virus infection. NPJ Vaccines (2020) 5. doi: 10.1038/s41541-020-0206-5
32. Laursen NS, Friesen RHE, Zhu X, Jongeneelen M, Blokland S, Vermond J, et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science (2018) 362:598–602. doi: 10.1126/science.aaq0620
33. Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, et al. A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nat Commun (2021) 12:1722. doi: 10.1038/s41467-021-21954-2
34. Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science (2019) 366:499–504. doi: 10.1126/science.aay0678
35. Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, et al. Structure and function analysis of an antibody recognizing all influenza a subtypes. Cell (2016) 166:596–608. doi: 10.1016/j.cell.2016.05.073
36. Del Rosario JMM, da Costa KAS, Asbach B, Ferrara F, Ferrari M, Wells DA, et al. Exploiting pan influenza a and pan influenza b pseudotype libraries for efficient vaccine antigen selection. Vaccines (2021) 9:1520. doi: 10.3390/vaccines9070741
37. Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza a viruses. Cell (2016) 166:609–23. doi: 10.1016/j.cell.2016.06.043
38. Carnell G, Giotis S, Grehan K, Ferrara F. The bat influenza H17N10 can be neutralized by broadly-neutralizing monoclonal antibodies and its neuraminidase can facilitate viral egress. Science (2020) 368(6497):1335–40. doi: 10.1101/499947
39. Chen Y-Q, Lan LY-L, Huang M, Henry C, Wilson PC. Hemagglutinin stalk-reactive antibodies interfere with influenza virus neuraminidase activity by steric hindrance. J Virol (2019) 93:e01526–18. doi: 10.1128/JVI.01526-18
40. Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, et al. Analysis of anti-influenza virus neuraminidase antibodies in children, adults, and the elderly by ELISA and enzyme inhibition: Evidence for original antigenic sin. MBio (2017) 8:e02281-16. doi: 10.1128/mBio.02281-16
41. da Costa KAS, Del Rosario JMM, Ferrari M, Vishwanath S, Asbach B, Kinsley R, et al. (B/Victoria and B/Yamagata) neuraminidase pseudotypes as tools for pandemic preparedness and improved influenza vaccine design. Vaccines (2022) 10:1520. doi: 10.3390/vaccines10091520
42. Sadanand S. A llama-derived antibody for flu. Nat Biotechnol (2019) 37:27. doi: 10.1038/nbt.4328
43. Brandenburg B, Koudstaal W, Goudsmit J, Klaren V, Tang C, Bujny MV, et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PloS One (2013) 8:e80034. doi: 10.1371/journal.pone.0080034
44. Hashimoto G, Wright PF, Karzon DT. Antibody-dependent cell-mediated cytotoxicity against influenza virus-infected cells. J Infect Dis (1983) 148:785–94. doi: 10.1093/infdis/148.5.785
45. Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol (2007) 178:2448–57. doi: 10.4049/jimmunol.178.4.2448
46. Cox F, Kwaks T, Brandenburg B, Koldijk MH, Klaren V, Smal B, et al. HA antibody-mediated FcγRIIIa activity is both dependent on FcR engagement and interactions between HA and sialic acids. Front Immunol (2016) 7. doi: 10.3389/fimmu.2016.00399
47. Winarski KL, Tang J, Klenow L, Lee J, Coyle EM, Manischewitz J, et al. Antibody-dependent enhancement of influenza disease promoted by increase in hemagglutinin stem flexibility and virus fusion kinetics. Proc Natl Acad Sci (2019) 116:15194–9. doi: 10.1073/pnas.1821317116
48. Ekiert DC, Bhabha G, Elsliger MA, Friesen RHE, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Sci (80-. ). (2009) 324:246–51. doi: 10.1126/science.1171491
49. Lefranc M-P, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J. Immunoglobulins: 25 years of immunoinformatics and IMGT-ONTOLOGY. Biomolecules (2014) 4:1102–39. doi: 10.3390/biom4041102
50. Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet (2013) 92:530–46. doi: 10.1016/j.ajhg.2013.03.004
51. Chen F, Tzarum N, Wilson IA, Law M. V H 1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design. Curr Opin Virol (2019) 34:149–59. doi: 10.1016/j.coviro.2019.02.004
52. Throsby M, van den Brink E, Jongeneelen M, Poon LLM, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory b cells. PloS One (2008) 3:e3942. doi: 10.1371/journal.pone.0003942
53. Phillips AM, Lawrence KR, Moulana A, Dupic T, Chang J, Johnson MS, et al. Binding affinity landscapes constrain the evolution of broadly neutralizing anti-influenza antibodies. Elife (2021) 10:e71393. doi: 10.7554/eLife.71393
Keywords: broadly neutralizing antibodies, influenza virus, pan-influenza protection, CR9114, correlate of protection, antiviral therapies, immunization, vaccination
Citation: Beukenhorst AL, Frallicciardi J, Koch CM, Klap JM, Phillips A, Desai MM, Wichapong K, Nicolaes GAF, Koudstaal W, Alter G and Goudsmit J (2022) The influenza hemagglutinin stem antibody CR9114: Evidence for a narrow evolutionary path towards universal protection. Front. Virol. 2:1049134. doi: 10.3389/fviro.2022.1049134
Received: 20 September 2022; Accepted: 11 October 2022;
Published: 02 November 2022.
Edited by:
Sanjeev Kumar, International Centre for Genetic Engineering and Biotechnology, IndiaReviewed by:
Hyesun Jang, J. Craig Venter Institute, United StatesCopyright © 2022 Beukenhorst, Frallicciardi, Koch, Klap, Phillips, Desai, Wichapong, Nicolaes, Koudstaal, Alter and Goudsmit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacopo Frallicciardi, amFjb3BvLmZyYWxsaWNjaWFyZGlAbGV5ZGVubGFicy5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.