- Department of Health Sciences, Carleton University, Ottawa, ON, Canada
Understanding the host-virus interactions helps to decipher the viral replication strategies and pathogenesis. Viruses have limited genetic content and rely significantly on their host cell to establish a successful infection. Viruses depend on the host for a broad spectrum of cellular RNA-binding proteins (RBPs) throughout their life cycle. One of the major RBP families is the heterogeneous nuclear ribonucleoproteins (hnRNPs) family. hnRNPs are typically localized in the nucleus, where they are forming complexes with pre-mRNAs and contribute to many aspects of nucleic acid metabolism. hnRNPs contain RNA binding motifs and frequently function as RNA chaperones involved in pre-mRNA processing, RNA splicing, and export. Many hnRNPs shuttle between the nucleus and the cytoplasm and influence cytoplasmic processes such as mRNA stability, localization, and translation. The interactions between the hnRNPs and viral components are well-known. They are critical for processing viral nucleic acids and proteins and, therefore, impact the success of the viral infection. This review discusses the molecular mechanisms by which hnRNPs interact with and regulate each stage of the viral life cycle, such as replication, splicing, translation, and assembly of virus progeny. In addition, we expand on the role of hnRNPs in the antiviral response and as potential targets for antiviral drug research and development.
1 Introduction
The regulation of gene expression in eukaryotes is highly regulated (1) and the RNA is the central molecule of posttranscriptional regulation of gene expression (2). The primary transcripts of RNA polymerase II are historically referred to as heterogeneous nuclear RNAs (hnRNAs), also known as pre-mRNAs (3). These hnRNAs are associated with proteins in large complexes that structurally resemble the histones in the nucleus (4). The regulatory RNA binding proteins (RBPs) that associate with hnRNAs are known as heterogeneous nuclear ribonucleoproteins (hnRNPs) (2). hnRNPs constitute a large family of RBPs that bind with nascent pre-mRNAs/mRNA and are involved in the RNA metabolism and regulate gene expression (2). In addition to regulation of transcription, hnRNPs regulate RNA splicing and maturation, RNA packaging and transport, translational control, RNA silencing, DNA repair, and telomere biogenesis (5, 6). hnRNPs are also involved in the maintenance of transcriptome integrity, by protecting endogenous pre-mRNA transcripts from mis-splicing of intronic cryptic splicing signals (7). Moreover, hnRNPs function as dynamics and unstable interaction proteins in nuclear and cytoplasmic cellular processes. Many of the hnRNP proteins undergo constant nuclear-cytoplasmic shuttling through nuclear pores (5). Not surprisingly, due to their RNA metabolism and the nucleocytoplasmic shuttling activities, many hnRNPs are associated with the viral life cycle. Indeed, from the initial time of the hnRNP discovery, the association of hnRNPs with viral nucleic acid (mainly RNA) was observed. Even though the association between herpes virus and HeLa cells’ ribonucleoproteins was shown in a paper published in 1963 (8), the term “hnRNP” was officially used almost 20 years later, in a 1980 paper that demonstrated the association between adenovirus-2 DNA and hnRNP (the identity of this RBP was not known at the time, only that it had a relative molecular mass between 25,000 and 200,000) (9). The historical biochemical characterization of the hnRNPs was thoroughly reviewed previously (2, 10). In this review, we will describe and discuss the structural and functional diversity of hnRNPs as they relate to their regulatory role in distinct steps of the viral life cycle, including viral RNA transcription, pre-mRNA splicing, viral RNA and protein translocation, translational regulation, viral mRNA and particles packaging, and release.
1.1 Structure and function of hnRNPs
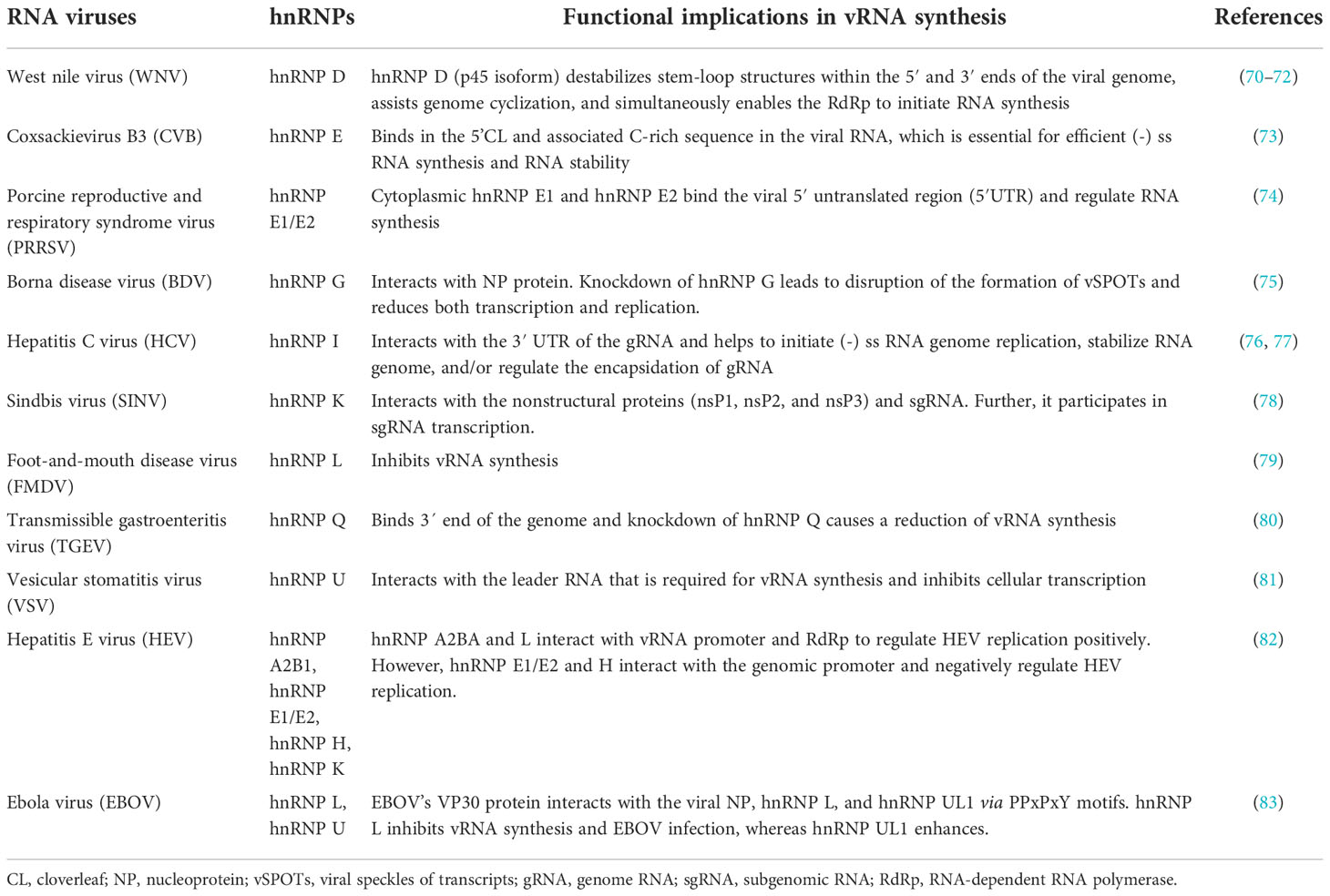
The hnRNP family is comprised of 20 major hnRNPs, named from hnRNP A1 to U, with molecular weights ranging from 34 to 120 kDa (5, 11, 12). Most hnRNPs are commonly known by their designated hnRNP names, but few of them, are also known by their alternative names, as summarized in Table 1. Among these hnRNPs, the hnRNP A/B sub-family of proteins (comprised of A0, A1, A2/B1, and A3) contributes to the regulation of the majority of cellular RNAs (15). In addition to the major hnRNPs, there are many minor hnRNPs such as HuR, PABP1, RNPS1, SRm160, and SRp20 [reviewed in (12)]. Unlike major hnRNPs, these minor hnRNPs are characterized either as unstably binding hnRNAs, or only associated with a subset of hnRNAs (5, 12, 13, 16).
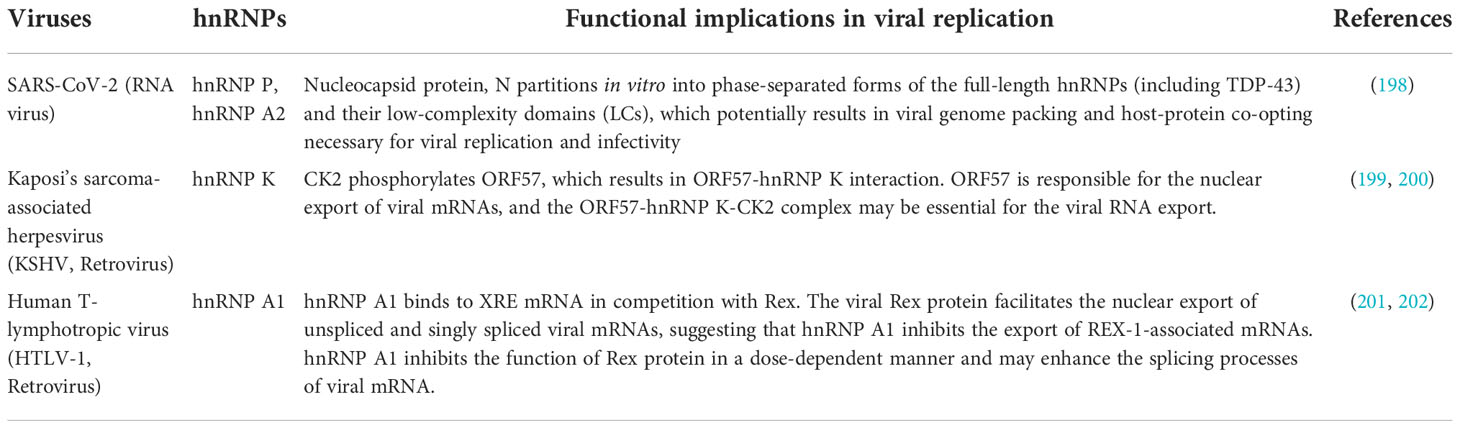
The binding of hnRNPs to nucleic acids is carried out by multiple protein domains (12). Most hnRNPs contain classical RNA-Binding Domains (RBDs) such as RNA recognition motif (RRM), glycine-arginine-rich (GAR) domains or arginine-glycine-glycine rich (RGG) boxes, hnRNP K-homology (KH) domain, DEAD-box helicase domain, dsRNA motif (DSRM), and zinc fingers (ZF) domain (17). RRMs play a crucial role in binding the RNA sequences (18). The RRM (the second RRM in some proteins), the external β-strands, the loops, and the C- and N-termini, can have a high RNA-binding affinity and specificity, as well as extreme structural versatility when they interact with RNA (19). This makes the RRM-containing proteins capable of performing numerous biological functions (19). In addition to RRM-containing hnRNPs, five hnRNPs (hnRNP E 1-4, hnRNP K) contain KH domains (5). The KH domains cooperatively bind polypyrimidine or C-rich regions in RNA, ssDNA, or dsDNA with high affinity and specificity (20). Moreover, there are numerous protein partners that interact specifically with the KH domain proteins (20). As a result, hnRNPs can influence many events in the cell at the transcriptional, post-transcriptional, and translational levels (21). hnRNP A/B, hnRNP P, hnRNP D, hnRNP H, and hnRNP U also contain a low-complexity domain (LCD) typically in the C-terminus region, which is also known as an intrinsically disordered region (IDR) or prion-like domain (PrLD) (22, 23). The PrLD encourages liquid-liquid phase separation as well as fibrillation by self-association (24). The activity of these interactions is thought to facilitate transient and diverse multivalent de-mixing of proteins and/or RNA (25). PrLDs/IDRs have also been shown to bind RNA (26–28). Since hnRNPs contain either RBDs or both RBDs and IDRs, they can interact with RNA in both specific and nonspecific ways.
hnRNPs have been reported to undergo various post-translational modifications (PTMs), such as phosphorylation, glycosylation, s-nitrosylation, methylation, acetylation, ubiquitination, and SUMOylation. These PTMs are essential for cellular localization, protein-protein interactions, and nucleic acid binding of hnRNPs, in addition to regulating viral RNA processing and virus replication (29–31). Some hnRNPs are strictly nuclear [e.g., hnRNP C and U (32)], whereas others remain associated with the mRNA as it passes through nuclear pores and therefore undergo constant nucleocytoplasmic movement (e.g., members of hnRNP A, B, E, D, I, and K group) (12). The subcellular localization is controlled by discreet sequence signals within hnRNPs. For example, the hnRNP C and U contain nuclear localization signal (NLS) sequence and are confined to the nucleus. hnRNP A1 harbours noncanonical nucleocytoplasmic shuttling (NS) domains called M9, and hnRNP K contains both an NLS (N-terminal) and an NS (termed KNS, K nuclear shuttling) domain (32–34). Of note, hnRNP A1 was identified as the first cargo of Transportin-1 (Trn-1), which is a well-characterized nuclear import receptor (35, 36). Trn-1 also imports other proteins of the hnRNP family such as hnRNP A/B, hnRNP D, hnRNP F, hnRNP H, hnRNP M (37). A schematic structural diagram of major hnRNPs, along with their size, major RNA binding domains, and localization signals are shown in Figure 1 and Table 1.
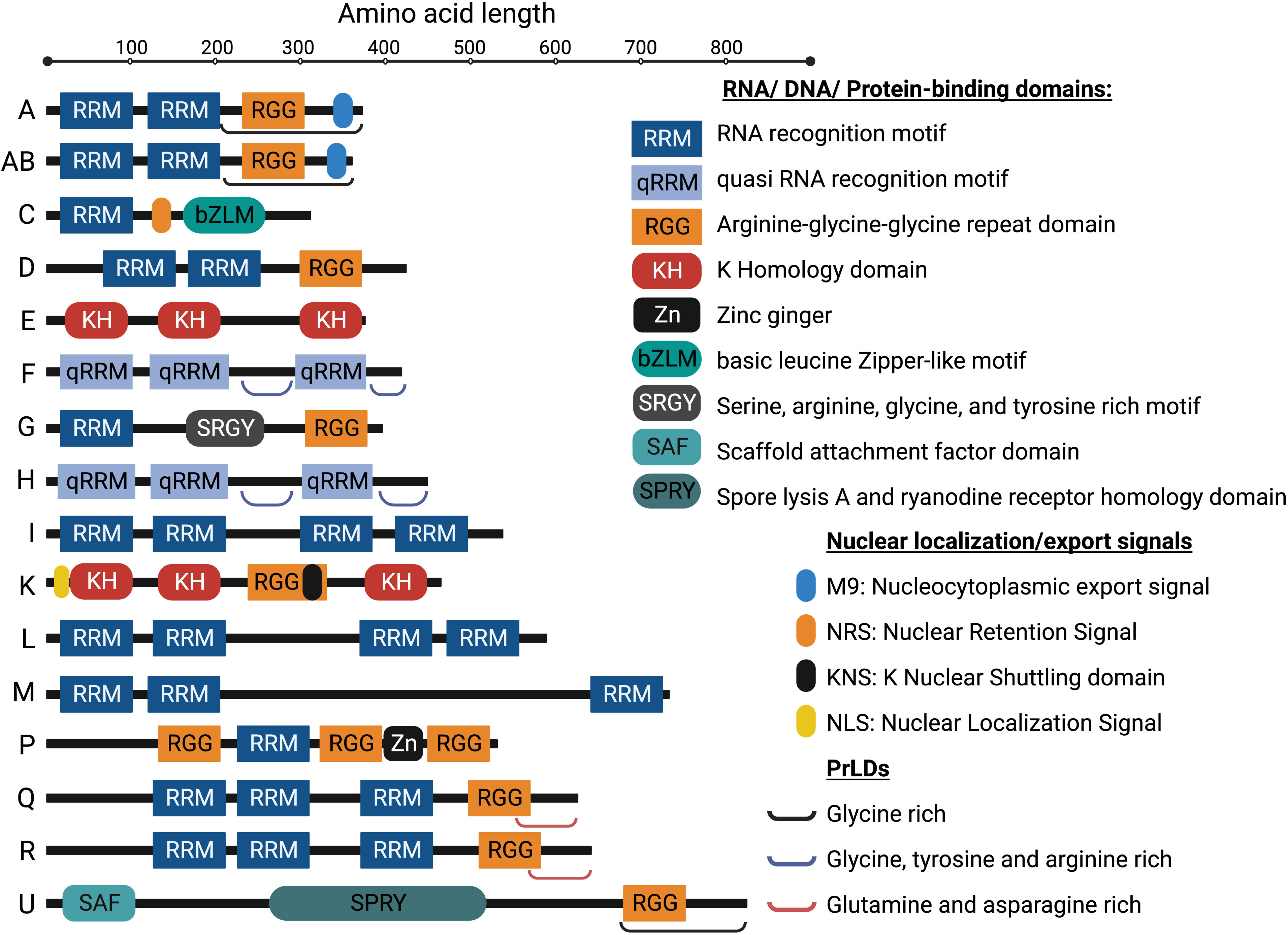
Figure 1 Schematic diagram of structures of the representative isoforms of the major hnRNP proteins. PrLDs, Prion-like domains (aka intrinsically disordered regions (IDRs)). The image was created with BioRender.com.
1.2 Brief overview of viruses and viral lifecycle
Pathogenic viruses continuously emerge and re-emerge, which have become a major threat to public health for millennia. Understanding the molecular mechanism of the viral life cycle is important for the development of diagnostic tests and effective antiviral therapies. The process of viral life cycle can be divided into six major stages: attachment, penetration, uncoating, genome replication and expression (including vRNA transcription and viral protein synthesis), assembly, and virion release from the host cell (Figure 2) (39). Viruses generate new infectious particles by assembling preformed components. Following entry into host, viruses must rapidly generate numerous copies of genome and package them into virions (39). To do so, viruses must express their genes as functional mRNAs as early as possible during infection, and subsequently instruct the cellular translation machine to create viral proteins (40).
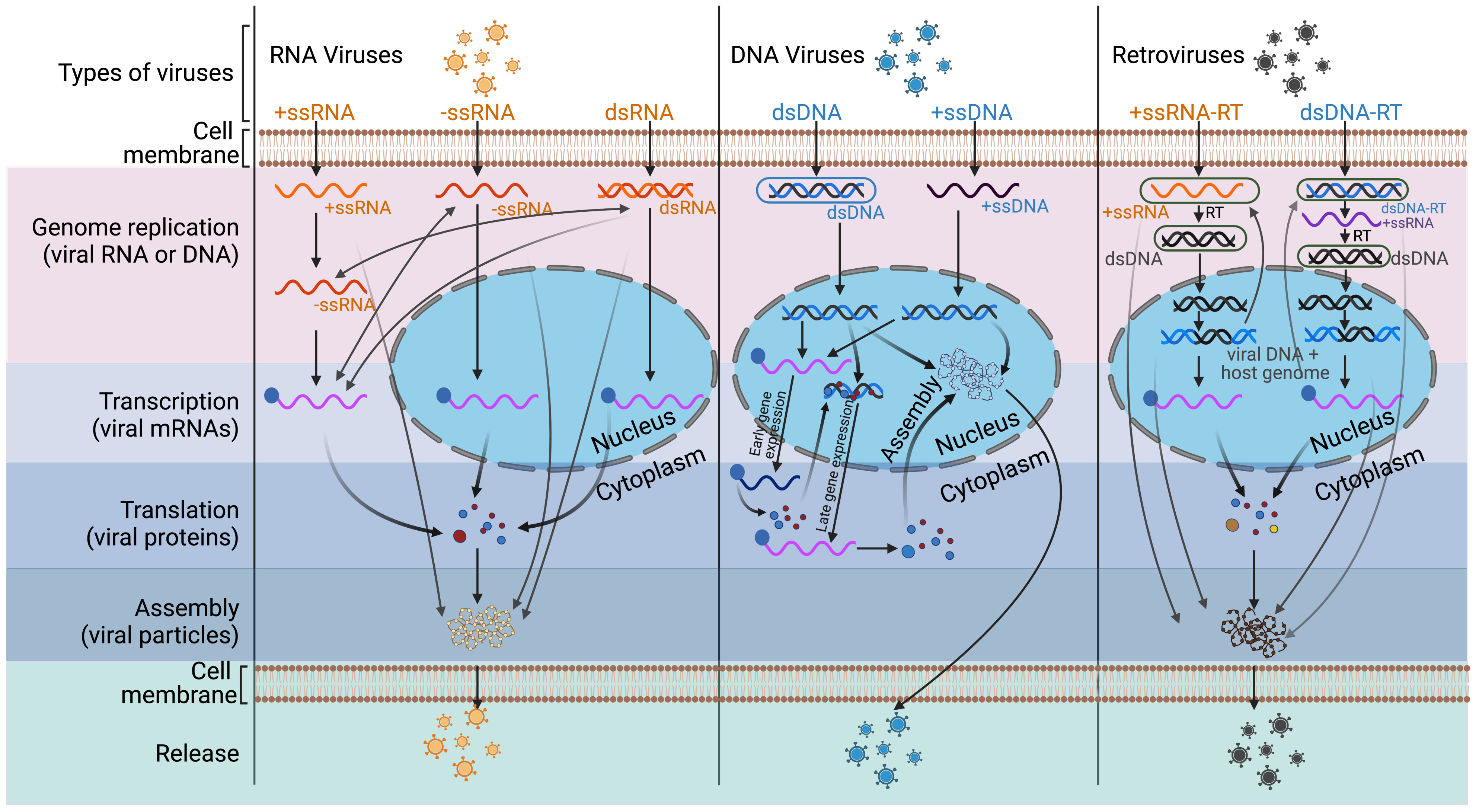
Figure 2 Life cycles of RNA viruses, DNA viruses, and retroviruses. This diagram illustrates the steps of the viral life cycle of different types of viruses. Further, unlike RNA or retroviruses, DNA viruses have a different replication compartment within the nucleus or cytoplasm, as described in (38). +ssRNA (CVB, DENV, EMCV, EV-A71, FMDV, HAV, HCV, HEV, HRV, JEV, MHV, PEDV, PRRSV, PV, SARS-CoV-2, SINV, SVV, TGEV, WNV), -ssRNA (BDV, BEFV, EBOV, IAV, JUNV, NDV, NiV, RABV, VSV), dsRNA (ARV), ssDNA (PPV), dsDNA (EBV, HCMV, HPV, HSV, KSHV), ssRNA-RT (HIV, HTLV, RSV), and dsDNA-RT (HBV) viruses are featured in this review. See text for details. RT, reverse transcriptase. The image was created with BioRender.com.
Based on the type of viral genome and mode of transcription, viruses are categories into RNA viruses, DNA viruses, and retroviruses. These viruses are further subcategorised into double-stranded (ds) or single-stranded (ss), and positive-strand (+) or negative strand (-) viruses, as shown in the Figure 2 (41). In RNA viruses, (+) ssRNA acts as mRNA, and one of the RNA strands of the dsRNA or (-) ssRNA acts as a template for viral mRNA or (+) ssRNA synthesis. The mRNA or (+) ssRNA is then recognized by the host cell translation machinery (42). Similarly, in DNA viruses, dsDNA is transcribed into viral mRNA. However, ssDNA is first converted to dsDNA intermediate, which is further transcribed into mRNA. For efficient viral replication, a high degree of regulation is required to switch from DNA transcription to replication when virion packaging occurs. The transcription begins with early genes, such as polymerase and regulatory proteins, followed by the initiation of viral DNA replication once these genes are transcribed. Finally, the transcription ends with late genes that code for the capsid and envelope (38). In retroviruses, a reverse transcription step precedes the replication of the genome and the transcription of DNA. In RNA retroviruses, virion enzyme reverse transcribes (+) ssRNA into dsDNA intermediates upon infection (43). Once a virus genome has been integrated into the host genome, that integrated virus genome is transcribed into mRNA and is translated by the host synthesis machinery (44). Like in DNA viruses, the transcription and genome replication are also highly regulated in retroviruses for efficient viral replication (45).
2 The roles of hnRNPs in the virus life cycle
Viruses depend on the host cells to replicate and complete their life cycle. To do so, viruses hijack and make use of the host molecular components (46). In addition, viruses use complex strategies to improve their ability to disrupt host cell responses, such as viruses target mRNA export factors and nucleoporins, and block nuclear export of host mRNAs that encode antiviral proteins (47). The nuclear trafficking of RBPs is inhibited by viruses, which could make them accumulate in the cytoplasm, affecting the cellular homeostasis while promoting viral replication and pathogenesis (46). Moreover, disruption of nucleocytoplasmic transport impairs cellular antiviral response by affecting the nuclear import of signal transducers and activators of transcription, which are involved in the production of interferon (IFN) (48). Dicker et al. (49) have compared virion proteomes of ten different human RNA viruses and discovered a pool of cellular RBPs, including some hnRNPs, which have important functions in viral particle formation and infectivity and were incorporated into viral particles (49). The hnRNPs are considered as one of the most important RBPs family for the functions in the viral life cycle (50). Several members of the hnRNP family form functional complexes with viral RNAs and proteins (31). A study on proteome-wide scale analysis of viral RNA-host cell interaction identified 12 hnRNPs in the core vRNA interactome, which makes these proteins usual suspects in vRNA interactome analysis (51). Viral nucleic acids or proteins alter the hnRNPs and vice-versa in infected cells. The changes in alterations to the nucleic acids/proteins may have different functional implications according to different phases of infection. The major functions of the hnRNPs in the virus life cycle are shown in Table 1.
In this section, we review the known functions of hnRNPs in each step of viral life cycle. In general, although viruses usurp hnRNPs to accomplish their biological goals, hnRNPs also negatively control many viral functions and steps of the viral life cycle and therefore act as critical parts of the cellular antiviral strategy. As defined by this text, the term “viral replication” or “viral life cycle” includes the whole process, from the entry of the virus into the host cell to the release of progeny viruses.
2.1 hnRNPs in viral genome replication and transcription
Viruses depend on the host to supply tools for their DNA/RNA replication and transcription. New viral RNA complementary synthesis from an RNA template is described by the term “vRNA synthesis”, whereas mRNA synthesis from an RNA or DNA template is described by the term “vRNA transcription”. In this text, the term “viral genome replication” includes both vRNA synthesis as well as viral DNA replication. In this section, we review the roles of hnRNPs in viral genome replication and vRNA transcription in RNA, DNA, and retroviruses.
RNA viruses: One of the best-studied RNA viruses is human poliovirus (PV). During PV infection, hnRNP C binds to the 3′ end of the PV (-) ssRNA through its RRM domain and interacts with the PV replication protein 3CD (52). This binding further recruits 3CD to the (-) ssRNA template (52). The chaperone activity of hnRNP C helps to fold (-) ssRNA into a replication-compatible conformation, which promotes RNA synthesis (52). Additionally, in the replication initiation complex at the 5′ end, PV encoded RNA-dependent RNA polymerase (RdRp) is activated by the cleavage of 3CD. hnRNP E2 directly interacts with PABP bound to the 3’ poly(A) sequence and carries activated RdRp to its 3’end (53, 54). Further, hnRNP C interacts with both 5’- and 3’-end sequences of PV (-) ssRNA, enabling the convergence of 5’- and 3’-end sequences, which facilitates (+) ssRNA synthesis (55). On the other hand, upon the binding to viral RNAs, hnRNP E2 is cleaved by the PV-encoded RdRp precursor (3CD) or protease (3C) proteins and, as a consequence, loses one of its three RNA-binding sites (as explained in the translation section below) (56, 57). In the cleaved form of hnRNP E2, two RNA-binding sites are retained, allowing the protein to bind to the cloverleaf (CL) structure located at the 5’ untranslated region (UTR) of PV (+) ssRNA (56, 57). This makes PV RNA replication possible, as the poly(A)-binding proteins (PABPs) unite the 3’ and 5’ ends of the viral RNA (Figure 3B) (56, 57).
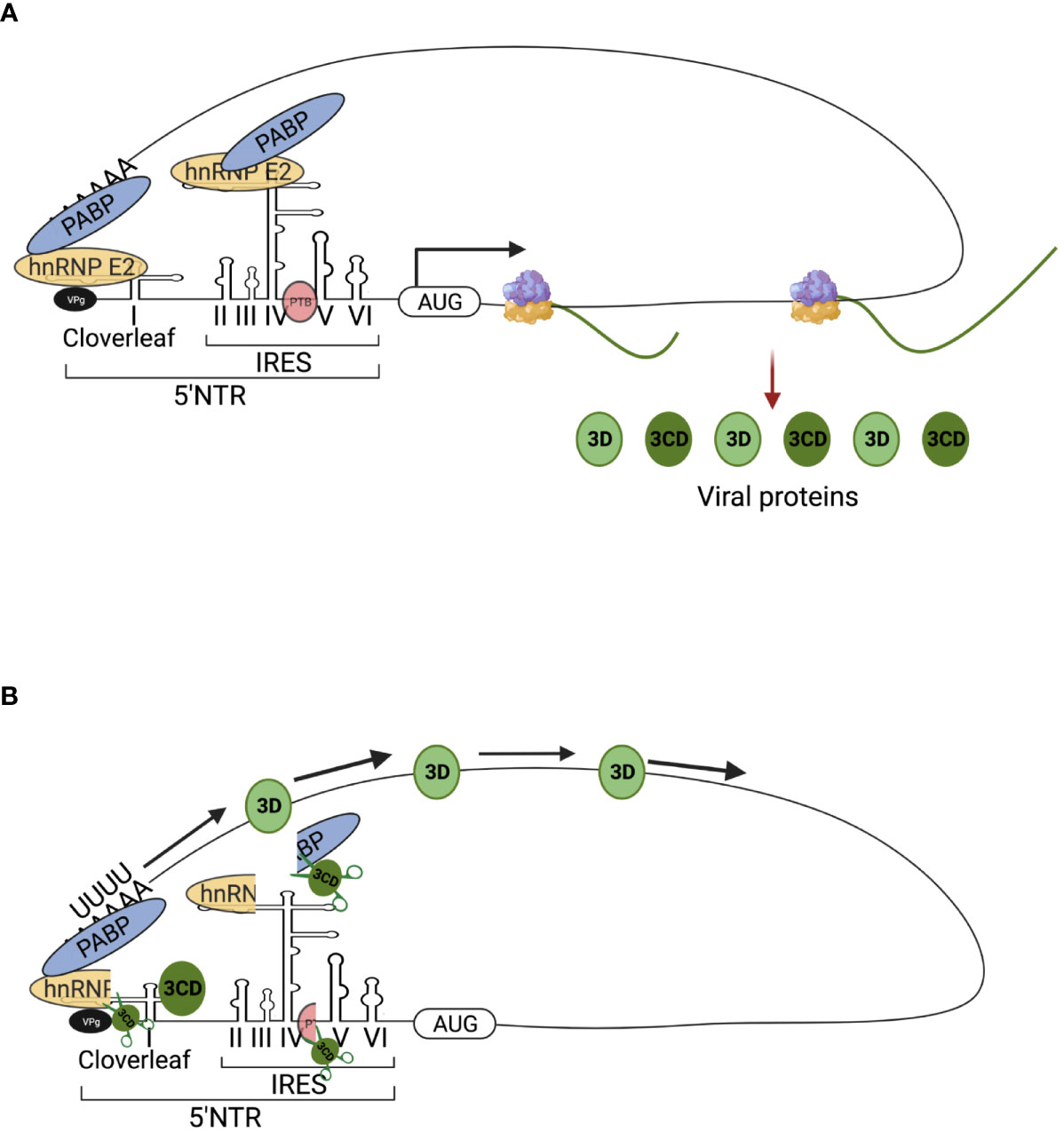
Figure 3 Schematic figures of involvement of hnRNP E (PCBP) in poliovirus (PV) translation and replication. The viral replication proteins may repress translation and switch to viral RNA synthesis, where hnRNP E is involved in the two roles of PV translation and replication. (A) PV translation: Full-length hnRNP E2 (PCBP 2) binds to stem-loop IV of IRES, which is necessary for cap-independent translation of the PV genome. The translation produces viral proteins, such as 3C/CD protease, which are required for subsequent switching of translation to replication. (B) Switching of translation to replication: The 3C/CD proteases cleavage hnRNP E2 (PCBP 2) and PTB, resulting in translation inhibition and template clearance from ribosomes. The cleaved hnRNP E2 (PCBP 2) binds to stem-loop I and forms a functional ternary complex with 3CD viral proteinase, which permits the initiation of RNA replication. The image was created with BioRender.com.
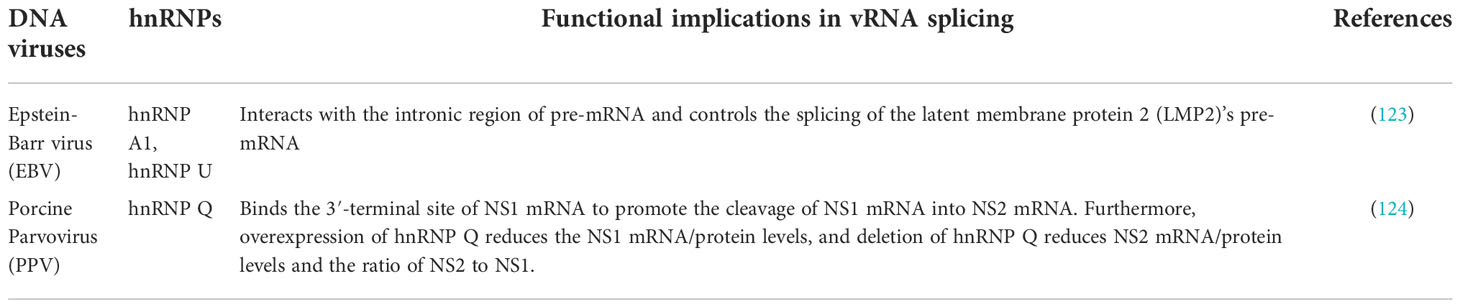
For other RNA viruses, including coronavirus and its prototype mouse hepatitis virus (MHV), hnRNP A1 specifically binds to the leader and intergenic sequences in MHV (-) ssRNA (58, 59). MHV’s RNA synthesis is stimulated by overexpression of wild-type hnRNP A1 while its replication is inhibited by the expression of a dominant-negative mutant hnRNP A1 (carrying a 75 amino acid deletion from the C-terminus) (60). It is evident from an in vitro reconstitution assay that the hnRNP A1 interacts with the leader and intergenic sequences and form a RNP complex which may participate in MHV RNA transcription (61). Further, hnRNP A1 interacts with MHV’s nucleocapsid protein (N), which suggests that hnRNP A1 might recruit the viral N protein to MHV transcription sites (62). Moreover, N protein recruitment to replication-transcription complexes (RTCs), via binding to non-structural protein 3 (nsp3), is a critical step for optimal MHV viral RNA synthesis (63). hnRNP I is also strongly associated with (+) ss leader sequence and with the 5’ UTR of (-) ssRNA (64). RNA affinity purification identified the interaction between hnRNP Q and the 5’ NCR of MHV genomic RNA and 5’ UTR of antigenomic RNA (65). This interaction is not limited to in vitro, but also to MHV-infected cells that overexpressed hnRNP Q (65). Moreover, an hnRNP Q mutant lacking the C terminus interferes with viral replication but does not affect viral translation which indicates that the hnRNP Q participates in the synthesis of RNA (65). In SARS CoV infection, hnRNP A1’s C terminal region has a high affinity and directly binds to the SARS’s N protein, suggesting that the interaction facilitates viral transcription and replication (66–68). In addition, hnRNP A2/B1 along with ILF3, QKI, and SFPQ interact with the SARS-CoV-2 genome and promote RNA synthesis (69). There are other examples of the involvement of hnRNPs in viral RNA synthesis used by other RNA viruses, and they are summarized in Table 2.
DNA viruses: Unlike RNA viruses where there are many interactions between hnRNPs and viruses, there are only two main examples in DNA viruses. In a hepatitis B virus (HBV), APOBEC3B (A3B) protein inhibits HBV expression by inhibiting the binding of hnRNP K to the enhancer II of HBV (84). A3B also directly suppresses HBV’s S gene promoter activity (84). Enhancer II of HBV plays an important role in activating the transcription of the HBV gene (85). HBV expression may be inhibited by A3B at the transcription level by reducing hnRNP K’s interactions with the enhancer II of the virus (84). According to this model, A3B and hnRNP K may form a super-complex that may affect hnRNP K’s binding to the enhancer II of HBV by changing its conformation (84). In another DNA virus, Kaposi’s sarcoma-associated herpesvirus (KSHV), the interaction of RNA polymerase II (RNAP II) with hnRNP P negatively affects serine-2 phosphorylation of RNAP II’s C-terminal domain at the replication and transcription activator gene (RTA) of KSHV, resulting in decreased nascent RNA synthesis (86). Further, knockdown of hnRNP P increases transcription of RTA, and enhances expression of KSHV lytic genes (86).
hnRNPs are also associated with viral DNA replication. Lytic replication is a major step of herpesvirus pathogenesis (87). Immunocompromised individuals are often at risk of life-threatening diseases from the lytic phase of human cytomegalovirus (HCMV) infection (88), and also the lytic phase contributes to cancer cell evolution during chronic infections of KSHV (87). Several hnRNPs are associated with lytic DNA replication of herpesviruses. A herpesvirus’s lytic DNA replication begins at an origin (oriLyt) and requires trans-acting elements (89). In addition to its role in promoting lytic DNA synthesis, oriLyt is also thought to play a role in regulating the transition between lytic and latent phases (89). In case of HCMV, hnRNP K is required for the efficient lytic DNA replication by interacting with the oriLyt DNA and virally encoded UL84 protein (89). RNAi knockdown of hnRNP K prevents amplification of oriLyt in a transient assay (89). Similarly, several hnRNPs such as hnRNP A1, hnRNP A1/B1, hnRNP A3, hnRNP C, hnRNP D, hnRNP G, hnRNP K and hnRNP U were shown to be associated with the KSHV oriLyt DNA in a mass spectrometry study (90).
Retroviruses: Human immunodeficiency virus (HIV) is the well-studied retrovirus with many demonstrated interactions between hnRNPs and the viral RNA synthesis. HIV’s pathogenicity factor, Nef forms a protein complex with Lck kinase and nPKC subfamily (PKC δ and θ) proteins that form a complex called NAKC (Nef-associated kinase complex), which is required to increase viral replication and infectivity (91). The hnRNP K interacts with Nef and nucleates Nef-interacting kinases, including Lck, PKC δ, and PI-3 kinase, leading to the activation of Lck and Erk1/2 (92). Thus, hnRNP K may mediate transcriptional de-repression by coordinated membrane signaling with Erk1/2, and HIV targets it to enable Tat-dependent transcription (92). Another hnRNP, hnRNP G regulates HIV-1 repressive trimethylation of histone H3 lysine 9 (H3K9me3) by binding to proviral DNA at the LTR downstream region (93). This results in the blockage of the recruitment of the phosphorylated RNA polymerase II and substantially impediment of transcription, which helps the virus to maintain its latency (93). Another study showed hnRNP A2/B1 as a HIV’s long terminal repeat (LTR)-G-quadruplex-interacting protein that functions as a transcription activator (94). Moreover, silencing of the hnRNP A2/B1 in cells decreases the LTR activity (94).
2.2 hnRNPs control post-transcriptional regulation of viral RNA
hnRNP proteins control viral mRNA splice site selection, promote viral RNA polyadenylation, and stabilize viral RNA in a variety of ways.
RNA viruses: hnRNP K helps to splice influenza A virus (IAV)’s M mRNA segments by binding M mRNA downstream of the M2 5’ splice sites and promoting U1 snRNP recruitment (95). A mutation in either or both of the hnRNP K and NS1-BP-binding sites results in a mis-spliced M segment and diminished replication of the IAV (95). Further, M1 mRNA segment is properly spliced to yield M2 mRNA by NS1-BP, without affecting either M4 or NS mRNA segments. hnRNP K plays a role in mediating the interaction of NS1-BP and M RNA (96). Another study showed that nuclear speckles of hnRNP K increase in IAV-infected cells, which could influence the accessibility of hnRNP K to host transcripts, thereby enhancing the replication of IAV (97).
In case of a Rous sarcoma virus (RSV), a negative regulator of splicing (NRS) element is located in the pre-mRNA of the gag gene, which inhibits RNA splicing (98). hnRNP H, serine/arginine-rich (SR), and U1/U11 snRNPs bind to the NRS element and inhibit splicing by causing a decoy 5′ splicing site selection, subsequently promoting polyadenylation (99, 100).
hnRNPs also stabilize PV mRNA. PV mRNA does not possess a 5’-terminal cap structure (101), and uncapped mRNAs are rapidly degraded by 5’ exonuclease (102). Yet, PV mRNA is very stable despite the absence of a 5’-terminal cap in HeLa S10 translation-replication reaction (103). PV’s genome possesses a 5’ cloverleaf structure that binds to hnRNP E, and this binding stabilizes PV (+) ssRNA (Figure 3A) (103, 104). The mutant hnRNP E protein that is unable to bind to the cloverleaf structure causes PV (+) ssRNA to degrade rapidly (103). Since in-vitro 5’ 7-methylguanosine capped PV (+) ssRNA can bypass the requirement for hnRNP E in stability assays, it is likely that binding of hnRNP E to PV (+) ssRNA blocks XRN1 5’-to-3’ exonuclease-mediated RNA decay (103, 104). In another example, rabies virus glycoprotein mRNA transcripts steal host hnRNP E2, which leads to increased transcript abundance and stability (105).
DNA viruses: hnRNP A1 is shown to regulate human papillomavirus (HPV) splicing and polyadenylation (Figure 4). HPV life cycle progression occurs in conjunction with host epithelial cell differentiation (106). The expression of nuclear hnRNP A1 increases during differentiation of HPV-16 infected epithelial cells, suggesting that hnRNP A1 is necessary for appropriate alternative splicing for late transcript synthesis (107). Further, hnRNP A1 also directly interacts with the HPV-16 late regulatory element (LRE) in the differentiated HPV-16-infected cells (107). Several post-transcriptional mechanisms have been identified as part of the LRE’s regulation of late gene expression. The HPV-16 genome is known to contain four 5’ splice donor sites (SDs) (e.g., SD3632) and seven 3’ splice acceptor sites (SAs) (e.g., SA409, SA742, SA2709, SA5639) (108, 109). A splicing silencer element (SSE) interacts with hnRNP A1 and hnRNP A2 to inhibit splicing at the SA409 to modulate E6 and E7 mRNA transcripts (Figure 4) (108, 109). Another hnRNP, hnRNP G interacts with splicing enhancer element (SEE), and this interaction promotes splicing to SA2709 and enhances E2 mRNA production through splicing factor U2AF65 (Figure 4) (110). The splice sites SD3632 and SA5639 are responsible for balancing the production of HPV 16 L1 and L2 transcripts. A splicing silencer element suppresses SD3632, allowing polyadenylation at an early polyadenylation signal (pAE). In particular, SD3632 and SA5639 are found to be “hot spots” for RBPs including hnRNPs (Figure 4) (111). The hnRNP A2/B1, hnRNP D, and hnRNP L bind to the SSE of the SD3632 to modulate L1 and L2 transcripts (112–114). In addition, overexpression of hnRNP C reduces suppression of the splice site SD3632 in an HPV-16 early 3′-UTR-dependent manner (115). Similarly, to modulate L1 and L2 mRNA, hnRNPs such as hnRNP A1, A2/B1, and hnRNP L bind to the SSE at the SA5639 that is located immediately upstream of the AUG sequence at Late 1 (L1) mRNA (114, 116, 117). Further, binding of hnRNP A1 to the SSE suppresses the use of the SA5639, and results into the inhibition of late mRNAs splicing by influencing SSE, preventing L1 expression prematurely (118, 119). This hnRNP A1 inhibition of HPV 16 mRNAs makes the virus infection undetectable by the host immune surveillance and allows for viral persistence (116). Moreover, hnRNP L binds to SD3632 and SA5639 in response to Akt-dependent phosphorylation (114).
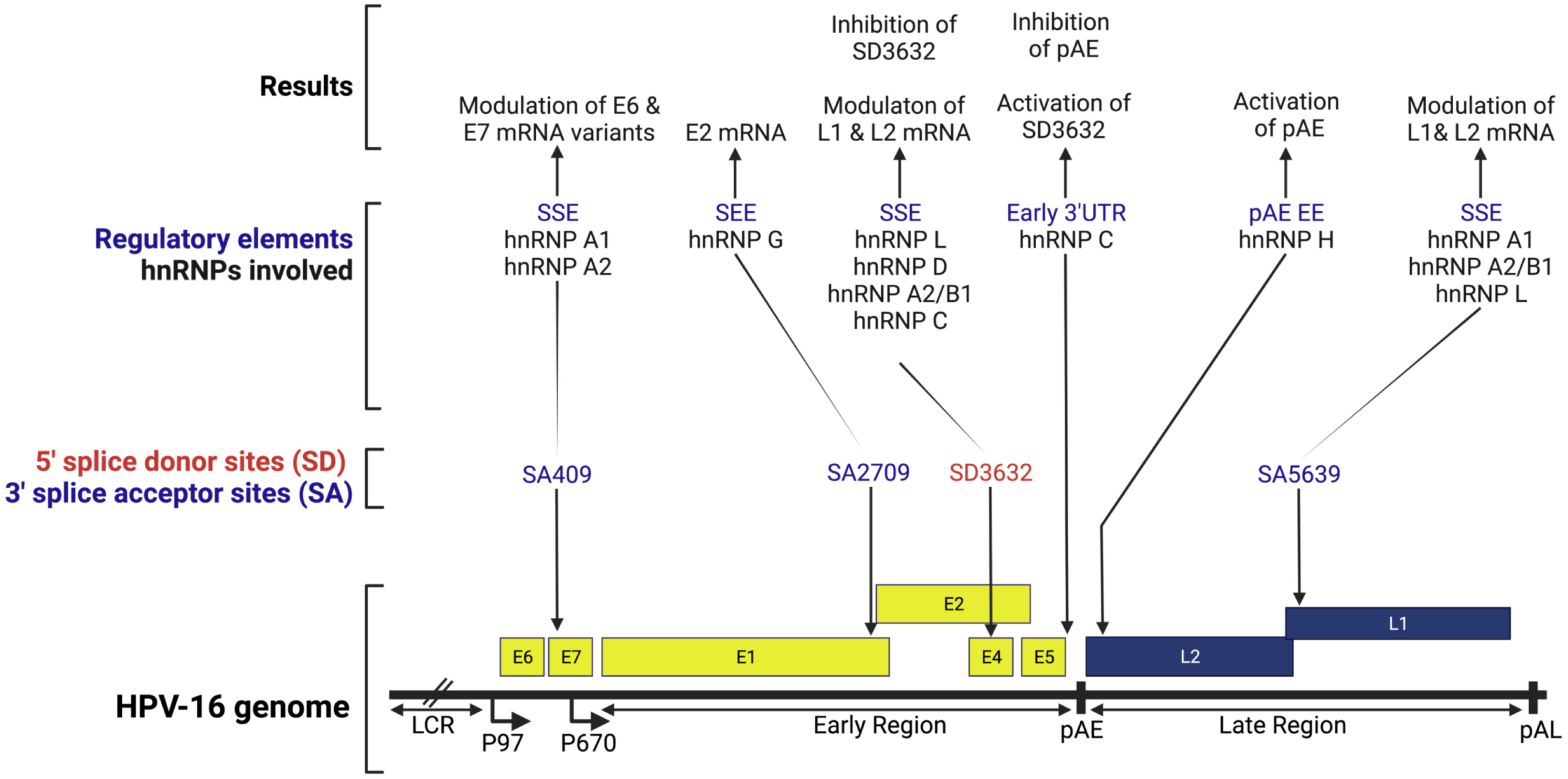
Figure 4 Schematic representation of the HPV-16 genome. This figure shows splice sites and the interfering and interacting positions of hnRNP proteins in the HPV-16 splicing sites. LCR, long control region; p97, early promoter; p670, late promoter; Early genes (E1, E2, and E4-7); late genes (L1 & L2); pAE, early polyadenylation signal; pAL, late polyadenylation signal; SSE, splicing silencer element; SEE, splicing enhancer element; EE, enhancer element. The image was created with BioRender.com.
In addition to the above mechanism, hnRNP D inhibits the splicing of HPV-16 E1/E2 and E6/E7-mRNAs, thereby causing E1-and E6-mRNAs to contain introns, respectively (120). As a result, the cytoplasmic levels of intron retained HPV-16 mRNAs increases in the presence of hnRNP D (120). hnRNP A1 promotes HPV-16 E6 exon exclusion, whereas Brm and Sam68 promote exon inclusion (121). hnRNP I also blocks splicing inhibitory elements located at the downstream and upstream of HPV-16’s major late 5’ splicing site (SD), thereby enhancing gene expression in the late stage (122). Moreover, some of the examples of hnRNPs on the viral RNA splicing used by other DNA viruses are summarized in Table 3.
In case of HPV polyadenylation, an early polyadenylation signal (pAE) plays a significant role in the transition from the early to late stages of viral lifecycles (125). An GGG motif located 174 nucleotides downstream of the pAE is known to act as a signal for hnRNP H to downregulate the late gene L2 (125). As a result of this hnRNP H binding, polyadenylation at pAE is enhanced, inhibiting L2 mRNA synthesis (125). It is necessary to read-through to the late region of L2 mRNA and polyadenylate the late transcripts at the late polyadenylation signal (pAL) in order to synthesize L2 mRNA (Figure 4) (125). L1 mRNA captures hnRNP H and releases the inhibitory effects on L2 at the late infection stage (125, 126). This late gene autoregulation mechanism promotes the production of viral capsid protein (125, 126). In addition, in the splice site SD3632, activation of the DNA damage response increases binding of hnRNP C and inhibits polyadenylation at pAE (Figure 4) (127).
Similarly, in the case of Epstein-Barr virus (EBV), hnRNPs helps to compensate the inefficient processing of the mRNA through polyadenylation process (128). Inefficient cleavage and polyadenylation of EBV DNA polymerase (pol), which contains a noncanonical polyadenylation signal, UAUAAA, is thought to be regulated by the EBV early proteins SM and M (128). hnRNP C1/C2 and hnRNP A1/A2 interact with SM suggesting that the hnRNPs participate in the pre-mRNA 3′ processing of the intronless pol transcript and compensate the inefficient processing of the mRNA (128).
Retroviruses: Several hnRNPs regulate HIV mRNA splicing. Following infection, short spliced mRNAs are produced that encode the viral regulatory proteins Tat, Rev, and Nef, which are required in the early stages of viral infection (129, 130). In response to their expressions, the partially spliced mRNA coding for Vif, Vpr, Vpu, and Env are produced and exported to the nucleus (129, 130). Lastly, the 9 kb transcript encodes both viral structural proteins, Gag and Pol (129, 130). Virus maturation and multiplication depend heavily on the control and coordination of these splicing activities. This post-transcriptional regulation is mediated by proteins Rev and Tat, and by four alternative 5’ splice sites and eight alternative 3’ splice acceptor sites (Figure 5) (130).
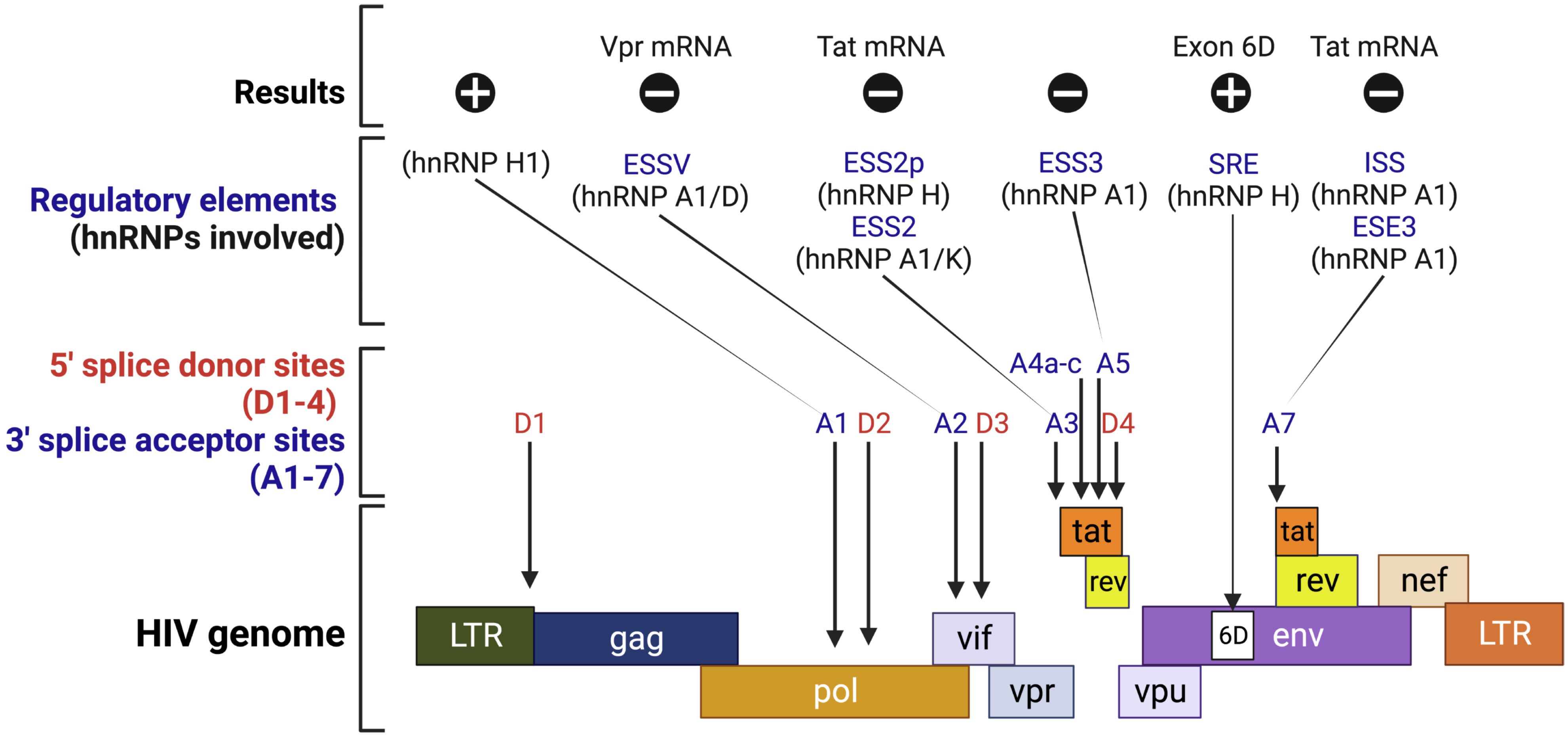
Figure 5 Schematic representation of the HIV genome. This figure shows splice sites and the interfering and interacting positions of hnRNP proteins in the HIV splicing sites. , positive regulation; ϴ, negative regulation; ESS, exonic splicing silencer; ESE, exonic splicing enhancer; ISS, intronic splicing silencer; SRE, splicing regulatory element. The image was created with BioRender.com.
A few hnRNPs modulate Tat protein expression by interfering with enhancer splicing elements (ESEs) or interacting with the exonic and intron splicing silencer (ESS and ISS) elements. hnRNP A/B bind the Tat exon 2 ESE to repress splicing, while SC35 binds to the ESE to activate it (131). Both hnRNP A1 and SC35 have overlapping binding sites within the ESE/ESS9 (131). The hnRNP A1 binds to the ESS and directly covers the SC35 binding site, inhibiting the splicing of the upstream intron (131, 132). Additionally, hnRNP A1 has also been shown to compete at ESE3/(GAA)3 with ASF/SF2 (131). Consequently, the ratio of ASF/SF2 to hnRNP A1 determines whether ESE3/(GAA)3 is activated or repressed at site A7 (133, 134). Inhibition of A3 splicing occurs when hnRNP A1 and hnRNP K bind to ESS2 (131, 135). The UAG triplets in ESS2 are required for hnRNP A1 binding (136). The hnRNP H also involves in splicing inhibition by binding to the ESS2p element, which results in the inhibition of splicing at 3′-splice site A3 (137). In addition, hnRNP A1 inhibits Tat splicing by binding with ISS, which is not a component of ESS2 and prevents entry of U2 snRNP (133, 138). hnRNP A/B proteins also inhibit splicing at 3′ splice site A2, which is responsible for the production of Vpr mRNA (139). hnRNP D also binds to ESSV and inhibits splicing (140). The hnRNP A/B protein binding at ESSV inhibits U2AF65 binding to the repressed 3′ splice site and interferes with the splicing efficiency of the 3′ splice site (139).
Interestingly, hnRNP H acts positively on the splicing of exon 6D of HIV mRNA, by interacting with CGGA and enabling the assembly of the U1 snRNP onto exon 6D (135, 141). Another study revealed that hnRNP H1 plays an important role in splicing enhancement by binding to its target elements (142). Depleted or mutated hnRNP H1 leads to diminished Vif expression and decreased replicative fitness, resulting from the uncontrollable splice acceptor A1 (142). hnRNP F also serves as a modulator of splicing events, indicating that hnRNPs are crucial factors for the regulatory pathways of each viral mRNA (143).
2.3 hnRNPs are needed for viral protein synthesis
Viruses are fully dependent on the translation system of their host cells to produce the proteins that are essential for viral replication (40). Therefore, viruses recruit host ribosomes to translate viral mRNAs, which also concurrently suppresses host translation. Virus families are characterized by mRNAs that have a range of structures at their 5’ (capped, not capped, cap-substituted) and 3’ (polyadenylated or not) ends and carry various cis-acting sequences (such as internal ribosome entry sites, IRES) features that determine many aspects of viral mRNA translation (144).
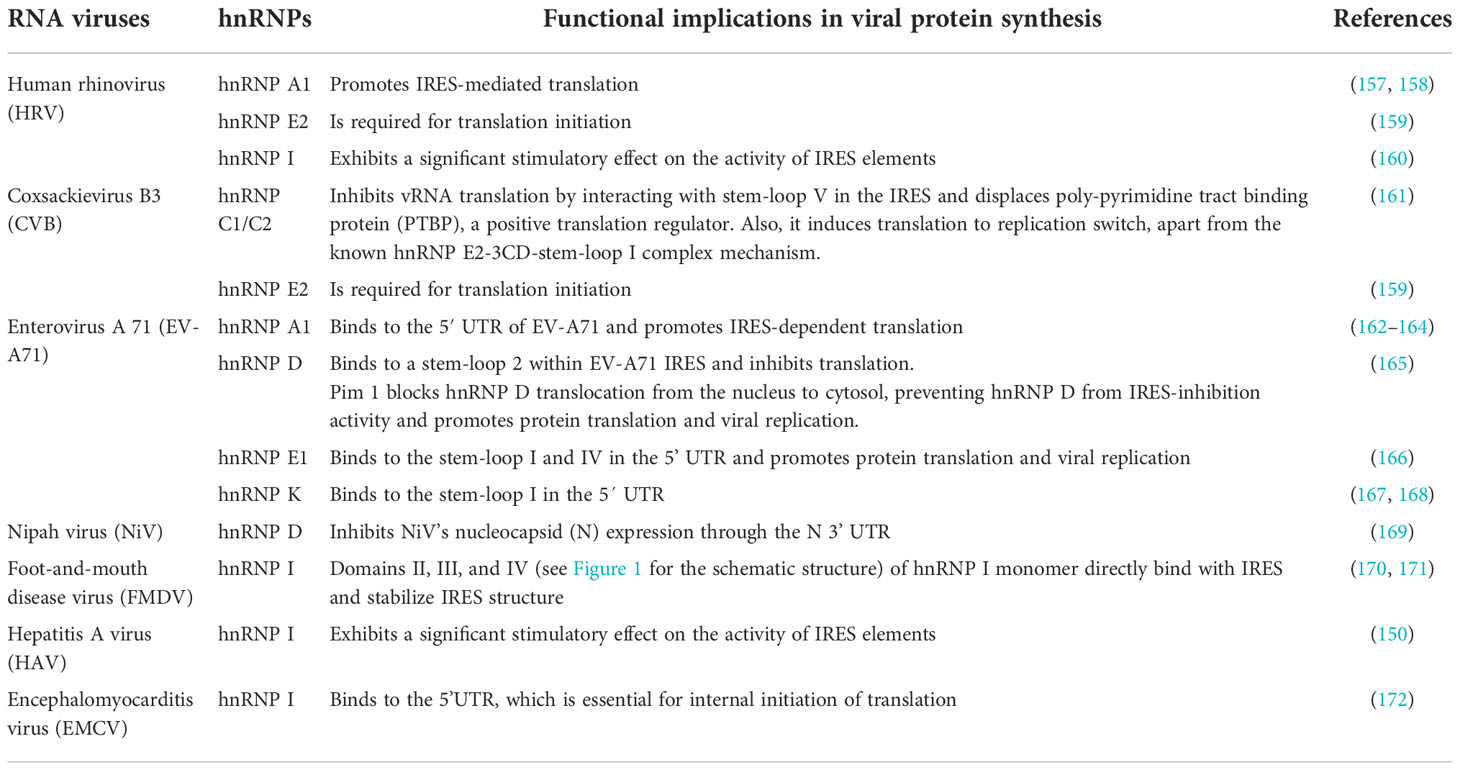
RNA viruses: Most hnRNPs positively regulate virus mRNA translation. The IRES sequences of (+) ssRNA viruses drive translation of viral proteins (145). IRESs were initially discovered in picornaviruses where they have been shown to directly recruit ribosomes to viral mRNA (Figure 3A) (146). Several hnRNPs bind to the IRES of different viruses and act as IRES-trans acting factors (ITAFs) that assist with the translation of viral mRNAs.
During hepatitis C virus (HCV) virus infection, the majority of hnRNPs promote translation of HCV viral mRNA by binding to the 5′ UTR sequence (6). hnRNP D interacts with the stem-loop II, and its overexpression promotes HCV IRES-dependent translation, while hnRNP D knockdown inhibits the HCV IRES activity (147, 148). Additionally, hnRNP D sequestration with an interacting DNA polymer inhibits HCV mRNA translation in an in vitro system, reinforcing its role in the translation process (148). Other hnRNPs, such as hnRNP A1, E, I, L, and Q also stimulate the translation of HCV RNAs. hnRNP A1 binds to the cis-acting elements which are present in both the 5’ NTRs and the 3’ NTRs of HCV RNA (149). Similarly, hnRNP I interacts with NS5B, blocking the binding of NS5B to the 3’NCR, which positively affects the translation of HCV RNAs in vivo (150–152). hnRNP E plays a role in the circularization of HCV RNA through interactions with the 5’- and 3’UTRs for RNA translation and replication (153). hnRNP L bind to the 3’ border region of the HCV IRES and enhances mRNA translation (154). hnRNP Q assists the proper positioning of the 40S ribosomal subunit on HCV mRNA in order to facilitate IRES-dependent translation, which occurs through the interaction(s) between hnRNP Q and r-proteins of the 40S ribosomal subunit (155, 156). Additional roles of hnRNPs in the viral protein synthesis of RNA viruses are summarized in Table 4.
DNA viruses: In HPV-16, hnRNPs control the translation of the late genes (HPV-16 genome is shown in Figure 4) (111). The hnRNPs E1, E2, and K bind to the HPV-16 L2 mRNA in a sequence-specific manner, inhibiting its translation (173). During the L2 translation inhibition, hnRNP K is phosphorylated on multiple tyrosines by c-Src, which inhibits its binding to RNA (174). In another DNA virus, KSHV, hnRNP I interacts with an IRES RNA within a vCyclin (latently expressed gene) that controls the expression of a downstream ORF encoding an inhibitor of apoptosis viral FLICE inhibitory protein (vFLIP) (175).
Retroviruses: In the HIV infected cells, hnRNP A1 is redistributed into the cytoplasm during late HIV infection and increases IRES-mediated translation (176). Knock-down of hnRNP A1 and A2 in HeLa cells transfected with a bicistronic dual-luciferase construct containing Gag under the control of the HIV-1 IRES decreases Gag synthesis (176). Similarly, depletion of hnRNP D expression decreases the synthesis of HIV-1 Gag and Env structural proteins (177). In addition, hnRNP D relocalizes to the cytoplasm upon HIV-1 infection and is associated with Gag protein (177). Moreover, the four isoforms of hnRNP D have differential effects on HIV-1 Gag expression, in which p45 and p42 isoforms increase viral Gag synthesis while p40 and p37 suppress it (177). It is subsequently confirmed by selective deletion of p45 and p42 that relative abundance of hnRNP D isoforms likely contribute to cell types’ ability to replicate HIV-1 (177). In another study, overexpression of hnRNP E1, but not hnRNP E2, lead to a reduction in Rev production, primarily through the suppression of viral RNA translation, and the C- terminal end of hnRNP E1 is required for this effect (178).
A retroviral IRES’s activity is not only influenced by the recruited ITAFs, but also by the post-translational modifications to these proteins (179). hnRNP A1 serves as an ITAF for HTLV-1 and MMTV IRESs, with MMTV IRES showing a higher dependence on hnRNPA1 than those of HTLV-1 or HIV-1 (179). In addition, HIV-1 and HTLV-1 IRES exhibit equally responsive IRES-activity to hnRNP A1 and its phosphorylation mutants S4A/S6A, S4D/S6D, and S199A/D (179). It appears, however, that the S4D/S6D mutant stimulates MMTV-IRES activity significantly higher than the wild type hnRNP A1 (179). The PRMT5-induced symmetrical di-methylation of arginine residues of hnRNP A1 enhances the stimulation activity of the hnRNP A1 on HIV-1 and HTLV-1 IRES, while decreasing the stimulation activity on MMTV IRES. (179).
2.4 hnRNPs switch vRNA from translation to replication
When cells are infected with (+) ssRNA viruses, the viral genomic (+) ssRNA serves not only as template for translation but also for genome replication (180). Although the two processes, translation and replication, rely on the same strand of (+) ssRNA, their path to completion is opposite (180). The viral replication proteins may repress translation and switch to viral RNA synthesis for a variety of reasons, one being that viral genome (+) ssRNAs must avoid degradation by mRNA decay pathways after translation in order to continue replication (180). The host RBPs, including hnRNPs, contribute to the regulation of this process by interacting with cis-elements in the viral (+) ssRNA (54, 181).
In PV genome replication and translation, hnRNP E2 has two functions, which makes it an ideal mediator of the switch from translation to replication (Figures 3A, B) (54, 181–183). During mid-to-late infection, the viral protease 3C/3CD cleaves hnRNP E2 to facilitate PV’s genome replication (57; 184). A full-length hnRNP E2, which contains three RNA binding domains (KH-domains) (Figure 1), binds selectively to the stem-loop IV of IRES (as shown in Figure 3A) (185, 186). However, proteolytic cleavage of the linker sequence between KH2 and KH3 results in the truncated hnRNP E2 protein being devoid of a KH3 domain, which blocks its binding to the IRES and inhibits PV translation (57; 184). Interestingly, in hnRNP E2-depleted extract, truncated hnRNP E2 still rescues PV (+) ssRNA replication (57). With the similar mechanism, hnRNP K plays dual role in foot-and-mouth disease virus (FMDV) translation and RNA replication (187). hnRNP K’s KH2 and KH3 domains bind directly to FMDV IRES domains II, III, and IV, inhibiting IRES-mediated translation by interfering with the recognition of another positive ITAF, hnRNP I (187). The viral protease, 3C, however, antagonizes the inhibition of hnRNP K through cleavage at the Glu-364 residue of hnRNP K during FMDV infection (187). This result indicates that the N-terminal cleavage product is capable of inhibiting IRES activity while the C-terminal cleavage product promotes FMDV replication (187).
2.5 hnRNPs participate in vRNA trafficking and genome packaging
hnRNP A2/B1 binds NS1 and NS2 mRNAs of IAV and inhibits the nuclear export of the NS1 mRNA (188). Conversely, knockdown of hnRNP A2/B1 promotes the transport of NS1 mRNA to the cytoplasm in infected cells and results in increased levels of NS1 (vRNA, mRNA, and protein) consequently leading to enhanced virus replication (188). Therefore, the hnRNP A2/B1 inhibits the replication of IAV in host cells possibly by suppressing nucleocytoplasmic translocation of NS1 mRNA (188). In addition, nucleoprotein (NP) cooperates with hnRNP A/B to further restrict viral mRNA nuclear export by acting as an RNA retention factor for IAV mRNA (189). An additional consequence of IAV NP’s interaction with hnRNP A/B is that it interrupts the interaction between ALY and UAP56 (RNA helicase), leading to repression of the ALY-viral mRNA binding, followed by inhibition of viral mRNA binding for NXF1 (mRNA transport receptor) (189).
For retroviruses, viral RNA trafficking from nucleus to cytoplasm is the major event in the viral lifecycle. In HIV, hnRNPs facilitate translocation of viral RNAs. Several spliced and unspliced RNA molecules of HIV exist in the nucleus after transcription and splicing (6). hnRNP D may assist in HIV RNA translocation, because loss of hnRNP D reduces the accumulation of HIV-1 unspliced and singly spliced RNAs in the cytoplasm but does not affect viral RNA abundance (177). HIV-1 Vpr and Gag mRNAs contain cis-acting RNA trafficking sequences (hnRNP A2RE-1 and A2RE-2) that are responsible for cytoplasmic RNA trafficking (190, 191). hnRNP A2 and both A2REs bind to form a complex in vitro, which is required for the nuclear export of RNA in oligodendrocyte (190, 191). RNA transport mediated by the A2RE requires both microtubules and hnRNP A2 (190, 191). As a consequence, the replication of A2RE-2 mutant viruses is significantly compromised and genome RNA encapsidation is reduced (191). hnRNP A2 is involved in carrying HIV-1 RNA to the microtubule-organizing center (MTOC) and then transporting it to the plasma membrane for virion assembly (177, 192, 193). Although inhibiting the localization of genomic RNA at the MTOC as a result of hnRNP A2 knockdown does not impact Gag synthesis, it adversely affects viral production and infection (192).
Similarly, hnRNP A2B1 and hnRNP C can retain the genomic RNA in the nucleus. hnRNP A2B1 participates in the nuclear retention of the HIV-1 genomic RNA in the absence of Rev as well as in the release of the genomic RNA from cytoplasmic ribonucleoprotein (RNP) complexes (194), but the exact mechanism is unknown. hnRNP C binds to the nuclear retention sequence (NRS) within the intron of unspliced RNA and mediates nuclear retention of the RNA (195). hnRNP U also controls HIV-1 mRNA nuclear export by targeting the 3′ long terminal repeat (3′LTR) and blocking HIV-1 mRNA accumulation in the cytoplasm (196). Interestingly, HIV-1 also increases hnRNP A1 expression and promotes its relocation to the cytoplasm. The cytoplasmic translocation of hnRNP A1 is escorted by the viral genomic RNA and is synergistically induced by Rev (197). During the late phases of HIV-1 replication, hnRNP A1 and viral genomic RNA colocalize in the cytoplasm, supporting a post nuclear function of hnRNP A1 (176). Additional examples of hnRNPs involvement on viral RNA trafficking and packaging are summarized in Table 5.
2.6 hnRNPs involve in viral egress
Viral egress involves capsid assembly and release, where hnRNPs play a role in either one or both steps. In dengue virus (DENV), vimentin interacts with hnRNP C1/C2, hnRNP K, and NS1 protein to form a complex, and the dissociation of the complex by acrylamide treatment reduces viral release (203). The vimentin-hnRNP K-NS1 complex was also studied in Japanese encephalitis virus (JEV) infection, in which knocking down vimentin and hnRNP K reduces viral replication, whereas over-expression of vimentin and hnRNP K enhances viral replication (204). Moreover, vimentin was also shown to interact with hnRNP K following JEV infection or after overexpression of NS1 (204).
In contrast, hnRNP E1 normally limits virion assembly and secretion, which prevents the escape of viral genomes packaged into virions that contribute to viral RNA accumulation in infected cells (205). Similarly, hnRNP K suppresses HCV particle production. The hnRNP K protein is recruited to the virion assembly sites in proximity of lipid droplets and the hnRNP K colocalizes with core protein and HCV plus-strand RNA (206).
Viruses also utilize hnRNPs for controlling the apoptosis process for the release of mature virions as an indirect mechanism. EV71 releases its virus through hnRNP A1 via an apoptotic axis involving 3C protease, hnRNP A1, Apaf-1, and caspase 3 (207). When EV71 infection is present, or 3C protease is expressed, hnRNP A1 is cleaved, which affects its binding to apaf-1 IRES (207). This allows IRES-dependent synthesis of Apaf-1, and subsequent activation of caspase-3, and apoptosis (207). Conversely, hnRNP K suppresses bovine ephemeral fever virus (BEFV) replication by degrading viral α3 gene and thus inhibits apoptosis induced by α3 gene (208). Moreover, BEFV infection degrades hnRNP K protein in BHK-21 cells, which is mediated by viral activation of caspase 3 (208). However, the hnRNP K has a supportive role in the HSV-1 virion egress (209). Infectious particles are released by the exocytic pathway after newly assembled HSV-1 particles acquire a secondary envelope at the trans-Golgi network (209). The downregulation of hnRNP K leads to a blockage of virus particles leaving the cell, and the virion release is crucial for viral propagation (209). Similarly, the hnRNP A2B1 supports the egress of exosomes and HSV-1 from infected cells (210).
2.7 Nucleocytoplasmic shuttling of hnRNPs to control viral replication
Viral nucleic acids and/or proteins encounter hnRNPs in these cellular compartments and alter their subcellular localization to promote viral replication. The Ebola virus (EBOV) prevents hnRNP C1/2 from entering the nucleus (211). VP24 from the EBOV binds to NPI-1 subfamily karyopherin α (KPNA) nuclear import proteins, which prevents KPNA from interacting with phospho-STAT1 (211). This results in the inhibition of nuclear import of the hnRNP C1/C2-KPNA-VP24 complex (211). Similarly, hnRNP A1, hnRNP K, and hnRNP C1/C2 are relocalized to the cytoplasm during vesicular stomatitis virus (VSV) infection (212). Recently, it was shown that open-reading frame 6 (ORF6) protein of SARS-CoV-2 can directly manipulate localization and functions of nucleoporins by disrupting nuclear rim of nucleoporins, RAE1 and NUP98 (213). This disruption results into aberrant nucleocytoplasmic trafficking and leads to nuclear accumulation of hnRNP A1 (213). Similarly, in case of junin virus (JUNV), hnRNP A1 interacts with the N protein of JUNV in acutely infected cells, and upregulating N protein induces cytoplasmic re-localization of hnRNP A1 (214). JUNV as well as DENV-2 induce hnRNP K cytoplasmic translocation to favor viral particle production (214, 215).
Many hnRNPs relocalize to the cytoplasm upon rotavirus infection. Rotavirus replicates in the cytoplasm of infected cells in viroplasms (VMs, unique inclusion bodies induced by the virus), which are nucleated by two essential virus nonstructural proteins, NSP2 and NSP5 (216). NSP2 and NSP5 interact directly with the hnRNPs (which are translocated to cytoplasm from the nucleus) in an RNA independent manner and sequester themselves in the viroplasms of infected cells, thus creating favorable conditions for virus replication (216). In another example, Seneca Valley virus (SVV) 3C protease (SVV 3C) cleaves hnRNP A1 and this cleaved hnRNP A1 translocates to the cytoplasm, which is utilized by SVV to aid viral replication (217).
2.8 hnRNPs interact with viral proteins to control viral replication
hnRNPs interact with the viral proteins to control the viral gene expression or translocation of the viral proteins. hnRNP A1 interacts with multiple proteins of different viruses. hnRNP A1 interacts with p17 protein of avian reovirus (ARV) to facilities its nuclear import and export (218). hnRNP A1 also interacts with nucleoprotein (NP) of IAV and co-localize in both nucleus and mitochondria of mammalian cells that are either transfected with NP or infected with IAV (219). Similarly, hnRNP A1 interacts with nucleocapsid (N) protein of porcine epidemic diarrhea virus (PEDV) and enhances the PEDV infection (220). During infection with DENV, hnRNP C1/C2 and hnRNP I accumulate in the cytoplasm and colocalizes with NS1 and NS2 viral proteins (221, 222). DENV core protein is also shown to interact with hnRNP K by coimmunoprecipitation analysis (221). Moreover, silencing of hnRNP I inhibits virus replication, while overexpression of hnRNP I enhances DENV replication (223). hnRNP H also interacts with DENV non-structural 1 protein to promote the viral replication (224). During IAV infection, the central BACK domain of cellular protein NS1-BP interacts directly with hnRNP K and hnRNP I and with the influenza virulence factor, NS1 protein (225). In addition, proteomic analysis revealed that hnRNP M and hnRNP H1 are part of the complexes formed by the IAV polymerase (226). hnRNP H interacts with Newcastle disease virus (NDV) V protein to promote viral replication (227). Rev protein, which is a key regulator of HIV-1 gene expression, also interacts with hnRNP A1, hnRNP Q, hnRNP K, hnRNP R, and hnRNP U (228). This interaction of Rev with functionally diverse hnRNPs is proposed to be involved in HIV replication and virus-host cell interactions (228).
2.9 hnRNPs involvement in viral immune modulation
The presence of infection triggers innate immune responses against viruses in mammalian cells. The cells detect viral dsRNA by means of cytoplasmic receptors, such as the retinoic acid-inducible gene I (RIG-I) and the melanoma differentiation-associated gene 5 (MDA5), both of which interact with the adaptor mitochondrial antiviral signaling (MAVS) (229). The MAVS integrates signaling from RIG-I-like receptors (RLRs) and mediates the downstream activation of TANK-binding kinase 1 (TBK1)/IκB kinase ε, eventually leading to the induction of type I IFNs, pro-inflammatory cytokines, and multiple IFN-stimulated genes (229–232). This results in pacing cells in an antiviral state (229). hnRNP E1 and E2 are synergetic feedback inhibitors of MAVS, which facilitate the degradation of the MAVS protein by recruiting Atrophin 1-interacting protein 4 (AIP4, E3 ubiquitin ligase) to MAVS-containing complex after viral infection (233). This mechanism is considered as the fine tuning of antiviral innate immunity for the prevention of excessive harmful immune responses (232, 233). In addition, hnRNP M was shown to act as a negative regulator of RLRs-mediated signaling, where hnRNP M interacts with RIG-I and MDA5 and impairs the binding of the RLRs to viral RNA, leading to the inhibition of RNA virus-triggered innate immune responses (234).
hnRNP U (a nuclear matrix protein functions as a nuclear viral dsRNA sensor for both DNA and RNA viruses. As a response to viral dsRNA, hnRNP U oligomerizes and activates the distal enhancers of antiviral genes, including Interferon beta 1 (IFNB1) (235). Also, hnRNP U is required for the activation of super-enhancers, which initiate robust immune gene expression for antiviral defense (235, 236). Another study showed NLS domain of hnRNP U interacts with bunyavirus NP and recognizes the bunyavirus genomic RNA in the cytoplasm, which activates STING-TBK1 signaling axis against the virus infection (237). In addition, hnRNP U was shown to interact with a IVRPIE (Inhibiting IAV Replication by Promoting IFN and ISGs Expression, an lncRNA) (238). The expression of IVRPIE significantly inhibits IAV replication in A549 cells (238). Additionally, IVRPIE positively regulates interferon β1 transcription and several vital interferon-stimulated genes (ISGs) (238).
hnRNP A2B1 recognizes and binds to viral DNA in the nucleus during herpes simplex virus-1 (HSV-1) infection (239). The binding is followed by homodimerization and demethylation of hnRNP A2B1 which leads to the release of HSV-1 DNA (239). Subsequently, hnRNP A2B1 translocates to the cytosol and activates TBK1-IRF3 signaling pathway to initiate cytokine production, thus enabling innate immunity through type I interferon signaling for virus elimination, ultimately leading to the inhibition of HSV-1 replication (239). During an innate immune response to HSV-1 infection, hnRNP A2B1 dissociates from fat mass and obesity-associated protein (FTO), leading to the promotion of N6-methyladenosine (m6A) modification, nucleocytoplasmic trafficking, and translation of cGAS, STING, and IFI16 mRNAs to enhance IFN-I production (239).
3 Implications and conclusions
In this review we have highlighted numerous functions and mechanisms by which hnRNPs impact the viral life cycle (summarized in Figure 6). hnRNPs are important emerging targets for antiviral drug research and development. The discovery of novel therapeutic options against important human and animal viral pathogens can be advanced by understanding how cellular RBPs, including hnRNPs, regulate virus infection (49). For example, in addition to acting as a host-protective mechanism, hnRNP A1 may also hold potential as an antiviral target to develop therapy for human rhinovirus (HRV) and IAV (157, 158, 219). Similarly, hnRNP E2 has the inhibitory effects on VSV replication (240). Expression of many cancer-related genes, including therapy-resistant genes, is altered by aberrant SR proteins and hnRNPs in cervical cancer. As a result, SR proteins and hnRNPs make good drug targets for cervical cancer, and research is underway to develop hnRNPs and SR targeting strategies to reduce expression of oncogenic factors while simultaneously inhibiting HPV replication (241, 242). hnRNP P modulates HIV and global gene transcription, and viral latency (243). Therefore, the hnRNP P can be a potential target for future therapies that are intended to reactivate HIV from its latent state (243). For virus-associated human pathologies, hnRNP A1 binds both structured and unstructured RNAs with affinities that vary by several orders of magnitude (244). Therefore, disrupting the hnRNP A1’s RNA-binding functions is the most promising approach for developing new therapeutics (244).
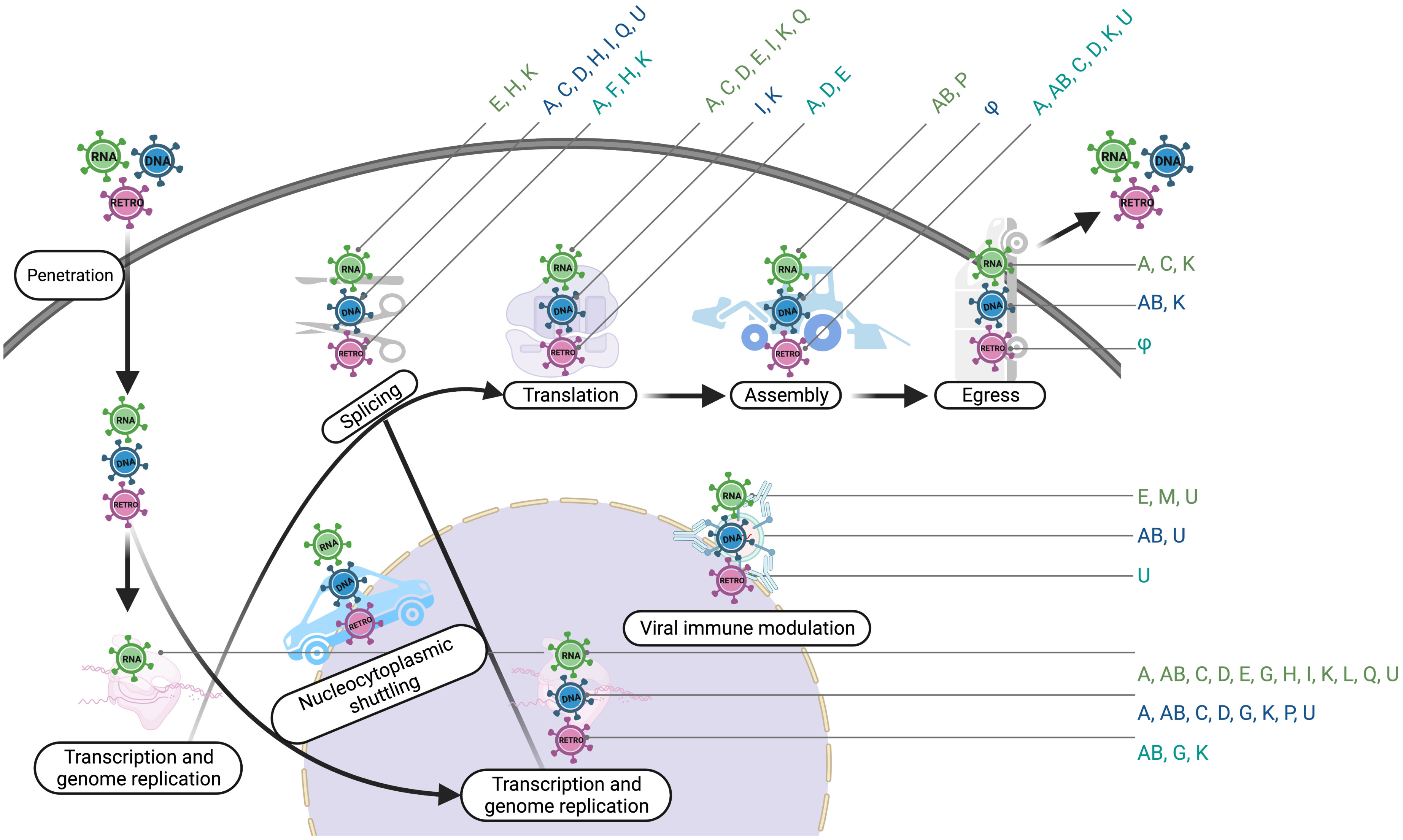
Figure 6 hnRNPs participate in different steps of viruses. This diagram illustrates the involvement of hnRNPs in the life cycle of different types of viruses. Transcription and genome replication include vRNA synthesis/transcription and viral DNA replication; Splicing includes vRNA splicing, vRNA polyadenylation, and vRNA stability; Assembly includes vRNA trafficking and genome packaging; φ, Not Known. The image was created with BioRender.com.
Some drugs that act on hnRNPs are already available on the market. For example, Apigenin inhibits hnRNP A2 dimerization and affects the alternative splicing of many mRNAs (245). In addition, it inhibits the interaction of the hnRNP A1 and A2 with the enterovirus A71 (EV-A71) RNA, inhibiting its translation (246, 247). VPC-80051 targets the RRM of hnRNP A1 and alters hnRNP A1 spicing activity (248). In addition, quercetin impairs the ability of hnRNP A1 to shuttle between the nucleus and the cytoplasm by binding to the C-terminal region (249). DZNep inhibits the recruitment of hnRNP A1 to N6-methyl adenosine (m6A)-modified SARS-CoV-2 RNA, inhibiting the synthesis of the viral genome (250). Targeting the methylome with DZNep could be of great benefit to treating SARS-CoV-2 infection (250). Another compound, DMA-135 changes the structure of the EV-A71 RNA and stabilizes a ternary complex with the hnRNP D protein, repressing translation (251). The DMA-135 and its mechanism of action could be broadly applied to other viral RNA structural elements (251). Similarly, an approved anticancer drug, Idarubicin (IDR) impairs binding between the EV-A71 IRES RNA and hnRNP A1, and IDR and similar drugs could spearhead the development of novel antiviral therapies directed at the EV-A71 IRES RNA (252). miR-555 reduces levels of hnRNP C1/C2 required for PV replication in the infected cells, suggesting miR-555 can be incorporated into developing antiviral therapies against PV (253). For viral diseases that lack therapies and for newly emerging viruses, hnRNPs are especially attractive targets due to their role in viral life cycles (6). In addition, some of the studies of hnRNPs suggest that these proteins could also provide prognostic value for various infections. For example, hnRNP A1 overexpression has diagnostic value in distinguishing between HBV-related hepatocellular carcinoma (HCC) and non-HCC liver tissue (254, 255).
Author contributions
This manuscript was co-written by KB and MH. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) [Discovery Grants, RGPIN-2020-04731 to MH]; Alexander Graham Bell Canada Graduate Scholarships Doctoral Award (to KB). Funding for open access charge: NSERC and Carleton University.
Acknowledgments
Thanks to our colleagues for their constructive and spirited discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell (2009) 136:731–45. doi: 10.1016/j.cell.2009.01.042
2. Dreyfuss G, Matunis MJ, Pinolroma S, Burd CG. hnRNP proteins and the biogenesis of messenger-RNA. Annu Rev Biochem (1993) 62:289–321. doi: 10.1146/annurev.bi.62.070193.001445
3. Sommerville J, Scheer U. Structural organization of nascent transcripts and hnRNA molecules in amphibian oocytes. Mol Biol Rep (1981) 7:53–6. doi: 10.1007/BF00778733
4. Miller OL, Bakken AH. Morphological studies of transcription. Acta Endocrinol (Copenh) (1972) 168:155–77. doi: 10.1530/acta.0.071s155
5. Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol (2002) 3:195–205. doi: 10.1038/nrm760
6. Wan Q, Song D, Li H, He M-L. Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development. Signal Transduct Target Ther (2020) 5:125. doi: 10.1038/s41392-020-00233-4
7. Das U, Nguyen H, Xie J. Transcriptome protection by the expanded family of hnRNPs. RNA Biol (2019) 16:155–9. doi: 10.1080/15476286.2018.1564617
8. Love R, Wildy P. Cytochemical studies of the nucleoproteins of HeLa cells infected with herpes virus. J Cell Biol (1963) 17:237–54. doi: 10.1083/jcb.17.2.237
9. Gattoni R, Stevenin J, Jacob M. Comparison of the nuclear ribonucleoproteins containing the transcripts of adenovirus-2 and HeLa cell DNA. Eur J Biochem (1980) 108:203–11. doi: 10.1111/j.1432-1033.1980.tb04713.x
10. Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol (1986) 2:459–98. doi: 10.1146/annurev.cb.02.110186.002331
11. Piñol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev (1988) 2:215–27. doi: 10.1101/gad.2.2.215
12. Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA (2010) 16:1449–62. doi: 10.1261/rna.2254110
13. Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol (1999) 11:363–71. doi: 10.1016/S0955-0674(99)80051-9
14. Frank L, Rippe K. Repetitive RNAs as regulators of chromatin-associated subcompartment formation by phase separation. J Mol Biol (2020) 432:4270–86. doi: 10.1016/j.jmb.2020.04.015
15. Thibault PA, Ganesan A, Kalyaanamoorthy S, Clarke J.-P. W. E., Salapa HE, Levin MC. hnRNP A/B proteins: An encyclopedic assessment of their roles in homeostasis and disease. Biol (Basel) (2021) 10:712. doi: 10.3390/biology10080712
16. Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol (2005) 12:645–53. doi: 10.1038/nsmb961
17. Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J (2010) 430:379–92. doi: 10.1042/bj20100396
18. Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Sci (1994) 265:615–21. doi: 10.1126/science.8036511
19. Maris C, Dominguez C, Allain FH-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J (2005) 272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x
20. Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J (2008) 275:2712–26. doi: 10.1111/j.1742-4658.2008.06411.x
21. Nazarov IB, Bakhmet EI, Tomilin AN. KH-domain Poly(C)-binding proteins as versatile regulators of multiple biological processes. Biochemistry (2019) 84:205–19. doi: 10.1134/S0006297919030039
22. March ZM, King OD, Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res (2016) 1647:9–18. doi: 10.1016/j.brainres.2016.02.037
23. Batlle C, Yang P, Coughlin M, Messing J, Pesarrodona M, Szulc E, et al. hnRNPDL phase separation is regulated by alternative splicing and disease-causing mutations accelerate its aggregation. Cell Rep (2020) 30:1117–1128.e5. doi: 10.1016/j.celrep.2019.12.080
24. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell (2015) 163:123–33. doi: 10.1016/j.cell.2015.09.015
25. Hyman AA, Weber CA, Juelicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol (2014) 30:39–58. doi: 10.1146/annurev-cellbio-100913-013325
26. Basu S, Bahadur RP. A structural perspective of RNA recognition by intrinsically disordered proteins. Cell Mol Life Sci (2016) 73:4075–84. doi: 10.1007/s00018-016-2283-1
27. Castello A, Fischer B, Frese CK, Horos R, Alleaume A-M, Foehr S, et al. Comprehensive identification of RNA-binding domains in human cells. Mol Cell (2016) 63:696–710. doi: 10.1016/j.molcel.2016.06.029
28. Jaervelin AI, Noerenberg M, Davis I, Castello A. The new (dis)order in RNA regulation. Cell Communication Signaling (2016) 14:9. doi: 10.1186/s12964-016-0132-3
29. Clarke JP, Thibault PA, Salapa HE, Levin MC. A comprehensive analysis of the role of hnRNP A1 function and dysfunction in the pathogenesis of neurodegenerative disease. Front Mol Biosci (2021) 8:659610. doi: 10.3389/fmolb.2021.659610
30. Velázquez-Cruz A, Baños-Jaime B, Díaz-Quintana A, la Rosa MA, Díaz-Moreno I. Post-translational control of RNA-binding proteins and disease-related dysregulation. Front Mol Biosci (2021) 8:658852. doi: 10.3389/fmolb.2021.658852
31. Wang J, Sun D, Wang M, Cheng A, Zhu Y, Mao S, et al. Multiple functions of heterogeneous nuclear ribonucleoproteins in the positive single-stranded RNA virus life cycle. Front Immunol (2022) 13:989298. doi: 10.3389/fimmu.2022.989298
32. Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: A novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J (1997) 16:3587–98. doi: 10.1093/emboj/16.12.3587
33. Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell (1995) 83:415–22. doi: 10.1016/0092-8674(95)90119-1
34. Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol (1995) 129:551–60. doi: 10.1083/jcb.129.3.551
35. Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell (1996) 86:985–94. doi: 10.1016/S0092-8674(00)80173-7
36. Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci (1997) 110:1325–31. doi: 10.1242/jcs.110.11.1325
37. Mboukou A, Rajendra V, Kleinova R, Tisné C, Jantsch MF, Barraud P. Transportin-1: A nuclear import receptor with moonlighting functions. Front Mol Biosci (2021) 8:638149. doi: 10.3389/fmolb.2021.638149
38. Schmid M, Speiseder T, Dobner T, Gonzalez RA. DNA Virus replication compartments. J Virol (2014) 88:1404–20. doi: 10.1128/JVI.02046-13
39. Rampersad S, Tennant P. Replication and expression strategies of viruses. Viruses (2018), 55–82. doi: 10.1016/B978-0-12-811257-1.00003-6
40. Walsh D, Mohr I. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol (2011) 9:860–75. doi: 10.1038/nrmicro2655
41. Flint SJ, Racaniello VR, Rall GF, Hatziioannou T, Skalka AM. Principles of virology, volume 1: molecular biology. Washington, DC, USA: John Wiley & Sons with publisher ASM Press (2020).
42. Payne S. Introduction to RNA viruses. Viruses (2017), 97–105. doi: 10.1016/B978-0-12-803109-4.00010-6
43. Hu W-S, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med (2012) 2:a006882. doi: 10.1101/cshperspect.a006882
44. Suzuki Y, Craigie R. The road to chromatin - nuclear entry of retroviruses. Nat Rev Microbiol (2007) 5:187–96. doi: 10.1038/nrmicro1579
45. Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology (2009) 6:8. doi: 10.1186/1742-4690-6-8
46. Dimitrov DS. Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol (2004) 2:109–22. doi: 10.1038/nrmicro817
47. Kuss SK, Mata MA, Zhang L, Fontoura BMA. Nuclear imprisonment: viral strategies to arrest host mRNA nuclear export. Viruses (2013) 5:1824–49. doi: 10.3390/v5071824
48. Romanelli MG, Morandi C. Importin alpha binds to an unusual bipartite nuclear localization signal in the heterogeneous ribonucleoprotein type I. Eur J Biochem (2002) 269:2727–34. doi: 10.1046/j.1432-1033.2002.02942.x
49. Dicker K, Järvelin AI, Garcia-Moreno M, Castello A. The importance of virion-incorporated cellular RNA-binding proteins in viral particle assembly and infectivity. Semin Cell Dev Biol (2021) 111:108–18. doi: 10.1016/j.semcdb.2020.08.002
50. Garcia-Moreno M, Jaervelin AI, Castello A. Unconventional RNA-binding proteins step into the virus-host battlefront. WIREs-RNA (2018) 9:e1498. doi: 10.1002/wrna.1498
51. Iselin L, Palmalux N, Kamel W, Simmonds P, Mohammed S, Castello A. Uncovering viral RNA-host cell interactions on a proteome-wide scale. Trends Biochem Sci (2022) 47:23–38. doi: 10.1016/j.tibs.2021.08.002
52. Brunner JE, Nguyen JHC, Roehl HH, Ho T, Swiderek KM, Semler BL. Functional interaction of heterogeneous nuclear ribonucleoprotein c with poliovirus RNA synthesis initiation complexes. J Virol (2005) 79:3254–66. doi: 10.1128/JVI.79.6.3254-3266.2005
53. Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell (2001) 7:581–91. doi: 10.1016/S1097-2765(01)00205-2
54. Li Z, Nagy PD. Diverse roles of host RNA-binding proteins in RNA virus replication. RNA Biol (2011) 8:305–15. doi: 10.4161/rna.8.2.15391
55. Ertel KJ, Brunner JE, Semler BL. Mechanistic consequences of hnRNP c binding to both RNA termini of poliovirus negative-strand RNA intermediates. J Virol (2010) 84:4229–42. doi: 10.1128/JVI.02198-09
56. Walter BL, Parsley TB, Ehrenfeld E, Semler BL. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol (2002) 76:12008–22. doi: 10.1128/JVI.76.23.12008-12022.2002
57. Perera R, Daijogo S, Walter BL, Nguyen JHC, Semler BL. Cellular protein modification by poliovirus: The two faces of Poly(rC)-binding protein. J Virol (2007) 81:8919–32. doi: 10.1128/JVI.01013-07
58. Zhang XM, Lai MM. Regulation of coronavirus RNA transcription is likely mediated by protein-RNA interactions. Adv Exp Med Biol (1995) 380:515–21. doi: 10.1007/978-1-4615-1899-0_82
59. Li HP, Zhang XM, Duncan R, Comai L, Lai MMC. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci (1997) 94:9544–9. doi: 10.1073/pnas.94.18.9544
60. Shi ST, Huang PY, Li HP, Lai MMC. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J (2000) 19:4701–11. doi: 10.1093/emboj/19.17.4701
61. Zhang XM, Li HP, Xue WM, Lai MMC. Formation of a ribonucleoprotein complex of mouse hepatitis virus involving heterogeneous nuclear ribonucleoprotein A1 and transcription-regulatory elements of. Virology (1999) 264:115–24. doi: 10.1006/viro.1999.9970
62. Wang YC, Zhang XM. The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo. Virology (1999) 265:96–109. doi: 10.1006/viro.1999.0025
63. Cong Y, Ulasli M, Schepers H, Mauthe M, V’kovski P, Kriegenburg F, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol (2020) 94:e01925–19. doi: 10.1128/JVI.01925-19
64. Huang PY, Lai MMC. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3 ` untranslated region, thereby altering RNA conformation. J Virol (1999) 73:9110–6. doi: 10.1128/JVI.73.11.9110-9116.1999
65. Choi KC, Mizutani A, Lai MMC. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J Virol (2004) 78:13153–62. doi: 10.1128/JVI.78.23.13153-13162.2004
66. Jiang XS, Tang LY, Dai J, Zhou H, Li SJ, Xia QC, et al. Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches - implications for cellular responses to virus infection. Mol Cell Proteomics (2005) 4:902–13. doi: 10.1074/mcp.M400112-MCP200
67. Luo HB, Chen Q, Chen J, Chen KX, Shen X, Jiang HL. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett (2005) 579:2623–8. doi: 10.1016/j.febslet.2005.03.080
68. Hussain S, Perlman S, Gallagher TM. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J Virol (2008) 82:7212–22. doi: 10.1128/JVI.02406-07
69. Labeau A, Fery-Simonian L, Lefevre-Utile A, Pourcelot M, Bonnet-Madin L, Soumelis V, et al. Characterization and functional interrogation of the SARS-CoV-2 RNA interactome. Cell Rep (2022) 39:110744. doi: 10.1016/j.celrep.2022.110744
70. Friedrich S, Schmidt T, Geissler R, Lilie H, Chabierski S, Ulbert S, et al. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol (2014) 88:11586–99. doi: 10.1128/JVI.01283-14
71. Friedrich S, Schmidt T, Schierhorn A, Lilie H, Szczepankiewicz G, Bergs S, et al. Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45. RNA (2016) 22:1574–91. doi: 10.1261/rna.055269.115
72. Friedrich S, Engelmann S, Schmidt T, Szczepankiewicz G, Bergs S, Liebert UG, et al. The host factor AUF1 p45 supports flavivirus propagation by triggering the RNA switch required for viral genome cyclization. J Virol (2018) 92:e01647–17. doi: 10.1128/JVI.01647-17
73. Sharma N, Ogram SA, Morasco BJ, Spear A, Chapman NM, Flanegan JB. Functional role of the 5’ terminal cloverleaf in coxsackievirus RNA replication. Virology (2009) 393:238–49. doi: 10.1016/j.virol.2009.07.039
74. Beura LK, Dinh PX, Osorio FA, Pattnaik AK. Cellular Poly(C) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1 beta and support viral replication. J Virol (2011) 85:12939–49. doi: 10.1128/JVI.05177-11
75. Hirai Y, Honda T, Makino A, Watanabe Y, Tomonaga K. X-Linked RNA-binding motif protein (RBMX) is required for the maintenance of borna disease virus nuclear viral factories. J Gen Virol (2015) 96:3198–203. doi: 10.1099/jgv.0.000273
76. Chung RT, Kaplan LM. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3 ` terminus of hepatitis c viral RNA. Biochem Biophys Res Commun (1999) 254:351–62. doi: 10.1006/bbrc.1998.9949
77. Aizaki H, Choi KS, Liu M, Li Y, Lai MMC. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J BioMed Sci (2006) 13:469–80. doi: 10.1007/s11373-006-9088-4
78. Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology (2007) 367:212–21. doi: 10.1016/j.virol.2007.05.008
79. Sun C, Liu M, Chang J, Yang D, Zhao B, Wang H, et al. Heterogeneous nuclear ribonucleoprotein l negatively regulates foot-and-mouth disease virus replication through inhibiting viral RNA synthesis by interacting with IRES in 5′ untranslated region. J Virol (2020) 94:e00282–20. doi: 10.1128/JVI.00282-20
80. Galan C, Sola I, Nogales A, Thomas B, Akoulitchev A, Enjuanes L, et al. Host cell proteins interacting with the 3’ end of TGEV coronavirus genome influence virus replication. Virology (2009) 391:304–14. doi: 10.1016/j.virol.2009.06.006
81. Gupta AK, Drazba JA, Banerjee AK. Specific interaction of heterogeneous nuclear ribonucleoprotein particle U with the leader RNA sequence of vesicular stomatitis virus. J Virol (1998) 72:8532–40. doi: 10.1128/JVI.72.11.8532-8540.1998
82. Pingale KD, Kanade GD, Karpe YA. Heterogeneous nuclear ribonucleoproteins participate in hepatitis e virus replication. J Mol Biol (2020) 432:2369–87. doi: 10.1016/j.jmb.2020.02.025
83. Batra J, Mori H, Small GI, Anantpadma M, Shtanko O, Mishra N, et al. Non-canonical proline-tyrosine interactions with multiple host proteins regulate Ebola virus infection. EMBO J (2021) 40:e105658. doi: 10.15252/embj.2020105658
84. Zhang W, Zhang X, Tian C, Wang T, Nguyen Sarkis PT, Fang Y, et al. Cytidine deaminase APOBEC3B interacts with heterogeneous nuclear ribonucleoprotein K and suppresses hepatitis b virus expression. Cell Microbiol (2008) 10:112–21. doi: 10.1111/j.1462-5822.2007.01020.x
85. Ng LFP, Chan M, Chan SH, Cheng PCP, Leung EHC, Chen WN, et al. Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis b virus replication. PloS Med (2005) 2:673–83. doi: 10.1371/journal.pmed.0020163
86. Dunker W, Song Y, Zhao Y, Karijolich J. FUS negatively regulates kaposi’s sarcoma-associated herpesvirus gene expression. Viruses (2018) 10:359. doi: 10.3390/v10070359
87. Flemington EK. Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol (2001) 75:4475–81. doi: 10.1128/JVI.75.10.4475-4481.2001
88. Steininger C, Puchhammer-Stöckl E, Popow-Kraupp T. Cytomegalovirus disease in the era of highly active antiretroviral therapy (HAART). J Clin Virol (2006) 37:1–9. doi: 10.1016/j.jcv.2006.03.005
89. Kagele D, Rossetto CC, Tarrant MT, Pari GS. Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt. Virology (2012) 424:106–14. doi: 10.1016/j.virol.2011.12.010
90. Wang Y, Li H, Tang Q, Maul GG, Yua Y. Kaposi’s sarcoma-associated herpesvirus ori-lyt-dependent DNA replication: involvement of host cellular factors. J Virol (2008) 82:2867–82. doi: 10.1128/JVI.01319-07
91. Wolf D, Giese SI, Witte V, Krautkraemer E, Trapp S, Sass G, et al. Novel (n)PKC kinases phosphorylate nef for increased HIV transcription, replication and perinuclear targeting. Virology (2008) 370:45–54. doi: 10.1016/j.virol.2007.08.015
92. Wolf D, Witte V, Clark P, Blume K, Lichtenheld MG, Baur AS. HIV Nef enhances tat-mediated viral transcription through a hnRNP-k-nucleated signaling complex. Cell Host Microbe (2008) 4:398–408. doi: 10.1016/j.chom.2008.08.013
93. Ma L, Jiang Q-A, Sun L, Yang X, Huang H, Jin X, et al. X-Linked RNA-binding motif protein modulates HIV-1 infection of CD4(+) T cells by maintaining the trimethylation of histone H3 lysine 9 at the downstream region of the 5’ long terminal repeat of HIV proviral DNA. mBio (2020) 11:e03424–19. doi: 10.1128/mBio.03424-19
94. Scalabrin M, Frasson I, Ruggiero E, Perrone R, Tosoni E, Lago S, et al. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci Rep (2017) 7:45244. doi: 10.1038/srep45244
95. Thompson MG, Munoz-Moreno R, Bhat P, Roytenberg R, Lindberg J, Gazzara MR, et al. Co-Regulatory activity of hnRNP K and NS1-BP in influenza and human mRNA splicing. Nat Commun (2018) 9:2407. doi: 10.1038/s41467-018-04779-4
96. Tsai P-L, Chiou N-T, Kuss S, Garcia-Sastre A, Lynch KW, Fontoura BMA. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza a virus RNA splicing. PloS Pathog (2013) 9:e1003460. doi: 10.1371/journal.ppat.1003460
97. Thompson MG, Dittmar M, Mallory MJ, Bhat P, Ferretti MB, Fontoura BM, et al. Viral-induced alternative splicing of host genes promotes influenza replication. Elife (2020) 9:e55500. doi: 10.7554/eLife.55500
98. Fogel BL, McNally MT. A cellular protein, hnRNP h, binds to the negative regulator of splicing element from rous sarcoma virus. J Biol Chem (2000) 275:32371–8. doi: 10.1074/jbc.M005000200
99. Fogel BL, McNally LM, McNally MT. Efficient polyadenylation of rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acid Res (2002) 30:810–7. doi: 10.1093/nar/30.3.810
100. Wilusz JE, Beemon KL. The negative regulator of splicing element of rous sarcoma virus promotes polyadenylation. J Virol (2006) 80:9634–40. doi: 10.1128/JVI.00845-06
101. Nomoto A, Kitamura N, Golini F, Wimmer E. The 5’-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc Natl Acad Sci U.S.A. (1977) 74:5345–9. doi: 10.1073/pnas.74.12.5345
102. Decker CJ, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci (1994) 19:336–40. doi: 10.1016/0968-0004(94)90073-6
103. Murray KE, Roberts AW, Barton DJ. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA (2001) 7:1126–41. doi: 10.1017/S1355838201010044
104. Kempf BJ, Barton DJ. Poly(rC) binding proteins and the 5 ` cloverleaf of uncapped poliovirus mRNA function during de novo assembly of polysomes. J Virol (2008) 82:5835–46. doi: 10.1128/JVI.01513-07
105. Palusa S, Ndaluka C, Bowen RA, Wilusz CJ, Wilusz J. The 3’ untranslated region of the rabies virus glycoprotein mRNA specifically interacts with cellular PCBP2 protein and promotes transcript stability. PloS One (2012) 7:e33561. doi: 10.1371/journal.pone.0033561
106. Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, et al. The biology and life-cycle of human papillomaviruses. Vaccine (2012) 30 Suppl 5:F55–70. doi: 10.1016/j.vaccine.2012.06.083
107. Cheunim T, Zhang J, Milligan SG, McPhillips MG, Graham SV. The alternative splicing factor hnRNP A1 is up-regulated during virus-infected epithelial cell differentiation and binds the human papillomavirus type 16 late regulatory element. Virus Res (2008) 131:189–98. doi: 10.1016/j.virusres.2007.09.006
108. Ajiro M, Tang S, Doorbar J, Zheng Z-M. Serine/arginine-rich splicing factor 3 and heterogeneous nuclear ribonucleoprotein A1 regulate alternative RNA splicing and gene expression of human papillomavirus 18 through two functionally distinguishable cis elements. J Virol (2016) 90:9138–52. doi: 10.1128/JVI.00965-16
109. Zheng Y, Jönsson J, Hao C, Shoja Chaghervand S, Cui X, Kajitani N, et al. Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and hnRNP A2 inhibit splicing to human papillomavirus 16 splice site SA409 through a UAG-containing sequence in the E7 coding region. J Virol (2020) 94:e01509–20. doi: 10.1128/JVI.01509-20
110. Hao C, Zheng Y, Jönsson J, Cui X, Yu H, Wu C, et al. hnRNP G/RBMX enhances HPV16 E2 mRNA splicing through a novel splicing enhancer and inhibits production of spliced E7 oncogene mRNAs. Nucleic Acids Res (2022) 50:3867–91. doi: 10.1093/nar/gkac213
111. Kajitani N, Schwartz S. Role of viral ribonucleoproteins in human papillomavirus type 16 gene expression. Viruses (2020) 12:1110. doi: 10.3390/v12101110
112. Rush M, Zhao X, Schwartz S. A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression. J Virol (2005) 79:12002–15. doi: 10.1128/JVI.79.18.12002-12015.2005
113. Li X, Johansson C, Glahder J, Mossberg A-K, Schwartz S. Suppression of HPV-16 late L1 5’-splice site SD3632 by binding of hnRNP d proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs. Nucleic Acids Res (2013) 41:10488–508. doi: 10.1093/nar/gkt803
114. Kajitani N, Glahder J, Wu C, Yu H, Nilsson K, Schwartz S. hnRNP l controls HPV16 RNA polyadenylation and splicing in an akt kinase-dependent manner. Nucleic Acids Res (2017) 45:9654–78. doi: 10.1093/nar/gkx606
115. Dhanjal S, Kajitani N, Glahder J, Mossberg A-K, Johansson C, Schwartz S. Heterogeneous nuclear ribonucleoprotein c proteins interact with the human papillomavirus type 16 (HPV16) early 3’-untranslated region and alleviate suppression of HPV16 late L1 mRNA splicing. J Biol Chem (2015) 290:13354–71. doi: 10.1074/jbc.M115.638098
116. Zhao XM, Rush M, Schwartz S. Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J Virol (2004) 78:10888–905. doi: 10.1128/JVI.78.20.10888-10905.2004
117. Zhao X, Rush M, Carlsson A, Schwartz S. The presence of inhibitory RNA elements in the late 3’-untranslated region is a conserved property of human papillomaviruses. Virus Res (2007) 125:135–44. doi: 10.1016/j.virusres.2006.12.012
118. Zhao XM, Oberg D, Rush M, Fay J, Lambkin H, Schwartz S. A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J Virol (2005) 79:4270–88. doi: 10.1128/JVI.79.7.4270-4288.2005
119. Zhao X, Schwartz S. Inhibition of HPV-16 L1 expression from L1 cDNAs correlates with the presence of hnRNP A1 binding sites in the L1 coding region. Virus Genes (2008) 36:45–53. doi: 10.1007/s11262-007-0174-0
120. Cui X, Hao C, Gong L, Kajitani N, Schwartz S. HnRNP d activates production of HPV16 E1 and E6 mRNAs by promoting intron retention. Nucleic Acids Res (2022) 50:2782–806. doi: 10.1093/nar/gkac132
121. Rosenberger S, Arce JD-C, Langbein L, Steenbergen RDM, Roesl F. Alternative splicing of human papillomavirus type-16 E6/E6{*}early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc Natl Acad Sci (2010) 107:7006–11. doi: 10.1073/pnas.1002620107
122. Somberg M, Zhao X, Froehlich M, Evander M, Schwartz S. Polypyrimidine tract binding protein induces human papillomavirus type 16 late gene expression by interfering with splicing inhibitory elements at the major late 5 ` splice site, SD3632. J Virol (2008) 82:3665–78. doi: 10.1128/JVI.02140-07
123. Kumarasinghe N, Moss WN. Analysis of a structured intronic region of the LMP2 pre-mRNA from EBV reveals associations with human regulatory proteins and nuclear actin. BMC Res Notes (2019) 12:33. doi: 10.1186/s13104-019-4070-1
124. Chen S, Miao B, Chen N, Chen C, Shao T, Zhang X, et al. SYNCRIP facilitates porcine parvovirus viral DNA replication through the alternative splicing of NS1 mRNA to promote NS2 mRNA formation. Vet Res (2021) 52:73. doi: 10.1186/s13567-021-00938-6
125. Oberg D, Fay J, Lambkin H, Schwartz S. A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP h. J Virol (2005) 79:9254–69. doi: 10.1128/JVI.79.14.9254-9269.2005
126. Zheng Z-Z, Sun Y-Y, Zhao M, Huang H, Zhang J, Xia N-S, et al. Specific interaction between hnRNP h and HPV16 Li proteins: Implications for late gene auto-regulation enabling rapid viral capsid protein production. Biochem Biophys Res Commun (2013) 430:1047–53. doi: 10.1016/j.bbrc.2012.12.042
127. Nilsson K, Wu C, Kajitani N, Yu H, Tsimtsirakis E, Gong L, et al. The DNA damage response activates HPV16 late gene expression at the level of RNA processing. Nucleic Acids Res (2018) 46:5029–49. doi: 10.1093/nar/gky227
128. Key SCS, Yoshizaki T, Pagano JS. The Epstein-Barr virus (EBV) SM protein enhances pre-mRNA processing of the EBV DNA polymerase transcript. J Virol (1998) 72:8485–92. doi: 10.1128/JVI.72.11.8485-8492.1998
129. Stoltzfus CM. Chapter 1. regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv Virus Res (2009) 74:1–40. doi: 10.1016/S0065-3527(09)74001-1
130. Dlamini Z, Hull R. Can the HIV-1 splicing machinery be targeted for drug discovery? HIV AIDS (Auckl) (2017) 9:63–75. doi: 10.2147/HIV.S120576
131. Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem (2004) 279:10077–84. doi: 10.1074/jbc.M312743200
132. del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol (1999) 19:251–60. doi: 10.1128/MCB.19.1.251
133. Damgaard CK, Tange TO, Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA (2002) 8:1401–15. doi: 10.1017/S1355838202023075
134. Marchand V, Mereau A, Jacquenet S, Thomas D, Mougin A, Gattoni R, et al. A janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J Mol Biol (2002) 323:629–52. doi: 10.1016/S0022-2836(02)00967-1
135. Caputi M, Zahler AM. SR proteins and hnRNP h regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J (2002) 21:845–55. doi: 10.1093/emboj/21.4.845
136. Hallay H, Locker N, Ayadi L, Ropers D, Guittet E, Branlant C. Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 tat exon 2 splicing site. J Biol Chem (2006) 281:37159–74. doi: 10.1074/jbc.M603864200
137. Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlan C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of tat mRNA and binds protein hnRNP h. J Biol Chem (2001) 276:40464–75. doi: 10.1074/jbc.M104070200
138. Asai K, Platt C, Cochrane A. Control of HIV-1 env RNA splicing and transport: investigating the role of hnRNP A1 in exon splicing silencer (ESS3a) function. Virology (2003) 314:229–42. doi: 10.1016/S0042-6822(03)00400-8
139. Domsic JK, Wang YB, Mayeda A, Krainer AR, Stoltzfus CM. Human immunodeficiency virus type 1 hnRNP A/B-dep endent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol Cell Biol (2003) 23:8762–72. doi: 10.1128/MCB.23.23.8762-8772.2003
140. Hillebrand F, Peter JO, Brillen A-L, Otte M, Schaal H, Erkelenz S. Differential hnRNP d isoform incorporation may confer plasticity to the ESSV-mediated repressive state across HIV-1 exon 3. Biochim Biophys Acta Gene Regul Mech (2017) 1860:205–17. doi: 10.1016/j.bbagrm.2016.12.001
141. Jablonski JA, Buratti E, Stuani C, Caputi M. The secondary structure of the human immunodeficiency virus type 1 transcript modulates viral splicing and infectivity. J Virol (2008) 82:8038–50. doi: 10.1128/JVI.00721-08
142. Kutluay SB, Emery A, Penumutchu SR, Townsend D, Tenneti K, Madison MK, et al. Genome-wide analysis of heterogeneous nuclear ribonucleoprotein (hnRNP) binding to HIV-1 RNA reveals a key role for hnRNP H1 in alternative viral mRNA splicing. J Virol (2019) 93:e01048–19. doi: 10.1128/JVI.01048-19
143. Jablonski JA, Caputi M. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J Virol (2009) 83:981–92. doi: 10.1128/JVI.01801-08
144. Walsh D, Mathews MB, Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol (2013) 5:a012351. doi: 10.1101/cshperspect.a012351
145. Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol (2009) 19:267–76. doi: 10.1016/j.sbi.2009.03.005
146. Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol (2005) 6:318–27. doi: 10.1038/nrm1618
147. Bokinsky G, Nivon LG, Liu S, Chai G, Hong M, Weeks KM, et al. Two distinct binding modes of a protein cofactor with its. J Mol Biol (2006) 361:771–84. doi: 10.1016/j.jmb.2006.06.048
148. Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-Binding protein hnRNP d modulates internal ribosome entry site-dependent translation of. J Virol (2008) 82:12082–93. doi: 10.1128/JVI.01405-08
149. Kim CS, Seol SK, Song O-K, Park JH, Jang SK. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis c virus replication. J Virol (2007) 81:3852–65. doi: 10.1128/JVI.01311-06
150. Gosert R, Chang KH, Rijnbrand R, Yi MY, Sangar DV, Lemon SM, et al. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol Cell Biol (2000) 20:1583–95. doi: 10.1128/MCB.20.5.1583-1595.2000.
151. Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. Role of la autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology (2005) 335:72–86. doi: 10.1016/j.virol.2005.02.009
152. Fontanes V, Raychaudhuri S, Dasgupta A. A cell-permeable peptide inhibits hepatitis c virus replication by sequestering IRES transacting factors. Virology (2009) 394:82–90. doi: 10.1016/j.virol.2009.08.012
153. Wang L, Jeng K-S, Lai MMC. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis c virus (HCV) 5 ` untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol (2011) 85:7954–64. doi: 10.1128/JVI.00339-11
154. Hwang B, Lim JH, Hahm B, Jang SK, Lee S-W. hnRNP l is required for the translation mediated by HCV IRES. Biochem Biophys Res Commun (2009) 378:584–8. doi: 10.1016/j.bbrc.2008.11.091
155. Kim JH, Paek KY, Ha SH, Cho SC, Choi K, Kim CS, et al. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis c virus polyprotein initiation codon. Mol Cell Biol (2004) 24:7878–90. doi: 10.1128/MCB.24.18.7878-7890.2004
156. Park SM, Paek KY, Hong KY, Jang CJ, Cho S, Park JH, et al. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acid Res (2011) 39:7791–802. doi: 10.1093/nar/gkr509
157. Cammas A, Pileur F, Bonnal S, Lewis SM, Lévêque N, Holcik M, et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell (2007) 18:5048–59. doi: 10.1091/mbc.E07-06-0603
158. Courteau L, Crasto J, Hassanzadeh G, Baird SD, Hodgins J, Liwak-Muir U, et al. Hexokinase 2 controls cellular stress response through localization of an RNA-binding protein. Cell Death Dis (2015) 6:e1837. doi: 10.1038/cddis.2015.209
159. Walter BL, Nguyen JHC, Ehrenfeld E, Semler BL. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA (1999) 5:1570–85. doi: 10.1017/S1355838299991483
160. Hunt SL, Jackson RJ. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA (1999) 5:344–59. doi: 10.1017/S1355838299981414
161. Dave P, George B, Balakrishnan S, Sharma DK, Raheja H, Dixit NM, et al. Strand-specific affinity of host factor hnRNP C1/C2 guides positive to negative-strand ratio in coxsackievirus B3 infection. RNA Biol (2019) 16:1286–99. doi: 10.1080/15476286.2019.1629208
162. Lin J-Y, Shih S-R, Pan M, Li C, Lue C-F, Stollar V, et al. hnRNP A1 interacts with the 5’ untranslated regions of enterovirus 71 and sindbis virus RNA and is required for viral replication. J Virol (2009) 83:6106–14. doi: 10.1128/JVI.02476-08
163. Levengood JD, Tolbert M, Li M-L, Tolbert BS. High-affinity interaction of hnRNP A1 with conserved RNA structural elements is required for translation and replication of enterovirus 71. RNA Biol (2013) 10:1136–45. doi: 10.4161/rna.25107
164. Leong SY, Ong BKT, Chu JJH. The role of misshapen NCK-related kinase (MINK), a novel Ste20 family kinase, in the IRES-mediated protein translation of human enterovirus 71. PloS Pathog (2015) 11:e1004686. doi: 10.1371/journal.ppat.1004686
165. Zhou F, Wan Q, Lu J, Chen Y, Lu G, He M-L. Piml impacts enterovirus A71 replication and represents a potential target in antiviral therapy. iScience (2019) 19:715+. doi: 10.1016/j.isci.201908.008
166. Luo Z, Dong X, Li Y, Zhang Q, Kim C, Song Y, et al. PolyC-binding protein 1 interacts with 5’-untranslated region of enterovirus 71 RNA in membrane-associated complex to facilitate viral replication. PloS One (2014) 9:e87491. doi: 10.1371/journal.pone.0087491
167. Lin J-Y, Li M-L, Huang P-N, Chien K-Y, Horng J-T, Shih S-R. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5’ untranslated region and participates in virus replication. J Gen Virol (2008) 89:2540–9. doi: 10.1099/vir.0.2008/003673-0
168. Shih S-R, Stollar V, Li M-L. Host factors in enterovirus 71 replication. J Virol (2011) 85:9658–66. doi: 10.1128/JVI.05063-11
169. Hino K, Sato H, Sugai A, Kato M, Yoneda M, Kai C. Downregulation of nipah virus n mRNA occurs through interaction between its 3’ untranslated region and hnRNP d. J Virol (2013) 87:6582–8. doi: 10.1128/JVI.02495-12
170. Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol (1997) 71:8330–9. doi: 10.1128/JVI.71.11.8330-8339.1997
171. Song YT, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, et al. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA (2005) 11:1809–24. doi: 10.1261/rna.7430405
172. Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. Rna (1995) 1:924–38.
173. Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and Poly(rC)-binding proteins 1 and 2. J Biol Chem (1998) 273:22648–56. doi: 10.1074/jbc.273.35.22648
174. Ostareck-Lederer A, Ostareck DH, Cans C, NEubauer G, Bomsztyk K, Superti-Furga G, et al. C-src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol (2002) 22:4535–43. doi: 10.1128/MCB.22.13.4535-4543.2002
175. Bieleski L, Hindley C, Talbot SJ. A polypyrimidine tract facilitates the expression of kaposi’s sarcoma-associated herpesvirus vFLIP through an internal ribosome entry site. J Gen Virol (2004) 85:615–20. doi: 10.1099/vir.0.19733-0
176. Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J Biol Chem (2009) 284:31350–62. doi: 10.1074/jbc.M109.048736
177. Lund N, Milev MP, Wong R, Sanmuganantham T, Woolaway K, Chabot B, et al. Differential effects of hnRNP D/AUF1 isoforms on HIV-1 gene expression. Nucleic Acid Res (2012) 40:3663–75. doi: 10.1093/nar/gkr1238
178. Woolaway K, Asai K, Emili A, Cochrane A. HnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression. Retrovirology (2007) 4:28. doi: 10.1186/1742-4690-4-28
179. Barrera A, Ramos H, Vera-Otarola J, Fernández-Garciá L, Angulo J, Olguín V, et al. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res (2020) 48:10479–99. doi: 10.1093/nar/gkaa765
180. Lloyd RE. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology (2015) 479–480:457–474. doi: 10.1016/j.virol.2015.03.001
181. Nagy PD, Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol (2012) 10:137–49. doi: 10.1038/nrmicro2692
182. Gamarnik A V, , Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev (1998) 12:2293–304. doi: 10.1101/gad.12.15.2293
183. Ogram SA, Spear A, Sharma N, Flanegan JB. The 5 ` CL-PCBP RNP complex, 3 ` poly(A) tail and 2A(pro) are required for optimal translation of poliovirus RNA. Virology (2010) 397:14–22. doi: 10.1016/j.virol.2009.11.006
184. Gamarnik AV, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5 ` untranslated region of the poliovirus genome. J Virol (2000) 74:2219–26. doi: 10.1128/JVI.74.5.2219-2226.2000
185. Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of Poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol (1997) 71:6243–6. doi: 10.1128/JVI.71.8.6243-6246.1997
186. Sean P, Nguyen JHC, Semler BL. The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. Virology (2008) 378:243–53. doi: 10.1016/j.virol.2008.05.007
187. Liu W, Yang D, Sun C, Wang H, Zhao B, Zhou G, et al. hnRNP K is a novel ITAF that negatively regulates foot-and-mouth disease virus translation and replication and is antagonized by viral 3C protease. J Virol (2020) 94:e00803–20. doi: 10.1128/JVI.00803-20
188. Wang Y, Zhou J, Du Y. hnRNP A2/B1 interacts with influenza a viral protein NS1 and inhibits virus replication potentially through suppressing NS1 RNA/protein levels and NS1 mRNA nuclear export. Virology (2014) 449:53–61. doi: 10.1016/j.virol.2013.11.009
189. Wang X, Lin L, Zhong Y, Feng M, Yu T, Yan Y, et al. Cellular hnRNPAB binding to viral nucleoprotein inhibits flu virus replication by blocking nuclear export of viral mRNA. iScience (2021) 24:102160. doi: 10.1016/j.isci.2021.102160
190. Mouland AJ, Xu HB, Cui HY, Krueger W, Munro TP, Prasol M, et al. RNA Trafficking signals in human immunodeficiency virus type 1. Mol Cell Biol (2001) 21:2133–43. doi: 10.1128/MCB.21.6.2133-2143.2001
191. Beriault V, Clement JF, Levesque K, LeBel C, Yong X, Chabot B, et al. A late role for the association of hnRNP A2 with the HIV-1 hnRNP A2 response elements in genomic RNA, gag, and vpr localization. J Biol Chem (2004) 279:44141–53. doi: 10.1074/jbc.M404691200
192. Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, et al. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic (2006) 7:1177–93. doi: 10.1111/j.1600-0854.2006.00461.x
193. Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res (2006) 4:43–55. doi: 10.2174/157016206775197655
194. Gordon H, Ajamian L, Valiente-Echeverria F, Levesque K, Rigby WF, Mouland AJ. Depletion of hnRNP A2/B1 overrides the nuclear retention of the HIV-1 genomic RNA. RNA Biol (2013) 10:1714–25. doi: 10.4161/rna.26542
195. Suh D, Seguin B, Atkinson S, Ozdamar B, Staffa A, Emili A, et al. Mapping of determinants required for the function of the HIV-1 env nuclear retention sequence. Virology (2003) 310:85–99. doi: 10.1016/S0042-6822(03)00073-4
196. Valente ST, Goff SP. Inhibition of HIV-1 gene expression by a fragment of hnRNP U. Mol Cell (2006) 23:597–605. doi: 10.1016/j.molcel.2006.07.021
197. Najera I, Krieg M, Karn J. Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J Mol Biol (1999) 285:1951–64. doi: 10.1006/jmbi.1998.2473
198. Perdikari TM, Murthy AC, Ryan VH, Watters S, Naik MT, Fawzi NL. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J (2020) 39:e106478. doi: 10.15252/embj.2020106478
199. Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci (2003) 28:419–24. doi: 10.1016/S0968-0004(03)00142-7
200. Malik P, Clements JB. Protein kinase CK2 phosphorylation regulates the interaction of kaposi’s sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acid Res (2004) 32:5553–69. doi: 10.1093/nar/gkh876
201. Dodon MD, Hamaia S, Martin J, Gazzolo L. Heterogeneous nuclear ribonucleoprotein A1 interferes with the binding of the human T cell leukemia virus type 1 Rex regulatory protein to its response element. J Biol Chem (2002) 277:18744–52. doi: 10.1074/jbc.M109087200
202. Kress E, Baydoun HH, Bex F, Gazzolo L, Dodon MD. Critical role of hnRNP Al in HTLV-1 replication in human transformed T lymphocytes. Retrovirology (2005) 2:8. doi: 10.1186/1742-4690-2-8
203. Kanlaya R, Pattanakitsakul S, Sinchaikul S, Chen S-T, Thongboonkerd V. Vimentin interacts with heterogeneous nuclear ribonucleoproteins and dengue nonstructural protein 1 and is important for viral replication and release. Mol Biosyst (2010) 6:795–806. doi: 10.1039/b923864f
204. Wang P, Liu X, Li Q, Wang J, Ruan W. Proteomic analyses identify intracellular targets for Japanese encephalitis virus nonstructural protein 1 (NS1). Virus Res (2021) 302:198495. doi: 10.1016/j.virusres.2021.198495
205. Cousineau SE, Rheault M, Sagan SM. Poly(rC)-binding protein 1 limits hepatitis c virus virion assembly and secretion. Viruses (2022) 14:291. doi: 10.3390/v14020291
206. Poenisch M, Metz P, Blankenburg H, Ruggieri A, Lee J-Y, Rupp D, et al. Identification of HNRNPK as regulator of hepatitis c virus particle production. PloS Pathog (2015) 11:e1004573. doi: 10.1371/journal.ppat.1004573
207. Li M-L, Lin J-Y, Chen B-S, Weng K-F, Shih S-R, Calderon JD, et al. EV71 3C protease induces apoptosis by cleavage of hnRNP A1 to promote apaf-1 translation. PloS One (2019) 14:e0221048. doi: 10.1371/journal.pone.0221048
208. Jiang H, Hou P, He H, Wang H. Cell apoptosis regulated by interaction between viral gene alpha 3 and host heterogeneous nuclear ribonucleoprotein K facilitates bovine ephemeral fever virus replication. Vet Microbiol (2020) 240:108510. doi: 10.1016/j.vetmic.2019.108510
209. Schmidt T, Striebinger H, Haas J, Bailer SM. The heterogeneous nuclear ribonucleoprotein K is important for herpes simplex virus-1 propagation. FEBS Lett (2010) 584:4361–5. doi: 10.1016/j.febslet.2010.09.038
210. Zhou X, Wang L, Zou W, Chen X, Roizman B, Zhou GG. hnRNPA2B1 associated with recruitment of RNA into exosomes plays a key role in herpes simplex virus 1 release from infected cells. J Virol (2020) 94:e00367–20. doi: 10.1128/JVI.00367-20
211. Shabman RS, Gulcicek EE, Stone KL, Basler C.F. The Ebola virus VP24 protein prevents hnRNP C1/C2 binding to karyopherin α1 and partially alters its nuclear import. J Infect Dis (2011) 204:S904–10. doi: 10.1093/infdis/jir323
212. Kneller ELP, Connor JH, Lyles DS. hnRNPs relocalize to the cytoplasm following infection with vesicular stomatitis virus. J Virol (2009) 83:770–80. doi: 10.1128/JVI.01279-08
213. Kato K, Ikliptikawati DK, Kobayashi A, Kondo H, Lim K, Hazawa M, et al. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem Biophys Res Commun (2021) 536:59–66. doi: 10.1016/j.bbrc.2020.11.115
214. Maeto CA, Knott ME, Linero FN, Ellenberg PC, Scolaro LA, Castilla V. Differential effect of acute and persistent junin virus infections on the nucleo-cytoplasmic trafficking and expression of heterogeneous nuclear ribonucleoproteins type a and b. J Gen Virol (2011) 92:2181–90. doi: 10.1099/vir.0.030163-0
215. Brunetti JE, Scolaro LA. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a host factor required for dengue virus and junin virus multiplication. Virus Res (2015) 203:84–91. doi: 10.1016/j.virusres.2015.04.001
216. Dhillon P, Tandra VN, Chorghade SG, Namsa ND, Sahoo L, Rao CD. Cytoplasmic relocalization and colocalization with viroplasms of host cell proteins, and their role in rotavirus infection. J Virol (2018) 92:e00612–18. doi: 10.1128/JVI.00612-18
217. Song J, Wang D, Quan R, Liu J. Seneca Valley virus 3C(pro) degrades heterogeneous nuclear ribonucleoprotein A1 to facilitate viral replication. Virulence (2021) 12:3125–36. doi: 10.1080/21505594.2021.2014681
218. Chiu H-C, Huang W-R, Wang Y-Y, Li J-Y, Liao T-L, Nielsen BL, et al. Heterogeneous nuclear ribonucleoprotein A1 and lamin A/C modulate nucleocytoplasmic shuttling of avian reovirus p17. J Virol (2019) 93:e00851–19. doi: 10.1128/JVI.00851-19
219. Kaur R, Batra J, Stuchlik O, Reed MS, Pohl J, Sambhara S, et al. Heterogeneous ribonucleoprotein A1 (hnRNPA1) interacts with the nucleoprotein of the influenza a virus and impedes virus replication. Viruses (2022) 14:199. doi: 10.3390/v14020199
220. Li Z, Zeng W, Ye S, Lv J, Nie A, Zhang B, et al. Cellular hnRNP A1 interacts with nucleocapsid protein of porcine epidemic diarrhea virus and impairs viral replication. Viruses-Basel (2018) 10:127. doi: 10.3390/v10030127
221. Chang CJ, Luh HW, Wang SH, Lin HJ, Lee SC, Hu S.T. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) interacts with dengue virus core protein. DNA Cell Biol (2001) 20:569–77. doi: 10.1089/104454901317094981
222. Noisakran S, Sengsai S, Thongboonkerd V, Kanlaya R, Sinchaikul S, Chen S-T, et al. Identification of human hnRNP C1/C2 as a dengue virus NS1-interacting protein. Biochem Biophys Res Commun (2008) 372:67–72. doi: 10.1016/j.bbrc.2008.04.165
223. Azael Agis-Juarez R, Galvan I, Medina F, Daikoku T, Padmanabhan R, Ludert JE, et al. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in vero cells. J Gen Virol (2009) 90:2893–901. doi: 10.1099/vir.0.013433-0
224. Diwaker D, Mishra KP, Ganju L, Singh SB. Dengue virus non-structural 1 protein interacts with heterogeneous nuclear ribonucleoprotein h in human monocytic cells. Asian Pac J Trop Med (2016) 9:112–8. doi: 10.1016/j.apjtm.2016.01.015
225. Zhang K, Shang G, Padavannil A, Wang J, Sakthivel R, Chen X, et al. Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression. Proc Natl Acad Sci (2018) 115:E12218–27. doi: 10.1073/pnas.1818012115
226. Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, et al. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics (2008) 8:2077–88. doi: 10.1002/pmic.200700508
227. Tong L, Chu Z, Gao X, Yang M, Adam FEA, Theodore DWP, et al. Newcastle Disease virus V protein interacts with hnRNP H1 to promote viral replication. Vet Microbiol (2021) 260:109093. doi: 10.1016/j.vetmic.2021.109093
228. Hadian K, Vincendeau M, Maeusbacher N, Nagel D, Hauck SM, Ueffing M, et al. Identification of a heterogeneous nuclear ribonucleoprotein-recognition region in the HIV rev protein. J Biol Chem (2009) 284:33384–91. doi: 10.1074/jbc.M109.021659
229. Rehwinkel J, Gack MU. RIG-i-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol (2020) 20:537–51. doi: 10.1038/s41577-020-0288-3
230. Clément J-F, Meloche S, Servant MJ. The IKK-related kinases: from innate immunity to oncogenesis. Cell Res (2008) 18:889–99. doi: 10.1038/cr.2008.273
231. Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol (2010) 20:4–22. doi: 10.1002/rmv.633
232. Zhou X, You F, Chen H, Jiang Z. Poly(C)-binding protein 1 (PCBP1) mediates housekeeping degradation of mitochondrial antiviral signaling (MAVS). Cell Res (2012) 22:717–27. doi: 10.1038/cr.2011.184
233. You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, et al. PCBP2 mediates degradation of via the HECT ubiquitin ligase AIP4. Nat Immunol (2009) 10:1300–8. doi: 10.1038/ni.1815
234. Cao P, Luo W-W, Li C, Tong Z, Zheng Z-Q, Zhou L, et al. The heterogeneous nuclear ribonucleoprotein hnRNPM inhibits RNA virus-triggered innate immunity by antagonizing RNA sensing of RIG-i-like receptors. PloS Pathog (2019) 15:e1007983. doi: 10.1371/journal.ppat.1007983
235. Cao L, Liu S, Li Y, Yang G, Luo Y, Li S, et al. The nuclear matrix protein SAFA surveils viral RNA and facilitates immunity by activating antiviral enhancers and super-enhancers. Cell Host Microbe (2019) 26:369–384.e8. doi: 10.1016/j.chom.2019.08.010
236. Cao L, Luo Y, Guo X, Liu S, Li S, Li J, et al. SAFA facilitates chromatin opening of immune genes through interacting with anti-viral host RNAs. PloS Pathog (2022) 18:e1010599. doi: 10.1371/journal.ppat.1010599
237. Liu B-Y, Yu X-J, Zhou C-M. SAFA initiates innate immunity against cytoplasmic RNA virus SFTSV infection. PloS Pathog (2021) 17:e1010070. doi: 10.1371/journal.ppat.1010070
238. Zhao L, Xia M, Wang K, Lai C, Fan H, Gu H, et al. A long non-coding RNA IVRPIE promotes host antiviral immune responses through regulating interferon β1 and ISG expression. Front Microbiol (2020) 11:260. doi: 10.3389/fmicb.2020.00260
239. Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science (2019) 365:eaav0758. doi: 10.1126/science.aav0758
240. Dinh PX, Beura LK, Panda D, Das A, Pattnaik AK. Antagonistic effects of cellular Poly(C) binding proteins on vesicular stomatitis virus gene expression. J Virol (2011) 85:9459–71. doi: 10.1128/JVI.05179-11
241. Cerasuolo A, Buonaguro L, Buonaguro FM, Tornesello ML. The role of RNA splicing factors in cancer: Regulation of viral and human gene expression in human papillomavirus-related cervical cancer. Front Cell Dev Biol (2020) 8:474. doi: 10.3389/fcell.2020.00474
242. Francies FZ, Bassa S, Chatziioannou A, Kaufmann AM, Dlamini Z. Splicing genomics events in cervical cancer: Insights for phenotypic stratification and biomarker potency. Genes (Basel) (2021) 12:130. doi: 10.3390/genes12020130
243. Krasnopolsky S, Marom L, Victor RA, Kuzmina A, Schwartz JC, Fujinaga K, et al. Fused in sarcoma silences HIV gene transcription and maintains viral latency through suppressing AFF4 gene activation. Retrovirology (2019) 16:16. doi: 10.1186/s12977-019-0478-x
244. Levengood JD, Tolbert BS. Idiosyncrasies of hnRNP A1-RNA recognition: Can binding mode influence function. Semin Cell Dev Biol (2019) 86:150–61. doi: 10.1016/j.semcdb.2018.04.001
245. Arango D, Morohashi K, Yilmaz A, Kuramochi K, Parihar A, Brahimaj B, et al. Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc Natl Acad Sci U.S.A. (2013) 110:E2153–62. doi: 10.1073/pnas.1303726110
246. Lv X, Qiu M, Chen D, Zheng N, Jin Y, Wu Z. Apigenin inhibits enterovirus 71 replication through suppressing viral IRES activity and modulating cellular JNK pathway. Antiviral Res (2014) 109:30–41. doi: 10.1016/j.antiviral.2014.06.004
247. Zhang W, Qiao H, Lv Y, Wang J, Chen X, Hou Y, et al. Apigenin inhibits enterovirus-71 infection by disrupting viral RNA association with trans-acting factors. PloS One (2014) 9:e110429. doi: 10.1371/journal.pone.0110429
248. Carabet LA, Leblanc E, Lallous N, Morin H, Ghaidi F, Lee J, et al. Computer-aided discovery of small molecules targeting the RNA splicing activity of hnRNP A1 in castration-resistant prostate cancer. Molecules (2019) 24:763. doi: 10.3390/molecules24040763
249. Ko C-C, Chen Y-J, Chen C-T, Liu Y-C, Cheng F-C, Hsu K-C, et al. Chemical proteomics identifies heterogeneous nuclear ribonucleoprotein (hnRNP) A1 as the molecular target of quercetin in its anti-cancer effects in PC-3 cells. J Biol Chem (2014) 289:22078–89. doi: 10.1074/jbc.M114.553248
250. Kumar R, Khandelwal N, Chander Y, Nagori H, Verma A, Barua A, et al. S-adenosylmethionine-dependent methyltransferase inhibitor DZNep blocks transcription and translation of SARS-CoV-2 genome with a low tendency to select for drug-resistant viral variants. Antiviral Res (2022) 197:105232. doi: 10.1016/j.antiviral.2021.105232
251. Davila-Calderon J, Patwardhan NN, Chiu L-Y, Sugarman A, Cai Z, Penutmutchu SR, et al. IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex. Nat Commun (2020) 11:4775 doi: 10.1038/s41467-020-18594-3
252. Hou H-Y, Lu W-W, Wu K-Y, Lin C-W, Kung S-H. Idarubicin is a broad-spectrum enterovirus replication inhibitor that selectively targets the virus internal ribosomal entry site. J Gen Virol (2016) 97:1122–33. doi: 10.1099/jgv.0.000431
253. Shim B-S, Wu W, Kyriakis CS, Bakre A, Jorquera PA, Perwitasari O, et al. MicroRNA-555 has potent antiviral properties against poliovirus. J Gen Virol (2016) 97:659–68. doi: 10.1099/jgv.0.000372
254. Gunaseelan S, Wong KZ, Min N, Sun J, Ismail NKBM, Tan YJ, et al. Prunin suppresses viral IRES activity and is a potential candidate for treating enterovirus A71 infection. Sci Transl Med (2019) 11:eaar5759. doi: 10.1126/scitranslmed.aar5759
Keywords: hnRNPs, viruses, viral lifecycle, ribonucleoprotein, translation, splicing
Citation: Bhattarai K and Holcik M (2022) Diverse roles of heterogeneous nuclear ribonucleoproteins in viral life cycle. Front. Virol. 2:1044652. doi: 10.3389/fviro.2022.1044652
Received: 14 September 2022; Accepted: 28 November 2022;
Published: 22 December 2022.
Edited by:
Hernan Garcia-Ruiz, University of Nebraska-Lincoln, United StatesReviewed by:
Yuexiu Zhang, The Ohio State University, United StatesDirk H. Ostareck, University Hospital RWTH Aachen, Germany
Copyright © 2022 Bhattarai and Holcik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Holcik, bWFydGluLmhvbGNpa0BjYXJsZXRvbi5jYQ==
 Krishna Bhattarai
Krishna Bhattarai Martin Holcik
Martin Holcik