- 1Laboratory of Virus-Host Coevolution, Institute for Frontier Life and Medical Sciences, Kyoto University, Kyoto, Japan
- 2Department of Molecular Life Science, Tokai University School of Medicine, Isehara, Japan
- 3Research and Development Section, Anicom Specialty Medical Institute Inc., Kanagawa, Japan
- 4Labratory of Molecular Diagnostics and Therapeutics, The United Graduate School of Veterinary Medicine, Yamaguchi University, Yamaguchi, Japan
Endogenous retroviruses (ERVs) are remnants of ancestral retroviruses that infected host germ cells in the past. Most ERVs are thought to be non-functional elements, but some ERVs retain open reading frames (ORFs) capable of expressing proteins. The proteins encoded by ERV-ORFs have potential roles in oncogenesis; however, studies on mammals other than humans and mice are limited. Here, we identified ERV-derived genes expressed in canine oral malignant melanoma (OMM). We identified 11 ERV-derived genes in our OMM samples. Differential expression gene analysis revealed that four ERV-derived genes (PEG10, LOC102155597, and two newly identified genes) were upregulated in OMM compared to healthy tissues. PEG10 is a conserved long terminal repeat (LTR)-type retrotransposon-derived gene among mammals and is involved in human cancers. LOC102155597 is a retroviral env gene conserved in Carnivora. This Env protein harbors an immunosuppressive domain, implying the potential adverse effects on the immune system. While the production of viral particles from ERVs has been reported in human and mouse melanoma, we found no ERV-derived genes having the potential to produce viral particles. These results provide insights into the different and conserved features of ERV-derived genes in mammalian melanoma.
Introduction
Endogenous retroviruses (ERVs) are remnants of ancient retroviral infections to germ cells and have accumulated in all mammalian genomes (1). Retroviruses have three structural and enzymatic genes: gag gene encoding the major structural protein; pol gene encoding a polyprotein consisting of multiple proteins including protease, reverse transcriptase, RNase H, and integrase; env gene encoding the envelope protein. In addition, some retroviruses encode regulatory and/or accessory genes that control viral gene expression and/or suppress host defense mechanisms. Most ERVs have lost their open reading frames (ORFs) due to the accumulation of mutations. However, some genes derived from ERVs are expressed as proteins in host cells when the ORFs are still intact because they are young ERVs, or when the ORFs are domesticated as de novo host genes (1).
In human melanoma, aberrant expression of ERVs has been reported to be associated with oncogenesis (2). Human ERV-K (HERV-K) is actively expressed in human melanoma, and their protein expression, as well as particle formation, have been reported (3, 4). ERV3-1, an env gene of HERV-R, is also reported to be expressed in colorectal cancers (5) and acute myelogenous leukemia (6). In mice, active expression of an ERV called MelARV has been reported in B16/BL6 melanoma cells (7, 8). Knockdown of MelARV reduces the aggressiveness of B16 melanoma in transplanted mice (9). They proposed a functional model in which Env protein of MelARV regulates regulatory T cells (9). Thus, aberrantly expressed ERV genes may be a promising therapeutic target for melanoma. However, the association between melanoma and ERVs in mammals other than humans and mice remains unresolved.
Long terminal repeat (LTR)-type retrotransposons that mainly correspond to ERVs comprise 8 and 10% of the human and mouse genomes, respectively (10, 11). The overall proportion of ERVs as well as their lineages varies greatly among mammalian species (12). In the dog genome, for example, a proportion of genomic regions corresponding to ERVs is relatively low at 4.85% (13). According to a comprehensive study identifying ERV-derived ORFs (ERV-ORFs) in the genome, the number of canine ERV-ORFs is one-third of those in humans (14). Therefore, the expressed ERV-derived genes in canine melanoma are supposed to be fewer than in human and mouse melanoma; however, comprehensive characterization of ERVs expressed in canine melanoma has not been performed.
Canine oral malignant melanoma (OMM) is a highly metastatic tumor in dogs, spreading to the lung, lymph nodes, and liver, and shows a grave prognosis (15). Surgical resection and radiation are first-line treatments for OMM cases with no metastasis; however, there are no effective systemic treatments after metastasis (15). Canine cancers, including OMM, have been proposed as research models for human cancer studies (16, 17). On the contrary, the applications of the cancer therapies developed in humans to dogs are also underway, such as the administration of immune checkpoint inhibitors (18, 19). These studies have shown the effectiveness of strategies targeting genes conserved in humans and dogs. Though most ERVs are not conserved in humans and dogs, considering the active expression of ERVs in human and mouse melanoma, it is plausible that ERVs, including canine-specific ones, are expressed in canine OMM. Since cancer-specific ERVs may not be detectable in normal tissues, de novo transcripts in OMM samples, in addition to the reference transcripts, are required for the comprehensive gene expression analysis. Here, we made a catalog of transcripts retaining the ERV-ORFs that are expressed in canine OMM samples and conducted differential gene expression analyses. Since ERVs are involved in oncogenesis in humans and mice, these canine ERVs have the potential to be new therapeutic targets and markers. This study also will provide a model for studying ERVs' common roles in melanoma across mammalian species.
Materials and Methods
RNA Sequencing
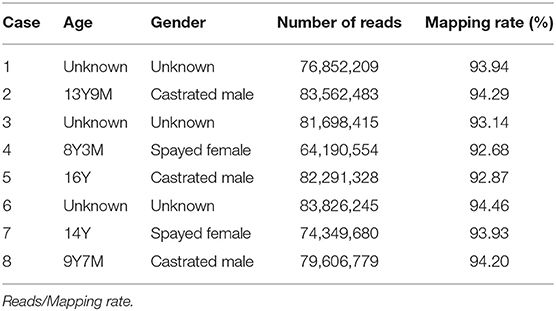
Canine OMM samples (n = 8) were obtained following surgical resection as treatment at Animal Medical Center, Yamaguchi University (Table 1). The bleed of all dogs used in this study was Dachshund. The patient's owners were informed before sample collection. Total RNA was isolated from fresh OMM from eight dogs using RNA Premium Kit (FastGene #FG-81050). We collected mRNA by NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB #E7490). Strand-specific libraries were generated by NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB #E7420) and indexed by NEBNext Multiplex Oligos for Illumina (NEB #E7335). All libraries were sequenced on Illumina NextSeq 500. We also obtained transcriptome sequencing data of canine OMM and oral healthy tissues (8 and 3 samples, respectively) from NCBI sequence read archives (https://www.ncbi.nlm.nih.gov/sra) (20) to confirm our RNA-seq analyses. Accession numbers and read mapping results were summarized in Supplementary Table 1.
Transcript Assembly
All paired-end FASTQ files were trimmed, and their sequence qualities were checked using Trim Galore (version 0.6.4) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Then, the filtered short reads were aligned to the canine reference genome (un-masked canFam3 genome from UCSC genome browser: https://hgdownload.soe.ucsc.edu/goldenPath/canFam3/bigZips/) using HISAT2 (version 2.1.0) (21). For each RNA-seq sample, we conducted transcript assemblies using StringTie2 (version 2.1.1) (21) with “–rf” options to specify strandedness. Resultant GTF files obtained from the eight canine OMM samples and the canine RefSeq GTF file (canFam3, downloaded from UCSC genome browser on June 15, 2021) were merged using StringTie2 with “–merge” option to generate a merged GTF file. The “-m 500 -f 0.05” options were combined to improve the specificity of the transcript assembly. Each melanoma transcript identical to a RefSeq transcript was allocated to the RefSeq nomenclature. The resultant transcript assemblies are available in Supplementary File 1 (a compressed GTF file).
Detection of ERV-Derived Genes
A BED file of canine ERV-ORFs was downloaded from gEVE (version 1.1) (http://geve.med.u-tokai.ac.jp/) (14). In gEVE, pol genes that were thought to be derived from long interspersed nuclear elements (LINEs) were included (14). To remove LINE-derived sequences, ERV-ORFs which were annotated as “LINE” by RepeatMasker and/or “YP_073558.1” or “NP_048132.1” by BLASTP against the NCBI Viral Genome Database were removed. Transcripts in the merged GTF file overlapped with ERV-ORFs were identified using BEDtools intersect with “-s” option to consider strandedness (22). Forty-three ERV-derived genes detected at this step were listed in Supplementary Table 2. All ERV-derived genes were manually checked using Integrative Genomics Viewer (version 2.4.9) (23). Transcripts per kilobase million (TPM) was calculated using StringTie2 with “-e –rf” options using the merged GTF file. Eleven ERV-derived genes with the mean TPM > 1 were listed including Canis-env2 (Table 2). Differentially expressed genes were identified and visualized with an MA plot and a volcano plot using DESeq2 (version 1.26.0) (24) in R (version 3.6.1).
RT-PCR
Total RNA was extracted from cultured OMM cells from four animals (Supplementary Table 3), which were maintained in Dulbecco's modified Eagle medium (Sigma-Aldrich #D5796) supplemented with 10% heat-inactivated fetal bovine serum (Gibco #10270), penicillin (100 IU/ml) and streptomycin (100 μg/ml) (Nacalai tesuque # 09367-34). To synthesize the cDNA, RNA was reverse transcribed using Verso cDNA Synthesis kit (Thermo Fisher Scientific #AB1453B). For negative controls, samples without reverse transcriptase were prepared. PCR was performed using KOD One (TOYOBO #KMM-101). Primer pairs used for NMG-1 and NMG-4 were 5′-ACCCGCTGACTATGACTCAGGAAC-3′ (forward) and 5′-AACAGTCTTTGCCTCTGCTGTCAG-3′ (reverse), and 5′-AGGCACTCCTCCACGCCACTACTAG-3′ (forward) and 5′-TCAAGTCTGTGCTTATGTAGGGACCAG-3′ (reverse), respectively. For loading control, GAPDH was amplified with a primer pair, 5′-AAGGTCGGAGTGAACGGATTTGG-3′ (forward) and 5′-CGGTTGCTGTAGCCAAATTCATTG-3′ (reverse). After preheating at 94°C for 2 min, thermal cycling reaction (98°C for 10 s and 68°C for 10 s) was repeated 35 times (for NMG-1 and NMG-4) or 30 times (for GAPDH). The sequences of the amplicons were checked by the Sanger sequencing (Fasmac Co., Ltd.).
Evolutionally Analyses of Canis-Env2
To collect Canis-env2 orthologs in Carnivora genomes, coding sequences of Canis-env2 in the dog was searched using Blat (25) in the following represented species in the UCSC genome browser (https://genome.ucsc.edu): Southern Sea otter, enhLutNer1 (Jun. 2019); Ferret, musFur1 (Apr. 2011); Hawaiian monk seal, neoSch1 (Jun. 2017); panda, ailMel1 (Dec. 2009); and Cat, felCat9 (Nov. 2017). The hits with the highest similarity scores were confirmed to be located between BTN2A1 and BTN1A1. To conduct the following evolutionary analyses of Canis-env2, we used MEGA-X (26). We first aligned nucleotide sequences of Canis-env2 genes by MUSCLE program with “Align codons” option (27), and pairwise p-distance values of amino acids, which are proportions of amino acid sites at which the two sequences to be compared are different, were calculated. The numbers of non-synonymous substitutions (dN) and synonymous substitutions (dS) per site were estimated by Nei-Gojobori method with Jukes-Canter correlation (28).
Results
Identification of Transcriptionally Active ERV-Derived Genes in Canine OMM
We conducted RNA-seq analyses using canine oral malignant melanoma (OMM) cases (n = 8). To identify ERV-derived genes from these sequence data, we constructed a pipeline to identify ERV-ORFs expressed in canine melanoma (Figure 1A). First, we constructed de novo transcript assemblies with reference to the RefSeq gene coordinates using StringTie2. In this procedure, transcripts expressed from the overlapped locus were given a single gene name. In total, we obtained 106,209 transcripts from the 8 OMM RNA-seq samples, and these transcripts were organized into 34,292 genes, of which 3,035 genes were not reported in the reference gene annotation (Figure 1B). Next, we extracted genes whose transcripts were overlapped with ERV-ORFs in the gEVE database (http://geve.med.u-tokai.ac.jp/) (14) (Figure 1C). As a result, we obtained 43 genes, of which five genes were newly identified (Figure 1D, Supplementary Table 2).
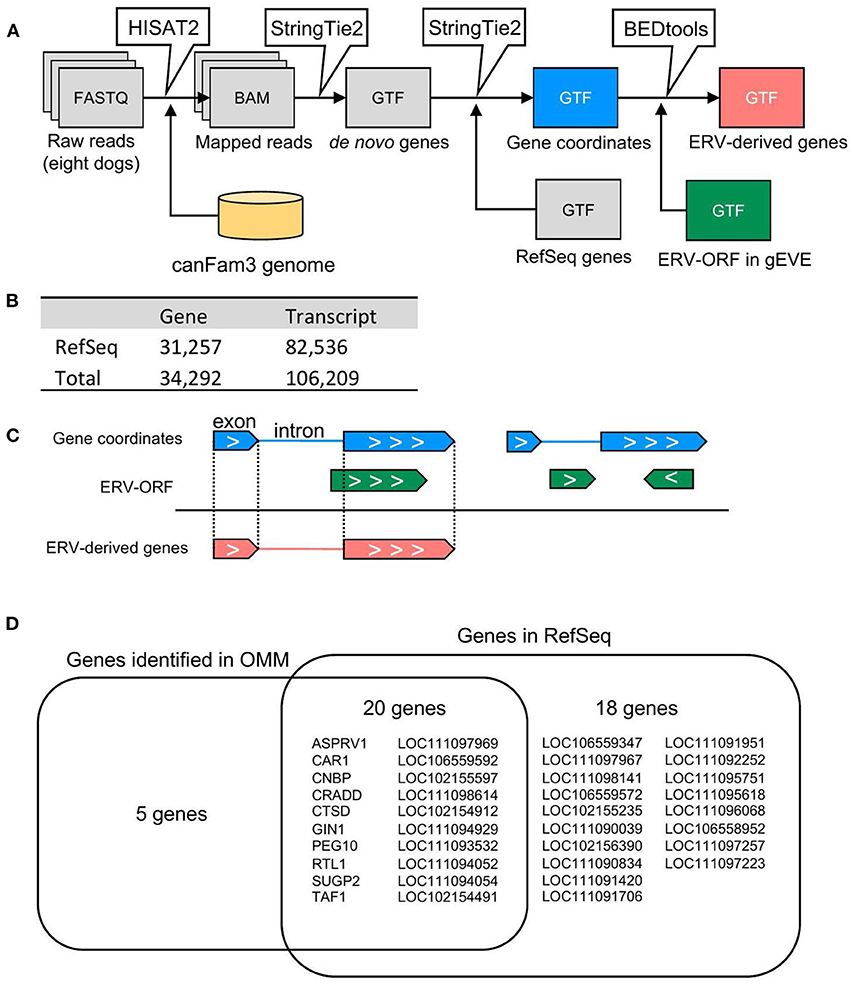
Figure 1. Identification of ERV-derived genes from canine OMM. (A) Schematic representation of the pipeline for detection of ERV-derived genes. (B) The numbers of genes and transcripts identified in the RNA-seq analysis. (C) The extraction method for ERV-derived genes. Genes of which exon(s) were overlapped with the same coordinates annotated in the gEVE database were defined as ERV-derived genes. Note that genes whose intron(s) were overlapped with the gEVE annotation, or genes overlapped with the gEVE annotation in reverse direction were not regarded to ERV-derived genes. (D) Names of ERV-derived genes. Five ERV-derived genes were not found in the RefSeq genes. Twenty ERV-derived genes were annotated in the RefSeq database and expressed in the OMM samples. Eighteen ERV-derived genes were RefSeq genes, but those were not expressed in the OMM samples.
Characterization of ERV-Derived Genes
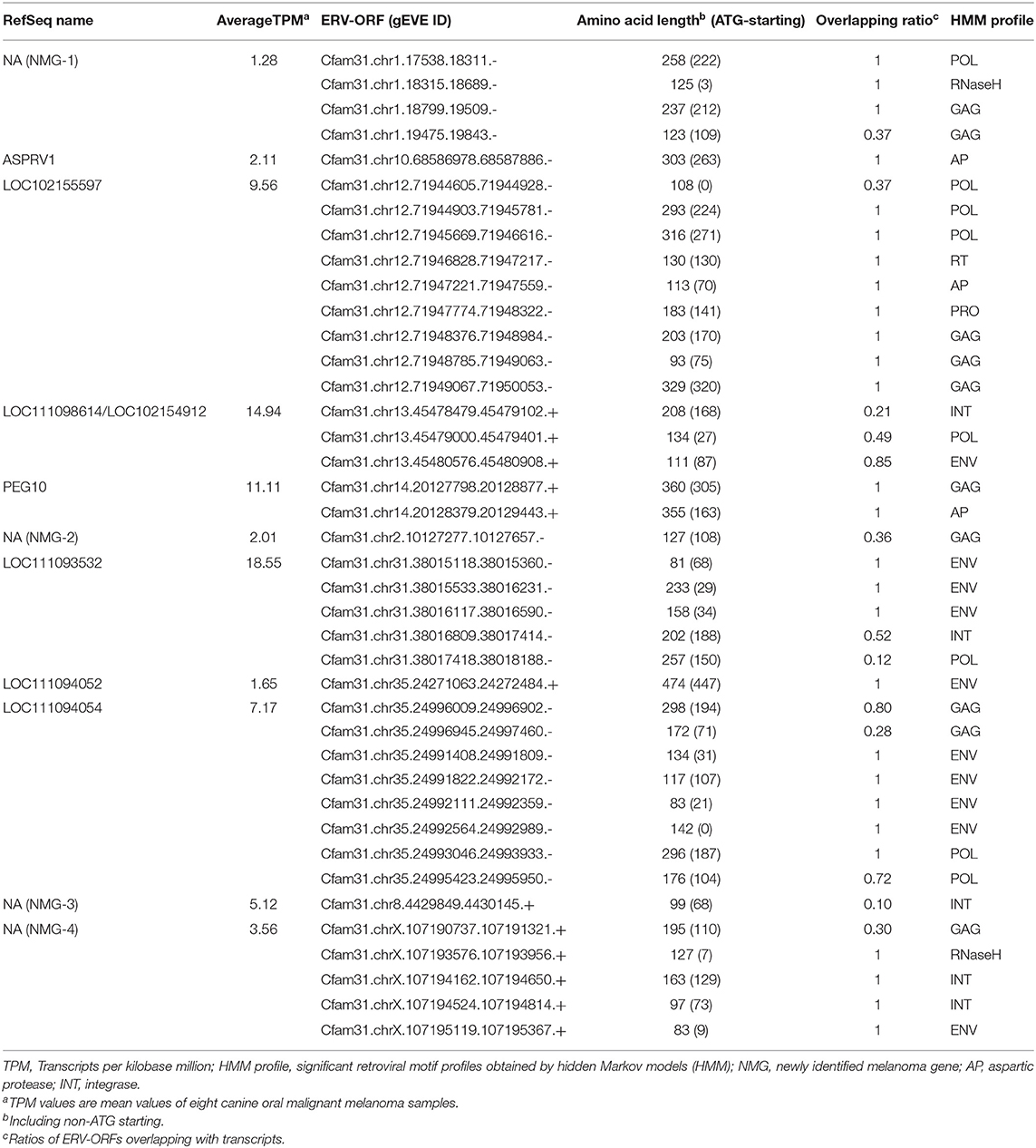
Next, we examined the structures of these 43 genes overlapped with ERV-ORFs. In nine genes, ERV-ORFs were identical to full or partial coding sequences of host genes: ASPRV1, CAR1, CNBP, CTSD, GIN1, PEG10, RTL1, SUGP2, and TAF1 (Figure 2). In these genes, four genes—ASPRV1, CAR1, PEG10, and RTL1 — were known to originate from ERVs or LTR-type retrotransposons and mammalian or Carnivora-specific genes. ASPRV1 codes an aspartic protease conserved in mammals and has roles in skin maintenance (29). PEG10 and RTL1 are gag-pol genes derived from Ty3/Gypsy retrotransposon and were involved in placental development (30, 31). CAR1, also known as syncytin-Car1, is a retroviral env gene conserved in Carnivora (32). Syncytin-Car1 protein is involved in the cell fusion of syncytiotrophoblast during placentation (32). We used these four definite retroviral genes for further analysis. The remaining five genes—CNBP, CTSD, GIN1, SUGP2, and TAF1—consist of several exons, and one exon identified as containing ERV-ORFs (Figure 2A). Orthologs of these exons were found in at least chickens, mice, and humans and may be derived from ERVs before the divergence of birds and mammals. This possibility has been particularly suggested for GIN1, where its integrase domain shows a high similarity to the GIN element, an animal-specific DNA transposon that encodes a Gypsy/Ty3 retrotransposon-like integrase (33). On the other hand, to the best of our knowledge, no evidence has been reported to indicate that CNBP, CTSD, SUGP2, and TAF1 were derived from transposons. It is also plausible that these genes independently acquired protein motifs similar to ERVs. Since it is beyond the focus of this study to make conclusions, we did not regard these five genes as ERV-derived genes in this study. Two splicing variants of CRADD were overlapped with two ERV-ORFs, although they were in the 5′ UTR. Therefore, CRADD was also removed for further analyses. Others were uncharacterized genes in the RefSeq database (i.e., genes whose names start with LOC), including non-coding genes or newly identified non-RefSeq genes. To identify sufficiently expressing genes, we collected genes that were expressed in the eight canine OMM samples with an average TPM > 1. Finally, we identified 11 ERV-derived genes, including the newly identified melanoma genes 1 to 4 (NMG-1 to 4), which were expressed in canine oral melanoma (Table 2). Since NMG-1 to 4 overlapped with RepeatMasker track of an ERV group, CfERV1-int (Figure 2B; Supplementary Figure 1), these genes are presumed to be derived from ERVs.
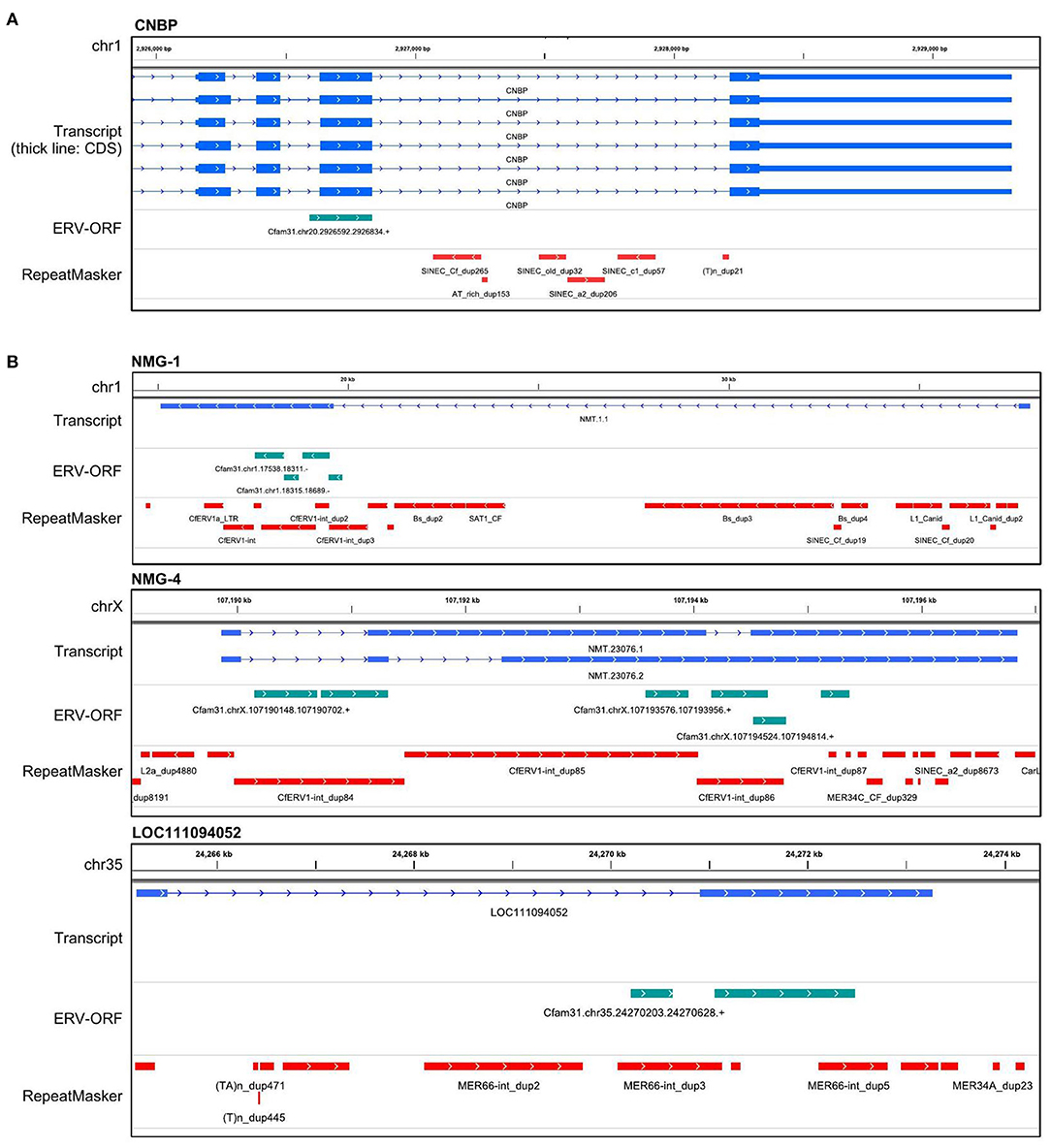
Figure 2. Gene structures of ERV-derived genes. (A) An example of an ERV-derived gene, CNBP, whose single exon overlapped with an ERV-ORF. The transcript track displays the coordinates generated by StringTie2. The ERV-ORF track displays the coordinates of ERV-ORFs obtained from gEVE version 1.1. RepeatMasker track displays the coordinates annotated by RepeatMasker downloaded from the UCSC genome browser. (B) Examples of other ERV-derived genes. Structures of the other ERV-derived genes were in Supplementary Figure 1.
Identification of Upregulated ERV-Derived Genes in Canine OMM
Next, we identified the ERV-derived genes that showed higher expression levels in the OMM samples than those in healthy tissues. Although we did not conduct the transcriptome sequencing of control samples, we utilized publicly available RNA-seq data of eight canine OMM samples and three canine oral healthy samples, all of which were obtained from the SRA database (20) (Supplementary Table 1). Then, we found that four ERV-derived genes were significantly upregulated (PEG10, NMG-1, NMG-4, and LOC102155597), and two ERV-derived genes were down-regulated (ASPRV1 and LOC10215559) in OMM (adjusted p < 0.05) (Figures 3A,B, Supplementary Table 4). TPMs of these four upregulated genes varied among melanoma samples, but their expression levels were low in all healthy tissues (Figure 3C). PEG10 was reported as an important gene in several species of human cancers, such as hepatocellular carcinoma (34), breast cancer (35), lung cancer (36), and neuroendocrine prostate cancer (37). NMG-1 and NMG-4 are newly identified genes, and their expressions were experimentally verified by RT-PCR (Figure 3D). NMG-1 gene has a single transcript isoform that is composed of two exons, and its second exon was overlapped with four ERV-ORFs annotated by gEVE database (Figure 2B). NMG-4 gene has two transcripts, and both are composed of three exons. LOC102155597 was overlapped with an Env-like ERV-ORF of 447 amino acid length (Figure 2B). This env gene was previously designated as Canis-env2 (32). We found that Canis-env2 homologs located between BTN2A1 and BTN1A1 among six Carnivora species available in UCSC genome browser, and they were presumed to be orthologs (Figure 4A). We found that the coding sequences of Canis-env2 orthologs were under purifying selection, suggesting that the proteins are functionally important (Figure 4B). This Canis-Env2 protein retains the signal peptide, the furin cleavage site, and the immunosuppressive domain, but not the transmembrane (TM) domain (Figure 4C). Together, we successfully identified newly identified or uncharacterized retroviral coding genes expressed in canine OMM.
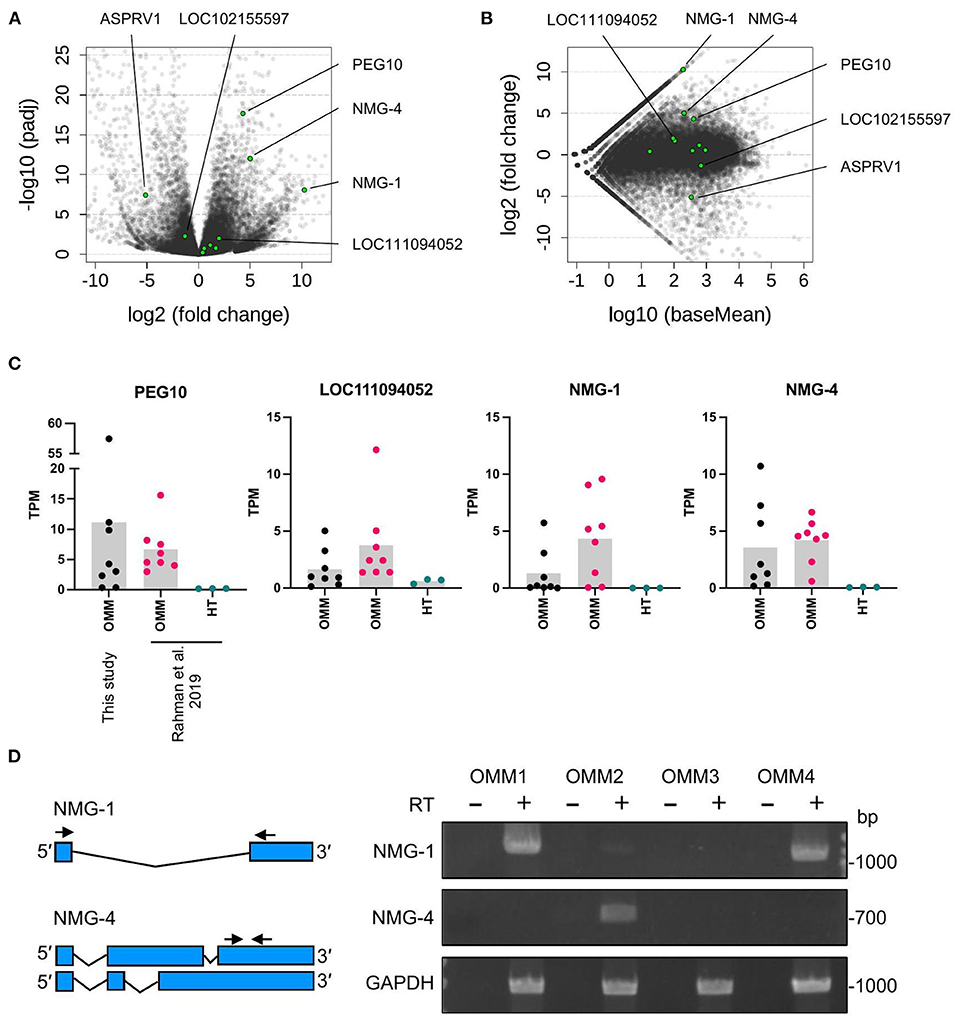
Figure 3. Differentially expressed ERV-derived genes. (A) Volcano plot representing fold changes and adjusted p-values (padj) between OMM and healthy oral tissues obtained from same sites in different individuals. Each dot represents one gene. Green dots indicate ERV-derived genes, and gray dots indicate the other genes. Names of significantly changed genes (padj < 0.05) are also represented. (B) MA plot representing fold changes and expression levels (baseMean). (C) TPM values of individual samples. Bars represent the means. OMM, oral malignant melanoma; HT, healthy tissue. (D) Agarose gels showing the RT-PCR products of NMG-1, NMG-4, and GAPDH that were amplified from OMM cell lines from four dogs with the primers shown in the left panel. Negative controls without the reverse transcriptase (RT) were included to confirm the absence of DNA contamination.
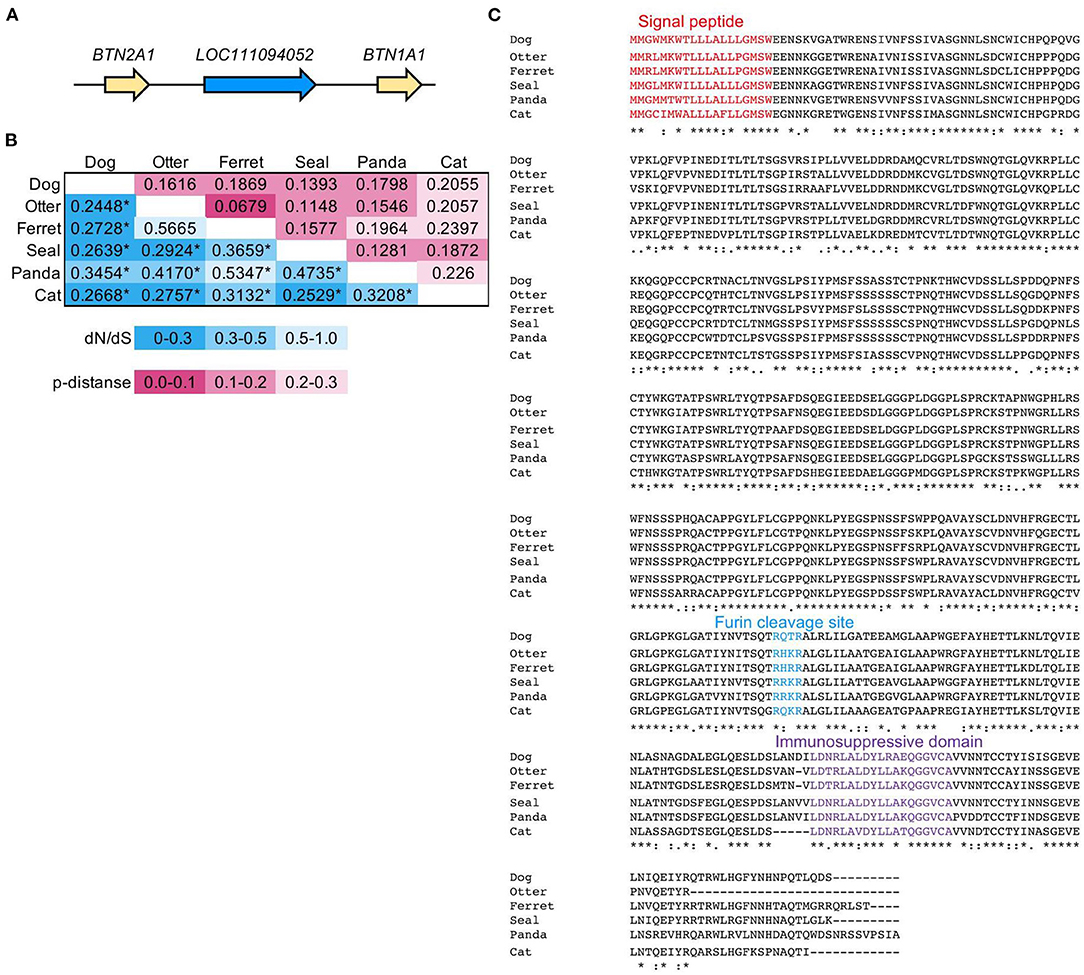
Figure 4. Evolutionally conservation of an Env protein coded in LOC111094052. (A) Synteny block of LOC111094052 in canFam3. (B) Purifying selection on the Env protein, Canis-Env2, coded in LOC111094052. Pairwise dN/dS values and amino acid p-distances of Canis-Env2 orthologs from six Carnivora species were represented in the double-entry table. Statistical significance of selection was estimated by the codon-based test of neutrality using Nei-Gojobori method [27]. *p < 0.01. (C) Amino acid sequence alignment of Canis-env2 orthologs. Colored amino acids represent characteristic domains. Signal peptide was detected using SignalP (version 5.0) (https://services.healthtech.dtu.dk/service.php?SignalP-5.0). Furin cleavage site has the RXXR motif. Immunosuppressive domain was identified by comparison to the CKS-17 peptide (NH2-LQNRIRGLDLLFLKEGGL-COOH) from the immunosuppressive domain conserved in retroviral Env proteins (38).
Discussion
In this study, we identified and characterized ERV-derived genes that are expressed in canine OMM samples (Figures 1, 2). Among them, four ERV-derived genes are upregulated in melanoma compared to healthy tissues (Figure 3). Though ERV-derived viral particles have been found in melanoma in humans and mice (3, 7), we could not find such ERV-derived genes that could potentially produce viral particles. Since this study is based on the canine reference genome (canFam3), we could not detect newly generated genes in OMM genomes. Therefore, the relationship between polymorphisms and ERVs in the genome of canine OMM remains to be investigated. Even with these limitations, evolutionarily conserved as well as expressed ERV-derived genes were newly identified in canine OMM in this study (Figure 4).
PEG10 is upregulated in several human cancers (39). Comprehensive expression analysis revealed that PEG10 is upregulated, especially in hepatocellular carcinoma and breast carcinoma (39). In hepatocellular carcinoma, PEG10 is involved in the TGF-β1-triggered epithelial-mesenchymal transmission, progressing metastasis (34). PEG10 interacts with SIAH1 and suppresses apoptosis of hepatocellular carcinoma (40). Though a correlation between PEG10 and human melanoma has not been reported yet, considering the upregulation of PEG10 in canine OMM, this gene may be involved in tumor development in various mammalian species.
NMG-1 and NMG-4 are two ERV-derived genes that were newly identified in this study. Both retained fragmented ORFs and overlapped with CfERV1-int, a canine-specific ERV family. Therefore, these two genes are relatively new ERVs and may be under being disrupted in the canine genome by mutation. Since their expressions were suppressed to very low levels in healthy samples, they may become a valuable marker for OMM. Future studies on the correlation of the expression levels with malignancy are needed.
LOC111094052 is an Env-coding gene, conveniently named Canis-env2 in the previous study (32). Canis-env2 orthologs were identified in all mammals belonging to the four major lineages of the order Carnivora (Figure 4A). Pairwise dN/dS values among orthologs were <1 (Figure 4B), and their amino acid sequences were conserved (Figure 4C), suggesting the functional importance of the protein. Therefore, Canis-env2 has been under co-option as a host gene in the Carnivora. While the function of Canis-Env2 protein is still unknown, the structure of Canis-Env2 is very similar to a primate-specific Env protein, HEMO, which lost the TM domain by protease cleavage (41). In addition, env-panMars, an env gene conserved in marsupials, also lost the TM domain due to a stop codon before the TM domain. HEMO, Env-panMars, and Canis-Env2 retain immunosuppressive domains and may be involved in cancer immunity. Tumors expressing Env can avoid immune rejection when transplanted into mice (42). MCA205 tumor cells expressing ERV-FRD1 Env (Syncytin-2) and ERV3 Env suppress the immune rejection in mice depending on the immunosuppressive domains. Syncytin-2 and ERV3 are expressed in the placenta, and their immunosuppressive activities are thought to be conserved for fetal-maternal immunosuppression (43). Thus, aberrant expression of Canis-Env2 may have a role in suppressing immune systems against OMM.
Our current study has some limitations: the small number of samples as well as the lack of well-designed control groups. These are partially due to the inevitable circumstances that our sample collection was dependent on the hospital surgeries for domesticated dogs. Furthermore, the expression of the transcripts and proteins identified in our RNA-seq analysis were not immunohistologically validated. However, our analysis clearly indicated that four ERV-derived genes (PEG10, LOC111094052, and two fragmented-ORF genes) was expressed in canine OMM. These results indicate that even in dogs with relatively low ERV copy numbers, ERV-derived genes similar to those of humans and mice are expressed.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Experiment Ethics Committee of Yamaguchi University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
KK and TMiy designed the study. AS, MY, and TMiz performed experiments. KK performed the computational analyses, with supervision by SN. KK, SN, TMiz, and TMiy prepared the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by JSPS KAKENHI (Grant Numbers 20J22607 to KK, 19K22365 to SN, TMiz, and TMiy, 20K06775 to SN and TMiy, and 20H03150 to SN and TMiy).
Conflict of Interest
MY was employed by the company Anicom Specialty Medical Institute Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2021.785678/full#supplementary-material
References
1. Johnson WE. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. (2019) 17:355–70. doi: 10.1038/s41579-019-0189-2
2. Singh S, Kaye S, Gore ME, McClure MO, Bunker CB. The role of human endogenous retroviruses in melanoma. Br J Dermatol. (2009) 161:1225–31. doi: 10.1111/j.1365-2133.2009.09415.x
3. Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. (2003) 63:8735–41.
4. Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. (2005) 65:4172–80. doi: 10.1158/0008-5472.CAN-04-2983
5. Lee SH, Kang YJ, Jo JO, Ock MS, Baek KW, Eo J, et al. Elevation of human ERV3-1 env protein expression in colorectal cancer. J Clin Pathol. (2014) 67:840–4. doi: 10.1136/jclinpath-2013-202089
6. Nakagawa S, Kawashima M, Miyatake Y, Kudo K, Kotaki R, Ando K, et al. Expression of ERV3-1 in leukocytes of acute myelogenous leukemia patients. Gene. (2021) 773:145363. doi: 10.1016/j.gene.2020.145363
7. Li MF, Muller J, Xu F, Hearing VJ, Gorelik E. Inhibition of melanoma-associated antigen expression and ecotropic retrovirus production in B16BL6 melanoma cells transfected with major histocompatibility complex class I genes. Cancer Res. (1996) 56:4464–74.
8. Pothlichet J, Mangeney M, Heidmann T. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. Int J Cancer. (2006) 119:1869–77. doi: 10.1002/ijc.22066
9. Mangeney M, Pothlichet J, Renard M, Ducos B, Heidmann T. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res. (2005) 65:2588–91. doi: 10.1158/0008-5472.CAN-04-4231
10. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. (2001) 409:860–921. doi: 10.1038/35057062
11. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. (2002) 420:520–62. doi: 10.1038/nature01262
12. Platt RN, Vandewege MW, Ray DA. Mammalian transposable elements and their impacts on genome evolution. Chromosom Res. (2018) 26:25–43. doi: 10.1007/s10577-017-9570-z
13. Wang C, Wallerman O, Arendt ML, Sundström E, Karlsson Å, Nordin J, et al. A novel canine reference genome resolves genomic architecture and uncovers transcript complexity. Commun Biol. (2021) 4:1–11. doi: 10.1038/s42003-021-01698-x
14. Nakagawa S, Takahashi MU. gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database. (2016) 2016:1–8. doi: 10.1093/database/baw087
15. Prouteau A, André C. Canine melanomas as models for human melanomas: clinical, histological, and genetic comparison. Genes. (2019) 10. doi: 10.3390/genes10070501
16. Khanna C, Lindblad-Toh K, Vail D, London C, Bergman P, Barber L, et al. The dog as a cancer model. Nat Biotechnol. (2006) 24:1065–6. doi: 10.1038/nbt0906-1065b
17. Rowell JL, McCarthy DO, Alvarez CE. Dog models of naturally occurring cancer. Trends Mol Med. (2011) 17:380–8. doi: 10.1016/j.molmed.2011.02.004
18. Maekawa N, Konnai S, Takagi S, Kagawa Y, Okagawa T, Nishimori A, et al. A canine chimeric monoclonal antibody targeting PD-L1 and its clinical efficacy in canine oral malignant melanoma or undifferentiated sarcoma. Sci Rep. (2017) 7:1–12. doi: 10.1038/s41598-017-09444-2
19. Igase M, Nemoto Y, Itamoto K, Tani K, Nakaichi M, Sakurai M, et al. A pilot clinical study of the therapeutic antibody against canine PD-1 for advanced spontaneous cancers in dogs. Sci Rep. (2020) 10:1–16. doi: 10.1038/s41598-020-75533-4
20. Rahman MM, Lai Y, Husna A, Chen H, Tanaka Y, Kawaguchi H, et al. Transcriptome analysis of dog oral melanoma and its oncogenic analogy with human melanoma. Oncol Rep. (2019) 43:16–30. doi: 10.3892/or.2019.7391
21. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. (2016) 11:1650–67. doi: 10.1038/nprot.2016.095
22. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. (2010) 26:841–2. doi: 10.1093/bioinformatics/btq033
23. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. (2011) 29:24–6. doi: 10.1038/nbt.1754
24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:1–21. doi: 10.1186/s13059-014-0550-8
25. Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. (2002) 12:656–64. doi: 10.1101/gr.229202
26. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
27. Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–97. doi: 10.1093/nar/gkh340
28. Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. (1986) 3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410
29. Matsui T, Miyamoto K, Kubo A, Kawasaki H, Ebihara T, Hata K, et al. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol Med. (2011) 3:320–33. doi: 10.1002/emmm.201100140
30. Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. (2006) 38:101–6. doi: 10.1038/ng1699
31. Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. (2008) 40:243–8. doi: 10.1038/ng.2007.51
32. Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Veron G, Mulot B, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci USA. (2012) 109:E432–41. doi: 10.1073/pnas.1115346109
33. Marín I. GIN transposons: Genetic elements linking retrotransposons and genes. Mol Biol Evol. (2010) 27:1903–11. doi: 10.1093/molbev/msq072
34. Zhang M, Sui C, Dai B, Shen W, Lu J, Yang J. PEG10 is imperative for TGF-β1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Oncol Rep. (2017) 37:510–8. doi: 10.3892/or.2016.5282
35. Li X, Xiao R, Tembo K, Hao L, Xiong M, Pan S, et al. PEG10 promotes human breast cancer cell proliferation, migration and invasion. Int J Oncol. (2016) 48:1933–42. doi: 10.3892/ijo.2016.3406
36. Deng X, Hu Y, Ding Q, Han R, Guo Q, Qin J, et al. PEG10 plays a crucial role in human lung cancer proliferation, progression, prognosis and metastasis. Oncol Rep. (2014) 32:2159–67. doi: 10.3892/or.2014.3469
37. Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. (2015) 12:922–36. doi: 10.1016/j.celrep.2015.07.012
38. Cianciolo GJ, Copeland TD, Oroszlan S, Snyderman R. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Science. (1985) 230:453–5. doi: 10.1126/science.2996136
39. Xie T, Pan S, Zheng H, Luo Z, Tembo KM, Jamal M, et al. PEG10 as an oncogene: expression regulatory mechanisms and role in tumor progression. Cancer Cell Int. (2018) 1–10. doi: 10.1186/s12935-018-0610-3
40. Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. (2003) 63:3043–8.
41. Heidmann O, Béguin A, Paternina J, Berthier R, Deloger M, Bawa O, et al. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc Natl Acad Sci USA. (2017) 114:E6642–51. doi: 10.1073/pnas.1702204114
42. Mangeney M, Heidmann T. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc Natl Acad Sci USA. (1998) 95:14920–5. doi: 10.1073/pnas.95.25.14920
Keywords: dog, endogenous retrovirus, oral malignant melanoma, protein-coding gene, RNA-Seq
Citation: Kitao K, Sumiyoshi A, Nakagawa S, Matsumoto Y, Mizuno T and Miyazawa T (2021) Systematic Identification of Endogenous Retroviral Protein-Coding Genes Expressed in Canine Oral Malignant Melanoma. Front. Virol. 1:785678. doi: 10.3389/fviro.2021.785678
Received: 29 September 2021; Accepted: 03 November 2021;
Published: 24 November 2021.
Edited by:
Jie Cui, Institut Pasteur of Shanghai (CAS), ChinaCopyright © 2021 Kitao, Sumiyoshi, Nakagawa, Matsumoto, Mizuno and Miyazawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takayuki Miyazawa, dGFrYXZldEBpbmZyb250Lmt5b3RvLXUuYWMuanA=
 Koichi Kitao
Koichi Kitao Aoi Sumiyoshi1
Aoi Sumiyoshi1 So Nakagawa
So Nakagawa Yuki Matsumoto
Yuki Matsumoto Takayuki Miyazawa
Takayuki Miyazawa