
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol., 29 November 2021
Sec. Emerging and Reemerging Viruses
Volume 1 - 2021 | https://doi.org/10.3389/fviro.2021.783407
This article is part of the Research TopicTranslational Virology in PregnancyView all 12 articles
 Nicole N. Haese1
Nicole N. Haese1 Hannah Smith2
Hannah Smith2 Kosiso Onwuzu3
Kosiso Onwuzu3 Craig N. Kreklywich1
Craig N. Kreklywich1 Jessica L. Smith1
Jessica L. Smith1 Michael Denton1
Michael Denton1 Nicholas Kreklywich1
Nicholas Kreklywich1 Aaron D. Streblow2
Aaron D. Streblow2 Antonio E. Frias2,4
Antonio E. Frias2,4 Terry K. Morgan4,5
Terry K. Morgan4,5 Alec J. Hirsch1,3
Alec J. Hirsch1,3 Benjamin N. Bimber1,3
Benjamin N. Bimber1,3 Victoria H. J. Roberts2
Victoria H. J. Roberts2 Daniel N. Streblow1,3*
Daniel N. Streblow1,3*Zika virus (ZIKV) is an arthropod-borne Flavivirus that can also be transmitted vertically from infected mother to fetus. Infection of the fetus during pregnancy can lead to congenital malformations and severely impact fetal brain development causing a myriad of diseases now labeled Congenital Zika Syndrome (CZS). The mechanisms by which ZIKV crosses the placenta into the fetal circulation and the extent of ZIKV-induced changes remain unclear. We have previously shown that ZIKV infection of pregnant rhesus macaques results in abnormal oxygen transport across the placenta which may promote uterine vasculitis and placental villous damage. Changes in immune cell frequencies and activation status were also detected, as were distinct changes in the proportions of CD14+ cell subsets with an altered ratio of classical to non-classical CD14+ monocyte cells in both the maternal decidua and placental villous from ZIKV-infected animals compare to uninfected controls. In the current study, we performed single cell RNA sequencing on CD14+ cells isolated from the decidua of animals that were ZIKV infected at 31, 51, or 115 days of gestation (where term is ~168 days) compared to pregnant, time-matched uninfected controls. Bioinformatic analysis identified unique transcriptional phenotypes between CD14+ cells of infected and uninfected animals suggesting a distinct and sustained difference in transcriptomes between infected and uninfected CD14+ cells derived from the decidua. The timing of ZIKV infection had no effect on the CD14+ cell transcriptional profiles. Interestingly, ZIKV infection caused changes in expression of genes in pathways related to cellular stress and metabolism as well as immune response activation. Type 1 interferon response genes (ISGs) were among those that were differentially expressed following infection and these included members of the ISG12 family, IFI27 and IFI6. These ISGs have been recently described as effectors of the IFN response to flaviviruses. Supplementing our animal findings, in CD14+ cells isolated from human placenta, ZIKV infection similarly induced the expression of IFI27 and IFI6. Overall, our results showed that ZIKV infection during pregnancy induces the stable expression of antiviral genes within CD14+ cells of the placenta, which may provide an immune shield to protect the placenta from further infection and damage.
Zika virus (ZIKV) is a flavivirus most commonly transmitted by the bite from an infected Aedes mosquito. Unique to ZIKV as a flavivirus, vertical transmission can occur from an infected mother to the fetus, and through sexual contact. ZIKV has emerged in recent years as a significant threat to humans primarily because of the effects it has on neonates. Historically, in healthy individuals, infection is most often either asymptomatic or results in mild symptoms. However, in the most recent epidemic, evidence indicates that ZIKV can cause neurological sequelae such as Guillain-Barre Syndrome in adults (1, 2). Prenatal ZIKV exposure was first associated with an increased risk for severe microcephaly in infants (3, 4). It is now clear that infection during pregnancy can result in a range of presentations with variable severity referred to as Congenital Zika Syndrome (CZS), divided into mutually exclusive categories: (1) brain abnormalities and/or microcephaly and (2) neural tube defects, eye abnormalities, or consequences of central nervous system dysfunction among fetuses or infants without evidence of other brain abnormalities or microcephaly (3, 5). The most severe phenotype appears to be associated with exposure during the first trimester (6), although confirmed CZS after exposure during all 3 trimesters has been reported. In fact, nearly half of the infants exposed to Zika in utero manifest abnormalities at birth or have symptoms during the first year of life (4, 7).
The severity and lasting effects of ZIKV infection on the developing fetus and pregnancy outcomes has attracted attention as a research topic. However, it is unknown what role placental dysfunction plays in ZIKV infection outcome. ZIKV RNA and/or antigen have been detected in multiple cell types within the placenta of human, non-human primate (NHP) and mouse models of infection (8, 9). For example, in human placenta from mothers infected with ZIKV, evidence of viral infection has been detected in the chorionic villi, placenta macrophages and histocytes in the intervillous space (9, 10). In a single pregnant pigtail macaque (Macaca nemestrina) infected with ZIKV, several aspects of CZS were characterized along with inflammation at the maternal-fetal interface, including mild decidual perivascular inflammation (not unusual in human decidua) and placental acute chorioamnionitis (11). Our group has shown that ZIKV infection of pregnant rhesus macaques (Macaca mulata), regardless of the severity of fetal effects, results in abnormal oxygen transport within the fetus with a dramatic effect on placental oxygen reserve (8). We hypothesize that these findings may be related to alterations in placental cell immune activation as well as a decrease in the ratio of classical (CD14+CD16-) to non-classical monocytes (CD14-CD16+) and an increase in intermediate monocytes (CD14+CD16+) in both the decidua and villous from ZIKV infected dams compared to uninfected controls (8).
Monocytes are a key immunomodulating cell during pregnancy these cells are critical for placental development and fruition (12). Infection of these cells is likely to alter their antiviral state and alter dissemination of the virus. For example, studies with human samples have shown ZIKV infects macrophages of the placenta inducing production of anti-viral factors including type 1 interferons (IFNs) (13). Beyond the placenta, human CD14+ monocytes are a primary site of ZIKV infection in pregnant and non-pregnant individuals (14). Analysis of human PBMCs infected in vitro, and PBMCs isolated from Zika virus positive Nicaraguan patients, demonstrated that CD14+CD16+ cells are the main targets of infection and this population of cells is expanded during infection (15).
Studies thus far support placenta CD14+ cells as an important site of infection and antiviral response during ZIKV infection. However, the impact of maternal ZIKV infection on CD14+ cell gene expression in the placenta and the effect that expression specific changes have on downstream cellular functions and viral infection is unknown. Viral infection is a dynamic process driven by an interplay between cellular pathways and viral mechanisms. In the current study, we performed single cell RNA sequencing on CD14+ cells isolated at 135 days of gestation (G) from the decidua of animals that were ZIKV infected at either G31, G51, or G115 and the results were compared to pregnant, time-matched uninfected controls. This method provided an unbiased characterization of transcriptional changes in individual CD14+ cells with insights into unique molecular signatures and discovery of specific cell functions. The overall objective of our study was to build upon our previous data from our NHP model of ZIKV infection during pregnancy by taking a targeted approach to determining immune cell characterization and function. Our data reveal the upregulation of multiple gene pathways involved in translation control, cellular stress and growth regulation, inflammation, and innate immunity that we suggest contribute to the antiviral response generated by CD14+ placenta cells following in utero ZIKV exposure.
Animal samples used in this study were collected from previous studies performed in compliance with local and national animal welfare bodies and in strict accordance with Institutional Animal Care and Use Committee (IACUC) protocols. Rhesus macaque studies were performed in a bio-containment facility at the Oregon National Primate Research Center (ONPRC), which is accredited by the Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. In our previous studies the dams and fetuses were humanely euthanized using a method that is consistent with the recommendation of the American Veterinary Medical Association.
Zika virus isolate PRVABC59 received from the Centers for Disease Control (CDC) (16). PRVABC59 was passaged twice on C6/36 cells (ATCC CRL-1660) this working stock was concentrated by ultracentrifugation through a 20% sorbitol cushion, titered on confluent monolayers of Vero cells (ATCC CRL-1586), and sequenced and described previously (17). All cells were cultured in Dulbecco's modified eagle medium (DMEM) containing penicillin-streptomycin-glutamine and 5–10% fetal bovine serum (FBS) and cultured at 37°C.
For our in vitro studies, CD14+ cells were isolated from experimentally infected rhesus macaque placenta and full-term human placenta that were processed as described below.
At the time of necropsy, ~0.5–1g samples of rhesus macaque maternal decidua and fetal villous tissue were carefully dissected to avoid cross contamination (n = 3 infected and uninfected). Decidua and villous samples were collected separately into 5 mL of HBSS supplemented with 2% FBS and 10 mM Hepes (HBSS +). Tissues were dissected into a fine slurry using forceps prior to being digested with 50 mg collagenase in 35 mL HBSS + for 30 min at 37°C with continuous rocking.
De-identified Human full-term placentas were obtained following non-laboring cesarian section (c-section) from OHSU patients (IRB# 15196). Multiple ~0.5–1g samples of full thickness placenta were collected into HBSS+. Tissues were dissected into a fine slurry prior to being digested with 0.25% trypsin and 0.2% DNase in HBSS+ for 30 min at 37°C while rocking. Trypsin digested tissues were washed with HBSS + and further digested with 50 mg collagenase in 35 mL HBSS +, for 1 h at 37°C, with rocking.
Digested rhesus macaque and human placenta were washed with HBSS + and flushed through a 70 μM filter (Falcon). The cells present in the flow through were pelleted by centrifugation at 500 × g. Red blood cells were lysed with 5 mL BD Pharm Lyse lysing buffer (BD Biosciences) for 5 min followed by the addition of 10 mL HBSS+ to stop lysis. After a final wash in HBSS +, cells were resuspended in 5 mL HBSS +. Cells in 5 mL HBSS+ was centrifuged over lymphocyte separation medium (Corning) for 45 min at 3,000 rpm (1,459 x g) to isolate placental mononuclear cells. For magnetic bead isolation using MACS MicroBead Technology (Miltenyi Biotec), mononuclear were resuspended in MACS buffer at a concentration of approximately 1 x 107 cells/mL. Prior to magnetic separation, cells were passed through a 70 μM filter, to remove cell clumps. All cell types were isolated using a magnetic isolation method with species-specific reagents (MACS, Miltenyi Biotec). Approximately 1 x 107 cells were incubated with magnetic beads coated with anti-CD14 (Miltenyi Biotech) for 15 min at 4°C. The cells were then washed and resuspended in 500 μL MACS buffer before being loaded onto a LD magnetic separation column in the presence of a magnetic field. After sample loading the column was washed with 2 mL of MACS buffer. The magnetically labeled CD14+ cells retained on the column were eluted by removing the column from the magnetic field and flushing with 1 mL MACS buffer using the provided plunger. The eluted cells were pelleted and resuspended in RPMI supplemented with 10% FBS and the purity of cells was demonstrated by flow cytometry (see below).
Flow cytometry was used to demonstrate purity of CD14+ isolated cells. 1 x 104 cells were sampled from the 1 mL of cells eluted after magnetic separation (positive fraction) and from the column flow through during separation (negative fraction). Cells were stained with a CD14 specific antibody (HCD14 10 μg/mL Biolegend) and cellular surface staining was quantified using an LSRII instrument (BD Bioscience). Flow cytometry data was analyzed using FlowJo Software (TreeStar).
Rhesus macaque placenta derived CD14+ cells were counted using a hemocytometry, and the cell number was normalized to a concentration of 1,000 cells/μL in chilled media. The cells were loaded into a 10x Genomics Chromium instrument (10x Genomics) and processed using the Single Cell 5′ kit Version 1.1 following the manufacturer's protocol. Generation of gene expression libraries was performed using manufacturer's instructions. To multiplex samples, cell hashing was performed using the MULTI-Seq lipid labeling system (18, 19). Libraries were sequenced using Illumina chemistry on either Novaseq or HiSeq instruments (Illumina). Raw sequence data are available under BioProject PRJNA639805 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA639805.
The resulting sequence data were processed using cell ranger software (10x Genomics, version 4.0). Data were aligned to the MMul_10 genome build (release 98, assembly ID GCA_003339765.3), using Ensembl gene annotations (Build 10.103). The raw count matrix was imported directly into R and analyzed using the Seurat software package, version 4.0.2 (20). Briefly, the EmptyDrops algorithm was used to select droplets containing viable transcriptomes (21), followed by filtering on UMI count (discarding cells >20,000), feature count (discarding cells with <200 or >5,000). Passing cells were merged across samples and normalized using Seurat's “LogNormalize” method. Feature counts are divided by the total counts for that cell, multiplied by a scale factor, and natural-log transformed using log1p. The top 2,000 variable features were selected using Seurat's FindVariableFeatures function, using the “vst” method. Dimensionality reduction was performed with PCA followed by tSNE on the first 12 components. Differential expression was calculated using FindMarkers, using a Wilcoxon Rank Sum test, and then filtered using adjusted p < 0.001 and average log fold-change >0.5. Data were visualized using the heatmap R package (1.0.12). Pathway analysis of the differentially regulated genes was performed using Qiagen's Ingenuity Pathway Analysis (IPA), Gene Ontology (Go) Term analysis, and String analysis.
RNA from in vitro infected placental samples was isolated using TRIzol reagent (Invitrogen) as previously described (8, 17, 22). Isolated RNA was quantified using a Nanodrop spectrophotometer, diluted to a concentration of 100 ng/μl and DNAse treated with ezDNase (Invitrogen). Single-stranded cDNA was generated using 1 μg of total RNA, random hexamers and Superscript IV reverse transcriptase (Invitrogen, Carlsbad, CA). qRT-PCR was performed with Taqman Fast Advanced Master Mix using the following cycling conditions: 95° for 2 min followed by 40 cycles of two steps with the first at 95° for 1 s followed by 60° for 20 s (Applied Biosystems, Foster City, CA). Forward and reverse primers directed against ZIKV and rhesus GAPDH were used at 300 nM in the reaction and the probe at 150 nM (Primer/Probe sequences are listed in Table 1). The results from each assay were analyzed with Applied Biosystem's QuantStudio 7 Flex Real Time PCR System software. Expression was normalized across all samples using the Rhesus macaque housekeeping gene GAPDH. The sensitivity of this qRT-PCR assay is <100 copies (8, 17, 22).
Total rhesus placenta RNA was used to generate cDNA by qRT-PCR using methods described above. The gene for rhesus IFI27 containing an in frame C'terminal HiBiT tag was amplified from cDNA and the product was ligated into pGEM-Teasy in E. coli DH5? (Promega). Individual colonies were screened for inserts and plasmid DNA was digested with restriction digested with EcoRI and HindIII and cloned into pCDNA-3.1(-). Resultant clones were sequenced verified and plasmid DNA was produced using a Qiagen midi-prep kit (Qiagen) and quantified using a Nanodrop spectrophotometer.
An 80% confluent flat bottom 96-well plate of HEK293 cells was transfected with 100 μg/well of the DNA constructs described above using lipofectamine 2,000 (Life Technologies) in replicates of ten. After overnight incubation at 37°C with 5% CO2, the supernatant was removed and fresh media containing ZIKV [multiplicity of infection (MOI) =1] was added. At 72 h post infection (hpi), supernatants were collected in a 96-well plate and stored at −80°C. ZIKV virus in supernatant was quantified using a focus forming assay. Stored samples were thawed and serial dilutions of supernatant were plated in 96-well plates seeded with Vero cells, allowed to adsorb for 1h, followed by overlay with 0.5% carboxymethyl-cellulose (CMC; Sigma). At 30 hpi, cells were fixed with 4% paraformaldehyde, washed twice with PBS and blocked/ permeabilized for 1 h in PBS supplemented with 2% normal goat serum (NGS; Sigma) and 0.4% triton X-100. Cells were then washed twice with PBS followed by incubation with 0.3μg/ml anti-flavivirus monoclonal antibody 4G2 (23) in PBS supplemented with 2% NGS for 1h. The plates were washed twice with PBS and then incubated with anti-mouse IgG-horseradish peroxidase (Santa Cruz Biotech) for 1 h, after which they were washed twice with PBS. ZIKV foci were visualized by incubation with the Vector VIP peroxidase substrate kit (Vector Labs) according to manufacturer's specifications and counted using an ELIspot reader (AID).
Statistical analysis of single cell sequencing data is described in Single-cell RNA-seq and Data Analysis. All other statistical analysis was performed using Prism v6 software (GraphPad Software, Inc). For gene expression experiments, data was analyzed by either a Student's t-test or a two-way ANOVA with secondary analysis to determine significance.
In our previous study, pregnant rhesus macaques (RMs) were infected with ZIKV at G31, G51, or G115 followed by cesarean section delivery at G135, at which time maternal and fetal tissues and the placenta were collected and processed for further analysis (8). Uninfected control RMs from ongoing studies underwent similar delivery and tissue collection at G135. Our previous analysis of these tissues confirmed the presence of persistent ZIKV nucleic acids in the placenta and other tissues of the RM. We also described substantial virus specific changes in the innate and adaptive immune responses within the placenta. Of primary interest to us was the increased activation of CD14+ cells and changes in monocyte subset proportions reported in both the decidua and villous (8). To further understand the role of CD14+ cells in the response to ZIKV infection during pregnancy, single cell sequencing was used to identify unique transcriptional signatures in CD14+ cells isolated from the maternal decidua of RM infected with ZIKV during gestation (Figure 1A). To isolate CD14+ cells, previously collected single cell RM decidua samples were bound to CD14+ labeled magnetic beads and processed through a separation column, resulting in a >90% pure cell population (Figure 1B).
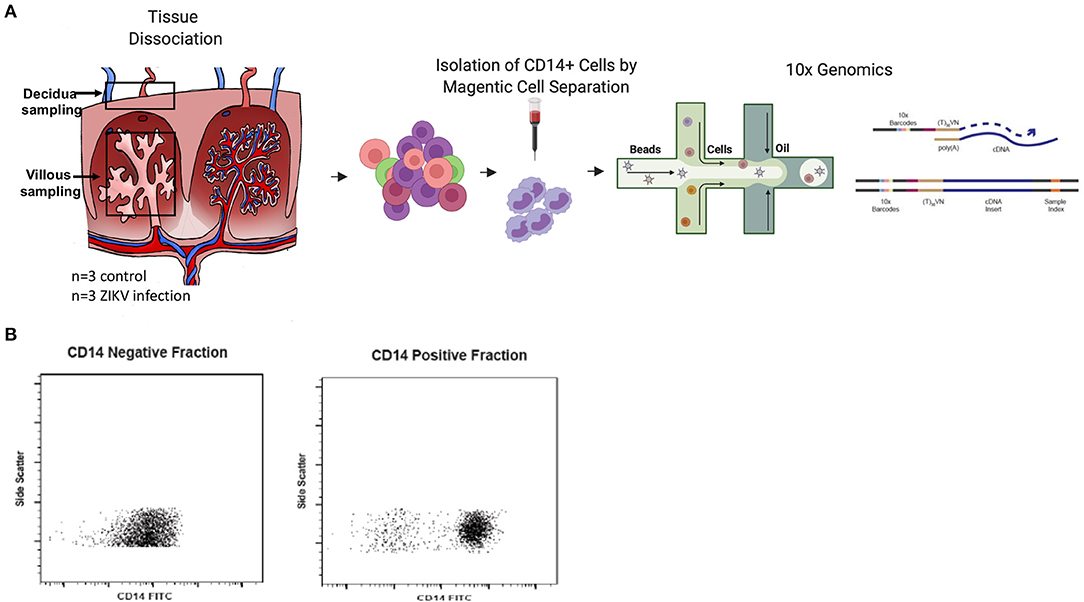
Figure 1. Experimental design. (A) Decidua samples were collected after C-section delivery at G135 of pregnant dams either infected with ZIKV at G31, G51, or G115 or time matched uninfected control dams (n = 3). Tissues were dissociated and processed to single cells. For isolation of CD14+ cells, cells were positively selected using CD14+ microbeads and magnetic cell separation. Isolated cells were loaded into a 10x Genomics Chromium instrument and libraries were sequenced using Illumina chemistry. Resulting sequence data was processed using cell ranger software and further analyzed in the Seurat software package of R. (B) Both the CD14+ positive cells and negative flowthrough cells were analyzed by flow cytometry to determine purity of the CD14+ cell fraction.
To examine the state of placental monocytes/macrophages in ZIKV-infected RM, we isolated CD14+ cells and performed single cell RNA-seq (scRNA-seq). Using single-cell transcriptomes, we first performed unsupervised clustering to determine whether there were particular transcriptional states over-represented in cells from ZIKV-infected RM that subdivided into five phenotypic clusters (Figure 2A). We found clearly distinct phenotypes between the ZIKV and uninfected animals, with relatively homogenous transcriptional phenotypes within each cohort regardless of duration of animal infection (Figures 2B–D). Notably, the majority of CD14+ cells from ZIKV-infected animals formed two clusters (1 and 3, Figure 2C). To determine transcriptional differences between ZIKV-infected and control samples, we identified differentially expressed genes (DEGs) between each cluster (Figure 2E). Cluster 1, which represents 80.8% of cells from ZIKV-infected RM, shows a strong signal of immune activation, including up-regulation of chemokines CCL4L1/MIP-1-B and CCL2, MHC class II (Mamu-DPA), and dual-specificity phosphatase genes (DUSP1/2). This signature also includes ENSMMUG00000054210, an uncharacterized lncRNA. A subset of CD14+ cells from ZIKV-infected animals did not display this immune activation signature; however, these cells showed strong activation of genes involved in oxidative phosphorylation (Figure 2E, Cluster 3). CD14+ cells from ZIKV-infected RM also displayed a near universal down-regulation of a gene module including HMOX1, which is a macrophage modulator associated with anti-inflammatory M2 macrophages (24). Overall, we found expression profiles from infected animals did not significantly overlap with those from uninfected animals suggesting a distinct and sustained difference in transcriptomes between CD14+ cells derived from the decidua of animals infected during gestation.
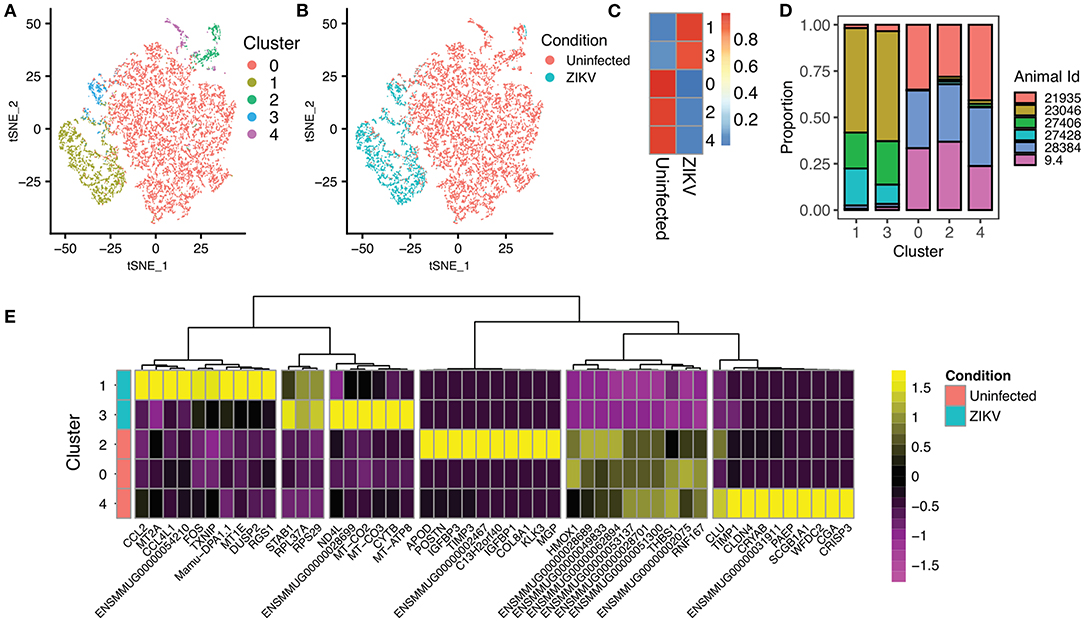
Figure 2. Single-cell RNA sequencing of CD14+ cells from the placenta of ZIKV and uninfected RM. (A) Unsupervised clustering of single-cell transcriptomes from 11,752 CD14+ placental monocytes. Cells are colored based on cluster assignment. (B) Identical dimensionality reduction as (A), with cells colored by the ZIKV of the RM, with cells from ZIKV-infected RM colored blue, and cells from uninfected controls colored red. (C) The heat map illustrates the proportion of cells from ZIKV vs. uninfected RM in each cluster. There is very little overlap between cells from ZIKV-infected and uninfected animals. (D) Contribution of samples to each cluster. (E) Heatmap showing the top differentially expressed genes for each cluster. Clusters 1 and 3 (top) are primarily from ZIKV-infected animals. Clusters 0, 2, 4, and 5 are primarily from uninfected RM.
Gene ontology enrichment analysis (GO) and String: Functional Protein Association Networks online analysis tools were used to identify enriched biological processes and gene nodes within the significantly differentially expressed genes (DEGs) from ZIKV vs uninfected placenta samples (Figures 3A,B). The top enriched processes identified by these two analyses fit into the following broad categories: (1) translation control (e.g., EIF2 signaling; Regulation of eIF4 and p70S6k); (2) stress response (e.g., Mitochondrial dysfunction; HIF1α signaling; Sirtuin signaling); (3) control of cellular growth (e.g., mTOR signaling; HER-2 signaling); (4) inflammatory responses (e.g., Granulocyte adhesion and diapedesis; Antigen processing); (5) signaling pathways mediated through cytokines/chemokines (IL-17 signaling; IL-10 signaling; OX40 signaling); and (6) innate immunity and antiviral responses (e.g., Role of PKR in interferon induction; Communication between innate and adaptive immune cells) (Figure 3B). Ingenuity pathway analysis (IPA) also demonstrated a clear relationship for interferon signaling in placenta derived CD14+ cells with nodes formed around IRF1 and IFNα (Figure 4). Among the genes upregulated in tissues from CD14+ cells of decidua from infected RM were type 1 IFN stimulated genes (ISGs) from the ISG12 family IFI6 and IFI27, which have been recently described as effectors of the IFN response to flaviviruses (25) (Figure 4). To confirm the upregulation of the ISG12 family during ZIKV infection of the placenta, we directly measured expression of IFI27 by qRT-PCR in CD14+ cells isolated from the villous and decidua of RMs infected with ZIKV at various time points during gestation. These samples were from additional, previously published, animal cohorts to bolster sample size (n = 6) (8, 22). IFIT3, a well-studied type 1 IFN ISG, was used as a positive control. CD14+ cells from the decidua or villous of ZIKV infected RM expressed higher levels of IFI27 compared to cells from placentas from uninfected controls (Figures 5A,B). The villous showed higher expression of both IFI27 and IFIT3 compared to the decidua, suggesting a more robust IFN response in villous tissue. These data confirm ZIKV infection during gestation induces IFI27 expression in the decidua and villous. Importantly, we did not see the same dysregulation of IFI27 in peripheral blood mononuclear cells from infected animals compared to uninfected controls indicating a specific and persistent response to infection by cells in the placenta (Figure 5C).
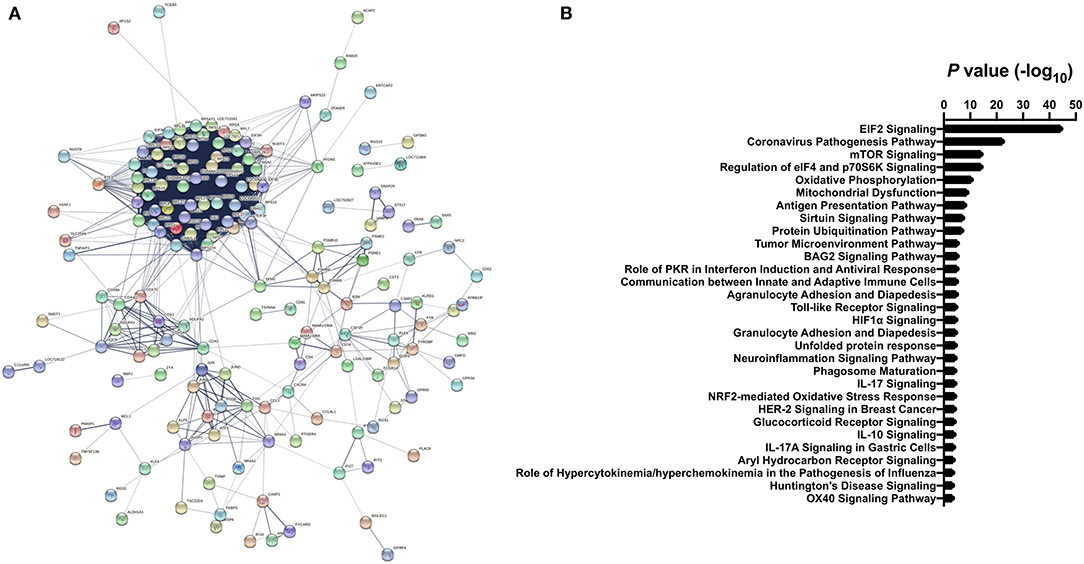
Figure 3. Differential gene expression analysis of CD14+ villous cells (A) STRING analysis of significant differentially expressed genes from ZIKV infected vs. uninfected clusters. Lines with different colors represent the existence of different types of evidence applied to predict associates between proteins. (B) The top Gene Ontology (GO) terms from differentially expressed genes represented as -log10 (P value). Differentially expressed genes between ZIKV infected and uninfected clusters were identified using a Wilcoxon Rank Sum and filtered using adjusted p < 0.001 and average fold-change >0.5.
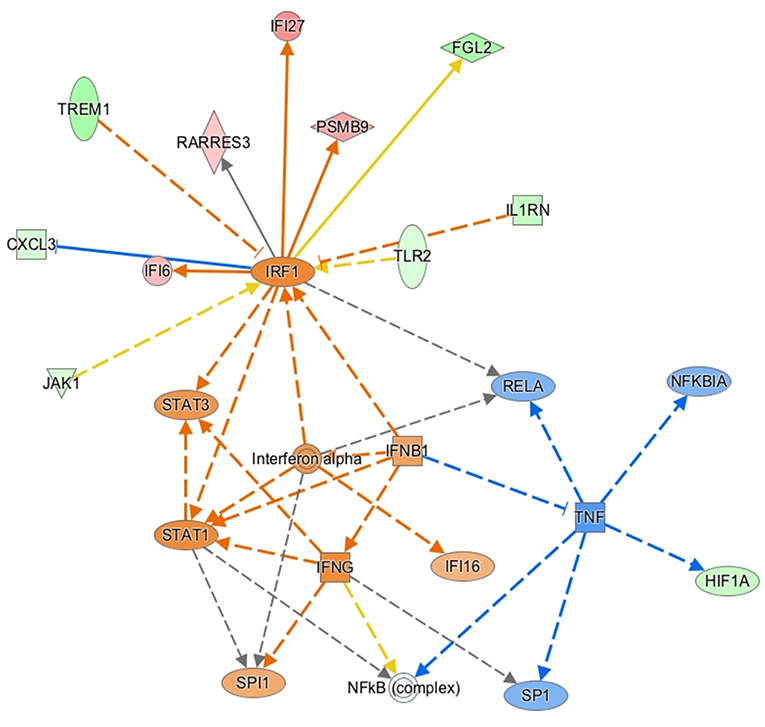
Figure 4. Ingenuity pathway core analysis was used to identify enrichment in biological processes (ZIKV Infected vs. Uninfected). Statistically significant alterations in differentially expressed genes from SC-RNAseq analysis of decidua CD14+ cells are represented by labeled shapes depending on their molecular function. Expression and predicted activation levels determined by fold-change between ZIKV infected vs. uninfected are presented using the following convention on color scales with increasing saturation associated with a greater absolute fold-change value: red = increased detection in CD14+ cells from ZIKV infected decidua; green = decreased detection; orange = predicted activation. Predicted relationships are indicated by arrow-heads and blue lines to indicate predicted inhibition; yellow lines indicate inconsistent findings that make it difficult to predict outcomes of interactions.
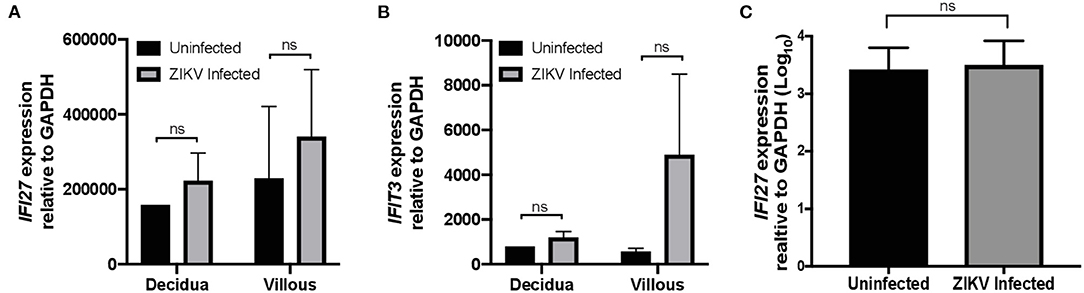
Figure 5. IFI27 is highly expressed in CD14+ decidua and villous cells after ZIKV infection. Validation of increased expression of IFI27 in CD14+ decidua and villous cells collected from pregnant dams infected with ZIKV and uninfected controls from multiple study cohorts (n = 6 per condition). RNA was isolated and gene expression analyzed by qRT-PCR of (A) IFI27 and (B) control IFIT3 relative to GAPDH. (C) RNA was isolate from the PBMCs of dams infected with ZIKV and uninfected controls from multiple study cohorts (n = 12 per condition) and analyzed for IFI27 gene expression using GAPDH to normalize the data across samples. Student's t-test was used to determine statistical significance of expression data obtained from infected and uninfected animals.
To validate ZIKV infection-induced expression of IFI27 in placental monocyte/macrophages, we explored gene expression in ZIKV naïve cells from RM and human placenta that we infected with ZIKVPRABC in vitro. For this experiment, CD14+ cells were isolated from the decidua and villous of control RM using the methods described above. In the human placenta, the division between decidua and villous is not as distinct and consequently is difficult to definitively isolate these cell types, therefore whole thickness samples from human placenta were digested followed by CD14+ cell isolation using antibody-conjugated magnetic beads. Isolated CD14+ cells were infected with ZIKVPRABC in vitro at 0.5 or 5 FFU/cell. Cells were collected into Trizol reagent at 48hpi or 72hpi, RNA was isolated gene expression analyzed by qRT-PCR of ZIKV, IFI27, and IFIT3. CD14+ cells from both RM and human placenta tissues were readily infected with ZIKV (Figures 6A, 7A). ZIKV infection resulted in a MOI dependent increase in the level of IFI27 and IFIT3 transcripts in both RM and human CD14+ placenta cells (Figures 6B,C and 7B,C). These results confirm the ability of ZIKV infection to induce IFI27 and IFIT3 as well as the transferability of the model to a human infection scenario.
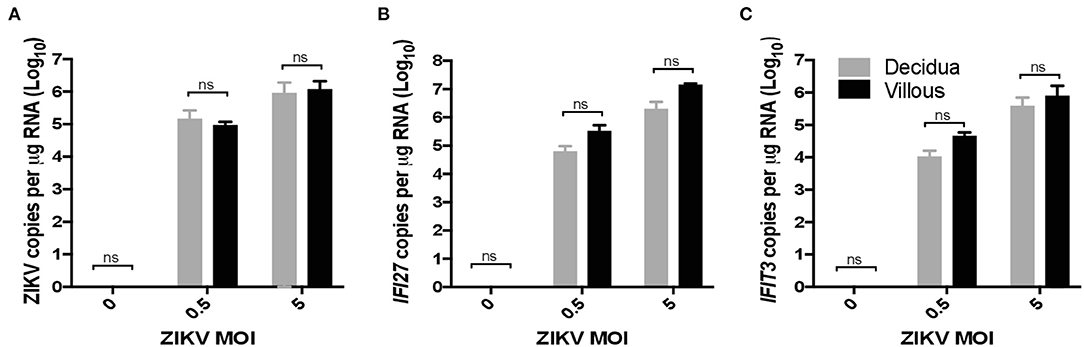
Figure 6. ZIKV infection in vitro induces IFI27 expression in rhesus macaque decidua and villous. CD14+ cells were isolated from the decidua and villous of uninfected control rhesus macaque and infected with ZIKV (MOIs = 0.5 and 5 FFU/cells) at 72hpi supernatants were collected and analyzed by qRT-PCR for expression of (A) ZIKV (B) IFI27 (C) IFIT3 using species specific primers. Student's t-test was used to determine statistical significance.
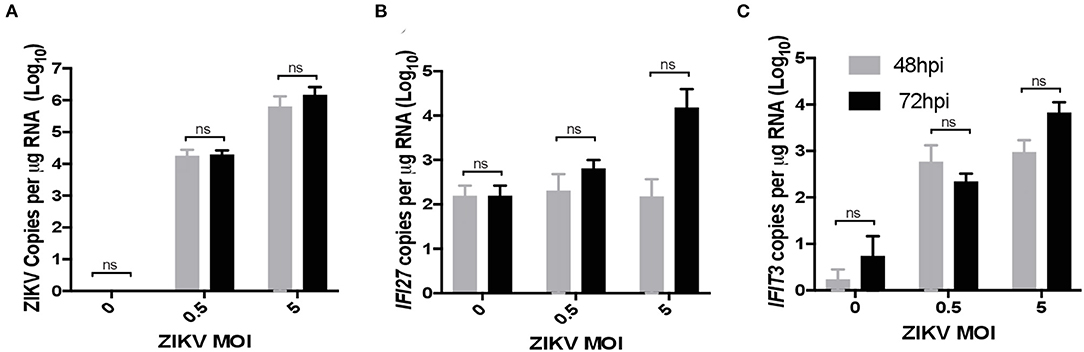
Figure 7. ZIKV infection in vitro induces IFI27 expression in human placenta. CD14+ cells were isolated from de-identified human full-term placentas and infected with ZIKV (MOIs = 0.5 and 5 FFU/cells) at 48 and 72 hpi supernatants were collected and analyzed by qRT-PCR for expression of (A) ZIKV (B) IFI27 (C) IFIT3. A two-way ANOVA with secondary testing was used to determine statistical significance for these data.
To confirm the role of IFI27 as an antiviral factor against ZIKV, we constructed a plasmid expressing rhesus IFI27 to allow exogenous expression of the ISG in ZIKV-infected cells. IFI27 was transfected into 293 cells and expression from the plasmid was confirmed by western blot (Figure 8A). HEK293 cells were transfected with an IFI27 expressing or control plasmid one day prior to infection with ZIKV. Supernatants were collected 3dpi and infectious virus was quantified by focus forming assay. In this system, exogenous expression of IFI27 significantly decreased the production of infectious virus by nearly a log (Figure 8B), confirming that IFI27 plays a role as an antiviral factor against ZIKV.
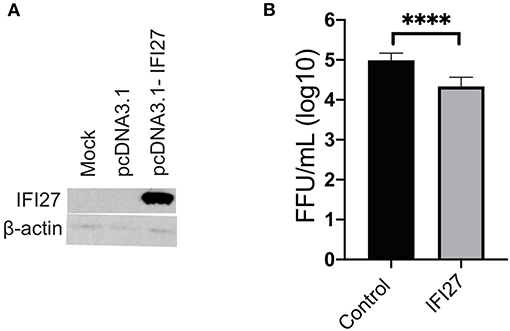
Figure 8. IFI27 overexpression reduces ZIKV replication in vitro. (A) To confirm IFI27 protein expression 100 μg of pcDNA3.1 or pcDNA3.1-IFI27 DNA was transfected into HEK293 cells. At 7 h post-transfection cell lysates were analyzed for IFI27 or actin by western blotting. (B) HEK293 cells were transfected with 100 μg of a control vector constitutively expressing GFP or pcDNA3.1-IFI27 24 h post-transfection cells were infected with ZIKV (MOI = 1). At 72 hpi supernatants were collected and virus quantified by focus forming assay. P values were determined by paired student's t-test ****p < 0.0001, all other comparison were not significant.
The rhesus macaque model of in utero ZIKV infection recapitulates many of the hallmarks of the human disease caused by this neurotropic flavivirus including fetal neuropathy and demise. We previously showed that ZIKV infection in Rhesus macaques during pregnancy induces placental damage associated with increased vascular damage that leads to altered nutrient perfusion across placental membranes, as well as increased inflammation and remodeling (8). Placental monocytes and macrophages (especially of the classical lineage) are key cells for normal placental vascular development and actively patrol the placenta for pathogenic invaders as the first line of defense. We previously reported dramatic alterations in the activation profiles for both monocyte/macrophages and CD4+ T cells that were consistent with ongoing inflammatory processes with possible correlations with ZIKV persistence. This long-term cellular activation was not observed for either dendritic cell or CD8+ T cell populations indicating that there is a unique effect of ZIKV infection on placenta immunopathology. Since macrophages are a key cell type in placental immunobiology and development, we interrogated the changes to the placenta macrophage population following infection. Herein, we utilized a single cell sequencing approach to further investigate the CD14+ monocyte/macrophage cell population to identify transcriptional signatures in cells isolated from the maternal decidua of ZIKV infected animals. Our objective was to identify mechanistic pathways that are dysregulated in the placenta following ZIKV infection. Our study findings reveal significantly altered gene expression in placental macrophages isolated from ZIKV-infected animals and demonstrate the sustained upregulation of macrophage functions even in the absence of positive viral detection indicating a prolonged response to viral exposure. This may be an adaptive response to put up an immune shield to prevent further infection or disease in the placenta and to limit the adverse impacts on fetal growth and development.
Specifically, in the samples analyzed in this study, the DEGs in CD14+ villous cells of ZIKV infected RM were strongly associated with eIF2 signaling, eIF4 signaling, mTOR signaling, oxidative phosphorylation, and mitochondria dysfunction. eIF2, EIF4, and mTOR are regulators of protein translation and the cellular stress unfolded protein response (UPR). These pathways are tightly interrelated with inflammatory pathways leading to production of proinflammatory molecules (26). The mitochondrion also plays a role in regulating the antiviral immune response, a function that viruses can regulate to evade the immune response, leading to host cell damage through changes in ROS and cell metabolism (27–29). Altered bioenergetics and mitochondrial dysfunction of peripheral blood monocytes have been described for other viral infections in connection to changes in CD14/CD16 monocyte population distribution (30).
Our results are in accordance with a recent a study by Lum et al. using placenta from pregnant women infected with ZIKV during the first, second, or third trimester. Only ZIKV infection during the first trimester resulted in a gene expression profile that was different from healthy controls, and the resulting DEGs were associated with eIF2 signaling, oxidative phosphorylation, and mitochondrial dysfunction, which is very consistent with DEG found in our study. Additionally, there was a distinct relationship between the degree of infection and level of CD14+ monocytes in the patients (31). While in our study we did not see an effect of infection gestational age on the transcriptional program in CD14+ cells, this could be a result of the fact that we looked at this cell type specifically at only G135 or possibly due to the RM model or ZIKV virus strain used for infection. The similar DEGs between the studies do suggest the importance of these conserved macrophage-centric pathways in ZIKV infection on the immunopathology of the placenta. Our findings implicate that CD14+ cells play an important role in regulating the antiviral response through multiple pathways following infection with ZIKV. Notably flaviviruses possess multiple mechanisms to modulate aspects of innate immunity that may affect infection of cells including CD14+ monocytes (32–38). Future experiments will focus on understanding the role of these interactions in placental pathology and effects on outcomes of ZIKV infection during pregnancy.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: National Center for Biotechnology Information (NCBI) BioProject, https://www.ncbi.nlm.nih.gov/bioproject, PRJNA639805.
The animal study was reviewed and approved by OHSU Institutional Animal Care and Use Committee.
NH, DS, AH, and VR: designed, coordinated, and implemented NHP experiments. NH, DS, AH, VR, BB, and AF: reviewed the data, aided in experiment designed, and drafted and edited manuscript. AH and JS: provided and prepared viral stocks and performed plaque assays. NH and HS: performed rhesus macaque and human cell isolation. BB and KO: performed 10x genomics statistical analysis. NK, MD, HS, and CK: performed cloning and gene expression quantification. All authors contributed to the article and approved the submitted version.
This work was supported by National Institutes of Health P51 ODO11092 (Haigwood), National Institutes of Health R01 HD096741 (Streblow).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Paixao ES, Barreto F, Teixeira Mda G, Costa Mda C, Rodrigues LC. history, epidemiology, and clinical manifestations of Zika: A systematic review. Am J Public Health. (2016) 106:606–12. doi: 10.2105/AJPH.2016.303112
2. Brasil P, Sequeira PC, Freitas AD, Zogbi HE, Calvet GA, De Souza RV, et al. Guillain-Barre syndrome associated with Zika virus infection. Lancet. (2016) 387:1482. doi: 10.1016/S0140-6736(16)30058-7
3. Reynolds MR, Jones AM, Petersen EE, Lee EH, Rice ME, Bingham A, et al. Vital signs: update on Zika virus-associated birth defects and evaluation of all U.S. infants with congenital Zika virus exposure - U.S. Zika pregnancy registry, 2016. MMWR Morb Mortal Wkly Rep. (2017). 66:366–73. doi: 10.15585/mmwr.mm6613e1
4. Brasil P, Nielsen-Saines K. More pieces to the microcephaly-Zika virus puzzle in Brazil. Lancet Infect Dis. (2016) 16:1307–9. doi: 10.1016/S1473-3099(16)30372-3
5. Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, et al. Birth defects among fetuses and infants of US women with evidence of possible zika virus infection during pregnancy. JAMA. (2017) 317:59–68. doi: 10.1001/jama.2016.19006
6. Molnar Z, Kennedy S. Neurodevelopmental disorders: Risks of Zika virus during the first trimester of pregnancy. Nat Rev Neurol. (2016) 12:315–6. doi: 10.1038/nrneurol.2016.71
7. Soares De Souza A, Moraes Dias C, Braga FD, Terzian AC, Estofolete CF, Oliani AH, et al. Fetal infection by Zika virus in the third trimester: report of 2 cases. Clin Infect Dis. (2016) 63:1622–5. doi: 10.1093/cid/ciw613
8. Hirsch AJ, Roberts VHJ, Grigsby PL, Haese N, Schabel MC, Wang X, et al. Zika virus infection in pregnant rhesus macaques causes placental dysfunction and immunopathology. Nat Commun. (2018) 9:263. doi: 10.1038/s41467-017-02499-9
9. Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, et al. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell. (2016) 165:1081–91. doi: 10.1016/j.cell.2016.05.008
10. Noronha L, Zanluca C, Azevedo ML, Luz KG, Santos CN. Zika virus damages the human placental barrier and presents marked fetal neurotropism. Mem Inst Oswaldo Cruz. (2016) 111:287–93. doi: 10.1590/0074-02760160085
11. Nguyen SM, Antony KM, Dudley DM, Kohn S, Simmons HA, Wolfe B, et al. Highly efficient maternal-fetal Zika virus transmission in pregnant rhesus macaques. PLoS Pathog. (2017) 13:e1006378. doi: 10.1371/journal.ppat.1006378
12. Faas MM, Spaans F, De Vos P. Monocytes and macrophages in pregnancy and pre-eclampsia. Front Immunol. (2014) 5:298. doi: 10.3389/fimmu.2014.00298
13. Quicke KM, Bowen JR, Johnson EL, Mcdonald CE, Ma H, O'Neal JT, et al. Zika virus infects human placental macrophages. Cell Host Microbe. (2016) 20:83–90. doi: 10.1016/j.chom.2016.05.015
14. Foo SS, Chen W, Chan Y, Bowman JW, Chang LC, Choi Y, et al. Asian Zika virus strains target CD14(+) blood monocytes and induce M2-skewed immunosuppression during pregnancy. Nat Microbiol. (2017) 2:1558–70. doi: 10.1038/s41564-017-0016-3
15. Michlmayr D, Andrade P, Gonzalez K, Balmaseda A, Harris E. CD14(+)CD16(+) monocytes are the main target of Zika virus infection in peripheral blood mononuclear cells in a paediatric study in Nicaragua. Nat Microbiol. (2017) 2:1462–70. doi: 10.1038/s41564-017-0035-0
16. Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor Ldel C. Phylogeny of zika virus in western hemisphere, 2015. Emerg Infect Dis. (2016) 22:933–5. doi: 10.3201/eid2205.160065
17. Hirsch AJ, Smith JL, Haese NN, Broeckel RM, Parkins CJ, Kreklywich C, et al. Zika Virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLoS Pathog. (2017) 13:e1006219. doi: 10.1371/journal.ppat.1006219
18. Mcginnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, et al. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods. (2019) 16:619–26. doi: 10.1038/s41592-019-0433-8
19. Boggy G, Bimber BN. cellhashR: An R package designed to demultiplex cell hashing data. (2021). Available online at: https://github.com/bimberlab/cellhashr
20. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. (2018) 36:411–20. doi: 10.1038/nbt.4096
21. Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, participants in the 1st human cell, Atlas J, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. (2019). 20:63. doi: 10.1186/s13059-019-1662-y
22. Steinbach RJ, Haese NN, Smith JL, Colgin LMA, Macallister RP, Greene JM, et al. A neonatal nonhuman primate model of gestational Zika virus infection with evidence of microencephaly, seizures and cardiomyopathy. PLoS ONE. (2020) 15:e0227676. doi: 10.1371/journal.pone.0227676
23. Henchal EA, Gentry MK, Mccown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. (1982) 31:830–6. doi: 10.4269/ajtmh.1982.31.830
24. Naito Y, Takagi T, Higashimura Y. Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch Biochem Biophys. (2014) 564:83–8. doi: 10.1016/j.abb.2014.09.005
25. Lucas TM, Richner JM, Diamond MS. The interferon-stimulated gene Ifi27l2a restricts west nile virus infection and pathogenesis in a cell-type- and region-specific manner. J Virol. (2015) 90:2600–15. doi: 10.1128/JVI.02463-15
26. Kolattukudy PE, Niu J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ Res. (2012) 110:174–89. doi: 10.1161/CIRCRESAHA.111.243212
27. Jin HS, Suh HW, Kim SJ, Jo EK. Mitochondrial control of innate immunity and inflammation. Immune Netw. (2017) 17:77–88. doi: 10.4110/in.2017.17.2.77
28. Zhang L, Qin Y, Chen M. Viral strategies for triggering and manipulating mitophagy. Autophagy. (2018) 14:1665–73. doi: 10.1080/15548627.2018.1466014
29. Zuo H, Wan Y. Metabolic reprogramming in mitochondria of myeloid cells. Cells. (2019) 9. doi: 10.3390/cells9010005
30. Gibellini L, De Biasi S, Paolini A, Borella R, Boraldi F, Mattioli, et al. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia. EMBO Mol Med. (2020) 12:e13001. doi: 10.15252/emmm.202013001
31. Lum FM, Narang V, Hue S, Chen J, Mcgovern N, Rajarethinam R, et al. Immunological observations and transcriptomic analysis of trimester-specific full-term placentas from three Zika virus-infected women. Clin Transl Immunology. (2019) 8:e01082. doi: 10.1002/cti2.1082
32. Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. (2016) 19:882–90. doi: 10.1016/j.chom.2016.05.009
33. Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. (2016) 17:1766–75. doi: 10.15252/embr.201642627
34. Chan YK, Gack MU. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol. (2016) 17:523–30. doi: 10.1038/ni.3393
35. Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, et al. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc Natl Acad Sci U S A. (2018) 115:E6310–8. doi: 10.1073/pnas.1803406115
36. Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. (2005) 437:1167–72. doi: 10.1038/nature04193
37. Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol. (2017) 2:17037. doi: 10.1038/nmicrobiol.2017.37
Keywords: Zika virus, pregnancy, monocyte, macrophage, interferon
Citation: Haese NN, Smith H, Onwuzu K, Kreklywich CN, Smith JL, Denton M, Kreklywich N, Streblow AD, Frias AE, Morgan TK, Hirsch AJ, Bimber BN, Roberts VHJ and Streblow DN (2021) Differential Type 1 IFN Gene Expression in CD14+ Placenta Cells Elicited by Zika Virus Infection During Pregnancy. Front. Virol. 1:783407. doi: 10.3389/fviro.2021.783407
Received: 26 September 2021; Accepted: 08 November 2021;
Published: 29 November 2021.
Edited by:
Kristina M. Adams Waldorf, University of Washington, United StatesReviewed by:
Yoel Sadovsky, Magee-Womens Research Institute, United StatesCopyright © 2021 Haese, Smith, Onwuzu, Kreklywich, Smith, Denton, Kreklywich, Streblow, Frias, Morgan, Hirsch, Bimber, Roberts and Streblow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel N. Streblow, c3RyZWJsb3dAb2hzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.