- 1Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
- 2Faculty of Medicine and Health Sciences, University of Nottingham, Nottingham, United Kingdom
- 3School of Science and Technology, Nottingham Trent University, Nottingham, United Kingdom
- 4Department of Clinical Microbiology, Kings Mill Hospital, Sutton-in-Ashfield, United Kingdom
During the COVID-19 pandemic, countries with robust population-based asymptomatic testing were generally successful in controlling virus spread, hence reducing hospitalizations and deaths. This effectiveness inspired widespread asymptomatic surveillance for COVID-19/SARS-CoV-2 globally. Polarized vaccination programs, coupled with the relatively short-lived immunity vaccines provide, mean that reciprocal cross-border exchanges of each new variant are likely, as evidenced by Delta and Gamma, and asymptomatic testing will be required for the foreseeable future. Reliance on nasopharyngeal swabs contributes to “testing fatigue” arising due to difficulties in standardizing administration, unpleasantness, and inappropriateness of use in younger people or individuals with special needs. There has also been erosion in confidence of testing due to variable and/or poor accuracy of lateral flow devices to detect COVID-19. Here, we question why saliva-based PCR assays are not being used more widely, given that standardization is easy and this non-invasive test is suitable for everyone, providing high sensitivity and accuracy. We reflect on our experience with the University of Nottingham COVID-19 Asymptomatic Testing, where (as of October 2021) 96,317 samples have been processed by RT-qPCR from 23,740 repeat saliva donors, yielding 465 positive cases. We challenge myths that saliva is difficult to process, concluding that it is an undervalued resource for both asymptomatic and symptomatic detection of SARS-CoV-2 genomes to an accuracy of >99% and a sensitivity of 1–10 viral copies/μl. In July 2021, our data enabled Nottingham to become the first UK University to gain accreditation and the first UK institute to gain this accolade for saliva.
Introduction
Since the first reports of SARS-CoV-2 infections in late 2019, there has been an emerging acceptance of the need to co-exist with the virus in our communities. Outbreak control will be critical, requiring large-scale testing for the foreseeable future, well beyond mass vaccination programs. General population surveillance provides valuable real-time data on infection rates, spread, and demographics (1). The importance of monitoring new variants, prevalence, and capacity for evasion of immunity is highlighted by Delta and Gamma variants (2), and now Delta+. By August 2021, 70% of the 3 billion vaccines produced were delivered in just 10 countries vs. 1% in the developing world. New variants will emerge from what the World Health Organization has dubbed a “two-tier pandemic”, hence perpetuating cycles of reinfection1.
For SARS-CoV-2 detection, public testing schemes typically rely on nasopharyngeal swabs for lateral flow or polymerase chain reaction tests (LFTs or PCR, respectively). With high specificity and sensitivity (circa 95–99%), PCR approaches are the mainstay of COVID-19 tests, employing extraction of RNA followed by reverse transcriptase quantitative PCR (RT-qPCR) or loop mediated isothermal amplification (LAMP). LFTs detect epitopes in the viral spike protein, giving sensitivities of 5%−70% relative to RT-qPCR detection of the SARS-CoV-2 genome (3, 4). Detection limit of LFTs is circa 100 viral copies/μl (5), 10- to 100-fold less sensitive than PCR approaches.
Discussion continues on the relative merits of each approach regarding cost, labor, route of deployment, and result turnaround. However, a common issue is the use of nasopharyngeal swabs (6). A pervasive error exists in the failure to reach the correct nasopharynx target site, even when performed by trained medical experts (7, 8). Inexperienced or self-administered operators have LFT sensitivities of sub-50%, with uncertainty on how far “up” or “back” the swab should go or at what “angle” and for “how long”. At best, this causes discomfort because the swab is wedged against the middle turbinate (7), dissuading regular repeat testing. At worst, false-negative results lead to relaxed behaviors that amplify virus transmission. Indeed, numerous people with overt COVID-19 symptoms report that they have tested negative via multiple LFTs over consecutive days but positive by RT-qPCR approaches.
Overlooked Benefits of Saliva for SARS-COV-2 Detection
The purpose of this Perspective is to prompt discussion and highlight saliva-based direct RT-qPCR detection of SARS-CoV-2 as an alternative method, thus far overlooked for mainstream testing. Saliva samples avoid the issues of invasive, qualitative nasopharyngeal swabs by easy provision, less variability and more reliability because volumes of 100 μl are acceptable. Anecdotally, samples provided in the morning before eating food, brushing teeth, or using oral hygiene products, such as mouth wash, provide high-quality samples. Our Asymptomatic Testing Service2. (University ethics approval committee approval FMHS 96-0920) is currently evaluating if simplified collection routes benefit sensitive communities, such as special education, dementia patients, homeless hostels, or victims of sexual abuse, where any perceived penetration is unwelcome.
Other benefits of direct saliva-based RT-qPCR detection are as follows: (i) Reduced risk of infection to the staff conducting the sampling, since self-harvesting is easy. (ii) Sample stability; at least 20 days at 4°C without viral transport medium (see Figure 1). (iii) No RNA-extraction step, decreasing testing time and cost. (iv) Possibility of sample pooling to increase throughput and reduce cost. (v) Not competing with other diagnostic schemes, hence increasing capacity without additional demand on manufacturers. (vi) Fewer components, reducing supply chain issues, the importance of which was highlighted during the worldwide shortage of nasopharyngeal swabs (9). (vii) High specificity/sensitivity comparable with nasopharyngeal swab-based detection (3, 6, 10–12).
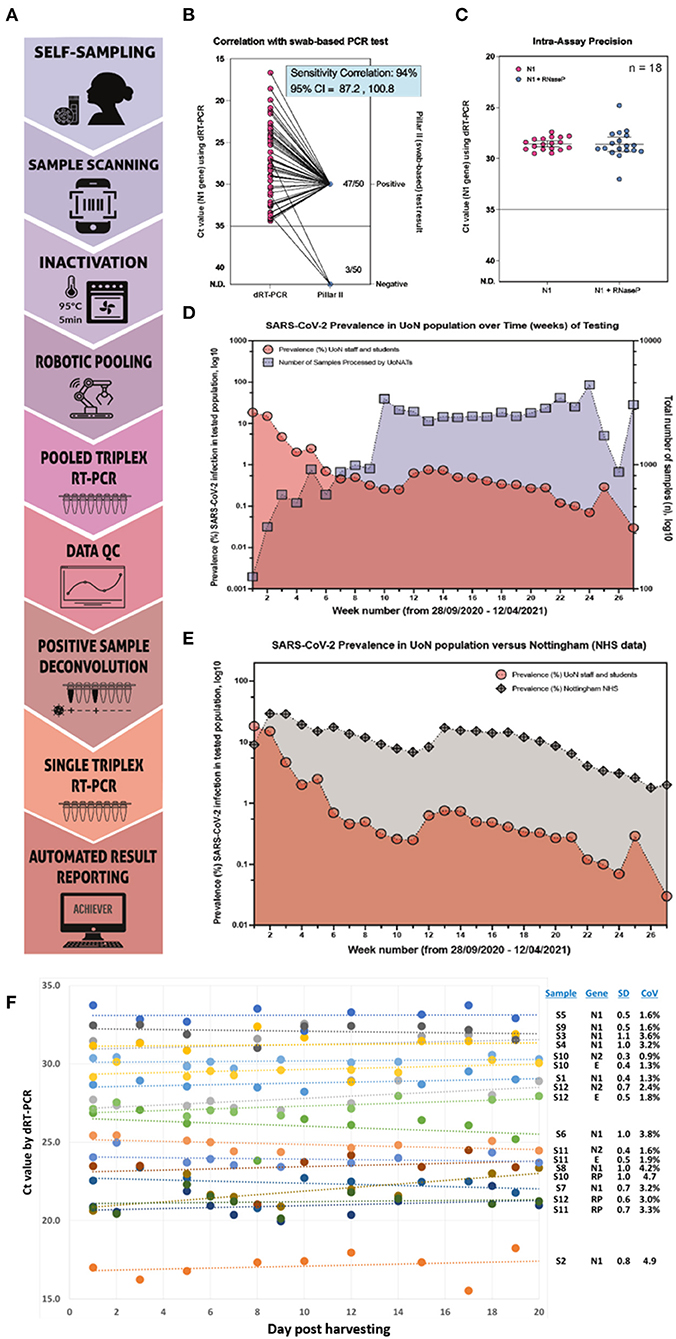
Figure 1. Saliva-based RT-qPCR detection of SARS-CoV-2 is a reliable diagnostic method. (A) A Stepwise testing process: (i) Self-sampling of saliva into a barcoded vial; (ii) Donor scans barcode using their personal devices (e.g., smartphone) and enter information to secure database; (iii) Batch inactivation and lysis for 5 min at 95°C in an oven; (iv) Robot-assisted two-way pooling into 96- or 384-well plates; (v) Direct RT-qPCR to target genes; (vi) Data analysis; (vii) Deconvolution of positive samples to allow (viii) re-testing of original saliva as single samples in a new RT-qPCR cycle; (ix) Automated reporting via Achiever Medical LIMS, Interactive software. (B) Correlation of positive results from our saliva test vs. a UK government-approved swab-based testing service known as a “Pillar II” test. Of 50 patients identified as positive using our testing method, 94% tested positive via Pillar II. (C) Assay reproducibility shown as the plotted Ct values for a positive sample tested repeatedly over 14 days using four different qPCR machines by four different operators (n = 18). (D) Plotted prevalence (% of detected positive samples over total samples tested) over time (weeks, red circles) against the number of total samples processed each week (blue squares). (E) Plotted prevalence (% of detected positive samples over total samples tested) in our cohort over time (weeks, red circles) and the prevalence (%) of positive samples detected by the local public healthcare system (gray diamonds). The data for the prevalence (%) of positive SARS-CoV-2 diagnostic tests were sourced from a UK government site. (F) Stability of saliva was evaluated via regular repeat analysis of 12 samples stored at 4°C for up to 20 days via RT-qPCR amplifying viral genes N1 (n = 9), N2 (n = 3), or E (n = 3), or the internal control human gene, RP (RNAseP; n = 3). High stability was observed over the time course, with all standard deviations (SD) being ≤1.1 and coefficients of variation (CoV) being <5%. Dataset consistency was confirmed using Kolmogorov–Smirnov normality test (normality test passed, p < 0.05).
Underscoring these benefits, from November 2020 to February 2021, the University of Nottingham provided free access to undergraduate students (typically 18–22-year-olds) of LFTs using nasopharyngeal swabs (provided by the UK Government) and RT-qPCR testing using saliva collection aids (provided by our Service). Circa 20,000 tests were completed, with evidence citing ease, reliability, and confidence in the result for why there was a preference toward saliva tests (13).
Why saliva is underutilized is unclear, though it may be due to historical reasons and paucity of data in the literature on accuracy. Indeed, nearly a year into the pandemic (late 2020), the Infectious Diseases Society of America guidelines stated that “saliva as the sole sample source for COVID-19 diagnosis cannot be recommended due to a paucity of studies” (14). However, other studies have shown high sensitivity of SARS-CoV-2 detection using saliva, wherein there was a higher correlation with care worker-collected nasopharyngeal swabs than self-sampled anterior nasal swabs (15–17). Another factor is the small sample size and unclear saliva sampling methodology, no doubt being an underlying cause of the conflict on the level of correlation with nasopharyngeal swab3.
Within our own service, the issues associated with saliva as a diagnostic sample are modest relative to benefits. For example, while we overcame potential background fluorescence issues by using double-quenched probes from IDT (internal ZEN quencher at 9 base pairs from 5' end; IowaBlack Quencher at 3' end), the level of complementarity between the N1 and E primer/probe sets (and/or amplicons) caused aberrant amplification curves, which could only be overcome switching to a combination of N2 and E. We have also found that sequencing the viral genome from saliva can be a challenge, possibly due to fragmented viral RNA.
Impurities and inhibitors within saliva may be problematic but can be overcome if processed in conjunction with compatible RT-qPCR reaction mixes, as explained below. It is true that pooling presents the greatest challenge to sensitivity due to a skewed ratio of impurities vs. viral genomes per unit volume. While we have found the limit of detection in single saliva samples is <1 viral genome/μl, the maximum sensitivity in eight samples combined in two-way pools reduced to 4 viral genomes/μl. Impact on samples of samples with medium to low viral loads is negligible, but when Ct values exceed 30, the error rate of detection in pooled samples reduces accuracy to <99%. Although this level is required by UK regulatory agencies for diagnosis of SARS-CoV-2, saliva is an attractive sample, especially when mass surveillance is needed during easing of social distancing and travel bans.
An Evidence-Based Pipeline for SARS-COV-2 Detection Using Saliva
The simple, streamlined pipeline we use is in Figure 1A, while Figures 1B–E show data generated between September 2020 and July 2021 from regular testing among University of Nottingham staff, students, and support services. At time of writing (October 2021), circa 96,317 samples were processed from 23,740 unique donors, yielding 465 positive test results. Key aspects are as follows.
Harvesting
Donors are provided with a Ziploc bag containing a collection tube (dual linear and QR bar codes; Brooks Ltd, product [65-7643]), tissue, paper straw (cut to lengths of ~5 cm; purchased via Amazon from IntrinsicPaperStraws.com, item Black 6 × 140 mm), and a stepwise guide4. To avoid exogenous contaminants, donors are requested to provide a saliva sample in the morning before they have eaten, brushed their teeth, or used oral hygiene products, consistent with the guidelines from the European Center for Disease Prevention and Control (ECDC) (see footnote 3).
Inactivation
Saliva samples are oven-baked to a target temperature of 95°C/5 min to inactivate and lyse virus, hence simplifying safety procedures and bypassing the need for toxic chemicals and/or RNA extraction. While, at least in our experience, heating causes swabs and/or viral transport medium to become more viscous, with saliva, the effect is to increase sample fluidity. This is critical, yet often overlooked because of compatibility with downstream liquid handling processes.
Pooling, Then RT-qPCR
When prevalence of infection is low (<6%), samples are configured into two-way pools of six to eight samples per pool for one-step RT-qPCR with Center for Disease Control (CDC) primer-probes for the N and/or E genes. Critical points are as follows: (i) Quantabio UltraPlex 1-Step ToughMix RT-qPCR Reagents, designed for use with samples containing high levels of potential inhibitory factors. (ii) Positive pooled samples are deconvoluted and confirmed via single, unpooled tests. If prevalence exceeds 6%, the complexity of deconvolution becomes prohibitive and the process pipeline defaults to single, non-pooled testing.
In support of saliva in surveillance and diagnosis of SARS-CoV-2 infection, we provide illustrative data from the University of Nottingham Asymptomatic Testing Service. Figure 1B shows that of samples identified as positive in our assay, 94% agreement correlation (95% CI 87.2–100.8) existed with a hospital-accredited swab-based qPCR service. The 6% differential might be explained by saliva being a more consistent sample to harvest, as explored above. Also, 1–5 days elapsed between positive saliva result and the swab provision; hence, viral load may have reduced.
Saliva tests showed high intra-assay precision after repeat testing of the same positive samples over 14 days between four different operators and four different qPCR machines (Figure 1C). The high concordance required to satisfy the regulators and achieve accreditation (see below) suggests that heat inactivation has little or no negative effect on sensitivity and, anecdotally, may increase sensitivity in some cases. This is possibly because there is no loss of viral RNA, which occurs to varying degrees when using extraction procedures. In Figures 1D,E, an increased number of samples were tested from people on campus associates with reducing prevalence rates in staff and students (1D) and with 10–100-fold lower infection rates than the surrounding geographical area of Nottingham (1E, data from UK government). These data suggest that early detection is breaking transmission chains, even in high-population zones such as student halls of residence.
Discussion
Combined with the few data available in the literature, our work on SARS-CoV-2 RT-qPCR detection points to saliva as an undervalued resource. Via this Perspective, we seek to promote discussion around the potential for a missed opportunity to achieve COVID-19 surveillance and outbreak control. The perception that saliva is difficult to work with can be overcome by simple modifications, such as by heating and using one-step inhibitor-resistant RT-qPCR. To assist with appropriate harvesting approaches, the European Center for Disease Prevention and Control (ECDC) recently published (17) a technical report “Considerations for the use of saliva as sample material for COVID-19 testing”. Although concluding that saliva sample collection is easy, non-invasive, acceptable for repeat testing, and can be performed by non-healthcare professionals, ECDC noted that performance of RT-qPCR tests has variously reported both higher and lower sensitivity for saliva samples compared with nasopharyngeal swabs. In part, heterogeneity is likely to reflect differences in sampling techniques, sampling times, and the type of population being tested, which the ECDC technical report explores (17).
Within the guidance from ECDC is the need to provide a sample into a “collection container, upon waking up, before brushing teeth and eating”. We came to the same conclusions early on in the UoN Testing Service because saliva samples of various consistencies and viscosities slowed down testing and processing time. Sample provision before eating is likely to be one factor in the high accuracy, sensitivity, and consistency observed within the UoN Testing Service and in other laboratories, which have reported that saliva has offered greater sensitivity than nasopharyngeal swabs for diagnosis of asymptomatic and mild COVID-19 infection (11). Retrospective studies have provided similar findings. Guillaume and colleagues (12) surveyed 385 references, which yielded 16 unique studies that were identified for quantitative synthesis. Eight peer-reviewed studies and eight preprints were included in the meta-analyses (5,922 unique patients), with a conclusion that diagnostic accuracy of saliva is similar to that of nasopharyngeal swabs.
Other benefits include ease of donation, minimal invasiveness, high-sensitivity testing, and accurate reporting. Stability of saliva as a source material is also high (Figure 1F), wherein regular analysis via RT-qPCR to N1, N2, E, and/or RNAseP of the same samples stored at 4°C for up to 20 days showed standard deviations (SD) of ≤ 1.1 and coefficients of variation (CoV) of <5%. These attributes meant that saliva samples were preferred over nasopharyngeal swabs within our cohorts, and we expect the same to be true for communities with special considerations; hence, the assay will increase inclusivity.
In July 2021, the University of Nottingham became the first university in the UK and the first institution in the UK to gain accreditation status from the oversight body, UKAS (UK Accreditation Service). This permitted results from our testing service to be reported directly to the government organization, Public Health England, thereby requiring donors who are positive for SARS-CoV-2 to follow national laws. At the time of writing (October 2021), all datasets in the form of research and protocols manuscripts are being prepared to give detailed information on the use of a triplex testing via CDC primers N2 and E, along with an internal control of RNAseP, in SARS-CoV-2 detection in saliva following direct heat inactivation. In these articles, we will draw on evidence from circa 100,000 samples tested. This will include data required for UKAS accreditation showing >99% concordance of 400 samples (250 negative and 150 positive), most of which were twinned swab and saliva hospital samples. We will provide evidence for analytical specificity, analytical sensitivity (limit of detection), limit of quantification, diagnostic specificity, diagnostic sensitivity, precision, sample stability, repeatability, reproducibility, range/linearity/accuracy, robustness (control of known interference), and low coefficients of variation of ≤ 5.3% even in the most viscous saliva samples.
Receiving accreditation from UKAS means that we can assist other institutions to gain this accolade and both accelerate and broaden their own testing programs. In parallel, the US Food & Drug Administration (FDA) approved Emergency Use Authorization (EUA) for “SalivaDirect™” (18). Various saliva RT-qPCR tests are in development or in the process of regulatory approval through the FDA EUA process or the CE Marking process in the European Union, including Rutgers University, OraSure Technologies/DNA Genotek, University of Illinois Champagne, and others. In instances where community surveillance requires pooling of large numbers of samples (10 or more per pool), there is the potential that viscosity may cause pipetting errors or reduce the sensitivity of SARS-CoV-2 detection, especially when using direct RT-qPCR approaches on samples with low viral loads (17). These issues can be overcome by extracting viral RNA prior to pooling and analysis but may not be needed due to differences in regulatory bodies. In the UK, regulations permit only a maximum of four samples to be pooled for diagnostic purposes for SARS-CoV-2.
Saliva may not be a one-size-fits-all solution. While various companies now offer saliva-based antigen or antibody tests, the ECDC suggests that the current limited evidence does not support the use of this sample material in this way and further clinical validation studies are needed on the different available tests (17). Nevertheless, even if this stance does not change for protein-based testing, the acceptability and ease of saliva as a donor sample coupled with approval as a diagnostic for SARS-CoV-2 genomes by multiple regulatory bodies, including the FDA, ECDC, and UKAS, is positive. This is likely to assist with sustained regular repeat testing over long periods, which will be essential to detect emergence of new variants during this two-tier pandemic. Thus, to conclude, saliva is presented as a suitable first-line diagnostic test to survey and control infection rates among populations in a more efficient and less invasive manner, complementing other testing strategies and improving our ability to control infectious events in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by University of Nottingham Ethics Committee, reference number FMHS 96-0920. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CD is lead of the programme and SP-G lead on data harvesting and first draft of the manuscript. All authors were involved in writing and contributing to the manuscript, and running the asymptomatic testing service.
Funding
This work was funded by the University of Nottingham, to support the COVID-19 Asymptomatic Testing Service, and Medical Research Council Urgency Award MC_PC_20027 “COVID-19 in university settings”.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://www.nature.com/articles/d41586-021-01390-4
2. ^http://www.nottingham.ac.uk/coronavirus/university-testing-service/index.aspx
3. ^https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-use-saliva-sample-material-testing.pdf
4. ^https://www.nottingham.ac.uk/coronavirus/university-testing-service/how-to-provide-your-sample/how-to-get-tested.aspx
References
1. Wu SL, Mertens AN, Crider YS, Nguyen A, Pokpongkiat NN, Djajadi S, et al. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat Commun. (2020) 11:4507. doi: 10.1038/s41467-020-18272-4
2. Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da https://pubmed.ncbi.nlm.nih.gov/?term=da$+$Silva$+$Filipe$+$A&cauthor_id=33621484 Silva Filipe A, Wojcechowskyj JA, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 184:1171–87.e20. doi: 10.1016/j.cell.2021.01.037
3. Wise J. CCovid-19: Safety of lateral flow tests questioned after they are found to miss half of cases. BMJ. 371:m4744. doi: 10.1136/bmj.m4744
4. Koller G, Morrell AP, Galão RP, Pickering S, MacMahon E, Johnson J, et al. More than the eye can see: shedding new light on sars-cov-2 lateral flow device-based immunoassays. ACS Appl Mater Interfaces. (2021) 22:25694–700. doi: 10.1021/acsami.1c04283
5. Peto T. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine. (2021) 36:100924. doi: 10.1016/j.eclinm.2021.100924
6. Torjesen I. Covid-19: How the UK is using lateral flow tests in the pandemic. BMJ. (2021) 372:1–3. doi: 10.1136/bmj.n287
7. Higgins TS, Wu AW, Ting JY. SARS-CoV-2 nasopharyngeal swab testing-false-negative results from a pervasive anatomical misconception. JAMA Otolaryngol Head Neck Surg. (2020) 146:993–4. doi: 10.1001/jamaoto.2020.2946
8. Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results, Expert Review of Molecular Diagnostics. (2020) 5:453–454. doi: 10.1080/14737159.2020.1757437
9. Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C https://pubmed.ncbi.nlm.nih.gov/?term=Cant%C3%BA-Cuevas+MA&cauthor_id=32703816 Cantú-Cuevas MA, Barón-Olivares H, V OD Saliva sampling its direct lysis, an excellent option to increase the number of SARS-CoV-2 diagnostic tests in settings with supply shortages. J Clin Microbiol. (2021) 58. doi: 10.1128/JCM.01659-20
10. Yee R, Truong TT, Pannaraj PS, Eubanks N, Gai E, Jumarang J, et al. Saliva is a promising alternative specimen for the detection of SARS-CoV-2 in children and adults. J Clin Microbiol. (2021) 59:e02686–20. doi: 10.1128/JCM.02686-20
11. Teo AKJ, Choudhury Y, Tan IB, Cher CY, Chew SH, Wan ZY, et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Scientific Rep. (2021) 11:3134. doi: 10.1038/s41598-021-82787-z
12. Butler-Laporte G, Lawandi A, Schiller I, Yao M, Dendukuri N, McDonald EG, et al. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med. (2021) 181:353–60. doi: 10.1001/jamainternmed.2020.8876
13. Blake H, Knight H, Jia R, Corner J, Morling JR, Denning C, et al. Students' views towards sars-cov-2 mass asymptomatic testing, social distancing and self-isolation in a university setting during the covid-19 pandemic: A qualitative study. Int J Environ Res Public Health. (2021) 18:4182. doi: 10.3390/ijerph18084182
14. Landry ML, Criscuolo j, Peaper DR. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. (2020) 130:104567. doi: 10.1016/j.jcv.2020.104567
15. Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, et al. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. (2020) 58: e01824–20. doi: 10.1128/JCM.01824-20
16. Warsi I, Khurshid Z, Shazam H, Umer MF, Imran E, Khan MO, et al. Saliva exhibits high sensitivity and specificity for the detection of SARS-COV-2. Diseases. (2021) 9:38. doi: 10.3390/diseases9020038
17. Azzi L, Carcano G, Gianfagna F, Grossi P https://pubmed.ncbi.nlm.nih.gov/?term=Gasperina+DD&cauthor_id=32298676 Gasperina DD, Genoni A Saliva is a reliable tool to detect SARS-CoV-2. J Infect. (2020) 81:e45–50. doi: 10.1016/j.jinf.2020.04.005
Keywords: SARS-CoV-2, lateral flow, polymerase chain reaction, COVID-19, nasopharyngeal swab, saliva, asymptomatic testing
Citation: Pijuan-Galito S, Tarantini FS, Tomlin H, Jenkins H, Thompson JL, Scales D, Stroud A, Tellechea Lopez A, Hassall J, McTernan PG, Coultas A, Arendt-Tranholm A, Reffin C, Hill I, Lee I-n, Wu S, Porte J, Chappell J, Lis-Slimak K, Kaneko K, Doolan L, Ward M, Stonebridge M, Ilyas M, McClure P, Tighe P, Gwynne P, Hyde R, Ball J, Seedhouse C, Benest AV, Petrie M and Denning C (2021) Saliva for COVID-19 Testing: Simple but Useless or an Undervalued Resource? Front. Virol. 1:778790. doi: 10.3389/fviro.2021.778790
Received: 17 September 2021; Accepted: 26 October 2021;
Published: 15 December 2021.
Edited by:
Seshadri Vasan, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Paul Desmond Slowey, Oasis Diagnostics, United StatesErik Albert Karlsson, Institut Pasteur du Cambodge, Cambodia
Copyright © 2021 Pijuan-Galito, Tarantini, Tomlin, Jenkins, Thompson, Scales, Stroud, Tellechea Lopez, Hassall, McTernan, Coultas, Arendt-Tranholm, Reffin, Hill, Lee, Wu, Porte, Chappell, Lis-Slimak, Kaneko, Doolan, Ward, Stonebridge, Ilyas, McClure, Tighe, Gwynne, Hyde, Ball, Seedhouse, Benest, Petrie and Denning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chris Denning, Y2hyaXMuZGVubmluZ0Bub3R0aW5naGFtLmFjLnVr
 Sara Pijuan-Galito1
Sara Pijuan-Galito1 Jamie Louise Thompson
Jamie Louise Thompson Philip G. McTernan
Philip G. McTernan Andy Coultas
Andy Coultas Joanne Porte
Joanne Porte Patrick McClure
Patrick McClure Chris Denning
Chris Denning