- 1Hebei Key Laboratory of the Bohai Sea Fish Germplasm Resources Conservation and Utilization, Beidaihe Central Experiment Station, Chinese Academy of Fishery Sciences, Qinhuangdao, China
- 2College of Animal Science and Veterinary Medicine, Shenyang Agricultural University, Shenyang, China
- 3College of Science and Engineering, Flinders University, Bedford Park, SA, Australia
- 4Panjin Guanghe Crab Industry Co. Ltd., Panjin, China
- 5Key Laboratory of Breeding and Propagation of Chinese Mitten Crab, Ministry of Agriculture and Rural Affairs, Panjin, Liaoning, China
- 6Liaoning Panjin Wetland Ecosystem National Observation and Research Station, Shenyang, China
Introduction: To support the sustainable development of rice and aquaculture industries, various rice-animal coculture systems have been developed. One such system, the rice-crab coculture system (RCC), has been practiced for decades in northern China. However, studies on the crab physiological status in RCC remain limited. Microorganisms play a crucial role in aquaculture by influencing animal nutrition, health, nutrient cycling, water quality, and environmental impact. Research on the gut and environmental microbiota in RCC is scarce.
Methods: This study compared the growth performance, immune and digestive enzyme activities of crabs between RCC and traditional pond farming system (PF). In addition, the microbiota in crab guts, water, and sediment from both systems was investigated using 16S rRNA gene sequencing.
Results: Crabs in RCC exhibited superior growth performance and higher enzymatic activities, including acid phosphatase (ACP), alkaline phosphatase (AKP), lipase (LPS), and trypsin (TRY). Significant differences were observed in microbiota composition across crab gut, water, and sediment samples, respectively. RCC crabs had a lower abundance of Bacteroidota and a higher abundance of Firmicutes in their gut microbiota. The RCC environment was enriched with beneficial bacteria such as Rhizobiales, Methylococcales, KD4-96, C39, Xanthomonadales, and Nitrosomonadaceae. Microbial function predictions confirmed enhanced methanotrophy and nitrogen fixation in the RCC.
Discussion: The RCC enhances the growth rate and immune capability of crabs. Crabs from RCC consume more animal-based nutrition, which results in distinct differences in gut microbiota composition and higher levels of LPS and TRY compared to those in PF. Additionally, RCC supports environmentally beneficial bacteria that contribute to greenhouse gas reduction, carbon and nitrogen fixation, organic matter decomposition, and ammonia oxidation, benefiting both the crabs and their ecosystem. These findings enhance our understanding of crab physiology and microbial communities in RCC and PF systems.
1 Introduction
The Chinese mitten crab (Eriocheir sinensis) is one of the most important freshwater crustaceans in China due to its commercial and ecological value (1, 2). Chinese mitten crab cultivation is practiced widespread across China (3), with annual production reaching 7.76 million tons in 2020 (4). The primary adult crab culture methods of Chinese mitten crabs include traditional pond farming system (PF) and rice-crab coculture system (RCC) (3). The former can cause severe environmental issues, such as eutrophication, greenhouse gas emissions, and acidification (5, 6). In contrast, RCC is considered a more sustainable method (7). Similar to other rice-animal coculture systems, RCC is not only more profitable than rice monoculture systems but also reduces the use of pesticides and fertilizers (8). Consequently, the Chinese government has promoted this culture system for many years (3). In 2019, China's rice-crab coculture area was ~138,600 hm2, and the annual yield reached 61,800 tons (9, 10). Previous research has investigated the effect of stocking density on the yield and nutritional quality of crabs in the RCC, suggesting that an appropriate density can significantly affect crab growth rate, gonadosomatic index, and meat yield (11–13). The positive ecological impact of RCC has also been confirmed in some studies. For example, with the same fertilizer N application, RCC improved rice yield compared to rice monoculture (RM) and also increased crab yield compared to crab monoculture when provided the same amount of feed (14). Moreover, RCC enhances nutrient levels in the soil and rice compared to RM (15). Additionally, through optimizing crab density and surplus N from crab feed, RCC can significantly reduce the emission of methane (CH4) and nitrous oxides (N2O), which are potent greenhouse gases (16). However, studies on the physiological status of crabs are limited.
Microorganisms play an essential role in aquaculture systems. They are closely related to the nutrition and health status of the culture animals and the productivity, nutrient cycling, water quality, and environmental impact of the effluent (17, 18). As one of the most significant aquaculture species, numerous comparative studies on the gut microbiota of Chinese mitten crabs, focusing on factors such as genetic background, developmental stage, habitat, feedstuff, health status, and environmental conditions, have been reported (19–25). However, there are very limited studies on environmental microorganisms in Chinese mitten crab culture ecosystems (23, 26), especially in RCC.
This study aimed to compare the growth performance, immune and digestive enzyme activities, and gut microbiota of crabs cultured in PF and RCC systems. Additionally, we analyzed environmental microorganisms in water and sediment to assess the differences in these systems. These findings expand our understanding of the physiological status of crabs and microbial communities in ecosystems in PF and RCC systems.
2 Materials and methods
2.1 Experimental design
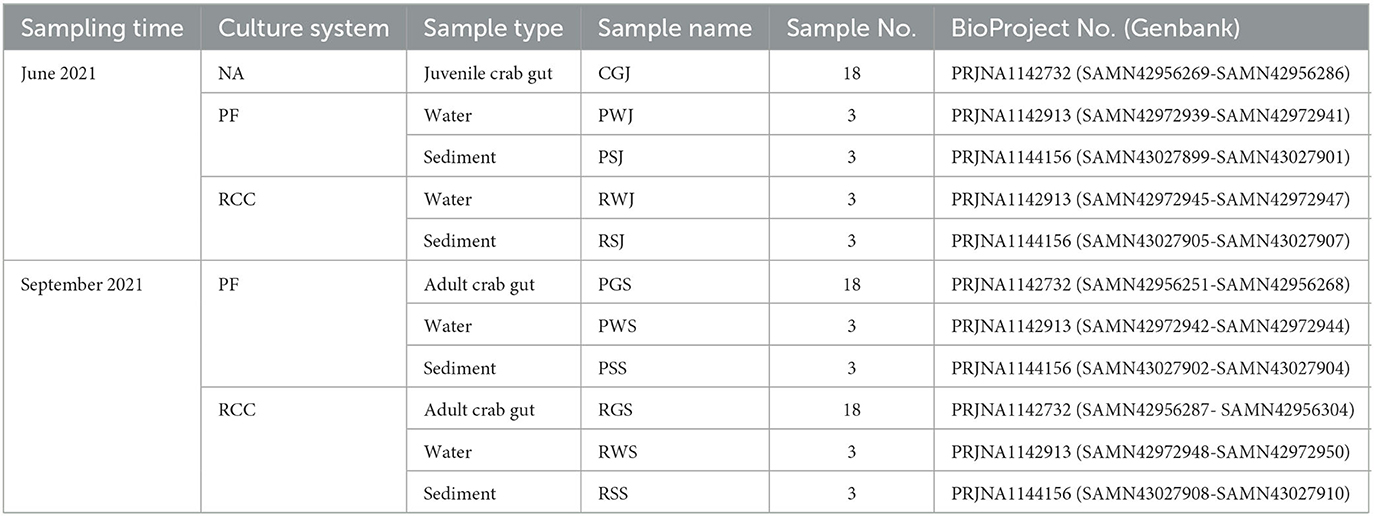
The experiments were conducted in Panjin Guanghe Crab Industry Co., Ltd. in Panjin City, China (N 40°54′, E121°52′). The rice field preparation followed the methods of Li et al. (11). Prior to the experiment, the juvenile crabs were kept in the wintering ponds. Two weeks after rice seedlings were transplanted, the juvenile crabs pooled from wintering ponds with an average weight (mean ± SD) of 10 ± 0.2 g were transferred to the experimental ponds (three replicates, approximate size 0.1 ha) and rice fields (three replicates, approximate size 0.15 ha) at a density of 2500 crabs per pond or rice field in June 1st, 2021. Before being transferred to the plots, the robust crabs (showing no disease and visually assessed healthy intestine and hepatopancreas) were randomly collected from pooled juvenile crabs. Water and sediment samples were collected from three experimental ponds and rice fields, respectively (Table 1).
The management of the two culture systems was identical. During the experimental period, crabs were fed twice daily with a commercial pelleted feed (Wellhope Aquatic Feed Co. Ltd., Shenyang, China). The feeding amount was adjusted by 5%–10% based on the size and population of the crabs. Water was added when the water level in the pond or rice field was significantly reduced, with no drainage occurring during the experiment.
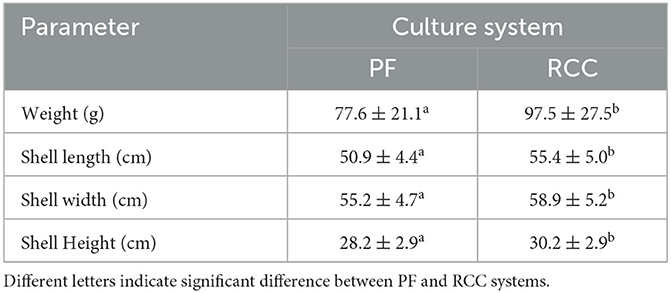
Crabs from both culture systems were harvested in September 1st, 2021. After the harvest, robust crabs, water, and sediment were collected from each experimental pond and rice field for further analysis (Table 1). 100 crabs from each pond and rice field were randomly collected to measure their carapace length, width, height and weight to evaluate their growth performances followed the methods described by Xue et al. (27).
2.2 Sample collection
2.2.1 Gut and hepatopancreas samples
The crabs were temporarily raised in the same tank under continuous aeration at ~20°C in the lab. After 24-h fast, six individuals were randomly collected from each group. We rinsed the crab's shell surfaces with sterile water and wipe the surfaces using 75% ethanol. The gut and hepatopancreas were dissected immediately in a sterile environment. Approximately 1.5 cm of mid and hindgut from each individual crab was collected as one replicate for gut microbiota sequencing. Meanwhile, three individual hepatopancreas samples were pooled as one replicate for enzyme activity assessment. Both gut and hepatopancreas replicates were placed in 2 mL cryogenic vials, submerged in liquid nitrogen, and stored at −80°C for subsequent analyses.
2.2.2 Water and sediment samples
In each experimental pond and rice field, water and sediment samples were collected from six spots (four corners and two middle spots of the long sides) and then pooled as one biological replicate. Water samples were collected from 5 cm below the water surface using sterile bottles. In each pond or rice field, 500 mL of water from each sample was filtered through a 0.22 μm polycarbonate membrane filter (Millipore, Germany). The filters were then stored at −80°C in 2 mL cryogenic vials for DNA extraction. Sediment samples were taken using plastic pipes (diameter 2.5 cm), and filtered through a 2 mm filter to remove large debris. At last, ~2 g sediment for each pond or rice field was placed into a 2 mL cryogenic vial at −80°C for DNA extraction.
2.3 Analysis of enzyme activities
Three hepatopancreas replicates (each replicate pooled from the hepatopancreas of three crabs) tissue was used for enzyme activity assessment. The assessment included two immune-related enzymes [acid phosphatase (ACP) and alkaline phosphatase (AKP)] and three digestive enzymes [amylase (AMS), lipase (LPS), and trypsin (TRY)] by using assay kits (Nanjing Jiancheng Bioengineering Institute, China) according to the manufacturer's protocols. Detailed methods and equipment for assessing the above enzymes and total protein content (TP) are shown in Supplementary Table S1.
2.4 Microbiota sequencing
Microbial community genomic DNA of crab gut, water and sediment was extracted with an E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) following the manufacturer's instructions. DNA concentration and purity were measured by NanoDrop 2000 (Thermo Scientific, Wilmington, USA). The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GACTACHVGGGTWTCTAAT-3′). The PCR reaction mixture included 4 μL 5 × FastPfu buffer, 2 μL 2.5 mM dNTPs, 0.8 μL each primer (5 μM), 0.4 μL Fast Pfu polymerase, 10 ng of template DNA, and ddH2O to a final volume of 20 μL. PCR amplification cycling conditions for gut samples were as follows: initial denaturation at 95°C for 3 min, followed by 29 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 45 s, and single extension at 72°C for 10 min, and end at 4°C. For water and sediment samples, the annealing temperature was 55°C and the number of PCR cycles was 27, while other conditions were the same for gut samples. The PCR product was extracted from 2% agarose gel and purified using the PCR Clean-Up Kit (YuHua, Shanghai, China) according to manufacturer's instructions and quantified using Qubit 4.0 (Thermo Fisher Scientific, USA). Purified amplicons were pooled in equimolar and paired-end sequenced on an Illumina MiSeq PE300 platform (Illumina, San Diego, USA) under the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under the BioProject number PRJNA1142732, PRJNA1142913, and PRJNA1144156 (Table 1).
2.5 Data processing and statistical analyses
After sequencing, the raw 16S rRNA gene sequencing reads were demultiplexed and quality-filtered by fastp version 0.20.0 (28). The reads were merged by FLASH 1.2.11 (29) with the same criteria as described in a previous study (30). Operational taxonomic units (OTUs) were clustered using UPARSE version 7.1 (31) with a 97% similarity cutoff, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier version 2.2 (32) against the 16S rRNA database (Silva SSU 138) using a confidence threshold of 0.7. After removing chloroplast and mitochondria sequences, the Chao and Shannon indices for α-diversity were calculated by Mothur 1.30.2 (33). β-diversity was determined through Bray-Curtis dissimilarity matrices and displayed using principal coordinate analysis (PCoA). To identify differences between groups, we conducted an Analysis of similarities (ANOSIM) using Bray-Curtis dissimilarity. A linear discriminant analysis (LDA) effect size (LEfSe) was performed to present the enrichment of microbial communities within the different samples (34). Functional profiling was inferred from the relative abundance of 16S rRNA sequences using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) (35). Additionally, the Functional Annotation of Prokaryotic Taxa (FAPROTAX) (36) was used to predict the ecological relevant functions of bacterial community in water and sedimental samples.
All of these analyses were carried out on the Majorbio I-Sanger Cloud Platform (37). One-way ANOVA with Waller-Duncan test was performed using the SPSS 20.0 software package (SPSS Inc., Chicago, USA) to estimate the differences of Shannon and Chao indices within different sample types or ecosystems, and hepatopancreas enzymatic activity level among crabs in June (CJ), crabs in September from PF (PS) and RCC (RS). Significance was declared at P < 0.05. Additionally, One-way ANOVA was employed to assess differences in relative abundance in KEGG pathways (Level 3) and ecological functions within gut, water, and sediment samples. P-values were corrected using the False Discovery Rate (FDR) method, with significance was set at corrected P-values < 0.05.
3 Results
3.1 Growth performance and enzymatic activities in Chinese mitten crabs
The growth parameters are shown in Table 2. Crabs from RCC were significantly higher in all parameters compared to crabs from PF, indicating they have a better growth performance.
The enzyme activities among crabs in June (CJ), crabs in September from PF (PS) and RCC (RS) are shown in Figure 1. The amylase (AMS) activity of crabs under both culture systems in September (PS and RS) did not differ significantly, but they were significantly higher than those of the CJ group. Lipase (LPS) and trypsin (TRY) activities in RS were significantly higher than in CJ and PS, while the latter two showed no significant difference. Acid phosphatase (ACP) and alkaline phosphatase (AKP) activities in CJ was significantly lower than in PS and RS, whereas in RS, they were significantly higher than in PS.
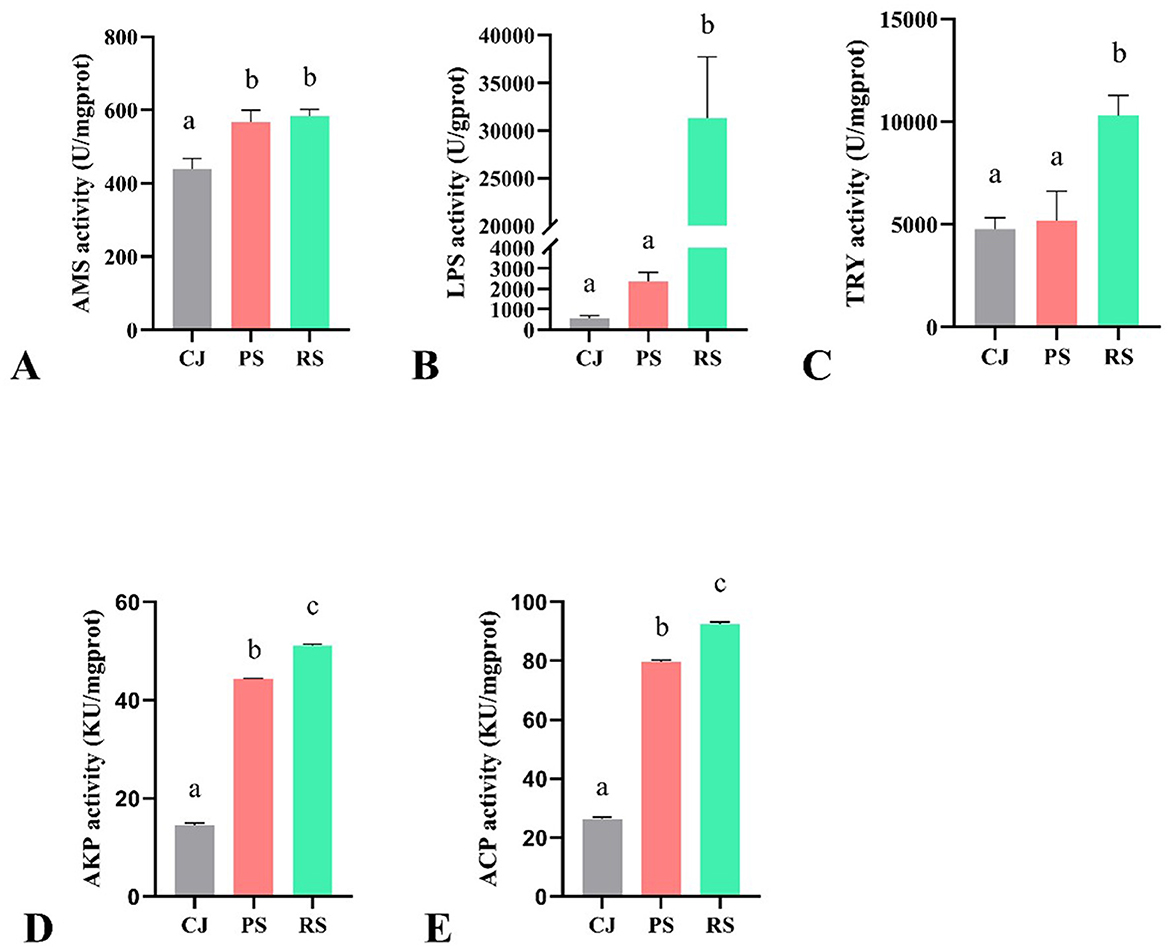
Figure 1. Enzyme activity of Chinese mitten crabs under two culture systems. (A) AMS; (B) LPS; (C) TRY; (D): AKP; (E) ACP. CJ, crabs in June; PS, crabs under PF in September; RS, crabs under RCC in September; KU, King's unit. Different lowercase letters indicate significant differences (P < 0.05).
3.2 Overview of 16s rRNA sequencing data
A total of 1,501,890 OTUs were obtained, with an average of 56,976, 46,197, and 57,184 OTUs in the gut, water, and sediment microbial communities, respectively. The α-diversity indices, including the Chao and Shannon indices, were calculated from the OTUs of each group (Table 3), the group abbreviations are explained in Table 1.
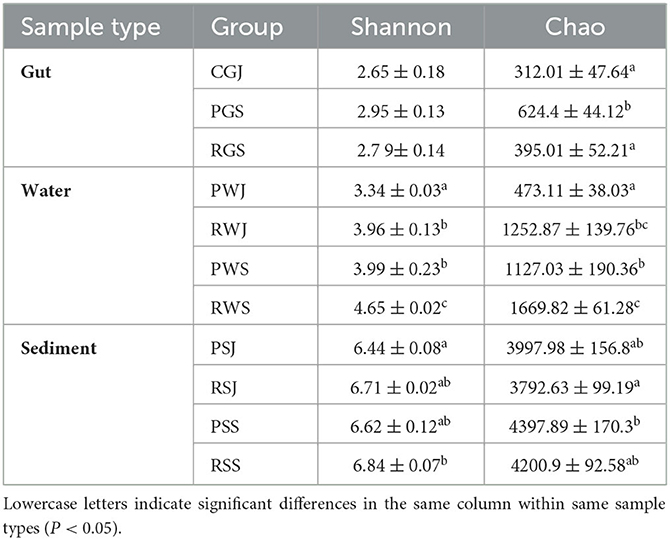
Table 3. α-diversity indices of gut, water and sediment samples from two different culture systems of Chinese mitten crabs.
In the gut microbial community, there were no significant differences in the Shannon index among these three groups, although the values of PGS and RGS were higher than for CGJ. The Chao index was the highest in PGS (624.40 ± 44.12), which was significantly higher than those observed in CGJ (312.01 ± 47.64) and RGS (395.01 ± 52.21).
Regarding the water microbial community, compared to PF, RCC had higher Shannon and Chao indices in June and September, respectively. The Shannon indices significantly increased in both PF [3.34 ± 0.03 (PWJ) to 3.99 ± 0.23 (PWS)] and RCC [3.96 ± 0.13 (RWJ) to 4.65 ± 0.02 (RWS)]. A similar pattern was also observed in Chao indices in the water microbial community.
Concerning the sediment microbial community, Shannon and Chao indices were also higher in September than in June in both systems, but the increases were not statistically significant. At two sampling times, higher Shannon indices were observed in RCC [6.71 ± 0.02 (RSJ) vs. 6.44 ± 0.08 (PSJ), 6.84 ± 0.07 (RSS) vs. 6.71 ± 0.02 (PSS), while higher Chao indices were observed in PF [3997.98 ± 156.8 (PSJ) vs. 3792.63 ± 99.19 (RSJ), 4397.89 ± 170.3 (PSS) vs. 4200.9 ± 92.58 (RSS)], although the differences were not significant.
In the comparison of the α-diversity of gut, water, and sediment microbial communities in each ecosystem, the Shannon and Chao indices were significantly higher in the sediment, followed by the water, with the gut having the lowest indices. Except the PF in June, the Shannon and Chao indices of water were significantly higher than those of the gut (Figure 2).
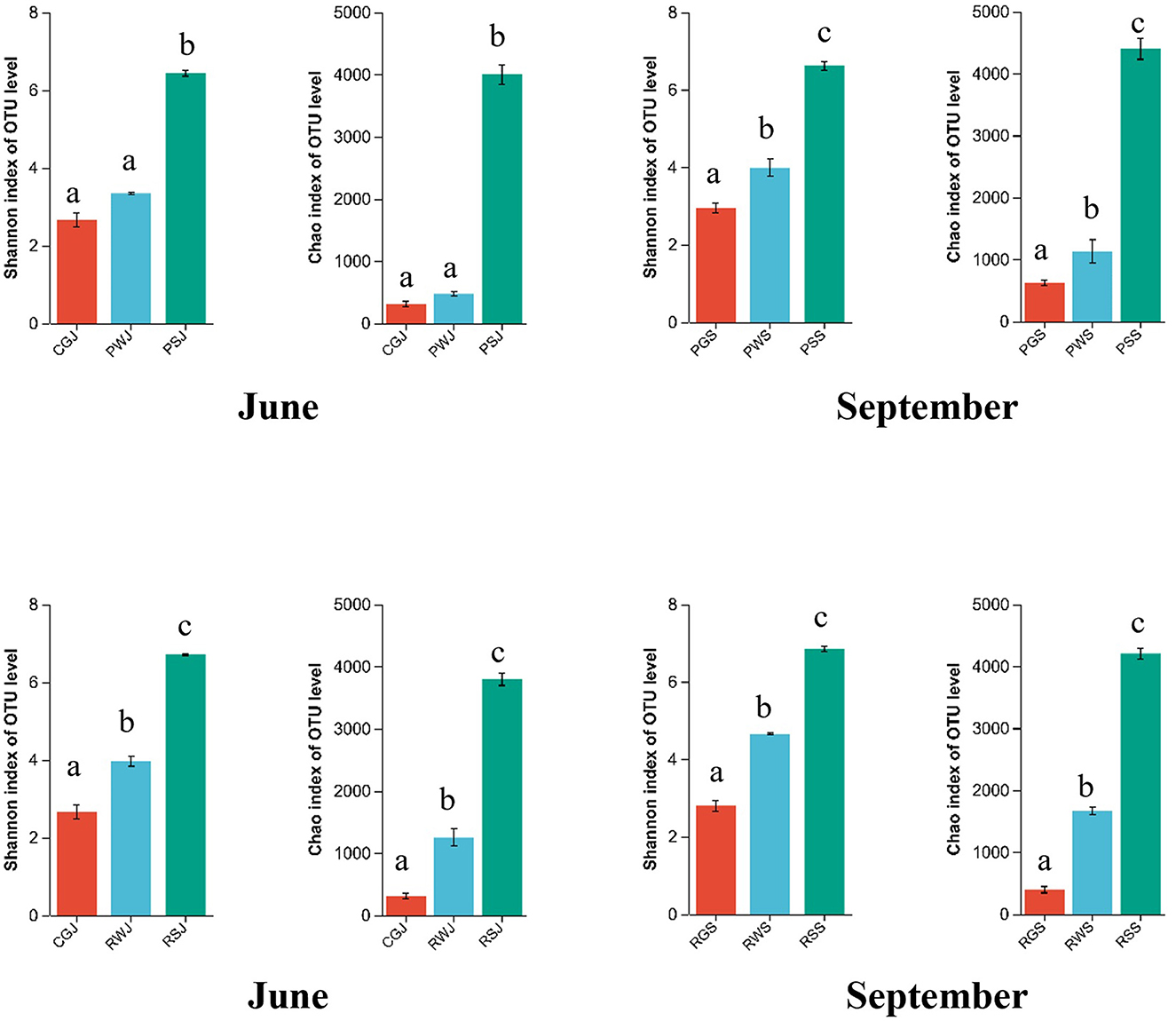
Figure 2. Shannon and Chao indices of gut, water, and sediment within the same ecosystem. Lowercase letters indicate significant difference (P < 0.05).
3.3 Differences in microbiota between the two culture systems
Differences in bacterial community compositions of intestines, water, and sediment between two culture systems in different months were visualized by conducting the PCoA based on Bray-Curtis distances. ANOSIM analysis and β-diversity analysis through PCoA indicated significant differences in gut microbiota (R = 0.3634, P = 0.001; Figure 3A), water microbiota (R = 0.7531, P = 0.001; Figure 3B), and sediment microbiota (R = 0.7438, P = 0.002; Figure 3C).
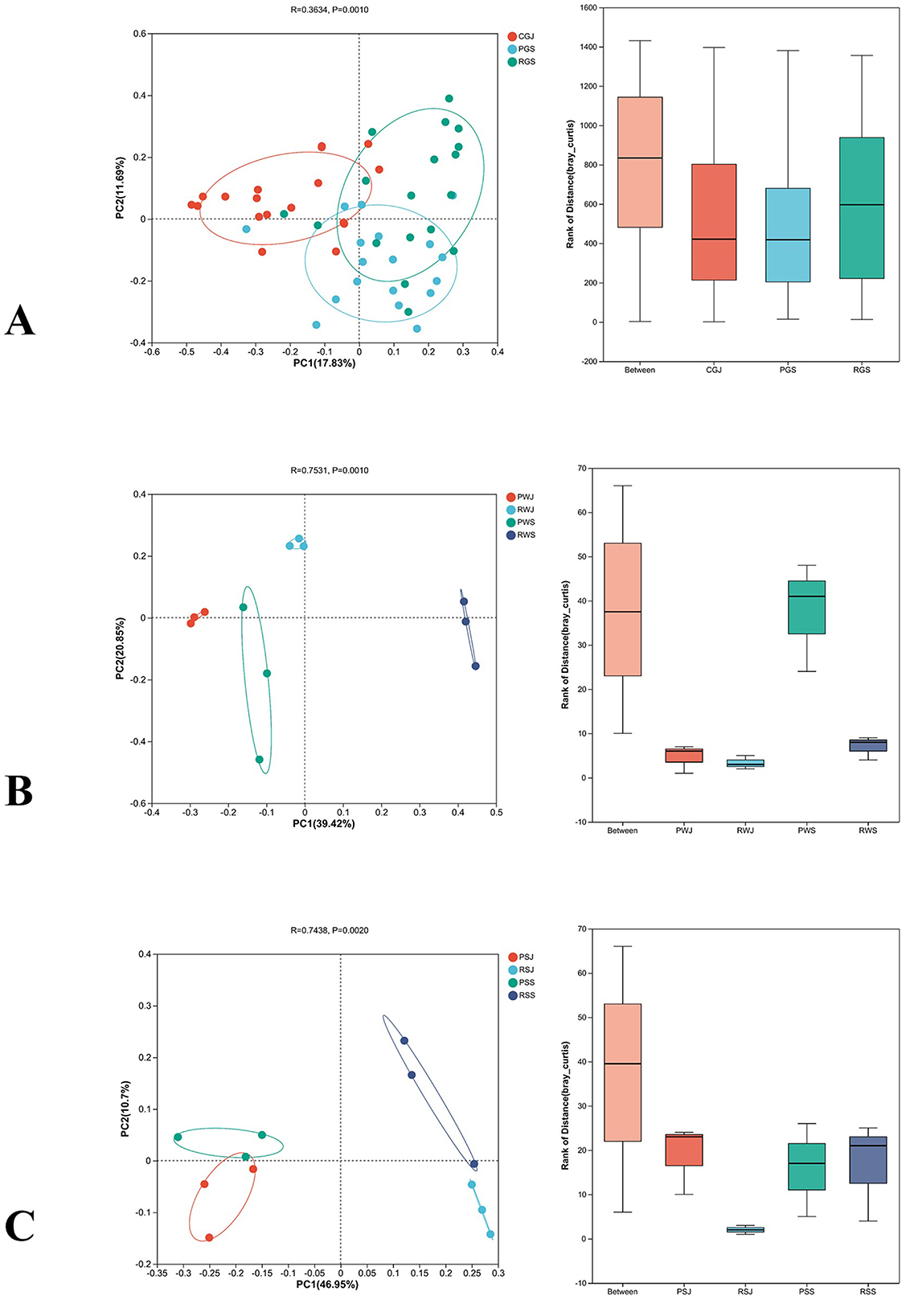
Figure 3. β-diversity comparison of the gut (A), water (B) and sediment microbiota (C) between two culture systems in June and September at the OTU level.
3.4 Microbiota compositions in the two culture systems
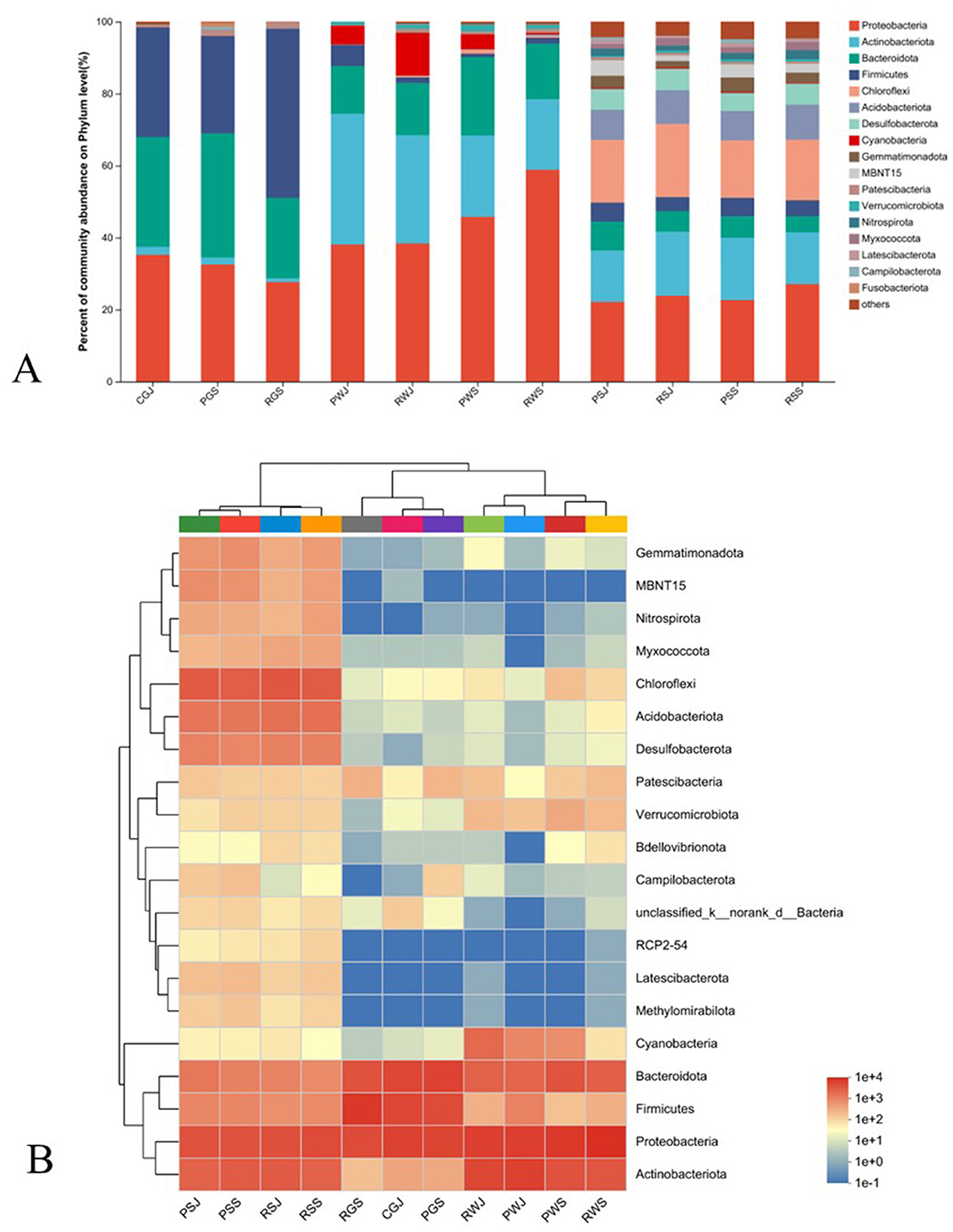
The relative abundance of bacterial community in different samples at the phylum level is shown in Figure 4A. The community heatmap suggests apparent divergence among gut, water, and sediment samples (Figure 4B).
In the crab gut, Proteobacteria (27.64%–35.24%), Bacteroidota (22.34%–34.49%), and Firmicutes (27.07%–47.04%) were the three most dominant phyla in all three gut groups. Figure 5 shows the microorganisms with significant differential abundance between CGJ and PGS, CGJ and RGS, and PGS and RGS. Compared with CGJ, PGS had a higher abundance in two phyla (Patescibacteria and Fusobacteriota), three classes (Saccharimonadia, Clostridia, and Fusobacterila), six orders, ten families and 11 genera. CGJ was richer in four orders (Aeromonadales, Burkholderiales, Erysipelotrichales, and Lactobacillales), five families, and five genera. Between CGJ and RGS, RGS had a significantly higher abundance in two phyla (Firmicutes and Patescibacteria), two classes (Saccharimonadia and Clostridia), five orders, seven families and nine genera. CGJ had a higher abundance in the one phylum (Proteobacteria), one class (Alphaproteobacteria), five orders, six families, and six genera. In the comparison between PGS and RGS, PGS was rich in three phyla (Bacteroidota, Fusobacteriota, and Campilobacterota), four classes, eight orders, ten families, and eight genera, but lower in Firmicutes (phylum level), Bacilli (class level), Entomoplasmatales, Lactobacillales, and Mycoplasmatales (order level), three families, and four genera.
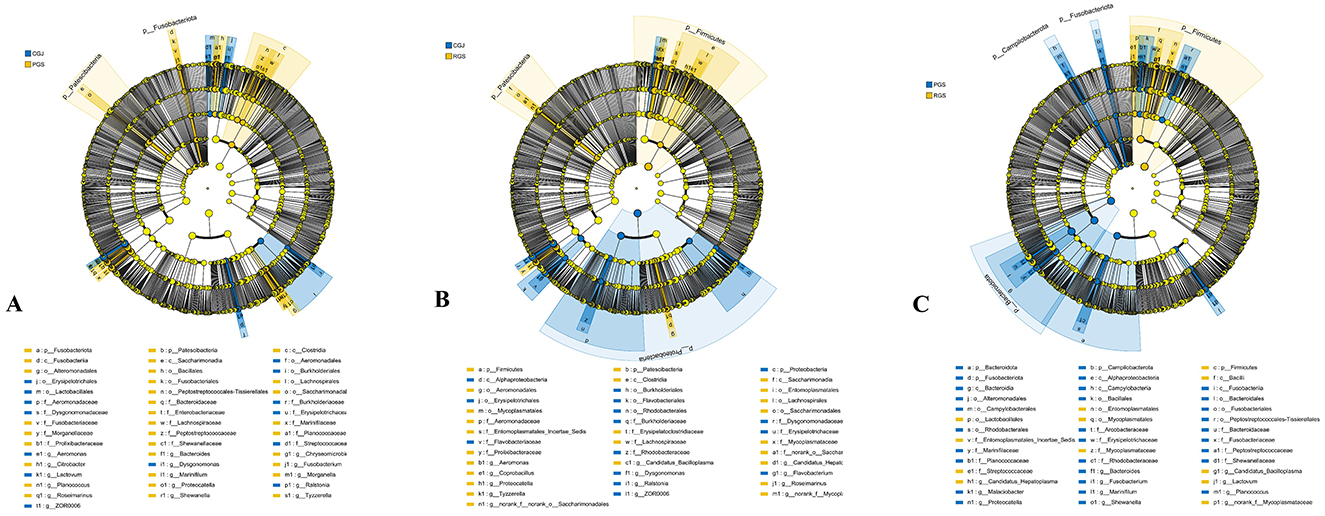
Figure 5. The linear discriminant analysis EffectSize (LEfSe) displaying the differences in the gut microbiota between CGJ and PGS (A), CGJ and RGS (B), and PGS and RGS (C). LDA score > 3.5.
Regarding the water microbial community, Proteobacteria (38.06%–58.84%), Actinobacteriota (19.60%–36.33%), Bacteroidota (13.28%–21.72%), and Cyanobacteria (0.39%–11.82%) were the four most dominant phyla in all samples (Figure 4). In June, PWJ had a higher abundance of Actinobacteriota but a lower abundance of Patescibacteria and Cyanobacteria compared with RWJ (Figure 6A). In the comparison between PWS an RWS, PWS was rich in Cyanobacteria (phylum level), Cyanobacteriia (class level), Synechococcales, Sphingobacteriales, SAR11_clade, and unclassified_Actinobacteria (order level), while RWS was rich in Rhizobiales, Caulobacterales, Corynebacteriales, Methylococcales, Xanthomonadales, and Pem15 (order level; Figure 6B). In the PF, a higher abundance of Bacteroidota and Chloroflexi were observed in PWS, while a higher abundance of Firmicutes and Actinobacteriota were found in PWJ (Figure 6C). In the RCC, compared with RWJ, RWS had a higher abundance of Proteobacteria but a lower abundance of Actinobacteriota and Cyanobacteria (Figure 6D).
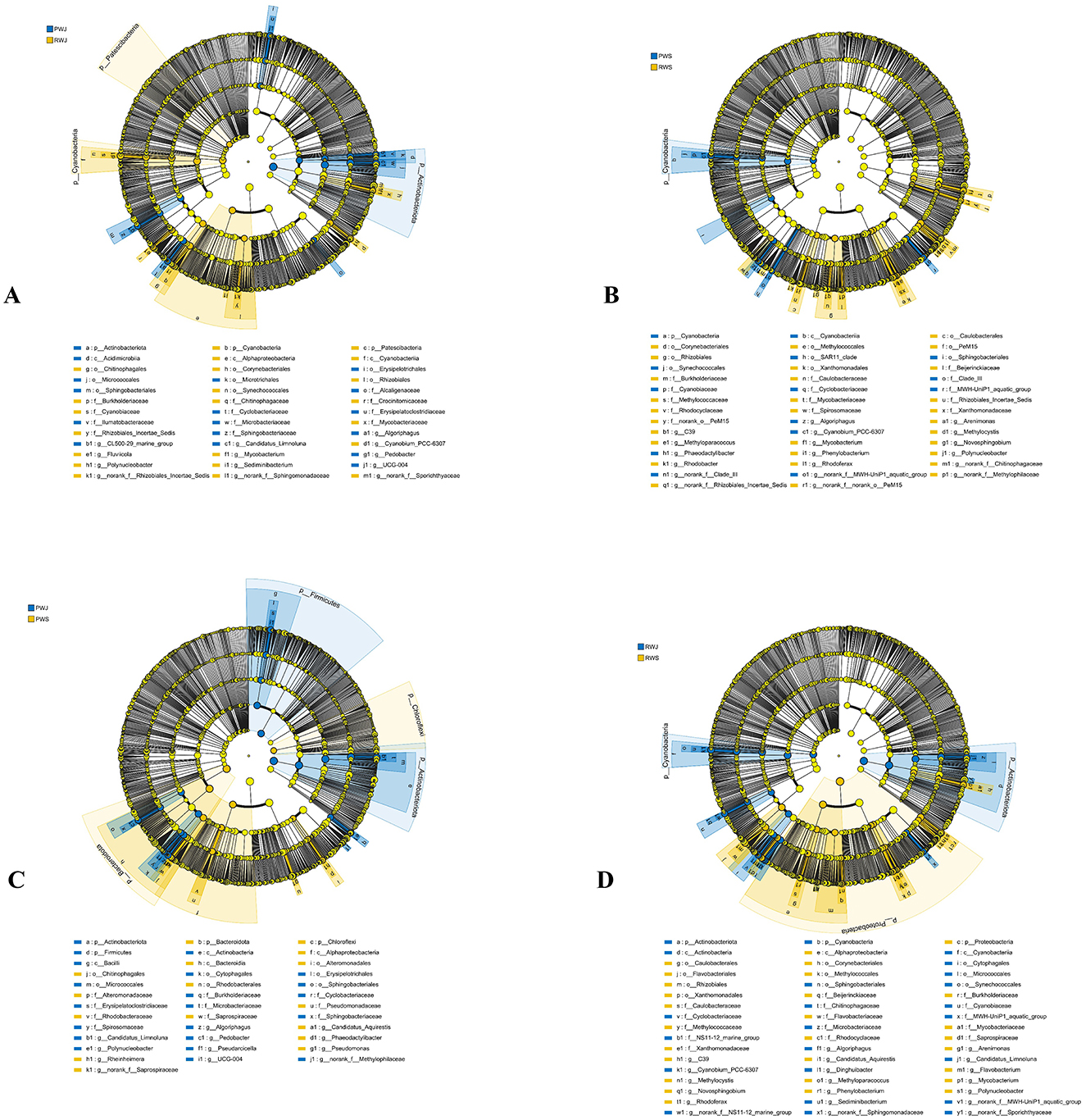
Figure 6. The linear discriminant analysis EffectSize (LEfSe) displaying the differences in the water between PWJ and RWJ (A), PWS and RWS (B), PWJ and PWS (C), and RWJ and RWS (D). LDA score > 3.5.
In the sediment samples, Proteobacteria (22.14%–27.07%), Chloroflexi (15.94%–20.34%), Actinobacteriota (14.29%–17.78%), and Acidobacteriota (8.16%–9.62%) were the most dominant phyla. Bacteroidota, Desulfobacterota, and Firmicutes also had higher relative abundance (3.89%–8.03%) than those low-abundance phyla (Figure 4A). In June, RSJ was rich in phyla of Actinobacteriota and Myxococcota, while PSJ was rich in phyla of MBNT15, Nitrospirota, Campilobacterota, and Gemmatimonadota (Figure 7A). Compared with RSS, PSS was rich in MBNT15 (phylum level), Bacilli, Acidimicrobiia, Desulfuromonadia, MB-A2-108, and BD2-11 terrestrial group (class level), and eight orders, whilst RSS was rich in KD4-96, Thermoleophilia and Syntrophobacteria (class level), Rhizobiales, Syntrophobacterales, and unclassified KD4-96 (order level; Figure 7B). In the PF, PSS had a higher abundance of MB-A2-108 and Thermoleophilia (class level) but a lower abundance in Flavobacteriales (order level) when compared with PSJ (Figure 7C). In the RCC, RSS had a greater abundance of Nitrospirota (phylum level), Acidobacteriae, Thermodesulfovibrionia, and Thermoleophilia (class level) but a lower abundance in Actinobacteriota (phylum level), Anaerolineae, and Actinobacteria (class level), as shown in Figure 7D.
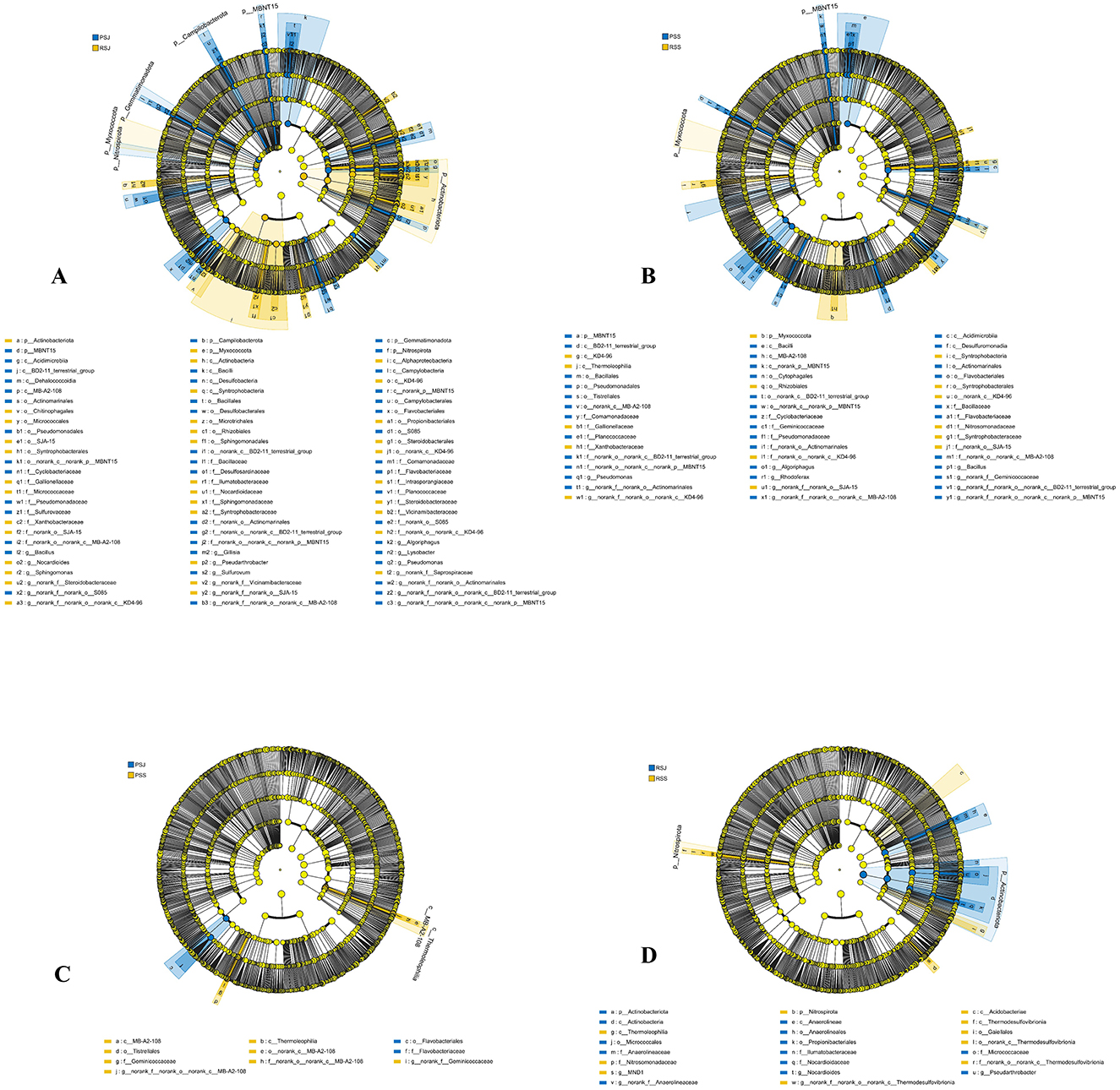
Figure 7. The linear discriminant analysis EffectSize (LEfSe) displaying the differences in the sediment between PSJ and RSJ (A), PSS and RSS (B), PSJ and PSS (C), and RSJ and RSS (D), LDA score > 3.5.
3.5 Microbial function prediction
The relative abundance of predicted function pathways (top 30) based on the KEGG annotation at level 3 was shown in Figure 8A. The most abundant four KEGG pathways at level 3 were metabolic pathways, biosynthesis of secondary metabolites, microbial metabolism in diverse environments, and biosynthesis of amino acids. Among gut samples, significant difference was observed in Glycolysis/gluconeogenesis, Amino sugar and nucleotide metabolism, Oxidative phosphorylation, Purine metabolism, and Ribosome. Specifically, when compared with RGS, PGS had higher abundance in oxidative phosphorylation but lower abundance in the other four pathways (Figure 8B). As for water samples, only two pathways (Fatty acid biosynthesis and Sulfur metabolism) showed significantly different abundances and had the same pattens. RWS had the highest abundances, while PWJ had the lowest abundances in these two pathways (Figure 8C). In sediment samples, no significant different pathway was identified based on KEGG pathways.
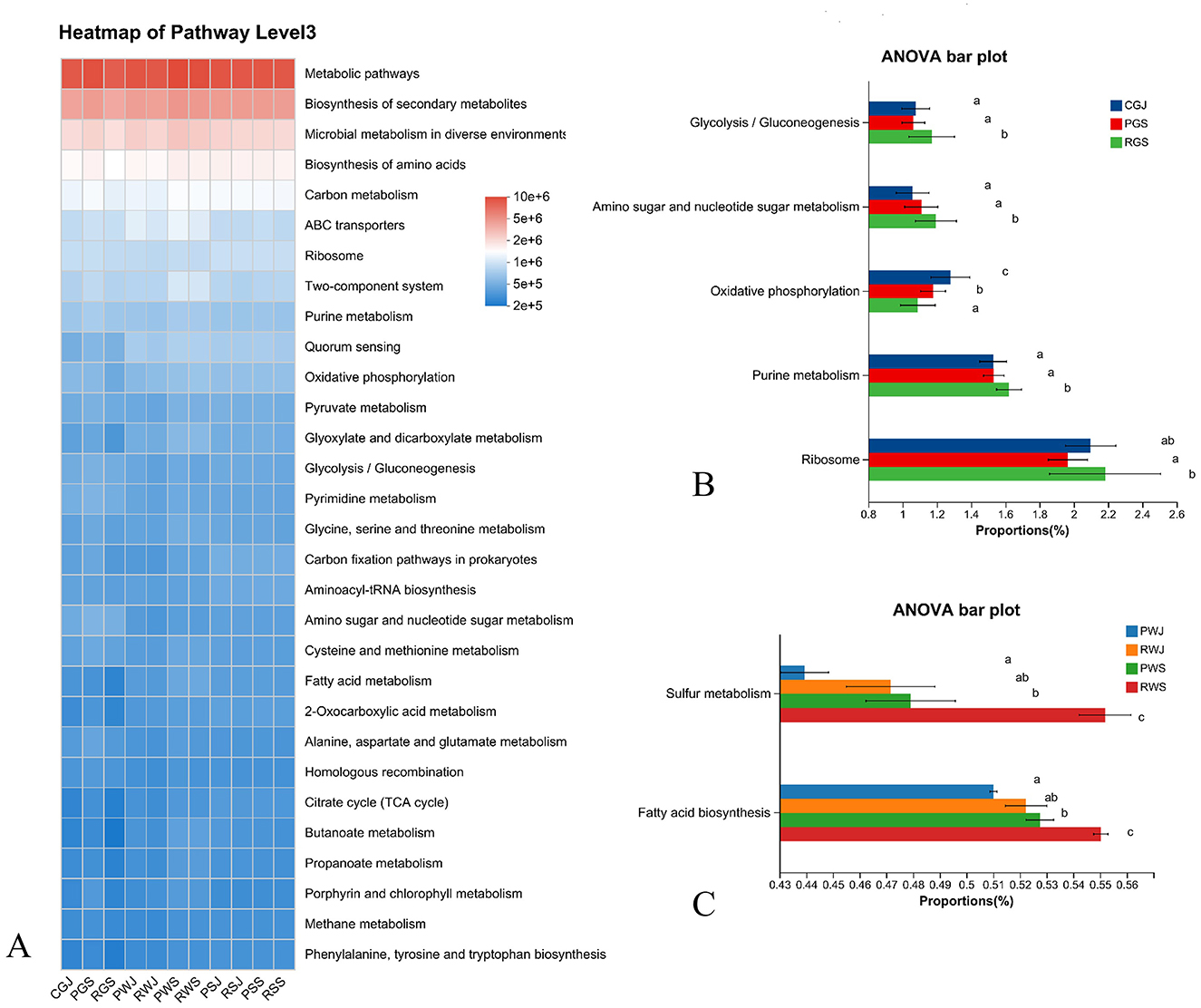
Figure 8. Predicted microbial functions (KEGG pathways, level 3) in gut, water, and sediment across two crab culture systems. (A) Heatmap of top 30 predicted functions, (B) Significant different pathways in gut samples (corrected P < 0.05), (C) Significant different pathways in water samples (corrected P < 0.05). Different letters indicate significant differences.
The ecologically relevant functions annotated by FAPROTAX are shown in Figure 9. In the water samples, functions related to Phototrophy, Photoautotrophy, Cyanbacteria, Oxygenic photoautotrophy, Photoheterotrophy, Hydrocarbon degradation, Methanotrophy, and Intracellular parasites showed significantly differences (Figure 9B). Generally, the abundances of the first four functions were the highest in RWJ. For Phototrophy, there were no significant differences in abundances among RWJ, PWS, and RWS. For the other three functions, the abundances in RWS were significantly lower than in RWJ and PWS. For the last four functions, the abundances in RWS were significantly higher than in PWJ, RWJ, and PWS, and the abundances among the latter three did not differ significantly. In comparisons among sediment samples, significant differences in abundance were observed in six functions, including Aromatic compound degradation, Hydrocarbon degradation, Nitrogen fixation, Methanotrophy, Ureolysis, and Sulfite respiration, as shown in Figure 9C. Overall, for all six functions, the abundances from samples of RCC (RSJ and RSS) were higher than those from PF (PSJ and PSS). In June, the abundances were significantly higher in RSJ compared to PSJ. In September, the abundances in RSS were higher than in PSS, although the differences were not significant for Aromatic compound degradation, Ureolysis, and Sulfite respiration.
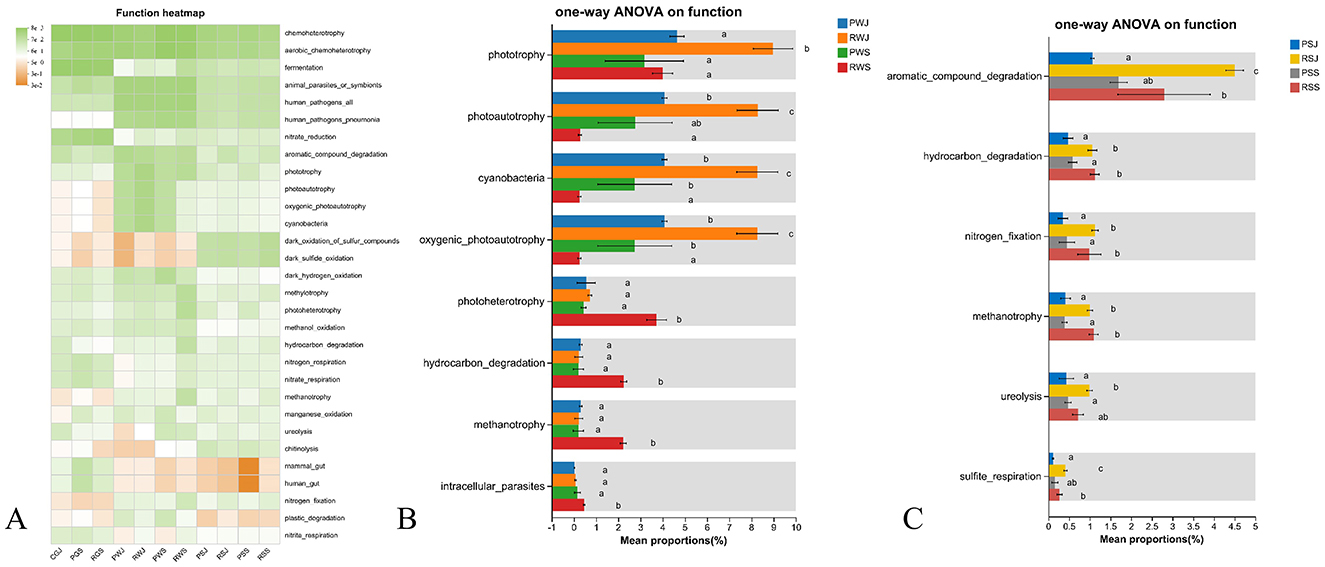
Figure 9. Predicted ecological relevant functions in microbial communities from water, and sediment across two crab culture systems. (A) Heatmap of top 30 predicted ecological relevant functions, (B) Significant different pathways among water samples (corrected P < 0.05), (C) Significant different pathways among sediment samples (corrected P < 0.05). Different letters indicate significant differences.
3.6 Correlation analysis between gut and environmental microbiota
The Venn diagram exhibits the overlap of microbial OTUs among crab gut, water and sediment in June and September in two culture systems (Figure 10). In June, the gut unique OTUs accounted 62.01% and 63.83% in PGJ and RGJ, respectively. When the crabs were harvested in September, the amount of gut unique OTUs decreased to 52.60% and 45.92% in PGS and RGS, respectively.
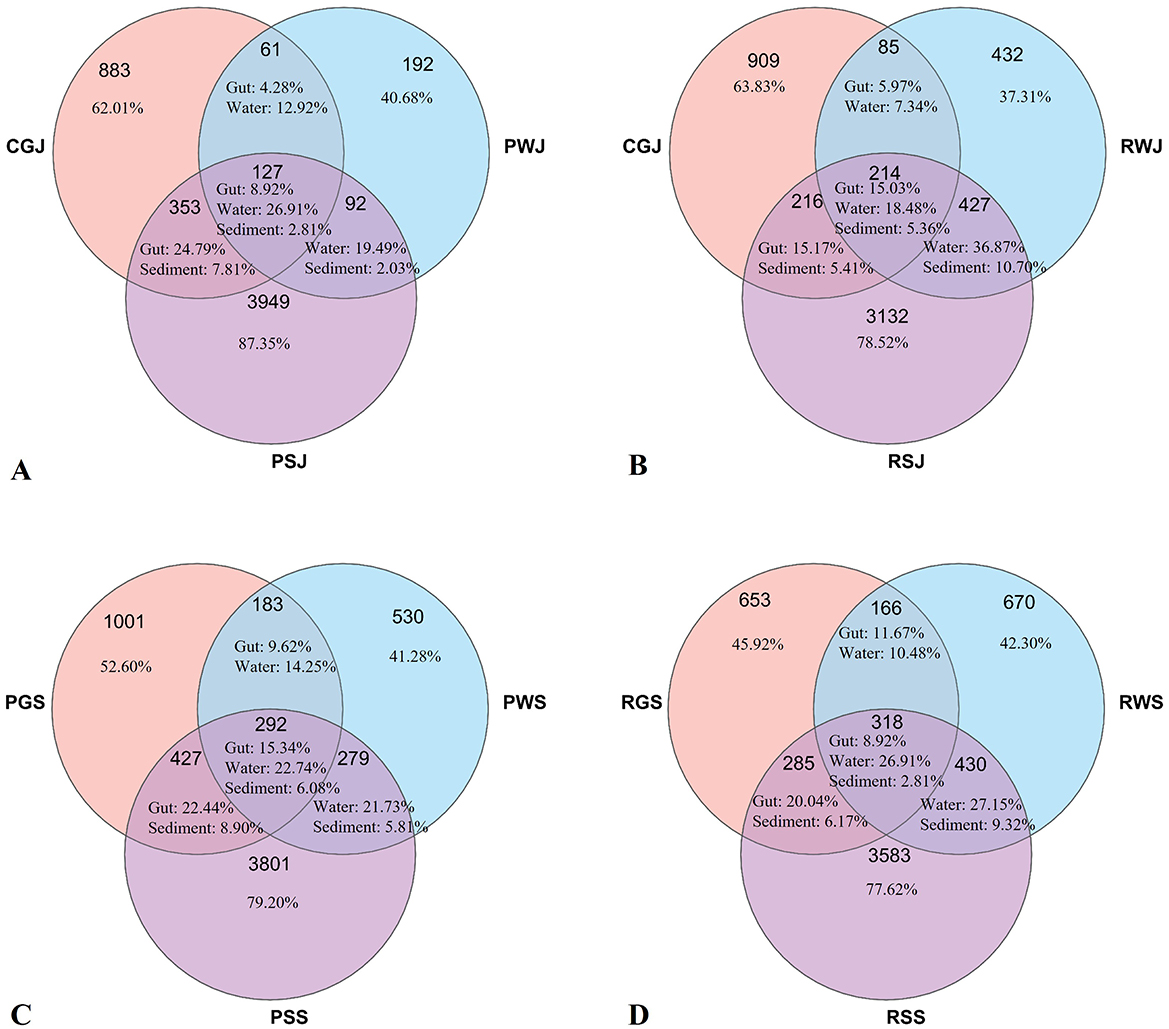
Figure 10. Venn diagram displaying the unique and shared OTU numbers among gut and environmental microbiota in two culture systems in June and September. (A) PF in June. (B) RCC in June. (C) PF in September. (D) RCC in September.
4 Discussion
4.1 Differences in growth performance and enzymatic activities between two culture systems
In this study, the growth performance of crabs from the RCC was significantly better than crabs from the PF. In a previous study in tilapia (Oreochromis niloticus), better growth performance was also observed in fish from fish-rice coculture system compared to monoculture system (38), and the authors attribute it to superior water quality and additional nutrient acquisition in the fish-rice coculture system. In this study, before the release of the crabs, dissolved oxygen in the RCC was significantly higher than in the PF, while ammonia levels were significantly lower, as shown in the Supplementary Table S2. However, these differences were not significant in September. Chinese mitten crabs are omnivores, which have a highly diversified food resources including detritus, macrophytes, algae, fish, Arthropoda, Oligochaeta, Mollusca, and other aquatic invertebrates (39–41). Previous literature indicated that rice-animal coculture systems usually have higher biodiversity and more stable community structure (42, 43). For example, compared to rice monoculture system, the biodiversity of benthic and plankton fauna are more dynamic in rice-fish coculture system (44), and individual spider number is significantly greater in the rice-crab coculture system (45). Hu et al. investigated the dynamics of plankton structure in PF and RCC systems in Panjin City (46), which is the same region to our study. Significantly higher species richness, density, average biomass, and Shannon index in phytoplankton and zooplankton were observed in the RCC than PF. Aquatic animals in the rice-animal coculture system usually play a significant role in pest control (47). For example, common carps significantly reduce the abundance of stemboring moths, chironomid midges, and rice planthoppers in the rice fields (48, 49). Yellow finless eels (Monopterus albus) and loaches (Misgurnus spp.) significantly reduced the herbivore insect abundance by 24.07% (50). Due to its omnivorous behavior, the pest control of Chinese mitten crabs in rice fields is predictable, although the direct evidence is lack so far. Therefore, the higher biodiversity in the rice fields may provide crabs with extra nutrients, which contributes to the better growth performance of crabs from the RCC in this study.
The hepatopancreas of crustaceans is a key organ involved in digestion, detoxification, and immunity (51, 52). To assess the digestive and immunity capacities of crabs from two culture systems, we investigated the hepatopancreas' enzymatic activity of three digestive enzymes and two immune-related enzymes. Digestive enzymes represent a crucial link between the nutrients in the diet and those incorporated into the body for further use in basal metabolism and growth (53). In the present study, the levels of TRY and LPS in RS were significantly higher than that of PS, and the latter had similar levels to the CJ. Previous studies suggest that a moderate increase in protein and lipids in the diets contributes to higher levels of TRY and LPS in Chinese mitten crabs (54, 55). Therefore, the extra nutrition, probably animal-type food, in the RCC may contribute to this result. ACP and AKP are important hydrolases that play a significant role in digesting extracellular invaders and have a close relationship with phagocytosis (56, 57). They are considered sensitive immune parameters in crustaceans (58). In this study, higher ACP and AKP activities were observed in RS compared to PS. In the comparison of crayfish between rice-crayfish coculture and pond culture systems, ACP and AKP levels were higher in the former system (59), which aligns with our study. Therefore, crabs from RCC have higher levels of non-specific immune enzymes and are healthier compared to traditional PF.
4.2 α-diversity of gut and environmental microbiota
In each ecosystem, the highest α-diversity indices were observed in sediment microbiota compared to water and gut microbiota. The same results have been reported in previous literature across various aquaculture systems of crustacean species, including Chinese mitten crabs (23, 26), Kuruma prawns (Penaeus japonicus) (60, 61), tropical shrimp (Penaeus notialis) (62), and Pacific white shrimp (Litopenaeus vannamei) (63, 64). It is likely due to the abundant organic matter accumulated from aquaculture animal feces, food residuals, and other materials that serve as nutrients to support microorganism growth (61, 65).
4.3 Differences in gut microbiota between two systems
In the comparison between juvenile (June) and adult (September) crabs, PGS had significantly higher Chao index than CGJ and RGS, whereas no significant difference was observed between CGJ and RGS. As for Shannon indices, CGJ, PGS, and RGS did not differ significantly. Comparable Shannon indices between juvenile and adult aquaculture species have been reported in previous studies in Chinese mitten crabs (23), Pacific white shrimp (66), and common carps (Cyprinus carpio) (67).
Between the adult crabs from PF and RCC systems, although PGS and RGS owned similar Shannon indices, their β-diversity differed significantly. Specifically, compared with PGS, RGS was rich in Firmicutes (47.05% vs. 27.13%) but lower in Bacteroidota (22.38% vs. 34.34%). The gut microbiota of aquatic animals is evidently closely with their diets (68–70). In a comparison of the gut microbiota of Chinese mitten crabs fed plant- and animal-type diet, lower abundance of Bacteroidota and higher abundance of Firmicutes were observed in the group fed by animal-type diet when compared with plant-type diet (21). In this study, the same artificial diet was applied in ponds and rice fields. Therefore, as previously mentioned, the higher biodiversity in the rice fields may provide crabs with extra nutrients, especially the animal-type diet, which contributes to the lower abundance of Bacteroidota and higher abundance of Firmicutes in this study.
4.4 Differences in water microbiota between two systems
In the comparison of water microbiota between PF and RCC systems prior to the release of juvenile crabs in June, the abundance of cyanobacteria in RWJ (11.82%) was significantly higher than in PWJ (5.22%). This is probably due to fertilizer application in the rice field before transplanting the rice seedlings. By the harvest season in September, the abundance of cyanobacteria in RWS decreased to 0.39%, significantly lower than that in PWS (4.12%). Since the water was not released until the harvest season, the microbial community in September is more important compared to that in June. Cyanobacteria can potentially cause algal blooms and produce cyanotoxins, which is a significant concern for managing water quality in aquaculture systems (71, 72). A lower abundance of cyanobacteria in RCC indicated less impact on the environment after release. Order SAR11 bacteria (Alphaproteobacterial) also had a higher abundance in PWS compared with RWS, which plays a role in converting fixed carbon to atmospheric CO2 (73).
Due to the presence of rice, RWS is unsurprisingly rich in the Rhizobiales order compared to PWS. Members of the order Rhizobiales are beneficial in oxidizing nitrogen and methane (74, 75). Genera of Rhizobiales Incertae Sedis and Methylocystis had a higher abundance in RWS than PWS. The former can utilize a wide range of carbohydrates and is involved in nitrogen fixation (76, 77). Methylocystis spp. play an important role in reducing methane emission into the atmosphere from methanogenic wetlands (78). In addition to Methylocystis spp., the order Methylococcales also had higher abundance in RWS. Methylococcales members are aerobic methanotrophs–bacteria that can metabolize methane (79, 80). RWS also had higher abundance in Arenimonas spp. (gammaproteobacterial order Xanthomonadales), which can catalyze a variety of enzymes and play an essential role in the decomposition of organic matter. They enhance plant growth by improving the utilization rate of nutrients (81). The genus C39 (gammaproteobacterial family Rhodocyclaceae) had a higher abundance in RWS. Its high abundance has been detected in the water of plant and fish farming systems (82), herbivorous grass carp (Ctenopharyngodon idella) and carnivorous southern catfish (Silurus meridionalis) coculture systems (83), and rice–crayfish coculture systems (84). C39 possesses versatile metabolic capabilities, contributing to the removal of total nitrogen and total phosphorus (85).
4.5 Differences in sediment microbiota between two systems
In this study, significant differences in microbial community abundance were observed before release and during the harvest season. At phylum level, phyla of MBNT15 and Myxococcota had higher abundance in PF and RCC, respectively. MBNT15 bacteria are obligate anaerobes and usually thrive in anoxic sediment (86). In contrast, Myxococcota species are predominantly aerobic soil-dwelling microorganisms (87). The difference in the abundances of these two phyla between PF and RCC may reflect variations in the dissolved oxygen statuses of their sediments. In the Desulfobacterota phylum, species harbor sulfur-cycling bacteria (88). PF was rich in the Desulfuromonadia class, while RCC was rich in the Syntrophobacteria class. All members of the Desulfuromonadia class are mesophilic, whereas members of the Syntrophobacteria are either mesophilic or moderately thermophilic (89). The lower water depth in the rice field likely contributes to temperature increases in water and sediment, further affecting the abundance differences in Desulfuromonadia and Syntrophobacteria between the two systems.
Chloroflexi phylum had a high abundance in the sediment of PF and RCC, which is in line with previous studies (23, 90). Although the abundance of Chloroflexi did not differ significantly between two systems, higher abundance of KD4-96 class was observed in RCC than PF. KD4–96 species are proposed to be involved in C fixation by assimilating CO2 (91, 92). In the study on the rice-crayfish system, Huang et al. found that the relative abundance of Firmicutes phylum in the ponds without rice was significantly higher than that of the rice field (93), and relevant studies have confirmed that crops can significantly reduce the relative abundance of Firmicutes in sediment (94). In this study, the abundance of Firmicutes was slightly higher in PF compared with RCC, but not statistically significant. In Firmicutes, however, PF had a higher abundance of Bacilli class in PF compared to RCC. Some Bacillus species can colonize the rhizosphere or endophytic tissues of rice plants, benefiting rice cultivation by improving rice yield and resistance to diseases and wind (95, 96). Therefore, rice seeds or seedlings treated with Bacillus spp. can enhance germination, growth, and resistance to many pathogens (97–99). Whether the abundance of Bacillus spp. is sufficient in the rice fields in this study needs further research in the future.
Before the crab release in June, the abundance of the Nitrosomonadaceae family did not differ significantly between PF and RCC. However, by September, its abundance in RCC was significantly higher than in PF. Nitrosomonadaceae bacteria oxidize ammonia to nitrate (100), which is an important process in aquaculture systems (101) that helps maintain a better environment for Chinese mitten crabs. As expected, the abundance of the Rhizobiales order was higher in the RCC, as high abundance of Rhizobiales in the rice field has been reported by Kim and Liesack (102). Members of the Rhizobiales order play a beneficial role in nitrogen and methane oxidation, providing absorbable nutrients for plants and supporting sustain nutrient cycles within the field (75).
4.6 Microbial function potentials
Among gut samples, five metabolism-related KEGG pathways were significantly different in their abundance. For Glycolysis/gluconeogenesis, Amino sugar and nucleotide sugar metabolism, and Purine metabolism, abundances in RGS were significantly higher than in CGJ and PGS. A metagenomics analysis of gut microbiota of red swamp crayfish (Procambarus clarkii) revealed higher abundances of genes related to above-mentioned pathways in crayfish from rice fields compared to those from ponds (103). These results indicated that both red swamp crayfish and Chinese mitten crabs from the RCC could have better capabilities in those metabolism pathways. In this study, the abundance for Oxidative phosphorylation was lower in RGS than in CGJ and PGS. Although the increased function of oxidative phosphorylation has been associated with providing more free energy to host animal (104), it is also linked to higher redox potentials (105) and even a pathogenesis-related microbial function in Pacific white shrimp (106).
In terms of ecologically relevant functions in the water samples, the increases in Phototrophy, Photoautotrophy, and Oxygenic photoautotrophy in RWJ were attributed to its higher abundance in cyanobacteria, as previously mentioned. Compared to other samples, RWS had higher levels of Hydrocarbon degradation, Methanotrophy, and Intracellular parasites. Here, hydrocarbon mainly refers to methane, based on relative abundances. Methane, a potent greenhouse gas, is the second-largest contributor to climate warming after carbon dioxide (CO2). Both Chinese mitten crab aquaculture ponds and rice fields are significant contributors to atmospheric greenhouse gas (e.g., CO2, CH4, and N2O) (107, 108). Previous studies indicate that rice-animal coculture systems, such as rice-duck, rice-fish, and rice-frog can effectively alleviate greenhouse gas emissions compared to rice monoculture system (109). Although this advantage is less significant in the RCC (109), PF significantly increases the methane emission compared to rice monoculture field (108). In this study, the abundant methanotrophs (e.g., Rhizobiales, Methylocystis spp.) in RWS contributed to the higher methanotrophy function in RCC. This suggests that RCC system may reduce the methane emission compared with PF system, though a direct comparison is lacking.
In the sediment samples, similar to water samples, Hydrocarbon degradation and Methanotrophy functions were enhanced in RCC than in PF, which can be attributed to the higher abundance of Rhizobiales in the RCC. The high abundance of Rhizobiales also likely contributed to the increased function of Nitrogen fixation in RCC compared to PF. The function of Aromatic compound degradation was also increased in the RCC. Some aromatic compounds (e.g., benzoate, phenylpropionate and phenylacetate) are intermediates in the methanogenic degradation of rice straw and soil organic matter (110). Therefore, it is not surprising that sediment samples from the RCC had an increased capacity for this function. Similarly, urea fertilization in the rice field increased the Ureolysis function in RCC, and the increase in the abundance of soil ureolytic bacterial in the rice fields has been confirmed by previous study (111).
4.7 Correlation between gut and environmental microbiota
Previous studies on shrimp indicate that the gut microbiota of Kuruma prawns and Pacific white shrimp is closely associated with environmental microbiota (61, 64, 112). For example, the unique OTU number of gut microbiota of Kuruma prawns accounts for as low as 15.67% of the total gut OTU number. In this study, however, unique OTU numbers account for ~50% in either PF or RCC in September. This finding is similar to the previous studies on Chinese mitten crabs (26) and suggests that gut microbiota of crabs is not as closely related to the environment as that of shrimp. The reasons contributing to this difference between Chinese mitten crabs and Kuruma prawns and Pacific white shrimp require further investigation.
5 Conclusions
This study compared the growth performance, immune and digestive enzyme activities of crabs, as well as the microbiota in crab gut, water, and sediment between the RCC and PF systems. The results showed that RCC crabs exhibited better growth performance, and higher enzymatic activities of ACP and AKP, suggesting that RCC enhances the growth rate and immune response in Chinese mitten crabs. Significant differences were observed in the microbial communities of crab gut, water, and sediment samples between these two systems. RGS was characterized by an increased abundance of Firmicutes and a decreased abundance of Bacteroidota, suggesting that RCC crabs acquired additional nutrients, especially from animal-based diets, which was confirmed by their elevated LPS and TRY levels. In the surrounding environment, RCC was rich in various environmentally beneficial bacteria such as Rhizobiales, Methylococcales, KD4-96, C39, Xanthomonadales, and Nitrosomonadaceae. These bacteria play important roles in greenhouse gas reduction, carbon and nitrogen fixation, organic matter decomposition, and ammonia oxidation, respectively. The microbial function predictions also suggested that the water and sediment samples in the RCC had increased methanotrophy functions, with the sediment showing enhanced functions in nitrogen fixation, aromatic compound degradation. Conversely, PWS contained higher level of Cyanobacteria, which requires monitoring due to their potential to cause harmful algal blooms. Additionally, RCC sediment had a lower abundance of Bacillus species compared with sediment of PF, and these bacteria are considered beneficial for rice. Further investigation is needed to determine whether the abundance of Bacillus spp. is sufficient in the rice fields in this study.
This study deepens the understanding of the physiological status of crabs and expands our knowledge in RCC and PF ecosystems from the perspective of microbial communities. It provides insights into the advantages of RCC over traditional PF, particularly in promoting crab growth and improving environmental sustainability.
However, as this study was conducted in the Liaohe drainage during a single production cycle, other factors (e.g., location and climate) may influence microbial community. Long-term studies and research in different locations are needed to fully understand the microbiota dynamic in these two aquaculture systems. Additionally, we used 16S rRNA sequencing in study, and metagenomic analyses in the subsequent studies can provide deeper insight into microbial functions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1142732; https://www.ncbi.nlm.nih.gov/, PRJNA1142913; https://www.ncbi.nlm.nih.gov/, PRJNA1144156.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of Shenyang Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XZ: Data curation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. MN: Investigation, Methodology, Writing – original draft. NS: Methodology, Resources, Writing – original draft. YZhan: Investigation, Methodology, Writing – original draft. MS: Investigation, Methodology, Writing – original draft. CL: Investigation, Methodology, Writing – original draft. QJ: Investigation, Methodology, Writing – original draft. HW: Resources, Supervision, Writing – original draft. YL: Resources, Supervision, Writing – original draft. QH: Resources, Supervision, Writing – original draft. YZhao: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing, Writing – original draft. XL: Conceptualization, Funding acquisition, Resources, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research work was supported by grant from Applied Basic Research Program of Liaoning Science and Technology Department (2023JH2/101300129), National Key Research and Development Program of China (2023YFD2401802), and Major Science and Technology Special Project of Liaoning Province (2024JH1/11700009).
Conflict of interest
NS and XL were employed by Panjin Guanghe Crab Industry Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1571454/full#supplementary-material
References
1. Wang J, Xu P, Zhou G, Li X, Lu Q, Liu X, et al. Genetic improvement and breeding practices for Chinese mitten crab, Eriocheir sinensis: aquaculture of Chinese mitten crab. J World Aquacult Soc. (2018) 49:292–301. doi: 10.1111/jwas.12500
2. Cui Z, Liu Y, Yuan J, Zhang X, Ventura T, Ma KY, et al. The Chinese mitten crab genome provides insights into adaptive plasticity and developmental regulation. Nat Commun. (2021) 12:2395. doi: 10.1038/s41467-021-22604-3
3. Cheng Y, Wu X, Li J. Chinese mitten crab culture: current status and recent progress towards sustainable development. In:Gui J-F, Tang Q, Li Z, Liu J, De Silva SS, , editors. Aquaculture in China. New York City: Wiley (2018). p. 197–217
5. Poore J, Nemecek T. Reducing food's environmental impacts through producers and consumers. Science. (2018) 360:987–92. doi: 10.1126/science.aaq0216
6. Liu X, Wu H, Wang Y, Liu Y, Zhu H, Li Z, et al. Comparative assessment of Chinese mitten crab aquaculture in China: Spatiotemporal changes and trade-offs. Environ Pollut. (2023) 337:122544. doi: 10.1016/j.envpol.2023.122544
7. Xu Q, Wang X, Xiao B, Hu K. Rice-crab coculture to sustain cleaner food production in Liaohe River Basin, China: an economic and environmental assessment. J Clean Prod. (2019) 208:188–98. doi: 10.1016/j.jclepro.2018.10.092
8. Cui J, Liu H, Wang H, Wu S, Bashir MA, Reis S, Sun Q, Xu J, Gu B. Rice-animal co-culture systems benefit global sustainable intensification. Earths Future. (2023) 11:e2022EF002984. doi: 10.1029/2022EF002984
9. Bao J, Jiang H, Li X. Thirty years of rice-crab coculture in China—Research progress and prospects. Rev Aquac. (2022) 14:1597–612. doi: 10.1111/raq.12664
10. Yuan J, Liao C, Zhang T, Guo C, Liu J. Advances in ecology research on integrated rice field aquaculture in China. Water. (2022) 14:2333. doi: 10.3390/w14152333
11. Li X, Dong S, Lei Y, Li Y. The effect of stocking density of Chinese mitten crab Eriocheir sinensis on rice and crab seed yields in rice–crab culture systems. Aquaculture. (2007) 273:487–93. doi: 10.1016/j.aquaculture.2007.10.028
12. Wang X, Tjale PM, Yao Q, Zhang D-M, Zhang B-Y, Chen Y, et al. Comparison of the growth performance and nutritional qualities of Chinese mitten crab (Eriocheir sinensis) with different stocking densities in rice-crab culture systems. Aquac Rep. (2021) 20:100761. doi: 10.1016/j.aqrep.2021.100761
13. Zhang B, Fang W, Zhu R, Wang N, Yao Q, Liu H, et al. Comparative study on growth index and nutritional quality of female Chinese mitten crab Eriocheir sinensis selected at different growth periods in rice-crab culture systems. Aquacult Nutr. (2023) 2023:1–15. doi: 10.1155/2023/4805919
14. Hu L, Guo L, Zhao L, Shi X, Ren W, Zhang J, et al. Productivity and the complementary use of nitrogen in the coupled rice-crab system. Agr Syst. (2020) 178:102742. doi: 10.1016/j.agsy.2019.102742
15. Bashir MA, Wang H, Pan J, Khoshnevisan B, Sun W, Zhai L, et al. Variations in soil nutrient dynamics and their composition in rice under integrated rice-crab co-culture system. J Clean Prod. (2021) 281:125222. doi: 10.1016/j.jclepro.2020.125222
16. Khoshnevisan B, Bashir MA, Sun Q, Pan J, Wang H, Xu Y, et al. Optimal rice-crab co-culture system as a new paradigm to air-water-food nexus sustainability. J Clean Prod. (2021) 291:125936. doi: 10.1016/j.jclepro.2021.125936
17. Ding X, Jin F, Xu J, Zhang S, Chen D, Hu B, et al. The impact of aquaculture system on the microbiome and gut metabolome of juvenile Chinese softshell turtle (Pelodiscus sinensis). iMeta. (2022) 1:e17. doi: 10.1002/imt2.17
18. Moriarty DJW. The role of microorganisms in aquaculture ponds. Aquaculture. (1997) 151:333–49. doi: 10.1016/S0044-8486(96)01487-1
19. An H-E, Malik A, Lee J, Mun M-H, Lee KH, Yoo HY, et al. Comparison of bacterial community composition in gut of Chinese mitten crabs from three distinct rivers in Korea. Fishes. (2024) 9:144. doi: 10.3390/fishes9040144
20. Cao Y, Zhang L, Yang Y, Li J, Luan X, Xia X, et al. Proteome and gut microbiota analysis of Chinese mitten crab (Eriocheir sinensis) in response to Hepatospora eriocheir infection. Aquaculture. (2024) 582:740572. doi: 10.1016/j.aquaculture.2024.740572
21. Chen X, Lu D, Li Z, Yue W, Wang J, Jiang X, Han H, Wang C. Plant and animal-type feedstuff shape the gut microbiota and metabolic processes of the Chinese mitten crab Eriocheir sinensis. Front Vet Sci. (2021) 8:589624. doi: 10.3389/fvets.2021.589624
22. Shao C, Zhao W, Li N, Li Y, Zhang H, Li J, et al. Gut microbiome succession in Chinese mitten crab Eriocheir sinensis during seawater–freshwater migration. Front Microbiol. (2022) 13:858508. doi: 10.3389/fmicb.2022.858508
23. Wang C, Zhou Y, Lv D, Ge Y, Li H, You Y. Change in the intestinal bacterial community structure associated with environmental microorganisms during the growth of Eriocheir sinensis. Microbiologyopen. (2019) 8:e00727. doi: 10.1002/mbo3.727
24. Zhang R, Zhao Z, Li M, Luo L, Wang S, Guo K, et al. Effects of saline-alkali stress on the tissue structure, antioxidation, immunocompetence and metabolomics of Eriocheir sinensis. Sci Total Environ. (2023) 871:162109. doi: 10.1016/j.scitotenv.2023.162109
25. Zhu X, Zhao Y, Sun N, Li C, Jiang Q, Zhang Y, et al. Comparison of the gut microbiota and untargeted gut tissue metabolome of Chinese mitten crabs (Eriocheir sinensis) with different shell colors. Front Microbiol. (2023) 14:1218152. doi: 10.3389/fmicb.2023.1218152
26. Sun Y, Han W, Liu J, Huang X, Zhou W, Zhang J, et al. Bacterial community compositions of crab intestine, surrounding water, and sediment in two different feeding modes of Eriocheir sinensis. Aquac Rep. (2020) 16:100236. doi: 10.1016/j.aqrep.2019.100236
27. Xue J, Liu H, Jiang T, Chen X, Yang J. Shape variation in the carapace of Chinese mitten crabs (Eriocheir sinensis H. Milne Edwards, 1853) in Yangcheng Lake during the year-long culture period. Eur Zool J. (2022) 89:217–28. doi: 10.1080/24750263.2022.2038290
28. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. (2018) 34:i884–90. doi: 10.1093/bioinformatics/bty560
29. Magoc T Salzberg SL FLASH FLASH fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
30. Zhu Z, Ma X, Wu J, Xiao Z, Wu W, Ding S, et al. Altered gut microbiota and its clinical relevance in mild cognitive impairment and Alzheimer's disease: shanghai aging study and shanghai memory study. Nutrients. (2022) 14:3959. doi: 10.3390/nu14193959
31. Edgar RC UPARSE highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
32. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
33. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
34. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. (2011) 12:R60. doi: 10.1186/gb-2011-12-6-r60
35. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. (2020) 38:685–8. doi: 10.1038/s41587-020-0548-6
36. Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. (2016) 353:1272–7. doi: 10.1126/science.aaf4507
37. Ren Y, Yu G, Shi C, Liu L, Guo Q, Han C, et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta. (2022) 1:e12. doi: 10.1002/imt2.12
38. He J, Feng P, Lv C, Lv M, Ruan Z, Yang H, et al. Effect of a fish–rice co-culture system on the growth performance and muscle quality of tilapia (Oreochromis niloticus). Aquacult Rep. (2020) 17:100367. doi: 10.1016/j.aqrep.2020.100367
39. Jin G, Xie P, Li Z. Food habits of two-year-old Chinese mitten crab (Eriocheir sinensis) stocked in Lake Bao'an, China. J Freshw Ecol. (2003) 18:369–75. doi: 10.1080/02705060.2003.9663972
40. Koester M, Frenzel C, Becker G, Sahm R. Food spectrum of the Chinese mitten crab (Eriocheir sinensis): insights from the Lower River Rhine comparing stable isotope mixing models and genetic gut content analyses. AI. (2022) 17:560–76. doi: 10.3391/ai.2022.17.4.06
41. Mao Z, Gu X, Zeng Q. Food sources and trophic relationships of three decapod crustaceans: insights from gut contents and stable isotope analyses. Aquac Res. (2016) 47:2888–98. doi: 10.1111/are.12739
42. Ge L, Sun Y, Li Y, Wang L, Guo G, Song L, et al. Ecosystem sustainability of rice and aquatic animal co-culture systems and a synthesis of its underlying mechanisms. Sci Total Environ. (2023) 880:163314. doi: 10.1016/j.scitotenv.2023.163314
43. Ji Z, Zhao L, Zhang T, Dai R, Tang J, Hu L, Chen X. Coculturing rice with aquatic animals promotes ecological intensification of paddy ecosystem. J Plant Ecol. (2023) 16:rtad014. doi: 10.1093/jpe/rtad014
44. Li F, Gao J, Xu Y, Nie Z, Fang J, Zhou Q, et al. Biodiversity and sustainability of the integrated rice-fish system in Hani terraces, Yunnan province, China. Aquacult Rep. (2021) 20:100763. doi: 10.1016/j.aqrep.2021.100763
45. Ma X, Che X, Wang J, Sang H. The structure of spider communities in crab paddies and conventional paddies. Chin J Eco-Agricult. (2019) 27:1157–62. doi: 10.13930/j.cnki.cjea.181068
46. Hu Q, Mei J, Li X, Li P. Effects of crab culture in paddy field and pond on plankton in the culture environment. J Shenyang Agricult Univ. (2023) 54:555–62.
47. Yu L, Li C, Zhang Y, Guo X, Cao N, Guo S, et al. Residue monitoring of propiconazole in the rice–crab co-culture field and its toxicity and bioaccumulation to Eriocheir sinensis. Front Environ Sci. (2022) 10:848348. doi: 10.3389/fenvs.2022.848348
48. Halwart M, Litsinger JA, Barrion AT, Viray MC, Kaule G. Efficacy of Common Carp and Nile Tilapia as biocontrol agents of rice insect pests in the Philippines. Int J Pest Manag. (2012) 58:330–46. doi: 10.1080/09670874.2012.705918
49. Xie J, Hu L, Tang J, Wu X, Li N, Yuan Y, Yang H, Zhang J, Luo S, Chen X. Ecological mechanisms underlying the sustainability of the agricultural heritage rice–fish coculture system. Proc Natl Acad Sci USA. (2011) 108:E1381–7. doi: 10.1073/pnas.1111043108
50. Wan N, Li S, Li T, Cavalieri A, Weiner J, Zheng X, et al. Ecological intensification of rice production through rice-fish co-culture. J Clean Prod. (2019) 234:1002–12. doi: 10.1016/j.jclepro.2019.06.238
51. Chen Y, Huang X, Wang J, Li C. Effect of pure microcystin-LR on activity and transcript level of immune-related enzymes in the white shrimp (Litopenaeus vannamei). Ecotoxicology. (2017) 26:702–10. doi: 10.1007/s10646-017-1802-7
52. Wang X, Huang Z, Wang C, Qi C, Gu Z, Li E, et al. comparative study on growth and metabolism of Eriocheir sinensis juveniles under chronically low and high ph stress. Front Physiol. (2020) 11:885. doi: 10.3389/fphys.2020.00885
53. Santos JF, Soares KLS, Assis CRD, Guerra CAM, Lemos D, Carvalho LB. Bezerra RS. Digestive enzyme activity in the intestine of Nile tilapia (Oreochromis niloticus L.) under pond and cage farming systems. Fish Physiol Biochem. (2016) 42:1259–74. doi: 10.1007/s10695-016-0215-5
54. Cui Y, Ma Q, Limbu SM, Du Z, Zhang N, Li E, et al. Effects of dietary protein to energy ratios on growth, body composition and digestive enzyme activities in Chinese mitten-handed crab, Eriocheir sinensis. Aquac Res. (2017) 48:2243–52. doi: 10.1111/are.13061
55. Li X, Li Z, Liu J, Murphy BR. Growth, precocity, enzyme activity and chemical composition of juvenile Chinese mitten crab, Eriocheir sinensis, fed different dietary protein-to-energy ratio diets. Aquac Res. (2012) 43:1719–28. doi: 10.1111/j.1365-2109.2011.02981.x
56. Hong Y, Huang Y, Yan G, Pan C, Zhang J. Antioxidative status, immunological responses, and heat shock protein expression in hepatopancreas of Chinese mitten crab, Eriocheir sinensis under the exposure of glyphosate. Fish Shellfish Immunol. (2019) 86:840–5. doi: 10.1016/j.fsi.2018.12.020
57. Zhou L, Chu L, Du J, Nie Z, Cao L, Gao J, et al. Oxidative stress and immune response of hepatopancreas in Chinese mitten crab Eriocheir sinensis under lipopolysaccharide challenge. Comp Biochem Physio Part C Toxicol Pharmacol. (2023) 263:109495. doi: 10.1016/j.cbpc.2022.109495
58. Hong Y, Yang X, Yan G, Huang Y, Zuo F, Shen Y, et al. Effects of glyphosate on immune responses and haemocyte DNA damage of Chinese mitten crab, Eriocheir sinensis. Fish Shellfish Immunol. (2017) 71:19–27. doi: 10.1016/j.fsi.2017.09.062
59. Wang J, Ye J, Zhang Z, An Z, Wang T, Dong X. Comparison of the nutrient value, nonspecific immunity, and intestinal microflora of red swamp crayfish (Procambarus clarkii) in different culture modes. Aquacult Rep. (2023) 31:101683. doi: 10.1016/j.aqrep.2023.101683
60. Song M, Pan L, Zhang M, Huang F, Gao S, Tian C. Changes of water, sediment, and intestinal bacterial communities in Penaeus japonicus cultivation and their impacts on shrimp physiological health. Aquac Int. (2020) 28:1847–65. doi: 10.1007/s10499-020-00562-9
61. Zhou L, Qu Y, Qin JG, Chen L, Han F, Li E. Deep insight into bacterial community characterization and relationship in the pond water, sediment and the gut of shrimp (Penaeus japonicus). Aquaculture. (2021) 539:736658. doi: 10.1016/j.aquaculture.2021.736658
62. Dabadé DS, Wolkers-Rooijackers JCM, Azokpota P, Hounhouigan DJ, Zwietering MH, Nout MJR, et al. Bacterial concentration and diversity in fresh tropical shrimps (Penaeus notialis) and the surrounding brackish waters and sediment. Int J Food Microbiol. (2016) 218:96–104. doi: 10.1016/j.ijfoodmicro.2015.11.013
63. Hou D, Huang Z, Zeng S, Liu J, Weng S, He J. Comparative analysis of the bacterial community compositions of the shrimp intestine, surrounding water and sediment. J Appl Microbiol. (2018) 125:792–9. doi: 10.1111/jam.13919
64. Huang F, Pan L, Song M, Tian C, Gao S. Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Appl Microbiol Biotechnol. (2018) 102:8585–98. doi: 10.1007/s00253-018-9229-5
65. Jia B, Li Y, Zi X, Gu X, Yuan H, Jeppesen E, et al. Nutrient enrichment drives the sediment microbial communities in Chinese mitten crab Eriocheir sinensis culture. Environ Res. (2023) 223:115281. doi: 10.1016/j.envres.2023.115281
66. Fan J, Chen L, Mai G, Zhang H, Yang J, Deng D, et al. Dynamics of the gut microbiota in developmental stages of Litopenaeus vannamei reveal its association with body weight. Sci Rep. (2019) 9:734. doi: 10.1038/s41598-018-37042-3
67. Tao L, Chai J, Liu H, Huang W, Zou Y, Wu M, et al. Characterization and dynamics of the gut microbiota in rice fishes at different developmental stages in rice-fish coculture systems. Microorganisms. (2022) 10:2373. doi: 10.3390/microorganisms10122373
68. Sun Y, Han W, Liu J, Liu F, Cheng Y. Microbiota comparison in the intestine of juvenile Chinese mitten crab Eriocheir sinensis fed different diets. Aquaculture. (2020) 515:734518. doi: 10.1016/j.aquaculture.2019.734518
69. Tanaka R, Ootsubo M, Sawabe T, Ezura Y, Tajima K. Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture-independent techniques. Aquaculture. (2004) 241:453–63. doi: 10.1016/j.aquaculture.2004.08.032
70. Yang G, Bao B, Peatman E, Li H, Huang L, Ren D. Analysis of the composition of the bacterial community in puffer fish Takifugu obscurus. Aquaculture. (2007) 262:183–91. doi: 10.1016/j.aquaculture.2006.11.031
71. Dang C, Wang J, He Y, Yang S, Chen Y, Liu T, et al. Rare biosphere regulates the planktonic and sedimentary bacteria by disparate ecological processes in a large source water reservoir. Water Res. (2022) 216:118296. doi: 10.1016/j.watres.2022.118296
72. Jin L, Ding A, Lin J, Wu X, Ji G. Dynamics of phytoplankton communities and environmental drivers in Chinese mitten crab aquaculture ponds: highlighting the need for Cyanobacteria control. Water. (2024) 16:1688. doi: 10.3390/w16121688
73. Bolaños LM, Tait K, Somerfield PJ, Parsons RJ, Giovannoni SJ, Smyth T, et al. Influence of short and long term processes on SAR11 communities in open ocean and coastal systems. ISME Commun. (2022) 2:116. doi: 10.1038/s43705-022-00198-1
74. Garrido-Oter R, Nakano RT, Dombrowski N, Ma K-W, McHardy AC, Schulze-Lefert P. Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe. (2018) 24:155–67.e5. doi: 10.1016/j.chom.2018.06.006
75. Juliyanti V, Itakura R, Kotani K, Lim SY, Suzuki G, Chong CW, et al. Rahman S. Comparative analysis of root associated microbes in tropical cultivated and weedy rice (Oryza spp.) and temperate cultivated rice. Sci Rep. (2024) 14:9656. doi: 10.1038/s41598-024-60384-0
76. Liu X, Lu X, Zhao W, Yang S, Wang J, Xia H, et al. The rhizosphere effect of native legume Albizzia julibrissin on coastal saline soil nutrient availability, microbial modulation, and aggregate formation. Sci Total Environ. (2022) 806:150705. doi: 10.1016/j.scitotenv.2021.150705
77. Zhang J, Ge Z, Ma Z, Huang D, Zhang J. Seasonal changes driving shifts of aquatic rhizosphere microbial community structure and the functional properties. J Environ Manage. (2022) 322:116124. doi: 10.1016/j.jenvman.2022.116124
78. Guo K, Glatter T, Paczia N, Liesack W. Asparagine uptake: a cellular strategy of Methylocystis to combat severe salt stress. Appl Environ Microbiol. (2023) 89:e00113–23. doi: 10.1128/aem.00113-23
79. Deng Y, Gui Q, Dumont M, Han C, Deng H, Yun J, et al. Methylococcaceae are the dominant active aerobic methanotrophs in a Chinese tidal marsh. Environ Sci Pollut Res. (2019) 26:636–46. doi: 10.1007/s11356-018-3560-3
80. Orata FD, Meier-Kolthoff JP, Sauvageau D, Stein LY. Phylogenomic analysis of the gammaproteobacterial methanotrophs (order Methylococcales) calls for the reclassification of members at the genus and species levels. Front Microbiol. (2018) 9:3162. doi: 10.3389/fmicb.2018.03162
81. Shi H, Lu L, Ye J, Shi L. Effects of two Bacillus velezensis microbial inoculants on the growth and rhizosphere soil environment of Prunus davidiana. IJMS. (2022) 23:13639. doi: 10.3390/ijms232113639
82. Eck M, Sare AR, Massart S, Schmautz Z, Junge R, Smits THM, et al. Exploring bacterial communities in aquaponic systems. Water. (2019) 11:260. doi: 10.3390/w11020260
83. Zhang Z, Li D, Xu W, Tang R, Li L. Microbiome of co-cultured fish exhibits host selection and niche differentiation at the organ scale. Front Microbiol. (2019) 10:2576. doi: 10.3389/fmicb.2019.02576
84. Wang Y, Wang C, Chen Y, Zhang D, Zhao M, Li H, et al. Microbiome analysis reveals microecological balance in the emerging rice–crayfish integrated breeding mode. Front Microbiol. (2021) 12:669570. doi: 10.3389/fmicb.2021.669570
85. Wang P, You G, Gao Y, Chen J, Wang X, Wang C. Agricultural practices and ditch size drive microbial community assembly and mediate N- and P-transformation in multistage drainage networks of paddy fields: Insights from a large-scale irrigation district in eastern China. J Environ Manage. (2024) 350:119625. doi: 10.1016/j.jenvman.2023.119625
86. Chen Y-J, Leung PM, Wood JL, Bay SK, Hugenholtz P, Kessler AJ, et al. Metabolic flexibility allows bacterial habitat generalists to become dominant in a frequently disturbed ecosystem. ISME J. (2021) 15:2986–3004. doi: 10.1038/s41396-021-00988-w
87. Murphy CL, Yang R, Decker T, Cavalliere C, Andreev V, Bircher N, et al. Genomes of novel Myxococcota reveal severely curtailed machineries for predation and cellular differentiation. Appl Environ Microbiol. (2021) 87:e0170621 doi: 10.1101/2021.07.06.451402
88. Bell E, Lamminmäki T, Alneberg J, Qian C, Xiong W, Hettich RL, et al. Active anaerobic methane oxidation and sulfur disproportionation in the deep terrestrial subsurface. ISME J. (2022) 16:1583–93. doi: 10.1038/s41396-022-01207-w
89. Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, Wagner M, et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol. (2020) 70:5972–6016. doi: 10.1099/ijsem.0.004213
90. Jiang X, Ma H, Zhao Q, Yang J, Xin C, Chen B. Bacterial communities in paddy soil and ditch sediment under rice-crab co-culture system. AMB Expr. (2021) 11:163. doi: 10.1186/s13568-021-01323-4
91. Beulig F, Heuer VB, Akob DM, Viehweger B, Elvert M, Herrmann M, et al. Carbon flow from volcanic CO2 into soil microbial communities of a wetland mofette. ISME J. (2015) 9:746–59. doi: 10.1038/ismej.2014.148
92. Qin L, Tian W, Freeman C, Jia Z, Yin X, Gao C, Zou Y, Jiang M. Changes in bacterial communities during rice cultivation remove phenolic constraints on peatland carbon preservation. ISME Commun. (2024) 4:ycae022. doi: 10.1093/ismeco/ycae022
93. Huang J, Li J, Zhou W, Cheng Y, Li J. Effect of different rice transplanting patterns on microbial community in water, sediment, and Procambarus clarkii intestine in rice-crayfish system. Front Microbiol. (2023) 14:1233815. doi: 10.3389/fmicb.2023.1233815
94. Zhang X, Xu M, Shi F. Impact of typical agricultural land use on the characteristics of soil microbial communities in the Nyingchi region of southeastern Tibet. J Agro-Environ Sci. (2020) 39:331–42.
95. Rajer FU, Samma MK, Ali Q, Rajar WA, Wu H, Raza W, et al. Gao X. Bacillus spp.-mediated growth promotion of rice seedlings and suppression of bacterial blight disease under greenhouse conditions. Pathogens. (2022) 11:1251. doi: 10.3390/pathogens11111251
96. Tu C-K, Huang W-D, Wang P-H, Lin W-L, Chen H-Y, Rau S-T, et al. The rice endophytic bacterium Bacillus velezensis LS123N provides protection against multiple pathogens and enhances rice resistance to wind with increase in yield. Biol Control. (2024) 192:105507. doi: 10.1016/j.biocontrol.2024.105507
97. Ali Q, Ayaz M, Mu G, Hussain A, Yuanyuan Q, Yu C, et al. Revealing plant growth-promoting mechanisms of Bacillus strains in elevating rice growth and its interaction with salt stress. Front Plant Sci. (2022) 13:994902. doi: 10.3389/fpls.2022.994902
98. Zhou H, Ren Z, Zu X, Yu X, Zhu H, Li X, et al. Efficacy of plant growth-promoting bacteria Bacillus cereus YN917 for biocontrol of rice blast. Front Microbiol. (2021) 12:684888. doi: 10.3389/fmicb.2021.684888
99. Zhu F, Wang J, Jia Y, Tian C, Zhao D, Wu X, et al. Bacillus subtilis GB519 promotes rice growth and reduces the damages caused by rice blast fungus Magnaporthe oryzae. PhytoFrontiersTM. (2021) 1:330–8. doi: 10.1094/PHYTOFR-12-20-0041-R
100. Prosser JI, Head IM, Stein LY. The family Nitrosomonadaceae. In:Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, , editors. The Prokaryotes. Berlin, Heidelberg: Springer (2014). p. 901–18.
101. Preena PG, Rejish Kumar VJ, Singh ISB. Nitrification and denitrification in recirculating aquaculture systems: the processes and players. Rev Aquacult. (2021) 13:2053–75. doi: 10.1111/raq.12558
102. Kim Y, Liesack W. Differential assemblage of functional units in paddy soil microbiomes. PLoS ONE. (2015) 10:e0122221. doi: 10.1371/journal.pone.0122221
103. Chen X, Fan L, Qiu L, Dong X, Wang Q, Hu G, et al. Metagenomics analysis reveals compositional and functional differences in the gut microbiota of red swamp crayfish, procambarus clarkii, grown on two different culture environments. Front Microbiol. (2021) 12:735190. doi: 10.3389/fmicb.2021.735190
104. Dehler CE, Secombes CJ, Martin SAM. Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci Rep. (2017) 7:13877. doi: 10.1038/s41598-017-13249-8
105. Xu R, Li Q, Wang H, Su Y, Zhu W. Reduction of redox potential exerts a key role in modulating gut microbial taxa and function by dietary supplementation of pectin in a pig model. Microbiol Spectr. (2023) 11:e03283–22. doi: 10.1128/spectrum.03283-22
106. Deris ZM, Iehata S, Gan HM, Ikhwanuddin M, Najiah M, Asaduzzaman M, et al. Understanding the effects of salinity and Vibrio harveyi on the gut microbiota profiles of Litopenaeus vannamei. Front Mar Sci. (2022) 9:974217. doi: 10.3389/fmars.2022.974217
107. Chen Y, Hua X, Li S, Zhao J, Yu H, Wang D, Yang J, Liu L. Aromatic compound 2-acetyl-1-pyrroline coordinates nitrogen assimilation and methane mitigation in fragrant rice. Curr Biol. (2024) 34:3429–38.e4. doi: 10.1016/j.cub.2024.06.063
108. Yuan J, Xiang J, Liu D, Kang H, He T, Kim S, et al. Rapid growth in greenhouse gas emissions from the adoption of industrial-scale aquaculture. Nat Clim Chang. (2019) 9:318–22. doi: 10.1038/s41558-019-0425-9
109. Wang C, Shi X, Qi Z, Xiao Y, Zhao J, Peng S, et al. How does rice-animal co-culture system affect rice yield and greenhouse gas? A meta-analysis. Plant Soil. (2023) 493:325–40. doi: 10.1007/s11104-023-06233-x
110. Glissmann K, Hammer E, Conrad R. Production of aromatic compounds during methanogenic degradation of straw in rice field soil. FEMS Microbiol Ecol. (2005) 52:43–8. doi: 10.1016/j.femsec.2004.10.004
111. Sun R, Li W, Hu C, Liu B. Long-term urea fertilization alters the composition and increases the abundance of soil ureolytic bacterial communities in an upland soil. FEMS Microbiol Ecol. (2019) 95:e0170621. doi: 10.1093/femsec/fiz044
Keywords: Eriocheir sinensis, gut microbiota, environmental microbiota, culture system, 16S rRNA, growth performance, enzyme activity
Citation: Zhu X, Nie M, Sun N, Zhang Y, Sun M, Li C, Jiang Q, Wei H, Li Y, Hu Q, Zhao Y and Li X (2025) Comparative analysis of crab growth performance, enzyme activity, and microbiota between rice-crab coculture and pond farming systems. Front. Vet. Sci. 12:1571454. doi: 10.3389/fvets.2025.1571454
Received: 05 February 2025; Accepted: 03 March 2025;
Published: 19 March 2025.
Edited by:
Donghong Niu, Shanghai Ocean University, ChinaReviewed by:
Xing Wu, Chinese Academy of Sciences (CAS), ChinaHonglin Chen, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2025 Zhu, Nie, Sun, Zhang, Sun, Li, Jiang, Wei, Li, Hu, Zhao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingying Zhao, emhhb3l5QHN5YXUuZWR1LmNu; Xiaodong Li, bGl4aWFvZG9uZ0BzeWF1LmVkdS5jbg==
 Xiaochen Zhu
Xiaochen Zhu Miao Nie2
Miao Nie2 Yingdong Li
Yingdong Li Qingbiao Hu
Qingbiao Hu Yingying Zhao
Yingying Zhao Xiaodong Li
Xiaodong Li