
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 14 February 2025
Sec. Livestock Genomics
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1540613
This article is part of the Research Topic Advances in Livestock Genetics: Enhancing Breeding Practices and Improving Animal Health View all 5 articles
Long non-coding RNAs (lncRNAs) are RNA molecules exceeding 200 nucleotides in length. Recent studies have demonstrated their involvement in regulating gene expression and various biological processes. Among these, myogenesis and lipogenesis are particularly important because of their direct effects on muscle development and fat deposition in farm animals. These processes are crucial for determining meat quality, growth rates, and overall economic value in animal husbandry. Although the specific mechanisms through which lncRNAs influence these pathways are still under investigation, further research into their roles in muscle and fat development is crucial for optimizing farm animal breeding strategies. Here, we review the characteristics of lncRNAs, including their biogenesis, localization, and structures, with a particular focus on their association with myogenesis and adipogenesis. This review seeks to establish a theoretical foundation for enhancing farm animal production. In particular, focusing on lncRNAs may reveal how these molecules can enhance the economic traits of farm animals, thereby contributing to the optimization of farm animal breeding processes.
Farm animals are important as they can supply the basic nutritional needs of humankind, including meat, eggs, and milk. Additionally, animal products such as gelatin are widely used as food additives in confectionery, and farm animal waste serves as an excellent fertilizer. Optimizing farm animal production is crucial to meet the ever-growing demand for animal products. A key strategy would be to improve the amount and quality of farm animal products by understanding the molecular mechanisms of the key biological processes that govern animal reproduction and well-being. The mechanisms that regulate skeletal muscle growth, development, and fat deposition are critical determinants of meat yield and quality.
Numerous studies have attempted to elucidate the molecular mechanisms underlying various animal traits, especially skeletal muscle and adipose tissue development. For instance, myogenesis regulators such as MyoD, Myf5, Myogenin (MyoG), and MRF4 are crucial for skeletal muscle development, satellite cell activation, and regenerative myogenesis (1). In addition, genetic variations in fatty acid synthesis and deposition between different cattle breeds have been observed to influence beef marbling (2). Recent studies have shown that traits in farm animals, such as muscle (3–6) and fat development (7–11), are influenced not only by coding genes but also by the regulatory roles of long non-coding RNAs (lncRNAs).
LncRNAs are a type of non-coding RNA (ncRNA) molecule exceeding 200 nucleotides (nts) in length and are known to significantly influence various traits in farm animals. For example, lncRNA muscle growth promoting factor (lncMGPF) is a conserved lncRNA found in pigs, which promotes muscle growth and regeneration by enhancing HuR-mediated mRNA stability of myogenic regulators and acting as a molecular sponge for miR-135a-5p (12). LncCCPG1 in bovine adipose cells can alleviate the inhibition of lncSLC30A9 expression by miR-93 through miR-93 adsorption LncSLC30A9 inhibits cell proliferation by downregulating AKT expression and promotes bovine adipocyte differentiation through the recruitment of FOS proteins to the peroxisome proliferation-activated receptor gamma (PPARγ) promoter (8). A highly specific sheep enhanced muscularity Transcript lncRNA (lnc-SEMT) has been identified in sheep skeletal muscle tissue. Lnc-SEMT acts as a molecular sponge by antagonizing miR-125b to control IGF2 protein abundance and promote sheep myoblast differentiation in vitro (13). In chicken skeletal muscle, a lncRNA named myosin, heavy chain 1G (MYH1G)-antisense transcript (MYH1G-AS) has been identified. It promotes the transcription of SMAD4 by reducing the interaction between FGF18 and SMARCA5. This action activates the SMAD4-dependent pathway, thereby enhancing the proliferation of myoblasts (14). This review will examine recent progress in understanding the role of lncRNAs in skeletal muscle development and fat deposition in different farm animal species. It will also delve into their regulatory mechanisms at the epigenetic, transcriptional, and post-transcriptional levels.
There are several ways to form lncRNAs in living cells (15): (1): Protein-coding gene open reading frames (ORFs) can be cut and mutated to produce lncRNAs (15); (2) Chromatin rearrangement can cause two distant untranscribed fragments to concatenate, generating multi-exon lncRNAs (15); (3) Retro-transposition can produce lncRNAs (15); (4) The same sequence can be duplicated resulting in the production of lncRNAs with adjacent repeating sequences (15); (5) Insertion of transposable elements can produce functional lncRNAs (15); (6) Enhancer transcription can produce enhancer-associated lncRNAs (elncRNAs) (16); (7) The upstream region of a promoter can be transcribed in order to generate short-lived lncRNAs (17); (8) Excised intron-derived small nucleolar RNA (snoRNA)-ended can give rise to some lncRNAs (18, 19). LncRNAs, similar to protein-coding genes, have conserved core promoter sequences. However, because there are fewer overlapping motifs bound by TFs in lncRNA promoters (Figure 1A), the expression levels of lncRNAs are generally lower than those of protein-coding genes (20). The DNA core promoter initiates transcription, which results in the production of mRNAs and lncRNAs. The pre-mRNAs are transcribed by Pol II and must undergo 5′ capping, splicing and 3′ cleavage and polyadenylation. Therefore, the lncRNAs produced are frequently cleaved and prematurely terminated during co-transcription. Although the splicing mechanisms of lncRNAs resemble those of protein-coding genes, the splicing efficiency in lncRNAs is typically reduced (Figure 1B).
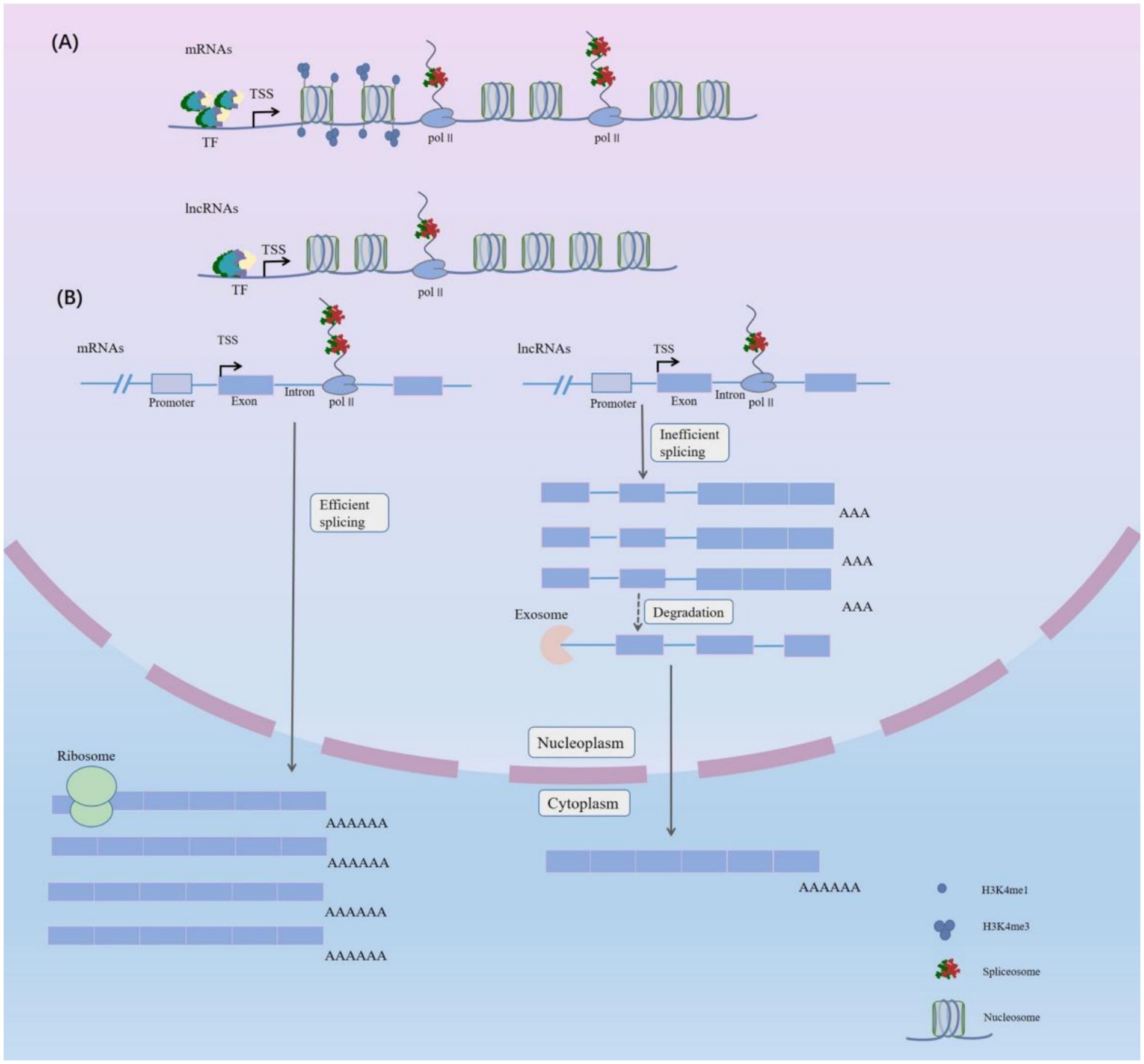
Figure 1. Distinguishing the Characteristics of Long Noncoding RNAs and mRNAs. (A) Compared with mRNAs, lncRNAs have fewer transcription factors (TFs) bound to their promoters. In addition, the splicing efficiency of lncRNAs is lower than that of mRNAs. mRNAs are associated with H3K4me1 (associated with enhancers) and H3K4me3 modifications (associated with promoters). In addition, lncRNAs are characterized by enrichment of H3K9me3 modifications at the promoter site, which correlates with tissue specificity. RNA Pol II, RNA polymerase II; TSS, transcription start site. (B) Unlike mRNAs, many lncRNAs transcribed by RNA polymerase II (Pol II) are processed inefficiently and most remain in the nucleus. Only a small proportion which are similar to mRNAs enters the cytoplasm, and some lncRNAs which are located in the nuclei are degraded by exosomes. While mRNAs are more abundant in the cytoplasm and these are invariably associated with ribosomes.
LncRNA can function through multiple ways to regulate gene expression. They can interact with proteins, RNAs, and DNAs, acting as guides (8), scaffolds (21), and bait (22) molecules in order to regulate transcription (Figure 2). This section investigates the functions of lncRNAs in the contexts of epigenetics, transcription, and post-transcription.
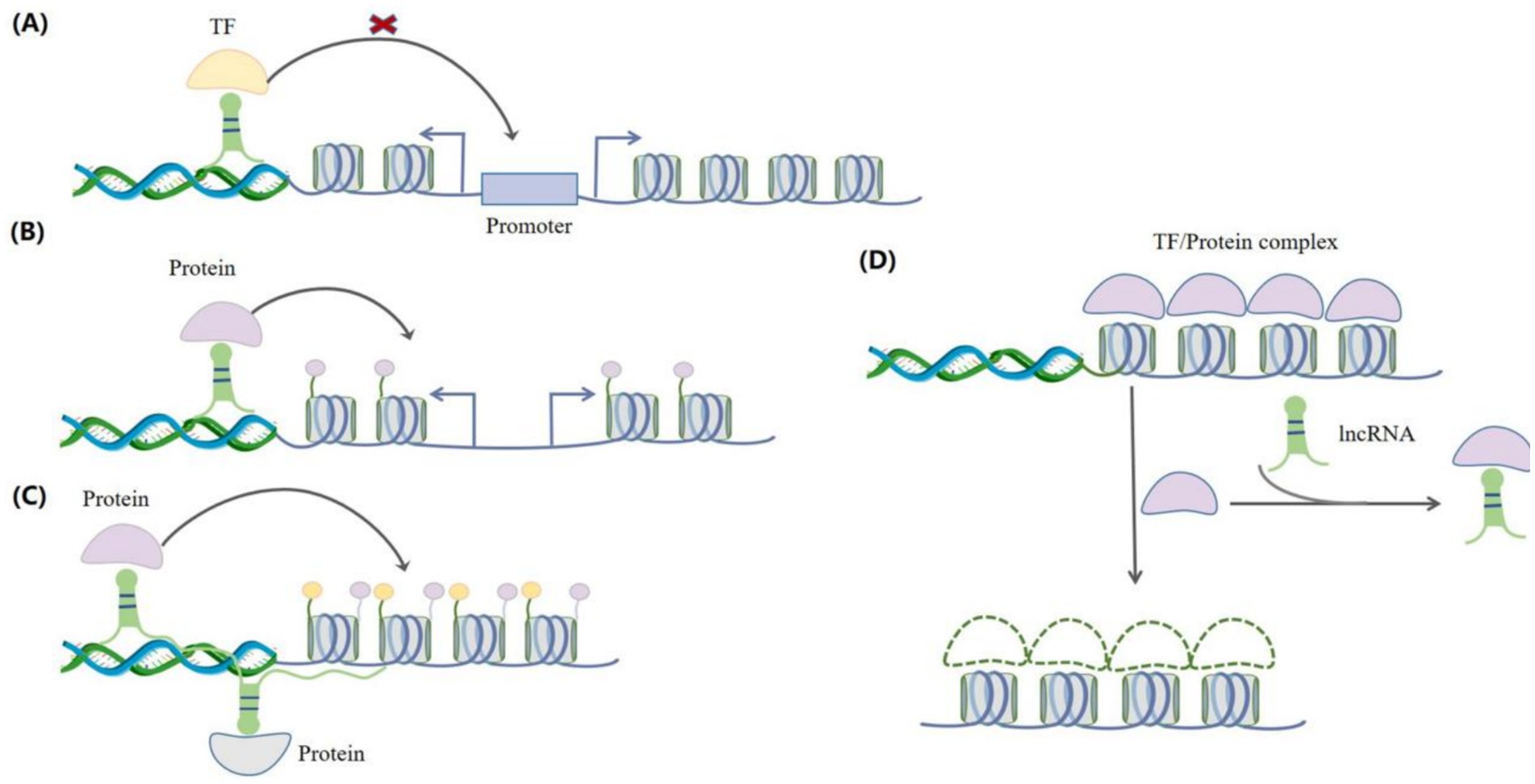
Figure 2. Interactions between lncRNAs and regulatory factors. (A) As guide molecules, lncRNAs can bind to transcription factors to prevent them from attaching to the promoters of target genes, thereby regulating gene expression. (B) As guide molecules, lncRNAs can recruit epigenetic regulators (such as proteins) to chromatin and regulate the expression of target genes. (C) As scaffold molecules, lncRNAs can recruit different protein complexes to regulate target gene expression. (D) LncRNAs can also act as decoys for transcription factors and proteins, preventing them from binding to chromatin, thereby regulating gene expression.
In eukaryotes, chromatin is highly folded and compressed, which reduces its ability to bind to TFs, promoters, and enhancers. Dynamic alterations of the chromatin structure can promote its accessibility. One of the key determinants of transcriptional activity is the state of the chromatin, and lncRNAs can regulate this parameter in several ways to mediate gene transcription and silencing. For instance, lncRNAs interact with proteins, histone-modifying enzymes, and chromatin-modifying complexes (23) and are involved in chromatin epigenetic regulation, such as chromatin remodeling (24), gene imprinting (25), and dosage compensation (26).
LncRNAs can organize chromatin domains to coordinate gene activation. LncRNAs can regulate gene expression by recruiting chromatin-modifying complexes, such as PRC2, G9a, and hnRNPK, to specific gene loci for chromatin remodeling. The HOX Transcript Antisense Intergenic RNA (lncRNA HOTAIR) recruits PRC2, leading to histone H3 lysine-27 trimethylation (H3K27me3) and transcriptional silencing of a ~ 40-kb region at the HOXD locus in various human fibroblasts cultured in vitro (24). Chromatin looping involves alterations in the three-dimensional structure of chromatin. LncRNAs promote the construction of chromatin loops by recruiting specific chromatin modification complexes (27, 28), which can alter the expression of adjacent genes (Figures 3A,B). LncRNA HOTTIP attracts the MLL1 protein’s histone lysine methyltransferase complex by directly binding to the WDR5 protein. This targets MLL1 to the HOXA site by chromatin looping, which then induces histone H3 lysine 4 trimethylation (H3K4me3) and promotes HOXA gene transcription (27).
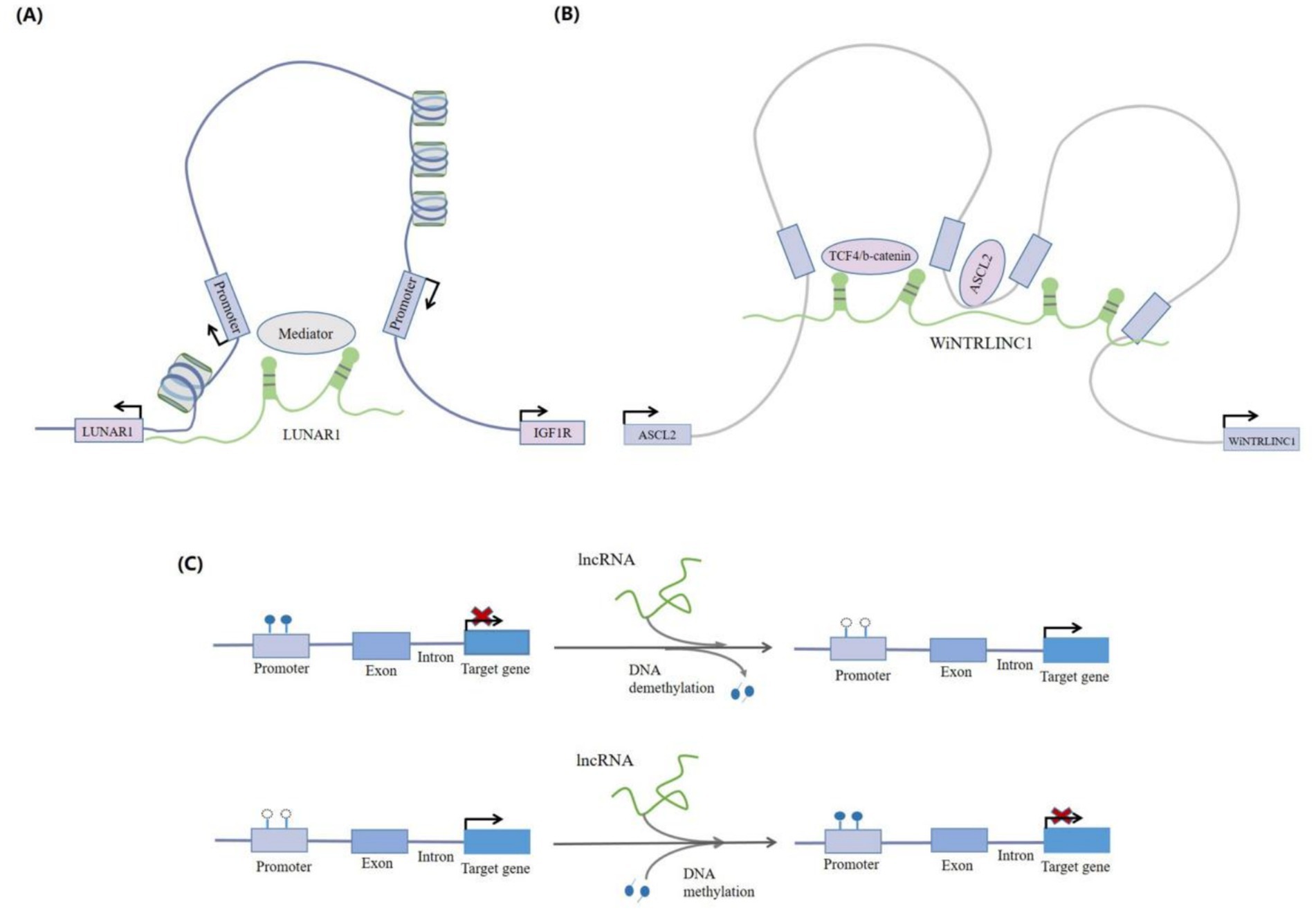
Figure 3. LncRNA-mediated chromatin regulation. (A) LncRNAs can interact with chromatin modification complexes and recruit them to target gene promoters in order to activate or repress the transcription of target genes. For example, the lncRNALUNAR1 can promote and interact with chromosome looping, and activate the transcription of IGF1R by recruiting the chromatin modification complex mediator to the promoter region of the IGF1R gene. (B) The lncRNA, WiNTRLINC1K, can bind to the TCF4/b-catenin complex and mediate the formation of chromatin loops between the promoter of WiNTRLINC1 and the regulatory domain of transcription factor, ASCL2. In turn, ASCL2 can bind to the WiNTRLINC1 promoter and positively regulate WiNTRLINC1 transcription. (C) LncRNAs can bi-directionally regulate DNA methylation levels. They can, not only promote DNA methylation in promoter regions (such as lncRNA Xist), but also play a role in DNA demethylation.
DNA methylation silencing is a common form of epigenetic regulation that suppresses gene transcription. LncRNAs often influence gene expression by modulating the methylation status of CpG islands within promoter regions. DNA methylation silencing is a common epigenetic regulatory mechanism that inhibits gene transcription. LncRNAs regulate DNA methylation in a bidirectional manner, promoting both DNA methylation and demethylation (Figure 3C). For instance, LncRNA CRNDE enhances NDRG2 expression through DNA methylation in B lymphocytes, thereby inhibiting cell proliferation and promoting apoptosis (29).
Gene transcription is a rigorous and complex biological process, and lncRNAs can regulate it through various mechanisms. The multidimensional mechanisms by which lncRNAs participate in gene regulation suggest that we can only learn more about how lncRNAs are used for gene regulation by elucidating the RNA sequences and structural elements that make the lncRNAs functional.
LncRNAs can modulate gene transcription by either recruiting TFs to target the gene promoter regions or by interfering with Pol II at the targeted loci. When lncRNAs are transcribed, they can interfere with TF binding to the promoter, thus inhibiting gene transcription. A case of this kind of transcriptional interference effect is when the lncRNA SRG1, upstream of the yeast gene SER3, straddles the SER3 promoter sequence during transcriptional elongation, sequestering Pol II from binding to the SER3. This results in the repression of SER3 transcription (30). LncRNAs can also influence gene transcription by functioning as co-transcriptional factors (co-TFs). For instance, the lncRNA Evf2 interacts with the TF DLX2, forming a complex that enhances the transcriptional activity of the Dlx-5/6 enhancer, thereby regulating the expression of Dlx5 and Dlx6 (31). LncRNAs can also influence gene transcription by hybridizing with DNA to form triple helices, recruiting transcriptional cofactors to target gene promoter regions. For example, the lncRNA Khps1 generates a DNA–RNA triplet with the upstream region of the SPHK1 enhancer, which in turn promotes the recruitment of the histone acetyltransferase p300/CBP to the SPHK1 promoter. This process activates SPHK1-eRNA transcription and enhances SPHK1 expression (32). Numerous studies have demonstrated that DNA elements like enhancers (33) and promoters (34) located at lncRNA sites, rather than lncRNA transcripts, play a regulatory role in gene transcription (Figure 4A). For instance, a DNase hypersensitive site on the Lockd locus promoter interacts with multiple TFs and regulates Cdkn1b gene expression by binding to the Cdkn1b promoter. A study demonstrated a significant reduction in Cdkn1b expression following the deletion of the 25 kb Lockd locus. However, the reduction of Lockd lncRNA transcripts did not affect the expression of Cdkn1b (34). In addition, lncRNAs can promote enhancer circularization and regulate gene transcription and expression by recruiting chromatin activation complexes to target gene promoters (Figure 4B). For example, the Colon Cancer Associated Transcript 1, the Long isoform (lncRNA CCAT1L), situated within a super-enhancer region, regulates MYC transcription by facilitating the long-range interaction between the MYC promoter and its enhancer (28).
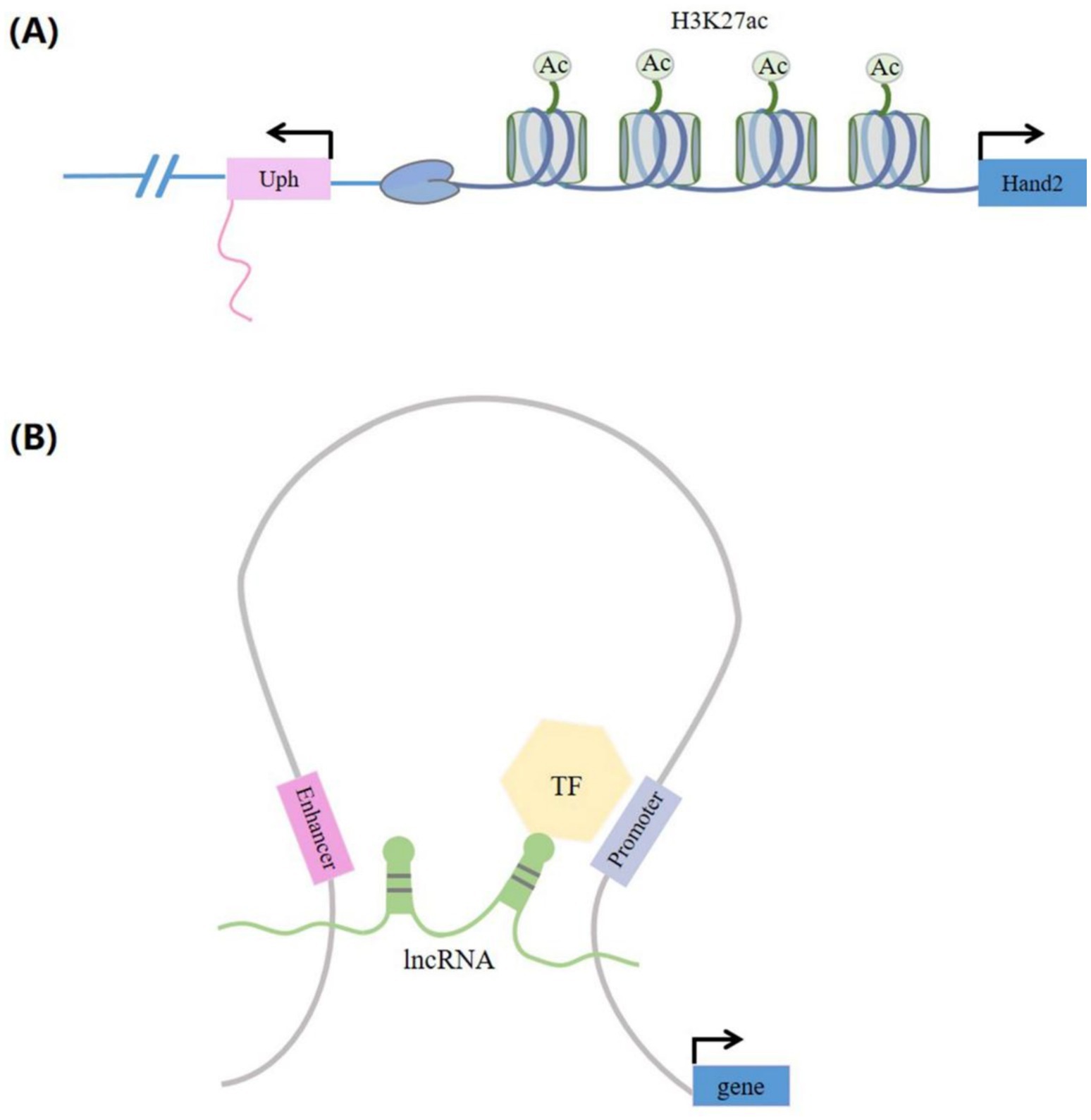
Figure 4. LncRNAs produced through enhancer-mediated gene activation. (A) When the lncRNAs were transcribed, the H3K27ac active enhancer tag in the Uph locus is guaranteed to activate transcription of the proximal Hand2 gene. (B) LncRNAs have the function of activating gene expression. LncRNAs can promote enhancer cyclization and are able to recruit chromatin-activated complexes (such as transcription factors) to the promoter of the protein coding genes. These are capable of regulating the transcription and expression of target genes. TF, transcription factors.
LncRNAs are crucial in the regulation of RNA splicing. Specifically, metastasis-associated lung adenocarcinoma transcript 1 (lncRNA-MALAT1) influences the alternative splicing of precursor mRNA by affecting the activity of serine/arginine-rich splicing factors (35). Additionally, lncRNAs can be cleaved and processed into smaller non-coding RNAs, such as miRNAs and piwi-interacting RNAs (piRNAs), which are involved in regulating gene expression after transcription.
Furthermore, LncRNAs can regulate RNA activity by interacting with proteins (36) and RNAs (37). For example, LncMyoD directly binds to IGF2-mRNA-binding protein 2 (IMP2) and negatively regulates IMP2-mediated translation of proliferation genes such as N-Ras and c-Myc (36). LncRNAs can regulate mRNA stability by absorbing miRNAs (Figure 5A). For example, intramuscular fat deposition-associated long noncoding RNA 1 (lncRNA IMFlnc1) binds to miR-199a, preventing it from degrading the target gene, caveolin-1 (CAV-1), and promoting adipogenesis (37). LncRNAs can influence mRNA stability and translation by binding to mRNAs through base pairing interactions (Figure 5B). For example, PU.1 AS lncRNA can form a sense-antisense RNA duplex with PU.1 mRNA, thereby inhibiting the translation of porcine PU.1 mRNAs (38).
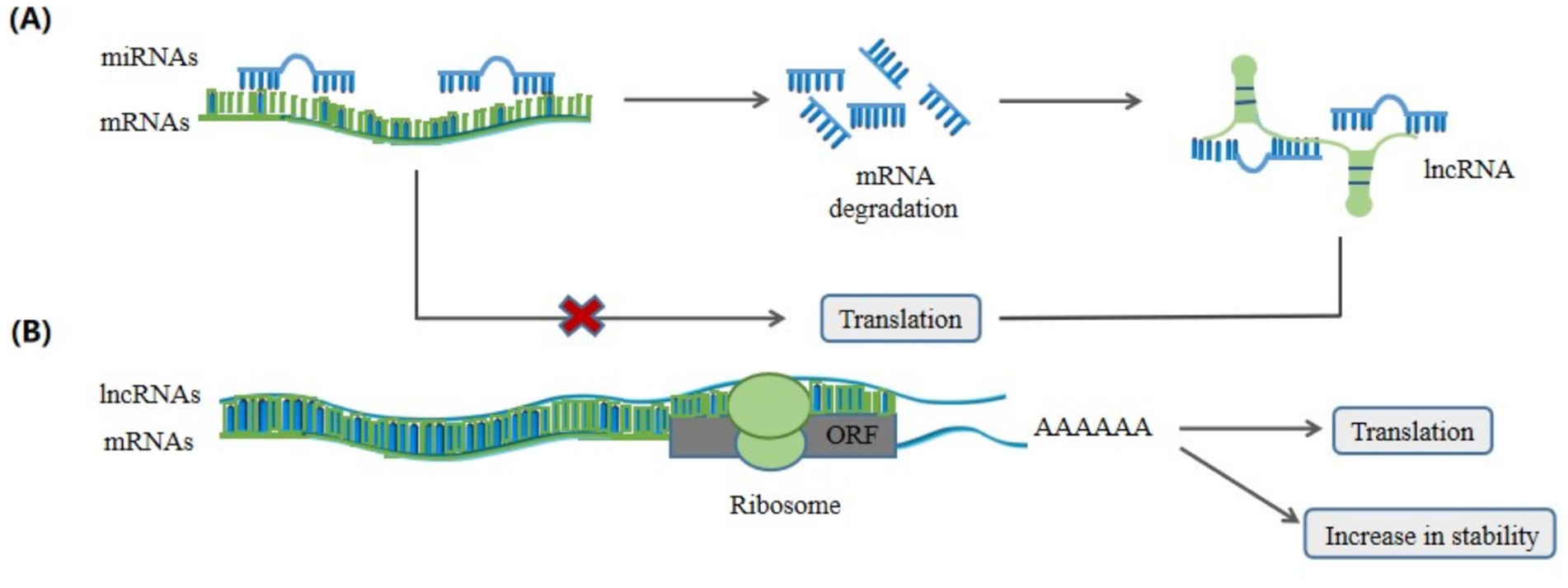
Figure 5. LncRNAs regulate mRNA degradation, stability and translation. (A) MiRNAs bind to mRNAs to regulate the degradation and translation of mRNAs, while lncRNAs can prevent miRNAs from degrading the target gene mRNAs and regulate mRNAs translation by absorbing to microRNAs (miRNAs). (B) Cytoplasmic lncRNAs can bind to mRNAs to form double-stranded RNAs in order to increase the stability and regulate the translation of the mRNAs.
Moreover, extensive research has shown that lncRNAs can encode small functional peptides. Most lncRNAs are classified as ncRNAs because they typically contain only small open reading frames (smORFs), ORFs with 100 codons or less, potentially able to be translated into proteins shorter than 100 aa before post-translational modifications (such as proteolytic cleavage) (39). However, recent research has indicated that lncRNAs can encode small peptides, which have been found to be critical for muscle growth (40) and relaxation (41, 42). By interacting with lysosomal v-type ATPase, the short peptide, SPAR, encoded by LINC00961, can limit muscle regeneration by selectively reducing the ability of mTORC1 to recognize amino acid stimulation (40).
Pigs, cattle, sheep and poultry are four key farm animals that provide abundant animal products for population worldwide, including meat, eggs and milk. LncRNAs have been demonstrated to influence economic traits in farm animals, though research on their role in these animals remains in the early stages. In this section, we review the recent studies that explore the functions and mechanisms of lncRNAs on myogenesis, lipogenesis in the aforementioned four farm animals.
Skeletal muscle is the main component of animal bodies, and its formation and structure directly influence meat yield and quality. Recent decades of research have underscored the vital role of lncRNAs in the development of skeletal muscle (Table 1).
NONCODE is a comprehensive knowledge database focusing on ncRNA genes. Among farm animals, this database1 contains the highest number of lncRNA transcripts in pigs, totaling 29,585. In recent years, researchers have studied the expression of several lncRNAs in porcine muscle tissues (11, 43–47). They have examined the differential expression of lncRNAs between muscle and other tissues (48), in cloned and normal breeding piglets (49), and in gene-edited and normal pigs (50). They have identified numerous lncRNAs linked to the development of porcine muscle in research on pig breeds with varying growth rates (43–46). Specifically, 1,407 differentially expressed lncRNAs (DELs) were found in the skeletal muscles of pigs with distinct muscle growth rates throughout their lifespans (43). Both RNA-seq and miRNA-seq techniques were used to analyze the expression of ncRNAs in the longissimus dorsi muscles (LDMs) of pigs (46). By predicting quantitative trait loci (QTL) for the DELs, it was observed that most of them were associated with muscle development (45).
The quality of pork, including color, water retention and tenderness, has been a concern for decades. Breed is a significant factor that can affect meat quality in various ways, such as total fiber count, fiber cross-sectional area (CSA), and the composition of fiber types in a specific muscle within the species. The differential expression of lncRNAs among different pig breeds may be crucial in accounting for differences in growth rates and meat quality among these breeds (44). In the study of lncRNAs in the longest muscle of pigs from the same breed at different life stages (11, 47), QTL mapping analysis of the DELs identified several loci associated with growth and meat quality traits (47). Since vertebrate skeletal muscle consists mainly of muscle fibers, the quality of fresh meat is closely linked to these fibers’ characteristics. Morphological features, including total fiber number (TNF) and (CSA), are key determinants of muscle mass and meat quality (51). In addition, the quality of fresh meat was closely correlated with the fiber type component (FTC) within muscle tissue (51). One study identified 92 differentially expressed lncRNAs in the muscles of fast-twitch Biceps femoris (Bf) and slow-twitch Soleus (Sol). The study found that lncRNA MSTRG.42019 is linked to fiber types in porcine skeletal muscle and associated with meat quality traits (52).
Meat production can be directly increased by increasing the cross-sectional area of muscle fibers (53, 54). Biological pathways that effectively increase meat yield include promoting cell proliferation, differentiation, and muscle cell hypertrophy, leading to increased muscle fiber cross-sectional area. Research has shown that lncRNAs influence the proliferation and differentiation of porcine muscle cells via diverse mechanisms, thereby regulating muscle development at the epigenetic level. LncMREF, a conserved lncRNA found in both humans and pigs, promotes myogenic differentiation and muscle regeneration through its interaction with Smarca5/p300 complexes. This interaction leads to the upregulation of key myogenesis regulators, including MyoD (55). Certain lncRNAs, such as MSTRG.59589, can impact biological processes by modulating the expression of adjacent genes. Specifically, lncRNA MSTRG.59589 increases PALLD expression and promotes the differentiation of porcine skeletal muscle satellite cells (PSCs) (5). As members of the ceRNA (competitive endogenous RNA) family, lncRNAs play a critical regulatory role in the growth of porcine muscle. For example, lncRNA maternally expressed gene 3 (lncRNA MEG3) can competitively bind to miR-423-5p, thereby upregulating serum response factor (SRF) expression and facilitating the differentiation of porcine skeletal muscle (6). Some lncRNAs are known to regulate muscle atrophy in pigs. Synaptopodin-2 (SYNPO2) intron sense-overlapping lncRNA (SYISL), a conserved lncRNA, has been shown to regulate myogenesis across mice, humans, and pigs. Additionally, SYISL can promote muscle atrophy by interacting with miR-23a-3p/miR-205-5p, and miR-103-3p (56). Interestingly, certain lncRNAs, such as H19, can influence pig muscle development through various mechanisms. Specifically, lncRNA H19 regulates the differentiation of PSCs via distinct pathways (57). H19 exhibits two distinct roles: it serves as a molecular sponge for miR-140-5p, inhibiting PSCs differentiation, and directly binds to the DBN1 protein to regulate their differentiation. Additionally, lncRNA H19 interacts directly with the DNA/RNA-binding protein TDP43 to promote PSCs differentiation (58).
The current NONCODE database2 shows that the number of known lncRNAs in cattle is second only to that in pigs, with a total of 23,515. Recent research has extensively identified lncRNAs in bovine muscle tissue. Huang et al. created the first extensive genome-wide catalog of bovine intergenic lncRNAs, identifying a total of 449 lncRNAs situated in intergenic regions (59). Since then, a series of studies have characterized lncRNAs in bovine muscle tissue and explored their functions.
Research has suggested that lncRNAs influence beef quality. Billerey et al. assessed lncRNA expression in LDMs and discovered that numerous lncRNAs are positioned within QTLs linked to meat quality (60). Since then, several studies have found that lncRNAs are located in bovine QTL chromosomal regions associated with muscle development (61, 62), and these are mainly linked to those of intramuscular fat (IMF), lean meat, and longissimus muscle regions and are associated with shear forces. One study found that the DELs in hard and tender beef tissues appeared to play important roles in physiological processes associated with meat quality (63). In addition, one study that analyzed bovine and buffalo meat, which had significant differences in shear forces and muscle fiber content, detected 2,161 DELs, which allowed the construction of co-expression and ceRNA networks (64).
Promoting cell proliferation and differentiation can increase meat production. Numerous studies have highlighted the critical role of lncRNAs in regulating the proliferation and differentiation of bovine skeletal muscle. For example, lncRNA H19 facilitates the differentiation of bovine skeletal muscle satellite cells (BSCs) by downregulating myogenic suppressor genes such as SIRT1 and FoxO1 (65). LncRNAs have been identified to regulate muscle production in cattle through a variety of pathways. LncRNAs can modulate muscle development in cattle through the adsorption of miRNAs. For instance, Liu et al. discovered and analyzed the lncRNAs in the LDM of Shandong Black and Luxi cattle breeds and found that LOC104975788 could compete with miR-133a for binding to Pax7. This allowed Pax7 expression to regulate skeletal muscle development (66). An additional instance is the binding of lncRNA MDNCR (67) to miR-133a, which stimulates GosB expression, leading to the differentiation of bovine myoblasts and reducing cell growth. LncRNA-MEG3 interacts with miRNA-135 and MEF2C to promote differentiation in bovine skeletal muscle (68). LncRNAs also influence bovine muscle development by modulating the expression of nearby genes, such as lnc403. Lnc403 is specifically expressed in bovine myoblasts and myotubes, where it suppresses the differentiation of BSCs by disrupting the expression of the neighboring gene, Myf6 (69). In addition, lncRNAs influence muscle development through interactions with proteins. Lnc23 promotes myogenesis in BSCs by binding to PFN1 protein and reducing its inhibitory effect on RhoA and Rac1 (70). Importantly, some lncRNAs such as IGF2 AS can regulate bovine myogenesis through more than one pathway. IGF2 AS acts as a complement to the IGF2 gene’s intronic area, subsequently influencing the stability and expression levels of IGF2 mRNA. Additionally, it interacts with the interleukin enhancer binding factor 3 protein to facilitate the proliferation and differentiation of bovine myoblasts.
In addition, some studies have explored lncRNAs in yak muscle. These studies revealed that some lncRNAs had varying expression levels between yaks and bovine-yak hybrids. These lncRNAs could regulate muscle growth and development in bovines through multiple signaling pathways (71). In yak embryos, Ma et al. discovered many DELs in muscle tissues at various developmental stages. Some regulatory elements, like IGF2 and Pax7, were incorporated into the co-expression networks of these lncRNAs along with their corresponding target genes (72). Furthermore, Huang et al. sequenced the transcriptomes of the LDMs from hybrids of cattle and yaks at different ages and identified 857 differentially expressed lncRNAs (73). In addition, this group also identified 791 DELs in cattle-yak and yak LDM samples and constructed six differentially expressed lncRNA-dominated ceRNA networks (71).
Compared to pigs and cattle, research on lncRNAs in sheep skeletal muscle is less extensive, with only a limited number of lncRNA transcripts identified. However, by analyzing the lncRNAs in the skeletal muscle of sheep at different developmental stages, a total of 4,738 lncRNAs were identified, including 997 that were DELs. Among these, lncGTL2 was highly expressed during the differentiation of skeletal muscle satellite cells (SCs) and was shown to promote myogenic differentiation in sheep by affecting the phosphorylation levels of PKA and CREB (74). Multiple studies have analyzed lncRNAs in the skeletal muscle of embryos at both gestational and postnatal stages. Li et al. identified 404 lncRNAs with differential expression between prenatal and postnatal stages of sheep skeletal muscle development, providing a detailed expression profile of these lncRNAs in the context of sheep skeletal muscle (75). Yuan et al. also identified several lncRNAs that regulate myogenic differentiation through interactions with miRNAs (76).
At present, research into the roles and mechanisms of lncRNAs in sheep muscle formation is still scarce. However, evidence indicates that lncRNAs could function as miRNA sponges, potentially playing a pivotal role in skeletal muscle development. The lncRNA CTTN-IT1 restores YAP1 expression by absorbing miR-29a, thereby promoting the proliferation and differentiation of satellite cells in sheep skeletal muscle (77).
Numerous studies, including those by Li et al., have highlighted the critical role of lncRNAs in muscle development in poultry, with Li et al. being the first to identify 281 novel lincRNAs in chicken skeletal muscle at different embryonic stages using RNA-seq technology. Notably, the lncRNA gga-lnc-0181 exhibits high expression levels in skeletal muscle, indicating its potential importance in muscle development (78). Since then, numerous studies have identified lncRNAs in chicken skeletal muscle at different embryonic stages (79–81). Comparing lncRNA expression in skeletal muscle tissues of border chickens with different muscle growth rates at various embryonic stages revealed that specific DELs may be essential in explaining these differences in growth rates (82). Recently, several DELs have been identified at various stages of chicken muscle development. For example, Li et al. investigated lncRNA expression in the breast muscles of both juvenile and laying chickens, indicating that these lncRNAs could be involved in breast muscle development via the MAPK signaling pathways (83). Furthermore, Ju et al. also found that lncRNAs play potential regulatory roles in oxidative and glycolytic muscle fibers in chickens (84).
Research has not only identified numerous lncRNAs in chicken muscle through sequencing but has also highlighted their crucial roles in regulating muscle development in poultry. These lncRNAs play a role in multiple processes, including the proliferation and differentiation of myoblasts, muscle fiber differentiation and transformation, and muscle atrophy. Similar to other species, some lncRNAs regulate skeletal muscle development by acting as miRNA sponges. For example, lncRNA-Six1 functions as a ceRNA by binding to miR-1611, thereby modulating the expression of the Six1 protein. This interaction influences both the proliferation and differentiation of chicken myoblasts and the transition between different muscle fiber types (4). LncIRS1 has been demonstrated to regulate muscle mass and fiber composition in living organisms. This lncRNA promotes the proliferation and differentiation of chicken myoblasts in vitro by activating the IGF1-PI3K/AKT signaling pathways through adsorption of miR-15 (85). Besides ceRNA mechanisms, some lncRNAs regulate gene expression through both cis and trans interactions. For example, the lncRNA Six1, which is highly expressed in chicken breast tissue, regulates the expression of the Six1 gene in cis. It encodes a micro-peptide that activates the Six1 gene, thereby enhancing skeletal muscle cell proliferation and promoting muscle growth in chickens (86). In addition, lncRNAs can interact with proteins to regulate muscle development in chickens. For example, epidermal differentiation protein containing cysteine histidine motifs 1(lncEDCH1) is a lncRNA that shows varying levels of expression between fat broilers and lean Chinese native breeds. The lncEDCH1 acts as a decoy to bind SERCA2 protein in order to increase its stability and its activity. This modulates Ca2+ homeostasis, promotes the slow-twitch phenotype, and helps reduce muscle atrophy (87). Additionally, lncRNA-FKBP1C, found to be differentially expressed between Bai Yinyan (WRR) and Xinghua (XH) chickens, binds to MYH1B to stabilize its protein, thereby influencing myoblast proliferation, differentiation, and the transformation of skeletal muscle fiber types (88).
Mammalian skeletal muscle is an important aspect of meat quality research, and there is a significant correlation between animal fat deposition and meat production traits. The directional deposition of fat determines the efficiency of feed utilization by farm animals. High subcutaneous fat content leads to poor meat quality, but IMF is crucial for regulating the tenderness, water retention, and flavor of the meat. In recent decades, a growing number of lncRNAs have been recognized for their significant roles in adipose tissue (Table 2). Research indicates that lncRNAs can regulate gene expression and signaling pathways associated with adipogenesis through various mechanisms.
For the past few years, reports have emerged about the discovery of lncRNAs in porcine intramuscular (PIM) tissue (89–91), dorsal (92, 93), and subcutaneous adipose tissues (94–96). LncRNAs are critical in porcine adipogenesis and can influence processes such as proliferation (97) and differentiation (98) of porcine adipocytes, which in turn affects meat quality. Inadequate IMF and excessive subcutaneous (SC) fat present the primary challenges to pork quality (99).
The content of intramuscular fat was positively associated with the flavor, tenderness, and juiciness of pork and was closely linked to overall pork quality (97, 100). LncRNAs were found to be involved in intramuscular lipogenesis in pigs. Zou et al. conducted transcriptome sequencing of porcine LDM at four developmental stages with varying IMF content, and identified 1,032 lincRNAs, among which 6 lincRNAs might be crucial for IMF development (91). Another investigation discovered 6 DELs associated with pathways related to fat deposition and lipid metabolism in the IMF of Jinhua and Landrace pigs (89). Additionally, the differentially expressed lnc_000414 has been shown to inhibit the proliferation of porcine intramuscular adipocytes in fat-type versus lean-type pigs (97).
SC fat accumulation correlates with lean carcass percentage (101); however, excessive deposition of subcutaneous fat can impair growth performance and reduce meat production efficiency (95). LncRNAs can also regulate subcutaneous fat deposition (94–96). For instance, Zhang et al. performed transcriptome sequencing on subcutaneous adipocytes from Jiaxing black pigs and Large White pigs, which exhibit substantial differences in subcutaneous fat deposition. They observed that several DEL target genes were implicated in the PI3K-Akt and MAPK signaling pathways, which are associated with fat formation and lipid metabolism (96).
Adipocyte maturation must go through two important steps: adipocyte proliferation and adipocyte differentiation. Currently, many lncRNAs are considered to be key regulators of porcine adipocyte proliferation and differentiation. For example, lncIMF4 inhibited adipogenesis in PIM pre-adipocytes by promoting lipolysis (102). LncRNAs regulate porcine lipogenesis through various mechanisms, including interactions with other RNAs. For instance, PU.1 AS lncRNAs, transcribed from the porcine PU.1 gene, can suppress PU.1 protein expression and promote porcine adipogenesis by forming sense-antisense duplexes with PU.1 mRNA (38). Furthermore, lncRNAs can also interact with proteins to regulate porcine fat development. LncMYOZ2 was shown to interact with adenosylhomocysteinase (AHCY) protein to regulate MYOZ2 expression and thus promote adipogenesis and differentiation in porcine pre-adipocytes (98). Several lncRNAs are known to regulate adipogenesis by acting as ceRNAs. The lncRNA IMFlnc1 promoted adipogenesis in PIM adipocytes by absorbing miR-199a, thereby upregulating the expression of the caveolin-1 gene in a similar way to some ceRNAs (37). In addition, lncIMF2 can promote adipogenesis in PIM pre-adipocytes by sponging miR-217 (103).
Tenderness, flavor, juiciness, and color are crucial parameters for assessing beef quality. The deposition of fat is related to the quality of beef. LncRNAs have been identified as essential regulators of lipogenesis. Recent research has revealed numerous lncRNAs linked to bovine adipose development by analyzing their expression across various developmental stages of adipose tissue (104, 105) and by contrasting them with muscle tissues (64).
The IMF content influences the degree of marbling and is regarded as a key factor impacting the sensory quality of beef (106). However, the function of lncRNAs in intramuscular fat deposition in cattle is not yet fully understood, with only a few studies addressing this aspect. Yang et al. performed comprehensive transcriptional sequencing and analyzed intramuscular preadipocytes at various differentiation stages in Qinchuan cattle, identifying 501 differentially expressed lncRNAs. In addition, they found that the lncRNAs’ target genes are linked to pathways related to lipogenesis and adipocyte differentiation. They proposed that some lncRNAs may absorb miRNAs and regulate lipogenesis (105). Another study identified lncRNAs in the longest back muscles of yaks with varying intramuscular fat content and made a similar finding (9). The function of lncRNAs in bovine intramuscular fat still requires further investigation.
Preadipocytes such as fibroblasts gradually develop into adipocytes, and adipocytes continue to accumulate and eventually form adipose tissue within. Adipogenesis occurs through the proliferation and differentiation of adipocytes. LncRNAs play a vital role in regulating the proliferation and differentiation of adipocytes. The novel lncRNA miR-221 host gene (MIR221HG), located in the transcripts of beef cattle, was identified as having differential expression during adipocyte differentiation in beef cattle, and its inhibition significantly increased adipocyte differentiation (107). Furthermore, the expression of lncFAM200B, which had higher levels in fat than in muscles, reduced cyclin D1 expression and notably suppressed the proliferation of bovine pre-adipocytes (108). LncRNAs have been shown to affect both the proliferation and differentiation of bovine adipocytes through multiple pathways. LncRNAs can influence the proliferation and differentiation of bovine adipocytes by cis-regulating gene expression. GLRX5 acts as a stimulator that enhances lipid droplet formation and the expression of adipogenic genes. Conversely, the bovine adipocyte differentiation–related lncRNA 1 (lncRNA BADLNCR1) suppresses bovine lipogenesis by downregulating GLRX5 expression (7). In addition, lncRNAs can also regulate cattle fat development as ceRNAs. For example, lncPRRX1 functions as a ceRNA to promote bovine myoblast proliferation by releasing CDC42 by competitively binding to miRNA-137 (109). The lncRNA, adipocyte differentiation-associated lncRNA (ADNCR), inhibits adipocyte differentiation by competing with miR-204 for binding, preventing it from inhibiting its target gene, SIRT1, which is a histone deacetylase (110). In buffalo adipocytes, lncRNA NDUFC2-AS promotes adipogenic differentiation by increasing the expression of C/EBP-α and THRSP (111). Sorting and assembly machinery component 50 (LncSAMM50) was also implicated to promote adipogenic differentiation of buffalo adipocytes by upregulating adipogenic markers in a 3 T3-L1 cell line in vitro (112). The regulation of lncRNAs in adipose tissue is intricate and encompasses various pathways. Therefore, evaluating their effects on adipocyte proliferation and differentiation requires consideration of their impact on signaling pathways as well.
At present, research on the regulatory functions of lncRNAs in sheep fat development is limited, with the majority of studies concentrating specifically on tail fat. Some DELs were identified by sequencing adipose tissue from different sheep breeds. Ma et al. performed high-throughput sequencing on tail adipose tissue from sheep breeds with varying levels of tail fat and identified 37 DELs (113). The study revealed that certain lncRNAs are involved in fatty acid metabolism and elongation, as well as in other pathways contributing to fat deposition. Another study identified 7 DELs in fat-tailed versus thin-tailed sheep, with target genes associated with fat development pathways. Notably, three of these lncRNAs were located within the QTLs linked to ‘tail fat deposition,’ indicating their potential role in lipid metabolism (114). Su et al. performed comprehensive transcriptome sequencing on tail tissues from sheep breeds with varying tail types, identifying 728 DELs. Among them, lncRNA-MSTRG.24995 directly affected tail fat deposition through the FASN gene, while lncRNA-MSTRG.36913 indirectly affected this process through the THRSP gene (115). In addition, Bao et al. investigated a DEL in the longissimus thoracis muscle of Tibetan sheep at four different developmental stages. This lncRNA was characterized as a trans-regulator of FASN and plays a role in regulating fatty acid metabolism throughout the growth and development of the animals (116). A recent study characterized lncRNAs in sheep intramuscular fat, finding that 61 lncRNAs were differentially expressed during fat deposition in Aohan fine wool sheep. The pathways associated with lipid accumulation were significantly enriched among the target genes of these lncRNAs (117).
Different from mammalian adipose cells, poultry adipose cells have limited capacity to generate fat. The body fat of broilers is mainly deposited in abdominal fat. Research has demonstrated that lncRNAs can modulate abdominal fat deposition in poultry. Jing et al. performed transcriptome sequencing on the abdominal adipose tissue of both fat and lean broilers, identifying 30 DELs. Among them, 16 lncRNAs were specifically expressed in adipose/lean cells (118).
The first fat to be deposited is intermuscular fat, and the amount of intermuscular fat determines the tenderness of the meat. Research has revealed that lncRNAs can modulate intermuscular fat accumulation in poultry. 7 DELs were identified during the differentiation of intramuscular pre-adipocytes, which may have significant roles in the development of intramuscular pre-adipocytes in chickens (119). LncRNA IMFNCR can also absorb miR-128-3p and miR-27b-3p, which increases the expression of PPARγ and promotes the differentiation of chicken intramuscular adipocytes (120).
Many lncRNAs have been discovered to regulate adipogenesis in chickens through various mechanisms. Some lncRNAs can influence gene expression through a cis-regulatory mechanism. For instance, adipocyte differentiation-associated lncRNA (lncAD) was identified to promote the expression of TXNRD1, thus promoting adipogenic differentiation and inhibiting the proliferation of chicken pre-muscular adipocytes (121). Other studies have also revealed that lncRNAs regulate fat development in chickens at an epigenetic level. For example, Chen et al. sequenced lncRNA in abdominal adipose tissue of broiler strains with different abdominal fat content, and found one DEL, lncPRDM16. The 5′-end functional element of lncPRDM16 is essential for it to inhibit the proliferation of adipocytes and regulate the activity of the PRDM16 promoter (122). Furthermore, some lncRNAs can also absorb miRNAs and release the expression of their target gene in order to regulate adipose formation in chickens. For example, Tian et al. identified 19,212 potential lncRNAs in the abdominal fat of chickens. MSTRG.25116.1 can absorb miR-1635, leading to increased FAAH expression, which is essential for adipogenic differentiation in chicken pre-adipocytes (119). LncRNA FNIP2 has been shown to accelerate chicken lipid synthesis through the release of FNIP2 by adsorption of miR-24-3p (123).
In this review, we discuss the latest developments in lncRNA research concerning the biogenesis, myogenesis, and lipogenesis of lncRNAs in farm animals, including pigs, cattle, sheep, and poultry. Our review highlights the growing understanding of lncRNAs and their significant impact on various biological processes affecting farm animal production. Advancements in modern molecular biology and next-generation sequencing technologies have led to the identification of an increasing number of lncRNAs associated with farm animal traits, including muscle and fat development (7–11). However, a comprehensive understanding of lncRNA functions and mechanisms in farm animals remains incomplete, with many lncRNAs and their roles still to be fully elucidated.
A major objective in farm animal production is to enhance meat yield and quality through the regulation of lncRNA expression. This involves developing strategies to harness lncRNAs, which are known to play a vital role in regulating muscle cell proliferation, differentiation, and atrophy. Techniques such as gene editing, RNA interference, and antisense oligonucleotides have been employed to overexpress or knock down specific lncRNAs, thereby influencing muscle hypertrophy and overall meat yield.
However, progress in lncRNA research in farm animals is limited by several factors. Firstly, there is a lack of data regarding the recognition and functional annotation of lncRNAs. Current lncRNA databases primarily cover humans and mice, with only a few providing expression profiles of lncRNAs in farm animals. The lncRNA sequences are generally less conserved among farm animal species, which can lead to recognition impairment due to insertions and deletions within these sequences, which may occur at the same position in the genome. Secondly, the annotation of lncRNAs in farm animals is far less complete than in humans and mice, both regarding the quantity of gene loci and the variety of alternative isomers identified. Therefore, there is a concerted and urgent need to accelerate the annotation of the non-coding regions of the farm animal genome. Thirdly, the focus is primarily on identifying lncRNAs, with a need for further research into their functions and regulatory mechanisms, particularly in cattle and sheep.
Future studies need to explore the functions and mechanisms of action of lncRNAs, as well as clarify their roles in myogenesis, adipogenesis, and other traits in farm animals. In research on the mechanisms of lncRNAs, there are fewer studies on their epigenetic and transcriptional roles, as well as their involvement in pre/during/post transcriptional processes. To explore the mechanisms of lncRNAs more effectively, further development and application of advanced techniques, such as domain-specific chromatin isolation by RNA purification and capture hybridization analysis coupled with RNA target mass spectrometry, are needed. Moreover, existing research on lncRNA functionality is primarily confined to in vitro studies involving farm animal cells or cell lines, with a limited number of in vivo investigations. Therefore, specific lncRNAs knockout models in vivo are required as their effects on the whole organism remain largely unknown. The potential of lncRNA knockout or overexpression for enhancing farm animal breeds still requires further investigation.
In conclusion, lncRNAs represent a promising frontier in farm animal research, with the potential to revolutionize animal breeding and production. The insights gained from studying lncRNAs in myogenesis and lipogenesis offer valuable opportunities for improving farm animal traits. Continued research, supported by technological advancements and interdisciplinary approaches, will be essential for fully realizing the potential benefits of lncRNAs in animal husbandry.
WL: Data curation, Writing – original draft. MC: Data curation, Writing – original draff, Writing – review & editing. YL: Visualization, Writing – review & editing. XL: Writing – review & editing, Visualization. HL: Data curation, Funding acquisition, Resources, Supervision, Writing – review & editing. JW: Data curation, Funding acquisition, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Specific Research Project of Guangxi for Research Bases and Talents (Guike AD23026212), Guangxi Natural Science Foundation (Grant No. 2024GXNSFBA010422 and 2024GXNSFFA010018), and National Natural Science Foundation of China (Grant No. 32302701, 32472863 and 32102514).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zammit, PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. (2017) 72:19–32. doi: 10.1016/j.semcdb.2017.11.011
2. Dinh, TT, Blanton, JR Jr, Riley, DG, Chase, CC Jr, Coleman, SW, Phillips, WA, et al. Intramuscular fat and fatty acid composition of longissimus muscle from divergent pure breeds of cattle. J Anim Sci. (2010) 88:756–66. doi: 10.2527/jas.2009-1951
3. Song, C, Yang, Z, Jiang, R, Cheng, J, Yue, B, Wang, J, et al. lncRNA IGF2 AS regulates bovine Myogenesis through different pathways. Mol Ther. (2020) 21:874–84. doi: 10.1016/j.omtn.2020.07.002
4. Ma, M, Cai, B, Jiang, L, Abdalla, BA, Li, Z, Nie, Q, et al. lncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 protein expression and Fiber type switching in chicken Myogenesis. Cells. (2018) 7:243. doi: 10.3390/cells7120243
5. Li, L, Cheng, X, Chen, L, Li, J, Luo, W, and Li, C. Long noncoding ribonucleic acid MSTRG.59589 promotes porcine skeletal muscle satellite cells differentiation by enhancing the function of PALLD. Front Genet. (2019) 10:10. doi: 10.3389/fgene.2019.01220
6. Cheng, X, Li, L, Shi, G, Chen, L, Fang, C, Li, M, et al. MEG3 promotes differentiation of porcine satellite cells by sponging miR-423-5p to relieve inhibiting effect on SRF. Cells. (2020) 9:449. doi: 10.3390/cells9020449
7. Cai, H, Li, M, Jian, W, Song, C, Huang, Y, Lan, X, et al. A novel lncRNA BADLNCR1 inhibits bovine adipogenesis by repressing GLRX5 expression. J Cell Mol Med. (2020) 24:7175–86. doi: 10.1111/jcmm.15181
8. Kang, Z, Zhang, S, Jiang, E, Wang, X, Wang, Z, Chen, H, et al. circFLT1 and lncCCPG1 sponges miR-93 to regulate the proliferation and differentiation of adipocytes by promoting lncSLC30A9 expression. Mol Ther. (2020) 22:484–99. doi: 10.1016/j.omtn.2020.09.011
9. Wang, H, Zhong, J, Zhang, C, Chai, Z, Cao, H, Wang, J, et al. The whole-transcriptome landscape of muscle and adipose tissues reveals the ceRNA regulation network related to intramuscular fat deposition in yak. BMC Genomics. (2020) 21:347. doi: 10.1186/s12864-020-6757-z
10. Wang, Z, Yin, Z-T, Zhang, F, Li, X-Q, Chen, S-R, Yang, N, et al. Dynamics of transcriptome changes during subcutaneous preadipocyte differentiation in ducks. BMC Genomics. (2019) 20:6055. doi: 10.1186/s12864-019-6055-9
11. Tan, Y, Gan, M, Shen, L, Li, L, Fan, Y, Chen, Y, et al. Profiling and functional analysis of Long noncoding RNAs and mRNAs during porcine skeletal muscle development. Int J Mol Sci. (2021) 22:503. doi: 10.3390/ijms22020503
12. Lv, W, Jin, J, Xu, Z, Luo, H, Guo, Y, Wang, X, et al. lncMGPF is a novel positive regulator of muscle growth and regeneration. J Cachexia Sarcopenia Muscle. (2020) 11:1723–46. doi: 10.1002/jcsm.12623
13. Wei, C, Wu, M, Wang, C, Liu, R, Zhao, H, Yang, L, et al. Long noncoding RNA Lnc-SEMT modulates IGF2 expression by sponging miR-125b to promote sheep muscle development and growth. Cell Physiol Biochem. (2018) 49:447–62. doi: 10.1159/000492979
14. Cai, B, Ma, M, Yuan, R, Zhou, Z, Zhang, J, Kong, S, et al. MYH1G-AS is a chromatin-associated lncRNA that regulates skeletal muscle development in chicken. Cell Mol Biol Lett. (2024) 29:9. doi: 10.1186/s11658-023-00525-x
15. Ponting, CP, Oliver, PL, and Reik, W. Evolution and functions of Long noncoding RNAs. Cell. (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
16. Statello, L, Guo, C-J, Chen, L-L, and Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2020) 22:96–118. doi: 10.1038/s41580-020-00315-9
17. Preker, P, Almvig, K, Christensen, MS, Valen, E, Mapendano, CK, Sandelin, A, et al. PROMoter uPstream transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. (2011) 39:7179–93. doi: 10.1093/nar/gkr370
18. Wu, H, Yang, L, and Chen, L-L. The diversity of Long noncoding RNAs and their generation. Trends Genet. (2017) 33:540–52. doi: 10.1016/j.tig.2017.05.004
19. Chen, X, He, L, Zhao, Y, Li, Y, Zhang, S, Sun, K, et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell. Discovery. (2017) 3:2017. doi: 10.1038/celldisc.2017.2
20. Mattioli, K, Volders, P-J, Gerhardinger, C, Lee, JC, Maass, PG, Melé, M, et al. High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res. (2019) 29:344–55. doi: 10.1101/gr.242222.118
21. Tsai, M-C, Manor, O, Wan, Y, Mosammaparast, N, Wang, JK, Lan, F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. (2010) 329:689–93. doi: 10.1126/science.1192002
22. Lambrou, GI, Hatziagapiou, K, and Zaravinos, A. The non-coding RNA GAS5 and its role in tumor therapy-induced resistance. Int J Mol Sci. (2020) 21:633. doi: 10.3390/ijms21207633
23. Ransohoff, JD, Wei, Y, and Khavari, PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. (2017) 19:143–57. doi: 10.1038/nrm.2017.104
24. Rinn, JL, Kertesz, M, Wang, JK, Squazzo, SL, Xu, X, Brugmann, SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
25. Kelley, RL, and Kuroda, MI. Noncoding RNA genes in dosage compensation and imprinting. Cell. (2000) 103:9–12. doi: 10.1016/S0092-8674(00)00099-4
26. Chen, CK, Blanco, M, Jackson, C, Aznauryan, E, Ollikainen, N, Surka, C, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. (2016) 354:468–72. doi: 10.1126/science.aae0047
27. Wang, KC, Yang, YW, Liu, B, Sanyal, A, Corces-Zimmerman, R, Chen, Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. (2011) 472:120–4. doi: 10.1038/nature09819
28. Xiang, J-F, Yin, Q-F, Chen, T, Zhang, Y, Zhang, X-O, Wu, Z, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. (2014) 24:513–31. doi: 10.1038/cr.2014.35
29. Yang, Z, Xu, F, Teschendorff, AE, Zhao, Y, Yao, L, Li, J, et al. Insights into the role of long non-coding RNAs in DNA methylation mediated transcriptional regulation. Front Mol Biosci. (2022) 9:9. doi: 10.3389/fmolb.2022.1067406
30. Martens, JA, Laprade, L, and Winston, F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. (2004) 429:571–4. doi: 10.1038/nature02538
31. Feng, J, Bi, C, Clark, BS, Mady, R, Shah, P, and Kohtz, JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. (2006) 20:1470–84. doi: 10.1101/gad.1416106
32. Postepska-Igielska, A, Giwojna, A, Gasri-Plotnitsky, L, Schmitt, N, Dold, A, Ginsberg, D, et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell. (2015) 60:626–36. doi: 10.1016/j.molcel.2015.10.001
33. Engreitz, JM, Haines, JE, Perez, EM, Munson, G, Chen, J, Kane, M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. (2016) 539:452–5. doi: 10.1038/nature20149
34. Paralkar Vikram, R, Taborda Cristian, C, Huang, P, Yao, Y, Kossenkov Andrew, V, Prasad, R, et al. Unlinking an lncRNA from its associated cis element. Mol Cell. (2016) 62:104–10. doi: 10.1016/j.molcel.2016.02.029
35. Tripathi, V, Ellis, JD, Shen, Z, Song, DY, Pan, Q, Watt, AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. (2010) 39:925–38. doi: 10.1016/j.molcel.2010.08.011
36. Gong, C, Li, Z, Ramanujan, K, Clay, I, Zhang, Y, Lemire-Brachat, S, et al. A Long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. (2015) 34:181–91. doi: 10.1016/j.devcel.2015.05.009
37. Wang, J, Chen, M-y, Chen, J-f, Ren, Q-l, Zhang, J-q, Cao, H, et al. LncRNA IMFlnc1 promotes porcine intramuscular adipocyte adipogenesis by sponging miR-199a-5p to up-regulate CAV-1. BMC Mol Cell Biol. (2020) 21:77. doi: 10.1186/s12860-020-00324-8
38. Wei, N, Wang, Y, Xu, RX, Wang, GQ, Xiong, Y, Yu, TY, et al. PU.1antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. Anim Genet. (2015) 46:133–40. doi: 10.1111/age.12275
39. Andrews, SJ, and Rothnagel, JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. (2014) 15:193–204. doi: 10.1038/nrg3520
40. Matsumoto, A, Pasut, A, Matsumoto, M, Yamashita, R, Fung, J, Monteleone, E, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. (2017) 541:228–32. doi: 10.1038/nature21034
41. Anderson Douglas, M, Anderson Kelly, M, Chang, C-L, Makarewich Catherine, A, Nelson Benjamin, R, McAnally John, R, et al. A micropeptide encoded by a putative Long noncoding RNA regulates muscle performance. Cell. (2015) 160:595–606. doi: 10.1016/j.cell.2015.01.009
42. Nelson, BR, Makarewich, CA, Anderson, DM, Winders, BR, Troupes, CD, Wu, F, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. (2016) 351:271–5. doi: 10.1126/science.aad4076
43. Gao, PF, Guo, XH, du, M, Cao, GQ, Yang, QC, Pu, ZD, et al. LncRNA profiling of skeletal muscles in large white pigs and Mashen pigs during development1,2. J Anim Sci. (2017) 95:4239–50. doi: 10.2527/jas2016.1297
44. Hou, X, Wang, L, Zhao, F, Liu, X, Gao, H, Shi, L, et al. Genome-wide expression profiling of mRNAs, lncRNAs and circRNAs in skeletal muscle of two different pig breeds. Animals. (2021) 11:169. doi: 10.3390/ani11113169
45. Huang, Z, Li, Q, Li, M, and Li, C. Transcriptome analysis reveals the long intergenic noncoding RNAs contributed to skeletal muscle differences between Yorkshire and Tibetan pig. Sci Rep. (2021) 11:126. doi: 10.1038/s41598-021-82126-2
46. Sun, J, Xie, M, Huang, Z, Li, H, Chen, T, Sun, R, et al. Integrated analysis of non-coding RNA and mRNA expression profiles of 2 pig breeds differing in muscle traits. J Anim Sci. (2017) 95:867. doi: 10.2527/jas2016.0867
47. Zhao, WJ, Li, ZJ, Liu, Q, Xie, S, Li, MX, Wang, Y, et al. Analysis of long intergenic non-coding RNAs transcriptomic profiling in skeletal muscle growth during porcine embryonic development. Sci Rep. (2021) 11:14. doi: 10.1038/s41598-021-94014-w
48. Chen, G, Cheng, X, Shi, G, Zou, C, Chen, L, Li, J, et al. Transcriptome analysis reveals the effect of Long intergenic noncoding RNAs on pig muscle growth and fat deposition. Biomed Res Int. (2019) 2019:1–15. doi: 10.1155/2019/2951427
49. Zou, C, Li, JX, Luo, WZ, Li, L, Hu, A, Fu, YH, et al. Transcriptome analysis reveals long intergenic non-coding RNAs involved in skeletal muscle growth and development in pig. Sci Rep. (2017) 7:8704. doi: 10.1038/s41598-017-07998-9
50. Wang, D, Pu, Y, Li, Y, Pan, D, Wang, S, Tian, W, et al. Comprehensive analysis of lncRNAs involved in skeletal muscle development in ZBED6-knockout Bama pigs. BMC Genomics. (2021) 22:593. doi: 10.1186/s12864-021-07881-y
51. Lee, SH, Joo, ST, and Ryu, YC. Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality. Meat Sci. (2010) 86:166–70. doi: 10.1016/j.meatsci.2010.04.040
52. Li, R, Li, B, Jiang, A, Cao, Y, Hou, L, Zhang, Z, et al. Exploring the lncRNAs related to skeletal muscle Fiber types and meat quality traits in pigs. Genes. (2020) 11:883. doi: 10.3390/genes11080883
53. Bernat, JL, and Ochoa, JL. Muscle hypertrophy after partial denervation: a human case. J Neurol Neurosurg Psychiatry. (1978) 41:719–25. doi: 10.1136/jnnp.41.8.719
54. Walters, J. Muscle hypertrophy and pseudohypertrophy. Pract Neurol. (2017) 17:369–79. doi: 10.1136/practneurol-2017-001695
55. Lv, W, Jiang, W, Luo, H, Tong, Q, Niu, X, Liu, X, et al. Long noncoding RNAlncMREFpromotes myogenic differentiation and muscle regeneration by interacting with the Smarca5/p300 complex. Nucleic Acids Res. (2022) 50:10733–55. doi: 10.1093/nar/gkac854
56. Jin, J, du, M, Wang, J, Guo, Y, Zhang, J, Zuo, H, et al. Conservative analysis of Synaptopodin-2 intron sense-overlapping lncRNA reveals its novel function in promoting muscle atrophy. J Cachexia Sarcopenia Muscle. (2022) 13:2017–30. doi: 10.1002/jcsm.13012
57. Li, J, Su, T, Zou, C, Luo, W, Shi, G, Chen, L, et al. Long non-coding RNA H19 regulates porcine satellite cell differentiation through miR-140-5p/SOX4 and DBN1. Front Cell Dev Biol. (2020) 8:8. doi: 10.3389/fcell.2020.518724
58. Li, J, Zhao, W, Li, Q, Huang, Z, Shi, G, and Li, C. Long non-coding RNA H19 promotes porcine satellite cell differentiation by interacting with TDP43. Genes. (2020) 11:259. doi: 10.3390/genes11030259
59. Huang, W, Long, N, and Khatib, H. Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim Genet. (2012) 43:674–82. doi: 10.1111/j.1365-2052.2012.02325.x
60. Billerey, C, Boussaha, M, Esquerré, D, Rebours, E, Djari, A, Meersseman, C, et al. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genomics. (2014) 15:499. doi: 10.1186/1471-2164-15-499
61. Choi, J-Y, Shin, D, Lee, H-J, and Oh, J-D. Comparison of long noncoding RNA between muscles and adipose tissues in Hanwoo beef cattle. Anim Cells Syst. (2018) 23:50–8. doi: 10.1080/19768354.2018.1512522
62. Liu, XF, Ding, XB, Li, X, Jin, CF, Yue, YW, Li, GP, et al. An atlas and analysis of bovine skeletal muscle long noncoding RNAs. Anim Genet. (2017) 48:278–86. doi: 10.1111/age.12539
63. Muniz, MMM, Simielli Fonseca, LF, Scalez, DCB, Vega, AS, Silva, DBS, Ferro, JA, et al. Characterization of novel lncRNA muscle expression profiles associated with meat quality in beef cattle. Evol Appl. (2022) 15:706–18. doi: 10.1111/eva.13365
64. Li, H, Huang, K, Wang, P, Feng, T, Shi, D, Cui, K, et al. Comparison of Long non-coding RNA expression profiles of cattle and Buffalo differing in muscle characteristics. Front Genet. (2020) 11:98. doi: 10.3389/fgene.2020.00098
65. Xu, X, Ji, S, Li, W, Yi, B, Li, H, Zhang, H, et al. LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/FoxO1. Cell Mol Biol Lett. (2017) 22:10. doi: 10.1186/s11658-017-0040-6
66. Liu, R, Han, M, Liu, X, Yu, K, Bai, X, and Dong, Y. Genome-wide identification and characterization of Long non-coding RNAs in longissimus dorsi skeletal muscle of Shandong black cattle and Luxi cattle. Front Genet. (2022) 13:399. doi: 10.3389/fgene.2022.849399
67. Li, H, Yang, J, Jiang, R, Wei, X, Song, C, Huang, Y, et al. Long non-coding RNA profiling reveals an abundant MDNCR that promotes differentiation of myoblasts by sponging miR-133a. Mol Ther. (2018) 12:610–25. doi: 10.1016/j.omtn.2018.07.003
68. Liu, M, Li, B, Peng, W, Ma, Y, Huang, Y, Lan, X, et al. LncRNA-MEG3 promotes bovine myoblast differentiation by sponging miR-135. J Cell Physiol. (2019) 234:18361–70. doi: 10.1002/jcp.28469
69. Zhang, XJ, Chen, MM, Liu, XF, Zhang, LL, Ding, XB, Guo, YW, et al. A novel lncRNA, lnc403, involved in bovine skeletal muscle myogenesis by mediating KRAS/Myf6. Gene. (2020) 751:144706. doi: 10.1016/j.gene.2020.144706
70. Chen, M, Zhang, L, Guo, Y, Liu, X, Song, Y, Li, X, et al. A novel lncRNA promotes myogenesis of bovine skeletal muscle satellite cells via PFN1-RhoA/Rac1. J Cell Mol Med. (2021) 25:5988–6005. doi: 10.1111/jcmm.16427
71. Huang, C, Ge, F, Ma, X, Dai, R, Dingkao, R, Zhaxi, Z, et al. Comprehensive analysis of mRNA, lncRNA, circRNA, and miRNA expression profiles and their ceRNA networks in the longissimus Dorsi muscle of cattle-yak and yak. Front Genet. (2021) 12:557. doi: 10.3389/fgene.2021.772557
72. Ma, X, Fu, D, Chu, M, Ding, X, Wu, X, Guo, X, et al. Genome-wide analysis reveals changes in polled yak Long non-coding RNAs in skeletal muscle development. Front Genet. (2020) 11:365. doi: 10.3389/fgene.2020.00365
73. Huang, C, Dai, R, Meng, G, Dingkao, R, Wang, X, Ren, W, et al. Transcriptome-wide study of mRNAs and lncRNAs modified by m6A RNA methylation in the longissimus Dorsi muscle development of cattle-yak. Cells. (2022) 11:654. doi: 10.3390/cells11223654
74. Chen, Q, Bao, J-J, Zhang, H-C, Huang, C, Zhao, Q, et al. LncRNA GTL2 regulates myoblast proliferation and differentiation via the PKA-CREB pathway in Duolang sheep. Zool Res. (2024) 45:1261–75. doi: 10.24272/j.issn.2095-8137.2024.125
75. Li, C-Y, Li, X, Liu, Z, Ni, W, Zhang, X, Hazi, W, et al. Identification and characterization of long non-coding RNA in prenatal and postnatal skeletal muscle of sheep. Genomics. (2019) 111:133–41. doi: 10.1016/j.ygeno.2018.01.009
76. Yuan, C, Zhang, K, Yue, Y, Guo, T, Liu, J, Niu, C, et al. Analysis of dynamic and widespread lncRNA and miRNA expression in fetal sheep skeletal muscle. PeerJ. (2020) 8:e9957. doi: 10.7717/peerj.9957
77. Wu, T, Wang, S, Wang, L, Zhang, W, Chen, W, Lv, X, et al. Long noncoding RNA (lncRNA) CTTN-IT1 elevates skeletal muscle satellite cell proliferation and differentiation by acting as ceRNA for YAP1 through absorbing miR-29a in Hu sheep. Front Genet. (2020) 11:843. doi: 10.3389/fgene.2020.00843
78. Li, T, Wang, S, Wu, R, Zhou, X, Zhu, D, and Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics. (2012) 99:292–8. doi: 10.1016/j.ygeno.2012.02.003
79. Li, Y, Jin, W, Zhai, B, Chen, Y, Li, G, Zhang, Y, et al. LncRNAs and their regulatory networks in breast muscle tissue of Chinese Gushi chickens during late postnatal development. BMC Genomics. (2021) 22:356. doi: 10.1186/s12864-020-07356-6
80. Li, Z, Ouyang, H, Zheng, M, Cai, B, Han, P, Abdalla, BA, et al. Integrated analysis of Long non-coding RNAs (LncRNAs) and mRNA expression profiles reveals the potential role of LncRNAs in skeletal muscle development of the chicken. Front Physiol. (2017) 7:687. doi: 10.3389/fphys.2016.00687
81. Pan, Z, Yang, C, Zhao, R, Jiang, X, Yu, C, and Li, Z. Characterization of lncRNA/circRNA-miRNA-mRNA network to reveal potential functional ceRNAs in the skeletal muscle of chicken. Front Physiol. (2022) 13:854. doi: 10.3389/fphys.2022.969854
82. Wu, P, Zhou, K, Zhang, J, Ling, X, Zhang, X, Li, P, et al. Transcriptome integration analysis at different embryonic ages reveals key lncRNAs and mRNAs for chicken skeletal muscle. Frontiers in veterinary. Science. (2022) 9:9. doi: 10.3389/fvets.2022.908255
83. Loor, JJ, Li, D, Li, F, Jiang, K, Zhang, M, Han, R, et al. Integrative analysis of long noncoding RNA and mRNA reveals candidate lncRNAs responsible for meat quality at different physiological stages in Gushi chicken. PLoS One. (2019) 14:5006. doi: 10.1371/journal.pone.0215006
84. Ju, X, Liu, Y, Shan, Y, Ji, G, Zhang, M, Tu, Y, et al. Analysis of potential regulatory LncRNAs and CircRNAs in the oxidative myofiber and glycolytic myofiber of chickens. Sci Rep. (2021) 11:20861. doi: 10.1038/s41598-021-00176-y
85. Li, Z, Cai, B, Abdalla, BA, Zhu, X, Zheng, M, Han, P, et al. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J Cachexia Sarcopenia Muscle. (2019) 10:391–410. doi: 10.1002/jcsm.12374
86. Cai, B, Li, Z, Ma, M, Wang, Z, Han, P, Abdalla, BA, et al. LncRNA-Six1 encodes a micropeptide to activate Six1 in Cis and is involved in cell proliferation and muscle growth. Front Physiol. (2017) 8:230. doi: 10.3389/fphys.2017.00230
87. Cai, B, Ma, M, Zhang, J, Wang, Z, Kong, S, Zhou, Z, et al. LncEDCH1 improves mitochondrial function to reduce muscle atrophy by interacting with SERCA2. Mol Ther. (2022) 27:319–34. doi: 10.1016/j.omtn.2021.12.004
88. Yu, J-a, Wang, Z, Yang, X, Ma, M, Li, Z, and Nie, Q. LncRNA-FKBP1C regulates muscle fiber type switching by affecting the stability of MYH1B. Cell death. Discovery. (2021) 7:463. doi: 10.1038/s41420-021-00463-7
89. Miao, Z, Wang, S, Zhang, J, Wei, P, Guo, L, Liu, D, et al. Identification and comparison of long non-conding RNA in Jinhua and landrace pigs. Biochem Biophys Res Commun. (2018) 506:765–71. doi: 10.1016/j.bbrc.2018.06.028
90. Zappaterra, M, Tan, L, Chen, Z, Teng, M, Chen, B, and Xu, H. Genome-wide analysis of mRNAs, lncRNAs, and circRNAs during intramuscular adipogenesis in Chinese Guizhou Congjiang pigs. PLoS One. (2022) 17:1293. doi: 10.1371/journal.pone.0261293
91. Zou, C, Li, L, Cheng, X, Li, C, Fu, Y, Fang, C, et al. Identification and functional analysis of Long intergenic non-coding RNAs underlying intramuscular fat content in pigs. Front Genet. (2018) 9:102. doi: 10.3389/fgene.2018.00102
92. Kumar, H, Srikanth, K, Park, W, Lee, S-H, Choi, B-H, Kim, H, et al. Transcriptome analysis to identify long non coding RNA (lncRNA) and characterize their functional role in back fat tissue of pig. Gene. (2019) 703:71–82. doi: 10.1016/j.gene.2019.04.014
93. Xing, K, Wang, K, Ao, H, Chen, S, Tan, Z, Wang, Y, et al. Comparative adipose transcriptome analysis digs out genes related to fat deposition in two pig breeds. Sci Rep. (2019) 9:12925. doi: 10.1038/s41598-019-49548-5
94. Feng, H, Liu, T, Yousuf, S, Zhang, X, Huang, W, Li, A, et al. Identification and analysis of lncRNA, miRNA and mRNA related to subcutaneous and intramuscular fat in Laiwu pigs. Front Endocrinol. (2023) 13:1460. doi: 10.3389/fendo.2022.1081460
95. Liu, X, Liu, K, Shan, B, Wei, S, Li, D, Han, H, et al. A genome-wide landscape of mRNAs, lncRNAs, and circRNAs during subcutaneous adipogenesis in pigs. J Anim Sci Biotechnol. (2018) 9:76. doi: 10.1186/s40104-018-0292-7
96. Zhang, D, Wu, W, Huang, X, Xu, K, Zheng, C, and Zhang, J. Comparative analysis of gene expression profiles in differentiated subcutaneous adipocytes between Jiaxing black and large white pigs. BMC Genomics. (2021) 22:361. doi: 10.1186/s12864-020-07361-9
97. Sun, Y, Chen, X, Qin, J, Liu, S, Zhao, R, Yu, T, et al. Comparative analysis of Long noncoding RNAs expressed during intramuscular adipocytes Adipogenesis in fat-type and lean-type pigs. J Agric Food Chem. (2018) 66:12122–30. doi: 10.1021/acs.jafc.8b04243
98. Yang, Y, Wu, Y, Ji, M, Rong, X, Zhang, Y, Yang, S, et al. The long non-coding RNA lncMYOZ2 mediates an AHCY/MYOZ2 axis to promote adipogenic differentiation in porcine preadipocytes. BMC Genomics. (2022) 23:700. doi: 10.1186/s12864-022-08923-9
99. Wu, W, Zhang, D, Yin, Y, Ji, M, Xu, K, Huang, X, et al. Comprehensive transcriptomic view of the role of the LGALS12 gene in porcine subcutaneous and intramuscular adipocytes. BMC Genomics. (2019) 20:509. doi: 10.1186/s12864-019-5891-y
100. Huang, W, Zhang, X, Li, A, Xie, L, and Miao, X. Genome-wide analysis of mRNAs and lncRNAs of intramuscular fat related to lipid metabolism in two pig breeds. Cell Physiol Biochem. (2018) 50:2406–22. doi: 10.1159/000495101
101. Grunert, KG, Bredahl, L, and Brunsø, K. Consumer perception of meat quality and implications for product development in the meat sector—a review. Meat Sci. (2004) 66:259–72. doi: 10.1016/S0309-1740(03)00130-X
102. Sun, Y, Cai, R, Wang, Y, Zhao, R, Qin, J, and Pang, W. A newly identified LncRNA LncIMF4 controls Adipogenesis of porcine intramuscular Preadipocyte through attenuating autophagy to inhibit lipolysis. Animals. (2020) 10:926. doi: 10.3390/ani10060926
103. Yi, X, He, Z, Tian, T, Kou, Z, and Pang, W. LncIMF2 promotes adipogenesis in porcine intramuscular preadipocyte through sponging MiR-217. Anim Biotechnol. (2023) 34:268–79. doi: 10.1080/10495398.2021.1956509
104. Jiang, R, Li, H, Huang, Y, Lan, X, Lei, C, and Chen, H. Transcriptome profiling of lncRNA related to fat tissues of Qinchuan cattle. Gene. (2020) 742:144587. doi: 10.1016/j.gene.2020.144587
105. Yang, X, Ma, X, Mei, C, and Zan, L. A genome-wide landscape of mRNAs, lncRNAs, circRNAs and miRNAs during intramuscular adipogenesis in cattle. BMC Genomics. (2022) 23:691. doi: 10.1186/s12864-022-08911-z
106. Stewart, SM, Gardner, GE, McGilchrist, P, Pethick, DW, Polkinghorne, R, Thompson, JM, et al. Prediction of consumer palatability in beef using visual marbling scores and chemical intramuscular fat percentage. Meat Sci. (2021) 181:108322. doi: 10.1016/j.meatsci.2020.108322
107. Li, M, Gao, Q, Tian, Z, Lu, X, Sun, Y, Chen, Z, et al. MIR221HG is a novel Long noncoding RNA that inhibits bovine adipocyte differentiation. Genes. (2019) 11:29. doi: 10.3390/genes11010029
108. Zhang, S, Kang, Z, Cai, H, Jiang, E, Pan, C, Dang, R, et al. Identification of novel alternative splicing of bovine lncRNA lncFAM200B and its effects on preadipocyte proliferation. J Cell Physiol. (2020) 236:601–11. doi: 10.1002/jcp.29887
109. Zhang, W, Sun, B, Zhao, Y, Raza, SHA, Li, Y, Wang, J, et al. Proliferation of bovine myoblast by LncPRRX1 via regulation of the miR-137/CDC42 axis. Int J Biol Macromol. (2022) 220:33–42. doi: 10.1016/j.ijbiomac.2022.08.018
110. Li, M, Sun, X, Cai, H, Sun, Y, Plath, M, Li, C, et al. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim Biophys. (2016) 1859:871–82. doi: 10.1016/j.bbagrm.2016.05.003
111. Huang, J, Zheng, Q, Wang, S, Wei, X, Li, F, and Ma, Y. High-throughput RNA sequencing reveals NDUFC2-AS lncRNA promotes Adipogenic differentiation in Chinese Buffalo (Bubalus bubalis L.). Genes. (2019) 10:689. doi: 10.3390/genes10090689
112. Zhu, R, Feng, X, Wei, Y, Guo, D, Li, J, Liu, Q, et al. lncSAMM50 enhances Adipogenic differentiation of Buffalo adipocytes with no effect on its host gene. Front Genet. (2021) 12:158. doi: 10.3389/fgene.2021.626158
113. Ma, L, Zhang, M, Jin, Y, Erdenee, S, Hu, L, Chen, H, et al. Comparative transcriptome profiling of mRNA and lncRNA related to tail adipose tissues of sheep. Front Genet. (2018) 9:365. doi: 10.3389/fgene.2018.00365
114. Bakhtiarizadeh, MR, and Salami, SA. Identification and expression analysis of Long noncoding RNAs in fat-tail of sheep breeds. G3. (2019) 9:1263–76. doi: 10.1534/g3.118.201014
115. Su, XH, He, HY, Fang, C, Liu, LL, and Liu, WJ. Transcriptome profiling of LncRNAs in sheep tail fat deposition. Anim Biotechnol. (2023) 34:900–10. doi: 10.1080/10495398.2021.2002882
116. Bao, G, Li, S, Zhao, F, Wang, J, Liu, X, Hu, J, et al. Comprehensive transcriptome analysis reveals the role of lncRNA in fatty acid metabolism in the longissimus Thoracis muscle of Tibetan sheep at different ages. Frontiers. Nutrition. (2022) 9:9. doi: 10.3389/fnut.2022.847077
117. Han, F, Li, J, Zhao, R, Liu, L, Li, L, Li, Q, et al. Identification and co-expression analysis of long noncoding RNAs and mRNAs involved in the deposition of intramuscular fat in Aohan fine-wool sheep. BMC Genomics. (2021) 22:98. doi: 10.1186/s12864-021-07385-9
118. Jing, Y, Cheng, B, Wang, H, Bai, X, Zhang, Q, Wang, N, et al. The landscape of the long non-coding RNAs and circular RNAs of the abdominal fat tissues in the chicken lines divergently selected for fatness. BMC Genomics. (2022) 23:45. doi: 10.1186/s12864-022-09045-y
119. Tian, W, Hao, X, Nie, R, Ling, Y, Zhang, B, Zhang, H, et al. Comparative transcriptome analysis reveals regulatory mechanism of Long non-coding RNAs during abdominal Preadipocyte Adipogenic differentiation in chickens. Animals. (2022) 12:1099. doi: 10.3390/ani12091099
120. Zhang, M, Li, F, Sun, J-w, Li, D-h, Li, W-t, Jiang, R-r, et al. LncRNA IMFNCR promotes intramuscular adipocyte differentiation by sponging miR-128-3p and miR-27b-3p. Front Genet. (2019) 10:10. doi: 10.3389/fgene.2019.00042
121. Zhang, M, Ma, X, Zhai, Y, Zhang, D, Sui, L, Li, W, et al. Comprehensive transcriptome analysis of lncRNAs reveals the role of lncAD in chicken intramuscular and abdominal Adipogenesis. J Agric Food Chem. (2020) 68:3678–88. doi: 10.1021/acs.jafc.9b07405
122. Chen, Y, Zhao, S, Ding, R, Li, H, Yang, C-X, and Du, Z-Q. Identification of a Long noncoding RNA (lncPRDM16) inhibiting Preadipocyte proliferation in the chicken. J Agric Food Chem. (2022) 70:1335–45. doi: 10.1021/acs.jafc.1c05554
123. Guo, L, Chao, X, Huang, W, Li, Z, Luan, K, Ye, M, et al. Whole transcriptome analysis reveals a potential regulatory mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 Axis in chicken Adipogenesis. Front Cell Dev Biol. (2021) 9:9. doi: 10.3389/fcell.2021.653798
124. Wang, L, Zhao, Y, Bao, X, Zhu, X, Kwok, YK, Sun, K, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. (2015) 25:335–50. doi: 10.1038/cr.2015.21
125. Zhou, L, Sun, K, Zhao, Y, Zhang, S, Wang, X, Li, Y, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. (2015) 6:10026. doi: 10.1038/ncomms10026
126. Zhou, Y, Cheunsuchon, P, Nakayama, Y, Lawlor, MW, Zhong, Y, Rice, KA, et al. Activation of paternally expressed genes and perinatal death caused by deletion of theGtl2gene. Development. (2010) 137:2643–52. doi: 10.1242/dev.045724
127. Yu, X, Zhang, Y, Li, T, Ma, Z, Jia, H, Chen, Q, et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nature. Communications. (2017) 8:16. doi: 10.1038/ncomms14016
128. Caretti, G, Schiltz, RL, Dilworth, FJ, di, M, Zhao, P, Ogryzko, V, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are Coregulators of MyoD and skeletal muscle differentiation. Dev Cell. (2006) 11:547–60. doi: 10.1016/j.devcel.2006.08.003
129. Mueller, AC, Cichewicz, MA, Dey, BK, Layer, R, Reon, BJ, Gagan, JR, et al. MUNC, a Long noncoding RNA that facilitates the function of MyoD in skeletal Myogenesis. Mol Cell Biol. (2023) 35:498–513. doi: 10.1128/MCB.01079-14
130. Mousavi, K, Zare, H, Dell’Orso, S, Grontved, L, Gutierrez-Cruz, G, Derfoul, A, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. (2013) 51:606–17. doi: 10.1016/j.molcel.2013.07.022
131. Cesana, M, Cacchiarelli, D, Legnini, I, Santini, T, Sthandier, O, Chinappi, M, et al. A Long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. (2011) 147:358–69. doi: 10.1016/j.cell.2011.09.028
132. Kallen Amanda, N, Zhou, X-B, Xu, J, Qiao, C, Ma, J, Yan, L, et al. The imprinted H19 LncRNA antagonizes Let-7 MicroRNAs. Mol Cell. (2013) 52:101–12. doi: 10.1016/j.molcel.2013.08.027
133. Zhu, M, Liu, J, Xiao, J, Yang, L, Cai, M, Shen, H, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nature. Communications. (2017) 8:718. doi: 10.1038/ncomms14718
134. Sun, X, Li, M, Sun, Y, Cai, H, Lan, X, Huang, Y, et al. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim Biophys Acta Rev Cancer. (2016) 1863:2835–45. doi: 10.1016/j.bbamcr.2016.08.014
135. Lu, L, Sun, K, Chen, X, Zhao, Y, Wang, L, Zhou, L, et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. (2013) 32:2575–88. doi: 10.1038/emboj.2013.182
136. Wang, J, Gong, C, and Maquat, LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. (2013) 27:793–804. doi: 10.1101/gad.212639.112
137. Wang, G-q, Wang, Y, Xiong, Y, Chen, X-C, Ma, M-l, Cai, R, et al. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. (2016) 6:1865. doi: 10.1038/srep21865
138. Cabianca Daphne, S, Casa, V, Bodega, B, Xynos, A, Ginelli, E, Tanaka, Y, et al. A Long ncRNA links copy number variation to a Polycomb/Trithorax epigenetic switch in FSHD muscular dystrophy. Cell. (2012) 149:819–31. doi: 10.1016/j.cell.2012.03.035
139. Jin, JJ, Lv, W, Xia, P, Xu, ZY, Zheng, AD, Wang, XJ, et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proceedings of the National Academy of Sciences (2018) 115:E9802–11. doi: 10.1073/pnas.1801471115,
140. Song, C, Wang, J, Ma, Y, Yang, Z, Dong, D, Li, H, et al. Linc-smad7 promotes myoblast differentiation and muscle regeneration via sponging miR-125b. Epigenetics. (2018) 13:591–604. doi: 10.1080/15592294.2018.1481705
141. Xiao, T, Liu, L, Li, H, Sun, Y, Luo, H, Li, T, et al. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Rep. (2021) 16:1006–8. doi: 10.1016/j.stemcr.2021.03.024
142. Zhu, E, Zhang, J, Li, Y, Yuan, H, Zhou, J, and Wang, B. Long noncoding RNA Plnc1 controls adipocyte differentiation by regulating peroxisome proliferator-activated receptor γ. FASEB J. (2019) 33:2396–408. doi: 10.1096/fj.201800739RRR
143. Sun, Y, Song, Y, Liu, C, and Geng, J. LncRNA NEAT1-MicroRNA-140 axis exacerbates nonalcoholic fatty liver through interrupting AMPK/SREBP-1 signaling. Biochem Biophys Res Commun. (2019) 516:584–90. doi: 10.1016/j.bbrc.2019.06.104
144. Nuermaimaiti, N, Liu, J, Liang, X, Jiao, Y, Zhang, D, Liu, L, et al. Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem Biophys Res Commun. (2018) 495:1878–84. doi: 10.1016/j.bbrc.2017.12.006
145. Zhang, X, Xue, C, Lin, J, Ferguson, JF, Weiner, A, Liu, W, et al. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci Transl Med. (2018) 10:987. doi: 10.1126/scitranslmed.aar5987
146. Huang, Y, Zheng, Y, Jin, C, Li, X, Jia, L, and Li, W. Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep. (2016) 6:897. doi: 10.1038/srep28897
147. Cui, X, You, L, Li, Y, Zhu, L, Zhang, F, Xie, K, et al. A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J. (2016) 30:4301–12. doi: 10.1096/fj.201600694R
148. Alvarez-Dominguez Juan, R, Bai, Z, Xu, D, Yuan, B, Lo Kinyui, A, Yoon Myeong, J, et al. De novo reconstruction of adipose tissue transcriptomes reveals Long non-coding RNA regulators of Brown adipocyte development. Cell Metab. (2015) 21:764–76. doi: 10.1016/j.cmet.2015.04.003
149. Hotamisligil, G, Bai, Z, Chai, X-r, Yoon, MJ, Kim, H-J, Lo, KA, et al. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. (2017) 15:76. doi: 10.1371/journal.pbio.2002176
150. Zhao, X-Y, Li, S, Wang, G-X, Yu, Q, and Lin, JD. A Long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. (2014) 55:372–82. doi: 10.1016/j.molcel.2014.06.004
151. Agoulnik, IU, Liu, S, Xu, R, Gerin, I, Cawthorn, WP, MacDougald, OA, et al. SRA regulates Adipogenesis by modulating p38/JNK phosphorylation and stimulating insulin receptor gene expression and downstream signaling. PLoS One. (2014) 9:416. doi: 10.1371/journal.pone.0095416
152. Huang, Y, Jin, C, Zheng, Y, Li, X, Zhang, S, Zhang, Y, et al. Knockdown of lncRNA MIR31HG inhibits adipocyte differentiation of human adipose-derived stem cells via histone modification of FABP4. Sci Rep. (2017) 7:131. doi: 10.1038/s41598-017-08131-6
153. Firmin, FF, Oger, F, Gheeraert, C, Dubois-Chevalier, J, Vercoutter-Edouart, A-S, Alzaid, F, et al. The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci Rep. (2017) 7:14087. doi: 10.1038/s41598-017-14570-y
154. Shang, G, Wang, Y, Xu, Y, Zhang, S, Sun, X, Guan, H, et al. Long non-coding RNA TCONS_00041960 enhances osteogenesis and inhibits adipogenesis of rat bone marrow mesenchymal stem cell by targeting miR-204-5p and miR-125a-3p. J Cell Physiol. (2018) 233:6041–51. doi: 10.1002/jcp.26424
155. Chen, Y, Li, K, Zhang, X, Chen, J, Li, M, and Liu, L. The novel long noncoding RNA lncRNA-Adi regulates adipogenesis. Stem Cells Transl Med. (2020) 9:1053–67. doi: 10.1002/sctm.19-0438
156. Ming, Y, and Liu, ZP. Overexpression of lncRNA-NEF regulates the miR-155/PTEN axis to inhibit adipogenesis and promote osteogenesis. Kaohsiung J Med Sci. (2021) 37:930–9. doi: 10.1002/kjm2.12423
Keywords: lncRNAs, biogenesis, farm animals, myogenesis, lipogenesis
Citation: Liu W, Chen M, Liu Y, Li X, Li H and Wang J (2025) Understanding lncRNAs: key regulators of myogenesis and lipogenesis in farm animals. Front. Vet. Sci. 12:1540613. doi: 10.3389/fvets.2025.1540613
Received: 06 December 2024; Accepted: 27 January 2025;
Published: 14 February 2025.
Edited by:
Carrie S. Wilson, Agricultural Research Service (USDA), United StatesReviewed by:
Paolo Zambonelli, University of Bologna, ItalyCopyright © 2025 Liu, Chen, Liu, Li, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bGlodWkzODc2QDE2My5jb20=; Jian Wang, amlhbndhbmdAZ3h1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.