
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Vet. Sci. , 11 February 2025
Sec. Veterinary Infectious Diseases
Volume 12 - 2025 | https://doi.org/10.3389/fvets.2025.1523698
Bovine mastitis is a major problem with huge economic losses in dairy farming worldwide. One of the most common pathogens is Staphylococcus aureus, which is highly contagious and often spread during milking. A sanitation of a dairy herd can be challenging particularly in terms of diagnostics, because of intermittent shedding of Staphylococcus aureus in milk. The observation of intermittent shedding of Staphylococcus aureus in longitudinal studies and applied detection methods were reviewed in this study. Categorization of detection methods is used to describe the basic influence of intermittent shedding on sensitivity of diagnostic of each category. The laboratory diagnostic methods evaluated have a wide range regarding the detection limit (40 cfu/mL−106 cfu/mL). A low detection limit is essential for the detection of even chronically infected cows with intermittent shedding of the pathogen. The literature overview shows that only a few studies (n = 6) examined occurrence of intermittent shedding of Staphylococcus aureus in milk at cow level. A detection-free period of ≤0.5–1 d was only observed in 3 studies.
Bovine mastitis is the most common disease in dairy herds and leads to major economic losses in dairy farms (1). Staphylococcus (S.) aureus is one of the most frequently occurring mastitis pathogens, causes huge costs (2) and can lead to chronical intramammary infections. In a recent prevalence study from Lower Saxony, Germany, S. aureus was detected in bulk milk in 18.3% of the investigated dairy herds (3). Diagnosis is particularly difficult as the pathogen is shed cyclically, and only small quantities of the pathogen are excreted at times (4, 5). Intermittent shedding patterns and the resulting diagnostic challenges have also been investigated in other mastitis pathogens such as Prototheca spp. and Mycoplasma spp. (6, 7). S. aureus has different pathogenic properties to circumvent the immune response of the host (8), e.g., the ability to form a biofilm is suggested to be a major factor (9), although biofilm formation in vivo has rarely been investigated (10). While pathogen quantities of 55,000 cfu/mL are excreted in severe cases of mastitis (11), the excretion of S. aureus in milk is partially below the detection limit of standard laboratory diagnostics (12–14). False negative results due to such intermittent shedding can prevent the successful sanitation of a dairy, therefore research should prioritize on early detection to control the disease through treatment and management (15). The aim of this mini review is to obtain an overview on the occurrence of intermittent S. aureus-shedding in milk and to evaluate the sampling strategies and diagnostic possibilities.
The excretion of S. aureus in milk can differ greatly from day to day and even from two infected quarters of the same cow (5). Sears et al. (4) were able to show that the detection rate of S. aureus could be increased from 75 % to 94 % and respectively 98 % following a second and third sample of the quarter. The time of milk sampling and handling also was found to be relevant: the likelihood of detecting subclinical S. aureus-infection is higher in fresh milk samples taken before milking than in samples taken after milking (16). Villanueva et al. (17) suspected that freezing milk samples could destroy bacterial cell aggregates, which would improve the sensitivity of BC. Such a positive effect could not be confirmed in comparison with fresh pre-milking samples in a study by Godden et al. (16). The freezing of milk samples seems to have different effects on pathogens, whereby no effect could be confirmed especially for S. aureus (18). A study with centrifuged milk samples showed that cultures from the sediment of quarter milk samples can increase the number of positive results by up to 145.5 % (19). Furthermore, Mahmmod et al. (20) demonstrated that pre-sampling procedures (cleaning, disinfecting and discarding first milked streaks) significantly reduced the likelihood of false-positive S. aureus results by eliminating colonies from the skin and teat canal.
Hence, for detecting pathogens in milk different sample types can be used. Quarter milk sample (QMS) are the standard method used for the detection of intramammary infections (IMI) (21). Maisano et al. (22) suggested that the serial sampling of composite milk samples is an alternative to a few QMS. To assess the infection status of a herd, bulk milk samples could be analyzed for the prevalence of S. aureus (23, 24). Britten (13) recommended the targeted use of selective media in general for composite and bulk milk samples to improve sensitivity. However, these results do not allow any statement to be made about the prevalence of the pathogen in individual cows.
In mastitis diagnostics bacterial culture (BC) is standard for detection of bovine IMI. For S. aureus, different selective media have been reported. The use of Baird Parker agar (BP), Vogel-Johnson (VJ), Champman agar (CHAP), CHROMagar S. aureus (CHROM) and chromID S. aureus (SAID) were compared by Graber et al. (27). BP, VJ, and CHAP are used for diagnostic of bovine originated isolates whereas the CHROM and SAID have been evaluated for use of S. aureus-isolates from human origin (27). However, in this study the authors concluded that the specificity of the different selective media is unsatisfactory, mainly because of the similar reactions and occurrence of non-aureus staphylococci (27). In food industry BP is used for enumeration of gram-positive staphylococci, i.e. S. aureus (28). Baird and Lee (29) rated BP as standard medium for enumeration of S. aureus. Artursson et al. (30) investigated eight methods for the isolation of S. aureus from bovine milk samples varying different culture volumes, enrichment, incubation and freezing methods, as well as sedimentation and use of Mastistrip cassette (SVA, Uppsala, Sweden). They concluded that pre-incubation of milk without additives at 37°C for 18 hours increased the number of positive udder quarters by 50%. Middleton et al. (31) stated that in routine mastitis diagnostics with BC, the standard application volume is 10 μl and therefore the minimum detection limit is 100 cfu/mL (see Table 1). Walker et al. (32) demonstrated that the inoculum size of 0.1 mL was found to be the most accurate size for detecting a S. aureus-infection. In addition, the growth of only one colony forming unit (cfu) was classified as sufficient for a positive result (32, 33). However, it was found that the sensitivity of a single sample can be up to 90 % if all cultures (including mixed cultures) are considered positive for S. aureus at a threshold of 1 cfu/0,01mL (34). Furthermore, mixed infections and low pathogen excretion can complicate the diagnosis of S. aureus using BC (35). More evidence could also be obtained for mixed cultures through standardized thresholds and definitions as described by Dohoo et al. (34). The suitable use of culture improvement methods can significantly increase the sensitivity of detection of mastitis organisms in milk (13). The definition of an infection or a positive finding in the BC ranges from ≥1cfu/0.1 mL (32), ≥2 cfu/0,1/mL (12), ≥3 cfu/0.1 mL (36) to ≥1 cfu/0.01 mL (5, 33). These different definitions have a major influence on studies on the occurrence of intermittent shedding. Consequently, the authors had different opinions whether there was cyclical intermittent shedding, in which cultural detection is not possible (4, 37), or rather only low shedding, which is detectable with a larger inoculum (32).
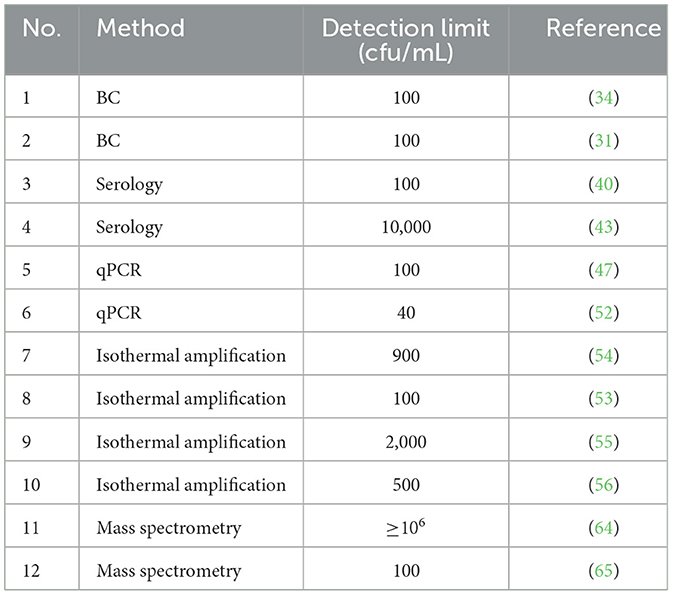
Table 1. Detection methods and reported detection limits in the diagnosis of Staphylococcus aureus from milk.
Several rapid immunological tests for the detection of S. aureus in milk have been developed. ELISA rapid test for the detection of antigen-specific IgG was investigated by Yang and Laven (38) and El- Rashidy et al. (39) for the detection of S. aureus in milk samples. The sensitivity of the tests was found to be 94 % (31) and 86 % (32), but the detection limit is not reported. Another Biosensing method was used to detect S. aureus from milk by binding to the Fc fragment of human IgG on bio-functionalized beads and detected using antibodies labeled by fluorescence markers. Here, the detection limit in milk was 100 cfu/mL (40). In another ELISA-based technique, the enzyme thermostable nuclease (TNase) produced by S. aureus was detected. In this method, a combination of immunomagnetic separation and ELISA (IMS-ELISA) was developed and tested on composite milk samples from 444 cows. The detection limit was approximately 105 S. aureus per mL milk (41). The sensitivity was considered to be limited, and it was stated that TNase is not specific for S. aureus as also other coagulase-positive staphylococci (S. intermedius and S. hyicus), as well as some coagulase-negative staphylococci produce TNase (41, 42). A further rapid test based on immunology, an immunochromatographic strip test (ICS) was developed and reported with a detection limit of 104 cfu/mL (43). In all 3 studies (40, 41, 43) that determined a detection limit used spiked milk samples. Antibody tests are not dependent on the shedding pattern of the bacterium and in addition fast and relatively cheap (44). Many serological detection methods for S. aureus are rapid tests, but with a detection limit that might be unsatisfactory for the detection of subclinical infections (43). Furthermore, it must be noted that there might be a discrepancy between antibodies and the actual amount of pathogens excreted (45).
There are many different approaches in molecular diagnostics for example thermocycle methods as conventional PCR or qPCR as well as isothermal methods are used. Many protocols apply for direct DNA-extraction from milk samples (46–48). The selected protocol and performance of DNA extractions has a major influence on the result, for example gram-positive bacteria such as S. aureus, extraction is challenging, as these bacteria often remain in the cream fraction (49). In conventional PCR each protocol defines a specific number of amplification cycles before the determination the qPCR runs a determination after each cycle. Hence, the result is not only negative or positive but allows also for graduation of positive results (50). Cederlöf et al. (51) using Ct-value cut offs and proposed that low PCR-Ct-values could be defined as “truly/strongly infection” whereas high Ct-values could be defined as “S. aureus- positive cow.” In studies with qPCR, detection limits of 40 cfu/mL (52) and 100 cfu/mL (47) were determined. In opposite to conventional PCR and qPCR, isothermal methods amplify in constant temperature and, hence, do not need to apply a thermocycler. The detection limits for isothermal method (see Table 1) range from 1 × 102 cfu/mL to 2 × 103 cfu/mL in milk (53–56). Studer et al. (36) compared the sensitivity of the qPCR protocol of Graber et al. (57) with the sensitivity of classical BC in the examination of chronically infected S. aureus quarter milk samples. They summarized that the sensitivity was 92.9% for qPCR and 21.4% for BC, with a low pathogen shedding rate, and concluded that one sample per quarter examined with qPCR was sufficient to obtain a definitive result. Nevertheless, the sensitivity of PCR diagnostics is also dependent on the shedding pattern and requires investments in equipment that usually exceeds those of BC (38). Furthermore, a PCR assay detects the DNA of viable and non-viable bacteria, whereas BC only detects viable bacteria (58). Consequently, PCR could detect dead DNA from infections that have already subsided (13). Such cases have hardly been investigated and the relevance of PCR-positive and culture-negative results should be focused on future research (13).
In the early years of the use of mass spectrometry in mastitis research, it was based on single protein analysis (59–61). Hettinga et al. (62) attempted to link the analysis of volatile bacterial metabolites to certain mastitis pathogens. Barreiro et al. (63) started to use mass spectrometry to detect complete mastitis pathogens and considered the species identification of mastitis isolates using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI TOF MS) for faster identification than by biochemical methods. Barreiro et al. (64) developed a protocol to investigate the opportunity of direct detection of bacteria in milk without previous BC using MALDI TOF MS. Experimentally contaminated skim milk identified adequate (score ≥2) S. aureus with concentrations ≥106 cfu/mL (see Table 1). Sauerbrey (65) demonstrated a detection limit of 102 cfu/mL with spiked skim milk in a validation test (see Table 1), but bacteria identification from raw milk samples was not possible without doing BC beforehand. The use of MALDI TOF MS for identification of mastitis pathogen is time consuming with previous BC. The approaches of Barreiro et al. and Sauerbrey (64, 65) to identify the pathogens directly from milk are promising. However, this current detection limit is not satisfactory as only clinical mastitis with extremely high bacterial shedding can be detected.
In the future, the use of AI might also be an option to identify bacteria genera and species based on colony morphology, although there is still limited literature available. Garcia et al. (25) compared an AI-based plate reading application with MALDI-TOF MS on clinical mastitis-causing pathogens and had difficulties to differentiate non-aureus Staphylococci from S. aureus. In addition, the colonies within Staphylococcus spp. are quite similar in morphology, which makes differentiation and thus diagnosis quite difficult (26). Currently this is not an option for the detection of intermittent bacterial species such as S. aureus, as the AI cannot recognize anything unless there is any growth on the plate.
In general, enrichment processes are applied before microbial cultures and initiated in specific sampling situations (21). Keefe (14) assumed that the level of S. aureus-excretion may be below the detection limit of conventional BC and stated that enrichment media could be a helpful tool to increase sensitivity in case of low S. aureus-shedding. Also in human medicine for isolation of S. aureus from swab sample selective enrichment broths were used (66, 67). Furthermore, enrichment media for S. aureus were used to investigate the food safety of milk powder and compared with direct plating at selective Baird Parker medium and Hauschild pork plasma fibrinogen medium (68). In this study, the enrichment medium (Giolitti Cantoni broth with Tween 80) did not achieve better results than direct plating on Baird Parker medium but compared to direct plating on Hauschild medium it did (68). Only a few studies have investigated the potential of enrichment broths for more sensitive bovine mastitis diagnostics with different results (30, 69, 70): Thurmond et al. (70) showed in an experiment with composite milk samples that pre-enrichment of milk samples in Brain heart fusion increases the probability of isolating S. aureus by 1.6 times. Artursson et al. (30) could not demonstrate positive effect on the isolation of S. aureus with nutrient broth containing 10% horse serum in analyses of subclinical quarter milk samples. In some studies an enrichment of the milk was done before PCR to carry out more sensitive diagnostics (71–73). Further studies with different enrichment media for the detection of subclinical S. aureus-infections could provide further insights.
Intermittent shedding of S. aureus in bovine milk samples at cow level was investigated in six longitudinal studies shown in Table 2 (4, 5, 12, 33, 36, 39). Despite an extensive literature search from 1980 to 2024, only these six studies provided information on the shedding of S. aureus at cow level, of which five had detailed information on intermittent shedding.
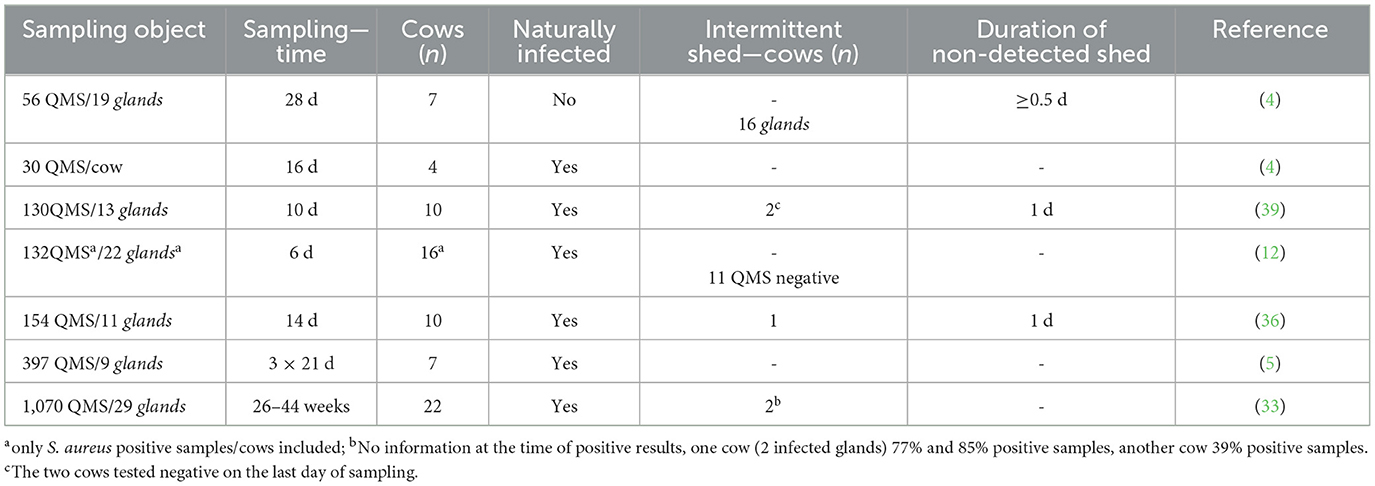
Table 2. Comparison of longitudinal studies investigating intermittent shedding of Staphylococcus aureus in bovine milk samples.
The six studies differ slightly in scope of sample, with 4–22 S. aureus infected cows being analyzed, all examined QMS using BC as detection method. Studer et al. (36) additionally made use of qPCR (see Table 2). Sears et al. (4) investigated naturally and artificially infected cows with S. aureus in an experimental model. In four of the longitudinal studies, the duration of sampling ranged from 6 d to 28 d with at least one sample taken daily. In a study by Walker et al. (5) samples are taken three times on 21 consecutive days during lactation. In another study (33) quarter milk samples are taken weekly over a period of 26–44 weeks. In those studies (5, 33) the bacterial genome was additionally analyzed using pulsed field electrophoresis. Consequently, it should be noted that the six studies differ greatly in terms of study design, which makes comparability difficult.
In three studies, undetectable intervals (see Table 2; “Duration of non-detected shed”) were not investigated in detail, only three studies mention undetectable intervals of ≥0.5 d or 1 d (4, 36, 39). Walker et al. (5) determined that 97.5% of the samples were positive. Unfortunately, the duration of non-detected shed and the number of those cows was not reported. Similar to the study by Buelow et al. (12) in which 11 negative QMS are mentioned, but the number of cows and time interval were not discussed.
Walker et al. (33) compared the results of their study with those of Sears et al. (4) and concluded that naturally infected mammary glands excrete S. aureus in a more consistent pattern than experimentally infected mammary glands. In addition it was found that S. aureus strains have different affinities for the mammary gland, which is why the strain selection might achieve different results (33, 74). However, Sears et al. and Walker et al. (4, 33) agreed on the definition of different types of shedding, so-called low shedding pattern (≤ 10 cfu/0.01 mL) and high shedding pattern (≥20 cfu/0.01 mL). Furthermore, Walker et al. and Studer et al. (33, 36) both observed sinusoidal shedding pattern, i.e., an alternation between low and high S. aureus shedding over time. Studer et al. (36) suggested that this is a result of a synchronized process between the pathogen and the immune system. However, Walker et al. (33) noted that the duration and amplitude of each pattern varied, so that no consistent pattern was observed between or within cows. In a few longitudinal studies transmission of S. aureus was examined on herd level and focusing on the transmission of S. aureus examining the different genotypes (75, 76). Sommerhäuser (75) concluded that the persistence of the pathogen in the udder tissue is more likely in herd sub-suspensions than a temporary cure and subsequent reinfection by the same S. aureus-type. On the other hand Wente et al. (77) investigated recurrent clinical mastitis and stated that S. aureus showed the highest recurrence rate (27 % of all S. aureus cases).
In summary, the occurrence of intermittent shedding of Staphylococcus aureus and the need for sensitive diagnostics is to be investigated in further longitudinal studies. Particular attention should be paid in future research to the duration of undetectable shedding. As this duration was reported as very short in the studies by Sears et al. and Studer et al. (4, 36), close monitoring should also be carried out in future research. Furthermore, whole genome sequencing of the isolates could provide further insights into microevolution in the host in order to determine the persistence of the bacterium in the udder or a re-infection. It is therefore important that a consistent terminology is established to characterize IMI over time (78). Furthermore, there is no standardized definition for the diagnosis of an IMI with S. aureus (32). For BC a sample could already be considered positive if 1 cfu grows in an inoculum of 0.1 mL, as stated by Walker et al. (32, 33). For molecular diagnostics, the subdivision into infected and positive depending on the Ct-value, which was proposed by Cederlöf et al. (51), could be useful. Definitions need to be clarified to enhance research in the field of intermittent shedding of S. aureus in bovine mastitis. Regarding the detection limit, the serological detection and direct MALDI TOF MS studies require further investigations to achieve detection even with low shedding.
LM: Data curation, Investigation, Methodology, Validation, Visualization, Writing – original draft. NK: Supervision, Writing – review & editing. JB: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
BP, baird parker; BC, bacterial culture; cfu, colony forming unit; ELISA, enzyme-linked immunosorbent assay; IMI, intramammary infection; QMS, quarter milk samples.
1. Ruegg PL. A 100-year review: mastitis detection, management, and prevention. J Dairy Sci. (2017) 100:10381–97. doi: 10.3168/jds.2017-13023
2. Sørensen LP, Mark T, Sørensen MK, Østergaard S. Economic values and expected effect of selection index for pathogen-specific mastitis under Danish conditions. J Dairy Sci. (2010) 93:358–69. doi: 10.3168/jds.2009-2506
3. Kortstegge J, Krömker V. Prevalence of contagious mastitis pathogens in bulk tank milk from dairy farms in Lower Saxony, Germany. Hygiene. (2024) 26:122–34. doi: 10.3390/hygiene4020009
4. Sears PM, Smith BS, English PB, Herer PS, Gonzalez RN. Shedding pattern of Staphylococcus aureus from bovine intramammary infections. J Dairy Sci. (1990) 73:2785–9. doi: 10.3168/jds.S0022-0302(90)78964-3
5. Walker JB, Rajala-Schultz PJ, Walker WL, Mathews JL, Gebreyes WA, DeGraves FJ. Variation in daily shedding patterns of Staphylococcus aureus in naturally occurring intramammary infections. J Vet Diagn Invest. (2011) 23:1114–22. doi: 10.1177/1040638711425587
6. Tenhagen BA, Hille A, Schmidt A, Heuwieser W. Development of cell content and shedding of Prototheca spp. in milk from infected udder quarters of cows. Dtsch Tierarztl Wochenschr. (2005) 112:44–8.
7. Okella H, Tonooka K, Okello E. A systematic review of the recent techniques commonly used in the diagnosis of Mycoplasma bovis in dairy cattle. Pathogens. (2023) 19:1178. doi: 10.3390/pathogens12091178
8. Schukken YH, Günther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, et al. Host-response patterns of intramammary infections in dairy cows. Vet Immunol Immunopathol. (2011) 144:270–89. doi: 10.1016/j.vetimm.2011.08.022
9. Kerro Dego O, Vidlund J. Staphylococcal mastitis in dairy cows. Front Vet Sci. (2024) 11:1356259. doi: 10.3389/fvets.2024.1356259
10. Pedersen RR, Krömker V, Bjarnsholt T, Dahl-Pedersen K, Buhl R, Jørgensen E. Biofilm research in bovine mastitis. Front Vet Sci. (2021) 8:656810. doi: 10.3389/fvets.2021.656810
11. Krebs I, Zhang Y, Wente N, Leimbach S, Krömker V. Severity of clinical mastitis and bacterial shedding. Pathogens. (2023) 12:1098. doi: 10.3390/pathogens12091098
12. Buelow KL, Thomas CB, Goodger WJ, Nordlund KV, Collins MT. Effect of milk sample collection strategy on the sensitivity and specificity of bacteriologic culture and somatic cell count for detection of Staphylococcus aureus intramammary infection in dairy cattle. Prevent Vet Med. (1996) 26:1–8. doi: 10.1016/0167-5877(95)00518-8
13. Britten AM. The role of diagnostic microbiology in mastitis control programs. Vet Clini North Am. (2012) 28:187–202. doi: 10.1016/j.cvfa.2012.03.006
14. Keefe G. Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet Clini North Am. (2012) 28:203–16. doi: 10.1016/j.cvfa.2012.03.010
15. Rainard P, Foucras G, Fitzgerald JR, Watts JL, Koop G, Middleton JR. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound Emerg Dis. (2018) 65:149–65. doi: 10.1111/tbed.12698
16. Godden SM, Jansen JT, Leslie KE, Smart NL, Kelton DF. The effect of sampling time and sample handling on the detection of Staphylococcus aureus in milk from quarters with subclinical mastitis. Can Vet J. (2002) 43:38–42.
17. Villanueva MR, Tyler JW, Thurmond MC. Recovery of Streptococcus agalactiae and Staphylococcus aureus from fresh and frozen bovine milk. J Am Vet Med Assoc. (1991) 198:1398–400. doi: 10.2460/javma.1991.198.08.1398
18. Schukken YH, Grommers FJ, Smit JA, Vandegeer D, Brand A. Effect of freezing on bacteriologic culturing of mastitis milk samples. J Dairy Sci. (1989) 72:1900–6. doi: 10.3168/jds.S0022-0302(89)79309-7
19. Zecconi A, Piccinini R, Zepponi A, Ruffo G. Recovery of Staphylococcus aureus from centrifuged quarter milk samples. J Dairy Sci. (1997) 80:3058–63. doi: 10.3168/jds.S0022-0302(97)76273-8
20. Mahmmod YS, Klaas IC, Nielsen SS, Katholm J, Toft N. Effect of presampling procedures on real-time PCR used for diagnosis of intramammary infections with Staphylococcus aureus in dairy cows at routine milk recordings. J Dairy Sci. (2013) 96:2226–33. doi: 10.3168/jds.2012-6059
21. German Veterinary Association. Guideline for the Laboratory Diagnostic of Mastitis. Geneva: GVA Service (2018).
22. Maisano AM, Luini M, Lorenzi V, Bolzoni L, Romanò A, Spelta C, et al. Diagnostic value of composite milk sample vs single quarter milk sample for the detection of Staphylococcus aureus intra-mammary infections in cattle. Prev Vet Med. (2019) 167:80–4. doi: 10.1016/j.prevetmed.2019.03.026
23. Haran KP, Godden SM, Boxrud D, Jawahir S, Bender JB, Sreevatsan S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from minnesota dairy farms. J Clin Microbiol. (2012) 50:688–95. doi: 10.1128/JCM.05214-11
24. Patel K, Godden SM, Royster EE, Crooker BA, Johnson TJ, Smith EA, et al. Prevalence, antibiotic resistance, virulence and genetic diversity of Staphylococcus aureus isolated from bulk tank milk samples of US dairy herds. BMC Genomics. (2021) 22:367. doi: 10.1186/s12864-021-07603-4
25. Garcia BLN, Martins CMDMR, Porto LF, Nobrega DB, Dos Santos MV. Accuracy of an AI-based automated plate reading mobile application for the identification of clinical mastitis-causing pathogens in chromogenic culture media. Sci Rep. (2024) 14:1208. doi: 10.1038/s41598-023-50296-w
26. Garcia BLN, Fidelis CE, Freu G, Granja BDM, Dos Santos MV. Evaluation of chromogenic culture media for rapid identification of gram-positive bacteria causing mastitis. Front Vet Sci. (2021) 8:662201. doi: 10.3389/fvets.2021.662201
27. Graber HU, Pfister S, Burgener P, Boss R, Meylan M, Hummerjohann J. Bovine Staphylococcus aureus: Diagnostic properties of specific media. Res Vet Sci. (2013) 95:38–44. doi: 10.1016/j.rvsc.2013.02.023
28. ISO 6888-1:2021. Part 1: Method using Baird-Parker agar medium. In: Microbiology of the Food Chain — Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus aureus and other species). Geneva: International Organization for Standardization.
29. Baird RM, Lee WH. Media used in the detection and enumeration of Staphylococcus aureus. Int J Food Microbiol. (1995) 26:15–24. doi: 10.1016/0168-1605(93)E0028-P
30. Artursson K, Nilsson-Öst M, Persson Waller K. An improved method to culture Staphylococcus aureus from bovine milk. J Dairy Sci. (2010) 93:1534–8. doi: 10.3168/jds.2009-2544
31. Middleton JR, Fox LK, Pighetti G, Petersson-Wolfe C. Laboratory Handbook on Bovine Mastitis. New Prague, MN: National Mastitis Council Inc. (2017).
32. Walker JB, Rajala-Schultz PJ, DeGraves FJ. The effect of inoculum volume on the microbiologic detection of naturally occurring Staphylococcus aureus intramammary infections. J Vet Diagn Invest. (2010) 22:720–4. doi: 10.1177/104063871002200508
33. Walker JB, Walker WL, DeGraves FJ, Mathews JL, Gebreyes WA, Rajala-Schultz PJ. Staphylococcus aureus shedding pattern throughout lactation in dairy cows with naturally occurring intramammary infection. JAVMA. (2013) 242:1410–8. doi: 10.2460/javma.242.10.1410
34. Dohoo IR, Smith J, Andersen S, Kelton DF, Godden S. Diagnosing intramammary infections: Evaluation of definitions based on a single milk sample. J Dairy Sci. (2011) 94:250–61. doi: 10.3168/jds.2010-3559
35. Gomes AFN, De Castro FDFA, Silva MR, Lange CC, Ribeiro JB, Guimarães ADS, et al. Interference of Streptococcus agalactiae blitz therapy in Staphylococcus aureus microbiological diagnosis in subclinical bovine mastitis. Vet Sci. (2024) 11:233. doi: 10.3390/vetsci11060233
36. Studer E, Schaeren W, Naskova J, Pfaeffli H, Kaufmann T, Kirchhofer M, et al. A longitudinal field study to evaluate the diagnostic properties of a quantitative real-time polymerase chain reaction–based assay to detect Staphylococcus aureus in milk. J Dairy Sci. (2008) 91:1893–902. doi: 10.3168/jds.2007-0485
37. Daley MJ, Oldham ER, Williams TJ, Coyle PA. Quantitative and qualitative properties of host polymorphonuclear cells during experimentally induced Staphylococcus aureus mastitis in cows. Am J Vet Res. (1991) 52:474–9. doi: 10.2460/ajvr.1991.52.03.474
38. Yang DA, Laven RA. Performance of the S taphGold ELISA test in determining subclinical Staphylococcus aureus infections in dairy cows using a Gaussian mixture model. Vet Med. (2022) 8:1632–9. doi: 10.1002/vms3.785
39. El-Rashidy AA, Fox LK, Gay JM. Diagnosis of Staphylococcus aureus intramammary infection by detection of specific antibody titer in milk. J Dairy Sci. (1992) 75:1430–5. doi: 10.3168/jds.S0022-0302(92)77897-7
40. Juronen D, Kuusk A, Kivirand K, Rinken A, Rinken T. Immunosensing system for rapid multiplex detection of mastitis-causing pathogens in milk. Talanta. (2018) 178:949–54. doi: 10.1016/j.talanta.2017.10.043
41. Yazdankhah SP, Sølverød L, Simonsen S, Olsen E. Development and evaluation of an immunomagnetic separation-ELISA for the detection of Staphylococcus aureus thermostable nuclease in composite milk. Vet Microbiol. (1999) 67:113–25. doi: 10.1016/S0378-1135(99)00035-8
42. Park CE, Serrano ADM, Landgraf M, Huang JC, Stankiewicz Z, Rayman MK, et al. survey of microorganisms for thermonuclease production. Can J Microbiol. (1980) 26:532–5. doi: 10.1139/m80-089
43. Nagasawa Y, Kiku Y, Sugawara K, Yabusaki N, Oono K, Fujii K, et al. Rapid Staphylococcus aureus detection from clinical mastitis milk by colloidal gold nanoparticle-based immunochromatographic strips. Front Vet Sci. (2020) 6:504. doi: 10.3389/fvets.2019.00504
44. Fabres-Klein MH, Aguilar AP, Silva MP, Silva DM, Ribon AOB. Moving towards the immunodiagnosis of staphylococcal intramammary infections. Eur J Clin Microbiol Infect Dis. (2014) 33:2095–104. doi: 10.1007/s10096-014-2181-0
45. Bartlett PC, Erskine RJ, Gaston P, Sears PM, Houdijk HW. Enzyme-linked immunosorbent assay and microbiologic culture for diagnosis of Staphylococcus aureus intramammary infection in cows. J Food Protect. (1996) 59:6–10. doi: 10.4315/0362-028X-59.1.6
46. Cremonesi P, Castiglioni B, Malferrari G, Biunno I, Vimercati C, Moroni P, et al. Technical note: improved method for rapid DNA extraction of mastitis pathogens directly from milk. J Dairy Sci. (2006) 89:163–9. doi: 10.3168/jds.S0022-0302(06)72080-X
47. Cressier B, Bissonnette N. Assessment of an extraction protocol to detect the major mastitis-causing pathogens in bovine milk. J Dairy Sci. (2011) 94:2171–84. doi: 10.3168/jds.2010-3669
48. Kim CH, Khan M, Morin DE, Hurley WL, Tripathy DN, Kehrli M, et al. Optimization of the PCR for detection of Staphylococcus aureus NUC gene in bovine milk. J Dairy Sci. (2001) 84:74–83. doi: 10.3168/jds.S0022-0302(01)74454-2
49. Schwenker JA, Friedrichsen M, Waschina S, Bang C, Franke A, Mayer R, et al. Bovine milk microbiota: Evaluation of different DNA extraction protocols for challenging samples. Microbiol. Open. (2022) 11:e1275. doi: 10.1002/mbo3.1275
50. Mahmmod YS, Klaas IC, Enevoldsen C, DNA. carryover in milk samples from routine milk recording used for PCR-based diagnosis of bovine Staphylococcus aureus mastitis. J Dairy Sci. (2017) 100:5709–16. doi: 10.3168/jds.2016-12330
51. Cederlöf SE, Toft N, Aalbaek B, Klaas IC. Latent class analysis of the diagnostic characteristics of PCR and conventional bacteriological culture in diagnosing intramammary infections caused by Staphylococcus aureus in dairy cows at dry off. Acta Vet Scand. (2012) 54:65. doi: 10.1186/1751-0147-54-65
52. Boss R, Naskova J, Steiner A, Graber HU. Mastitis diagnostics: quantitative PCR for Staphylococcus aureus genotype B in bulk tank milk. J Dairy Sci. (2011) 94:128–37. doi: 10.3168/jds.2010-3251
53. Tie Z, Chunguang W, Xiaoyuan W, Xinghua Z, Xiuhui Z. Loop-mediated isothermal amplification for detection of Staphylococcus aureus in dairy cow suffering from mastitis. J Biomed Biotechnol. (2012) 2012:1–5. doi: 10.1155/2012/435982
54. Sheet OH, Grabowski NT, Klein G, Abdulmawjood A. Development and validation of a loop mediated isothermal amplification (LAMP) assay for the detection of Staphylococcus aureus in bovine mastitis milk samples. Mol Cellular Probes. (2016) 30:320–5. doi: 10.1016/j.mcp.2016.08.001
55. Griffioen K, Cornelissen J, Heuvelink A, Adusei D, Mevius D, Van Der Wal FJ. Development and evaluation of 4 loop-mediated isothermal amplification assays to detect mastitis-causing bacteria in bovine milk samples. J Dairy Sci. (2020) 103:8407–20. doi: 10.3168/jds.2019-18035
56. Heng P, Liu J, Song Z, Wu C, Yu X, He Y. Rapid detection of Staphylococcus aureus using a novel multienzyme isothermal rapid amplification technique. Front Microbiol. (2022) 13:1027785. doi: 10.3389/fmicb.2022.1027785
57. Graber HU, Casey MG, Naskova J, Steiner A, Schaeren W. Development of a highly sensitive and specific assay to detect Staphylococcus aureus in bovine mastitic milk. J Dairy Sci. (2007) 90:4661–9. doi: 10.3168/jds.2006-902
58. Koskinen MT, Wellenberg GJ, Sampimon OC, Holopainen J, Rothkamp A, Salmikivi L, et al. Field comparison of real-time polymerase chain reaction and bacterial culture for identification of bovine mastitis bacteria. J Dairy Sci. (2010) 93:5707–15. doi: 10.3168/jds.2010-3167
59. Hogarth CJ, Fitzpatrick JL, Nolan AM, Young FJ, Pitt A, Eckersall PD. Differential protein composition of bovine whey: a comparison of whey from healthy animals and from those with clinical mastitis. Proteomics. (2004) 4:2094–100. doi: 10.1002/pmic.200300723
60. Napoli A, Aiello D, Di Donna L, Prendushi H, Sindona G. Exploitation of endogenous protease activity in raw mastitic milk by MALDI-TOF/TOF. Anal Chem. (2007) 79:5941–8. doi: 10.1021/ac0704863
61. Taverna F, Negri A, Piccinini R, Zecconi A, Nonnis S, Ronchi S, et al. Characterization of cell wall associated proteins of a Staphylococcus aureus isolated from bovine mastitis case by a proteomic approach. Vet Microbiol. (2007) 119:240–7. doi: 10.1016/j.vetmic.2006.09.007
62. Hettinga KA, Van Valenberg HJF, Lam TJGM, Van Hooijdonk ACM. Detection of mastitis pathogens by analysis of volatile bacterial metabolites. J Dairy Sci. (2008) 91:3834–9. doi: 10.3168/jds.2007-0941
63. Barreiro JR, Ferreira CR, Sanvido GB, Kostrzewa M, Maier T, Wegemann B, et al. Short communication: identification of subclinical cow mastitis pathogens in milk by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Dairy Sci. (2010) 93:5661–7. doi: 10.3168/jds.2010-3614
64. Barreiro JR, Gonçalves JL, Braga PAC, Dibbern AG, Eberlin MN, Veiga Dos Santos M. Non-culture-based identification of mastitis-causing bacteria by MALDI-TOF mass spectrometry. J Dairy Sci. (2017) 100:2928–34. doi: 10.3168/jds.2016-11741
65. Sauerbrey K. Möglichkeiten und Grenzen der Mastitisdiagnostik mittels MALDI-TOF MS-Analytik und molekularbiologischen Methoden [Internet]. Munich: Ludwig-Maximilians-Universität München (2015). Available at: https://edoc.ub.uni-muenchen.de/id/eprint/17991 (accessed 26 June, 2024).
66. Nilsson P, Ripa T. Staphylococcus aureus throat colonization is more frequent than colonization in the anterior nares. J Clin Microbiol. (2006) 44:3334–9. doi: 10.1128/JCM.00880-06
67. Mernelius S, Löfgren S, Lindgren PE, Matussek A. The role of broth enrichment in Staphylococcus aureus cultivation and transmission from the throat to newborn infants: results from the Swedish hygiene intervention and transmission of S. aureus study. Eur J Clin Microbiol Infect Dis. (2013) 32:1593–8. doi: 10.1007/s10096-013-1917-6
68. Chopin A, Malcolm S, Jarvis G, Asperger H, Beckers HJ, Bertona AM, et al. ICMSF methods studies XV comparison of four media and methods for enumerating Staphylococcus aureus in powdered milk. J Food Prot. (1985) 48:21–8. doi: 10.4315/0362-028X-48.1.21
69. Innes AG. An enrichment broth and differential medium for the isolation of mastitis organisms from the milk of distant herds. J Comp Pathol Therapeut. (1953) 63:136–42. doi: 10.1016/S0368-1742(53)80015-9
70. Thurmond MC, Tyler JW, Luiz DM, Holmberg CA, Picanso JP. The effect of pre-enrichment on recovery of Streptococcus agalactiae, Staphylococcus aureus and mycoplasma from bovine milk. Epidemiol Infect. (1989) 103:465–74. doi: 10.1017/S0950268800030879
71. Gillespie BE, Oliver SP. Simultaneous detection of mastitis pathogens, Staphylococcus aureus, Streptococcus uberis, and Streptococcus agalactiae by multiplex real-time polymerase chain reaction. J Dairy Sci. (2005) 88:3510–8. doi: 10.3168/jds.S0022-0302(05)73036-8
72. Sunagar R, Deore SN, Deshpande PV, Rizwan A, Sannejal AD, Sundareshan S, et al. Differentiation of Staphylococcus aureus and Staphylococcus epidermidis by PCR for the fibrinogen binding protein gene. J Dairy Sci. (2013) 96:2857–65. doi: 10.3168/jds.2012-5862
73. Sartori C, Boss R, Ivanovic I, Graber HU. Development of a new real-time quantitative PCR assay for the detection of Staphylococcus aureus genotype B in cow milk, targeting the new gene adlb. J Dairy Sci. (2017) 100:7834–45. doi: 10.3168/jds.2017-12820
74. Smith EM, Green LE, Medley GF, Bird HE, Fox LK, Schukken YH, et al. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J Clin Microbiol. (2005) 43:4737–43. doi: 10.1128/JCM.43.9.4737-4743.2005
75. Sommerhäuser J, Kloppert B, Wolter W, Zschöck M, Sobiraj A, Failing K. The epidemiology of Staphylococcus aureus infections from subclinical mastitis in dairy cows during a control programme. Vet Microbiol. (2003) 96:91–102. doi: 10.1016/S0378-1135(03)00204-9
76. Anderson KL, Lyman RL. Long-term persistence of specific genetic types of mastitis-causing Staphylococcus aureus on three dairies. J Dairy Sci. (2006) 89:4551–6. doi: 10.3168/jds.S0022-0302(06)72504-8
77. Wente N, Grieger AS, Klocke D, Paduch JH, Zhang Y, Leimbach S, et al. Recurrent mastitis–persistent or new infections? Vet Microbiol. (2020) 244:108682. doi: 10.1016/j.vetmic.2020.108682
Keywords: Staphylococcus aureus, bovine mastitis, intermittent shedding, S. aureus diagnostic, S. aureus mastitis
Citation: Mues L, Kemper N and Blumenberg JA (2025) Occurrence and diagnostic of intermittent shedding of Staphylococcus aureus in bovine mammary infection. Front. Vet. Sci. 12:1523698. doi: 10.3389/fvets.2025.1523698
Received: 06 November 2024; Accepted: 20 January 2025;
Published: 11 February 2025.
Edited by:
Csaba Varga, University of Illinois at Urbana–Champaign, United StatesReviewed by:
Rima Shrestha, University of Illinois at Peoria, United StatesCopyright © 2025 Mues, Kemper and Blumenberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Anna Blumenberg, amJsdW1lbmJlcmdAZW1haWwudW5pLWtpZWwuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.