- 1Hebei Collaborative Innovation Center for Eco-Environment, Hebei Key Laboratory of Animal Physiology, Biochemistry and Molecular Biology, College of Life Sciences, Hebei Normal University, Shijiazhuang, Hebei, China
- 2Hebei Research Center of the Basic Discipline Cell Biology, Ministry of Education Key Laboratory of Molecular and Cellular Biology, Shijiazhuang, Hebei, China
- 3Department of Zoology, Shaheed Benazir Bhutto University, Sheringal, Upper Dir, Khyber Pakhtunkhwa, Pakistan
Species of Heterakis (Ascaridida: Heterakoidea) are commonest nematode parasites occurring in the alimentary canal of wild and domestic birds, which are of major socio-economic importance, due to some Heterakis species causing Heterakidosis in wild birds and poultry. In the present study, a new species of Heterakis, H. pucrasia sp. n., was described using integrated methods based on specimens collected from the koklass pheasant Pucrasia macrolopha (Lesson) (Galliformes: Phasianidae) in Pakistan. The complete mitochondrial genome of H. pucrasia sp. n. was sequenced and annotated for the first time to enrich the mitogenomic data, and reveal the pattern of mitogenomic evolution of the family Heterakidae. Moreover, phylogenetic analyses of the orders Ascaridida, Spirurida, Oxyurida and Rhigonematida based on the amino acid sequences of 12 protein coding genes (PCGs) of mitochondrial genomes, revealed that the order Ascaridida is not monophyletic, and the superfamily Heterakoidea has a closer affinity with Rhigonematida + Oxyurida + Spirurida, than the superfamily Ascaridoidea in Ascaridida. The present findings enriched the global species composition of heterakid nematodes and their mitogenomic data, and also provided novel insight on the phylogenetic relationships between Heterakoidea and its related groups.
1 Introduction
The koklass pheasant Pucrasia macrolopha (Lesson) (Galliformes: Phasianidae) is the only species of the monotypic genus Pucrasia, which is mainly distributed in China, Pakistan, India, Afghanistan and Nepal (1–3). The koklass pheasant is mostly herbivorous, mainly feeding on pine nuts, pine shoots, bamboo shoots, fruits and seeds, but it is highly insectivorous during the breeding season.1 However, our present knowledge of nematode parasites of the koklass pheasant remains very limited. To date, only Heterakis gallinarum (Schrank, 1788) has been reported from this bird (4).
The genus Heterakis (Ascaridida: Heterakoidea) is an important group of zooparasitic nematodes, including approximately 49 nominal species distributed globally, which mainly parasitize wild and domestic birds, but also occasionally occur in the alimentary canal of mammals (5–7). Some species of Heterakis (i.e., H. gallinarum, H. dispar and H. isolonche) can cause Heterakidosis in wild birds and poultry (8, 9). However, current knowledge of the global species composition of Heterakis nematodes remains far from complete. Furthermore, the molecular identification, population genetics and phylogeny of Heterakis spp. remain in their infancy (9–12), due to the limited genetic data available, especially the mitochondrial genomes. To date, only H. gallinarum, H. beramporia and H. dispar have been reported for the complete mitochondrial genomes in the genus Heterakis (13, 14).
In the present study, some Heterakis nematodes were collected from the koklass pheasant P. macrolopha in Pakistan. Multiple methods were used to study the specimens, including light and scanning electron microscopy, Assembled Species by Automatic Partitioning (ASAP) and Bayesian inference (BI) analyses, in order to exactly identify these specimens to species level. Moreover, the complete mitochondrial genome of the present material was sequenced and annotated to enrich the mitogenomic data and investigate the pattern of mitogenomic evolution of the family Heterakidae. Phylogenetic analyses were also performed in order to assess the systematic position of the superfamily Heterakoidea in the order Ascaridida, and the evolutionary relationships of the four orders Ascaridida, Spirurida, Oxyurida and Rhigonematida based on the amino acid sequences of 12 protein coding genes (PCGs) of mitogenomes using maximum likelihood (ML) and Bayesian inference (BI), respectively.
2 Methods
2.1 Parasite collection
During a helminthological survey of Pakistani birds in 2023, a single dead koklass pheasant P. macrolopha was dissected for parasites in the Sahoor Top Sheringal Upper Dir (35.2691° N, 72.0627° E), and a total of 23 nematode specimens were collected from the ceca of this bird. The specimens were fixed and stored in 80% ethanol until study.
2.2 Morphological observation
For light microscopy, nematode specimens were cleared in glycerin, then examined and photographed using a Nikon® optical microscope (Nikon ECLIPSE Ni-U, Nikon Corporation, Tokyo, Japan). For scanning electron microscopy (SEM), the anterior and posterior ends of 1 male and 1 female specimens, and the mid-body (vulval region) of 2 female specimens were transferred to 4% formaldehyde solution, post-fixed in 1% OsO₃, dehydrated via an ethanol series and acetone, and critical point dried. The specimens were coated with gold and examined using a Hitachi S-4800 scanning electron microscope (Hitachi Ltd., Tokyo, Japan) at an accelerating voltage of 20 kV. Measurements (minimum, maximum, followed by mean in parentheses) are given in micrometers (μm), unless otherwise stated. Type specimens were deposited at the College of Life Sciences, Hebei Normal University, Hebei Province, China.
2.3 Molecular procedures
The mid-body of three female specimens were randomly selected for molecular analysis. Genomic DNA from each individual was extracted using a Column Genomic DNA Isolation Kit (Shanghai Sangon, China) according to the manufacturer’s instructions, and stored at −20°C for further study. The primers and cycling conditions for amplifying the nuclear small ribosomal subunit (18S), large ribosomal subunit (28S) and internal transcribed spacer (ITS), and mitochondrial cytochrome c oxidase subunit 1 (cox1), cytochrome c oxidase subunit 2 (cox2) and small subunit ribosomal RNA gene (12S) by polymerase chain reaction (PCR) were provided in Supplementary Table S1. All PCR reactions were performed in 50 μL consisting of 10 mM Tris HCl at pH 8.4, 50 mM KCl, 3.0 mM MgCl₂, 250 μM of each dNTP, 50 pmol of each primer and 1.5 U of Taq polymerase (Takara Bio Inc., Kusatsu, Shiga, Japan) in a thermocycler (model 2,720; Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). PCR products were checked on GoldView-stained 1.5% agarose gel and purified using the Column PCR Product Purification Kit (Shanghai Sangon, China). Sequencing of each sample was carried out for both strands using a DyeDeoxyTerminator Cycle Sequencing Kit (v.2, Applied Biosystems, California, USA). The ITS1-5.8S-ITS2 region was assembled using GetOrganelle v1.7.7.0 (15) based on genomic data sequenced by Illumina NovaSeq 6,000 platform. The 18S, 28S, ITS, cox1, cox2 and 12S sequences obtained herein were deposited in the National Center for Biotechnology Information (NCBI) database.2
2.4 Species delimitation
The Assembled Species by Automatic Partitioning (ASAP) (16) was used for species delimitation of Heterakis spp. based on different genetic markers (i.e., ITS, cox1, cox2, and 12S sequences). Ascaridia columbae was chosen as the out-group. The ASAP analyses were performed using an online ASAP web server3 under the Kimura (k8o) ts/tv 2.0 model. The group with the lowest score from ASAP was considered to be optimal.
2.5 Mitochondrial genome sequencing, assembly, and annotation
The mid-body of 1 female specimen was used for mitogenome sequencing. A total of 50 GB of clean genomic data was generated using the Pair-End 150 sequencing method on the Illumina NovaSeq 6,000 platform by Novogene (Tianjin, China). The complete mitochondrial genome was assembled using GetOrganelle v1.7.7.0 (15). Protein coding genes (PCGs), rRNAs and tRNAs were annotated using the MITOS2 web server4 and MitoZ v3.6 (17). The open reading frame (ORF) of each PCG was confirmed manually using the web version of the ORF finder.5 The “lost” tRNA genes ignored by both MitoS2 and MitoZ (17), were identified using BLAST-based on a database of existing tRNA sequences of nematodes. The secondary structures of tRNAs were predicted using ViennaRNA module (18), by MitoS2 and RNAstructure v6.4 (19), followed by manual correction. MitoZ v3.6 was used to visualize and depict gene element features (17). Base composition, amino acid usage, and relative synonymous codon usage (RSCU) were calculated using Python script, which refers to the Codon Adaptation Index (CAI) (20). The total length of the bases included ambiguous bases. Base skew analysis was used to describe the base composition. The complete mitochondrial genome obtained herein was deposited in the GenBank database (see footnote 2).
2.6 Phylogenetic analyses
Phylogenetic analyses of Heterakis spp. were performed based ITS, cox1, cox2 and 12S sequence data, using Bayesian inference (BI) with MrBayes v3.2.7a (21). Ascaridia columbae was chosen as the out-group. Phylogenetic analyses of Ascaridida, Spirurida, Oxyurida and Rhigonematida were conducted based on concatenated amino acid (AA) sequences of the 12 PCGs using maximum likelihood (ML) with IQ-TREE v2.2.2.7 (22), and Bayesian inference (BI) with MrBayes v3.2.7a (21). Caenorhabditis elegans (Rhabditida: Rhabditidae) was chosen as the outgroup. The ingroup included 56 species representing 11 superfamilies of these four orders. Detailed information on the representatives of Ascaridida, Spirurida, Oxyurida and Rhigonematida included in the present phylogenetic analyses was provided in Supplementary Table S2. Genes were aligned separately with MAFFT v7.526 using the iterative refinement method of E-INS-I (23). Ambiguous sites and poorly aligned positions were eliminated using BMGE v1.12 (m = BLOSUM90, h = 0.5) (24). The aligned and eliminated sequences were concatenated into a matrix using PhyloSuite v1.2.3 (25). The mtMet+F+R4 model for ML inference, and the JTT+I+G model for BI were identified as the optimal nucleotide substitution models, respectively. Reliabilities for ML inference was tested using 1,000 bootstrap replications and Bayesian information criterion for BI analysis was run for 5 × 106 Markov Chain Monte Carlo (MCMC) generations. In the ML tree, bootstrap support (BS) values ≥90 were considered to constitute strong nodal support, whereas BS values ≥70 and < 90 were considered to constitute moderate nodal support. In the BI tree, Bayesian posterior probability (BPP) values ≥0.90 were considered to constitute strong nodal support, whereas BPP values≥0.85 and < 0.90 were considered to constitute moderate nodal support.
3 Results
3.1 Description of Heterakis pucrasia sp. n. (Figures 1–4)
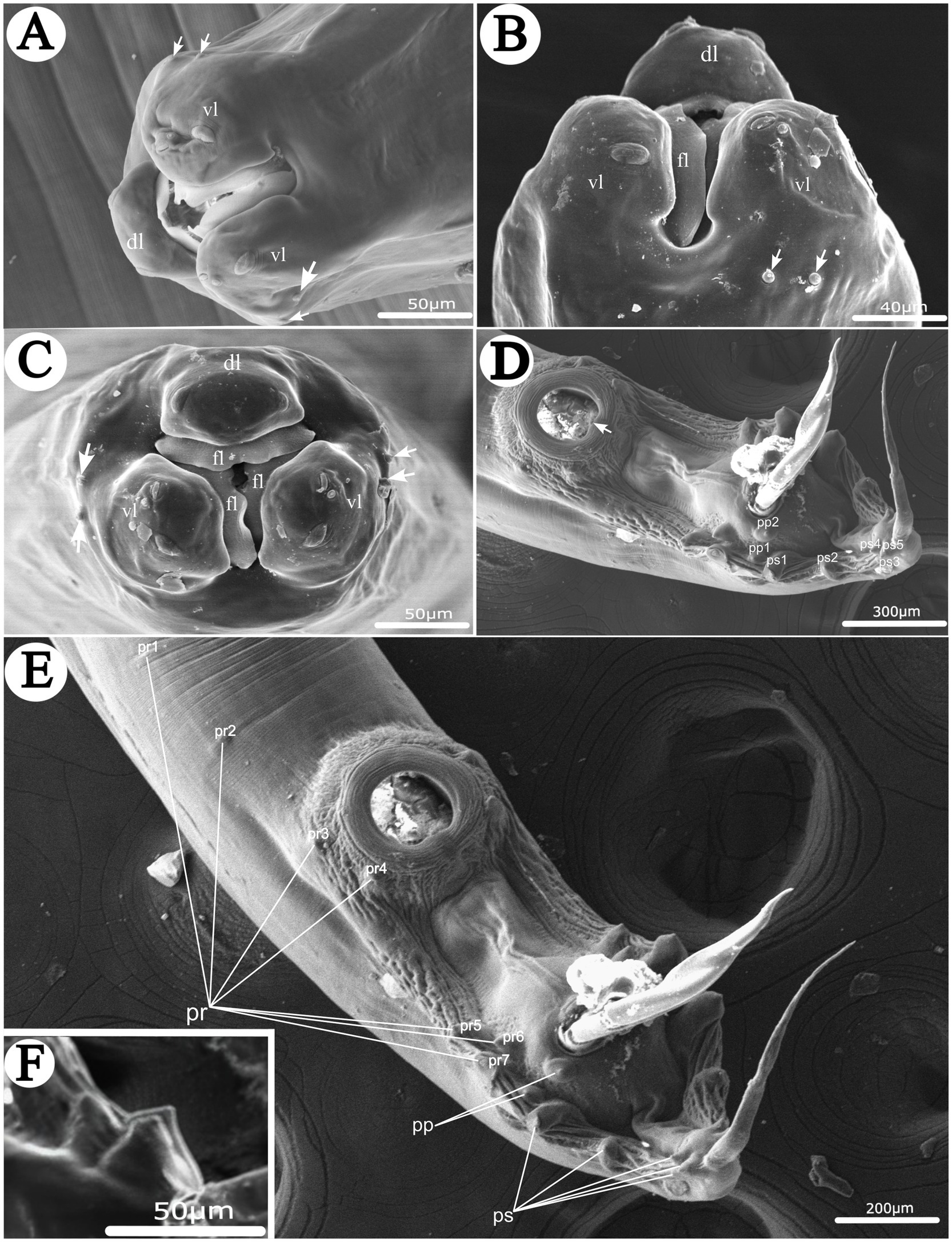
Figure 1. Scanning electron micrographs of Heterakis pucrasia sp. n. from Pucrasia macrolopha in Pakistan. (A) Cephalic extremity of male (cephalic papillae arrowed), subapical view. (B) Cephalic extremity of female (cephalic papillae arrowed), ventral view. (C) Cephalic extremity of female (cephalic papillae arrowed), apical view. (D) Posterior end of male (single medio-ventral sessile papilla on posterior margin of precloacal sucker arrowed), ventral view. (E) Posterior end of male, ventral view. (F) Magnified image of last 3 pairs of postcloacal papillae. dl, dorsal lip; vl, ventrolateral lip; fl, inner flanges on each lip; pr, precloacal papillae; pp., paracloacal papillae; ps, postcloacal papillae.
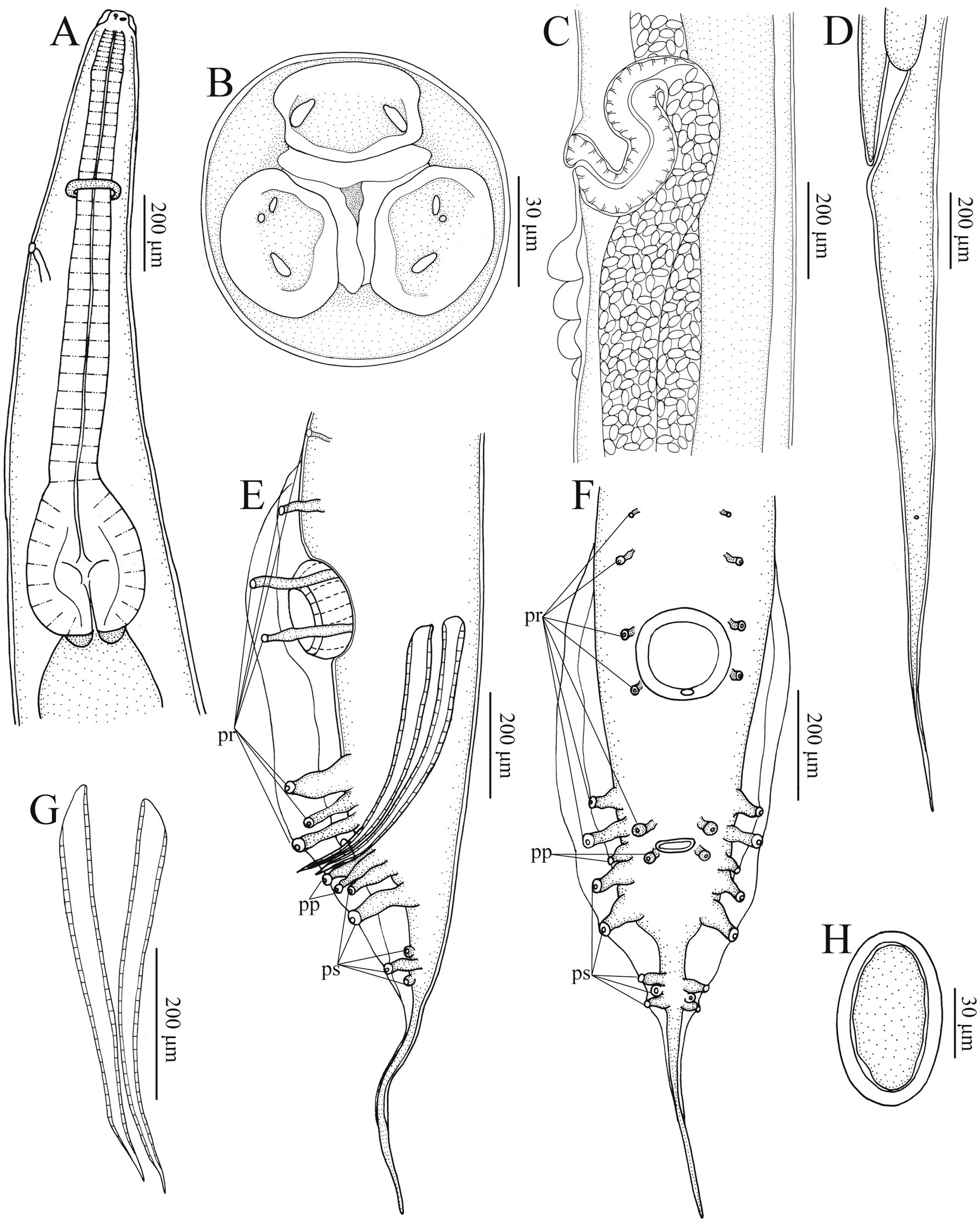
Figure 2. Heterakis pucrasia sp. n. from Pucrasia macrolopha in Pakistan. (A) Anterior part of female, sublateral view. (B) Cephalic extremity of female, apical view. (C) Region of vulva, lateral view. (D) Tail of female, lateral view. (E) Posterior end of male, lateral view. (F) Posterior end of male, ventral view. (G) Spicules. (H) Egg. pr, precloacal papillae; pp., paracloacal papillae; ps, postcloacal papillae.

Figure 3. Photomicrographs of Heterakis pucrasia sp. n. from Pucrasia macrolopha in Pakistan. (A) Anterior part of female, ventral view. (B) Cephalic extremity of female, sublateral view. (C) Posterior end of male, lateral view. (D) Posterior end of male, ventral view. (E) Eggs. (F) Region of vulva, lateral view.
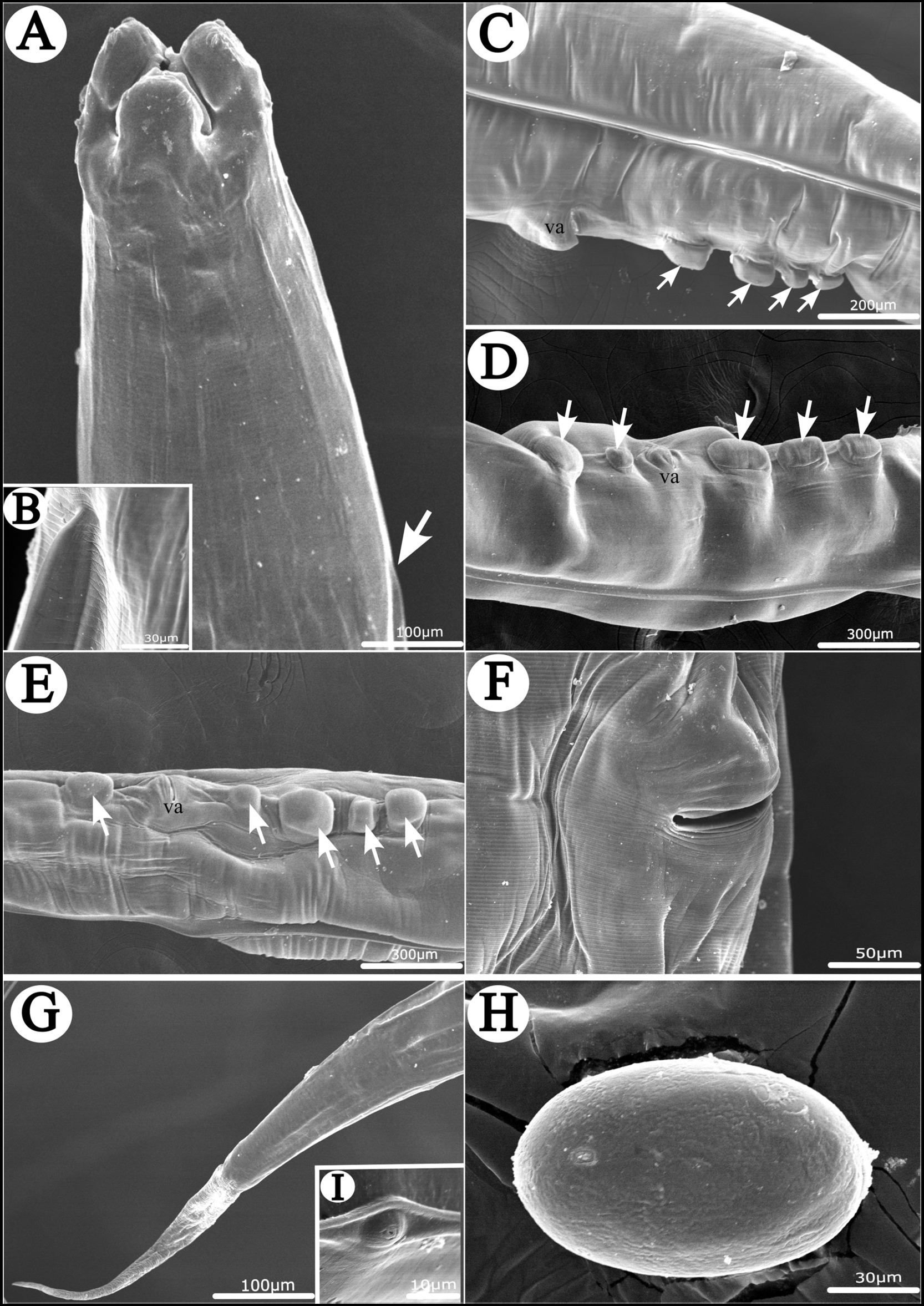
Figure 4. Scanning electron micrographs of Heterakis pucrasia sp. n. from Pucrasia macrolopha in Pakistan, female. (A) Anterior part of body (starting position of lateral ala arrowed), dorsal view. (B) Magnified image of starting position of lateral ala. (C–E) Region of vulva showing different number and arrangement of cuticular protrusions at region of vulva (cuticular protrusions arrowed), lateral or subventral view. (F) Magnified image of vulva. (G) Tail tip. (H) Magnified image of egg. (I) Magnified image of phasmid. va, vulva.
General: Medium-sized, whitish nematodes with finely transversely striated cuticle. Maximum width at about region of mid-body. Cephalic extremity with three roughly trapeziform lips, approximately equal in size; each lip possesses remarkable inner flanges (Figures 1A–C, 2A,B, 3A,B, 4A). Dorsal lip with one pair of large double labial papillae; ventrolateral lips each with single double labial papilla, small labial papilla, and amphid (Figures 1A–C, 2B, 3A). One pair of cephalic papillae present at base of each ventro-lateral lip (Figures 1A–C). Interlabia absent. Esophagus muscular, divided into anterior short pharynx, cylindrical corpus, inconspicuous isthmus and terminating posterior bulb with valves (Figures 2A, 3A,B). Nerve ring at about 1/5 of esophageal length. Excretory pore just posterior to nerve ring (Figure 2A). Lateral alae well-developed, starting from some distance posterior to base of ventrolateral lips extending to caudal region (Figures 3A, 4A,B). Tails of both sexes conical, with long filamentous tips (Figures 1D–F, 3C,D, 4G).
Male [based on 2 mature specimens] Body 9,100–9,390 (9,245 ± 145) long, maximum width 480–510 (495 ± 15). Dorsal and ventrolateral lips almost equal in size, 24–29 (27 ± 2) long, 53–58 (56 ± 2) wide. Esophagus 1,370–1,420 (1,395 ± 25) long in total length (including posterior bulb), representing 10.7–11.7 (11.2) % of body length; pharynx + corpus + isthmus 1,000–1,060 (1,030 ± 30) long, size of posterior bulb 360–370 (365 ± 5) × 290–300 (295 ± 5). Nerve-ring and excretory pores 390–390 (385 ± 5) and 550–680 (615 ± 65) from the cephalic extremity, respectively. Posterior end of body distinctly curves ventrally. Spicules short, alate, nearly equal in length, distal end pointed, 550–570 (560 ± 10) long, representing 5.80–6.26 (6.06) % of body length (Figures 1D,E, 2E,G, 3C,D). Gubernaculum absent. Precloacal sucker well developed, circular, 190–200 (195 ± 5) in diameter, 310–350 (330 ± 20) from its center to cloaca; single medio-ventral sessile papilla present at posterior margin of precloacal sucker (Figures 1D,E, 2E,F, 3C,D). Caudal papillae 14 pairs in total, including 7 pairs of precloacal papillae (1st pair small, sessile; 2nd pair being pedunculate, located at about starting position of caudal alae; 3rd and 4th pairs being pedunculate, located at about level of anterior and posterior margin of sucker, respectively; 5th–7th pairs large, close to each, slightly anterior to cloaca, 5th and 6th pairs located ventrolaterally, being pedunculate, 7th pair located ventrally, being sessile), 2 pairs of paracloacal papillae (one pair located ventrolaterally, being pedunculate; the other located ventrally, being sessile) and 5 pairs of postcloacal pedunculate papillae (anterior 2 pairs large; posterior 3 pairs small, grouped together) (Figures 1D–F, 2E,F, 3C,D). Tail 650–660 (655 ± 5) long (Figures 1D–F, 2E,F, 3C,D). Phasmids not observed.
Female [based on 10 gravid specimens] Body 10,317–14,220 (12,135 ± 1,527) long, maximum width 420–560 (500 ± 41). Dorsal and ventrolateral lips almost equal in size, 24–39 (32 ± 4) long, 48–58 (55 ± 3) wide. Esophagus 1,430–1,600 (1,532 ± 56) long in total length (including posterior bulb), representing 11.3–13.9 (12.6) % of body length; pharynx + corpus + isthmus 1,010–1,220 (1,152 ± 59) long, size of posterior bulb 280–310 (296 ± 8) × 280–310 (296 ± 8). Nerve ring 370–430 (399 ± 21) and excretory pore 580–680 (619 ± 27) from cephalic extremity. Vulva slit-like, with slightly protruded vulval lip (Figures 2C, 3F, 4C–F), 4,470–6,995 (5,860 ± 907) from cephalic extremity, representing 43.3–53.5 (48.2) % of body length. Four or five cuticular protrusions located at region of vulva in various arrangement types (Figures 2C, 3F, 4C–E). Vagina muscular, uteri didelphic, opistodelphic. Eggs oval, thick-shelled, 68–72 (71 ± 2) × 39–43 (43 ± 1) (n = 20) in size (Figures 2H, 3E, 4H). Tail 1,310–1,680 (1,517 ± 128) long (Figures 2D, 3G). Phasmids at about posterior 1/3 of the tail (Figures 2D, 4I).
Type host: Koklass pheasant Pucrasia macrolopha (Lesson) (Galliformes: Phasianidae).
Type locality: Sahoor Top Sheringal Upper Dir (35.2691° N, 72.0627° E), Pakistan.
Site in host: Ceca.
Prevalence and intensity: Single koklaas pheasant examined with an intensity of 23 specimens.
Type specimens: Holotype: 1 male (HBNU–N–B20240426YL); allotype: 1 female (HBNU–N–B20240427YL); paratypes: 1 complete male and 12 complete females (HBNU–N–R20240428YL), 1 incomplete male (only anterior and posterior end, mid-body for molecular analysis) and 7 incomplete females (only anterior and posterior end, mid-body for molecular analysis) (HBNU–N–B20240428YL); deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China.
To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN), details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:C5319C09-F4DB-4CCA-99C6-02DE2B88B41C. The LSID for the new name Heterakis pucrasia sp. n. is urn:lsid:zoobank.org:act:9CAFD43C-072A-4294-A06B-AC97025AC838.
Etymology: The specific name refers to the generic name of the host type.
3.2 Genetic characterization
3.2.1 Partial 18S region
Three 18S sequences of H. pucrasia sp. n. obtained herein are 781 bp long, with no nucleotide divergence detected. In the genus Heterakis, the 18S sequences are available in GenBank for H. gallinarum (DQ503462, MK844591, MW073433, MW073432 and MW073553), Heterakis sp. (AF083003) and H. spumosa (MH571872). Pairwise comparison of the 18S sequences of H. pucrasia sp. n. with that of H. gallinarum, Heterakis sp. and H. spumosa available in GenBank showed 0.40% (H. gallinarum) to 5.72% (Heterakis sp.) of nucleotide divergence.
3.2.2 Partial 28S region
Three 28S sequences of H. pucrasia sp. n. obtained herein are 1,071 bp long, with no nucleotide divergence detected. In the genus Heterakis, only H. gallinarum (LC777441) and H. spumosa (MH571869) with the partial 28S sequence are available in GenBank. Pairwise comparison of the 28S sequences of H. pucrasia sp. n. with that of H. gallinarum and H. spumosa available in GenBank showed 11.1% (H. gallinarum) to 11.3% (H. spumosa) of nucleotide divergence.
3.2.3 Partial ITS (ITS-1+5.8S+ITS-2) region
Single ITS sequence of H. pucrasia sp. n. obtained herein is 954 bp long. In the genus Heterakis, the ITS sequences are available in GenBank for H. gallinarum (MW408729–MW412732, MW403874, MW661165, MW561275, MF066721–MF066726, MF403056, LC592776–LC592805, LC592808, KT310099–KT310157, OQ848045, JQ995320, OR619556, AJ876757), H. indica (LC592806–LC592809), H. dispar (OM530142–OM530145, OM530147, MF319969), H. isolonche (KM212953), Heterakis sp. (ON244646–ON244647), H. spumosa (LC389877, JX845278), H. beramporia (LC592731–LC592733, KU529974, OL470970–OL470973) and H. dahomensis (JX845277). Pairwise comparison of the ITS sequence of H. pucrasia sp. n. with that of Heterakis spp. available in GenBank showed 8.47–8.82% (H. dispar) to 12.2–18.2% (H. beramporia) of nucleotide divergence.
3.2.4 Partial cox1 region
Three cox1 sequences of H. pucrasia sp. n. obtained herein are 682 bp long, with no nucleotide divergence detected. In the genus Heterakis, the cox1 sequences are available in GenBank for H. gallinarum (KP308308–KP308363, MF066712–MF066720, LC592865, NC_029839, KU529973), H. indica (LC592869–LC592874), H. dispar (NC_042411, MK024389), H. isolonche (MN732844–MN732847, FJ009625–FJ009627), Heterakis sp. (MN737038–MN737040, MN732849–MN732852), H. spumosa (LC626017) and H. beramporia (LC592900–LC592902, NC_029838, KU529972). Pairwise comparison of the cox1 sequence of H. pucrasia sp. n. with that of Heterakis spp. available in GenBank showed 9.20% (H. gallinarum) to 18.1% (H. isolonche) nucleotide divergence.
3.2.5 Partial cox2 region
Three cox2 sequences of H. pucrasia sp. n. obtained herein are 519 bp long, with no nucleotide divergence detected. In the genus Heterakis, the cox2 sequences are available in GenBank for H. gallinarum (MF114536–MF114591, NC_029839), H. beramporia (NC_029838) and H. dispar (NC_042411). Pairwise comparison of the cox2 sequences of H. pucrasia sp. n. with that of Heterakis spp. available in GenBank showed 10.6–11.0% (H. gallinarum) to 11.2% (H. dispar) nucleotide divergence.
3.2.6 Partial 12S region
Three 12S sequences of H. pucrasia sp. n. obtained herein are 486 long, with no nucleotide divergence. In the genus Heterakis, the 12S sequences are available in GenBank for H. gallinarum (KR080329, KR153881–KR153937, NC_029839.1), H. dispar (NC_042411), H. beramporia (NC_029838) and H. spumosa (MT135066, MT135065). Pairwise comparison of the 12S sequences of H. pucrasia sp. n. with that of Heterakis spp. available in GenBank showed 8.27% (H. gallinarum) to 13.5–13.6% (H. spumosa) nucleotide divergence.
3.3 Species delimitation
The results of ASAP and BI analyses based on the ITS, cox1, cox2 and 12S sequences, all supported that H. pucrasia sp. n. represents a separated species from its congeners (Figures 5, 6). However, the optimal results of ASAP and BI analyses based on the ITS sequences did not support the species partition of H. dispar and H. isolonche, and H. spumosa and H. dahomensis (Figure 5). The results of BI analyses based on the ITS and 12S sequences showed that H. pucrasia sp. n. is a sister to H. dispar, but the BI results based on the cox1 and cox2 sequences displayed H. pucrasia sp. n. and H. gallinarum have a closer relationship than H. dispar (Figure 6).
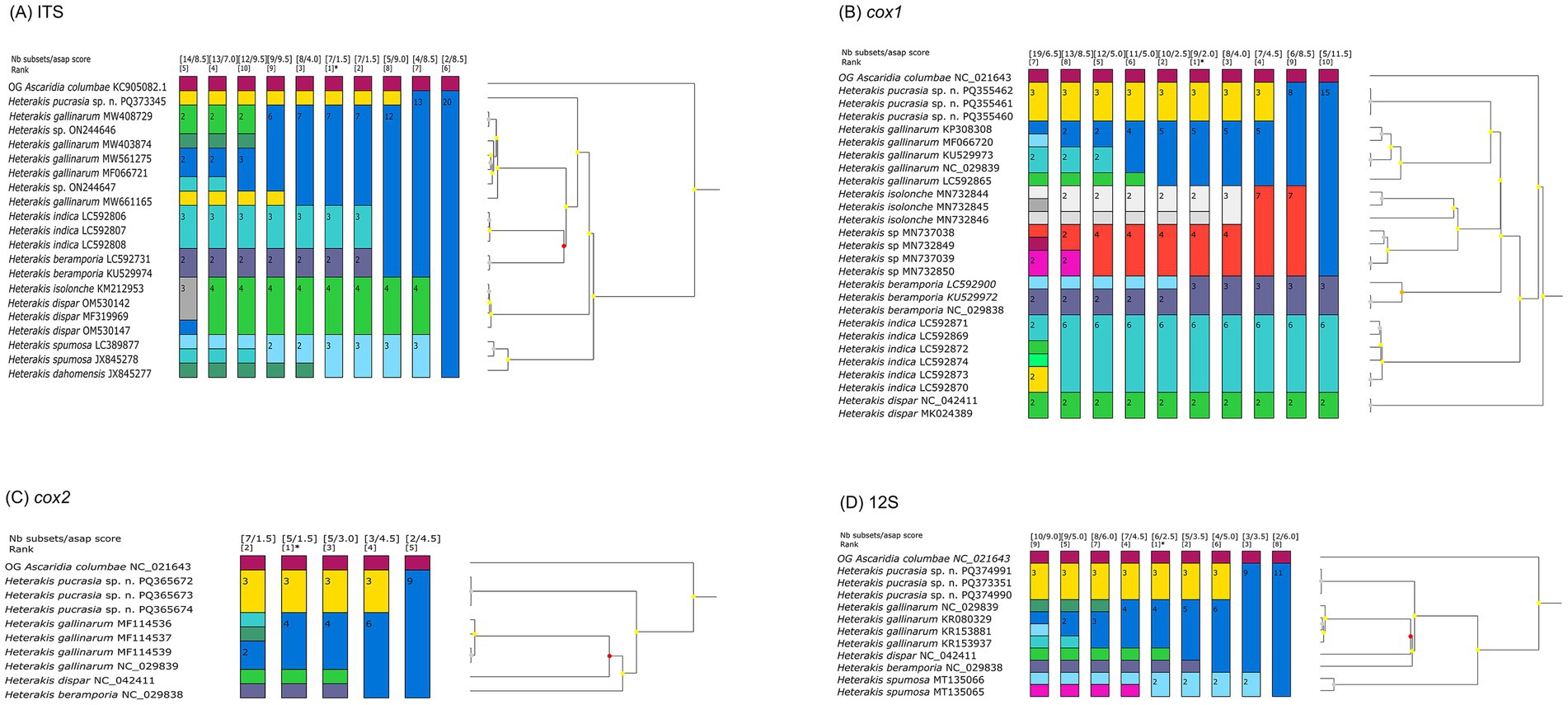
Figure 5. Assemble species by automatic partitioning (ASAP) analyses of Heterakis spp. based on different genetic markers. Asterisk representing the optimal result recommended by ASAP. Ascaridia columbae was chosen as the out-group (OG). (A) ASAP results based on ITS data. (B) ASAP results based on cox1 data. (C) ASAP results based on cox2 data. (D) ASAP results based on 12S data.
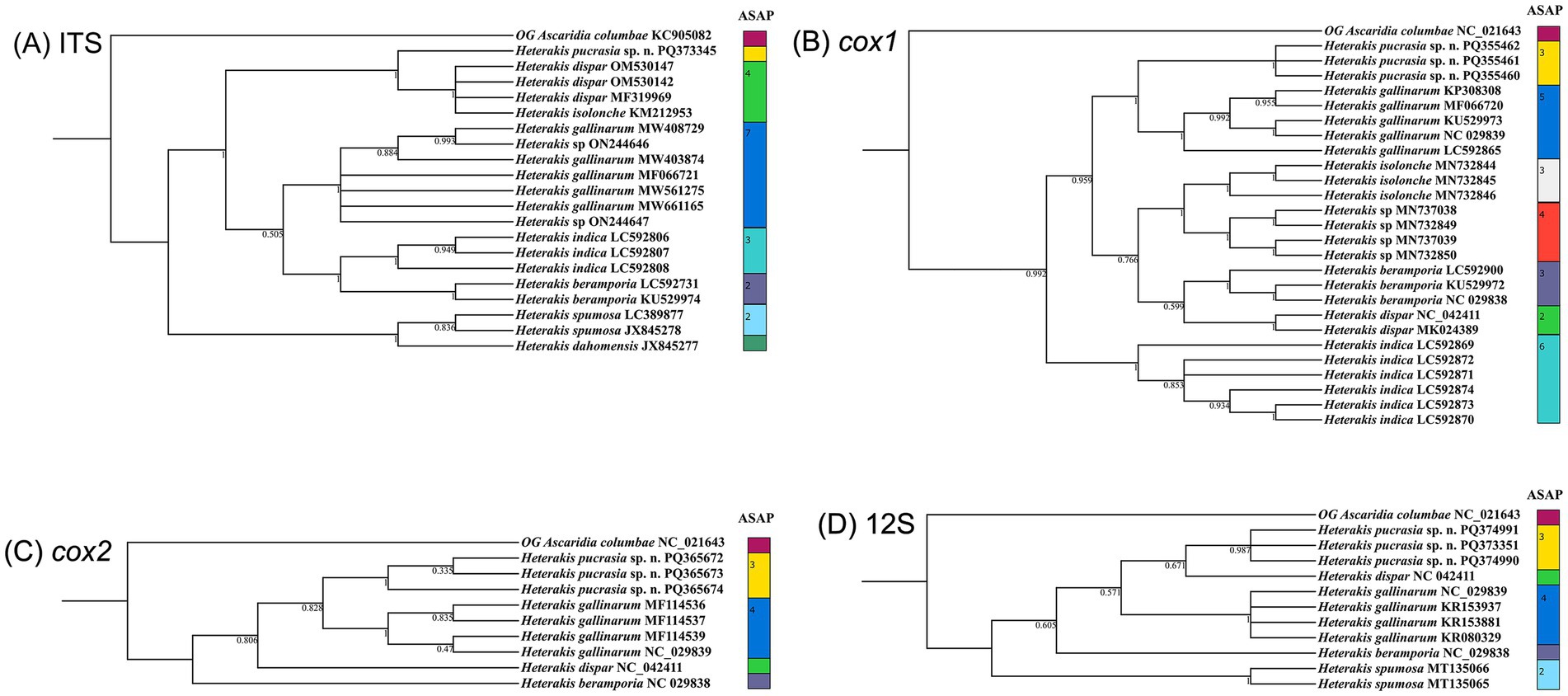
Figure 6. Comparative results of Bayesian inference (BI) and ASAP of Heterakis spp. based on different genetic markers. Ascaridia columbae was chosen as the out-group (OG). (A) BI results based on ITS data. (B) BI results based on cox1 data. (C) BI results based on cox2 data. (D) BI results based on 12S data.
3.4 Characterization of complete mitochondrial genome
The mitogenome of H. pucrasia sp. n. is 13,978 bp in length, containing 36 genes, including 12 PCGs (missing atp8) (cox1–3, cytb, nad1–6, nad4L and atp6), 22 tRNA genes and 2 rRNA genes (rrnL and rrnS) (Figure 7; Supplementary Table S1). All genes are transcribed from the same DNA strand. Two non-coding regions (LNCR and SNCR) are present in the mitogenome of H. pucrasia sp. n. (LNCR is 619 bp, between tRNA-Cys and tRNA-Asn; SNCR is 123 bp, between nad4 and tRNA-Met) (Figure 7). The overall A+T content in the mitogenome of H. pucrasia sp. n. is 68.9%, displaying a strong nucleotide compositional bias toward A+T (Supplementary Table S2). The nucleotide content of the mitogenome of H. pucrasia sp. n. was provided in Supplementary Table S2.
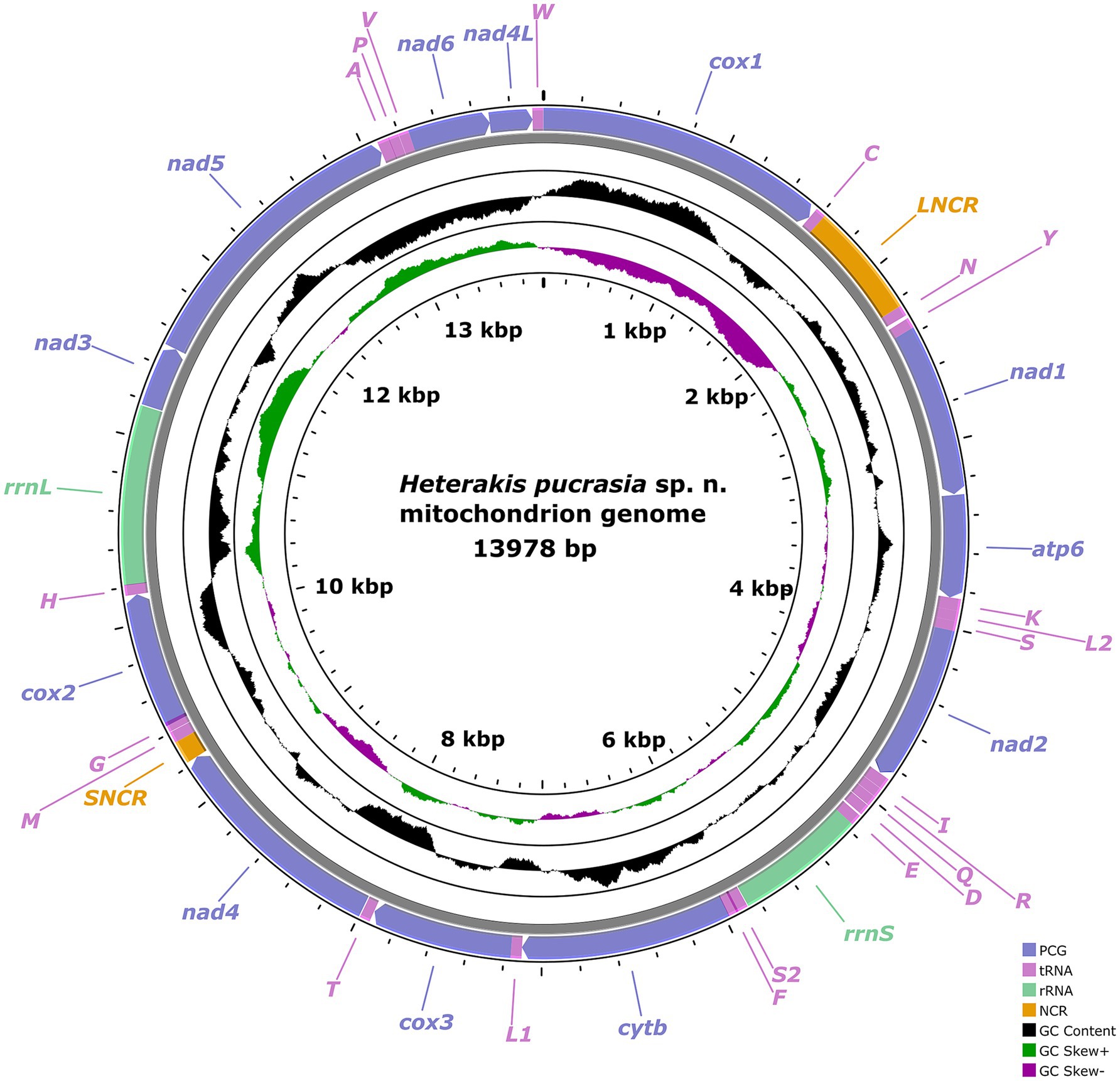
Figure 7. Gene map of the mitochondrial genome of Heterakis pucrasia sp. n. All 22 tRNA genes are nominated by the one-letter coding with numbers differentiating each of the two tRNAs, serine and leucine.
The 12 PCGs of the mitogenome of H. pucrasia sp. n. has 10,277 bp (excluding termination codons), ranged in size from 226 bp (nad4L) to 1,563 bp (cox1), which encoded 3,427 amino acids. Among the 12 PCGs, TTG is the most common start codon used for five genes (nad1, nad2, nad3, nad5 and nad6), followed by ATT for three genes (cox2, cox3 and nad4L), and ATG for cox1 and cytb and ATA for atp6 and nad4. TAA is the most commonly used termination codon for seven genes (cox3, nad1, nad2, nad4, nad5, nad6, and cytb), followed by TAG for cox1, nad3, cox2 and atp6, and the incomplete stop codon TA for nad4L (Table 1). The components and usage of codons in the mitogenome of H. pucrasia sp. n. were provided in Table 1 and Figure 8. The length of 22 tRNAs in the mitogenome of H. pucrasia sp. n. was provided in Table 1.
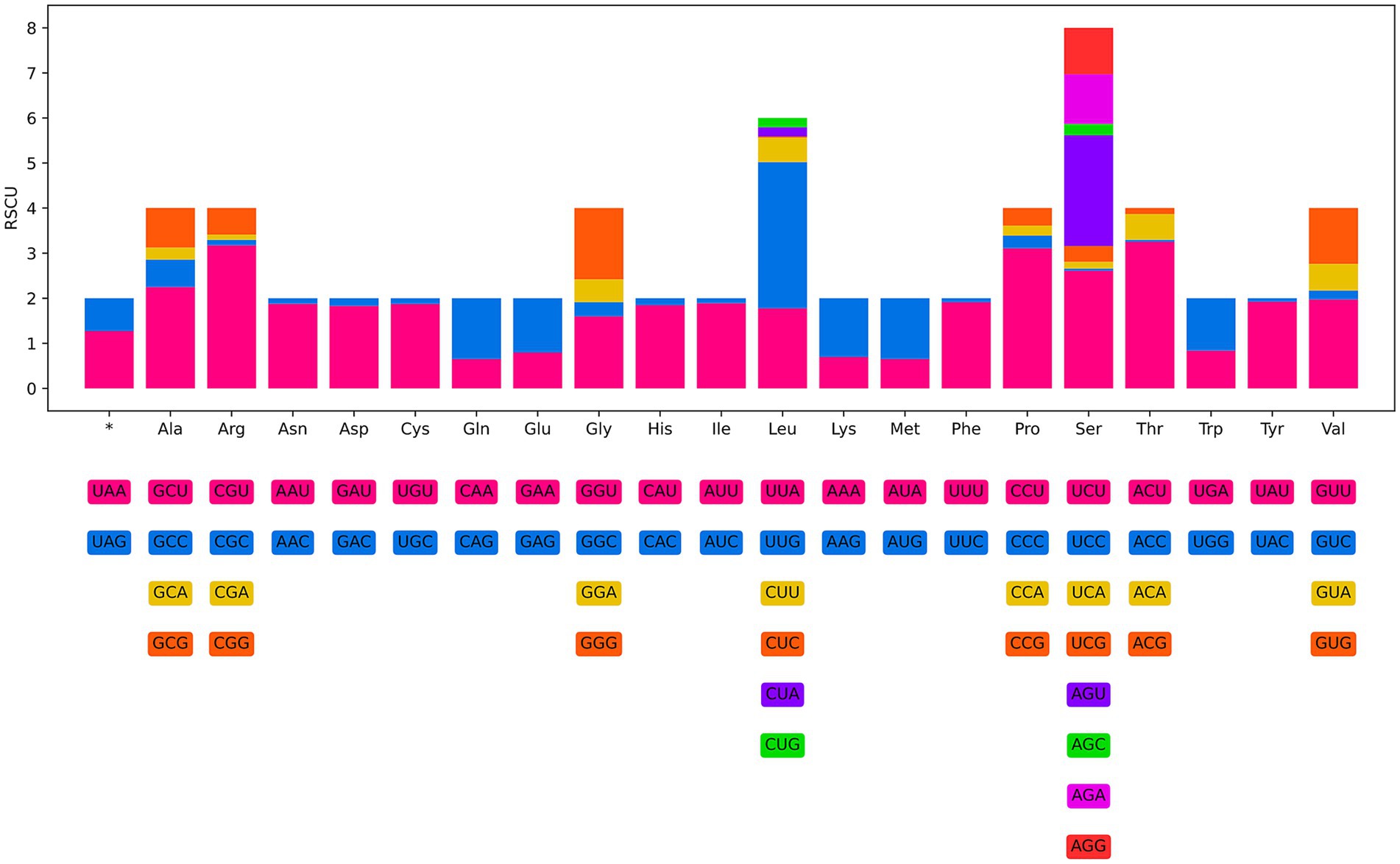
Figure 8. Relative synonymous codon usage (RSCU) of Heterakis pucrasia sp. n. The codon families (in alphabetical order) indicated below the horizontal axis. Values at the top of each bar representing amino acid usage in percentage.
The 36 gene arrangements in the mitogenome of H. pucrasia sp. n. are in the following order: cox1, tRNA-Cys, tRNA-Asn, tRNA-Tyr, nad1, atp6, tRNA-Lys, tRNA-Leu2, tRNA-Ser1, nad2, tRNA-Ile, tRNA-Arg, tRNA-Gln, tRNA-Asp., tRNA-Glu, rrnS, tRNA-Ser2, tRNA-Phe, cytb, tRNA-Leu1, cox3, tRNA-Thr, nad4, tRNA-Met, tRNA-Gly, cox2, tRNA-His, rrnL, nad3, nad5, tRNA-Ala, tRNA-Pro, tRNA-Val, nad6, nad4L, and tRNA-Trp (Figure 7).
3.5 Phylogenetic analyses
Phylogenetic results based on the concatenated amino acid sequences of the 12 protein-coding genes of mitogenomes using BI and ML methods, are nearly identical, which showed the representatives of Ascaridida, Spirurida, Oxyurida and Rhigonematida divided into six large clades (Clade I-VI) (Figure 9). Clade I included species of the superfamily Ascaridoidea. Clade II included two representatives of the superfamily Seuratoidea (Pingus sinensis and Cucullanus robustus). Clade III contained species of Ascaridia and Heterakis, representing the superfamily Heterakoidea. The single representative Rhigonema thysanophora comprised the Clade IV, representing the order Rhigonematida. Clade V included six species of the order Oxyurida. Clade VI contained 12 species of the order Spirurida (Table 2).

Figure 9. Phylogenetic results based on the concatenated amino acid sequences of 12 PCGs of mitogenomes of Ascaridida, Spirurida, Oxyurida and Rhigonematida. Caenorhabditis elegans (Rhabditida: Rhabditidae) was chosen as the out-group. Bootstrap values ≥70 and Bayesian posterior probability values ≥0.85 were shown in the phylogenetic trees.
4 Discussion
In the genus Heterakis, there have been about 38 nominal species reported from birds (26, 27). However, the validity of some species remains under debate. Among these species parasitic in birds, the new species is similar to the following species by having nearly equal spicule length with 0.50–1.00 mm, including H. alata Schneider, 1866, H. skrjabini Cram, 1927, H. papillosa (Bloch, 1782), H. macroura Linstow, 1883, H. dispar (Schrank, 1790) and H. psophiae Travassos, 1913. Heterakis pucrasia sp. n. differs from H. psophiae by having much larger precloacal sucker (0.19–0.20 mm in diameter in H. pucrasia vs only 0.09 mm in diameter in the latter) and longer male tail (0.65–0.66 mm in the new species vs 0.35 mm in H. psophiae) (28, 29). Heterakis papillosa is a poorly known heterakid nematode reported from Otis tarda, O. tetrax and Tetrastes bonasia in Europe and Asia (28, 30), which is very similar to the new species in the lengths of body and esophagus, the morphology and size of precloacal sucker, and the morphology and lengths of spicules and tail. However, the new species can be distinguished from H. papillosa by more caudal papillae (12 pairs in H. pucrasia vs 10 pairs in H. papillosa) and the different position of vulva (vulva 4.47–6.95 mm from cephalic extremity, representing 43.3–53.5% of body length in the new species vs vulva 6.29–8.12 mm from cephalic extremity, representing about 62.2% of body length in H. papillosa).
The new species differs from H. dispar by having distinctly longer male tail (0.65–0.66 mm in H. pucrasia vs about 0.32 mm in H. dispar), smaller female body (10.3–14.2 mm long in the new species vs 15.0–23.0 mm long in H. dispar) and the different position of vulva (the length of vulva from cephalic extremity representing 43.3–53.5% of body length in the new species vs representing about 56.3% of body length in H. dispar) (28, 29).
With the larger male body (17.0–27.8 mm), smaller posterior esophageal bulb (0.18–0.28 × 0.21–0.23 mm) and shorter male tail (0.28–0.37 mm), H. alata can be easily distinguished from the new species (male 9.10–9.39 mm long), posterior esophageal bulb 0.36–0.37 × 0.29–0.30 mm, male tail 0.65–0.66 mm (28, 31).
Heterakis skrjabini is also different from H. pucrasia sp. n. by having male tail lacking long filamentous tip (vs the presence of long filamentous tip in male in the new species), relatively shorter esophagus of female (esophageal length representing only 4.50% of body length in H. skrjabini vs representing 11.3–13.9% of body length in H. pucrasia) and the different position of the vulva (length of the vulva from cephalic extremity representing 17.7% of body length in H. skrjabini vs representing 43.3–53.5% of body length in the new species). Heterakis macroura is a common nematode parasite from birds of Tetraogallus.
Barus and Sonin (32) listed H. altaica Spaul, 1929 and H. lahulensis Soota & Chaturvedi, 1969 as synonyms of H. macroura. Heterakis pucrasia sp. n. differs from H. macroura by having more caudal papillae (14 pairs in the new species vs 12 pairs in H. macroura), more cuticular protrusions at region of vulva (presence of 4 or 5 cuticular protrusions in H. pucrasia vs usually presence of 2 cuticular protrusions in the latter) and smaller body of female (10.3–14.2 mm long in the new species vs 15.0–20.2 mm long in H. macroura) (28, 29).
Comparative mitogenome analysis revealed that the size and overall A+T content in the mitogenome of H. pucrasia sp. n. (13,978 bp in length, 68.9% overall A+T content) are similar to that of H. gallinarum, H. beramporia and H. dispar (13973–14,012 bp in length, and 69.8–71.3% overall A+T content in the latter three species) (13, 14). Moreover, the composition and gene arrangement of the mitogenome of H. pucrasia sp. n. are accordant with that of H. gallinarum, H. beramporia and H. dispar. To date, a total of 63 types of gene arrangement in the mitogenomes of Nematoda have been reported (33). The gene arrangement of the new species belongs to the GA1 type proposed by Yatawara et al. (34), which is identical to that of heterakoid species, including H. gallinarum, H. beramporia, H. dispar, Ascaridia columbae, A. galli and Ascaridia sp. (13, 14, 35).
Blaxter et al. (36) and Holterman et al. (37) provided phylogenies of the phylum Nematoda based on 18S sequence data, and preliminary revealed the evolutionary relationships of the four orders Ascaridida, Spirurida, Oxyurida and Rhigonematida. However, the phylogenetic relationships of these four orders, together with some their included superfamilies and families, remain uncertain (38). The present phylogenetic results showed that the current order Ascaridida is not monophyletic, which agreed well with some previous phylogenies (13, 35, 37–42). To date, only some molecular phylogenies tried to investigate the systematic position of the superfamily Heterakoidea, but most of phylogenetic results showed the Heterakoidea has a close relationship with the order Rhigonematida (37, 40–42); in contrast, some other results indicated the Heterakoidea has a close affinity with Ascaridoidea (38, 43, 44) or Oxyurida (39) or Rhabditomorpha + Ascaridomorpha (13, 35). Interestingly, the present phylogenetic results displayed the Heterakoidea clustered together with the representatives of Rhigonematida + Oxyurida + Spirurida, which are different from all of the previous studies (37, 38, 40–44). The present results suggested that the traditional superfamily Heterakoidea could be elevated as the infra-order Heterakomorpha. Although the present study provided novel insight into the systematic position of the Heterakoidea, it is yet an unsolved mystery of the phylogenetic position of the Heterakoidea. However, a more rigorous molecular phylogenetic study that includes broader representatives of the Heterakoidea, especially species of the families Aspidoderidae and Kiwinematidae, is needed to further solve the deep phylogenetic relationships between the Heterakoidea and its related groups.
5 Conclusion
A new species of Heterakis, H. pucrasia sp. n. was described using integrated methods based on specimens collected from the koklass pheasant P. macrolopha in Pakistan. The complete mitochondrial genome of H. pucrasia sp. n. was sequenced and annotated for the first time to enrich the mitogenomic data and reveal the pattern of mitogenomic evolution of Heterakidae. Mitogenomic phylogenetic results revealed that the order Ascaridida is not monophyletic, and suggested the erection of the infraorder Heterakomorpha for the traditional superfamily Heterakoidea.
Data availability statement
The nuclear and mitochondrial DNA sequences of Heterakis pucrasia sp. n. obtained in the present study were deposited in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) under the accession numbers: PQ373346, PQ373350, PQ358945 (18S); PQ358944, PQ358946, PQ358947 (28S); PQ373345 (ITS); PQ355460-PQ355462 (cox1); PQ374991, PQ373351, PQ374990 (12S); PQ365672-PQ365674 (cox2); PQ389430 (mitogenome).
Ethics statement
This study was conducted under the protocol of Hebei Normal University. All applicable national and international guidelines for the protection and use of animals were followed. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MY: Conceptualization, Data curation, Formal analysis, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. Y-NS: Formal analysis, Software, Writing – review & editing. H-XC: Investigation, Validation, Writing – review & editing. AK: Data curation, Writing – review & editing. LL: Funding acquisition, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Grant No. 32170442), and the Key Development Foundation of Hebei Normal University (L2024ZD17) for Liang Li.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1519092/full#supplementary-material
Footnotes
1. ^https://avibase.bsc-eoc.org/species.jsp?avibaseid=6383E214
2. ^http://www.ncbi.nlm.nih.gov
3. ^https://bioinfo.mnhn.fr/abi/public/asap/
References
1. Miller, JR. Survey of Western Tragopan, Koklass pheasant, and Himalayan Monal populations in the great Himalayan National Park, Himachal Pradesh, India. Indian Birds. (2010) 6:60–5.
2. Badrulislam, KA, Khalil, S, Hussain, M, Saqib, Z, Altaf, J, Hadi, R, et al. Habitat suitability modelling of Koklass pheasant (Pucrasia macrolopha) in moist temperate forest. PLoS One. (2024) 19:e0296921. doi: 10.1371/journal.pone.0296921
3. Shansaz, U, Fazili, MF, and Bhat, BA. Distribution and threats to two threatened pheasants in the brein nishat conservation reserve, Kashmir, India. Uttar Pradesh J Zool. (2021) 42:88–94. doi: 10.9734/bpi/nvbs/v5/2776E
4. Lu, X-S, Fu, B-Z, Shi, X-Q, Zhou, Z-Y, and Hu, Q-H. Notes on the nematodes of wild birds in Shanghai zoo. J Agric Sci. (1990) 11:53–5.
5. Skryabin, K, Shikhobalova, N, and Mozgovoi, A. Descriptive catalogue of parasitic nematodes. Volume 2. Oxyurata and Ascaridata. Moscow: Akademiya Nauk SSSR Publishers. (1951). 631 pp.
6. Smales, L. Heterakis fieldingi n. sp. (Nematoda: Heterakoidea) from the Australian water-rat, with a review of heterakids occurring in mammals. Syst Parasitol. (1996) 35:127–32. doi: 10.1007/BF00009821
7. Upadhyay, SK. Morphotaxometry of a new roundworm Heterakis equispiculis n. sp.(Nematoda: Heterakidae) from rodents of Bundelkhand region at Uttar Pradesh, India. Proc Zool Soc. (2019) 72:171–9. doi: 10.1007/s12595-017-0252-9
8. Menezes, RC, Tortelly, R, Gomes, DC, and Pinto, RM. Nodular typhlitis associated with the nematodes Heterakis gallinarum and Heterakis isolonche in pheasants: frequency and pathology with evidence of neoplasia. Mem Inst Oswaldo Cruz. (2003) 98:1011–6. doi: 10.1590/S0074-02762003000800005
9. Bobrek, K, Hildebrand, J, Urbanowicz, J, and Gaweł, A. Molecular identification and phylogenetic analysis of Heterakis dispar isolated from geese. Acta Parasitol. (2019) 64:753–60. doi: 10.2478/s11686-019-00112-1
10. Bobrek, K, Gaweł, A, and Urbanowicz, J. Molecular analysis of the Heterakis dispar population in domestic geese based on the ITS1−5.8 rRNA-ITS2 fragment. Animals. (2022) 12:e926. doi: 10.3390/ani12070926
11. Ribas, A, Bellocq, J, Ros, A, Ndiaye, P, and Miquel, J. Morphometrical and genetic comparison of two nematode species: H. Spumosa and H. Dahomensis (Nematoda, Heterakidae). Acta Parasitol. (2013) 58:389–98. doi: 10.2478/s11686-013-0156-4
12. Abdel-Gaber, R, Kamel, R, Maher, S, and Fergani, Y. Morphological and molecular studies of the nematode parasite Heterakis gallinarum (Heterakidae) infecting the cattle egret Bubulcus ibis (Ardeidae). Arq Bras Med Vet Zoot. (2023) 75:1096–106. doi: 10.1590/1678-4162-13052
13. Wang, B-J, Gu, X-B, Gu, X-B, Yang, G-Y, Wang, T, Lai, W-M, et al. Mitochondrial genomes of Heterakis gallinae and Heterakis beramporia support that they belong to the infraorder Ascaridomorpha. Infect Genet Evol. (2016) 40:228–35. doi: 10.1016/j.meegid.2016.03.012
14. Gao, J-F, Hou, M-R, Wang, W-F, Gao, Z-Y, Zhang, X-G, Lu, Y-X, et al. The complete mitochondrial genome of Heterakis dispar (Ascaridida: Heterakidae). Mitochondrial DNA B. (2019) 4:1630–1. doi: 10.1080/23802359.2019.1574627
15. Jin, J-J, Yu, W-B, Yang, J-B, Song, Y, De Pamphilis, CW, Yi, T-S, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. (2020) 21:1–31. doi: 10.1186/s13059-020-02154-5
16. Puillandre, N, Brouillet, S, and Achaz, G. ASAP: assemble species by automatic partitioning. Mol Ecol Resour. (2021) 21:609–20. doi: 10.1111/1755-0998.13281
17. Meng, G, Li, Y, Yang, C, and Liu, S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. (2019) 47:e63. doi: 10.1093/nar/gkz173
18. Gruber, AR, Bernhart, SH, and Lorenz, R. The ViennaRNA web services. Methods Mol Biol. (2015) 1269:307–26. doi: 10.1007/978-1-4939-2291-8_19
19. Reuter, JS, and Mathews, DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. (2010) 11:1–9. doi: 10.1186/1471-2105-11-129
20. Lee, BD. Python implementation of codon adaptation index. J Open Res Softw. (2018) 3:905. doi: 10.21105/joss.00905
21. Ronquist, F, Teslenko, M, Van Der Mark, P, Ayres, DL, Darling, A, Höhna, S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. (2012) 61:539–42. doi: 10.1093/sysbio/sys029
22. Minh, BQ, Hahn, MW, and Lanfear, R. New methods to calculate concordance factors for phylogenomic datasets. Mol Biol Evol. (2020) 37:2727–33. doi: 10.1093/molbev/msaa106
23. Katoh, K, and Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. (2010) 26:1899–900. doi: 10.1093/bioinformatics/btq224
24. Criscuolo, A, and Gribaldo, S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. (2010) 10:210–21. doi: 10.1186/1471-2148-10-210
25. Zhang, D, Gao, F, Jakovlić, I, Zou, H, Zhang, J, Li, WX, et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. (2020) 20:348–55. doi: 10.1111/1755-0998.13096
26. Inglis, WG. The evolution, host relationships and classification of the nematode superfamily Heterakoidea. Bull Br Mus Nat Hist. (1967) 15:1–28. doi: 10.5962/bhl.part.27515
27. Smales, LR. Two new species of the genus Heterakis (Heterakidae) from the genera Echymipera, Isoodon and Perameles (Peramelidae), bandicoots from Papua New Guinea and Australia. Acta Parasitol. (2023) 68:782–92. doi: 10.1007/s11686-023-00706-w
28. Cram, EB. Bird parasites of the nematode suborders Strongylata, Ascaridata, and Spirurata. Bull US Natl Mus. (1927) 140:1–465. doi: 10.5479/si.03629236.140.1
29. Skryabin, K, Shikhobalova, N, and Lagodovskaya, E. Principles of nematoíogy In: KI Skryabin, editor. Oxyurata of animals and man. Part 2, Moscow: Izdatel ‘stvo Akademiya Nauk SSSR Publishers. vol. X (1961). 499 pp.
30. Li, H. Report on a collection of parasitic nematodes mainly from North China. Pt. III. Oxyuroidea. Chin Med J. (1933) 47:1307–25.
31. Vicente, JJ, Pinto, RMP, and Noronha, D. Remarks on six species of heterakid nematodes parasites of Brazilian Tinamid birds with description of a new species. Mem Inst Oswaldo Cruz. (1993) 88:271–8. doi: 10.1590/S0074-02761993000200015
32. Barus, V, and Sonin, M. Survey of nematodes parasitizing hosts of the genus Tetraogallus Gray, 1834 (Galliformes). Folia Parasitol. (1978) 25:211–21.
33. Zeng, J-L, Chen, H-X, Ni, X-F, Kang, J-Y, and Li, L. Molecular phylogeny of the family Rhabdiasidae (Nematoda: Rhabditida), with morphology, genetic characterization and mitochondrial genomes of Rhabdias kafunata and R. Bufonis. Parasite Vector. (2024) 17:100. doi: 10.1186/s13071-024-06201-z
34. Yatawara, L, Wickramasinghe, S, Rajapakse, R, and Agatsuma, T. The complete mitochondrial genome of Setaria digitata (Nematoda: Filarioidea): mitochondrial gene content, arrangement and composition compared with other nematodes. Mol Biochem Parasitol. (2010) 173:32–8. doi: 10.1016/j.molbiopara.2010.05.004
35. Liu, G-H, Shao, R, Li, J-Y, Zhou, D-H, Li, H, and Zhu, X-Q. The complete mitochondrial genomes of three parasitic nematodes of birds: a unique gene order and insights into nematode phylogeny. BMC Genomics. (2013) 14:1–13. doi: 10.1186/1471-2164-14-414
36. Blaxter, ML, de, P, Garey, JR, Liu, LX, Scheldeman, P, Vierstraete, A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. (1998) 392:71–5. doi: 10.1038/32160
37. Holterman, M, van der Wurff, A, van den Elsen, S, van Megen, H, Bongers, T, Holovachov, O, et al. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol Biol Evol. (2006) 23:1792–800. doi: 10.1093/molbev/msl044
38. Ahmed, M, and Holovachov, O. Twenty years after De Ley and Blaxter—how far did we progress in understanding the phylogeny of the phylum Nematoda? Animals. (2021) 11:3479. doi: 10.3390/ani11123479
39. Nadler, S, Carreno, R, Mejía-Madrid, H, Ullberg, J, Pagan, C, Houston, R, et al. Molecular phylogeny of clade III nematodes reveals multiple origins of tissue parasitism. Parasitology. (2007) 134:1421–42. doi: 10.1017/S0031182007002880
40. Kim, T, Kim, J, Cho, S, Min, GS, Park, C, Carreno, RA, et al. Phylogeny of Rhigonematomorpha based on the complete mitochondrial genome of Rhigonema thysanophora (Nematoda: Chromadorea). Zool Scr. (2014) 43:289–303. doi: 10.1111/zsc.12047
41. Choudhury, A, and Nadler, SA. Phylogenetic relationships of Cucullanidae (Nematoda), with observations on Seuratoidea and the monophyly of Cucullanus, Dichelyne and Truttaedacnitis. J Parasitol. (2016) 102:87–93. doi: 10.1645/15-806
42. Gendron, EM, Qing, X, Sevigny, JL, Li, H, Liu, Z, Blaxter, M, et al. Comparative mitochondrial genomics in Nematoda reveal astonishing variation in compositional biases and substitution rates indicative of multi-level selection. BMC Genomics. (2024) 25:615. doi: 10.1186/s12864-024-10500-1
43. Meldal, BH, Debenham, NJ, De Ley, P, De Ley, IT, Vanfleteren, JR, Vierstraete, AR, et al. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol. (2007) 42:622–36. doi: 10.1016/j.ympev.2006.08.025
Keywords: parasite, bird, Heterakoidea, integrative taxonomy, mitochondrial genome, phylogeny, new species
Citation: Yousaf MA, Sun Y-N, Chen H-X, Khan AU and Li L (2025) Morphology, complete mitochondrial genome and molecular phylogeny of Heterakis pucrasia sp. n. (Nematoda: Ascaridida) from the koklass pheasant Pucrasia macrolopha (Lesson) (Galliformes: Phasianidae) in Pakistan. Front. Vet. Sci. 12:1519092. doi: 10.3389/fvets.2025.1519092
Edited by:
Ruediger Hauck, Auburn University, United StatesReviewed by:
Luis Garcia Prieto, National Autonomous University of Mexico, MexicoGeorge Sangster, Naturalis Biodiversity Center, Netherlands
Copyright © 2025 Yousaf, Sun, Chen, Khan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Li, bGlhbmdsaWFuZ2V4MzY5QDEyNi5jb20=
 Muhammad Amjad Yousaf
Muhammad Amjad Yousaf Yi-Nuo Sun
Yi-Nuo Sun Hui-Xia Chen
Hui-Xia Chen Asmat Ullah Khan
Asmat Ullah Khan Liang Li
Liang Li