- 1Department of Animal Sciences, Center for Nutrition and Pregnancy, North Dakota State University, Fargo, ND, United States
- 2Department of Agriculture and Natural Resources, University of Minnesota Crookston, Crookston, MN, United States
- 3Department of Veterinary Biomedical Sciences, University of Saskatchewan, Saskatoon, SK, Canada
- 4United States Department of Agriculture, Agriculture Research Service, U.S. Meat Animal Research Center, Clay Center, NE, United States
Introduction: Maternal nutrition during pregnancy critically influences offspring development and immune function. One-carbon metabolites (OCM) are epigenetic modifiers that may modulate antimicrobial peptide (AMP) expression, which is vital for innate immunity. This study investigated the effects of maternal nutrient restriction and OCM supplementation on mRNA expression of AMP in fetal and maternal lung, mammary gland, and small intestine of beef cattle.
Methods: Twenty-nine crossbred Angus beef heifers were synchronized for estrus and artificially inseminated. They were assigned to one of four treatments in a 2 × 2 factorial design: nutritional plane [control (CON) vs. restricted (RES)] and OCM supplementation [without OCM (−OCM) or with OCM (+OCM)]. Heifers on the CON diet were fed to gain 0.45 kg/day, while RES heifers were fed to lose 0.23 kg/day. Treatments were applied from day 0 to 63 of gestation, after which all heifers were fed a common diet to gain 0.45 kg/day until day 161 of gestation, when samples were collected. Quantitative RT-qPCR was used to assess mRNA expression of AMP.
Results: Nutritional plane had no effect (p ≥ 0.24) on mRNA expression of AMP in either the fetus or dams. However, the mRNA expression of cathelicidin5 (CATHL5; p = 0.07) and bovine neutrophil β-defensin5 (BNBD5; p = 0.07) in the fetal lung and mammary gland, respectively, was lower in the +OCM groups compared to the −OCM groups. In the maternal small intestine, the expression of enteric β-defensin (EBD) was lower (p = 0.01) in the +OCM groups compared to the −OCM groups. Additionally, in the maternal lung, there was a tendency (p = 0.06) for an interaction in CATHL5 mRNA expression, with the RES + OCM group showing greater expression compared to the CON + OCM (p = 0.07) and RES − OCM (p = 0.08) groups.
Discussion: Our findings suggest that while restricted maternal nutrition did not affect mRNA expression of AMP, OCM supplementation modulated AMP expression in both fetal and maternal tissues. Further research is needed to elucidate the mechanisms underlying OCM’s impact on AMP expression.
1 Introduction
Maternal nutrition during pregnancy plays a crucial role in influencing the health and development of offspring (1, 2). Factors such as drought-induced forage shortages and inadequate intake of essential minerals and supplements can disrupt maternal nutrition, leading to lasting effects on fetal growth, immune function, and metabolic regulation in livestock (3, 4). This phenomenon, known as developmental programming, suggests that stressors, including nutritional stress, during critical developmental windows can elicit both immediate and prolonged consequences in the offspring (5). A principal mechanism by which maternal nutrition can affect fetal development is through epigenetic modifications (6–8).
Central to this epigenetic control are one-carbon metabolites (OCM), which include methyl-donors such as folate, vitamin B12, methionine, and choline. These metabolites serve as vital epigenetic modifiers, influencing DNA methylation and regulating the expression of genes involved in various biological processes, including immune function (9–12). Dysregulation of these metabolites and the one-carbon metabolism pathway can lead to immune dysfunction (13, 14). Studies indicate increased maternal plasma concentrations of OCM are associated with greater expression of genes related to the immune system, histone modification, and RNA processing in offspring (15). Additionally, our previous research has shown that nutrient restriction in pregnant beef heifers can result in reduced concentrations of methionine, an essential OCM, in allantoic fluid and increased concentrations of homocysteine in maternal serum during early gestation (16). These alterations in OCM levels can subsequently alter metabolic pathways related to one-carbon metabolism and influence epigenetic modifications (10). This highlights the potential impact that altering OCM supply could have on the developing immune system.
Antimicrobial peptides (AMP), such as β-defensins (17), cathelicidins (18), and S100 proteins (19), are diverse molecules that serve as a first-line host innate defense mechanism. Typically short and positively charged, AMP interact with the negatively charged membranes of microbes, disrupting the membrane and causing microbial death (20, 21). They can also target other microbial components, such as DNA or proteins (22). AMP offers several advantages over traditional antibiotics. They are less likely to trigger resistance in bacteria, act much faster, and can work against a wider range of bacteria, fungi, and enveloped viruses. This makes them particularly effective against even those strains of bacteria that are resistant to multiple antibiotics (23, 24). AMP exhibit potent bactericidal activity against pathogens affecting the respiratory tract, intestine, and udder, such as Mannheimia haemolytica, Histophilus somni, Pasteurella multocida, Escherichia coli, and Staphylococcus aureus (25–29). Epigenetic pathways, including DNA methylation and histone modification, play key roles in regulating the expression and production of AMP (30, 31). Bovine respiratory disease (32), infectious diarrhea (33), and mastitis (34) are significant health concerns in cattle industry, contributing to substantial economic losses and health complications. The rise in antimicrobial resistance due to the overuse of synthetic antimicrobials further emphasizes the need to enhance the natural immune system to prevent diseases and combat pathogens (35, 36). Therefore, understanding the variables that affect the epigenetic control of AMP is essential for enhancing their expression and boosting the natural immunity of cattle. However, the influence of maternal nutrition and strategic OCM supplementation on AMP expression remains to be elucidated.
The objective of this study was to determine the effects of restricted maternal nutrition during early gestation, with or without OCM supplementation, on mRNA expression of AMP in the bovine fetal and maternal lung, mammary gland, and small intestine. We hypothesized that maternal nutrient restriction would reduce antimicrobial peptide expression, while one-carbon supplementation would at least partially ameliorate these effects. The data will provide insight into how the maternal nutritional level affects the innate immune system in the bovine fetus and dams, and whether strategic supplementation with methyl-donor molecules may support normal innate immunity maturation under conditions of poor maternal nutrition.
2 Materials and methods
2.1 Animal ethics
All methodologies and experiments adhered to relevant guidelines and regulations. The design of the experiment, the management of animals, and the tissue collection processes were approved by the Institutional Animal Care and Use Committee at North Dakota State University (IACUC #20220059) on October 26, 2022.
2.2 Animals, diet, and treatment
Seventy-two Angus heifers were transported from the Central Grasslands Research Extension Center (Streeter, ND, USA) to the Animal Nutrition and Physiology Center at North Dakota State University (Fargo, ND, USA). After a 14-day adaptation to the feeding system, heifers were subjected to a 7-day Select Synch + CIDR estrus synchronization protocol (37) and artificially inseminated 18 to 22 h following the detection of estrus. To explore the effects of maternal nutrition and OCM on fetal development during gestation while removing sex-specific effects (38, 39), only female-sexed semen from a specific bull (Connealy Maternal Made [ST Genetics, Navasota, TX, USA]) was used for insemination. Of these 72 heifers, 29 (~ 14 months of age) successfully conceived [CON − OCM (n = 7), CON + OCM (n = 8), RES − OCM (n = 7), and RES + OCM (n = 7)]. The average initial body weight was 436 ± 42 kg. Upon breeding, the heifers were allocated into four different dietary groups based on a 2 × 2 factorial design that considered the level of nutritional plane (control vs. restricted) and OCM supplementation (with or without OCM). The heifers were individually fed daily in an electronic head gate facility (American Calan; Northwood, NH) at 0800 h daily.
Heifers on the control (CON) diet were fed to achieve 100% of the National Academy of Sciences, Engineering, and Medicine (NASEM) (40) nutritional requirements aiming for an average daily gain (ADG) of 0.45 kg/day, though they actually gained 0.60 kg/day. This regimen intended to bring them to 80% of their mature body weight by calving. Conversely, heifers on the restricted (RES) intake regimen were fed less to achieve a weight loss of 0.23 kg/day, mimicking the natural production responses observed in heifers undergoing dietary and environmental changes in early gestation (40). Their diets consisted of a total mixed ration including corn silage, alfalfa hay, corn grain, mixed alfalfa/grass hay, and a vitamin/mineral premix (Trouw dairy VTM w/Optimins, Trouw Nutrition USA, Highland, IL, USA). Diet compositions are more fully described in (41). Diets were formulated based on initial body weight at breeding to contain 2.25 Mcal/kg ME, 9.75% CP, and 58.6% NDF. Heifers were weighed weekly, and their individual feed intake was adjusted throughout the study to achieve the targeted body weight gains. Heifers supplemented with OCM (+OCM) received daily doses of 7.4 g of rumen-protected methionine (Smartamine, Adisseo, Beijing, China) and 44.4 g of rumen-protected choline (ReaShure, Balchem Inc., New Hampton, NY, USA) in a corn carrier, following dosages from previous studies (42, 43). They also received weekly intramuscular injections of vitamin B12 (50 mg of cyanocobalamin/mL; MWI Animal Health, Boise, ID, USA) and folate (53.33 mg of folic acid/mL; Spectrum Chemical Mfg. Corp., New Brunswick, NJ, USA) supplements, aimed at delivering 320 mg of folic acid and 20 mg of vitamin B12 each week, following previously described methods (43, 44). In contrast, the non-OCM supplemented heifers (−OCM) were given only the corn carrier daily and received weekly saline injections intramuscularly. These treatment protocols continued until day 63 of gestation, after which all heifers were managed on the CON − OCM treatment targeting a daily gain of 0.45 kg for the remainder of the study.
Pregnancy confirmation occurred on day 35 via transrectal ultrasonography by detecting fetal heartbeats, and a subsequent ultrasound on day 63 assessed fetal sex (45), retaining only pregnant heifers with female fetuses for further study stages.
2.3 Sample collection and preparation
Heifers were slaughtered on day 161 of gestation. The gravid uterus was promptly removed, and the fetus was immediately separated from the placenta, and subsequently dissected. Given that the cranioventral area of the lung is predominantly affected in bovine respiratory disease (46), lung samples from both the fetus and the dam were collected from this region (left cranioventral). Reflecting findings that high levels of AMP mRNA are present in the ileum (47), samples from this region were obtained from a section 10 cm proximal to the ileocecal junction. Additionally, considering the involvement of the mammary gland parenchyma in mastitis (48) and the uniform expression of AMP across different quarters (17), mammary gland samples were taken from the parenchyma of the right forequarters. All samples were wrapped in aluminum foil, snap-frozen in liquid nitrogen, and stored at −80°C until further analysis.
2.4 RNA extraction
Total RNA was extracted using the RNeasy® Plus Universal Kit (Qiagen, Germantown, MA, USA), following the manufacturer’s protocol and tissues were lysed using the TissueLyser LT (Qiagen Germantown, MA, USA). The RNA concentration was determined using the Qubit® RNA BR Assay Kit (Thermo Fisher Scientific, MA, USA) and measuring it with a Qubit 3.0 Fluorometer (LifeTechnologies, Carlsbad, CA, USA).
2.5 cDNA synthesis
The RNA was reverse transcribed into cDNA utilizing the High-Capacity Reverse Transcription Kit (Thermo Fisher Scientific, MA, USA) following the manufacturer’s protocol. The final cDNA concentration was 100 ng/μl in a volume of 20 μl, and samples were stored at −20°C until use.
2.6 RT-qPCR
The target AMP genes were as follows: lingual antimicrobial peptide (LAP), tracheal antimicrobial peptides (TAP), enteric β-defensin (EBD), bovine neutrophil β-defensin4 (BNBD4), bovine neutrophil β-defensin5 (BNBD5), S100 calcium binding protein A7 (S100A7), and cathelicidin5 (CATHL5). For each gene, the primers were designed using NCBI/Primer-BLAST or sourced from previous literature.
Optimization was conducted to determine the optimum cDNA concentration and primer efficiencies in each tissue type (49). The amplification efficiency (E) of target genes was evaluated by plotting the cycle threshold (Ct) versus log concentration. The E was calculated using the equation , with acceptable efficiencies ranging between 90 and 110% (50, 51). Details of the primer sequences used in the qPCR analysis can be found in Table 1. The primer sets for two genes (S100A7 and TAP) fell outside the acceptable efficiency range, likely due to low expression levels, as they exhibited high Ct values at the highest cDNA concentrations. Consequently, the relative mRNA expression of S100A7 and TAP were deemed below our limit of detection and were not further evaluated in this study. All other primers demonstrated efficiency within the acceptable range (90–110%). The results of the amplification efficiency experiment, including slope, R2, and efficiency, are summarized in Supplementary Tables S1, S2.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin beta (ACTB) and hypoxanthine phosphoribosyltransferase 1 (HPRT1) were used as reference genes. We employed the geNorm tool to assess the stability of their expression across all tissue samples. The lower M-values calculated by geNorm indicate more stable expression (52), and in our study, reference genes were considered stable if their M-values were below 0.5 (53).
Quantitative real-time PCR was performed using a QuantStudio™ 3 System (Thermofisher Scientific, MA, USA), with a 10 μl reaction mixture comprising 2 μl of the diluted cDNA (10 ng), 5 μl of 2× SYBR Green Master Mix (Bio-Rad Laboratories, Hercules, CA), 0.5 μl each of 10 μM forward and reverse primers (final concentration of 500 nM), and 2 μl of nuclease-free water. The amplification protocol included an initial denaturation at 95°C for 20 s, followed by 40 cycles of denaturation at 95°C for 1 s and annealing/elongation at 60°C for 20 s. A dissociation melt curve was determined at the end of each run, involving a brief denaturation at 95°C for 1 s, annealing at 60°C for 20 s, and a final denaturation at 95°C for 1 s. All runs included a negative control (without cDNA) and were performed in triplicate on a 96-well reaction plate (Thermo Fisher Scientific, MA, USA). Relative gene expression levels were quantified using the 2-ΔΔCT method (54), employing GAPDH, ACTB, and HPRT1 as reference genes for normalization, and using the CON − OCM group as control (set to 1).
2.7 Statistical analysis
Data were analyzed using the PROC MIXED function of SAS v.9.4 software (SAS Institute Inc., Cary, NC, USA). The normality of the data was assessed both before the analysis and for the resulting residuals after the analysis using PROC UNIVARIATE (Shapiro–Wilk test), followed by the QQ plot statement. In cases where the normality assumption was not met, data were transformed using . Residuals were subsequently re-evaluated for normality using the same methods. The model’s fixed effects included the level of nutrition, OCM treatment, and the interaction between these factors, with each individual heifer serving as the random effect. In the absence of significant interactions, the main effects of maternal nutrition and OCM treatment were presented. Means were separated using the PDIFF function of SAS (Tukey–Kramer adjustment) and all results were reported as least squares means (LSMEANS) and standard error of the mean (SEM). After analysis, the LSMEANS and SEM were back transformed to the original scale. The largest SEM is reported. All plots were generated using ggplot2 v.3.4.1 in R Studio v.4.2.2. Statistical significance was set at a p-value ≤0.05, and tendencies were reported for 0.05 < p ≤ 0.1.
3 Results
3.1 Genes expression in fetal tissue
The mRNA expression of BNBD4 in the fetal lung was not influenced by either the nutritional plane × OCM interaction or the main effect of treatment (p > 0.28; Figure 1A; Table 2); however, there was a tendency (p = 0.07) for OCM supplementation to affect the mRNA expression of CATHL5, with expression being 16.5% lower in the +OCM groups (0.81 ± 0.06 relative fold) compared to the −OCM groups (0.97 ± 0.06 relative fold) (Figure 1B; Table 2).
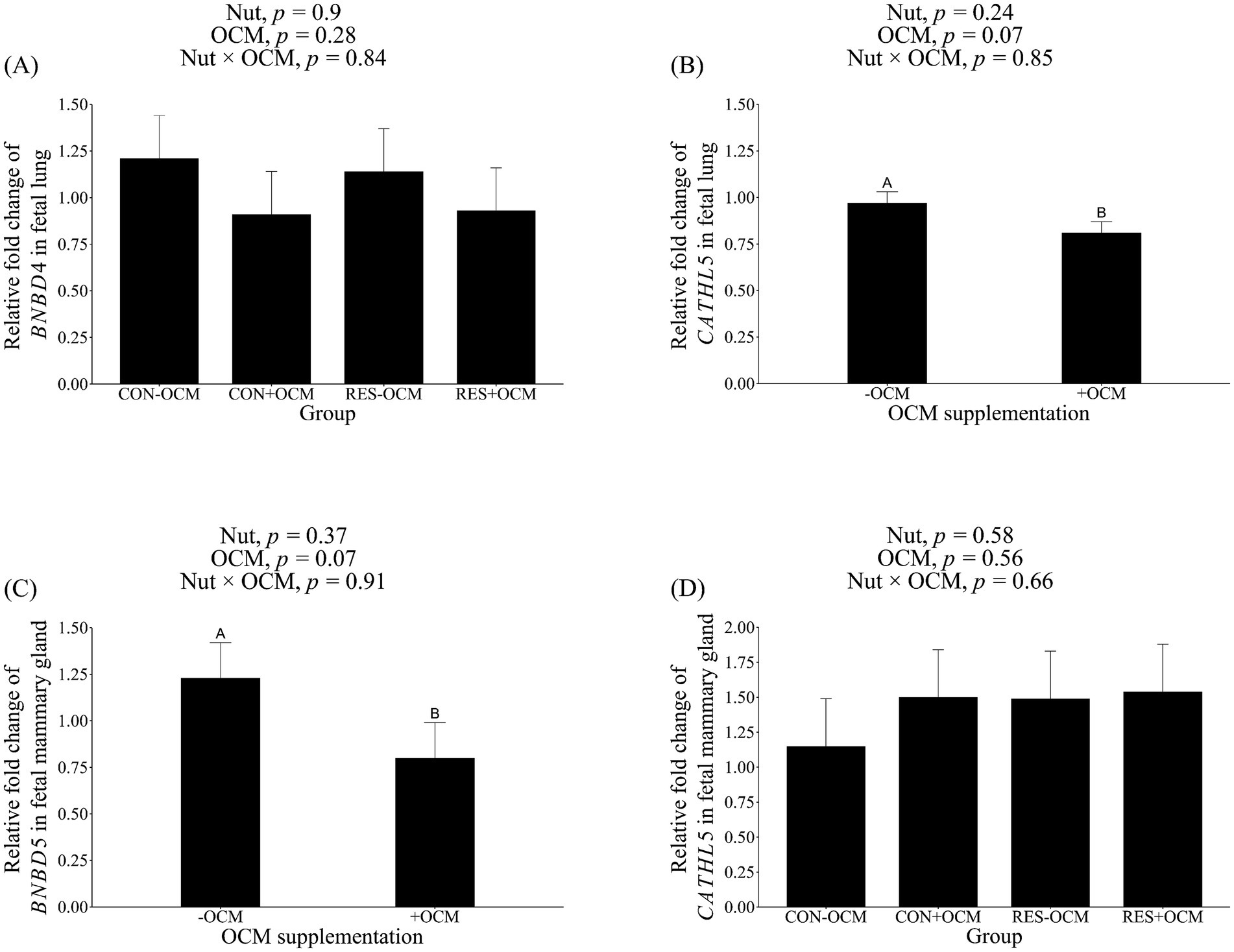
Figure 1. mRNA expression level of antimicrobial peptides in fetal tissues. (A) Bovine neutrophil β-defensin4 (BNBD4) in lung; (B) cathelicidin5 (CATHL5) in lung; (C) CATHL5 in mammary gland; (D) bovine neutrophil β-defensin5 (BNBD5) in mammary gland. Data presented as a 2-ΔΔCT-fold change normalized to GAPDH, ACTB, and HPRT1 as reference genes, and using the CON − OCM group as control. Different uppercase letters denote trends (0.05 < p ≤ 0.1).
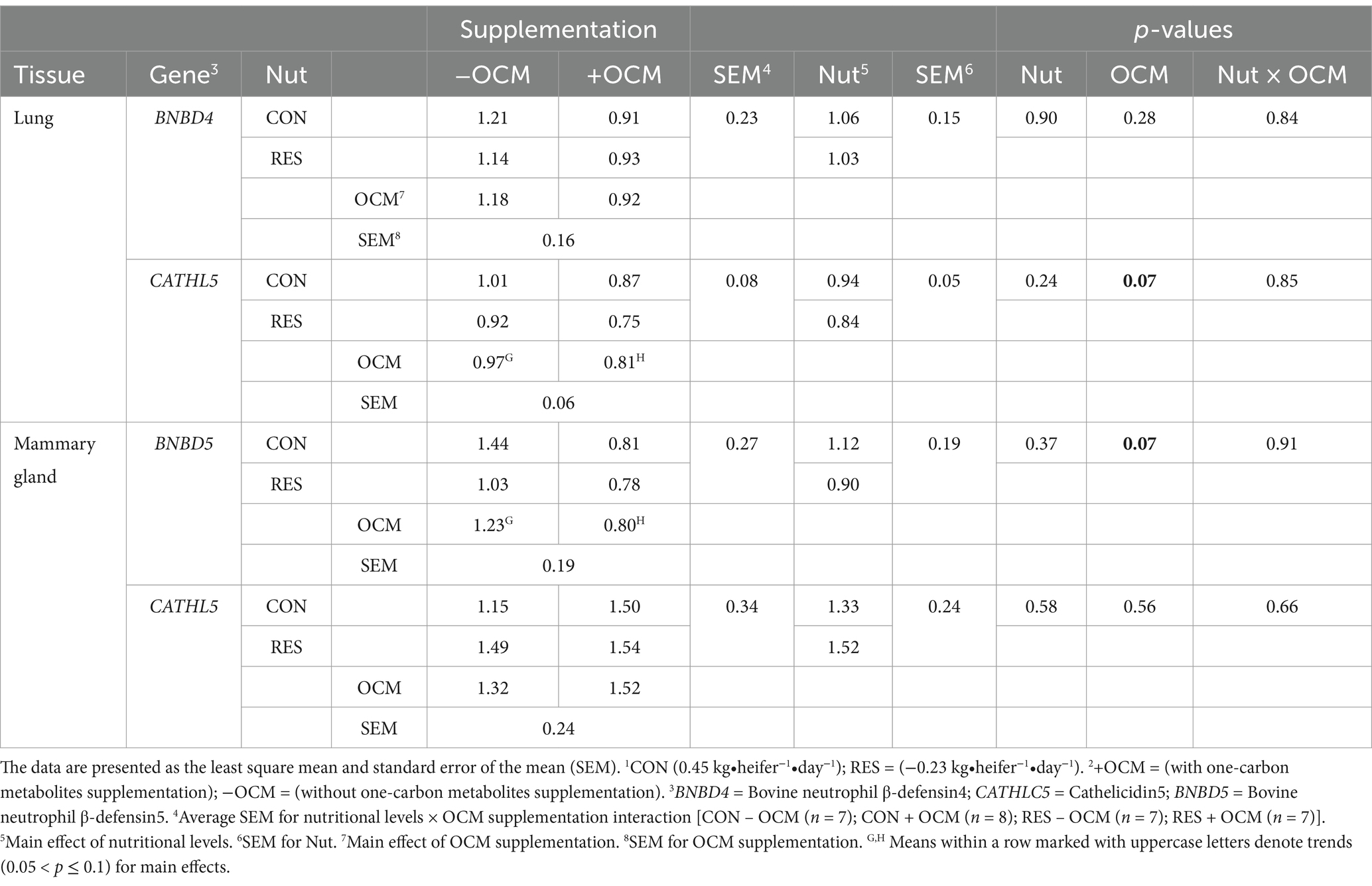
Table 2. The effect of maternal nutritional levels [Nut; control (CON) or restricted (RES)]1 and supplementation with one-carbon metabolites [OCM; with OCM (+OCM) or without (−OCM)]2 from day 0 to 63 of gestation on mRNA expression of antimicrobial peptides measured in fetal lung and mammary gland.
In the fetal mammary gland, the mRNA expression of BNBD5 tended (p = 0.07) to decrease by 35% in the +OCM groups (0.80 ± 0.19 relative fold) compared to the −OCM groups (1.23 ± 0.19 relative fold; Figure 1C; Table 2). No differences were observed in the mRNA expression of CATHL5 among the groups (p > 0.1; Figure 1D; Table 2).
3.2 Genes expression in maternal tissue
In the maternal lung, the mRNA expression of BNBD4 did not show differences among the groups (p > 0.1; Figure 2A; Table 3). Despite this, there was a tendency (p = 0.06) for an interaction for CATHL5 mRNA expression, with the RES + OCM group having higher expression compared to the CON + OCM (p = 0.07) and RES − OCM (p = 0.08) groups (Figure 2B; Table 3).
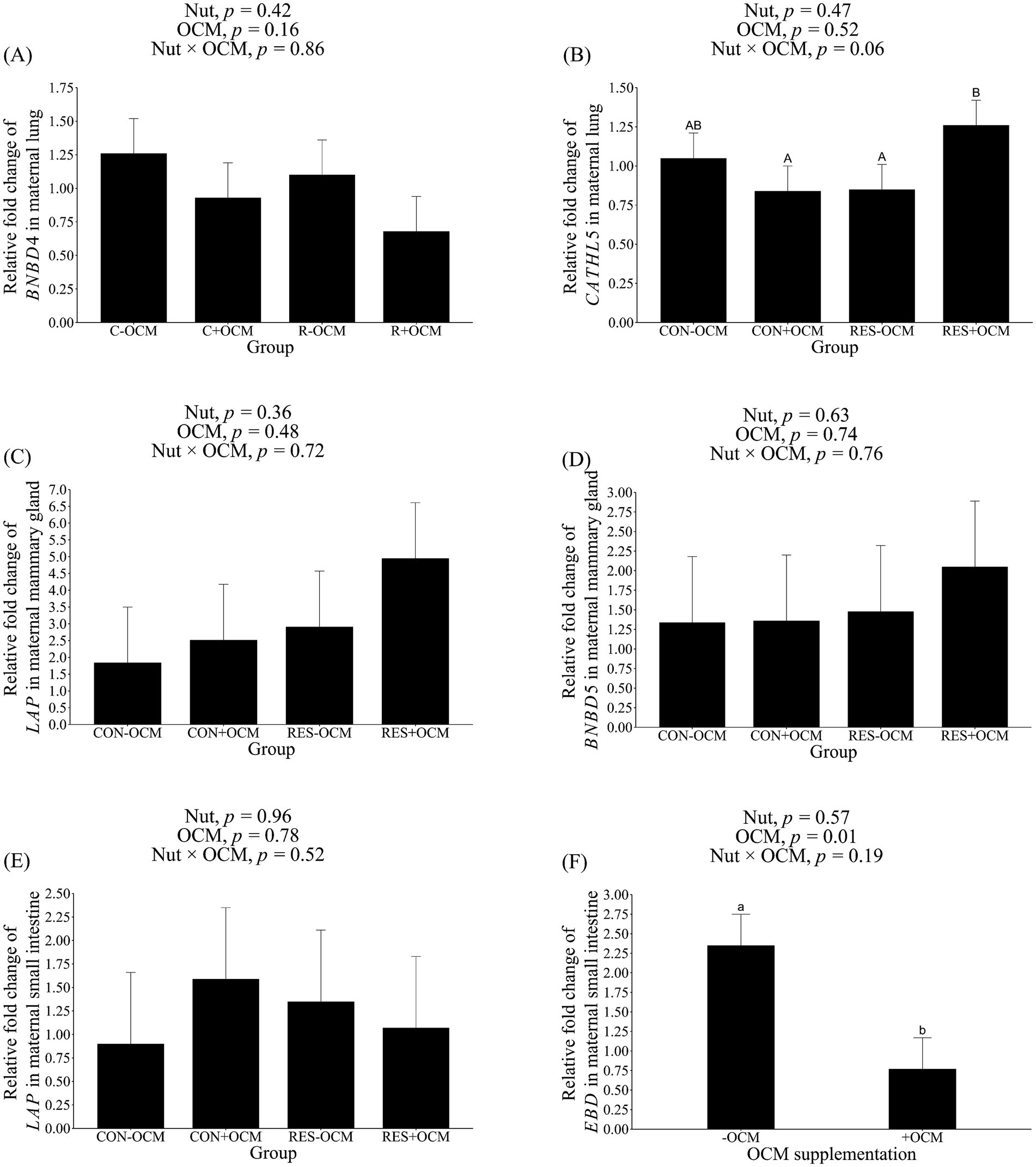
Figure 2. mRNA expression level of antimicrobial peptides in maternal tissues. (A) Bovine neutrophil β-defensin4 (BNBD4) in lung; (B) cathelicidin5 (CATHL5) in lung; (C) lingual antimicrobial peptide (LAP) in mammary gland; (D) bovine neutrophil β-defensin5 (BNBD5) in mammary gland; (E) LAP in small intestine; enteric β-defensin (EBD) in small intestine. Data presented as a 2-ΔΔCT-fold change normalized to GAPDH, ACTB, and HPRT1 as reference genes, and using the CON − OCM group as control. Different lowercase letters indicate significant differences (p ≤ 0.05), while uppercase letters denote trends (0.05 < p ≤ 0.1).
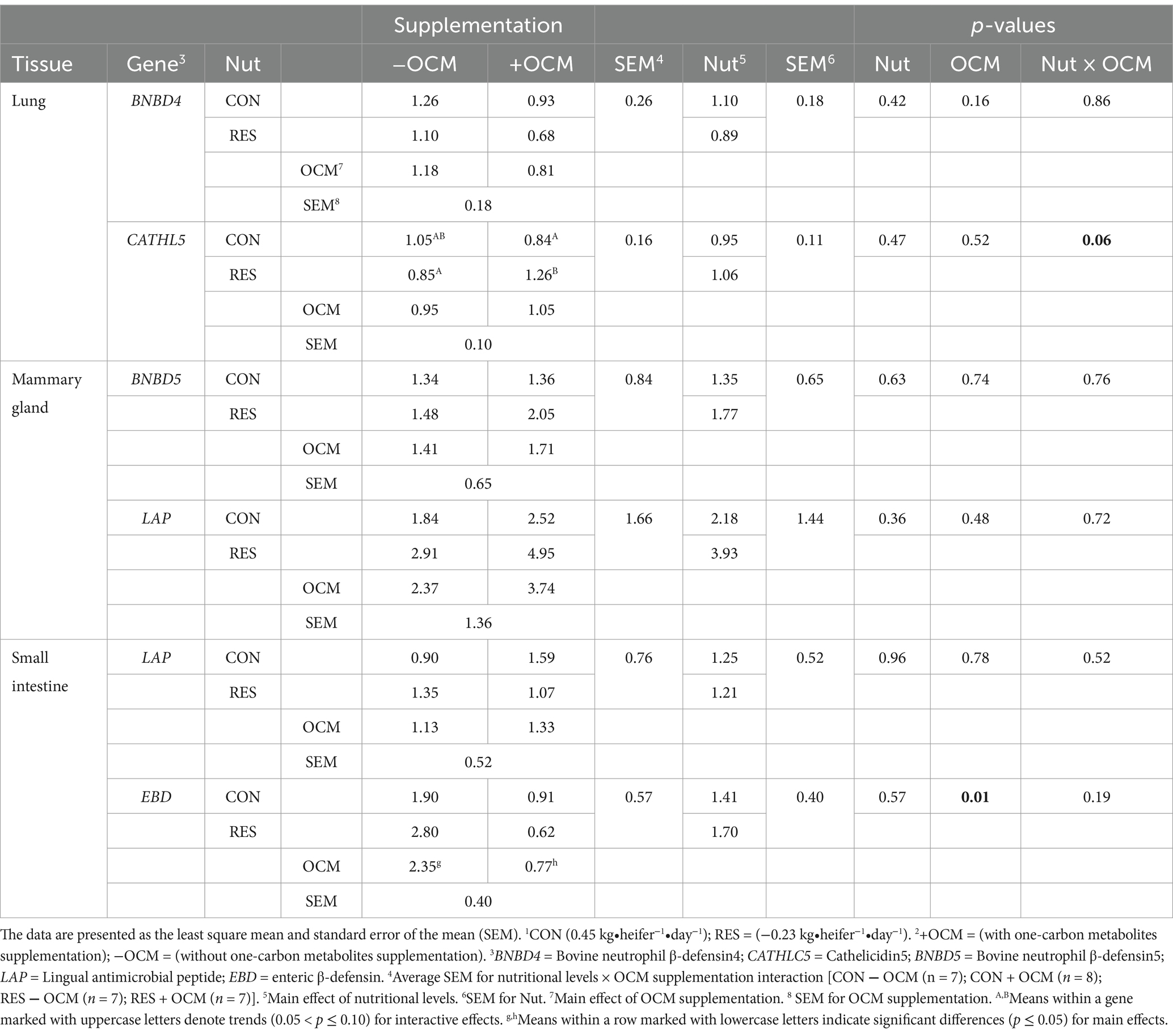
Table 3. The effect of maternal nutritional levels [Nut; control (CON) or restricted (RES)]1 and supplementation with one-carbon metabolites [OCM; with OCM (+OCM) or without (−OCM)]2 from day 0 to 63 of gestation on mRNA expression of antimicrobial peptides measured in maternal lung, mammary gland, and small intestine.
The mRNA expression of BNBD5 and LAP was assessed in the maternal mammary gland. No differences (p > 0.1) in the expression of these genes were observed (Figures 2C,D; Table 3).
In the maternal small intestine, the mRNA levels of LAP did not differ among the groups (p > 0.1; Figure 2E; Table 3). However, the expression of EBD was affected by OCM supplementation (p = 0.01), with the +OCM groups (0.77 ± 0.4 relative fold) showing a 67% lower expression compared to the −OCM groups (2.35 ± 0.4 relative fold; Figure 2F; Table 3).
4 Discussion
This study investigated the effects of maternal nutrient restriction during early gestation, with or without OCM supplementation, on the expression of AMP in the fetal and maternal lung, small intestine, and mammary gland tissues of beef cattle. Antimicrobial peptides play a crucial role in innate immunity, acting as a first line of defense against pathogens. To our knowledge, this is the first study to examine the impact of restricted maternal nutrition and OCM supplementation on AMP expression.
We aimed to assess the mRNA expression of several selected AMPs in bovine fetal and maternal tissues, based on previous research. Specifically, we chose representative members from three major AMP families—β-defensins, cathelicidins, and S100 proteins—that have been shown to be more highly expressed in these tissues compared to other family members. Most prior studies have been conducted in vitro, meaning that the influences of factors, such as the environment, have not been fully considered. Additionally, there is limited literature on AMP expression in bovine fetuses, which is critical for understanding the development of the innate immune system and how environmental factors might impact immunity across generations. To our knowledge, aside from studies on TAP in the lung (55) and EBD in the small intestine (56), no previous research has explored AMP expression in bovine fetal tissues.
To ensure the accuracy, reliability, and reproducibility of our RT-qPCR results, we validated primers and amplification efficiency within the CON − OCM group. Our findings revealed that S100A7 and TAP did not fall within the acceptable amplification efficiency range, likely due to low expression levels, as they exhibited high Ct values at the highest cDNA concentrations. Thus, we deemed the mRNA expression of these genes to be below our limit of detection. This could be attributed to several factors. Previous in vivo studies have demonstrated variation in the expression of AMPs in a single tissue analyzed from several animals of the same age group in cattle (55) and sheep (57, 58), which can account for this discrepancy.
Furthermore, previous studies in humans (59) and sheep (57, 58) models have demonstrated the developmental regulation of AMPs expression from fetus to neonate, with defensins showing increased expression during gestation. For instance, α-defensins in human fetal Paneth cells were detectable only after the 19th–24th week (60). It has also been shown that certain AMPs predominant in adulthood were not detectable in the fetus, both in human (59) and bovine models (55). Our data indicated low or absent expression of certain AMPs, such as in the fetal small intestine, aligning with findings that developmental regulation might only become apparent closer to term. It has been observed that the expression of certain β-defensins is lower in preterm humans than in term neonates and adults (60).
The birth delivery method can influence AMPs gene expression. Data indicate increased AMPs synthesis during vaginal delivery due to labor stress (60). In bovine neonates, LAP expression in the lung was lower following elective cesarean section compared to transvaginal delivery (61). Since our samples were collected from fetuses at 161 days of gestation without labor stress, the absence of certain gene expressions is logical.
Tissue-specific, or more precisely tissue-region-dependent, AMPs expression is another factor. For example, TAP is restricted to airway tissue, with the majority of expression in the trachea in cattle (55). In our study, lung samples were collected from the cranioventral region, where respiratory pathogens typically impact, and TAP was either not expressed or lowly expressed. This finding suggests that TAP is not constitutively expressed in the lung tissue of either mature cattle or their fetuses. Additionally, Tetens et al. showed a high level of S100A7 mRNA expression in the streak canal and the Rosette of Fürstenberg of the mammary gland; however, they could not detect S100A7 mRNA expression in the gland cisternal epithelium and udder parenchyma of healthy cattle (62). In agreement, our study found that S100A7 was either not expressed or lowly expressed in both fetal and maternal parenchyma of the mammary gland. Therefore, according to our findings and previous studies, it is conceivable that S100A7 is not constitutively expressed in the parenchyma of the mammary gland of both fetus and dam.
Inducible expression in response to stimuli is a characteristic of certain AMPs. In bovine studies, the inducible properties of β-defensins in the lung (63), mammary gland (64) and small intestine (56) have been properly shown, where AMPs expression was either low or nonexistent in the absence of stimuli. For example, LAP mRNA was significantly upregulated in the intestinal mucosa during chronic Mycobacterium paratuberculosis infection and in the bronchiolar epithelium during acute Pasteurella haemolytica infection, with low or absent signals in non-infected tissues (63). The absence of such stimuli in our study could explain the low or absent expression of certain AMPs.
4.1 Impact of maternal nutritional restriction on expression of AMP
Maternal undernutrition prior to parturition has been shown to interfere with the innate immune system of the ovine fetus and offspring (4, 65). However, to our knowledge, no studies have explored the effects of restricted maternal nutrition during pregnancy on the bovine fetal expression of AMP. Our data indicate that restricted maternal nutrition did not significantly affect the expression of AMP, neither in the fetus nor in the dams.
Several potential reasons may explain why the restricted nutritional plane did not independently result in significant changes in AMP expression. First, the severity and duration of the nutrient restriction imposed in this study may not have been sufficient to elicit significant changes in AMP expression. Although the extent of restricted nutrition was sufficient to assess impacts on gene expression associated with tissue metabolism, accretion, and function in our previous studies (66, 67), it may not have been severe or prolonged enough to affect AMP expression significantly.
Second, epigenetic adaptations might have occurred to maintain baseline AMP expression levels, potentially as a protective measure against nutrient restriction. It has been pointed out that the impact of prenatal events, such as undernutrition, on the immune system is not uniform. These events may lead to differential investments in various subsystems of the immune system, where some aspects might be downregulated or compromised, while others receive increased investment or remain unaffected (68, 69). In this context, it is possible that the body prioritized the maintenance of AMP expression over other immune functions in response to the prenatal nutritional stress experienced in our study. This differential investment could be an adaptive response, as it proposes that the immune system may shift its investment away from more energetically expensive specific immune defenses (like those involving specific antibodies and adaptive immune responses) and towards less costly nonspecific defenses (such as innate immune defenses) (68). This shift could help conserve resources while still providing adequate protection. It has been shown that undernutrition in ewes can upregulate specific genes, such as Cholesterol 25-Hydroxylase and MHC Class I Polypeptide-Related Sequence B, that modulate the innate immune response and inhibit the actions of receptors involved in cytotoxic activities, potentially beneficial for immunotolerance at the embryo-maternal interface (70).
Another factor is the variability in tissue response to stimuli. The AMP are constitutively expressed but can also be induced/increased in response to stimuli such as infection (17). Studies have shown that different energy levels during gestation do not affect the mRNA abundance of innate immune-related genes in weaned piglets under basal conditions; however, when challenged with lipopolysaccharide, the immune response was significantly higher in those born to dams under low-energy diets (71). Therefore, the lack of significant changes in AMP expression in the current study may be due to the absence of an immunological challenge. Hence, it is suggested that evaluating the expression of AMP in both dams and fetal tissues in response to different immunological challenges could provide further insights into the potential effects of maternal nutrition on the inducible expression of these innate immune effectors.
Additionally, maternal organisms may have evolved compensatory mechanisms to mitigate the effects of nutrient restriction on the developing fetus. These mechanisms could include alterations in nutrient partitioning, placental adaptation, or changes in maternal metabolism to ensure the provision of essential nutrients to the fetus, thereby minimizing the impact on fetal development (72, 73), including AMP expression.
Furthermore, sex-specific effects could play a role. Although the effects of sex were not explored in this study, there is strong evidence supporting sexual dimorphism in the innate immune system. Females tend to have a more effective and protective innate immune response, leading to better outcomes in the face of infections and immune challenges (9, 74). The mechanisms behind these observations may include evolutionary adaptations for higher reproductive potential (75) and hormonal differences between males and females (74).
4.2 Impact of OCM supplementation on expression of AMP
The supplementation of OCM during early gestation influenced the expression of AMP in both fetal and maternal tissues, indicating a modulatory role of these metabolites on the innate immune system.
In the fetal lung and mammary gland, the mRNA expression of CATHL5 and BNBD5, respectively, tended to decrease in the OCM-supplemented groups compared to the non-supplemented groups. Similarly, in the maternal small intestine, the mRNA levels of EBD were significantly lower in the OCM-supplemented groups. These findings demonstrate that the expression of AMP was suppressed by OCM supplementation.
One-carbon metabolism and its metabolites are essential for the proper regulation of immune function through epigenetic modifications and metabolic reprogramming, ensuring that immune cells can respond effectively to infections and stress (13, 14). For instance, a study by Sinclair et al. (9) demonstrated that a maternal diet low in B vitamins and methionine can lead to altered immune responses in ovine offspring, indicating potential negative impacts on the immune system (9). Although our results showed a decrease in the mRNA expression of AMP in response to OCM supplementation, this does not necessarily contradict previous findings. It is possible that the improved overall condition of the OCM-supplemented fetuses reduced the need for immune responses, thereby influencing the expression of these immune-related genes. Further research is warranted to evaluate the activation and function of AMP in response to OCM supplementation.
It is widely acknowledged that the expression of AMP increases during an inflammatory response (76, 77). The nuclear factor-kappa B (NF-κB) through toll-like receptor (TLR)-NF-κB pathway, and the mitogen-activated protein kinase (MAPK) signaling pathway play crucial roles in regulating the expression of AMP (17, 78). On the other hand, a deficiency in one-carbon metabolites has been shown to elevate homocysteine levels, leading to increased inflammation. In agreement, our previous research has shown that nutrient restriction in beef heifers can result in reduced concentrations of methionine in allantoic fluid and increased levels of homocysteine in maternal serum during early gestation (16). Conversely, OCM reduces inflammation in two ways. First, it increases levels of S-adenosylmethionine (SAM) while reducing S-adenosylhomocysteine (SAH). The ratio between these compounds (SAM/SAH) regulates most methyltransferases (79–82). Second, when SAM levels rise, they help decrease inflammation by reducing NF-κB production and suppressing both the NF-κB and MAPK pathways. This leads to lower levels of inflammatory markers (80, 83, 84). Therefore, it is likely that OCM supplementation attenuated AMP expression by exerting its anti-inflammatory properties through the suppression of the NF-κB and MAPK pathways. Future studies should evaluate these pathways in the context of OCM supplementation to further elucidate their role in regulating AMP expression.
5 Conclusion
In conclusion, our study provides valuable insights into the effects of maternal nutrient restriction and OCM supplementation during early gestation on the mRNA expression of AMP in fetal and maternal lung, small intestine, and mammary gland tissues of beef cattle. We evaluated some AMP genes in fetal tissues for the first time, which is crucial for understanding the development of the innate immune system in cattle. Additionally, our findings revealed that restricted maternal nutrition alone did not significantly alter AMP expression; however, OCM supplementation led to a decrease in the mRNA expression of CATHL5 and BNBD5 in fetal lung and mammary gland tissues, respectively, and a significant reduction in EBD expression in the maternal small intestine. Further research is warranted to elucidate the mechanisms by which OCM modulate AMP expression, potentially through exerting anti-inflammatory effects by suppressing the NF-κB and MAPK pathways.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee at North Dakota State University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MD: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Software, Validation, Writing – original draft, Writing – review & editing. PB: Methodology, Writing – review & editing. MH: Formal analysis, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. YE: Methodology, Writing – review & editing. JS: Methodology, Writing – review & editing. LK: Methodology, Writing – review & editing. KS: Methodology, Writing – review & editing. MA: Methodology, Writing – review & editing. LR: Methodology, Writing – review & editing. AW: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing. CD: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing. MC: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing. JC: Conceptualization, Funding acquisition, Methodology, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was funded by the USDA National Institute of Food Agriculture, USDA-NIFA-AFRI (#2018-07055) and the North Dakota State Board of Agricultural Research Education (#FARG090516).
Acknowledgments
The authors would like to express their gratitude to colleagues, including undergraduate and graduate students, as well as postdoctoral fellows, for their unwavering dedication. Mention of a trade name, proprietary product, or specific agreement does not constitute a guarantee or warranty by the USDA and does not imply approval of the inclusion of other products that may be suitable. USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1505427/full#supplementary-material
References
1. Caton, JS, Crouse, MS, McLean, KJ, Dahlen, CR, Ward, AK, Cushman, RA, et al. Maternal Periconceptual nutrition, early pregnancy, and developmental outcomes in beef cattle. J Anim Sci. (2020) 98:358. doi: 10.1093/jas/skaa358
2. Hammer, CJ, Caton, JS, Dahlen, CR, Ward, AK, Borowicz, PP, and Reynolds, LP. Dohad: a menagerie of adaptations and perspectives: large animal models of developmental programming: sustenance, stress, and sex matter. Reproduction. (2023) 165:F1–F13. doi: 10.1530/REP-22-0453
3. Funston, RN, Summers, AF, and Roberts, AJ. Alpharma beef cattle nutrition symposium: implications of nutritional management for beef cow-calf systems. J Anim Sci. (2012) 90:2301–7. doi: 10.2527/jas.2011-4568
4. Eckersall, PD, Lawson, FP, Kyle, CE, Waterston, M, Bence, L, Stear, MJ, et al. Maternal undernutrition and the ovine acute phase response to vaccination. BMC Vet Res. (2008) 4:1. doi: 10.1186/1746-6148-4-1
5. Caton, JS, Crouse, MS, Reynolds, LP, Neville, TL, Dahlen, CR, Ward, AK, et al. Maternal nutrition and programming of offspring energy requirements. Transl Anim Sci. (2019) 3:976–90. doi: 10.1093/tas/txy127
6. Reynolds, LP, Borowicz, PP, Caton, JS, Crouse, MS, Dahlen, CR, and Ward, AK. Developmental programming of fetal growth and development. Vet Clin North Am Food Anim Pract. (2019) 35:229–47. doi: 10.1016/j.cvfa.2019.02.006
7. Chmurzynska, A . Fetal programming: link between early nutrition, DNA methylation, and complex diseases. Nutr Rev. (2010) 68:87–98. doi: 10.1111/j.1753-4887.2009.00265.x
8. Wang, X, Lan, X, Radunz, A, and Khatib, H. Maternal nutrition during pregnancy is associated with differential expression of imprinted genes and DNA methyltranfereases in muscle of beef cattle offspring. J Anim Sci. (2015) 93:35–40. doi: 10.2527/jas.2014-8148
9. Sinclair, KD, Allegrucci, C, Singh, R, Gardner, DS, Sebastian, S, Bispham, J, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci USA. (2007) 104:19351–6. doi: 10.1073/pnas.0707258104
10. Zeisel, SH . The supply of choline is important for fetal progenitor cells. Semin Cell Dev Biol. (2011) 22:624–8. doi: 10.1016/j.semcdb.2011.06.002
11. Clare, CE, Brassington, AH, Kwong, WY, and Sinclair, KD. One-carbon metabolism: linking nutritional biochemistry to epigenetic programming of long-term development. Annu Rev Anim Biosci. (2019) 7:263–87. doi: 10.1146/annurev-animal-020518-115206
12. Zhao, H, Raines, LN, and Huang, SC. Carbohydrate and amino acid metabolism as hallmarks for innate immune cell activation and function. Cells. (2020) 9:562. doi: 10.3390/cells9030562
13. Vachharajani, V, and McCall, CE. Epigenetic and metabolic programming of innate immunity in Sepsis. Innate Immun. (2019) 25:267–79. doi: 10.1177/1753425919842320
14. Dang, S, Jain, A, Dhanda, G, Bhattacharya, N, Bhattacharya, A, and Senapati, S. One carbon metabolism and its implication in health and immune functions. Cell Biochem Funct. (2024) 42:e3926. doi: 10.1002/cbf.3926
15. Zhu, Y, Mordaunt, CE, Durbin-Johnson, BP, Caudill, MA, Malysheva, OV, Miller, JW, et al. Expression changes in epigenetic gene pathways associated with one-carbon nutritional metabolites in maternal blood from pregnancies resulting in autism and non-typical neurodevelopment. Autism Res. (2021) 14:11–28. doi: 10.1002/aur.2428
16. Crouse, MS, Greseth, NP, McLean, KJ, Crosswhite, MR, Pereira, NN, Ward, AK, et al. Maternal nutrition and stage of early pregnancy in beef heifers: impacts on hexose and aa concentrations in maternal and fetal fluids. J Anim Sci. (2019) 97:1296–316. doi: 10.1093/jas/skz013
17. Daneshi, M, Caton, JS, Caixeta, LS, Eftekhari, Z, and Ward, AK. Expression, regulation, and function of Beta-Defensins in the bovine mammary glands: current knowledge and future perspectives. Animals (Basel). (2023) 13:3372. doi: 10.3390/ani13213372
18. Young-Speirs, M, Drouin, D, Cavalcante, PA, Barkema, HW, and Cobo, ER. Host defense Cathelicidins in cattle: types, production, bioactive functions and potential therapeutic and diagnostic applications. Int J Antimicrob Agents. (2018) 51:813–21. doi: 10.1016/j.ijantimicag.2018.02.006
19. Singh, P, and Ali, SA. Multifunctional role of S100 protein family in the immune system: an update. Cells. (2022) 11:2274. doi: 10.3390/cells11152274
20. Chen, N, and Jiang, C. Antimicrobial peptides: structure, mechanism, and modification. Eur J Med Chem. (2023) 255:115377. doi: 10.1016/j.ejmech.2023.115377
21. Kaiser, V, and Diamond, G. Expression of mammalian Defensin genes. J Leukoc Biol. (2000) 68:779–84. doi: 10.1189/jlb.68.6.779
22. Kumar, R, Ali, SA, Singh, SK, Bhushan, V, Mathur, M, Jamwal, S, et al. Antimicrobial peptides in farm animals: an updated review on its diversity, function, modes of action and therapeutic prospects. Vet Sci. (2020) 7:206. doi: 10.3390/vetsci7040206
23. Mwangi, J, Hao, X, Lai, R, and Zhang, ZY. Antimicrobial peptides: new Hope in the war against multidrug resistance. Zool Res. (2019) 40:488–505. doi: 10.24272/j.issn.2095-8137.2019.062
24. Zhang, QY, Yan, ZB, Meng, YM, Hong, XY, Shao, G, Ma, JJ, et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil Med Res. (2021) 8:48. doi: 10.1186/s40779-021-00343-2
25. Wu, J, Wang, C, He, H, Hu, G, Yang, H, Gao, Y, et al. Molecular analysis and recombinant expression of bovine neutrophil Beta-Defensin 12 and its antimicrobial activity. Mol Biol Rep. (2011) 38:429–36. doi: 10.1007/s11033-010-0125-z
26. Diamond, G, Zasloff, M, Eck, H, Brasseur, M, Maloy, WL, and Bevins, CL. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a CDNA. Proc Natl Acad Sci USA. (1991) 88:3952–6. doi: 10.1073/pnas.88.9.3952
27. Yarus, S, Rosen, JM, Cole, AM, and Diamond, G. Production of active bovine tracheal antimicrobial peptide in milk of transgenic mice. Proc Natl Acad Sci USA. (1996) 93:14118–21. doi: 10.1073/pnas.93.24.14118
28. Bowdish, DM, Davidson, DJ, Scott, MG, and Hancock, RE. Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother. (2005) 49:1727–32. doi: 10.1128/AAC.49.5.1727-1732.2005
29. Glaser, R, Harder, J, Lange, H, Bartels, J, Christophers, E, and Schroder, JM. Antimicrobial psoriasin (S100a7) protects human skin from Escherichia Coli infection. Nat Immunol. (2005) 6:57–64. doi: 10.1038/ni1142
30. Ochoa-Zarzosa, A, Villarreal-Fernandez, E, Cano-Camacho, H, and Lopez-Meza, JE. Sodium butyrate inhibits Staphylococcus Aureus internalization in bovine mammary epithelial cells and induces the expression of antimicrobial peptide genes. Microb Pathog. (2009) 47:1–7. doi: 10.1016/j.micpath.2009.04.006
31. Kweh, MF, Merriman, KE, and Nelson, CD. Short communication: inhibition of DNA methyltransferase and histone deacetylase increases beta-defensin expression but not the effects of lipopolysaccharide or 1,25-Dihydroxyvitamin D(3) in bovine mammary epithelial cells. J Dairy Sci. (2019) 102:5706–12. doi: 10.3168/jds.2018-16141
32. Snyder, E, and Credille, B. Mannheimia Haemolytica and Pasteurella Multocida in bovine respiratory disease: how are they changing in response to efforts to control them? Vet Clin North Am Food Anim Pract. (2020) 36:253–68. doi: 10.1016/j.cvfa.2020.02.001
33. Cho, YI, and Yoon, KJ. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. (2014) 15:1–17. doi: 10.4142/jvs.2014.15.1.1
34. Krishnamoorthy, P, Goudar, AL, Suresh, KP, and Roy, P. Global and countrywide prevalence of subclinical and clinical mastitis in dairy cattle and buffaloes by systematic review and Meta-analysis. Res Vet Sci. (2021) 136:561–86. doi: 10.1016/j.rvsc.2021.04.021
35. Velazquez-Meza, ME, Galarde-Lopez, M, Carrillo-Quiroz, B, and Alpuche-Aranda, CM. Antimicrobial resistance: one health approach. Vet World. (2022) 15:743–9. doi: 10.14202/vetworld.2022.743-749
36. Mulchandani, R, Wang, Y, Gilbert, M, and Van Boeckel, TP. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLOS Glob Public Health. (2023) 3:e0001305. doi: 10.1371/journal.pgph.0001305
37. Lamb, GC, Dahlen, CR, Larson, JE, Marquezini, G, and Stevenson, JS. Control of the estrous cycle to improve fertility for fixed-time artificial insemination in beef cattle: a review. J Anim Sci. (2010) 88:E181–92. doi: 10.2527/jas.2009-2349
38. Schneider, JE, Brozek, JM, and Keen-Rhinehart, E. Our stolen figures: the Interface of sexual differentiation, endocrine disruptors, maternal programming, and energy balance. Horm Behav. (2014) 66:104–19. doi: 10.1016/j.yhbeh.2014.03.011
39. Gionbelli, TRS, Rotta, PP, Veloso, CM, Valadares Filho, SC, Carvalho, BC, Marcondes, MI, et al. Intestinal development of bovine Foetuses during gestation is affected by Foetal sex and maternal nutrition. J Anim Physiol Anim Nutr (Berl). (2017) 101:493–501. doi: 10.1111/jpn.12572
40. National Academies of Sciences E, Medicine . Committee on nutrient requirements of beef C. Nutrient requirements of beef cattle, vol. xvii. 8th rev. ed. Washington, D.C: National Academies Press (2016). 475 p.
41. Syring, JG, Crouse, MS, Entzie, YL, King, LE, Hirchert, MR, Ward, AK, et al. One-carbon metabolite supplementation increases vitamin B12, folate, and methionine cycle metabolites in beef heifers and fetuses in an energy dependent manner at day 63 of gestation. J Anim Sci. (2024) 102:102. doi: 10.1093/jas/skae202
42. Jacometo, CB, Zhou, Z, Luchini, D, Correa, MN, and Loor, JJ. Maternal supplementation with rumen-protected methionine increases Prepartal plasma methionine concentration and alters hepatic Mrna abundance of 1-carbon, methionine, and Transsulfuration pathways in neonatal Holstein calves. J Dairy Sci. (2017) 100:3209–19. doi: 10.3168/jds.2016-11656
43. Crouse, MS, Freetly, HC, Lindholm-Perry, AK, Neville, BW, Oliver, WT, Lee, RT, et al. One-carbon metabolite supplementation to heifers for the first 14 D of the estrous cycle alters the plasma and hepatic one-carbon metabolite Pool and methionine-folate cycle enzyme transcript abundance in a dose-dependent manner. J Anim Sci. (2023) 101:101. doi: 10.1093/jas/skac419
44. Gagnon, A, Khan, DR, Sirard, MA, Girard, CL, Laforest, JP, and Richard, FJ. Effects of intramuscular administration of folic acid and vitamin B12 on granulosa cells gene expression in postpartum dairy cows. J Dairy Sci. (2015) 98:7797–809. doi: 10.3168/jds.2015-9623
45. Lamb, G, Dahlen, C, and Brown, D. Reproductive ultrasonography for monitoring ovarian structure development, fetal development, embryo survival, and twins in beef cows. Prof Anim Sci. (2003) 19:135–43. doi: 10.15232/S1080-7446(15)31392-9
46. Wilkins, PA, Lascola, KM, Woolums, AR, Bedenice, D, Giguère, S, Boyle, AG, et al. Chapter 31 - diseases of the respiratory system. In: BP Smith, DC MetreVan, and N Pusterla, editors. Large Animal Internal Medicine (Sixth Edition). St. Louis (MO): Mosby (2020). p. 515–701.e42.
47. Zhu, HL, Zhao, XW, Wang, XZ, Qi, YX, Huang, DW, Cheng, GL, et al. Changes in expression of antimicrobial peptides and fc receptors in the small intestines of neonatal calves during the passive immunity period. J Dairy Sci. (2020) 103:9515–24. doi: 10.3168/jds.2019-18113
48. Constable, PD, Hinchcliff, KW, Done, SH, and Grünberg, W. Veterinary medicine: a textbook of the diseases of cattle, horses, sheep. Pigs and Goats: Elsevier Health Sciences (2016).
49. Ruiz-Villalba, A, van Pelt-Verkuil, E, Gunst, QD, Ruijter, JM, and van den Hoff, MJ. Amplification of nonspecific products in quantitative polymerase chain reactions (Qpcr). Biomol Detect Quantif. (2017) 14:7–18. doi: 10.1016/j.bdq.2017.10.001
50. Fraga, D, Meulia, T, and Fenster, S. Real-Time Pcr. Curr Prot Essent Lab Tech. (2014) 8:8. doi: 10.1002/9780470089941.et1003s08
51. Harshitha, R, and Arunraj, DR. Real-time quantitative Pcr: a tool for absolute and relative quantification. Biochem Mol Biol Educ. (2021) 49:800–12. doi: 10.1002/bmb.21552
52. Vandesompele, J, De Preter, K, Pattyn, F, Poppe, B, Van Roy, N, De Paepe, A, et al. Accurate normalization of real-time quantitative Rt-Pcr data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
53. Rocha-Martins, M, Njaine, B, and Silveira, MS. Avoiding pitfalls of internal controls: validation of reference genes for analysis by Qrt-Pcr and Western blot throughout rat retinal development. PLoS One. (2012) 7:e43028. doi: 10.1371/journal.pone.0043028
54. Livak, KJ, and Schmittgen, TD. Analysis of relative gene expression data using real-time quantitative Pcr and the 2− Δδct method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
55. Diamond, G, Jones, DE, and Bevins, CL. Airway epithelial cells are the site of expression of a mammalian antimicrobial peptide gene. Proc Natl Acad Sci USA. (1993) 90:4596–600. doi: 10.1073/pnas.90.10.4596
56. Tarver, AP, Clark, DP, Diamond, G, Russell, JP, Erdjument-Bromage, H, Tempst, P, et al. Enteric Beta-Defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium Parvum infection. Infect Immun. (1998) 66:1045–56. doi: 10.1128/IAI.66.3.1045-1056.1998
57. Huttner, KM, Brezinski-Caliguri, DJ, Mahoney, MM, and Diamond, G. Antimicrobial peptide expression is developmentally regulated in the ovine gastrointestinal tract. J Nutr. (1998) 128:297S–9S. doi: 10.1093/jn/128.2.297S
58. Meyerholz, DK, Gallup, JM, Grubor, BM, Evans, RB, Tack, BF, McCray, PB Jr, et al. Developmental expression and distribution of sheep Beta-Defensin-2. Dev Comp Immunol. (2004) 28:171–8. doi: 10.1016/S0145-305X(03)00105-8
59. Starner, TD, Agerberth, B, Gudmundsson, GH, and McCray, PB Jr. Expression and activity of Beta-Defensins and Ll-37 in the developing human lung. J Immunol. (2005) 174:1608–15. doi: 10.4049/jimmunol.174.3.1608
60. Agakidou, E, Agakidis, C, Kontou, A, Chotas, W, and Sarafidis, K. Antimicrobial peptides in early-life host defense, perinatal infections, and necrotizing enterocolitis-an update. J Clin Med. (2022) 11:5074. doi: 10.3390/jcm11175074
61. Mandic Havelka, A, Yektaei-Karin, E, Hultenby, K, Sorensen, OE, Lundahl, J, Berggren, V, et al. Maternal plasma level of antimicrobial peptide Ll37 is a major determinant factor of neonatal plasma Ll37 level. Acta Paediatr. (2010) 99:836–41. doi: 10.1111/j.1651-2227.2010.01726.x
62. Tetens, J, Friedrich, JJ, Hartmann, A, Schwerin, M, Kalm, E, and Thaller, G. The spatial expression pattern of antimicrobial peptides across the healthy bovine udder. J Dairy Sci. (2010) 93:775–83. doi: 10.3168/jds.2009-2729
63. Stolzenberg, ED, Anderson, GM, Ackermann, MR, Whitlock, RH, and Zasloff, M. Epithelial antibiotic induced in states of disease. Proc Natl Acad Sci USA. (1997) 94:8686–90. doi: 10.1073/pnas.94.16.8686
64. Petzl, W, Zerbe, H, Gunther, J, Yang, W, Seyfert, HM, Nurnberg, G, et al. Escherichia Coli, but not Staphylococcus Aureus triggers an early increased expression of factors contributing to the innate immune defense in the udder of the cow. Vet Res. (2008) 39:18. doi: 10.1051/vetres:2007057
65. Jones, AK, Hoffman, ML, Pillai, SM, McFadden, KK, Govoni, KE, Zinn, SA, et al. Gestational restricted- and over-feeding promote maternal and offspring inflammatory responses that are distinct and dependent on diet in sheep. Biol Reprod. (2018) 98:184–96. doi: 10.1093/biolre/iox174
66. Daneshi, M, Borowicz, PP, Entzie, YL, Syring, JG, King, LE, Safain, KS, et al. Influence of maternal nutrition and one-carbon metabolites supplementation during early pregnancy on bovine fetal small intestine vascularity and cell proliferation. Vet Sci. (2024) 11:146. doi: 10.3390/vetsci11040146
67. Crouse, MS, Caton, JS, Cushman, RA, McLean, KJ, Dahlen, CR, Borowicz, PP, et al. Moderate nutrient restriction of beef heifers alters expression of genes associated with tissue metabolism, accretion, and function in fetal liver, muscle, and cerebrum by day 50 of gestation. Transl Anim Sci. (2019) 3:855–66. doi: 10.1093/tas/txz026
68. McDade, TW . Life history, maintenance, and the early origins of immune function. Am J Hum Biol. (2005) 17:81–94. doi: 10.1002/ajhb.20095
69. Dantzer, R, Cohen, S, Russo, SJ, and Dinan, TG. Resilience and immunity. Brain Behav Immun. (2018) 74:28–42. doi: 10.1016/j.bbi.2018.08.010
70. de Brun, V, Loor, JJ, Naya, H, Grana-Baumgartner, A, Vailati-Riboni, M, Bulgari, O, et al. The presence of an embryo affects day 14 uterine transcriptome depending on the nutritional status in sheep. B. Immune system and uterine remodeling. Theriogenology. (2021) 161:210–8. doi: 10.1016/j.theriogenology.2020.12.008
71. Chen, Y, Mou, D, Hu, L, Zhen, J, Che, L, Fang, Z, et al. Effects of maternal low-energy diet during gestation on intestinal morphology, Disaccharidase activity, and immune response to lipopolysaccharide challenge in pig offspring. Nutrients. (2017) 9:1115. doi: 10.3390/nu9101115
72. Van Gronigen, CG, Storey, KM, Parmeley, LE, and Schulz, LC. Effects of maternal nutrient restriction during the Periconceptional period on placental development in the mouse. PLoS One. (2021) 16:e0244971. doi: 10.1371/journal.pone.0244971
73. Reynolds, LP, and Vonnahme, KA. Livestock as models for developmental programming. Anim Front. (2017) 7:12–7. doi: 10.2527/af.2017-0123
74. Marriott, I, and Huet-Hudson, YM. Sexual dimorphism in innate immune responses to infectious organisms. Immunol Res. (2006) 34:177–92. doi: 10.1385/IR:34:3:177
75. Trivers, RL, and Willard, DE. Natural selection of parental ability to vary the sex ratio of offspring. Science. (1973) 179:90–2. doi: 10.1126/science.179.4068.90
76. Son, GH, Lee, JJ, Kim, Y, and Lee, KY. The role of antimicrobial peptides in preterm birth. Int J Mol Sci. (2021) 22:8905. doi: 10.3390/ijms22168905
77. Kim, Y, Lee, KY, Lee, JJ, Tak, H, Park, ST, Song, JE, et al. Expression of antimicrobial peptides in the amniotic fluid of women with cervical insufficiency. Am J Reprod Immunol. (2022) 88:e13577. doi: 10.1111/aji.13577
78. Diamond, G, Kaiser, V, Rhodes, J, Russell, JP, and Bevins, CL. Transcriptional regulation of Beta-Defensin gene expression in tracheal epithelial cells. Infect Immun. (2000) 68:113–9. doi: 10.1128/IAI.68.1.113-119.2000
79. Dominguez-Lopez, I, Kovatcheva, M, Casas, R, Toledo, E, Fito, M, Ros, E, et al. Higher circulating vitamin B12 is associated with lower levels of inflammatory markers in individuals at high cardiovascular risk and in naturally aged mice. J Sci Food Agric. (2024) 104:875–82. doi: 10.1002/jsfa.12976
80. Detopoulou, P, Panagiotakos, DB, Antonopoulou, S, Pitsavos, C, and Stefanadis, C. Dietary choline and betaine intakes in relation to concentrations of inflammatory markers in healthy adults: the Attica study. Am J Clin Nutr. (2008) 87:424–30. doi: 10.1093/ajcn/87.2.424
81. Takahashi, Y, Soejima, Y, and Fukusato, T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. (2012) 18:2300–8. doi: 10.3748/wjg.v18.i19.2300
82. Mehta, AK, Singh, BP, Arora, N, and Gaur, SN. Choline attenuates immune inflammation and suppresses oxidative stress in patients with asthma. Immunobiology. (2010) 215:527–34. doi: 10.1016/j.imbio.2009.09.004
83. Lan, W, Wang, Z, Liu, J, and Liu, H. Methionyl-methionine exerts anti-inflammatory effects through the Jak2-Stat5-Nf-Kappab and Mapk signaling pathways in bovine mammary epithelial cells. J Agric Food Chem. (2020) 68:13742–50. doi: 10.1021/acs.jafc.0c05962
84. Deniz, E, Topcu, A, Ozturk, A, Ozturk, SD, Arpa, M, and Atak, M. The effects of vitamin B12 on the Tlr-4/Nf-Kappab signaling pathway in ovarian ischemia-reperfusion injury-related inflammation. Int Immunopharmacol. (2022) 107:108676. doi: 10.1016/j.intimp.2022.108676
85. Cormican, P, Meade, KG, Cahalane, S, Narciandi, F, Chapwanya, A, Lloyd, AT, et al. Evolution, expression and effectiveness in a cluster of novel bovine Beta-Defensins. Immunogenetics. (2008) 60:147–56. doi: 10.1007/s00251-007-0269-8
86. Tellez-Perez, AD, Alva-Murillo, N, Ochoa-Zarzosa, A, and Lopez-Meza, JE. Cholecalciferol (vitamin D) differentially regulates antimicrobial peptide expression in bovine mammary epithelial cells: implications during Staphylococcus Aureus internalization. Vet Microbiol. (2012) 160:91–8. doi: 10.1016/j.vetmic.2012.05.007
87. Dai, H, Wei, G, Wang, Y, Ma, N, Chang, G, and Shen, X. Sodium butyrate promotes lipopolysaccharide-induced innate immune responses by enhancing mitogen-activated protein kinase activation and histone acetylation in bovine mammary epithelial cells. J Dairy Sci. (2020) 103:11636–52. doi: 10.3168/jds.2020-18198
88. Merriman, KE, Kweh, MF, Powell, JL, Lippolis, JD, and Nelson, CD. Multiple beta-defensin genes are upregulated by the vitamin D pathway in cattle. J Steroid Biochem Mol Biol. (2015) 154:120–9. doi: 10.1016/j.jsbmb.2015.08.002
89. Kosciuczuk, EM, Lisowski, P, Jarczak, J, Krzyzewski, J, Zwierzchowski, L, and Bagnicka, E. Expression patterns of beta-defensin and cathelicidin genes in parenchyma of bovine mammary gland infected with coagulase-positive or coagulase-negative staphylococci. BMC Vet Res. (2014) 10:246. doi: 10.1186/s12917-014-0246-z
90. Tomasinsig, L, De Conti, G, Skerlavaj, B, Piccinini, R, Mazzilli, M, D'Este, F, et al. Broad-spectrum activity against bacterial mastitis pathogens and activation of mammary epithelial cells support a protective role of neutrophil Cathelicidins in bovine mastitis. Infect Immun. (2010) 78:1781–8. doi: 10.1128/IAI.01090-09
Keywords: β-defensins, cathelicidins, developmental programming, early pregnancy, epigenetic modifications, innate immunity, nutrition restriction, one-carbon metabolism
Citation: Daneshi M, Borowicz PP, Hirchert MR, Entzie YL, Syring JG, King LE, Safain KS, Anas M, Reynolds LP, Ward AK, Dahlen CR, Crouse MS and Caton JS (2024) Influence of maternal nutrition and one-carbon metabolites supplementation on bovine antimicrobial peptides in fetal and maternal tissues. Front. Vet. Sci. 11:1505427. doi: 10.3389/fvets.2024.1505427
Edited by:
Luciana Rossi, University of Milan, ItalyReviewed by:
Jon Schoonmaker, Purdue University, United StatesAlejandro Bielli, University of the Republic, Uruguay
Mohammed Ahmed Elmetwally, Mansoura University, Egypt
Copyright © 2024 Daneshi, Borowicz, Hirchert, Entzie, Syring, King, Safain, Anas, Reynolds, Ward, Dahlen, Crouse and Caton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mojtaba Daneshi, bW9qdGFiYS5kYW5lc2hpQG5kc3UuZWR1
 Mojtaba Daneshi
Mojtaba Daneshi Pawel P. Borowicz
Pawel P. Borowicz Mara R. Hirchert1
Mara R. Hirchert1 Kazi Sarjana Safain
Kazi Sarjana Safain Muhammad Anas
Muhammad Anas Lawrence P. Reynolds
Lawrence P. Reynolds Alison K. Ward
Alison K. Ward Carl R. Dahlen
Carl R. Dahlen Matthew S. Crouse
Matthew S. Crouse Joel S. Caton
Joel S. Caton