
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 13 November 2024
Sec. Animal Nutrition and Metabolism
Volume 11 - 2024 | https://doi.org/10.3389/fvets.2024.1498639
This article is part of the Research Topic Crosslinking of feed nutrients, microbiome and production in ruminants View all 16 articles
This study aimed to investigate the effects of fermented soybean meal (FSM) on milk production, blood parameters, and rumen fermentation and microbial community in dairy cows. In this study, 48 healthy Holstein cows (parity, 3.0 ± 0.6; days in milk, 86.0 ± 6.7) were used. Cows were randomly assigned into four groups (CON, T-200, T-400, and T-600) with 12 cows per group. Cows in CON were not supplemented with FSM. Cows in T-200, T-400, and T-600 were supplemented with 200, 400, and 600 g/head/day FSM, respectively. This study lasted 5 weeks (1-week adaptation and 4-week treatment). The results showed that FSM did not affect milk yield and milk components (p > 0.05). In the serum, FSM greatly increased prolactin (p < 0.01), and a dosage effect was observed. Aspartate aminotransferase and total protein were the highest in the T-400 (p < 0.05), and triglycerides was the lowest in T-200 (p < 0.05), and there was no difference for the 3 measurements between the other 3 groups (p > 0.05). In the rumen, FSM did not affect pH, microbial crude protein, acetate, propionate, butyrate, valerate, total volatile fatty acids and the ratio of acetate:propionate (p > 0.05), only changed NH3-N, isobutyrate and isovalerate (p < 0.05). The results of the rumen bacterial 16S rRNA genes sequencing showed that FSM decreased the richness (p < 0.05) and evenness (p < 0.05) of the bacterial communities. PCoA analysis showed that FSH altered the rumen bacterial community (ANOSIM, R = 0.108, p = 0.002). In the relative abundance of phyla, FSM increased Firmicutes (p = 0.015) and Actinobacteriota (p < 0.01) and Patescibacteria (p = 0.012), decreased Bacteroidota (p = 0.024). In the relative abundance of genera, FSM increased Christensenellaceae R-7 group (p = 0.011), Lactococcus (p < 0.01), Candidatus Saccharimonas (p < 0.01), Olsenella (p < 0.01), decreased Muribaculaceae_norank (p < 0.01). Conclusively, supplemented FSM altered the rumen fermentation parameters and bacterial community, and increased serum prolactin level in lactating Holstein cows. These findings may provide an approach to keep the peak of lactation in dairy cows.
Fermented soybean meal (FSM) is a high-quality plant protein source for animals, containing probiotics, digestive enzymes, bioactive peptides, antioxidants and providing immunomodulatory effects (1). Many studies reported that feeding FSM to animals (pigs, chicken and calves) showed positive effects with improved nutrient digestibility and intestinal health and production performance (2–4). Due to the ban on the use of antimicrobial growth promoters in animal production, the use of FSM in ruminants has attracted a great interest.
In the study of Kim et al. (5), FSM had been used in a calf starter and showed positive effects on the health and growth of calves. As demonstrated by Feizi et al. (4), FSM improved the starter intake in calves, and altered the rumen fermentation and microbiota. In another study of Rezazadeh et al. (6), feeding FSM helped calves adapt to weaning stress during cold weather. One study in lactating cows reported that feeding FSM increased milk protein yield, milk fat yield and fat corrected milk, and decreased milk somatic cell count (7). Studies also showed that feeding FSM alter the rumen fermentation parameters and rumen microbiota in lactating Holstein cows (7, 8). However, the results were not consistent. As reported by Wang et al. (8), FSM reduced total volatile fatty acid concentration, acetate to propionate ratio and increased propionate percentage. According to Amin et al. (7), FSM increased rumen pH, acetate percentage and acetate to propionate ratio. Most studies regarding the use of FSM in ruminants have been focused on calves, few studies investigated the lactating cows, especially for cows in early stage of lactation (1).
We hypothesized that feeding FSM could cause changes in the rumen fermentation and microbiota and blood parameters which could lead to improve in the milk performance of dairy cows. In this study, we aimed to explore the effects of feeding FSM on the milk performance, blood parameters, and rumen fermentation and bacterial community in dairy cows in the early stage of lactation. The results would offer a reference for the application of FSM in the dairy cow industry.
This experiment was conducted from November 2020 to December 2020 at Shanghai Jinshan Yinan Dairy Farm (Shanghai, China). In this study, 48 healthy Holstein cows in similar parity (3.0 ± 0.6) and lactation stages (86.0 ± 6.7 day in milk) and milk yield (41.0 ± 2.8 kg) were used. Cows were randomly assigned into 4 groups (CON, T-200, T-400, and T-600) with 12 cows per group. Cows in CON were not supplemented with FSM. Cows in T-200, T-400, and T-600 were supplemented with 200, 400, and 600 g/head/day FSM, respectively. This study lasted 5 weeks (1-week adaptation and 4-week treatment). FSM (yellow granular substance, fermented by Saccharomyces cerevisiae and Bacillus subtilis) used in this study was purchased from Shanghai Yuanyao Agriculture Co., Ltd. The basic diet used in this study was formulated based on NRC (2001) guidelines for lactating cows. The nutritional composition of FSM was shown in Supplementary Table S1. The ingredients and chemical composition of the basic diet were shown in Supplementary Table S2. All cows were housed in a tie stall barn, milked three times daily using a fully automated pipeline milking machine (02,30, 10,30, 16:30), and fed with total mixed ration three times daily (03,30, 10,30, 16:30), ensuring that cows had at least 20 h of free access to feed per day and free access to fresh water.
Milk yield was determined by a Tunisian flow-meter (JHF-G17, Sichuan Jinhaifeng Animal Husbandry Technology Co., Ltd., Sichuan, China). Milk samples were collected at the last 2 days in each week, and preserved with potassium dichromate, at 4°C. Milk samples collected from the morning, afternoon, and evening milking daily were mixed at a ratio of 4:3:3 before determining the milk composition using a near-infrared analyzer (MilkoScanTM 7 RM, Foss Electric, Denmark).
The blood samples were collected via the tail vein of the cows before morning feeding on the last day of the trial. The collected blood samples were immersed in warm water (37°C) for 10 min immediately before centrifuging at 3,500 r/min for 15 min. The supernatant was collected and stored at −20°C for the determination of serum biochemical indices.
The rumen content samples were collected at 4 h after morning feeding using an oral ruminal tube (Wuhan Kelibao Co., Ltd., Wuhan, China) on the last day of the trial. In order to avoid saliva contamination, the first 200 mL rumen fluid was discarded. A portion of the rumen content was stored in liquid nitrogen for the measuring the microbial community. Another portion was filtered through four layers of sterilized cheesecloth, and stored at −20°C for the determination of microbial crude protein, NH3-N, and volatile fatty acids.
The serum biochemical indices were measured using a fully automated biochemical analyzer (Vital Scientific NV, The Netherlands) following the standard procedure. Prolactin (PRL) is a milk-production hormone, was measured using an enzyme-linked immunosorbent assay kit (ELISA kit, Shanghai, China).
The pH value of rumen fluid was measured using a portable pH meter (HI 9024C; HANNA Instruments, Woonsock, RI). The concentration of NH3-N in rumen fluid was determined using a phenol sodium hypochlorite colorimetric method according to Weatherburn (9). The microbial crude protein (MCP) content in rumen fluid was determined using a Coomassie Brilliant Blue colorimetric method according to Makkar et al. (10). The volatile fatty acids (VFA) concentration in rumen fluid was determined by a gas chromatography (GC-2014B, Shimadzu, Japan) equipped with a capillary column (column temperature: 110°C, film thickness: 30 m × 0.32 mm × 0.25 μm) (11).
Rumen microbial genomic DNA was extracted using a phenol-chloroform extraction and cell lysis methods (12). The concentration of DNA was measured by a Nanodrop spectrophotometer (Nyxor Biotech; Paris, France) and stored at −80°C for further sequencing.
A pair of PCR primers was used to amplify the V3-V4 region of the rumen bacterial 16S rRNA genes (13). The primers were 341F (5-CCTAYGGGGRBGCASCAG-3) and 806R (5-GGACTACNNGGGTATCTAAT-3). The amplicons were sequenced on an Illumina MiSeq PE 300 platform (Illumina Inc., San Diego, California, United States) in a commercial laboratory (Shanghai Biozeron Technology Co., Ltd., Shanghai, China). The raw data were stored in the Sequence Read Archive (SRA) database, the accession number is PRJNA1162692.
Trimmomatic (v.0.33) software was used to trim adapters and low-quality sequences. FLASH (1.2.7) software was utilized to concatenate paired segments into a sequence (14). A software (QIIME2 v1.9.0) was used to process the raw Illumina fastq files (15). Bases with an average quality value below 20 were filtered. UPARSE software was used to classify sequences with a similarity level ≥ 97% into OTUs (16). The SILVA database was used to perform the taxonomic assignment of the representative OTU sequences (17). Principal coordinate analysis (PCoA) was conducted based on the Bray–Curtis metrics (18). The differences among groups was evaluated by ANOSIM using the vegan package in R.
A SPSS 20.0 software (SPSS Inc., Chicago, IL, United States) was used to analyze the data in this study.
The data (milk yield, and components) were analyzed with repeated measurements using a MIXED procedure, and adjusted with the data of adaption period as a covariate factor. The model included the fixed effects of treatment (CON, T-200, T-400, and T-600), time (week 1 to 4), treatment × time, and covariate. Time (week) was used as a repeated measurement with cows as the subject.
Data (rumen fermentation parameters, serum biochemical indices) were analyzed using the one-way ANOVA test. Significant difference between treatments was evaluated using Duncan’s test. Data on bacterial communities were analyzed using the nonparametric test (Kruskal-Wallis). Significance was declared at p < 0.05. All results were expressed as mean ± standard error.
As shown in Table 1, there were no effects of treatment (p > 0.05), time (p > 0.05), and treatment by time (p > 0.05) for milk yield, milk fat percentage, total milk solids, and somatic cell count. There were effects of time (p < 0.05), but not treatment (p > 0.05) and treatment by time (p > 0.05) for milk protein percentage, milk lactose percentage or milk urea nitrogen concentration.
As shown in Table 2, there were treatment effects for PRL (prolactin), AST (Aspartate aminotransferase), TP (Total Protein), TRIG (Triglycerides) in the serum (p < 0.05). PRL showed a dosage effect, and increased with the dosage increase of FSM (394.67, 493.81, 536.16, and 608.13 mIU/L, p < 0.01). AST was higher in T-400 (p < 0.05), and did not differ between the other 3 groups (p > 0.05). TP was higher in T-400 than that in CON and T-200 (p < 0.05), and T-400 did not differ with T-600 (p > 0.05). TRIG was lower in T-200 than that in CON and T-600 (p < 0.05), and T-200 did not differ with T-400 (p > 0.05). There were no treatment effects for T-SOD (superoxide dismutase), ALT (Alanine aminotransferase), ALB (Albumin), ALP (Alkaline Phosphatase), GLOB (Globulin), A/G (Albumin/ Globulin), ALP (Alkaline Phosphatase), CK (Creatine Kinase), LDH (Lactic Acid Dehydrogenase), HDL-C (High-density lipoprotein), LDL-C (Low-density lipoprotein), CREAT (Creatinine), TCHO (Total cholesterol), GLU (Glucose), and UA (uric acid) in the serum (p > 0.05).
As shown in Table 3, there were no treatment effects for rumen pH, microbial crude protein, acetate, propionate, butyrate, valerate, total volatile fatty acids and the ratio of acetate to propionate (p > 0.05). There were treatment effects for the concentration of NH3-N, isobutyrate, and isovalerate (p < 0.05). The concentration of NH3-N was higher in T-400 than the other 3 groups (p < 0.05). Isobutyrate was lower in T-200 and T-400 than that in CON (p < 0.05), but not differ with that in T-600 (p > 0.05). Isovalerate was lower in T-400 than that in CON and T-600 (p < 0.05), but not differ with that in T-200 (p > 0.05).
There were total of 2,128,625 high-quality sequences were obtained from 48 samples, with an average of 44,346 sequences for each sample. The rarefaction curve tended to flatten out, indicating that the sequencing depth were sufficient for analyzing the rumen bacterial communities (Supplementary Figure S1). PCoA based on the Bray Curtis metric algorithm showed that FSM altered the rumen bacterial community structure (ANOSIM: R = 0.108, p = 0.002) (Figure 1). Significant differences were observed between CON and T-400 (R = 0.074, p = 0.032); CON and T-600 (R = 0.109, p = 0.004); T-200 and T-400 (R = 0.076, p = 0.018); T-200 and T-600 (R = 0.103, p = 0.003). There were no differences between CON and T-200 (R = 0.042, p = 0.441); T-400 and T-600 (R = 0.039, p = 0.539).
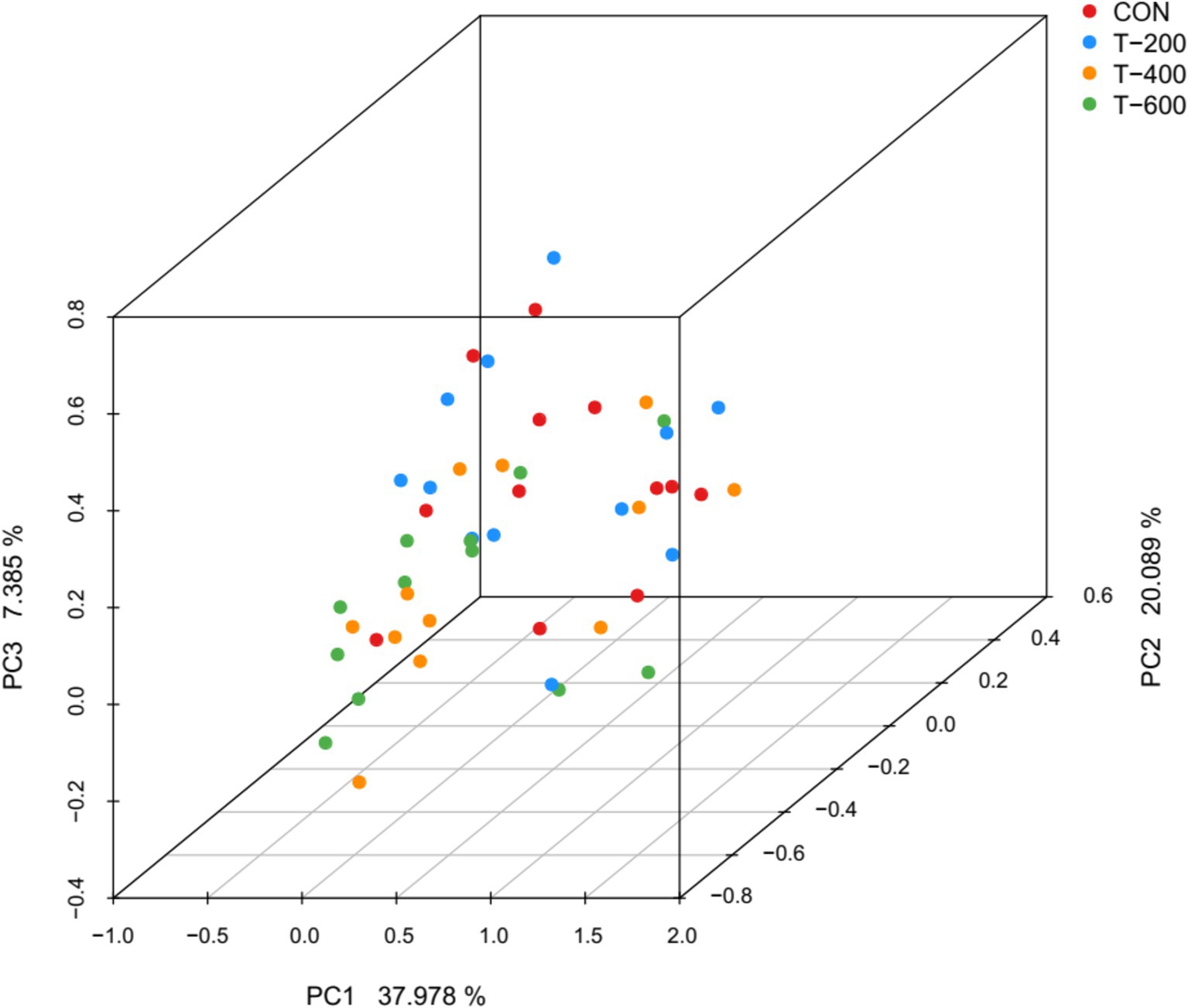
Figure 1. Principal coordinate analysis (PCoA) plot of ruminal bacterial communities based on the Bray Curtis metrics.
As shown in Table 4, there were treatment effects for the number of OTUs (p < 0.01), Chao 1 index (p = 0.043), and Shannon index (p = 0.028). The number of OTUs and Chao 1 and Shannon were lower in T-400 and T-600 than that in CON (p < 0.05). There were no treatment effects for Simpson (p = 0.051).
As shown the relative abundance of phyla in Table 5, there were treatment effects for Firmicutes (p = 0.015), Bacteroidota (p = 0.024), Actinobacterota (p < 0.01), and Patescibacteria (p = 0.012). Firmicutes was higher in T-400 and T-600 than that in CON and T-200 (p < 0.05). Bacteroidota was lower in T-400 and T-600 than that in CON and T-200 (p < 0.05).
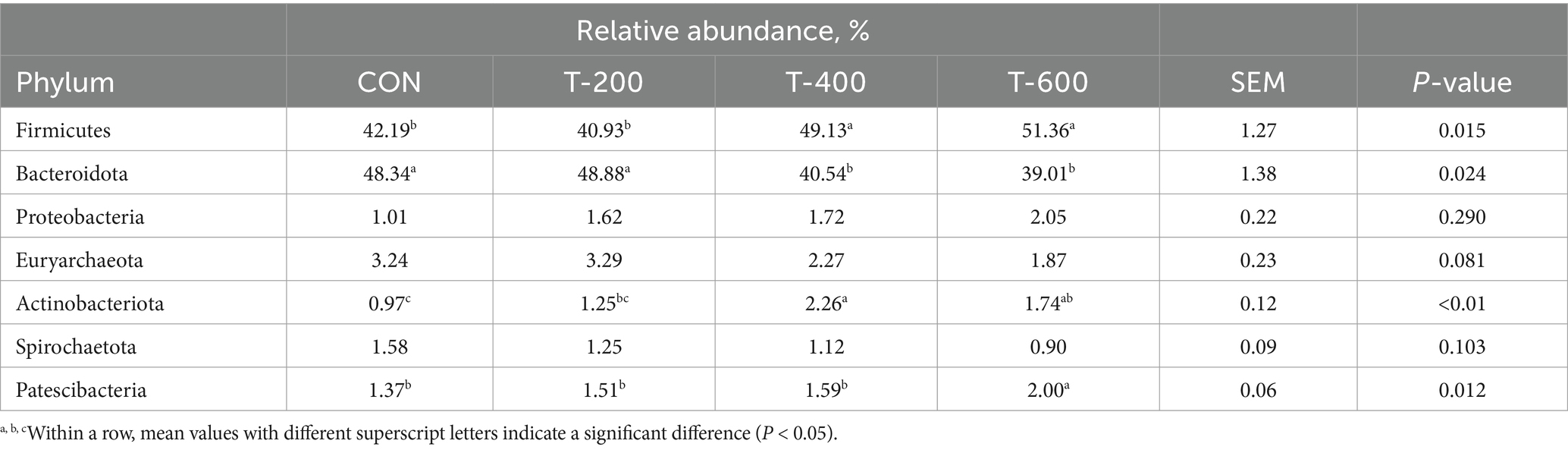
Table 5. Effects of feeding FSM on the relative abundance of phyla of rumen bacterial community in lactating cows.
Actinobacterota was higher in T-400 and T-600 than that in CON (p < 0.05). Patescibacteria was higher in T-600 than that in the other 3 groups (p < 0.05).
As shown the relative abundance of genera in Table 6, there were treatment effects for Muribaculaceae_norank (p < 0.01), Christensenellaceae R-7 group (p = 0.011), Lactococcus (p < 0.01), Candidatus Saccharimonas (p < 0.01), and Olsenella (p < 0.01). Muribaculaceae_norank was lower in T-400 and T-600 than that in CON (p < 0.05). Christensenellaceae R-7 group was higher in T-600 than that in CON and T-200 (p < 0.05). Lactococcus and Candidatus Saccharimonas were higher in T-600 than that in the other 3 groups (p < 0.05). Olsenella was higher in T-400 and T-600 than that in CON (p < 0.05).
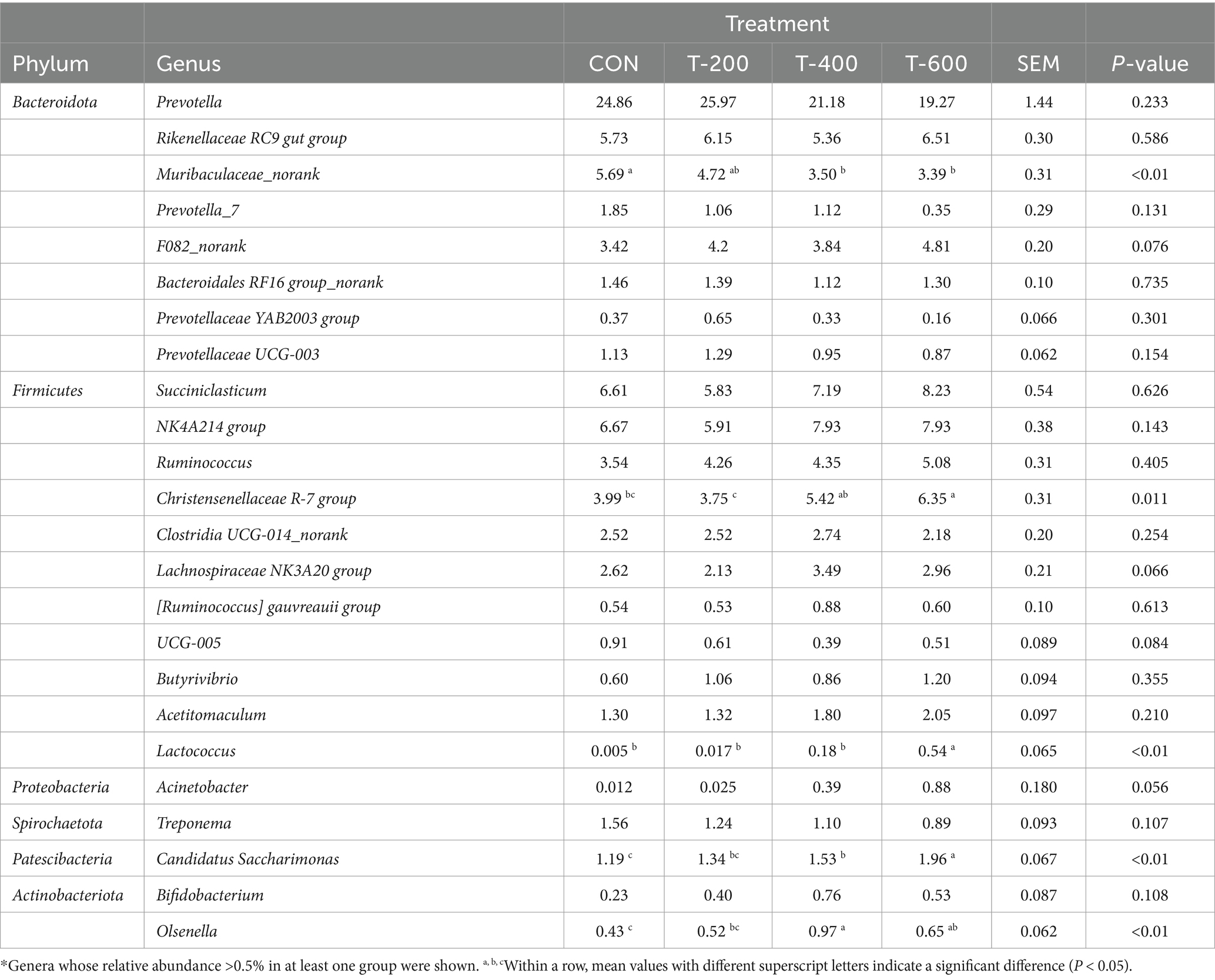
Table 6. Effects of feeding FSM on the relative abundance of genera of rumen bacterial community in lactating cows*.
FSM is a high-quality protein, containing probiotics, digestive enzymes, bioactive peptides, antioxidants, and low-antinutritional-factors. Feeding FSM would provide positive effects on dairy cows, especially for the cows in the early stage of lactation, during which cows suffer multiple stress. In this study, feeding FSM did not affect the milk yield and milk composition, just observed a numerical increase in milk yield, milk protein percentage and milk urea nitrogen. It is not consistent with a previous study. Amin et al. (7) reported that feeding FSM increased the milk protein yield, milk fat yield and fat corrected milk, and decreased milk somatic cell count in cows in early lactation stage (54 days in milk). The different results observed may be attributed to the varying dosages of FSM supplementation, the inoculum used, the composition of the basic diets, and the lactation stages of cows involved in the different studies (1). Studies regarding FSM on lactating cows are very few, thus more works are needed to elucidate the action mode of FSM in lactating cows.
Serum biochemical indices are indirect indicators of the health and metabolic status of livestock. Feeding FSM caused a little change in a few blood measurements, but caused a great increase in the serum prolactin concentration. These changes were not observed or not measured in the previous studies of Amin et al. (7) and Wang et al. (8). Prolactin is an important lactation hormone that plays a crucial role in promoting mammary gland development, milk synthesis, milk yield, and maintaining lactation (19). During milk synthesis, prolactin facilitates the absorption of glucose and amino acids, as well as the synthesis of milk lactose, fat, casein, and lactoglobulin (20). The mechanism underlying the increase of serum prolactin by feeding FSM is still unclear. In this study, FSM treatment only last 4 weeks, the effect of maintenance of lactation did not exhibited. Further studies would last 8 weeks or longer to explore the effect of FSM on the lactation maintenance in lactating cows. Nevertheless, the new finding might provide a new strategy for the utilization of FSM on lactating cows, especially in the early lactation stage.
Feeding FSM did not affect the concentration of acetate, propionate, butyrate, total volatile fatty acids in the rumen, only caused a little change in concentration of NH3-N, isobutyrate and isovalerate. Two previous studies reported that FSM changed the rumen fermentation parameters in lactating Holstein cows (7, 8). Wang et al. (8) reported that FSM reduced total volatile fatty acid concentration, acetate to propionate ratio and increased propionate percentage. Amin et al. (7) reported that FSM increased rumen pH, acetate percentage and acetate to propionate ratio. The results from these studies were not consistent. The underlying reasons remain to elucidate.
Feeding FSM caused changes in the rumen bacterial community. The previous studies also reported that feeding FSM modified the rumen bacterial communities in lactating Holstein cows (7, 8). However, the changes in these studies were not consistent. In the current study, feeding FSM decreased the number of OTUs, Chao 1 and Shannon indexes, and increased the relative abundance of Firmicutes and decreased the relative abundance of Bacteroidota in the phylum levels, which were not observed in the two previous studies. Amin et al. (7) reported that FSM enriched the genus of Muribaculaceae_norank, which were reduced in the current study. Both the current study and the study of Amin et al. (7) observed the enrichment of the genus of Christensenellaceae R-7 group by feeding FSM. The enrichment of the genera of Lactococcus, Candidatus Saccharimonas and Olsenella was not observed in the two previous studies. Fernando et al. (21) reported that Firmicutes were more adapted to fiber fermentation, while Bacteroidota was more effective in degrading starch. Wang et al. (22) reported that Actinobacteriota had the ability to degrade polysaccharides. It suggested that FSM enhanced the fiber fermentation ability in the rumen. Lagkouvardos et al. (23) reported that the genomes of Muribaculaceae contained a substantial and versatile set of carbohydrate-active enzymes, suggesting that the members in this family had the ability to degrade complex carbohydrates, the authors also stated that the fitness of Muribaculaceae species in degrading dietary carbohydrates most likely explains the decreased occurrence in the feeding trials using high-calories or carbohydrate-enriched diets. Lactococcus are homofermentative and are used for the production of L(+) lactic acid from glucose. In dairy industry, Lactococcus species are used majorly in the production of lactic acid from lactose, hydrolysis of casein, fat lipolysis by weak esterase activities, and citric acid fermentation (24). The enrichment of Lactococcus might be due to the enhancement of the fiber degradation or due to the nutrients provided by FSM. The enrichment of Candidatus Saccharimonas in the rumen was observed by feeding a Saccharomyces cerevisiae fermentation product in lactating Holstein cows (25). Tong et al. (26) reported that the abundance of the Candidatus Saccharimonas was positively correlated with the concentration of propionate in the rumen of lactating cows. Ranilla et al. (27) observed that an antioxidant (carvacrol) enriched Candidatus Saccharimonas in an in vitro trial. It suggested that the enrichment of Candidatus Saccharimonas might be associated with the antioxidant provided by FSM. The members of Olsenella could utilize starch and glycogen, producing lactate, acetate, and formate (28). Kim et al. (29) reported that the relative abundance of Olsenella was higher in the rumen of Holstein cows fed a high-grain diet. McLoughlin et al. (30) reported that the relative abundance of Olsenella in the rumen was positively associated with feed efficiency in sheep. Elolimy et al. (31) observed a higher relative abundance of Olsenella in the hindgut of Holstein heifer calves with high feed efficiency. However, Ellison et al. (32) found a higher abundance of Olsenella in the rumen of low feed efficient lambs fed a concentrate diet. It suggested that FSM increased the feed efficiency in the current study. Unfortunately, the feed efficiency was not determined in this study. It should be measured in the further study.
FSM is a high-quality plant protein source containing more than 50% crude protein. The study supplemented FSM directly into the diets without modifying the dietary protein levels across the various treatment groups. As a result, the dietary crude protein levels in the treatment groups increased by approximately 0.7 to 1.4% compared to the control group. A slight rise was observed in the numeric value of the milk protein percentage and milk urea nitrogen, but this increase was statistically insignificant. The further studies would adjust the dietary protein levels to be the same across all treatments.
Feeding FSM to lactating cows did not affect the milk performance, but increased the serum prolactin levels which would help cows maintain the lactation. Moreover, feeding FSM only caused a minor change in rumen fermentation parameters, but greatly alter the rumen microbiota, with the increase of Firmicutes, and decrease of Bacteroidota in the relative abundance. Though more work should be done to demonstrate the effects of FSM, these findings may provide an approach to keep the peak of lactation in dairy cows.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was approved by the Animal Care and Use Committee of Nanjing Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
JZ: Data curation, Investigation, Visualization, Writing – original draft. FG: Writing – original draft, Investigation. SH: Investigation, Writing – original draft. YM: Writing – review & editing, Investigation. SW: Writing – review & editing, Conceptualization. WJ: Writing – original draft, Writing – review & editing. SM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (32072755).
YM was employed by the Jiangsu Jiahui Biotechnology Co., Ltd. and SW was employed by the Shanghai Menon Animal Nutrition Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1498639/full#supplementary-material
1. Lambo, MT, Ma, H, Zhang, H, Song, P, Mao, H, Cui, G, et al. Mechanism of action, benefits, and research gap in fermented soybean meal utilization as a high-quality protein source for livestock and poultry. Anim Nutr. (2024) 16:130–46. doi: 10.1016/j.aninu.2023.10.003
2. Yuan, L, Chang, J, Yin, Q, Lu, M, Di, Y, Wang, P, et al. Fermented soybean meal improves the growth performance, nutrient digestibility, and microbial flora in piglets. Anim Nutr. (2017) 3:19–24. doi: 10.1016/j.aninu.2016.11.003
3. Jazi, V, Mohebodini, H, Ashayerizadeh, A, Shabani, A, and Barekatain, R. Fermented soybean meal ameliorates Salmonella Typhimurium infection in young broiler chickens. Poultry Sci. (2019) 98:5648–60. doi: 10.3382/ps/pez338
4. Feizi, LK, Zad, SS, Jalali, SAH, Rafiee, H, Jazi, MB, Sadeghi, K, et al. Fermented soybean meal affects the ruminal fermentation and the abundance of selected bacterial species in Holstein calves: a multilevel analysis. Sci Rep. (2020) 10:12062. doi: 10.1038/s41598-020-68778-6
5. Kim, MH, Yun, CH, Lee, CH, and Ha, JK. The effects of fermented soybean meal on immunophysiological and stress-related parameters in Holstein calves after weaning. J Dairy Sci. (2012) 95:5203–12. doi: 10.3168/jds.2012-5317
6. Rezazadeh, F, Kowsar, R, Rafiee, H, and Riasi, A. Fermentation of soybean meal improves growth performance and immune response of abruptly weaned Holstein calves during cold weather. Anim Feed Sci Technol. (2019) 254:114206. doi: 10.1016/j.anifeedsci.2019.114206
7. Amin, AB, Zhang, L, Zhang, J, and Mao, S. Fermented soybean meal modified the rumen microbiome to enhance the yield of milk components in Holstein cows. Appl Microbiol Biotechnol. (2022) 106:7627–42. doi: 10.1007/s00253-022-12240-2
8. Wang, Z, Yu, Y, Li, X, Xiao, H, Zhang, P, Shen, W, et al. Fermented soybean meal replacement in the diet of lactating Holstein dairy cows: modulated rumen fermentation and ruminal microflora. Front Microbiol. (2021) 12:625857. doi: 10.3389/fmicb.2021.625857
9. Weatherburn, MW . Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. (1967) 39:971–4. doi: 10.1021/ac60252a045
10. Makkar, H, Sharma, O, Dawra, R, and Negi, S. Simple determination of microbial protein in rumen liquor. J Dairy Sci. (1982) 65:2170–3. doi: 10.3168/jds.S0022-0302(82)82477-6
11. Wang, Y, Xu, L, Liu, J, Zhu, W, and Mao, S. A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front Microbiol. (2017) 8:2080. doi: 10.3389/fmicb.2017.02080
12. Mao, S, Zhang, R, Wang, D, and Zhu, W. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet Res. (2012) 8:237–13. doi: 10.1186/1746-6148-8-237
13. Behrendt, L, Larkum, AW, Trampe, E, Norman, A, Sørensen, SJ, and Kühl, M. Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J. (2012) 6:1222–37. doi: 10.1038/ismej.2011.181
14. Magoč, T, and Salzberg, SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
15. Campbell, BJ, Polson, SW, Hanson, TE, Mack, MC, and Schuur, EA. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol. (2010) 12:1842–54. doi: 10.1111/j.1462-2920.2010.02189.x
16. Edgar, RC . Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
17. Quast, C, Pruesse, E, Yilmaz, P, Gerken, J, Schweer, T, Yarza, P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
18. Mitter, EK, de Freitas, JR, and Germida, JJ. Bacterial root microbiome of plants growing in Oil Sands reclamation covers. Front Microbiol. (2017) 8:849. doi: 10.3389/fmicb.2017.00849
19. Lacasse, P, Ollier, S, Lollivier, V, and Boutinaud, M. New insights into the importance of prolactin in dairy ruminants. J Dairy Sci. (2016) 99:864–74. doi: 10.3168/jds.2015-10035
20. Kim, YJ . Pivotal roles of prolactin and other hormones in lactogenesis and the nutritional composition of human milk. Clin Exp Pediatr. (2020) 63:312–3. doi: 10.3345/cep.2020.00311
21. Fernando, SC, Purvis, H, Najar, F, Sukharnikov, L, Krehbiel, C, Nagaraja, T, et al. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl Environ Microbiol. (2010) 76:7482–90. doi: 10.1128/AEM.00388-10
22. Wang, C, Dong, D, Wang, H, Müller, K, Qin, Y, Wang, H, et al. Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol Biofuels. (2016) 9:1–17. doi: 10.1186/s13068-016-0440-2
23. Lagkouvardos, I, Lesker, TR, Hitch, TCA, Gálvez, EJC, Smit, N, Neuhaus, K, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. (2019) 7:28. doi: 10.1186/s40168-019-0637-2
24. Kim, W . The genus Lactococcus In: WH Holzapfel and BJB Wood, editors. Lactic acid bacteria: Biodiversity and Taxonomy John Wiley & Sons, Ltd. (2014) doi: 10.1002/9781118655252
25. Jiang, Q, Sherlock, DN, Elolimy, AA, Yoon, I, and Loor, JJ. Feeding a Saccharomyces cerevisiae fermentation product during a gut barrier challenge in lactating Holstein cows impacts the ruminal microbiota and metabolome. J Dairy Sci. (2024) 107:4476–94. doi: 10.3168/jds.2023-24147
26. Tong, J, Zhang, H, Yang, D, Zhang, Y, Xiong, B, and Jiang, L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS One. (2018) 13:e0198225. doi: 10.1371/journal.pone.0198225
27. Ranilla, MJ, Andrés, S, Gini, C, Biscarini, F, Saro, C, Martín, A, et al. Effects of Thymbra capitata essential oil on in vitro fermentation end-products and ruminal bacterial communities. Sci Rep. (2023) 13:4153. doi: 10.1038/s41598-023-31370-9
28. Göker, M, Held, B, Lucas, S, Nolan, M, Yasawong, M, Glavina Del Rio, T, et al. Complete genome sequence of Olsenella uli type strain (VPI D76D-27C). Stand Genomic Sci. (2010) 3:76–84. doi: 10.4056/sigs.1082860
29. Kim, YH, Nagata, R, Ohkubo, A, Ohtani, N, Kushibiki, S, Ichijo, T, et al. Changes in ruminal and reticular pH and bacterial communities in Holstein cattle fed a high-grain diet. BMC Vet Res. (2018) 14:310. doi: 10.1186/s12917-018-1637-3
30. McLoughlin, S, Spillane, C, Claffey, N, Smith, PE, O'Rourke, T, Diskin, MG, et al. Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front Microbiol. (2020) 11:1981. doi: 10.3389/fmicb.2020.01981
31. Elolimy, A, Alharthi, A, Zeineldin, M, Parys, C, and Loor, JJ. Residual feed intake divergence during the preweaning period is associated with unique hindgut microbiome and metabolome profiles in neonatal Holstein heifer calves. J Anim Sci Biotechnol. (2020) 11:13. doi: 10.1186/s40104-019-0406-x
Keywords: fermented soybean meal, cows, milk performance, serum biochemical indices, rumen fermentation, bacterial community
Citation: Zhang J, Guan F, Huang S, Ma Y, Wen S, Jin W and Mao S (2024) Fermented soybean meal modified the rumen microbiota and increased the serum prolactin level in lactating Holstein cows. Front. Vet. Sci. 11:1498639. doi: 10.3389/fvets.2024.1498639
Received: 19 September 2024; Accepted: 24 October 2024;
Published: 13 November 2024.
Edited by:
Shengguo Zhao, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Xiaodan Huang, Lanzhou University, ChinaCopyright © 2024 Zhang, Guan, Huang, Ma, Wen, Jin and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Jin, amlud2VpQG5qYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.