- 1Fujian Key Laboratory for Avian Diseases Control and Prevention, Fujian Academy of Agricultural Sciences, Institute of Animal Husbandry and Veterinary Medicine, Fujian Animal Diseases Control Technology Development Centre, Fuzhou, China
- 2School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Animal Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
Duck adeno-associated Virus (DAAV) is a novel pathogen that was recently discovered in ducks. To establish a molecular detection assay for DAAV for further epidemiological investigation and pathogenic mechanism. Here, we designed specific primers and probes according to the sequence characteristics of the newly discovered DAAV and then established a TaqMan real-time PCR method (TaqMan-qPCR) for the detection of DAAV. Our data showed that the established TaqMan-qPCR for detecting DAAV had high sensitivity, with the lowest detection limit of 29.1 copies/μL. No cross reaction was found with duck circovirus (DuCV), H9N2 subtype avian influenza virus (AIV), avian Tembusu virus (ATmV). duck hepatitis A virus 1 and 3 (DHAV-1 and DHAV-3), duck adenovirus A (DAdV-A), duck adenovirus 3 (DAdV-3), or duck enteritis virus (DEV). The repeatability was excellent, with the coefficients of variation of repeated intragroup and intergroup tests ranging from 0.12–0.21% and 0.62–1.42%, respectively. Seventy-eight clinical samples collected from diseased or deceased ducklings were tested. The results showed that the DAAV positive rate was 21.79%, and a triple infection (DAAV+MDPV+GPV) was found. These data provide technical support for further molecular epidemiological surveillance and pathogenic mechanism studies of DAAV infection.
1 Introduction
The Parvoviridae family consists of linear single-stranded DNA viruses with genomes ranging from ~4 to 6 kb (1). According to the latest classification of The International Committee on Taxonomy of Viruses (ICTV), the viruses of this family are divided into three subfamilies: Parvovirinae, which contain viruses that infect vertebrate hosts (2); Densovirinae, encompassing viruses that infect arthropods; and Hamaparvovirinae, which include viruses from both invertebrates and vertebrates (3). To date, there are 11 genera in Parvovirinae, of which adeno-associated virus (AAV) belongs to the genus Dependoparvovirus under Parvovirinae.
Adeno-associated viruses (AAVs) belong to the Parvoviridae-dependent Parvovirus genus. AAV is a nonenveloped, regular icosahedral, single-stranded DNA with a diameter of 20 to 26 nm and a genome size of 4.7 kb (4). As a replication-defective virus, AAV can replicate and proliferate only under the influence of helper viruses (such as adenovirus and herpes virus) (5). It has two large open reading frames (ORFs) with an inverted terminal repeat (ITR) flanking the open reading frames. The left Rep encodes nonstructural proteins that are important for virus packaging and replication. The right Cap encodes structural proteins (6, 7), which are responsible for the integration, replication, and assembly of viral particles. The infection range of AAV is not limited to dividing cells but also nondividing cells and shows a wide range of tissue tropism in vivo (8).
At present, AAV can be divided into three groups, most of which have been detected in mammals (9). Studies have also identified more than 150 viral variants (10, 11). Recently, a duck adeno-associated virus (designated DAAV) was first identified from Muscovy ducks in China (12). Then, DAAV was isolated in Fujian (FJFF001 strain, GenBank accession number: MW286836) in Southeast China using Muscovy duck embryo fibroblasts (MDCEFs). To date, there has been no report on the primers, probes and methods of real-time PCR for the detection of DAAV. Therefore, it is necessary to develop an accurate and efficient DAAV-specific molecular diagnostic platform to help us to better understand the prevalence and transmission of DAAV in waterfowl. This paper describes a novel TaqMan-based real-time polymerase chain reaction method (TaqMan-qPCR) for DAAV, which will help us for further molecular epidemiological surveillance and pathogenic mechanism research of DAAV infection.
2 Materials and methods
2.1 Viruses
Duck circovirus (DuCV), H9N2 subtype avian influenza virus (AIV), avian Tembusu virus (ATmV), duck hepatitis A virus 1 and 3 (DHAV-1 and DHAV-3), duck adenovirus A (DAdV-A), duck adenovirus 3 (DAdV-3), duck enteritis virus (DEV), goose parvovirus (GPV), Muscovy duck parvovirus (MDPV), and DAAV were stored at Fujian Key Laboratory for Avian Diseases Control and Prevention.
2.2 Clinical samples
A total of 78 dead ducklings’ samples (including the spleen, liver, kidney, and intestinal tissue mixture) were collected during 2022–2023 in Southeast China. All dead samples were handled in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of China, which were kindly provided by Professor Bin Jiang. All samples used in this study was obtained by Animal Disease Detection Center, Institute of Animal Husbandry and Veterinary Medicine, Fujian Academy of Agricultural Sciences, which is accredited in accordance with ISO/IEC 17025: 2017 General Requirements for the Competence of Testing and Calibration Laboratories (CNAS-CL01 Accreditation Criteria for the Competence of Testing and Calibration Laboratories) for the competence to undertake the service described in the schedule attached to this certificate (Registration No. CNAS L18448). The 78 samples (including the spleen, liver, kidney, and intestinal tissue mixture) were pooled and regarded as one sample. These samples were homogenized in phosphate-buffered saline (PBS) (20%, w/v). Viral DNA was extracted from duckling sample homogenates via the Magnetic Animal Tissue Genomic DNA Kit.
2.3 Primers and probe
According to the sequence characteristics of the novel duck adeno-associated virus (DAAV) gene, specific primer sets (DAAV-TqF and DAAV-TqR) and a probe (DAAV-TqP) for TaqMan-qPCR were designed using Primer Express 3. The primers and probe sequences (Table 1) were subsequently subjected to BLAST1 to verify their specificity and synthesized by Sangon Bioengineering Co., Ltd. (Sangon Biotech, Shanghai, China).
2.4 Standard plasmids
The EasyPure Viral DNA/RNA Kit (TransGen Biotech, Beijing, China) was used to extract the nucleic acid DNA of the DAAV-FJ strain (GenBank accession number: MW380871), and then the Cap gene fragments were amplified using DAAV-F and DAAV-R by conventional PCR (cPCR) technology (Table 1). After PCR, the PCR products (approximately 617 bp) were identified by electrophoresis on a 1.0% agarose gel (TransGen Biotech, Beijing, China) and then cloned and inserted into the pEASY-T1 vector according to the instructions of the pEASY-T1 Simple Cloning Kit (TransGen Biotech, Beijing, China). The obtained recombinant plasmids were sent to Sangon Bioengineering (Shanghai, China) Co., Ltd. for sequencing. The sequencing results were verified by BLAST analysis in NCBI. The positive recombinant plasmids (T-DAAV-Tq) were used as the positive standard of TaqMan-qPCR and quantified using an ND-2000c spectrophotometer (NanoDrop2000, Wilmington, United States). The copy number was calculated to be 2.91 × 1010 copies/μL according to a previous study (13), using the formula: Amount (copies/μL) = [DNA concentration (g/μL)/(plasmid length in bp × 660)] × 6.02 × 1023. Ten-fold dilutions of T-DAAV-Tq, ranging from 2.91× 109–2.91 × 100 copies/μL, were prepared using TE buffer (10 mmol/L Tris–HCl, 1 mmol/L EDTA). All aliquots of each dilution were stored at −80°C until use.
2.5 TaqMan-qPCR reaction
TaqMan-qPCR was prepared according to the instructions of the Premix Ex Taq™ (Probe qPCR) kit (TaKaRa Biotechnology, Dalian, China), and the total reaction volume was 25 μL. To determine the optimal primer amount of the reaction, 0.1–1.0 μL (10 μmol/L) was used for the reaction, and different annealing temperatures (ranging from 54 to 62°C) were set for the reaction to determine the optimal annealing temperature. Tenfold serial dilutions of T-DAAV-Tq (ranging from 2.91× 106–2.91 × 101 copies/μL), were used to generate the standard curve under the optimal reaction conditions using LightCycler 96 Instrument (Mannheim, Germany). The plasmid copy number logarithm was plotted against the corresponding Ct values (cycle number threshold), and the standard curve was obtained.
2.6 Sensitivity, specificity, and repeatability
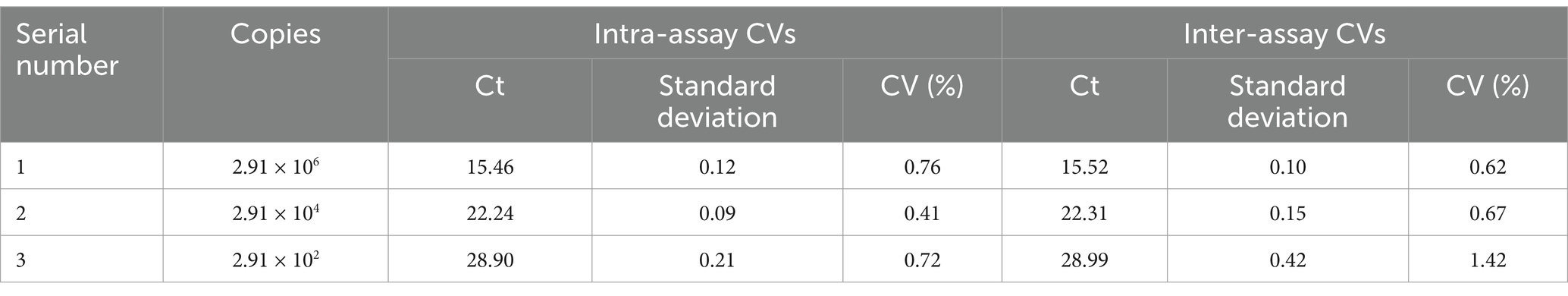
To evaluate the sensitivity, serial dilutions of T-DAAV-Tq (ranging from 2.91 × 103–2.91 × 100 copies/μL) were used as the template, and optimized TaqMan-qPCR was used for detection. To evaluate the specificity, nucleic acid DNA was extracted from DuCV, AIV, ATmV, DHAV-1, DHAV-3, DAdV-A, DAdV-3, DEV, GPV and MDPV. To evaluate the reproducibility, DAAV-positive standard samples with three different concentrations (2.91 × 106 copies/μL, 2.91 × 104 copies/μL, 2.91 × 102 copies/μL) were used as templates. Three replicates were performed for each concentration of plasmid, to determine the coefficient of variation (CV). The intragroup and intergroup with the coefficient of variation (CVs) for Ct values were calculated.
2.7 Clinical testing
A total of 78 samples were teste using the established TaqMan-qPCR method. The weight of each sample was 25 mg and was homogenized with sterile PBS in a mortar. Then, the suspensions were collected after centrifugation at 4,000 rpm at 4°C for 15 min. Nucleic acid was extracted from the supernatant using the Magnetic Animal Tissue Genomic DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Because GPV and MDPV also belong to the genus Dependoparvovirus under the Parvoviridae family, the GPV and MDPV were also detected in the above samples by the real-time PCR previously reported by us (14, 15), which will help us to learn about the co-infection situation of duck-origin related diseases under the Parvoviridae family.
3 Results
3.1 TaqMan-qPCR optimization
The optimal reaction system of 25 μL was optimized by the established TaqMan-qPCR assay as follows: Premix Ex Taq™ (Probe qPCR) master mix 12.5 μL, 0.4 μL each of DAAV-TqF and DAAV-TqR primers (10 μmol/L), and probe (DAAV-TqP, 5 μmol/L), template (1 μL), and sterilized ddH2O to a final volume of 25 μL. The optimized reaction conditions were as follows: 95°C for 120 s and 40 cycles of 95°C for 10 s and 60°C for 30 s.
3.2 Standard curve
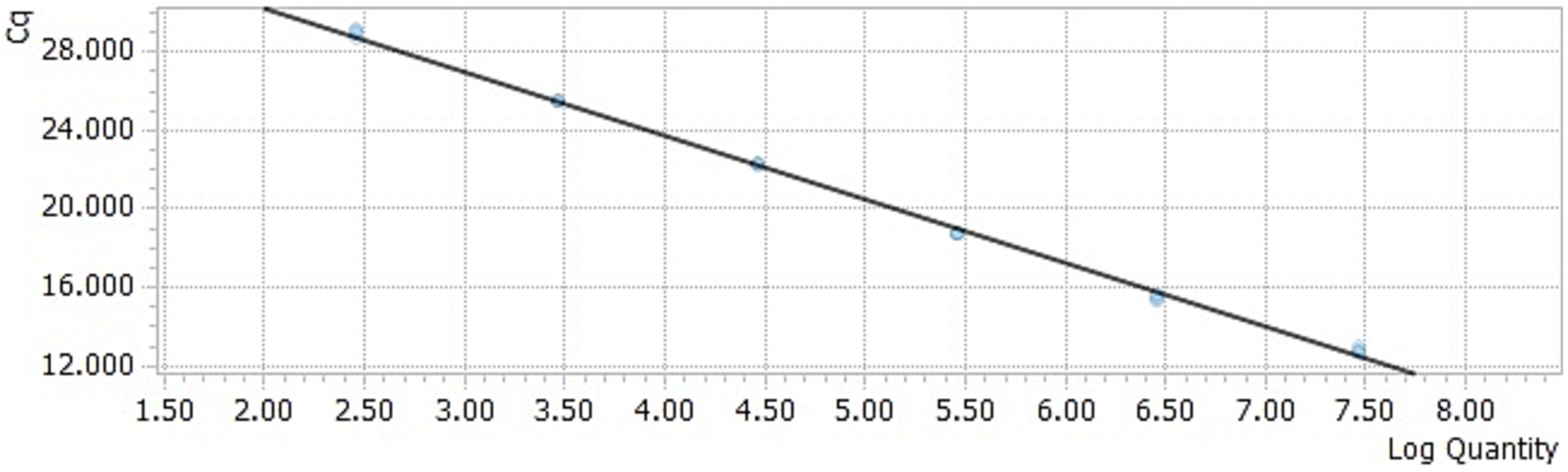
The DAAV real-time PCR standard curve was obtained by taking the common logarithm (lgC) of copy number in each concentration standard template as the horizontal coordinate and cycle threshold (Ct value) as the ordinate (Figures 1, 2). The slope of the standard curve was −3.2634, the Y-intercept was 36.787, the R2 was 0.9987, and the amplification efficiency was 99%, which indicated that the standard curve of the TaqMan-qPCR has a good linear relationship.
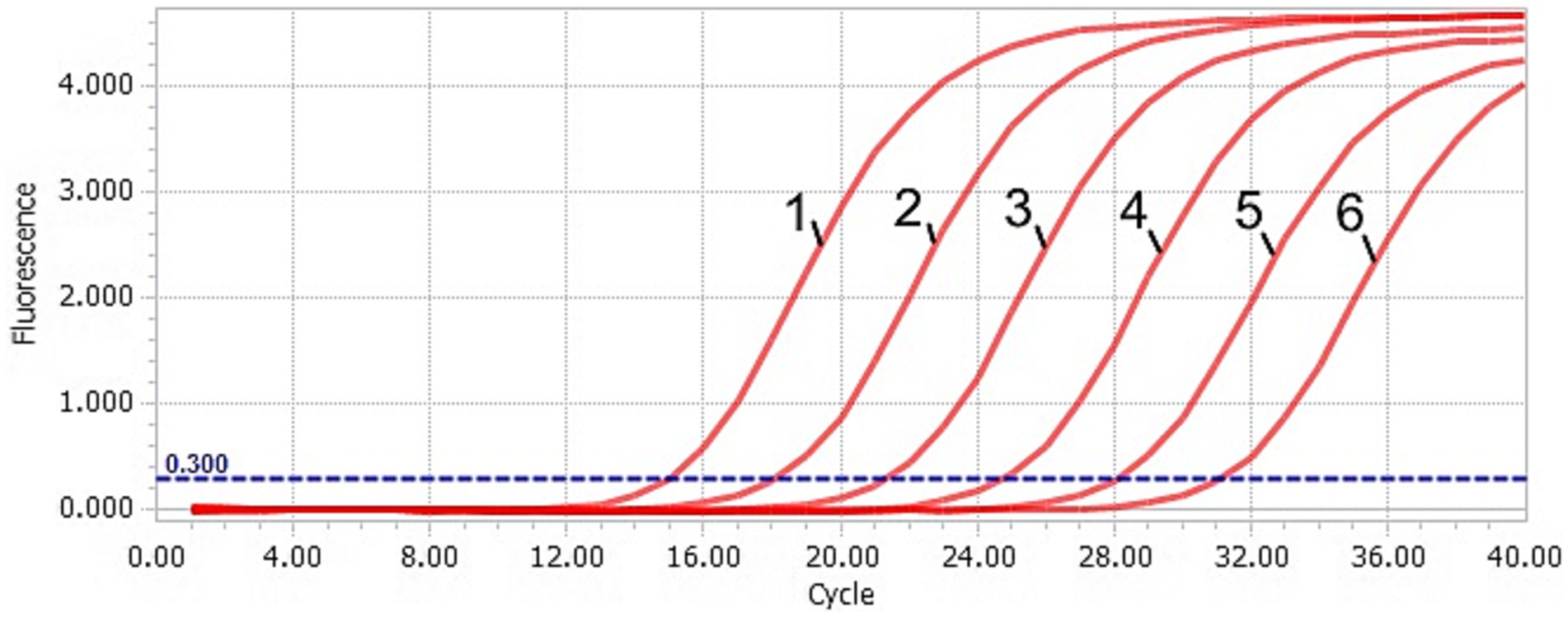
Figure 1. Amplification curve of the TaqMan-qPCR assay. 1–6: plasmid concentration ranging from 2.91 × 106–2.91 × 101 copies/μL.
3.3 Sensitivity, specificity, and repeatability
After TaqMan-qPCR amplification, the minimum detection limit of the assay was 2.91 × 101 copies/μL (29.1 copies/μL) (Figure 3). Using the viral DNA and cDNA as template (DuCV, AIV, ATmV, DHAV-1, DHAV-3, DAdV-A, DAdV-3, DEV, GPV and MDPV), the data showed only DAAV with a positive signal, while other templates followed with no response signal (Figure 4). Similar data were observed after three independent reactions were repeated. The results showed that TaqMan-qPCR was highly specific. The intragroup coefficient of variation of the assay was 0.12–0.21%, and the intergroup coefficient of variation was 0.62–1.42%. The results are shown in Table 2, demonstrating the good repeatability of the assay.
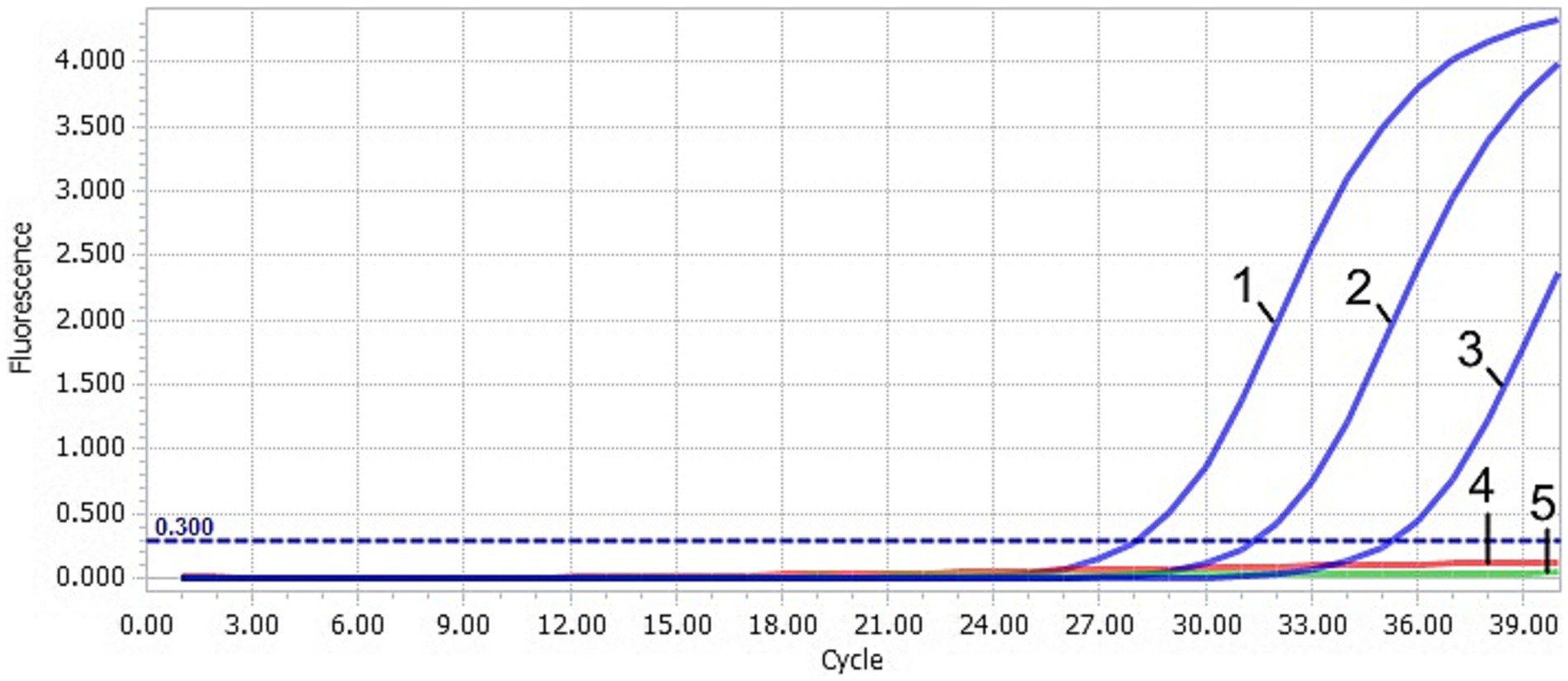
Figure 3. Sensitivity of the TaqMan-qPCR assay. 1–4: plasmid concentration ranging from 2.91 × 103–2.91 × 100 copies/μL. 5: negative control (ddH2O).
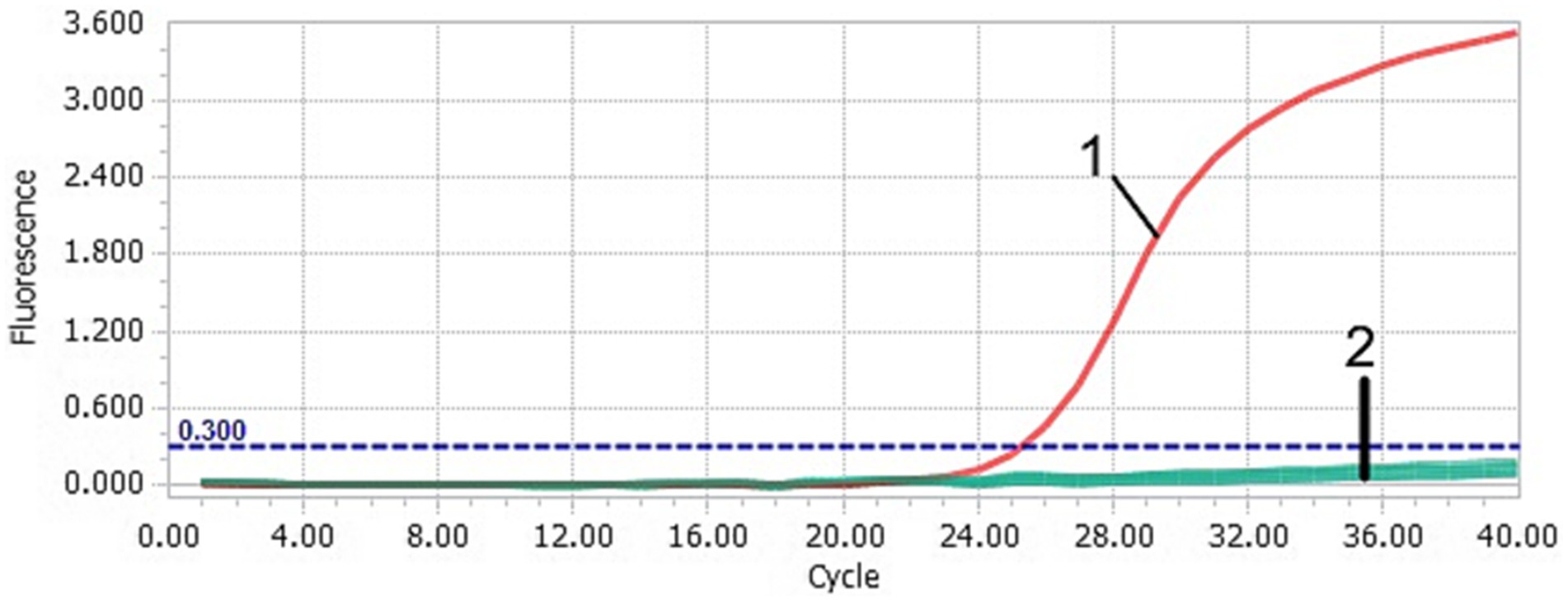
Figure 4. Specificity of the TaqMan-qPCR assay. 1: DAAV; 2: DuCV, AIV, ATmV, DHAV-1, DHAV-3, DAdV-A, DAdV-3, DEV, GPV, MDPV and ddH2O. These controls did not yield a positive fluorescent signal. It cannot be effectively distinguished by the naked eye.
3.4 Clinical samples evaluation
3.4.1 Duck adeno-associated virus evaluation
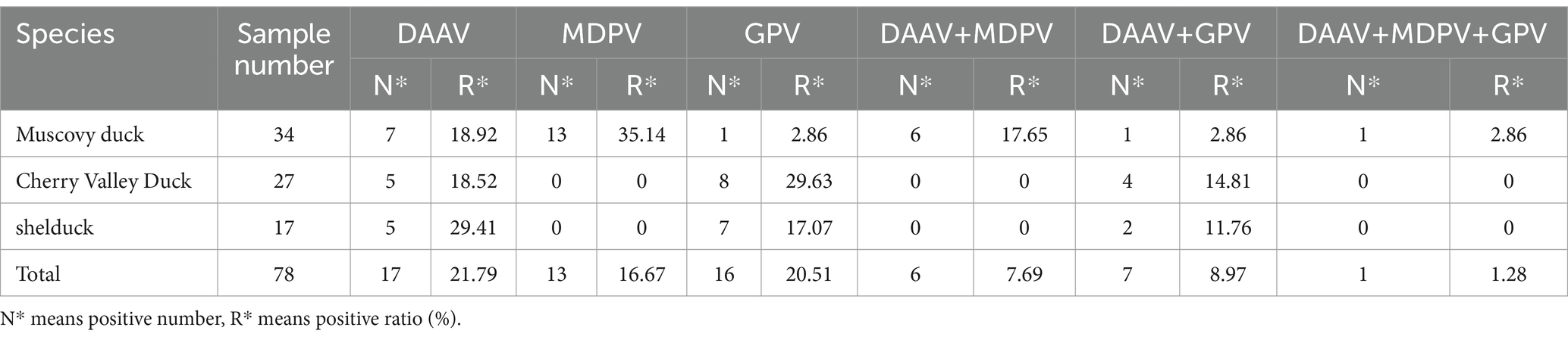
The 78 collected clinical samples were tested using the established TaqMan-qPCR assay (Table 3). For DAAV, 17 samples tested positive, with a positive ratio of 21.79% (17/78). Moreover, the TaqMan-qPCR-positive samples were also verified by cPCR method (described at Section2.4, using DAAV-F and DAAV-R), 14 of 17 were tested with cPCR-positive. To confirm the presence of DAAV in cPCR-positive samples, the expected product sizes were cloned (described at Section2.4). For each PCR product, three colonies were selected for Sanger sequencing in both directions at Sangon (Shanghai, China). In terms of sequence identity, the identified DAAV-cPCR-positive viruses showed nucleotide of 100% with FJFF001 (GenBank No. MW286836).
3.4.2 Muscovy duck parvovirus and GPV coinfection evaluation
For MDPV, 13 samples tested positive, with a positive ratio of 16.67% (13/78). For GPV, 16 samples tested positive, with a positive ratio of 20.51% (16/78). Moreover, the DAAV and MDPV coinfection positive ratio was 7.69% (6/78), the DAAV and GPV coinfection positive ratio was 8.97% (7/78), and the DAAV, MDPV and GPV triple infection rate was 1.28% (1/78), with the copy number of 8.75 × 103 copies/μL(for DAAV), 3.92 × 104 copies/μL(for MDPV), and 7.09 × 103copies/μL(for GPV), respectively.
4 Discussion
In recent years, the emergence of new duck viral diseases has caused enormous losses to waterfowl breeding in China. Therefore, the exploration of unknown duck-origin viruses is conducive to the prevention and control of waterfowl diseases. Adeno-associated virus (AAV), a kind of single-stranded DNA-defective virus, is the smallest type of animal virus due to its simple genetic structure and small size (16, 17). AAV is widely used in the construction of gene expression vectors and gene therapy because of its good safety, wide host range and low immunogenicity (18, 19). A novel duck adeno-associated virus (DAAV) was identified during the virological investigation of waterfowl disease in the Muscovy ducks. Due to limited information, the specific pathogenicity of DAAV remains to be fully understood, and accurate measurement of viral loads in different tissues of DAAV-infected ducks could allow researchers to more fully understand the relationship between this virus and the onset or progression of the disease.
Fluorescence PCR is widely used in the diagnosis of clinical diseases due to its high sensitivity and good specificity (20). Real-time fluorescence PCR technology based on TaqMan fluorescence-labeled probes is the most widely used in clinical diagnosis in China (14, 15, 21–23). The TaqMan fluorescent probe is labeled with a fluorescent reporter group at the 5′ end and a quenching agent at the 3′ end. Based on this principle, we analyzed the DAAV Cap gene and established a TaqMan-based real-time PCR assay for the detection of DAAV.
In the present study, the establishment and evaluation of the TaqMan-qPCR for fast, sensitive and accurate detection of DAAV was used. Our data were analytically specific and sensitive and presented excellent intra- and inter-assay CVs (both less than 1.50%), demonstrating that the established TaqMan-qPCR assay is a reliable and reproducible platform. For clinical evaluation, the positive rate of DAAV, MDPV and GPV were 21.79, 16.67 and 20.51%, respectively. The coinfection rates of DAAV+MDPV and DAAV+GPV was 7.69 and 8.97%, respectively; moreover, we also found a triple infection (DAAV+MDPV+GPV) in Muscovy duck. The role of DAAV and its coinfection with GPV and MDPV needs further study.
5 Conclusion
In conclusion, our study provides a DAAV detection platform using the TaqMan-qPCR assay with good sensitivity, specificity, and repeatability. This platform will help us further molecular epidemiological surveillance and pathogenesis studies.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
All samples were handled in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SC: Investigation, Methodology, Writing – original draft. YC: Investigation, Writing – original draft. MZ: Conceptualization, Investigation, Writing – original draft. WZ: Investigation, Writing – original draft. HF: Investigation, Writing – review & editing. YH: Investigation, Writing – review & editing. LC: Methodology, Writing – review & editing. CW: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by grants from the Fujian Science and Technology Program (grant no. 2020 J06029), China Agriculture Research System (grant no. CARS-42-19), ‘5511’ Collaborative Innovation Project of Fujian Academy of Agricultural Sciences (XTCXGC2021012) and Research and Technology Program of Fujian Academy of Agricultural Sciences (grant nos. YCZX202412, DWHZ2024-13, and CXTD2021005). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Cotmore, SF, Agbandje-McKenna, M, Chiorini, JA, Mukha, DV, Pintel, DJ, Qiu, J, et al. The family Parvoviridae. Arch Virol. (2014) 159:1239–47. doi: 10.1007/s00705-013-1914-1
2. Meyer, NL, and Chapman, MS. Adeno-associated virus (AAV) cell entry: structural insights. Trends Microbiol. (2022) 30:432–51. doi: 10.1016/j.tim.2021.09.005
3. Souza, WMD, Romeiro, MF, Fumagalli, MJ, Modha, S, de Araujo, J, Queiroz, LH, et al. Chapparvoviruses occur in at least three vertebrate classes and have a broad biogeographic distribution. J Gen Virol. (2017) 98:225–9. doi: 10.1099/jgv.0.000671
4. Rabinowitz, J, Chan, YK, and Samulski, RJ. Adeno-associated virus (AAV) versus immune response. Viruses. (2019) 11:102. doi: 10.3390/v11020102
5. Meier, AF, Fraefel, C, and Seyffert, M. The interplay between adeno-associated virus and its helper viruses. Viruses. (2020) 12:662. doi: 10.3390/v12060662
6. Zhu, C, Wang, C, Wu, J, Ye, F, Lv, R, Hu, D, et al. Distribution and genetic diversity of adeno-associated viruses in bats from coastal areas of Southeast China. Sci Rep. (2020) 10:3725. doi: 10.1038/s41598-020-60721-z
7. Hull, JA, Mietzsch, M, Chipman, P, Strugatsky, D, and McKenna, R. Structural characterization of an envelope-associated adeno-associated virus type 2 capsid. Virology. (2022) 565:22–8. doi: 10.1016/j.virol.2021.09.010
8. MacLoughlin, RJ, Higgins, BD, Devaney, J, O’Toole, D, Laffey, JG, and O’Brien, T. Aerosol-mediated delivery of AAV2/6-IkappaBalpha attenuates lipopolysaccharide-induced acute lung injury in rats. Hum Gene Ther. (2015) 26:36–46. doi: 10.1089/hum.2014.053
9. Pillay, S, Meyer, NL, Puschnik, AS, Davulcu, O, Diep, J, Ishikawa, Y, et al. An essential receptor for adeno-associated virus infection. Nature. (2016) 530:108–12. doi: 10.1038/nature16465
10. Mietzsch, M, Jose, A, Chipman, P, Bhattacharya, N, Daneshparvar, N, McKenna, R, et al. Completion of the AAV structural atlas: serotype capsid structures reveals clade-specific features. Viruses. (2021) 13:101. doi: 10.3390/v13010101
11. Yao, Y, Wang, J, Liu, Y, Qu, Y, Wang, K, Zhang, Y, et al. Variants of the adeno-associated virus serotype 9 with enhanced penetration of the blood–brain barrier in rodents and primates. Nat Biomed Eng. (2022) 6:1257–71. doi: 10.1038/s41551-022-00938-7
12. Su, X, Liu, J, Zhou, Q, Zhang, X, Zhao, L, Xie, Q, et al. Isolation and genetic characterization of a novel adeno-associated virus from Muscovy ducks in China. Poult Sci. (2017) 96:3867–71. doi: 10.3382/ps/pex235
13. Yun, JJ, Heisler, LE, Hwang, II, Wilkins, O, Lau, SK, Hyrcza, M, et al. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic Acids Res. (2006) 34:e85. doi: 10.1093/nar/gkl400
14. Wan, C, Chen, C, Cheng, L, Chen, H, Fu, Q, Shi, S, et al. Specific detection of Muscovy duck parvovirus infection by TaqMan-based real-time PCR assay. BMC Vet Res. (2018) 14:267. doi: 10.1186/s12917-018-1600-3
15. Wan, C, Chen, C, Cheng, L, Liu, R, Shi, S, Fu, G, et al. Specific detection and differentiation of classic goose parvovirus and novel goose parvovirus by TaqMan real-time PCR assay, coupled with host specificity. BMC Vet Res. (2019) 15:389. doi: 10.1186/s12917-019-2090-7
16. György, B, Fitzpatrick, Z, Crommentuijn, HM, Mu, D, and Casey, A. MaguireNaturally, enveloped AAV vectors for shielding neutralizing antibodies and robust gene delivery in vivo. Biomaterials. (2014) 35:7598–609. doi: 10.1016/j.biomaterials.2014.05.032
17. Rapti, K, and Grimm, D. Adeno-associated viruses (AAV) and host immunity - a race between the hare and the hedgehog. Front Immunol. (2021) 12:753467. doi: 10.3389/fimmu.2021.753467
18. Li, X, Le, Y, Zhang, Z, Nian, X, Liu, B, and Yang, X. Viral vector-based gene therapy. Int J Mol Sci. (2023) 24:7736. doi: 10.3390/ijms24097736
19. Bulcha, JT, Wang, Y, Ma, H, Tai, P, and Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct Target Ther. (2021) 6:53. doi: 10.1038/s41392-021-00487-6
20. Zhang, D, Wu, J, Sun, J, Bai, C, Xu, F, Duan, P, et al. Establishment of TaqMan-based real-time PCR assay for rapid detection of duck circovirus. 3 Biotech. (2021) 11:470. doi: 10.1007/s13205-021-03021-1
21. Wang, J, Zhang, Y, Wang, J, Liu, L, Pang, X, and Yuan, W. Development of a TaqMan-based real-time PCR assay for the specific detection of porcine circovirus 3. J Virol Methods. (2017) 248:177–80. doi: 10.1016/j.jviromet.2017.07.007
22. Yu, J, Zou, J, Liu, X, Pan, Y, Mu, Y, Li, S, et al. TaqMan-probe-based multiplex real-time RT–qPCR for simultaneous detection of GoAstV, GPV, and GoCV. Poult Sci. (2023) 102:102396. doi: 10.1016/j.psj.2022.102396
Keywords: duck adeno-associated virus, DAAV, TaqMan, qPCR, epidemiological surveillance
Citation: Chen S, Chen Y, Zhang M, Zhang W, Fu H, Huang Y, Cheng L and Wan C (2024) Specific detection of duck adeno-associated virus using a TaqMan-based real-time PCR assay. Front. Vet. Sci. 11:1483990. doi: 10.3389/fvets.2024.1483990
Edited by:
Francisco Ruben Carvallo Chaigneau, Virginia Tech, United StatesReviewed by:
Manish Muhuri, Voyager Therapeutics, Inc., United StatesStephanie Michelle Todd, Virginia Tech, United States
Copyright © 2024 Chen, Chen, Zhang, Zhang, Fu, Huang, Cheng and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunhe Wan, Y2h1bmhld2FuQDEyNi5jb20=
†These authors have contributed equally to this work
 Shuyu Chen
Shuyu Chen YuYi Chen
YuYi Chen Mengyan Zhang
Mengyan Zhang Wenyu Zhang
Wenyu Zhang Huanru Fu
Huanru Fu Yu Huang
Yu Huang Longfei Cheng
Longfei Cheng Chunhe Wan
Chunhe Wan