- 1Department of Veterinary Internal Medicine, College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
- 2Department of Infectious Disease, College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
Bacterial urinary tract infections (UTIs) are prevalent in dogs and necessitate antibiotic intervention. However, the emergence of multidrug-resistant (MDR) bacteria poses significant challenges to antibiotic therapy. Although fosfomycin has been demonstrated to achieve and maintain high concentrations in urine, suggesting its potential for treating UTIs in dogs, its efficacy and the resistance profiles of urinary pathogens from canine UTIs remain elusive. Therefore, this study was conducted to investigate the antibiotic susceptibility of bacterial pathogens isolated from companion dogs with UTIs, with a particular focus on their susceptibility and resistance to fosfomycin. A total of 70 isolates from urine samples were analyzed, of which Escherichia coli (n = 18), Proteus mirabilis (n = 9), Klebsiella pneumoniae (n = 5), and Staphylococcus pseudintermedius (n = 5) were predominant. Resistance to erythromycin was most prevalent (94.59%), followed by clindamycin (91.89%) and ampicillin (78.37%), whereas the lowest resistance rate was observed for amikacin (5.40%). Resistance to fosfomycin was observed in 15 out of the 37 predominant isolates (40.54%), including all K. pneumoniae isolates (100%). All isolates, except 4 E. coli strains, were categorized as MDR (33 out of 37; 89.18%). The resistance rates for amoxicillin/clavulanic acid and trimethoprim-sulfamethoxazole, which are common first-line antibiotics for canine UTIs, were 48.64 and 56.75%, respectively. Whole-genome sequencing of K. pneumoniae isolates, which exhibited high resistance to fosfomycin, revealed multiple antibiotic resistance genes, with chromosomal fosA present in all isolates. Among the 27 dogs with recurrent infection included in this study, 2 were administered fosfomycin, resulting in clinical remission, as evidenced by negative urine culture tests. Overall, this study is the first to demonstrate the importance of assessing fosfomycin resistance profile for optimal treatment of canine UTIs, particularly in cases involving MDR strains.
1 Introduction
Bacterial urinary tract infections (UTIs) are prevalent in small veterinary practices, affecting approximately 14% of companion dogs during their lifetime (1). Antibiotics are pivotal in the treatment of UTIs, with selection dependent on the antibiotic susceptibility profiles of the uropathogens (2). However, empirical antibiotic therapy is frequently initiated in veterinary clinics during the interim period awaiting susceptibility test results (3). Amoxicillin is commonly selected as the initial treatment, followed by trimethoprim-sulfadiazine (3). Notably, the misuse and overuse of antibiotics, particularly in cases of empirical administration without prior antibiotic susceptibility testing, have contributed to the emergence of multidrug-resistant (MDR) bacteria in both human and veterinary medicine.
The increasing prevalence of MDR bacteria in both human and veterinary medicine has significantly limited the range of antibiotics available for the treatment of UTIs. Notably, fosfomycin, discovered in 1969, has emerged as a viable alternative antibiotic. Recently, it has become a common choice for primary treatment in humans, particularly for uncomplicated UTIs and infections caused by MDR bacteria (4–6).
Fosfomycin is eliminated via glomerular filtration and exhibits higher concentrations in urine than in plasma, which enhances its effectiveness against urinary pathogens (7–9). Moreover, fosfomycin is effective against MDR bacteria, such as methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, such as Klebsiella pneumoniae, and carbapenemase-producing Enterobacteriaceae (10, 11). Previous studies involving canine models have demonstrated that fosfomycin achieves and maintains high concentrations in urine, suggesting its potential for treating UTIs in dogs (12, 13). However, the efficacy of fosfomycin and the resistance profiles of urinary pathogens from canine UTIs remain largely unexplored.
This study was undertaken to investigate the antibiotic susceptibility and resistance profiles of major bacterial species isolated from companion dogs with UTIs, with a specific focus on their susceptibility and resistance to fosfomycin. Furthermore, we sought to examine the presence of antibiotic resistance genes in K. pneumoniae, which exhibited high resistance to fosfomycin, using whole-genome sequencing (WGS).
2 Materials and methods
2.1 Sampling
Between September 2019 and September 2022, a total of 221 clinical samples were collected from various lesions in dogs at the Veterinary Medical Teaching Hospital of Konkuk University in Seoul, South Korea. These samples included 52 urine specimens and were obtained from diverse anatomical regions, such as the skin, eyes, pleuroperitoneal effusion, gastrointestinal tract, and urogenital areas. All samples were promptly transported to the NosVet Laboratory (Gyeonggi-do, South Korea) and analyzed within 3–4 h for the isolation of causative agents through antibiotic susceptibility testing. The study protocol was reviewed and approved by the Institutional Animal Care and Use Committee (KU24059). Owners provided written informed consent for their dogs to participate in the study.
2.2 Bacterial isolation and identification
A total of 420 bacterial isolates were obtained from clinical samples, with 70 isolates derived from 52 urine samples. For bacterial isolation from urine samples, 50 μL aliquots of the samples were inoculated onto 2 blood agar plates and incubated at 37°C overnight under aerobic and anaerobic conditions. For anaerobic cultivation, the blood plate was placed in an anaerobic jar with an anaerobic gas pack. Identification testing was performed if at least 1 colony was detected on the blood agar plate; the colonies were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (ASTA, Gyeonggi-do, South Korea). Among the 70 isolates from urine samples (Supplementary Table 1), the 4 predominant bacterial species (n = 37), isolated from 27 dogs, included Escherichia coli (n = 18), Proteus mirabilis (n = 9), K. pneumoniae (n = 5), and Staphylococcus pseudintermedius (n = 5). These species were subjected to further analysis.
2.3 Canine patient characteristics
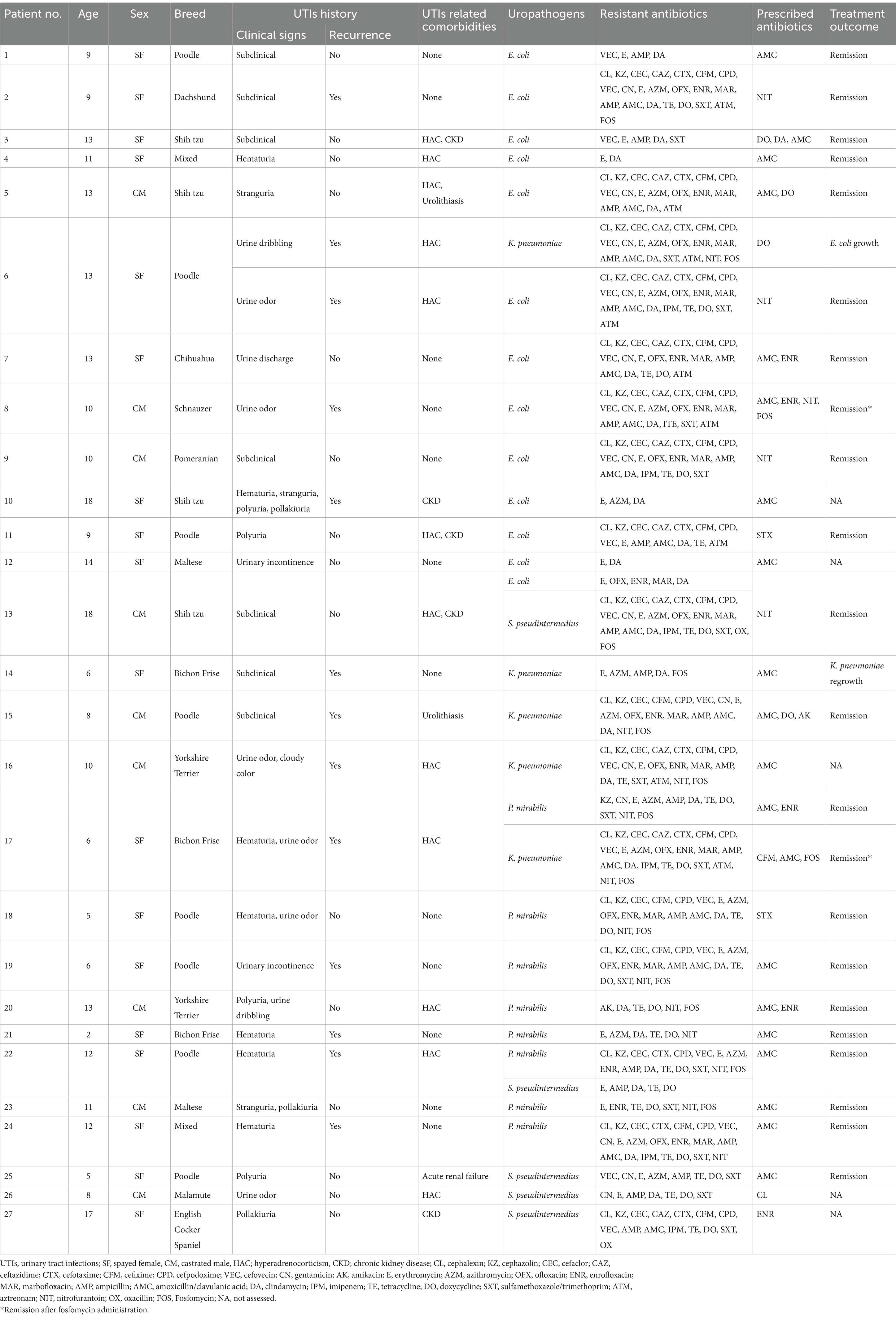
Detailed information about the 27 dogs included in this study is presented in Table 1. The median age of the dogs was 10 years, with an age range from 2 to 17 years. Clinical signs of UTIs were observed in 20 dogs (74.07%), and 12 dogs (44.44%) had a history of recurrent infections. Concurrent conditions potentially affecting UTIs were identified as follows: hyperadrenocorticism in 10 dogs (37.03%), chronic kidney disease in 4 dogs (14.81%), urolithiasis in 2 dogs (7.40%), and acute renal failure in 1 dog (3.70%). Conditions unrelated to UTIs are not listed.
2.4 Antibiotic susceptibility test
Susceptibility testing for 25 antibiotics was performed using the Kirby-Bauer disk diffusion method, following the interpretive criteria recommended by the Clinical and Laboratory Standards Institute (CLSI) for consensus interpretation (14).
Susceptibility to fosfomycin was assessed using the agar dilution method, adhering to the CSLI and European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (14, 15). Minimum inhibitory concentrations (MICs) were determined using Mueller Hinton agar supplemented with 25 mg/L glucose-6-phosphate (Sigma Aldrich Co., South Korea), with fosfomycin trometamol (Sigma Aldrich Co., South Korea) tested in 2-fold dilutions ranging from 1 mg/L to 1,024 mg/L. The experiments were performed in triplicate, and results were interpreted using CLSI and EUCAST breakpoints/MICs. For Enterobacterales and Staphylococcus spp., MIC results were interpreted according to EUCAST guidelines, whereas for E. coli, interpretations were based on CLSI standards owing to their specificity and higher reliability (16, 17).
2.5 Whole-genome sequencing
Among the 37 bacterial isolates analyzed in this study, K. pneumoniae isolates (n = 5), which exhibited high resistance to fosfomycin, were subjected to WGS. Genomic DNA was isolated from pure cultures of K. pneumoniae isolates using the MagNA Pure 96 DNA and Viral NA Small Volume Kit on the MagNA Pure 96 instrument (Roche Applied Sciences, Germany), following the manufacturer’s instructions. DNA concentration was determined using a Qubit BR dsDNA assay kit (Invitrogen, Carlsbad, CA), and DNA samples at a concentration of 0.2 ng/μL were used for library preparation using the Illumina Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA), following the procedures described in our previous study (18). The library pool, comprising 500 μL of 10 pM libraries, was loaded into the MiniSeq High Output Reagent cartridge (300 cycles) (Illumina). Paired FASTQ files were generated via base-calling from the raw read data obtained from Illumina sequencing.
2.6 Whole-genome sequencing analysis
Raw reads were trimmed using Bbduk1 (quality score [Q] > 20; minimum length > 50). Subsequently, the trimmed reads were assembled de novo using SPAdes 3.15.5 (19) with default settings in Geneious Prime 10 software. Contigs with coverage less than 5× and sizes below 300 bases were eliminated from the assembly. MLST 2.0 (Multi-Locus Sequence Typing) was used to determine the sequence types of the isolates. The presence of acquired antimicrobial resistance genes and chromosomal mutations in the gyrA, gyrB, parC, and parE genes was assessed using ResFinder 4.12, applying a threshold of 90% and a minimum length of 60% with the assembled contigs.
3 Results
3.1 Antimicrobial treatments and clinical outcomes
Antibiotics were prescribed based on antibiotic susceptibility test results for all dogs (Table 1). Empirically prescribed antibiotics were maintained if the isolates were sensitive and replaced if resistance was detected. Amoxicillin/clavulanic acid (AMC) was the most frequently prescribed antibiotic, administered to 20 of the 27 dogs following susceptibility confirmation. Among the 37 bacterial strains isolated, susceptibility testing revealed a resistance rate of 48.64% (18/37) to AMC. AMC alone resulted in UTI remission in 8 cases. However, for 2 out of the 27 dogs with recurrent infections, no viable antibiotic options were available based on susceptibility test results. Despite initial effectiveness, the empirical prescriptions did not prevent recurrent infections, depleting available oral antibiotic options. In these cases, fosfomycin was administered at 40 mg/kg orally every 8 h for 2 weeks. Subsequent remission was achieved, as indicated by negative urine culture results and improved clinical signs (Table 1).
3.2 Antimicrobial resistance
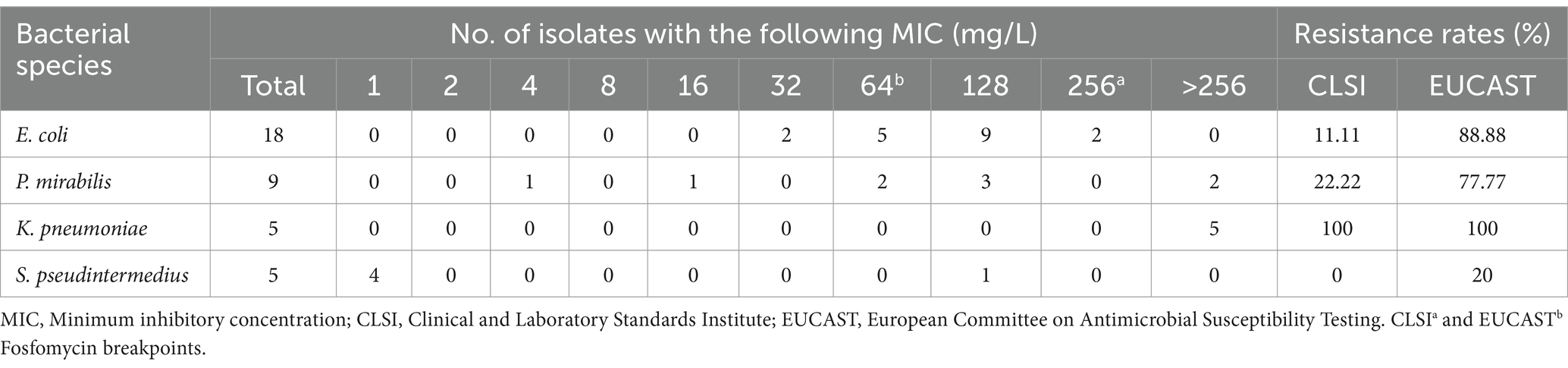
The antibiotic resistance profiles for the 37 isolates are presented in Table 2, and the MIC values for fosfomycin by species are presented in Table 3. The highest observed resistance was against erythromycin (94.59%), followed by clindamycin (91.89%) and ampicillin (78.37%), whereas the lowest resistance rate was observed for amikacin (5.40%) (Table 2). Resistance rates for AMC and trimethoprim-sulfamethoxazole—common initial antibiotics for canine UTIs—were notably high at 48.64 and 56.75%, respectively (Table 2). Resistance rates against third-generation cephalosporins ranged from 40.54 to 64.86%. Among the antibiotics used for UTIs in dogs, the lowest resistance rate (40.54%) was observed for ceftazidime and nitrofurantoin, whereas the highest resistance rate was observed for cefovecin (64.86%) (Table 2). All isolates, except 4 E. coli strains, were categorized as MDR (89.18%) (data not shown). Two S. pseudintermedius isolates were resistant to oxacillin and categorized as methicillin-resistant S. pseudintermedius (MRSP).
The antibiotics to which the isolates exhibited 100% resistance, categorized by species, are as follows: erythromycin and clindamycin for E. coli; tetracycline, doxycycline, and nitrofurantoin for P. mirabilis; erythromycin, ampicillin, clindamycin, and nitrofurantoin for K. pneumoniae; and ampicillin, tetracycline, and doxycycline for S. pseudintermedius.
Resistance to fosfomycin was observed in 15 out of the 37 isolates (40.54%) based on our criteria. Specifically, resistance was observed in 100% of K. pneumoniae, 77.77% of P. mirabilis, 20% of S. pseudintermedius, and 11.11% of E. coli isolates (Table 2). However, following CLSI and EUCAST breakpoints, the resistance rates for E. coli were 11.11 and 88.88%, respectively; the resistance rates for P. mirabilis were 22.22 and 77.77%, respectively; all K. pneumoniae isolates (100%) exhibited resistance to fosfomycin according to both criteria; and the resistance rates for S. pseudintermedius were 0 and 20%, respectively (Table 3).
All K. pneumoniae isolates exhibited high levels of resistance to fosfomycin, with MIC values of >256 mg/L (Table 3). One isolate (K. pneumoniae_45) demonstrated intermediate susceptibility to imipenem while showing resistance to all other 24 antibiotics tested; therefore, it was classified as extensively drug-resistant (Table 2).
3.3 Whole-genome sequencing analysis
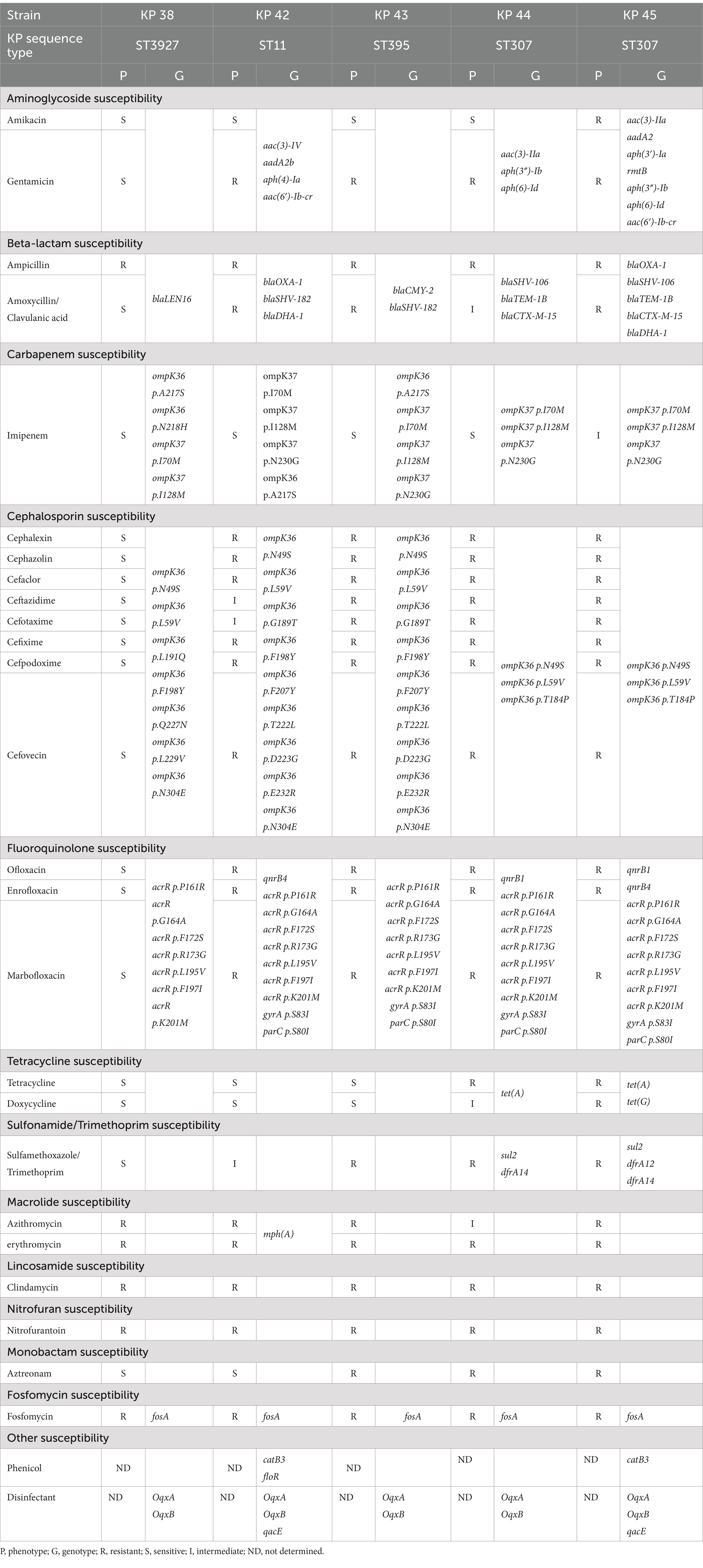
The 5 K. pneumoniae isolates were subjected to WGS analysis to determine their sequence types (ST) and identify antibiotic resistance genes (Table 4). The isolates were classified as ST 307 (n = 2), ST 3927 (n = 1), ST 11 (n = 1), and ST 395 (n = 1) (Table 4).
The isolates harbored multiple antibiotic resistance genes. All isolates carried the fosA gene and exhibited chromosomal mutations in acrR, ompK37, ompK36, parC, and gyrA (Table 4). The 2 ST 307 isolates exhibited a higher number of antibiotic resistance genes than the other ST isolates. These isolates harbored genes conferring resistance to aminoglycoside, beta-lactam, tetracycline, sulfonamide/trimethoprim, fosfomycin, and disinfectants (Table 4). However, none of the isolates harbored genes for resistance to lincosamide, nitrofuran, or monobactam (Table 4).
ESBL genes were also confirmed through WGS, with 4 out of the 5 K. pneumoniae isolates testing positive for ESBLs. The BlaSHV gene was identified in all 4 samples, whereas blaTEM and blaCTX-M co-existed in the ST 307 isolates.
4 Discussion
In this study, we investigated the antibiotic resistance profiles of UTI-causing bacteria isolated from dogs, focusing on their resistance to fosfomycin, and evaluated the clinical application and treatment response in dogs. Fosfomycin is currently recommended for treating UTIs, particularly severe UTIs caused by MDR Enterobacteriaceae in humans (4). Common uropathogens, including MDR isolates, have been demonstrated to exhibit high susceptibility to fosfomycin (20–23). Sabharwal and Sharma et al. reported that isolates with high levels of antimicrobial resistance, including E. coli and P. mirabilis, exhibited the highest susceptibility to fosfomycin (6). Additionally, Sreenivasan et al. identified fosfomycin as the sole oral antibiotic with significant in vitro antimicrobial efficacy against Enterobacteriaceae isolates (24). Consequently, the renewed interest in fosfomycin has prompted an increase in the number of studies investigating its susceptibility and resistance (25).
Variations in susceptibility and resistance rates were observed upon comparing fosfomycin susceptibility test results based on CLSI and EUCAST criteria. According to CLSI breakpoints, the rates of E. coli, P. mirabilis, K. pneumoniae, and S. pseudintermedius resistance to fosfomycin were 11.11, 22.22, 100, and 0%, respectively. Conversely, the resistance rates were 88.88, 77.77, 100, and 20%, respectively, based on EUCAST breakpoints (Table 3). Similarly, in a previous study, susceptibility rates for E. coli, Klebsiella spp., and other Enterobacterales were 92.9, 92.1, and 100% based on CLSI breakpoints. Conversely, susceptibility rates of 85.7, 86.9, and 92.9%, respectively, were reported based on EUCAST breakpoints (26, 27). The discrepancies between these criteria complicate the interpretation of fosfomycin susceptibility and resistance results when relying solely on agar dilutions. Therefore, additional genetic testing is warranted to achieve more accurate results, and establishing standardized criteria to reduce the variability in breakpoints between the two institutions is necessary for further investigation into fosfomycin susceptibility.
Studies on the susceptibility of MRSP to fosfomycin are limited. DiCicco et al. (28) analyzed 31 MRSP strains from dogs and reported a 77% susceptibility rate to fosfomycin according to EUCAST criteria. In our study, 80% (4 of 5) of S. pseudintermedius strains were susceptible to fosfomycin according to EUCAST criteria. Among the 5 strains, 2 were identified as MRSP, 1 of which exhibited resistance to fosfomycin. Further research is required to explore the mechanisms of fosfomycin resistance in S. pseudintermedius despite its anticipated efficacy against MDR strains.
Klebsiella pneumoniae is globally recognized as a significant disease-causing pathogen (29). Given the increasing incidence of K. pneumoniae infections in humans worldwide, the potential for transmission and resistance gene transfer in veterinary medicine cannot be overlooked. In a 2009 study conducted in South Korea, the susceptibility rate of K. pneumoniae from human patients to fosfomycin was 95.2% (30). However, this rate declined to 61.9% in a study by Cho et al., conducted from 2011 to 2013, indicating an increase in resistance (31). In the present study, all 5 K. pneumoniae isolates exhibited high-level resistance to fosfomycin. WGS revealed that the fosA gene was present in the chromosomal DNA of all K. pneumoniae isolates (data not shown). In a previous study by Huang et al., 80% of K. pneumoniae human isolates exhibited resistance to fosfomycin, and chromosomal fosA was identified in 98.8% of cases (32). The fosA gene can be located on either the bacterial chromosome or on a plasmid, with chromosomal fosA being widely recognized as a major determinant of high-level resistance to fosfomycin in various gram-negative species. This chromosomal presence represents a key contributor to the observed high-level resistance (33, 34).
All dogs from which K. pneumoniae was isolated in our study had recurrent UTIs and were repeatedly exposed to multiple antibiotics. While the median duration of antibiotic treatment for all dogs was 2 months, those with K. pneumoniae infections received treatment for a longer duration of 3.5 months, indicating prolonged antibiotic use. Fosfomycin was prescribed to 2 dog patients owing to the lack of alternative oral antibiotic options, resulting in remission in both cases. Despite agar dilution results indicating resistance to fosfomycin in 1 dog, treatment efficacy was observed, suggesting a possible discrepancy between in vitro and in vivo efficacy.
This study had a few limitations. First, the small sample size of bacterial isolates, which were collected from a single center, restricted the generalizability of the findings. Additionally, the unequal distribution of bacterial strains included in the analysis posed challenges in accurately comparing their resistance to fosfomycin. Furthermore, differences in MIC breakpoints for fosfomycin using the agar dilution method resulted in inconsistent results, rendering precise interpretation challenging. Finally, owing to the lack of clinical data on fosfomycin in veterinary medicine, the discussion predominantly relied on comparisons with fosfomycin studies conducted in humans.
In veterinary medicine, the MIC breakpoint for fosfomycin susceptibility testing remains undefined. Although MIC results provide in vitro susceptibility data, actual responses in vivo may differ from expected outcomes. The prediction of in vivo results in dogs is further complicated by the limited understanding of fosfomycin pharmacodynamics and the absence of extensive clinical trials. However, in cases where alternative antibiotic treatments are limited, fosfomycin could be considered an effective alternative, particularly when susceptibility is corroborated by sufficient evidence from urine bacterial culture and quantitative susceptibility testing.
5 Conclusion
To our knowledge, this study is the first to emphasize the importance of evaluating resistance to antibiotics, including fosfomycin, for the effective management of canine UTIs, particularly in cases involving MDR pathogens. This study revealed high levels of antibiotic resistance among UTI pathogens in dogs and highlighted the efficacy of fosfomycin against certain species, such as E. coli, P. mirabilis, and S. pseudintermedius, especially against MDR isolates. However, K. pneumoniae exhibited the highest level of resistance to fosfomycin, harboring multiple antibiotic resistance genes. In conclusion, this study provides key insights into antibiotic resistance in canine UTIs and the potential role of fosfomycin in the treatment of these infections. While fosfomycin shows promise as a treatment option for certain UTI-causing species, the high resistance observed in K. pneumoniae isolates is concerning. Therefore, caution is advised against its indiscriminate use owing to the risk of escalating resistance and transmission between animals and humans.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding author. Paired-end reads of K. pneumoniae isolates from this study have been deposited in the National Center for Biotechnology Information (NCBI) under the Bioproject accession number PRJNA956693.
Ethics statement
The animal studies were approved by Institutional Animal Care and Use Committee (KU24059). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
D-EL: Writing – original draft. J-YH: Writing – original draft. S-WK: Methodology, Writing – original draft. D-YL: Methodology, Writing – original draft. J-HK: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the KU Research Professor Program of Konkuk University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1455021/full#supplementary-material
Abbreviations
AMC, Amoxicillin/clavulanic acid; CLSI, Clinical and Laboratory Standards Institute; ESBL, Extended-spectrum β-lactamase; MDR, Multidrug resistant; MIC, Minimum inhibitory concentration; MLST, Multi Locus Sequence Typing; MRSP, Methicillin-resistant Staphylococcus pseudintermedius; UTIs, Urinary tract infections; WGS, Whole genome sequencing.
Footnotes
References
1. Byron, JK . Urinary tract infection. Vet Clin North Am Small Anim Pract. (2019) 49:211–21. doi: 10.1016/J.CVSM.2018.11.005
2. Olin, SJ, and Bartges, JW. Urinary tract infections treatment/comparative therapeutics. Vet Clin North Am Small Anim Pract. (2022) 52:581–608. doi: 10.1016/j.cvsm.2022.01.002
3. Weese, JS, Blondeau, J, Boothe, D, Guardabassi, LG, Gumley, N, Papich, M, et al. International Society for Companion Animal Infectious Diseases (ISCAID) guidelines for the diagnosis and management of bacterial urinary tract infections in dogs and cats. Vet J. (2019) 247:8–25. doi: 10.1016/j.tvjl.2019.02.008
4. Bader, MS, Loeb, M, and Brooks, AA. An update on the management of urinary tract infections in the era of antimicrobial resistance. Postgrad Med. (2017) 129:242–58. doi: 10.1080/00325481.2017.1246055
5. Hendlin, D, Stapley, EO, Jackson, M, Wallick, H, Miller, AK, Wolf, FJ, et al. Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science. (1969) 166:122–3. doi: 10.1126/science.166.3901.122
6. Sabharwal, ER, and Sharma, R. Fosfomycin: an alternative therapy for the treatment of UTI amidst escalating antimicrobial resistance. J Clin Diagn Res. (2015) 9:DC06–9. doi: 10.7860/JCDR/2015/15227.6951
7. Kuiper, SG, Dijkmans, AC, Wilms, EB, Kamerling, IMC, Burggraaf, J, Stevens, J, et al. Pharmacokinetics of fosfomycin in patients with prophylactic treatment for recurrent escherichia coli urinary tract infection. J Antimicrob Chemother. (2020) 75:3278–85. doi: 10.1093/jac/dkaa294
8. Patel, SS, Balfour, JA, Bryson, HM, and Tromethamine, F. Fosfomycin Tromethamine. Drugs. (1997) 53:637–56. doi: 10.2165/00003495-199753040-00007
9. Zhanel, GG, Walkty, AJ, and Karlowsky, JA. Fosfomycin: a first-line oral therapy for acute uncomplicated cystitis. Can J Infect Dis Med Microbiol. (2016) 2016:2082693. doi: 10.1155/2016/2082693
10. Falagas, ME, Athanasaki, F, Voulgaris, GL, Triarides, NA, and Vardakas, KZ. Resistance to fosfomycin: mechanisms, frequency and clinical consequences. Int J Antimicrob Agents. (2019) 53:22–8. doi: 10.1016/j.ijantimicag.2018.09.013
11. Tulara, NK . Nitrofurantoin and fosfomycin for extended spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae. J Glob Infect Dis. (2018) 10:19–21. doi: 10.4103/jgid.jgid_72_17
12. Harada, K, Shimizu, T, Kawaguchi, K, Furuhashi, T, and Ishihara, G. Urinary pharmacokinetic and pharmacodynamic profiles of fosfomycin against extended-spectrum β-lactamase-producing escherichia coli with canine ex vivo modeling: a pilot study. Antibiotics. (2020) 9:230. doi: 10.3390/antibiotics9050230
13. Jariyapamornkoon, N, Patthanachai, K, and Suanpairintr, N. Plasma and urine pharmacokinetics of oral fosfomycin tromethamine in dogs. Vet Sci. (2023) 10:391. doi: 10.3390/vetsci10060391
14. Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing. Pennsylvania: Clinical and Laboratory Standards Institute (2021).
15. EUCAST . The European committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 13.0 (2023). Available at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_50_Breakpoint_Table_01.pdf (Accessed June 28, 2023).
16. Karlowsky, JA, Lagacé-Wiens, PRS, Laing, NM, Baxter, MR, Adam, HJ, and Zhanel, GG. Susceptibility of clinical isolates of Escherichia coli to fosfomycin as measured by four in vitro testing methods. J Clin Microbiol. (2020) 58:e01306–20. doi: 10.1128/JCM.01306-20
17. Karlowsky, JA, Baxter, MR, Walkty, AJ, Lagacé-Wiens, PRS, Bay, D, Adam, HJ, et al. In vitro activity of fosfomycin against bacterial pathogens isolated from urine specimens in Canada from 2007 to 2020: CANWARD surveillance study. J Antimicrob Chemother. (2022) 77:3035–8. doi: 10.1093/jac/dkac275
18. Hyeon, JY, Helal, ZH, Polkowski, R, Vyhnal, K, Mishra, N, Kim, J, et al. Genomic features of Salmonella enterica subspecies houtenae serotype 45:g,z51: isolated from multiple abdominal abscesses of an African fat-tailed gecko, United States, 2020. Antibiotics. (2021) 10:1322. doi: 10.3390/antibiotics10111322
19. Bankevich, A, Nurk, S, Antipov, D, Gurevich, AA, Dvorkin, M, Kulikov, AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:455–77. doi: 10.1089/cmb.2012.0021
20. Banerjee, S, Sengupta, M, and Sarker, TK. Fosfomycin susceptibility among multidrug-resistant, extended-spectrum beta-lactamase-producing, carbapenem-resistant uropathogens. Indian J Urol. (2017) 33:149–54. doi: 10.4103/iju.IJU_285_16
21. Mittal, S, Sharma, M, and Chaudhary, U. Fosfomycin use in multi drug resistant uropathogenic Escherichia coli. Infect Disord Drug Targets. (2015) 15:196–201. doi: 10.2174/1871526515666150916141907
22. Patwardhan, V, and Singh, S. Fosfomycin for the treatment of drug-resistant urinary tract infections: potential of an old drug not explored fully. Int Urol Nephrol. (2017) 49:1637–43. doi: 10.1007/s11255-017-1627-6
23. Behera, B, Mohanty, S, Sahu, S, and Praharaj, A. In vitro activity of fosfomycin against multidrug-resistant urinary and nonurinary gram-negative isolates. Indian J Crit Care Med. (2018) 22:533–6. doi: 10.4103/ijccm.IJCCM_67_18
24. Sreenivasan, S, Kali, A, Pravin Charles, MV, and Kunigal, S. Evaluation of in vitro susceptibility of fosfomycin among Enterobacteriaceae isolates from urine cultures: a study from Puducherry. J Lab Physicians. (2019) 11:249–52. doi: 10.4103/jlp.jlp_27_19
25. Aghamali, M, Sedighi, M, Zahedi bialvaei, A, Mohammadzadeh, N, Abbasian, S, Ghafouri, Z, et al. Fosfomycin: mechanisms and the increasing prevalence of resistance. J Med Microbiol. (2019) 68:11–25. doi: 10.1099/jmm.0.000874
26. Bir, R, Mohapatra, S, Kumar, A, Arif, N, Tyagi, S, Gautam, H, et al. Comparison of in-vitro susceptibility of fosfomycin against drug resistant uropathogens by various susceptibility testing methods. Res Sq. (2022) 1:84. doi: 10.21203/RS.3.RS-414884/V1
27. Bir, R, Mohapatra, S, Kumar, A, Arif, N, Tyagi, S, Ak, AP, et al. Genomic analysis of fosfomycin resistance in multi-drug resistant uropathogens and comparison of in-vitro susceptibility methods uropathogens. Iran J Microbiol. (2022) 14:636–44. doi: 10.18502/ijm.v14i5.10956
28. DiCicco, M, Weese, S, Neethirajan, S, Rousseau, J, and Singh, A. Fosfomycin susceptibility of canine methicillin-resistant Staphylococcus pseudintermedius isolates. Res Vet Sci. (2014) 96:251–3. doi: 10.1016/j.rvsc.2014.02.004
29. Juan, CH, Chuang, C, Chen, CH, Li, L, and Lin, YT. Clinical characteristics, antimicrobial resistance and capsular types of community-acquired, healthcare-associated, and nosocomial Klebsiella pneumoniae bacteremia. Antimicrob Resist Infect Control. (2019) 8:1. doi: 10.1186/s13756-018-0426-x
30. Lee, SY, Park, YJ, Yu, JK, Jung, S, Kim, Y, Jeong, SH, et al. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother. (2012) 67:2843–7. doi: 10.1093/jac/dks319
31. Cho, YH, Jung, SI, Chung, HS, Yu, HS, Hwang, EC, Kim, SO, et al. Antimicrobial susceptibilities of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in health care-associated urinary tract infection: focus on susceptibility to fosfomycin. Int Urol Nephrol. (2015) 47:1059–66. doi: 10.1007/s11255-015-1018-9
32. Huang, L, Cao, M, Hu, Y, Zhang, R, Xiao, Y, and Chen, G. Prevalence and mechanisms of fosfomycin resistance among KPC-producing Klebsiella pneumoniae clinical isolates in China. Int J Antimicrob Agents. (2021) 57:106226. doi: 10.1016/j.ijantimicag.2020.106226
33. Ito, R, Mustapha, MM, Tomich, AD, Callaghan, JD, McElheny, CL, Mettus, RT, et al. Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. MBio. (2017) 8:e00749-17. doi: 10.1128/mBio.00749-17
Keywords: antibiotic susceptibility test, dog, fosfomycin, Klebsiella pneumoniae, multidrug resistance, urinary tract infection
Citation: Lee D-E, Hyeon J-Y, Kang S-W, Lee D-Y and Kim J-H (2024) Antibiotic efficacy and resistance patterns of urinary tract infection-causing bacteria in dogs and resistome of multidrug-resistant Klebsiella pneumoniae via whole genome sequencing in South Korea. Front. Vet. Sci. 11:1455021. doi: 10.3389/fvets.2024.1455021
Edited by:
Carmel T. Mooney, University College Dublin, IrelandReviewed by:
Sunghyun Yoon, National Center for Toxicological Research (FDA), United StatesTariq Jamil, Friedrich-Loeffler-Institute, Germany
Copyright © 2024 Lee, Hyeon, Kang, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung-Hyun Kim, anVuZ2h5dW5Aa29ua3VrLmFjLmty
†These authors have contributed equally to this work
 Da-Eun Lee
Da-Eun Lee Ji-Yeon Hyeon
Ji-Yeon Hyeon Seok-Won Kang2
Seok-Won Kang2 Dong-Yeop Lee
Dong-Yeop Lee Jung-Hyun Kim
Jung-Hyun Kim