- 1College of Life Science, Nanyang Normal University, Nanyang, China
- 2Department of Zoology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 3Department of Anatomy, Faculty of Veterinary Science, University of Agriculture, Faisalabad, Pakistan
- 4Department of Veterinary Medicine, Southwest University, Chongqing, China
Introduction: Intestinal dysfunction poses a severe problem by preventing the digestion and absorption of nutrients. The gut, being the most vital organ for these processes, plays a crucial role in ensuring our body receives the nutrients it needs. We explored the mitigating effect of Morchella esculenta polysaccharides (MEP) on intestinal injury induced by lipopolysaccharides (LPS) through the modulation of intestinal flora.
Methods: For this purpose, Kunming mice (KM) were divided into three groups, namely, PC, PM, and PY. Group PY was treated with MEP, while groups PM and PY were induced with LPS.
Results: The results showed that weight loss in the PM group was significantly greater than that in the PY group (P < 0.05), and the organ indexes of the lung and spleen in the PM group were significantly higher than those in the PC (P < 0.01) and PY (P < 0.05) groups. LPS caused severe injuries in KM mice in the PM group, characterized by broken villi. However, MEP treatment could alleviate this damage in the PY group, resulting in relatively intact villi. The serum analysis showed that tumor necrosis factor alpha (TNF-ɑ) (P < 0.01), interleukin 6 (IL-6) (P < 0.01), and 3,4-methylenedioxyamphetamine (MDA) (P < 0.05) levels were significantly higher in the PM group, while IL-10 (P < 0.001), superoxide dismutase (SOD) (P < 0.01) and glutathione peroxidase (GSH-Px) (P < 0.01) were significantly lower in that group. Interestingly, supplementation with MEP could lower the levels of TNF-ɑ, IL-10, IL-6, MDA while increasing the levels of superoxide dismutase (SOD) (P < 0.01) and GSH-Px. The gut microbiota analysis yielded 630,323 raw reads and 554,062 clean reads, identifying 3,390 amplicon sequencing variants (ASVs). One phylum and five genera were notably different among animal groups, including Escherichia_Shigella, Limosilactobacillus, unclassified_Geminicoccaceae, unclassified_Rhodobacteraceae, and Parabacteroides (P. distasonis).
Discussion: In conclusion, we found that MEP could mitigate the intestinal damage caused by LPS by modulating the inflammatory response, oxidative resistance, and intestinal flora of KM mice. Our results may provide insights into novel treatment options for intestine-related diseases.
Introduction
The gut is the most important organ for the digestive absorption of nutrients. Various enzymes in the villi contribute to digestion, while intestinal epithelial cells combined with tight junction proteins promote absorption. Intestinal integrity is crucial for host health, and compromised intestines have been reported in individuals with colitis as well as those affected by infectious bacterial or viral intestinal diseases (1, 2). Gut flora refers to the microorganisms inhabiting the intestines, including commonly known bacteria, viruses, and fungi, as well as millions of archaea and protozoa (3, 4). In individuals, the intestinal microbiota is positively related to the host's nutrition and xenobiotic metabolism, immunity, and body homeostasis (3, 5, 6). Intestinal epithelial cells (IECs) and the intestinal microbiota function together as a protective barrier against toxins and pathogens (7). An imbalance in the structure of gut flora may lead to metabolic disorders in the host (8), and dysbiosis is commonly observed in various diseases, including lung disease, kidney disease, and diarrhea (9–11). There is an urgent need for novel and effective therapies for treating gastrointestinal diseases.
The well-known endotoxin lipopolysaccharide is distributed in the external panniculus of Gram-negative microbes, guaranteeing its bacterial structure and function integrity (12). The bacteria release LPS after multiplication, death, and lysis. The released lipopolysaccharides (LPS) cause not only an inflammatory response but also fever, septic shock, diarrhea, organ damage, and even serious physiological effects (13, 14). Among these complications, sepsis is a common disease with high mortality, which has limited therapeutic options for its treatment (13). Additionally, dysbiosis caused by LPS has been reported in several investigations (15–17). Polysaccharides consist of numerous polymer carbohydrates that have important biological functions, such antioxidant effects, antitumor activities, anti-microbial properties, and the ability to regulate the immune system (18, 19). Morchella esculenta (L.) Pers is a well-known edible and medicinal fungus, which is recorded in the Chinese medicine masterpiece Compendium of Materia Medica (also known as Pen-tsao Kan-mu) as having the effect of “benefiting the intestines and stomach, helping food digestion and absorption”. Polysaccharides from Morchella esculenta (L.) Pers (MEP) are highly valued healthcare products known for their biological activities, including oxidation resistance (20), anti-inflammatory effects, and regulatory flora properties (43). However, there is only a limited understanding regarding the effect of MEP on mice challenged with LPS. Hence, we performed this trial to understand the remission effect of MEP on animals induced by LPS through the modulation of intestinal flora.
Materials and methods
Animals
A total of 36 Kunming (KM) mice, aged 4 weeks (21.50 ± 0.67 g), with equal numbers of male and female animals were used in this study. They were obtained from the Experimental Animal Center at Yangzhou University (Yangzhou, China). All KM mice were divided into three groups, namely PC (the control group), PM (the model group), and PY (the MEP group), after acclimatization for 3 days. The mice in the PY group were supplemented with MEP (100 mg/kg) via intragastric administration for 14 days, while KM mice in the other groups were given the same volume of normal saline. On the 15th day, the KM mice in the PM and PY groups were intraperitoneally induced with LPS (10 mg/kg, Acmec Biochemical Technology Co., Ltd., Shanghai, China). Then, all KM mice were euthanized the following day to collect serum, organs, and large intestines (21). The daily weight of KM mice and organs was documented and the organ index was calculated.
Extraction and purification of MEP
First, 500 g of Morchella esculenta (L.) Pers was decolored and defatted with 95% ethanol (2,000 mL) twice under reflux for 1 h each time. After reflux, ethanol was removed, and the residue was dried at 60°C. The dried residue was extracted by boiling water (4,000 mL) twice for 1.5 h each time. Finally, the extract was filtered, combined, and concentrated. After cooling to room temperature, the concentration was adjusted to 500 mL by deionized water and then 90% ethanol (v/v) was added until the total percentage of ethanol in the solution reached 80%. The mixture was kept at 4°C overnight and then filtered to give the total crude Morchella esculenta (L.) Pers polysaccharides (MEPct). The MEPct solution (0.1 g/mL) was prepared using deionized water, and the proteins in the MEPct solution were sequentially removed five times using the Sevag method. The deproteined solution was concentrated to 500 mL under vacuum and then freeze-dried at −20°C for 1 week to obtain the total purified MEP.
Histopathological analysis
Large intestine tissues obtained from KM mice were fixed in paraformaldehyde for further hematoxylin and eosin (H&E) staining (Pinuofei Biological, Wuhan, China). Subsequently, the pathological examination was carried out using a microscope (Olympus BX46, Japan) following the methodology of a previous study (22).
Antioxidant ability and inflammatory factor in KM mice
Serum samples obtained from KM mice were tested for indexes of antioxidant abilities and inflammatory factors [tumor necrosis factor alpha (TNF-ɑ), interleukin 6 (IL-6), IL-10, IL-1β, superoxide dismutase (SOD), total antioxidant capacity (T-AOC), methylenedioxyamphetamine (MDA), and glutathione peroxidase (GSH-Px)] using assay kits provided by the Jiancheng Bioengineering Research Institute and Solarbio Life Science, following the previously described methods (23).
Rectal flora sequencing and analysis
A total of nine rectum samples from KM mice were used for DNA extraction, and amplification of the V3–V4 variable region of the 16S rRNA gene was conducted (24). The generated products were then processed for library construction using the Hieff NGS Ultima Pro DNA Library Prep Kit (Yeasen, China) and sequenced via the Bioyi Biotechnology Illumina NovaSeq platform (Wuhan, China). The sequenced raw data were subjected to quality control via Trimmomatic (v0.33) (25), Cutadapt (1.9.1) (26), and QIIME2 (27). Then, the filtered data were used for amplicon sequence variant (ASV) analysis (28) and taxonomy annotation.
Statistical analysis
Data regarding KM mice were presented as means ± standard deviation and analyzed using SPSS version 26.0. Alpha and beta diversity analyses were performed by employing QIIME (29, 30). Significant differences among the three KM animal groups were determined by analysis of variance (ANOVA), linear discriminant analysis (30), and Metastats (31). The network analysis of the intestinal flora in KM mice was performed using Spearman's rank correlation analysis (32). A p-value of < 0.05 was considered statistically significant.
Results
The effect of MEP on weight and organ indexes in KM mice induced by LPS
Although the mean body weight of the KM mice was nearly the same in the three groups, the weight loss in PM mice was dramatically higher than that in PY mice (P < 0.05) (Figure 1A). The organ indexes of the lung and spleen of KM mice in the PM group were higher than those in the PC (P < 0.01) and PY (P < 0.05) groups (Figure 1B). LPS caused severe injuries in KM mice in the PM group, characterized by broken villi, while MEP could alleviate this damage in the PY group with relatively intact villi (Figure 1C).
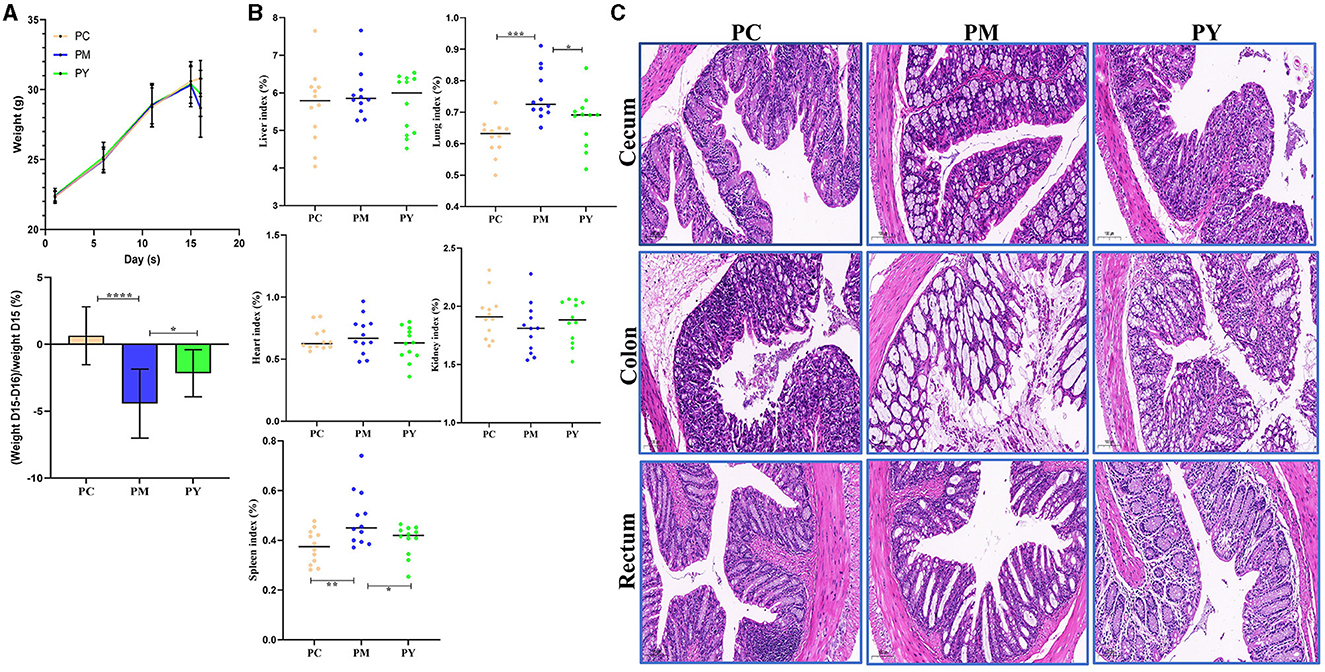
Figure 1. MEP-remitted damages in KM mice induced by LPS. (A) Body weight, (B) organ index, (C) pathological analysis. Scale bar 50 μm. Data are shown as mean ± SEM (n = 12). Significance is marked as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
The effect of MEP on oxidation resistance and inflammatory response in KM mice induced by LPS
Serum analysis showed that TNF-ɑ (P < 0.01), IL-6 (P < 0.01) and MDA (P < 0.05) were significantly higher in PM mice, while IL-10 (P < 0.001), SOD (P < 0.01), and GSH-Px (P < 0.01) were markedly lower in that group. Interestingly, supplementation with MEP decreased TNF-ɑ, IL-10, IL-6, and MDA levels and increased SOD (P < 0.01) and GSH-Px levels (Figure 2).
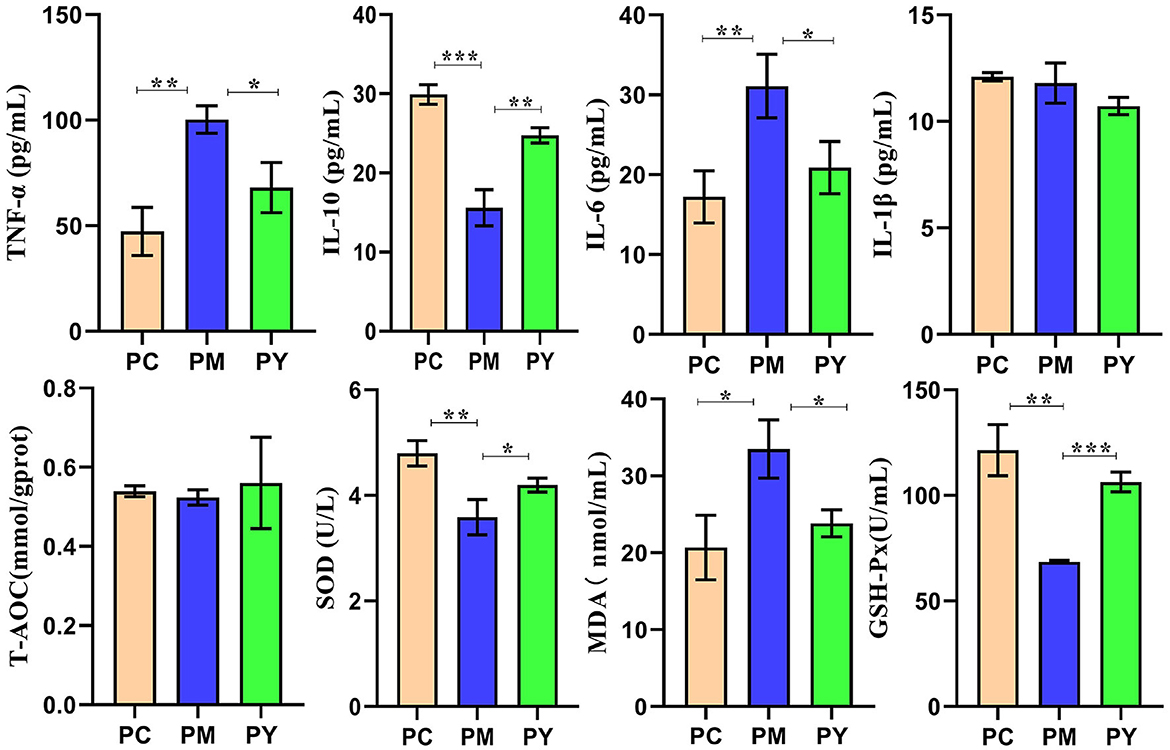
Figure 2. MEP promoted oxidation resistance and decreased inflammatory response in KM mice challenged by LPS. Data are exhibited as mean ± SEM (n = 3). Significance is marked as *P < 0.05, **P < 0.01, and ***P < 0.001.
The effect of MEP on gut flora of KM mice induced by LPS
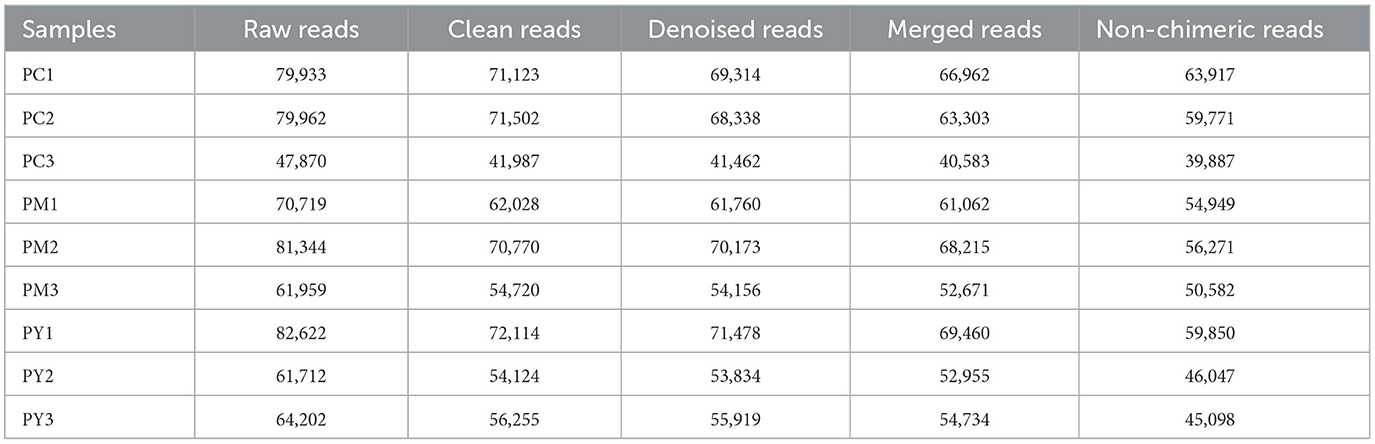
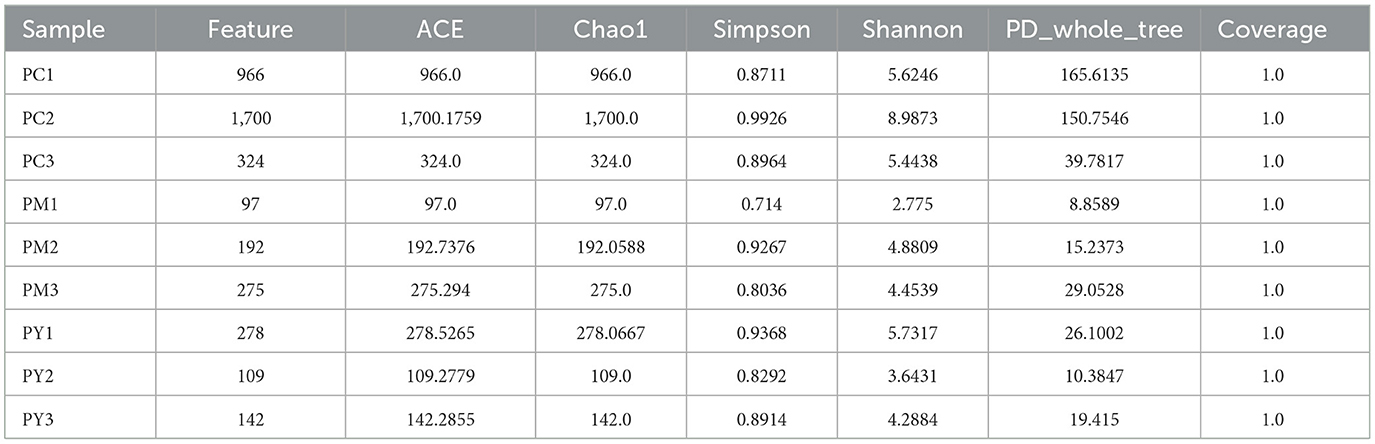
Over 47,870 raw and 41,987 filtered reads were found in each sample of KM mice (Table 1), with 3,390 ASVs found in the three animal groups (Figure 3A). The curves of Shannon index, rarefaction, and rank abundance in KM mice were flat and displayed saturated curves (Figures 3B–D), demonstrating that the sequencing depth was sufficient in the current experiment. No marked difference in the α-diversity was observed between the groups (Table 2, Figure 3E).
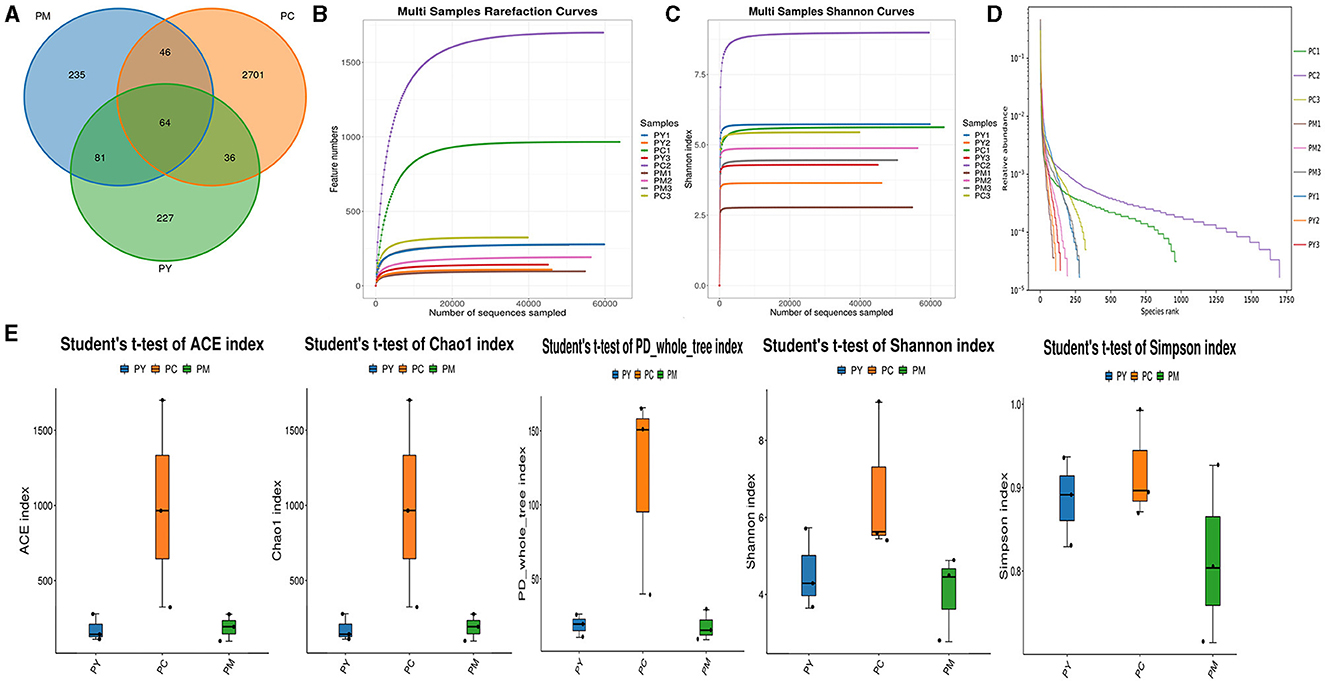
Figure 3. Intestinal flora structure and α-diversity analysis. (A) Venn map, (B) rarefaction curve, (C) Shannon index curve, (D) rank abundance curve, and (E) Statistical analysis of α-diversity.
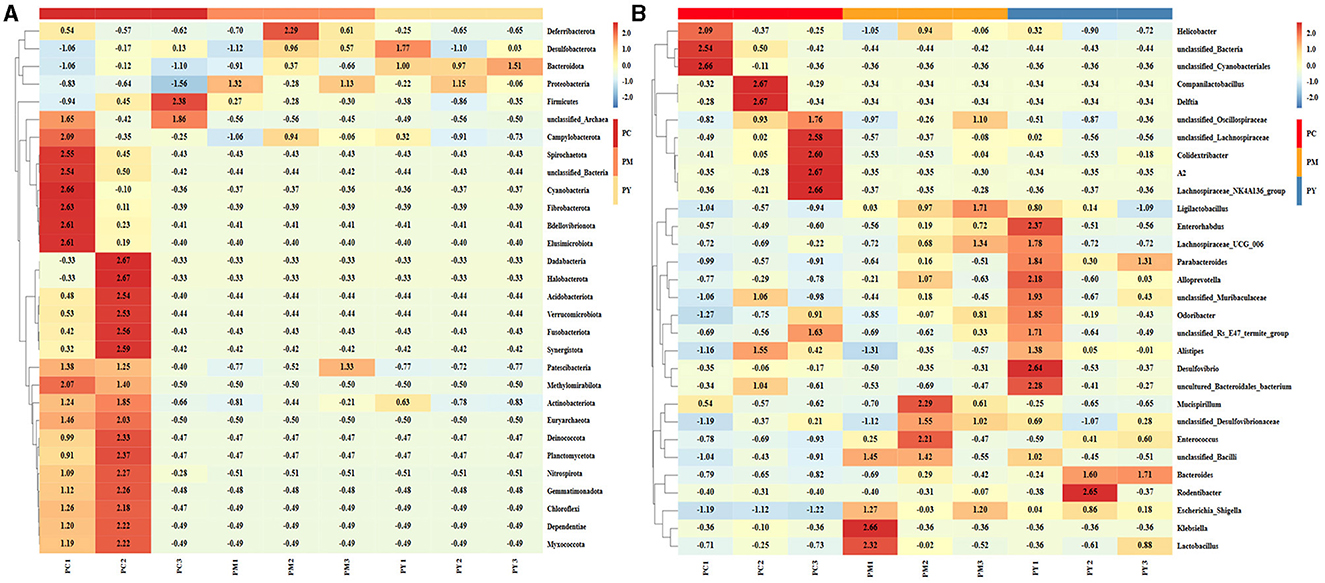
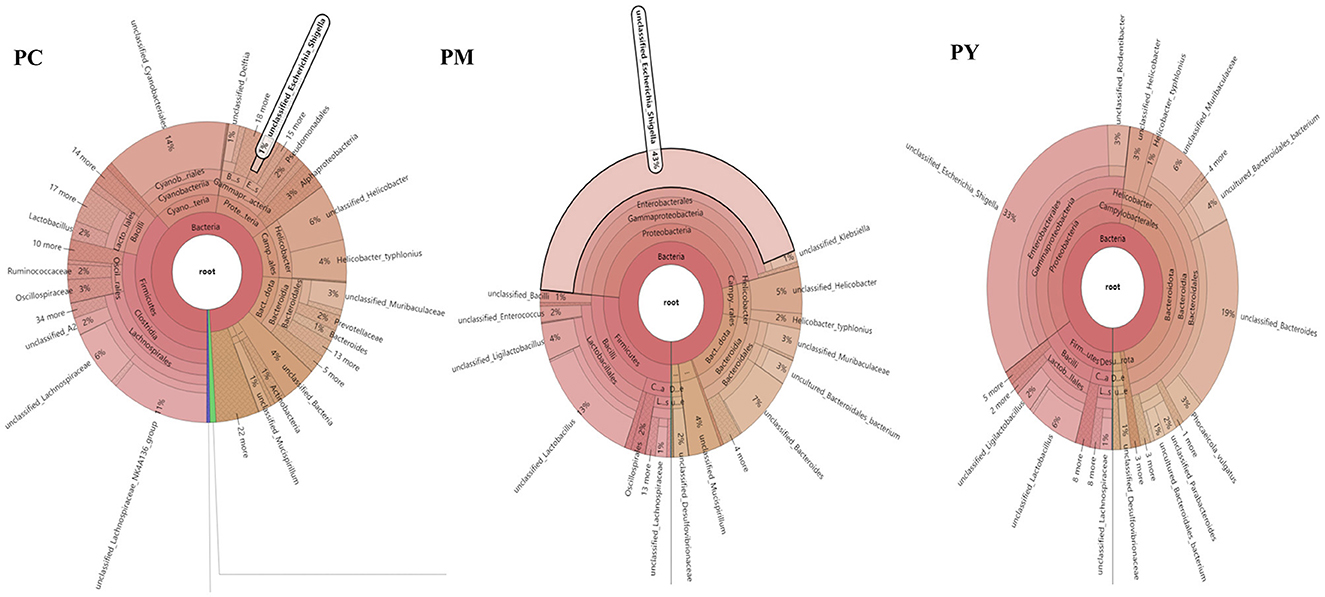
The predominant phyla in different groups were as follows: Firmicutes (phylum Firmicutes) (37.97%), Cyanobacteria (14.54%), and Proteobacteria (12.85%) in the PC group; Proteobacteria (44.88%), Firmicutes (26.36%), and Bacteroidota (15.71%) in the PM group; and Bacteroidota (39.70%), Proteobacteria (35.98%), and Firmicutes (16.24%) in the PY group (Figure 4A). At the class level, Clostridia (28.51%), Cyanophyceae (14.50%), and Bacteroidia (10.32%) were mainly observed in the PC group, while Gammaproteobacteria, Bacilli (Bacillus subtilis), and Bacteroidia were predominantly found in the PM (44.88, 20.58, and 15.71%) and PY (35.98, 39.70, and 11.45%) groups (Figure 4B). At the order level, Lachnospirales (21.67%), Cyanobacteriales (14.36%), and Campylobacterales (10.77%) were predominantly observed in the PC group, while Enterobacterales, Lactobacillales, and Bacteroidales were the dominant orders in the PM (44.87, 18.90, and 15.71%) and PY (35.97, 39.70, and 10.03%) groups (Figure 4C). At the family level, Lachnospiraceae (21.65%), unclassified_Cyanobacteriales (14.24%), and Helicobacteraceae (10.72%) were primarily dominant in the PC group, while Enterobacteriaceae, Lactobacillaceae, and Bacteroidaceae were the main families in the PM (44.30, 17.10, 7.35%) and PY (33.07, 8.86, 22.32%) groups (Figure 4D). At the genus level, unclassified_Cyanobacteriales (14.24%), Lachnospiraceae_NK4A136_group (12.04%), and Helicobacter (10.72%) were predominantly found in the PC group, while Escherichia_Shigella, Lactobacillus, and Bacteroides were the main genera in the PM (43.04, 13.23, and 7.34%) and PY (33.07, 6.64, and 22.32%) groups (Figure 4E). The heat map showed a higher abundance of phyla of unclassified_Archaea, Acidobacteriota, Nitrospirota, and Chloroflexi in the PC group, Proteobacteria in the PM group, and Bacteroidota in the PY group (Figure 5A). At the genus level, a higher abundance of Lachnospiraceae_NK4A136_group in the PC group, Ligilactobacillus and Escherichia_Shigella in the PM group, and Parabacteroides, Bacteroides, and Escherichia_Shigella in the PY group were observed (Figure 4B). The Krona analysis indicated that the main genera in each group were as follows: unclassified_Cyanobacteriales, unclassified_ Lachnospiraceae_NK4A136_group, unclassified_Lachnospiraceae, and unclassified_Helicobacter in the PC group; unclassified_Escherichia_Shigella, unclassified_Lactobacillus, and unclassified_Bacteroides in the PM group; and unclassified_Escherichia_Shigella, unclassified_Muribaculaceae, and unclassified_Lactobacillus in the PY group, respectively (Figure 5).
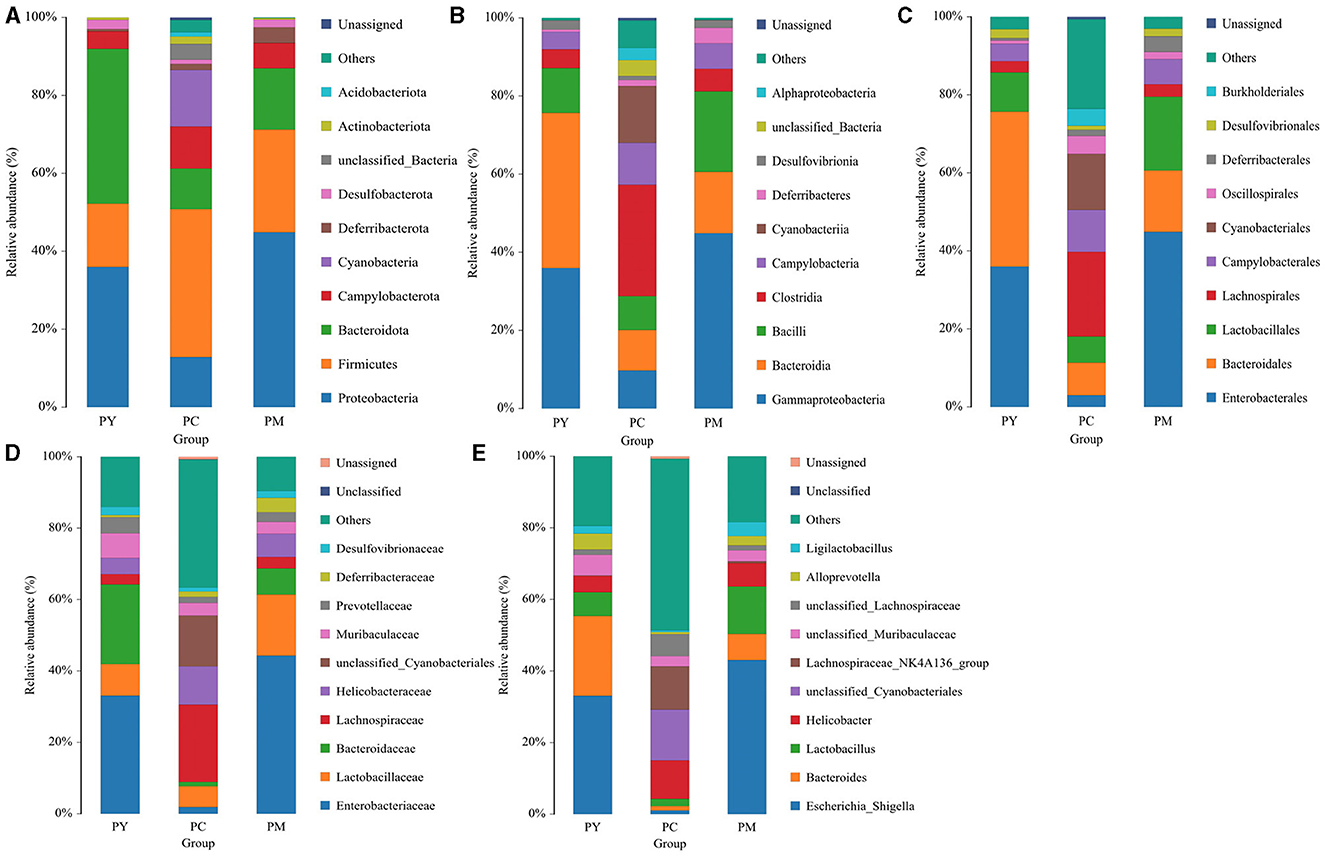
Figure 4. Comparing intestinal flora of KM mice in different taxa: (A) phylum, (B) class, (C) order, (D) family, and (E) genera.
Beta diversity analysis of intestinal flora
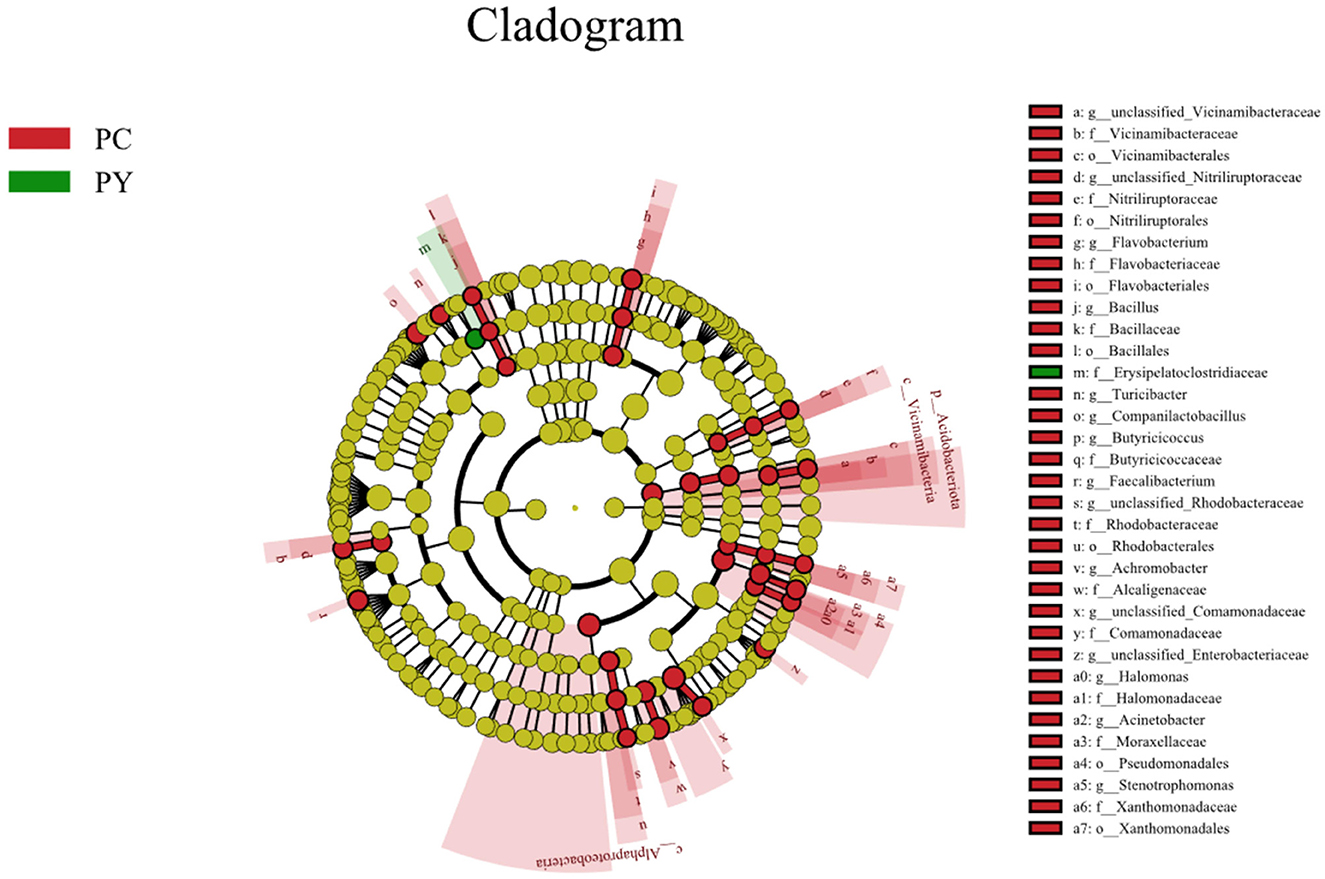
Beta diversity analysis showed that LPS-challenged KM mice were significantly distant from PC mice. However, the distance between PY and PC mice was shorter than that between PM and PC mice (Figure 6). The linear discriminant analysis effect size (LEfSe) revealed that taxa o_Pseudomonadales (P < 0.05), f_Moraxellaceae (P < 0.05), f_Butyricicoccaceae (P < 0.05), g_Butyricicoccus (P < 0.05), f_Alcaligenaceae (P < 0.05), g_Achromobacter (P < 0.05), g_unclassified_Enterobacteriaceae (P < 0.05), p_Acidobacteriota (P < 0.05), c_Vicinamibacteria (P < 0.05), o_Vicinamibacterales (P < 0.05), f_Vicinamibacteraceae (P < 0.05), g_unclassified_Vicinamibacteraceae (P < 0.05), o_Nitriliruptorales (P < 0.05), f_Nitriliruptoraceae (P < 0.05), g_unclassified_Nitriliruptoraceae (P < 0.05), o_Flavobacteriales (P < 0.05), f_Flavobacteriaceae (P < 0.05), g_Flavobacterium (P < 0.05), o_Bacillales (P < 0.05), f_Bacillaceae (P < 0.05), g_Bacillus (P < 0.05), g_Turicibacter (P < 0.05), g_Companilactobacillus (P < 0.05), g_Faecalibacterium (P < 0.05), c_Alphaproteobacteria (P < 0.05), o_Rhodobacterales (P < 0.05), f_Rhodobacteraceae (P < 0.05), g_unclassified_Rhodobacteraceae (P < 0.05), f_Comamonadaceae (P < 0.05), g_unclassified_Comamonadaceae (P < 0.05), f_Halomonadaceae (P < 0.05), g_Halomonas (P < 0.05), g_Acinetobacter (P < 0.05), o_Xanthomonadales (P < 0.05), f_Xanthomonadaceae (P < 0.05), and g_Stenotrophomonas (P < 0.05) were higher in the PC group, while f_Erysipelatoclostridiaceae (P < 0.05) was higher in the PY group (Figure 7). An ANOVA showed that Bacteroidota was significantly higher in the PY group (P < 0.05) (Figure 8A). At the genus level, the abundance of Escherichia_Shigella in the PC group was markedly lower compared to the PM (P < 0.01) and PY (P < 0.05) groups. Limosilactobacillus (P < 0.01), unclassified_Geminicoccaceae (P < 0.0001), and unclassified_Rhodobacteraceae (P < 0.05) were significantly higher in the PC group. Parabacteroides in the PY group was significantly higher compared to the PM (P < 0.05) and PC (P < 0.01) groups (Figure 8).
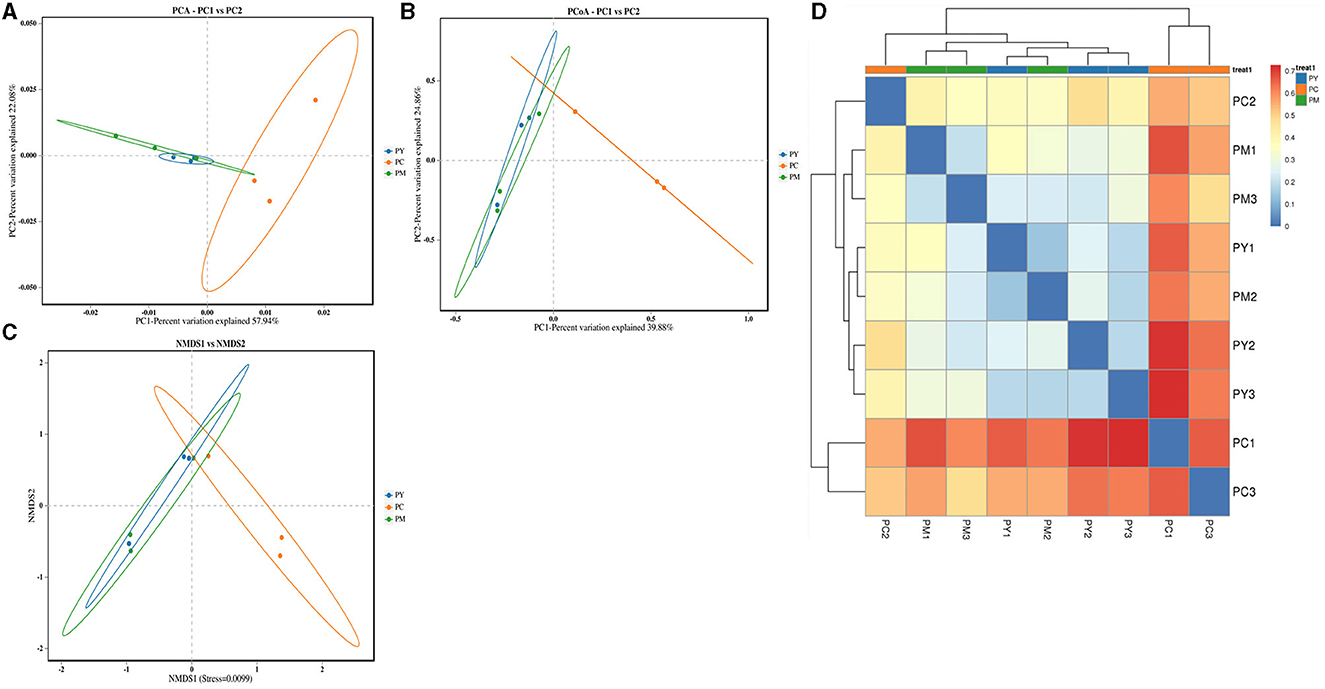
Figure 6. Beta diversity analysis of intestinal flora: (A) PCA, (B) PCoA, (C) NMDS, and (D) clustering heat map.
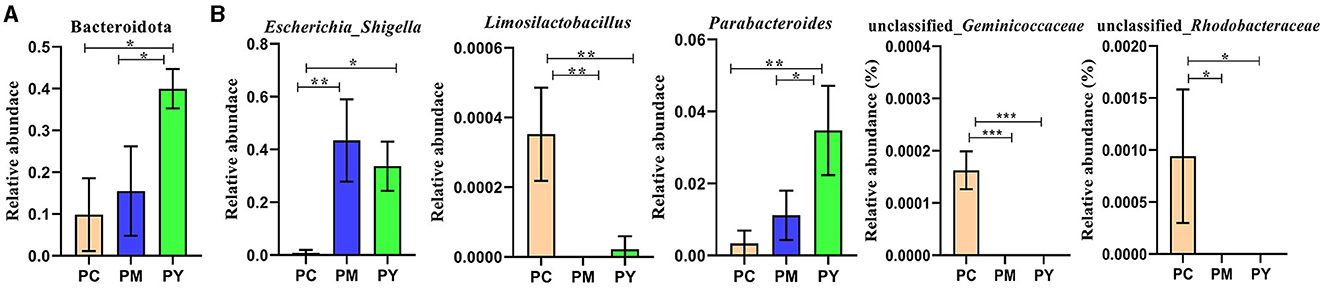
Figure 8. ANOVA of intestinal flora: (A) phylum and (B) genera. Significance is presented as *p < 0.05, **p < 0.01, and ***p < 0.001; data are presented as mean ± SEM (n = 3).
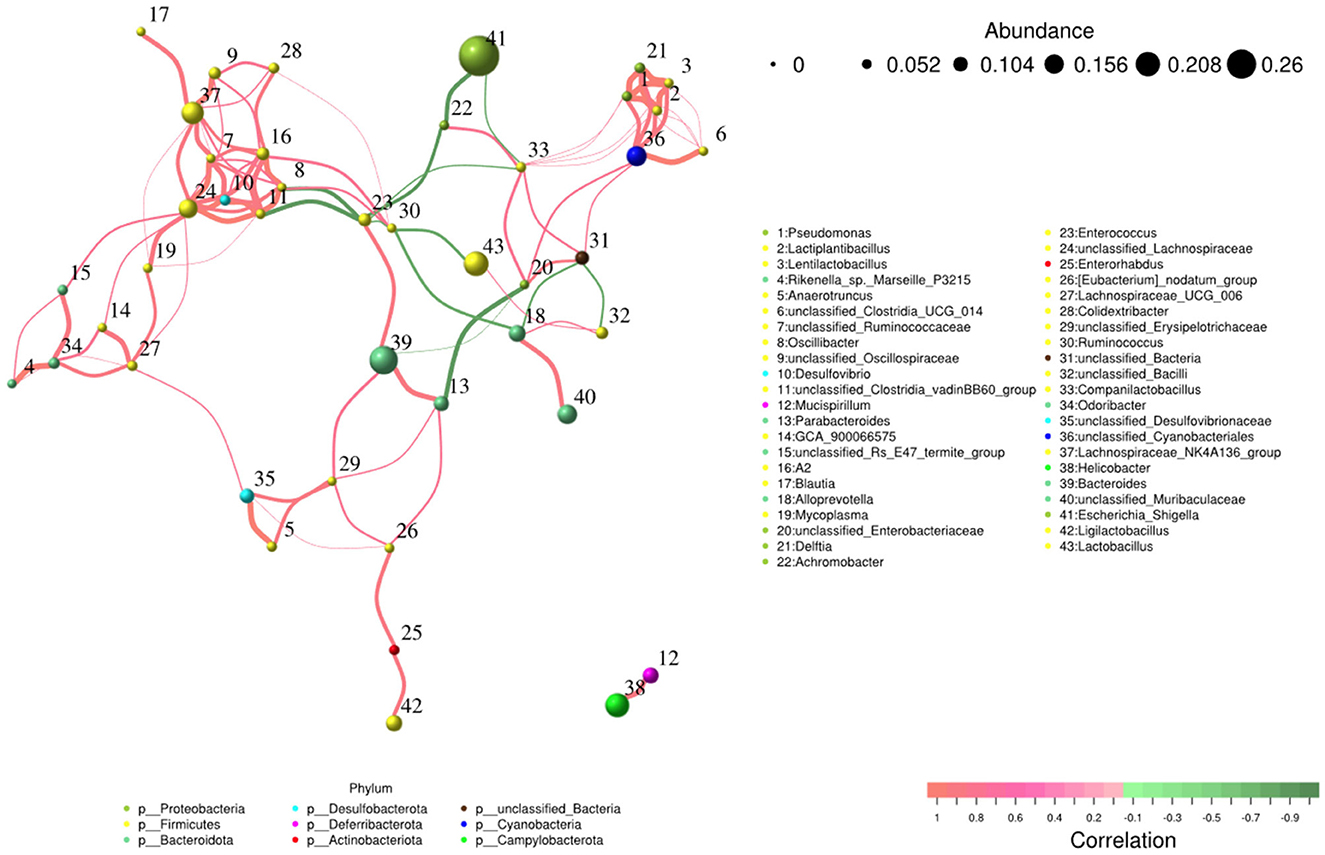
Network and function analysis of intestinal flora
The network analysis revealed that Escherichia_Shigella genus negatively contributed to the gut flora, while Lachnospiraceae_NK4A136_group, Bacteroides, unclassified_Lachnospiraceae, and unclassified_Cyanobacteriales genera made positive contributions to the gut flora (Figure 9).
Discussion
Plant and microorganism extracts are natural products with various biological activities such as immunoregulation, anti-inflammatory effects, oxidative stress reduction, and liver protection (33). In this study, we found that MEP could alleviate body weight loss in KM mice induced by LPS by maintaining the gut integrity (Figure 1). Consistent with previous studies reporting splenomegaly in animals treated with LPS (34, 35), a higher spleen index was observed in the PM group. However, MEP significantly decreased it (P < 0.05) (Figure 1B). Previous studies have also found an association between splenomegaly and IL-6 and TNF-α (35). Therefore, we examined the levels of inflammatory factors in the serum of KM mice. Similar to previous studies demonstrating higher levels of TNF-α and IL-6 and lower levels of IL-10 in LPS-induced animals (36, 37), we observed comparable results for these pro-inflammatory and anti-inflammatory cytokines in this study. Interestingly, MEP significantly reduced inflammation by notably decreasing TNF-α and IL-6 levels and increasing IL-10 levels in PY mice. The antioxidant system and reactive oxygen species are the two side effects of host health and their imbalance is associated with the disease (38). Oxidative damage is a well-known consequence of LPS exposure (39, 40), and SOD, GSH-Px, and MDA are indicators affected by LPS (16). Higher levels of MDA and lower levels of SOD and GSH-Px in PM animals were consistent with previous results (16, 40). However, MEP could reduce oxidative damage by regulating these enzymes in the PY group (Figure 2).
In addition to inflammatory responses and oxidative injuries, dysbiosis was also observed in animals challenged with LPS (15, 16, 23). Next, we performed the gut microbiota analysis of KM mice and obtained 630,323 raw reads and 55,462 clean reads. These reads were identified with 3,390 ASVs, and 63 ASVs were shared among the KM mouse groups (Figure 3). No marked difference in α-diversity was observed between the KM groups, which is consistent with the findings in people receiving synbiotics (24) and liver-damaged mice (23). However, this is contrary to the observations in antibiotic-treated mice (21) and obese animals (44). LPS altered the abundance of bacteria across different taxa, and MEP were able to partially restore the microbiota structure in KM mice (Figures 4, 5, 10). In a healthy host, the primary phyla of microbiota are Firmicutes and Bacteroidetes (3). In this study, MEP increase the abundance of those two phyla in the PY group (55.96%) compared with the PM group (42.06%). Similar results were found at the genus level; LPS increase the abundance of Escherichia_Shigella (43.04%), while KM mice treated with MEP had a lower abundance of this genus. The proteobacteria Escherichia Shigella is a conditioned pathogen leading to intestine diseases (45). Furthermore, we explored the significant variations in phyla and genera among KM groups, detecting one phylum and five genera (Figure 8). These genera were Escherichia_Shigella, Limosilactobacillus, unclassified_Geminicoccaceae, unclassified_Rhodobacteraceae, and Parabacteroides. The network analysis revealed that Escherichia_Shigella was an important genus that negatively interacted with the gut flora (Figure 9). Species of Limosilactobacillus have been commercialized for use in probiotics (41), and a previous study found a lower abundance of Limosilactobacillus in heat-stressed birds (42). Interestingly, KM mice supplemented with MEP increased the abundance of this genus. These results suggest that MEP could regulate the intestinal microbiota in KM mice. However, due to species and strain differences, all the results and findings in this study require further research on other animal strains and species.
Conclusion
In conclusion, our study demonstrates that MEP have the potential to alleviate intestinal damage induced by LPS in KM mice. By modulating inflammatory responses, enhancing oxidation resistance, and restoring intestinal flora, MEP show promise as a therapeutic agent in mitigating intestinal-related diseases. These findings offer valuable insights into the development of novel treatment options aimed at preserving gut health and combating the detrimental effects of LPS-induced damage.
Data availability statement
All raw data from ICR animals was deposited in the NCBI Sequence Read Archive under accession number: PRJNA1073062.
Ethics statement
The animal study was approved by the Ethics Committee of Southwest University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YZ: Conceptualization, Data curation, Formal analysis, Methodology, Software, Writing – original draft. RQ: Data curation, Formal analysis, Software, Writing – review & editing. ZZ: Data curation, Formal analysis, Software, Writing – review & editing. MA: Writing – review & editing. SN: Writing – review & editing. SD: Conceptualization, Methodology, Supervision, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received support from the Research on Germplasm Enhancement Project of Edible Fungi (Lentinus edodes as the Main Type) Industry in the Water Source Conservation Area of the South to North Water Diversion (Z20221343035) and the Fundamental Research Funds for the Central Universities (SWU-KT22012).
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R191), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1446924/full#supplementary-material
References
1. Quansah E, Gardey E, Ramoji A, Meyer-Zedler T, Goehrig B, Heutelbeck A, et al. Intestinal epithelial barrier integrity investigated by label-free techniques in ulcerative colitis patients. Sci Rep. (2023) 13:2681. doi: 10.1038/s41598-023-29649-y
2. Bron PA, Kleerebezem M, Brummer R, Cani PD, Mercenier A, Macdonald TT, et al. Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr. (2017) 117:93–107. doi: 10.1017/S0007114516004037
3. Jandhyala SM. Role of the normal gut microbiota. World J Gastroenterol. (2015) 21:8787. doi: 10.3748/wjg.v21.i29.8787
4. Jabeen Z, Bukhari SA, Malik SA, Hussain G, Kamal S. Improved gut microbiota escalates muscle function rehabilitation and ameliorates oxidative stress following mechanically induced peripheral nerve injury in mice. Pak Vet J. (2023) 43:707–13. doi: 10.29261/pakvetj/2023.098
5. Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. (2010) 330:831–5. doi: 10.1126/science.1191175
6. Tang J, Xu L, Zeng Y, Gong F. Effect of gut microbiota on LPS-induced acute lung injury by regulating the TLR4/NF-kB signaling pathway. Int Immunopharmacol. (2021) 91:107272. doi: 10.1016/j.intimp.2020.107272
7. Sánchez De Medina F, Romero-Calvo I, Mascaraque C, Martínez-Augustin O. Intestinal inflammation and mucosal barrier function. Inflamm Bowel Dis. (2014) 20:2394–404. doi: 10.1097/MIB.0000000000000204
8. Wang Z, Li Y, Liao W, Huang J, Liu Y, Li Z, et al. Gut microbiota remodeling: a promising therapeutic strategy to confront hyperuricemia and gout. Front Cell Infect Microb. (2022) 12:935723. doi: 10.3389/fcimb.2022.935723
9. Ma P, Wang M, Wang Y. Gut microbiota: a new insight into lung diseases. Biomed Pharmacother. (2022) 155:113810. doi: 10.1016/j.biopha.2022.113810
10. Bhargava S, Merckelbach E, Noels H, Vohra A, Jankowski J. Homeostasis in the gut microbiota in chronic kidney disease. Toxins. (2022) 14:648. doi: 10.3390/toxins14100648
11. Li Y, Xia S, Jiang X, Feng C, Gong S, Ma J, et al. Gut microbiota and diarrhea: an updated review. Front Cell Infect Microb. (2021) 11:625210. doi: 10.3389/fcimb.2021.625210
12. Gorman A, Golovanov AP. Lipopolysaccharide structure and the phenomenon- of low endotoxin recovery. Eur J Pharm Biopharm. (2022) 180:289–307. doi: 10.1016/j.ejpb.2022.10.006
13. Sun Y, Shang D. Inhibitory effects of antimicrobial peptides on lipopolysaccharide-induced inflammation. Mediat Inflamm. (2015) 2015:167572 doi: 10.1155/2015/167572
14. Zhang J, Xue J, Xu B, Xie J, Qiao J, Lu Y. Inhibition of lipopolysaccharide induced acute inflammation in lung by chlorination. J Hazard Mater. (2016) 303:131–6. doi: 10.1016/j.jhazmat.2015.10.024
15. Candelli M, Franza L, Pignataro G, Ojetti V, Covino M, Piccioni A, et al. Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int J Mol Sci. (2021) 22:6242. doi: 10.3390/ijms22126242
16. Xu B, Yan Y, Yin B, Zhang L, Qin W, Niu Y, et al. Dietary glycyl-glutamine supplementation ameliorates intestinal integrity, inflammatory response, and oxidative status in association with the gut microbiota in LPS-challenged piglets. Food Funct. (2021) 12:3539–51. doi: 10.1039/D0FO03080E
17. Lozano CP, Wilkens LR, Shvetsov YB, Maskarinec G, Park S, Shepherd JA, et al. Associations of the Dietary Inflammatory Index with total adiposity and ectopic fat through the gut microbiota, LPS, and C-reactive protein in the Multiethnic Cohort–Adiposity Phenotype Study. Am J Clin Nutr. (2022) 115:1344–56. doi: 10.1093/ajcn/nqab398
18. Zeng P, Li J, Chen Y, Zhang L. The structures and biological functions of polysaccharides from traditional Chinese herbs. Prog Mol Biol Transl. (2019) 423–44. doi: 10.1016/bs.pmbts.2019.03.003
19. Sun B, Yu S, Zhao D, Guo S, Wang X, Zhao K. Polysaccharides as vaccine adjuvants. Vaccine. (2018) 36:5226–34. doi: 10.1016/j.vaccine.2018.07.040
20. Li S, Tang D, Wei R, Zhao S, Mu W, Qiang S, et al. Polysaccharides production from soybean curd residue viaMorchella esculenta. J Food Biochem. (2019) 43:e12791. doi: 10.1111/jfbc.12791
21. Pan H, Chen X, Wang P, Peng J, Li J, Ding K. Effects of Nemacystus decipiens polysaccharide on mice with antibiotic associated diarrhea and colon inflammation. Food Funct. (2023) 14:1627–35. doi: 10.1039/D1FO02813H
22. Wang S, Cao Z, Wu Q, AI MHA, Dong H. A comparative analysis and verification of differentially expressed miRNAs could provide new insights for the treatment of endometritis in yaks. Pak Vet J. (2023) 43:486–92. doi: 10.29261/pakvetj/2023.061
23. He X, Hao P, Wang Y, Wu C, Yin W, Shahid MA, et al. Swertia bimaculata moderated liver damage in mice by regulating intestine microbiota. Ecotox Environ Safe. (2023) 263:115223. doi: 10.1016/j.ecoenv.2023.115223
24. Sergeev IN, Aljutaily T, Walton G, Huarte E. Effects of synbiotic supplement on human gut microbiota, body composition and weight loss in obesity. Nutrients. (2020) 12:222. doi: 10.3390/nu12010222
25. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. (2018) 34:i884–90. doi: 10.1093/bioinformatics/bty560
26. Kalabiska I, Annar D, Keki Z, Borbas Z, Bhattoa HP, Zsakai A. The oral microbiome profile of water polo players aged 16–20. Sports. (2023) 11:216. doi: 10.3390/sports11110216
27. Rai SN, Qian C, Pan J, Rai JP, Song M, Bagaitkar J, et al. Microbiome data analysis with applications to pre-clinical studies using QIIME2: statistical considerations. Genes Dis. (2021) 8:215–23. doi: 10.1016/j.gendis.2019.12.005
28. Schloss PD. Amplicon sequence variants artificially split bacterial genomes into separate clusters. mSphere. (2021) 6:e0019121. doi: 10.1128/mSphere.00191-21
29. Wells PM, Sprockett DD, Bowyer RCE, Kurushima Y, Relman DA, Williams FMK, et al. Influential factors of saliva microbiota composition. Sci Rep. (2022) 12:18894. doi: 10.1038/s41598-022-23266-x
30. Aivelo T, Lemoine M, Tschirren B. Elevational changes in bacterial microbiota structure and diversity in an arthropod-disease vector. Microb Ecol. (2022) 84:868–78. doi: 10.1007/s00248-021-01879-5
31. Zhao M, Du S, Li Q, Chen T, Qiu H, Wu Q, et al. High throughput 16SrRNA gene sequencing reveals the correlation between Propionibacterium acnes and sarcoidosis. Resp Res. (2017) 18:28. doi: 10.1186/s12931-017-0515-z
32. Xie F, Xu H, Zhang J, Liu X, Kou B, Cai M, et al. Dysregulated hepatic lipid metabolism and gut microbiota associated with early-stage NAFLD in ASPP2-deficiency mice. Front Immunol. (2022) 13:974872. doi: 10.3389/fimmu.2022.974872
33. Ekiert HM, Szopa A. Biological activities of natural products. Molecules. (2020) 25:5769. doi: 10.3390/molecules25235769
34. Zhang J, Ma L, Wan X, Shan J, Qu Y, Hashimoto K. (R)-Ketamine attenuates LPS-induced endotoxin-derived delirium through inhibition of neuroinflammation. Psychopharmacology. (2021) 238:2743–53. doi: 10.1007/s00213-021-05889-6
35. Wu Y, Zhang Y, Xie B, Abdelgawad A, Chen X, Han M, et al. RhANP attenuates endotoxin-derived cognitive dysfunction through subdiaphragmatic vagus nerve-mediated gut microbiota–brain axis. J Neuroinflamm. (2021) 18:300. doi: 10.1186/s12974-021-02356-z
36. Reyes-Díaz A, Mata-Haro V, Hernández J, González-Córdova A, Hernández-Mendoza A, Reyes-Díaz R, et al. Milk fermented by specific lactobacillus strains regulates the serum levels of IL-6, TNF-α and IL-10 cytokines in a LPS-stimulated murine model. Nutrients. (2018) 10:691. doi: 10.3390/nu10060691
37. Qiu Y, Zhang J, Liu Y, Ma H, Cao F, Xu J, et al. The combination effects of acetaminophen and N-acetylcysteine on cytokines production and NF-κB activation of lipopolysaccharide-challenged piglet monon-uclear phagocytes in vitro and in vivo. Vet Immunol Immunop. (2013) 152:381–8. doi: 10.1016/j.vetimm.2013.01.013
38. Yang X, Li L, Xue Y, Zhou X, Tang J. Flavonoids from Epimedium pubescens: extraction and mechanism, antioxidant capacity and effects on CAT and GSH-Px of Drosophila melanogaster. PeerJ. (2020) 8:e8361. doi: 10.7717/peerj.8361
39. Zhou J, Peng Z, Wang J. Trelagliptin alleviates lipopolysaccharide (LPS)-induced inflammation and oxidative stress in acute lung injury mice. Inflammation. (2021) 44:1507–17. doi: 10.1007/s10753-021-01435-w
40. Li C, Ma D, Chen M, Zhang L, Zhang L, Zhang J, et al. Ulinastatin attenuates LPS-induced human endothelial cells oxidative damage through suppressing JNK/c-Jun signaling pathway. Biochem Bioph Res Commun. (2016) 474:572–8. doi: 10.1016/j.bbrc.2016.04.104
41. Ksiezarek M Grosso F Ribeiro TG and Peixe L. Genomic diversity of genus Limosilactobacillus. Microb Genom. (2022) 8:847. doi: 10.1099/mgen.0.000847
42. Goel A, Kim B, Ncho C, Jeong C, Gupta V, Jung J, et al. Dietary supplementation of shredded, steam-exploded pine particles decreases pathogenic microbes in the cecum of acute heat-stressed broilers. Animals. (2021) 11:2252. doi: 10.3390/ani11082252
43. Liu B, Yu L, Zhai Q, Li M, Li L, Tian F, et al. Effect of water-soluble polysaccharides from Morchella esculenta on high-fat diet-induced obese mice: changes in gut microbiota and metabolic functions. Food Funct. (2023) 14:5217–31. doi: 10.1039/D3FO00574G
44. Liu GM, Lu JJ, Sun WX, Jia G, Zhao H, Chen XL, et al. Dietary alpha-ketoglutarate enhances intestinal immunity byTh17/Treg immune response in piglets after lipopolysaccharide challenge. J Anim Sci. (2023) 101:skad213. doi: 10.1093/jas/skad213
Keywords: Morchella esculenta polysaccharides, LPS, intestinal flora, rectal microbiota, intestinal dysfunction
Citation: Zhang Y, Qiu R, Zhang Z, Almutairi MH, Nawaz S and Dong S (2024) Effect of Morchella esculenta polysaccharides on the rectal microbiota of mice challenged with lipopolysaccharides. Front. Vet. Sci. 11:1446924. doi: 10.3389/fvets.2024.1446924
Received: 10 June 2024; Accepted: 16 August 2024;
Published: 19 September 2024.
Edited by:
Isa Ozaydin, Kafkas University, TürkiyeReviewed by:
Ali Raza Jahejo, Guangxi University, ChinaRanran Hou, Qingdao Agricultural University, China
Copyright © 2024 Zhang, Qiu, Zhang, Almutairi, Nawaz and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiqi Dong, dGN2bUBzd3UuZWR1LmNu
 Yingjun Zhang1
Yingjun Zhang1 Reng Qiu
Reng Qiu Mikhlid H. Almutairi
Mikhlid H. Almutairi Shiqi Dong
Shiqi Dong