- 1Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture and Rural Affairs, Shanghai Ocean University, Shanghai, China
- 2Science and Technology Service Center, Ministry of Science and Technology, Shanghai Mugao Biotechnology Co., Ltd., Shanghai, China
- 3Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 4Department of Pediatric, Shanghai Sixth People's Hospital, Shanghai, China
Bone morphogenetic proteins (BMPs) play an important biological role in pearl biomineralization in pearl mussels. In this study, based on the genome data of the triangular sail mussel (Hyriopsis cumingii), the genome-wide identification and bioinformatic analysis of BMP gene family were performed, and the expression pattern of the BMP genes was investigated by the insertion experiments. The results showed that a total of 12 BMP gene family members (BMP2a/2b, BMP3, BMP5a/5b, BMP7a/7b/7c, BMP9, BMP10a/10b, and BMP11) were identified, which were unevenly distributed on chromosome 3/14/18, encoding 169–583 amino acids, with molecular weights ranging from 19.32 to 65.99 kDa. BMP2a, BMP7b, and BMP10a were distributed, respectively, in the cytoplasm, endoplasmic reticulum and mitochondria, others were distributed in the nucleus. qRT-PCR results showed the significant tissue specificity in BMPs gene expression. The HcBMPs were differentially expressed in the mantle and visceral mass, and the expression level was higher in the visceral mass. The expressing trends of HcBMPs were not consistent between the mantle and visceral mass insertion, suggesting that HcBMPs may perform different functions. We also found that insertion surgery in the mantle and visceral mass significantly alters the expression profiling of the BMP gene family. Insertion of the mantle induced the biomineralization function of BMP2a, BMP7a, and BMP7b, while BMP3 and BMP10b played opposite roles in visceral mass insertion. Visceral mass insertion could suppress BMP9 expression at 5 d and BMP5b expression at 90 d after insertion This work lays the foundation and data support for the preliminary elucidation of regulatory role and mechanism of HcBMPs in the pearl-cultivating process of mantle and visceral mass.
1 Introduction
The shell of the mussel serves as its exoskeleton, which can protect the soft internal tissues and visceral organs (1), and is a product of its biomineralization (2). In addition, pearls are formed by the secretion of an organic matrix from the mantle of pearl mussel, which forms calcium carbonate and undergoes biomineralization. They are identical in composition to that of the shell, but are structurally different (3), and the formation process is regulated at the genetic level. The triangular sail mussel (Hyriopsis cumingii) is the most important freshwater pearl-breeding mussel in China (4), and the pearls cultured are one of the typical representatives of biomineralization products. When the mussel is stimulated by artificially inserted nucleus and pieces of donor mantle, the epithelial cells on the outer side of the mantle at the insertion point are stimulated by the foreign substances. They will interact with the mantle pieces, proliferate continuously and gradually enclose the inserted nucleus, forming a sac-like structure called the pearl sac, which continuously secretes organic macromolecules and ions deposited on the surface of the nucleus (5). At present, the common nucleus insertion parts of H. cumingii are mantle and visceral mass, the mantle is relatively thin, so the nucleus is easy to penetrate and fall off, and the rate of pearl formation is low (6). The visceral mass, as a potential site for cultivating round and large pearls, is characterized by its large size and multiple insertion sites, which can form high-quality pearls with good luster, large particles and round shapes (7), but there have been fewer studies on the regulatory mechanisms of H. cumingii after the insertion of nucleus into the mantle and visceral mass. Therefore, the study of the different response mechanisms of H. cumingii after the insertion surgery of mantle and visceral masses is of great theoretical and practical significance for production.
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-beta (TGF-β) superfamily and are synthesized as secreted signaling factors with additional prepeptide sequences (8). BMPs have attracted much attention in higher organisms, and are associated with bone formation and differentiation, regulation of reproduction, cell proliferation, migration and differentiation (9–11). BMPs are osteogenic inducers that play an important role in bone formation, limb formation, osteoblast differentiation and embryonic kidney development. Jiang et al. (12) observed the effects on zebrafish bone metabolism after inhibition of BMP2a expression by knockdown. The conclusion showed that zebrafish BMP2 was down-regulated by knockdown technique, it was visualized that zebrafish had delayed bone development, and BMP2 played a crucial role in zebrafish bone metabolism. By cloning the BMP4 gene of Siniperca chuatsi and analyzing its expression, Cao et al. (13) demonstrated that BMP4 is involved in the regulation of jaw remodeling, and the development of jaw remodeling is correlated with its feeding behaviors at different stages. In higher organisms, BMP11 can inhibit the progression of atherosclerosis by activating TGF-β/Smad2/3, AMPK/eNOS signaling pathways and inhibiting JNK and NF-κB signaling pathways (14). In addition, BMPs are involved in the biomineralization process of mollusks. BMP3 is involved in the shell formation of Pinctada fucata, as well as calcium ion metabolism in the gills (15). BMP10 is involved in the formation of shells and pearls of H. cumingii, which plays an important role in the calcium carbonate-mediated biomineralization reactions and may be involved in calcium ion uptake and metabolism (16).
In the present study, we identified the BMP gene family members in H. cumingii from the genome database and used bioinformatics methods to study their phylogenetic relationships, motif composition, gene structure and chromosomal localization. Moreover, qRT-PCR was used to examine the relative expression of BMP genes before and after the insertion to the mantle and visceral masses. This study provides a theoretical basis for further studies on the different roles played by the BMP gene family in the mantle and visceral mass of H. cumingii after insertion.
2 Materials and methods
2.1 Experimental animals and sample collection
Experimental animals: One hundred healthy 2-year-old H. cumingii of uniform body size (length: 12 ± 2 cm, width: 7 ± 1 cm, height: 2.5 ± 0.8 cm, weight: 225.23 ± 30.77 g) purchased from the aquaculture farm in Jinhua City, Zhejiang Province, China were randomly selected and temporarily cultivated in the laboratory. Twenty were assigned to the control group without insertion and 80 were assigned to the experimental group, small pieces of donor mantle (2 mm × 2 mm) and 3 mm diameter nucleus were inserted in the posterior part of the host mantle and visceral mass (17). After the insertion surgery, the mussels were temporarily cultivated in a laboratory tank with 100 L circulating water, and the water temperature was maintained at about 26°C. Chlorella was fed twice a day.
Sample collection: Six mussels were collected at five time points (0, 5, 20, 50, and 90 d), respectively. The mussels were killed by cutting off adductor muscle, 80–100 mg of samples were collected from the insertion sites of mantle and visceral mass immediately, and were frozen by liquid nitrogen and stored in a refrigerator at −80°C.
2.2 Genome-wide identification of BMP genes
The whole genome sequencing data of the H. cumingii (GCA_028554795.2) had been obtained by laboratory in the early stage (18). The Hidden Markov Model file of BMP genes (PF00019 and PF00688) was downloaded from the Pfam protein family database1 and the amino acid sequences of the genome of H. cumingii were searched by hmmsearch function of the TBtools software. Subsequently, structural domain validation was performed using NCBI-CDD2 and SMART3. The BMP gene family of H. cumingii was finally confirmed and named based on NCBI-BLAST4 results, combined with genome-wide annotation. The number of amino acids (aa), molecular weight (MW), isoelectric point (pI), stability index and grand average of hydrophilicity (GRAVY) of BMP family proteins were predicted by ExPASy5. Subcellular localization of BMP gene family members was predicted using the Euk-mPLoc2.06. The secondary structures of BMP family proteins were analyzed using the PSORT II7.
2.3 Phylogenetic analysis of BMP gene family
Amino acid sequences of typical species (Homo sapiens, Danio rerio, Crassostrea gigas, Crassostrea virginica, and Mizuhopecten yessoensis) were obtained from the NCBI database8 and saved in fasta format. All protein accession numbers and sequences used for the analysis were shown in Supplementary Table S1. The amino acid sequences were imported into MEGA 11 software and the multiple sequence alignment of full-length proteins among species was assessed using MUSCLE. An phylogenetic tree was constructed using the Neighbor-Joining (NJ) method and the bootstrap replications were 1,000. The phylogenetic tree was visually improved using iTOL9.
2.4 Gene structure analysis
The conserved domains and motifs of the BMP gene family members were predicted using the NCBI-CDD and MEME10, respectively. The maximum number of motifs was set 10. The BMP protein sequences of the H. cumingii were subjected to multiple sequence comparison analysis using the MEGA 11 software (19), and the phylogenetic tree was constructed by the Maximum Likelihood (ML) method (bootstrap replications of 1,000). The visualization of the gene structure of BMP family members were applied by TBtools software (v 2.096) (20) based on the genome annotation file.
2.5 Chromosomal location analysis
BMP genes were mapped to chromosomes based on the chromosomal position provided in the H. cumingii genome database. Chromosomal location of 12 BMP genes was visualized by TBtools software (v 2.096).
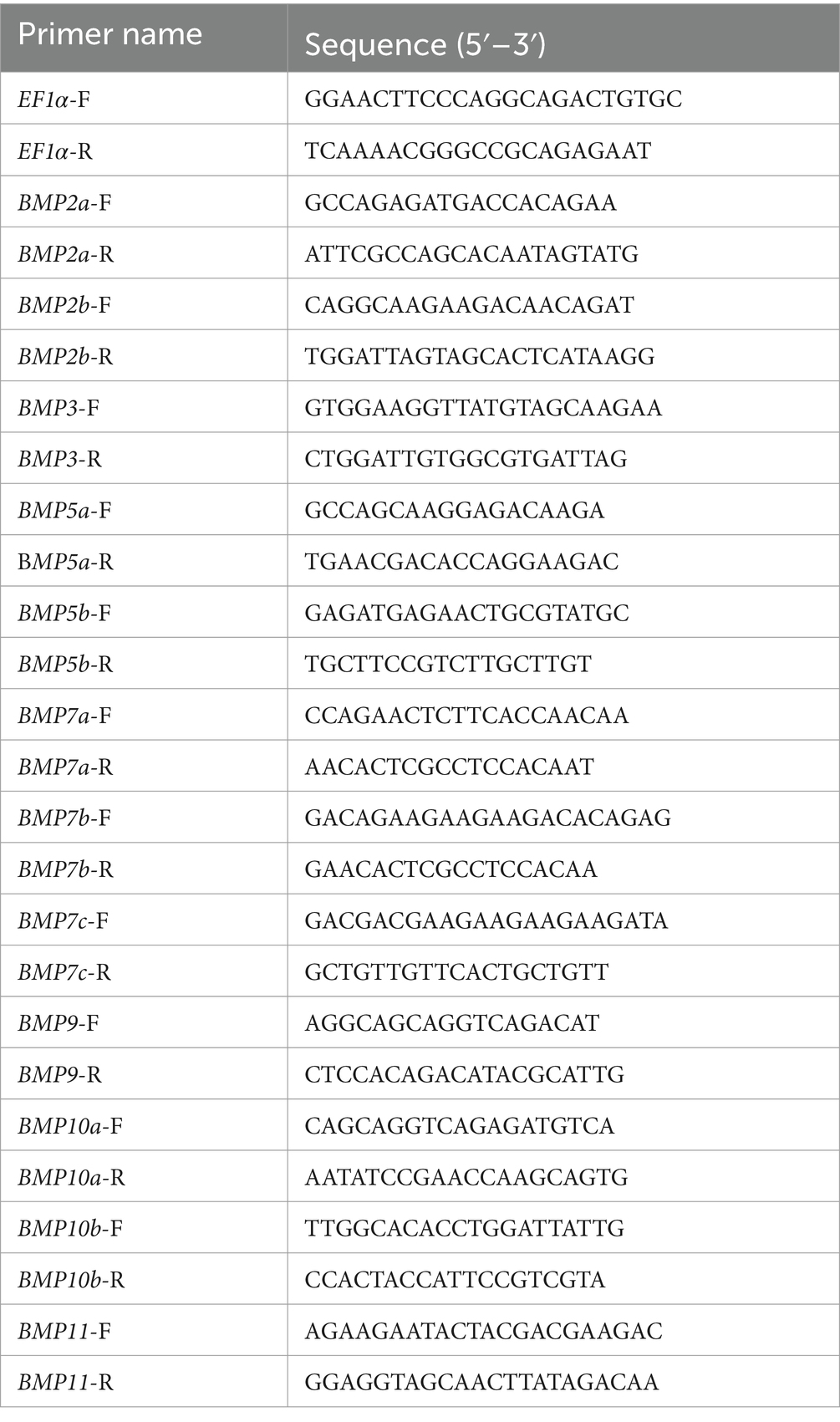
2.6 Quantitative real-time PCR (qRT-PCR) of BMP gene family
Total RNA was extracted from the mantle and visceral mass of each mussel at five time points by using the Trizol Reagent (TaKaRa, Tokyo, Japan). The quality of RNA was tested by 1% agarose gel electrophoresis, and the concentration of total RNA and A260/A280 ratio were examined on a NanoDrop 2000 (Thermo, Waltham, MA, USA). cDNA was synthesized using a Evo M-MLV Reverse Transcription Mix Kit (Accurate Biology, Hunan, China) according to the manufacturer’s protocol. Specific primers for qRT-PCR were designed using Premier Primer 5.0 (Table 1). Follow the instructions of TB Green® Premix Ex Taq™ (TaKaRa, Tokyo, Japan) for qRT-PCR. The amplification was performed in triplicate on a Bio-Rad CFX96 (Bio-Rad, Hercules, CA, USA), which was consistent with the method reported by Shen et al. (21). Relative gene expression data was analyzed using the 2−ΔΔCt method to obtain the expression of each sample relative to the internal reference gene EF1α. All reactions were performed in three replicates using six biological samples. t-test and one-way ANOVA were performed using SPSS 27.0. The means among various groups were compared by Duncan’s multiple range tests. The results are presented as the means ± SEM, and the statistical significance is represented by p < 0.05.
3 Results
3.1 Identification of BMP genes in H. cumingii
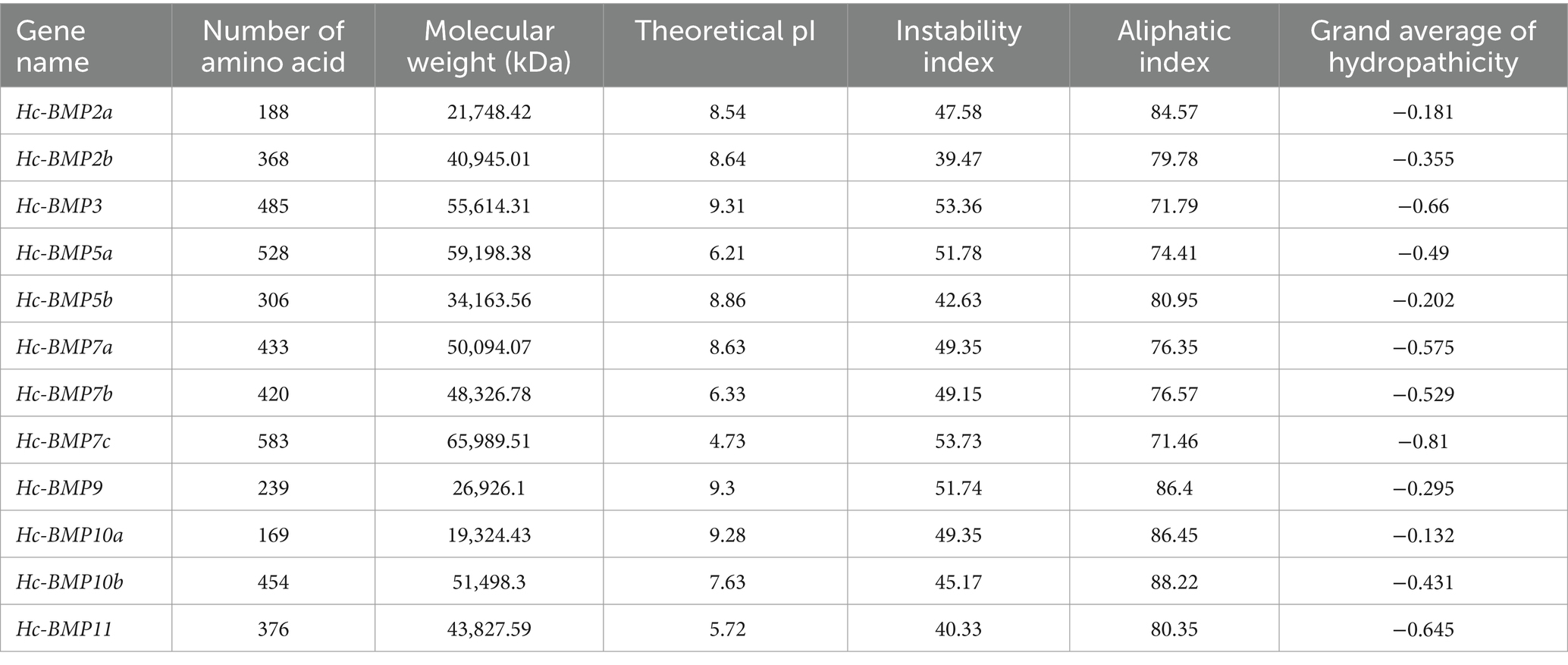
As shown in Table 2, 12 BMP gene family members, named BMP2a, BMP2b, BMP3, BMP5a, BMP5b, BMP7a, BMP7b, BMP7c, BMP9, BMP10a, BMP10b, BMP11 were identified within the genome of H. cumingii in this study. The length of ORF (open reading frames) ranged from 510 to 1,752 bp. The longest sequence was Hc-BMP7c, encoding 583 amino acids, and the shortest sequence was Hc-BMP10a, encoding 169 amino acids. The corresponding protein MW ranged from 19.32 to 65.99 kDa, and the pI ranged from 4.73 to 9.31. Four BMPs proteins (BMP5a, BMP7b, BMP7c, and BMP11) were acidic proteins (pI < 7.5), and the rest of BMPs proteins were alkaline proteins (pI > 7.5). All of the 12 BMPs proteins had an instability index greater than 40 and the GRAVY of proteins were all negative, indicating that the family proteins were unstable hydrophilic proteins.
The subcellular localization and secondary structures of BMP gene family proteins were analyzed, as shown in Table 3. BMP2a was distributed in the cytoplasm, BMP7b was distributed in the endoplasmic reticulum and BMP10a was distributed in the mitochondria, while the rest of BMPs were distributed in the nucleus. The results of secondary structure analysis showed that 12 BMPs proteins were dominated by random coil and α-helix structures, with a percentage ranging from 62.34 to 81.91%, while β-turn accounted for the lowest percentage (2.13–11.30%).
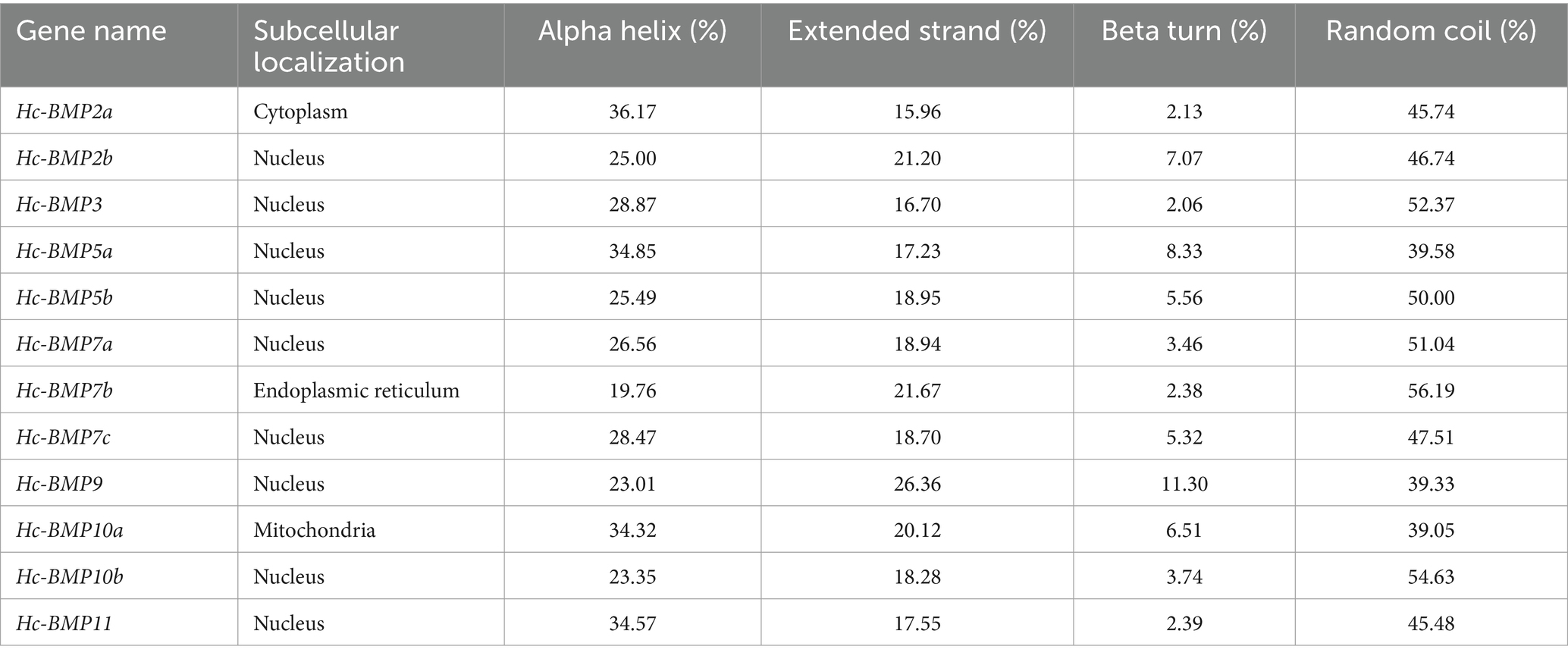
Table 3. The subcellular localization and secondary structure prediction of BMP proteins in H. cumingii.
3.2 Phylogenetic analysis of the BMP genes
To investigate the phylogenetic relationships of BMP genes in H. cumingii, a phylogenetic tree was constructed (Figure 1) using BMP amino acid sequences from H. sapiens, D. rerio, C. gigas, C. virginica, and M. yessoensis. BMP2a, BMP2b, BMP5a, BMP5b, and BMP7c were clustered into one branch by themselves, BMP9 and BMP10a were clustered into one branch, all of which were phylogenetically distant from other higher organisms and shellfish. BMP3 and BMP10b were clustered into one branch with C. gigas, C. virginica, and M. yessoensis, which indicated that BMP3 and BMP10b are phylogenetically closely related between H. cumingii and other shellfish. BMP7a and BMP7b were clustered with C. gigas and C. virginica, which were phylogenetically close to each other. BMP11 was clustered with BMP1 of M. yessoensis and its phylogenetic relationship needs to be further investigated.
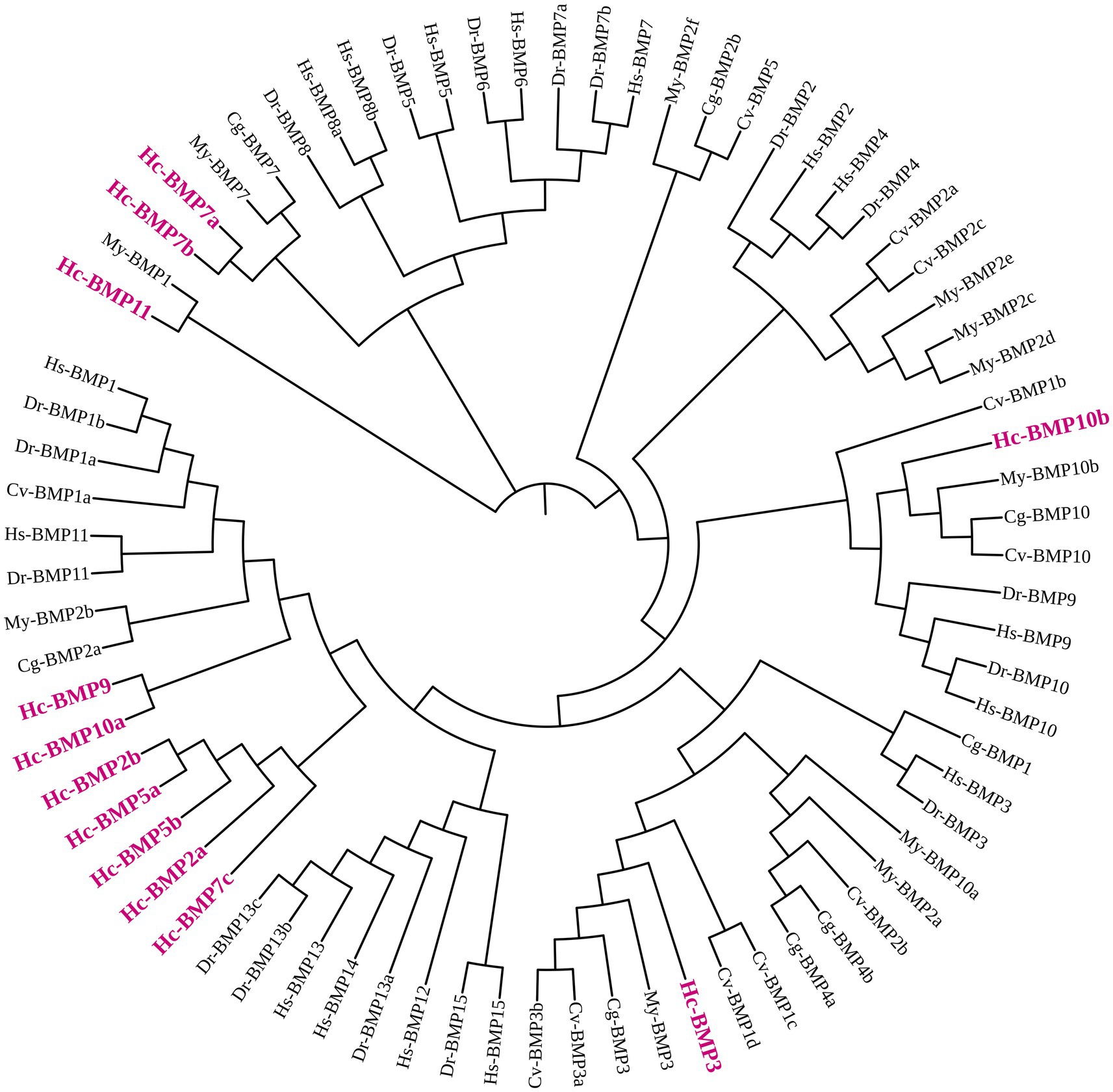
Figure 1. Phylogenetic tree of the BMP gene family in H. cumingii (Hc) and representative species. H. sapiens (Hs), D. rerio (Dr), C. gigas (Cg), C. virginica (Cv), and M. yessoensis (My). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates.
3.3 Gene structures analysis of the BMP genes
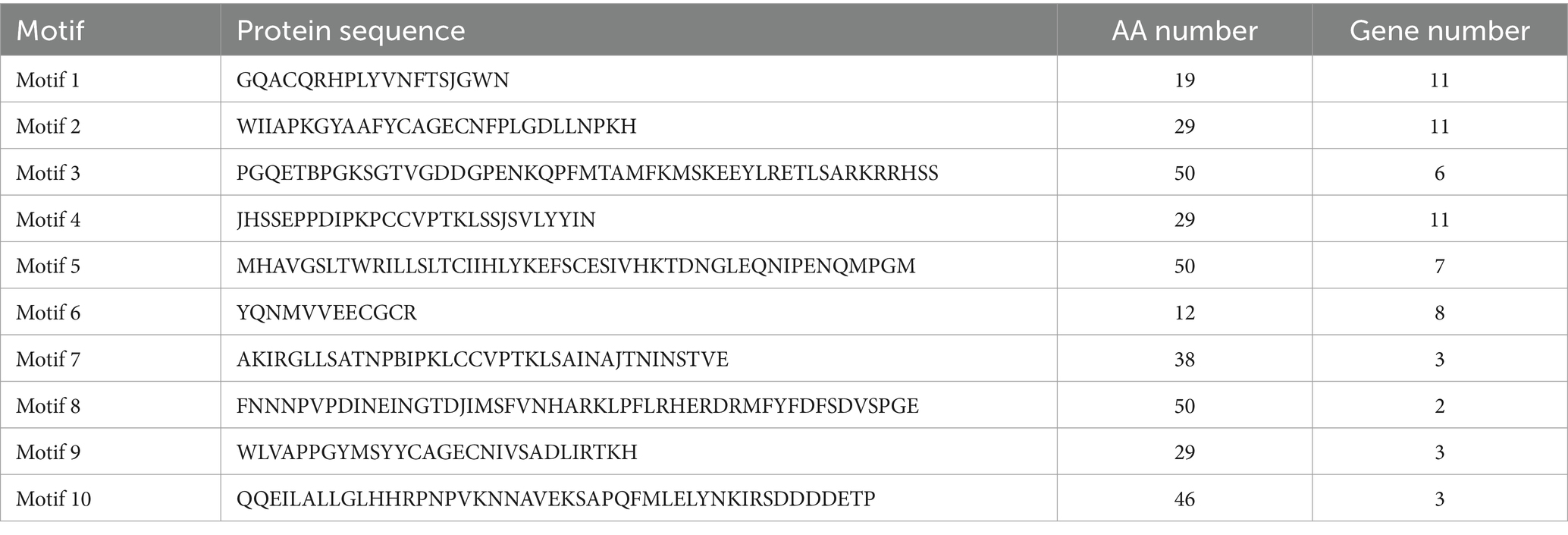
Analysis of the conserved motifs of BMP proteins (Figure 2A; Table 4) showed that the amino acid sequence length of motifs ranged from 12 to 50 aa. Motif 1, Motif 2, and Motif 4 were highly conserved, which were shared by 11 BMP genes, followed by Motif 6, and fewer family members contained Motif 7, Motif 8, Motif 9, and Motif 10. The distribution of conserved motifs was similar among BMP genes with near positions in the phylogenetic tree.
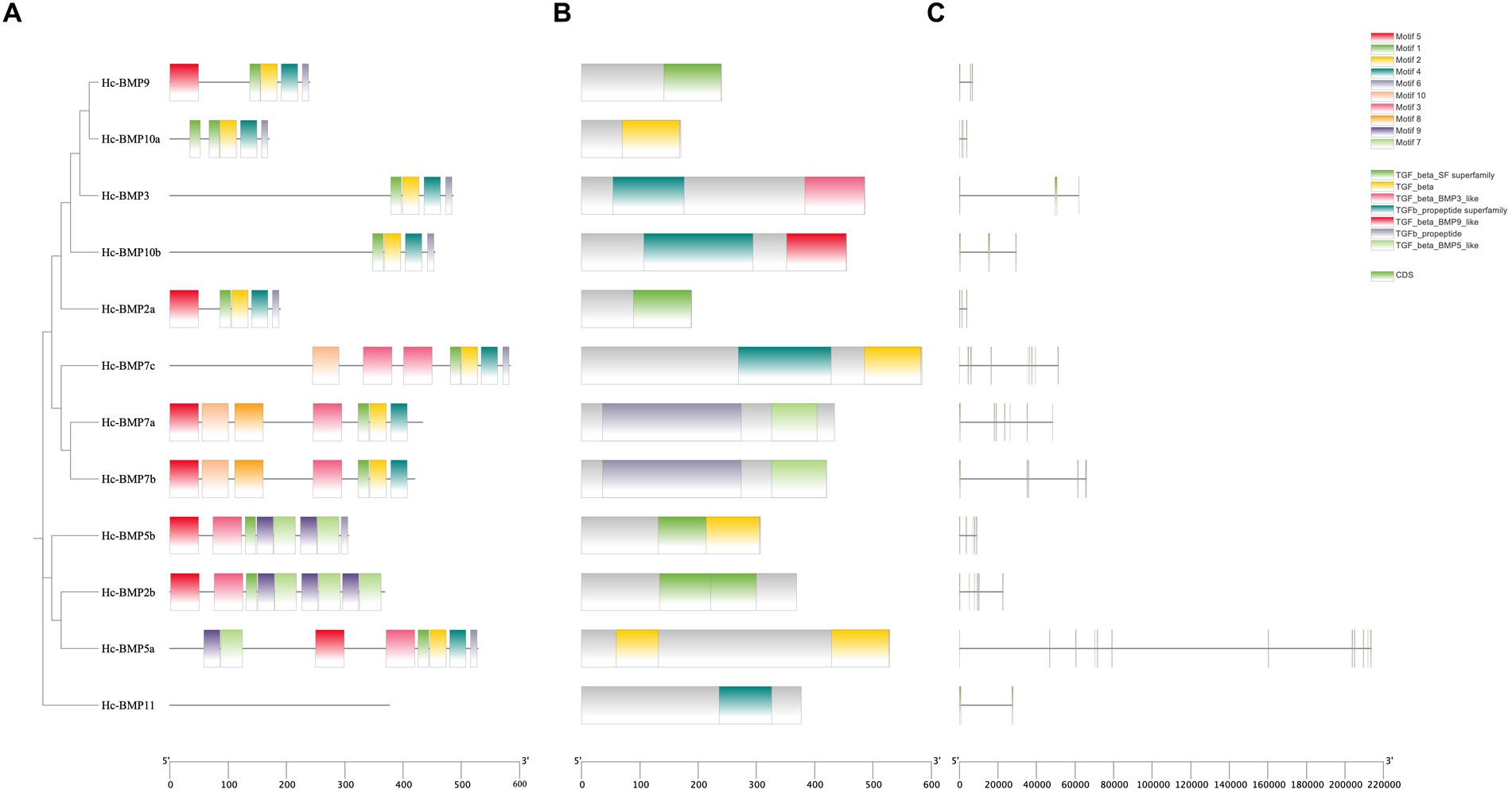
Figure 2. Phylogenetic tree, conserved motifs, conserved domains and gene structures of BMP genes in H. cumingii. (A) phylogenetic tree and conserved motifs, (B) conserved domains, and (C) gene structures.
Analysis of the conserved domains of BMP genes (Figure 2B) showed that most of BMP genes possessed the TGF-β SF superfamily and TGF-β superfamily. The distribution of conserved domains was similar among BMP genes with near positions in the phylogenetic tree. As shown in the figure, Motif 1, Motif 2, Motif 3, and Motif 4 are involved in the formation of the TGF-β SF superfamily, and Motif 1, Motif 2, Motif 4, and Motif 6 are involved in the formation of the TGF-β superfamily. These conserved motifs may perform the functions of the corresponding structural domains.
Analysis of the gene structure of BMP genes (Figure 2C) showed that the total length of the different genes varied, with BMP2a being the shortest and BMP5a being the longest. In addition, similarity of gene structure among BMP proteins with near positions in the phylogenetic tree can be seen from the figure.
3.4 Chromosomal localization
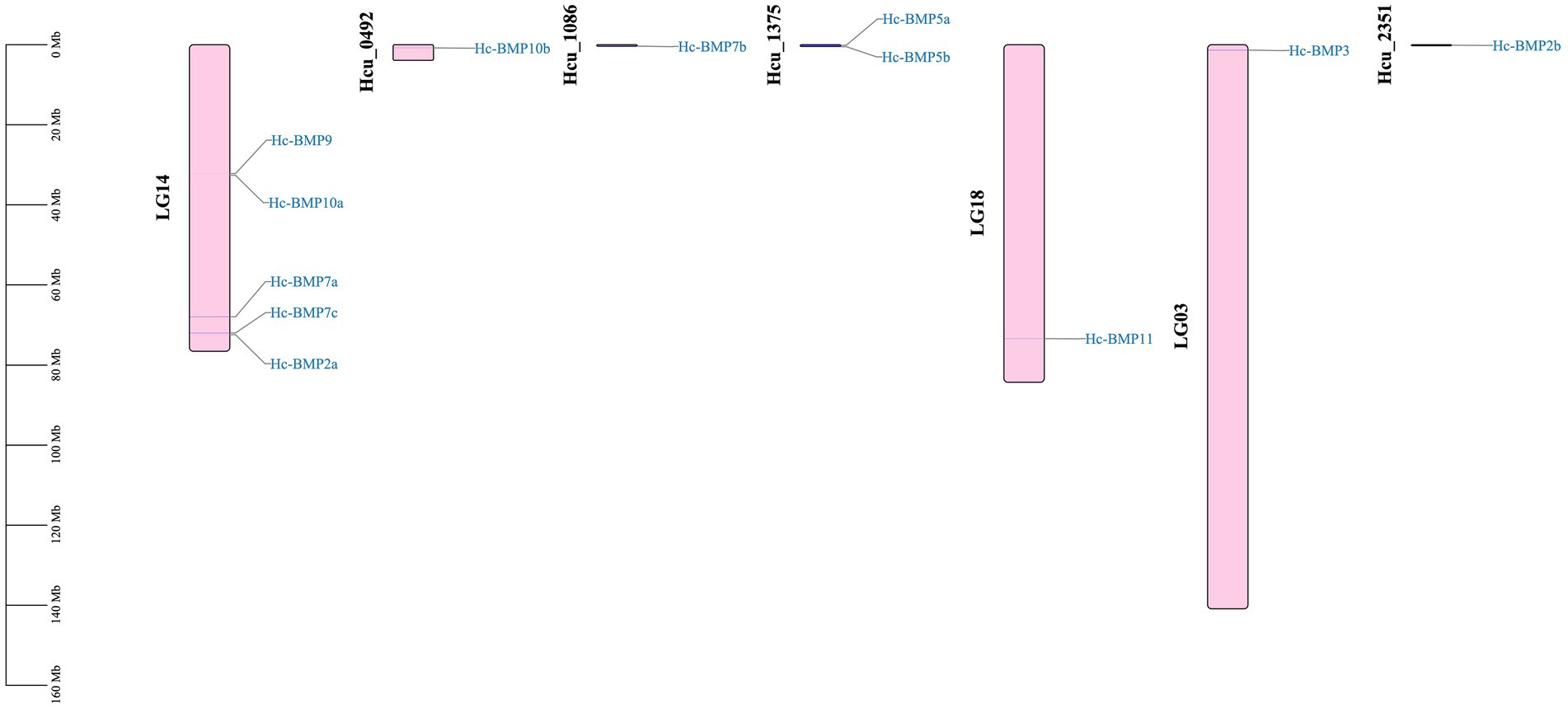
The localization of the BMPs gene family on the chromosomes is shown in Figure 3. The number of chromosomes in the H. cumingii is 2n = 38. Seven genes (BMP2a, BMP3, BMP7a, BMP7c, BMP9, BMP10a, and BMP11) were located to four different chromosomes, and five genes (BMP2b, BMP5a, BMP5b, BMP7b, and BMP10b) were not localized on chromosomes. Chromosome 14 contained five BMP genes, and chromosomes 18 and 3 contained one BMP gene, respectively. Meanwhile, BMP9 and BMP10a were located in the middle part of chromosome 14, while the other family members were distributed on both ends of the chromosomes. The scale is in megabases (Mb).
3.5 Expression analysis
qRT-PCR was performed to detect the expression of 12 BMP genes in the mantle and visceral mass of H. cumingii, which were not inserted. As shown in Figure 4, except for BMP2b, BMP5b, and BMP10b, the expression of the other BMPs genes in the mantle and visceral mass were significantly different (p < 0.05), this research found significant tissue specificity in BMPs gene expression. Among them, the expression of BMP2a, BMP5a, BMP7c, and BMP9 were significantly higher in the visceral mass than in the mantle (p < 0.001), and their expression in the visceral mass were 1.84 times, 3.68 times, 3.86 times and 4.74 times higher than in the mantle, respectively. In contrast, BMP3, BMP10a and BMP11 expression in the mantle were significantly higher than in the visceral mass, especially the expression of BMP10a in the mantle was 2.37 times higher than in the visceral mass.
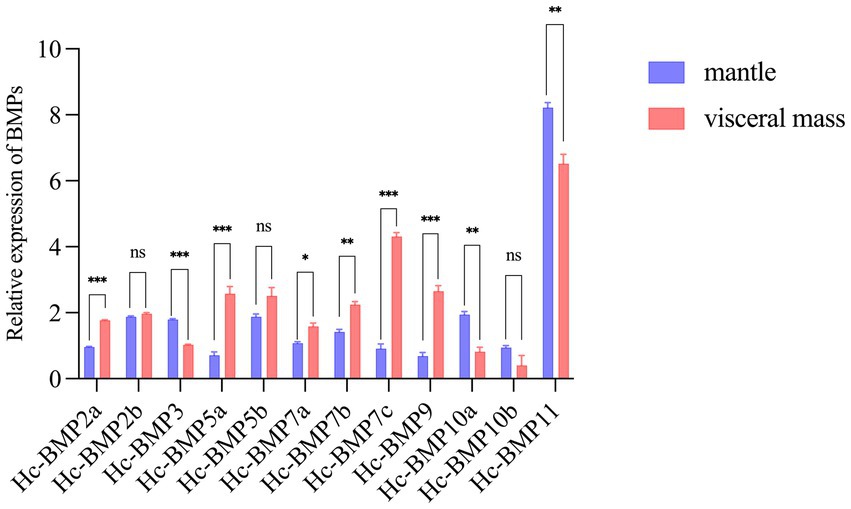
Figure 4. Relative expression of HcBMPs in the mantle and visceral mass without insertion. *p < 0.05, **p < 0.01, ***p < 0.001, ns means not significant.
qRT-PCR was performed to detect the expression changes of 12 BMP genes at 5, 20, 50, and 90 d after the insertion surgery. As shown in Figure 5, the expression of all 12 BMP family members was significantly different (p < 0.05) after the mantle and visceral mass were inserted. Compared with the control group, BMP genes showed different expression patterns during the insertion of nucleus for pearl cultivating, except for BMP7c, BMP10a, and BMP11. Among them, the expression of BMP2a was significantly higher in mantle than in visceral mass at 5 d, unlike the control and other time points, and the expression trends of BMP7a and BMP2a were basically the same. The expression of BMP3 was higher in the visceral mass at 20 d, and the expression pattern of BMP10b was opposite to that of the control at 5 d and 20 d, with the expression in visceral mass significantly higher than that of mantle. In the early stage, the of BMP5a and BMP5b were both significantly higher in visceral mass than in mantle, while at 90 d BMP5a showed no significant difference between the two, and BMP5b showed significantly higher expression in mantle than in visceral mass. BMP7b had significantly higher expression in mantle than in visceral mass at 50 d and 90 d, unlike the early stage. In addition, the expression pattern of BMP2b and BMP9 changed greatly after insertion, BMP2b showed no significant difference between the expression of mantle and visceral mass in the control group, while the expression in mantle was higher at all time points after insertion except for the higher expression in visceral mass at 20 d, and BMP9 expression was significantly higher in mantle than in visceral mass at all time points after insertion, in contrast to the control group, except at 50 d when there was no significant difference between the two.
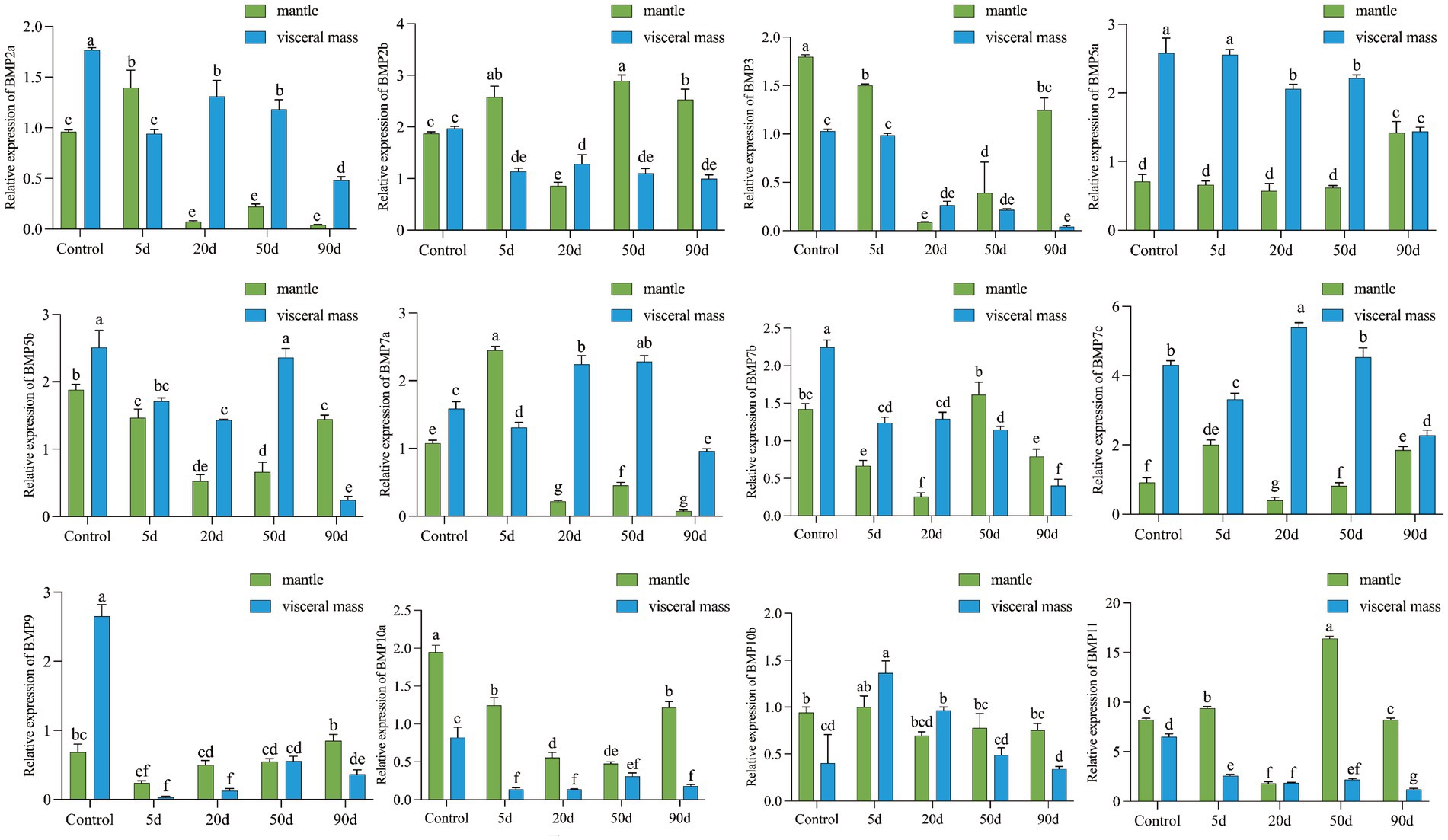
Figure 5. Relative expression of HcBMPs in the mantle and visceral mass after insertion. Different letters indicate significant differences between groups (p < 0.05).
4 Discussion
BMPs are key factors in the biomineralization of pearl mussels, but their roles in pearl culture are less reported. Because of the functional diversity of BMP gene family, in-depth studies have been conducted in several species, with 44 BMP genes identified from Cyprinus carpio (22), 11 BMP genes from Chinese soft-shell turtle (Pelodiscus sinensis) (23), 26 BMP genes were identified from Cynoglossus semilaevis (24), and 18 BMP genes from Hypophthalmichthys nobilis (25). In this study, 12 BMP gene family members were identified from the genome of H. cumingii, including two BMP2 members, one BMP3 member, two BMP5 members, three BMP7 members, one BMP9 member, two BMP10 members and one BMP11 member. The BMP gene family identified in this study is complete, which lays the foundation for subsequent functional and evolutionary analyses. Analysis of the physicochemical properties of the BMP gene family members showed that the ORF lengths were shorter and the corresponding protein molecular weights were smaller compared to those of other species (26). Moreover, the pI of most BMP proteins was greater than 7.5, proving that they may be a class of basic proteins. Subcellular localization revealed that most of the BMPs were present in the nucleus, but also in the mitochondria, cytoplasm, and endoplasmic reticulum, which is inconsistent with the findings of BMPs from Rachycentron canadum (27), where most of the BMPs exist outside the cell. Structural analysis demonstrated that their secondary structures contain a higher number of α-helices and random coil, suggesting that α-helix and random coil have a greater influence on the function of BMP family proteins of H. cumingii.
By constructing a phylogenetic tree of H. cumingii with higher organisms (human, zebrafish) and other shellfish (Pacific oyster, American oyster, and scallop), it was found that the H. cumingii BMPs sequences were distantly phylogenetically related to the higher organisms and none of them clustered into a single branch, whereas some of the BMP genes clustered into a single branch with other shellfish, and some of them clustered into a single branch on their own. Many literatures (28–30) suggest that the evolutionary relationship of BMP genes in molluscs is closer to that of other molluscs and more distantly related to higher organisms (human and zebrafish), which is consistent with the results of present study. However, the phylogenetic analyses of BMP gene family in other species (H. nobilis, R. canadum, C. carpio, and T. dalaica) all classified the BMP gene family into five subfamilies (22, 25, 31, 32): BMP2/4/16, BMP5/6/7/8, BMP9/10, BMP12/13/14, and BMP1/3/11/15, but the phylogenetic analysis carried out in the present study could not come to this conclusion, suggesting that the phylogenetic pattern of BMP gene family in H. cumingii is worthy of further in-depth study. Statistical analysis of the conserved motifs in the BMP gene family revealed that Motif 1, Motif 2, and Motif 4 were the most conserved, 11 genes contained these motifs, suggesting that they may be the key factors in the functioning of structural domains of BMPs. Members of different branches have different motif compositions, and these conserved motifs may be the main reason why different BMP genes are involved in different biological functions. Structural domain analysis showed that most of BMP gene family members in this study have the TGF-β SF superfamily and TGF-β superfamily structural domain, which has been proven to be related to the formation and repair of shells. Both BMP3 and BMP10 of Pinctada martensi have the TGF-β structural domain, which is associated with damage repair of shells (15), BMP2 and BMP10 of H. cumingii have also been demonstrated to be related to the formation of the prismatic layer and pearl layer of shells as well as calcium metabolism (16). TGF-β of H. cumingii also possesses this structural domain, which was found to be related to immune response and wound repair (33). The above results all suggest that the BMP gene family may play an important role in the biomineralization process of pearl mussels.
Many studies have shown that nucleus insertion to H. cumingii causes changes in the expression of relevant biomineralization genes in response to wound repair (34–36). Since most of the BMP genes showed significant differences of expression between mantle and visceral mass, it is hypothesized that these genes perform different functions in the mantle and visceral mass. We suggested that BMPs have different mechanisms of action after insertion surgery of the mantle and visceral mass, so the insertion surgery was performed. We focused on the differences of BMP expression in mantle and visceral mass after nucleus insertion for pearl cultivating. Except for BMP7c, BMP10a, and BMP11, the expression pattern of other BMP genes in mantle and visceral mass after insertion were changed. The trend of BMP2a and BMP7a expression change before and after insertion in mantle and visceral mass were basically the same, the expression of both genes in mantle significantly increased at 5 d, which indicated that these two genes played a role 5 days after mantle insertion. But BMP2a and BMP7a expression decreased significantly in the late stage of mantle insertion, which may be suppressed during the pearl formation process. Interestingly, the phylogenetic tree showed that their evolutionary relationship was not close, which might be due to the fact that their action mechanisms during pearl cultivating were similar, whereas the expression of BMP7b increased significantly at the late stage of mantle insertion. BMP7a and BMP7b may perform different functions after mantle and visceral mass insertion. Meanwhile, Lin (37) demonstrated that BMP7 was involved in the formation of nacreous layer and prismatic layer of H. cumingii, and Fan et al. (38) found that BMP7 may play an important role in the shell formation and repair process of P. martensi, suggesting that BMP7 can indeed participate in the biomineralization of mantle of the pearl mussel. We suppose that BMP2 and BMP7 play an important role in the biomineralization of mantle and that the difference in the roles of two homologous genes, BMP7a and BMP7b, is of interest. There are literatures (15) suggesting that the short-term expression of BMP3 in P. martensi increased significantly during damage repair of shells, which was presumed to be involved in the damage repair of shells. Duan (16) showed that the expression of BMP10 in H. cumingii mantle increased significantly after 7 days of mantle insertion. Both indicated that BMP3 and BMP10 play important roles in the biomineralization of mantle. In our study, we found that BMP3 and BMP10a were expressed at higher levels and with similar expression trends after mantle insertion, suggesting that they play an important role in pearl cultivating by mantle insertion. However, the specific mechanism still needs to be further investigated. Both BMP5a and BMP5b were expressed at higher levels after visceral mass insertion, with BMP5b expression being suppressed after 90 days of visceral mass insertion. This finding implies that BMP5a and BMP5b play a role in pearl cultivating by visceral insertion. But there is less literature available on mollusc BMP5, and only in higher organisms has it been shown to be related to bone formation and trauma repair (39, 40). Therefore, the function of BMP5 in the pearl cultivating of H. cumingii requires further study. BMP9 expression was significantly decreased 5 days after nucleus insertion in the visceral mass, and it may be suppressed in the short-term inflammatory and biomineralization. BMP9 was also not found to be studied in mollusks, but higher organisms’ studies have shown that its expression induces osteoblast differentiation, which is found to be related to bone formation and repair (41, 42). BMP9 may perform opposite functions in higher organisms and shellfish, and the specific mechanisms need to be further investigated. The differential expression patterns of BMP in mantle and visceral mass of the mussel after insertion indicate that the BMP gene family plays different roles in the pearl-cultivating process of mantle and visceral mass, some of which are promoted and some of which are suppressed. A more in-depth study of the above genes can provide a theoretical basis for shortening the pearl-culture cycle of H. cumingii and improving the quality of pearls.
5 Conclusion
In this study, bioinformatics analysis of BMP genes was performed based on genome of H. cumingii. A total of 12 BMP genes were identified, which were distributed on four chromosomes. The phylogenetic tree showed that HcBMPs are evolutionarily close to other shellfish, but evolutionarily distant from higher organisms (human and zebrafish). qRT-PCR results showed that the HcBMPs were differentially expressed in the mantle and visceral mass, and the expression level was higher in the visceral mass. This research found significant tissue specificity in BMPs gene expression. Further insertion surgery in the mantle and visceral mass significantly alters the expression profiling of the BMP gene family. BMP2a, BMP5a, BMP5b, BMP7a, and BMP7c had higher expression after visceral mass insertion, whereas BMP2b, BMP3, BMP9, and BMP10a had higher expression after mantle insertion, suggesting that HcBMPs may play different roles during the insertion of nucleus in the mantle and visceral mass, respectively. The present study lays the foundation and data support for the preliminary elucidation of regulatory role and mechanism of HcBMPs in the pearl-cultivating process of mantle and visceral mass.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Shanghai Ocean university. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YC: Methodology, Writing – original draft, Writing – review & editing. SL: Conceptualization, Writing – review & editing. YY: Investigation, Methodology, Writing – original draft. JS: Investigation, Writing – original draft. XC: Writing – original draft, Software. XY: Visualization, Writing – original draft. XX: Supervision, Writing – review & editing. XB: Conceptualization, Project administration, Supervision, Writing – review & editing, Funding acquisition, Resources. WL: Conceptualization, Project administration, Supervision, Writing – review & editing, Resources.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2022YFD2400105), the earmarked fund for China Agriculture Research System of MOF and MARA (CARS-49), the National Natural Science Foundation of China (No. 31201991).
Conflict of interest
SL was employed by Shanghai Mugao Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1445594/full#supplementary-material
Footnotes
2. ^https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
3. ^http://smart.embl-heidelberg.de
4. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
5. ^http://web.expasy.org/compute_pi/
6. ^http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/
7. ^https://psort.hgc.jp/form2.html
References
1. Kong, W . Research on cell culture and biomineralization roles of mantle tissue cells from the pearl oyster Pinctada facata. [Master’s thesis]. Beijing: Tsinghua University (2015).
2. Dauphin, Y . Structure and composition of the septal nacreous layer of Nautilus macromphalus L. (Mollusca, Cephalopoda). Zoology. (2006) 109:85–95. doi: 10.1016/j.zool.2005.08.005
3. Addadi, L, Joester, D, Nudelman, F, and Weiner, S. Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chem. (2006) 37. doi: 10.1002/chin.200616269
4. Li, W, Yang, J, Feng, S, Li, X, Chen, Y, Shen, X, et al. Analysis of proteomic differences and immune factors between Hyriopsis cumingii mantle and pearl sac. J Shanghai Ocean Univ. (2022) 31:344–54. doi: 10.12024/jsou.20210603491
5. Wang, A, Yan, B, and Ye, L. Pearl sacs culture in vitro and production of pearls by culturing pearl sacs in the pearl oysters (Pinctada martensi). Guangxi Sci. (2000) 7:70–4. doi: 10.13656/j.cnki.gxkx.2000.01.022
6. Liang, F, Xie, S, and Lin, W. Progress on pearl production from Hyriopsis cumingii. Trans Oceanol Limnol. (2015) 2:113–8. doi: 10.13984/j.cnki.cn37-1141.2015.02.016
7. Shi, Z, Xie, X, and He, X. Study on the bioloical characteristic of part culturing pearl in visceral mass of Hyriopsis cumingii. Biotechnol Bull. (2008) 3:178–81. doi: 10.13560/j.cnki.biotech.bull.1985.2008.03.042
8. Bragdon, B, Moseychuk, O, Saldanha, S, King, D, Julian, J, and Nohe, A. Bone morphogenetic proteins: a critical review. Cell Signal. (2011) 23:609–20. doi: 10.1016/j.cellsig.2010.10.003
9. Cheng, H, Jiang, W, Phillips, FM, Haydon, RC, Peng, Y, Zhou, L, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). Urol Oncol Semin Orig Invest. (2004) 22:79–80. doi: 10.1016/j.urolonc.2003.12.008
10. Deng, ZL, Sharff, KA, Tang, N, Song, WX, Luo, J, Luo, X, et al. Regulation of osteogenic differentiation during skeletal development. Front Biosci-Landmark. (2008) 13:2001–21. doi: 10.2741/2819
11. Luu, HH, Song, WX, Luo, X, Manning, D, Luo, J, Deng, ZL, et al. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. (2010) 25:665–77. doi: 10.1002/JOR.20359
12. Jiang, Y, Wang, L, Zhu, G, and Xu, Y. Effects on osteoblasts by bone morphgenetic protein 2a knocked down in zebrafish. Chin J Osteoporosis Bone Miner Res. (2019) 12:165–71. doi: 10.3969/j.issn.1674-2591.2019.02.010
13. Cao, X, Ma, C, Xia, X, Zhang, R, Chen, X, and Zhao, J. Cloning and expression analysis of jaw remodeling developmental gene Bmp4 in Siniperca chuatsi. Genomics Appl Bio. (2020) 39:5075–83. doi: 10.13417/j.gab.039.005075
14. Mei, W, Xiang, GD, Li, YX, Li, H, Xiang, LW, Lu, JY, et al. GDF11 protects against endothelial injury and reduces atherosclerotic lesion formation in Apolipoprotein E-null mice. Mol Ther. (2016) 24:1926–38. doi: 10.1038/mt.2016.160
15. Zhou, D . Moleculer cloning and preliminary functional studies of BMP3 and BMP10 gene from pearl oyster, Pinctada fucata. [Master’s thesis]. Shanghai: Shanghai Ocean University (2016).
16. Duan, S . Comparison of growth and pearling performance of Hyriopsis cumingii and Cristaria plicata after graft operation and identification and functional research of BMP genes. [Master’s thesis]. Shanghai: Shanghai Ocean University (2021).
17. Li, W, Huang, K, Li, Q, Qi, X, Shang, C, and Zhou, Z. Effects of different tissue sites insert-nucleus on pearl-sac formation in visceral mass of Hyriopsis cumingii. J Fishery Sci Chin. (2014) 21:1098–107. doi: 10.3724/SP.J.1118.2014.01098
18. Bai, Z, Lu, Y, Hu, H, Yuan, Y, Li, Y, Liu, X, et al. The first high-quality genome assembly of freshwater pearl mussel Sinohyriopsis cumingii: new insights into pearl biomineralization. Int J Mol Sci. (2024) 25:3146. doi: 10.3390/ijms25063146
19. Kumar, S, Stecher, G, Li, M, Knyaz, C, and Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
20. Chen, C, Chen, H, Zhang, Y, Thomas, HR, Frank, MH, He, Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. (2020) 13:1194–202. doi: 10.1016/j.molp.2020.06.009
21. Shen, X, Chen, Y, Jia, L, He, W, Chen, X, Chen, Y, et al. Transcriptome and digital gene expression analysis reveal immune responses of mantle and visceral mass pearl culturing in Hyriopsis cumingii. Front Mar Sci. (2023) 10:1251251. doi: 10.3389/fmars.2023.1251251
22. Chen, L, Dong, C, Kong, S, Zhang, J, Li, X, and Xu, P. Genome wide identification, phylogeny, and expression of bone morphogenetic protein genes in tetraploidized common carp (Cyprinus carpio). Gene. (2017) 627:157–63. doi: 10.1016/j.gene.2017.06.020
23. Lei, L, Zhu, J, Chen, C, Wang, Y, Wu, C, Qi, M, et al. Genome-wide identification, evolution and expression analysis of bone morphogenetic protein (BMP) gene family in Chinese soft-shell turtle (Pelodiscus sinensis). Front Genet. (2023) 14:1109478. doi: 10.3389/fgene.2023.1109478
24. Shi, R, Li, X, Xu, X, Chen, Z, Zhu, Y, and Wang, N. Genome-wide analysis of BMP/GDF family and DAP-seq of YY1 suggest their roles in Cynoglossus semilaevis sexual size dimorphism. Int J Mol. (2023) 253:127201. doi: 10.1016/j.ijbiomac.2023.127201
25. Wu, Y, Zhou, X, Chen, G, Yu, X, and Tong, J. Genome wide identification, phylogeny, and expression of bone morphogenetic protein genes in bighead carp (Hypophthalmichthys nobilis). Acta Hydrobiol Sin. (2024) 48:1085–1094. doi: 10.7541/2024.2023.0314
26. Ma, Y, Xiao, Y, Xiao, Z, Wu, Y, Zhao, H, Gao, G, et al. Genome-wide identification, characterization and expression analysis of the BMP family associated with beak-like teeth in Oplegnathus. Front Genet. (2022) 13:938473. doi: 10.3389/fgene.2022.938473
27. Ma, Q, Yang, YS, Mao, FF, Zhou, QL, Wang, LY, and Chen, G. Genome-wide identification, phylogeny and expression analysis of the gene family associated with development and skeleton deformity in cobia (Rachycentron canadum). Aquacult Rep. (2023) 31:101644. doi: 10.1016/j.aqrep.2023.101644
28. Ai, J, Li, Z, Liu, J, and Shen, Y. Molecular cloning and expression analysis of BMP-2 cDNA in Haliotis diversicolor supertexta. Mar Sci. (2018) 42:107–15. doi: 10.11759/hykx20180310001
29. Feng, L, Guo, H, Li, X, Yu, Q, Hu, X, Zhang, L, et al. Cloning and expression analysis of bone Morphgenetic protein 2 gene of Chlamys farreri. Period Ocean Univ China. (2013) 43:48–55. doi: 10.16441/j.cnki.hdxb.2013.12.008
30. Jiang, M, Zhang, Q, Meng, Y, Bai, K, Zhang, X, Nong, Z, et al. Molecular cloning and expression analysis of BMP7 gene of Crassostrea hongkongensis. J Guangdong Ocean Univ. (2023) 43:129–36. doi: 10.3969/j.issn.1673-9159.2023.04.016
31. Mao, F . Genome-wide identification and Expressionprofiling of the bone morphogenetic proteins gene family, osteologicalontogeny in larval and juvenile Rachycentron canadum. [Master’s thesis]. Guangdong: Guangdong Ocean University (2021).
32. Zhang, Y, Yu, J, Han, R, Ma, Z, Zhang, M, Li, Y, et al. Genome-wide identification and structural analysis of the BMP gene family in Triplophysa dalaica. BMC Genomics. (2024) 25:194. doi: 10.1186/s12864-024-10049-z
33. Yi, P . The role of TGF-β and its type I receptor gene in immunity stress and wound repair of Hyriopsis cumingii. [Master’s thesis]. Nanchang: Nanchang University (2020).
34. Jin, C, Li, JL, and Liu, XJ. Teosin, a novel basic shell matrix protein from induces calcium carbonate polycrystal formation. Int J Mol Sci. (2020) 150:1229–37. doi: 10.1016/j.ijbiomac.2019.10.134
35. Luo, W, Jiang, R, Ren, G, and Jin, C. Hic12, a novel acidic matrix protein promotes the transformation of calcite into vaterite in. Comp Biochem Physiol, Part B: Biochem Mol Biol. (2022) 261:110755. doi: 10.1016/j.cbpb.2022.110755
36. Zhang, X, Xia, Z, Liu, X, and Li, J. The novel matrix protein hic7 of hyriopsis cumingii participates in the formation of the shell and pearl. Comp Biochem Physiol B Biochem Mol Biol. (2021) 256:110640. doi: 10.1016/j.cbpb.2021.110640
37. Lin, J . Cloning and expression analysis of genes involved in the pearl formation of Hyriopsis cumingii. [Master’s thesis]. Shanghai: Shanghai Ocean University (2014).
38. Fan, S, Zhou, D, Liu, B, Deng, Z, Guo, Y, and Yu, D. Molecular cloning and expression analysis of BMP7b from Pinctada fucata. South China Fisheries Sci. (2018) 14:121–6. doi: 10.3969/j.issn.2095-0780.2018.01.016
39. Heller, IS, Guenther, CA, Meireles, AM, Talbot, WS, and Kingsley, DM. Characterization of mouse Bmp5 regulatory injury element in zebrafish wound models. Bone. (2022) 155:116263. doi: 10.1016/j.bone.2021.116263
40. Snelling, SJ, Hulley, PA, and Loughlin, J. BMP5 activates multiple signaling pathways and promotes chondrogenic differentiation in the ATDC5 growth plate model. Growth Factors. (2010) 28:268–79. doi: 10.3109/08977191003752296
41. Bing, C, Ling-Jiao, K, Jin-Xia, L, Lu, S, Cai, Z, and Jin-Hua, W. BMP9 induces osteogenic/odontogenic differentiation of ISCAP through the Smad pathway. China Biotechnol. (2016) 36:16–22. doi: 10.13523/j.cb.20160803
Keywords: Hyriopsis cumingii, bone morphogenetic proteins, identification, expression analysis, pearl culture
Citation: Chen Y, Liu S, Yao Y, Sun J, Chen X, Yu X, Xuan X, Bian X and Li W (2024) Identification and expression profiling of the bone morphogenetic protein gene family based on pearl culture in mantle and visceral mass of Hyriopsis cumingii. Front. Vet. Sci. 11:1445594. doi: 10.3389/fvets.2024.1445594
Edited by:
Ran Di, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Minglin Wu, Anhui Academy of Agricultural Sciences, ChinaKeyi Ma, Chinese Academy of Fishery Sciences, China
Copyright © 2024 Chen, Liu, Yao, Sun, Chen, Yu, Xuan, Bian and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangli Bian, Qmlhbl94aWFuZ19saUAxNjMuY29t; Wenjuan Li, d2psaUBzaG91LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Yiwen Chen
Yiwen Chen Shijun Liu2†
Shijun Liu2† Yuanbin Yao
Yuanbin Yao Wenjuan Li
Wenjuan Li